Abstract
Domain dynamics of the chicken gizzard smooth muscle myosin catalytic domain (heavy chain Cys-717) and regulatory domain (regulatory light chain Cys-108) were determined in the absence of nucleotides using saturation-transfer electron paramagnetic resonance. In unphosphorylated synthetic filaments, the effective rotational correlation times, τr, were 24 ± 6 μs and 441 ± 79 μs for the catalytic and regulatory domains, respectively. The corresponding amplitudes of motion were 42 ± 4° and 24 ± 9° as determined from steady-state phosphorescence anisotropy. These results suggest that the two domains have independent mobility due to a hinge between the two domains. Although a similar hinge was observed for skeletal myosin (Adhikari and Fajer (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 9643–9647. Brown et al. (2001) Biochemistry 40, 8283–8291), the latter displayed higher regulatory domain mobility, τr = 40 ± 3 μs, suggesting a smooth muscle specific mechanism of constraining regulatory domain dynamics. In the myosin monomers the correlation times for both domains were the same (~4 μs) for both smooth and skeletal myosin, suggesting that the motional difference between the two isoforms in the filaments was not due to intrinsic variation of hinge stiffness. Heavy chain/regulatory light chain chimeras of smooth and skeletal myosin pinpointed the origin of the restriction to the heavy chain and established correlation between the regulatory domain dynamics with the ability of myosin to switch off but not to switch on the ATPase and the actin sliding velocity. Phosphorylation of smooth muscle myosin filaments caused a small increase in the amplitude of motion of the regulatory domain (from 24 ± 4° to 36 ± 7°) but did not significantly affect the rotational correlation time of the regulatory domain (441 to 408 μs) or the catalytic domain (24 to 17 μs). These data are not consistent with a stable interaction between the two catalytic domains in unphosphorylated smooth muscle myosin filaments in the absence of nucleotides.
Myosins are a superfamily of motor proteins that convert the chemical energy derived from ATP hydrolysis to mechanical energy by generating movement or force. Myosin II, the major protein from muscle, consists of two globular head domains and a coiled-coil tail or rod domain. At physiological ionic strength, myosin forms thick filaments through intermolecular interactions of the tail domains. The globular heads have two distinct domains: a catalytic domain (CD)1 with ATP- and actin-binding properties; and a RD consisting of an α-helix and associated ELC and RLC (1). According to the current working model of muscle contraction during the ATPase cycle, the CD of myosin interacts with the actin filament generating a torque that is transmitted to the thick filament by rotation of the RD (for review see ref 2).
In skeletal muscle, actomyosin ATPase activity is switched on by Ca2+ binding to the thin filament, but in smooth muscle the activity is switched on by phosphorylation of SMM on the RLC at Ser-19. Although skMM can be phosphorylated in vivo, phosphorylation does not regulate the ATPase, but modulates the sensitivity of the muscle to Ca2+ (3). SkMM does not have a stable “OFF” state whereas unphosphorylated SMM has a low actin-activated ATPase and does not support actin movement. Biochemical studies of SMM suggest that interhead and head–S2 interactions play a critical role in the regulatory switch. Two head domains are required for regulation, as single-headed myosin cannot switch off (4). Filament assembly is not required as heavy meromyosin (HMM), a fragment of SMM lacking the filament assembly domain, is fully regulated (5). An HMM molecule with two RDs and only one CD also cannot switch off, suggesting that both CDs are required for regulation (6).
The structural details of the regulatory switch are not known, as an atomic-resolution structure for a two-headed myosin is not available. In smooth HMM phosphorylation increases the sedimentation coefficient (7), stabilizes the heads extended away from the rods at the expense of heads bent back toward the rod (7), increases proteolytic susceptibility of the head–rod junction (8), increases the mobility of the active site and the RLC (9), and increases the distribution of distances between the two heads (10, 11). Together these findings point to an immobilized head–rod junction, with closer heads, and a more compact overall structure for the unphosphorylated versus the phosphorylated state. Studies with calcium-regulated scallop HMM show similar trends induced by calcium binding (12). The two RLC can be cross-linked in the unphosphorylated, but not phosphorylated smooth HMM (13), and phosphorylation causes a large structural rearrangement of the RLC N-terminus (14) to a more exposed and nonpolar environment (15) consistent with recent EPR studies (16). Cryo-EM image reconstructions of nucleotide-bound smooth HMM ordered on a lipid monolayer have visualized contacts between the two CDs that are abolished upon phosphorylation (17, 18). These contacts were not observed for phosphorylated HMM.
Less is known about the structural changes upon SMM phosphorylation in the physiologically relevant filamentous state. SMM filaments have a side-polar structure, as opposed to the bipolar structure of skMM filaments (19, 20). In the absence of ATP, both unphosphorylated and phosphorylated SMM filaments have a disordered head appearance by microscopy, but upon addition of ATP the unphosphorylated SMM heads appear to become ordered on the filament surface (21). Interestingly, vertebrate striated (3, 22) and invertebrate muscles displaying thick filament regulation (22–24) may also undergo similar structural changes from a relaxed (presence of ATP) to an active condition (RLC phosphorylation). The structural basis of these observations has recently been uncovered in nucleotide-bound unphosphorylated tarantula myosin filaments (25), which are also regulated by phosphorylation. The actin-binding surface of one CD contacts the converter region of the other CD, consistent with the above-mentioned studies with HMM. This inter-CD contact is proposed to prevent actin binding of one head and converter rotation of the other head that is necessary for force generation (18). Whether this interaction is stable and required to maintain myosin in the OFF state is a matter of debate (11).
The inter-CD interactions in both smooth muscle HMM and tarantula filaments, as described above, were observed in the presence of ATP. However, it is well documented that phosphorylation can induce structural changes that are independent of ATP-binding at the active site. Structural differences between the phosphorylation states of smooth HMM have been detected in the absence of the motor domains using both cross-linking (13, 14) and fluorescence anisotropy (9). Nucleotides have little effect upon the distribution of the relative head-to-head positions of both phosphorylated and unphosphorylated SMM at low ionic strength as determined by atomic force microscopy (11). Large structural differences between the conformation of the RLC N-terminal domain are observed in the absence of nucleotides (15, 16), although nucleotide binding can enhance the structural differences (15). Two measures that are consistent with partial release of cross-bridges from the vertebrate skMM filament surface by phosphorylation, the phosphorylation-induced decrease in the chemical cross-linking rate between the S2 and the filament surface and the increase in the proteolytic susceptibility of the HMM-LMM junction, are observed without nucleotides (26).
The objective of this study was to use ST-EPR and steady-state phosphorescence anisotropy to characterize RD and CD mobility of both SMM and skMM filaments in the microsecond time domain. This was a logical first step in the study of SMM filament dynamics in light of our prior work on skMM filament dynamics (27–30). ST-EPR and steady-state phosphorescence anisotropy are complementary to EM methods (mentioned above) in that the nature of the helical order on the filament surface can be deduced, that is, static versus dynamic. The experiments in this study were performed in the absence of nucleotide (rigor) because SMM filaments, unlike those of skMM, will disassemble into a mixture of monomers and filaments in the presence of ATP. The current study is a prerequisite to a study of nucleotide effects. Since phosphorylation effects have previously been noted under rigor conditions (see above), we also examined the effects of phosphorylation.
