Abstract
Testicular germ cell tumors (TGCTs) share germline ancestry but diverge phenotypically and clinically as seminoma (SE) and nonseminoma (NSE), the latter including the pluripotent embryonal carcinoma (EC) and its differentiated derivatives, teratoma (TE), yolk sac tumor (YST), and choriocarcinoma. Epigenomes from TGCTs may illuminate reprogramming in both normal development and testicular tumorigenesis. Herein we investigate pure-histological forms of 130 TGCTs for conserved and subtype-specific DNA methylation, including analysis of relatedness to pluripotent stem cell (ESC, iPSC), primordial germ cell (PGC), and differentiated somatic references. Most generally, TGCTs conserve PGC-lineage erasure of maternal and paternal genomic imprints and DPPA3 (also known as STELLA); however, like ESCs, TGCTs show focal recurrent imprinted domain hypermethylation. In this setting of shared physiologic erasure, NSEs harbor a malignancy-associated hypermethylation core, akin to that of a diverse cancer compendium. Beyond these concordances, we found subtype epigenetic homology with pluripotent versus differentiated states. ECs demonstrate a striking convergence of both CpG and CpH (non-CpG) methylation with pluripotent states; the pluripotential methyl-CpH signature crosses species boundaries and is distinct from neuronal methyl-CpH. EC differentiation to TE and YST entails reprogramming toward the somatic state, with loss of methyl-CpH but de novo methylation of pluripotency loci such as NANOG. Extreme methyl-depletion among SE reflects the PGC methylation nadir. Adjacent to TGCTs, benign testis methylation profiles are determined by spermatogenetic proficiency measured by Johnsen score. In sum, TGCTs share collective entrapment in a PGC-like state of genomic-imprint and DPPA3 erasure, recurrent hypermethylation of cancer-associated targets, and subtype-dependent pluripotent, germline, or somatic methylation.
Malignant testicular germ cell tumors (TGCTs) are the most common malignancy among young men, and the incidence continues to increase (Sheikine et al. 2012; Trabert et al. 2015). Also referred to as type II TGCTs (Oosterhuis and Looijenga 2005), they are divided histologically and clinically into seminoma (SE) and nonseminoma (NSE). NSEs are further distinguished by variable composition of the histological elements embryonal carcinoma (EC), teratoma (TE), yolk sac tumor (YST), and choriocarcinoma, typically as mixed tumors but occasionally in pure form. TE, YST, and choriocarcinoma are considered to be differentiated derivatives of the pluripotent progenitor, EC. TGCTs’ association with developmental gonadal defects supports the prevailing model by which they originate from a multipotent stem cell arising in the germline, which fails to disable pluripotency to mature into a spermatogonium (McGlynn and Cook 2009; Sheikine et al. 2012). On the other hand, recent studies identifying reversion to pluripotency in the germline (Nettersheim et al. 2015; Oliveros-Etter et al. 2015) question whether TGCTs entail a continuation of, versus reversion to, functional pluripotency.
SE and the various forms of NSE originate from a common pathologic germ cell precursor, historically referred to as Carcinoma In Situ (CIS) (Skakkebaek 1972), Intratubular Germ Cell Neoplasia Unclassified (IGCNU), or, most recently, Germ Cell Neoplasia In Situ (GCNIS) according to the World Health Organization classification 2016 (Berney et al. 2016). This progenitor shares many characteristics of a primordial germ cell (PGC)/gonocyte, such as mRNA/miRNA/protein expression (including the coexpression of POU5F1 [also known as OCT3/4] and SOX17) (Looijenga et al. 2003; de Jong et al. 2008), as well as global CpG methylation erasure, features also found in invasive SE (Port et al. 2005; Netto et al. 2008; Wermann et al. 2010; Al-Hussain et al. 2015). However, while PGCs express multiple biomarkers of pluripotency shared with blastocyst inner cell mass stem cells, they are normally unipotent to produce gametogenic stem cells, and the spawning of differentiated somatic elements in this lineage is pathologic. Elucidating the underlying basis of such pathologic TGCT functional pluripotency remains a key question, particularly as the GCNIS predecessor reportedly lacks genetic anomalies, such as gain of 12p (e.g., i(12p)), that characterize the invasive state.
It is well established that distinct epigenetic profiles exemplified by DNA methylation signify pluripotent as well as divergent somatic lineage states (Krausz et al. 2012; Nazor et al. 2012; Reinius et al. 2012; Yuen et al. 2013; Anton et al. 2014; Court et al. 2014; He et al. 2014). The germ lineage also bears a unique developmentally regulated state, with transient erasure of genomic imprints that ensues from a post-inner cell mass wave of methylome erasure (Tada et al. 1998), followed by the establishment of male or female gametic epigenetic sexual dimorphism required for physiological reproduction (Gkountela et al. 2015; Guo et al. 2015; Tang et al. 2015). Thus a recognized molecular distinction between PGC and precursor and downstream states is the erasure of all genomic imprints.
The status of parental imprints in TGCTs is of fundamental importance to the investigative biology and pathology of the germ cell lineage. Perturbation of imprint balance drives several pathologies epitomized by female ovarian germ cell tumors (Linder et al. 1975; Kajii and Ohama 1977; Surani et al. 1984; Surti et al. 1990; Devriendt 2005; Oosterhuis and Looijenga 2005; McFadden and Robinson 2006). TGCTs have been less explored as imprinting models, perhaps because they originate early on in the male germline and their imprint status is less established, with reports ranging from generally erased to relatively intact (Verkerk et al. 1997; Looijenga et al. 1998; Sievers et al. 2005; Rijlaarsdam et al. 2015). Recent reports of maintained imprints in TGCTs would place their derivation temporally closer to inner cell mass precursor cells (Rijlaarsdam et al. 2015). These uncertainties may be due in part to measuring methylated allele frequencies in the TGCT histophenotypic amalgam of malignant cells and nontumorous elements, including lymphocytes.
Beyond the question of imprint status, whether or not TGCT subtypes harbor core and/or subtype-dependent DNA methylation signatures, including those that are pathogenic, physiologic, or descendant from PGC ancestors, is not known. Many of the surface and other markers now used to identify ESCs were originally discovered in EC (Andrews 2002), yet its epigenetic territory has been less explored. Prior studies based on immunohistochemistry and targeted approaches have reported that SE is relatively more CpG-hypomethylated than the more-differentiated NSE (Netto et al. 2008; Minami et al. 2010; Wermann et al. 2010), yet a genome-scale high-resolution methylation analysis of pure forms is required to further elucidate the epigenomic programming in TGCTs, particularly as it relates to pluripotent and differentiated states.
In the present study, we perform genome-wide DNA methylation analysis of TGCT pure forms, including a relatedness analysis to a wide variety of high-quality reference methylomes from pluripotent and somatic differentiated states. The overall goals were to seek insight into PGC tumorigenesis and also to explore TGCT utility as a model system for the study of epigenetic programming in general.
Results
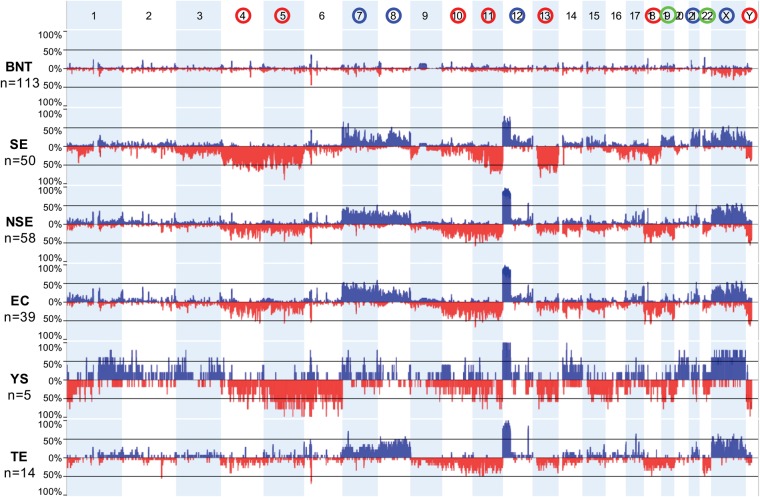
Series description
An initial set of 130 individual pure TGCTs (61 SEs, 43 ECs, 20 TEs, six YSTs) were selected, along with 128 matched benign neighboring testes (BNTs) that were annotated for histological Johnsen-score (JS) measure of spermatogenesis and absence of GCNIS from dissected foci (Supplemental Tables S1, S2). Quality control (QC) of array data included evaluation for proper tumor/normal source matching based upon built-in single-nucleotide polymorphisms (SNPs) on the methylation arrays (Supplemental Fig. S1). Next we performed somatic copy number aberration (SCNA) analysis for presence of anticipated anomalies (see Methods), typified by tumor-specific gain of the short arm of Chromosome 12, which is independent of subtype histology (Atkin and Baker 1982; Gibas et al. 1986; van Echten et al. 1995). Based on these data, 108 of the 130 assayed tumors and 113 of 128 BNTs met inclusion criteria for further analysis (Methods) (Supplemental Fig. S2). In addition to gain of 12p in individual samples, SCNAs were consistent with available historical data (van Echten et al. 1995; Faulkner et al. 2000; Looijenga et al. 2000; Skotheim and Lothe 2003; McIntyre et al. 2007; Litchfield et al. 2015), notably recurrent pan-TGCT gains of Chromosomes 7, 8, 22, and X, and losses of Chromosomes 4, 5, 10, 11, 13, 18, and Y (Fig. 1); also evident are the relative gains of Chromosomes 19 and 22 in SEs relative to NSEs (van Echten et al. 1995; Summersgill et al. 1998). The determination of SCNA in TGCTs from methylation data was validated on an independent public data set (Supplemental Fig. S1). Following these various QC validations for inclusion of original data from 108 TGCTs and 113 BNTs, multiple pluripotent, PGC, differentiated somatic, and diverse cancer reference sample data sets from the public domain were also included (Supplemental Tables S1, S2).
Figure 1.
Somatic copy number aberration (SCNA) profiles in TGCT subtypes. (Top) Blue and red circles around chromosome numbers indicate recurrent shared SCNA gains and losses, respectively; green circles, chromosomes relatively gained in SE versus NSE. (Next three panels) Aggregate view of BNTs, SEs, and NSEs. (Bottom three panels) NSEs were further subtyped as EC, YST, and TE. SCNAs were computed from 450K-methylation array signal intensities (see Methods). Tumor inclusion criteria required the presence of gain of the short arm of Chromosome 12p. For abbreviations used, see text.
The SNPs also enabled evaluation of allelic heterozygosity status in TGCTs. Historically, the recurrent genome-wide loss of allelic polymorphism in the female germ cell tumor ovarian teratoma has been interpreted as tumor origin from failed meiosis II or endoreduplication of a mature ovum, and we validated our ability to detect genome-wide loss of heterozygosity (LOH) in this context (Supplemental Fig. S1). We measured, in contrast with ovarian teratoma, equivalent autosomal SNP heterozygosity for tumor/normal pairs in TGCTs, including a validation data set (Methods) (Supplemental Fig. S1), consistent with an origin from somatic/premeiotic cells and thus underscoring the prevailing view that TEs, and male TGCTs in general, are biologically distinct from female OTEs with regards to meiosis.
