SUMMARY
Understanding individual susceptibility to drug-induced cardiotoxicity is key to improving patient safety and preventing drug attrition. Human induced pluripotent stem cells (hiPSCs) enable the study of pharmacological and toxicological responses in patient-specific cardiomyocytes (CMs), and may serve as preclinical platforms for precision medicine. Transcriptome profiling in hiPSC-CMs from seven individuals lacking known cardiovascular disease-associated mutations, and in three isogenic human heart tissue and hiPSC-CM pairs, showed greater inter-patient variation than intra-patient variation, verifying that reprogramming and differentiation preserve patient-specific gene expression, particularly in metabolic and stress-response genes. Transcriptome-based toxicology analysis predicted and risk-stratified patient-specific susceptibility to cardiotoxicity, and functional assays in hiPSC-CMs using tacrolimus and rosiglitazone, drugs targeting pathways predicted to produce cardiotoxicity, validated inter-patient differential responses. CRISPR/Cas9-mediated pathway correction prevented drug-induced cardiotoxicity. Our data suggest that hiPSC-CMs can be used in vitro to predict and validate patient-specific drug safety and efficacy, potentially enabling future clinical approaches to precision medicine.
Keywords: Induced pluripotent stem cells, cardiomyocytes, personalized drug safety and efficacy, precision medicine
INTRODUCTION
Precision medicine is an emerging approach with great promise for disease prevention and treatment that takes into account individual variability in genes, environment, and lifestyle for each person (Collins and Varmus, 2015). It has rapidly become a major socioeconomic and healthcare focus, drawing attention to the need for developing appropriate research platforms to realize the promise of personalized patient care in the future. The advent of hiPSC reprogramming technologies (Takahashi et al., 2007; Yu et al., 2007) has not only circumvented ethical issues surrounding the use of human embryonic stem cells (hESCs), but has also provided patient-specific cells that act as new platforms for the derivation of differentiated cell types which are ideal for disease modelling research, patient-oriented drug discovery, and toxicology studies (Matsa et al., 2014). In parallel, technological revolution in the “omics” field has enabled cost effective assembly of personalized and dynamic genetic information, providing unprecedented mechanistic insights into physiological states and disease susceptibility (Ozsolak and Milos, 2011). The combination of “omics” and hiPSC technologies provides unique opportunities for building preclinical functional readout systems that could serve as a solid foundation for precision medicine (Chen et al., 2016; Matsa et al., 2016).
Unpredicted drug-induced cytotoxicity has led to thousands of preventable hospitalizations and deaths. In addition, the resulting drug attrition has caused the pharmaceutical industry billions of dollars over the past decade (Laustriat et al., 2010). Over 30% of drugs entering clinical trials are withdrawn due to safety concerns, while the costs of drug discovery have climbed dramatically without a corresponding increase in the numbers of new drugs being released to the market (Scannell and Bosley, 2016). These problems point to the urgent need for developing more appropriate human in vitro models for pre-clinical patient-specific testing of drug candidates. While the use of reprogramming technologies holds great promise for generation of such models, in some studies, divergent reprogramming or differentiation strategies, and chromosomal aberrations acquired during reprogramming and in vitro expansion, have been shown to cause phenotype variation in hiPSCs and their derivatives. (Martins-Taylor and Xu, 2012; Sgodda and Cantz, 2013; Tapia and Schöler, 2016). Such findings call into question the ability of differentiated hiPSC derivatives to reflect the genetic identity of the individual. Therefore, comprehensive comparison of inter- and intra-patient genetic heterogeneity is required to validate the hiPSC technology. Importantly, the ability of hiPSC derivatives from individuals lacking known mutations associated with a relevant disease to respond differentially to drug treatment has not been verified. Addressing this challenge will be critical for validating future applications of hiPSC derivatives in personalized drug discovery and toxicology studies.
In this study, we used RNA sequencing to assess the variation in gene expression signatures between the hiPSC-CMs of two separate hiPSC clones derived from five females (F1-F5, Table S1) and two males (M1-M2). With the use of rigorous methodology standardization, we determined that hiPSC reprogramming and subsequent cardiac differentiation can preserve patient-specific gene expression signatures. This finding was confirmed by transcriptomic comparison of isogenic human heart tissues and hiPSC-CMs from three patients (P1-P3). We also used comprehensive toxicology analysis based on gene expression profiles to show that patient-specific hiPSC-CM susceptibility to drug-induced cardiotoxicity can be computationally predicted, functionally validated, and rescued in vitro. Our study, therefore, represents a step forward for deploying hiPSC-CMs clinically in precision medicine.
RESULTS
Derivation and characterization of hiPSC-CMs
hiPSC lines were first generated from the skin fibroblasts of 5 females (Fig. 1a, S1a) using a non-integrating Sendai virus method. hiPSCs satisfied all characterization criteria for pluripotency (Fig. S1b-c), had normal karyotypes (Fig. S1d), and showed close correlation in SNP profiles to the primary skin fibroblasts they were derived from (Fig. S1e). In addition, hiPSC clones for each individual did not show significant differences in copy number variants. hiPSCs were differentiated to CMs using a small molecule-based methodology (Fig. S2a). Differentiation had over 80% efficiency based on TNNT2+ flow cytometry, following a metabolic selection process in glucose-deprived media (Fig. S2b). hiPSC-CMs also demonstrated regular structural and electrophysiological characteristics at baseline (Fig. S2c-e, Table S2).
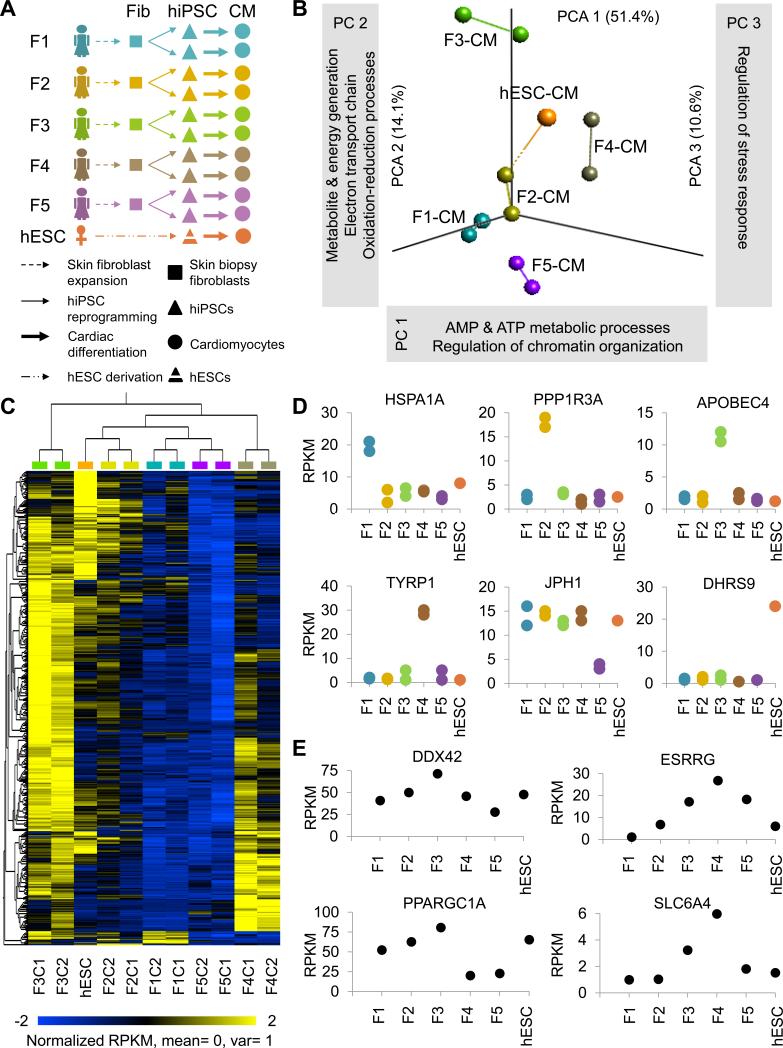
Figure 1. Inter- and intra-patient variation analysis in hiPSC-CMs. See also Figures S1 and S2, and Tables S1-S4.
(a) Schematic outline of study design. Skin fibroblasts were collected from 5 female volunteers, reprogrammed to hiPSCs, and two hiPSC clones per patient were differentiated to CMs for RNA sequencing, pathway analysis, and drug testing. (b) Principal component analysis (PCA) with computation of closest neighboring samples and (c) hierarchical clustering showing closer correlation between hiPSC-CMs from hiPSC clones of the same individual (intra-patient correlation), rather than between individuals (inter-patient variation). (d) The most highly differentially expressed genes for each line were conserved in hiPSC-CMs from both hiPSC clones analyzed. (e) Gene ontology (GO) clusters least conserved in individuals’ hiPSC-CMs were associated with ATP metabolism and chromatin organization (PC1), redox reactions and the electron transport chain (PC2), and regulation of stress response (PC3). Among the most highly differentially expressed genes for each principal component were DDX2 (PC1), ESRRG (PC1), PPARGC1A (PC2), and SLC6A4 (PC3).
Inter- and intra-patient variation in hiPSC-CMs
We next utilized a bioinformatics approach to analyze the variation in transcriptome signatures between hiPSC-CMs derived from the gender-matched female individuals, and from different hiPSC clones of the same individual. Quality scores were high for all RNA sequencing data (Q > 30), reflecting a low probability of base-calling errors (P < 0.001), equivalent to > 99.9% accuracy. Correlation between averaged gene expression values from all clones of each line was high (R = 0.84-0.94; Fig. S2f), reflecting the purity of hiPSC-CM.