We observed differential dynamics between the RD and CD of SMM, indicating that there is a flexible interdomain hinge, as we had previously shown for skMM (27). The SMM RD was found to be considerably less mobile than the skMM RD. This relative motional restriction was limited to the RD as the CD of the two isoforms had similar mobility. The intrinsic stiffness of the hinge between the RD and CD appeared to be similar in monomeric myosin. The differences between the isoforms are defined by the HC rather than the RLC as suggested by chimeric myosin mobilities. Restricted RD dynamics in SMM correlated with the ability to adopt the switched-off, but not the switched-on, state. Phosphorylation did not significantly affect the RD correlation time but caused a small increase in the amplitude of RD motion that might be related to the release of enzymatic inhibition.
MATERIALS AND METHODS
Protein Purification
SMM and RLC were purified from frozen chicken gizzards (31, 32), and skMM was purified from the back muscle of adult white New Zealand rabbits (33). A single cysteine mutant of skeletal RLC, in which Cys-125 was replaced with an arginine residue, leaving a single Cys-154 (RLC154), was expressed in Escherichia coli and purified by ion exchange on Q-Sepharose (Amersham Biotech) (30).
Spin Labeling, RLC Exchange, and Phosphorylation
The labeling of CD of the SMM was carried out at 22 °C for 1 h in 20 mM Tris, 0.4 M KCl, 2 mM MgCl2, pH 7.5 with the equimolar concentrations of InVSL (indane dione spin label, courtesy of K. Hideg, U. Pecs, Hungary) and protein. The unreacted spin label was removed by dialysis against myosin filament buffer (50 mM KCl, 0.1 mM EGTA, 10 mM MgCl2, 15 mM Tris, pH 7.5). Labeling with fluorescent probes under similar conditions resulted in modification of Cys-717 with some labeling of light chains and the myosin rod (34). To quantify and localize domain labeling we digested spin-labeled myosin with papain and compared the extent of labeling in myosin and subfragment 1. The initial ratio of 0.69 spin label per head in myosin decreased to 0.59 in S1. Thus, the labeling of the rod was about 15%. The extent of labeling of the light chains was estimated by removal of the RLC (with trifluoroperazine) or both light chains (with ammonium chloride and TFP) (35). The 95% removal of light chains resulted in the 15% loss of the signal, suggesting that no more than 16% of the spin label was on the light chains. Thus, ~70% of the InVSL was localized to the CD.
Chicken gizzard regulatory light (gRLC) was labeled at Cys-108 as previously described (31). To ensure complete reduction of all sulfhydryls, the gRLC was incubated for 2 h at room temperature in 6 M guanidine HCl, 10 mM DTT, and 100 mM Tris, pH 8.0. After dialysis to remove DTT (100 mM NaCl, 2 mM EDTA, and 20 mM MOPS, pH 7.0), a 5-fold excess of InVSL was added and allowed to react at 22 °C for 2–4 h. Unreacted label was removed by extensive dialysis at 4 °C. Skeletal RLC154 and skeletal myosin SH1 (707) were labeled as described previously (27, 29, 30). Labeling levels for CD of SMM, smooth RLC108, skeletal RLC154, and skeletal SH1 are 0.65–0.7, 0.9–1.0, 0.6–0.9, and 0.35 labels per myosin head for RLC.
Labeled RLC was exchanged onto SMM at 10-fold molar excess over myosin (2 mg/mL), in 500 mM KCl, 2 mM ATP, 30 mM HEPES, pH 7.5, 10 mM EDTA buffer at 42 °C for 30 min. The exchange was terminated by addition of 10 mM MgCl2 followed by dialysis against filament buffer. Thiophosphorylation of the exchanged SMM was accomplished by adding 30 μg/mL myosin light chain kinase, 4 μg/mL calmodulin, 0.5 mM ATPγS (Sigma) in 50 mM NaCl, 2.5 mM MgCl2, 2.5 mM CaCl2, for 1 h at pH 7.0, 25 °C (36). Thiophosphorylation levels were found to be higher than 90%, as verified by 8 M urea gel electrophoresis. The steady-state actin-activated ATPase activity of RLC108-labeled SMM was assayed as a rate of Pi formation measured with malachite green. The reaction was carried out in 50 mM KCl, 15 mM Tris, pH 7.5, 5 mM MgCl2, 0.1 mM EGTA, and 1 mM ATP at 25 °C in the presence of 25 μM actin (13). The activity of SMM was 3 nmol min–1 mg–1 for unphosphorylated myosin, with 10-fold increase upon thiophosphorylation (33 nmol min–1 mg–1). As found before, the RLC exchanges or modification (at Cys-108) had little effect on ATPase regulation (13). Activities of SMM labeled on the CD were measured by single turnover of formycin triphosphate in the presence of 5 and 12.5 μM actin, 100 μM ATP, 0.4 mM MgCl2, 10 mM MOPS (pH 7.0), 150 mM NaCl, 0.1 mM EGTA at 25 °C as described in ref 6 except that the emission passed through a P70 filter ThermoCorion. Data were collected for 2000 s and 500 s for the unphosphorylated and phosphorylated samples, respectively, and were fit to a biexponential model (6). Figure 1 shows the calculated rates and amplitudes of the single turnover ATPase measurements (6). The control values show a fast phase that is not seen in fully regulated samples, which is likely to be due to age of the samples as previously shown (15) and the fact that they were prepared in the absence of DTT. The values for unphosphorylated and phosphorylated InVSL-labeled myosins are similar to control values except that the faster of the two phases represents a greater fraction of the total amplitude. Therefore, although the presence of the InVSL probe alters the kinetics somewhat, there is no evidence that the InVSL-labeled fraction of the sample is unregulated. A sufficient level of regulation remains to examine the effects of phosphorylation on domain dynamics.
Figure 1.
Actin-activated single turnover rates and amplitudes of unlabeled (control) and InVSL-labeled SMM. Single turnover rates were fit to a double exponential model. The faster of the two derived rates is shown in solid black bars, and the slower is shown as cross-hatched bars. The respective amplitudes for each derived rate are shown as gray bars and open bars. The unphosphorylated samples (left) and phosphorylated samples (right) were measured at two actin concentrations (5 and 12.5 μM). Each value is the average of at least 3 measurements per sample. Variations in both rates and amplitudes were approximately ±5%. Total amplitudes of all measurements were similar.
EPR and ST-EPR Experiments
Mobility in filaments was measured in 10 mM MgCl2, 15 mM Tris pH 7.5, 0.1 mM EGTA, and 50 mM KCl, and 6S extended monomeric myosin was measured in 0.5 M KCl, 10 mM MOPS pH 7.0, 0.1 mM EDTA. EPR experiments were performed on a Bruker EMX spectrometer (Bruker Inc., Billerica, MA) using a TE102 cavity. Conventional EPR spectra were recorded at a microwave power of 0.032 G microwave field and a modulation field of 2.5 G, while saturation transfer EPR spectra (ST-EPR, second harmonic absorption, out-of-phase) were obtained at a saturating microwave field of 0.25 G with modulation amplitude of 5 G. All experiments were performed at 4 °C. The effective τr was determined from the diagnostic ratio of intensities, L″/L, of ST-EPR spectra calibrated using the empirical equation τr = 10(L″/L+0.21)/0.39 obtained from a standard curve of spin-labeled hemoglobin tumbling in media of known viscosity (87.55% w/w, glycerol/water mixture, at a range of temperatures between –30 to +20 °C) (37). The rigid limit of the spin label was determined from the spectrum of protein immobilized by cross-linking to preactivated DITC glass beads (Sigma Chemical Co., St. Louis, MO). The cross-linking was carried out overnight, at an equimolar ratio of protein and DITC beads at pH 7.0 with unreacted protein removed by repeated washing and sedimentation of the glass beads.
