Abstract
The ability of microbes to secrete bioactive chemical signals into their environment has been known for over a century. However, it is only in the last decade that imaging mass spectrometry has provided us with the ability to directly visualize the spatial distributions of these microbial metabolites. This technology involves collecting mass spectra from multiple discrete locations across a biological sample, yielding chemical ‘maps’ that simultaneously reveal the distributions of hundreds of metabolites in two dimensions. Advances in microbial imaging mass spectrometry summarized here have included the identification of novel strain- or coculture-specific compounds, the visualization of biotransformation events (where one metabolite is converted into another by a neighboring microbe), and the implementation of a method to reconstruct the 3D subsurface distributions of metabolites, among others. Here we review the recent literature and discuss how imaging mass spectrometry has spurred novel insights regarding the chemical consequences of microbial interactions.
Keywords: imaging mass spectrometry, MALDI-TOF, molecular networking, microbial interactions, microbial coculture
This review highlights recent advances in the applications of imaging mass spectrometry to assist in both identifying and directly visualizing the distributions of specialized metabolites in complex bacterial populations.
Graphical Abstract Figure.
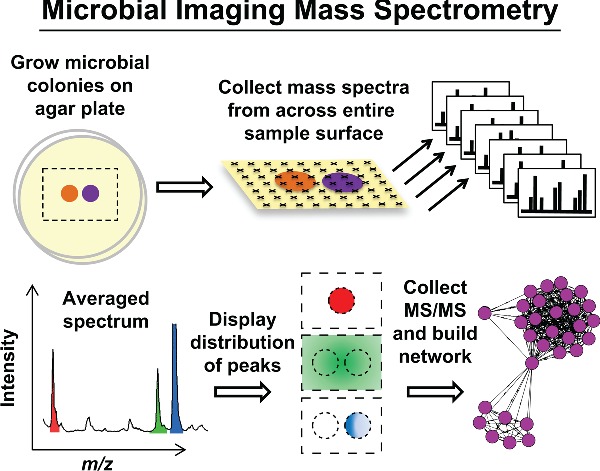
This review highlights recent advances in the applications of imaging mass spectrometry to assist in both identifying and directly visualizing the distributions of specialized metabolites in complex bacterial populations.
INTRODUCTION
For decades, humans have been reaping the therapeutic benefits of microbially produced specialized metabolites (formerly called secondary metabolites). However, we are only just beginning to understand and appreciate how interkingdom, intergenera and interspecies interactions mediated by microbial specialized metabolites and other secreted molecules (including quorum-sensing compounds and virulence factors) lead to the expression of compounds that are not readily detected in monoculture laboratory conditions. Methodological advances in deep sequencing, genome mining and bacterial gene network analysis have provided tantalizing insights into the extensive networks of chemical communication occurring between microbial populations. The capability to directly visualize the distributions of such metabolic signals using imaging mass spectrometry (IMS) has further expanded our appreciation of the complex interactions shaping microbial communities, and will be the subject of this review.
IMS has been in use in pathology settings since the late 1990's (Caprioli, Farmer and Gile 1997). However, its active application to microbial samples grown on agar began less than a decade ago, led by the Dorrestein lab's adaptation of matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) IMS for this purpose (Yang et al. 2009, 2012). While the specifics vary, the overall concept of microbial IMS involves collecting hundreds of mass spectra from across the 2D surface of a microbial colony or coculture grown on agar, in a raster or grid-like pattern (Yang et al. 2012). This permits the spatial distributions of the chemical signatures of microbes to be directly visualized and correlated with biological phenotypes of interest. Many dozens of metabolites can be visualized simultaneously in a single IMS sample. IMS has chiefly facilitated the investigation of the consequences of microbial interspecies interactions. It allows for the observation of previously undetected, coculture-specific metabolites and permits the identification of the metabolite-producing microbes. Previous reviews have discussed the technical aspects of IMS sample preparation and its applications in microbiology (Watrous and Dorrestein 2011; Gonzalez et al. 2012; Fang and Dorrestein 2014). Our intent in this review is to discuss how IMS has been applied to address small molecule production during bacterial coculture interactions, with an emphasis on papers from the last 4 years. We have structured the review by grouping studies based on the ionization or mass spectrometry detection method used.
MALDI-IMS
Using MALDI-IMS as a stand-alone analytical technique has led to a deeper understanding of how specialized metabolites regulate bacterial behavior and how the metabolites themselves are regulated during bacterial interactions. This section will discuss how MALDI-IMS provides useful insights into microbial interactions that would be undetectable using non-imaging methods.
One advantage of MALDI-IMS is its compatibility with microbial phenotypic assays. Commercially available software makes it straightforward to create overlays of the diffusion patterns of specialized metabolites (i.e. m/z peaks) in agar with the visual images of the bacterial colonies that secreted them. This permits the direct correlation of biological phenomena with their putative causative signals. Our research group recently demonstrated the utility of this approach (Bleich et al. 2015). Bacillus cereus secretes a molecule that activates biofilm gene expression in B. subtilis 3610 (Shank et al. 2011). In order to identify the biofilm-activating molecule produced by B. cereus, we took advantage of the fact that a positive result in our bioassay generates a distinct, 2D distribution of fluorescent B. subtilis colonies on agar (Bleich et al. 2015). We surmised that the metabolite of interest must then be physically present in the areas where we saw activation of the B. subtilis fluorescent reporter. Using MALDI-IMS, we identified m/z peaks exhibiting spatial distributions that overlapped with the fluorescence, and thus represented putative biofilm-stimulating molecules. This method allowed us to predict, and then confirm, that a group of thiazolyl peptide antibiotics, the thiocillins, were the metabolites produced by B. cereus that stimulated biofilm gene expression in B. subtilis (Bleich et al. 2015). Thus, in this example, MALDI-IMS allowed a phenotype-to-chemotype connection to be rapidly and definitively made, and the biologically relevant molecule to be identified.