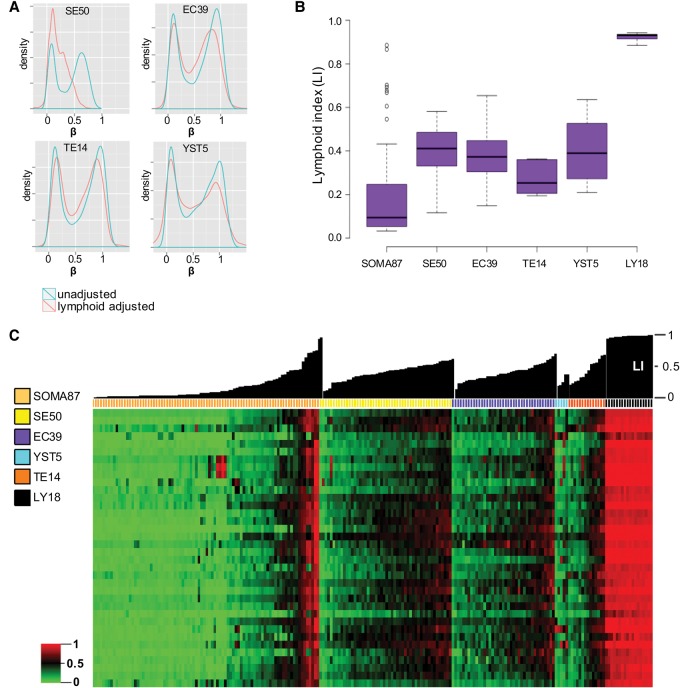
Lymphoid-compensated TGCT global methylation densities
A plot of raw global methylation density in SEs and the three histological subtypes of NSEs showed a relatively more profound hypomethylation in SEs, as expected based on earlier reports (Fig. 2A, blue methylation density distribution curves; Netto et al. 2008; Wermann et al. 2010); however, raw methylation levels were incompletely erased, particularly in SEs, where we anticipated near-total erasure. Because lymphoid infiltrates are a prominent parahistological component typically associated with SEs but potentially underappreciated in NSEs as well (Supplemental Fig. S3), we applied the principle of lymphoid cell–specific methylated allele measurement (Killian et al. 2011) to empirically calculate a lymphoid cell fraction of each TGCT sample (see Methods).
Figure 2.
Lymphoid cell fraction in TGCT samples. (A) Genome-wide methylation β-density distribution in TGCT subtypes, without and after correction for lymphoid infiltrates (blue and red traces, respectively) (see Methods). Notably, the lymphoid adjustment further exposes global methylation erasure in SEs, substantially shifting the peak methylation β-density closer to zero. (B) Box plots of lymphoid infiltrate levels in various TGCTs and reference sample groups as measured by lymphoid-specific methylation frequencies (i.e., LI; see Methods) reveal comparable levels across TGCT subtypes. (C) Heatmap of LS36 variable module (see Methods) and TGCT subtypes, with reference somatic tissues and purified lymphocytes (samples ordered by group and then by increasing LI); individual sample LI plotted above heatmap. The numbers of specimens investigated are indicated in the sample color legend. For abbreviations used, see text.
The resultant lymphoid index (LI) revealed that not only SEs but also NSEs manifest a substantial median LI (Fig. 2B,C). As controls for leukocyte-specific allelic methylation, nonlymphoid somatic cell cultures and TGCT cell lines had low-level methylation, while reference leukocytes were high (Fig. 2; Supplemental Fig. S3). To better gauge tumor cell–intrinsic cytosine methyl erasure, we next derived a simple equation to adjust observed target methylation values to LI (see Methods), which revealed a sizable shift toward global erasure in SEs (Fig. 2A, red trace on global methylation density plot). EC manifests a more modest left shift toward erasure, while TE and YST global methylation densities are unshifted, indicating somatic levels as measured in lymphoid cells (Fig. 2A). In sum, TGCT subtypes run the gamut of global methylation density, where lymphoid infiltrates may confound detection of tumor cell–intrinsic methylation erasure.
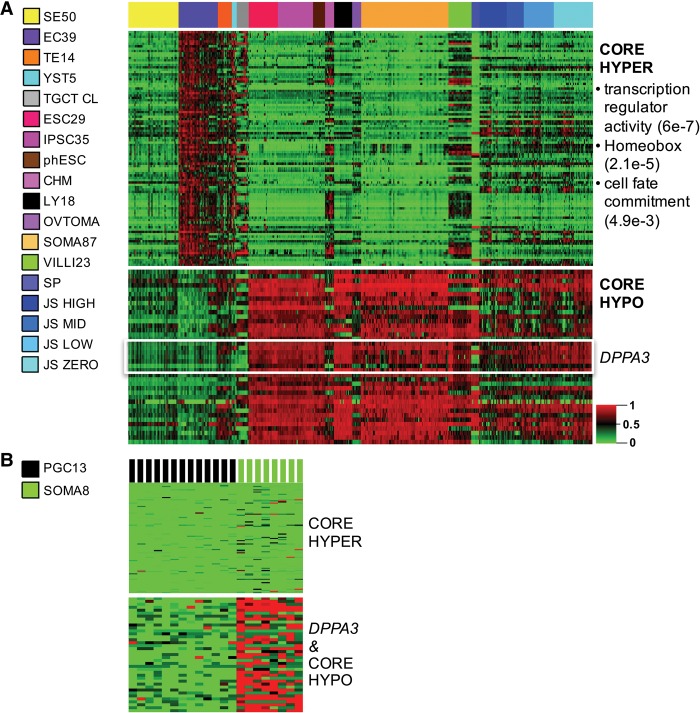
TGCT core methylation
Because of their global methylation erasure, we excluded SEs from the discovery of core hyper- and hypomethylation modules in TGCTs. Subsequently, we identified a core hypermethylation target module in the NSE group (EC, TE, and YST) methylome relative to benign pluripotent and somatic references (Fig. 3; Supplemental Table S3). Here, significant hypermethylation common to both pluripotent and differentiated NSE forms but absent from benign counterparts is potentially malignancy related. Gene ontology (GO) analyses on the top 100 hypermethylated targets (mapped to 44 genes) revealed significant enrichment for homeobox and cell fate commitment genes (Fig. 3A; Supplemental Table S3, “core hyper”). Moreover, as expected, these TGCT core hypermethylated targets are demethylated in PGC (Fig. 3B). We detected hypermethylation of this TGCT hypermethylation module broadly across a compendium of cancers arising in numerous anatomic sites (Supplemental Fig. S4A), suggesting that these are recurrent de novo general malignancy-related methylations.
Figure 3.
Core TGCT methylation. The nonseminomas (ECs, TEs, and YSTs) were collectively compared with reference samples. (A) Heatmap of the TGCT core hypermethylation module (“core hyper,” n = 100 targets) (Supplemental Table S3) across the four TGCT subtypes and multiple references. SEs show erasure for the otherwise TGCT-hypermethylated module, as shown in this unclustered heatmap. Functional annotation of core hypermethylated genes using the DAVID Functional Annotation Clustering Tool revealed significant enrichment for transcription regulatory, homeobox, and cell fate commitment genes (FDR shown in parentheses at right). Among the “core hypo” targets (Supplemental Table S3), seven of 39 targets map to DPPA3. (B) PGCs are erased for both core hyper- and hypomethylated targets, while embryonic somatic tissues serve as an internal control and show inverse methylation of the modules relative to TGCTs. For abbreviations used, see text.
Interestingly, it has been recently reported that certain noncoding piRNAs and their modifiers are involved in testis development and show DNA methylation aberrations in TGCTs (Ferreira et al. 2014), in particular the hypermethylation of PIWIL1, PIWIL2, PIWIL4, and TDRD1 in NSEs relative to SEs and benign testes. However, these genes were not present in our identified NSE-specific core hypermethylation module. Further inspection of our data found relative erasure of these genes in sperm, BNTs (high-JS BNTs in particular), and SEs compared to NSEs (Supplemental Fig. S5A), as was reported. At the same time, we found relatively high methylation of these genes (as represented on Illumina 450K array) broadly across the benign references, both pluripotent and differentiated, as well as BNTs with a low JS (Supplemental Fig. S5A). Like SEs and sperm, PGCs are demethylated at these genes, while differentiated tissues are methylated (Supplemental Fig. S5B). Thus, hypermethylation of these genes is not specifically malignancy related and is commonly found outside of SEs, PGCs, and spermatogenesis. piRNA gene hypermethylation is therefore associated with the expression of both benign and malignant differentiated phenotypes and their corresponding pluripotent precursors during embryonic, fetal, and adult life.
In the direction of erasure, we identified a core hypomethylation space in NSEs versus an expanded reference panel, including trophoblast and other lineages (Fig. 3; Supplemental Table S3); as anticipated, these core hypomethylated targets were also erased in SEs and PGCs (Fig. 3B). The singular most uniformly and robustly hypomethylated locus is the promoter region of DPPA3 (also known as STELLA), a well-known TGCT- and PGC-expressed maternal-effect gene (Bowles et al. 2003; von Meyenn and Reik 2015), whose corresponding methylation dynamics in human are nevertheless not well characterized. Therefore, we broadly analyzed DPPA3 methylation across TGCTs and reference pluripotent, PGC, gamete, and somatic differentiated references. Eleven 450K-methylation array targets within a 5-kbp window around the DPPA3 transcription start site demonstrate DPPA3 methyl-vacancy in PGC and male germline derivatives including TGCTs (SEs and NSEs) and sperm, while conversely DPPA3 is comparatively hypermethylated in physiologic embryonic tissues and differentiated derivatives, as well as the female germ cell tumor ovarian teratoma (false-discovery rate [FDR] = 0.05) (Supplemental Fig. S6). Interestingly, the hypomethylated DPPA3 promoter interval does not harbor a CpG island (CGI); ENCODE (Encyclopedia of DNA Elements Project) annotations for this domain as viewed in the UCSC Genome Browser reveal CTCF transcription factor binding, as well as DNase hypersensitivity (DHS) uniquely in ESC among lineages (Gerstein et al. 2012). Importantly, our reanalysis of recent PGC whole-genome bisulfite sequencing (WGBS) data and public database TGCT data demonstrates significant erasure of DPPA3 here as well (Fig. 3B; Supplemental Fig. S7A, respectively). In sum, DPPA3 status in TGCTs is most consistent with the retention of both gene expression and methyl-vacant epigenotype from the PGC progenitor, irrespective of histologic subtype of the invasive derivative. This TGCT property more firmly implicates the PGC or PGC-like cell as its origin. As presented above already and in more detail further below, this DPPA3 erasure/expression in TGCTs persists in the setting of de novo and maintenance methylation at other loci, both malignancy related and physiologic.
