Abstract
Kinase inhibitor resistance often involves upregulation of poorly understood “bypass” signaling pathways. Here, we show that extracellular proteomic adaptation is one path to bypass signaling and drug resistance. Proteolytic shedding of surface receptors, which can provide negative feedback on signaling activity, is blocked by kinase inhibitor treatment and enhances bypass signaling. In particular, MEK inhibition broadly decreases shedding of multiple receptor tyrosine kinases (RTKs) including HER4, MET, and most prominently AXL, an ADAM10 and ADAM17 substrate, thus increasing surface RTK levels and mitogenic signaling. Progression-free survival of melanoma patients treated with clinical BRAF/MEK inhibitors inversely correlates with RTK shedding reduction following treatment, as measured non-invasively in blood plasma. Disrupting protease inhibition by neutralizing TIMP1 improves MAPK inhibitor efficacy, and combined MAPK/AXL inhibition synergistically reduces tumor growth and metastasis in xenograft models. Altogether, extracellular proteomic rewiring through reduced RTK shedding represents a surprising mechanism for bypass signaling in cancer drug resistance.
Keywords: mitogen activated protein kinase (MAPK), tissue inhibitor of metalloproteinase (TIMP), a disintegrin and metalloproteinase (ADAM), Kras mutation
Introduction
Mere overexpression of a drug’s target often fails to predict efficacy, in part due to “bypass” signaling whereby inhibition of one signaling pathway leads to compensatory signaling through alternative routes. Previous work has largely focused on how intracellular processes such as gene expression changes and mutations contribute to bypass signaling, heterogeneous drug responses, and drug resistance. For example, targeting mitogen-activated-protein-kinase (MAPK) signaling through MEK or B-RAF inhibition leads to increased transcription of multiple receptor tyrosine kinases (RTKs), which then provide alternative pro-growth and pro-survival signals that circumvent the intended inhibitor effects (1–3). However, genetic and gene expression changes account for only a fraction of observed bypass signaling (4, 5). For instance, only half of melanoma patients receiving B-RAF inhibitor therapy exhibit mutations in known B-RAF resistance genes upon emergence of drug resistance; a large fraction of drug resistance arises without defined genetic or epigenetic explanation; and the functional consequences of common genetic or translational alterations often still depend on the activity of signaling through RAS-CRAF and other pathways (6). Importantly, these observations carry substantial implications in the clinic, where many strategies for designing and monitoring an individual’s therapeutic course rely largely on genetic or transcriptional information.
This work investigates several cancer types where bypass signaling is evident (1, 2, 7, 8), and focuses primarily on malignant melanoma and triple-negative breast cancer (TNBC), a subtype of breast cancer with poor prognosis, no approved targeted therapies, and which is classified by low expression of estrogen receptor, progesterone receptor, and HER2 (9). MEK inhibition (MEKi) represents one promising therapeutic strategy, as MAPK signaling is dysregulated in many cancers, including TNBC and melanoma (9, 10), and clinical trials have recently been completed or are ongoing in several cancers including TNBC (9). As a treatment strategy, MAPK inhibition (MAPKi) has been most successful in melanoma, with three such inhibitors gaining FDA approval since 2011. However, resistance to MAPKi develops in most patients within a year (11). More generally, MAPKi carries broad importance given many other relevant drug targets including various RTKs are upstream of the MAPK pathway and indirectly affect its activity.
Relative to tumor gene expression changes, little attention has been paid to how the tumor-derived extracellular proteome changes in response to targeted kinase inhibitors, and how such changes impact bypass signaling and drug efficacy. Of central importance, the ‘A Disintegrin and Metalloproteinases’ (ADAMs) ADAM10 and ADAM17 are widely known as the principal “sheddases” of the cell surface responsible for shedding ectodomains of hundreds of transmembrane substrates, including many growth factors, cytokines, adhesion molecules, and metalloproteinases involved in the processes described above. ADAM10 is required for activation of the Notch signaling pathway, while ADAM17 is needed for TNFα cleavage, and both ADAM10−/− and ADAM17−/− mice are not viable (12). ADAM10 and ADAM17 are particularly overexpressed in many cancers including breast cancer and melanoma (13, 14), with activities governed by frequently dysregulated MAPK signaling (10, 15). Furthermore, ADAM10 and ADAM17 have been considered promising drug targets for their part in shedding EGF-family growth factor ligands from the surface of cancer cells, a process that mediates ErbB-family receptor mitogenic signaling in an autocrine manner (13, 16, 17). Unfortunately, metalloproteinase inhibitors, including a second-generation inhibitor with specificity towards ADAM10 and ADAM17 (INCB7839, Incyte), have failed in clinical trials despite promising initial results (18). These failures can largely be attributed to a poor understanding of how the broad activity of ADAMs, and metalloproteinases in general, integrate to influence overall tumor behavior (19).
Here, we identify differential extracellular proteolytic shedding as a major post-translational mechanism of bypass signaling that complements other pathways of drug resistance. Proteolytic shedding of surface receptors, which can provide negative feedback on signaling network activity, is dramatically reduced upon inhibition of kinase pathways such as the MAPK pathway. Decreased RTK proteolysis consequently leads to surface RTK accumulation and increased signaling through other pathways that support mitogenesis. Thus, we hypothesized that RTK proteolysis could (a) be non-invasively monitored in patients; (b) enable early detection of drug resistance in the clinic; and (c) guide the design of combination therapies that forestall disease progression. Indeed, we found that circulating RTKs were detectable at elevated levels in a subset of patients, and that shed RTK levels accurately predicted clinical MAPKi response better than mere RTK gene expression. We tested two strategies to enhance MAPKi efficacy: (1) modulating ADAM10 through neutralizing its cognate inhibitor “tissue inhibitor of metalloproteinases 1” (TIMP1), and (2) inhibiting the RTK AXL, a key protease substrate that we observed to be upregulated following MAPKi. In several orthotopic animal models, combination therapies exhibited synergistic effects on tumor growth, metastasis, and survival. Altogether, our findings demonstrate that extracellular proteomic rewiring through reduced proteolytic receptor shedding represents a significant and targetable mechanism for bypass signaling in acquired cancer drug resistance.
Results
MAPKi causes a global reduction in circulating RTKs
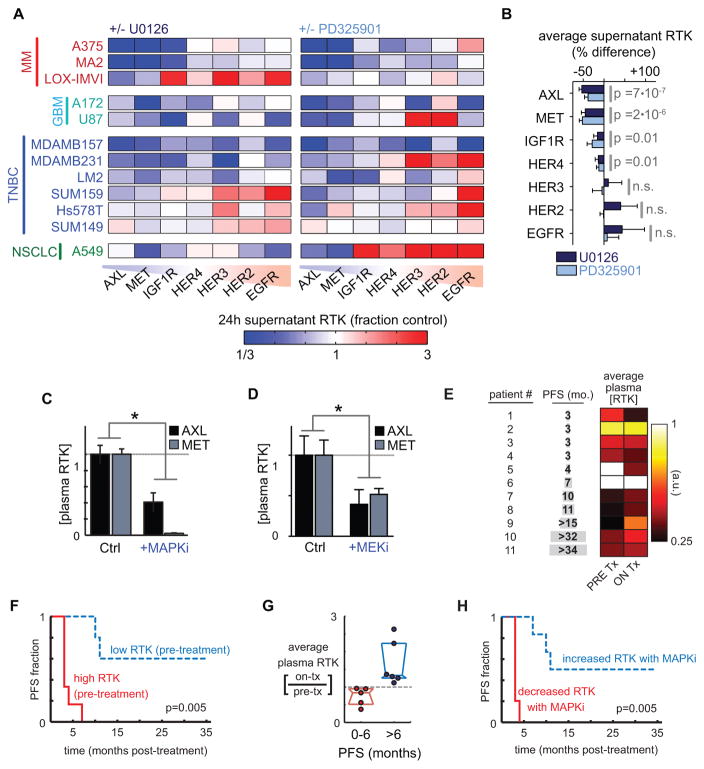
RTK signaling mediates drug resistance (1, 7, 20), and the release of RTK ectodomains from the cell surface has become an increasingly appreciated regulator of signaling activity in contexts of cancer metastasis (21), antibody therapies (22), and in other invasive diseases (15). However, little is known regarding how extracellular RTK levels change in response to targeted kinase inhibitor treatment, and how such changes influence drug efficacy in cancer. To study these effects, we first measured how MEKi impacts the supernatant accumulation of seven key RTKs that have been implicated in drug resistance: the four ErbB-family receptors (epidermal growth factor receptor EGFR, HER2, HER3, and HER4); insulin-like growth factor receptor 1 (IGF1R); hepatocyte growth factor receptor (HGFR/MET); and the TAM-family RTK member, AXL. Using two MEK inhibitors U0126 and PD325901 (Fig 1A, Fig. S1A validates reduction in pErk), we examined a panel of 12 cell lines from several cancer types where bypass signaling has been implicated, including malignant melanoma (MM), TNBC, non-small cell lung cancer (NSCLC), and glioblastoma multiforme (GBM). We surprisingly found that only decreased (rather than increased) supernatant RTK accumulation was consistently and significantly observed across the panel of measurements (Fig. 1A–B). Most prominently, supernatant AXL and MET decreased by roughly 50% in nearly every cell line. We confirmed by ultracentrifugation that AXL and MET were unassociated with supernatant microvesicles (Fig. S1B). Supernatant accumulation of both RTKs could be blocked by broad-spectrum metalloproteinase inhibition (MPi) using batimastat (BB94), suggesting their accumulation resulted from proteolytic release off the cell surface (Fig. S1C). EGFR, which is not a suspected metalloproteinase substrate, behaved in stark contrast to MET and AXL: in the supernatant, EGFR i) did not consistently decrease in response to MEKi (Fig. 1A–B) ii) was substantially associated with microvesicles (Fig. S1B), and iii) did not decrease in response to MPi (Fig. S1C). Thus, we find that MEKi treatment induces significant changes in supernatant RTK levels, which contains a combination of free proteolytically shed soluble receptor (as with AXL and MET), and surface receptor on microvesicles (as with EGFR), with the significant effects of MEKi dominated by the former (Fig. 1A–B).
Figure 1. MEKi broadly reduces extracellular release of protein ectodomains and correlates with drug resistance in melanoma patients.
A) MEKi with U0126 and PD325901 reduces supernatant accumulation of soluble RTKs (measured by ELISA) in multiple cancer cell lines (n≥2). B) Change in supernatant RTKs after treatment with MEKi, averaged across all cell lines shown in A (pooled two-tailed t-test; n=22). C–D) Circulating plasma levels of soluble AXL and MET decrease after (C) MAPKi treatment (10 mg/kg vemurafenib + 1 mg/kg trametinib) in LOX-IMVI melanoma xenografts (p=0.004, n=4, pooled two-tailed t-test), or (D) MEKi treatment (1 mg/kg PD325901) in orthotopic LM2 breast cancer xenografts (p=0.0036, n≥4, pooled two-tailed t-test). E) Plasma from melanoma patients was assayed for soluble RTK levels before and on treatment with dual BRAFi/MEKi therapy; heatmap shows the average of 6 RTK levels (Fig. S1G for full dataset). F) Kaplan-Meier analysis based on average pre-treatment RTK levels (see E; p=0.005; two-tailed log-rank test; total n=11). G) Circulating RTK levels decrease in melanoma patients with short progression free survival (PFS) after MAPKi treatment (median +/− IQR of RTK levels, averaged as in E). H) Kaplan-Meier analysis based on the change in RTK levels with MAPKi therapy initiation (p=0.005; two-tailed log-rank test; total n=11).