Another advantage of mass spectrometry in studying bacterial interactions is the ability to discriminate strain-specific metabolites and how they vary in response to coculture. This capacity is particularly relevant due to growing interest in studying natural microbial isolates from environmental samples or animal hosts. The lab-adapted (‘domesticated’) bacterial strains typically studied in microbiology labs may not exhibit traits identical to those of their wild relatives (Aguilar et al. 2007). IMS can ascertain such metabolite differences between related bacteria that may influence specific interactions. An example of this was demonstrated in a comparative study of two Pseudomonas aeruginosa strains: the lab-adapted PAO1 and the human-host-adapted DK2-P2M24-2003 (DK2). These strains are phenotypically distinct in how they interact with Staphylococcus aureus strain JE2: PAO1 suppresses the growth of JE2, while DK2 coexists with JE2 (Frydenlund Michelsen et al. 2015). MALDI-IMS revealed that the metabolic profiles of these two P. aeruginosa strains differed when they were grown adjacent to JE2: pyocyanin (an antimicrobial) and several rhamnolipids were produced by PAO1 but not by DK2 in coculture (Phelan et al. 2014; Frydenlund Michelsen et al. 2015). JE2 also generated at least two additional compounds when grown with DK2 that were not present during its interactions with PAO1 (Frydenlund Michelsen et al. 2015). This differential metabolite production was proposed to be due to DK2's less adversarial interaction with JE2. Particularly intriguing was the lack of the PQS molecule or the antimicrobial HQNO from the DK2 strain during growth with JE2. Instead, DK2 produced more of a PQS precursor, HHQ. This difference in metabolite production was linked to decreased transcription of the pqsH gene, and the resulting low levels of PQS were hypothesized to be the reason why JE2 did not kill DK2 (Frydenlund Michelsen et al. 2015). In this study, then, MALDI-IMS not only revealed important variability in the metabolic profiles of bacterial strains isolated from different ecological habitats, but also permitted their chemotypic differences to be tied to a single gene that likely explains their distinct coculture phenotypes.
MALDI-IMS can also be used to rapidly identify metabolic differences between closely related strains, as recently shown in a comparative study of Lysobacter strains grown in coculture with the fungus Rhizoctonia solani (de Bruijn et al. 2015). The intragenus comparisons between these Lysobacter strains, whether in monoculture or fungal coculture, exemplify how phylogenetically related species can display distinct metabolic profiles, and highlights the importance of examining metabolic exchange not only in lab strains, but also in ‘wild’ microbial isolates. Comparing the metabolic profiles of these Lysobacter strains also permitted the identification of some specialized metabolites that were predicted from genome-mining studies (de Bruijn et al. 2015).
Another major advantage of IMS is its ability to indicate (via spatial distributions) which organism is producing a specific metabolite of interest within interspecies interactions. The relevance of this was brought to the fore in a recent paper by Moree et al. (2012) examining the interkingdom interactions between P. aeruginosa and Aspergillus fumigatus, pathogens that frequently coexist in the lungs of cystic fibrosis patients (Amin, Dupuis and Aaron 2010; Mowat et al. 2010). Their use of MALDI-IMS revealed a unique chemical interaction that would have been overlooked by non-imaging analysis (Moree et al. 2012). They used MALDI-TOF and MALDI-FT-ICR imaging as well as MS/MS networking (see below) to explore how the metabolites produced by these pathogens influence each other (Moree et al. 2012). In coculture, four P. aeruginosa phenazines (PCN, PCA, PYO, and 1-MP) were highly abundant across the sample surface, without regard for the A. fumigatus colony. In contrast, three phenazines (1-HP, 5-MPCA, and phenazine-1-sulfate) exhibited unusual spatial distributions, being more abundant in and around the A. fumigatus colony, despite Aspergillus lacking the biosynthesis genes required to produce these compounds (Moree et al. 2012). This led to the hypothesis that A. fumigatus was biotransforming some of these metabolites. Moree et al. demonstrated that A. fumigatus was transforming the PCA secreted by P. aeruginosa into 1-HP, which was being further transformed into 1-MP and phenazine-1-sulfate. The generation of 1-HP elicited the production of two siderophores by A. fumigatus (Moree et al. 2012). Without IMS, this unique biotransformation event may easily have been erroneously attributed to P. aeruginosa directly stimulating A. fumigatus to produce siderophores rather than A. fumigatus actively self-eliciting this response.
This is not the only example where IMS has revealed metabolic biotransformation events. Bacillus subtilis 3610 inhibits aerial hyphae development in streptomyces via the production of an antibiotic, surfactin (Straight, Willey and Kolter 2006), but Streptomyces sp. Mg1 appears to be resistant to this effect (Hoefler et al. 2012). Hoefler et al. (2012) further inspected the interaction between strains 3610 and Mg1. They observed that the m/z peaks corresponding to surfactin decreased in proximity to the Mg1 colony. Such metabolite voids can indicate microbial digestion or biotransformation, and indeed there was a concomitant increase in new peaks in these regions that corresponded to the surfactin ions +18 m/z, a mass shift indicative of hydrolysis. Hoefler et al. (2012) went on to demonstrate that Mg1 secretes a surfactin hydrolase (which opens the macrolide ring of surfactin), and that the resulting linear, hydrolyzed form of surfactin is no longer able to block aerial hyphae development in Mg1. Additional examples of how IMS can inform our understanding of how microbes alter and transform their surrounding metabolic environments can be found in a recent review by Silva and Northen (2015) on exometabolomics.
