SUMMARY
Inherited germline polymorphisms can cause gene expression levels in normal tissues to differ substantially between individuals. We present an analysis of the genetic architecture of normal adult skin from 470 genetically unique mice, demonstrating the effect of germline variants, skin tissue location, and perturbation by exogenous inflammation or tumorigenesis on gene signaling pathways. Gene networks related to specific cell types and signaling pathways including Sonic Hedgehog (Shh), Wnt, Lgr family stem cell markers, and keratins differed at these tissue sites, suggesting mechanisms for the differential susceptibility of dorsal and tail skin to development of skin diseases and tumorigenesis. The Pten tumor suppressor gene network is rewired in premalignant tumors compared to normal tissue, but this response to perturbation is lost during malignant progression. We present a software package for eQTL network analysis and demonstrate how network analysis of whole tissues provides insights into interactions between cell compartments and signaling molecules.
Graphical abstract
eTOC
The demands placed on human skin vary by physical location on the body, producing location specificity in cellular composition, signaling pathways, and response to perturbation, including differential susceptibility to inflammation and disease. Quigley et al. show how skin gene networks respond to perturbation by genetic variation, inflammation, and tumorigenesis.
INTRODUCTION
The skin is the largest human organ, forming an essential barrier against environmental insults, including physical and chemical exposures. Skin is the tissue of origin of the commonest form of cancer in Caucasian populations (Diepgen and Mahler, 2002), as well as a host of other diseases ranging from relatively common inflammatory conditions such as atopic dermatitis, to rare life-threatening conditions, such as the skin fragility syndrome Epidermolysis Bullosa. Studies both in mice and in humans have uncovered the underlying genetic basis of many skin diseases, and have led to pioneering discoveries in tissue transplantation, regeneration, and stem cell biology (Blanpain and Fuchs, 2006). Keratinocytes, the most common cell type in skin, produce several closely related families of proteins with distinctive locations and functions (Fuchs, 1995; Schneider et al., 2009). Keratins were initially characterized as structural proteins that form the cytoskeletal architecture (Steinert et al., 1985), but they can also function in signaling pathways in skin in response to tissue perturbation (Arwert et al., 2012; Gu and Coulombe, 2007; Paramio and Jorcano, 2002).
Skin morphology, function, and tumor susceptibility vary in different parts of the body (Rinn et al., 2008). To withstand physical stress, the soles of human feet, mouse paws, and mouse tails have thicker and tougher epidermal layers than dorsal skin. Mouse dorsal skin, but not tail skin, is highly sensitized to squamous papilloma development induced by chemical initiators and promoters of carcinogenesis (Schweizer and Marks, 1977b). Exposing Ptch1+/− mice to ionizing radiation produces basal cell carcinomas (BCCs) in dorsal skin (Wang et al., 2011). In contrast, activation of the hedgehog pathway by oncogenic smoothened (Smo) driven by widely expressed Krt14-Cre results in development of BCCs preferentially in mouse tail skin (Youssef et al., 2012). Similarly, over-expression of Gli2 using a Krt5 promoter also led to development of BCCs in the tail skin (Grachtchouk et al., 2000). The underlying basis of these site-specific phenotypes is not presently understood.
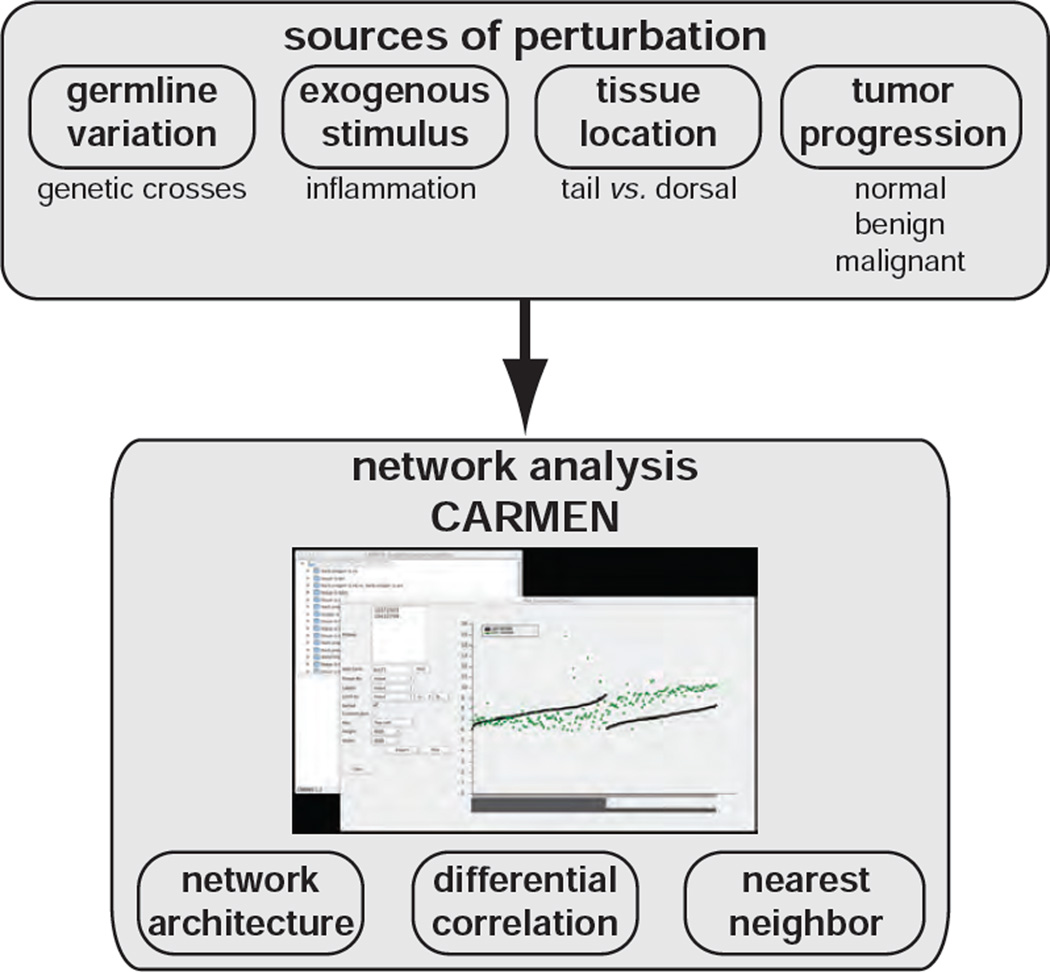
In this study we identify significant differences in the network architecture of signaling pathways between mouse dorsal and tail skin using gene expression quantitative trait locus (eQTL) and differential correlation analysis. We performed this analysis in a cohort of 470 genetically distinct animals produced by crossing FVB/N and Mus spretus mice, two highly divergent strains. We analyzed differential gene expression networks after stimulation of inflammation and epithelial proliferation using the tumor promoter TPA, or in tumors induced by sequential treatment with Dimethylbenzanthracene (DMBA) and TPA. We also present CARMEN, a user-friendly software package for network analysis. We demonstrate how this systems genetics strategy identifies the genes and pathways that are engaged in response to different forms of perturbation. These resources provide opportunities for generation and testing of many specific hypotheses related to skin biology.
RESULTS
Gene expression networks reconstruct the cellular composition of the skin
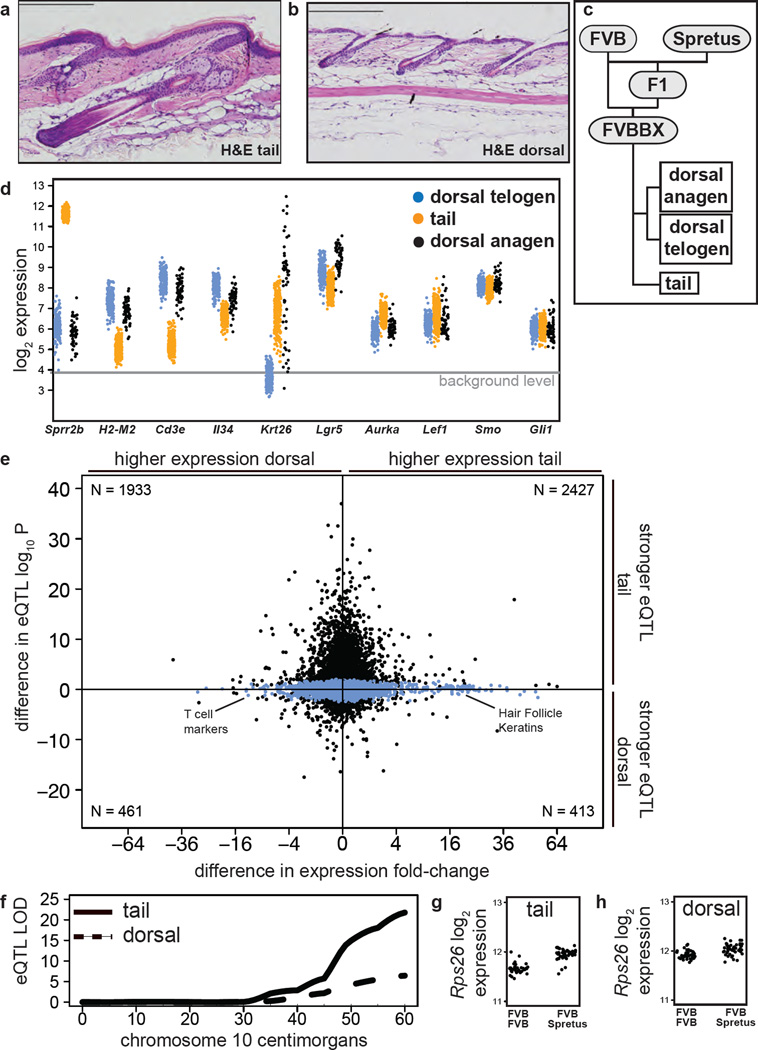
Adult mouse dorsal skin in the resting (telogen) phase has thin cornified and epithelial layers and small, densely packed hair follicles, while mouse tail skin has a thick and scaled cornified envelope layer and large, sparsely placed anagen hair follicles (Schweizer and Marks, 1977a) (Figure 1a, 1b). To investigate the genetic basis of the skin site-specific biological differences, we measured gene expression in 222 dorsal skins and 248 tail skins from a backcross of eight-week-old Mus musculus (FVB/N) and Mus spretus mice, referred to as FVBBX mice (Figure 1c, Methods). Tail skins had significantly higher expression of genes expressed in the outer cornified layer of the skin such as small proline receptor 2b (Sprr2b, Figure 1d). Individual hair follicles cycle through periods of growth (anagen) and destruction (catagen), with an intervening period of quiescence (telogen) (Stenn and Paus, 2001). Hair follicle cycling affects the skin’s physical structure and its susceptibility to experimental models of carcinogenesis (Mancuso et al., 2006; Miller et al., 1993). In the first two months after a mouse is born, its dorsal hair follicles progress synchronously through this cycle in a temporally predictable manner (Muller-Rover et al., 2001). Expression of hair-follicle inner-root-sheath specific keratins such as Krt26 was at or below background levels in 77% of the dorsal skin samples (Figure 1d), indicating that follicles in these skins were in telogen. All tail samples expressed hair-follicle-specific keratins at levels above background, consistent with follicles in tail being primarily in the anagen phase (Figure 1a, d).
