Summary
An influential striatal model postulates that neural activities in the striatal direct and indirect pathways promote and inhibit movement, respectively. Normal behavior requires coordinated activity in the direct pathway to facilitate intended locomotion and indirect pathway to inhibit unwanted locomotion. In this striatal model, neuronal population activity is assumed to encode locomotion relevant information. Here, we propose a novel encoding mechanism for the dorsal striatum. We identified spatially compact neural clusters in both the direct and indirect pathways. Detailed characterization revealed similar cluster organization between the direct and indirect pathways, and cluster activities from both pathways were correlated with mouse locomotion velocities. Using machine-learning algorithms, cluster activities could be used to decode locomotion relevant behavioral states and locomotion velocity. We propose that neural clusters in the dorsal striatum encode locomotion relevant information, and that coordinated activities of direct and indirect pathway neural clusters are required for normal striatal controlled behavior.
Introduction
Basal ganglia circuits transform activity in the cerebral cortex to control motor learning, habit formation, and action selection based on desirable outcomes (Balleine et al., 2007; Cisek and Kalaska, 2010; Graybiel, 2004; Hikosaka et al., 2000; Mink, 1996; Wichmann and DeLong, 2003; Yin and Knowlton, 2006). These effects on behavior are mediated by two parallel striatal circuits involving two subtypes of medium spiny neurons (MSN): the direct pathway neurons that express the dopamine D1 receptor (D1-MSN), and the indirect pathway neurons that express the dopamine D2 receptor (D2-MSN) (Gerfen et al., 1990). Activities in the direct and indirect pathways have been proposed to promote and inhibit movement respectively, and imbalances in their relative activities are thought to account for movement disorders such as Parkinson’s disease (Albin et al., 1989; Alexander and Crutcher, 1990; DeLong, 1990; Gerfen et al., 1990). Recent studies demonstrated concurrent activation of both direct and indirect pathway neurons during action initiation (Cui et al., 2013; Isomura et al., 2013; Jin et al., 2014; Tecuapetla et al., 2014), supporting an updated dual circuit striatal model arguing that activities of the direct pathway neurons promote intended behaviors and activities of the indirect pathway neurons inhibit competing behaviors (Brown, 2007; Chan et al., 2005; Hikosaka et al., 2000; Mink, 2003; Nambu, 2008).
In previous models of striatal function, population neural activities of the direct and indirect pathways are assumed to carry locomotion relevant information. However, how exactly the dorsal striatal neurons encode locomotion remains unknown. Identifying such encoding mechanisms requires advanced techniques that allow examination of the relationship between spatially and temporally coordinated neural activity and locomotion. To address this challenge, we developed a custom miniature fluorescent microscope (miniScope) to concurrently record calcium activities of hundreds of striatal neurons from freely moving mice. Detailed computational analysis showed the existence of spatially compact neural clusters in both direct and indirect pathways. Cluster activities correlated with mouse locomotion velocities, and cluster dynamics were distinct at different behavioral states. Using machine-learning algorithms, we were able to more accurately predict mice behavioral states and locomotion velocities using neural cluster activities than using randomly selected subset of single neurons or population MSN activities. We propose that dorsal striatal neural clusters encode locomotion relevant information, and that spatially and temporally coordinated neural cluster activities of direct and indirect pathways are required for normal striatal-controlled behavior.
Results
Imaging Calcium Activities of Striatal Medium Spiny Neurons in Freely Moving Mice
We developed a custom miniScope system (Figure 1A, also see Supplemental Experimental Procedures for details), based on a previously developed mobile imaging system (Park et al., 2011). We injected D1-Cre and D2-Cre mice (Gerfen et al., 2013; Gong et al., 2007) with Cre-dependent AAV-GCaMP6s (Chen et al., 2013) to selectively label D1- or D2- MSN in dorsal striatum (Figure S1A). Subsequently, we implanted a gradient index (GRIN) lens into the dorsal striatum, and mounted the miniScope above the GRIN lens (Figure 1B). We placed the mice in an open field, acquired GCaMP6 fluorescence from dorsal striatum through the miniScope, and concurrently recorded mouse locomotor activity using a video-recording system (Movie S1). Individual active neurons were detected using a Spatio-Temporal Gradient Matching (STGM, see Supplemental Experimental Procedures) cell identification algorithm, and calcium transients were extracted using an Annular Region Subtraction method (ARS, see Supplemental Experimental Procedures) (Figure 1C, Figure S1B–F, and Movie S2). We thoroughly compared our STGM/ARS algorithms with the recently published constrained nonnegative matrix factorization (CNMF) framework (Pnevmatikakis et al., 2016), and concluded that the two methods performed similarly (Figure S2 and S3). For the rest of the manuscript, we used only STGM/ARS for our calculations.
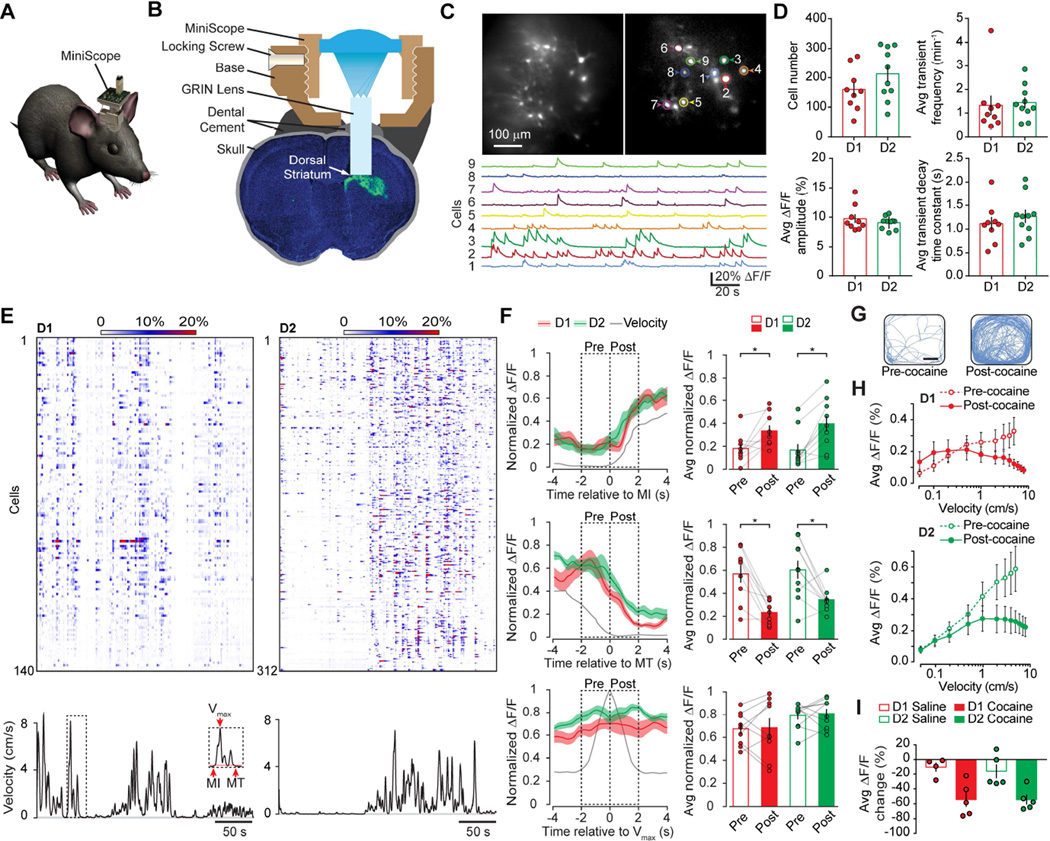
Figure 1. Dorsal Striatal D1- and D2-MSN Population Activity Displayed Similar Relationship with Locomotion in Freely Moving Mice.
