Abstract
All positive-strand RNA [(+)RNA] viruses replicate their RNA on intracellular membranes, often in association with spherular invaginations of the target membrane. For brome mosaic virus, we previously showed that such spherules serve as compartments or mini-organelles for RNA replication and that their assembly, structure, and function have similarities to the replicative cores of retrovirus and double-stranded RNA virus virions. Some other (+)RNA viruses conduct RNA replication in association with individual or clustered double-membrane vesicles, appressed double membranes, or other structures whose possible relationships to the spherular invaginations are unclear. Here we show that modulating the relative levels and interactions of brome mosaic virus replication factors 1a and 2a polymerase (2apol) shifted the membrane rearrangements associated with RNA replication from small invaginated spherules to large, karmellae-like, multilayer stacks of appressed double membranes that supported RNA replication as efficiently as spherules. Spherules were induced by expressing 1a, which has functional similarities to retrovirus virion protein Gag, or 1a plus low levels of 2apol. Double-membrane layers were induced by 1a plus higher levels of 2apol and were suppressed by deleting the major 1a-interacting domain from 2apol. The stacked, double-membrane layers alternated with spaces that, like spherule interiors, were 50–60 nm wide, connected to the cytoplasm, and contained 1a and 2apol. These and other results suggest that seemingly diverse membrane rearrangements associated with RNA replication by varied (+)RNA viruses may represent topologically and functionally related structures formed by similar protein–protein and protein–membrane interactions and interconverted by altering the balances among those interactions.
Positive-strand RNA [(+)RNA] viruses are the largest genetic class of viruses and include many pathogens, such as the severe acute respiratory syndrome (SARS) coronavirus, hepatitis C virus, and potential bioterrorism agents. Such (+)RNA viruses encapsidate messenger-sense genomic RNAs and replicate those genomes through negative-strand RNA intermediates. The RNA replication complexes of (+)RNA viruses invariably form on intracellular membranes, usually in association with vesiculation or other membrane rearrangements. Different (+)RNA viruses use distinct but usually specific membranes, ranging from the outer membranes of the endoplasmic reticulum (ER), later or mixed compartments of the secretory pathway, endosomes, mitochondria, and other organelles (1–7).
Many (+)RNA viruses, including alphaviruses, nodaviruses, bromoviruses, and many others form RNA replication complexes at virus-induced, vesicular invaginations of specific intracellular membranes (4–6, 8–11). One such virus is brome mosaic virus (BMV), a member of the alphavirus superfamily of human, animal, and plant viruses. BMV encodes two proteins that direct viral RNA replication in its natural plant hosts or yeast. Viral replication factor 1a contains a C-terminal helicase domain and a self-interacting N-terminal domain with m7G methyltransferase and covalent m7GMP-binding activities required for viral RNA capping (12–14). Viral replication factor 2a polymerase (2apol) contains a central polymerase domain and an N-terminal domain that interacts with the 1a helicase domain (15, 16). Replication factor 1a localizes to outer perinuclear ER membranes, where it induces membrane lipid synthesis and 50- to 60-nm vesicular invaginations or spherules, connected by means of necks to the cytoplasm, that serve as compartments or miniorganelles for RNA replication (6, 17–19). 1a also recruits viral RNA templates and 2aPol to these compartments, which concentrate replication factors, link successive replication steps, and protect double-stranded RNA intermediates from host RNA interference and IFN responses (6, 16, 20–22). The roles of 1a, 2aPol, and certain cis RNA signals in assembly and function of these intracellular spherular BMV RNA replication complexes parallel those of Gag-, Pol-, and RNA-packaging signals in the membrane-enveloped replicative cores of retrovirus virions (6). These roles also show similarities to double-stranded RNA virus virion cores, suggesting functional and possible evolutionary links among these three virus classes (6).
Although all (+)RNA viruses assemble their replication complexes on membranes, RNA replication by some (+)RNA viruses induce apparently distinct membrane rearrangements involving alternate vesicle types, appressed membranes, or both. Flavivirus replication factors and double-stranded RNA replication intermediates colocalize in packets of 50- to 100-nm vesicles enclosed in a second bounding membrane (1). Replication of the 12.5-kb arterivirus RNA localizes to 80-nm double-membrane vesicles (2), and the related coronaviruses replicate their >30-kb genomes in association with 200- to 350-nm double-membrane vesicles (23). Poliovirus RNA replication is associated with clusters of 150- to 300-nm, double-membrane-bounded vesicles (3, 24). RNA replication has been proposed to occur in the space between these clustered vesicles (25).
Here we show that modulating the relative levels of BMV replication factors 1a and 2apol, and, thus, the balance of their homotypic and heterotypic interactions, shifts the structure of membrane rearrangements associated with RNA replication from small spherular invaginations, as is found in natural infections by bromoviruses and many other (+)RNA viruses (4–6, 8–11), to large stacks of karmellae-like, appressed double-membrane layers that also support efficient viral RNA replication. We also show that 1a–2apol interaction motifs are critical for inducing this new membrane rearrangement. The results suggest that alternate membrane morphologies associated with RNA replication by various (+)RNA viruses may embody common, underlying principles of architecture and assembly. The results also have implications for other features of RNA replication, such as the frequent down-regulation of polymerase expression in (+)RNA and reverse-transcribing viruses.
