Abstract
Label-free quantitative MS based on the Normalized Spectral Abundance Factor (NSAF) has emerged as a straightforward and robust method to determine the relative abundance of individual proteins within complex mixtures. Here, we present Morpheus Spectral Counter (MSpC) as the first computational tool that directly calculates NSAF values from output obtained from Morpheus, a fast, open-source, peptide-MS/MS matching engine compatible with high-resolution accurate-mass instruments. NSAF has distinct advantages over other MS-based quantification methods, including a higher dynamic range as compared to isobaric tags, no requirement to align and re-extract MS1 peaks, and increased speed. MSpC features an easy to use graphic user interface that additionally calculates both distributed and unique NSAF values to permit analyses of both protein families and isoforms/proteoforms. MSpC determinations of protein concentration were linear over several orders of magnitude based on the analysis of several high-mass accuracy datasets either obtained from PRIDE or generated with total cell extracts spiked with purified Arabidopsis 20S proteasomes. The MSpC software was developed in C# and is open sourced under a permissive license with the code made available at http://dcgemperline.github.io/Morpheus_SpC/.
Keywords: Bioinformatics, Label-free quantification, LC-MS/MS, Morpheus, proteomics
Quantification of individual polypeptides within complex mixtures by MS is an extremely useful tool to understand proteomic changes in organisms during growth and development, and after environmental perturbation [1]. While a number of MS/MS strategies have been developed to measure protein abundance, including Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC), labeling with isobaric tags, and Absolute Quantification of proteins (AQUA) [2-5], label-free quantification (LFQ) have become increasingly popular given their simplicity and low cost [1, 6]. One LFQ strategy infers abundance from the number of observed peptide spectra matches (PSMs). For these PSM-based approaches, changes in protein abundance can be generated artifactually when total PSMs differ among samples and because longer proteins tend to produce more raw counts. For these reasons normalizing for both protein length and total PSMs is paramount. While this adjustment can be made in a number of ways; one of the most straight forward methods is to use Normalized Spectral Abundance Factor (NSAF), a length- and count-normalized measure for each protein [7]. Further improvements to the NSAF algorithm have been made by accounting for shared peptides in distributed NSAF (dNSAF), which distributes common PSMs among a family of isoforms/proteoforms based on the number of distinct PSMs observed for each isoform/proteoform, and unique NSAF (uNSAF), which ignores shared PSMs and only assigns distinct PSMs to each specific isoform/proteoform [8].
The Morpheus MS search engine was recently designed for high-resolution, accurate-mass data obtained from Orbitrap-based instruments to provide faster matching of spectra to peptides [9]. Unfortunately, no downstream automated tools are available to facilitate LFQ analysis, which can be quite challenging, if not impossible, to complete manually when accounting for shared peptides. To overcome this bottleneck, we developed Morpheus Spectral Counter (MSpC) as the first LFQ computational tool that integrates directly with Morpheus to calculate NSAF, dNSAF, uNSAF, and corrected PSM [10] values in complex protein samples. MSpC is fully automated, and only requires a Morpheus search summary file (summary.tsv) as input. The user interface (Supplemental Figure 1A) allows one to select the summary file and displays the raw MS/MS files that will be analyzed by MSpC. Some important features of MSpC are its ability to handle fractionation experiments as input, and the ability to whitelist proteins of interest in the output by specifying a csv file (see Tutorial). Options exist to specify global PSM and protein group FDR rates (thus avoiding increased FDRs when one analyzes many experiments at once), to output NSAF, dNSAF, and uNSAF values, to require a minimum number of unique peptides to quantify a protein, and to specify an output directory. A progress bar indicates completion of the analysis by MSpC.