A. Schematic demonstration of a miniScope carried by a mouse for in vivo imaging purpose. B. Schematic demonstration of mounting miniScope on mouse head. GRIN lens was implanted into the GCaMP6s AAV infected dorsal striatum (Green region) and anchored on the mouse skull using dental cement; miniScope base was mounted on the mouse skull using dental cement; miniScope body was secured to miniScope base through a locking screw. C. Representative fluorescent images of medium spiny neurons labeled with GCaMP6s at two different time points (Upper panels, scale bar: 100 µm). Regions of interest (ROIs) were drawn around 9 representative neurons. Traces at lower panel represented calcium transients from ROIs with matched colors. D. Quantifications of identified active neuron number (top left panel), calcium transient frequency (top right panel), calcium transient amplitude (bottom left panel), and decay time constant (bottom right panel) for D1- (Red) and D2- (Green) MSN. Each dot on the plot represented the averaged result from one mouse, and histogram bar represented the mean value for all D1- or D2- mice, with error bars representing sem. E. D1- and D2- MSN displayed similar activity during locomotion. Top panels: rasterplots of calcium transients from 140 neurons of a representative D1-Cre mouse (Left) and from 312 neurons of a representative D2-Cre mouse (Right). Bottom panels: black traces indicated locomotion velocity of the mouse. Inset panel indicated motion initiation (MI), motion termination (MT), and maximum velocity of locomotion (Vmax). Horizontal scale bar: 50 seconds. F. D1- (Red) and D2- (Green) MSN displayed similar population activity during MI, MT, and Vmax. Left panels indicated population activity of D1- and D2- MSN, red trace representing averaged population activity from 9 D1-Cre mice; green trace representing averaged population activity from 10 D2-Cre mice; shadowed area on the traces representing sem; black trace in each plot represented mouse locomotion velocity; vertical dotted lines indicated the exact moment for the onset of MI, MT and Vmax. Right histogram plots quantified MSN population activity two seconds before and after MI, MT, and Vmax. Each dot on the plot represented the averaged result from one mouse, connected with gray lines for the “pre” and “post” values of each mouse. Histogram bar represented the mean value for all D1- or D2- mice, with error bars representing sem. G. Representative mouse locomotion traces in an open field before and after cocaine injection, indicating that cocaine substantially enhanced mouse locomotor activity. H. Cocaine altered relationships between mouse locomotion velocity and D1- (Red) and D2- (Green) MSN population activity. Open circles and dashed lines indicated before cocaine injections, solid circles and solid lines indicated after cocaine injections. I. Quantification of D1- and D2- MSN population activity changes during ambulation before and after saline and cocaine injections. Each dot on the plot represented the averaged result from one mouse, and histogram bar represented the mean value for all D1- or D2- mice, with error bars representing sem. Asterisk (*) represents statistical significance.
We identified similar numbers of active D1- and D2- MSN from each D1-Cre or D2-Cre mouse (D1-MSN: 160 ± 24 neurons, n = 9 mice; D2-MSN: 213 ± 28 neurons, n = 10 mice; p = 0.1823, Mann-Whitney test, Figure 1D, also refer to Table S1 for all statistical comparisons throughout the manuscript). Calcium activities from D1- and D2- MSN displayed similar frequency (D1-MSN: 1.3 ± 0.4 events per minute, n = 9 mice; D2-MSN: 1.4 ± 0.2 events per minute, n = 10 mice; p = 0.3154, Mann-Whitney test), similar amplitude (D1-MSN: 9.7% ± 0.7%, n = 9 mice; D2-MSN: 9.1% ± 0.3%, n = 10 mice; p = 0.9682, Mann-Whitney test), and similar decay time (D1-MSN: 1.1 ± 0.1 seconds, n = 9 mice; D2-MSN: 1.3 ± 0.1 seconds, n = 10 mice; p = 0.4470, Mann-Whitney test, Figure 1D). D1- and D2- MSN population activities were both high during ambulation and low during immobility (Figure 1E).
We then performed detailed comparisons for D1- and D2-MSN population activities (the averaged ΔF/F of all recorded neurons for each mouse) at various locomotion states, including prior to and post motion initiation (MI), prior to and post motion termination (MT), as well as prior to and post maximum velocity of locomotion (Vmax). We found that D1- and D2-MSN population activities displayed similar activity patterns during locomotion states MI and MT, with increased activities post MI, and decreased activities post MT (D1-MSN prior to and post MI: 0.18 ± 0.04 and 0.33 ± 0.05, p = 0.0273; D2-MSN prior to and post MI: 0.17 ± 0.05 and 0.40 ± 0.07, p = 0.0039, Figure 1F top panels; D1-MSN prior to and post MT: 0.57 ± 0.08 and 0.23 ± 0.03, p = 0.0078; D2-MSN prior to and post MT: 0.60 ± 0.08 and 0.34 ± 0.03, p = 0.0195, Figure 1F middle panels; n = 9 mice for D1-MSN; n = 10 mice for D2-MSN, Wilcoxon matched-pairs signed rank test). In contrast, D1- and D2- MSN population activity did not exhibit distinguishable patterns around Vmax (D1-MSN prior to and post Vmax: 0.67 ± 0.05 and 0.68 ± 0.08, p = 0.91; D2-MSN prior to and post Vmax: 0.79 ± 0.04 and 0.80 ± 0.04, p = 0.625, n = 9 mice for D1-MSN; n = 10 mice for D2-MSN, Wilcoxon matched-pairs signed rank test; Figure 1F bottom panels). Comparisons of D1- and D2- MSN population activities during each locomotion state did not reveal any statistical significance, except for a tendency of higher activity of D2- over D1- MSN prior to Vmax (Figure 1F; comparing D1- and D2-MSN, prior to MI, p = 0.3154; post MI, p = 0.4967; prior to MT, p = 0.9048; post MT, p = 0.0947; prior to Vmax, p = 0.0535; post Vmax, p = 0.2775; n = 9 mice for D1-MSN; n = 10 mice for D2-MSN, Mann-Whitney test).
Collectively, these results demonstrate that D1- and D2- MSN population activities during locomotion are overall similar, consistent with other recent reports (Cui et al., 2013; Isomura et al., 2013; Jin et al., 2014; Tecuapetla et al., 2014). These results also support the updated dual circuit striatal model arguing that activities of the direct pathway neurons promote intended behaviors and activities of the indirect pathway neurons inhibit competing behaviors (Brown, 2007; Chan et al., 2005; Hikosaka et al., 2000; Mink, 2003; Nambu, 2008).
Injection of the psychostimulant drug cocaine (20 mg/kg, i.p) substantially enhanced mouse locomotor activity (Figure 1G). Based on the updated dual circuit striatal model, we reasoned that D1- and D2- MSN population activity would be further increased with the enhanced locomotor activity. Before cocaine exposure, both D1- and D2- MSN displayed increased population activities with increasing mouse locomotion velocity (Figure 1H, open circles). However, after cocaine injection, population activities of D1- and D2- MSN during ambulation did not increase despite substantially increased mouse locomotion activity (Figure 1H, filled dots). We further calculated population activity changes post-injection versus pre-injection during ambulation, and compared cocaine injection to saline injection as control. We found that saline injection slightly suppressed D1 and D2- MSN population activities (D1-MSN: −17 ± 9%, n = 4 mice; D2-MSN: −22 ± 16%, n = 5 mice), likely reflecting acclimation to open field environment during post-injection period. Notably, cocaine injection did not increase either D1-or D2- MSN population activity, instead cocaine injection inhibited D1-MSN population activity and showed a tendency to inhibit D2-MSN population activity (D1-MSN cocaine injection: −68 ± 9%, n = 5 mice, p = 0.0317 compared to D1-MSN saline injection; D2-MSN cocaine injection: −69 ± 6%, n = 5 mice, p = 0.0556 compared to D2-MSN saline injection, Mann-Whitney test, Figure 1I). The observation that cocaine enhanced mouse locomotor activity but failed to increase D1- and D2- MSN population activities suggests that population activities of D1- or D2-MSN do not serve the purpose of encoding locomotion relevant information as suggested by previous striatal models. Together, our results revealed a more complex functional activity of the MSNs on locomotion than what could be predicted from the classical model, highlighting the need for a better neural encoding model for dorsal striatum to explain the functional role of D1-and D2- MSN on locomotion.
Spatially Compact Neural Clusters in both Direct and Indirect Pathways
Previous studies employing neural clustering approach identified cell assemblies that represented functional states of the striatal network (Adler et al., 2012; Bakhurin et al., 2016; Carrillo-Reid et al., 2008; Jaidar et al., 2010; Ponzi and Wickens, 2012). We therefore applied similar neural clustering approach (Humphries, 2011; Ozden et al., 2008) to our data and identified neural clusters of D1- or D2- MSN based solely on their calcium activity correlation (Figure S4). A representative standard deviation projection of GCaMP6 labeled active neurons from one single imaging session was shown in Figure 2A. We then anatomically mapped active neurons for all clusters identified from all the imaging sessions in each mouse, and found that neurons within each cluster displayed compact spatial distribution (Figure 2B). Diversity in the activity patterns among clusters was prominent (Figure 2C). For example, spatially adjacent clusters C1 and C4 (from the representative clusters shown in Figure 2B) displayed vastly different activity patterns (Figure 2C). Further characterization of neural clusters showed that the number of clusters per mouse was similar between D1- and D2- MSN (D1-MSN: 15 ± 3 clusters, n = 9 mice; D2-MSN: 17 ± 2 clusters, n = 10 mice; p = 0.4829, Mann-Whitney test; Figure 2D). The number of neurons per cluster was also similar between D1- and D2- MSN (D1-MSN: 10 ± 1 neurons, n = 9 mice; D2-MSN: 11 ± 1 neurons, n = 10 mice; p = 0.0947, Mann-Whitney test; Figure 2E). In addition, there was no difference between D1- and D2- MSN in average cell distance within a cluster (D1-MSN: 43.0 ± 5.3 µm, n = 9 mice; D2-MSN: 40.0 ± 1.9 µm, n = 10 mice; p = 0.7197, Mann-Whitney test; Figure 2F).