Principal component analysis (PCA; Fig. 1b) and hierarchical clustering (Fig. 1c) revealed a closer correlation in CMs derived from hiPSC clones of the same patient rather than from clones of different patients. Inter-patient variation was largely attributed to genes involved in metabolic and stress-response processes (Fig 1b). Importantly, the most significantly differentially expressed genes per cell line (Fig. 1d) showed similar expression patterns for both of the analyzed clones, reinforcing the observation that intra-patient variation in hiPSC-CMs is minimal. Intra-patient clone clustering was also evident amongst over 150 genes that are important for CM function, which included those encoding sarcomere proteins, ion channels, and calcium-handling associated genes (Fig S2g). This indicated that genes conferring functionality to CMs exhibit donor-specific gene expression signatures, suggesting that inter-patient variability could result in patient-specific responses to cardioprotective and cardiotoxic pharmacology.
Functional enrichment analysis revealed minimal intra-patient variation (Fig. 1b-d), which was attributed to differences in molecular functions such as transcriptional regulation, RNA processing and metabolism, and protein localization. A closer investigation into the functions of most differentially expressed genes per line also identified potential susceptibility to a variety of cardiovascular defects. For example, hiPSC-CMs derived from patient 1 (F1) had a significantly elevated expression of the protein-folding factor HSPA1A (Fig. 1d), which is a prognostic indicator of increased risk for coronary heart disease (He et al., 2009). Patient 2's (F2) hiPSC-CMs showed high expression levels of Protein Phosphatase 1, Regulatory Subunit 3A (PPP1R3A), an important regulator of glycogen metabolism. Since glycogen breakdown is the fastest source of energy in the heart, differences in PPP1R3A expression could lead to varied tolerance towards exercise and cardiac performance (Groop and Orho-Melander, 2008). Female patient 3 (F3) was distinct for high expression levels of Apolipoprotein B mRNA Editing Enzyme (APOBEC), a primary component of LDL particles that are key indicators of cardiovascular disease (Wilson et al., 1998). Further details are outlined in Table S3. Such indications, with further refinement and in combination with patients’ clinical history, could become informative for therapy.
GOs most responsible for inter-patient variation (principal component 1; PC1) involved the regulation of chromatin organization, as well as catabolic and metabolic processes such as ATP synthesis and breakdown (Table S4), as exemplified by differential expression of genes such as the ATP-dependent helicase DDX42 (Fig. 1e). Because tight coupling between ATP production and CM contraction is crucial for normal cardiac function (Ingwall and Weiss, 2004), a significant variance in ATP processing suggests potential differences in stress response among hiPSC-CMs from the 5 female patients analyzed. Significant differences were also observed in the estrogen receptor signaling pathway (e.g., Estrogen-Related Receptor Gamma gene ESRRG, Fig 1e), which is known to regulate processes involved in the development of cardiovascular disease, including the mitochondrial electron transport chain, reactive oxygen species (ROS) production, contractile protein function, and ion channel function during cardiac repolarization, through its interaction with the partnered co-factor peroxisome proliferator-activated receptor gamma coactivator 1α [PPARGC1A (Murphy, 2011)].
Perhaps not surprisingly, inter-patient variation in PC2 was attributed to genes involved in the mitochondrial electron transport chain, such as PPARGC1A (Fig 1e). In PC3, genes associated with serotonin transport [e.g., Solute Carrier Family 6 Member 4 (SLC6A4 or SERT1)] were amongst those accounting for inter-patient variation. Pharmaceuticals acting through serotonin (5-HT)-related pathways are widely utilized as antidepressants and appetite suppressants, but are often associated with significant adverse cardiovascular effects, including pulmonary hypertension, arrhythmias, and heart valve disease (Levy, 2006). It is likely that inter-patient variation in the expression of genes involved in serotonin transport could account for the observed side effects, and patient-specific hiPSC-CMs would be ideal platforms for further investigation.
Toxicology pathway analysis in hiPSC-CMs
Unraveling the causes of heterogeneity amongst hiPSC lines and their differentiated derivatives is essential for better understanding of their therapeutic applications. To further elucidate whether the transcriptomic analysis of hiPSC-CMs could predict patient-specific cardiotoxicity, we performed toxicology analysis using the Ingenuity Pathway Analysis (IPA) tools, which use the collective expression levels of interacting genes involved in certain pathways to predict the overall likelihood of adverse cellular function. Toxicology analysis using the average RPKM gene expression in CMs from both hiPSC clones of each patient revealed particularly interesting outcomes concerning the development of oxidative stress mediated through the nuclear factor erythroid 2-like 2 (NFE2L2 or NRF2) pathway. hiPSC-CMs from three lines had a low predicted risk for NRF2-mediated oxidative stress (F2, F3, and F4), whereas F1 and F5 had medium and high predicted risk, respectively (Fig. 2a).
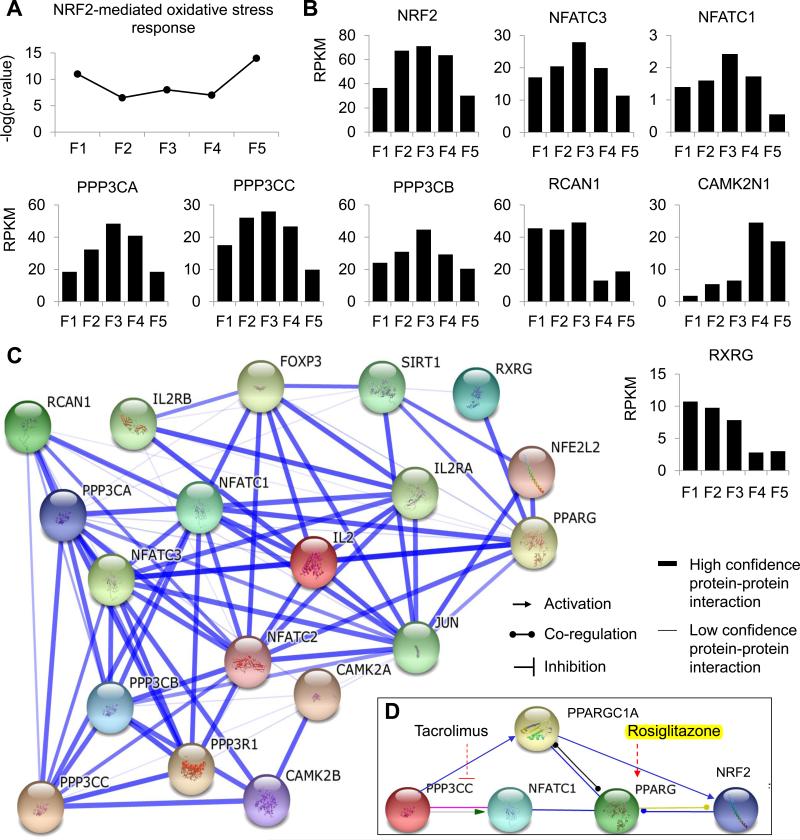
Figure 2. Toxicology pathway analysis. See also Table S3.
(a) Pathway analysis for prediction of risk towards cytotoxicity revealed a higher probability for NRF2-mediated oxidative stress in female line 5 (F5) CMs compared to other lines. (b) NRF2 as well as key genes in the NRF2 pathway were consistently expressed at significantly different levels only in F5. To further probe this complex pathway, (c) protein-protein interactions of the NRF2-pathway were identified using STRING analysis, and (d) inhibitors or agonists of key upstream pathway regulators (PPP3CC inhibitor, tacrolimus; PPARG agonist, rosiglitazone) were identified to investigate potential cardiotoxicity in F5, compared to lines predicted to suffer minimum NRF2-mediated oxidative stress.
The NRF2 pathway is a major mechanism in cellular defense against oxidative stress (Nguyen et al., 2009), and its activation controls the expression of genes involved in detoxification and elimination of ROS and other electrophilic species. In our data, low expression levels of NRF2 in F1 and F5 (Fig. 2b) suggested that hiPSC-CMs could exhibit functional abnormalities upon pharmacological challenge. To investigate this hypothesis and identify suitable drug targets, gene expression and protein-protein interactions of NRF2-associated genes were further investigated (Fig. 2c). It was identified that PPARGC1 (or PGC-1α), which was significantly down-regulated in F5 (Fig. 1e), is an upstream regulator of NRF2 (Ventura-Clapier et al., 2008) and is activated by the protein phosphatase calcineurin, a key regulator of CM ion channels and receptors, calcium homeostasis, fatty acid oxidation, and energy metabolism (Casademont and Miro, 2002; Ventura-Clapier et al., 2008). Interestingly, all three isozymes of calcineurin's catalytic subunit (PPP3CA, PPP3CB, and PPP3CC), as well as the calcineurin regulator RCAN2 were expressed at lower levels in F5 hiPSC-CMs compared to hiPSC-CMs from other female patients (Fig. 2b). At the same time, the calcium/calmodulin-dependent protein kinase II inhibitor CAMK2N1 (or CaMKIINα), a negative regulator of calcium homeostasis, was markedly upregulated in F5, thus supporting the greater potential for functional disruption in F5 hiPSC-CMs. Further interrogation of NRF2 interactions led to the observation that PPARGC1 is also a co-activator of PPARG (PPAR-γ), a receptor which becomes functional upon hetero-dimerization with Retinoid X nuclear Receptors (RXR), including the RXRG receptor that is down-regulated in F5 (Fig 2b). Notably, decreased PPARGC1 expression, as well as decreased expression or DNA binding of the PPAR-RXR complex, have been shown to play a significant role in the development of heart failure (Finck, 2007; Ventura-Clapier et al., 2008). Our pathway analysis, therefore, reinforced the potential heightened susceptibility of F5 hiPSC-CMs to functional disruption of the NRF2 pathway.