We next examined if reduced accumulation of extracellular RTK ectodomain was detectable in mice undergoing MAPKi therapy. We used two different xenograft tumor models: subcutaneous BRAF-mutant melanoma using LOX-IMVI cells, and orthotopic TNBC using the BRAF/KRAS mutant LM2 cells. With human-specific solution-phase immunoassays, we measured plasma levels of tumor-derived receptors in tumor-bearing mice following drug treatment. Indeed, MAPKi using combined MEKi (trametinib) and BRAFi (vemurafenib) treatment in the melanoma model decreased circulating levels of tumor-derived AXL and MET (Fig. 1C). Similarly, MEKi significantly reduced levels of the circulating AXL and MET in mice with mammary fat pad tumors (Fig. 1D).
To test whether this RTK ectodomain accumulation may be relevant to clinical pathology, we measured circulating levels of AXL, MET, HER2, and HER4 ectodomains in stage I and stage IV breast cancer patients along with healthy controls, and observed that AXL contributes to an overall pattern of increased RTKs in patients (Fig. S1D–F). The combined level of these four RTKs in a given serum sample increased significantly in breast cancer patients who have not been exposed to the inhibitors shown here, compared to healthy controls (Fig. S1D). In other words, roughly 50% of stage I (10/20) and stage IV (9/19) patient samples exhibited a “quadruple-positive” phenotype with increased levels of all four RTKs in circulation, compared to 5% (1/20) of healthy controls (Fig. S1E). Among “quadruple-positive” samples, circulating RTK levels in stage IV patients were 40% higher than levels from stage I patients (Fig. S1E; p=0.03, two-tailed t-test). The statistical significance of an elevated serum RTK signature was dependent upon inclusion of AXL measurements (Fig. S1F), indicating it as the most vital among the four. These data confirm that stage I and especially advanced metastatic stage IV diseases are associated with changes in RTK shedding that are detectable circulating in human patients.
Circulating RTK levels but not their mere tumor expression predict MAPKi resistance in melanoma patients
We next investigated whether reduced RTK ectodomain levels were observable in patients undergoing MAPKi therapy and if markers of RTK accumulation correlated with clinical outcomes. Plasma, rather than serum which was used in the breast cancer cohort, was collected from melanoma patients undergoing treatment with a combination of trametinib (MEKi) and dabrafenib (BRAFi), both before and while on treatment. As a surrogate marker of RTK shedding, we measured soluble levels of six RTKs known to be sheddase substrates: MET, HER2, HER4, and the three TAM receptors (AXL, MERTK, and TYRO3) using solution-phase antibody arrays (Fig. 1E, Fig. S1G). With this blood-based test, we found that patients showing high levels of circulating RTKs before MAPKi treatment exhibited rapid disease progression (Fig. 1F). Motivated by the hypothesis that reduced RTK shedding may lead to MAPKi resistance, we next examined (a) if circulating RTK levels changed with MAPKi treatment, and (b) if changes correlated with disease progression. We found that 5/11 patients, principally those with high initial circulating RTK levels, showed decreased circulating RTK levels upon initiation of MAPKi treatment (Fig. 1G), and that disease rapidly progressed in these patients (Fig. 1H). In fact, these circulating RTK changes were highly predictive of progression-free survival (PFS; p=0.005; n=11; two-tailed log-rank test) (Fig. 1H). In contrast, initial tumor response as measured by Response Evaluation Criteria for Solid Tumors (RECIST) failed to reliably predict PFS in a statistically significant manner (p=0.08; n=11; two-tailed log-rank test), as observed in other cancers (23), thus demonstrating the comparative superiority of circulating RTKs as predictive markers of disease progression.
We next examined whether mere RTK expression in the tumor, rather than levels of circulating RTKs, could similarly predict PFS in patients. In the same cohort used to assess circulating RTKs, total tumor AXL measured by immunohistochemistry of its intracellular C-terminus failed to correlate with PFS (Fig. S1H). Furthermore, in an independent dataset of 21 melanoma patients undergoing BRAFi therapy, RNA expression of the six RTKs measured here did not substantially predict PFS (Fig. S1I). Up-regulation of RTK RNA expression, measured by comparing matched gene expression before and after BRAFi therapy began, only mildly trended towards worse PFS but the difference was not significant (Fig. S1J; p=0.055, two-tailed log-rank test). These results, combined with the aforementioned TNBC and melanoma xenograft studies, show that circulating RTK levels (a) can be non-invasively, quantitatively, and longitudinally monitored in patients undergoing MAPKi treatment, (b) provide an early indication of MAPKi efficacy, (c) are more predictive of MAPKi efficacy than mere expression in the tumor, and (d) may consequently have utility as a patient selection criterion.
MEKi increases total and phosphorylated AXL on the cell surface
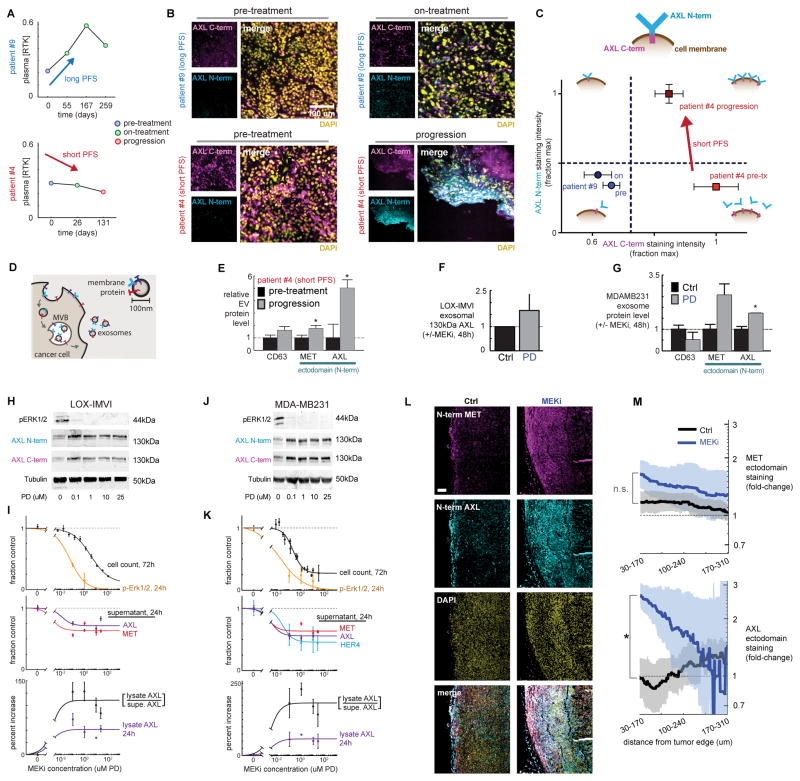
We next investigated the relationship between AXL levels in circulation and within the tumors of melanoma patients. We used samples from patients with similar initial tumor responses by RECIST (Fig. S2A), yet very different plasma RTK patterns and times of disease progression (Fig 2A). To simultaneously measure total tumor AXL and its ectodomain release from the tumor, we compared immunostaining by antibodies targeting either the AXL intracellular C-terminus (corresponding to total tumor AXL) or the N-terminus ectodomain (the latter was also used to detect circulating AXL in plasma). In one patient exhibiting relatively long PFS (patient #9), low plasma AXL levels (Fig. 2A, top row) corresponded to low tumor AXL levels pre-treatment (Fig. 2B, top row). A sustained increase in AXL and other RTKs was detected in plasma (Fig. 2A, top row) with MAPKi treatment initiation, and indeed C-terminus AXL expression was detected at higher levels in a subset of the tumor cells analyzed on treatment (Fig. 2B; S2B). In contrast, another patient showing rapid disease progression (patient #4) exhibited high pre-treatment AXL levels both in plasma and in the pre-treatment tumor (Fig. 2A–B, bottom row), but tumor AXL appeared to be shed at high levels: although AXL C-terminus was high in this tumor, ectodomain staining was low by comparison. Even though plasma AXL levels declined with MAPKi therapy in this patient (Fig. 2B, bottom row), AXL C-terminus staining remained high in the on-treatment tumor biopsy and tumor AXL ectodomain increased substantially (Fig. 2C). This evidence suggests that MAPKi-induced decrease in plasma RTK does not reflect decreased AXL expression in the tumor, but rather decreased release of the AXL ectodomain from the tumor.
Figure 2. Decreased circulating AXL correlates with increased AXL surface levels in the tumor after MEKi.
A) Longitudinal plasma RTK levels monitored in melanoma patients showing similar initial response to MAPKi (Fig. S2A) but dramatically different PFS. B) Immunofluorescence of AXL cytoplasmic domain (C-term) and ectodomain (N-term) from tumor biopsies from patients in A. C) Schematic of AXL measurements and quantification of data from B. D) Multivesicular bodies (MVB) generate exosomes containing membrane proteins resembling those on the cell surface. E) Levels of MET, AXL, and the exosome marker CD63 on EVs isolated from plasma from patient #4 pre-treatment and after disease progression (nPLEX; *p=0.008, n=8, two-tailed t-test). F) Levels of full-length AXL on exosomes isolated from LOX-IMVI cells in culture (western blot; n=3). G) Levels of MET, AXL, and the exosome marker CD63 on exosomes isolated from MDA-MB231 cells +/− MEKi treatment (nPLEX; *p=0.01, n≥2, two-tailed t-test). H) LOX-IMVI lysate western blot, probed with Abs for AXL ectodomain (N-term) and cytoplasmic (C-term) epitopes. I) Quantification of cell count, pERK1/2, and RTK levels in LOX-IMVI cells (n≥3). J) MDA-MB231 lysate western blot, probed with Abs for AXL ectodomain (N-term) and cytoplasmic (C-term) epitopes. K) Quantification of cell count, pERK1/2, and RTK levels in MDA-MB231 (n≥3). L) Representative immunofluorescence of LM2 primary TNBC tumors 21 days after PD325901 treatment, showing upregulation of AXL ectodomain near the tumor edge. Scale bar = 100μm. M) Mean (thick line) +/− SEM (shaded area) for staining intensity for MET and AXL measured within a 140 μm sliding window from the tumor edge (*p=0.013; n≥3 tumors per group).
Circulating tumor-derived extracellular vesicles (EVs) including exosomes can also be detected in patients, and surface proteins on such vesicles often correlate with membrane protein levels on the tumor (Fig. 2D) (24). We examined if changes in EV RTK levels could also explain the post-treatment decrease in circulating RTKs seen for patient #4. Using the recently developed nPLEX (nanoplasmonic exosome) assay to sensitively detect surface RTK ectodomains on EVs (24), we found that these EVs did not explain the overall circulating RTK decrease (Fig. 2E; Fig. S2C). In fact, EV AXL and MET ectodomain levels increased with MAPKi in patient #4, which is consistent with the observed tumor histology (Fig. 2C) and likely reflects intact RTK ectodomain on the tumor cell surfaces. In sum, these data demonstrate the divergence between the levels of circulating, soluble RTKs and RTK levels on cancer cell membranes following MAPKi in patients: decreased circulating RTK levels do not simply reflect decreased expression in the tumor.