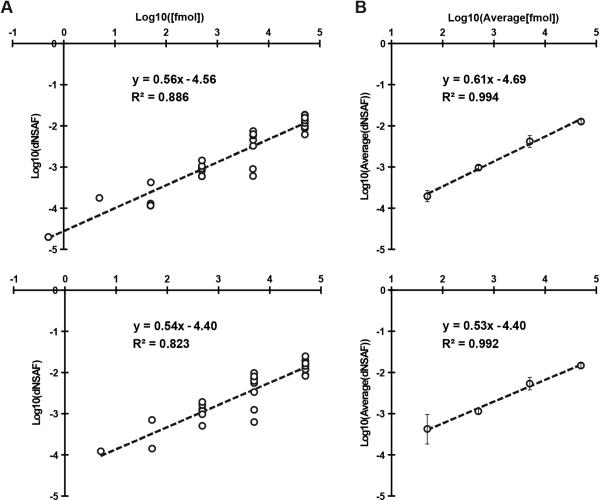
To validate the accuracy of MSpC, we analyzed two MS/MS datasets available in PRIDE that were previously generated by high-energy collision-induced dissociation using Thermo Q-Exactive Orbitrap instruments. Here, Xenopus egg (top, Figure 1) and embryo (bottom, Figure 1) extracts were spiked at a 4:1 ratio with the Universal Proteome Standard 2 (UPS2), a mix of 48 purified proteins at defined molar ratios of 0.5, 5, 50, 500, 5000, and 50,000, with each ratio containing a different set of 8 of the 48 proteins. As shown in Figure 1A, when the Morpheus/MSpC pipeline was used to calculate the average dNSAF value for each UPS2 protein, requiring only a single unique peptide to quantify, strong linear correlations (R2 = 0.886 and 0.823) were obtained across a 1,000 fold change in abundance (50 fmol to 50,000 fmol). In fact, the R2 values were similar to those obtained by others with PSM-based LFQ methods [11, 12]. This linear correlation was further strengthened when the dNSAF values were averaged for all UPS2 proteins within each of the concentration groups, with R2 values of 0.994 and 0.992 for the egg and embryo datasets, respectively (Figure 1B). Notably, the slope of the concentration series was significantly less than unity, showing that NSAF measurements are not appropriate for absolute quantification, which was expected given that NSAF is a relative value.
Figure 1.
Confirmation of MSpC accuracy by analysis of MS/MS datasets generated with the Universal Proteome Standard 2 (UPS2). The array of UPS2 standards were spiked into Xenopus laevis egg (Top) and embryo (Bottom) extracts at a range of concentrations. Following MS/MS analysis, dNSAF values for each protein was determined by Morpheus and MSpC. (A) A log-log plot of dNSAF versus concentration for each UPS2 protein detected across each fmol range. (B) A log-log plot of average dNSAF vs average concentration of each group of UPS2 proteins at each fmol range: (50, 500, 5000, and 50,000 fmol).
We also reprocessed the UPS2 dataset using the option of requiring a minimum of two unique peptides for quantification, which should improve stringency. This option provided only a minor improvement in overall linearity for the average UPS2 dNSAF values, but decreased linearity when each UPS2 protein was considered individually and removed some UPS2 proteins at low concentrations (compare Supplemental Figure 2A to Figure 1A). Consequently, caution should be exercised when selecting this option even though it might provide a slight improvement in stringency (see supplemental discussion in Supporting Information).