Materials and Methods
Yeast and Plasmids. Yeast strain YPH500 and culture conditions were as described in ref. 16. BMV 1a was expressed from the GAL1 promoter by using pB1YT3H (26). BMV 2apol was expressed from pB2CT15 (ADH1 promoter) (26), pB2YT5 (GAL1 promoter) (14), or pAON55 (CUP1 promoter), a pB2YT5 derivative with the GAL1 promoter replaced by that of CUP1. BMV RNA3 was expressed from pB3MS82, which encodes a full-length RNA3 derivative that does not express coat protein (14). Sec63-GFP was expressed from pJK59, kindly provided by J. Kahanab and P. Silver (Harvard Medical School, Cambridge, MA).
RNA and Protein Analysis. Cell fractionation, isolation of nuclei, RNA-dependent RNA polymerase reactions (6), and Northern (16) and Western blot analyses (17) were as described. RNA was purified from cell fractions by using Qiagen RNeasy columns.
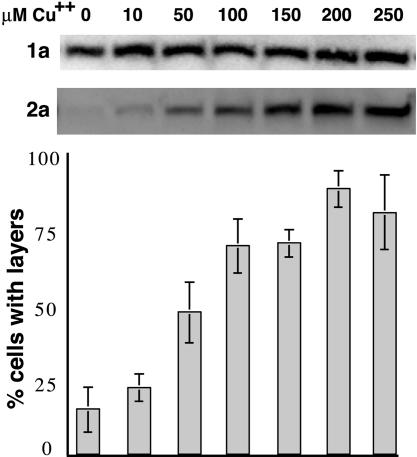
Microscopy. Confocal and electron microscopy (EM) were performed as described in refs. 6 and 16. Under EM, not all cell sections in populations expressing 1a or 1a plus 2apol show nuclei or perinuclear membrane rearrangements because of the limited cell region revealed by the random plane of sectioning, incomplete plasmid segregation and consequent absence of plasmids from significant fractions of continuously selected yeast populations (27), and other effects. Thus, the relative frequencies of perinuclear membrane layers versus spherules were calculated as the percentage of cell sections with BMV-induced perinuclear membrane rearrangements that showed layers or spherules, respectively (see Figs. 4 and 5).
Fig. 4.
Increasing 2apol levels promotes 1a/2apol-mediated induction of double-membrane layers. Western analysis (Top and Middle) showing nearly constant 1a expression from the GAL1 promoter and linearly increasing 2apol expression from the copper-inducible CUP1 promoter in yeast grown in medium with the indicated levels of CuSO4. For each CuSO4 concentration, the histogram indicates the frequency of EM-visualized cell sections showing BMV-induced perinuclear double-membrane layers, relative to cells showing BMV-induced perinuclear spherules. The results show averages over two independent experiments, and in each experiment 150- to 200-cell sections with BMV-induced membrane rearrangements were scored for each indicated CuSO4 concentration. Standard error bars are indicated.
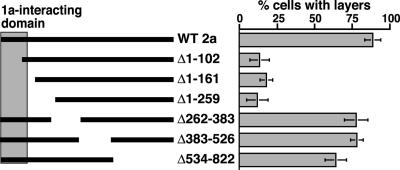
Fig. 5.
Deleting the N-proximal 1a-interacting domain of 2apol inhibits induction of double-membrane layers. Diagram of 2apol deletion derivatives with the indicated amino acids deleted. The shaded box indicates the 2apol segment directing high-affinity interaction with the 1a helicase-like domain (15, 16). The histogram indicates the frequency of EM-visualized cell sections that show BMV-induced perinuclear double-membrane layers relative to cells showing BMV-induced perinuclear spherules. The results show averages over two independent experiments, and in each experiment 150- to 200-cell sections with BMV-induced membrane rearrangements were scored for each 2apol deletion derivative. Standard error bars are indicated.
Results
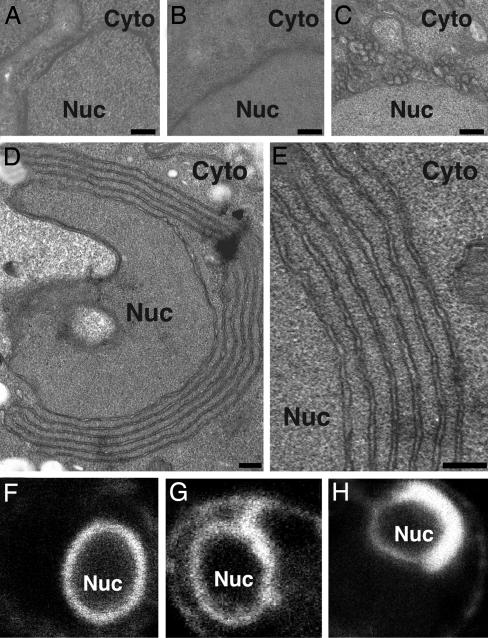
Increased 2apol Levels Alter 1a-Induced Membrane Rearrangements. BMV 1a protein associates as a peripheral membrane protein with the cytoplasmic face of the outer ER membrane and induces invaginations of this membrane into the ER lumen to form vesicles or spherules whose interiors are connected through narrow necks with the cytoplasm (6). These spherules are induced either in yeast expressing 1a alone or in yeast expressing 1a and 2apol from plasmids by using the stronger GAL1 and weaker ADH1 promoters, respectively (1aG+2aA yeast; Fig. 1C) (6). By contrast, in the great majority of yeast cells expressing both 2apol and 1a from plasmids by using the GAL1 promoter (1aG+2aG yeast), we discovered by EM that the perinuclear membrane did not form spherules but proliferated into a series of 2–7 appressed layers of double-membrane ER (Fig. 1 D and E). These karmellae-like, multilayer structures were formed by folding over continuous sheets of ramified, double-membrane ER with its enclosed lumen (Fig. 1D). The successive, double-membrane ER layers were separated by regular, 50- to 60-nm spaces (Fig. 1E), which at their ends were contiguous with the cytoplasm (Fig. 1D, top and bottom left). Yeast expressing 2apol from either the GAL1 or ADH1 promoters but lacking 1a lacked either layers or spherules and showed no detectable membrane changes from WT yeast (Fig. 1B). By contrast, yeast expressing 1a from either the GAL1 or ADH1 promoters but lacking 2apol contained perinuclear spherules but never layers (ref. 6 and results not shown).