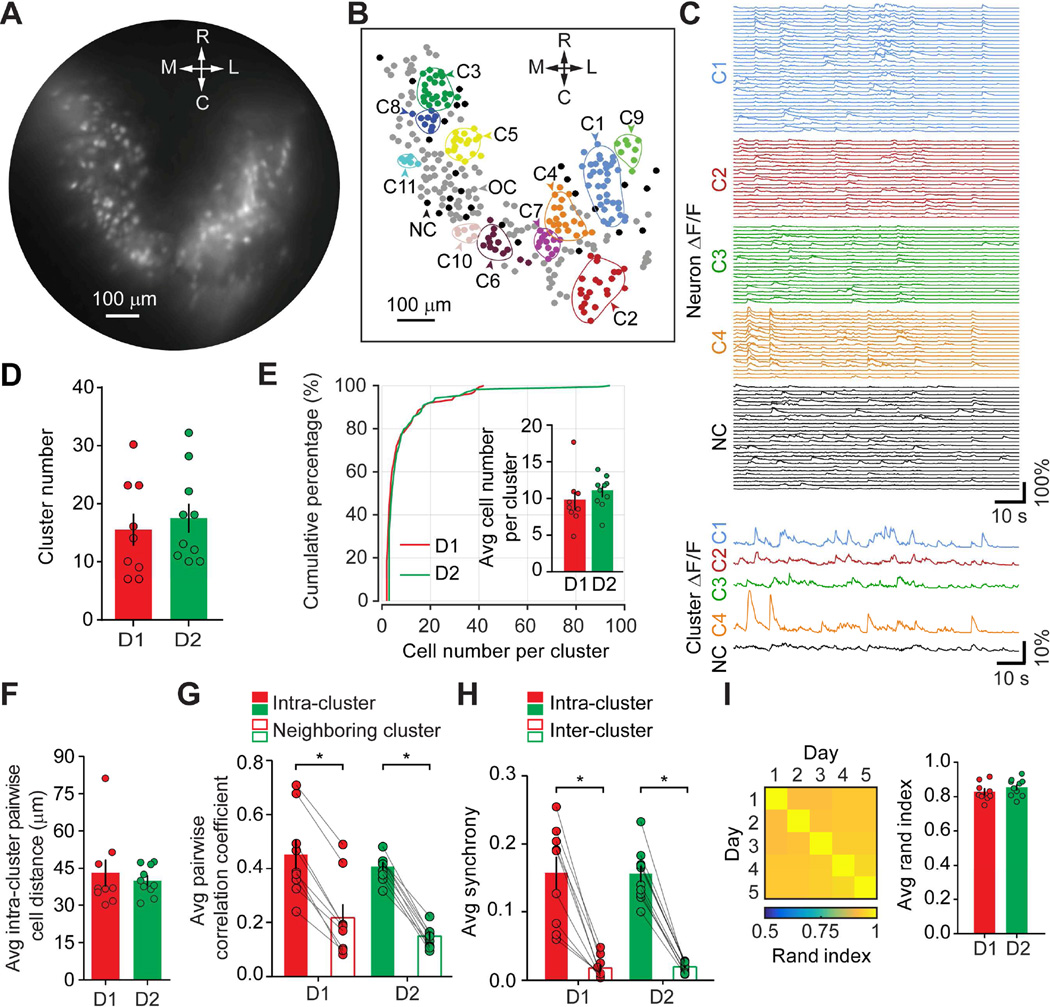
Figure 2. Cluster Analysis Revealed Spatially Compact Neural Clusters Within Dorsal Striatal D1- and D2- MSNs.
A. A representative standard deviation projection of 3000 GCaMP6 images of dorsal striatum from a representative D2-Cre mouse, scale bar 100 µm. B. Spatial distribution of clusters identified from all imaging sessions of a representative D2-Cre mouse. A total of 11 colored enclosed areas indicated 11 different neural clusters (C1 to C11), each containing more than 7 neurons (neurons within each cluster were labeled with the same color). Neurons in clusters containing less than 7 neurons were labeled in gray (OC), and un-clustered neurons were labeled in black (NC). Colored contour indicates the estimated boundary for each cluster. Scale bar 100 µm. Orientation of the map for both A and B: M, medial; L, Lateral; R, Rostral; C, Caudal. C. Individual neural calcium transient traces and mean cluster activity traces from four representative clusters (C1 to C4, colored); and individual calcium transient traces from un-clustered neurons (NC) and the averaged calcium trace from all the NC neurons (Black). D. Quantification of neural cluster numbers identified from each mouse dorsal striatum. Each dot on the plot represented the averaged result from one mouse, histogram bar represented the mean value for all D1- or D2- mice, with error bars representing sem (same for E through I histogram plots). E. Cumulative plot of cell numbers per cluster for both D1- and D2-MSN. Inset histogram bars were quantification of averaged cluster numbers. F. Quantification of the averaged cell distance within a cluster. G. Quantification of pair-wise neuron correlation coefficient within a cluster (Intra-cluster) or between neighboring clusters (Inter-cluster). H. Quantification of calcium activity synchrony within a cluster (Intra-cluster) or between clusters (Inter-cluster). In both G and H, gray lines connected the “Intra-” and “Inter-” cluster values from each mouse. I. Left panel was representative matrix of Rand index from a representative mouse showing clustering was consistent over five days. Each square in the matrix indicated the Rand index value calculated for the two days specified by the corresponding row and column. The range indicator below the matrix indicated the value of Rand index in the matrix. Right histogram bar plot quantified the average Rand index for D1- and D2- MSN clusters over five days. Asterisk (*) represents statistical significance.
Correlation analysis revealed that D1- and D2- MSN intra-cluster correlation was greater than inter-cluster correlation between adjacent neural clusters (D1-MSN: Intra 0.45 ± 0.05, Inter 0.21 ± 0.05, p = 0.0039, n = 9 mice; D2-MSN: Intra 0.40 ± 0.01, Inter 0.14 ± 0.01, p = 0.0020, n = 10 mice, Wilcoxon matched-pairs signed rank test, Figure 2G), while both intra- and adjacent inter- cluster correlation were similar between D1- and D2- MSN (Intra: p = 0.9048, Inter: p = 0.3562 between D1- and D2- MSN, Mann-Whitney test, Figure 2G). We also examined synchronous calcium activity, with simultaneously active neurons within a cluster categorized as intra-cluster synchrony, and simultaneously active neurons from different clusters categorized as inter-cluster synchrony. We found that intra-cluster synchrony was more prominent than inter-cluster synchrony for both D1- and D2- MSN (D1-MSN: Intra 0.155 ± 0.024, Inter 0.018 ± 0.005, p = 0.0039, n = 9 mice; D2-MSN: Intra 0.154 ± 0.012, Inter 0.019 ± 0.002, p = 0.0020, n = 10 mice, Wilcoxon matched-pairs signed rank test, Figure 2H), while both intra-cluster and inter-cluster synchrony were similar between D1- and D2- MSN (Intra: p = 0.7802, Inter: p = 0.268 between D1-MSN and D2-MSN, Mann-Whitney test, Figure 2H).
To determine the stability of these identified neural clusters, we calculated the Rand index of clusters for each mouse over 5 days (Figure 2I left panel). Rand index indicates the similarity between two clustering schemes, ranging from zero to one, with zero indicating that no pairs of cells is clustered in agreement in the two schemes, and one indicating that each pair of cells is clustered identically in the two schemes. We found that both D1- and D2- MSN clusters displayed consistently high value of Rand index over 5 days (D1-MSN: 0.83 ± 0.02, n = 9 mice; D2-MSN: 0.85 ± 0.02, n = 10 mice; p = 0.3154, Mann-Whitney test, Figure 2I right panel). This result indicates that D1- and D2- MSN clusters from the same mouse are stable over different days. Moreover, the overall similarities between D1- and D2- MSN neural clusters suggest that the organizations of neural clusters for the direct and indirect pathways in dorsal striatum are similar.
Neural Cluster Activity Dynamics at Different Behavioral States
We next explored dynamics of neural cluster activity (the averaged ΔF/F of all neurons within each cluster) under different behavioral states. If the neural clusters identified from the direct and indirect pathways in the dorsal striatum encode locomotion relevant information, individual cluster activity should correlate with mouse locomotion. We therefore analyzed the cross correlation for individual neural cluster activity with Vmax of mouse locomotion. We found that different clusters displayed distinct cross correlations with Vmax of mouse locomotion. For example, C1 and C4 were spatially adjacent clusters (from the representative clusters shown in Figure 2B), yet the peak activity of cluster C4 preceded Vmax, whereas that of cluster C1 lagged behind Vmax (Figure 3A and 3B). This observation indicates that cluster C1 and C4 were active at different phases of locomotion. We next calculated the averaged cross correlation between cluster activities and Vmax for each mouse. We found that the averaged cross correlation from each mouse exhibited a higher value than cross correlation predicted by pure chance (see Supplemental Experimental Procedures), and the overall averaged cross correlation was similar between D1- and D2-MSN clusters (D1-MSN: 0.41 ± 0.03, n = 9 mice; D2-MSN: 0.45 ± 0.02, n = 10 mice; p = 0.3562, Mann-Whitney test; Figure 3C). Moreover, cross correlation was stable over 5 consecutive days (Figure 3D). Together, these results demonstrate that neural cluster activities in the dorsal striatum correlated with mouse locomotion, and suggest that neural cluster activity encodes locomotion relevant information.