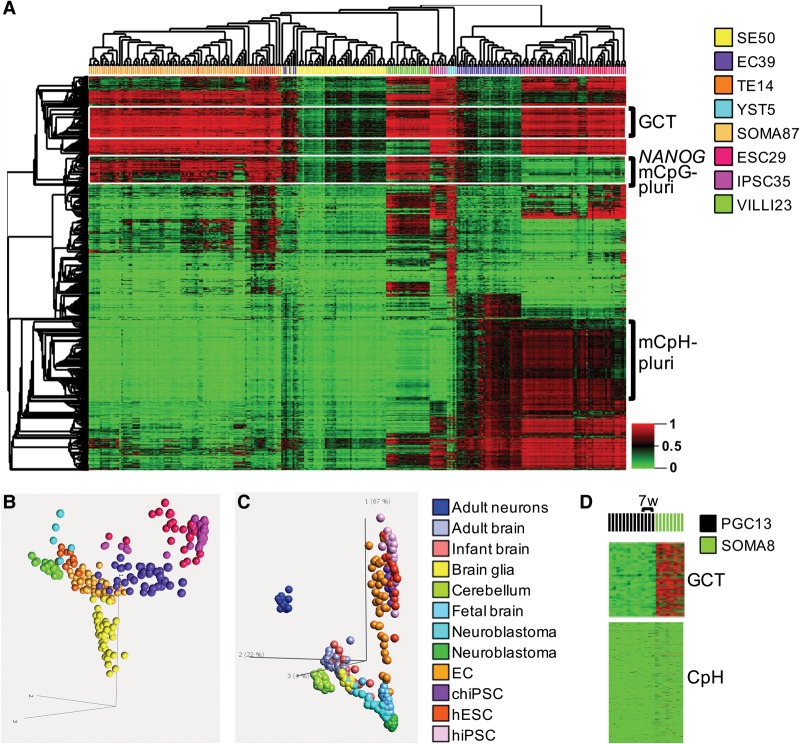
TGCT (NSE) subtype-specific methylation
Initial hierarchical clustering inclusive of both SE and NSE was dominated by the extensive relative hypomethylation of SE, which was subsequently excluded to better distinguish among NSE subtypes (Supplemental Fig. S8A,B). Multigroup difference analysis of NSE subtypes (EC, TE, YST) identified the top 2000 differentially methylated targets (Supplemental Table S3, “DMT2000”), unselected with respect to any of the benign pluripotent and differentiated reference specimens. Subsequently, we performed a relatedness analysis of TGCT subtypes (including SE) with a compendium of reference samples based upon these 2000 differentially methylated targets (DMTs) (Fig. 4A,B), which clearly showed that each unique tumor histology segregates with its phenotypic developmental counterpart. In particular, EC clustered with ESC/iPSC and TE with soma.
Figure 4.
Differentially methylated targets (DMTs) in TGCT subtypes. (A) The methylome profiles of the various TGCT histological subtypes recapitulate embryonic and extra-embryonic epigenomic differentiation. NSE multi-subtype DMTs (selected blinded to reference tissue profiles; see Methods) (see Supplemental Table S3) were used in hierarchical clustering of TGCT and reference GEO database samples representative of pluripotent (ESC), induced pluripotent (iPSC), somatic (SOMA), and extra-embryonic/trophoblastic lineage (placenta tissues and trophoblast cultures). Coclustering emerges for EC with ESC/iPSC, TE with somatic samples, and prominent CpG methylation of YST. The GCT cluster comprises 161 CpG targets uniquely hypomethylated in ECs among NSEs and references, and maps to 76 genes with significantly up-regulated expression in embryonal carcinoma. The “NANOG/PLURI” 141 target module maps to 76 genes, is cohypomethylated in EC/ESC, and includes NANOG, DNMT3B, BCOR, SMAD3, POU5F1, EYA1, FOXH1, HOXB1, HOXB3, LRP4, PBX1, SALL4, TFAP2A, TRIM71, and DPPA4, among others. A striking ESC-like CpH (non-CpG) hypermethylation process was identified in ECs, also present in iPSCs. (B) PCA plot further highlights TGCT-subtype/embryonic-lineage convergence. Color legend at right of A. (C) Pluripotency- versus neuron-signifying CpH methylation. PCA using 473 CpH target module (Methods) (Supplemental Table S3), including ECs and multiple different test groups indicated in color legend at right. PCA shows orthogonality of neuronal versus pluripotency methyl-CpH axes: ECs, and human (hiPSCs), and chimp (chiPSCs) track along the ESC/pluripotency axis, while post-natal cortex and cerebellum lay on the neuronal axis; minimal CpH reprogramming seen in neuroblastoma cell lines (green) and tissues (light blue). (D) Fetal somatic tissues and PGCs as early as 7 wk are erased of CpH methylation, while benign fetal somatic tissue, but not PGCs, maintains methylation at the GCT targets erased in ECs. The numbers of specimens investigated are indicated in the sample color legend. For abbreviations used, see text.
TGCT programming convergence with that of physiologic pluripotent versus differentiated lineages is evident. For example, we discovered high levels of CpH methylation (methyl-CpH) specifically in ECs (Fig. 4A). Since methyl-CpH has been reported in both pluripotent ESCs (Ziller et al. 2011) and terminally differentiated adult neurons (Guo et al. 2014), we further explored the distinction between pluripotent and neuronal methyl-CpH, and to what extent ECs potentially converge with one or the other. Difference analysis of methyl-CpH profiles of adult neurons versus ESCs identified 458 high-contrast DMTs (Methods) (Supplemental Table S3, “CpH458”); this module manifests uniform/recurrent cell-specific methylation in neurons versus pluripotent human ESCs (hESCs) (Supplemental Fig. S9A, upper left), and includes 252 and 206 CpH targets that are hypermethylated in hESCs versus adult neurons, respectively. By using this module, we then performed both principal component analysis (PCA) and unsupervised hierarchical clustering of a multitude of module-agnostic samples including iPSCs; ECs; fetal, infant and adult brain cortex; adult cerebellum; and neuroblastoma (Fig. 4C; Supplemental Fig. S9A). As a result, we first found that ECs and both human and chimpanzee iPSCs (Gallego Romero et al. 2015) are tightly clustered with hESCs in the heatmap and are orthogonal to the neuronal axis in the PCA, which instead associates with post-natal brain tissues. (Parenthetically, our exploration of methyl-CpH in the reference brain specimens resulted in localization of acquisition of this mark to the fetal-to-adult brain transition, during the first days of life, suggesting a potential biomarker utility for live birth and post-natal neural cell physiology worthy of future investigation.) Thus, the methyl-CpH profile of ECs is a pluripotency signature, which is convergent with that of ESCs/iPSCs and conserved across mammalian evolutionary boundaries and which is significantly distinct from that of the neuronal (post-natal) methyl-CpH signature. We next interrogated a cancer compendium of over 1000 cell lines from 14 distinct anatomic sites for their levels of pluripotency-specific methyl-CpH (mCpH-pluri; Methods) (Supplemental Table S3). We found no cancers outside of testis with EC levels of mCpH-pluri, including a multitude of tumors with a hypermethylator phenotype, as well as glial and neuronal cancers and neuroblastoma (Supplemental Fig. S9B).
General erasure of methyl-CpH in human PGCs relative to ESCs has been previously reported (Takashima et al. 2014). More recently, none of three methylome studies of human PGCs (von Meyenn and Reik 2015) reported methyl-CpH, yet still we revisited the PGC WGBS data (Guo et al. 2015), which confirmed the erasure of CpH hypermethylation at the earliest measured 7-wk time point (Fig. 4D), and therefore, CpH erasure convincingly coincides with PGC lineage divergence from pluripotent ancestor cells. Thus, all available data pertaining to the programming of pluripotency during early development and following its induction in vitro indicate that mCpH-pluri is a unique conserved feature of the functional pluripotent state, is a signifier that is absent in the post-primordial germline and soma, and is distinct from de novo methyl-CpH in post-natal neurons (mCpH-neuro). In our study, the analysis of numerous benign and malignant cells and tissues from post-embryonic time points only identified mCpH-pluri in invasive germ cell malignancy, namely, EC, and this property converged upon the ESC and iPSC state. While there are no CpH data directly available for GCNIS, we may infer its absence here as well, having demonstrated its erasure in the proximal ancestor PGCs and descendent SEs. Plus, GCNIS lacks expression of DNMT3B (Sperger et al. 2003; Skotheim et al. 2005; Sonne et al. 2009), thought to be coupled to DNA methylation in the pluripotential state (Chedin 2011; Tiedemann et al. 2014). The most parsimonious interpretation of these observations is that the prominent burst of mCpH-pluri in ECs, having already passed through an erased PGC/GCNIS ancestry, represents a reversion to/reactivation of pluripotency, with de novo apparition. As further evidence for such a recrudescence of pluripotency in ECs, we found that induction of pluripotent EC phenotypes from the seminomatous TCam-2 cell line following xenograft to murine testis (Nettersheim et al. 2015) spawns mCpH-pluri (Supplemental Fig. S9C), which we now interpret as a key molecular event previously unrecognized in that experiment. Thus, recurrent and specific mCpH-pluri elevation upon the stimulation of functional pluripotency in TGCT cell lines further validates our tissue-based discovery based upon data from archival pathology specimens. Importantly regarding cell line models of TGCTs, we found that in vivo EC tumors in our study demonstrate greater convergence with pluripotent states (ESC/iPSC) than ECs in vitro, driven in part by lower levels of mCpH-pluri in the malignant cell lines (Supplemental Fig. S9C), levels that are still, however, significantly elevated relative to differentiated states; mCpH-pluri reduction in this context may reflect an adaptation to or result of cell culture. Moreover, the lowest level of mCpH-pluri among undifferentiated samples is found in the nullipotent cultures (SE-like TCam-2 and EC-like 2102EP), while, as noted above, activation of potency in TCam-2 testis xenotransplants is coupled to mCpH-pluri elevation (Supplemental Fig. S9C).
Another noteworthy example of physiologic reprogramming found among the TGCT DMT2000 is the homologous methyl-vacancy between ECs and ESCs of a pluripotency gene space that is significantly enriched for functional annotation terms related to transcription regulation and embryonic morphogenesis (Fig. 4A “pluri,” Supplemental Fig. S10A; Supplemental Table S3). These genes included canonical pluripotency expressed genes such as DNMT3B, POU5F1, EYA1, PBX1, FOXH1, FOXK1, SMAD3, TFAP2A, TRIM71, SALL4, and NANOG (Adewumi et al. 2007; Biermann et al. 2007), several of which have been previously shown to have up-regulated expression in ECs as well (Alagaratnam et al. 2011). Notably this shared epigenomic space is not significantly associated with disease, consistent with a physiologic state. The prior finding of NANOG epigenotype-expression correlation in ECs (Nettersheim 2011) we now report is broadly extended to pluripotency loci in general. Many of the 76 genes in this cluster (Supplemental Table S3) have not yet been associated with pluripotency, and thus ECs are an extant tangible resource for identifying pluripotency-enabling genes and their associated epigenetic state.
Beyond ECs, we uncovered a large epigenomic space in YSTs that is uniquely programmed relative to any of the reference specimens (Fig. 4A). YSTs have been previously reported to harbor an overall hypermethylated profile (Amatruda et al. 2013; Furukawa et al. 2009), although we find strong lineage-specific hypomethylation as well. We now further interpret the YST methylome to have subtype-specific, differentiation-level programming with some unique homology with trophoblast, yet not entirely convergent with it. Future profiling of additional references, including the yolk sac itself, are needed to identify the true developmental counterpart of YSTs.
Regarding potentially pathologic methylation states in ECs, we identified a uniquely hypomethylated space relative to other NSEs (but absent in SEs) and any of the embryonic and somatic references (Fig. 4A; Supplemental Table S3, “GCT”). Notably, whereas many DMT subsets reported from myriad cancer studies may not map significantly onto expression-based GO terms, this EC-hypomethylation module returned significant functional annotation enrichment for genes up-regulated in ECs (Supplemental Fig. S10B), thus identifying a direct correlation between this particular significant EC hypomethylation space and positive gene expression. Despite such evidence, we identify coerasure of this module in PGCs (and SEs) as well (Fig. 4C), which counters the pathogenicity of this epigenomic state and instead implicates PGC ontogeny.