Fig. 1.
Two alternate membrane rearrangements are induced by 1a and 2apol. (A–E) Representative electron micrographs of yeast cells expressing no BMV components (A), GAL1 promoter-driven 2apol (B), GAL1 promoter-driven 1a and ADH1 promoter-driven 2apol (1aG+2aA) (C), or GAL1 promoter-driven 1a and 2aPol (1aG+2aG)(D and E). (F–H) Representative confocal fluorescence images of live yeast cells expressing Sec63p-GFP and no BMV components (F), 1aG+2aA (G), or 1aG+2aG (H). Nuc, nucleus; Cyto, cytoplasm. (Scale bars, 100 nm.)
Confocal imaging of live cells expressing a fusion of GFP to Sec63, an integral ER membrane protein, correlated well with the higher resolution ultrastructure obtained by EM. In cells not expressing BMV components, Sec63-GFP defined a perinuclear ER layer of relatively uniform thickness (Fig. 1F). Yeast expressing 1aG + 2aA, which contained perinuclear spherules (Fig. 1C), showed modest thickening of some perinuclear sections (Fig. 1G). In 1aG+2aG yeast, which contained predominantly BMV-induced double-membrane layers, large sections of the GFP-fluorescent perinuclear layer showed a strikingly greater thickening (Fig. 1H). Thus, the Sec63-GFP results confirmed that, in the presence of 1a, 2apol expression from the stronger GAL1 promoter induced alternate perinuclear membrane changes in live cells and not just in cells fixed for EM analysis.
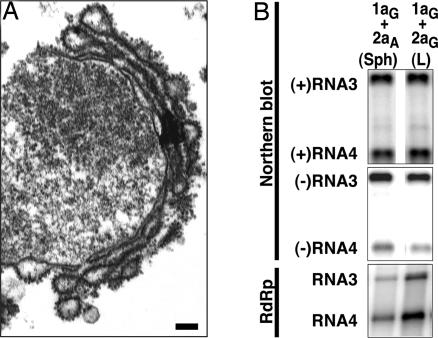
Double-Membrane Layers Contain 1a and 2apol and Support BMV RNA Replication. Yeast expressing BMV 1a, 2apol, and BMV genomic RNA3 support RNA3 replication, including production of negative-strand (–)RNA3 that is copied to dramatically amplify (+)RNA3 and to produce the subgenomic mRNA RNA4 (26, 28). Northern blot analysis (Fig. 2A) showed that, when RNA3 was expressed from a third plasmid, similar levels of RNA3 replication and subgenomic RNA4 production occurred in 1aG+2aA and 1aG+2aG yeast, which contained membrane spherules or layers, respectively (Fig. 1). Thus, BMV RNA replication can occur in association with two distinct membrane architectures.
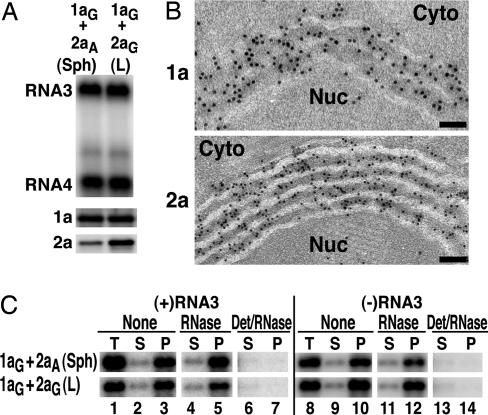
Fig. 2.