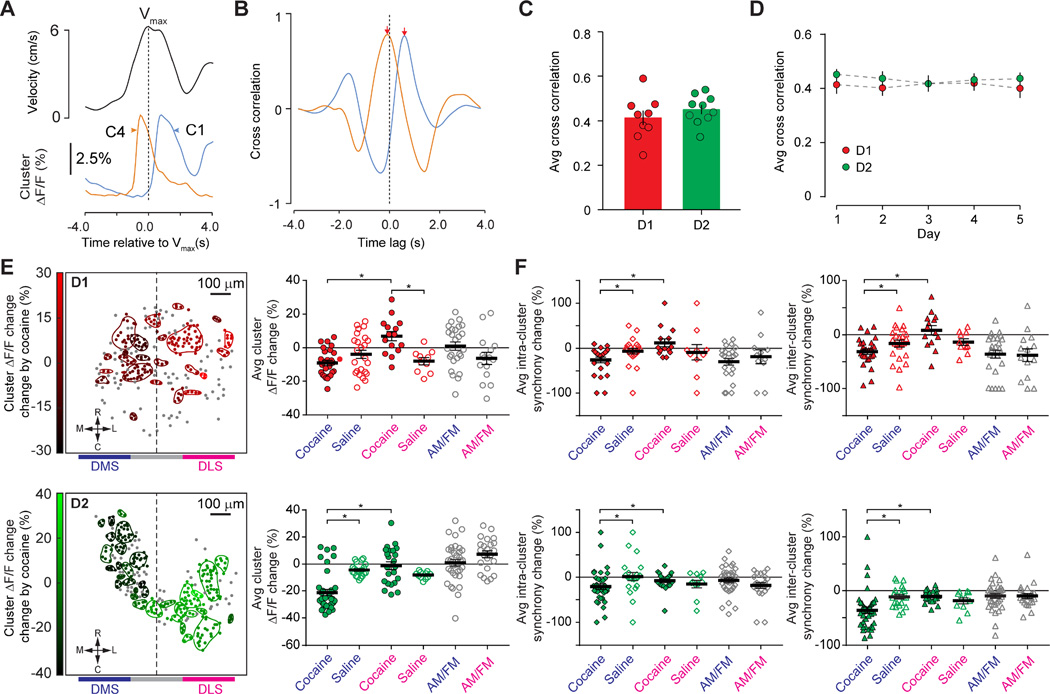
Figure 3. Cluster Activity Dynamics Under Different Behavior States.
A. Representative cluster activities during maximum velocity (Vmax) of locomotion. Upper panel: a representative mouse locomotion velocity peak; Lower panel: cluster activity of C1 and C4 (corresponding to clusters shown in Figure 2B). Vertical dash line indicated peak position of Vmax B. Cross correlation between cluster C1 and C4 activity with Vmax. Vertical dash line indicated zero time lag in the cross correlation. C. Quantification of cross-correlation between cluster activity and Vmax for D1- or D2- MSN from Day One. Each dot indicated the average result from one mouse on Day One, and histogram bars indicated the mean value for all D1- or D2- mice, with error bars representing sem. D. Reliability of cross-correlation over 5 days, X-axis represented days, Y-axis represented averaged cross correlation value for each day, each dot indicating the averaged value from all D1- or D2- mice on the corresponding day, and error bars representing sem. E. Left panels were representative maps for cluster activity change from a D1-Cre (top left panel) and D2-Cre mouse (bottom left panel). Orientation of the map: M, medial; L, Lateral; R, Rostral; C, Caudal. Colored contour indicated the estimated boundary for each cluster. Dots with the same color as the contour indicate the centroids of neurons in the cluster. Gray dots indicated un-clustered neurons. Range indicator on the left indicated activity change, with reduced activity towards black color, and increased activity towards brighter red (D1-MSN) or green (D2-MSN). Horizontal bar beneath panels indicated the approximate location of dorsomedial and dorsolateral striatum (DMS and DLS). Scale bar 100 µm. Right panels were quantifications of cluster activity change near DMS (Blue color) and DLS (Pink color). Y-axis represented activity change, with negative value representing activity suppression and positive value representing activity increase. Each dot indicated value from one cluster. Black horizontal line represented the average value from all clusters in either DMS or DLS, error bar represented sem. Cocaine: activity change following cocaine injection; Saline: activity change following saline injection; AM/FM: activity difference between ambulation and fine movement. F. Quantification of cluster synchrony change near DMS (Blue color) and DLS (Pink color). Left panels were intra-cluster synchrony change for D1-MSN (top) and D2-MSN (bottom), Right panels were inter-cluster synchrony change for D1-MSN (top) and D2-MSN (bottom). The definition for positive and negative values in Y-axis was the same as E. Each dot indicated value from one cluster. Black horizontal line represented the average value from all clusters in either DMS or DLS, error bar represented sem. Asterisk (*) represents statistical significance.
We next explored D1- and D2- MSN cluster activity changes following cocaine exposure. We calculated D1- and D2- MSN cluster activity changes after cocaine or saline injection versus before injection, at dorsomedial striatum (DMS) and dorsolateral striatum (DLS) respectively (Figure 3E). We found that cocaine enhanced D1-MSN cluster activity near DLS compared to saline injection controls (D1-MSN at DLS: cocaine: 7.0% ± 2.7%, n = 15 clusters from 5 mice, saline: −7.9% ± 2.1%, n = 10 clusters from 4 mice, p = 0.0005, Mann-Whitney test, Figure 3E upper right panel), whereas cocaine suppressed D2-MSN cluster activity near DMS compared to saline injection controls (D2-MSN at DMS: cocaine: −21.3% ± 2.4%, n = 37 clusters from 5 mice, saline: −4.4% ± 1.0%, n = 22 clusters from 5 mice, p < 0.0001, Mann-Whitney test, Figure 3E lower right panels). Cocaine did not change D1-MSN cluster activity near DMS or D2-MSN activity near DLS (D1-MSN at DMS: cocaine: −8.9% ± 1.6%, n = 26 from 5 mice, saline: −3.8% ± 2.4%, n = 24 clusters from 4 mice, p = 0.12; D2-MSN and at DLS: cocaine: −1.2% ± 2.9%, n = 23 clusters from 5 mice, saline: −8.3% ± 0.8%, n = 11 clusters from 5 mice, p = 0.21; Mann-Whitney test). When comparing activity changes between DMS and DLS in either D1- or D2-MSN clusters, cocaine elicited differential effect on DMS versus DLS cluster activity changes (D1-MSN: DMS: −8.9% ± 1.6%, n = 26 clusters from 5 mice, DLS: 7.0% ± 2.7%, n = 15 clusters from 5 mice, p < 0.0001; D2-MSN: DMS: −21.3% ± 2.4%, n = 37 clusters from 5 mice, DLS: −1.2% ± 2.9%, n = 23 clusters from 5 mice, p < 0.0001, Mann-Whitney test, Figure 3E right panels). This differential effect on DMS vs. DLS by cocaine could not be explained simply by higher locomotor activity under cocaine influence, because cluster activity changes between ambulation (AM, high locomotor activity) and fine movement (FM, low locomotor activity) under normal locomotion conditions did not display any difference between DMS and DLS (AM/FM D1-MSN: DMS: 1.0% ± 2.4%, n = 26 clusters from 5 mice, DLS: −6.2% ± 3.6%, n = 15 clusters from 5 mice, p = 0.0857; AM/FM D2-MSN: DMS: 1.0% ± 2.3%, n = 37 clusters from 5 mice, DLS: 7.2% ± 2.5%, n = 23 clusters from 5 mice, p = 0.0778, Mann-Whitney test, Figure 3E right panels).