Patients F1-F5 had not undergone any clinical procedures allowing isolation of heart tissue for direct comparison of gene expression in primary cardiomyocytes and hiPSC-CMs. To address this, we performed transcriptome profiling in an independent set of patients (P1-P3, Table S1) suffering with non-ischemic dilated cardiomyopathy (NICM) from which heart tissues were available (Fig. 3a), either following orthotopic heart transplantation (OHT) or placement of left ventricular assist devices (LVAD). These data showed that despite the differences in expression of certain cardiac maturation markers (Fig. 3b), over 1,500 genes had close correlation between isogenic left ventricular (LV) primary tissues and corresponding hiPSC-CMs (Fig. S3c and d). GOs with the most preserved expression profiles between LV tissues and hiPSC-CMs included metabolic processes, oxidative stress, and signal transduction pathways such as ERK/MAPK (Fig. S3e). These data supported the hypothesis that pathways involved in NRF2-mediated oxidative stress are preserved between patient-specific primary heart tissues and in vitro-derived hiPSC-CMs, prompting us to choose two drugs - tacrolimus (or FK-506, a calcineurin inhibitor) and rosiglitazone (a PPARG activator) - to assess whether hiPSC-CMs from lines with low, medium and high predicted risk for NRF2-mediated cardiotoxicity would show patient-specific drug responses in vitro (Fig. 2d).
Figure 3. Transcriptomic comparison of isogenic human left ventricular (LV) heart tissues vs. hiPSC-CMs.
(a) Schematic outline of study design for direct comparison of isogenic human heart tissue and hiPSC-CMs. LV tissue and skin fibroblasts were collected from 3 DCM patients. Fibroblasts were reprogrammed to hiPSCs, and differentiated to CMs for AmpliSeq Transcriptome sequencing and pathway analysis. (b) RPKM values of cardiac maturation markers from AmpliSeq RNA sequencing in isogenic LV tissue and fibroblast-derived hiPSC-CMs from three patients with non-ischemic dilated cardiomyopathy. Higher expression of markers associated with cardiac maturation (TNNI3 and MYH7) were observed in LV tissue vs. hiPSC-CMs, while markers associated with cardiac immaturity (TNNI1 and MYH6) were expressed at higher levels in hiPSC-CMs vs. LV tissue. (c) Principal component analysis (PCA) with computation of closest neighboring samples and (d) hierarchical clustering under multivariate analysis of variance (MANOVA) statistical analysis (P = 0.2) showed over 1,500 genes with preserved patient-specific expression in both LV tissue and hiPSC-CMs. (e) Gene ontology (GO) analysis identified that these genes were involved in metabolic processes, oxidative stress, and signal transduction (e.g., ERK/MAPK signaling).
Patient-specific cardiotoxic responses of hiPSC-CMs to pharmacology
Tacrolimus is an immunosuppressant drug used clinically to inhibit organ rejection post-transplantation. It acts via targeted inhibition of calcineurin, a calcium/calmodulin-dependent protein phosphatase, resulting in decreased production of interleukin-2 (IL2) by T cells, inhibiting their growth and proliferation (Musson et al., 2012). Calcineurin is also known as a potent and sufficient inducer of cardiac growth or hypertrophy under physiological or pathological conditions, which is thought to occur through downstream activation of NFAT (e.g., NFATC1, NFATC3, and NFATC4) signaling that is important for calcium handling in CMs (Leinwand, 2001). Tacrolimus has thus been investigated as an anti-hypertrophy drug (Sussman et al., 1998). However, contradictory data exist showing that tacrolimus can itself induce hypertrophy or other cardiotoxicity when used as an immunosuppressant in transplant patients (Zhang et al., 2012).
Optimal doses for in vitro experimentation with tacrolimus were selected based on cell survival and contractility assays performed in hiPSC-CMs from a healthy control female line (F6), which was unrelated to F1-F5 (Fig. S3, S4). Both assays were achieved using high throughput detection systems, for drug concentrations ranging from 1 pM to 1 mM, with 10-fold serial dilutions between. Drug concentrations that did not induce an increase in cell death over a 7-day period and had minimal effects on beating prevalence (% beating area in contractility analysis) compared to untreated controls were considered optimal for further functional assays. Functional characterization was performed in hiPSC-CMs derived from both hiPSC clones of patients F1-F5, and data from both clones were averaged to enhance statistical power.
When tested in low risk (F2, F3, F4), medium risk (F1), or high risk (F5) hiPSC-CMs, we noticed selected concentrations of tacrolimus ranging from 10 nM - 1 μM did not induce significant increases in cell death (Fig. 4a), changes in cell morphology or size (data not shown), beat rate (Fig. 4b), ROS/RNS production (Fig. 4c), or mitochondrial DNA (mtDNA) content measured as a ratio of nuclear to mitochondrial DNA copies using droplet digital PCR (ddPCR, Fig. 4d). This is consistent with literature showing that tacrolimus treatment does not lead to changes in ROS/RNS levels or mtDNA copy number (Rostambeigi et al., 2011). However, it is thought that tacrolimus can induce metabolic stress through uncoupling of mitochondrial oxidative phosphorylation by decreasing inner membrane fluidity (Simon et al., 2003) or inhibiting complex III and IV activity (Zini et al., 1998). The latter can in turn lead to decreased respiratory control ratio (RCR) and ATP depletion, which are often associated with cardiomyopathy (Marin-Garcia et al., 2001). Indeed, RNA-sequencing analysis in tacrolimus-treated F5 hiPSC-CMs detected reduced expression of cardiolipin synthase (CRLS1), which is a key molecule in the maintenance of mitochondrial membrane integrity, as well as Cytochrome C Oxidase Assembly Factor (COX20) and Cytochrome C (CYCS), which are central components of the electron transport chain and ATP production (Table S5). These changes were not observed in low risk F2 hiPSC-CMs.
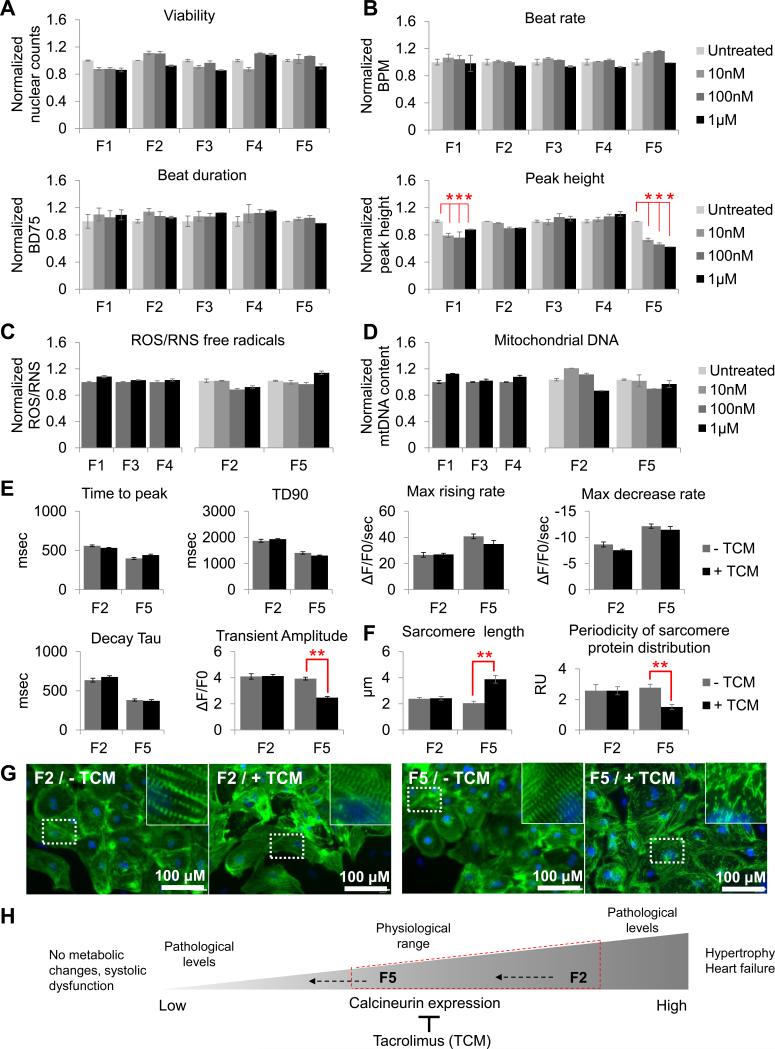
Figure 4. Inter-patient variation in tacrolimus effect. See also Figures S3-S7.