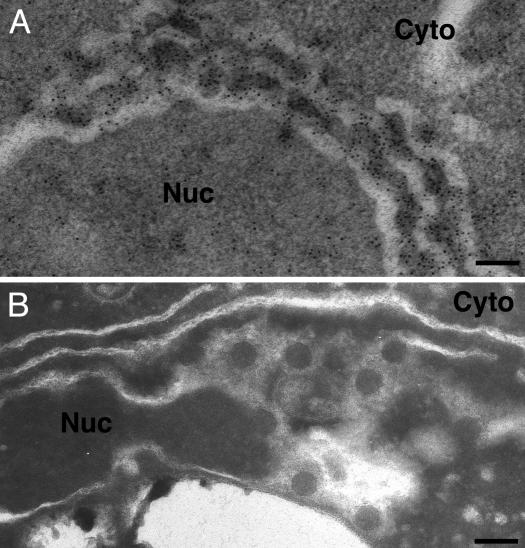
Double-membrane layers induced by 1a/2apol contain 1a and 2apol and support BMV RNA replication and 1a-induced membrane association and nuclease resistance of viral RNAs. (A) Northern and Western blot analyses of RNA3, RNA4, 1a and 2apol accumulation in 1aG+2aA yeast that contain perinuclear spherules (Sph), or 1aG+2aG yeast that contain double-membrane layers (L). (B) ImmunoGold EM localization of 1a (Upper) and 2apol (Lower) in 1aG+2aG yeast containing double-membrane layers. Replication factor 1a was localized with polyclonal anti-1a antiserum (17). Because anti-2a antibodies gave weak ImmunoGold labeling, 2apol was localized by using an anti-GFP monoclonal antibody (JL-8, Clontech) to detect a 2apol-GFP fusion that supports BMV RNA replication (16) and induced ultrastructural changes indistinguishable from WT 2apol (compare with Fig. 1D). Nuc, nucleus; Cyto, cytoplasm. (Scale bars, 100 nm.) (C) Northern blot analysis of cell fractionation extracts from 1aG+2aA yeast that contain spherules (upper row) and 1aG+2aG yeast that contain double-membrane layers (lower row). The analysis shows the distribution of (+)RNA3 and (–)RNA3 in total lysate (T), 20,000 × g membrane-depleted supernatant (S), or membrane-enriched pellet (P) fractions after one of the following treatments: no additional treatment (none), addition of 0.01 units/ml micrococcal nuclease/1 mM CaCl2 for 15 min at 30°C (RNase), or addition of 0.5% Nonidet P-40 for 15 min at 4°C followed by nuclease treatment (Det/RNase).
Yeast expressing 1aG+2aG accumulate 1a and 2apol in a 1a/2apol ratio of ≈25 (6). Western blot analysis confirmed that the levels of 1a accumulation were indistinguishable in 1aG+2aA and 1aG+2aG yeast (Fig. 2A). However, as expected, GAL1-promoted 2apol expression associated with membrane layers increased 2apol levels by >2-fold relative to the ADH1-promoted 2apol expression associated with spherules (Fig. 2A).
Confocal ImmunoGold EM and biochemical analyses showed previously that 1a localizes both itself and 2apol to the cytoplasmic face of the perinuclear ER membrane and that, in 1aG+2aA yeast, each BMV-induced spherule contains hundreds of 1a proteins and ≈10–15 2apol proteins (6, 17, 18, 29). ImmunoGold EM analysis showed that, in 1aG+2aG yeast, 1a and 2apol localized to the perinuclear membrane layers, with ≈80% of the ImmunoGold label in the cytoplasmic spaces between the double-membrane ER layers or on the cytoplasmic face of the outermost membrane layer (Fig. 2B). Some clustering of gold particles occurred, suggesting possible points of 1a and 2apol concentration within the layers (see also below).
Cells Containing Membrane Layers Sequester (+)RNA3 and (–)RNA3 Templates in a Membrane-Associated, Nuclease-Resistant State. In the absence of 2apol, 1a acts through a specific cis-acting RNA sequence to recruit RNA3 replication templates to a membrane-associated, nuclease-resistant state (6, 20, 21). This state appears to correspond to the interior of the 1a-induced spherules because, in 1aG+2aA yeast replicating RNA3, (+)RNA3 and (–)RNA3 templates and nascent RNA are retained in an indistinguishable, membrane-associated, nuclease-resistant state and ImmunoGold EM localizes BrUTP-labeled nascent RNA to spherules (6).
In 1aG+2aG yeast containing double-membrane layers, we found similar membrane association and protection of (+)RNA3 and (–)RNA3 (Fig. 2C). Samples of 1aG+2aA or 1aG+2aG yeast replicating RNA3 and containing spherules or layers, respectively, were treated with lyticase to remove cell walls and then lysed and centrifuged at 20,000 × g to yield a membrane-enriched pellet (P) and a cytosolic supernatant (S). In both cases, the majority of (+)RNA3 and (–)RNA3 was recovered in the membrane-containing pellet, with little RNA3 in the supernatant (Fig. 2C, lanes 1–3 and 8–10). This membrane-associated RNA3 was highly resistant to added nuclease (Fig. 2C, lanes 4–5 and 11–12) but became nuclease-susceptible after treatment with a membrane-disrupting, nonionic detergent (Fig. 2C, lanes 6–7 and 13–14). Thus, in association with both membrane architectures, (+)RNA3 and (–)RNA3 were found in a membrane-associated, nuclease-resistant, nonionic detergent-susceptible state.
Isolated Nuclei Retain Double-Membrane Layers, Viral (+)RNA and (–)RNA, and RNA-Dependent RNA Polymerase. To further characterize the perinuclear, double-membrane layers, nuclei were isolated from 1aG+2aG yeast. These nuclei retained double-membrane layers that were stable through nuclear isolation and EM analysis, suggesting relatively strong adhesion between the double-membrane layers (Fig. 3A). Similarly, nuclei isolated from 1aG+2aA yeast retain perinuclear spherules (6). Prior results show that 1a is required to direct RNA3 to nuclear membranes and that in the absence of 1a, RNA3 fractionates with the cytosol (6). Nuclei isolated from cells containing membrane layers or spherules contained similar levels of (+)RNA3, (–)RNA3, and RNA4 (Fig. 3B). Nuclei from both types of 1a- and 2apol-expressing yeast, but not from WT yeast, also incorporated radiolabeled ribonucleotides into BMV RNA replication intermediates (Fig. 3B) (6).