Labeling with Phosphorescent Probes
SH1 (Cys-717) of SMM was labeled with E-5-IA (Molecular Probes, OR) by incubation of myosin filaments (2 mg/mL) with a 4-fold molar excess of label for 30 min at 4 °C in 100 mM NaCl, 2 mM EDTA, 20 mM MOPS, pH 7.0. The filaments were dissolved in 1 mL of 0.5 M KCl, 2 mM EDTA, 10 mM MOPS, pH 7.0, and unconjugated probe was removed on Sephadex G-25 Fine (2 × 20 cm) followed by dialysis against 1 mM EDTA, 10 mM MOPS, pH 7.0 to re-form filaments.
RLC was labeled as described by Brown et al. (30) and modified for chicken gRLC. gRLC (~5 mg/mL) was incubated for 2 h at 22 °C in reducing buffer (6 M guanidine-HCl, 10 mM DTT, 2 mM EDTA, 50 mM Tris-HCl, pH 7.0), followed by dialysis against 100 mM NaCl, 2 mM EDTA, 50 mM Tris-HCl, pH 7.0 to remove DTT and denaturant. The sample was then reacted with a 3-fold molar excess of E-5-IA at 4 °C overnight. Unconjugated probe was removed by Sephadex G-25 gel filtration and extensive dialysis.
Phosphorescence Anisotropy Measurements
Phosphorescence anisotropy was measured with a Perkin Elmer LS50B fluorimeter equipped with a red-sensitive photomultiplier (R928, Hamamatsu), emission and excitation polarizers. The E-5-IA-labeled SMM filaments (2 mg/mL, 10 mM MgCl2, 0.1 mM EGTA and 50 mM KCl, 10 mM MOPS pH 7.5) were excited at 480 nm, and the emission intensity was monitored at 690 nm with a 1.5 ms integration time window and an initial delay of 0.04 ms to avoid fluorescence. To minimize the effects of instrumental drift we collected each of four polarizations (IVV, IVH, IHV, and IHH, where the first subscript denotes polarization of the excitation and the second subscript refers to the emitted light) for 5 s followed by rotation of polarizers. Three hundred cycles were required for the 10% accuracy in final anisotropy. Oxygen was removed from the sample before the phosphorescence measurement by addition of glucose oxidase (30 μg/mL), glucose (45 μg/mL), and catalase (20 μg/mL). The surface of the sample was flushed continuously with argon for 15–20 min prior to data collection to maintain an oxygen-free environment. Steady state phosphorescence anisotropy (r) was calculated from the emission of vertically (IVV) and horizontally (IVH) polarized light according to
| (1) |
where G is an instrumental correction factor (G = IHV/IHH). Initial anisotropy, r0, was estimated from a Perrin plot of the steady-state anisotropy as a function of solvent viscosity (38):
| (2) |
η is the viscosity of the solution, R is the gas constant, τ is the phosphorescence lifetime, T is the absolute temperature, and V is the volume of the molecule. The viscosity was varied from 1.004 to 2.642 cP by varying sucrose concentration between 5 and 25% (wt/wt) and the temperature between 5 and 20 °C (38). A plot of 1/r versus 1/η is linear for isotropic motion with an ordinate intercept corresponding to 1/r0. The cone half-angle (θc) of the probe motion was estimated from the ratio of the initial anisotropy and anisotropy in the absence of sucrose (39, 40):
| (3) |
RESULTS
InVSL as a Probe for Domain Motion
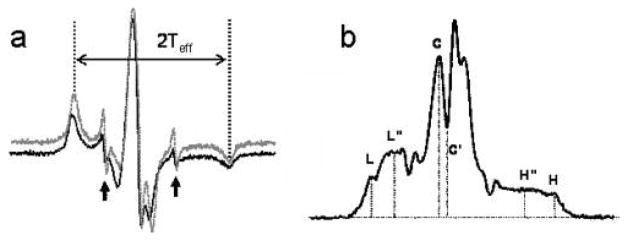
When external probes are used to report global or domain protein motions, it is necessary to ascertain that the probe does not move with respect to the protein. InVSL has been previously found to be fully immobilized on a number of proteins, e.g., skeletal myosin S1 and ELC (27, 28), RLC (29), troponin C and troponin I (41). To check the immobilization of InVSL on Cys-108 of gRLC we have cross-linked the InVSL-RLC to DITC-coated glass beads. Label immobilization was first determined in the nanosecond time scale with conventional EPR, which is sensitive to motions with τr = 0.01 to 100 ns. The conventional EPR spectrum of the cross-linked protein has a broad line shape with splitting, 2Teff, of 70.9 ± 0.2 G (Figure 2a, lower spectrum). The value of the splitting is at the rigid limit for a nitroxide spin label, indicating full immobilization on the nanosecond time scale. Equally immobilized was the spectrum of SMM exchanged with InVSL-RLC, 2Teff = 70.7 G (Figure 2a, upper spectrum). The small peaks near the center field (indicated by arrows in Figure 2a) arise due to free nitroxide cleaved from the protein in the retro-Michael reaction of InVSL. This unconjugated InVSL contributed less than 5% of the total intensity and did not affect interpretation of the spectra.
Figure 2.
(a) Conventional EPR spectra of InVSL-RLC exchanged onto SMM (upper spectrum) and bound to DITC glass beads (lower spectrum). (b) ST-EPR spectrum of InVSL-RLC bound to DITC glass beads.
Label immobilization is also required for measurements in the microsecond time scale of saturation transfer EPR (ST-EPR). The ST-EPR spectrum of DITC-immobilized InVSL-RLC is shown in Figure 2b. Typically, in a ST-EPR spectrum, the signal intensity at field positions L″, C′, and H″ decreases with the increasing motion while the intensity at L, C, and H remains constant. The diagnostic line height ratios L″/L, C′/C, and H″/H are used to estimate the effective correlation time, τr, by comparison to a standard curve from labeled hemoglobin tumbling in media of known viscosity (42). Here, we report τr calculated from the low-field region (L″/L), which is less influenced by spectral overlap than the central field (C′/C), and has higher signal-to-noise ratio than the high field region (H″/H). The ST-EPR spectrum of immobilized RLC has a L″/L of 1.6, which yields a τr greater than 1 ms, the rigid limit of ST-EPR. Thus, InVSL is rigidly attached to the RLC with no independent motion on the nanosecond (EPR) and the microsecond time scales (ST-EPR).
Dynamics in SMM: RD vs CD
The ST-EPR spectra of the CD and RD of unphosphorylated SMM filaments in the absence of nucleotide are shown in Figure 3. The ratio L″/L = 1 for the RD is much higher than that for the CD, L″/L = 0.52 (Table 1). These line shape parameters translate into an effective correlation time τr of ~441 μs for the RD and 24 μs for the CD (Table 2). In the case of the CD, 70% of the label was specifically localized to the CD and 30% nonspecifically to the ELC (in the RD) and the rod (see Materials and Methods). Since the RD and rod have slower motion than the CD, the values reported in Table 1 for the CD must somewhat underestimate the mobility. Therefore there is at least a ~20-fold difference in mobility between the CD and RD, which indicates the presence of a hinge between the two domains. For comparison, the corresponding hinge in the skMM filaments resulted only in ~4-fold difference in mobility.
FIGURE 3.
