Abstract
The thymus, an organ responsible for T cell development, is one of the more stress-sensitive tissues in the body. Stress, in the form of infections, radiation exposure, and steroids, impairs thymic epithelial cell (TEC) functions and induces the programmed cell death of immature thymocytes. MicroRNAs are small noncoding RNAs involved in tissue repair and homeostasis, with several supporting T cell development. We report that miR-205, an epithelial-specific miR, maintains thymopoiesis following inflammatory perturbations. Thus, the activation of diverse pattern recognition receptors in mice causes a more severe thymic hypoplasia and delayed T cell recovery when miR-205 is conditionally ablated in TECs. Gene expression comparisons in the TECs with/without miR-205 revealed a significant differential regulation of chemokine/chemokine receptor pathways, antigen processing components, and changes in the Wnt signaling system. This was partly a consequence of reduced expression of the transcriptional regulator of epithelial cell function, Forkhead Box N1 (Foxn1), and its two regulated targets, stem cell factor and ccl25, following stress. miR-205 mimics supplemented into miR-205-deficient fetal thymic organ cultures restored Foxn1 expression along with ccl25 and stem cell factor. A number of putative targets of miR-205 were up-regulated in TECs lacking miR-205, consistent with an important role for this miR in supporting T cell development in response to stress.
Keywords: development, lipopolysaccharide (LPS), microRNA (miRNA), pathogen-associated molecular pattern (PAMP), pattern recognition receptor (PRR), stress, stress response, T-cell biology
Introduction
The thymus provides a unique stromal niche for the development of T cells of the adaptive immune system (1, 2). This tissue is specified from the 3rd pharyngeal pouch during embryogenesis, when endothelial cells form a thymic epithelial cell (TEC)2 meshwork. These cells recruit hematopoietic stem cells and support their development into mature thymocytes through cell-cell interactions, chemokine gradients, and growth factors. TECs comprise two major subsets with distinct functions. Cortical TECs (cTECs) sustain the development and positive selection of immature thymocytes. Positive selection establishes the T cell receptor specificity of the developing thymocytes through interactions with the self-peptide/MHC molecules expressed by the TECs. After a repertoire of T cell receptor expressing single positive thymocytes (CD4+CD8− and CD4−CD8+) is generated, the thymocytes migrate into the medulla via chemokine gradients, where interactions with medullary TECs (mTECs) ensure the T cells are tolerant to self-peptide·MHC complexes (negative selection), which includes the expression of tissue-restricted antigens (TRAs) controlled by the transcriptional regulator, Aire (3). These mTECs, in conjunction with resident dendritic cells, also select for regulatory T cells.
The critical role of TECs is clearly revealed in patients with selected primary immunodeficiency diseases. A subgroup of individuals with 22q11.2 deletion syndrome (DiGeorge syndrome) have a peripheral T cell lymphopenia resulting from impaired specification and/or expansion of TECs (4, 5). Patients with mutations in Foxn1, the master transcription factor critical for TECs, have a severe combined immunodeficiency due to an ensuing thymic hypoplasia (6, 7). In normal individuals, the natural aging process contributes to a thymic atrophy (8, 9). The atrophy is linked to the reduced expression of Foxn1 and Wnt4, epithelial-to-mesenchymal transitions, adipogenesis, and fibrosis (10–12). Notably, enforced expression of Foxn1 in older mice attenuates the severity of this thymic atrophy (11, 13).
The thymus is exceedingly sensitive to stress, with cellular losses often reaching 90%. Infections, radiation exposure, trauma, and alcoholism each can elicit a thymic hypoplasia, reducing T cell output (14–17). Corticosteroids, produced following infections, and sex hormones such as testosterone, cause a thymic hypoplasia (16, 18, 19). The type of stress predicates whether TECs, thymocytes, and/or both populations are compromised (17). For example, activation of Toll-like receptor 3, MDA-5, and the RIG-I-like receptor pathways following viral infections generates a type I interferon (IFN) response that primarily targets TECs (20, 21). Toll-like receptor 4 driven responses, initiated by the detection of lipopolysaccharide (LPS) released by Gram− bacteria, triggers a programmed cell death that primarily affects immature CD4+CD8+ thymocytes.
MicroRNAs (miRs) are small noncoding RNAs that regulate stress responses and maintain cellular homeostasis in diverse organs (22, 23). The functional contribution of miRs in regulating TEC functions was revealed in mice harboring a conditional ablation of the miR-processing enzyme, Dicer (21, 23, 24). The failure to generate miRs in these TECs results in a progressive destruction of the thymic architecture (21, 23). The damage is exacerbated following stress responses involving type I IFNs (21, 23). Several of the stress responsive miRs identified in the thymus include miR-29a, miR-181d, miR-185, and miR-205 (14, 21, 25, 26). miR-29a is a key miR mitigating TEC involution because it targets IFN receptor α in the TECs, reducing their sensitivity to IFN (21). A miR-29a deficiency is not as damaging as that revealed with the loss of Dicer, suggesting that additional miRs are coupled to TEC functions.
A second candidate is miR-205. This miR is epithelial cell-specific, predominately expressed in the thymus, skin, stomach, tongue, and bladder (27–29). The deletion of miR-205 results in a partial postnatal lethality in mice (28, 29). miR-205 is highly expressed in both cortical and medullary TECs, with higher levels observed in the mTEC subsets (30). Recent reports comparing the stress damage in mice lacking miR-205 selectively in TECs demonstrated equivalent thymic involution and normal post-stress recovery upon low dose injections of poly(I:C), a dsRNA mimic that induces type I IFN (30). We previously reported that miR-205 is elevated in the thymic tissue in response to strong inflammatory perturbations (14). Consequently, we compared the thymic tissue damage in mice with/without miR-205 in TECs using high doses of poly(I:C), as well as stress induced by other pathogen-associated molecular patterns (PAMPs). We report that the miR-205 deficiency causes a more pronounced thymic involution, with compromised TEC functions. Gene expression comparisons revealed a differential expression of chemokines, cytokines, and components of the Wnt signaling pathway in miR-205-deficient TECs. Foxn1 expression was significantly reduced in these TECs following stress. Restoring miR-205 expression in fetal thymic organ culture with miR-205 mimics increased Foxn1 expression and two chemokines regulated by Foxn1, ccl25 and scf, indicating an essential regulatory role for miR-205 in TECs following stress.
Results
miR-205 Positively Regulates Thymic Cellularity following Stress
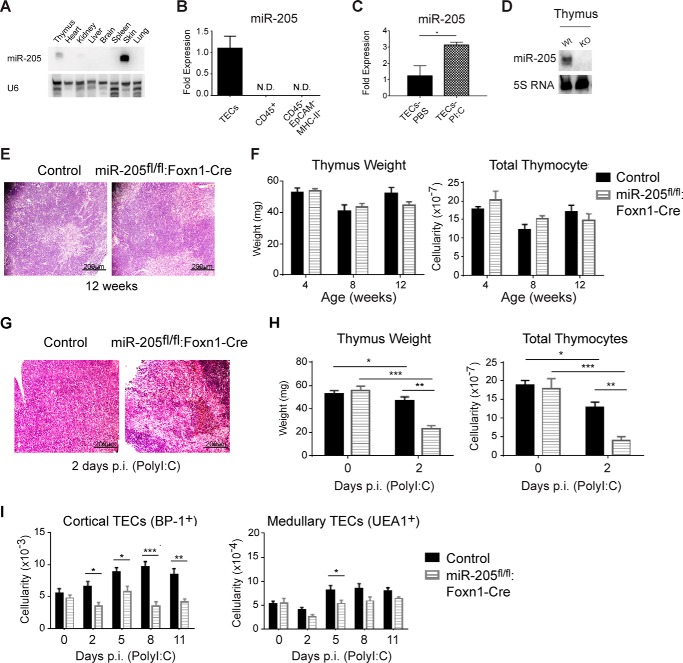
Northern blot analysis comparing different tissues revealed that miR-205 was predominantly expressed in the thymus and skin (Fig. 1A). In the thymus, miR-205 was selectively expressed in TECs, with no expression detected in the CD45+ hematopoietic cells, which comprise mostly thymocytes, or other thymic stromal cells (CD45−EpCAM−MHC class II−) (Fig. 1B). miR-205 is expressed in both cortical and medullary TECs, with higher levels noted in the medullary subset (30). It is up-regulated in TECs following inflammatory responses (Fig. 1C) (14). An initial characterization of human thymii, several from patients with 22q11.2 deletion syndrome, revealed diminished or absent miR-205 expression in severely hypoplastic tissues. Although suggesting a connection between miR-205 and thymopoiesis, the undefined nature of these hypoplastic tissues prevented confirmation of this hypothesis. To determine whether miR-205 contributes to thymopoiesis, mice were generated in which miR-205 was selectively removed in TECs by crossing miR-205 conditional knock-out mice with a Foxn1-Cre expressing line (miR205fl/fl:Foxn1-Cre mice) (supplemental Fig. S1 A) (27, 31). The elimination of miR-205 in the thymus was confirmed by Northern blot analysis (Fig. 1D). The elimination of miR-205 in TECs revealed a similar thymic appearance in control and miR205fl/fl:Foxn1-Cre mice (Fig. 1E). The thymic weight and cellularity in female age-matched miR205fl/fl:Foxn1-Cre mice was similar to littermate controls at 4, 8, and 12 weeks of age (Fig. 1F). The littermate controls included miR-205fl/fl, miR-205fl/+, miR-205fl/+:Foxn1-Cre, or Foxn1-Cre mice, and these were indistinguishable from wild type C57Bl/6 mice.