We next sought to understand how MEKi affects AXL surface levels and signaling activity in cultured melanoma (LOX-IMVI) and TNBC (MDA-MB231) cells. Consistent with the clinical melanoma findings, we observed that although MEKi reduced total supernatant RTK for both cell lines (Fig. 1A), these changes did not correlate with exosomal RTK levels within that same supernatant (Fig. 2F–G). MEKi did not impact exosome production or size in MDA-MB231 (Fig. S2D), and previously discussed control experiments revealed that the majority of supernatant MET and AXL from MDA-MB231 and LOX-IMVI cell culture was not exosome-anchored (Fig. S1B). We next examined how MEKi influenced protein levels on the cell surface and lysate. Changes in both surface and phosphorylated AXL were detectable by 3h post-treatment and further increased by 24h (Fig. S2E). However, MEKi did not similarly increase RNA levels of AXL (Fig. S2F–G), suggesting that surface AXL changes are not simply due to transcriptional regulation.
For a dose-response analysis of MEKi-induced AXL effects, we treated LOX-IMVI (Fig. 2H–I) and MDA-MB231(Fig. 2J–K) cells with increasing concentrations of the MEK inhibitors PD325901 and U0126; measured p-ERK1/2 as a readout of proximal MEKi efficacy; monitored cell survival; measured supernatant RTK ectodomain levels; and assessed levels of full-length intact AXL in cell lysates using antibodies targeted to either the ectodomain (N-terminus) or the intracellular domain (C-terminus) (also Fig. S2H–I). Reduction of supernatant RTK correlated with p-Erk1/2 decrease and anti-correlated with lysate levels of full-length AXL, which increased. Particularly with U0126 treatment, coordinated changes in p-Erk1/2, supernatant RTK, and lysate AXL all were observable at lower MEKi concentrations that did not substantially impact cell growth, therefore suggesting the RTK changes are not directly caused by cell death and/or apoptosis processes.
To parallel the clinical and in vitro observations, we next tested whether MEKi similarly increased tumor AXL in a TNBC xenograft model. In association with our data that circulating RTK levels decreased substantially with MEKi (Fig. 1D), we observed that MEKi concomitantly increased AXL in the primary tumor, particularly at tumor edges (Fig. 2L–M). Overall, MEKi-induced reduction in extracellular soluble RTKs corresponds with a contrasting accumulation on the membranes of cells and exosomes, with corresponding increases in AXL phosphorylation.
AXL mediates MEKi resistance, and co-treatment with AXLi synergistically reduces tumor growth, metastasis, and extends survival in mice
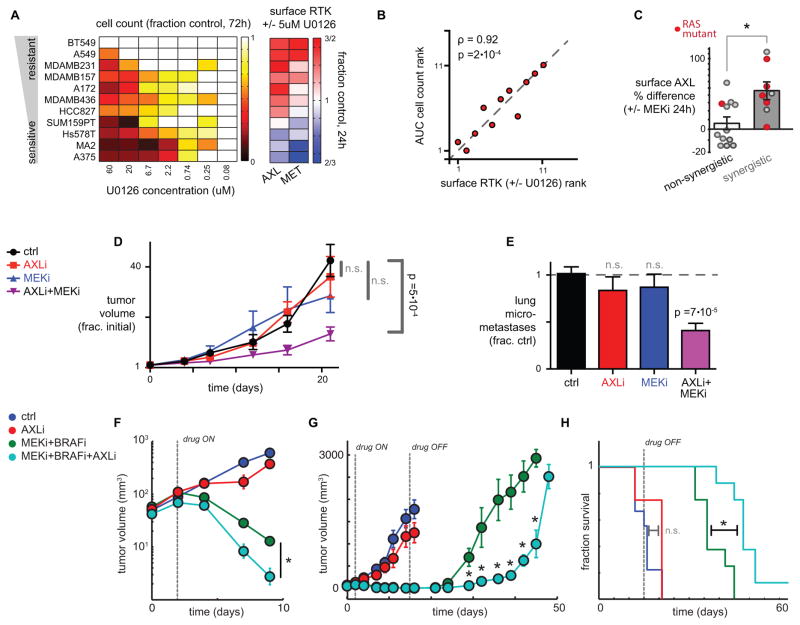
We next asked how increased RTK surface accumulation following MEKi relates to overall drug response. Across 11 cancer cell lines, we observed a significant correlation between MEKi resistance and surface changes in MET and AXL: cells showing RTK accumulation following MEKi were insensitive to MEKi as measured by proliferation across a dose-response (Fig. 3A–B). From these data, we hypothesized that MEKi-induced AXL upregulation would also correlate with synergistic response to combined MEKi/AXLi treatment. To test this, we quantified MEKi/AXLi synergistic response using a model of bliss independence across 10 cell lines and correlated the results to changes in surface AXL (Fig. S3A–C). Consistent with our model, cell lines displaying synergistic responses to combined AXLi/MEKi treatment also showed corresponding upregulation of surface AXL following MEKi, in comparison to cell lines displaying non-synergistic responses (Fig. 3C, Fig. S3C). Notably, cell lines displaying synergistic response were also enriched for RAS mutation, which is possibly related to a decreased reliance on signaling through proteolytically shed EGF-ligands for MAPK activity, and which is clinically relevant due to the frequent assessment of RAS mutation in patient tumors (Fig. 3C; Fig. S3D). Indeed, RAS-mutant MDA-MB231 cells, among others, were resistant to treatment with an anti-EGFR antibody that blocks ligand binding (mAb225, non-humanized cetuximab; Fig. S3E), and Kras mutation is a contra-indication for cetuximab therapy in the colorectal cancer setting due to lack of efficacy in those patients (25).
Figure 3. Combination MEKi and AXLi synergistically reduce tumor growth and metastasis in mouse models of melanoma and TNBC.
A) Cell counts and surface AXL and MET changes (measured by live-cell immunostaining; n=3) after 24h treatment with MEKi in 11 different cell lines. B) Correlation between cell count and surface RTK expression after MEKi treatment, ranked by order across cell lines in A (ρ = Spearman correlation, p-value from exact permutation test). C) U0126 and PD325901 increase surface AXL (measured by live-cell immunostaining) more in cell lines showing synergistic inhibition of proliferation from dual AXLi/MEKi (p=0.01, two-tailed t-test). Dots represent data from 10 cell lines and two drug combinations (see Fig. S3). Treatments in KRAS or HRAS mutant cell lines are shown in red, and are significantly enriched among the synergistic responses (p=0.02, Fisher’s exact test). D) Dual AXLi/MEKi reduces tumor growth more than either treatment individually (1 mg/kg PD325901; 30 mg/kg R428), in LM2 TNBC xenografts (*two-tailed t-test, n≥7). E) Dual AXLi/MEKi reduces metastasis after 21 days of treatment, corresponding to D (*two-tailed t-test, n≥7). F–H) AXLi (30 mg/kg R428) co-treatment synergistically increases efficacy of BRAFi/MEKi (1 mg/kg PD0325901 with 10 mg/kg vemurafinib) in LOX-IMVI xenografts by enhancing initial tumor shrinkage (F; p=0.031, two-tailed t-test), delaying tumor recurrence (G; p=0.002, two-tailed t-test), and extending survival (H; p=0.03, two-tailed log-rank test); n≥8 animals per group for all. Note AXLi alone fails to significantly affect tumor growth and animal survival.
We next tested the synergistic efficacy of dual MEKi/AXLi treatment in two mouse xenograft models, using cancer cell lines that showed increased surface AXL following MEKi. First, we employed orthotopic TNBC xenografts using the highly lung-metastatic derivative of MDA-MB231, LM2. Combined MEKi/AXLi treatment reduced both tumor growth (Fig. 3D) and metastasis (Fig. 3E) more than either treatment alone, with significant super-additive synergistic effects in tumor growth reduction (p=0.015; two-way ANOVA interaction term; n≥7). In melanoma LOX-IMVI xenografts, we hypothesized that MEKi+BRAFi (trametinib+vemurafenib) combined with the AXL inhibitor R428 (AXLi) might extend progression free survival (PFS) by compensating for drug resistance arising via AXL-mediated bypass signaling. Using an in vitro proliferation/cytotoxicity assay, AXLi combined with either MEKi or BRAFi showed synergistic effects in LOX-IMVI (Fig. S3F). In the LOX-IMVI xenograft, addition of AXLi to the BRAFi/MEKi treatment regimen led to an enhanced initial tumor response (Fig. 3F), delayed tumor recurrence after the initial treatment course ended (Fig. 3G), and extended median overall survival by roughly 50% more than MAPKi treatment alone (Fig. 3H). In contrast, AXLi treatment by itself had no significant impact on overall survival, thus demonstrating that the interaction between MAPKi and AXLi is synergistic (Fig. 3H). Overall, these results provide evidence that AXL mediates bypass signaling in response to BRAFi/MEKi treatment, contributes to drug resistance, and is therapeutically targetable using combination treatment regimens.
MEKi-induced AXL and MET upregulation is consistent with decreased proteolytic receptor shedding
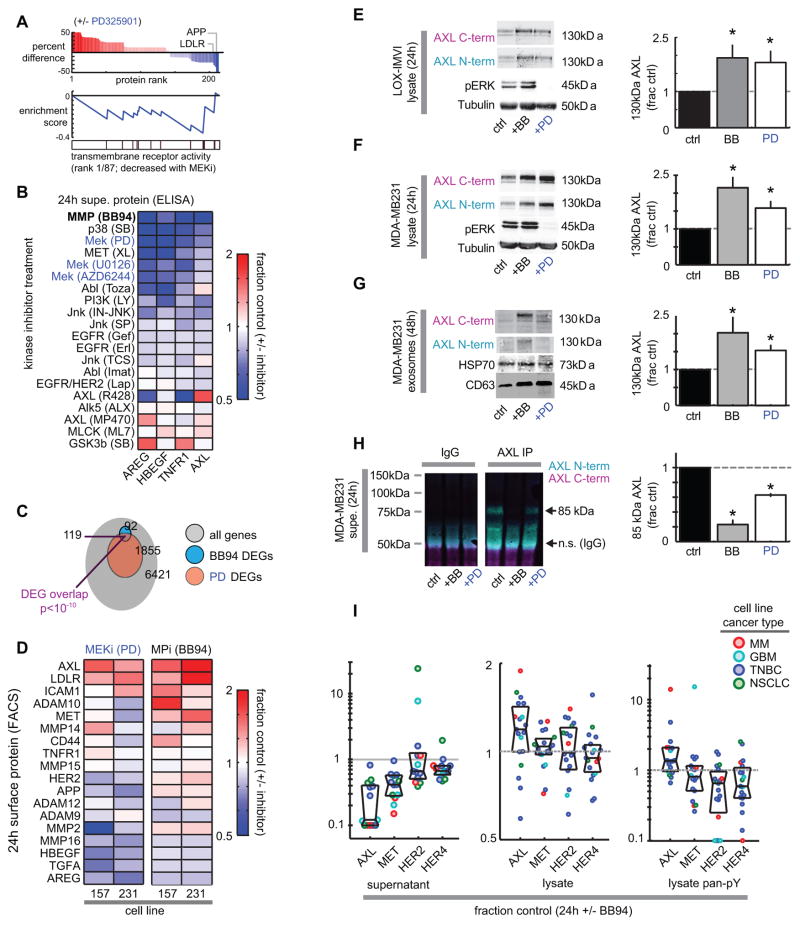
We next investigated the mechanism by which AXL and MET accumulate on the cell surface following MEKi, while simultaneously decreasing levels in the supernatant and in circulation. We first took a proteomic approach to look for global patterns in how the tumor-derived extracellular proteome changes in response to targeted kinase inhibitor treatment, using antibody microarrays to screen 1000 proteins for differential supernatant accumulation following MEKi in the TNBC MDA-MB231 cell line. Gene set enrichment analysis (26) of the ~200 proteins exhibiting significantly altered levels in the supernatant indicated that MEKi reduced transmembrane receptor ectodomain abundance (Fig. 4A) while increasing secreted (not proteolytically shed) cytokine levels (Fig. S4A). Among proteins that were depleted with MEKi from the supernatant, the top-ranked “transmembrane receptor activity” gene-set (Fig. 4A) comprised various known sheddase substrates including amyloid precursor protein (APP) (27) and low-density lipoprotein receptor (LDLR) (28). These results combined with the significantly decreased supernatant accumulation of RTKs including known sheddase substrates MET and HER4 (Fig. 1A–B) collectively implicate reduced proteolytic activity as a potentially key effect of MEKi.