Figure 1. Synchronized expression of major signaling pathways in dorsal skin.
(a,b) Hematoxylin and eosin stain of (a) tail skin and (b) dorsal skin from an eight-week-old mouse. Scale bars indicate 500 µM. (c) Outline of FVBBS breeding scheme and tissues obtained. (d) Gene expression levels of key genes in tail, dorsal anagen, and dorsal telogen skin. (e) Scatter plot comparing the difference in eQTL effect size for each probeset in tail and dorsal samples (y-axis) with the difference in mean expression levels in tail and dorsal samples (x-axis). Probes with significant eQTL in either tail or dorsal skin drawn in black; otherwise drawn in blue. The genes with the largest increase in eQTL strength tended to have the smallest change in gene expression levels. (f) Statistical strength of the eQTL for Rps26 in tail skin (solid line) and dorsal skin (dashed line) on chromosome 10. Expression of Rps26 in (g) tail skin and (h) dorsal skin separated by genotype at Chromosome 10, 118 Mb. F/F: homozygous FVB/N; F/S: heterozygous FVB/N / Spret/Ei.
Expression of mitotic markers such as Aurora kinase A (Aurka) was significantly higher in tail than in dorsal telogen or anagen skin (Figure 1d), consistent with elevated proliferation rates. In agreement with prior observations of site-specific immune responses in the skin (Bergstresser et al., 1980; Nabors and Farrell, 1994; Tong et al., 2014), tail skin showed dramatically reduced expression of markers of tissue-resident Langerhans cells such as Il34 (Wang et al., 2012), genes of the Major Histocompatibility Complex (MHC) such as H2-m2, and T cell surface markers such as Cd3e (Figure 1d). The highest expression of these genes in dorsal was during telogen phase (Figure 1d), confirming measurements made by Paus and colleagues (Paus et al., 1998).
Genetic influence on energy and metabolism gene expression is stronger in tail skin
We hypothesized that the genetic architectures of tail and dorsal skin would differ in ways that reflect their distinct forms and functions. To test this idea, we performed an expression Quantitative Trait Locus (eQTL) study in the 89 FVBBX mice where we had both dorsal skin and tail skin from the same animal. Gene expression data were corrected for strain-specific SNP effects on microarray hybridization (Methods). Using matched tissues reduced non-genetic variation and provided equivalent statistical power in both conditions. Surprisingly, we identified 4,753 genes with significant eQTL in tail skin but only 1,710 genes with significant eQTL in dorsal skin.
One possible explanation for this result would be if cell populations present preferentially in tail skin were the source of additional significant eQTL in tails. However, the number of genes expressed above background levels was similar in tail and dorsal skin and only a small number of genes had nearly-complete tissue-specific expression. Although most genes were significantly differentially expressed between matched tail and dorsal skin, the effect size was usually very small (median fold-change 6% higher in tail, Figure 1e). Genes with large location-specific differences in expression did not tend to have large differences in eQTL strength. Intermediate filament keratins in the hair follicle network such as Krt26 had the highest relative mean expression levels in tail skin (Figure 1e), reflecting the fact that most matched dorsal skin samples were in telogen. Many of the genes with highest relative expression in dorsal skin were T cell surface markers such as Cd3e (Figure 1d). However, the eQTL strength of these hair follicle keratins and T cell markers was similar in both tail and dorsal skin (Figure 1e). There was not a significant correlation between the difference in gene expression levels and the difference in eQTL strength. The peaks of almost all eQTL mapped to the same locus in tail and dorsal skin samples, and after increasing the number of samples in the tail eQTL analysis from 89 to 248 by adding additional unmatched FVBBX animals, we did not identify more than a handful of genes with eQTL exclusively in dorsal skin. These results suggest the tissue locations differed in linkage strength but not in their genetic architecture.
Gene Ontology enrichment analysis indicated that genes with the largest increase in eQTL strength (−log10P increase ≥ 10) were significantly enriched for roles in the mitochondria, cellular metabolism, translation, and energy production. These genes usually had very small differences in expression levels. One example was Rps26, a ribosomal protein mutated in Diamond-Blackfan Anemia, which can affect TP53 transactivation (Cui et al., 2014; Doherty et al., 2010). Rps26 was expressed at nearly identical levels in tail and dorsal skin but was under much stronger genetic control in tail skin (LODtail 21.8 vs. LODdorsal 6.4, Figure 1f–h). We interpret this to mean that the increase in tail eQTL strength was not simply the result of cell populations present in the tail skin and absent in dorsal skin, or the result of higher expression levels producing stronger linkage signals. Rather, these differences reflect differences in the activity of genes linked to protein production and cellular metabolism. This was concordant with the observation that levels of mitotic genes such as Aurora Kinase A are expressed at constitutively higher levels in tail skin (Figure 1d).
Conserved and tissue-specific networks in dorsal and tail skins
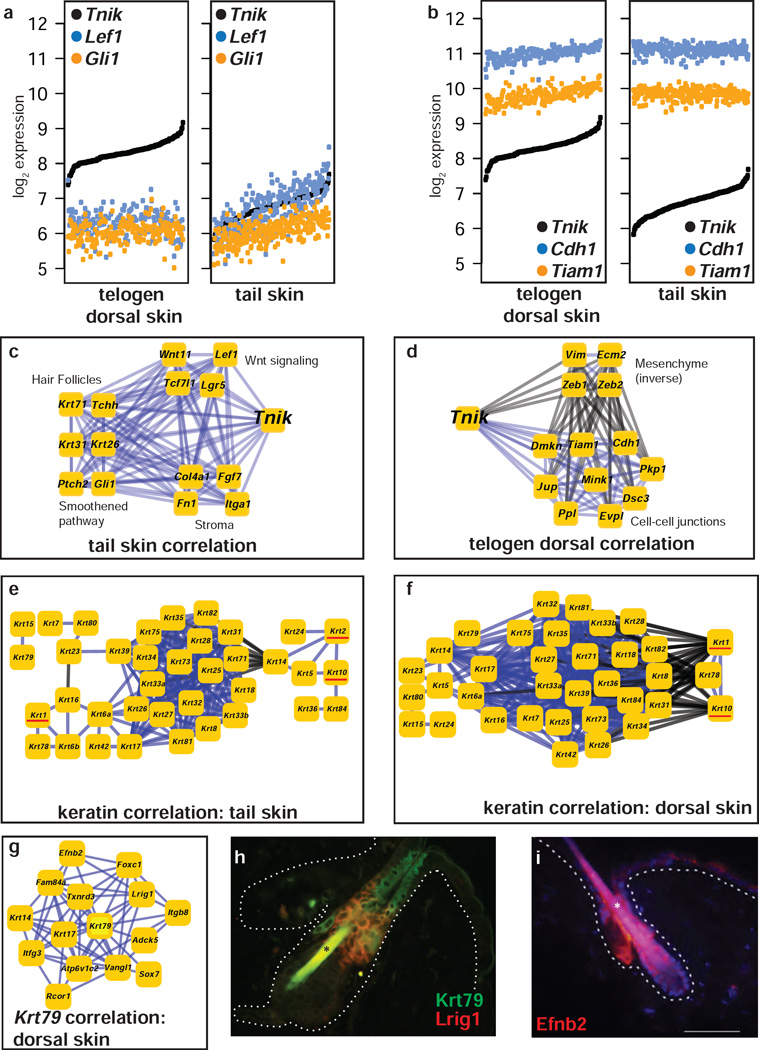
Signals from the Wnt and hedgehog pathways organize the hair placode during mouse embryonic development, and Wnt activation from the dermal papillae is required for follicle down-growth and cycling (Chiang et al., 1999; Gat et al., 1998; Huelsken et al., 2001; Kishimoto et al., 2000; St-Jacques et al., 1998). Expression of the Wnt-associated transcription factor Lef1 (DasGupta and Fuchs, 1999) was modestly higher in tail skin and dorsal anagen skin than in dorsal telogen, but expression levels of the Hedgehog pathway genes Smo and Gli1 were nearly identical between these tissues (Figure 1d). Gene expression correlation analysis can reveal features of signaling networks that are not detectable by measuring differential expression levels between tissues. Correlation was among the first statistical tools to be applied to microarray data to identify sets of genes which act in concert (Chu et al., 1998). In contrast, relatively few studies have exploited how the differences in correlation between contrasting conditions can elucidate the changes in how genes work together under different biological conditions. Several groups have developed formal methods to analyze differential correlation (reviewed in (de la Fuente, 2010)). Some methods determine whether pre-specified gene sets are differently correlated in two conditions (Braun et al., 2008; Choi and Kendziorski, 2009), while others discover gene sets directly from the data (Watson, 2006). We used CARMEN to calculate differential correlation between tail and dorsal telogen tissues (Methods). We then calculated pair-wise differential correlation genome-wide and ranked each gene by the number of significant differential correlations between the two tissue conditions. The 100 genes with the largest number of rewired correlations was enriched for genes with key roles in Wnt signaling such as Lef1, Fzd1, Tnik, Rspo3, and Lgr5, as well as genes with key roles in hair follicle stem cell biology such as Lgr5, Msx2, Lhx2, Hoxb13, and Dlx3 (Supplementary Table 1). The second-most-frequently rewired gene, TRAF2 and NCK interacting kinase (Tnik), activates Wnt target genes in intestinal crypts (Mahmoudi et al., 2009) and has a role in Wnt-driven colorectal tumorigenesis (Shitashige et al., 2010). In tail skin but not dorsal telogen skin, expression of Tnik was significantly correlated with expression of Lef1 and Gli1 (Figure 2a), whereas in dorsal but not tail skin Tnik expression was correlated with expression of adherens junctions genes such as e-cadherin (Cdh1) and T-cell lymphoma invasion and metastasis 1 (Tiam1) (Figure 2b). Plotting these gene networks (Figure 2c, 2d), allows the identification of gene pathway rewiring that would not be detectable from analysis of gene expression levels alone.