FIGURE 1.
Mice lacking miR-205 in thymic epithelial cells exhibit a more severe thymic stress response. A, Northern blots with a probe specific for miR-205 reveals its predominant expression in the thymus and skin versus the heart, kidney, liver, brain, spleen, and lung. U6 was used as a RNA loading control. B, real-time PCR experiments were used to evaluate the expression of miR-205 in sorted thymic epithelial cells (CD45−EpCAM+MHCII+) compared with isolated thymocytes (CD45+) and other stromal cells (CD45−EpCAM−MHCII−). C, real-time PCR revealed the expression of miR-205 in sorted thymic epithelial cells (CD45−EpCAM+MHCII+) before and following a systemic inflammatory response induced by poly(I:C) for 5 days. D, the elimination of miR-205 in thymic epithelial tissue was confirmed by Northern blots of RNA miR-205fl/fl:Foxn1-Cre mice compared with sibling controls. 5S RNA was used as a RNA loading control. E, histochemical analysis of the indicated thymii isolated from female mice 12 weeks of age was compared using H&E staining of 5-μm thymus sections (×10, scale bar 200 μm). F, thymus weight and total thymic cellularity was compared in the control and miR-205fl/fl:Foxn1-Cre female at 4, 8, and 12 weeks of age. No statistically significant differences were observed. G, histochemical analysis of the indicated thymii processed 2 days after a 2nd poly(I:C) injection was compared using H&E staining of 5-μm thymus sections (×10, scale bar 200 μm). H, thymus weight and total thymic cellularity were compared in the control and miR-205fl/fl:Foxn1-Cre mice at day 0 or 2 days following treatment of poly(I:C). I, the total number of cortical (EpCAM+MHCII+UEA1−BP-1+) and medullary (EpCAM+MHCII+UEA1+BP-1−) TECs was compared between the indicated mice, either untreated (day 0) or 2, 5, 8, and 11 days post-poly(I:C) injection (gating not shown). *, p < 0.04; **, p < 0.006; ***, p < 0.0002; ****, p < 0.00001 (Student's t test). Data are representative of mean ± S.E. from at least 3 mice per group. All mice used for this experiment were female.
Recent work characterizing the stress role of miR-205 in the thymus revealed that the tissue damage in mice lacking miR-205 in TECs is comparable with control mice when using low doses of poly(I:C), ranging from 50 to 250 μg (30). We obtained similar results using these identical doses. Because the generation of a more severe thymic hypoplasia requires strong inflammatory insults, we exposed the miR205fl/fl:Foxn1-Cre mice to higher doses of poly(I:C) (supplemental Fig. S1B) (20, 21). A dose of 750 μg of poly(I:C) elicited a significant reduction in thymic cellularity in normal mice. Using this dose, the thymic architecture, weight, and cellularity were more severely disrupted in the miR-205fl/fl:Foxn1-Cre female mice, relative to littermate controls (Foxn1-Cre or miR-205fl/+:Foxn1-Cre) (Fig. 1, G and H). We next enumerated the number of cortical and medullary TECs at days 0, 2, 5, and 8, and 11 days post-poly(I:C) (Fig. 1I). In littermate controls, the number of cTECs increased at each time point examined following stress (Fig. 1H). In the miR-205fl/fl:Foxn1-Cre mice, the cTEC numbers failed to increase, with statistically significant reductions compared with littermate controls noted at 2, 5, 8, and 11 days post-injection (Fig. 1H). Medullary TEC numbers were reduced relative to controls uniquely at day 5 (Fig. 1H). Similar findings were revealed in male mice, indicating that miR-205 is required for optimal TEC function and recovery post-stress, independent of sex. By comparing the cTEC and mTEC subsets defined by the expression of MHC class II high and low, a similar reduction in each subpopulation was noted, indicating that all TEC subgroups are impacted equivalently (supplemental Fig. S1D).
To assess the impact of the TEC changes in response to poly(I:C) on thymocyte development, the percentage of CD4−CD8−, CD4+CD8+, and mature CD4+CD8− and CD4−CD8+ thymocytes was compared (Fig. 2A). The total number of thymocytes and CD4+CD8+ subset were significantly reduced in the miR-205 TEC-deficient mice at days 2 and 8 p.i. (Fig. 2B). The single positive CD4 and CD8 subsets were reduced at early and late time points (days 2, 8, and 11 p.i.), revealing that the developmental recovery of the SP subset was delayed in the miR-205fl/fl:Foxn1-Cre mice (Fig. 2B). This was not attributed to an altered TEC architecture as cytokeratin 5 (medulla) and cytokeratin 8 (cortex) staining were comparable between the controls and miR-205-deficient thymii (supplemental Fig. S2B). However, the number of F4/80-positive tangible body macrophages, a reflection of increased uptake of dying cells, appeared slightly higher in the mice lacking miR-205 in TECs 2 days p.i. poly(I:C) (supplemental Fig. S2B). Taken together, our findings indicate that miR-205 positively regulates both cortical and medullary TEC functions following interferon-triggered inflammatory responses.
FIGURE 2.
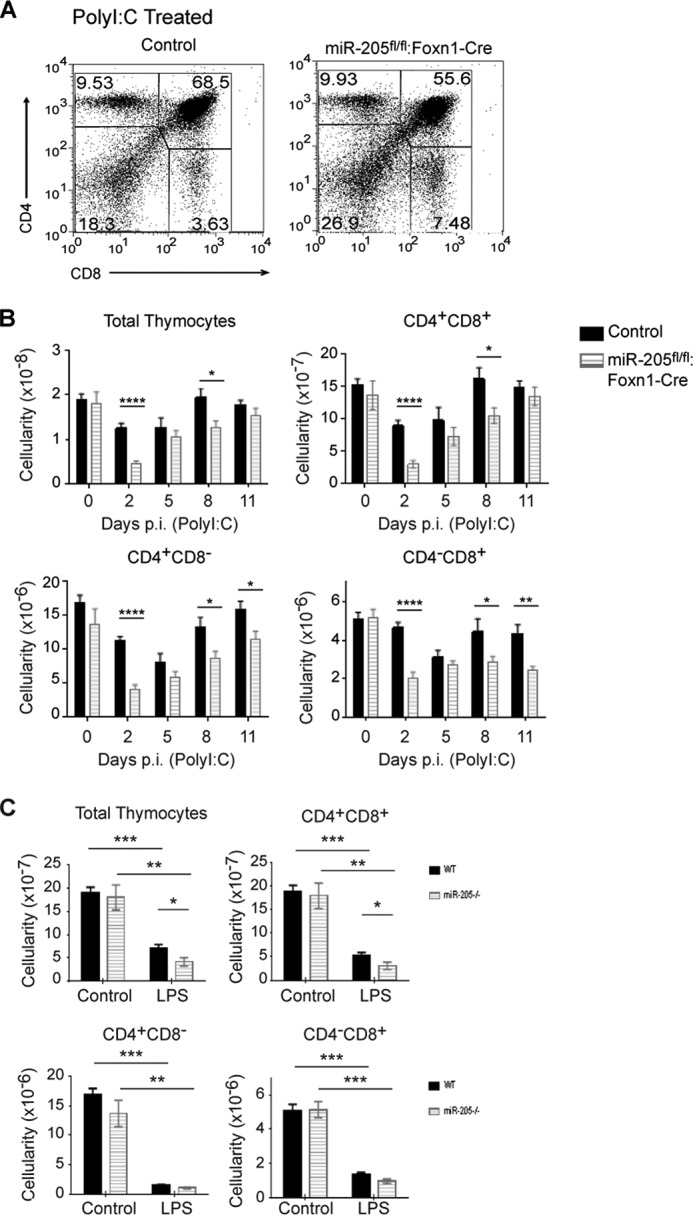
Stress induces a greater loss of all thymocyte subsets in mice lacking miR-205 in thymic epithelial cells. A, the percent of various thymocyte subsets following poly(I:C) treatment was determined by flow cytometry analysis staining with antibodies against CD4 and CD8 cell surface markers. B, the total thymic cellularity and the number of CD4+CD8+, CD4+CD8− and CD4−CD8+ thymocytes were compared between untreated (day 0) and poly(I:C)-treated mice 2, 5, 8, and 11 days post-injection. C, the total thymic cellularity and subset cellularity in untreated and LPS-treated sibling controls and miR-205fl/fl:Foxn1-Cre was calculated at 7 days post-injection. The effects of LPS on thymocyte subsets were determined by flow cytometric analysis of thymocytes stained with mAbs that detect the CD4 and CD8 cell surface markers. The thymii from the indicated mice were isolated 7 days post-LPS injection. *, p < 0.04; **, p < 0.006; ***, p < 0.0002; ****, p < 0.00001 (Student's t test). Data are representative of mean ± S.E. from at least 5 mice per group. All mice used for the poly(I:C) experiment were female. The LPS experiments were performed with male and female mice.