When bacterial colonies are grown on agar, they deposit their metabolites in three dimensions, both diffusing away from the colony as well as down into the substrate. The IMS data discussed so far have described the metabolic and phenotypic interactions of bacterial colonies in two dimensions, effectively collapsing the 3D distributions of metabolites within the agar into a 2D visual image. Investigating the secretion profiles of subsurface metabolites in three dimensions was first investigated by Watrous et al. (2013b). They established a method of growing bacteria on thick agar, sectioning it into vertical slices, collecting 2D IMS data from each slice, and then using that data to build a 3D reconstruction of the original sample (Watrous et al. 2013b). The utility of this 3D MALDI-IMS technique was demonstrated by looking at the undersurface distributions of metabolites within the hyphal growth of Candida albicans when grown with P. aeruginosa. This unique perspective revealed some metabolites that had dome-like distributions matching those of the regions with hyphae, some that had doughnut-like distributions near the surface of the agar that did not penetrate to the full depth of the hyphae, and some that had hollow-shell distributions that followed the outer curve of hyphal growth deep under the surface (Watrous et al. 2013b). While the biological ramifications of these unusual and distinctive metabolic distributions remain unclear, this adaptation of MALDI-IMS allows detection of previously unseen metabolic distributions that may impact microbial interactions.
A main challenge in imaging microbial interactions on agar is that the irregular thickness of the agar and granularity of matrix crystals can decrease mass resolution (Hoffmann and Dorrestein 2015). Initially, the most common method of applying MALDI matrix to agar samples was to sieve dry matrix over them (Yang et al. 2012), a method shown to be highly suitable and successful when imaging small molecules in tissue samples (Goodwin et al. 2010; Trimpin et al. 2010). On agar, however, this approach leads to many metabolites being inadequately extracted, leading to weak detection or broad, poorly resolved mass peaks (Hoffmann and Dorrestein 2015). Hoffmann and Dorrestein (2015) adapted a matrix-spraying technique compatible with bacterial colonies grown on agar that, compared to dry-matrix sieving, gives images with higher spatial resolution and less background noise, while still ensuring the agar does not flake off in the high vacuum of a MALDI instrument. When tested on a colony of Sorangium cellulosum, the peak corresponding to the metabolite pellasoren exhibited a >5-fold increase in intensity in the sprayed sample compared with the sieved sample (Hoffmann and Dorrestein 2015). Many other known metabolites, such as microsclerodermin M, exhibited greater peak reproducibility using the spray method of matrix application, an improvement that would better support targeted compound identification (Hoffmann and Dorrestein 2015). Future methodological developments in matrix sample preparation for MALDI-IMS will only continue to drive the field forward. Methods of performing MS analysis on agar samples without the need for MALDI matrix have also been developed (for instance nanoDESI and REX-NIMS) and are discussed below.
MALDI-IMS WITH LC-MS/MS
As described above, MALDI-IMS can provide a variety of insights into microbial interspecies interactions; however, its low mass resolution makes it better suited for studies of known compounds rather than unguided metabolomics. Combining MALDI-IMS with methods such as liquid chromatography (LC) and tandem MS/MS fragmentation, however, can provide additional information about unknown metabolites, facilitating their chemical identification. Several recent publications have coupled MALDI-IMS with LC-MS/MS to explore how specialized metabolite production is regulated in P. aeruginosa (Phelan et al. 2014; Frydenlund Michelsen et al. 2015; Phelan, Fang and Dorrestein 2015). Phelan, Fang and Dorrestein (2015) investigated how the macrolide antibiotic azithromycin (AZM) impacted specialized metabolite production in P. aeruginosa when delivered at levels below the minimum inhibitory concentration. Although not strictly examining an interspecies interaction, this study is important in that it describes an unexpected response to subinhibitory concentrations of an antimicrobial molecule, many of which are produced by other microbes. It was previously proposed that AZM inhibited specialized metabolite biosynthesis in P. aeruginosa by altering quorum sensing (Tateda et al. 2001; Nalca et al. 2006). The MALDI-IMS data initially supported this view, indicating that increasing AZM concentrations led to decreases in the production of quorum-sensing signals and siderophores (Phelan, Fang and Dorrestein 2015). However, upon quantifying bacterial cell numbers and metabolite concentrations (via LC-MS/MS), it became clear that metabolite production actually increased in AZM-sensitive strains treated with AZM (Phelan, Fang and Dorrestein 2015). Thus, despite fewer bacteria surviving AZM treatment, those that do survive increase their metabolite production (Phelan, Fang and Dorrestein 2015). [This phenotype did not hold for AZM-resistant strains, which showed negligible differences over increasing doses of AZM (Phelan, Fang and Dorrestein 2015).] These results highlight the need to consider both bacterial growth rates and metabolite concentrations when drawing conclusions from MALDI-IMS data.