Figure 2. Keratin correlation networks in tail and dorsal skin.
(a, b) Gene expression levels of (a) Tnik, Lef1, and Gli1 and (b) Tnik, Cdh1, and Tiam1 in mouse dorsal telogen and tail skin, sorted by increasing Tnik expression. (c, d) Gene expression correlation of genes differentially expressed with Tnik expression in (c) tail skin and (d) telogen dorsal skin. (e,f) Gene expression correlation networks for all keratins in (e) tail skin and (f) dorsal skin; Krt1, Krt2, Krt10 underlined where present. Edges connect genes (boxes) with significantly genes expression; black edges are inverse correlation, blue edges direct correlation. (g) Gene expression correlation network for Krt79. (h, i) Immunofluorescence image for antibodies against (h) Lrig1 (red) and Krt79 (green) in FVB mice and (i) Efnb2.
Keratin gene expression networks in dorsal and tail skin
Many skin disorders are caused by germline alterations in genes encoding keratins, resulting in hyperproliferation, epidermal barrier defects, or defective control of epidermal permeability (Lane and McLean, 2004). We hypothesized that network expression analysis could identify rewired gene expression pathways associated with differing susceptibility to disease models. Mutations in Krt1 and Krt10 are linked to epidermolytic hyperkeratosis and ichthyosis, rare conditions which result in blistering and thickening of skin (Cheng et al., 1992; Rothnagel et al., 1992). The palmoplantar variety of hyperkeratosis particularly affects the palms and soles of the feet. The Dsk2 mutation at Krt2, a model for epidermolysis bullosa, results in hyperkeratosis in mouse footpads, ears, and tails but not dorsal skin (Fitch et al., 2003; Rentrop et al., 1987). Correlation analysis of keratin gene expression in dorsal and tail skin (Figure 2e, 2f, Krt1, Krt2, Krt10 underlined) provided a rationale for the functional significance of tail-specific expression of Krt2 (Rentrop et al., 1987). Expression of type II keratin Krt1 and type I keratin Krt10, while strongly correlated in dorsal skin, showed no mRNA relationship in tail skin. Although both Krt10 and Krt1 were highly expressed at the mRNA level in tails, Krt10 was significantly correlated only with Krt2. In agreement with these data, germline deletion of Krt2 in the mouse causes acanthosis, hyperkeratosis and inflammation of the ear and tail, with accumulation of aggregates of Krt10 (Fischer et al, 2014). Correlation network analysis would correctly have predicted that in thickened skin locations such as the tail, Krt2 substitutes for roles normally played by Krt1 in barrier function and inflammation.
We further analyzed the mRNA expression network for a keratin that is localized within a specific compartment of the skin. We calculated a correlation network for expression of the type II keratin Krt79 (Figure 2g), which is expressed in cells that line the infundibulum and co-localizes in part with expression of Krt17 (Veniaminova et al., 2013). The expression network for Krt79 included, in addition to Krt17, the stem cell marker Leucine-rich repeats and immunoglobulin-like domains 1 (Lrig1). Importantly, Lrig1 is a transmembrane protein and a marker of a stem cell that maintains the infundibulum and sebaceous gland, but not the hair follicle nor the interfollicular epidermis (Page et al., 2013; Veniaminova et al., 2013). Immunohistochemical analysis confirmed that Krt79 and Lrig1 protein are both localized within the infundibulum region, but primarily in adjacent cells rather than in exactly the same cell population (Figure 2h).
Expression of Efnb2 was also significantly correlated with expression of Krt79 in this network. Since very little is known about possible functions and localization of Efnb2 in skin, we carried out an immunohistochemical analysis of Efnb2 expression, and found that it is also expressed in the infundibulum and sebaceous gland, in a pattern that overlaps with but is more extensive than Krt79 and Lrig1 (Figure 2i). We conclude that analysis of expression networks derived from this heterogeneous mouse population can provide novel information on the localization and possible functional relationships between genes expressed in complex whole tissues in vivo. It is however important to note that many of the statistically significant genes within the Krt79 network may be expressed outside the infundibulum. While it is possible that significantly correlated genes are expressed in the same cells within a tissue compartment, or interact within the same signalling pathway, it is also possible that RNA correlations reflect paracrine relationships between adjacent cell types, or a hierarchical relationship within a cell lineage.
Network changes in response to inflammatory agents
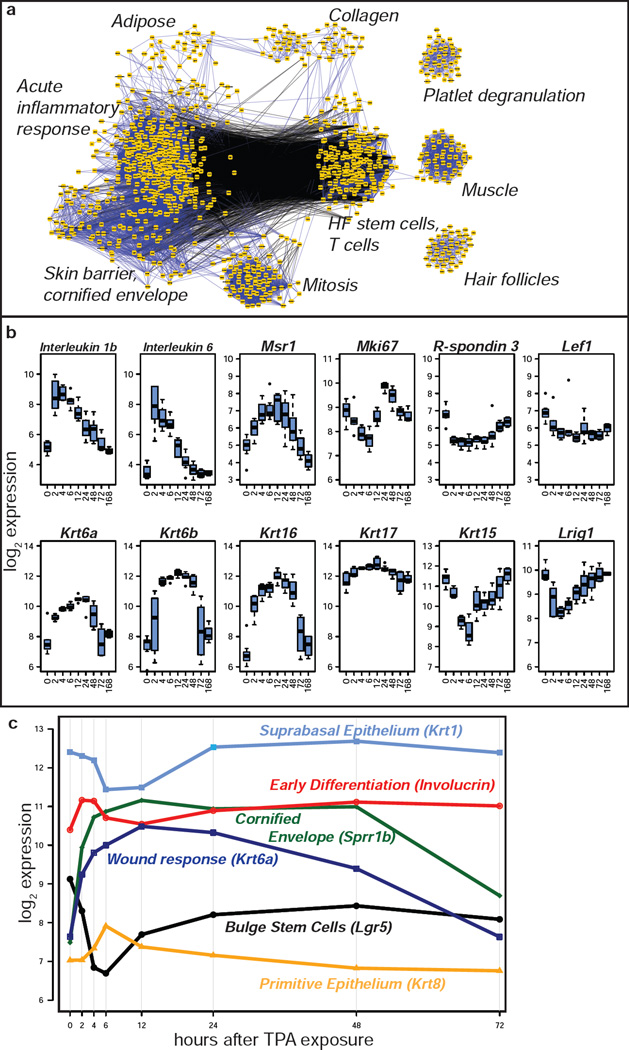
It is also possible to visualize network correlations using samples from inbred mice subjected to a strong perturbation such as induction of inflammation. To model epithelial response to inflammation, we treated dorsal skin of inbred FVB mice with the phorbol ester TPA, which results in edema and an influx of inflammatory cells into the skin (Furstenberger and Marks, 1983; Schlingemann et al., 2003). TPA treatment has been observed to induce differentiation of basal keratinocytes (Reiners and Slaga, 1983). On induction of a wound stimulus, cells positive for the Wnt-responsive stem cell markers Lgr5 and Lgr6 can give rise to progeny that migrate into the epidermis and may contribute to the wound healing process (Kasper et al., 2011). Correlation network analysis of gene expression across a one week time course after a single treatment with TPA (Figure 3a, Supplementary File 1), revealed the coordinated response of cytokines, keratins, and stem cell markers over time.
Figure 3. Keratin correlation and TPA response networks.
(a) Gene expression correlation network for genes with significantly correlated response in dorsal skin to TPA exposure. (b) Box plots of gene expression in dorsal skin at baseline (0 hours) and at time points from two hours to one week after TPA treatment. Number of animals for each timepoint was: 0 hr: 9, 2 hr: 6, 4 hr: 4, 6 hr: 7, 12 hr: 6, 24 hr: 7, 48 hr: 8, 72 hr: 4, 1 wk: 4. (c) Mean expression levels of Krt1, Involucrin, Sprr1b, Krt6a, Lgr5, and Krt8, in the TPA treatment experiment, with the X-axis plotted to scale for number of hours since treatment.
Expression of the immediate response interleukins Il1b and Il6 increased 8 to 16-fold two hours after TPA treatment (Figure 3b). The “acute inflammatory response” motif in Figure 3a includes Il1b as well as other known early response genes such as Tgfa, Junb, and Nfkb2. Expression of macrophage surface markers such as macrophage scavenger receptor 1 (Msr1) peaked at a five-fold increase after 12 hours, consistent with the influx of macrophages into the treated skin. Within the “Mitosis” network, expression of markers of the M phase of the cell cycle such as Mki67 decreased immediately after treatment only to rebound to higher than baseline levels by 24 hours, in agreement with earlier observations of early decrease in DNA synthesis after TPA treatment, followed by entry into S-phase after 12–18 hours (Balmain et al., 1977). Expression of the proliferation-associated keratins Krt6a, Krt6b, Krt16, and Krt17 increased significantly after TPA treatment, and these genes were linked to the “Skin Barrier and Cornified Envelope” module. Expression of the Wnt signaling agonist Rspo3 and Wnt effector Lef1 was significantly decreased by TPA treatment, with Rspo3 expression gradually recovering after acute inflammatory stimulus (Glinka et al., 2011).