To determine whether miR-205 mitigates thymic stress in response to other pattern recognition receptors, mice were injected with a Toll-like receptor 4 agonist, LPS. LPS causes a severe thymic atrophy, primarily as a result of immature CD4+CD8+ thymocyte cell death (14, 16, 19). A statistically significant impaired recovery of the thymocytes occurred in the miR-205fl/fl:Foxn1-Cre mice by day 7 (Fig. 2C). However, the overall change in the thymic subsets and architecture was not as severe as that noted with poly(I:C) (Fig. 2C and data not shown). This indicates that the ability of miR-205 to mitigate thymic tissue damage depends on which PRR pathway in involved. The more severe damage to the thymus in the miR-205fl/fl:Foxn1-Cre mice following inflammatory perturbations prompted us to assess whether relocating these mice from a specific pathogen-free facility to a conventional facility would further establish a role for miR-205 in stress responses. The miR-205fl/fl:Foxn1-Cre female mice exhibited a significantly decreased thymic weight and cellularity compared with the controls after 8 weeks in the conventional facility (supplemental Fig. S2C). These experiments confirm that miR-205 significantly mitigates thymic involution to various microbial-mediated responses.
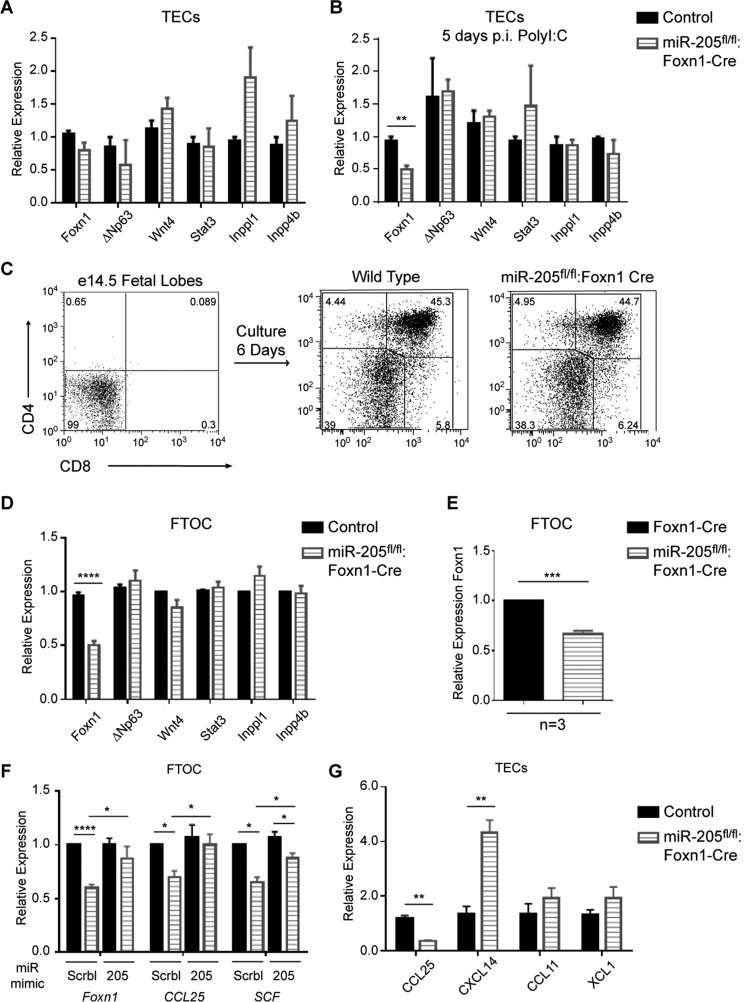
miR-205 Maintains Foxn1 Levels in TECs
To identify mRNAs affected by the absence of miR-205 in TECs, gene expression comparisons were made between TECs (CD45−EpCAM+MHCII+) sorted from 8-week-old sex-matched miR-205fl/fl:Foxn1-Cre and control mice. Quantitative RT-PCR (qPCR) with primers specific for genes important in thymopoiesis revealed no significant differences in the expression of Wnt4, ΔNp63, Stat3, or Inppl1 and Inpp4b, the latter two confirmed targets of miR-205 in skin epithelia (Fig. 3A) (28). Interestingly, the level of Foxn1, a master transcriptional regulator of TECs, was slightly decreased at the 8-week time point, but this did not reach significance (Fig. 3A). As the effects of a miR-205 TEC-selective deficiency were most obvious following stress, the gene expression comparisons were repeated with TECs sorted 5 days following poly(I:C) injections. This time point was chosen because miR-205 expression is at its peak, and the percentage of MHC-II high and low mTECs and cTECs was equivalent between the miR-205fl/fl:Foxn1-Cre and control (supplemental Fig. S1, C and D). Quantitative RT-PCR revealed a statistically significant 2-fold reduction in Foxn1 (Fig. 3B). ΔNp63, Wnt4, Stat3, Inppl1, and Inpp4b levels were again unchanged. To independently confirm the changes in Foxn1 expression, fetal thymic organ cultures (FTOC) were prepared from E14.5 embryos from wild type and miR-205fl/fl:Foxn1-Cre mice. At E14.5, cTECs are at the highest percent relative to the smaller numbers of medullary TECs and CD4−CD8− thymocytes (Fig. 3C and data not shown). Following 6 days of culture, the percentage of emerging CD4+CD8+ and single positive subsets was similar between the mice (Fig. 3C). Quantitative RT-PCR revealed a statistically significant 2-fold decrease in Foxn1 expression (Fig. 3D). This reduction was not due to the insertion of Cre in the distal exon of Foxn1, as Foxn1 levels were 2-fold higher in Foxn1-Cre mice compared with the miR-205fl/fl:Foxn1-Cre line (Fig. 3E). Wnt4, ΔNp63, Stat3, and the inhibitors of the PI3K pathway were not affected (Fig. 3D). E14.5 thymic lobes from miR-205fl/fl:Foxn1-Cre mice and control littermates were subsequently incubated with cholesterol stabilized control or miR-205 mimics. miR-205 mimics restored Foxn1 levels to that noted in control samples (Fig. 3F). Noteworthy, miR-205 mimics also increased the levels of ccl25 and stem cell factor (scf) (Fig. 3F). Previous work has established that these two genes are transcriptionally up-regulated by Foxn1 in knock-out and reconstitution experiments (32, 33). Of note, Foxn1 levels were not increased in the control cultures supplemented with the miR-205 mimics, suggesting that a maximum threshold of Foxn1 expression may exist (Fig. 3F). Other chemokines, such as cxcl14 were increased in the absence of miR-205, whereas others (ccl11 and xcl1) were unchanged (Fig. 3G). miR-205 mimics also reduced Frk, Inppl1, and Phlda3 levels in the cultures, with each a confirmed target of this miR in skin epithelia (supplemental Fig. S3A) (28). In summary, the data indicate that miR-205 is necessary to maintain normal levels of Foxn1 in TECs, which in turn transcriptionally regulates ccl25 and scf.