(a) 7-day treatment with 10 nM - 1 μM tacrolimus in F1-F5 hiPSC-CMs did not affect cell viability. (b) Video-based contractility analysis in hiPSC-CMs also showed no changes in beat rate or beat duration (BD75). However, after 7 days of treatment with tacrolimus, peak height decreased in high risk F5 hiPSC-CMs and medium risk F1 hiPSC-CMs (P < 0.05), but not in low risk F2, F3, or F4 hiPSC-CMs. (c) Total production of free radicals (ROS/RNS) and (d) mitochondrial DNA (mtDNA) content were not affected. (e) Calcium imaging revealed a reduced transient amplitude with 1 μM tacrolimus specifically in F5, designating contractile dysfunction. This was accompanied with (g) disrupted sarcomeric organization, (f) significantly decreased periodicity of sarcomere protein distribution, and seemingly increased sarcomere length in tacrolimus-treated F5-CMs, but not in low risk F2 hiPSC-CMs. (h) Correlation between our data and published literature, suggesting that abrogated calcineurin signaling coupled with calcineurin inhibition by tacrolimus can increase susceptibility to systolic dysfunction. Y-axis represents data normalized to the untreated samples for each cell line. RU, relative units; BPM, beats per minute; BD75, 75% of beat duration; TCM, tacrolimus.
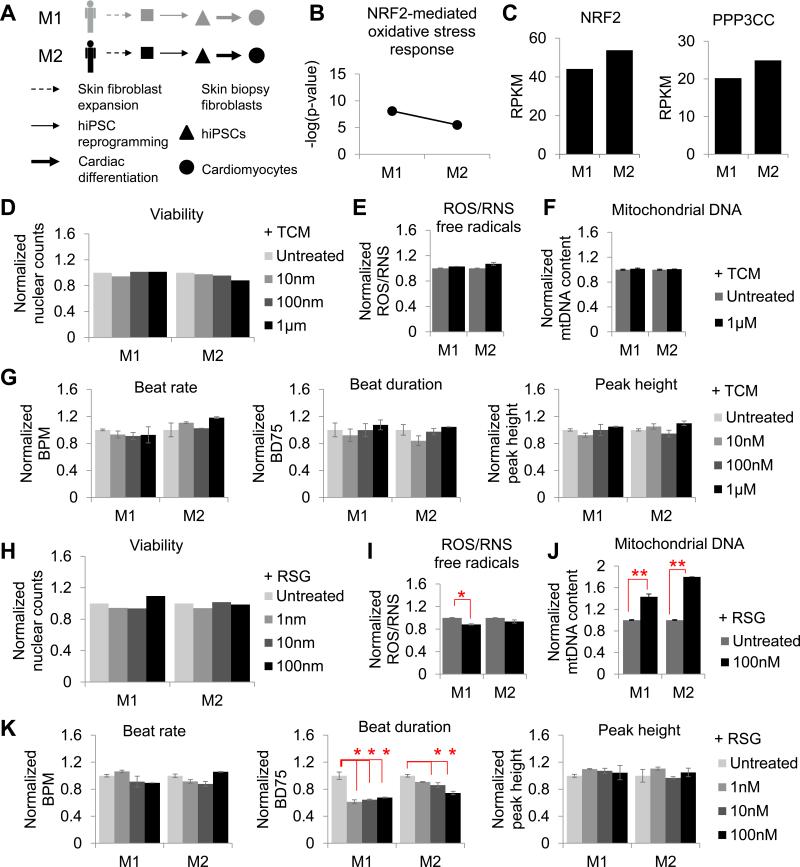
In addition, several key molecules of the ERK/MAPK signaling pathway involved in calcium homeostasis were downregulated in F5 hiPSC-CMs, including CAQS2, CALM2, PKA, and CAMKII (Table S5). In accordance, calcium handling and contractility assays showed a significant decrease in transient amplitude (P < 0.005; Fig. 4e) and contraction/relaxation amplitude in F5 hiPSC-CMs and, to a lesser extent in F1 CMs, but not in F2, F3, or F4 hiPSC-CMs (Fig. 4b). These patient-specific changes were accompanied by sarcomere disarray only in F5, as seen by immunofluorescence staining with cardiac α-actinin (Fig. 4g). Periodicity of sarcomere protein distribution was also significantly decreased in F5, and greater sarcomere length was observed as a result of decreased periodicity (Fig. 4f). To exclude the possibility that observed responses to tacrolimus were gender-specific, hiPSC-CMs from two additional male patients (M1 and M2) lacking known mutations associated to cardiovascular disease were generated (Fig. S5a, S2b and e, and Tables S1 and S2), and shown to have a low risk for NRF2-mediated oxidative stress using transcriptome sequencing (Fig. S5b), as well as high expression of genes involved in NRF2-signaling (Fig. S5c) and functional responses similar to low-risk female hiPSC-CMs (Fig. S5d-g).
Decreased calcium transient amplitude and sarcomeric disarray are characteristic of systolic dysfunction, and are often observed in dilated or hypertrophic cardiomyopathy patients (Frazier et al., 2011). Therefore, this experimental evidence suggests that patients with deficiencies in calcineurin-NRF2 signaling can experience pathological reduction in calcineurin levels upon tacrolimus treatment, leading to patient-specific drug-induced cardiotoxicity (Fig. 4h).
The second drug selected for investigation of patient-specific responses in hiPSC-CMs was rosiglitazone, a member of the thiazolidinedione (TZD) class of drugs, which has been marketed as a type II anti-diabetic drug due to its ability to increase fatty acid responsiveness to glucose by enhancing PPARγ activity (Liu et al., 2012). It has been suggested that rosiglitazone can also improve cardiac efficiency in heart failure, possibly by regulating genes involved in myocardial fatty acid oxidation and utilization, lactate uptake and oxidation, and glucose oxidation. (Fillmore and Lopaschuk, 2013). However, conflicting evidence from the “Rosiglitazone Evaluated for Cardiac Outcomes and Regulation of Glycaemia in Diabetes” (RECORD) clinical trial showed that rosiglitazone can be detrimental to heart function in heart failure and diabetic patients (Mahaffey et al., 2013), likely due to increased oxidative stress and mitochondrial dysfunction (He et al., 2014). Major safety concerns regarding rosiglitazone led the European Medicines Agency (EMA) to withdraw this drug from the market; in the USA, the Food and Drug Administration (FDA) allows rosiglitazone to be sold only following prescription from certified doctors and patients’ informed consent regarding associated risks (Mishra et al., 2014). It is unclear why rosiglitazone can be beneficial in some patients, and yet cardiotoxic in others.
For our experiments, optimal rosiglitazone doses were selected as described above for tacrolimus, and ranged from 1 nM - 100 nM (Fig. S3, S4). These doses did not induce cell death (Fig. 5a), changes in CM morphology and size (data not shown), or sarcomere integrity (Fig 5b-c), but did induce a decrease in ROS/RNS production in F1, F2, F3, and F4 after treatment for 7 days (P < 0.05, Fig. 5d). These observations confirm existing findings that PPARG activation can stimulate anti-oxidant pathways (Potenza et al., 2009). mtDNA content was increased in F2, F3, and F4 (Fig. 5e), in line with evidence showing that PPARG and subsequent NRF2 activation lead to mitochondrial biogenesis in heart tissue (Jornayvaz and Shulman, 2010; Lehman et al., 2000). Concurrently, calcium imaging and contractility analysis indicated an increase in contractility in F1, F2, F3, and F4 in the form of reduced contraction/relaxation periods after 7 days of rosiglitazone treatment (Fig. 5f), as well as decreased time to peak (P = 0.0003), transient duration (TD90, P = 0.00005), and decay Tau (P = 0.000073) of calcium transients (Fig. 5g). Maximum rising rate (P = 0.0004) and maximum decrease rate (P = 0.002) were also reduced in low risk hiPSC-CMs. To exclude the possibility that observed responses to rosiglitazone were gender-specific, hiPSC-CMs from M1 and M2 male patients were assessed and shown to have similar functional responses to low risk female hiPSC-CMs (Fig. S5h-k). Taken together, these data support studies in rat neonatal and adult CMs in which therapeutic levels of rosiglitazone were observed to enhance contractility due to decreased time to peak and maximum rising time, and faster return to resting calcium levels (Shah et al., 2005).
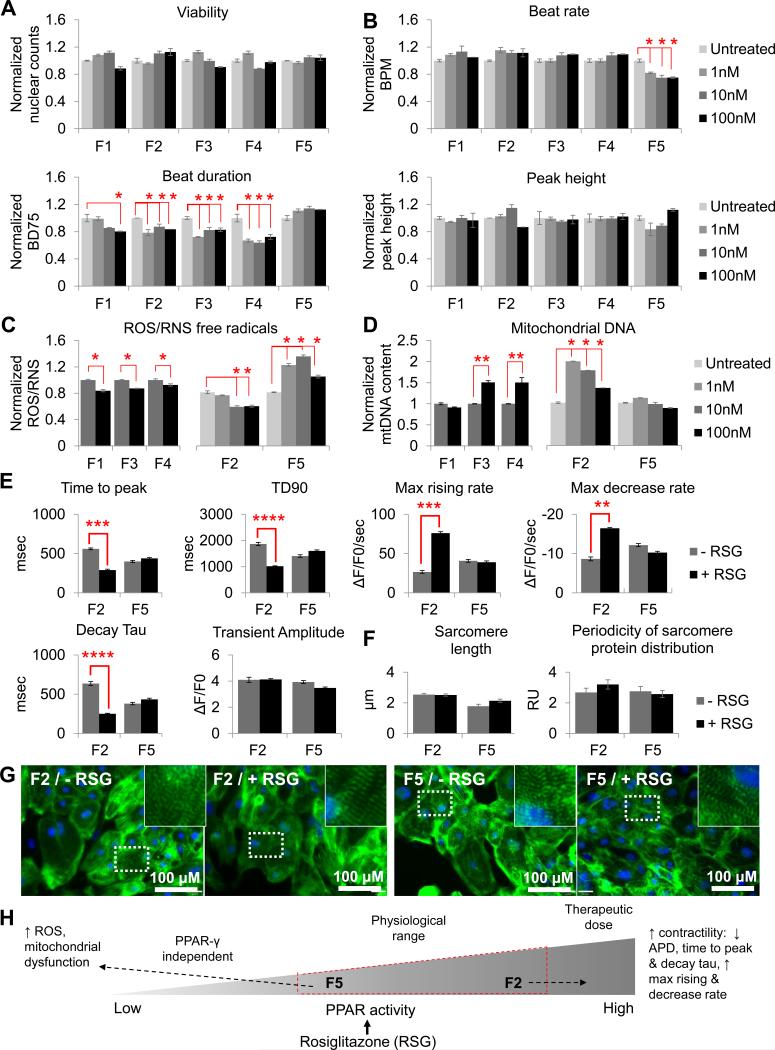
Figure 5. Inter-patient variation in rosiglitazone effect. See also Figures S3-S7.