Fig. 3.
Isolated nuclei retain perinuclear, double-membrane layers, BMV RNA templates, and BMV-specific RNA-dependent RNA polymerase (RdRp) activity. (A) Representative electron micrograph of a nucleus isolated from 1aG+2aG yeast and bearing 1a/2apol-induced, double-membrane layers. (Scale bar, 100 nm.) (B) Northern blot analysis of (+)- and (–)-strand RNA3 and RNA4 and BMV-specific RNA-dependent RNA polymerase activity found in preparations of nuclei isolated from 1aG+2aA yeast that contain perinuclear spherules (Sph) or 1aG+2aG yeast containing double-membrane layers (L).
2apol Levels Modulate Layer Formation. To further explore the relationship between double-membrane layers and 2apol levels (Fig. 2A), the copper-responsive, linearly inducible CUP1 promoter was used to regulate 2apol levels in cells coexpressing 1a from the standard GAL1-promoted 1a expression plasmid. As shown in Fig. 4, increasing [Cu2+] in the medium progressively increased 2apol accumulation, whereas 1a levels remained essentially constant. EM analysis showed that the percentage of cells with perinuclear membrane layers increased in parallel with 2apol (Fig. 4), whereas the percentage of cells with perinuclear spherules declined correspondingly. In the absence of added Cu2+, little 2apol was detected, but a low percentage of cells contained membrane layers. However, when the CUP1-2apol plasmid was omitted, no membrane layers were seen at any [Cu2+] concentration tested, confirming that 2apol was required to induce layers and that Cu2+ alone did not promote layering.
Deleting a 1a-Interactive, N-Terminal 2apol Domain Inhibits Membrane Layer Formation. BMV 1a and 2apol interact in vitro and in vivo through the 1a C-terminal helicase domain and 2apol amino acids 50–113 (15, 16). To test the contribution of this 1a/2apol interaction to 1a- and 2apol-dependent induction of perinuclear membrane layers, we constructed a series of 2apol deletions and assayed their ability to induce layers when expressed from the GAL1 promoter in yeast coexpressing 1a (Fig. 5). Western blot analysis with multiple monoclonal antibodies against various regions of 2apol to visualize all deletion derivatives showed that each derivative accumulated to levels close to WT 2apol (results not shown). Deletions overlapping the polymerase domain but retaining the 1a-interactive domain (amino acids 262–383, 385–526, or 534–822) induced double-membrane layers at frequencies approaching those of WT (64–78%). By contrast, all deletions overlapping the N-proximal, 1a-interactive 2apol domain, including deleting the first 102, 161, or 259 amino acids, drastically reduced the frequency of perinuclear membrane layers (12–18%) relative to WT 2apol (92%). The low, residual frequency of membrane layering in the absence of the N-proximal 2apol domain may depend on lower affinity interaction between 1a and the central polymerase domain of 2apol (30).
Internal Structures Between Double-Membrane Layers. Although the BMV-induced double-membrane layers showed relatively smooth contours in most EM images of fixed cells (Fig. 1 D and E), most isolated nuclei (Fig. 3A) and a smaller percentage of fixed cells (Figs. 2B and 6A) showed perinuclear membrane layers with a distinctly ruffled or corrugated morphology. Thus, in at least some cases, the intermembrane space was not uniform but possessed some underlying variation or structure. Moreover, in rare instances, oblique sections of the membrane layers exposing significant areas of intermembrane space revealed the presence of spheres whose 50- to 60-nm diameters corresponded closely to those of spherules and to the average spacing of ER membrane layers (Fig. 6B). The possible relationship of membrane ruffling and these spherical structures to the underlying architecture of the intermembrane space is considered further in Discussion.
Fig. 6.
Underlying structure in the cytoplasm-connected spaces between double-membrane layers in 1aG+2aG yeast. See Results for further comments. Nuc, nucleus; Cyto, cytoplasm. (Scale bar, 100 nm.)
Discussion
As noted in the Introduction, the universal membrane association of (+)RNA virus RNA replication appears crucial to replication complex assembly and function, yet RNA replication by different (+)RNA viruses induces varied membrane rearrangements including invaginations, double-membrane vesicles, and layered membranes. The results presented here suggest that such apparently distinct morphologies may share underlying structural features. Specifically, modulating the relative expression or interactions of BMV RNA replication proteins 1a and 2apol switched the membrane rearrangements associated with RNA replication from the spherular invaginations (spherules) found in natural infections by bromoviruses, the related alpha-viruses, and many other (+)RNA viruses (4–6, 8–11) to stacked layers of double ER membranes (Fig. 1). Like spherules, these stacked, double-membrane layers were the sites of 1a and 2apol accumulation, protected viral RNA templates from nuclease, and supported RNA replication (Figs. 2 and 3). Below we discuss relevant interactions of 1a and 2apol, the underlying structure of membrane spherules and layers, and possible relationships to other (+)RNA viruses.
Modulation of Replication-Associated Membrane Rearrangements by 2apol. In the absence of other viral factors, 1a induces membrane spherules but never layers, whereas 2apol alone induces no membrane rearrangements (Fig. 1B and ref. 6). Formation of double-membrane layers required coexpressing 1a and 2apol, was progressively favored by increasing 2apol levels (Fig. 4), and was inhibited by deleting the major 1a-interacting domain from 2apol (Fig. 5). Double-membrane layers also were promoted by reducing 1a expression relative to 2apol. When 1a was expressed from the weaker ADH1 promoter, cells formed layers when 2apol was expressed from either the GAL1 or ADH1 promoter (unpublished results).