Genomic imprinting in TGCT subsets
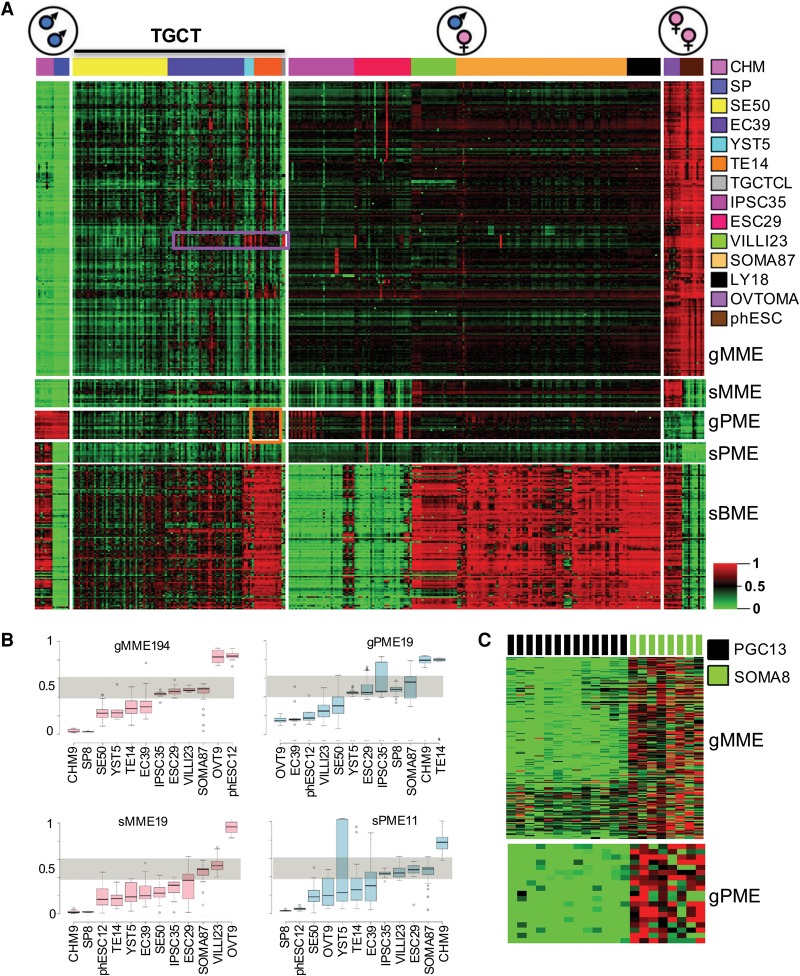
To investigate genomic imprinting in our TGCT samples, robust imprinted methylation modules including distinct germline and secondary maternal and paternal imprints (gMME, sMME, gPME, and sPME) (Okae et al. 2014) were empirically derived from reference tissue panels assayed on the same 450K platform (see Methods) (see Supplemental Tables S2, S3; Supplemental Fig. S11). The corresponding heatmap demonstrates an overall pattern of biparental germline and secondary imprint erasure in TGCTs relative to somatic tissues, gametes, and parthenogenetic and androgenetic specimens (Fig. 5A). Adjustment of observed TGCT tissue methylation imprints to lymphoid cell infiltrates (Methods) (Fig. 5B; Supplemental Table S3) enhanced the detection of generalized imprint erasure of gMME in all four TGCT subtypes and also erasure of gPME in three of four subtypes. Visual inspection of the heatmap (Fig. 5B) reveals that, like ESCs and TGCTs in vitro, TGCTs in vivo manifest focalized recurrent imprinted domain hypermethylation, as evidenced by HM13 among NSEs, and subtype-dependent hypermethylation of H19 in TEs (Fig. 5A, purple and orange boxes, respectively). This locoregional recurrent gMME and gPME hypermethylation variation is not to be attributed to lymphoid infiltrate, which would instead produce a broader, pan-imprint methylation. Distinct from NSEs, SEs were more uniformly, globally erased of imprints; the shared GCNIS ancestry of SEs and NSEs, coupled with global imprint erasure in SEs, suggests that the focal hypermethylation of imprinted loci in NSEs arises de novo following complete erasure. Notably, sMMEs and sPMEs were both erased across all TGCT subtypes, including in TE with its H19 gDMR hypermethylation (Fig. 5B), suggesting that hypermethylation of erased gDMRs may not engender secondary/somatic sDMR methylation, and further supporting the idea of global imprint erasure in TGCT at the outset. Our imprinted gene findings were validated in an independent TGCT data set (Supplemental Fig. S7). Finally, analysis of gMME194 and gPME19 in a compendium of 1028 cancer cell lines from 14 distinct anatomic sites demonstrated pan-imprint erasure to be unique to TGCTs (Supplemental Fig. S4B).
Figure 5.
Analysis of imprint erasure in TGCT subtypes. (A) 2D heatmap of imprinted target methylation in TGCTs and references. The heatmap conveys general erasure of maternal and paternal germline (gMME, gPME) and secondary/somatic (sMME, sPME) imprinted methylation, along with differentiation-associated biallelic somatic de novo methylation (sBME), particularly evident in TEs. Like ESCs and TGCTs in vitro, TGCTs show focalized recurrent hypermethylation affecting imprinted loci, such as HM13 in NSEs and H19 in TEs (purple and orange boxes, respectively). (B) Box plots of imprint erasure in TGCT subtypes following adjustment to account for lymphoid infiltrates (see Methods). gMMEs are erased in all subtypes; interestingly, gPMEs are erased in three of four TGCT subtypes, while as noted in A, TEs manifest hypermethylation of H19. Both sMMEs and sPMEs are erased in TGCT subtypes, including TEs. (Nonadjusted measures are provided in Supplemental Fig. S12.) (C) Reference WGBS data show gMME and gPME imprint erasure in PGCs, with maintenance in fetal soma. For abbreviations used, see text.
Beyond parental imprints, we further evaluated the de novo somatic differentiation-coupled methylation activity of TGCT subtypes. This was done by first identifying a pan-soma hypermethylation module relative to the ESCs (module termed sBME; see Methods) (see Supplemental Table S3; Supplemental Fig. S11E). sBMEs demonstrated maximal levels in the TE and YST subtypes (Fig. 5A). Thus, to summarize, histologic differentiation in NSEs is accompanied by subtype-specific, physiologic CpG and CpH de novo methylation, from which the imprint- and DPPA3-erased background largely escapes.
BNT methylation
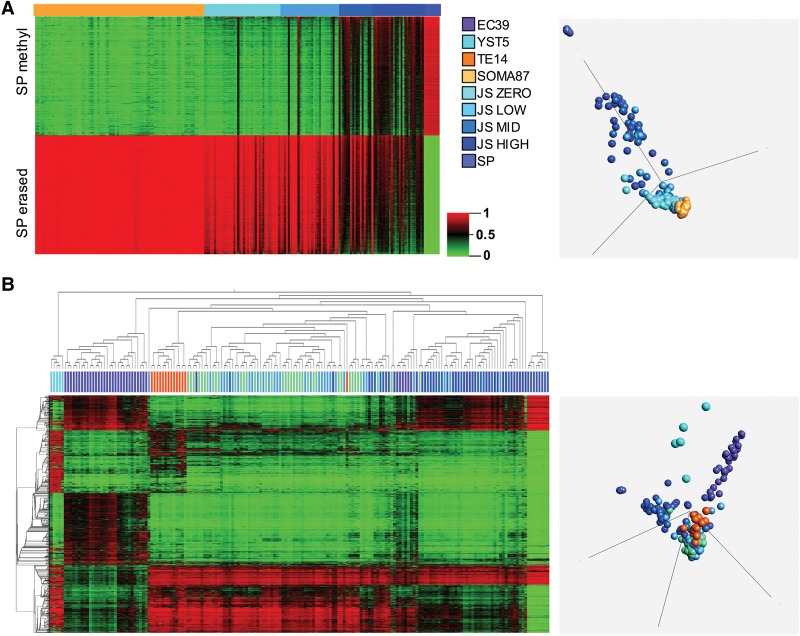
A range of BNT histology was sampled and profiled, including the seminiferous tubule with spermatogenesis in varying stages of maturation as measured by JS. This allowed us to ask whether the BNT methylation profile correlated with spermatogenetic proficiency. BNTs demonstrated a relatively sperm-like or soma-like profile when plotted with somatic reference tissues and sperm (SP) using CpG targets either germline erased or methylated in SP (Fig. 6A). PCA showed a strong relationship between JS and proximity to SP versus soma (Fig. 6A). BNT clustering with NSE and SP demonstrated that JS-zero and JS-low samples segregate with TEs, while JS-high samples have SP-like methylation (Fig. 6B), further consistent with somatic versus gamete-specific methylation as a function of JS. No TGCT subtype reprogrammed in the direction of mature spermatogenesis (Fig. 6B).
Figure 6.
Benign neighboring testis (BNT) methylation profile is determined by spermatogenic proficiency. (A, left) Unclustered heatmap of BNT annotated for Johnsen score (JS) of zero (n = 39), low (n = 30), intermediate (n = 17), or high (n = 27), with reference sperm (SP, n = 8) and somatic tissues/cultures (SOMA, n = 87). 450K array methylation variables were preselected for the top 1000 hyper- and hypomethylated targets in SP relative to SOMA by fold-change (SP erased: SOMA > SP fold-change = 1.915, n = 998 targets; SP methylated: SP > SOMA fold-change = 1.685, n = 998 targets). (Right) PCA on same targets. BNT methylome resembles a two-component mixture of SP and SOMA, with proximity to SP versus SOMA correlated to JS. (B, left) 2D hierarchical clustering of NSE and BNT samples and DMT2000 variables. Note clustering of TE with zero- and low-JS BNT and of high-JS BNT with SP. (Right) PCA of same. NSEs are orthogonally methylated to SPs. For abbreviations used, see text.
We also explored BNT imprinting as a function of JS (Supplemental Fig. S12). A JS of zero (i.e., absent germ cells) had somatic-level g/sMME and g/sPME, while high-JS testis showed g/sMME erasure but supra- and infra-somatic gPME and sPME methylation, respectively (i.e., approaching homogeneous gDMR paternalization). Low- and mid-JS testis gPME levels were in between somatic and paternalized. The finding of sPME erasure as a function of increasing JS further validated the secondary PME module. Thus, the hypermethylation of gPME and the erasure of gMME, sMME, and sPME provide complementary measures of spermatogenesis within testis parenchyma. Erasure of somatic biparental methylation (sBME) was significantly coupled to rising JS as well (Supplemental Fig. S12).
Discussion
Our study offers new insights into the epigenomic machinations of the TGCT microcosm, illuminating multiple convergences with normal embryogenesis, achieved through the focus on pure tumor forms and informed by analysis of relatedness to pluripotent, primordial and mature germ cell, and differentiated reference specimens. Broadly speaking, TGCT methylomes comprise an anachronistic, anatomically misplaced, genetic and epigenetically anomalous development circuitry, nevertheless replete with striking conservation of pluripotency and differentiation programs (Fig. 7).