7-day treatment with 1 nM - 100 nM rosiglitazone in F1-F5 hiPSC-CMs did not affect (a) cell viability, (b) sarcomere length or periodicity of sarcomere protein distribution, or (c) sarcomeric organization. (d) Total production of free radicals (ROS/RNS) decreased in F1-F4 following 7 days of drug treatment, whereas in F5 ROS/RNS levels increased significantly (P < 0.05). (e) Mitochondrial DNA content also increased in F2-F4, but not in F1 or F5. (f) Beat rate and peak height were not affected, whereas beat duration (BD75) decreased at 7-days post-treatment with F1 showing the most moderate response (P < 0.05), and F5 showing no response. (g) Calcium imaging revealed that F5 did not respond to the beneficial effects of rosiglitazone towards improved contractility, as seen in F2 (decreased time to peak, TD90, and decay Tau; increased max rising and decay rates). (h) Correlation between our data and published literature, suggesting that reduced PPAR abundance encourages the PPAR-γ agonist, rosiglitazone, to bind non-specifically, thus increasing susceptibility to mitochondrial dysfunction. Y-axis represents data normalized to the untreated samples for each cell line. RU, relative units; BPM, beats per minute; BD75, 75% of beat duration; RSG, rosiglitazone.
In contrast to low risk hiPSC-CMs, F5-CMs showed no beneficial changes in contractility (Fig. 5f and g) in response to rosiglitazone treatment. Furthermore, a significant increase in ROS/RNS production was observed after 7 days of treatment (1 nM, P = 0.029; 10 nM, P = 0.014; 100 nM, P = 0.015; Fig. 5d), while there was also a lack of mitochondrial biogenesis in F5-CMs (Fig. 5e), consistent with impaired PPARG/NRF2 signaling. In support of functional data, RNA-sequencing analysis showed that rosiglitazone treatment in F5 hiPSC-CMs reduced the expression of the ROS scavenger superoxide dismutase 2 (SOD2), and increased the expression of NADPH oxidase 4 (NOX4) that is involved in ROS synthesis (Table S5).
It should also be noted that F1 was shown by RNA-sequencing to have an intermediate susceptibility to cardiomyocyte dysfunction (Fig. 2a). hiPSC-CMs from this patient had markedly low expression of NRF2 and the calcineurin sub-units PPPP3CA, PPP3CB, and PPP3CC. These gene expression profiles likely contributed to the increased predicted susceptibility for cardiotoxicity compared to F2, F3, and F4. At the same time, F1 hiPSC-CMs also had high expression of the PPARG co-factor RXRG and low expression of the calcium/calmodulin-dependent protein kinase II inhibitor CAMK2N1. The latter are in contract to F5, and could explain the lower susceptibility of F1 to cardiotoxicity compared to F5. The moderate susceptibility of F1 hiPSC-CMs to cardiotoxicity was also shown experimentally (Fig. 4 and 5).
The cardiovascular effects of PPARγ agonists have been contradictory (Paolini et al., 2014). Our study now suggests that inter-patient variation in pathways regulating NRF2-mediated oxidative stress could account for this phenomenon. Originally thought to specifically bind PPARγ receptors and act as an agonist, rosiglitazone has subsequently been found to bind to several off-target molecules such as ion channels and proteins involved in glucose homeostasis, synaptic transduction, and mitochondrial energy production (Hoffmann et al., 2012), which could cause cardiotoxicity independently of PPARγ. Indeed, C57BL/6 mice with CM-specific PPARγ deficiency as well as mice treated with the PPARγ antagonist GW9662 developed cardiotoxicity involving oxidative stress-induced energy deficiency, mitochondrial dysfunction, and left ventricular systolic dysfunction, after treatment with rosiglitazone (He et al., 2014). Therefore, it is possible that due to impairment of the PPARG-NRF2 signaling pathway in F5, rosiglitazone was able to bind primarily to its off-target PPARγ-independent pathways, which are associated with cardiotoxicity (Fig. 5h).
To investigate whether drug-induced toxicity was specific to hiPSC-CMs, we also treated primary skin fibroblasts and hiPSC-derived endothelial cells from all male and female patients with tacrolimus and rosiglitazone. Optimal doses for in vitro experimentation with tacrolimus and rosiglitazone in fibroblasts and hiPSC-ECs were selected based on cell survival assays performed in cells from a healthy control female line (F6) that was unrelated to F1-F5 (Fig. S3). Fibroblasts showed no changes in cell viability, total production of ROS/RNS, or mtDNA content (data not shown), likely due to their low metabolic demand and low nuclear to mtDNA ratio (1: 403.8±43) compared to hiPSC-CMs (1: 998.9±174). hiPSC-ECs (Fig. S6a-b) had a high nuclear to mtDNA ratio (1: 885.6.8±153) and exhibited patient-specific cytotoxicity patterns. Notably, tacrolimus treatment in F5 hiPSC-ECs did not induce cell death, or ROS/RNS and mtDNA changes (Fig. S6c-e), but resulted in a reduced expression of genes associated with EC function, including CD31, CD144, and eNOS (Fig. S6f). Gene expression was not affected in hiPSC-ECs from other patients following tacrolimus treatment. Rosiglitazone treatment also resulted in a reduced expression of genes associated with EC function in F5 hiPSC-ECs, whereas gene expression was increased in hiPSC-ECs from other patients (Fig. S6g). In addition, F5 hiPSC-ECs showed increased levels of ROS/RNS (Fig. S7i) and no changes in mitochondrial biogenesis (Fig. S7j), in contrast to hiPSC-ECs from other patients where ROS/RNS levels were decreased or unaltered (Fig. S7i) and mtDNA content was increased (Fig. S7j). These data suggested that hiPSCs derivatives could be used to study patient-specific drug-induced cytotoxicity in a pleiotropic manner, and that patient-specific drug effects may arise from genomic variations that exert effects on multiple cell types.
Correction of patient-specific cardiotoxicity through CRISPR/Cas9-mediated genome editing
Whole exome sequencing in primary skin fibroblasts from F1-F5 and M1-M2 patients identified 43-54 potentially pathogenic known variants in each patient (Table S6), consistent with the number of deleterious non-synonymous variants observed in human genetic variation studies from large populations (The 1000 Genomes Project Consortium, 2012). Of these, approximately 10% were unique to patient F5 and had a low (0.01–0.05) minor allele frequency (MAF) that is indicative of a higher likelihood of association to disease risk (Table S7). However, none of the variants were associated with metabolic dysfunction, oxidative stress, or aberrant response to tacrolimus or rosiglitazone pharmacotherapy (dbSNP database; http://www.ncbi.nlm.nih.gov/SNP/). In addition, no potentially deleterious SNPs were identified in over 150 genes that are important for CM function, which included those that encode sarcomere proteins, ion channels, and calcium-handling associated genes, or in genes identified to have abnormal expression in F5 hiPSC-CMs following toxicology pathway analysis (Fig. 2b). This does not exclude the possibility that novel variants not listed in the dbSNP database could have accounted for the drug-induced cytotoxicity observed in patient F5.