We summarize the kinetics of epithelial response to TPA exposure using markers of stem, basal, suprabasal, and terminally differentiated cells in Figure 3c. Bulge stem cell markers (Lgr5) drop immediately by four-fold and slowly recover to baseline levels by 72 hours. Early differentiation markers (involucrin) increased to a peak at two hours, followed by markers of the primitive epithelium (Krt8), which increase to peak at six hours and revert to homeostatic levels by 12 hours. The suprabasal epithelium (Krt1) shows the inverse trend, decreasing at six hours and recovering by 24 hours. Markers of the cornified envelope (Sprr1b) and keratin wound response (Krt6a) increase to a peak at 12 hours and are still significantly elevated 48 hours after treatment. These observations are compatible with an immediate differentiation response to TPA by primitive cells and suprabasal cells, followed by an increase in basal cell production to replenish the suprabasal cell population between 12 and 24 hours after treatment. The early drop in mitotic gene expression suggested the increase in involucrin and Sprr1b expression was the result of a direct induction of a differentiation response rather than a consequence of proliferation. Our observations are compatible with earlier morphological and biochemical observations of the effects of TPA on mouse skin, which showed an early induction of differentiation markers and a change in the balance of basal and suprabasal cells following TPA treatment (Reiners and Slaga, 1983).
Keratin gene networks linked to Pachyonichia Congenita
Germline mutations in Krt6a, Krt6b, Krt16 and Krt17 are all linked to the rare skin disease pachyonichia congenita (PC), a congenital disorder characterized by blisters, thickened nails, and hyperkeratosis (Bowden et al., 1995; McLean et al., 1995). Network analysis revealed strong correlations between these mRNA levels of these genes under normal and inflammatory conditions, suggesting coordinated functions in skin architecture. The type II keratins Krt6a and Krt6b are thought to heterodimerize with the type I keratin Krt16, but in normal dorsal skin, expression of Krt16 was significantly correlated with expression of Krt6a but not Krt6b (Figure 2f) suggesting preferential heterodimerization between Krt16 and Krt6a compared to Krt6b, while in normal tail skin expression of Krt16 was linked to expression of both Krt6a and Krt6b (Figure 2e). To our knowledge there are currently no antibodies that can reliably distinguish Krt6a from Krt6b to address the question of differential binding under these conditions. After TPA treatment, the Krt16 expression profile was significantly correlated with that of Krt6b, as well as Krt6a, and barrier/inflammation genes such as S100a8, Lce3a, and small proline receptors Sprr2e and Sprr2d (Supplementary Table 2). There was also significant differential correlation between expression of Krt16 and S100a8, Defb3, and other genes induced by TPA exposure when comparing untreated FVBBX tail skin to dorsal skin (Supplementary Table 3). The tail-specific correlation of Krt16 with other genes induced during wound response, as well as elevated constitutive expression of Krt16 in normal tail, was compatible with elevated keratinocyte proliferation in tail skin and was reminiscent of the wound response seen in TPA-treated dorsal skin. These observations are consistent with previous analysis of data from the tail skins of aged (>1 year old) tumor-bearing mice, which implicated Krt16 in regulation of innate immune responses to barrier dysfunction (Lessard et al., 2013).
Network analysis of Krt17 in normal tail skin, in contrast to the analysis of Krt16, showed no significant correlations with S100a8 or other markers of acute inflammatory responses, but was linked to expression of hair follicle keratins (Supplementary Table 4, Supplementary Table 5, Figure 2e, 2f). In dorsal skin, Krt17 was correlated with a number of epidermal stem cell markers (e.g. Sox9, Dlx3, Tbx1) as well as Krt6a, but only weakly with Krt16. All of the genes that play causative roles in Pachyonichia Congenita (Krt6a, Krt6b, Krt16 and Krt17) were however significantly correlated with each other after treatment of skin with TPA, indicating that these genes form part of a coordinated network response to inflammation and tissue regeneration.
Smoothened activity is rewired between tail and dorsal skin
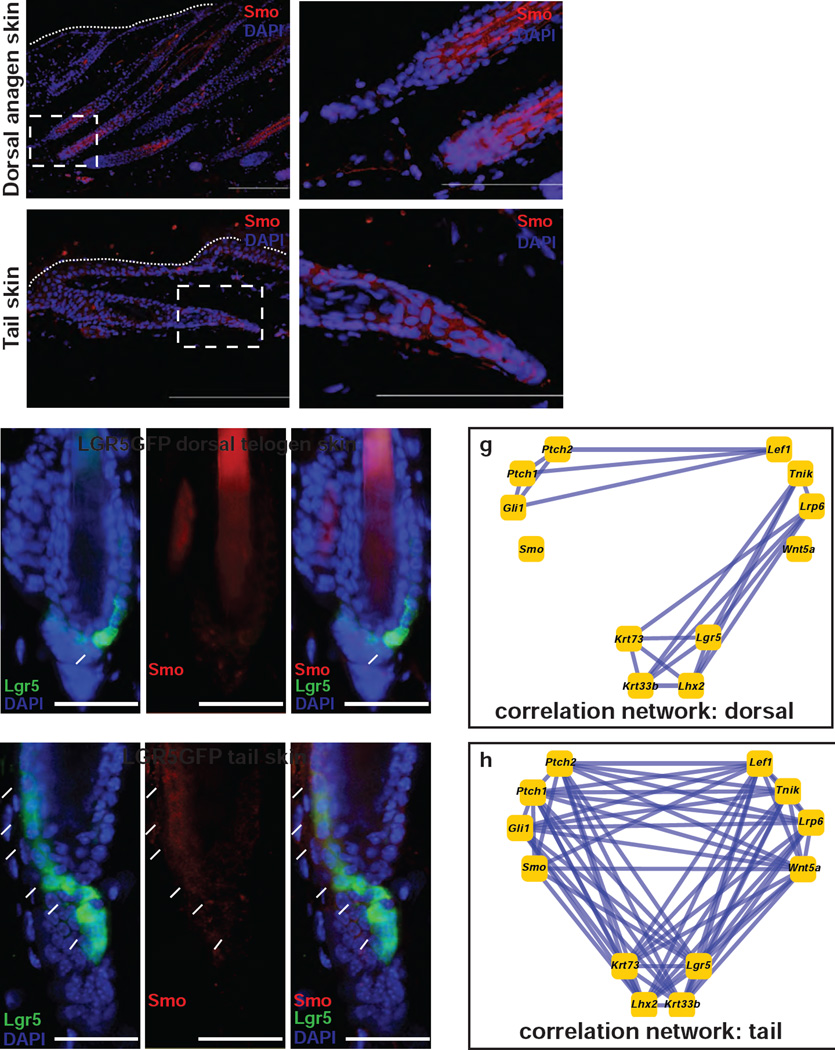
Skin tumorigenesis studies have shown differences in susceptibilty of tail and dorsal skin to induction of basal cell carcinomas (BCCs) by activation of the Shh signaling pathway. We hypothesized that tissue-specific regulation of Smoothened is responsible for the observation that models of BCC which activate Smoothened only produce tumors on the tail skin. It has previously been reported that Smo protein is absent in tail follicles (Wang et al., 2011). We first performed immunostaining for Smo protein with the antibody used by Wang and colleagues (Wang et al., 2011). In agreement with their results, we detected cytoplasmic Smo protein at the inner root sheath of dorsal anagen follicles (Figure 4a, 4b). In contrast to the previous report, we also detected Smo protein at the base of tail follicles (Figure 4c, 4d). Specificity of Smo staining was confirmed by staining Smo induced by the Ptch+/− radiation model (Supplementary Figure 1). We then co-stained for Smo protein and EGFP in mice expressing EGFP at the endogenous Lgr5 locus. Dorsal telogen skin showed Lgr5-positive cells at the dermal papilla, but no Smo protein, as expected (Figure 4e). Tail skin showed co-localization of Lgr5 and Smo protein in cells between the dermal papilla and bulge region (Figure 4f), thus confirming that Smo is in fact expressed in tail skin during anagen.
Figure 4. Rewired hedgehog networks in tail and dorsal hair follicles.
Correlation networks for key Hedgehog, Wnt, and Hair Follicle genes in (c) dorsal and (d) tail skin. Lines connect significantly correlated gene pairs. (a–d) Mouse skin immunostained with an anti-SMO antibody (red) and counter-stained with DAPI (blue). SMO protein was present in (a) dorsal anagen follicles [(b) higher magnification of dashed box] and (c) tail follicles [(d) higher magnification] most prominently near the dermal papillae. Lower/Higher magnification scale bar indicate 500/200 µM. (e, f) (e) LGR5-EGFP dorsal telogen mouse skin and (f) tail skin immunostained with an anti-SMO antibody (red) and anti-EGFP antibody (green) and counter-stained with DAPI (blue). Scale bar is 50 µM, images taken at 40×. (g, h) Correlation networks for key Hedgehog, Wnt, and Hair Follicle genes in (g) dorsal and (h) tail skin, illustrating the tail skin-specific correlation between expression of Smo and expression of the Wnt and Hair Follicle networks. Lines connect significantly correlated gene pairs.