An interesting consideration is what happens if one specialized metabolite is removed from a metabolic network. Phelan et al. (2014) tackled this question by observing the consequences of disrupting a single gene in P. aeruginosa (phzF2, which is necessary for the production of five related phenazines). After visualization by MALDI-IMS, LC-MS/MS was used to quantify how metabolite production of the phzF2 mutant in monoculture compared to that of the wild-type parent. This comparison revealed that, as expected, production of four phenazines was drastically decreased in the phzF2 mutant compared to wild type, while a fifth (PCN) was unexpectedly increased (Phelan et al. 2014). Unpredictably, non-phenazine metabolites were also impacted by the phzF2 mutation. Pyoverdine and quorum-sensing molecules were decreased in the mutant strain, while levels of pyochelin and several rhamnolipids were increased (Phelan et al. 2014). Thus, disrupting a single phenazine biosynthesis gene led to global metabolic changes in P. aeruginosa metabolite production. Such alterations also impacted interspecies interactions: the phzF2 mutant stimulated Aspergillus fumigatus to increase the production of a siderophore compared to when cocultured with wild-type P. aeruginosa (Phelan et al. 2014). This study demonstrates how MALDI-IMS and LC-MS/MS can directly report on the cascading metabolic consequences of removing a single specialized metabolite from a complicated regulatory pathway.
One of the most understated benefits of IMS is the hypothesis-generating potential of its unbiased studies. This is exemplified in one of the most ambitious applications of MALDI-IMS to date: reconstructing in three dimensions a complete map of the detectable metabolites, peptides and proteins present on human skin as well as the bacterial strains residing at each sampling site (Bouslimani et al. 2015) This study used multiple mass spectrometry techniques (including ultra-performance LC/quadrupole time-of-flight and MALDI-TOF MS/MS) to analyze metabolic samples from the surface skin of both a male and female subject, and 16S rRNA gene sequencing to determine the bacterial communities present. Additionally, LC-MS/MS was performed on 34 bacterial strains from different genera and a selection of beauty products to generate a library of ‘known’ metabolic signatures (Bouslimani et al. 2015). Correlating the localization of particular bacteria with particular molecular species generated testable hypotheses. For example, their correlation analysis indicated that Propionibacterium spp. spatially colocalized with 491 different molecular features (Bouslimani et al. 2015). This led to the hypothesis that Propionibacteria could hydrolyze compounds such as triolein, a triacylglyceride, into oleic acid. By confirming this hypothesis they established that the bacteria colonizing skin actively contribute to, and alter, the unique chemical composition of human skin (Bouslimani et al. 2015). Another example was observed in the groin regions of the participants, where 8122 molecular species were correlated with seven bacterial genera in the male subject, and 9922 unique molecular species were correlated with six bacterial genera in the female groin (Bouslimani et al. 2015). Innumerable other hypothesis are likely to be generated by this massive dataset, leading to advances in our understanding of the complex interactions occurring within the microbial communities on human skin and the specialized metabolites they produce.
ELECTROSPRAY IONIZATION IMS
Combining MALDI-IMS with electrospray ionization (ESI) techniques offers better mass-resolving power than MALDI alone, and thus provides increased confidence in identifying observed metabolites. This is particularly important when trying to distinguish two metabolites that have the same nominal mass, and where fractional mass is necessary to distinguish them. Ye et al. (2013) combined MALDI-TOF/TOF IMS with ESI for this reason when examining the differential expression of metabolites on the root and nodules of the legume Medicago truncatula. To verify MALDI-observed metabolite profiles, they performed ESI ultra-high resolution quadrupole time-of-flight (UHR-Q-TOF) mass spectrometry with and without reverse-phase LC (Ye et al. 2013). Many of the metabolite predictions made based on the MALDI-IMS data were accurate, but its broad peaks hid some metabolites. For example, GABA and choline have theoretical masses of 104.0706 and 104.1070 Da, respectively. Their MALDI-TOF/TOF masses were both 104.10 Da, but they could be distinguished through UHR-Q-TOF analysis as having masses of 104.0706 and 104.1071 Da (Ye et al. 2013). This study also demonstrated that many metabolites were only able to be visualized when specific MALDI matrices were used (Ye et al. 2013). Finally, this study highlights the value of spatial imaging during metabolic profiling: the distributions of numerous metabolites varied dramatically across the root and nodule structures of the legume (Ye et al. 2013). Here, ESI was not necessary for the imaging process itself, but was vital to the ultimate identification of the metabolites being imaged.
In other cases, ESI is employed as the imaging method itself. Desorption ESI (DESI), developed in the early 2000's (Takáts et al. 2004), and nanospray DESI (nanoDESI), developed in 2010 (Roach, Laskin and Laskin 2010) can both be performed at ambient pressure, opening up the possibility of using ESI for IMS while overcoming some of the sample preparation issues that microbial agar samples present. NanoDESI allows for the analysis of bacterial colonies by mass spectrometry without requiring dehydration or matrix application (Watrous and Dorrestein 2011; Watrous et al. 2012). By scanning a bacterial sample with the nanoDESI probe in a grid-like fashion, images can be generated from the resulting m/z peaks (Watrous et al. 2013a). Watrous et al. applied this approach to a colony of Streptomyces coelicolor, which secretes a known repertoire of specialized metabolites. Some of these compounds, such as phosphocholine and prodiginine, were indeed detected in a location-specific manner (Watrous et al. 2013a). After the verification of this technique, Watrous et al. applied nanoDESI IMS to Shewanella oneidensis MR-1 and B. subtilis 3610, both in monoculture and as a coculture biofilm. They detected a large number of MR-1-specific signals with nanoDESI that had not previously been attributed to Shewanella species, including riboflavin, putrebactin (a siderophore) and heme (Watrous et al. 2013a). Data from the mixed biofilm revealed that MR-1 putrebactin biosynthesis was inhibited in coculture, as was 3610's production of plipastatin (an antifungal cyclic lipopeptide) (Watrous et al. 2013a). Finally, they detected two coculture-specific metabolites (riboflavin and protoporphyrin IX) along the interacting edge of the colonies (Watrous et al. 2013a). The main advantage of nanoDESI is that it permits the direct sampling of a bacterial colony surface without additional sample preparation. Thus, nanoDESI—as a technique that does not require endpoint sample preparation—can theoretically be used to monitor bacterial colonies over time (Watrous et al. 2012); however, since it is probable that the solvent added to the colony during data collection will alter bacterial cell physiology, any resampling should be limited to other, distinct and previously undisturbed locations within the colony.