Replication factor 1a interacts with ER membranes and itself (12, 29) and is present at hundreds of copies per spherule (6), suggesting that 1a may induce spherules by forming a virion-like shell (Fig. 7A) (6). At least two nonexclusive mechanisms could explain how increasing 2apol expression relative to 1a induces membrane layering rather than spherules. First, similar perinuclear layers of double ER membranes, termed karmellae, and other organized smooth ER double-membrane arrays, including whorls, sinusoidal arrays, and crystalloid ER, are formed when certain ER-associated proteins like 3-hydroxy-3-methylglutaryl CoA reductase are expressed above a threshold level (31). Such membrane stacking requires interaction between the cytoplasmic domains of the inducing membrane proteins but, when the responsible membrane protein is expressed at high levels, even low-affinity, dynamic interactions suffice (31).
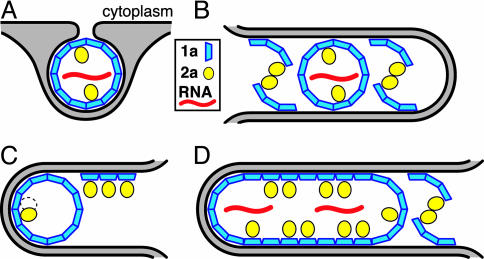
Fig. 7.
Models for 2apol modulation of 1a-dependent membrane rearrangements. See Discussion for further comments. (A) Invagination of a spherule (≈50 nm in diameter) into the outerperinuclear ER membrane by 1a formation of a membrane-enveloped, capsid-like shell with 1a recruitment of 2apol and viral RNA. Bold lines depict ER membranes and gray shading depicts the ER lumen. (B) High 2apol concentrations may promote zippering together of double-membrane layers by 1a–2a complexes. Replication factor 1a might participate as monomers or multimers (hexamers or pentamers). Self-interaction of 2apol might be direct or mediated by RNA or 2apol-interacting host proteins (33). As suggested by Fig. 6, spherule cores may be trapped in such layers. (C) High 2apol concentrations may block 1a–1a curvature needed to form spherules by steric hindrance between 2apol factors bound to adjacent 1a factors (left side), tending to restrict 1a to planar lattices (right side). (D) Between the stacked double-membrane layers, extended planar lattices of 1a with bound 2apol might provide a local replication complex environment similar to spherule interiors and may be closed by occasional regions of 1a curvature.
At sufficient levels, 2apol may induce stable layer formation by mediating similar zippering interactions between opposing membranes. Clustering of 2apol in vivo (16) suggests that 2apol interacts with itself, and the related poliovirus 3D polymerase self-interacts to form planar or tubular lattices (32). In keeping with the dependence of membrane layering on 2apol interaction with 1a (Fig. 5), 1a–2apol complexes might zipper together opposing membranes by means of 2apol–2apol interactions (Fig. 7B). Such 2apol–2apol interactions may be direct or mediated by RNA or 2apol-interacting host proteins (33). Most proteins inducing organized ER membrane arrays induce a cytoplasmic space of 8–11 nm between the stacked layers of double-membrane ER (31). The ≈50-nm spacing between double-membrane layers induced by 1a and 2apol suggests bridging by larger complexes, such as whole-spherule cores (Fig. 6), 1a hexamers or pentamers (see below), 2apol dimers or multimers, or protein-RNA complexes. Spherule cores trapped in these layers (Figs. 6 and 7B) could support RNA replication.
Alternatively or in addition, 2apol overexpression might promote ER membrane layering by inhibiting 1a formation of spherules. Like the CA subunit of HIV Gag (34), 1a may assemble on membranes as a planar hexameric lattice, into which pentamer defects are introduced to generate curvature and invaginate the spherule replication compartments. The 1a–2apol complex, which parallels many aspects of retroviral Gag-Pol fusion proteins, is incorporated into spherules at a 1a/2apol ratio of ≈25, similar to the Gag/Gag-Pol virion ratio of ≈20 (6). Gag-Pol overexpression interferes with HIV virion assembly (35), possibly because the virion interior, to which Pol is localized, lacks room for Pol on more than a fraction of Gags (36). Similarly, 2apol association with adjacent 1a replication factors may sterically block the 1a–1a curvature needed to form spherules (Fig. 7C). Thus, above a threshold density, 2apol binding might tend to restrict 1a to planar lattices on ER membranes (Fig. 7 C and D). The resulting membranes may be linked by occasional 1a–2apol bridges as noted above (Fig. 7B), and/or by occasional regions of curvature that allow the extended 1a lattice to close (Fig. 7D). The environment between such 1a and 1a–2apol bearing lattices would be locally similar to spherule interiors and might support RNA replication.