Our data show that Smo protein is expressed in both dorsal and tail skin, and furthermore that the average level of Smoothened mRNA expression is nearly identical in skin from both sites (Figure 1d). However, we found 676 genes with significant differential correlation with Smoothened in these tissues (P < 0.05, Bonferroni correction, see Methods). Gene Ontology enrichment analysis indiated significant activation of correlation in tails between expression of Smoothened and both canonical Wnt receptor signaling (P < 6 × 10−5) and the Hair Follicle cycle (P < 2 × 10−5). We plotted correlation between key genes in these networks, illustrating that while there was a constitutive association between expression levels of Lef1 and Hedgehog genes Gli1 and Ptch1 in both compartments, the association between Smo and the Wnt and hair follicle gene networks was restricted to tail skin (Figure 4g, 4h). We conclude that constitutive expression of a tail network encompassing Smo and components of the Wnt-Lef1 signaling pathway may explain the development of BCCs preferentially in the tail in spite of widespread targeting of mutant Smo to multiple body sites including dorsal skin using the Krt14 promoter (Wong and Reiter, 2011; Youssef et al., 2010).
Rewiring of tumor suppressor gene networks during premalignancy
We then applied differential correlation analysis to identify how relationships between genes are altered during tumorigenesis, analyzing previously published gene expression measurements on normal skin, benign tumors, and malignant tumors (Quigley et al., 2009; Quigley et al., 2011). Skin tumors were induced on mice using a two-stage skin carcinogenesis protocol as previously described; briefly: tumors were initiated using DMBA, which induces oncogenic mutations in Hras1 (Quintanilla et al., 1986), and were then promoted with tetradecanoyl-phorbol acetate (TPA), resulting in benign papillomas, some of which progress to malignant invasive carcinomas.
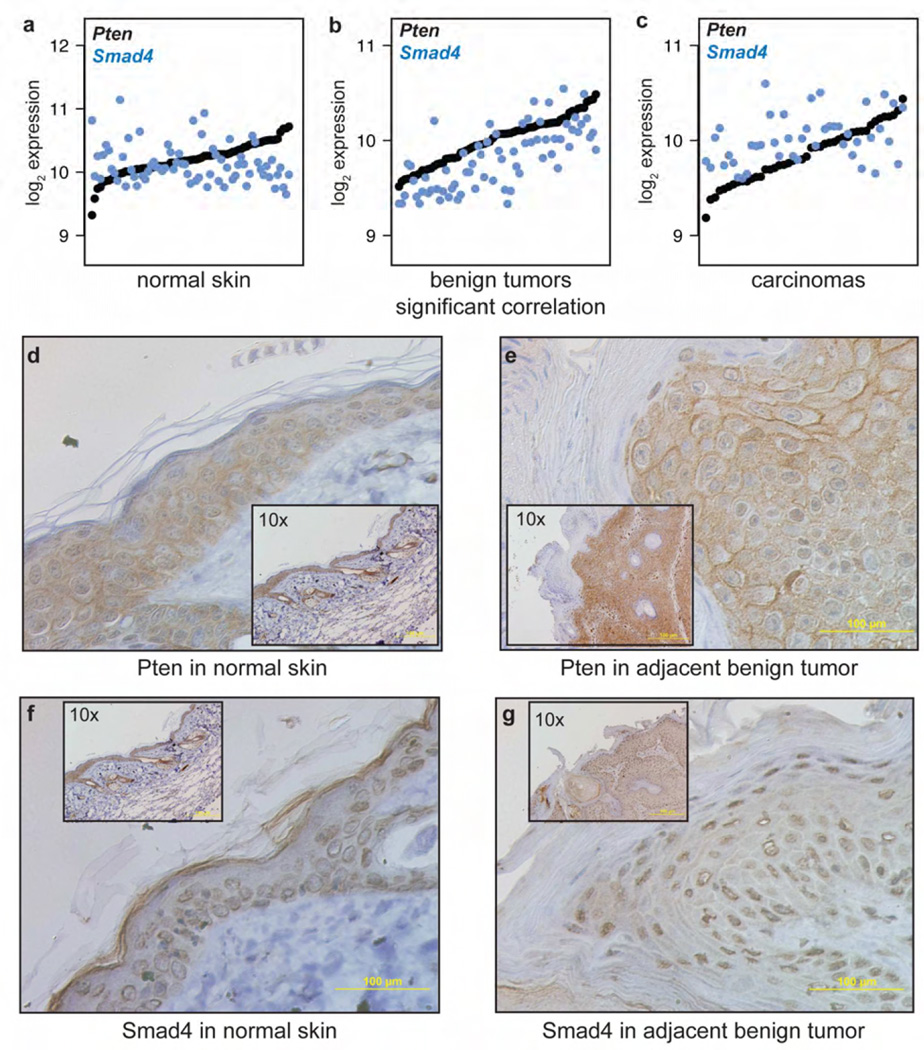
The most frequently differentially correlated genes when comparing normal skin to benign tumors were the ribosomal protein Rps7 and Phosphatase and tensin homolog deleted on chromosome 10 (Pten). Both genes were robustly expressed in skin, papillomas, and carcinomas, with mean change in expression levels between matched normal skin and papillomas of less than 20%. Analysis based on differential expression would not suggest that these genes are of special interest during tumorigenesis. However, these genes are both linked to tumor suppressor pathways potentially activated in epithelial tumors. Rps7 binds directly to Mdm2, inhibiting its ability to degrade p53 (Zhu et al., 2009). Pten is an antagonist of the PI3-kinase signaling pathway which is frequently inactivated in epithelial tumors (Salmena et al., 2008). Mice with heterozygous knockouts of Pten develop spontaneous tumors; mice with heterozygous or homozygous knockout of Pten targeted to their keratinocytes show a dose-dependent increase in spontaneous skin tumor formation and susceptibility to DMBA/TPA treatment (Suzuki et al., 2003). We have previously shown that 70% of the carcinomas that arise in Pten+/− mice after DMBA/TPA treatment do not have a Hras mutation, but all of these tumors lost their wild-type Pten allele. Wild-type animals treated with DMBA/TPA did not lose Pten copies or develop somatic Pten mutations in papilloma genomic DNA (Mao et al., 2004). These previous data identified a specific, mutually exclusive relationship between Hras mutations and Pten signaling in mouse skin. This relationship is supported by the observation that across the whole genome, Pten was the third-most-frequently “rewired” genes in papillomas carrying Hras mutations. Several of the genes that acquire correlations with Pten in papillomas are involved in alternative tumor suppressor pathways (Supplementary Table 6) suggesting that this rewiring is part of a concerted response to perturbation induced by activation of Ras signaling.
In many cases, the significant correlations with Pten that were acquired in premalignant papillomas were subsequently lost during malignant progression to carcinomas. This was the case with Pten and the TGF-beta signaling mediator SMAD family member 4 (Smad4), which is a well-characterized tumor suppressor gene both in mouse and human tumors (Figure 5a–c). Keratinocyte-specific inactivation of Smad4 blocks hair follicle differentiation and cycling and leads to invasive squamous cell carcinomas, while deletion of both Smad4 and Pten results in accelerated tumorigenesis (Yang et al., 2005). Moreover, Smad4 and Pten synergize to prevent pancreatic ductal adenocarcinoma (Ding et al., 2011; Xu et al., 2010).
Figure 5. Tumor suppressor gene rewiring during tumorigenesis.
(a–c) Expression of Pten and Smad4 in normal skin, benign tumors, and malignant carcinomas, showing significant correlation in benign tumors but not normal skin or malignant carcinomas. (d–g) Immunohistochemistry of (d, f) adjacent normal skin and (e, g) benign tumors shown at 40× (insets at 10×).
Pten and Smad4 proteins are expressed in the epithelial layer of normal dorsal skin with diffuse cytoplasmic staining (Figure 5d, 5f). In contrast, in papillomas Pten protein is expressed at moderately elevated levels in the membranes of epithelial cells, a location compatible with its known activity on the cell membrane (Figure 5e), while Smad4 protein is expressed at moderately higher levels in the nucleus. Our results suggest that even in the absence of inactivating mutations in Pten (McCreery et al., 2015) genetic networks are rewired to activate tumor suppressor pathways in benign tumors as a response to activated Ras signaling. Carcinomas however no longer show this activated response pattern, suggesting that alternative genetic or epigenetic mechanisms lead to loss of these growth controlling pathways during tumor progression.
DISCUSSION
Human skin is a self renewing tissue that has evolved specific functions according to location in the body. These functions may require the presence of specific cell types in particular locations, or gene expression patterns that determine responses to local perturbations or exogenous insults (Rinn et al., 2008). In the mouse, a similar heterogeneity in function is determined by site-specific requirements such as physical stress, barrier function or immune responses. Previous studies have shown that susceptibility to development of skin tumors is dependent on location. Transgenic approaches in which keratin gene promoters were used to drive expression of oncogenes in skin demonstrated preferential induction of tumors at specific sites, in spite of widespread expression of the gene promoters used. For example, expression of mutant Hras under the control of a keratin 5 promoter led to squamous tumors of the back, and sometimes ventral skin (Wakabayashi et al., 2007), whereas activation of mutant Smo in cells expressing keratin 14 gave rise to basal cell carcinomas (BCCs) mainly in tail or ear skin (Wong and Reiter, 2011; Youssef et al., 2012). Chemical carcinogenesis studies also showed that dorsal, but not tail skin, is susceptible to squamous tumor development (Schweizer and Marks, 1977b).
In the present study we have explored the basis for these site specific functional differences using gene expression network analysis of dorsal and tail skins from a genetically heterogeneous mouse population. Randomized inheritance of polymorphic variants between the strongly divergent Mus musculus and Mus spretus (Dejager et al., 2009) produced high variance in gene expression between normal individuals. Measuring the co-variation of these genes allowed us to visualize the network architecture of normal skin in two physically and functionally distinct locations.
Analysis of genetic control of gene expression in age- and gender-matched mice showed that the statistical strength of eQTLs calculated in 89 tail skins was much higher than that of eQTLs calculated from the same number of dorsal skins. Surprisingly, the stronger eQTLs were not enriched for roles in either hair follicle control or in cell populations such as Langerhans cells that differ in frequency between tail and dorsal. We found genes with stronger eQTLs in tail had roles in ribosomal structure, translation, and energy metabolism. We hypothesize that the apparently constitutive anagen at eight weeks of age in tail skin plays an important part in explaining this result and is a key difference between tail and dorsal skin overall. While most dorsal tissue samples at this timepoint were in telogen, each tail sample contained at least some anagen activity. The constitutive anagen and increased proliferative activity of tail skin may allow for clearer dissection of genetic differences between the parental strains.