We next explored if changes of D1- and D2- MSN cluster synchrony (as defined in Figure 2H) also reflect dorsal striatal network changes under cocaine influence. We calculated D1- and D2- MSN intra- and inter-cluster synchrony changes after cocaine or saline injection versus before injection at DMS and DLS, respectively. We found that compared to saline injection controls, cocaine reduced intra- and inter- cluster synchrony for both D1- and D2-MSN only at the DMS (D1-MSN Intra- at DMS: cocaine: −25.9% ± 6.2%, n = 24 cluster from 5 mice, saline: −6.4% ± 6.1%, n = 23 clusters from 4 mice, p = 0.0159, Figure 3F upper left panel; D2-MSN Intra- at DMS: cocaine −21.0% ± 6.1%, n = 37 clusters from 5 mice, saline: 1.4% ± 9.1%, n = 21 clusters from 5 mice, p = 0.0077, Figure 3F lower left panel; D1-MSN Inter- at DMS: cocaine: −31.0% ± 5.1%, n = 26 clusters from 5 mice, saline: −16.2% ± 6.5%, n = 24 clusters from 4 mice, p = 0.0481, Figure 3F upper right panel; D2-MSN Inter- at DMS: cocaine: −36.1% ± 5.9%, n = 37 clusters from 5 mice, saline: −11.3% ± 4.0%, n = 22 clusters from 5 mice, p < 0.0001, Figure 3F lower right panel. All pairwise comparisons were based on Mann-Whitney test). We did not observe difference of intra- or inter-cluster synchrony change at DLS for either D1- or D2- MSN between cocaine and saline controls (D1-MSN Intra- at DLS: cocaine: 12.2% ± 8.8%, n = 14 clusters from 5 mice, saline: −8.9% ± 17%, n = 10 clusters from 4 mice, p = 0.1334; D2-MSN Intra- at DLS: cocaine: −7.9% ± 3.9%, n = 23 clusters from 5 mice, saline: −15.2% ± 8.1%, n = 11 clusters from 5 mice, p = 0.67; D1-MSN Inter- at DLS: cocaine: 8.3% ± 8.8%, n = 14 clusters from 5 mice, saline: −13.4% ± 6.2%, n = 10 clusters from 4 mice, p = 0.0841; D2-MSN Inter- at DLS: cocaine: −10.6% ± 2.3%, n = 23 clusters from 5 mice, saline: −18.7% ± 5.3%, n = 11 clusters from 5 mice, p = 0.3256; Mann-Whitney test; Figure 3F). When comparing synchrony changes between DMS and DLS in either D1- or D2- MSN clusters, cocaine elicited differential effect on DMS versus DLS synchrony change (D1-MSN Intra-cluster: DMS: −25.9% ± 6.2%, n = 24 clusters from 5 mice, DLS: 12.2% ± 8.8%, n = 14 clusters from 5 mice, p = 0.0009, Figure 3F upper left panel; D2-MSN Intra-cluster: DMS: −21.0% ± 6.1%, n = 37 clusters from 5 mice, DLS: −7.9% ± 3.9%, n = 23 clusters from 5 mice, p = 0.0058, Figure 3F lower left panel; D1-MSN Inter-cluster: DMS: −31.0% ± 5.1%, n = 26 clusters from 5 mice, DLS: 8.3% ± 8.8%, n = 14 clusters from 5 mice, p = 0.0003, Figure 3F upper right panel; D2-MSN Inter-cluster: DMS: −36.1% ± 5.9%, n = 37 clusters from 5 mice, DLS: −10.6% ± 2.3%, n = 23 clusters from 5 mice, p < 0.0001, Figure 3F lower right panel. All pairwise comparisons were based on Mann-Whitney test). We did not observe similar differential DMS versus DLS synchrony change when comparing ambulation vs. fine movement under normal locomotion conditions (AM/FM Intra- synchrony changes for D1- MSN at DMS and DLS: −29.7% ± 7.4% and −18.6 ± 15.4%, n = 24 and 11 clusters from 5 mice, p = 0.6111; AM/FM Intra- synchrony changes for D2- MSN at DMS and DLS: −7.0% ± 5.2% and −18.4 ± 6.3%, n = 35 and 23 clusters from 5 mice, p = 0.148; AM/FM Inter- synchrony changes for D1- MSN at DMS and DLS: −35.8% ± 7.4% and −37.7 ± 11.8%, n = 26 and 14 clusters from 5 mice, p = 0.7207; AM/FM Inter- synchrony changes for D2- MSN at DMS and DLS: −9.7% ± 4.3% and −9.2 ± 4.6%, n = 36 and 23 clusters from 5 mice, p = 0.5839; Mann-Whitney test, Figure 3F).
Together, these results demonstrated that D1- and D2-MSN neural cluster activity and synchrony dynamics displayed distinctive spatial patterns under cocaine influence that was different from the activity and synchrony dynamics under normal locomotion conditions, and suggest that striatal network dynamics are different between normal locomotion behavior states and under cocaine influence. These results further suggest that neural cluster activity dynamics reflect striatal network states more accurately than population activity of MSN.
Neural Cluster Activities Perform Better in Behavior Decoding
To further test the hypothesis that neural cluster activities encode locomotion relevant information, we performed behavioral decoding experiments to predict mouse behavioral states using neural cluster activities (the averaged ΔF/F of all neurons within each cluster), activities of randomly selected subset of single neurons (the number of randomly selected neurons was equal to the number of clusters for each animal, in order to match the number of inputs for each decoding experiment using cluster activities, see Supplemental Experimental Procedures for details), or population activities (the averaged ΔF/F of all recorded neurons for each mouse). As a proof-of-concept experiment, we identified four different mouse behavioral states: three behavioral state classifications for normal locomotion (ambulation, fine movement, and immobility), and one behavioral state classification representing locomotion under cocaine influence. We trained a machine-learning algorithm using part of the behavior and MSN activity data, and subsequently used the rest of the data (not previously seen by the algorithm) to test the performance of the trained algorithm for behavioral state prediction (Figure 4A). We used decoding accuracy to quantify the performance of the algorithm trained by D1- or D2- MSN activities described above. Decoding accuracy ranged from 0.5 to 1.0, with 0.5 representing prediction by random guessing (pure chance), and 1.0 representing 100% accuracy. We statistically compared the decoding accuracy between those using cluster activities and those using activities from randomly chosen single neurons, or between those using cluster activities and those using population activities of MSN.
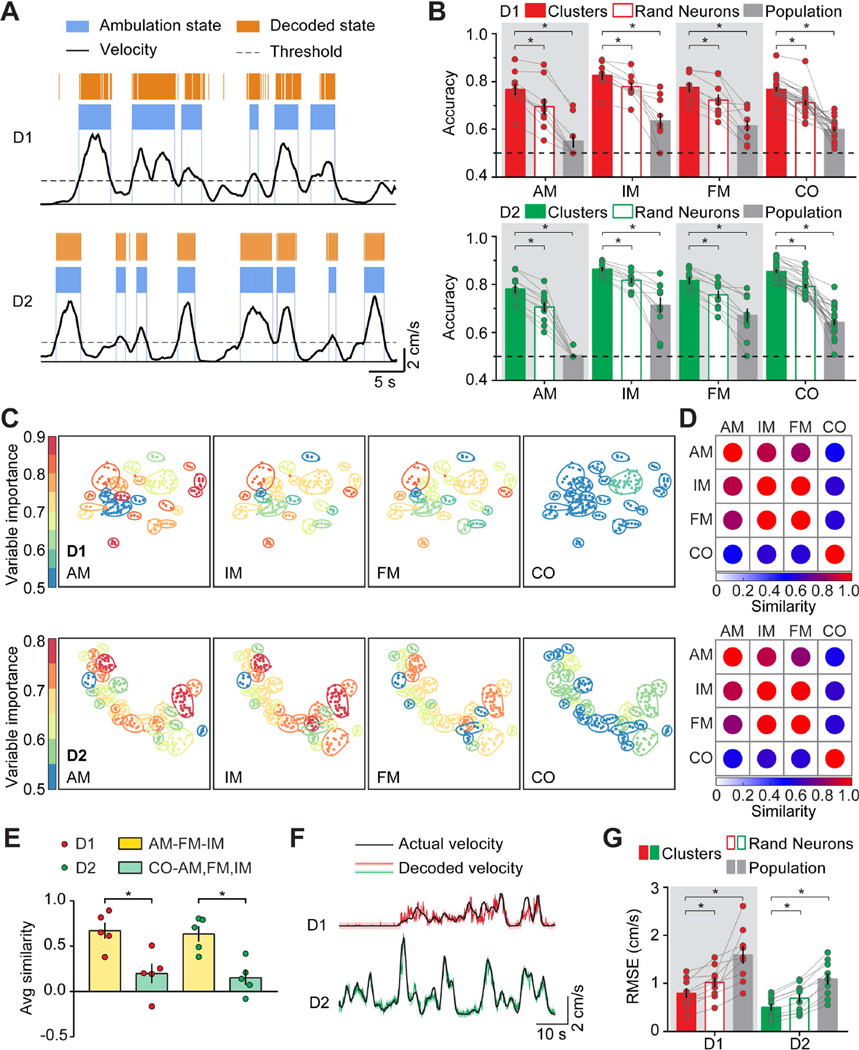
Figure 4. MSN Cluster Activities Perform Better in Behavior Decoding.