Pol Down-Regulation in (+)RNA Viruses and Retroviruses. Because 2apol levels and interactions dramatically affect BMV replication complex ultrastructure, it is notable that bromoviruses and many other (+)RNA viruses have multiple mechanisms to reduce polymerase expression, accumulation, and interaction. Like retroviruses, many (+)RNA viruses, including alphaviruses, coronaviruses, tobamoviruses, and others, use translational frame-shift or read-through to reduce polymerase expression 10- to 20-fold relative to upstream factors related to BMV 1a, and this regulation is linked to replicative fitness (37, 38). BMV, which encodes 1a and 2apol on separate genomic RNAs, inhibits 2apol translation at initiation (39). BMV and alphaviruses also regulate polymerase stability (40, 41). Bromovirus 1a–2apol interaction is further down-regulated by competing intramolecular 1a–1a interaction and 2apol phosphorylation (12, 42). Picornaviruses and some other (+)RNA viruses express polymerase at levels equimolar with other replication factors. Some of these viruses, such as the picornavirus-like potyviruses, sequester large amounts of excess polymerase away from RNA replication in self-assembled nuclear inclusions (43).
Relation to Other (+)RNA Viruses. The ability to experimentally modulate 1a–2apol interactions to switch the ultrastructure of functional BMV RNA replication complexes between small vesicular invaginations and extensive double-membrane layers suggests that seemingly diverse membrane rearrangements associated with RNA replication by varied (+)RNA viruses may represent related structures formed by similar protein–protein and protein–membrane interactions (Fig. 7). Similarly, retrovirus Gag proteins use a common set of underlying interactions to assemble sheets, tubes, cones, or isometric shells under various circumstances (34).
The varied membrane rearrangements associated with (+)RNA virus replication share some similarities. Flavivirus RNA replication localizes to packets of 50- to 100-nm vesicles surrounded by a second membrane (1). These vesicle packets appear similar to EM views of spherules invaginated into ER and mitochondrial lumens by BMV and nodaviruses, respectively, when sectioned perpendicular to the direction of invagination (refs. 6 and 9 and unpublished results). Picornavirus, coronavirus and arterivirus RNA synthesis occurs in or on double-membrane vesicles (2, 3, 23). Although BMV spherules typically are invaginated into a dilated perinuclear ER lumen and are tightly wrapped only by the outer ER membrane, the inner ER membrane constitutes a second bounding membrane surrounding the spherule, making them also double-membrane structures (Fig. 7A). Arterivirus double-membrane vesicles are thought to form similarly to spherules by invagination of appressed ER membranes (2). Poliovirus double-membrane-bounded vesicles appear to be formed by ER membranes wrapping around a portion of cytoplasm, which is topologically equivalent to invaginating a portion of ER membrane and its surrounding cytoplasm into the ER lumen (ref. 3; compare with Fig. 7A). Some EM sections of poliovirus double-membrane vesicles reveal a narrow neck at which, as for spherules, the inner and outer membranes are continuous and inner vesicle contents connect with the cytoplasm (44).
Like 1a/2apol-induced double-membrane layers, picornavirus double-membrane vesicles cluster by interaction of surface membranes carrying viral RNA replication factors (24, 25, 32). Two of these factors, 2C and 2BC, induce ordered ER-membrane arrays, including stacked membrane layers, whorls, and crystalloid ER (45, 46). Similarly, nodavirus and tymovirus RNA replication occurs in association with BMV- and alphavirus-like spherules invaginated into the outer membranes of mitochondria and chloroplasts, respectively, and the replication factor-bearing surface membranes of these modified organelles cluster like poliovirus vesicle rosettes (9, 47). Retargeting nodavirus RNA replication protein A from mitochondria to ER also induces karmellae-like double-membrane layers that, just as for BMV, support RNA replication (48, 49).
Acknowledgments
We thank Natalie Pellechia for technical assistance, Randall Massey and Ben August (University of Wisconsin Medical School Electron Microscopy Facility) for assistance with EM, and Ann Palmenberg for comments on the manuscript. This work was supported by National Institutes of Health Grant GM35072. P.A. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations: BMV, brome mosaic virus; (+)RNA, positive-strand RNA; (–)RNA, negative-strand RNA; ER, endoplasmic reticulum; EM, electron microscopy; 2apol, 2a polymerase.