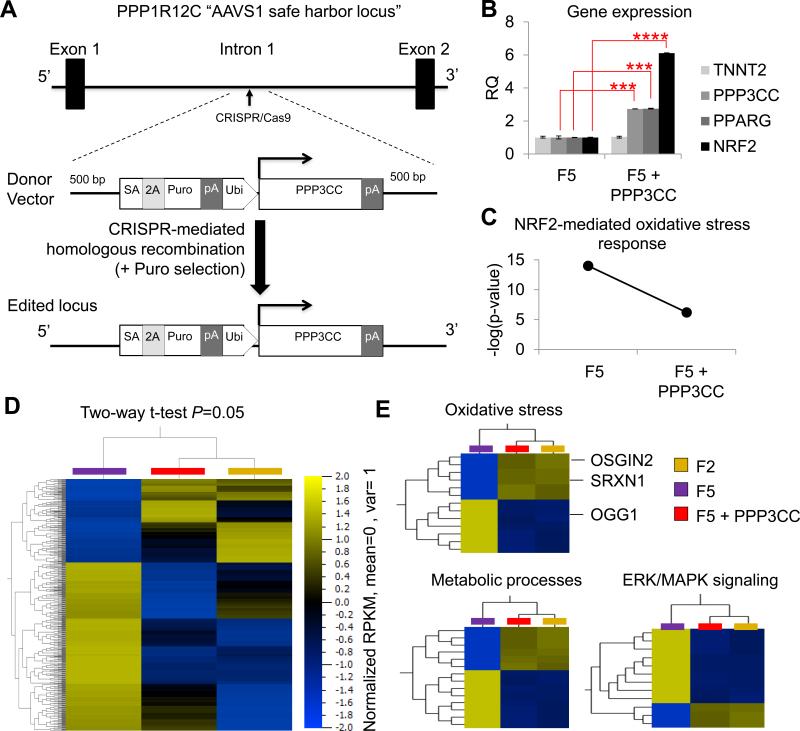
Nevertheless, in the absence of a known causative SNP in F5, phenotypic rescue was performed via CRISPR/Cas9-mediated transgenic overexpression of PPP3CC (Fig. 6a) which is an upstream activator of both PPARG and NRF2 (Long et al., 2007). In line with current literature, PPP3CC overexpression led to upregulation of PPARG and NRF2 in genome edited F5 hiPSC-CMs (henceforth referred to as F5 + PPP3CC, Fig. 6b). Nuclear to mtDNA ratio also increased in F5 + PPP3CC hiPSC-CMs (1: 1,596) compared to F5 CMs (1: 1,189). When analyzed at the transcriptome level, F5 + PPP3CC hiPSC-CMs had reduced probability for NRF2-mediated oxidative stress compared to parental F5 hiPSC-CMs (Fig. 6c). Hierarchical clustering under statistical analysis (two-way t-test, P = 0.05) showed that thousands of genes had similar gene expression levels in F2 and F5 + PPP3CC hiPSC-CMs, whereas F5 hiPSC-CMs had a markedly different gene expression pattern (Fig. 6d). The biggest variance between the two groups was found in genes involved with metabolic processes (GO: 0008152), oxidative stress (GO: 0006979), and ERK/MAPK signaling (GO: 0000165, Fig. 6e), consistent with the biological roles of calcineurin, PPARG, and NRF2 signaling (Trachootham et al., 2008). Examples of genes that were expressed at lower levels in F5 compared to F2 and F5 + PPP3CC included the oxidoreductases OSGIN2 and SRXN, whereas genes that are typically upregulated following exposure to ROS (e.g., OGG1) were expressed at higher levels in F5 compared to F2 and F5 + PPP3CC (Fig. 6e).
Figure 6. Generation and characterization of genome-edited F5 hiPSC-CMs. See also Figure S7 and Tables S5-S7.
(a) Schematic diagram of the CRISPR/Cas9 experimental strategy used to over-express the PPP3CC gene in the AAVS1 safe harbor locus of F5 hiPSCs. (b) Quantitative real-time PCR analysis of PPP3CC, PPARG, NRF2, and TNNT2 gene expression in F5 hiPSC-CMs and CRISPR/Cas9-mediated genome edited F5 hiPSC-CMs overexpressing PPP3CC (F5 + PPP3CC) from the ubiquitin promoter inserted in the AAVS1 safe harbor locus. (c) Pathway analysis for prediction of risk towards cytotoxicity revealed a reduced probability for NRF2-mediated oxidative stress in F5 + PPP3CC hiPSC-CMs compared to parental F5 hiPSC-CMs. (d) Hierarchical clustering of AmpliSeq RNA-sequencing data under two-way t-test (P = 0.05) showed that many genes had similar gene expression levels in F2 and genome edited F5 + PPP3CC hiPSC-CMs, while F5 hiPSC-CMs had a distinctly different gene expression pattern. (e) Gene ontology (GO) analysis identified that the most differentially expressed genes were involved in metabolic processes, oxidative stress, and signal transduction (e.g., ERK/MAPK signaling). SA, splice acceptor; Puro, puromycin resistance gene; Ubi, ubiquitin promoter; pA, polyadenylation signal.
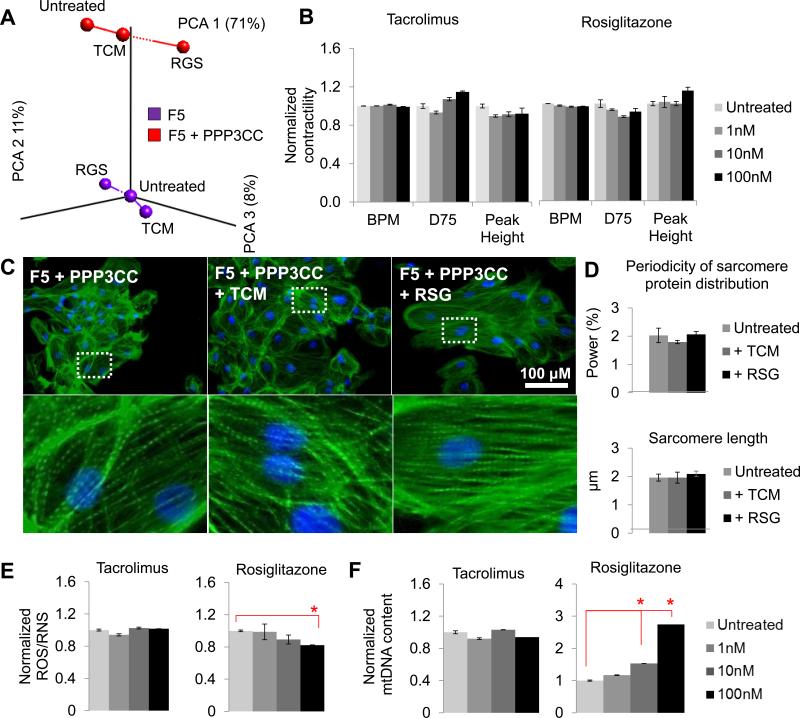
PCA analysis of RNA-sequencing data in F5 and F5 + PPP3CC hiPSC-CMs treated with 1 μM tacrolimus or 100 nM rosiglitazone for 7 days showed distinct responses between the two cell lines (Fig. 7a). Hierarchical clustering of RNA-sequencing data under a two-way t-test (P = 0.05) showed that many genes had similar RPKM fold changes compared to untreated samples in F2 and F5 + PPP3CC hiPSC-CMs, whereas F5 hiPSC-CMs had a distinctly different drug response. Not surprisingly, the most significant GO terms involved in differential drug response included metabolic processes, oxidative stress, and ERK/MAPK signaling (Fig. S7a-d). Functional characterization confirmed that over-expression of PPP3CC in F5 enabled phenotypic rescue; 7-day tacrolimus treatment had no deleterious effects in hiPSC-CM contractility (Fig. 7b), or sarcomere organization (Fig. 7c-d) in F5 + PPP3CC hiPSC-CMs. In addition, 7-day rosiglitazone treatment led to a significant decrease in ROS/RNS production (Fig. 7e) and enrichment in mtDNA content (Fig. 7f), without any deleterious effects in contractility (Fig. 7b). This is line with the rosiglitazone-induced activation of PPARG, which is involved in mitochondrial biogenesis (Jornayvaz and Shulman, 2010; Lehman et al., 2000). Taken together, these data suggest that NRF2 deficiency is an underlying mechanism for susceptibility towards drug-induced cardiotoxicity.
Figure 7. Drug-induced cytotoxicity rescued in F5 hiPSC-CMs through genome editing See also Table S5.
(a) Principal component analysis of AmpliSeq RNA-sequencing data in F5 and F5 + PPP3CC hiPSC-CMs showed cell line-specific responses with tacrolimus or rosiglitazone treatment. Over-expression of PPP3CC in F5 enabled phenotypic rescue; 7-day tacrolimus treatment had no deleterious effects in (b) contractility, (c, d) sarcomere organization, (e) ROS/RNS production, or (f) mitochondrial DNA (mtDNA) content in hiPSC-CMs. In addition, 7-day rosiglitazone treatment led to (e) significant decrease in ROS/RNS production and (f) enrichment in mitochondrial content, without any deleterious effects in (b) contractility or (c, d) sarcomere organization. Y-axis represents data normalized to the undertreated sample for the F5 + PPP3CC cell line. RQ; relative quantification; RU, relative units; BPM, beats per minute; BD75, 75% of beat duration; TCM, tacrolimus; RSG, rosiglitazone. See also Figure S7.
DISCUSSION
Understanding the level and patterns of gene expression variation between individuals is of paramount importance for biomedical research (Storey et al., 2007). Previous publications have used RNA sequencing data to show that genetic differences between individual donors were the major cause of transcriptional variation between pluripotent hiPSC lines, whereas other contributors such as cell type of origin and reprograming strategy had minimal effects on transcriptional heterogeneity (Choi et al., 2015; Rouhani et al., 2014). However, transcriptional variation in relevant differentiated derivatives of these hiPSC lines was not investigated. Our data show that rigorous standardization of hiPSC reprogramming and cardiac differentiation can preserve inter-individual variation in hiPSC differentiated derivatives with high fidelity, without increasing intra-individual variation, thus eliminating previously reported intra-clone variation in mouse hiPSC-CMs (Kaichi et al., 2010).
We also found that NRF2 pathway irregularities are implicated in clinically relevant drug-related side effects, including tacrolimus- and rosiglitazone-induced cardiovascular damage. The patients included in this study (F1-F5 and M1-M2) lacked known mutations associated with cardiovascular diseases and did not undergo cardiac biopsies, LVAD placement, or other procedures that would make isolation of heart tissue possible. They also had not been treated with tacrolimus or rosiglitazone to allow direct comparisons between in vitro and in vivo data. Nevertheless, our data from three unrelated patients (P1-P3) whose LV tissues and hiPSC-CMs were both available showed preserved patient-specific gene expression in metabolic, oxidative stress, and ERK/MAPK pathways (Fig. 3). This strengthens the ability of hiPSC-CMs to reflect clinically relevant perturbations regarding drug-induced cytotoxicity associated with these pathways.