Combining nanoDESI with MALDI-TOF IMS has proved to be a formidable combination to identify specialized metabolites from bacterial colony interactions. Traxler et al. (2013) grew S. coelicolor adjacent to five other actinobacterial strains (four different Streptomyces strains and one Amycolatopsis strain) that altered the S. coelicolor colony phenotype to varying degrees. These cocultures were then subjected to nanoDESI MS/MS and MALDI-TOF imaging. Networking the MS/MS data (see below) resulted in 629 different nodes (many of which represent highly structurally related metabolites) and facilitated compound identification (Traxler et al. 2013). Using their networking data in conjunction with predictions from antiSMASH—a program that identifies secondary metabolite biosynthetic genes in genome sequences (Medema et al. 2011)—they concluded that the coculture interactions induced S. coelicolor to produce the antibiotics actinorhodin and prodigiosin along with several desferrioxamine siderophores, many of which were novel acyl side chain variants (Traxler et al. 2013). Additional experiments indicated that the production of the siderophore amychelin by Amycolatopsis sp. AA4 caused competition for iron and thus stimulated S. coelicolor to produce 12 novel acyldesferrioxamine species (Traxler et al. 2013). This paper clearly illustrates how a single bacterial species can generate unique metabolic profiles depending on its neighboring interaction partner. Methodologically, this study also demonstrates how ESI-IMS and MS/MS networking can enhance our understanding of bacterial interspecies interactions without the need for MALDI matrix application or high vacuum.
COMBINING IMS WITH OTHER TECHNIQUES
As noted above, both matrix application and surface abnormalities in the agar and bacterial colony can affect MALDI-TOF performance due to the uneven surface. Louie et al. (2013) overcame this problem of an uneven biomolecule-absorbing surface by developing REX-NIMS (replica-extraction-transfer-nanostructure-initiator mass spectrometry). The NIMS approach itself does not require matrix application, instead relying on a nanostructured surface to trap initiator compounds under the sample (Woo et al. 2008). The REX protocol allows specialized metabolites and other small molecules to be extracted from an agar surface into an extraction gel, which is then ‘stamped’ onto a NIMS-chip surface for MALDI-MS analysis (Louie et al. 2013). The REX method transfers a very thin and even layer of gel onto the NIMS chip, more analogous to a tissue slice. This eliminates both the agar and matrix components from the sample data, thus increasing mass resolution and allowing for better spatial resolution of metabolites from interacting colonies. Louie et al. (2013) tested this protocol by coculturing Shewanella oneidensis and Pseudomonas stutzeri on solid agar before performing REX-NIMS. They showed that they could perform MALDI-TOF/TOF on the NIMS chip and identify spatially distinct m/z peaks that overlapped with where the bacterial colonies were prior to REX. Further applications of this technique with bacterial agar samples should provide data with better mass and spatial resolution than currently attainable with traditional agar-based MALDI-IMS.
The spatial resolution constraints of MALDI- and DESI-IMS effectively limit their utility to examining macroscopic bacterial colonies. By integrating secondary ion mass spectrometry (SIMS) with MALDI-IMS, Lanni et al. (2014b) combined many of the advantages of both approaches, even though it also required a sample preparation method that limits the detection range of the MALDI data to <1000 Da. They examined P. aeruginosa biofilms, which are poorly structured but contain microenvironments of interest. After using the coarser-grained MALDI-IMS data to identify areas that contained m/z peaks of interest, they investigated these specific small regions via SIMS, taking advantage of its high spatial resolution capabilities, (Lanni et al. 2014b). In this way, both quinolones and rhamnolipids were identified within the biofilm despite not being evenly dispersed throughout these structures (Lanni et al. 2014b). Another important facet of this study was their utilization of the laser-ablation marks resulting from MALDI data collection as spatial fiduciary marks to guide the subsequent collection of SIMS data (Lanni et al. 2014b). This clever procedure should prove useful in poorly structured samples where there are no clear sample boundaries to guide SIMS localization. This unique approach combining MALDI- and SIMS-IMS expands our ability to visualize the fine details of both intra- and interspecific interactions.
Although the majority of microbial IMS studies to date have focused on pairwise bacterial interactions, it is inevitable that growing interest in microbial communities will stimulate IMS investigations of mixed-species communities. Distinguishing which bacteria are producing which specialized metabolite will be a particularly acute problem in these more complex populations. A recent study coupling MALDI-IMS with fluorescence in situ hybridization (FISH) proposes a potential solution to this problem by attempting to correlate metabolite production with cells of known phylogenetic identify (Kaltenpoth, Strupat and Svatos 2016). Specifically, they imaged the surface of beewolf wasp cocoons in search of two antibiotics, piericidin A1 and B1, known to be produced by Streptomyces philanthi. Although varying concentrations of these antibiotics were observed across the sample surface, they appeared at high concentrations in the same locations (Kaltenpoth, Strupat and Svatos 2016). By overlaying the MALDI-IMS images with data from FISH probes for S. philanthi, these compounds are seen to be most highly abundant in regions where S. philanthi is present (Kaltenpoth, Strupat and Svatos 2016). Although this approach is limited by the requirement that the mass spectrometry method must preserve sufficient DNA for subsequent FISH analysis, advances in the spatial resolution attainable by MALDI-IMS or, alternatively, the use of nanoDESI, may provide future improvements to this method.
