Abstract
An essential step in the signal transduction pathway of Escherichia coli is the control of the protein kinase activity of CheA by the chemotaxis receptor proteins. This control requires the participation of the CheW protein. Although the biochemical nature of the coupling between the receptors and the kinase is unknown, it is likely that CheW interacts with the receptors and with CheA. In this communication, we report direct measurement of a physical interaction between CheW and CheA. We utilized the equilibrium column chromatography method of Hummel and Dreyer to show that CheW and CheA exhibit reversible binding with the stoichiometry of two CheW monomers per CheA dimer. CheW was found to exist as monomers and CheA was found to exist as dimers by equilibrium analytical ultracentrifugation. The dissociation constant for the CheW-CheA interaction (in 160 mM KCl/5 mM MgCl2, pH 7.4 at 4 degrees C) was determined to be in the physiologically relevant range of 17 microM. No evidence for cooperativity in the association of CheW with CheA was found.
Full text
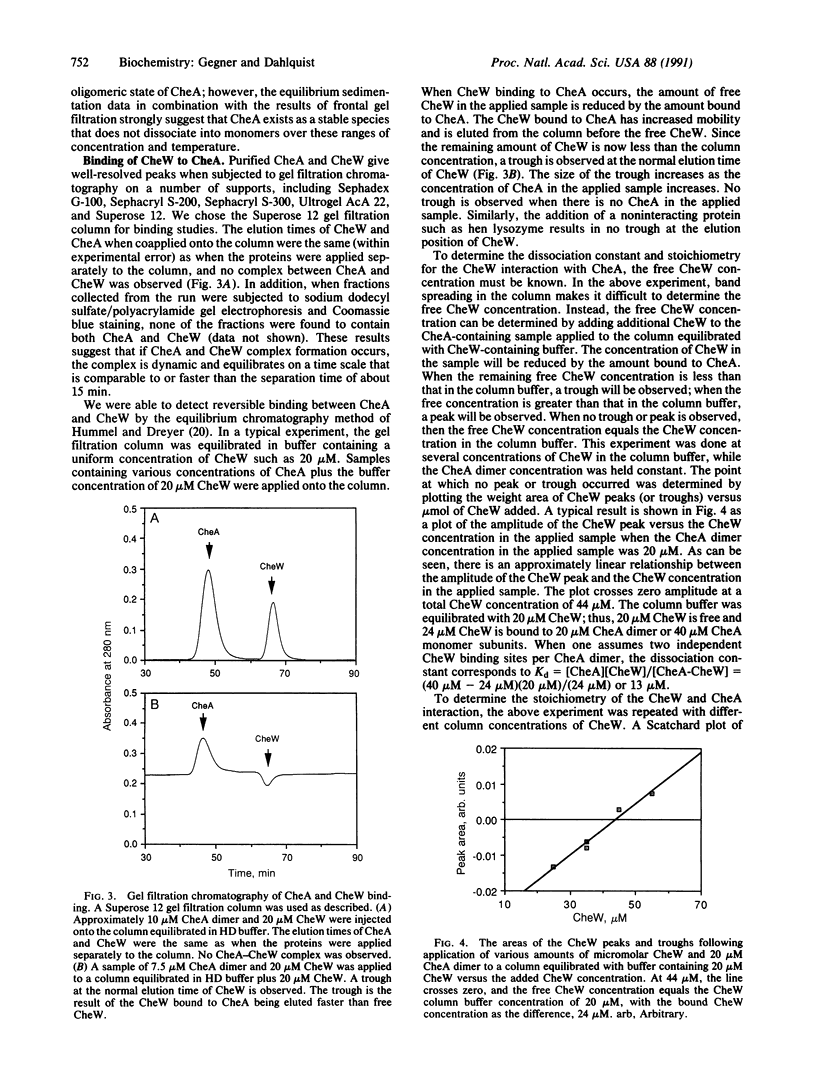
PDF


Selected References
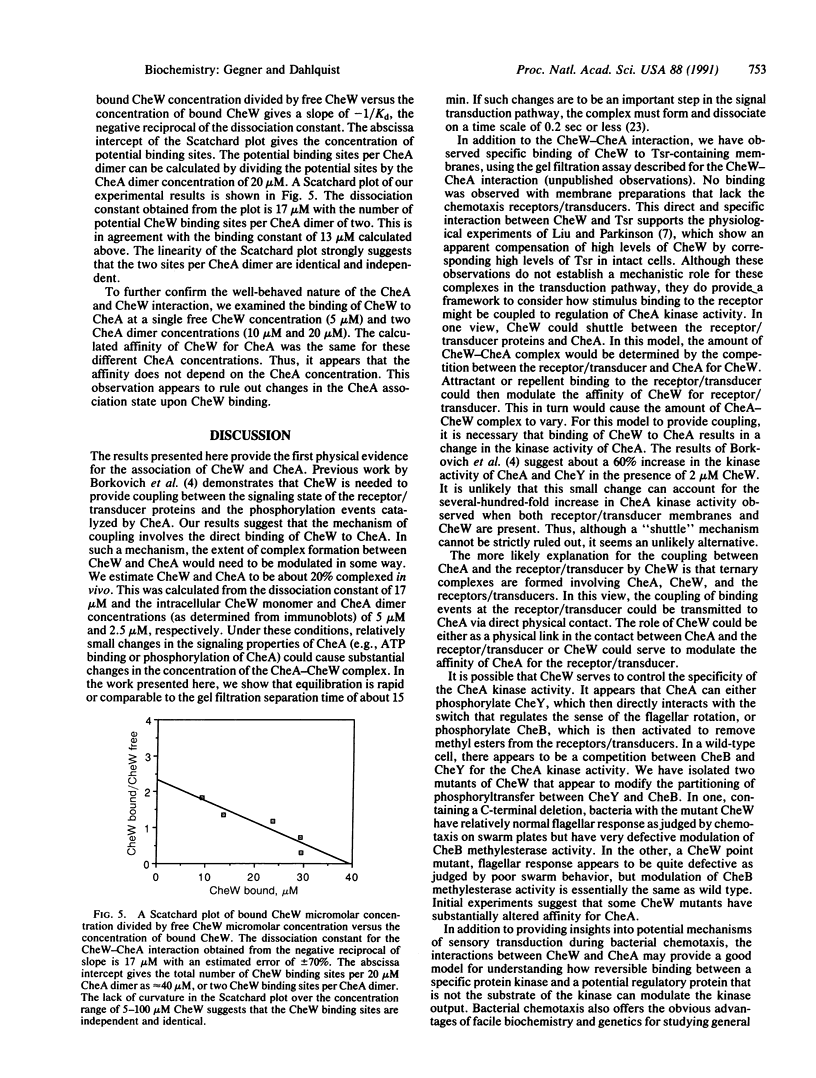
These references are in PubMed. This may not be the complete list of references from this article.