Differential cancer susceptibility in dorsal and tail skin
There is considerable debate over the cellular origins of skin BCCs, with some studies favoring the interfollicular epidermis (Wong and Reiter, 2011; Youssef et al., 2010) the hair follicle bulge region (Wang et al, Cancer Cell), or both locations (Grachtchouk et al., 2011). Direct comparison of these results is complicated by the differences in the mouse models used to induce BCCs by targeting oncogene expression or tumor suppressor loss to specific subpopulations of cells in the skin.. We set out to address a slightly different question, which is why expression of activated Smo using a Krt14 promoter which is universally active in interfollicular basal cells of both dorsal and tail skin, gives rise to BCCs mainly in the tail skin (Wong and Reiter, 2011; Youssef et al., 2010). Our expression analysis identified a network of genes linked to Smo expression in tail, but not dorsal skin, that included components of the Wnt/Lef1, and Shh signaling pathways. While many questions remain to be addressed regarding the cellular origins of skin BCCs and SCCs, we propose that mutant Smo activated by K14-Cre may have a stronger effect in tail keratinocytes because of the specific wiring of the Smo gene expression pattern in this tissue. Analyses of other Shh signaling components may reveal tissue- or cell type-specific patterns that could explain the diverse phenotypes observed when carcinomas are induced by different driver genes in sub-populations of skin target cells (Peterson et al., 2015). Loss of Ptch1 can also lead to BCC formation, but primarily in dorsal rather than tail skin (Wang et al., 2011). The explanation of this phenotype is unknown, but could be due to differential wiring of the Ptch1 gene network in dorsal and tail skin, or to differences in levels of expression of alternative genes, such as Ptch2, that can compensate for loss of Ptch1 (Adolphe et al., 2014). Indeed, while Ptch1 mRNA levels are similar in dorsal and tail skins, Ptch2 is expressed at a significantly higher level in the tails (data not shown), suggesting that compensation may explain the relative lack of tail BCCs when Ptch1 is deleted in this tissue.
Network changes during development of premalignancy
Mouse models of skin cancer initiated by the carcinogen DMBA display a high specificity for activation of the Hras gene by mutation at codon 61 (Bizub et al., 1986; Quintanilla et al., 1986). This event can be circumvented in mice lacking expression of the Pten tumor suppressor gene in keratinocytes (Mao et al., 2004), suggesting a connection between the Hras and Pten signaling pathways. It was therefore of interest that an unbiased screen for differentially correlated genes in normal skin and Hras-mutant papillomas from the same backcross mouse population (Quigley et al., 2011) identified Pten as one of the top rewired genes, in spite of minimal changes in mRNA expression levels during tumor development. In premalignant papillomas, Pten is correlated with a set of genes implicated in alternative tumor suppressor pathways including the Trp53 (Rps26) and Tgfβ (Smad4) pathways. These correlations are however disrupted in malignant carcinomas, suggesting that this concerted suppressor network response to Hras mutation has to be removed to permit tumor progression. The mechanisms involved are not presently clear, but do not appear to include direct mutations in Pten as shown by exome analysis of benign and malignant skin tumors (McCreery et al., 2015).
These observations demonstrate how network analysis of gene expression can reveal pathway engagement not obvious from differential expression analysis (Figure 6). Methods such as correlation analysis that identify relationships between genes in the same compartment or across compartments in intact tissues are a useful tool to understand the interactions that underlie normal tissue homeostasis, responses to damage, and development of progressive diseases including neoplasia.
Figure 6. Summary of network analysis and perturbations presented in this study.
EXPERIMENTAL PROCEDURES
Animal breeding and tissue collection
FVBBX mice were generated by breeding [(FVB/N × SPRET/Ei) × FVB/N]. Mice were housed in standard conditions, fed ad libitum, and sacrificed at 8 weeks of age. Animals were housed and treated in accordance with the regulations and protocols stipulated by the UCSF Institutional Animal Care and Use Committee (IACUC). For TPA treatment, the dorsal skins of FVB/N, Spret/Ei, and FVB/N × Spret/Ei mice were shaved and two days later treated with a single dose of TPA (200 ml of 10−4 M solution in acetone) or vehicle alone. Mice were sacrificed at 0, 2, 4, 6, 24, 48, and 72 hours, and after one week. Tissue was flash-frozen in liquid nitrogen and stored at −80° C.
Microarray analysis
RNA was isolated using TRIzol (Invitrogen) according to the manufacturer’s instructions. Residual genomic DNA was removed by DNase treatment (Ambion). RNA quality was assessed using a Bioanalyzer (Agilent). Gene expression was quantified using Affymetrix MoGene ST 1.1 arrays hybridized on an Affymetrix GeneTitan instrument. Batch effects were reduced using ComBat (Johnson et al., 2007). Affymetrix MoGene arrays were normalized using the oligo package (Carvalho and Irizarry, 2010) and a probe database prepared for FVBBX mice to avoid probes which intersect known FVB/N or Spret/Ei SNPs (Quigley, 2015). Probesets expressed below background levels (determined by expression of Y chromosome genes in female animals) or which did not map to Refseq-annotated locations for genes were discarded. Multiple probesets assigned to the same gene were collapsed to one signal using the rule: if two or more probesets had correlation > 0.8 these were combined by their mean, and other probesets discarded. If no two probesets had correlation > 0.8, the probeset with the highest variance was used and other probesets discarded. Normal/Papilloma/Carcinoma data were downloaded from GEO (accession GSE21264). Raw FVBBX microarray data are available for download at GEO (accession GSE52650).
Statistical methods
Spearman rank correlation 5% genome-wide significance was established by permutation (Churchill and Doerge, 1994). Statistical calculations were performed in the R statistical environment (R Core Team, 2012) and CARMEN. Differential expression was tested with SAM (Tusher et al., 2001). Differential correlation was calculated in CARMEN using Fisher’s transformation of Spearman rank correlation to determine the significance of a change in correlation between two conditions. Multiple test correction was performed by applying a Bonferroni correction to the number of gene pairs considered. Correlation networks were displayed with Cytoscape 3.2 (Shannon et al., 2003). Genotypes were measured with 240 custom-designed Applied Biosystems Taqman probes that distinguish the FVB/N and Spret/Ei genomes, measured using the Wafergen platform (www.wafergen.com). Genotypes were called using the manufacturer's software. Erroneously called genotypes were corrected and missing genotype data were imputed using R/QTL package (Broman et al., 2003). eQTL analysis was performed using CARMEN as described in (Quigley et al., 2009). CARMEN software and documentation are available at http://davidquigley.com/carmen.html with source code available at https://github.com/DavidQuigley/QuantitativeGenetics. CARMEN is written in C++ using the wxWidgets and Boost libraries (http://wxwidgets.org, http://boost.org). For each gene, association with genotype was tested within a chromosome by linear regression, with gene-wise significance assessed by permutation testing and genome-wide significance tested using permutation P values as input to the qvalue package (Storey and Tibshirani, 2003). eQTL LOD plots were generated with R/QTL (Broman et al., 2003). Differential correlation significance was assessed by Bonferroni correction of the Spearman correlation P value for the number of gene pairs considered.
Tissue staining
For Lrig1 and Krt79 staining, 5 µm skin sections were deparaffinised and antigen retrieval was performed in 10 mM sodium citrate solution (pH 6) or in Trilogy pre-treatment solution (Cell Marque). Sections were blocked for one hour at room temperature with 10% donkey serum (Abcam) diluted in PBS containing 0.3% Triton X-100 and incubated overnight with primary antibodies against Lrig1 (goat, R&D) or Krt79 (rabbit, Abcam) at 4 °C. After washing in PBS, sections were incubated with blocking solution containing an Alexa 555 conjugated donkey anti-goat/rabbit secondary antibody (Molecular Probes) and DAPI for one hour at room temperature. After another PBS wash slides were mounted using Citifluor. For other staining, tissues were fixed overnight in 4% paraformaldehyde (PFA) and embedded in paraffin. Hematoxylin and eosin stains were prepared by the UCSF Gladstone core facility with a Leica ST 5020 multistainer. For immunofluorescence, slides were de-pariffinized with citrisolve. Antigen retrieval was performed with citrate. Slides were blocked in 10% goat serum (Invitrogen 50062Z) with 0.3% Triton. The primary antibody was rabbit anti-Smo (1:200, Abcam) overnight, followed by secondary antibody conjugated with Alexa Fluor 555 (1:500, Invitrogen) and DAPI (Vectashield, Vector Labs).
Supplementary Material
Highlights.
Gene expression networks reconstruct the cellular composition of a complex tissue.
Genetic influence on gene expression varies by tissue location in the skin.
Smoothened pathway activity is rewired between tail and dorsal skin.
Gene expression networks are rewired in premalignant tumors and again in carcinoma.
Acknowledgments
AB acknowledges support from NCI grants CA084244-15 and CA141455-01, and support from Darrin Stuart and Nancy Pryer of the Novartis Institutes for Biomedical Research. We thank Ervin Epstein for sharing BCC tissue samples, and Peter Vuong and Douglas Chin for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
AB designed and oversaw the experiments. DAQ performed the genetic and genomic analyses and wrote CARMEN. RdR and DC performed animal husbandry. GH, KH, DW, PV contributed to genotyping and DNA/RNA extraction. FP, JS, and RrR performed the TPA time course experiment. CW, PH and EK performed immunofluorescence. AK contributed Basal Cell Carcinoma model data. DAQ and AB wrote the manuscript.