MS/MS NETWORKING
Even when unbiased IMS analyses show intriguing metabolite patterns and unique MS peaks, the greatest obstacle to furthering research in this field is the chemical identification of the metabolites of interest, initially identifiable solely by their m/z peak. This limitation has been somewhat alleviated by the application of MS/MS networking, peptidogenomics, and the concepts of molecular and gene cluster families (Watrous et al. 2012; Nguyen et al. 2013). Molecular families (MFs) are defined as “structurally related molecules based on their mass spectral fragmentation patterns” (Nguyen et al. 2013) and gene cluster families (GCFs) as “biosynthetic gene clusters that show similar gene cluster organization with a high degree of sequence similarity” (Nguyen et al. 2013). Integrating these ideas with MS/MS networking allows unknown m/z peaks to be associated with the MS/MS fragmentation signatures of known specialized metabolites from previously sequenced genomes.
In an application of this method, Nguyen et al. (2013) subjected 42 bacilli and 18 pseudomonads to nanoDESI and obtained their MS/MS spectra. These MS/MS data were used to generate a molecular network. In such networks, nodes represent consensus MS/MS spectra of parent ions at distinct m/z values, while the connections between the nodes indicate their spectral similarity to each other (Watrous et al. 2012). Thus, groups of closely related nodes can be designated as MFs (Nguyen et al. 2013). Nguyen et al. then focused on the MS/MS spectra that contained mass shifts indicative of amino acids, suggesting they were generated by peptidic compounds (either ribosomally or non-ribosomally generated). The resulting peptide sequence tags allowed them to perform peptidogenomics and identify genes in public genome databases that were predicted to generate such peptides. In this way, the MFs in their network were linked to known GCFs (Nguyen et al. 2013).
In their dataset, 3339 out of the 4311 nodes (78% of the spectra) were unique to either the bacilli or pseudomonads (Nguyen et al. 2013). These nodes were further grouped into 121 MFs that contained clusters of three or more nodes. A large cluster consisting of 78 nodes was identified as a GCF-MF pair for surfactin, a biosurfactant produced by the well-studied Bacillus subtilis 3610 strain (Nguyen et al. 2013). Thus, if an unsequenced bacillus strain generates a MS/MS spectrum that clusters into this GCF-MF, the source metabolite could be identified as a surfactin family member. As a proof of principle of this approach, Nguyen et al. examined two Pseudoalteromonas isolates that produced antibiotics that killed B. subtilis 3610. The MS/MS data collected from these antibiotics did not match those from any known metabolite, but they generated MS/MS spectra that clustered with nodes identified as originating from a bromoalterochromide. By genome mining all publically available Pseudoalteromonas genomes, a non-ribosomal peptide synthetase gene cluster was identified in Pseudomonas piscicida that was predicted to produce an antimicrobial bromoalterochromide (Nguyen et al. 2013). As noted throughout this review, MS/MS networking has facilitated the identification of specialized metabolites in a variety of studies (Moree et al. 2012; Watrous et al. 2012; Nguyen et al. 2013; Traxler et al. 2013; Frydenlund Michelsen et al. 2015), and will likely prove an extremely useful tool for the bacterial specialized metabolite community going forward.
CONCLUSIONS
Over the last few years, IMS has generated not only novel findings, but also has helped to stimulate a plethora of innovative hypotheses about microbial metabolic exchange. This review has delved into the recent microbial IMS literature to emphasize how this technique has been used to answer questions about the metabolic consequences of microbial interactions. In addition to the originally developed agar-based MALDI-TOF IMS, a variety of newer techniques are being applied to push the boundaries of resolving, detecting and identifying specialized metabolites, and correlate them with the organism generating them. Not covered here are multiple other exciting approaches in specialized metabolite imaging, including confocal Raman microscopy (Lanni et al. 2014a; Menezes et al. 2015) and laser desorption postionization mass spectrometry (Bhardwaj et al. 2013; Akhmetov, Bhardwaj and Hanley 2015). Future work will certainly expand upon the pairwise interactions typically being examined now, working towards defining the metabolic interactions occurring within complex multispecies communities. We anticipate that natural microbial communities consisting of small numbers of coexisting microbes, such as those seen in established symbioses (Aylward et al. 2014; Murfin et al. 2015), carnivorous plants (Adlassnig, Peroutka and Lendl 2011; Krieger and Kourtev 2011), or fermented foods (Wolfe and Dutton 2015) may prove to be ideal models for future IMS explorations.
Acknowledgments
This work was supported by the National Institute of General Medical Sciences, National Institutes of Health [grant number GM112981 to E.A.S]. N.M.S. and this work was supported by the Training, Workforce Development & Diversity division of the National Institute of General Medical Sciences, National Institutes of Health [grant number K12GM000678].
Conflict of interest. None declared.