FIGURE 3.
miR-205 positively regulates Foxn1 expression. A, RNA was prepared from purified TECs (CD45−EpCAM+MHCII+), isolated from thymii from 8-week-old control and miR-205fl/fl:Foxn1-Cre mice. Quantitative RT-PCR was used to assess the expression of Foxn1, ΔNp63, Wnt4, Stat3, Inppl1, and Inpp4b. B, RNA was isolated from CD45−EpCAM+MHCII+ sorted TECs 5 days p.i. of poly(I:C) from the indicated mice. Foxn1, ΔNp63, Wnt4, Stat3, Inppl1, and Inpp4b levels were compared using qPCR. C, FTOC were established for a 6-day period. CD4 and CD8 cell surface expression was measured by flow cytometry in E14.5 fetal thymic lobes at day 0 and after 6 days of culture using the indicated mice. D, quantitative RT-PCR was used to assess the expression of Foxn1, ΔNp63, Wnt4, Stat3, Inppl1, and Inpp4b in FTOCs from control and miR-205fl/fl:Foxn1-Cre mice after 6 days of culture. Control samples were normalized to one, with the relative mRNA expression calculated with 5 independently isolated samples. E, quantitative RT-PCR was used to compare Foxn1 expression in the Foxn1-Cre mice relative to the miR-205fl/fl:Foxn1-Cre line. Foxn1-Cre was normalized to one, and data were calculated from three independent experiments. F, FTOCs from the indicated mice were cultured in the presence of control miR mimics or miR-205 mimics. Six days post-culture, gene expression comparisons were undertaken with qRT-PCR. Control samples were normalized to one with the relative expression of the genes calculated using at least 3 independently isolated samples/gene. G, the relative expression of ccl25, cxcl14, ccl11, and xcl1 was determined by qRT-PCR with RNA isolated from sorted TECs as described in A. In A–F, relative expression was calculated by ΔΔCT normalized to the endogenous peptidylpropyl isomerase A levels, with 3 or more independent experiments performed in triplicate. *, p < 0.04; **, p < 0.006; ***, p < 0.0002; ****, p < 0.00001 (Student's t test). Data are representative of mean ± S.E. from at least 3 independent RNA samples.
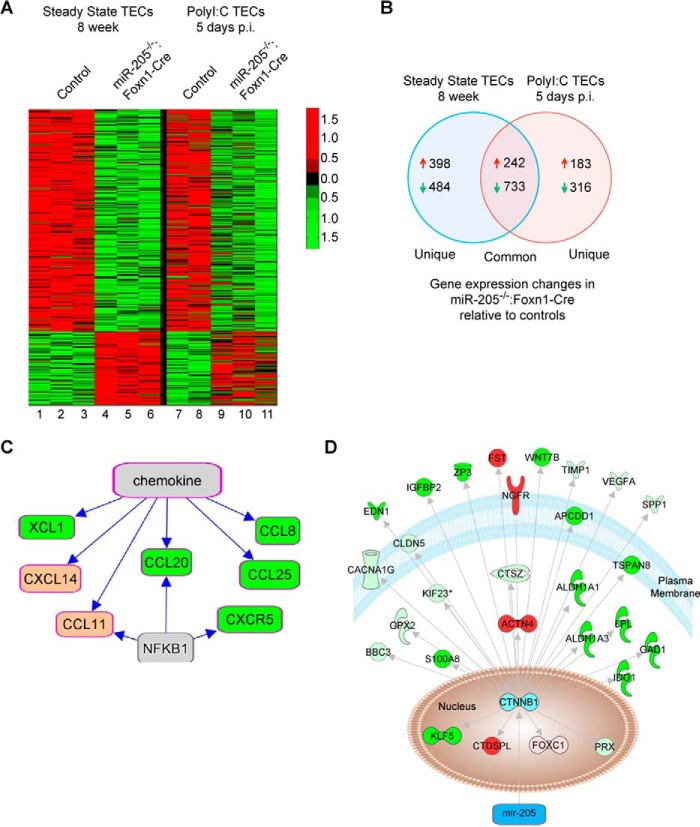
miR-205 Regulates Several Pathways Involved in Cell-Cell Communication and TEC Maintenance
To elucidate mechanisms whereby miR-205 supports TEC functions and Foxn1 expression, microarray gene expression comparisons were performed with TECs (CD45−EpCAM+MHCII+) sorted from the control mice and those lacking miR-205 in TECs (Fig. 4A). With 26,000 genes probed on the array, 242 genes were up- and 733 were down-regulated >1.5-fold in miR-205fl/fl:Foxn1-Cre mice at both steady state and in response to poly(I:C) (Fig. 4B, supplemental Table S1). Comparing TECs from 8-week-old mice, 398 genes were increased and 484 genes decreased in the absence of miR-205 (Fig. 4B). Following inflammatory perturbations, 183 up- and 315 down-regulated genes were differentially regulated in the miR-205fl/fl:Foxn1-Cre mice relative to controls (Fig. 4B, supplemental Table S2). As miRs primarily target the 3′ untranslated regions of mRNAs for degradation, microRNA prediction programs were used to identify genes up-regulated in the absence of miR-205. There were 109 putative targets of miR-205, with 19 having 3 or more miR-205 binding sites (Table 1, supplemental Table S3). In the stressed TECs, 69 of the 183 up-regulated genes were predicted targets of miR-205 (supplemental Table S3). Although several of the putative targets have roles in thymopoiesis, a substantial number remain uncharacterized, revealing many new candidates with a potential to regulate Foxn1 expression.
FIGURE 4.
The putative targets of miR-205 in TECs affect the expression of multiple chemokine regulated pathways. A, TECs were sorted from the thymii of 2 to 3 independent groups of control (lanes 1–3, 7, and 8) and miR-205fl/fl:Foxn1-Cre mice (lanes 4–6 and 9–11) from 8-week-old mice (lanes 1–6) or 5 days after a 2nd poly(I:C) treatment (lanes 7–11). RNA was isolated and used for gene expression comparisons. A heat map was used to indicate the up- (red) and down- (green) regulated genes from the indicated mice. B, Venn diagram demonstrating the number of up- and down-regulated genes that are common and unique to the steady state and stressed miR-205fl/fl:Foxn1-Cre TECs. C, Ingenuity Pathway Analysis was used to identify the most significant chemokine/chemokine receptor pathways differentially regulated in the absence of miR-205. D, Ingenuity Pathway Analysis of the genes increased (red) or decreased (green) coupled to the β-Catenin signaling pathway in miR-205-deficient TECs. Light green indicates genes uniquely down in stressed or 8 week TECs, whereas pink indicates genes uniquely up in 8 week or stressed TECs.
TABLE 1.
miR-205 predicted target genes up-regulated in thymic epithelial cells lacking miR-205
| Gene Name | miR-205 binding sites | Steady state fold ↑ | Stressed fold ↑ | Function |
|---|---|---|---|---|
| SULF1 | 5 | 1.64 | 1.53 | Sulfatase inhibits heparin-dependent growth factor signaling |
| KLHL29 (KBTBD9) | 4 | 1.62 | 1.97 | Unknown |
| Tmem169 (A830020B06RIK) | 3 | 1.85 | 1.62 | Unknown |
| Crtc3 (2610312F20RIK) | 3 | 1.66 | 1.63 | Transcriptional coactivator for CREB1 |
| Fam189b (1110013L07RIK) | 3 | 2.05 | 2.16 | Unknown |
| STC1 | 3 | 2.24 | 1.51 | Stimulates phosphate reabsorption |
| PPM1L | 3 | 3.83 | 2.06 | Inhibits apoptosis signal-regulating kinase 1 |
| Cnot1 (6030411K04RIK) | 3 | 2.15 | 1287.25 | CCR4-NOT complex scaffolding component |
| PLOD2 | 3 | 3.85 | 1.44 | Hydroxylates lysyl residues in collagen-like peptides |
| BMPR1A | 3 | 2.91 | 249.7 | BMP-2 and BMP-4 receptor |
| ANAPC5 | 3 | 2.12 | 1.42 | E3 ubiquitin ligase, controls mitosis and G1 cell cycle progression |
| ACTN4 | 3 | 4 | 1.5 | Epithelial cell tight junction assembly |
| SDK2 | 3 | 2 | 1.38 | Guides neuronal axons |
| BC021891 (Mlk4, KIAA1804) | 3 | 4.23 | 321.5 | Negatively regulates TLR4 |
| SEZ6L | 4 | 1.71 | 1.42 | ER functions in neurons |
| Phldb1 | 3 | 1.94 | 2.54 | Unknown |
| SATB1 | 3 | 4.84 | 2.22 | Mediates Xist RNA X inactivation |
| MYLK | 4 | 2.22 | 1.56 | Smooth muscle contration |
| Ninl (4930519N13RIK) | 6 | 1.63 | 1.69 | Regulates microtubule organization |
Ingenuity pathway analyses of the differentially expressed genes revealed a number of chemokines down-regulated (Fig. 4C). Others were increased in both experimental settings, and some uniquely in either steady state or stressed conditions (Table 2). These chemokines have essential roles in guiding T cell trafficking in the thymus (Table 2) (32, 34, 35). Cxcl14 was up-regulated >3.7-fold in the miR-205-deficient TECs (Table 2). Cxcl14 antagonizes the function of cxcl12, which is required for thymocyte selection and trafficking in the cortex (32, 35).