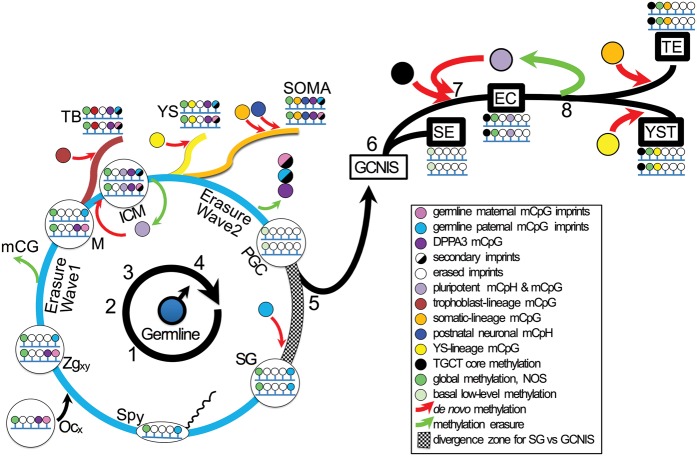
Figure 7.
Male germline programming to TGCTs. (1) A sperm with a Chromosome Y (Spy) fertilizes an oocyte (Ocx) to spawn a male zygote (Zgxy) with heteroparental germline imprints. DPPA3 methylation from the egg. (2) Cell divisions and post-zygotic genome-wide CpG methylation (mCpG) erasure en route to the morula (M), with persistent imprints and DPPA3 methylation. (3) Morula progression to blastocyst, with lineage-respective methylation of trophoblast (TB) and inner cell mass (ICM). Prominent pluripotency-signifying methyl-CpH (mCpH-pluri) in ICM stem cells is lost as they exit from pluripotency as yolk sac (YS) and somatic (SOMA) derivatives. Secondary imprints appear in TB and ICM. Differentiation-coupled mCpG, and post-natal neuron-specific CpH (mCpH-neuro). (4) Germline-specific near-total erasure wave eliminates imprints and DPPA3 methylation in the primordial germ cell (PGC) state. (5) PGCs either proceed to spermatogonia (germ cells of later spermatogenesis) with acquisition of paternal imprints and continuation of the physiologic cycle, or they pathologically exit the cycle to be first recognized as germ cell neoplasia in situ (GCNIS) within testicular seminiferous tubules. (6) GCNISs progress to invasive TGCT along two major branches. Seminoma (SEs) are nullipotent and propagate the erased genome of PGCs. (7) De novo methylation of a malignancy program and mCpH-pluri accompany reversion to pluripotency and characterize embryonal carcinoma (EC). (8) EC differentiation to teratoma (TE) or yolk sac tumor (YST) entails loss of mCpH-pluri and convergent reprogramming toward physiologic ICM differentiated derivatives. Global methylation not otherwise specified (NOS) may be found throughout, except in PGCs and SEs, which nevertheless have detectable basal methylation levels. For abbreviations used, see text and legend above.
First, a PGC-like state of imprinted gene and DPPA3 erasure resides at the collective TGCT core. Other than PGC and derivative male germline cells and TGCTs, all other tested tissues harbored a methylated DPPA3, including ESCs and iPSCs. It is already known that ECs express high levels of DPPA3 (Wongtrakoongate et al. 2013). DPPA3 has been recently demonstrated to protect parental imprints from erasure during post-fertilization demethylation; whether DPPA3 more generally also protects erased imprints in high-expressing PGCs and TGCTs requires further study. Furthermore, it may be that DPPA3 still functions in PGCs to enable silencing at critical sites like active retrotransposons, a function that carries over into TGCTs; such a function is supported by the proficiency of EC cultures to silence reporter genes introduced by retrotransposition (Garcia-Perez 2010). Functional analysis of the effects of stifling DPPA3 in EC cultures (currently more technically feasible than in human PGC lines, none of which have been reported) would be informative for these questions.
Next, in divergence from PGCs, a generalized malignancy-coupled hypermethylation core emerges in ECs and is propagated in its differentiated derivatives. And then, subtype-specific methylation signatures, including pluripotency-coupled methyl-CpH (Ziller et al. 2011; Arand et al. 2012) in ECs, illuminate an unprecedented molecular measure of operational activation of pluripotency in a human cancer. This pluripotency-specific signature (mCpH-pluri) was found to be evolutionarily conserved in nonhuman primates, re-enacted in induced- and reversion-to-pluripotency experimental data sets, and significantly distinct from a neuronal signature (mCpH-neuro). It is remarkable that this circuitry is invoked and maintained in a human malignancy in vivo. Previously it may have been argued that PGCs aberrantly retain functional pluripotency during testicular tumorigenesis; our study strongly implicates a post-PGC reversion process that re-enables pluripotency coupled to methyl-CpH renewal, a mark subsequently erased as ECs differentiate to YST and TE derivatives. In other words, the most parsimonious explanation of the restriction of a specific methyl-CpH profile to pluripotent states and its absence in PGC, SE, YST, and TE is that this program in ECs is invoked during divergence from GCNIS, which warrants future study. Moreover, further study of the pathway to pluripotency in ECs could inform novel routes to induced pluripotency in vitro.
Considering that pluripotency genes are reportedly protected from methylation throughout the male germline cycle (Leitch and Smith 2013), then ECs are still in the germline cycle, while derivative TEs and YSTs have exited from it, culminating with end-game methylation of the pluripotency circuit. The implication for other malignancies that may have passed through a similar pluripotent intermediate stage, but without a retained pluripotent state in the differentiated tumor mass, is that mCpH-pluri will likely be departed. On the other hand, it will be exciting to apply enhanced methods with low limits of detection to the search of additional cancer types for rare cells with mCpH-pluri.
The collective multilineage manifestations of TGCTs, taken together with core hypo- and hypermethylation modules and somatic subtype-dependent methylation, is consistent with antecedent physiologic global erasure in a PGC ancestor, followed by acquisition of mCpH-pluri and the malignancy-associated core hypermethylation module en route to ECs and, finally, establishment of differentiated derivative–specific methylation, the final two steps analogous to that of embryonic somatic de novo methylation (for review, see Monk 2015). The strong coexpression of DNMT3B and mCpH-pluri in both ECs and benign pluripotent states further supports the model whereby mCpH-pluri is established de novo, and then upon differentiation to YST and TE, its maintenance is neglected by the more CpG-philic DNMT1 maintenance methyltransferase (Sperger et al. 2003; Almstrup et al. 2005; Biermann et al. 2007; Chedin 2011; Tiedemann et al. 2014). This may be relevant for subtype-targeted treatment because elevated DNMT3B expression in ECs has been linked to exquisite sensitivity to demethylating agents (Beyrouthy et al. 2009; Biswal et al. 2012; Wongtrakoongate 2015). Interestingly, DNMT3A would conceivably be capable of methyl-CpH in ECs; yet, as this methyltransferase plays a specialized role in neurons and germ cell methylation, including gDMR de novo paternalization in the latter (Chedin 2011), the greater prominence of DNMT3B in ECs, coupled with mCpH-pluri, further distances the early developmental origin of TGCTs from germ cells downstream from de novo imprint paternalization. It would be interesting to explore pluripotent, neuronal, and possibly other cell types harboring specific methyl-CpH profiles for codependency upon DNMT3A versus -3B expression. Currently there is no well-accepted primary biological function for methyl-CpH (He and Ecker 2015). The null hypothesis here is that non-CpG methylation is an inert, transient byproduct of methyltransferase activity targeted to CpG sites. On the other hand, broad conservation of mCpH-pluri across mammalian benign and malignant states may favor a more selectionist argument. We expect that TGCTs, and ECs in particular, will be valuable informants for further investigation of this peculiar and mysterious mark.
The revival of methyl-CpH during GCNIS to EC development is coupled with reprogramming of CpG targets as well, with both modifications convergent upon physiologic pluripotent states. Importantly, germline and somatic imprints remain globally erased in this process, a finding relevant for future clinical utility of iPSC technology because it underscores the limitations of induced pluripotency to correct or restore imprinting defects present in somatic donor cells, including PGCs.
A recent study reported relatively somatic-level imprints in TGCTs, thus placing the developmental timing of tumorigenesis prior to, or very early after, germline specification and prior to extensive parental imprint erasure (Rijlaarsdam et al. 2015). To elucidate with high confidence the status of imprints in TGCTs, we employed lymphoid-compensated tumor tissue measurements of platform-validated germline and somatic maternal and paternal imprinted loci, which demonstrated generalized erasure of all imprints in the tumor cells. At the same time, we identified within the NSE lineage recurrent subtype-dependent hypermethylation, affecting H19 in TEs and HM13 across NSE subtypes. Recently it was reported that in vitro reversion to pluripotency of PGCs results in locus-specific hypermethylation of imprinted genes (Oliveros-Etter et al. 2015), a process we found played out in vivo in TGCTs. SEs had the most uniform and complete imprint erasure. Because cultures of pluripotent ECs/ESCs/iPSCs, including parthenogenetic derivatives, are prone to recurrent hypermethylation affecting imprinted genes, SEs therefore represent the most consistent biparental imprint-erased resource. These intriguing biological distinctions were only observable through analysis of pure forms of TGCTs.
Thus TGCTs occupy a singular niche in the spectrum of imprint states, stemming from the unique and transient allelic parental amnesia found in PGCs, whose upstream ancestors have heteroparental imprints, while downstream spermatogonial stem cells are paternalized (Sievers et al. 2005). TGCT entrapment in a state of imprint erasure is reminiscent of mouse 11.5–12.5 days post coitus (dpc) PGC clones (Lee et al. 2002), suggesting that imprints are already erased at the time of tumor initiation; however, we cannot exclude the interesting idea that completion of this process occurs following tumor initiation, as a form of continuation of PGC biology. The resistance of erased DMRs to de novo methylation through subsequent genome-wide programming of pluripotency and differentiation underscores a fundamental property of imprints whereby they are generally perpetuated in their state at the outset of clonal expansion. As discussed above, the suppression of imprint de novo methylation across TGCTs during tumor growth and differentiation may relate to continued expression of DPPA3, a core gene that is also passed over by methyltransferase as somatic differentiation-coupled reprogramming is established.
While female germ cell tumors have figured prominently in imprinting evolution theory (Haig 1994, 1996; Varmuza and Mann 1994), male GCTs have been less explored. Maternalized and paternalized genes are thought to drive somatic and trophoblast differentiation, respectively, as a result of inter-parental conflict over asymmetrical, mother-taxing investments in offspring. In this regard, it is interesting that testicular TE—defined by somatic histogenesis—features the exceptional hypermethylation of the H19 gDMR, just as in hydatidiform mole, the prototypical paternalized differentiation state (Fig. 5). Prior reports from murine pluripotent embryonic germ cell (EGC) models report a similar process of hypermethylation of H19 upon differentiation to soma (Shovlin et al. 2008). On the other hand, it may be more correct to refer to H19 hypermethylation in TEs as pseudopaternalization, since it is nonphysiologic and does not engender secondary imprinting in our study. More generally, as our analysis of gPME was limited by 450K array content to the H19 DMR, broader genomic methylation profiling of TEs is needed to assess hypermethylation of other foci of paternal imprinting. For now, in the context of the Haig hypothesis for the evolution of genomic imprinting (Haig and Graham 1991), the rationale for recurrent H19 methylation selectively among pure TGCTs with the histology of TE is curious and merits future attention.