REFERENCES
- Adlassnig W, Peroutka M, Lendl T. Traps of carnivorous pitcher plants as a habitat: composition of the fluid, biodiversity and mutualistic activities. Ann Bot. 2011;107:181–94. doi: 10.1093/aob/mcq238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar C, Vlamakis H, Losick R, et al. Thinking about Bacillus subtilis as a multicellular organism. Curr Opin Microbiol. 2007;10:638–43. doi: 10.1016/j.mib.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhmetov A, Bhardwaj C, Hanley L. Laser desorption postionization mass spectrometry imaging of biological targets. Method Mol Biol. 2015;1203:185–94. doi: 10.1007/978-1-4939-1357-2_18. [DOI] [PubMed] [Google Scholar]
- Amin R, Dupuis A, Aaron SD. The effect of chronic infection with Aspergillus fumigatus on lung function and hospitalization in patients with cystic fibrosis. Chest. 2010;137:171–6. doi: 10.1378/chest.09-1103. [DOI] [PubMed] [Google Scholar]
- Aylward FO, Suen G, Biedermann PHW, et al. Convergent bacterial microbiotas in the fungal agricultural systems of insects. mBio. 2014;5 doi: 10.1128/mBio.02077-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj C, Moore JF, Cui Y, et al. Laser desorption VUV postionization MS imaging of a cocultured biofilm. Anal Bioanal Chem. 2013;405:6969–77. doi: 10.1007/s00216-012-6454-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleich R, Watrous JD, Dorrestein PC, et al. Thiopeptide antibiotics stimulate biofilm formation in Bacillus subtilis. P Natl Acad Sci USA. 2015;112:3086–91. doi: 10.1073/pnas.1414272112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouslimani A, Porto C, Rath CM, et al. Molecular cartography of the human skin surface in 3D. P Natl Acad Sci USA. 2015;112:e2120–9. doi: 10.1073/pnas.1424409112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem. 1997;69:4751–60. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- de Bruijn I, Cheng X, de Jager V, et al. Comparative genomics and metabolic profiling of the genus Lysobacter. BMC Genomics. 2015;16 doi: 10.1186/s12864-015-2191-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Dorrestein PC. Emerging mass spectrometry techniques for the direct analysis of microbial colonies. Curr Opin Microbiol. 2014;19:120–9. doi: 10.1016/j.mib.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydenlund Michelsen C, Hossein Khademi SM, Krogh Johansen H, et al. Evolution of metabolic divergence in Pseudomonas aeruginosa during long-term infection facilitates a proto-cooperative interspecies interaction. ISME J. 2015 doi: 10.1038/ismej.2015.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez DJ, Xu Y, Yang Y-L, et al. Observing the invisible through imaging mass spectrometry, a window into the metabolic exchange patterns of microbes. J Proteomics. 2012;75:5069–76. doi: 10.1016/j.jprot.2012.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin RJA, Scullion P, MacIntyre L, et al. Use of a solvent-free dry matrix coating for quantitative matrix-assisted laser desorption ionization imaging of 4-bromophenyl-1,4-diazabicyclo(3.2.2)nonane-4-carboxylate in rat brain and quantitative analysis of the drug from laser microdissected tissue regions. Anal Chem. 2010;82:3868–73. doi: 10.1021/ac100398y. [DOI] [PubMed] [Google Scholar]
- Hoefler BC, Gorzelnik KV, Yang JY, et al. Enzymatic resistance to the lipopeptide surfactin as identified through imaging mass spectrometry of bacterial competition. P Natl Acad Sci USA. 2012;109:13082–7. doi: 10.1073/pnas.1205586109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann T, Dorrestein PC. Homogeneous matrix deposition on dried agar for MALDI imaging mass spectrometry of microbial cultures. J Am Soc Mass Spectr. 2015;26:1959–62. doi: 10.1007/s13361-015-1241-8. [DOI] [PubMed] [Google Scholar]
- Kaltenpoth M, Strupat K, Svatos A. Linking metabolite production to taxonomic identity in environmental samples by (MA) LDI-FISH. ISME J. 2016;10:527–31. doi: 10.1038/ismej.2015.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger JR, Kourtev PS. Bacterial diversity in three distinct sub-habitats within the pitchers of the northern pitcher plant, Sarracenia purpurea. FEMS Microbiol Ecol. 2011;79:555–67. doi: 10.1111/j.1574-6941.2011.01240.x. [DOI] [PubMed] [Google Scholar]
- Lanni EJ, Masyuko RN, Driscoll CM, et al. Correlated imaging with C-60-SIMS and confocal raman microscopy: visualization of cell-scale molecular distributions in bacterial biofilms. Anal Chem. 2014a;86:10885–91. doi: 10.1021/ac5030914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanni EJ, Masyuko RN, Driscoll CM, et al. MALDI-guided SIMS: multiscale imaging of metabolites in bacterial biofilms. Anal Chem. 2014b;86:9139–45. doi: 10.1021/ac5020222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie KB, Bowen BP, Cheng X, et al. “Replica-extraction-transfer” nanostructure-initiator mass spectrometry imaging of acoustically printed bacteria. Anal Chem. 2013;85:10856–62. doi: 10.1021/ac402240q. [DOI] [PubMed] [Google Scholar]
- Medema MH, Blin K, Cimermancic P, et al. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011;39:w339–46. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menezes RC, Kai M, Krause K, et al. Monitoring metabolites from Schizophyllum commune interacting with Hypholoma fasciculare combining LESA-HR mass spectrometry and Raman microscopy. Anal Bioanal Chem. 2015;407:2273–82. doi: 10.1007/s00216-014-8383-6. [DOI] [PubMed] [Google Scholar]