TABLE 2.
Chemokines differentially expressed in miR-205fl/fl:Foxn1-Cre thymic epithelial cells compared to controls
| Gene name | Steady state fold Δ | Stress fold Δ | Function |
|---|---|---|---|
| CCL8 | ↓2.07 | ↓2.12 | Attracts dendritic cells to the thymic cortex |
| CCL20 | ↓2.11 | ↓2.44 | Recruits DN1 thymocytes to the subcapsular zone and outer cortex |
| CCL25 | ↓4.29 | ↓4.02 | Recruits thymocyte precursors |
| XCL1 | ↓1.96 | ↓2.13 | Recruits dendritic cells into the thymic medulla. Regulated by AIRE |
| CXCL14 | ↑4.82 | ↑3.71 | Inhibits Cxcl12-Cxcr4-mediated chemotaxis |
| CCL11 (Eotaxin) | ↑1.95 | ↑1.68 | In the thymic medulla, function unknown |
| CCL2 | ↑1.67 | None | Elevations impair T cell development. |
| CXCL2 | None | ↓2.42 | Recruits neutrophils to stressed thymus |
| CXCL13 | None | ↓2.27 | Recruits B cells |
Twenty-nine distinct transcription factor-regulated pathways were down-modulated 2-fold or more when miR-205 was eliminated in TECs, whereas 6 pathways were increased (supplemental Fig. S3 B) (36–39). AIRE (autoimmune regulator) is highly expressed in medullary TECs and controls the expression of TRAs to prevent the development of autoimmunity (37). A number of TRAs were differentially expressed in the miR-205fl/fl:Foxn1-Cre TECs relative to controls. Yet, AIRE mRNA levels remained unchanged, suggesting that miR-205 regulates mRNAs that cooperate with AIRE (supplemental Tables S1 and S2) (40). Several β-Catenin-regulated genes, including TRAs were dysregulated in the miR-205-deficient TECs (Fig. 4D). As β-Catenin mRNA levels are unchanged in miR-205fl/fl:Foxn1-Cre TECs, miR-205 likely influences genes in complementary pathways.
Male Mice Lacking miR-205 in TECs Develop a Thymic Hypoplasia
In our initial studies of the mice housed in a pathogen-free facility, miR205fl/fl:Foxn1-Cre male mice exhibited an accelerated loss in thymic weight and cellularity at 8 and 12 weeks of age (Fig. 5A). By 24 weeks, the thymic cellularity in the control mice reached those noted in the knock-out line. With the exception of one time point, the cTEC and mTEC numbers were similar (Fig. 5B). As with the stressed female mice, the overall percentage of the different thymocyte subsets was similar to littermate controls (supplemental Fig. S4, A–C). The accelerated thymic hypoplasia impacted the peripheral lymphoid populations, as a statistically significant reduction in the number of CD4+CD8− and CD4−CD8+ T cells was noted in the spleens and lymph nodes of the miR-205fl/fl:Foxn1-Cre mice (supplemental Fig. S5, A and B). As testosterone produced in males reduces their thymic cellularity relative to females, we examined the contribution of sex hormones in the context of miR-205. Male mice were castrated and subsequently given dihydrotestosterone (5-DHT). The thymus weight increased similarly in the miR-205fl/fl:Foxn1-Cre castrated male mice and littermate controls (Fig. 5C). In addition, the subsequent reduction in thymic cellularity seen after DHT treatment was equivalent whether or not miR-205 was present in TECs (Fig. 5C). These findings indicate that other types of stress are affecting the male mice selectively.
FIGURE 5.
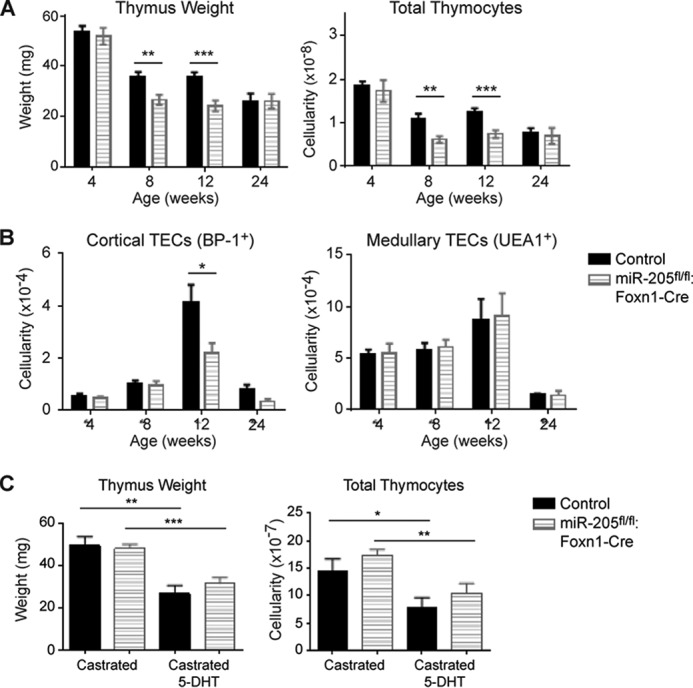
Male mice lacking miR-205 in TECs exhibit a testosterone-independent thymic hypoplasia relative to littermate controls. A, the thymus weight and total thymic cellularity was compared in littermate controls and the miR-205fl/fl:Foxn1-Cre males at 4, 8, 12, and 24 weeks of age. B, the total number of cortical (EpCAM+MHCII+UEA1−BP-1+) and medullary (EpCAM+MHCII+UEA1+BP-1−) TECs was compared between the indicated male mice at 4, 8, 12, and 24 weeks of age. C, 4-week-old miR-205fl/fl:Foxn1-Cre male mice and littermate controls were surgically castrated. Three to 4 weeks later, the mice received a 21-day slow release dihydrotestosterone pellet. At this time point, the thymic weight and cellularity were compared in the castrated mice versus those that received a subsequent increase in testosterone (DHT). *, p < 0.04; **, p < 0.006; ***, p < 0.0006. Data are representative of mean ± S.E. from at least 3 mice per group.
Discussion
MicroRNAs are critical regulators of cellular homeostasis, modulating tissue damage in many different organ systems following acute and chronic stress. The thymus is one of the more stress responsive organs, undergoing cellular losses that can approach 90% (17, 18, 41). miR-29a is one of the first individual miRs identified as playing a key role in TEC functions, in part via targeting of the interferon α receptor. We report herein that a deficiency of miR-205 in TECs results in an increased thymic involution in response to particular inflammatory mediators.
The most pronounced damage to the TECs lacking miR-205 occurred after an inflammatory response mediated by type I interferon. This is evidenced by the lack of cTEC expansion in mice lacking miR-205 in TECs following high dose poly(I:C) injections. Gene expression comparisons revealed a number of mRNA changes in the TECs that accounts for the more severe hypoplasia in the absence of miR-205. Included in this list are the reduced levels of many chemokines necessary for thymocyte recruitment and trafficking within the cortex and medulla (Table 2). Thus, miR-205 partly supports thymopoiesis by maintaining and/or re-establishing chemokine gradients following stress (42, 43). The localization of the cTECs and mTECs were not disrupted following stress, rather the thymocytes were mislocalized. Although many genes targeted by miR-205 could explain the reduced chemokine levels, the diminished expression of Foxn1 represents the most plausible explanation. Foxn1 transcriptionally activates the expression of the chemokines, scf, ccl25, cxcl12, and delta ligand like 4 (dll4). An important connection between miR-205 and Foxn1 was confirmed in fetal thymic organ culture experiments in which the addition of miR-205 mimics restored Foxn1 expression. In the presence of miR-205 mimics, scf and ccl25 increased. Both scf and ccl25 are positively regulated by Foxn1, as reported elsewhere (32, 33). As miRs principally function by targeting the 3′ untranslated regions of mRNAs, leading to the degradation of the latter, miR-205 likely binds and promotes the degradation of an mRNA that suppresses Foxn1 expression. Nineteen up-regulated genes had 3 or more predicted miR-205 binding sites. We are currently assessing whether any of these targeted mRNAs could impact Foxn1. It is interesting to note that many of the defined targets of miR-205 in skin epithelia were not the principal targets in TECs. As some were targets in FTOC when nanomolar concentrations of miR-205 mimics were added, the genes regulated by a miR are both cell type- and miR concentration-dependent.