References
- 1.Westaway, E., Mackenzie, J., Kenney, M., Jones, M. & Khromykh, A. (1997) J. Virol. 71, 6650–6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pedersen, K. W., van der Meer, Y., Roos, N. & Snijder, E. J. (1999) J. Virol. 73, 2016–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suhy, D. A., Giddings, T. H., Jr., & Kirkegaard, K. (2000) J. Virol. 74, 8953–8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Froshauer, S., Kartenbeck, J. & Helenius, A. (1988) J. Cell Biol. 107, 2075–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hatta, T., Bullivant, S. & Matthews, R. E. (1973) J. Gen. Virol. 20, 37–50. [DOI] [PubMed] [Google Scholar]
- 6.Schwartz, M., Chen, J., Janda, M., Sullivan, M., den Boon, J. & Ahlquist, P. (2002) Mol. Cell 9, 505–514. [DOI] [PubMed] [Google Scholar]
- 7.Ahlquist, P., Noueiry, A. O., Lee, W. M., Kushner, D. B. & Dye, B. T. (2003) J. Virol. 77, 8181–8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kujala, P., Ahola, T., Ehsani, N., Auvinen, P., Vihinen, H. & Kaariainen, L. (1999) J. Virol. 73, 7805–7811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller, D. J., Schwartz, M. D. & Ahlquist, P. (2001) J. Virol. 75, 11664–11676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim, K. S. (1977) J. Gen. Virol. 35, 535–543. [Google Scholar]
- 11.Hatta, T. & Francki, R. I. B. (1981) J. Gen. Virol. 53, 343–346. [Google Scholar]
- 12.O'Reilly, E. K., Wang, Z., French, R. & Kao, C. C. (1998) J. Virol. 72, 7160–7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kong, F., Sivakumaran, K. & Kao, C. (1999) Virology 259, 200–210. [DOI] [PubMed] [Google Scholar]
- 14.Ahola, T., den Boon, J. A. & Ahlquist, P. (2000) J. Virol. 74, 8803–8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kao, C. C. & Ahlquist, P. (1992) J. Virol. 66, 7293–7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, J. & Ahlquist, P. (2000) J. Virol. 74, 4310–4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Restrepo-Hartwig, M. & Ahlquist, P. (1996) J. Virol. 70, 8908–8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Restrepo-Hartwig, M. & Ahlquist, P. (1999) J. Virol. 73, 10303–10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, W. M. & Ahlquist, P. (2003) J. Virol. 77, 12819–12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janda, M. & Ahlquist, P. (1998) Proc. Natl. Acad. Sci. USA 95, 2227–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sullivan, M. & Ahlquist, P. (1999) J. Virol. 73, 2622–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahlquist, P. (2002) Science 296, 1270–1273. [DOI] [PubMed] [Google Scholar]
- 23.Gosert, R., Kanjanahaluethai, A., Egger, D., Bienz, K. & Baker, S. C. (2002) J. Virol. 76, 3697–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Egger, D., Pasamontes, L., Bolten, R., Boyko, V. & Bienz, K. (1996) J. Virol. 70, 8675–8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Egger, D., Teterina, N., Ehrenfeld, E. & Bienz, K. (2000) J. Virol. 74, 6570–6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janda, M. & Ahlquist, P. (1993) Cell 72, 961–970. [DOI] [PubMed] [Google Scholar]
- 27.Christianson, T. W., Sikorski, R. S., Dante, M., Shero, J. H. & Hieter, P. (1992) Genes 110, 119–122. [DOI] [PubMed] [Google Scholar]
- 28.Ishikawa, M., Janda, M., Krol, M. A. & Ahlquist, P. (1997) J. Virol. 71, 7781–7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.den Boon, J., Chen, J. & Ahlquist, P. (2001) J. Virol. 75, 12370–12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smirnyagina, E., Lin, N. S. & Ahlquist, P. (1996) J. Virol. 70, 4729–4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snapp, E. L., Hegde, R. S., Francolini, M., Lombardo, F., Colombo, S., Pedrazzini, E., Borgese, N. & Lippincott-Schwartz, J. (2003) J. Cell Biol. 163, 257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lyle, J. M., Bullitt, E., Bienz, K. & Kirkegaard, K. (2002) Science 296, 2218–2222. [DOI] [PubMed] [Google Scholar]
- 33.Tomita, Y., Mizuno, T., Diez, J., Naito, S., Ahlquist, P. & Ishikawa, M. (2003) J. Virol. 77, 2990–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ganser, B. K., Cheng, A., Sundquist, W. I. & Yeager, M. (2003) EMBO J. 22, 2886–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shehu-Xhilaga, M., Crowe, S. M. & Mak, J. (2001) J. Virol. 75, 1834–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coffin, J. M., Hughes, S. H. & Varmus, H. E. (1997) (Cold Spring Harbor Lab. Press, Plainview, NY).
- 37.Ishikawa, M., Meshi, T., Motoyoshi, F., Takamatsu, N. & Okada, Y. (1986) Nucleic Acids Res. 14, 8291–8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, G. & Rice, C. M. (1989) J. Virol. 63, 1326–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noueiry, A. O., Chen, J. & Ahlquist, P. (2000) Proc. Natl. Acad. Sci. USA 97, 12985–12990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishikawa, M., Diez, J., Restrepo-Hartwig, M. & Ahlquist, P. (1997) Proc. Natl. Acad. Sci. USA 94, 13810–13815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.deGroot, R. J., Rümenapf, T., Kuhn, R. J., Strauss, E. G. & Strauss, J. H. (1991) Biochemistry 88, 8967–8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim, S. H., Palukaitis, P. & Park, Y. I. (2002) EMBO J. 21, 2292–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Restrepo, M. A., Freed, D. D. & Carrington, J. C. (1990) Plant Cell 2, 987–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schlegel, A., Giddings, J. T., Ladinsky, M. & Kirkegaard, K. (1996) J. Virol. 70, 6576–6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cho, M. W., Teterina, N., Egger, D., Bienz, K. & Ehrenfeld, E. (1994) Virology 202, 129–145. [DOI] [PubMed] [Google Scholar]
- 46.Teterina, N. L., Bienz, K., Egger, D., Gorbalenya, A. E. & Ehrenfeld, E. (1997) Virology 237, 66–77. [DOI] [PubMed] [Google Scholar]
- 47.Prod'homme, D., Le Panse, S., Drugeon, G. & Jupin, I. (2001) Virology 281, 88–101. [DOI] [PubMed] [Google Scholar]
- 48.Miller, D. J., Schwartz, M. D., Dye, B. T. & Ahlquist, P. (2003) J. Virol. 77, 12193–12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rubino, L., Di Franco, A. & Russo, M. (2000) J. Gen. Virol. 81, 279–286. [DOI] [PubMed] [Google Scholar]