Figure 4. MEKi-induced RTK changes are consistent with decreased proteolytic receptor shedding.
A) Top bar graph: Differentially detected supernatant proteins from MDA-MB231 treated with MEKi (PD325901) for 24h, measured by Ab-microarray (n=4). Middle: Enrichment score (ES) trace for top-ranked gene-set of proteins depleted with MEKi; vertical bars below trace indicate location of proteins in the top-ranked gene-set. B) Diverse kinase inhibitors (labeled as drug target followed by drug name) affect supernatant concentrations in MDA-MB231 (n≥2; ELISA), after normalizing to cell count. C) Venn diagram of differentially expressed genes (DEGs) from RNA microarray analysis of MDA-MB231 treated with PD325901 or BB94 for 24h (q=FDR-corrected p-value; n≥2). D) Surface level changes of sheddase substrates following MPi or MEKi in two TNBC cell lines (live-cell immunostaining; n=3). E–F) Intact lysate AXL from LOX-IMVI (E) and MDA-MB231 cells (F), as detected using Abs probing N-term or C-term epitopes (n=4). G) Full-length exosomal AXL and exosome markers CD63 and HSP70, isolated MDA-MB231 supernatant (n=6). H) The immunoprecipitated shed AXL fragment from supernatant of MDA-MB231 cells decreases in abundance with BB94 or PD320951 treatment (n=3). For E–H, *p<0.05, two-tailed t-test. I) Supernatant, lysate total and phospho-RTK levels following MPi; each dot represents one of 19 cell lines tested (see Fig. S4K–L).
We next compared the supernatant effects of MEKi with broad-spectrum metalloproteinase inhibition (MPi) using batimastat (BB94), and found similar patterns of widely decreased sheddase substrates amphiregulin (AREG), heparin-binding epidermal growth factor (HBEGF), tumor necrosis factor receptor 1 (TNFR1), and AXL following inhibitor treatment (Fig. 4B). AREG, HBEGF, and TNFR1 have been largely associated with ADAM17 cleavage (13, 17) but all may also be shed by ADAM10 depending on the context (15, 29–32). Reduced sheddase substrate accumulation was not an exclusive effect of direct MEKi: among 19 clinical and pre-clinical inhibitors targeting diverse signaling pathways and RTKs, roughly 80% (15/19) inhibited substrate accumulation to some degree. Nonetheless, MEKi exhibited some of the strongest effects (Fig. 4B). p38, PI3K, and c-JUN N-terminal kinase (JNK) inhibitors also significantly reduced sheddase-substrate accumulation, consistent with previous reports describing their effects on ADAM activity (15, 33). In contrast to supernatant decreases, surface levels of sheddase-substrates AXL and TNFR1 correspondingly increased in response to several other inhibitors, but most significantly with MPi and MEKi (Fig. S4B). Kinetics of surface and phosphorylated AXL following MPi (Fig. S4C) closely resembled MEKi changes (Fig. S2D), with observable increases by 3h post-treatment and continued accumulation by 24h. To further compare the effects of MEKi and MPi, we profiled gene expression changes using RNA microarrays following either of the two treatments. Results show substantial overlap in the transcriptional responses arising from MEKi and MPi, suggesting a shared mechanism of action (Fig. 4C; Fig. S4D). However, gene set enrichment analysis indicated MEKi, but not MPi, induced growth arrest (Fig. S4E); in fact, MPi did not elicit any significant gene-set enrichment. Overall, these results show that reduced metalloproteinase activity is a surprisingly prominent effect of MEKi. Furthermore, these data implicate MAPK signaling as one of the key pathways broadly regulating supernatant sheddase-substrate accumulation, with effects similar to those seen when proteolytic shedding is directly inhibited.
We next compared the effects of MPi and MEKi on RTK changes in levels in cell lysate, cell surface, and in exosomes. Focusing on RTKs and key sheddase substrates identified in the Ab-microarray, we first analyzed changes in surface levels of proteins (Fig. 4D). In two TNBC cell lines (MDA-MB231 and MDA-MB157), we measured a panel of 18 sheddase-substrates on the cell surface in response MPi and MEKi, and found significant correlation between changes with MEKi compared to MPi (Pearson’s correlation = 0.56; p=0.0003, two-tailed t-test). Results show AXL as one of the most significantly upregulated membrane proteins (Fig. 4D). Across a panel of 12 cell lines, we observed a modest yet statistically significant correlation between changes in surface AXL and MET in response to either MEKi or MPi, such that cells are more likely to exhibit increased surface AXL or MET following MEKi if they also showed increased levels following MPi (Fig. S4F). Gene expression helps explains why some sheddase-substrates actually decrease on the cell surface following MPi; for example, HBEGF and AREG expression decrease following BB94 treatment, and their levels correspondingly decrease on the cell surface (Fig. S4G). In contrast, AXL expression does not significantly change with either MPi or MEKi, and in LOX-IMVI (Fig. 4E) and MDA-MB231 cells (Fig. 4F) treatment with both MPi and MEKi significantly increase levels of full-length intact lysate AXL (see Fig. S4H–I for confirming pErk reduction). Similarly, we found increased intact AXL levels in CD63+ exosomes isolated from supernatant of the same cells (Fig. 4G). To confirm the hypothesis that this is due to changes in AXL proteolysis, we measured levels of ectodomain AXL receptor fragment in the supernatant of treated cells. Indeed, MPi and MEKi both decreased the accumulation of an AXL ectodomain fragment in the supernatant (Fig. 4H), and full-length intact AXL protein was not detectable to any similar degree compared to the ectodomain fragment in the supernatant (Fig. 4H). No substantial increases in cell death, apoptosis, or autophagy were observed with BB94 or PD325901 at the concentrations tested, suggesting these processes are not involved in the AXL phenotype described above (Fig. S4J). Taken together, these data provide further evidence that MEKi effects on RTK processing are highly consistent with reduced proteolysis by metalloproteinases.
To more broadly assess the impact of direct MPi on RTK signaling, we next examined MPi-induced changes in total, phosphorylated, and supernatant RTK levels across a panel of 19 diverse cell lines. MPi widely decreased AXL, MET, HER2, and HER4 in the supernatant, and increased total and phospho-RTK levels in many cases, most significantly and consistently for AXL and MET (Fig. 4I; Fig. S4K–L). Overall, these data show that proteolytic RTK shedding substantially impacts RTK levels and signaling in many cells, especially for AXL.
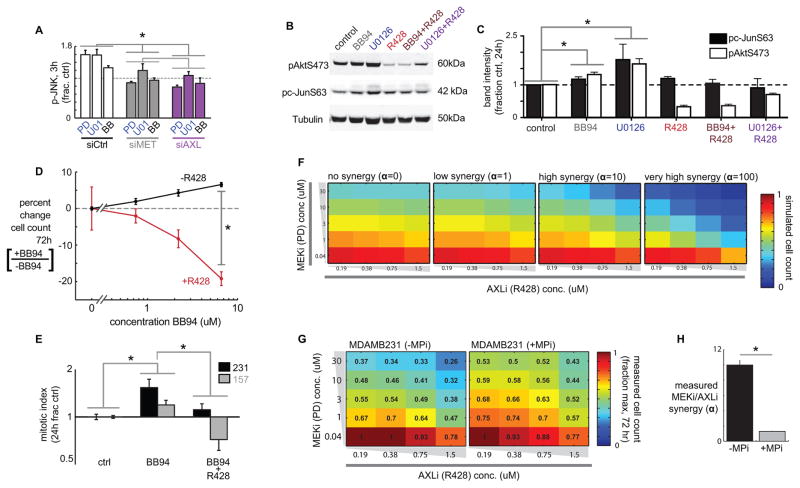
MPi and MEKi cause AXL-dependent bypass JNK/cJUN signaling
We next tested how AXL accumulation after MEKi or MPi leads to corresponding changes in downstream signaling events. At the RTK level, MEKi and MPi increase co-immunoprecipitation of AXL with MET and HER2, which has been associated with RTK co-localization, signaling cross-talk, and AXL transactivation (Fig. S5A) (34). Downstream of receptor signaling, we found that although MEKi reduced pErk, it increased phosphorylation in other signaling pathways (Fig. S1A). Previous reports have implicated signaling of JNK and its substrate cJUN in MAPKi resistance (35), and here we found p-cJUN consistently increased with MEKi treatment more than 5 other canonical signaling pathways averaged across 8 cell lines; p-Akt was also consistently elevated (Fig. S1A). In response to MPi, JNK and cJUN phosphorylation were among the strongest correlates with fluctuating surface AXL and p-AXL levels compared to 21 other signaling measurements across 13 cell lines (Fig. S5B–C), suggesting that accumulation of cell surface AXL following MPi leads to increased AXL phosphorylation and downstream signaling through the JNK/cJUN pathway. To test whether the observed JNK/cJUN signaling in fact depends on AXL activity, we monitored p-JNK following MPi or MEKi in the presence or absence of various AXL perturbations. After 2h MPi treatment, compensatory increase in phosphorylation occurs more in p-JNK than 4 other key phosphoproteins, and this increase can be blocked by co-treatment with AXLi using R428 (Fig. S5D). After 3h treatment with either MPi or MEKi, we found that compensatory increase in p-JNK can be eliminated by siRNA targeting either AXL, or to a lesser extent, MET (Fig. 5A; siRNA validation Fig. S5E). By 24h post-treatment, compensatory increases in p-cJUN and p-Akt are likewise blocked by co-treatment with AXLi using R428 (Fig. 5B–C). Compensatory JNK signaling in response to MAPKi leads to a synergistic response to dual JNK and MAPK inhibition (35), and our data suggest that sheddase activity is a key feedback component driving this synergistic interaction. To test this model, we measured the degree of drug synergy as a response to combination MAPKi and JNKi treatment, in the presence or absence of MPi, and found that the degree of synergy between JNKi and MAPKi is strongly reduced in the absence of metalloproteinase activity (Fig. S5F–G).
Figure 5. MPi increases proliferation in an AXL-dependent manner and blocks MEKi/AXLi synergy.