Recent advances in molecular biology are beginning to elucidate the transcriptional events governing NRF2-mediated metabolic and oxidative stress in CMs, and how these are associated with disease and drug response. It is known that increased levels of free radicals and decreased ATP availability due to increased mitochondrial DNA (mtDNA) damage or mitochondrial uncoupling can lead to sarcomeric damage, or excitation and calcium handling abnormalities (Austin and St-Pierre, 2012), as observed in our study (Fig. 4 and 5). Unraveling the intricate mechanistic relationship between metabolic or oxidative stress and heart failure in future studies could lead to improved pharmacological approaches and patient care. We also show that cardiotoxicity risk as exemplified by responses to tacrolimus and rosiglitazone can be computationally predicted and functionally validated in hiPSC-CMs from patients without known mutations related to cardiovascular diseases. Finally, we studied drug-induced cytotoxicity in isogenic hiPSC-CMs and hiPSC-ECs, which suggests that hiPSCs could potentially be utilized to investigate cell type-specific toxicity as well as patient-specific cytotoxicity in future studies.
Even though organismal physiology is more complex than in vitro cell biology, our data and methodology provide a proof of principle and a foundation for building a deeper mechanistic understanding of natural inter-patient variation in gene-expression levels that will be useful for interpreting variations in drug efficacy and drug-induced cytotoxicity. Further work in this field includes investigating the presence and impact of novel polymorphisms (e.g., small deletions or insertions, and single nucleotide polymorphisms) on drug treatment outcomes. Indeed, several PharmGKB variant annotations have been identified for tacrolimus (e.g., in ABCB1, CYP3A4, and CYP3A5) and rosiglitazone (e.g., in CYP2C8, LPIN1, PAX4, PPARG, and PARGC1A). The combination of hiPSCs and modern genome editing technologies (Wilson and Wu, 2015) should facilitate studies into this branch of precision medicine. Currently, hiPSC reprogramming followed by lineage-directed differentiation represent the most efficient and least invasive strategy for isolating functional human cells (e.g., CMs) for such in vitro experimentation.
In summary, our data demonstrate that by combining consistent and reproducible reprogramming, with controlled differentiation strategies, it is feasible to preserve the patient-specific identity of hiPSC-CMs. Lines generated from the same individual revealed that gene expression signatures were more similar to each other than to those of hiPSC-CMs generated from different individuals. With further refinement and in combination to patients’ clinical history, these findings may lead to the use of hiPSC-CMs for predicting and functionally validating personalized drug responses in the future.
EXPERIMENTAL PROCEDURES
hiPSC generation, culture, and differentiation
Details on the isolation of patient tissue, as well as the generation, culture, characterization and differentiation of hiPSCs have been previously published (Burridge et al., 2016), and can be found in Supplementary Methods.
RNA sequencing
RNA was extracted from frozen pellets of hiPSC-CMs using the QIAGEN RNeasy kit, under the manufacturer's instructions. Libraries for RNA sequencing were prepared from 100 ng RNA, with cDNA sheared to an average size of 300 bps. Fragments were repaired and polyadenylated, and Illumina adapters were ligated to their ends. Ten PCR cycles using primers specific for Illumina adapters were performed to amplify each sample to a suitable working concentration. Library quality was verified using the Agilent High Sensitivity DNA Kit on Agilent's 2100 Bioanalyzer, and 260 bp fragments were isolated by gel extraction. Fragments were sequenced using paired end reads (2×100 bp) on the Illumina HiSeq platform, with an average of 30 million reads per sample. Reads were aligned to the human reference genome (hg19) with Tophat software v2.0.9 and normalized to ERCC spike-in controls using a loess regression function in the affy package v1.42.3 in R (Gautier et al., 2004). An RPKM cut-off of >1 was applied. See also Supplemental Materials for AmpliSeq Transcriptome analysis. All transcriptome data is publically available via the NCBI Gene Expression Omnibus (GEO) database, under accession number GSE83668.
Bioinformatics analysis
Principal component and hierarchical clustering heat map analysis were performed with Omics Explorer v3.2 software (Qlucore). Enrichment analyses of GO terms represented in each PCA component were performed using the goseq package in R (Young et al., 2010). Inter-line comparisons were conducted by comparing the RPKM value of each gene, and genes significantly up- or down-regulated in each group were identified by variance of RPKM values. PANTHER Classification System software and Qlucore GSEA was used to identify molecular functions contributing to intra-line variation, and toxicology pathway analysis was performed using Ingenuity Pathway Analysis (IPA) software from QIAGEN. Protein-protein interaction pathways were built using IPA software and STRING online tools v10. See also Supplemental Materials for AmpliSeq Transcriptome analysis.
Drug treatments
Tacrolimus (FK 506) and rosiglitazone (both from Tocris Bioscience) were dissolved to a final concentration of 100 mM in DMSO, aliquoted, and stored at −20 °C. 10-fold serial dilutions ranging from 1 mM to 1 pM were prepared fresh each day, in CM culture media. For treatments longer than 24 h, fresh drugs were applied daily.
Supplementary Material
Acknowledgments
We thank funding support from the National Institutes of Health R01 HL113006, NIH R01 HL123968, NIH R01 HL130020, NIH R01 HL126527, NIH R01 HL128170, Burroughs Wellcome Foundation Innovation in Regulatory Science Award (JCW); CIRM Center of Excellence for Stem Cell Genomics GC1R-06673-A, NIH P01 GM099130 (MPS); NHLBI Progenitor Cell Biology Jump Start Award, American Heart Association 16BGIA27790017, Stanford Cardiovascular Institute Seed Grant (EM); NIH K99 HL121177, AHA 14BGIA20480329 (PWB); and Winston Chen Stanford Graduate Fellowship (KY).
Footnotes
Author contributions:
E.M. conceived, performed, and interpreted the experiments, and wrote the manuscript; P.W.B. performed hiPSC reprogramming and provided hiPSC lines; K.Y. and J.H.A. performed bioinformatics analysis; V.T. performed genome editing; J.M.C. performed RNA-seq library preparations; H.W. performed calcium imaging; P.S. performed patch-clamp electrophysiology; C.H. and N. Sayed performed EC differentiation; N. Shao performed GEO data curation; N.A.W. and A.S.C. performed droplet digital PCR; I.K. performed next-generation sequencing and provided experimental advice; J.D.G. and M.P.S. provided experimental advice; and J.C.W. provided experimental advice, manuscript review, and funding support.
Conflict of Interest Disclosures:
Dr. Joseph Wu is co-founder and member of scientific advisory board for Stem Cell Theranostics.