A. Mouse ambulation decoding using neural cluster activity data from D1- (Top panel) and D2- (Bottom panel) MSN. Mouse locomotor activity traces were shown at the bottom (Black traces). Ambulation was defined as locomotion velocity higher than 2 cm/s. Blue segments indicated actual ambulation period of mouse; Orange segments indicated prediction of mouse ambulation based on neural cluster activity. Horizontal scale bar: 5 seconds, vertical scale bar: 2 cm/s. B. Histogram showing accuracy of behavior state decoding based on cluster activity (Red filled bars on top panel, D1-MSN; Green filled bars on bottom panel, D2-MSN), randomly selected neuron activities (Red unfilled bars on top panel, D1-MSN; Green unfilled bars on bottom panel, D2-MSN) and population activity (Gray bars on top, D1-MSN; Gray bars on bottom, D1-MSN). Each dot on the plot represented the averaged result from one mouse, and histogram bar represented the mean value for all D1- or D2-mice, with error bars representing sem. AM: ambulation; IM: immobility; FM: fine movement; CO: cocaine. Dotted line at 50% accuracy indicated binary prediction by pure chance. C. Neural cluster map from representative D1-Cre (top panels) and D2-Cre (bottom panels) mouse indicating variable importance value of cluster decoding in four different behavior state decoding. Color on each cluster indicated variable importance value for the cluster in specified behavior decoding experiment, as indicated by the range indicator at the left. D. Similarity matrix for representative D1-Cre (top panel) and D2-Cre (bottom panel) mouse showing similarity of cluster variable importance between any two behavior decoding experiments. Color for the dot in the matrix indicated similarity value, as indicated by the range indicator at the bottom of the matrix. E. Quantification of averaged similarity value between the three normal locomotion behavior states (ambulation, immobility, and fine movement) and the cocaine injection behavior state. Red dots indicated D1-Cre mice, and green dots indicated D2-Cre mice. Each dot on the plot represented the averaged result from one mouse, and histogram bar represented the mean value for all D1- or D2- mice, with error bars representing sem. Yellow histogram bars indicated similarity scores between three normal locomotion behavior states, and green histogram bars indicate similarity scores between normal locomotion states and cocaine injection state. F. Representative mouse locomotion velocity decoding using cluster activity. Black traces indicated the actual mouse locomotion velocity; Red and green traces indicated predicted locomotion velocity based on D1- and D2-MSN cluster activities respectively. Horizontal scale bar: 10 seconds; Vertical scale bar: 2cm/s. G. Quantification of the root-mean-square error (RMSE) between predicted velocity and actual velocity for decoding error based on cluster (Red filled bars, D1-MSN; Green filled bars, D2-MSN), randomly selected neuron activities (Red unfilled bars, D1-MSN; Green unfilled bars, D2-MSN) and population activity (Gray bar at the left, D1-MSN; Gray bar at the right, D2-MSN). Lower RMSE value indicated lower prediction error therefore better prediction. Each dot on the plot represented the averaged result from one mouse, histogram bar represented the mean value for all D1- or D2- mice, with error bars representing sem. Red dots represented D1-Cre mice and green dots represented D2-Cre mice. Asterisk (*) represents statistical significance.
We found that decoding using cluster activities consistently yielded higher accuracies than those using activities of randomly selected subset of single neurons, as well as those using population activities. For ambulation decoding, decoding accuracies using cluster activity from D1- and D2-MSN were 0.77 ± 0.03 and 0.78 ± 0.02, whereas those using random neurons were 0.70 ± 0.03 and 0.71 ± 0.02, and those using D1- and D2-MSN population activity were 0.55 ± 0.03 and 0.505 ± 0.005 (comparing cluster and random neurons: for D1-MSN, p = 0.0039, n = 9 mice; for D2- MSN, p = 0.0020, n = 10 mice, Wilcoxon matched-pairs signed rank test; comparing cluster and population: for D1-MSN, p = 0.0039, n = 9 mice; for D2- MSN, p = 0.0020, n = 10 mice, Wilcoxon matched-pairs signed rank test, Figure 4B). For immobility decoding, decoding accuracies using cluster activity from D1- and D2-MSN were 0.83 ± 0.02 and 0.86 ± 0.01, whereas those using random neurons were 0.78 ± 0.02 and 0.82 ± 0.01, and those using D1- and D2-MSN population activity were 0.63 ± 0.03 and 0.71 ± 0.03 (comparing cluster and random neurons: for D1-MSN, p = 0.0039, n = 9 mice; for D2- MSN, p = 0.0020, n = 10 mice, Wilcoxon matched-pairs signed rank test; comparing cluster and population: for D1-MSN, p = 0.0039, n = 9 mice; for D2- MSN, p = 0.0020, n = 10 mice, Wilcoxon matched-pairs signed rank test, Figure 4B). For fine movement decoding, decoding accuracies using cluster activity from D1- and D2-MSN were 0.78 ± 0.02 and 0.82 ± 0.02, whereas those using random neurons were 0.72 ± 0.02 and 0.76 ± 0.02, and those using D1- and D2-MSN population activity were 0.61 ± 0.02 and 0.67 ± 0.03 (comparing cluster and random neurons: for D1-MSN, p = 0.0039, n = 9 mice; for D2- MSN, p = 0.0020, n = 10 mice, Wilcoxon matched-pairs signed rank test; comparing cluster and population: for D1-MSN, p = 0.0039, n = 9 mice; for D2- MSN, p = 0.0020, n = 10 mice, Wilcoxon matched-pairs signed rank test, Figure 4B). For cocaine decoding, decoding accuracies using cluster activity from D1- and D2-MSN were 0.77 ± 0.01 and 0.86 ± 0.01, whereas those using random neurons were 0.71 ± 0.01 and 0.79 ± 0.01, and those using D1- and D2- MSN population activity were 0.60 ± 0.01 and 0.64 ± 0.01 (comparing cluster and random neuron: for D1-MSN, p < 0.0001, n = 25 experiments from 5 mice; for D2-MSN, p < 0.0001, n = 25 experiments from 5 mice, Wilcoxon matched-pairs signed rank test; comparing cluster and population: for D1-MSN, p < 0.0001, n = 25 experiments from 5 mice; for D2-MSN, p < 0.0001, n = 25 experiments from 5 mice, Wilcoxon matched-pairs signed rank test, Figure 4B).
Together, these results suggest that clusters of similarly firing MSNs are more effective at encoding behaviorally relevant information than randomly selected subset of single neurons or population activities of D1- or D2- MSN.
We next calculated variable importance values, which measured weighted contribution for each cluster in specific decoding experiment, for all the neural clusters in the decoding experiment. We constructed decoding maps based on the variable importance value of neural clusters, and found that decoding maps for normal locomotion behavioral states (ambulation, fine movement, and immobility) appeared similar to one another, whereas that of cocaine injection state was vastly different (Figure 4C). We calculated and compared similarity scores among the three normal locomotion behavior states and the cocaine behavioral state (Figure 4D), and found that for D1- and D2-MSN clusters, the averaged similarity scores among the three normal locomotion states were 0.67 ± 0.09 and 0.64 ± 0.08 respectively, whereas the averaged similarity scores between normal locomotion states and cocaine state were 0.20 ± 0.11 and 0.15 ± 0.08 (comparing normal and cocaine state: for D1-MSN, p = 0.0159, n = 5 mice; for D2-MSN, p = 0.0159, n = 5 mice; Mann-Whitney test, Figure 4E). These results suggest that under cocaine influence, organization and connectivity of striatal cluster network dynamics is different from that of normal locomotion.
Given the similarity of decoding maps for all three normal locomotion behavior states, we further explored the possibility of predicting mouse locomotion velocity using cluster activities, activities of randomly selected subset of neurons matching the number of clusters, or population activities of D1- and D2- MSN. Both D1- and D2- MSN cluster activities performed well in predicting mouse locomotion velocity (Figure 4F). We further calculated and compared prediction errors using cluster activities, activities of randomly selected subset of neurons matching the number of clusters, and population activities from D1- and D2- MSN. Prediction error was calculated as root-mean-square error (RMSE) and reflected the differences between decoded velocity and actual mouse locomotion velocity, with lower value representing better prediction. We found that prediction errors using D1- and D2- MSN cluster activity were 0.80 ± 0.10 and 0.50 ± 0.07, whereas those using random neurons were 1.02 ± 0.11 and 0.69 ± 0.09, and those using D1- and D2-MSN population activities were 1.59 ± 0.19 and 1.09 ± 0.11 (comparing cluster and random neurons: for D1-MSN, p = 0.0039, n = 9 mice; for D2- MSN, p = 0.0020, n = 10 mice, Wilcoxon matched-pairs signed rank test; comparing cluster and population: for D1-MSN, p = 0.0039, n = 9 mice; for D2-MSN, p = 0.0020, n = 10 mice; Wilcoxon matched-pairs signed rank test, Figure 4G). We also noted from our results that randomly selected subset of single neurons outperformed averaged activities of the entire recorded MSN population in all our behavior decoding experiments (Figure 4B, 4G, and Table S1). These results suggest that for studies of neural encoding mechanisms in the brain, recording techniques allowing single neuron resolution will be more informative than the commonly used bulk recoding techniques such as fiber photometry.
These results demonstrated that locomotion velocity prediction based on cluster activity yielded lower prediction error therefore higher prediction accuracy than that based on randomly selected subset of single neurons or population activities of D1- or D2- MSN, consistent with our results from behavioral state decoding experiments (Figure 4B). Together, our results suggest that neural clusters activities in the dorsal striatum encode locomotion relevant information.
Discussion
In this study, we demonstrated that population activities of direct and indirect pathway neurons during locomotion were overall similar, providing additional evidence to support several recent studies showing concurrent activation of direct and indirect pathway neurons during behavior (Cui et al., 2013; Isomura et al., 2013; Jin et al., 2014; Tecuapetla et al., 2014). These data support an updated dual circuit striatal model arguing that normal behavior requires coordinated activities of the direct pathway neurons to promote intended behaviors and activities of the indirect pathway neurons to inhibit competing behaviors (Brown, 2007; Chan et al., 2005; Hikosaka et al., 2000; Mink, 2003; Nambu, 2008). We further took advantage of the single cell resolution that the miniScope system offers, in combination with advanced computational analysis, and identified spatially compact neural clusters in both the direct and indirect pathways of the dorsal striatum. Detailed characterization revealed that cluster organizations between direct and indirect pathways were overall similar. Cluster activities correlated with mouse locomotion, and cluster activity dynamics reflects striatal network states more accurately than population activity of MSN. Moreover, cluster activities outperformed MSN population activities in predicting mouse behavioral states as well as mouse locomotion velocities. Based on our results, we propose that neural clusters in the direct and indirect pathways encode locomotion relevant information, and postulate that spatially and temporally coordinated neural cluster activities of direct and indirect pathways are required for locomotion and likely other striatal-controlled behaviors.