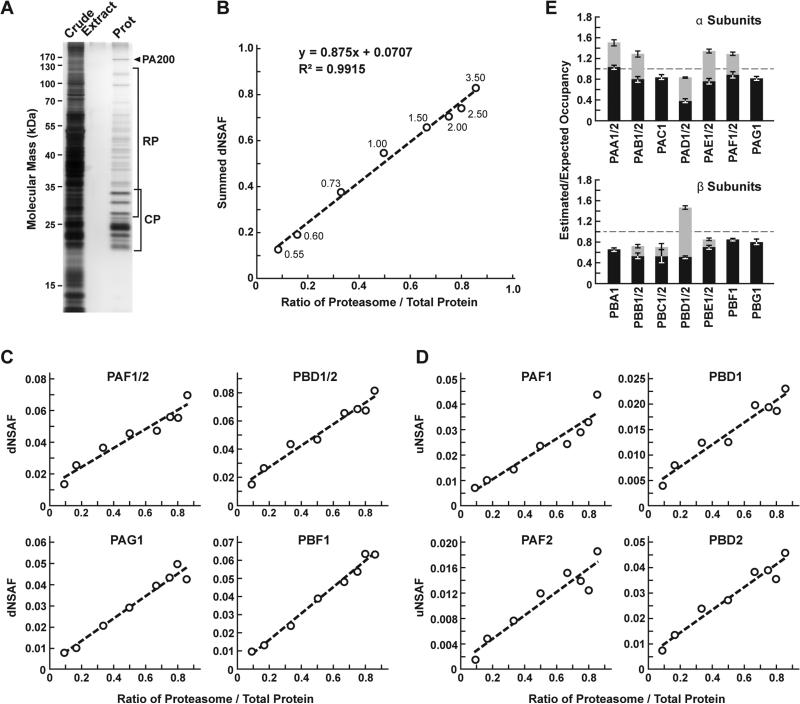
To demonstrate the utility and accuracy of MSpC as applied to our work, we analyzed 20S proteasomes isolated from Arabidopsis thaliana. This particle contains multiple subunits assembled in stoichiometric amounts, with many subunits encoded by two paralogous genes of sufficient amino acid identity (typically >90% [13]) such that discrimination between paralogs can be challenging using LFQ approaches [14]. To simulate changes in 20S proteasome abundance, we added varying amounts of trypsinized proteasomes (0.05 μg to 3 μg) to a fixed amount of trypsinized E. coli lysate (0.5 μg) to generate proteasome/lysate ratios of ~0.091, 0.167, 0.333, 0.500, 0.667, 0.750 0.800, 0.857. The digests were then subjected to MS/MS and the dNSAF value for each subunit along with the uNSAF value for individual isoforms were calculated by the Morpheus/MSpC pipeline (see Supplemental Methods). The data from this experiment are deposited in PRIDE with ID PXD003002. As shown in Figure 2, MSpC provided an excellent determination for the overall abundance of 20S proteasomes within a complex mixture, along with a good reflection of the abundance of individual subunits and their isoforms. When the dNSAF values for all subunits for the Arabidopsis 20S proteasome including their isoforms (representing 14 distinct subunits, 10 of which exist as isoform pairs) were summed, a very close approximation of the dNSAF/actual abundance was obtained (slope=0.875) with a very strong linear correlation (R2 = 0.99) over ~10-fold range in protein abundance.
Figure 2.
Confirmation of MSpC accuracy by analysis of MS/MS datasets generated with affinity purified Arabidopsis 20S proteasomes spiked into a total cell lysate from E. coli. Following MS/MS analysis, the dNSAF and uNSAF values for each subunit/isoform were determined by Morpheus and MSpC. (A) A silver-stained SDS-PAGE gel of 20S proteasome samples affinity purified from 10-d-old Arabidopsis seedlings. The crude seedling extract (CE), sample buffer (SB), and affinity-purified 20S proteasome samples (Prot) are shown. (B) Quantification of trypsinized 20S proteasomes when mixed at varying ratios with trypsinized total protein lysates from E. coli. The spiked samples were subjected to MS/MS followed by data analysis with the Morpheus and MSpC. dNSAF values for each proteasome subunit were averaged across three technical replicates, then summed to obtain an estimate of abundance for the 20S proteasome, and plotted against their known ratios. The total protein load is listed at each point in μg. (C and D) dNSAF and uNSAF values determined from the data in panel B for individual subunits (C) and their isoforms (D) for several subunits of the 20S proteasome. (E) Quantification accuracy of the Morpheus/MSpC pipeline for determining of the amount of each α and β subunit of the 20S proteasome. Single subunit isoforms are in black, whereas subunits having two isoforms are shown in black and grey to reflect the contributions of isoforms 1 and 2 respectively. Each bar represents the average of eight technical replicates (± SE). The dashed line represents the expected value of one assuming an equal stoichiometry of each subunit within the particle.