A) AXL and MET siRNAs reduce compensatory p-JNK signaling, measured following 3h drug treatment and 48h siRNA treatment in MDA-MB231 (*p=0.014, pooled t-test, n=2 reps/cond; siRNA validation, Fig. S5E). B–C) Representative western blots (B) and quantification (C) show enhanced p-Akt and p-cJUN following 24h treatment with either MPi or MEKi, which is blocked in the presence of AXLi (*p<0.05, two-tailed t-test, n≥3). D) MPi using BB94 enhances proliferation in the absence of R428, but decreases proliferation when R428 is present (p=0.02; n≥4, two-tailed t-test). E) BB94 increases mitotic index, measured by FACS cell cycle analysis, which is blocked by AXLi using R428 (bars denote p<0.05; n≥4 total reps; pooled two-tailed t-test). F) Computed simulations to illustrate how increasing the Loewe synergy term α theoretically affects cell survival (shown by heatmaps) in response to varying combinations of AXLi and MEKi co-treatment. G) Cells were treated with varying combinations of MEKi and AXLi for 72h, in the presence or absence of BB94, after which cell count was measured (shown as numbers and heatmap as a fraction of the max cell count for each plate). H) Data from combination drug treatments (G) were fit to a model of Loewe synergy, yielding the synergy interaction term α, which significantly decreased with MPi (*p<0.05, two-tailed jackknife test, n=20 measurements over n=2 reps).
MPi drives AXL-dependent cell proliferation and blocks response synergy to dual MEKi and AXLi
We next evaluated the direct effect of MPi on cell proliferation. MPi using BB94 caused a dose-dependent increase in cell growth in the absence of AXLi; however, in the presence of AXLi using R428, MPi actually caused a dose-dependent decrease in cell growth (Fig. 5D). Furthermore, MPi increased the mitotic index of two TNBC cell lines, and this increase was also blocked by R428 (Fig. 5E). Next, we directly tested the role of proteolytic activity in mediating MEKi/AXLi synergy. We treated cells with combinations of MEKi and AXLi in the presence or absence of MPi (using BB94), and then fit the resulting cell growth measurements to a computational model of Loewe synergy, as done above with MAPKi and JNKi. This model fits the response data to a parameter α that quantifies the degree to which drugs act in a synergistic, super-additive manner where the effect of both drugs combined is greater than what would be expected if the effects were independent and additive (simulated in Fig. 5F). This analysis revealed that synergistic interactions between MEKi and AXLi were substantially reduced in the absence of metalloproteinase activity (Fig. 5G–H), suggesting that proteolytic activity significantly contributes to AXL-dependent MEKi resistance.
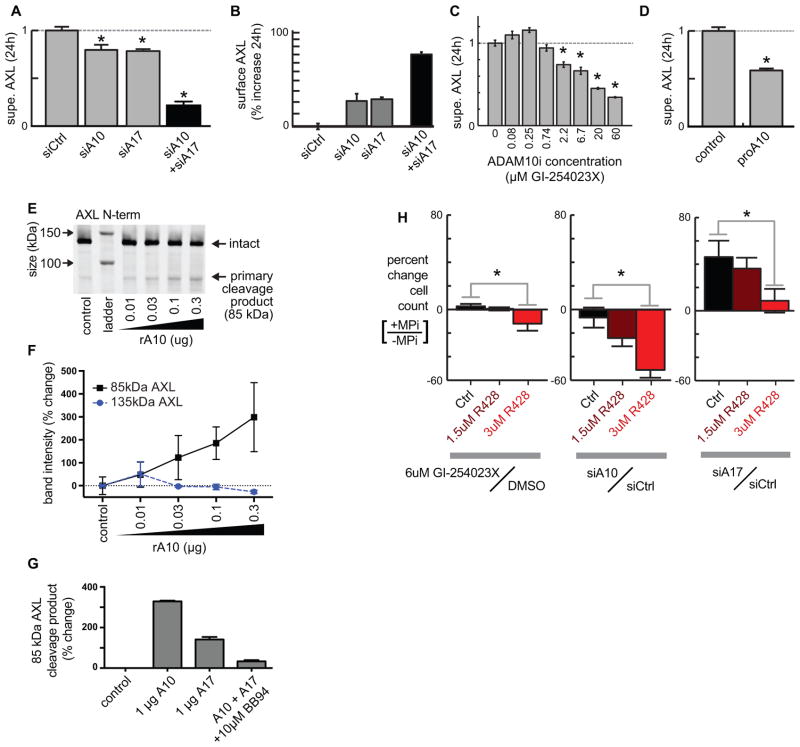
ADAM10 and ADAM17 proteolytically shed AXL to downregulate mitogenic signaling activity
We next investigated which sheddases cleave AXL directly, focusing on ADAM17 as a principal sheddase and ADAM10 because AXL and MET shedding are closely correlated throughout our data, and MET is a previously-known ADAM10 substrate (15, 21, 36). ADAM17 and ADAM10 knockdown using pooled small interfering RNA (siRNA; validation, Fig. S6A) decreased supernatant AXL accumulation (Fig. 6A) and enhanced its levels on the cell surface in both MDA-MB231 (Fig. 6B) and MDA-MB157 cell lines (Fig. S6B). Combined ADAM10 and ADAM17 knockdown yielded even greater effects, underscoring a role for both (Fig. 6A–B). Stable short hairpin RNA (shRNA) knockdown of either ADAM10 or ADAM17 also caused AXL accumulation on the surface of LOX-IMVI melanoma cells (Fig. S6C–D) and decreased supernatant AXL accumulation (Fig. S6E). ADAM10 inhibition with the ADAM10-selective inhibitor GI-254023X caused a dose-dependent decrease in supernatant AXL ectodomain (Fig. 6C). Similarly, treatment with the specific ADAM10 inhibitor proA10 (37) reduced supernatant AXL (Fig. 6D).
Figure 6. ADAM10 and ADAM17 regulate cell surface AXL and alter cell growth in an AXL-dependent manner.
A–B) ADAM-targeted siRNAs (validation: Fig. S6A) reduce supernatant AXL (A; ELISA measurement) and increase surface AXL (B; live-cell immunostaining) compared to non-targeted siRNA (*p<0.05; n≥2) in MDA-MB231. C) The specific ADAM10 inhibitor GI-254023X decreases supernatant AXL from MDA-MB231 (*p<0.05; n=3). D) The specific ADAM10 inhibitor proA10 reduces supernatant AXL compared to the vehicle control, measured by ELISA using MDA-MB231 (*p<0.05; n≥2). E–F) Recombinant ADAM10 cleaves recombinant AXL in a dose-response fashion, shown by AXL immunoblot (E) and corresponding quantification (F; *p<0.05; two-tailed t-test; n=2). G) Recombinant ADAM10 and ADAM17 cleave recombinant AXL, and cleavage is blocked by MPi using BB94 (n=2). H) In MDA-MB231 cells, ADAM10 inhibition or knockdown by siRNA only decreases proliferation in the presence of AXLi using R428. ADAM17 siRNA only induces proliferation in the absence of AXLi. SiRNA results were measured 24h after AXLi, 72h after transfection, and GI-254023X results were measured after 72h treatment (*p<0.05; n≥4; two-tailed t-test).
We examined the ability of recombinant ADAM10 to cleave purified AXL protein, and show by western blot that ADAM10 cleaves recombinant AXL (Fig. 6E–F) in a dose-dependent manner into ectodomain fragments of roughly the same size (~85kDa) as found in cell supernatant (Fig. 4H). As a control, we confirmed these cleavage products were reduced in the presence of MPi (Fig. 6G). We also confirmed the ability of ADAM17 to cleave both recombinant AXL (Fig. 6G) and full-length AXL that had been immunopurified from cell lysate (Fig. S6F–G).
Specific ADAM10 inhibition also affected cell proliferation in an AXL-dependent manner. Treatment with either GI-254023X or ADAM10 siRNA knockdown had minimal impact on cell growth in the absence of AXLi (Fig. 6H), and stable ADAM10 knockdown using shRNA had minimal impact on the growth of LOX-IMVI xenograft tumors (Fig. S6H). However, when AXL signaling was inhibited by R428, ADAM10 inhibition was effective in reducing cell growth (Fig. 6H). Similar trends were observed with ADAM17 siRNA knockdown (Fig. 6H). These combined results are consistent with BB94 effects on proliferation (Fig. 5D–E) and demonstrate AXL upregulation as significantly counteracting the anti-proliferative capacity of metalloproteinase inhibitors.
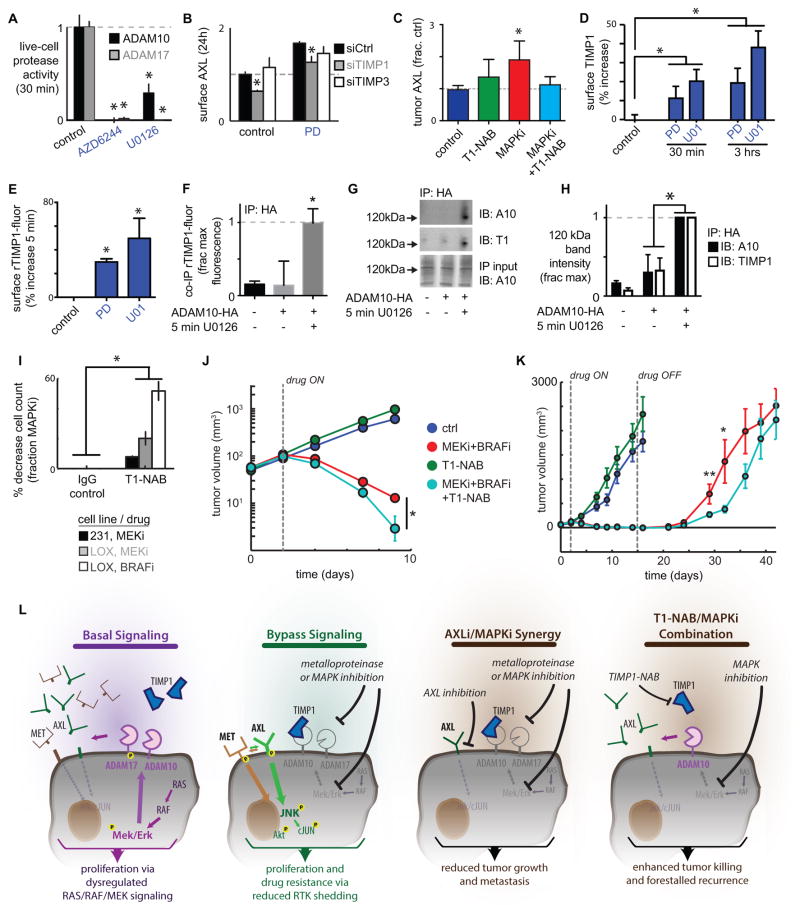
MEKi dynamically enhances cell surface TIMP1 association with ADAM10, thus reducing ADAM10 activity and causing drug resistance
We next investigated the mechanism through which MAPKi decreases AXL shedding. MEKi significantly reduced ADAM10 and ADAM17 catalytic activities in a live-cell assay (38) (Fig. 7A), without affecting their cell surface levels (Fig. S7A), suggesting direct regulation of the protease activities themselves. As further evidence of direct protease regulation, ADAM17 phosphorylation at a site previously associated with activity (33) decreased with MEKi (Fig. S7B–C). The regulated binding of ADAMs with Tissue Inhibitor of Metalloproteinases (TIMPs) including TIMP1 and TIMP3 has been implicated as a mechanism for governing protease activity, and we next tested the role of TIMPs on AXL shedding. SiRNA knockdown of TIMP1 but not TIMP3 mitigated MEKi-induced AXL accumulation on the surface (Fig. 7B; knockdown validation, Fig. S7D). In a melanoma xenograft model, we tested the in vivo effect of TIMP1 on AXL shedding by using a TIMP1 neutralizing antibody (T1-NAB) that binds TIMP1 and competitively prevents it from associating with proteases. Co-treatment with T1-NAB blocked tumor AXL accumulation after MAPKi treatment (Fig. 7C). Given that knockdown of TIMP3, which preferentially inhibits ADAM17, had no effect on AXL shedding, and because TIMP1 preferentially inhibits ADAM10, our data suggest a more prominent role for TIMP1 and ADAM10 regulating MEKi-driven AXL shedding.