A recent report demonstrated that miR-205 is not required for thymus recovery in response to low dose inflammation (30). We confirmed these findings and report herein that much higher doses of poly(I:C) were required to elicit a thymic hypoplasia that distinguished control littermates from the mice lacking miR-205 in TECs. Furthermore, environmental factors, such as transferring mice to a conventional facility, had a more severe impact in the thymic hypoplasia. This finding is consistent with our observation that differential PAMP exposure is more damaging in mice lacking miR-205 in TECs. Unlike PAMP-mediated effects, irradiation did not reveal any differences between control littermates and the miR-205 TEC-deficient mice. This agrees with a previous report (30). Sex differences connected to the absence of miR-205 were uncovered when comparing male versus female mice. The male mice, even when housed in a specific pathogen-free facility, exhibited thymic hypoplasia during the developmental stages when testosterone levels increase. Our experiments suggest that the contribution of miR-205 was not directly due to testosterone sensitivity even though miR-205 is reported to target the androgen receptor in the prostate (44). Thus, gene expression comparison of TECs ± miR-205 did not reveal differences in androgen receptor expression, either at steady state or following stress. These experiments suggest changes in inflammatory mediators are likely affecting the male mice, which may arise during in-fighting.
The successful use of miR-205 mimics in FTOC to restore normal Foxn1 expression reveals a new therapeutic avenue for improving T cell output in patients with decreased thymic functionality. This is particularly relevant in situations involving a chronic, interferon-dependent, inflammatory response. miR mimics are entering phase II clinical trials, increasing the possibility that miR-205 mimics could restore thymopoiesis in settings where T cell output has been reduced or compromised, such as in patients undergoing chemoablative therapies, and for the elderly.
Experimental Procedures
Mice
The animal studies were approved for use in this study by the Institutional Animal Care and Use Committee (IACUC) at the University of Texas Southwestern Medical Center (APN numbers 2010-0053 and 2015-101247). Mice were housed in both a specific pathogen-free facility and conventional room at University of Texas Southwestern Medical Center. The MIR205TM conditional knock-out mice were generated by the International Knock-out Mouse Consortium and acquired from the Jackson Lab (MGI:2676880) (27). Because the MIR205TM line (MGI:2676880) was initially provided on a mixed genetic background, the founder mice were backcrossed onto C57Bl/6 mice for 7–9 generations. This included crossing the mice with the C57Bl/6 pGK1-FlpO recombinase line to remove the LacZ cassette. The FlpO-recombinase mice were acquired from the Mutant Mouse Regional Resource Centers (C57Bl/6, MGI:4415609) (45). The miR-205fl/fl and miR-205fl/+ progeny were subsequently crossed with a Cre recombinase line in which Cre is expressed in a thymic epithelial tissue-specific manner using the Foxn1 3′ untranslated region, internal ribosome entry site Cre line (MGI:3760775). This results in the specific elimination of miR-205 in thymic epithelial cells starting at E11.25 (31). All progeny mice were further backcrossed onto C57Bl/6-derived lines for at least 4 generations to ensure all experiments were performed with conditional knock-out mice and sibling littermates on a C57Bl/6 background of >7 generations. Experiments included sibling controls, and no differences in these mice were observed when comparing MiR205fl/+:Foxn1-Cre, MiR205fl/fl, MiR205+/+:Foxn1-Cre, Foxn1-Cre, or wild type C57Bl/6 lines.
Thymic Epithelial Cell Isolation and Purification
Thymic epithelial cells were isolated from individual thymic lobes by digestion in LiberaseTM (Roche Applied Science) in the presence of DNase I (Roche) as described (46, 47). The cells were stained with antibodies against CD45 (Tonbo Scientific), MHC II (I-A/I-E) (Tonbo Scientific), EpCAM (eBioscience), BP-1 (eBioscience), and UEA-1 (Vector Laboratories). Samples were analyzed on FACSCantoII (BD Bioscience), with cell sorting done with a FACSAria (BD Bioscience). CD45−EpCAM+MHCII+ were collected and used for RNA isolation. Isolation of stromal cells was done by gating on CD45−EpCAM−MHCII− cells. FlowJo software (Tree Star Inc.) was used to analyze flow data. Thymic epithelial subsets were analyzed by selecting CD45−EpCAM+MHCII+ with either UEA1+BP1− for medullary TECs or BP1+UEA1− for cortical TECs. MHCII high and low cells were used to discriminate between the two cortical and two medullary TEC subsets. Thymocyte subsets were analyzed by cell surface expression of CD4 and/or CD8.
RNA Isolation and Analysis
Total RNA was isolated from intact thymic tissue homogenized in Qiazol (Qiagen), and processed using a miRNeasy Kit according to the manufacturer's instructions (Qiagen). Northern blot analysis was performed using 15–20 μg of RNA as previously described, using STARFireTM microRNA detection assays (IDT DNA Technologies) (14). RNA was prepared from the TECs, enriched using the cell sorter FACSAria (BD Bioscience), with the Ambion RNAaqueous micro kit (Invitrogen). All RNA samples were treated with DNase (Turbo DNase, Ambion), then cleaned and concentrated using a RNA clean and concentrator (TM-5, Zymo Research). cDNA was generated using ABI High Capacity cDNA synthesis kit (Life Technologies). qPCR was performed using Maxima SYBR Green (Thermo Scientific) on an ABI 7300 qPCR machine (Applied Biosystems). Experiments were done in triplicate with at least three independent samples per group. Relative expression of the genes analyzed was calculated as previously described (14). Student's t tests were performed between the wild type and miR-205-deficient samples with the wild type control normalized to one.
Fetal Thymic Organ Culture
Wild type C57B6 mice and miR-205fl/fl:Foxn1-Cre mice were used for timed pregnancies. Fetal thymus lobes were isolated at embryonic day 14.5 (E14.5) and cultured atop a nitrocellulose membrane in RPMI media (Cellgro) containing 20% fetal calf serum (Hyclone), and standard concentrations of penicillin, streptomycin, β-mercaptoethanol, l-glutamine, and non-essential amino acids at 37 °C in 5% CO2. Fetal lobes were harvested 6 days after culture. RNA was isolated using the miRVana kit (Ambion). MicroRNA mimics were purchased from Dharmacon (Thermo Scientific). miR-205 and control mimics were added at days 1 and 4 of FTOC, using a final concentration of 50 nm, delivered in Accell media (Thermo Scientific).
Immunofluorescence and Immunohistochemistry
Thymic tissues were fixed overnight in 4% paraformaldehyde in PBS at 4 °C. They were then dehydrated in an ethanol gradient of 25, 50, 75, and 100% (diluted in PBS). After washing in xylene, the tissues were embedded in paraffin and sectioned (4–6 μm thick). Slides were de-paraffinized in xylene and rehydrated using a descending ethanol gradient (100, 95, 90, 80, 70, and 40% ethanol). Antigen retrieval was performed for 30 min at 95 °C in Tris-EDTA buffer (10 mm Tris base, 1 mm EDTA, 0.05% Tween 20, pH 9.0). Slides were blocked in CAS Block (Invitrogen) for 1 h prior to addition of anti-cytokeratin 5 (Abcam), anti-cytokeratin 8 (Abcam), anti-Ki67 (BD Pharmingen), EpCAM (Thermo Scientific), and/or anti-Active Caspase-3 (BD Pharmingen) overnight. Secondary antibodies (Invitrogen) were used according to the manufacturer's instructions. The slides were stained with DAPI (Molecular Probes) prior to being mounted with Prolong Gold anti-fade reagent (Invitrogen). Images were taken on a Leica TCS SP5 confocal microscope and images were analyzed using ImageJ software. H&E-stained sections were imaged on an Axiovert 200M inverted fluorescent microscope.