Finally, the inclusion of a large number of BNTs in our study revealed that the spermatogenesis score of testicular parenchyma is well measured by DNA methylation profiles. This provides a rationale for future use of DNA methylation in evaluating seminiferous competency in the clinical evaluation of both benign and malignant testicular urologic pathology. The ability to evaluate spermatogenesis using methylation measures may facilitate understanding the link between TGCTs and infertility.
Methods
Study design
Original data from 130 individual cases of predominantly pure Type II TGCTs (43 ECs, 61 seminoma SEs, 20 TEs, six YSTs) and adjacent BNT parenchyma from 128 TGCT cases were initially included in this study (124 cases had paired T/N. There were four unpaired BNTs and six unpaired SEs). Individual cores were punched from formalin-fixed paraffin-embedded (FFPE) tissue under histological guidance based on a hematoxylin and eosin (H&E)–stained parallel section. In addition, the histological composition was supported by immunohistochemical staining for the proteins POU5F1, SOX17, SOX2, AFP, and CGB5 (also known as HCG), which specify various subtype elements, i.e., SEs, ECs, YSTs, and TEs. Moreover, the presence of spermatogenesis (JS) (Johnsen 1970) and the precursor lesion of Type II TGCTs (GCNIS) were evaluated. Additional original data included nine female ovarian teratomas, which were used as references for imprinting analyses (Supplemental Tables S1, S2). Additional reference samples were downloaded from GEO (Supplemental Tables S1, S2).
DNA methylation assay and processing
DNA extraction and genome-wide DNA methylation assays using the Infinium 450K Infinium beadarray were performed as previously described (Killian et al. 2013). Briefly, microdissected FFPE tissue was lysed in a cocktail containing mineral oil (for deparaffinization), proteinase K, and ATL lysis buffer (Qiagen); resultant lysates were sodium bisulfite treated and filter purified using Zymo reagents. Purified converted DNAs were further treated with FFPE DNA restoration solution (Illumina) and then input to the standard protocol Infinium 450K-methylation beadarray assay.
Infinium 450K-methylation array image files (.idat) were imported to GenomeStudio software (Illumina) using the methylation module. Cy3/Cy5 color channel normalization and background subtraction were performed according to manufacturer's instructions. Sample methylation β values (which approximate the percent methylation at targeted genomic cytosine loci) were computed using the GenomeStudio methylation analysis function. Beadarray data inclusion criteria included review of the nonpolymorphic probes QC dashboard in GenomeStudio and demonstration of red & green signal intensities substantially above background, following the manufacturer's technical support guidelines. β values were exported from GenomeStudio for subsequent statistical analyses and visualization using Qlucore Omics Explorer v.3 software (QOE).
Additional QC was performed on all sample microarray profiles to ensure continuity of sample identity and to empirically determine adequacy of malignant or benign cellularity in the DNA extractions derived from the needle core punches (based on which the profiles were generated). First, specimen identity was evaluated by tumor/normal matching for paired samples based on the 65 vendor-annotated SNPs represented within the 450K BeadChip array (Supplemental Fig. S1). Pearson correlation was maximal among T/N pairs, and hierarchical clustering appropriately matched the annotated T/N pairs. In addition, homozygosity for X-linked SNPs was observed in all samples, consistent with a male sex of the cases. On the autosomes, equivalent SNP heterozygous allele frequencies for T/N pairs were observed, thereby supporting the origin of Type II TGCT from a somatic/premeiotic cell (Supplemental Fig. S1). The 22 tumors without 12p gain were generally devoid of SCNA, consistent with inadequate tumor cell frequency, and were therefore excluded from further investigation (Supplemental Fig. S2). Among the BNT samples, 15 of 128 demonstrated SCNAs, including gain of 12p (Supplemental Fig. S2). Because 12p gain indicates cryptic aberrant cell contamination, perhaps from the deeper part of the cores, these were excluded from subsequent analyses as well, yielding 113 individual “flat genome” BNT samples for further analyses.
Genomic copy number derivation from methylation array data
The “FinalReport” files generated from Illumina Genome Studio were processed by using R packages “lumi” and/or “methylumi,” similar to previously described (Feber et al. 2014). Briefly, the total intensities from both methylated and unmethylated probes were calculated and then normalized by using a simple scaling normalization method (SSN implemented in lumi R package). Taking the median values of the same probes from all benign testis samples generated the “reference” intensities in the CGH experiment. A linear regression model was used to normalize the log2-transformed total intensities against the GC content of each target as well as the target types from the 450K-methylation array, and the relative log2 intensities against the reference, analogous to the log2Ratio in the array CGH, were derived for further processing. In order to reduce the noise level, the analogous log2Ratio values from every four adjacent probes sorted by chromosome positions on the same chromosome were merged by taking the mean value, and the resulting data were then uploaded into Nexus (BioDiscovery) software for further segmentation and visualization.
In sum, there were two main sample array data inclusion criteria: presence in tumor or absence in BNT of Chromosome 12p gain, and passing red/green signal intensity metrics on the GenomeStudio nonpolymorphic control probe QC window according to Illumina technical guidelines. This yielded 108 of 130 TGCTs and 113 of 128 BNTs passing QC. Analysis of historical SCNA was performed on TGCT subtypes in Nexus.
Samples were annotated into various reference and germ cell tumor groups (Supplemental Tables S1, S2). The between-group significance of each 450K-methylation cytosine target variable (i.e., q-value, a FDR corrected P-value) was computed in QOE, which uses a t-test for two-group comparison and F-test for multigroup comparison. Rank regression was also performed with QOE. Box plots were generated online (http://boxplot.tyerslab.com/), as were Kernel density estimation (KDE) plots (http://www.wessa.net/). Imprinted gene, somatic, lymphoid-specific, and additional target modules were identified as described below.
Lymphoid compensation
First, a lymphoid cell–specific target hypermethylation module termed the LS36 (see Methylation Modules below) was identified and utilized to calculate an average lymphoid-specific methylated allele frequency designated the lymphoid index (LI) of each specimen (note that LS36 refers to the set of 36 lymphoid-lineage-specific hypermethylated targets, while LI refers to the average methylated allele frequency of those targets within a sample). Not only SEs, but also all NSE subtypes manifest substantial median lymphoid infiltrates (i.e., LI) (Fig. 2B). The heatmap of LS36 targets with overlying plot of individual sample LI (Fig. 2C) further illustrates the range of LI across TGCT subtypes.
Methylation modules
LS36. This module is a robust set of 36 lymphoid-specific hypermethylation targets (Supplemental Table S3) that fit the general model: lymphocyte β = 1; nonlymphoid specimens β = 0. Specifically, two-group comparison of CD4/8/19+ cells (N = 18) versus a collection of somatic, nonlymphoid specimens (n = 87), ESCs (n = 29), iPSCs (n = 35), sperm (n = 8), and placenta/hydatidiform mole (n = 23/9) (β fold-change > 1.7, FDR < 2.3 × 10−42). The LI of a sample is then the average β value of these 36 targets.
TGCT_CORE_HYPER100. This module seeks targets where pan-TGCT (EC, TE, YST, total n = 58) methylation β = 1; somatic and pluripotent reference samples (n = 87 and 29, respectively) β = 0. Module targets (nonlymphoid calibrated) are specified by t-test of TGCT versus REFS (FDR = 3 × 10−35; fold-change +1.31 in TGCT group) (Fig. 3; Supplemental Table S3).
TGCT_CORE_HYPO39. General model: pan-TGCT (EC, TE, YST, total n = 58) methylation β = 0; reference samples (somatic, pluripotent, benign testis, total n = 343) β = 1. Specification: FDR = 0.05; fold-change 1.25 in references group (Fig. 3; Supplemental Table S3).
TGCT DMT2000. High-variance, subtype-significant differential-methylation module. The top 2000 differentially methylated targets among NSE samples were defined by variance = 0.5, ANOVA F-test FDR = 5.4 × 10−6 (nonlymphoid calibrated) (Fig. 4; Supplemental Table S3). EC_HYPO161: cluster subset of the DMT2000 of 161 CpG targets uniquely hypomethylated in ECs versus NSEs and references. EC_ESC COHYPO141: cluster subset of the DMT2000 of 141 targets cohypomethylated in ECs/ESCs.
gMME. This module is a robust set of 194 maternal germline methylated/imprinted targets that fit the general model: androgenetic samples = 0; biparental samples = 0.5; and gynogenetic samples = 1. The MME194 are specified by rank regression analysis on three sample groups ordered androgenetic < biparental < gynogenetic (sample group annotations indicated in Supplemental Fig. S11; Supplemental Table S3) and adjusting the FDR to 1.6 × 10−48 (Fig. 5; Supplemental Fig. S11A).
gPME. This module is a robust set of 19 paternal germline methylated/imprinted targets that fit the general model: partheno-/gynogenetic samples β = 0; biparental samples β = 0.5; and androgenetic samples β = 1. The PME19 are specified by rank regression analysis on three sample groups ordered gynogenetic < biparental < androgenetic (sample group annotations indicated in Supplemental Fig. S11; Supplemental Table S3) and adjusting the FDR to 2 × 10−28. (The FDR here is less stringent than MME194 because the ESCs/iPSCs included in the biparental group are intrinsically prone to hypermethylation of H19, and thus, there is greater variance in the biparental group for gPME target methylation) (Fig. 5; Supplemental Fig. S11B).
sMME. This module is a robust set of 19 secondary maternal methylated/imprinted targets that fit the general model: gamete (M&F) β = 0; biparental samples β = 0.5; and gynogenetic samples β = 1. The sMME19 are specified by rank regression analysis on three sample groups ordered gamete < biparental < gynogenetic (sample group annotations indicated in Supplemental Fig. S11; Supplemental Table S3) and adjusting the FDR to 5 × 10−38 (Fig. 5; Supplemental Fig. S11D).
sPME. This module is a robust set of 11 secondary paternal-allele methylated/imprinted targets that fit the general model: gamete (M&F) β = 0; biparental samples β = 0.5; and androgenetic samples β = 1. The sPME11 are specified by rank regression analysis on three sample groups ordered gamete < biparental < androgenetic (sample group annotations indicated in Supplemental Fig. S11; Supplemental Table S3), adjusting the FDR to 2 × 10−34 and selecting targets hemimethylated in biparental samples (Fig. 5; Supplemental Fig. S11D).
sBME. This module is a robust set of 100 pan-somatic, biparental CpG hypermethylated targets that fit the general model: gamete (M&F) & ESC β = 0; somatic differentiated samples β = 1 (sample group annotations indicated in Supplemental Fig. S11; Supplemental Table S3). The sBME is defined by two-group comparison with fold-change = 1.5 and FDR = 0.5 (Fig. 5; Supplemental Fig. S11E).
CpH_pluri_vs_neuron458. CpH DMT between ESCs (n = 19) and purified adult neurons (Supplemental Table S3; Supplemental Fig. S9). After filtering for intersection of 2208 (of 3091 array total) CpH targets across human and chimpanzee GEO data sets, then q = 0.05, fold-change = 1.25. Of these 458 targets, 256 are hypermethylated in ESCs and 202 in neurons.
mCpH pluri473. CpH targets hypermethylated in pluripotency (ESCs/iPSCs, n = 48) relative to neurons (n = 12). After filtering for intersection of 2208 (of 3091 array total) CpH targets across human and chimpanzee GEO data sets, then q = 0.05, fold-change = 1.2, yielding 473 targets (Supplemental Table S3).