ST-EPR spectra of the catalytic and RD of SMM in filaments. 70% of the spin probe is attached to the CD in panel a and 100% to Cys-108 of RLC in the RD in panel b.
Table 1.
Saturation Transfer EPR Line-Height Ratios for SkMM and SMM
| sample | regulatory domain
|
catalytic domain
|
||
|---|---|---|---|---|
| unphos | phos | unphos | phos | |
| smMM filamenta | 1.02 ± 0.03 (11)b | 1.00 ± 0.01 (3) | 0.52 ± 0.04 (7) | 0.47 ± 0.06 (2) |
| skMM filamenta | 0.61 ± 0.01 (3) | 0.38 ± 0.03 (3) | ||
| smMM monomer (6S) | 0.23 ± 0.02 (3) | 0.19 ± 0.02 (4) | ||
| skMM monomerc | 0.23 ± 0.02 (3) | 0.23 ± 0.02 (3) | ||
SMM filaments are side-polar, skMM filaments are bipolar.
Number of independent measurements.
Data from ref 27.
Table 2.
Motional Parameters for SkMM and SMM
| sample | motional param | regulatory domain
|
catalytic domain
|
buffer | ref | ||
|---|---|---|---|---|---|---|---|
| UPa | Pa | UP | P | ||||
| SMM filamentb | corr time (μs) | 441 ± 79 (11)c (Cys 108 RLC) | 408 ± 54 (3) | 24 ± 6 (7) (Cys 717) | 17 ± 7 (2) | 10 mM MgCl2, 15 mM Tris pH 7.5, 0.1 mM EGTA, 50 mM KCl, 4 °C | this work |
| cone angle | 24 ± 9° (Cys 108 RLC) | 36 ± 7° | 42 ± 4° (Cys 717) | 10 mM MgCl2, 10 mM MOPS pH 7.5, 0.1 mM EGTA, 50 mM KCl, 4 °C | this work | ||
| skMM filamentb | corr time (μs) | 40 ± 3 (3) (Cys 154 RLC) | 10 ± 2 (3) (Cys 707) | 40 mM KCl, 10 mM MOPS, pH 7.0, 4 °C | this work | ||
| 36 ± 12 (Cys 177 ELC) | 6.9 ± 1.5 (Cys 707) | 40 mM KCl, 10 mM MOPS, pH 7.0, 4 °C | 27 | ||||
| 26.2 ± 2.0 (4) (Cys 177 ELC) | 16.9 ± 8 (4) | 50 mM MOPS, 0.1 mM DTT, pH 7.0, 4 °C | 28 | ||||
| cone angle | 29 ± 8° (Cys 177 ELC) | 51 ± 5° (Cys 707) | 120 mM KCl, 5 mM MgCl2, 1 mM EGTA, 25 mM MOPS, pH 7.0, 12 °C | 30 | |||
| 27 ± 6° (Cys 154 RLC) | 10 mM MgCl2, 15 mM Tris pH 7.5, 0.1 mM EGTA, 50 mM KCl, 4 °C | this work | |||||
| SMM monomer (6S) | corr time (μs) | 4.4 ± 0.5 (3) (Cys 108 RLC) | 3.4 ± 0.2 (4) (Cys 717) | 0.5 M KCl, 10 mM MOPS pH 7.0, 0.1 mM EDTA, 4 °C | this work | ||
| skMM monomer | corr time (μs) | 4.1 ± 0.5 (3) (Cys 154 RLC) | 4.4 ± 0.6 (3) (Cys 707) | 0.6 M KCl, 20 mM MOPS, pH 7.0 | 27 | ||
| skMM rods | corr time (μs) | 31000 μs (rod domain) | 40 mM KCl, 10 mM MOPS pH 7.0, 4 °C | 27 | |||
UP, unphosphorylated; P, phosphorylated.
SMM are side-polar, skMM filaments are bipolar.
Number in parentheses is the number of independent measurements.
In the monomeric form, the SMM CD and RD domains have essentially the same mobility (Table 2), which was also observed for skMM (27, 30). The similarity of the correlation times for the two monomeric isoforms suggests that the intrinsic hinge stiffness is similar and cannot account for the large difference between the two isoforms in filaments. Note that the difference is also not due to the head–tail interaction of the hairpin-folded 10S monomer, as our conditions (no ATP, high ionic strength) favor an extended 6S monomer (43).
RD Dynamics of Different Isoforms: SMM vs skMM
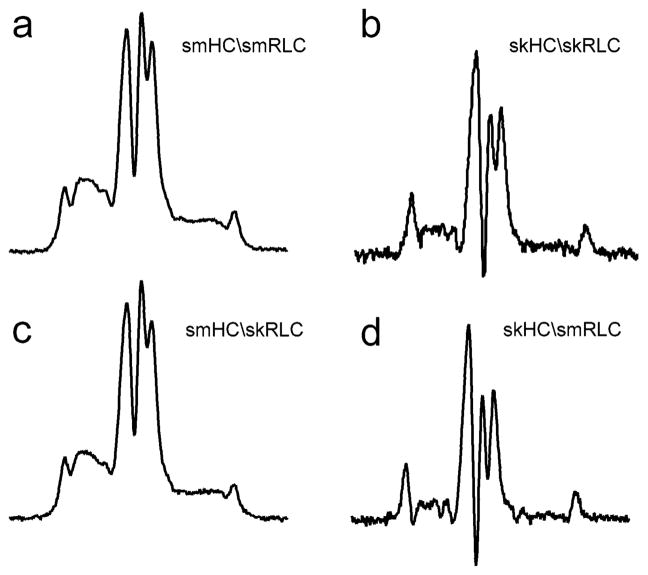
The RD dynamics of skMM filaments has been previously determined by ST-EPR with spin labels on the ELC (27) and RLC (29) (Table 2). The rotational correlation times were similar, implying that the RD is fairly rigid, thus allowing force transmission from the CD (29). The mobility of the RD of SMM is much slower than that of skMM as implied by higher intensity at diagnostic L″, C′, and H″ positions, see Figure 4a,b. The effective correlation times were 441 μs for SMM and 40 μs for skMM (Table 2). In contrast, the CD mobilities were relatively similar between the two isoforms (Table 2).
Figure 4.
ST-EPR spectra of the smooth and skeletal RLC reconstituted into a smooth or skeletal HC background: (a) smooth RLC on smooth HC; (b) skeletal RLC on skeletal HC; (c) chimera of skeletal RLC and smooth HC; and (d) chimera of smooth RLC and skeletal HC. Estimated rotational correlation times (τr) are given in Table 3.
Differences between the ST-EPR correlation times can arise from different orientations of the spin label with respect to the anisotropic diffusion axis (44). For example, if the z-axis of the nitroxide coincides with the molecular rotation axis, the resulting correlation time will be much higher (slower rate) than if it is perpendicular. This has to be considered whenever labels on different residues are compared. In this case, the labeled RLC residues are Cys-154 for skMM and Cys-108 for SMM. The different steric environments in the two light chains might orient the label at different angles with respect to the myosin. To check for such a possibility, we substituted the smooth RLC with a skeletal RLC on the SMM HC. The ST-EPR spectrum of this chimera (smHC/skRLC) in Figure 4c has the same line shape as the spectrum from native smHC/smRLC filaments in Figure 4a. Thus, the observed difference between SMM and skMM is not the result of different label orientations at sites of smRLC108 and skRLC154 but reflects a genuine difference in protein dynamics. Furthermore, the results suggest that the RLC mobility is dictated by the HC rather than the RLC. Whatever the mechanism that immobilizes RLC in SMM, the same mechanism appears to persist with the skeletal RLC exchanged onto SMM. A reverse chimera with smooth RLC exchanged onto skMM (skHC/smRLC) has a line shape similar to the native skHC/skRLC spectrum (Figure 4d and Figure 4b). Thus, the following characterizes the large difference in RD mobility between the two myosin isoforms in filaments: (1) the difference is only for the RD; the CDs have similar mobility; (2) the difference is not due to intrinsic stiffness of the hinge; (3) it is an intrinsic property of the myosin molecule, not a result of different spin label orientations and labeled sites; and (4) the RD dynamics is dictated by the type of HC.