Induction of Stress Responses
Four to 5-week-old mice were injected intraperitoneally (i.p.) with an increasing dosage of two injections spaced 3 days apart of 125, 250, 500, or 750 μg of polyinosinic:polycytidylic acid (poly(I:C)) (250 μl) (Sigma). Although no differences were seen at low doses, a thymic hypoplasia was consistently revealed at 750 μg. Subsequent experiments were performed with this dose. Thymii were harvested at days 2, 5, 8, and 11 days following the second injection. LPS (Sigma) was injected i.p. at a single dose of 100 μg.
Microarray Analysis
RNA was isolated from CD45−EpCAM+MHCII+ sorted TECs from control or miR-205fl/fl:Foxn1-Cre mice (n = 5 pooled thymii per group) at 8 weeks (steady state) or 5 days p.i. poly(I:C) (stressed). DNase treatments were used to remove contaminating genomic DNA followed by RNA clean and concentrator (Zymo Research). Two rounds of amplification for cDNA synthesis were performed. The cDNAs were applied to a MouseWG-6 version 2.0 Expression Bead Chip (triplicate samples/group). The amplification of cDNA and gene expression comparisons were performed by the University of Texas Southwestern Genomics and Microarray Core Facility. Data analysis was performed as described elsewhere (48).
Patient Samples
Informed consent was obtained for human studies under a protocol approved by the Institutional Review Board at University of Texas Southwestern Medical Center (STU-072010-003). The cardiothoracic group at Children's Health, Dallas, TX, identified patients with a likely diagnosis of DiGeorge/22q11.2 deletion syndrome. The thymus was obtained from individuals undergoing restorative cardiac surgery. The thymus size was variable from patient to patient. Samples were taken and processed for histological analyses. Thymic tissue sections were prepared and stained with hematoxylin and eosin by the Molecular Pathology Core at University of Texas Southwestern Medical Center. Larger fragments were used for RNA isolation. In brief, the samples were homogenized in 0.7 ml of Qiazol Lysis Reagent with a hand-held tissue homogenizer or Dounce homogenizer. RNA was extracted from these samples with a miRNeasy Mini Kit following the guidelines provided by the manufacturer (Qiagen). RNA was quantitated on a NanoDrop 2000 Spectrophotometer.
Single Cell Suspension of Lymphocytes and Flow Cytometry
Thymus, lymph nodes, and spleen were isolated and single cell suspensions generated. The mAbs used for staining and flow cytometry were performed as previously described (14). Samples were analyzed on a FACSCaliber (BD Bioscience) or FACSCantoII (BD Bioscience) and data were analyzed via FlowJo (Tree Star Inc.).
Castration and 5-DHT Treatment of Male Mice
Male mice were castrated at 4 weeks of age prior to the onset of sexual maturation. Slow release 5-DHT pellets (Innovative Research of America) were inserted under the skin 4 weeks post-castration as previously described (18). Thymocyte and TEC cellularity was assessed 21 days after insertion of slow release 5-DHT pellets as described above.
Statistical Analysis
All data were graphed and analyzed using GraphPad Prism software. Statistical analysis was performed using Student's t test. p values of <0.05 were considered significant. All graphical data are represented as mean ± S.E.
Author Contributions
A. H., M. d. l. M., and N. v. O. wrote the manuscript. A. H., J. M., Q. D., and N. v. O. performed the experiments. A. H., M. d. l. M., I. D., O. C., and N. v. O. designed the experiments, analyzed the data, and/or interpreted the findings. J. F. and K. G. participated in the isolation of normal and hypoplastic human thymii from patients.
Supplementary Material
Acknowledgments
We thank Angela Mobley and members of the flow cytometry core for assistance in cell sorting. We appreciate the suggestions provided by the graduate students and post-doctoral fellows in the Cleaver and Olson laboratories (Department of Molecular Biology at University of Texas Southwestern Medical Center). In addition, we sincerely appreciate the critical insights and suggestions from Drs. Joshua Mendell and Lora Hooper, both Howard Hughes Medical Institute investigators, at the University of Texas Southwestern Medical Center.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 AI114523 (to N. v. O.) and F31 AI110140 (to A. H.), and grants from the Children's Medical Center Research Foundation (to N. v. O., and M. d. l. M.), the Beecherl Grant from the University of Texas Southwestern Medical Center (to N. v. O.), and the Jeffery Model Foundation (to M. d. l. M.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Figs. S1–S5 and Tables S1–S3.
- TEC
- thymic epithelial cell
- cTEC
- cortical TEC
- mTEC
- medullary TEC
- TRA
- tissue-restricted antigens
- PAMP
- pathogen-associated molecular pattern
- miR
- microRNA
- qPCR
- quantitative PCR
- FTOC
- fetal thymic organ culture
- scf
- stem cell factor
- AIRE
- autoimmune regulator
- 5-DHT
- 5-dihydrotestosterone
- p.i.
- post-injection
- E
- embryonic day.
References
- 1. Anderson G., Lane P. J., and Jenkinson E. J. (2007) Generating intrathymic microenvironments to establish T-cell tolerance. Nat. Rev. Immunol. 7, 954–963 [DOI] [PubMed] [Google Scholar]
- 2. Anderson G., and Takahama Y. (2012) Thymic epithelial cells: working class heroes for T cell development and repertoire selection. Trends Immunol. 33, 256–263 [DOI] [PubMed] [Google Scholar]
- 3. Takaba H., Morishita Y., Tomofuji Y., Danks L., Nitta T., Komatsu N., Kodama T., and Takayanagi H. (2015) Fezf2 orchestrates a thymic program of self-antigen expression for immune tolerance. Cell 163, 975–987 [DOI] [PubMed] [Google Scholar]
- 4. Li B., Li J., Devlin B. H., and Markert M. L. (2011) Thymic microenvironment reconstitution after postnatal human thymus transplantation. Clin. Immunol. 140, 244–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gennery A. R. (2012) Immunological aspects of 22q11.2 deletion syndrome. Cell. Mol. Life Sci. 69, 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Adriani M., Martinez-Mir A., Fusco F., Busiello R., Frank J., Telese S., Matrecano E., Ursini M. V., Christiano A. M., and Pignata C. (2004) Ancestral founder mutation of the nude (FOXN1) gene in congenital severe combined immunodeficiency associated with alopecia in Southern Italy population. Ann. Hum. Genet. 68, 265–268 [DOI] [PubMed] [Google Scholar]
- 7. Romano R., Palamaro L., Fusco A., Giardino G., Gallo V., Del Vecchio L., and Pignata C. (2013) FOXN1: a master regulator gene of thymic epithelial development program. Front. Immunol. 4, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yager E. J., Ahmed M., Lanzer K., Randall T. D., Woodland D. L., and Blackman M. A. (2008) Age-associated decline in T cell repertoire diversity leads to holes in the repertoire and impaired immunity to influenza virus. J. Exp. Med. 205, 711–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gui J., Zhu X., Dohkan J., Cheng L., Barnes P. F., and Su D. M. (2007) The aged thymus shows normal recruitment of lymphohematopoietic progenitors but has defects in thymic epithelial cells. Int. Immunol. 19, 1201–1211 [DOI] [PubMed] [Google Scholar]
- 10. Dixit V. D. (2010) Thymic fatness and approaches to enhance thymopoietic fitness in aging. Curr. Opin. Immunol. 22, 521–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen L., Xiao S., and Manley N. R. (2009) Foxn1 is required to maintain the postnatal thymic microenvironment in a dosage-sensitive manner. Blood 113, 567–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Varecza Z., Kvell K., Talabér G., Miskei G., Csongei V., Bartis D., Anderson G., Jenkinson E. J., and Pongracz J. E. (2011) Multiple suppression pathways of canonical Wnt signalling control thymic epithelial senescence. Mech. Ageing Dev. 132, 249–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bredenkamp N., Nowell C. S., and Blackburn C. C. (2014) Regeneration of the aged thymus by a single transcription factor. Development 141, 1627–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Belkaya S., Silge R. L., Hoover A. R., Medeiros J. J., Eitson J. L., Becker A. M., de la Morena M. T., Bassel-Duby R. S., and van Oers N. S. (2011) Dynamic modulation of thymic microRNAs in response to stress. PloS One 6, e27580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Billard M. J., Gruver A. L., and Sempowski G. D. (2011) Acute endotoxin-induced thymic atrophy is characterized by intrathymic inflammatory and wound healing responses. PloS One 6, e17940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gruver A. L., and Sempowski G. D. (2008) Cytokines, leptin, and stress-induced thymic atrophy. J. Leukocyte Biol. 84, 915–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dooley J., and Liston A. (2012) Molecular control over thymic involution: from cytokines and microRNA to aging and adipose tissue. Eur. J. Immunol. 42, 1073–1079 [DOI] [PubMed] [Google Scholar]