REFERENCES
- Adolphe C, Nieuwenhuis E, Villani R, Li ZJ, Kaur P, Hui CC, Wainwright BJ. Patched 1 and patched 2 redundancy has a key role in regulating epidermal differentiation. J Invest Dermatol. 2014;134:1981–1990. doi: 10.1038/jid.2014.63. [DOI] [PubMed] [Google Scholar]
- Arwert EN, Hoste E, Watt FM. Epithelial stem cells, wound healing and cancer. Nat Rev Cancer. 2012;12:170–180. doi: 10.1038/nrc3217. [DOI] [PubMed] [Google Scholar]
- Balmain A, Alonso A, Fischer J. Histone phosphorylation and synthesis of DNA and RNA during phases of proliferation and differentiation induced in mouse epidermis by the tumor promoter 12-O-tetradecanoyl-phorbol-13-acetate. Cancer Res. 1977;37:1548–1555. [PubMed] [Google Scholar]
- Bergstresser PR, Fletcher CR, Streilein JW. Surface densities of Langerhans cells in relation to rodent epidermal sites with special immunologic properties. J Invest Dermatol. 1980;74:77–80. doi: 10.1111/1523-1747.ep12519909. [DOI] [PubMed] [Google Scholar]
- Bizub D, Wood AW, Skalka AM. Mutagenesis of the Ha-ras oncogene in mouse skin tumors induced by polycyclic aromatic hydrocarbons. Proc Natl Acad Sci U S A. 1986;83:6048–6052. doi: 10.1073/pnas.83.16.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C, Fuchs E. Epidermal stem cells of the skin. Annu Rev Cell Dev Biol. 2006;22:339–373. doi: 10.1146/annurev.cellbio.22.010305.104357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden PE, Haley JL, Kansky A, Rothnagel JA, Jones DO, Turner RJ. Mutation of a type II keratin gene (K6a) in pachyonychia congenita. Nat Genet. 1995;10:363–365. doi: 10.1038/ng0795-363. [DOI] [PubMed] [Google Scholar]
- Braun R, Cope L, Parmigiani G. Identifying differential correlation in gene/pathway combinations. BMC Bioinformatics. 2008;9:488. doi: 10.1186/1471-2105-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26:2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Syder AJ, Yu QC, Letai A, Paller AS, Fuchs E. The genetic basis of epidermolytic hyperkeratosis: a disorder of differentiation-specific epidermal keratin genes. Cell. 1992;70:811–819. doi: 10.1016/0092-8674(92)90314-3. [DOI] [PubMed] [Google Scholar]
- Chiang C, Swan RZ, Grachtchouk M, Bolinger M, Litingtung Y, Robertson EK, Cooper MK, Gaffield W, Westphal H, Beachy PA, et al. Essential role for Sonic hedgehog during hair follicle morphogenesis. Dev Biol. 1999;205:1–9. doi: 10.1006/dbio.1998.9103. [DOI] [PubMed] [Google Scholar]
- Choi Y, Kendziorski C. Statistical Methods for Gene Set Co-expression Analysis. Bioinformatics. 2009 doi: 10.1093/bioinformatics/btp502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui D, Li L, Lou H, Sun H, Ngai SM, Shao G, Tang J. The ribosomal protein S26 regulates p53 activity in response to DNA damage. Oncogene. 2014;33:2225–2235. doi: 10.1038/onc.2013.170. [DOI] [PubMed] [Google Scholar]
- DasGupta R, Fuchs E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development. 1999;126:4557–4568. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- de la Fuente A. From 'differential expression' to 'differential networking' - identification of dysfunctional regulatory networks in diseases. Trends Genet. 2010;26:326–333. doi: 10.1016/j.tig.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Dejager L, Libert C, Montagutelli X. Thirty years of Mus spretus: a promising future. Trends Genet. 2009;25:234–241. doi: 10.1016/j.tig.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Diepgen TL, Mahler V. The epidemiology of skin cancer. Br J Dermatol. 2002;146(Suppl 61):1–6. doi: 10.1046/j.1365-2133.146.s61.2.x. [DOI] [PubMed] [Google Scholar]
- Ding Z, Wu C-J, Chu GC, Xiao Y, Ho D, Zhang J, Perry SR, Labrot ES, Wu X, Lis R, et al. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature. 2011;470:269–273. doi: 10.1038/nature09677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty L, Sheen MR, Vlachos A, Choesmel V, O'Donohue MF, Clinton C, Schneider HE, Sieff CA, Newburger PE, Ball SE, et al. Ribosomal protein genes RPS10 and RPS26 are commonly mutated in Diamond-Blackfan anemia. Am J Hum Genet. 2010;86:222–228. doi: 10.1016/j.ajhg.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch KR, McGowan KA, van Raamsdonk CD, Fuchs H, Lee D, Puech A, Herault Y, Threadgill DW, Hrabe de Angelis M, Barsh GS. Genetics of dark skin in mice. Genes Dev. 2003;17:214–228. doi: 10.1101/gad.1023703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E. Keratins and the skin. Annu Rev Cell Dev Biol. 1995;11:123–153. doi: 10.1146/annurev.cb.11.110195.001011. [DOI] [PubMed] [Google Scholar]
- Furstenberger G, Marks F. Growth stimulation and tumor promotion in skin. J Invest Dermatol. 1983;81:157s–162s. doi: 10.1111/1523-1747.ep12540971. [DOI] [PubMed] [Google Scholar]
- Gat U, DasGupta R, Degenstein L, Fuchs E. De Novo hair follicle morphogenesis and hair tumors in mice expressing a truncated beta-catenin in skin. Cell. 1998;95:605–614. doi: 10.1016/s0092-8674(00)81631-1. [DOI] [PubMed] [Google Scholar]
- Glinka A, Dolde C, Kirsch N, Huang YL, Kazanskaya O, Ingelfinger D, Boutros M, Cruciat CM, Niehrs C. LGR4 and LGR5 are R-spondin receptors mediating Wnt/beta-catenin and Wnt/PCP signalling. EMBO reports. 2011;12:1055–1061. doi: 10.1038/embor.2011.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grachtchouk M, Mo R, Yu S, Zhang X, Sasaki H, Hui CC, Dlugosz AA. Basal cell carcinomas in mice overexpressing Gli2 in skin. Nat Genet. 2000;24:216–217. doi: 10.1038/73417. [DOI] [PubMed] [Google Scholar]
- Grachtchouk M, Pero J, Yang SH, Ermilov AN, Michael LE, Wang A, Wilbert D, Patel RM, Ferris J, Diener J, et al. Basal cell carcinomas in mice arise from hair follicle stem cells and multiple epithelial progenitor populations. J Clin Invest. 2011;121:1768–1781. doi: 10.1172/JCI46307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu LH, Coulombe PA. Keratin function in skin epithelia: a broadening palette with surprising shades. Curr Opin Cell Biol. 2007;19:13–23. doi: 10.1016/j.ceb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Huelsken J, Vogel R, Erdmann B, Cotsarelis G, Birchmeier W. beta-Catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell. 2001;105:533–545. doi: 10.1016/s0092-8674(01)00336-1. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Kasper M, Jaks V, Are A, Bergstrom A, Schwager A, Svard J, Teglund S, Barker N, Toftgard R. Wounding enhances epidermal tumorigenesis by recruiting hair follicle keratinocytes. Proc Natl Acad Sci U S A. 2011;108:4099–4104. doi: 10.1073/pnas.1014489108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto J, Burgeson RE, Morgan BA. Wnt signaling maintains the hair-inducing activity of the dermal papilla. Genes Dev. 2000;14:1181–1185. [PMC free article] [PubMed] [Google Scholar]
- Lane EB, McLean WH. Keratins and skin disorders. J Pathol. 2004;204:355–366. doi: 10.1002/path.1643. [DOI] [PubMed] [Google Scholar]
- Lessard JC, Pina-Paz S, Rotty JD, Hickerson RP, Kaspar RL, Balmain A, Coulombe PA. Keratin 16 regulates innate immunity in response to epidermal barrier breach. Proc Natl Acad Sci U S A. 2013;110:19537–19542. doi: 10.1073/pnas.1309576110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi T, Li VS, Ng SS, Taouatas N, Vries RG, Mohammed S, Heck AJ, Clevers H. The kinase TNIK is an essential activator of Wnt target genes. EMBO J. 2009;28:3329–3340. doi: 10.1038/emboj.2009.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso M, Leonardi S, Tanori M, Pasquali E, Pierdomenico M, Rebessi S, Di Majo V, Covelli V, Pazzaglia S, Saran A. Hair cycle-dependent basal cell carcinoma tumorigenesis in Ptc1neo67/+ mice exposed to radiation. Cancer Res. 2006;66:6606–6614. doi: 10.1158/0008-5472.CAN-05-3690. [DOI] [PubMed] [Google Scholar]
- Mao J-H, To MD, Perez-Losada J, Wu D, Del Rosario R, Balmain A. Mutually exclusive mutations of the Pten and ras pathways in skin tumor progression. Genes Dev. 2004;18:1800–1805. doi: 10.1101/gad.1213804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCreery MQ, Halliwill KD, Chin D, Delrosario R, Hirst G, Vuong P, Jen KY, Hewinson J, Adams DJ, Balmain A. Evolution of metastasis revealed by mutational landscapes of chemically induced skin cancers. Nat Med. 2015;21:1514–1520. doi: 10.1038/nm.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean WH, Rugg EL, Lunny DP, Morley SM, Lane EB, Swensson O, Dopping-Hepenstal PJ, Griffiths WA, Eady RA, Higgins C, et al. Keratin 16 and keratin 17 mutations cause pachyonychia congenita. Nat Genet. 1995;9:273–278. doi: 10.1038/ng0395-273. [DOI] [PubMed] [Google Scholar]
- Miller SJ, Wei ZG, Wilson C, Dzubow L, Sun TT, Lavker RM. Mouse skin is particularly susceptible to tumor initiation during early anagen of the hair cycle: possible involvement of hair follicle stem cells. J Invest Dermatol. 1993;101:591–594. doi: 10.1111/1523-1747.ep12366045. [DOI] [PubMed] [Google Scholar]
- Muller-Rover S, Handjiski B, van der Veen C, Eichmuller S, Foitzik K, McKay IA, Stenn KS, Paus R. A comprehensive guide for the accurate classification of murine hair follicles in distinct hair cycle stages. J Invest Dermatol. 2001;117:3–15. doi: 10.1046/j.0022-202x.2001.01377.x. [DOI] [PubMed] [Google Scholar]