Figure 7. MEKi reduces sheddase activity via increased TIMP1 association, and TIMP1 neutralization enhances MAPKi efficacy.
A) MEKi reduces ADAM10 and ADAM17 catalytic activities in MDA-MB231, directly measured using the live-cell PrAMA assay (*p<0.05; n=4). B) Live-cell immunostaining shows knockdown of TIMP1 but not TIMP3 reduces surface AXL in MDA-MB231, 24h after PD325901 treatment and 72h after transfection (*p<0.05; n=3; knockdown validation: Fig. S7D). C) Co-treatment with a TIMP1 neutralization antibody (T1-NAB) blocks tumor AXL accumulation following BRAFi/MEKi treatment in the LOX-IMVI melanoma xenograft model, shown by immunofluorescence quantification (*p=0.02; n≥2). D) Live-cell immunostaining shows MEKi increases surface TIMP1 in MDA-MB231. E) Flow cytometry shows 5 min MEKi treatment increases rTIMP1-fluor binding to the cell surface (*p<0.05; n=4). F) Significant co-IP of ADAM10-HA and rTIMP-fluor is only detected in bulk anti-HA IP lysate from cells transfected with ADAM10-HA and treated with U0126 (*p<0.05; n=3; pooled two-tailed t-test). G–H) The 120kDa ADAM10 dimerization band (39) and co-IP with rTIMP1 increases with 5 min MEKi (*p<0.05; n>3). I) 24h pre-treatment with a TIMP1 neutralization antibody (T1-NAB) followed by co-treatment with PD325901 (MEKi) or vemurafenib (BRAFi) lead to enhanced reduction in cell count at 72h, normalized to the effect-size of BRAFi or MEKi alone (*p=0.03, pooled two-tailed t-test, n=18 total reps). J–K) T1-NAB co-treatment synergistically increases BRAFi/MEKi efficacy in the LOX-IMVI xenograft model by enhancing initial tumor shrinkage (J; p=0.014, two-way ANOVA interaction term, total n=46) and delaying tumor recurrence (K; p=0.04, two-tailed t-test, n≥8 per group). L) Overview schematic of RTK shedding as a mechanism of MAPKi resistance.
We found that MEKi continually enhanced the binding of TIMP1 to the cell surface from 30 min (Fig. 7D) to 24h (Fig. S7E). In contrast, TIMP1 supernatant levels actually declined after MEKi (Fig. S7F), indicating enhanced TIMP1 surface levels do not simply reflect enhanced TIMP1 secretion. We thus hypothesized that MEKi induced a rapid change on the cell surface that enhanced TIMP1 binding. To test this, we first treated cells for 5 min with MEKi, rinsed, and incubated cells on ice with a fixed amount of fluorescently-tagged recombinant TIMP1 (rTIMP1-fluor). MEKi enhanced rTIMP1-fluor binding to the cell surface by nearly 50% (Fig. 7E), supporting the idea that MEKi treatment leads to increased accumulation of TIMP1 on the cell surface.
We next evaluated the role of ADAM10 in regulating MEKi-driven TIMP1 cell surface accumulation. Using siRNA, we found that TIMP1 accumulation on the cell surface was dependent upon ADAM10 expression (Fig. S7G; knockdown validation S6A). To directly observe interaction between TIMP1 and ADAM10, we over-expressed transgenic HA-tagged ADAM10 by 2-fold (Fig. S7H), treated cells for 5 min with MEKi, rinsed, and again incubated cells on ice with a fixed amount of rTIMP1-fluor. Proteins associating with ADAM10 were co-immunopurified (co-IP) using EGS crosslinking and anti-HA agarose resin. To sensitively detect rTIMP1-fluor co-IP, bulk immunoprecipitate was analyzed by fluorometry and found to exhibit detectable rTIMP1 fluorescence only after MEKi (Fig. 7F), further suggesting that MEKi induces ADAM10-driven cell surface TIMP1 accumulation.
We then investigated how MEKi dynamically influences interactions between ADAM10 and TIMP1. ADAM10 and ADAM17 dimerization has been associated with direct activity regulation (39, 40), and we hypothesized that MEKi elicited increased ADAM10 dimerization and association with TIMP1. Previously, immunoblots of EGS-crosslinked lysate have shown ADAM10 forms a dimerization band at roughly 120kDa (39), and here we found that this band increases with MEKi and co-immunostains for TIMP1 (Fig. 7G–H), indicating TIMP1-ADAM10 interaction. Thus, MEKi broadly reduces proteolytic shedding by enhanced TIMP1 association with ADAM10, negatively regulating its activity.
We next examined the role of TIMP1 in influencing the therapeutic response to MAPKi. We co-treated cells with T1-NAB and either MEKi or BRAFi. While T1-NAB had no detectable effect on cell growth in the absence of MAPKi, it enhanced MAPKi sensitivity by up to 50% (Fig. 7I; Fig. S7I). For more clinical relevance, we next examined the effect of T1-NAB co-treatment in a melanoma xenograft model undergoing a combined MEKi/BRAFi inhibitor regimen similar to those used in the clinic. T1-NAB treatment alone did not significantly reduce tumor growth (Fig. 7J; p=0.56, two-way ANOVA independent effect term, total n=46). In contrast, T1-NAB significantly enhanced the ability of MEKi/BRAFi to reduce tumor size by an additional 70% (Fig. 7J; note log-scale). Furthermore, T1-NAB extended the time to tumor recurrence after MEKi/BRAFi treatment had ended (Fig. 7K). Recurrent outgrown tumors from this study did not exhibit altered AXL levels (Fig. S7J), which is in contrast to tumors analyzed while on drug treatment (Fig. 7C), and suggests that effects on AXL shedding largely reverted during the roughly 3 weeks after treatment ended. Nevertheless, these results provide evidence that reduced proteolytic AXL shedding via TIMP1 association leads to blunted MAPKi efficacy.
Discussion
We have elucidated a new targetable mechanism of bypass cancer cell signaling with implications for the design and monitoring of cancer therapies (Fig. 7L). Inhibition of multiple signaling pathways, particularly ERK signaling through MEK1/2, reduces proteolytic RTK shedding and leads to enhanced mitogenic signaling through bypass kinase pathways including JNK. Numerous examples of increased RTK signaling activity have been observed following targeted kinase inhibitor treatment, often with little mechanistic explanation outside of transcriptional upregulation (1, 4, 5, 41). MEK and PI3K kinase inhibition have been shown to enhance the signaling of sheddase substrates including HER2 (3, 5) and AXL (1, 42), and we show here that inhibition of MEK, BRAF, and to some extent PI3K, p38, and JNK (as in Fig. 4B) reduce RTK shedding. The direct inhibition of RTKs also gives rise to bypass signaling. For example, enhanced AXL signaling mediates resistance to the EGFR/HER2 inhibitor lapatinib, even in the absence of AXL transcriptional upregulation (4). Although transcriptional and chromosomal reprogramming affect bypass signaling (2), these processes often fail to fully explain signaling network dynamics following drug treatment; this is especially true for AXL, which often exhibits little transcriptional upregulation despite sharply enhanced activity (1, 4). Here, we offer reduced proteolytic RTK shedding as a likely explanatory mechanism.
The promiscuous nature of metalloproteinases has made it difficult to anticipate the overall effects of perturbing their activities, especially in response to kinase inhibition (17, 19). Ectodomain shedding can be regulated through substrate-specific pathways (43), and in this work we find that substrate-specific regulation, for example through differential substrate accumulation on the cell-surface, can only be explained partially by transcriptional regulation (Fig. S4G). Nonetheless, substantial evidence suggests that MEKi inhibits ADAM catalytic activities themselves (Fig. 7), and we highlight an unappreciated role for TIMP1 association in this context. Consequently, MEKi essentially leads to a global reduction of ectodomain shedding across a wide range of substrates (Fig. 1; Fig. 4A). Sheddase substrates, such as EGF-ligands, have been studied for their role in autocrine growth factor signaling and have motivated protease inhibitor development (particularly targeting ADAM10 and ADAM17) for clinical use (44). This work suggests that past metalloproteinase inhibitor clinical trials may have failed partly due to unanticipated compensatory signaling feedback from unshed RTKs.
In general, we find that AXL and MET shedding downregulate signaling activity by limiting the accumulation of full-length, signaling-competent RTK on the cell surface. Ligand-dependent receptor activation is an important signaling feature, particularly in the context of receptor shedding (15), and is relevant for future studies. Nonetheless, AXL and the other RTKs studied here exhibit significant ligand-independent activity (34, 41), which amplifies as they accumulate on the cell surface following protease downregulation. Clinically, AXL upregulation often occurs without apparent dysregulation of its ligand Gas6, and roughly half of observed AXL bypass signaling acts independently of Gas6 in drug-resistant cell lines (41). Receptor shedding also results in the generation of inhibitory “decoy” receptors that both compete for binding of free extracellular ligand and block cell-surface dimerization between signaling-competent receptors; decoy functions have been therapeutically exploited for multiple receptors, including MET (45) and AXL (46). Here we show that kinase inhibition simultaneously increases full-length RTK on the cell surface while decreasing decoy receptor levels in the extracellular supernatant or in circulation.
Diminished RTK shedding likely complements other bypass signaling mechanisms. Many RTKs reported as transcriptionally upregulated in response to kinase inhibition are themselves sheddase substrates, including PDGFRβ (1, 8), VEGFR2 (1), and CD44 (47). In the context of MAPKi, AXL repeatedly surfaces in genome-wide screens as a top candidate for rescuing drug sensitivity upon transgenic overexpression (48). Reduced RTK shedding has the potential to amplify the effects of transcriptional upregulation by increasing the fraction of total RTK that remains intact on the cell surface. Within the tumor microenvironment, stromal-derived growth factors, cytokines, and extracellular matrix contribute to drug resistance (20, 49–52). Here we find that receptors impacted by reduced RTK shedding are also implicated in tumor-stroma ligand interactions, with a prominent example being MET activation by stromal-derived hepatocyte growth factor (HGF) (51). Of note, extracellular HGF and Gas6 release occurs through secretion rather than metalloproteinase shedding. Consequently, reduced RTK shedding has the capacity to amplify pro-survival and pro-metastatic tumor-stroma interactions, and previous work has demonstrated RTK shedding as a modifier of ligand-dependent receptor activation (15).