Effects of Phosphorylation
The ST-EPR spectra of un-phosphorylated and thiophosphorylated SMM filaments are shown in Figure 5, and their line height ratios are in Table 1. There was no significant difference in the ST-EPR spectra for the unphosphorylated (441 μs for the RD) versus the phosphorylated state (408 μs). The extent of phosphorylation was better than 90% as judged by gel electrophoresis (inset Figure 5), and the actin-activated ATPase was partially regulated (Figure 1), ruling out a trivial explanation for the observed similarity.
Figure 5.
Overlaid ST-EPR spectra of thiophosphorylated (gray dotted line) and unphosphorylated (black solid line) SMM filaments. The spin label InVSL was attached to RLC Cys-108.
The absence of a large effect of phosphorylation on the RD motion might be ascribed to changes in the amplitude of motion rather than the rate. ST-EPR spectra are not sensitive to the amplitude of motion if the amplitude is large enough >30° (45). Thus, to look for amplitude effects we have utilized phosphorescence anisotropy, a technique that is capable of differentiating rate and amplitude effects. The ratio of final and initial anisotropy is proportional to the amplitude of restricted motion (39, 40). The final and initial anisotropy values were obtained from samples in aqueous and sucrose solutions by extrapolation of Perrin plots to infinite viscosity, Figure 6 (46). For unphosphorylated SMM, the half-cone angle θc through which the probe rotates was 42 ± 4° for the CD and 24 ± 9° for RLC. These values are similar to the amplitudes for skMM, 51 ± 5° and 27 ± 6° for CD and RLC, respectively (30) (Table 2). The phosphorylation of SMM increased the amplitude of the RD to 36 ± 7°, a 1.5-fold increase with respect to the unphosphorylated state.
Figure 6.
Perrin plots of phosphorescence emission anisotropy as a function of solvent viscosity: SH1 (●), unphosphorylated RLC (○), and phosphorylated RLC (▼) in myosin filaments.
There was little effect of phosphorylation on the SMM CD correlation time, which decreased from 24 ± 6 to 17 ± 7 μs (Table 2). The SMM CD, whether in the phosphorylated or unphosphorylated state, displays dynamics similar to skMM CDs (τr = 10 μs) for which there is no evidence for intramolecular interactions. We expect that the CD motion should accelerate by more than a factor of 2 if phosphorylation were to release a stable intramolecular interaction between the two SMM CD. This is because noninteracting heads should diffuse independently with half the mass of dimerized heads and, more importantly, the coupling between the CD and RD should decrease, thereby increasing the independent motion of the CD and RD. A 2-fold decrease in correlation time would result in a decrease of the L″/L ratio by 0.12, well above the sensitivity and experimental error of the measurement. Therefore our data show that phosphorylation does not cause a major change in the intramolecular interactions between the two SMM CD under these conditions. The single turnover ATPase data in Figure 1, showing regulated activity, excludes the possibility that the SH1-labeled SMM is permanently switched on and does not form the inhibited complex.
DISCUSSION
We have characterized the CD and RD dynamics of SMM in filaments (the physiologically relevant state found in muscle tissue) with ST-EPR and phosphorescence anisotropy. We found that the CD mobility is faster and with larger amplitude than the RD mobility thus implying a hinge between the two domains. Furthermore, the motion of the unphosphorylated and phosphorylated RD is far more (~20-fold) restricted in SMM than in skMM. This restriction is dictated by the HC, as the mobility of HC/RLC chimeras consisting of smooth and skeletal myosin isoforms correlated with the HC origin, not with RLC origin. The observed RD immobilization might be functionally relevant because phosphorylation increases the amplitude of motion, which in turn might enhance contact of the heads with actin.
A Flexible Hinge Exists between the CD and RD in SMM
Our data provide strong support for a hinge between the SMM CD and RD. Rayment et al. (1) proposed this hinge from the crystal structure of skeletal S1. Head flexibility afforded by this hinge is an essential property of cross-bridges, allowing strain energy to be stored and dissipated gradually as proposed by Huxley and Simmons (47). The intradomain flexibility has been observed by EM and single-particle analysis (48), phosphorescence anisotropy (30) and ST-EPR (27). The comparison of crystal structures obtained with various nucleotide analogues suggests that the point of flexibility is localized between the converter domain (within the CD) and the RD (49, 50). This report is the first direct study of SMM dynamics to show that the two domains have independent mobility. It follows that the various RD conformations observed in crystals of SMM heads might reflect the dynamic nature of the RD rather than static conformations induced by the nucleotide state.
Mechanism of RD Motional Restriction in Filaments
Both the RD and CD of SMM and skMM myosin are more restricted in filaments than in 6S monomers (Table 2). This is due to dampening of the head motion by the larger filament structure. However, the transition from monomers to filaments in SMM slows down the RD to a much greater extent than the CD (100-fold vs 7-fold change). The corresponding changes for skMM were 10-fold and 2-fold restriction of the RD and CD mobility.
The rotational correlation time of the skMM rod in filaments is ~1 ms (L″/L = 1.4) (27), in a similar motional regime as the SMM RD. The similarity between the RD and the rod domain motion most likely reflects their interaction (Table 3). This interaction seems to be so strong that phosphorylation is insufficient to fully displace the RD from the filament surface under these conditions. Many lines of evidence suggest that phosphorylation weakens the interaction of S2 with the filament surface and that for many different myosin isoforms, including smooth and skeletal, causes a transition from a relatively ordered filament to one in which the heads appear to be disordered. Here we observe relatively small effects of phosphorylation on RD dynamics. This suggests that the disordered heads seen in the phosphorylated state remain tightly coupled to the motion of the filament backbone perhaps by restriction of the hinge at the head–tail junction, or head–head interaction. However, since the RD motion is much slower than the CD motion, a mechanism that selectively restricts the RD is required.
Table 3.
Comparison of Dynamics and Functions of Myosin and Myosin Chimeras
| myosin | τr of regulatory domaina (μs) | in vitro motility rate UP/Pb (μm s−1) | actin-activated ATPase UP/P (s−1) | regulation UP/P |
|---|---|---|---|---|
| smHC/smRLC | 441 ± 79 (11) | ~0/0.8 ± 0.2c | 0.05/0.57c 0.005/0.2d |
OFF/ON |
| smHC/skRLC | 321 ± 114 (3) | ~0/~ 0c | 0.03/0.06c | OFF/OFF |
| skHC/skRLC | 40 ± 3 (3) | 8.8 ± 1.4e | 1.28 f | ON/ON |
| skHC/smRLC | 36.6 ± 5 (3) | 7.6 ± 1.07e | 1.7/1.8d | ON/ON |
Under ionic conditions used in this work, SMM forms side-polar filaments (20, 51) in which the cross-bridges have the same polarity along one side of the filament with the reverse polarity on the opposite side. This is in contrast to the bipolar filament formed by skMM with helical arrangement of heads with a central bare zone of polarity reversal (52). It is possible that the mobility difference between SMM and skMM filaments is due to different filament packing. However, this is unlikely as the heads in the side-polar filaments protrude from the filament surface while the heads are splayed on the surface in bipolar filaments (53). Thus, the heads in SMM filaments should be more mobile than those of skMM, which was not observed.