The neural clusters identified in this manuscript may contribute to the formation of the cell assemblies identified in previous studies (Adler et al., 2012; Bakhurin et al., 2016; Carrillo-Reid et al., 2008; Jaidar et al., 2010; Ponzi and Wickens, 2012). Computation modeling of striatal networks (Ponzi and Wickens, 2012) has predicted that a real striatal network at its characteristic sparse connectivity level should display a connectivity distribution following the power-law distribution that were scale-free and capable of self-organizing (Barabasi and Albert, 1999). In this network, cell assemblies would form spontaneously and fire sequentially in a behavior relevant time scale (Ponzi and Wickens, 2012). Experimentally, dynamic striatal cell assemblies have recently been identified to display sequential activity in a behavior relevant manner (Adler et al., 2012; Bakhurin et al., 2016), suggesting that these cell assemblies likely carry behavior relevant information. Here we speculate that spatial and temporal coordination of neural cluster activities leads to the formation of different cell assemblies that encode distinct striatal network states. Alterations in cluster activity and synchrony dynamics allow transition between different cell assemblies that define different striatal network states, which correspond to specific behavioral states. It is worth noting that from an ex vivo study for Parkinson’s disease model, striatal network was locked into a dominant network state after dopamine deletion (Jaidar et al., 2010), consistent with the notion that abnormal higher striatal network synchrony contributed to Parkinson’s disease pathology (Costa et al., 2006). Therefore, under the pathological conditions of the Parkinson’s disease, insufficiency of dopamine in the dorsal striatum may entrain the striatal network into a dominant state, preventing proper transitions between different cell assembly states that are required for locomotion initiation or termination. Conversely, here in our study, under cocaine influence, excessive dopamine far beyond physiological level may lead to a different striatal network state that is distinct from normal locomotion conditions. In our behavior decoding experiment, same clusters contributed differently in different behavior decoding experiment (e.g., cocaine state versus normal locomotion states in Figure 4C–E). The weighted contributions from individual clusters led to different combinations of clusters that formed distinctive decoding maps, likely resembling the cell assemblies that were identified from previous studies, which may represent the specific functional state of the striatal network for a particular behavior state.
Although currently we do not know the exact mechanism driving the spatially compact organization of neural clusters in dorsal striatum, previous studies on synchronous cell assemblies in striatal networks offer some clues. Computation modeling predicted that striatal neurons would spontaneously form cell assemblies with background synaptic inputs (Humphries et al., 2009). Consistent with this computational prediction, ex vivo experiments using striatal slices demonstrated that bath application of NMDA enhanced glutamatergic transmission and revealed cell assemblies that exhibited spontaneous transitions between different network states, while blocking all glutamatergic transmissions abolished synchronous cell assembly activities (Carrillo-Reid et al., 2008). These results suggest that glutamatergic transmissions likely drive cell assembly formation. Recent in vivo experiments demonstrated that at rest, striatal neurons displayed resting state correlations that were spatially clustered, and the resting state network dynamics resembled behavior-modulated network dynamics (Bakhurin et al., 2016). Correlated activities of both spontaneous and behavior relevant network dynamics suggest a common structural basis to support the network dynamics, which could be attributed to common glutamatergic inputs (Cohen and Kohn, 2011). Together, these results support the role of glutamatergic transmissions driving cell assembly organization. By contrast, blocking GABAergic transmissions entrained cell assemblies into a preferred state (Carrillo-Reid et al., 2008), suggesting that GABAergic transmission is not required for synchronous cell assembly formation, but instead facilitates transitions between different cell assembly states. Computational modeling also predicted that striatal neurons could form cell assemblies spontaneously without dopaminergic inputs (Humphries et al., 2009). In an ex vivo experiment with dopamine depleted brain slices, after striatal neurons were locked into a dominant state, application of dopamine agonist was able to uncouple neurons from the dominant state, suggesting that dopamine also facilitated transitions between different cell assembly states (Jaidar et al., 2010). Taken together, evidence in the literature suggests that glutamatergic transmission drives striatal cell assembly formation, and may also be responsible for functional organization of neural cluster formation via synaptic plasticity mechanisms, whereas GABAergic and dopaminergic transmissions would modulate cluster activity and synchrony and shape transitions between different functional states.
While single neurons are likely the basic information-encoding units in the dorsal striatum, averaging signals from spatially compact clusters of similarly firing MSNs could give rise to a more reliable striatal code. Thus, it appears that neurons within a cluster could compensate for each other’s coding errors. Therefore, it is possible that downstream structures utilize average signals from MSN clusters to refine striatal-dependent control of movement, and that striatal disorders in which clusters are disrupted may lead to a less refined striatal code and consequently movement deficits. Our results revealed spatially compact neural clusters that may contribute to the functional cell assembly organization in the direct and indirect pathways. These findings make an important connection between the dual circuit model of the basal ganglia and the cell assembly functional model of striatum. Our results from behavior decoding experiments showing that cluster activities performed better than population activities for both D1- and D2- MSN, strongly suggest that neural cluster activities of D1- and D2- MSN encode locomotion relevant information. It is conceivable that D1-MSN clusters encode information that facilitates locomotion, and D2-MSN clusters encode information that inhibits unwanted locomotion, and coordination of D1- and D2- MSN clusters form cell assemblies that guide locomotion sequences. Notably, beyond the opposite aspect of locomotion regulation proposed by the longstanding model of striatal direct and indirect pathway, such opposing roles for direct and indirect pathway neurons have also been suggested in reward and punishment (Kravitz et al., 2012), in goal-directed behavior (Shan et al., 2014; Sippy et al., 2015; Tai et al., 2012), and in mechanisms influencing primary motor cortex (Oldenburg and Sabatini, 2015). We therefore speculate that similar coordinated activities of neural clusters in the direct and indirect pathways of dorsal striatum may also encode other striatal-controlled behaviors. Future studies applying in vivo imaging techniques to simultaneously record the activities from both direct and indirect pathway neural clusters will be important to test this hypothesis.
Methods
All experiments were conducted in accordance with the guidelines of Institutional Animal Care and Use Committee, the Intramural Research Program, National Institute on Drug Abuse, National Institutes of Health. Transgenic mice expressing Cre recombinase under the control of the dopamine D1 receptor (D1-Cre, FK150 line, C57BL/6J congenic, Gensat, RRID:MMRRC_036916-UCD) or dopamine D2 receptor (D2-Cre, ER44 line, C57BL/6J congenic, Gensat, RRID:MMRRC_032108-UCD) were used in the experiments. To image GCaMP6 fluorescence in dorsal striatum (DS), we first injected AAV1.CAG.Flex.GCaMP6s.WPRE.SV40 (University of Pennsylvania Vector Core) into the dorsal striatum. The AAV viruses were injected using the stereotactic coordinates (A/P: −0.93 mm, M/L: +1.8 mm, D/V: −3.46 mm, with 30° angle shift to caudate). One week after viral injection, a 1-mm diameter gradient index (GRIN) lens (GRINTECH GmBH) was directly implanted in the mouse brain right above the dorsal striatum under anesthesia with ketamine/xylazine (ketamine:100mg/kg. xylazine:15mg/kg). Two weeks after the GRIN lens implantation, the miniScope base was mounted onto the mouse head. We conducted a 5-day open-field test with a 34 cm×40 cm×20 cm chamber for all D1-Cre and D2-Cre mice. All the behavior tests were done in the light cycle. The calcium images were processed and analyzed using custom scripts in MATLAB. Clusters of neurons were identified based solely on their neural activity information (?F/F), through a clustering algorithm based on the meta k-means algorithm. All reported sample numbers represent biological replicates. All data were presented as mean ± sem unless otherwise stated. Detailed methods are in Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
Research was supported by NIH/NIDA/IRP. We would like to thank the Genetically-Encoded Neuronal Indicator and Effector (GENIE) Project and the Janelia Research Campus of the Howard Hughes Medical Institute for generously allowing the use of GCaMP6 in our research. We would like to thank Dr. Yavin Shaham of NIDA IRP for providing intellectual input on the data analysis and the write-up of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
DTL and YL conceptualized the project. DTL, YL, RC, GB, BL, LZ designed the experiments. GB and EC developed data acquisition hardware and software as well as data analysis software. BL and DTL designed the microscope and behavior recording system. LZ performed all surgical procedures. LZ and BL performed data acquisition. GB, BL, and RC performed data analysis under RC supervision, with inputs from DTL, YL, LZ, and CRG. All the authors contributed to the preparation of the manuscript.