When each 20S proteasome subunit was analyzed individually, a strong linear response was also obtained (R2 > 0.90) for a majority of subunits (Figure 2C and Supplemental Table 1). For example, reasonably accurate concentration plots were obtained for the PAF (α6) and PBD (β4) subunits that are encoded by the PAF1/2 and PBF1/2 gene pairs, and for the PAG (α7) and PBF (β6) subunits that are encoded by single PAG1 and PBF1 genes (R2 from 0.94 to 0.99). Even when we calculated uNSAF values for individual isoforms added to the E. coli lysate, strong linear responses were obtained (e.g., the PAF1/PAF2 and PBD1/PBD2 pairs) with robust correlations (R2 from 0.89 to 0.95) (Figure 2D). Taken together, MSpC worked well for relative LFQ analysis of a multi-subunit complex and its individual subunits and isoforms within a complex proteomic mixture.
The Morpheus/MSpC pipeline also allowed us to calculate the respective incorporation of each paralog in the complex (see Supplemental Methods). As shown in Figure 2E, these estimated/expected occupancies were close to unity for most subunits within both the α and β rings of the 20S proteasome. The only strong deviation was for PBD1/2 (β4), which had a greater dNSAF value relative to other β subunits across the experiments analyzed (Supplemental Table 1). The calculations for uNSAF values also estimated the relative proportion of each isoform within the complex for those subunits expressed from paralogous genes. The data obtained are similar to prior studies of the complex involving quantitative top-down proteomic analysis of purified proteasome samples using ultra violet-intrinsic fluorescence to quantify tyrosine-containing subunits [15]. However, our MSpC analysis provided a more complete picture as several subunit isoforms were difficult to quantify by fluorescence either because they lacked tryosine, or because their fluorescence peaks overlapped with those of other subunits/isoforms. Notably , the protein isoform ratios measured here agree well with the expression ratios for the paralogous genes [14], suggesting that the protein isoform abundance generally reflects the relative transcriptional activity of the gene pair. We consistently estimated slightly more α ring subunits (PAA-PAG) versus β ring subunits (PBA-PBG) in the final MSpC calculations (Figure 2E). This deviation could represent enhanced detection of α ring versus β ring subunits, or more likely that purification via the tagged α ring subunit PAG1 also isolated assembly intermediates comprised of only α ring subunits.
We compared the Morpheus and MSpC pipeline to the next most comparable open source, spectral-count-based LFQ pipeline, The Trans Proteomic Pipeline (TPP) [16] and ABACUS [10] using our datasets generated with the 20S proteasome/E. coli lysate mixture (Supplemental Table 1 and 2). Morpheus/MSpC slightly outperformed TPP/ABACUS by having a greater overall accuracy (average linearity of 0.88 compared to 0.84), and by having more subunits showing an R2 linear correlation greater than 0.9 (14/23 subunits for MSpC versus and 11/23 for ABACUS). In addition to this modest improvement, we note that the Morpheus/MSpC pipeline required significantly less intermediary steps, thus accelerating the data analysis. Some of the additional steps in TPP/ABACUS could be automated from the command-line, but it would likely be a challenge for the average user. Importantly, we found that the Morpheus/MSpC pipeline was faster. Timing tests using the proteasome/E. coli spike data generated here showed that the Morpheus/MSpC pipeline was 1.9-fold faster than the TPP/ABACUS pipeline (Figure 3). Such an improvement was expected given that Morpheus completes its searches on average 1.3 to 4.6 times faster than most other search engines available [9].
Figure 3.
MSpC combined with Morpheus works faster than TPP combined with ABACUS. Speed comparisons were performed for 12, 24, 48, and 96 raw MS/MS files generated with the 20S proteasome/E coli lysate samples analyzed in Figure 2. On average, Morpheus/MSpC finished the calculations 1.9 times faster than TPP/ABACUS over a ~ 10-fold range of dataset size.
Given its simplicity of use, speed, and open source nature, MSpC combined with Morpheus is clearly advantageous over other PSM-based LFQ approaches currently available. Moreover, by being open source, MSpC should allow others to extend its utility and to serve as a platform for integrating additional open source LFQ approaches into the Morpheus pipeline.