RD Dynamics Are Correlated to the Ability To Adopt the Switched-Off State
Table 3 shows a comparison of RD rotational dynamics, the actin sliding velocity, and the ATPase activities of SMM, skMM, and 2 HC/RLC chimeras. Slow RD rotational dynamics is characteristic of the two isoforms that exhibit slow in vitro motility rates and low ATPase activities in the unphosphorylated state (smHC/smRLC and smHC/skRLC). The two isoforms that cannot adopt a switched-off motility or ATPase in the unphosphorylated state exhibit much faster RD motion (skHC/skRLC and skHC/smRLC). Therefore the ability to adopt the switched-off state and slow RD motion are correlated. In contrast to RD dynamics, the CD dynamics does not appear to be correlated with the ability to adopt a switched-off state. Table 2 shows that the CD correlation times (24 and 10 μs) and cone angles (42° and 51°) are similar for the unphosphorylated SMM (OFF) and skMM (ON).
RD Dynamics Are Not Correlated to the Ability To Adopt the Switched-On State
A key question is whether the effects of phosphorylation upon the RD motion are correlated with activation of both the actin sliding motility and the ATPase activity. Even though SMM filaments are activated by phosphorylation, phosphorylation was not sufficient to increase the RD dynamics from an extremely slow regime (τr = 408 μs) to a fast regime as found in skMM (16.9 μs). This suggests that RD motion is not correlated with the ability of the molecule to be turned on by phosphorylation. However, we were able to detect effects of phosphorylation on the dynamics of the smooth RD and the CD (Table 2). The RD cone angle was increased by a factor of 1.5, and the CD correlation time was slightly shorter. For skeletal filaments, phosphorylation decreased the RD correlation time (16.9 μs) from the already low value for the unphosphorylated state (26.2 μs). Therefore, it appears that phosphorylation tends to increase the dynamic motion of both SMM and skMM to some extent; however, phosphorylation does not lead to the same dynamic behavior for these two turned-on molecules. Thus, RD dynamics is not correlated with the ability to adopt the ON state.
Structural Models of Heavy Meromyosin: Implications of Dynamics Studies
Cryo-EM images of unphosphorylated HMM in the presence of ATP suggest that one head of two-headed myosin binds to the partner head (17, 18). The binding is at the level of CDs with the first head docking its actin-binding domain into the converter domain of the second head. Such interacting heads were suggested to confer the “OFF” state as the first head cannot bind to actin while the second head cannot rotate the converter domain that might be necessary for product release. The head–head interaction was not observed in phosphorylated HMM (17, 18). If the head–head interaction observed in unphosphorylated HMM is sufficiently stable, the CD and RD would have a very restricted mobility in the unphosphorylated but not in the phosphorylated state. We do not observe this behavior. This does not necessarily mean that our findings are inconsistent with the Taylor model (17, 18). It is important to emphasize that our data were obtained in filaments, without ATP, while the EM model was from HMM with ATP. Therefore, our study may not be sensing the same state. However, others have shown that even in the absence of nucleotide there are phosphorylation-induced changes in smooth HMM (see introductory paragraphs). But it is clear that nucleotide at the active site biases the structure more toward an OFF state. Mazhari et al. (15) showed that the environment of the RLC N-terminus (phosphorylated or not) in the absence of nucleotides was intermediate between the nucleotide bound unphosphorylated and phosphorylated states in HMM. The fact that we see little effect of phosphorylation on the dynamics in the absence of nucleotides is consistent with the general trend of the Mazhari et al. data (15). Therefore, the Taylor model (17, 18) may represent a state that is more biased toward the OFF structure than the state that we are measuring in the absence of nucleotides. Several other studies also conclude that there are at least two conformations of myosin in equilibrium that are allosterically regulated by nucleotide, phosphorylation (15), or Ca2+ binding (54). Rosenfeld et al. (9) have shown that phosphorylation increases side chain or backbone RD motion (nanosecond time scale) of a smooth HMM-like construct lacking both CDs. Our result that phosphorylation modulates the motional restraint of the RD but does not abolish it is reasonably consistent with these studies. If RLC phosphorylation were to abolish the interhead or head–filament interactions and make SMM look like skMM (which does not have the “OFF” state), we would expect a ~10-fold increase in the rate of the RD motion. This was not observed.
The similarity of the RD mobility irrespective of the origin of the RLC (whether skeletal or smooth) implies that the restriction is not RLC specific. The difference in filament organization between smooth and skeletal myosin most likely persists in the chimera, such that the skeletal RLC exchanged into SMM HC are interacting with each other while there is no inter-RLC interaction within skMM HC.
In conclusion, the dynamics of the SMM RD is more restricted than that of skMM in filaments whereas the dynamics of the CDs are similar. The different RD dynamics was not due to a difference in the intrinsic hinge stiffness between the two isoforms, as the mobility of the monomers was the same. Formation of filaments introduced new interactions, most likely with the filament surface or with the other head. RD motional restriction was mediated by the HC as the dynamics of chimeric molecules correlated with the HC origin. Phosphorylation caused a small increase in the amplitude of RD motion without change in the rate of motion. Data for the CD suggest that there is probably not a static interaction of the two CDs in unphosphorylated SMM filaments in rigor. The CD is already quite mobile, and the mobility is only moderately increased by phosphorylation. Phosphorylation may increase the access of the head to actin in a native filament, but it does not release the strong interaction of the RD with the filament backbone and/or the RD of the other head.
Acknowledgments
We thank Drs. Louise Brown for the initial help with phosphorescence anisotropy and Olivia Henderson-Hall for technical assistance.
Footnotes
This material is based upon work supported by the National Science Foundation under Grant No. 0346650, the AHA GIA 0455236B to P.G.F., and NIH AR40917 to C.R.C. and TCMRC 90103 to H.-C.L. L.S. is a recipient of American Heart Graduate Fellowship.
Abbreviations: CD, catalytic domain; RD, regulatory domain; SMM, smooth muscle myosin; skMM, skeletal muscle myosin; HMM, heavy meromyosin; S1, myosin subfragment 1; RLC, regulatory light chain; RLC154, recombinant skeletal regulatory light chain with single cysteine at position 154; gRLC, native regulatory light chain purified from chicken gizzard; ELC, essential light chain; HC, heavy chain; EM, electron microscopy; ATP, adenosine 5′ triphosphate; ATPγS, adenosine-5′-O-(3-thiophosphate); DTT, dithiothreitol; EDTA, ethyl-enediamine tetraacetic acid aminoethyl ether; EGTA, ethylene-glycol-bis-N,N,N′,N′-tetraacetic acid; E-5-IA, eosin-5-iodoacetamide; HEPES, N-(2-hydroxyethyl)piperazine-N′-(2-ethanesulfonic acid); InVSL, 2-(oxyl-2,2,5,5-tetramethyl-3-pyrrolin-3-methynyl)indane-1,3-dione; MOPS, 3-(N-morpholino)propane-sulfonic acid; Tris, Tris (hydroxymethyl)-aminomethane; DITC, di-isothiocyanate.