Author Information
EC is a co-founder of TeraDeep Inc, Financial Conflicts of Interest are managed by Purdue University’s Conflict of Interest Committee. All other authors declare no competing financial interests.
Reference
- Adler A, Katabi S, Finkes I, Israel Z, Prut Y, Bergman H. Temporal convergence of dynamic cell assemblies in the striato-pallidal network. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32:2473–2484. doi: 10.1523/JNEUROSCI.4830-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albin RL, Young AB, Penney JB. The functional anatomy of basal ganglia disorders. Trends in neurosciences. 1989;12:366–375. doi: 10.1016/0166-2236(89)90074-x. [DOI] [PubMed] [Google Scholar]
- Alexander GE, Crutcher MD. Functional architecture of basal ganglia circuits: neural substrates of parallel processing. Trends in neurosciences. 1990;13:266–271. doi: 10.1016/0166-2236(90)90107-l. [DOI] [PubMed] [Google Scholar]
- Bakhurin KI, Mac V, Golshani P, Masmanidis SC. Temporal correlations among functionally specialized striatal neural ensembles in reward-conditioned mice. Journal of neurophysiology. 2016;115:1521–1532. doi: 10.1152/jn.01037.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balleine BW, Delgado MR, Hikosaka O. The role of the dorsal striatum in reward and decision-making. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27:8161–8165. doi: 10.1523/JNEUROSCI.1554-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabasi AL, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- Brown P. Abnormal oscillatory synchronisation in the motor system leads to impaired movement. Current opinion in neurobiology. 2007;17:656–664. doi: 10.1016/j.conb.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Carrillo-Reid L, Tecuapetla F, Tapia D, Hernandez-Cruz A, Galarraga E, Drucker-Colin R, Bargas J. Encoding network states by striatal cell assemblies. Journal of neurophysiology. 2008;99:1435–1450. doi: 10.1152/jn.01131.2007. [DOI] [PubMed] [Google Scholar]
- Chan CS, Surmeier DJ, Yung WH. Striatal information signaling and integration in globus pallidus: timing matters. Neuro-Signals. 2005;14:281–289. doi: 10.1159/000093043. [DOI] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisek P, Kalaska JF. Neural mechanisms for interacting with a world full of action choices. Annual review of neuroscience. 2010;33:269–298. doi: 10.1146/annurev.neuro.051508.135409. [DOI] [PubMed] [Google Scholar]
- Cohen MR, Kohn A. Measuring and interpreting neuronal correlations. Nature neuroscience. 2011;14:811–819. doi: 10.1038/nn.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa RM, Lin SC, Sotnikova TD, Cyr M, Gainetdinov RR, Caron MG, Nicolelis MA. Rapid alterations in corticostriatal ensemble coordination during acute dopamine-dependent motor dysfunction. Neuron. 2006;52:359–369. doi: 10.1016/j.neuron.2006.07.030. [DOI] [PubMed] [Google Scholar]
- Cui G, Jun SB, Jin X, Pham MD, Vogel SS, Lovinger DM, Costa RM. Concurrent activation of striatal direct and indirect pathways during action initiation. Nature. 2013;494:238–242. doi: 10.1038/nature11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong MR. Primate models of movement disorders of basal ganglia origin. Trends in neurosciences. 1990;13:281–285. doi: 10.1016/0166-2236(90)90110-v. [DOI] [PubMed] [Google Scholar]
- Gerfen CR, Engber TM, Mahan LC, Susel Z, Chase TN, Monsma FJ, Jr, Sibley DR. D1 and D2 dopamine receptor-regulated gene expression of striatonigral and striatopallidal neurons. Science. 1990;250:1429–1432. doi: 10.1126/science.2147780. [DOI] [PubMed] [Google Scholar]
- Gerfen CR, Paletzki R, Heintz N. GENSAT BAC cre-recombinase driver lines to study the functional organization of cerebral cortical and basal ganglia circuits. Neuron. 2013;80:1368–1383. doi: 10.1016/j.neuron.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Doughty M, Harbaugh CR, Cummins A, Hatten ME, Heintz N, Gerfen CR. Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27:9817–9823. doi: 10.1523/JNEUROSCI.2707-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graybiel AM. Network-level neuroplasticity in cortico-basal ganglia pathways. Parkinsonism & related disorders. 2004;10:293–296. doi: 10.1016/j.parkreldis.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Hikosaka O, Takikawa Y, Kawagoe R. Role of the basal ganglia in the control of purposive saccadic eye movements. Physiological reviews. 2000;80:953–978. doi: 10.1152/physrev.2000.80.3.953. [DOI] [PubMed] [Google Scholar]
- Humphries MD. Spike-train communities: finding groups of similar spike trains. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:2321–2336. doi: 10.1523/JNEUROSCI.2853-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries MD, Wood R, Gurney K. Dopamine-modulated dynamic cell assemblies generated by the GABAergic striatal microcircuit. Neural networks : the official journal of the International Neural Network Society. 2009;22:1174–1188. doi: 10.1016/j.neunet.2009.07.018. [DOI] [PubMed] [Google Scholar]
- Isomura Y, Takekawa T, Harukuni R, Handa T, Aizawa H, Takada M, Fukai T. Reward-modulated motor information in identified striatum neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:10209–10220. doi: 10.1523/JNEUROSCI.0381-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaidar O, Carrillo-Reid L, Hernandez A, Drucker-Colin R, Bargas J, Hernandez-Cruz A. Dynamics of the Parkinsonian striatal microcircuit: entrainment into a dominant network state. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:11326–11336. doi: 10.1523/JNEUROSCI.1380-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Tecuapetla F, Costa RM. Basal ganglia subcircuits distinctively encode the parsing and concatenation of action sequences. Nature neuroscience. 2014;17:423–430. doi: 10.1038/nn.3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravitz AV, Tye LD, Kreitzer AC. Distinct roles for direct and indirect pathway striatal neurons in reinforcement. Nature neuroscience. 2012;15:816–818. doi: 10.1038/nn.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mink JW. The basal ganglia: focused selection and inhibition of competing motor programs. Progress in neurobiology. 1996;50:381–425. doi: 10.1016/s0301-0082(96)00042-1. [DOI] [PubMed] [Google Scholar]
- Mink JW. The Basal Ganglia and involuntary movements: impaired inhibition of competing motor patterns. Archives of neurology. 2003;60:1365–1368. doi: 10.1001/archneur.60.10.1365. [DOI] [PubMed] [Google Scholar]
- Nambu A. Seven problems on the basal ganglia. Current opinion in neurobiology. 2008;18:595–604. doi: 10.1016/j.conb.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Oldenburg IA, Sabatini BL. Antagonistic but Not Symmetric Regulation of Primary Motor Cortex by Basal Ganglia Direct and Indirect Pathways. Neuron. 2015;86:1174–1181. doi: 10.1016/j.neuron.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozden I, Lee HM, Sullivan MR, Wang SS. Identification and clustering of event patterns from in vivo multiphoton optical recordings of neuronal ensembles. Journal of neurophysiology. 2008;100:495–503. doi: 10.1152/jn.01310.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Platisa J, Verhagen JV, Gautam SH, Osman A, Kim D, Pieribone VA, Culurciello E. Head-mountable high speed camera for optical neural recording. Journal of neuroscience methods. 2011;201:290–295. doi: 10.1016/j.jneumeth.2011.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnevmatikakis EA, Soudry D, Gao Y, Machado TA, Merel J, Pfau D, Reardon T, Mu Y, Lacefield C, Yang W, et al. Simultaneous Denoising, Deconvolution, and Demixing of Calcium Imaging Data. Neuron. 2016;89:285–299. doi: 10.1016/j.neuron.2015.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponzi A, Wickens J. Input dependent cell assembly dynamics in a model of the striatal medium spiny neuron network. Frontiers in systems neuroscience. 2012;6:6. doi: 10.3389/fnsys.2012.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Q, Ge M, Christie MJ, Balleine BW. The acquisition of goal-directed actions generates opposing plasticity in direct and indirect pathways in dorsomedial striatum. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2014;34:9196–9201. doi: 10.1523/JNEUROSCI.0313-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sippy T, Lapray D, Crochet S, Petersen CC. Cell-Type-Specific Sensorimotor Processing in Striatal Projection Neurons during Goal-Directed Behavior. Neuron. 2015;88:298–305. doi: 10.1016/j.neuron.2015.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai LH, Lee AM, Benavidez N, Bonci A, Wilbrecht L. Transient stimulation of distinct subpopulations of striatal neurons mimics changes in action value. Nature neuroscience. 2012;15:1281–1289. doi: 10.1038/nn.3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tecuapetla F, Matias S, Dugue GP, Mainen ZF, Costa RM. Balanced activity in basal ganglia projection pathways is critical for contraversive movements. Nature communications. 2014;5:4315. doi: 10.1038/ncomms5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichmann T, DeLong MR. Functional neuroanatomy of the basal ganglia in Parkinson’s disease. Advances in neurology. 2003;91:9–18. [PubMed] [Google Scholar]
- Yin HH, Knowlton BJ. The role of the basal ganglia in habit formation. Nature reviews Neuroscience. 2006;7:464–476. doi: 10.1038/nrn1919. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.