- Moree WJ, Phelan VV, Wu C-H, et al. Interkingdom metabolic transformations captured by microbial imaging mass spectrometry. P Natl Acad Sci USA. 2012;109:13811–6. doi: 10.1073/pnas.1206855109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowat E, Rajendran R, Williams C, et al. Pseudomonas aeruginosa and their small diffusible extracellular molecules inhibit Aspergillus fumigatus biofilm formation. FEMS Microbiol Lett. 2010;313:96–102. doi: 10.1111/j.1574-6968.2010.02130.x. [DOI] [PubMed] [Google Scholar]
- Murfin KE, Lee M-M, Klassen JL, et al. Xenorhabdus bovienii strain diversity impacts coevolution and symbiotic maintenance with Steinernema spp. nematode hosts. mBio. 2015;6:e00076. doi: 10.1128/mBio.00076-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalca Y, Jansch L, Bredenbruch F, et al. Quorum-sensing antagonistic activities of azithromycin in Pseudomonas aeruginosa PAO1: a global approach. Antimicrob Agents Ch. 2006;50:1680–8. doi: 10.1128/AAC.50.5.1680-1688.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DD, Wu C-H, Moree WJ, et al. MS/MS networking guided analysis of molecule and gene cluster families. P Natl Acad Sci USA. 2013;110:E2611–20. doi: 10.1073/pnas.1303471110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan VV, Fang J, Dorrestein PC. Mass spectrometry analysis of Pseudomonas aeruginosa treated with azithromycin. J Am Soc Mass Spectr. 2015;26:873–7. doi: 10.1007/s13361-015-1101-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan VV, Moree WJ, Aguilar J, et al. Impact of a transposon insertion in phzF2 on the specialized metabolite production and interkingdom interactions of Pseudomonas aeruginosa. J Bacteriol. 2014;196:1683–93. doi: 10.1128/JB.01258-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach PJ, Laskin J, Laskin A. Molecular characterization of organic aerosols using nanospray-desorption/electrospray ionization-mass spectrometry. Anal Chem. 2010;82:7979–86. doi: 10.1021/ac101449p. [DOI] [PubMed] [Google Scholar]
- Shank EA, Klepac-Ceraj V, Collado-Torres L, et al. Interspecies interactions that result in Bacillus subtilis forming biofilms are mediated mainly by members of its own genus. P Natl Acad Sci USA. 2011;108:E1236–43. doi: 10.1073/pnas.1103630108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva LP, Northen TR. Exometabolomics and MSI: deconstructing how cells interact to transform their small molecule environment. Curr Opin Biotech. 2015;34:209–16. doi: 10.1016/j.copbio.2015.03.015. [DOI] [PubMed] [Google Scholar]
- Straight PD, Willey JM, Kolter R. Interactions between Streptomyces coelicolor and Bacillus subtilis: role of surfactants in raising aerial structures. J Bacteriol. 2006;188:4918–25. doi: 10.1128/JB.00162-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takáts Z, Wiseman JM, Gologan B, et al. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science. 2004;306:471–3. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- Tateda K, Comte R, Pechere JC, et al. Azithromycin inhibits quorum sensing in Pseudomonas aeruginosa. Antimicrob Agents Ch. 2001;45:1930–3. doi: 10.1128/AAC.45.6.1930-1933.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traxler MF, Watrous JD, Alexandrov T, et al. Interspecies interactions stimulate diversification of the Streptomyces coelicolor secreted metabolome. mBio. 2013;4:e00459-1-13. doi: 10.1128/mBio.00459-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimpin S, Inutan ED, Herath TN, et al. Matrix-assisted laser desorption/ionization mass spectrometry method for selectively producing either singly or multiply charged molecular ions. Anal Chem. 2010;82:11–5. doi: 10.1021/ac902066s. [DOI] [PubMed] [Google Scholar]
- Watrous J, Roach P, Alexandrov T, et al. Mass spectral molecular networking of living microbial colonies. P Natl Acad Sci USA. 2012;109:E1743–52. doi: 10.1073/pnas.1203689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watrous J, Roach P, Heath B, et al. Metabolic profiling directly from the petri dish using nanoDESI imaging mass spectrometry. Anal Chem. 2013a;85:10385–91. doi: 10.1021/ac4023154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watrous JD, Dorrestein PC. Imaging mass spectrometry in microbiology. Nat Rev Microbiol. 2011;9:683–94. doi: 10.1038/nrmicro2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watrous JD, Phelan VV, Hsu C-C, et al. Microbial metabolic exchange in 3D. ISME J. 2013b;7:770–80. doi: 10.1038/ismej.2012.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe BE, Dutton RJ. Fermented foods as experimentally tractable microbial ecosystems. Cell. 2015;161:49–55. doi: 10.1016/j.cell.2015.02.034. [DOI] [PubMed] [Google Scholar]
- Woo H-K, Northen TR, Yanes O, et al. Nanostructure-initiator mass spectrometry: a protocol for preparing and applying NIMS surfaces for high-sensitivity mass analysis. Nat Protoc. 2008;3:1341–9. doi: 10.1038/nprot.2008.110. [DOI] [PubMed] [Google Scholar]
- Yang JY, Phelan VV, Simkovsky R, et al. Primer on Agar-Based Microbial Imaging Mass Spectrometry. J Bacteriol. 2012;194:6023–8. doi: 10.1128/JB.00823-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y-L, Xu Y, Straight P, et al. Translating metabolic exchange with imaging mass spectrometry. Nat Chem Biol. 2009;5:885–7. doi: 10.1038/nchembio.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Gemperline E, Venkateshwaran M, et al. MALDI mass spectrometry-assisted molecular imaging of metabolites during nitrogen fixation in the Medicago truncatula-Sinorhizobium meliloti symbiosis. Plant J. 2013;75:130–45. doi: 10.1111/tpj.12191. [DOI] [PubMed] [Google Scholar]