- 18. Olsen N. J., Olson G., Viselli S. M., Gu X., and Kovacs W. J. (2001) Androgen receptors in thymic epithelium modulate thymus size and thymocyte development. Endocrinology 142, 1278–1283 [DOI] [PubMed] [Google Scholar]
- 19. Berki T., Pálinkás L., Boldizsár F., and Németh P. (2002) Glucocorticoid (GC) sensitivity and GC receptor expression differ in thymocyte subpopulations. Int. Immunol. 14, 463–469 [DOI] [PubMed] [Google Scholar]
- 20. Vidalain P. O., Laine D., Zaffran Y., Azocar O., Servet-Delprat C., Wild T. F., Rabourdin-Combe C., and Valentin H. (2002) Interferons mediate terminal differentiation of human cortical thymic epithelial cells. J. Virol. 76, 6415–6424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Papadopoulou A. S., Dooley J., Linterman M. A., Pierson W., Ucar O., Kyewski B., Zuklys S., Hollander G. A., Matthys P., Gray D. H., De Strooper B., and Liston A. (2012) The thymic epithelial microRNA network elevates the threshold for infection-associated thymic involution via miR-29a mediated suppression of the IFN-α receptor. Nat. Immunol. 13, 181–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mendell J. T., and Olson E. N. (2012) MicroRNAs in stress signaling and human disease. Cell 148, 1172–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zuklys S., Mayer C. E., Zhanybekova S., Stefanski H. E., Nusspaumer G., Gill J., Barthlott T., Chappaz S., Nitta T., Dooley J., Nogales-Cadenas R., Takahama Y., Finke D., Liston A., Blazar B. R., Pascual-Montano A., and Holländer G. A. (2012) MicroRNAs control the maintenance of thymic epithelia and their competence for T lineage commitment and thymocyte selection. J. Immunol. 189, 3894–3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Khan I. S., Taniguchi R. T., Fasano K. J., Anderson M. S., and Jeker L. T. (2014) Canonical microRNAs in thymic epithelial cells promote central tolerance. Eur. J. Immunol. 44, 1313–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Belkaya S., and van Oers N. S. (2014) Transgenic expression of microRNA-181d augments the stress-sensitivity of CD4+CD8+ thymocytes. PloS one 9, e85274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Belkaya S., Murray S. E., Eitson J. L., de la Morena M. T., Forman J. A., and van Oers N. S. (2013) Transgenic expression of microRNA-185 causes a developmental arrest of T cells by targeting multiple genes including Mzb1. J. Biol. Chem. 288, 30752–30762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Park C. Y., Jeker L. T., Carver-Moore K., Oh A., Liu H. J., Cameron R., Richards H., Li Z., Adler D., Yoshinaga Y., Martinez M., Nefadov M., Abbas A. K., Weiss A., Lanier L. L., et al. (2012) A resource for the conditional ablation of microRNAs in the mouse. Cell Rep. 1, 385–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang D., Zhang Z., O'Loughlin E., Wang L., Fan X., Lai E. C., and Yi R. (2013) MicroRNA-205 controls neonatal expansion of skin stem cells by modulating the PI(3)K pathway. Nat. Cell Biol. 15, 1153–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Farmer D. T., Shariat N., Park C. Y., Liu H. J., Mavropoulos A., and McManus M. T. (2013) Partially penetrant postnatal lethality of an epithelial specific microRNA in a mouse knockout. PloS One 8, e76634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Khan I. S., Park C. Y., Mavropoulos A., Shariat N., Pollack J. L., Barczak A. J., Erle D. J., McManus M. T., Anderson M. S., and Jeker L. T. (2015) Identification of miR-205 as a microRNA that is highly expressed in medullary thymic epithelial cells. PloS One 10, e0135440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gordon J., Xiao S., Hughes B. 3rd, Su D. M., Navarre S. P., Condie B. G., and Manley N. R. (2007) Specific expression of lacZ and cre recombinase in fetal thymic epithelial cells by multiplex gene targeting at the Foxn1 locus. BMC Dev. Biol. 7, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Calderón L., and Boehm T. (2012) Synergistic, context-dependent, and hierarchical functions of epithelial components in thymic microenvironments. Cell 149, 159–172 [DOI] [PubMed] [Google Scholar]
- 33. Itoi M., Tsukamoto N., and Amagai T. (2007) Expression of Dll4 and CCL25 in Foxn1-negative epithelial cells in the post-natal thymus. Int. Immunol. 19, 127–132 [DOI] [PubMed] [Google Scholar]
- 34. Bunting M. D., Comerford I., Kara E. E., Korner H., and McColl S. R. (2014) CCR6 supports migration and differentiation of a subset of DN1 early thymocyte progenitors but is not required for thymic nTreg development. Immunol. Cell Biol. 92, 489–498 [DOI] [PubMed] [Google Scholar]
- 35. Bunting M. D., Comerford I., and McColl S. R. (2011) Finding their niche: chemokines directing cell migration in the thymus. Immunol. Cell Biol. 89, 185–196 [DOI] [PubMed] [Google Scholar]
- 36. Wallin J., Eibel H., Neubüser A., Wilting J., Koseki H., and Balling R. (1996) Pax1 is expressed during development of the thymus epithelium and is required for normal T-cell maturation. Development 122, 23–30 [DOI] [PubMed] [Google Scholar]
- 37. Liston A., Lesage S., Wilson J., Peltonen L., and Goodnow C. C. (2003) Aire regulates negative selection of organ-specific T cells. Nat. Immunol. 4, 350–354 [DOI] [PubMed] [Google Scholar]
- 38. Otero D. C., Baker D. P., and David M. (2013) IRF7 dependent IFNB production in response to RANKL promotes medullary thymic epithelial cell development. J. Immunol. 190, 3289–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liang C-C., You L-R., Yen J. J. Y., Liao N-S., Yang-Yen H-F., and Chen C-M. (2013) Thymic epithelial β-catenin is required for adult thymic homeostasis and function. Immunol. Cell Biol. 91, 511–523 [DOI] [PubMed] [Google Scholar]
- 40. Derbinski J., Gäbler J., Brors B., Tierling S., Jonnakuty S., Hergenhahn M., Peltonen L., Walter J., and Kyewski B. (2005) Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. J. Exp. Med. 202, 33–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dudakov J. A., Hanash A. M., Jenq R. R., Young L. F., Ghosh A., Singer N. V., West M. L., Smith O. M., Holland A. M., Tsai J. J., Boyd R. L., and van den Brink M. R. (2012) Interleukin-22 drives endogenous thymic regeneration in mice. Science 336, 91–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Halkias J., Melichar H. J., Taylor K. T., and Robey E. A. (2014) Tracking migration during human T cell development. Cell. Mol. Life Sci. 71, 3101–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Halkias J., Melichar H. J., Taylor K. T., Ross J. O., Yen B., Cooper S. B., Winoto A., and Robey E. A. (2013) Opposing chemokine gradients control human thymocyte migration in situ. J. Clin. Invest. 123, 2131–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hagman Z., Haflidadóttir B. S., Ceder J. A., Larne O., Bjartell A., Lilja H., Edsjö A., and Ceder Y. (2013) miR-205 negatively regulates the androgen receptor and is associated with adverse outcome of prostate cancer patients. Br. J. Cancer 108, 1668–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Birling M.-C., Dierich A., Jacquot S., Hérault Y., and Pavlovic G. (2012) Highly-efficient, fluorescent, locus directed cre and FlpO deleter mice on a pure C57BL/6N genetic background. Genesis 50, 482–489 [DOI] [PubMed] [Google Scholar]
- 46. Williams K. M., Mella H., Lucas P. J., Williams J. A., Telford W., and Gress R. E. (2009) Single cell analysis of complex thymus stromal cell populations: rapid thymic epithelia preparation characterizes radiation injury. Clin. Transl. Sci. 2, 279–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Seach N., Wong K., Hammett M., Boyd R. L., and Chidgey A. P. (2012) Purified enzymes improve isolation and characterization of the adult thymic epithelium. J. Immunol. Methods 385, 23–34 [DOI] [PubMed] [Google Scholar]
- 48. Dozmorov I., and Lefkovits I. (2009) Internal standard-based analysis of microarray data. Part 1: analysis of differential gene expressions. Nucleic Acids Res. 37, 6323–6339 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.