REFERENCES
- Austin S, St-Pierre J. PGC1alpha and mitochondrial metabolism--emerging concepts and relevance in ageing and neurodegenerative disorders. J Cell Sci. 2012;125:4963–4971. doi: 10.1242/jcs.113662. [DOI] [PubMed] [Google Scholar]
- Burridge PW, Diecke S, Matsa E, Sharma A, Wu H, Wu JC. Modeling Cardiovascular Diseases with Patient-Specific Human Pluripotent Stem Cell-Derived Cardiomyocytes. Methods Mol Biol. 2016;1353:119–130. doi: 10.1007/7651_2015_196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casademont J, Miro O. Electron transport chain defects in heart failure. Heart Fail Rev. 2002;7:131–139. doi: 10.1023/a:1015372407647. [DOI] [PubMed] [Google Scholar]
- Chen IY, Matsa E, Wu JC. Induced pluripotent stem cells: at the heart of cardiovascular precision medicine. Nature reviews Cardiology. 2016;13:333–349. doi: 10.1038/nrcardio.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Lee S, Mallard W, Clement K, Tagliazucchi GM, Lim H, Choi IY, Ferrari F, Tsankov AM, Pop R, et al. A comparison of genetically matched cell lines reveals the equivalence of human iPSCs and ESCs. Nat Biotechnol. 2015;33:1173–1181. doi: 10.1038/nbt.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillmore N, Lopaschuk GD. Targeting mitochondrial oxidative metabolism as an approach to treat heart failure. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2013;1833:857–865. doi: 10.1016/j.bbamcr.2012.08.014. [DOI] [PubMed] [Google Scholar]
- Finck BN. The PPAR regulatory system in cardiac physiology and disease. Cardiovasc Res. 2007;73:269–277. doi: 10.1016/j.cardiores.2006.08.023. [DOI] [PubMed] [Google Scholar]
- Frazier AH, Ramirez-Correa GA, Murphy AM. Molecular mechanisms of sarcomere dysfunction in dilated and hypertrophic cardiomyopathy. Prog Pediatr Cardiol. 2011;31:29–33. doi: 10.1016/j.ppedcard.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- Groop L, Orho-Melander M. New insights into impaired muscle glycogen synthesis. PLoS Med. 2008;5:e25. doi: 10.1371/journal.pmed.0050025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Tao H, Xiong H, Duan SZ, McGowan FX, Jr., Mortensen RM, Balschi JA. Rosiglitazone causes cardiotoxicity via peroxisome proliferator-activated receptor gamma-independent mitochondrial oxidative stress in mouse hearts. Toxicol Sci. 2014;138:468–481. doi: 10.1093/toxsci/kfu015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Guo H, Yang X, Zhang X, Zhou L, Cheng L, Zeng H, Hu FB, Tanguay RM, Wu T. Functional SNPs in HSPA1A gene predict risk of coronary heart disease. PLoS One. 2009;4:e4851. doi: 10.1371/journal.pone.0004851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann BR, El-Mansy MF, Sem DS, Greene AS. Chemical proteomics-based analysis of off-target binding profiles for rosiglitazone and pioglitazone: clues for assessing potential for cardiotoxicity. J Med Chem. 2012;55:8260–8271. doi: 10.1021/jm301204r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingwall JS, Weiss RG. Is the Failing Heart Energy Starved?: On Using Chemical Energy to Support Cardiac Function. Circulation Research. 2004;95:135–145. doi: 10.1161/01.RES.0000137170.41939.d9. [DOI] [PubMed] [Google Scholar]
- Jornayvaz FR, Shulman GI. Regulation of mitochondrial biogenesis. Essays in biochemistry. 2010;47:69–84. doi: 10.1042/bse0470069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaichi S, Hasegawa K, Takaya T, Yokoo N, Mima T, Kawamura T, Morimoto T, Ono K, Baba S, Doi H, et al. Cell line-dependent differentiation of induced pluripotent stem cells into cardiomyocytes in mice. Cardiovascular Research. 2010;88:314–323. doi: 10.1093/cvr/cvq189. [DOI] [PubMed] [Google Scholar]
- Laustriat D, Gide J, Peschanski M. Human pluripotent stem cells in drug discovery and predictive toxicology. Biochem Soc Trans. 2010;38:1051–1057. doi: 10.1042/BST0381051. [DOI] [PubMed] [Google Scholar]
- Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, Kelly DP. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. Journal of Clinical Investigation. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinwand LA. Calcineurin inhibition and cardiac hypertrophy: a matter of balance. Proc Natl Acad Sci U S A. 2001;98:2947–2949. doi: 10.1073/pnas.051033698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy RJ. Serotonin transporter mechanisms and cardiac disease. Circulation. 2006;113:2–4. doi: 10.1161/CIRCULATIONAHA.105.593459. [DOI] [PubMed] [Google Scholar]
- Liu T, Korantzopoulos P, Li G. Antioxidant therapies for the management of atrial fibrillation. Cardiovasc Diagn Ther. 2012;2:298–307. doi: 10.3978/j.issn.2223-3652.2012.10.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long YC, Glund S, Garcia-Roves PM, Zierath JR. Calcineurin Regulates Skeletal Muscle Metabolism via Coordinated Changes in Gene Expression. Journal of Biological Chemistry. 2007;282:1607–1614. doi: 10.1074/jbc.M609208200. [DOI] [PubMed] [Google Scholar]
- Mahaffey KW, Hafley G, Dickerson S, Burns S, Tourt-Uhlig S, White J, Newby LK, Komajda M, McMurray J, Bigelow R, et al. Results of a reevaluation of cardiovascular outcomes in the RECORD trial. Am Heart J. 2013;166:240–249. e241. doi: 10.1016/j.ahj.2013.05.004. [DOI] [PubMed] [Google Scholar]
- Marin-Garcia J, Goldenthal MJ, Moe GW. Mitochondrial pathology in cardiac failure. Cardiovasc Res. 2001;49:17–26. doi: 10.1016/s0008-6363(00)00241-8. [DOI] [PubMed] [Google Scholar]
- Martins-Taylor K, Xu RH. Concise review: Genomic stability of human induced pluripotent stem cells. Stem Cells. 2012;30:22–27. doi: 10.1002/stem.705. [DOI] [PubMed] [Google Scholar]
- Matsa E, Ahrens JH, Wu JC. Human Induced Pluripotent Stem Cells as a Platform for Personalized and Precision Cardiovascular Medicine. Physiological Reviews Accepted. 2016 doi: 10.1152/physrev.00036.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsa E, Burridge PW, J.C., W. Human stem cells for modeling heart disease and for drug discovery. Sci Transl Med. 2014;6 doi: 10.1126/scitranslmed.3008921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P, Singh SV, Verma AK, Srivastava P, Sultana S, Rath SK. Rosiglitazone induces cardiotoxicity by accelerated apoptosis. Cardiovasc Toxicol. 2014;14:99–119. doi: 10.1007/s12012-013-9234-y. [DOI] [PubMed] [Google Scholar]
- Murphy E. Estrogen signaling and cardiovascular disease. Circ Res. 2011;109:687–696. doi: 10.1161/CIRCRESAHA.110.236687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musson RE, Cobbaert CM, Smit NP. Molecular diagnostics of calcineurin-related pathologies. Clin Chem. 2012;58:511–522. doi: 10.1373/clinchem.2011.167296. [DOI] [PubMed] [Google Scholar]
- Nguyen T, Nioi P, Pickett CB. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J Biol Chem. 2009;284:13291–13295. doi: 10.1074/jbc.R900010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolini P, Pick D, Lapira J, Sannino G, Pasqualini L, Ludka C, Sprague LJ, Zhang X, Bartolotta EA, Vazquez-Hidalgo E, et al. Developmental and extracellular matrix-remodeling processes in rosiglitazone-exposed neonatal rat cardiomyocytes. Pharmacogenomics. 2014;15:759–774. doi: 10.2217/pgs.14.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potenza MA, Gagliardi S, De Benedictis L, Zigrino A, Tiravanti E, Colantuono G, Federici A, Lorusso L, Benagiano V, Quon MJ, et al. Treatment of spontaneously hypertensive rats with rosiglitazone ameliorates cardiovascular pathophysiology via antioxidant mechanisms in the vasculature. Am J Physiol Endocrinol Metab. 2009;297:E685–694. doi: 10.1152/ajpendo.00291.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostambeigi N, Lanza IR, Dzeja PP, Deeds MC, Irving BA, Reddi HV, Madde P, Zhang S, Asmann YW, Anderson JM, et al. Unique cellular and mitochondrial defects mediate FK506-induced islet beta-cell dysfunction. Transplantation. 2011;91:615–623. doi: 10.1097/TP.0b013e3182094a33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhani F, Kumasaka N, de Brito MC, Bradley A, Vallier L, Gaffney D. Genetic background drives transcriptional variation in human induced pluripotent stem cells. PLoS Genet. 2014;10:e1004432. doi: 10.1371/journal.pgen.1004432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell JW, Bosley J. When Quality Beats Quantity: Decision Theory, Drug Discovery, and the Reproducibility Crisis. PLoS One. 2016;11:e0147215. doi: 10.1371/journal.pone.0147215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgodda M, Cantz T. Small but Significant: Inter- and Intrapatient Variations in iPS Cell-based Disease Modeling. Mol Ther. 2013;21:5–7. doi: 10.1038/mt.2012.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah RD, Gonzales F, Golez E, Augustin D, Caudillo S, Abbott A, Morello J, McDonough PM, Paolini PJ, Shubeita HE. The antidiabetic agent rosiglitazone upregulates SERCA2 and enhances TNF-alpha- and LPS-induced NF-kappaB-dependent transcription and TNF-alpha-induced IL-6 secretion in ventricular myocytes. Cell Physiol Biochem. 2005;15:41–50. doi: 10.1159/000083637. [DOI] [PubMed] [Google Scholar]
- Simon N, Morin C, Urien S, Tillement JP, Bruguerolle B. Tacrolimus and sirolimus decrease oxidative phosphorylation of isolated rat kidney mitochondria. Br J Pharmacol. 2003;138:369–376. doi: 10.1038/sj.bjp.0705038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Madeoy J, Strout JL, Wurfel M, Ronald J, Akey JM. Gene-expression variation within and among human populations. Am J Hum Genet. 2007;80:502–509. doi: 10.1086/512017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman MA, Lim HW, Gude N, Taigen T, Olson EN, Robbins J, Colbert MC, Gualberto A, Wieczorek DF, Molkentin JD. Prevention of cardiac hypertrophy in mice by calcineurin inhibition. Science. 1998;281:1690–1693. doi: 10.1126/science.281.5383.1690. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tapia N, Schöler HR. Molecular obstacles to clinical translation of iPSCs. Cell Stem Cell. 2016 doi: 10.1016/j.stem.2016.06.017. In press. [DOI] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachootham D, Lu W, Ogasawara MA, Valle NR-D, Huang P. Redox Regulation of Cell Survival. Antioxidants & Redox Signaling. 2008;10:1343–1374. doi: 10.1089/ars.2007.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura-Clapier R, Garnier A, Veksler V. Transcriptional control of mitochondrial biogenesis: the central role of PGC-1alpha. Cardiovasc Res. 2008;79:208–217. doi: 10.1093/cvr/cvn098. [DOI] [PubMed] [Google Scholar]
- Wilson KD, Wu JC. Induced pluripotent stem cells. JAMA. 2015;313:1613–1614. doi: 10.1001/jama.2015.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson PW, D'Agostino RB, Levy D, Belanger AM, Silbershatz H, Kannel WB. Prediction of coronary heart disease using risk factor categories. Circulation. 1998;97:1837–1847. doi: 10.1161/01.cir.97.18.1837. [DOI] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yao L, Yu X, Ou J, Hui N, Liu S. A poor imitation of a natural process: A call to reconsider the iPSC engineering technique. Cell Cycle. 2012;11 doi: 10.4161/cc.22575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zini R, Simon N, Morin C, Thiault L, Tillement JP. Tacrolimus decreases in vitro oxidative phosphorylation of mitochondria from rat forebrain. Life Sci. 1998;63:357–368. doi: 10.1016/s0024-3205(98)00284-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.