The ability to assess RTK shedding in cancer patients using relevant biomarkers is essential for efficient clinical translation. A substantial proportion of molecular cancer diagnostics focuses on gene expression and/or genetic mutation analysis of tumors, is dependent on the presence of an accessible tumor, and is limited by factors such as tumor heterogeneity and post-translational regulation. We present RTK proteolysis as a mechanistic explanation for the discordance between gene expression and signaling activity, and provide evidence that receptor shedding can be non-invasively monitored in clinical samples following drug treatment. Although promising, the small cohort of patient samples examined here should be expanded to a study of both larger cohorts and a broader panel of sheddase substrates such as adhesion receptors, cytokines, and cytokine receptors. Importantly, RTK proteolysis may also reflect drug toxicity in addition to drug efficacy, and multiple physiological factors may influence altered circulating RTK levels. For instance, foretinib has been observed to cause elevated circulating MET in patients (53), which may be explained by its common liver toxicity (high AST/ALT) and the corresponding association of hepatotoxicity with elevated MET shedding (54). Changes may also be related to altered gene expression and known vascular effects of foretinib including hypertension. Nonetheless, the value of a biomarker that can predict resistance and shed light on a next potential line of therapy cannot be underestimated. Ultimately, circulating RTKs hold the potential to stand alone and to complement other diagnostic biomarkers in guiding targeted combination therapies, monitoring drug response, and non-invasively detecting the emergence of drug resistance.
Methods
Unless otherwise stated, reported replicates are from unique biological samples, statistical tests used the two-sided Student’s t-test for significance, and mean values are reported with error bars denoting standard error of the mean. With some explicitly stated exceptions, experiments used 10μM BB94, 3μM R428, 3μM PD325901, 5μM U0126, 10μM AZD6244, 15μM MP470, and 4μM pro-ADAM10. Please see Supplemental Experimental Procedures for further information regarding materials, experimental details, and computational analysis methods.
Cell Lines
The following cell lines were purchased directly from commercial or governmental repositories and immediately used for experiments in this manuscript: MDA-MB231 (ATCC, Jun. 2012); LOX-IMVI (NCI-DCTD repository, Mar. 2014); SUM149 and SUM102 (Asterand Biorepository, Nov. 2011); HCC827 (ATCC, Jan. 2012); BT549 (ATCC, Apr. 2010); BT20 (ATCC, Jan. 2012). Additional cell lines were procured from Massachusetts Institute of Technology (MIT), Harvard Medical School (HMS), and University of Michigan collaborating lab banks: MA2 and A375 cell lines (Richard Hynes, MIT, Jan. 2013); BT474, Hs578T, HCC38, MDAMB436, MDAMB468 (Mike Yaffe, MIT, Apr. 2011); A172 (Leona Samson, MIT, Jan. 2013); U87 (Dane Wittrup, MIT, May 2011); A549 (Linda Griffith, MIT, May 2011); SUM159 (Joan Brugge, HMS, Jan. 2011); LM2 (55) (Richard Hynes, MIT, Jan. 2013); SUM1315 (Stephen Ethier, Univ. Michigan, Dec. 2009). Cell lines were routinely tested for mycoplasma (Lonza MycoAlert) within 3 months of use and were not authenticated. All cell culture was performed according to the manufacturer’s guidelines.
Melanoma Patient Samples
Patients with metastatic melanoma containing BRAFV600E mutation (confirmed by genotyping) were enrolled on clinical trials for treatment with a BRAF inhibitor or combined BRAF + MEK inhibitor (dabrafenib 150mg b.i.d., trametinib 2mg q.d.) at Massachusetts General Hospital and were consented for blood and tissue acquisition per IRB-approved protocol, conducted in accordance with the Declaration of Helsinki. All patients (or legal representatives) gave written informed consent before enrollment. Blood was collected and tumor biopsies were performed pre-treatment (day 0), 10–14 days on treatment, and/or at time of progression if applicable. Multiple on treatment blood samples were collected over the course of therapy as available. Plasma was isolated immediately from blood samples using BD Vacutainer CPT tube with Sodium Citrate (BD 362761). Formalin-fixed tissue from each tissue biopsy was analyzed to confirm that viable tumor was present via hematoxylin and eosin (H&E) staining. Clinical response was assessed by RECIST (See supplemental experimental procedures).
In vivo tumor growth and metastasis assays
All animal experiments and husbandry were approved by the MIT Division of Comparative Medicine in accord with guidelines of the MIT-IACUC. For orthotopic mammary transplant assays, 6-week-old female NOD/SCID-gamma mice (JAX) were anesthetized by i.p. injection of 125–250 mg/kg body weight of Avertin (reconstituted in PBS), followed by i.p. injection of 100 μL of 12 μg/mL buprenorphine for analgesia. A small incision was made on the right flank, and 250,000 MDA-MB231-LM2 cells in 25 μL of HBSS were injected into the right #4 fat pad using a 25-μL Hamilton syringe. Mice received three additional i.p. injections of 100 μL of 12 μg/mL buprenorphine at 12h intervals following the surgery. Initial sample size was chosen based on previously published experiments with MDA-MB231-LM2 xenograft models (55), as well as previously published data with the MEK and AXL drugs of interest (56, 57). 20 days post-surgery, when tumor size was palpable, mice were ranked by tumor-size and semi-randomly divided into four groups of equal distribution in tumor size. Groups received one of four different drug treatments once daily for 21 days by oral gavage: vehicle (10% DMSO + 0.5% methylcellulose + 0.2% tween-80 in water), Axl inhibitor R428 at 30 mg/kg, PD0325901 at 1 mg/kg or a combination of both R428 at 30 mg/kg and PD0325901 at 1 mg/kg. Animals were sacrificed at the predetermined time of 21 days following initiation of drug treatment.
For the xenograft melanoma experiment, 7 week old female athymic nude mice (Taconic) were injected with 1×106 LOX-IMVI cells in 1:1 Matrigel:HBSS subcutaneously in each flank (58). 7 days post-cell injection, tumors were measured by calipers and mice were ranked by tumor size and semi-randomly divided into 6 groups of equal distribution in tumor size, with 10 mice per group. Groups received one of four different drug treatments once daily for 14 days by oral gavage: vehicle (10% DMSO + 0.5% methylcellulose + 0.2% tween-80 in water), Axl inhibitor R428 at 30 mg/kg, PD0325901 at 1 mg/kg and Vemurafinib at 10mg/kg or a combination of R428 at 30 mg/kg, PD0325901 at 1 mg/kg and Vemurafinib at 10mg/kg. To study the role TIMP1 in resistance to MAPKi, mice were treated with a TIMP1 neutralizing AB (AbD Serotec/Biorad) at 32 mg/kg IP (59) daily for 3 days prior to starting drug treatment and then every second day during drug treatment. Mice were sacrificed when overall tumor burden reached more than 3cm in diameter. Tumor size measurements, tissue processing, staining of tumors and exclusion criteria can be found in supplemental experimental procedures.
Supplementary Material
Statement of Significance.
Genetic, epigenetic, and gene expression alterations often fail to explain adaptive drug resistance in cancer. This work presents a novel post-translational mechanism of such resistance: kinase inhibitors, particularly targeting MAPK signaling, increase tumor cell surface receptor levels due to widely reduced proteolysis, allowing tumor signaling to circumvent intended drug action.
Acknowledgments
Grant Support: This work was supported by NIH grants U54-CA112967 and R01-CA096504 to D.A.L., and DoD award W81XWH-13-1-0031 to M.J.O.
We acknowledge support from the MIT Koch Institute Swanson Biotechnology Center and MIT BioMicroCenter core facilities. The authors express appreciation to Mike Hemann, Kevin Janes, and Katherine Yang for very helpful comments.
Footnotes
Accession Numbers: RNA microarray expression data can be found in the GEO repository under accession number GSE77868.
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
Author Contributions
M.A.M. helped design, perform, and analyze all experiments besides live animal work and clinical sample collection. M.J.O. measured melanoma patient samples; designed, performed, and analyzed the mouse xenograft experiments; performed in vitro cell cytotoxicity assays; and performed western blots. R.J.S., D.T.F, and K.T.F. designed and collected the melanoma patient samples, and performed immunohistochemistry. A.S.M. and S.J.W. performed phospho-signaling experiments, western blots, and soluble RTK measurements. J.T. helped with the mouse xenograft experiments. H.I., H.L., and R.W. helped design and perform exosome experiments. L.G.G. and F.B.G. were involved in the study design, data interpretation, and manuscript writing. M.A.M., M.J.O., and D.A.L. designed the study, analyzed the data, and wrote the paper. All authors discussed the results and commented on the manuscript.
References
- 1.Duncan JS, Whittle MC, Nakamura K, Abell AN, Midland AA, Zawistowski JS, et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell. 2012;149:307–21. doi: 10.1016/j.cell.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–7. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turke AB, Song Y, Costa C, Cook R, Arteaga CL, Asara JM, et al. MEK inhibition leads to PI3K/AKT activation by relieving a negative feedback on ERBB receptors. Cancer Res. 2012;72:3228–37. doi: 10.1158/0008-5472.CAN-11-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu L, Greger J, Shi H, Liu Y, Greshock J, Annan R, et al. Novel mechanism of lapatinib resistance in HER2-positive breast tumor cells: activation of AXL. Cancer Res. 2009;69:6871–8. doi: 10.1158/0008-5472.CAN-08-4490. [DOI] [PubMed] [Google Scholar]
- 5.Serra V, Scaltriti M, Prudkin L, Eichhorn PJ, Ibrahim YH, Chandarlapaty S, et al. PI3K inhibition results in enhanced HER signaling and acquired ERK dependency in HER2-overexpressing breast cancer. Oncogene. 2011;30:2547–57. doi: 10.1038/onc.2010.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Allen EM, Wagle N, Sucker A, Treacy DJ, Johannessen CM, Goetz EM, et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 2014;4:94–109. doi: 10.1158/2159-8290.CD-13-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turke AB, Zejnullahu K, Wu YL, Song Y, Dias-Santagata D, Lifshits E, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akhavan D, Pourzia AL, Nourian AA, Williams KJ, Nathanson D, Babic I, et al. De-repression of PDGFRβ transcription promotes acquired resistance to EGFR tyrosine kinase inhibitors in glioblastoma patients. Cancer Discov. 2013;3:534–47. doi: 10.1158/2159-8290.CD-12-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zardavas D, Baselga J, Piccart M. Emerging targeted agents in metastatic breast cancer. Nat Rev Clin Oncol. 2013;10:191–210. doi: 10.1038/nrclinonc.2013.29. [DOI] [PubMed] [Google Scholar]
- 10.Bartholomeusz C, Gonzalez-Angulo AM, Liu P, Hayashi N, Lluch A, Ferrer-Lozano J, et al. High ERK protein expression levels correlate with shorter survival in triple-negative breast cancer patients. Oncologist. 2012;17:766–74. doi: 10.1634/theoncologist.2011-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sullivan RJ, Flaherty KT. Resistance to BRAF-targeted therapy in melanoma. Eur J Cancer. 2013;49:1297–304. doi: 10.1016/j.ejca.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 12.Edwards DR, Handsley MM, Pennington CJ. The ADAM metalloproteinases. Mol Aspects Med. 2008;29:258–89. doi: 10.1016/j.mam.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGowan PM, Mullooly M, Caiazza F, Sukor S, Madden SF, Maguire AA, et al. ADAM-17: a novel therapeutic target for triple negative breast cancer. Ann Oncol. 2013;24:362–9. doi: 10.1093/annonc/mds279. [DOI] [PubMed] [Google Scholar]