- Borkovich K. A., Kaplan N., Hess J. F., Simon M. I. Transmembrane signal transduction in bacterial chemotaxis involves ligand-dependent activation of phosphate group transfer. Proc Natl Acad Sci U S A. 1989 Feb;86(4):1208–1212. doi: 10.1073/pnas.86.4.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourret R. B., Hess J. F., Simon M. I. Conserved aspartate residues and phosphorylation in signal transduction by the chemotaxis protein CheY. Proc Natl Acad Sci U S A. 1990 Jan;87(1):41–45. doi: 10.1073/pnas.87.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S. C., von Hippel P. H. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989 Nov 1;182(2):319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- HUMMEL J. P., DREYER W. J. Measurement of protein-binding phenomena by gel filtration. Biochim Biophys Acta. 1962 Oct 8;63:530–532. doi: 10.1016/0006-3002(62)90124-5. [DOI] [PubMed] [Google Scholar]
- Hess J. F., Oosawa K., Kaplan N., Simon M. I. Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell. 1988 Apr 8;53(1):79–87. doi: 10.1016/0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- Kehry M. R., Doak T. G., Dahlquist F. W. Stimulus-induced changes in methylesterase activity during chemotaxis in Escherichia coli. J Biol Chem. 1984 Oct 10;259(19):11828–11835. [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. D., Parkinson J. S. Role of CheW protein in coupling membrane receptors to the intracellular signaling system of bacterial chemotaxis. Proc Natl Acad Sci U S A. 1989 Nov;86(22):8703–8707. doi: 10.1073/pnas.86.22.8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A., Stock J. Phosphorylation of an N-terminal regulatory domain activates the CheB methylesterase in bacterial chemotaxis. J Biol Chem. 1989 Oct 15;264(29):17337–17342. [PubMed] [Google Scholar]
- Muchmore D. C., McIntosh L. P., Russell C. B., Anderson D. E., Dahlquist F. W. Expression and nitrogen-15 labeling of proteins for proton and nitrogen-15 nuclear magnetic resonance. Methods Enzymol. 1989;177:44–73. doi: 10.1016/0076-6879(89)77005-1. [DOI] [PubMed] [Google Scholar]
- Mutoh N., Simon M. I. Nucleotide sequence corresponding to five chemotaxis genes in Escherichia coli. J Bacteriol. 1986 Jan;165(1):161–166. doi: 10.1128/jb.165.1.161-166.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders D. A., Mendez B., Koshland D. E., Jr Role of the CheW protein in bacterial chemotaxis: overexpression is equivalent to absence. J Bacteriol. 1989 Nov;171(11):6271–6278. doi: 10.1128/jb.171.11.6271-6278.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segall J. E., Manson M. D., Berg H. C. Signal processing times in bacterial chemotaxis. Nature. 1982 Apr 29;296(5860):855–857. doi: 10.1038/296855a0. [DOI] [PubMed] [Google Scholar]
- Smith R. A., Parkinson J. S. Overlapping genes at the cheA locus of Escherichia coli. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5370–5374. doi: 10.1073/pnas.77.9.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart R. C., Roth A. F., Dahlquist F. W. Mutations that affect control of the methylesterase activity of CheB, a component of the chemotaxis adaptation system in Escherichia coli. J Bacteriol. 1990 Jun;172(6):3388–3399. doi: 10.1128/jb.172.6.3388-3399.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart R. C., Russell C. B., Roth A. F., Dahlquist F. W. Interaction of CheB with chemotaxis signal transduction components in Escherichia coli: modulation of the methylesterase activity and effects on cell swimming behavior. Cold Spring Harb Symp Quant Biol. 1988;53(Pt 1):27–40. doi: 10.1101/sqb.1988.053.01.007. [DOI] [PubMed] [Google Scholar]
- Stock A., Mottonen J., Chen T., Stock J. Identification of a possible nucleotide binding site in CheW, a protein required for sensory transduction in bacterial chemotaxis. J Biol Chem. 1987 Jan 15;262(2):535–537. [PubMed] [Google Scholar]
- YPHANTIS D. A. EQUILIBRIUM ULTRACENTRIFUGATION OF DILUTE SOLUTIONS. Biochemistry. 1964 Mar;3:297–317. doi: 10.1021/bi00891a003. [DOI] [PubMed] [Google Scholar]
- Zamyatnin A. A. Protein volume in solution. Prog Biophys Mol Biol. 1972;24:107–123. doi: 10.1016/0079-6107(72)90005-3. [DOI] [PubMed] [Google Scholar]