Supplementary Material
ACKNOWLEDGEMENTS
D.C.G. was supported by a grant from the U.S. Department of Energy Office of Science; Office of Basic Energy Sciences; Chemical Sciences, Geosciences, and Biosciences Division (DE-FG02-88ER13968) and a graduate training fellowship from the NIH (5 T32 GM 7133-37). M.S. and L.M.S were supported by a grant from the National Institute of Health/National Institute of General Medical Sciences (1P50HG004942). The authors thank Erin Gemperline, Richard S. Marshall, and Josh Coon for critical reading of the manuscript, and additionally thank Derek Bailey for a critical code review.
Abbreviations
- AQUA
Absolute QUAntification of proteins
- dNSAF
distributed Normalized Spectral Abundance Factor
- LFQ
Label-Free Quantification
- MSpC
Morpheus Spectral Counter
- NSAF
Normalized Spectral Abundance Factor
- PSM
Peptide Spectra Match
- TPP
Trans Proteomic Pipeline
- uNSAF
unique Normalized Spectral Abundance Factor
- UPS2
Universal Proteome Standard 2
- SILAC
Stable Isotope Labeling by Amino Acids in Cell Culture
Footnotes
The authors have declared no conflict of interest.
REFERENCES
- 1.Wong JW, Cagney G. An overview of label-free quantitation methods in proteomics by mass spectrometry. Methods Mol. Biol. 2010;604:273–283. doi: 10.1007/978-1-60761-444-9_18. [DOI] [PubMed] [Google Scholar]
- 2.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 3.Thompson A, Schafer J, Kuhn K, Kienle S, et al. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 4.Ross PL, Huang YN, Marchese JN, Williamson B, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. USA. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang B, VerBerkmoes NC, Langston MA, Uberbacher E, et al. Detecting differential and correlated protein expression in label-free shotgun proteomics. J. Proteome Res. 2006;5:2909–2918. doi: 10.1021/pr0600273. [DOI] [PubMed] [Google Scholar]
- 7.Zybailov B, Mosley AL, Sardiu ME, Coleman MK, et al. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J. Proteome Res. 2006;5:2339–2347. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Wen Z, Washburn MP, Florens L. Refinements to label free proteome quantitation: how to deal with peptides shared by multiple proteins. Anal. Chem. 2010;82:2272–2281. doi: 10.1021/ac9023999. [DOI] [PubMed] [Google Scholar]
- 9.Wenger CD, Coon JJ. A proteomics search algorithm specifically designed for high-resolution tandem mass spectra. J. Proteome Res. 2013;12:1377–1386. doi: 10.1021/pr301024c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fermin D, Basrur V, Yocum AK, Nesvizhskii AI. ABACUS: a computational tool for extracting and pre-processing spectral count data for label-free quantitative proteomic analysis. Proteomics. 2011;11:1340–1345. doi: 10.1002/pmic.201000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox J, Hein MY, Luber CA, Paron I, et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tu C, Li J, Sheng Q, Zhang M, Qu J. Systematic assessment of survey scan and MS2-based abundance strategies for label-free quantitative proteomics using high-resolution MS data. J. Proteome Res. 2014;13:2069–2079. doi: 10.1021/pr401206m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang P, Fu H, Walker J, Papa CM, et al. Purification of the Arabidopsis 26S proteasome: biochemical and molecular analyses revealed the presence of multiple isoforms. J. Biol. Chem. 2004;279:6401–6413. doi: 10.1074/jbc.M311977200. [DOI] [PubMed] [Google Scholar]
- 14.Book AJ, Gladman NP, Lee SS, Scalf M, et al. Affinity purification of the Arabidopsis 26S proteasome reveals a diverse array of plant proteolytic complexes. J. Biol. Chem. 2010;285:25554–25569. doi: 10.1074/jbc.M110.136622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russell JD, Scalf M, Book AJ, Ladror DT, et al. Characterization and quantification of intact 26S proteasome proteins by real-time measurement of intrinsic fluorescence prior to top-down mass spectrometry. PloS one. 2013;8:e58157. doi: 10.1371/journal.pone.0058157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deutsch EW, Mendoza L, Shteynberg D, Farrah T, et al. A guided tour of the Trans-Proteomic Pipeline. Proteomics. 2010;10:1150–1159. doi: 10.1002/pmic.200900375. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.