Adjustment of tumor tissue imprinted target methylation for lymphoid cell infiltrates was based on a two-component mixture model whereby
where ρ = sample lymphoid cell fraction (LI), β1 = average lymphoid cell target methylation in the 18 lymphoid reference samples, and β2 = tumor-intrinsic target methylation. Thus,
PGC WGBS data analysis
The BED files containing the chromosome positions of each cytosine “C” and also the methylation status (ratios equivalent to the β values) were downloaded from GEO (accession GSE63818). All positions that match exactly to the mapping positions on the Illumina H450K-methylation array were extracted and assigned the same H450K target ID. Then methylation modules from above were analyzed in the PGC bisulfite sequencing data using QOE.
Data access
DNA methylation array data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE74104.
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Program of the National Cancer Institute. We thank all contributors of high-quality data to public databases.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.201293.115.
References
- Adewumi O, Aflatoonian B, Ahrlund-Richter L, Amit M, Andrews PW, Beighton G, Bello PA, Benvenisty N, Berry LS, Bevan S, et al. 2007. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol 25: 803–816. [DOI] [PubMed] [Google Scholar]
- Alagaratnam S, Lind GE, Kraggerud SM, Lothe RA, Skotheim RI. 2011. The testicular germ cell tumour transcriptome. Int J Androl 34: e133–e150; discussion e150–e151. [DOI] [PubMed] [Google Scholar]
- Al-Hussain T, Bakshi N, Akhtar M. 2015. Intratubular germ cell neoplasia of the testis: a brief review. Adv Anat Pathol 22: 202–212. [DOI] [PubMed] [Google Scholar]
- Almstrup K, Hoei-Hansen CE, Nielsen JE, Wirkner U, Ansorge W, Skakkebaek NE, Rajpert-De Meyts E, Leffers H. 2005. Genome-wide gene expression profiling of testicular carcinoma in situ progression into overt tumours. Br J Cancer 92: 1934–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amatruda JF, Ross JA, Christensen B, Fustino NJ, Chen KS, Hooten AJ, Nelson H, Kuriger JK, Rakheja D, Frazier AL, et al. 2013. DNA methylation analysis reveals distinct methylation signatures in pediatric germ cell tumors. BMC Cancer 13: 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews PW. 2002. From teratocarcinomas to embryonic stem cells. Philos Trans R Soc Lond B Biol Sci 357: 405–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton L, Brown AG, Bartolomei MS, Elovitz MA. 2014. Differential methylation of genes associated with cell adhesion in preeclamptic placentas. PLoS One 9: e100148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arand J, Spieler D, Karius T, Branco MR, Meilinger D, Meissner A, Jenuwein T, Xu G, Leonhardt H, Wolf V, et al. 2012. In vivo control of CpG and non-CpG DNA methylation by DNA methyltransferases. PLoS Genet 8: e1002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkin NB, Baker MC. 1982. Specific chromosome change, i(12p), in testicular tumours? Lancet 2: 1349. [DOI] [PubMed] [Google Scholar]
- Berney DM, Looijenga L, Idrees M, Oosterhuis JW, Rajpert-De Meyts E, Ulbright TM, Skakkebaek NE. 2016. Germ cell neoplasia in situ (GCNIS): evolution of the current nomenclature for testicular pre-invasive germ cell malignancy. Histopathology 69: 7–10. [DOI] [PubMed] [Google Scholar]
- Beyrouthy MJ, Garner KM, Hever MP, Freemantle SJ, Eastman A, Dmitrovsky E, Spinella MJ. 2009. High DNA methyltransferase 3B expression mediates 5-aza-deoxycytidine hypersensitivity in testicular germ cell tumors. Cancer Res 69: 9360–9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biermann K, Heukamp LC, Steger K, Zhou H, Franke FE, Sonnack V, Brehm R, Berg J, Bastian PJ, Muller SC, et al. 2007. Genome-wide expression profiling reveals new insights into pathogenesis and progression of testicular germ cell tumors. Cancer Genomics Proteomics 4: 359–367. [PubMed] [Google Scholar]
- Biswal BK, Beyrouthy MJ, Hever-Jardine MP, Armstrong D, Tomlinson CR, Christensen BC, Marsit CJ, Spinella MJ. 2012. Acute hypersensitivity of pluripotent testicular cancer-derived embryonal carcinoma to low-dose 5-aza deoxycytidine is associated with global DNA damage-associated p53 activation, anti-pluripotency and DNA demethylation. PLoS One 7: e53003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles J, Teasdale RP, James K, Koopman P. 2003. Dppa3 is a marker of pluripotency and has a human homologue that is expressed in germ cell tumours. Cytogenet Genome Res 101: 261–265. [DOI] [PubMed] [Google Scholar]
- Chedin F. 2011. The DNMT3 family of mammalian de novo DNA methyltransferases. Prog Mol Biol Transl Sci 101: 255–285. [DOI] [PubMed] [Google Scholar]
- Court F, Tayama C, Romanelli V, Martin-Trujillo A, Iglesias-Platas I, Okamura K, Sugahara N, Simon C, Moore H, Harness JV, et al. 2014. Genome-wide parent-of-origin DNA methylation analysis reveals the intricacies of human imprinting and suggests a germline methylation-independent mechanism of establishment. Genome Res 24: 554–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong J, Stoop H, Gillis AJ, van Gurp RJ, van de Geijn GJ, Boer M, Hersmus R, Saunders PT, Anderson RA, Oosterhuis JW, et al. 2008. Differential expression of SOX17 and SOX2 in germ cells and stem cells has biological and clinical implications. J Pathol 215: 21–30. [DOI] [PubMed] [Google Scholar]
- Devriendt K. 2005. Hydatidiform mole and triploidy: the role of genomic imprinting in placental development. Hum Reprod Update 11: 137–142. [DOI] [PubMed] [Google Scholar]
- Faulkner SW, Leigh DA, Oosterhuis JW, Roelofs H, Looijenga LH, Friedlander ML. 2000. Allelic losses in carcinoma in situ and testicular germ cell tumours of adolescents and adults: evidence suggestive of the linear progression model. Br J Cancer 83: 729–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feber A, Guilhamon P, Lechner M, Fenton T, Wilson GA, Thirlwell C, Morris TJ, Flanagan AM, Teschendorff AE, Kelly JD, et al. 2014. Using high-density DNA methylation arrays to profile copy number alterations. Genome Biol 15: R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira HJ, Heyn H, Garcia del Muro X, Vidal A, Larriba S, Munoz C, Villanueva A, Esteller M. 2014. Epigenetic loss of the PIWI/piRNA machinery in human testicular tumorigenesis. Epigenetics 9: 113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa S, Haruta M, Arai Y, Honda S, Ohshima J, Sugawara W, Kageyama Y, Higashi Y, Nishida K, Tsunematsu Y, et al. 2009. Yolk sac tumor but not seminoma or teratoma is associated with abnormal epigenetic reprogramming pathway and shows frequent hypermethylation of various tumor suppressor genes. Cancer Sci 100: 698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego Romero I, Pavlovic BJ, Hernando-Herraez I, Zhou X, Ward MC, Banovich NE, Kagan CL, Burnett JE, Huang CH, Mitrano A, et al. 2015. A panel of induced pluripotent stem cells from chimpanzees: a resource for comparative functional genomics. eLife 4: e07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Perez JL, Morell M, Scheys JO, Kulpa DA, Morell S, Carter CC, Hammer GD, Collins KL, O'Shea KS, Menendez P, et al. 2010. Epigenetic silencing of engineered L1 retrotransposition events in human embryonic carcinoma cells. Nature 466: 769–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et al. 2012. Architecture of the human regulatory network derived from ENCODE data. Nature 489: 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibas Z, Prout GR, Pontes JE, Sandberg AA. 1986. Chromosome changes in germ cell tumors of the testis. Cancer Genet Cytogenet 19: 245–252. [DOI] [PubMed] [Google Scholar]
- Gkountela S, Zhang KX, Shafiq TA, Liao WW, Hargan-Calvopina J, Chen PY, Clark AT. 2015. DNA demethylation dynamics in the human prenatal germline. Cell 161: 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Shin JH, Shin J, Li H, Xie B, Zhong C, Hu S, Le T, Fan G, et al. 2014. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat Neurosci 17: 215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Yan L, Guo H, Li L, Hu B, Zhao Y, Yong J, Hu Y, Wang X, Wei Y, et al. 2015. The transcriptome and DNA methylome landscapes of human primordial germ cells. Cell 161: 1437–1452. [DOI] [PubMed] [Google Scholar]
- Haig D. 1994. Refusing the ovarian time bomb. Trends Genet 10: 346–347; author reply 348–349. [DOI] [PubMed] [Google Scholar]
- Haig D. 1996. Gestational drive and the green-bearded placenta. Proc Natl Acad Sci 93: 6547–6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D, Graham C. 1991. Genomic imprinting and the strange case of the insulin-like growth factor II receptor. Cell 64: 1045–1046. [DOI] [PubMed] [Google Scholar]
- He Y, Ecker JR. 2015. Non-CG methylation in the human genome. Annu Rev Genomics Hum Genet 16: 55–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Kang X, Du H, Song B, Lu Z, Huang Y, Wang D, Sun X, Yu Y, Fan Y. 2014. Defining differentially methylated regions specific for the acquisition of pluripotency and maintenance in human pluripotent stem cells via microarray. PLoS One 9: e108350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen SG. 1970. Testicular biopsy score count: a method for registration of spermatogenesis in human testes: normal values and results in 335 hypogonadal males. Hormones 1: 2–25. [DOI] [PubMed] [Google Scholar]
- Kajii T, Ohama K. 1977. Androgenetic origin of hydatidiform mole. Nature 268: 633–634. [DOI] [PubMed] [Google Scholar]
- Killian JK, Bilke S, Davis S, Walker RL, Jaeger E, Killian MS, Waterfall JJ, Bibikova M, Fan JB, Smith WI Jr, et al. 2011. A methyl-deviator epigenotype of estrogen receptor-positive breast carcinoma is associated with malignant biology. Am J Pathol 179: 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killian JK, Kim SY, Miettinen M, Smith C, Merino M, Tsokos M, Quezado M, Smith WI Jr, Jahromi MS, Xekouki P, et al. 2013. Succinate dehydrogenase mutation underlies global epigenomic divergence in gastrointestinal stromal tumor. Cancer Discov 3: 648–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krausz C, Sandoval J, Sayols S, Chianese C, Giachini C, Heyn H, Esteller M. 2012. Novel insights into DNA methylation features in spermatozoa: stability and peculiarities. PLoS One 7: e44479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Inoue K, Ono R, Ogonuki N, Kohda T, Kaneko-Ishino T, Ogura A, Ishino F. 2002. Erasing genomic imprinting memory in mouse clone embryos produced from day 11.5 primordial germ cells. Development 129: 1807–1817. [DOI] [PubMed] [Google Scholar]
- Leitch HG, Smith A. 2013. The mammalian germline as a pluripotency cycle. Development 140: 2495–2501. [DOI] [PubMed] [Google Scholar]