References
- 1.Rayment I, Rypniewski WR, Schmidt-Base K, Smith R, Tomchick DR, Benning MM, Winkelmann DA, Wesenberg G, Holden HM. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993;261:50–8. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 2.Geeves MA, Holmes KC. Structural mechanism of muscle contraction. Annu Rev Biochem. 1999;68:687–728. doi: 10.1146/annurev.biochem.68.1.687. [DOI] [PubMed] [Google Scholar]
- 3.Sweeney HL, Bowman BF, Stull JT. Myosin light chain phosphorylation in vertebrate striated muscle: regulation and function. Am J Physiol. 1993;264:C1085–95. doi: 10.1152/ajpcell.1993.264.5.C1085. [DOI] [PubMed] [Google Scholar]
- 4.Cremo CR, Sellers JR, Facemyer KC. Two heads are required for phosphorylation-dependent regulation of smooth muscle myosin. J Biol Chem. 1995;270:2171–5. doi: 10.1074/jbc.270.5.2171. [DOI] [PubMed] [Google Scholar]
- 5.Ellison PA, Sellers JR, Cremo CR. Kinetics of smooth muscle heavy meromyosin with one thiophosphorylated head. J Biol Chem. 2000;275:15142–51. doi: 10.1074/jbc.275.20.15142. [DOI] [PubMed] [Google Scholar]
- 6.Cremo CR, Wang F, Facemyer K, Sellers JR. Phosphorylation-dependent regulation is absent in a nonmuscle heavy meromyosin construct with one complete head and one head lacking the motor domain. J Biol Chem. 2001;276:41465–72. doi: 10.1074/jbc.M107103200. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki H, Stafford WF, 3rd, Slayter HS, Seidel JC. A conformational transition in gizzard heavy meromyosin involving the head-tail junction, resulting in changes in sedimentation coefficient, ATPase activity, and orientation of heads. J Biol Chem. 1985;260:14810–7. [PubMed] [Google Scholar]
- 8.Suzuki H, Kondo Y, Carlos AD, Seidel JC. Effects of phosphorylation, MgATP, and ionic strength on the rates of papain degradation of heavy and light chains of smooth muscle heavy meromyosin at the S1–S2 junction. J Biol Chem. 1988;263:10974–9. [PubMed] [Google Scholar]
- 9.Rosenfeld SS, Xing J, Cheung HC, Brown F, Kar S, Sweeney HL. Structural and kinetic studies of phosphorylation-dependent regulation in smooth muscle myosin. J Biol Chem. 1998;273:28682–90. doi: 10.1074/jbc.273.44.28682. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, Shao Z, Somlyo AP, Somlyo AV. Cryo-atomic force microscopy of smooth muscle myosin. Biophys J. 1997;72:1308–18. doi: 10.1016/S0006-3495(97)78777-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheng S, Gao Y, Khromov AS, Somlyo AV, Somlyo AP, Shao Z. Cryo-atomic force microscopy of unphosphorylated and thiophosphorylated single smooth muscle myosin molecules. J Biol Chem. 2003;278:39892–6. doi: 10.1074/jbc.M306094200. [DOI] [PubMed] [Google Scholar]
- 12.Frado LY, Craig R. Structural changes induced in scallop heavy meromyosin molecules by Ca2+ and ATP. J Muscle Res Cell Motil. 1992;13:436–46. [Google Scholar]
- 13.Wu X, Clack BA, Zhi G, Stull JT, Cremo CR. Phosphorylation-dependent structural changes in the regulatory light chain domain of smooth muscle heavy meromyosin. J Biol Chem. 1999;274:20328–35. doi: 10.1074/jbc.274.29.20328. [DOI] [PubMed] [Google Scholar]
- 14.Wahlstrom JL, Randall MA, Jr, Lawson JD, Lyons DE, Siems WF, Crouch GJ, Barr R, Facemyer KC, Cremo CR. Structural model of the regulatory domain of smooth muscle heavy meromyosin. J Biol Chem. 2003;278:5123–31. doi: 10.1074/jbc.M206963200. [DOI] [PubMed] [Google Scholar]
- 15.Mazhari SM, Selser CT, Cremo CR. Novel sensors of the regulatory switch on the regulatory light chain of smooth muscle Myosin. J Biol Chem. 2004;279:39905–14. doi: 10.1074/jbc.M407062200. [DOI] [PubMed] [Google Scholar]
- 16.Nelson WD, Blakely SE, Nesmelov YE, Thomas DD. Site-directed spin labeling reveals a conformational switch in the phosphorylation domain of smooth muscle myosin. Proc Natl Acad Sci U S A. 2005;102:4000–5. doi: 10.1073/pnas.0401664102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wendt T, Taylor D, Trybus KM, Taylor K. Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2. Proc Natl Acad Sci USA. 2001;98:4361–6. doi: 10.1073/pnas.071051098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wendt T, Taylor D, Messier T, Trybus KM, Taylor KA. Visualization of head–head interactions in the inhibited state of smooth muscle myosin. J Cell Biol. 1999;147:1385–90. doi: 10.1083/jcb.147.7.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu JQ, Harder BA, Uman P, Craig R. Myosin filament structure in vertebrate smooth muscle. J Cell Biol. 1996;134:53–66. doi: 10.1083/jcb.134.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tonino P, Simon M, Craig R. Mass determination of native smooth muscle myosin filaments by scanning transmission electron microscopy. J Mol Biol. 2002;318:999–1007. doi: 10.1016/S0022-2836(02)00191-2. [DOI] [PubMed] [Google Scholar]
- 21.Ikebe M, Ogihara S. Phosphorylation-dependent and ATP-induced changes in structural array in gizzard myosin filament bundles. J Biochem (Tokyo) 1982;92:1973–7. doi: 10.1093/oxfordjournals.jbchem.a134128. [DOI] [PubMed] [Google Scholar]
- 22.Levine RJ, Kensler RW, Yang Z, Sweeney HL. Myosin regulatory light chain phosphorylation and the production of functionally significant changes in myosin head arrangement on striated muscle thick filaments. Biophys J. 1995;68:224S. [PMC free article] [PubMed] [Google Scholar]
- 23.Vibert P, Craig R. Structural changes that occur in scallop myosin filaments upon activation. J Cell Biol. 1985;101:830–7. doi: 10.1083/jcb.101.3.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Craig R, Padron R, Kendrick-Jones J. Structural changes accompanying phosphorylation of tarantula muscle myosin filaments. J Cell Biol. 1987;105:1319–27. doi: 10.1083/jcb.105.3.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woodhead JL, Zhao FQ, Craig R, Egelman EH, Alamo L, Padron R. Atomic model of a myosin filament in the relaxed state. Nature. 2005;436:1195–9. doi: 10.1038/nature03920. [DOI] [PubMed] [Google Scholar]
- 26.Mrakovcic-Zenic A, Reisler E. Light-chain phosphorylation and cross-bridge conformation in myosin from vertebrate skeletal muscle. Biochemistry. 1983;22:525–30. doi: 10.1021/bi00272a001. [DOI] [PubMed] [Google Scholar]
- 27.Adhikari B, Hideg K, Fajer PG. Independent mobility of catalytic and regulatory domains of myosin heads. Proc Natl Acad Sci USA. 1997;94:9643–7. doi: 10.1073/pnas.94.18.9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adhikari BB, Somerset J, Stull JT, Fajer PG. Dynamic modulation of the regulatory domain of myosin heads by pH, ionic strength, and RLC phosphorylation in synthetic myosin filaments. Biochemistry. 1999;38:3127–32. doi: 10.1021/bi982553g. [DOI] [PubMed] [Google Scholar]