- 14.Lee SB, Schramme A, Doberstein K, Dummer R, Abdel-Bakky MS, Keller S, et al. ADAM10 is upregulated in melanoma metastasis compared with primary melanoma. J Invest Dermatol. 2010;130:763–73. doi: 10.1038/jid.2009.335. [DOI] [PubMed] [Google Scholar]
- 15.Miller MA, Meyer AS, Beste MT, Lasisi Z, Reddy S, Jeng KW, et al. ADAM-10 and -17 regulate endometriotic cell migration via concerted ligand and receptor shedding feedback on kinase signaling. Proc Natl Acad Sci U S A. 2013;110:E2074–83. doi: 10.1073/pnas.1222387110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Witters L, Scherle P, Friedman S, Fridman J, Caulder E, Newton R, et al. Synergistic inhibition with a dual epidermal growth factor receptor/HER-2/neu tyrosine kinase inhibitor and a disintegrin and metalloprotease inhibitor. Cancer Res. 2008;68:7083–9. doi: 10.1158/0008-5472.CAN-08-0739. [DOI] [PubMed] [Google Scholar]
- 17.Gooz M. ADAM-17: the enzyme that does it all. Crit Rev Biochem Mol Biol. 2010;45:146–69. doi: 10.3109/10409231003628015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newton RC, Bradley EC, Levy RS, Doval D, Bondarle S, Sahoo TP, et al. Clinical benefit of INCB7839, a potent and selective ADAM inhibitor, in combination with trastuzumab in patients with metastatic HER2+ breast cancer. J Clin Oncol. 2010;28 abstr 3025. [Google Scholar]
- 19.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487:505–9. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schelter F, Grandl M, Seubert B, Schaten S, Hauser S, Gerg M, et al. Tumor cell-derived Timp-1 is necessary for maintaining metastasis-promoting Met-signaling via inhibition of Adam-10. Clin Exp Metastasis. 2011;28:793–802. doi: 10.1007/s10585-011-9410-z. [DOI] [PubMed] [Google Scholar]
- 22.Feldinger K, Generali D, Kramer-Marek G, Gijsen M, Ng TB, Wong JH, et al. ADAM10 mediates trastuzumab resistance and is correlated with survival in HER2 positive breast cancer. Oncotarget. 2014;5:6633–46. doi: 10.18632/oncotarget.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takahashi R, Hirata H, Tachibana I, Shimosegawa E, Inoue A, Nagatomo I, et al. Early [18F]fluorodeoxyglucose positron emission tomography at two days of gefitinib treatment predicts clinical outcome in patients with adenocarcinoma of the lung. Clin Cancer Res. 2012;18:220–8. doi: 10.1158/1078-0432.CCR-11-0868. [DOI] [PubMed] [Google Scholar]
- 24.Im H, Shao H, Park YI, Peterson VM, Castro CM, Weissleder R, et al. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat Biotechnol. 2014;32:490–5. doi: 10.1038/nbt.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–65. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 26.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asai M, Hattori C, Szabó B, Sasagawa N, Maruyama K, Tanuma S, et al. Putative function of ADAM9, ADAM10, and ADAM17 as APP alpha-secretase. Biochem Biophys Res Commun. 2003;301:231–5. doi: 10.1016/s0006-291x(02)02999-6. [DOI] [PubMed] [Google Scholar]
- 28.Guo L, Eisenman JR, Mahimkar RM, Peschon JJ, Paxton RJ, Black RA, et al. A proteomic approach for the identification of cell-surface proteins shed by metalloproteases. Mol Cell Proteomics. 2002;1:30–6. doi: 10.1074/mcp.m100020-mcp200. [DOI] [PubMed] [Google Scholar]
- 29.Yang WS, Yu H, Kim JJ, Lee MJ, Park SK. Vitamin D-induced ectodomain shedding of TNF receptor 1 as a nongenomic action: D3 vs D2 derivatives. J Steroid Biochem Mol Biol. 2016;155:18–25. doi: 10.1016/j.jsbmb.2015.09.019. [DOI] [PubMed] [Google Scholar]
- 30.Lemjabbar H, Basbaum C. Platelet-activating factor receptor and ADAM10 mediate responses to Staphylococcus aureus in epithelial cells. Nat Med. 2002;8:41–6. doi: 10.1038/nm0102-41. [DOI] [PubMed] [Google Scholar]
- 31.Yan Y, Shirakabe K, Werb Z. The metalloprotease Kuzbanian (ADAM10) mediates the transactivation of EGF receptor by G protein-coupled receptors. J Cell Biol. 2002;158:221–6. doi: 10.1083/jcb.200112026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kasina S, Scherle PA, Hall CL, Macoska JA. ADAM-mediated amphiregulin shedding and EGFR transactivation. Cell Prolif. 2009;42:799–812. doi: 10.1111/j.1365-2184.2009.00645.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu P, Derynck R. Direct activation of TACE-mediated ectodomain shedding by p38 MAP kinase regulates EGF receptor-dependent cell proliferation. Mol Cell. 2010;37:551–66. doi: 10.1016/j.molcel.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyer AS, Miller MA, Gertler FB, Lauffenburger DA. The receptor AXL diversifies EGFR signaling and limits the response to EGFR-targeted inhibitors in triple-negative breast cancer cells. Sci Signal. 2013;6:ra66. doi: 10.1126/scisignal.2004155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fallahi-Sichani M, Moerke NJ, Niepel M, Zhang T, Gray NS, Sorger PK. Systematic analysis of BRAF(V600E) melanomas reveals a role for JNK/c-Jun pathway in adaptive resistance to drug-induced apoptosis. Mol Syst Biol. 2015;11:797. doi: 10.15252/msb.20145877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schelter F, Kobuch J, Moss ML, Becherer JD, Comoglio PM, Boccaccio C, et al. A disintegrin and metalloproteinase-10 (ADAM-10) mediates DN30 antibody-induced shedding of the met surface receptor. J Biol Chem. 2010;285:26335–40. doi: 10.1074/jbc.M110.106435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moss ML, Bomar M, Liu Q, Sage H, Dempsey P, Lenhart PM, et al. The ADAM10 prodomain is a specific inhibitor of ADAM10 proteolytic activity and inhibits cellular shedding events. J Biol Chem. 2007;282:35712–21. doi: 10.1074/jbc.M703231200. [DOI] [PubMed] [Google Scholar]
- 38.Miller MA, Barkal L, Jeng K, Herrlich A, Moss M, Griffith LG, et al. Proteolytic Activity Matrix Analysis (PrAMA) for simultaneous determination of multiple protease activities. Integr Biol (Camb) 2011;3:422–38. doi: 10.1039/c0ib00083c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu P, Liu J, Sakaki-Yumoto M, Derynck R. TACE activation by MAPK-mediated regulation of cell surface dimerization and TIMP3 association. Sci Signal. 2012;5:ra34. doi: 10.1126/scisignal.2002689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deng W, Cho S, Su PC, Berger BW, Li R. Membrane-enabled dimerization of the intrinsically disordered cytoplasmic domain of ADAM10. Proc Natl Acad Sci U S A. 2014;111:15987–92. doi: 10.1073/pnas.1409354111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Z, Lee JC, Lin L, Olivas V, Au V, LaFramboise T, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet. 2012;44:852–60. doi: 10.1038/ng.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Byers LA, Diao L, Wang J, Saintigny P, Girard L, Peyton M, et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin Cancer Res. 2013;19:279–90. doi: 10.1158/1078-0432.CCR-12-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dang M, Armbruster N, Miller MA, Cermeno E, Hartmann M, Bell GW, et al. Regulated ADAM17-dependent EGF family ligand release by substrate-selecting signaling pathways. Proc Natl Acad Sci U S A. 2013;110:9776–81. doi: 10.1073/pnas.1307478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duffy MJ, Mullooly M, O’Donovan N, Sukor S, Crown J, Pierce A, et al. The ADAMs family of proteases: new biomarkers and therapeutic targets for cancer. Clin Proteomics. 2011;8:9. doi: 10.1186/1559-0275-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michieli P, Mazzone M, Basilico C, Cavassa S, Sottile A, Naldini L, et al. Targeting the tumor and its microenvironment by a dual-function decoy Met receptor. Cancer Cell. 2004;6:61–73. doi: 10.1016/j.ccr.2004.05.032. [DOI] [PubMed] [Google Scholar]
- 46.Kariolis MS, Miao YR, Jones DS, Kapur S, Mathews II, Giaccia AJ, et al. An engineered Axl ‘decoy receptor’ effectively silences the Gas6-Axl signaling axis. Nat Chem Biol. 2014;10:977–83. doi: 10.1038/nchembio.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.To K, Fotovati A, Reipas KM, Law JH, Hu K, Wang J, et al. Y-box binding protein-1 induces the expression of CD44 and CD49f leading to enhanced self-renewal, mammosphere growth, and drug resistance. Cancer Res. 2010;70:2840–51. doi: 10.1158/0008-5472.CAN-09-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johannessen CM, Johnson LA, Piccioni F, Townes A, Frederick DT, Donahue MK, et al. A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature. 2013;504:138–42. doi: 10.1038/nature12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gilbert LA, Hemann MT. DNA damage-mediated induction of a chemoresistant niche. Cell. 2010;143:355–66. doi: 10.1016/j.cell.2010.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muranen T, Selfors LM, Worster DT, Iwanicki MP, Song L, Morales FC, et al. Inhibition of PI3K/mTOR leads to adaptive resistance in matrix-attached cancer cells. Cancer Cell. 2012;21:227–39. doi: 10.1016/j.ccr.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–4. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Obenauf AC, Zou Y, Ji AL, Vanharanta S, Shu W, Shi H, et al. Therapy-induced tumour secretomes promote resistance and tumour progression. Nature. 2015;520:368–72. doi: 10.1038/nature14336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shah MA, Wainberg ZA, Catenacci DV, Hochster HS, Ford J, Kunz P, et al. Phase II study evaluating 2 dosing schedules of oral foretinib (GSK1363089), cMET/VEGFR2 inhibitor, in patients with metastatic gastric cancer. PLoS One. 2013;8:e54014. doi: 10.1371/journal.pone.0054014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chalupský K, Kanchev I, Žbodáková O, Buryová H, Jiroušková M, Kořínek V, et al. ADAM10/17-dependent release of soluble c-Met correlates with hepatocellular damage. Folia Biol (Praha) 2013;59:76–86. [PubMed] [Google Scholar]
- 55.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–24. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoeflich KP, O’Brien C, Boyd Z, Cavet G, Guerrero S, Jung K, et al. In vivo antitumor activity of MEK and phosphatidylinositol 3-kinase inhibitors in basal-like breast cancer models. Clin Cancer Res. 2009;15:4649–64. doi: 10.1158/1078-0432.CCR-09-0317. [DOI] [PubMed] [Google Scholar]
- 57.Holland SJ, Pan A, Franci C, Hu Y, Chang B, Li W, et al. R428, a selective small molecule inhibitor of Axl kinase, blocks tumor spread and prolongs survival in models of metastatic breast cancer. Cancer Res. 2010;70:1544–54. doi: 10.1158/0008-5472.CAN-09-2997. [DOI] [PubMed] [Google Scholar]
- 58.Yang Y, Wang Y, Zeng X, Ma XJ, Zhao Y, Qiao J, et al. Self-control of HGF regulation on human trophoblast cell invasion via enhancing c-Met receptor shedding by ADAM10 and ADAM17. J Clin Endocrinol Metab. 2012;97:E1390–401. doi: 10.1210/jc.2012-1150. [DOI] [PubMed] [Google Scholar]
- 59.Stilley JA, Birt JA, Nagel SC, Sutovsky M, Sutovsky P, Sharpe-Timms KL. Neutralizing TIMP1 restores fecundity in a rat model of endometriosis and treating control rats with TIMP1 causes anomalies in ovarian function and embryo development. Biol Reprod. 2010;83:185–94. doi: 10.1095/biolreprod.109.083287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.