- Linder D, McCaw BK, Hecht F. 1975. Parthenogenic origin of benign ovarian teratomas. N Engl J Med 292: 63–66. [DOI] [PubMed] [Google Scholar]
- Litchfield K, Summersgill B, Yost S, Sultana R, Labreche K, Dudakia D, Renwick A, Seal S, Al-Saadi R, Broderick P, et al. 2015. Whole-exome sequencing reveals the mutational spectrum of testicular germ cell tumours. Nat Commun 6: 5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looijenga LH, Verkerk AJ, Dekker MC, van Gurp RJ, Gillis AJ, Oosterhuis JW. 1998. Genomic imprinting in testicular germ cell tumours. APMIS 106: 187–195; discussion 196–197. [DOI] [PubMed] [Google Scholar]
- Looijenga LH, Rosenberg C, van Gurp RJ, Geelen E, van Echten-Arends J, de Jong B, Mostert M, Wolter Oosterhuis J. 2000. Comparative genomic hybridization of microdissected samples from different stages in the development of a seminoma and a non-seminoma. J Pathol 191: 187–192. [DOI] [PubMed] [Google Scholar]
- Looijenga LH, Stoop H, de Leeuw HP, de Gouveia Brazao CA, Gillis AJ, van Roozendaal KE, van Zoelen EJ, Weber RF, Wolffenbuttel KP, van Dekken H, et al. 2003. POU5F1 (OCT3/4) identifies cells with pluripotent potential in human germ cell tumors. Cancer Res 63: 2244–2250. [PubMed] [Google Scholar]
- McFadden DE, Robinson WP. 2006. Phenotype of triploid embryos. J Med Genet 43: 609–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlynn KA, Cook MB. 2009. Etiologic factors in testicular germ-cell tumors. Future Oncol 5: 1389–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre A, Summersgill B, Lu YJ, Missiaglia E, Kitazawa S, Oosterhuis JW, Looijenga LH, Shipley J. 2007. Genomic copy number and expression patterns in testicular germ cell tumours. Br J Cancer 97: 1707–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami K, Chano T, Kawakami T, Ushida H, Kushima R, Okabe H, Okada Y, Okamoto K. 2010. DNMT3L is a novel marker and is essential for the growth of human embryonal carcinoma. Clin Cancer Res 16: 2751–2759. [DOI] [PubMed] [Google Scholar]
- Monk D. 2015. Germline-derived DNA methylation and early embryo epigenetic reprogramming: the selected survival of imprints. Int J Biochem Cell Biol 67: 128–138. [DOI] [PubMed] [Google Scholar]
- Nazor KL, Altun G, Lynch C, Tran H, Harness JV, Slavin I, Garitaonandia I, Muller FJ, Wang YC, Boscolo FS, et al. 2012. Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell Stem Cell 10: 620–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nettersheim D, Gillis A, Biermann K, Looijenga LH, Schorle H. 2011. The seminoma cell line TCam-2 is sensitive to HDAC inhibitor depsipeptide but tolerates various other chemotherapeutic drugs and loss of NANOG expression. Genes Chromosomes Cancer 50: 1033– 1042. [DOI] [PubMed] [Google Scholar]
- Nettersheim D, Jostes S, Sharma R, Schneider S, Hofmann A, Ferreira HJ, Hoffmann P, Kristiansen G, Esteller MB, Schorle H. 2015. BMP inhibition in seminomas initiates acquisition of pluripotency via NODAL signaling resulting in reprogramming to an embryonal carcinoma. PLoS Genet 11: e1005415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netto GJ, Nakai Y, Nakayama M, Jadallah S, Toubaji A, Nonomura N, Albadine R, Hicks JL, Epstein JI, Yegnasubramanian S, et al. 2008. Global DNA hypomethylation in intratubular germ cell neoplasia and seminoma, but not in nonseminomatous male germ cell tumors. Mod Pathol 21: 1337–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okae H, Chiba H, Hiura H, Hamada H, Sato A, Utsunomiya T, Kikuchi H, Yoshida H, Tanaka A, Suyama M, et al. 2014. Genome-wide analysis of DNA methylation dynamics during early human development. PLoS Genet 10: e1004868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveros-Etter M, Li Z, Nee K, Hosohama L, Hargan-Calvopina J, Lee SA, Joti P, Yu J, Clark AT. 2015. PGC reversion to pluripotency involves erasure of DNA methylation from imprinting control centers followed by locus-specific re-methylation. Stem Cell Reports 5: 337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosterhuis JW, Looijenga LH. 2005. Testicular germ-cell tumours in a broader perspective. Nat Rev Cancer 5: 210–222. [DOI] [PubMed] [Google Scholar]
- Port M, Schmelz HU, Stockinger M, Sparwasser C, Albers P, Pottek T, Abend M. 2005. Gene expression profiling in seminoma and nonseminoma. J Clin Oncol 23: 58–69. [DOI] [PubMed] [Google Scholar]
- Reinius LE, Acevedo N, Joerink M, Pershagen G, Dahlen SE, Greco D, Soderhall C, Scheynius A, Kere J. 2012. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS One 7: e41361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijlaarsdam MA, Tax DM, Gillis AJ, Dorssers LC, Koestler DC, de Ridder J, Looijenga LH. 2015. Genome wide DNA methylation profiles provide clues to the origin and pathogenesis of germ cell tumors. PLoS One 10: e0122146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikine Y, Genega E, Melamed J, Lee P, Reuter VE, Ye H. 2012. Molecular genetics of testicular germ cell tumors. Am J Cancer Res 2: 153–167. [PMC free article] [PubMed] [Google Scholar]
- Shovlin TC, Durcova-Hills G, Surani A, McLaren A. 2008. Heterogeneity in imprinted methylation patterns of pluripotent embryonic germ cells derived from pre-migratory mouse germ cells. Dev Biol 313: 674–681. [DOI] [PubMed] [Google Scholar]
- Sievers S, Alemazkour K, Zahn S, Perlman EJ, Gillis AJ, Looijenga LH, Gobel U, Schneider DT. 2005. IGF2/H19 imprinting analysis of human germ cell tumors (GCTs) using the methylation-sensitive single-nucleotide primer extension method reflects the origin of GCTs in different stages of primordial germ cell development. Genes Chromosomes Cancer 44: 256–264. [DOI] [PubMed] [Google Scholar]
- Skakkebaek NE. 1972. Possible carcinoma-in-situ of the testis. Lancet 2: 516–517. [DOI] [PubMed] [Google Scholar]
- Skotheim RI, Lothe RA. 2003. The testicular germ cell tumour genome. APMIS 111: 136–150; discussion 50–51. [DOI] [PubMed] [Google Scholar]
- Skotheim RI, Lind GE, Monni O, Nesland JM, Abeler VM, Fossa SD, Duale N, Brunborg G, Kallioniemi O, Andrews PW, et al. 2005. Differentiation of human embryonal carcinomas in vitro and in vivo reveals expression profiles relevant to normal development. Cancer Res 65: 5588–5598. [DOI] [PubMed] [Google Scholar]
- Sonne SB, Almstrup K, Dalgaard M, Juncker AS, Edsgard D, Ruban L, Harrison NJ, Schwager C, Abdollahi A, Huber PE, et al. 2009. Analysis of gene expression profiles of microdissected cell populations indicates that testicular carcinoma in situ is an arrested gonocyte. Cancer Res 69: 5241–5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperger JM, Chen X, Draper JS, Antosiewicz JE, Chon CH, Jones SB, Brooks JD, Andrews PW, Brown PO, Thomson JA. 2003. Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc Natl Acad Sci 100: 13350–13355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summersgill B, Goker H, Weber-Hall S, Huddart R, Horwich A, Shipley J. 1998. Molecular cytogenetic analysis of adult testicular germ cell tumours and identification of regions of consensus copy number change. Br J Cancer 77: 305–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani MA, Barton SC, Norris ML. 1984. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature 308: 548–550. [DOI] [PubMed] [Google Scholar]
- Surti U, Hoffner L, Chakravarti A, Ferrell RE. 1990. Genetics and biology of human ovarian teratomas. I. Cytogenetic analysis and mechanism of origin. Am J Hum Genet 47: 635–643. [PMC free article] [PubMed] [Google Scholar]
- Tada T, Tada M, Hilton K, Barton SC, Sado T, Takagi N, Surani MA. 1998. Epigenotype switching of imprintable loci in embryonic germ cells. Dev Genes Evol 207: 551–561. [DOI] [PubMed] [Google Scholar]
- Takashima Y, Guo G, Loos R, Nichols J, Ficz G, Krueger F, Oxley D, Santos F, Clarke J, Mansfield W, et al. 2014. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell 158: 1254–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WW, Dietmann S, Irie N, Leitch HG, Floros VI, Bradshaw CR, Hackett JA, Chinnery PF, Surani MA. 2015. A unique gene regulatory network resets the human germline epigenome for development. Cell 161: 1453–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedemann RL, Putiri EL, Lee JH, Hlady RA, Kashiwagi K, Ordog T, Zhang Z, Liu C, Choi JH, Robertson KD. 2014. Acute depletion redefines the division of labor among DNA methyltransferases in methylating the human genome. Cell Rep 9: 1554–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabert B, Chen J, Devesa SS, Bray F, McGlynn KA. 2015. International patterns and trends in testicular cancer incidence, overall and by histologic subtype, 1973–2007. Andrology 3: 4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Echten J, Oosterhuis JW, Looijenga LH, van de Pol M, Wiersema J, te Meerman GJ, Schaffordt Koops H, Sleijfer DT, de Jong B. 1995. No recurrent structural abnormalities apart from i(12p) in primary germ cell tumors of the adult testis. Genes Chromosomes Cancer 14: 133–144. [DOI] [PubMed] [Google Scholar]
- Varmuza S, Mann M. 1994. Genomic imprinting: defusing the ovarian time bomb. Trends Genet 10: 118–123. [DOI] [PubMed] [Google Scholar]
- Verkerk AJ, Ariel I, Dekker MC, Schneider T, van Gurp RJ, de Groot N, Gillis AJ, Oosterhuis JW, Hochberg AA, Looijenga LH. 1997. Unique expression patterns of H19 in human testicular cancers of different etiology. Oncogene 14: 95–107. [DOI] [PubMed] [Google Scholar]
- von Meyenn F, Reik W. 2015. Forget the parents: epigenetic reprogramming in human germ cells. Cell 161: 1248–1251. [DOI] [PubMed] [Google Scholar]
- Wermann H, Stoop H, Gillis AJ, Honecker F, van Gurp RJ, Ammerpohl O, Richter J, Oosterhuis JW, Bokemeyer C, Looijenga LH. 2010. Global DNA methylation in fetal human germ cells and germ cell tumours: association with differentiation and cisplatin resistance. J Pathol 221: 433–442. [DOI] [PubMed] [Google Scholar]
- Wongtrakoongate P. 2015. Epigenetic therapy of cancer stem and progenitor cells by targeting DNA methylation machineries. World J Stem Cells 7: 137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongtrakoongate P, Jones M, Gokhale PJ, Andrews PW. 2013. STELLA facilitates differentiation of germ cell and endodermal lineages of human embryonic stem cells. PLoS One 8: e56893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen RK, Chen B, Blair JD, Robinson WP, Nelson DM. 2013. Hypoxia alters the epigenetic profile in cultured human placental trophoblasts. Epigenetics 8: 192–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Muller F, Liao J, Zhang Y, Gu H, Bock C, Boyle P, Epstein CB, Bernstein BE, Lengauer T, et al. 2011. Genomic distribution and inter-sample variation of non-CpG methylation across human cell types. PLoS Genet 7: e1002389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.