- 29.Baumann BA, Hambly BD, Hideg K, Fajer PG. The regulatory domain of the myosin head behaves as a rigid lever. Biochemistry. 2001;40:7868–73. doi: 10.1021/bi002731h. [DOI] [PubMed] [Google Scholar]
- 30.Brown LJ, Klonis N, Sawyer WH, Fajer PG, Hambly BD. Independent movement of the regulatory and catalytic domains of myosin heads revealed by phosphorescence anisotropy. Biochemistry. 2001;40:8283–91. doi: 10.1021/bi010566f. [DOI] [PubMed] [Google Scholar]
- 31.Grand RJ, Perry SV. Preparation of the alkali and P light chains of chicken gizzard myosin. Amino acid sequence of the alkali light chain. Biochem J. 1983;211:267–72. doi: 10.1042/bj2110267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ikebe M, Hartshorne DJ. Effects of Ca2+ on the conformation and enzymatic activity of smooth muscle myosin. J Biol Chem. 1985;260:13146–53. [PubMed] [Google Scholar]
- 33.Margossian SS, Lowey S. Preparation of myosin and its subfragments from rabbit skeletal muscle. Methods Enzymol. 1982;85(Part B):55–71. doi: 10.1016/0076-6879(82)85009-x. [DOI] [PubMed] [Google Scholar]
- 34.Chandra TS, Nath N, Suzuki H, Seidel JC. Modification of thiols of gizzard myosin alters ATPase activity, stability of myosin filaments, and the 6–10 S conformational transition. J Biol Chem. 1985;260:202–7. [PubMed] [Google Scholar]
- 35.Baumann BA, Liang H, Sale K, Hambly BD, Fajer PG. Myosin regulatory domain orientation in skeletal muscle fibers: application of novel electron paramagnetic resonance spectral decomposition and molecular modeling methods. Biophys J. 2004;86:3030–41. doi: 10.1016/S0006-3495(04)74352-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adelstein RS, Klee CB. Purification of smooth muscle myosin light-chain kinase. Methods Enzymol. 1982;85(Part B):298–308. doi: 10.1016/0076-6879(82)85029-5. [DOI] [PubMed] [Google Scholar]
- 37.Fajer PG, Marsh D. Microwave and modulation field inhomogeneities and the effect of cavity Q in saturation transfer ESR spectra. Dependence on sample size. J Magn Reson. 1982;49:212–24. [Google Scholar]
- 38.Weast R. CRC Handbook of Chemistry and Physics. 63. CRC Press; Boca Raton, FL: 1982. [Google Scholar]
- 39.Kinosita K, Jr, Kawato S, Ikegami A. A theory of fluorescence polarization decay in membranes. Biophys J. 1977;20:289–305. doi: 10.1016/S0006-3495(77)85550-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kinosita K, Jr, Kawato S, Ikegami A. Dynamic structure of biological and model membranes: analysis by optical anisotropy decay measurement. Adv Biophys. 1984;17:147–203. doi: 10.1016/0065-227x(84)90027-3. [DOI] [PubMed] [Google Scholar]
- 41.Li HC, Hideg K, Fajer PG. The mobility of troponin C and troponin I in muscle. J Mol Recognit. 1997;10:194–201. doi: 10.1002/(SICI)1099-1352(199707/08)10:4<194::AID-JMR365>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 42.Hyde JS, Thomas DD. New EPR methods for the study of very slow motion: application to spin-labeled hemoglobin. Ann NY Acad Sci. 1973;222:680–92. doi: 10.1111/j.1749-6632.1973.tb15295.x. [DOI] [PubMed] [Google Scholar]
- 43.Olney JJ, Sellers JR, Cremo CR. Structure and function of the 10 S conformation of smooth muscle myosin. J Biol Chem. 1996;271:20375–84. doi: 10.1074/jbc.271.34.20375. [DOI] [PubMed] [Google Scholar]
- 44.Robinson BH, Dalton LR. Anisotropic rotational diffusion studied by passage saturation transfer electron paramagnetic resonance. J Chem Phys. 1980;72:1312–24. [Google Scholar]
- 45.Howard EC, Lindahl KM, Polnaszek CF, Thomas DD. Simulation of saturation transfer electron paramagnetic resonance spectra for rotational motion with restricted angular amplitude. Biophys J. 1993;64:581–93. doi: 10.1016/S0006-3495(93)81417-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lakowicz JR. Principles of fluorescence spectroscopy. 2. Plenum Press; New York: 1999. [Google Scholar]
- 47.Huxley AF, Simmons RM. Proposed mechanism of force generation in striated muscle. Nature. 1971;233:533–8. doi: 10.1038/233533a0. [DOI] [PubMed] [Google Scholar]
- 48.Burgess SA, Walker ML, White HD, Trinick J. Flexibility within myosin heads revealed by negative stain and single-particle analysis. J Cell Biol. 1997;139:675–81. doi: 10.1083/jcb.139.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dominguez R, Freyzon Y, Trybus KM, Cohen C. Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: visualization of the pre-power stroke state. Cell. 1998;94:559–71. doi: 10.1016/s0092-8674(00)81598-6. [DOI] [PubMed] [Google Scholar]
- 50.Houdusse A, Szent-Gyorgyi AG, Cohen C. Three conformational states of scallop myosin S1. Proc Natl Acad Sci USA. 2000;97:11238–43. doi: 10.1073/pnas.200376897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cross RA, Engel A. Scanning transmission electron microscopic mass determination of in vitro self-assembled smooth muscle myosin filaments. J Mol Biol. 1991;222:455–8. doi: 10.1016/0022-2836(91)90487-q. [DOI] [PubMed] [Google Scholar]
- 52.Stewart M, Kensler RW. Arrangement of myosin heads in relaxed thick filaments from frog skeletal muscle. J Mol Biol. 1986;192:831–51. doi: 10.1016/0022-2836(86)90032-x. [DOI] [PubMed] [Google Scholar]
- 53.Trybus KM, Lowey S. Assembly of smooth muscle myosin minifilaments: effects of phosphorylation and nucleotide binding. J Cell Biol. 1987;105:3007–19. doi: 10.1083/jcb.105.6.3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nyitrai M, Szent-Gyorgyi AG, Geeves MA. A kinetic model of the co-operative binding of calcium and ADP to scallop (Argopecten irradians) heavy meromyosin. Biochem J. 2002;365:19–30. doi: 10.1042/BJ20020099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trybus KM, Chatman TA. Chimeric regulatory light chains as probes of smooth muscle myosin function. J Biol Chem. 1993;268:4412–9. [PubMed] [Google Scholar]
- 56.Rajasekharan KN, Morita JI, Mayadevi M, Ikebe M, Burke M. Formation and properties of smooth muscle myosin 20-kDa light chain-skeletal muscle myosin hybrids and photocrosslinking from the maleimidylbenzophenone-labeled light chain to the heavy chain. Arch Biochem Biophys. 1991;288:584–90. doi: 10.1016/0003-9861(91)90240-j. [DOI] [PubMed] [Google Scholar]
- 57.Lowey S, Waller GS, Trybus KM. Skeletal muscle myosin light chains are essential for physiological speeds of shortening. Nature. 1993;365:454–6. doi: 10.1038/365454a0. [DOI] [PubMed] [Google Scholar]