- Nabors GS, Farrell JP. Site-specific immunity to Leishmania major in SWR mice: the site of infection influences susceptibility and expression of the antileishmanial immune response. Infection and immunity. 1994;62:3655–3662. doi: 10.1128/iai.62.9.3655-3662.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page ME, Lombard P, Ng F, Gottgens B, Jensen KB. The epidermis comprises autonomous compartments maintained by distinct stem cell populations. Cell stem cell. 2013;13:471–482. doi: 10.1016/j.stem.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paramio JM, Jorcano JL. Beyond structure: do intermediate filaments modulate cell signalling? Bioessays. 2002;24:836–844. doi: 10.1002/bies.10140. [DOI] [PubMed] [Google Scholar]
- Paus R, van der Veen C, Eichmuller S, Kopp T, Hagen E, Muller-Rover S, Hofmann U. Generation and cyclic remodeling of the hair follicle immune system in mice. J Invest Dermatol. 1998;111:7–18. doi: 10.1046/j.1523-1747.1998.00243.x. [DOI] [PubMed] [Google Scholar]
- Peterson SC, Eberl M, Vagnozzi AN, Belkadi A, Veniaminova NA, Verhaegen ME, Bichakjian CK, Ward NL, Dlugosz AA, Wong SY. Basal cell carcinoma preferentially arises from stem cells within hair follicle and mechanosensory niches. Cell stem cell. 2015;16:400–412. doi: 10.1016/j.stem.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley D. Equalizer reduces SNP bias in Affymetrix microarrays. BMC Bioinformatics. 2015;16:238. doi: 10.1186/s12859-015-0669-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley D, To M, Pérez-Losada J, Pelorosso F, Mao J, Nagase H, Ginzinger D, Balmain A. Genetic architecture of mouse skin inflammation and tumour susceptibility. Nature. 2009;458:505–508. doi: 10.1038/nature07683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley DA, To MD, Kim IJ, Lin KK, Albertson DG, Sjolund J, Perez-Losada J, Balmain A. Network analysis of skin tumor progression identifies a rewired genetic architecture affecting inflammation and tumor susceptibility. Genome biology. 2011;12:R5. doi: 10.1186/gb-2011-12-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintanilla M, Brown K, Ramsden M, Balmain A. Carcinogen-specific mutation and amplification of Ha-ras during mouse skin carcinogenesis. Nature. 1986;322:78–80. doi: 10.1038/322078a0. [DOI] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing (Vienna) 2012 [Google Scholar]
- Reiners JJ, Jr, Slaga TJ. Effects of tumor promoters on the rate and commitment to terminal differentiation of subpopulations of murine keratinocytes. Cell. 1983;32:247–255. doi: 10.1016/0092-8674(83)90515-9. [DOI] [PubMed] [Google Scholar]
- Rentrop M, Nischt R, Knapp B, Schweizer J, Winter H. An unusual type-II 70-kilodalton keratin protein of mouse epidermis exhibiting postnatal body-site specificity and sensitivity to hyperproliferation. Differentiation. 1987;34:189–200. doi: 10.1111/j.1432-0436.1987.tb00066.x. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Wang JK, Liu H, Montgomery K, van de Rijn M, Chang HY. A systems biology approach to anatomic diversity of skin. J Invest Dermatol. 2008;128:776–782. doi: 10.1038/sj.jid.5700986. [DOI] [PubMed] [Google Scholar]
- Rothnagel JA, Dominey AM, Dempsey LD, Longley MA, Greenhalgh DA, Gagne TA, Huber M, Frenk E, Hohl D, Roop DR. Mutations in the rod domains of keratins 1 and 10 in epidermolytic hyperkeratosis. Science. 1992;257:1128–1130. doi: 10.1126/science.257.5073.1128. [DOI] [PubMed] [Google Scholar]
- Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133:403–414. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- Schlingemann J, Hess J, Wrobel G, Breitenbach U, Gebhardt C, Steinlein P, Kramer H, Furstenberger G, Hahn M, Angel P, et al. Profile of gene expression induced by the tumour promotor TPA in murine epithelial cells. Int J Cancer. 2003;104:699–708. doi: 10.1002/ijc.11008. [DOI] [PubMed] [Google Scholar]
- Schneider MR, Schmidt-Ullrich R, Paus R. The hair follicle as a dynamic miniorgan. Curr Biol. 2009;19:R132–R142. doi: 10.1016/j.cub.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Schweizer J, Marks F. A developmental study of the distribution and frequency of Langerhans cells in relation to formation of patterning in mouse tail epidermis. J Invest Dermatol. 1977a;69:198–204. doi: 10.1111/1523-1747.ep12506298. [DOI] [PubMed] [Google Scholar]
- Schweizer J, Marks F. Induction of the formation of new hair follicles in mouse tail epidermis by the tumor promoter 12-O-tetradecanoylphorbol-13-acetate. Cancer Res. 1977b;37:4195–4201. [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shitashige M, Satow R, Jigami T, Aoki K, Honda K, Shibata T, Ono M, Hirohashi S, Yamada T. Traf2- and Nck-interacting kinase is essential for Wnt signaling and colorectal cancer growth. Cancer Res. 2010;70:5024–5033. doi: 10.1158/0008-5472.CAN-10-0306. [DOI] [PubMed] [Google Scholar]
- St-Jacques B, Dassule HR, Karavanova I, Botchkarev VA, Li J, Danielian PS, McMahon JA, Lewis PM, Paus R, McMahon AP. Sonic hedgehog signaling is essential for hair development. Curr Biol. 1998;8:1058–1068. doi: 10.1016/s0960-9822(98)70443-9. [DOI] [PubMed] [Google Scholar]
- Steinert PM, Steven AC, Roop DR. The molecular biology of intermediate filaments. Cell. 1985;42:411–420. doi: 10.1016/0092-8674(85)90098-4. [DOI] [PubMed] [Google Scholar]
- Stenn KS, Paus R. Controls of hair follicle cycling. Physiol Rev. 2001;81:449–494. doi: 10.1152/physrev.2001.81.1.449. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Itami S, Ohishi M, Hamada K, Inoue T, Komazawa N, Senoo H, Sasaki T, Takeda J, Manabe M, et al. Keratinocyte-specific Pten deficiency results in epidermal hyperplasia, accelerated hair follicle morphogenesis and tumor formation. Cancer Res. 2003;63:674–681. [PubMed] [Google Scholar]
- Tong PL, Roediger B, Kolesnikoff N, Biro M, Tay SS, Jain R, Shaw LE, Grimbaldeston MA, Weninger W. The Skin Immune Atlas: Three-Dimensional Analysis of Cutaneous Leukocyte Subsets by Multiphoton Microscopy. J Invest Dermatol. 2014 doi: 10.1038/jid.2014.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veniaminova NA, Vagnozzi AN, Kopinke D, Do TT, Murtaugh LC, Maillard I, Dlugosz AA, Reiter JF, Wong SY. Keratin 79 identifies a novel population of migratory epithelial cells that initiates hair canal morphogenesis and regeneration. Development. 2013;140:4870–4880. doi: 10.1242/dev.101725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi Y, Mao J-H, Brown K, Girardi M, Balmain A. Promotion of Hras-induced squamous carcinomas by a polymorphic variant of the Patched gene in FVB mice. Nature. 2007;445:761–765. doi: 10.1038/nature05489. [DOI] [PubMed] [Google Scholar]
- Wang GY, Wang J, Mancianti ML, Epstein EH., Jr Basal cell carcinomas arise from hair follicle stem cells in Ptch1(+/−) mice. Cancer Cell. 2011;19:114–124. doi: 10.1016/j.ccr.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Szretter KJ, Vermi W, Gilfillan S, Rossini C, Cella M, Barrow AD, Diamond MS, Colonna M. IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat Immunol. 2012;13:753–760. doi: 10.1038/ni.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M. CoXpress: differential co-expression in gene expression data. BMC Bioinformatics. 2006;7:509. doi: 10.1186/1471-2105-7-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SY, Reiter JF. Wounding mobilizes hair follicle stem cells to form tumors. Proc Natl Acad Sci U S A. 2011;108:4093–4098. doi: 10.1073/pnas.1013098108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Ehdaie B, Ohara N, Yoshino T, Deng C-X. Synergistic action of Smad4 and Pten in suppressing pancreatic ductal adenocarcinoma formation in mice. Oncogene. 2010;29:674–686. doi: 10.1038/onc.2009.375. [DOI] [PubMed] [Google Scholar]
- Yang L, Mao C, Teng Y, Li W, Zhang J, Cheng X, Li X, Han X, Xia Z, Deng H, et al. Targeted disruption of Smad4 in mouse epidermis results in failure of hair follicle cycling and formation of skin tumors. Cancer Res. 2005;65:8671–8678. doi: 10.1158/0008-5472.CAN-05-0800. [DOI] [PubMed] [Google Scholar]
- Youssef KK, Lapouge G, Bouvree K, Rorive S, Brohee S, Appelstein O, Larsimont JC, Sukumaran V, Van de Sande B, Pucci D, et al. Adult interfollicular tumour-initiating cells are reprogrammed into an embryonic hair follicle progenitor-like fate during basal cell carcinoma initiation. Nat Cell Biol. 2012;14:1282–1294. doi: 10.1038/ncb2628. [DOI] [PubMed] [Google Scholar]
- Youssef KK, Van Keymeulen A, Lapouge G, Beck B, Michaux C, Achouri Y, Sotiropoulou PA, Blanpain C. Identification of the cell lineage at the origin of basal cell carcinoma. Nat Cell Biol. 2010;12:299–305. doi: 10.1038/ncb2031. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Poyurovsky MV, Li Y, Biderman L, Stahl J, Jacq X, Prives C. Ribosomal protein S7 is both a regulator and a substrate of MDM2. Molecular cell. 2009;35:316–326. doi: 10.1016/j.molcel.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.