Abstract
Activation of telomerase is crucial for cells to gain immortality. Most normal human somatic cells have a limited proliferative life span, and expression of the rate-limiting telomerase catalytic subunit, known as human telomerase reverse transcriptase (hTERT), has been believed to be tightly repressed. This model of hTERT regulation is challenged by the recent identification of the induction of hTERT in normal cycling human fibroblasts during their transit through S phase. Here we show the small-molecule-based identification of the assembly and disassembly of E2F–pocket protein–histone deacetylase (HDAC) complex as a key mechanistic basis for the repression and activation of hTERT in normal human cells. A cell-based chemical screen was used to identify a small molecule, CGK1026, that derepresses hTERT expression. CGK1026 inhibits the recruitment of HDAC into E2F–pocket protein complexes assembled on the hTERT promoter. Chromatin immunoprecipitation analysis reveals dynamic alterations in hTERT promoter occupancy by E2F and pocket proteins according to the cell cycle-dependent regulation of hTERT. Dominant-negative or protein-knockout strategies to disrupt the assembly of E2F–pocket protein–HDAC complex derepress hTERT and telomerase activity. Taken together with the results on the regulatory function of these complexes in cellular senescence and tumorigenesis, our findings suggest that dynamic assembly of E2F–pocket protein–HDAC complex plays a central role in the regulation of hTERT in a variety of proliferative conditions (e.g., normal cycling, senescent, and tumor cells).
The catalytic subunit of telomerase, known as human telomerase reverse transcriptase (hTERT), is highly expressed in stem cells, germ cell lines, and most human tumors (1–3). The expression of hTERT induces telomerase activity, which prevents the mortality checkpoints evoked by programmed telomere shortening during each round of cell division. In contrast, insufficient hTERT is expressed in most mortal somatic cells for telomerase to maintain telomere length during cycles of chromosome replication (4, 5). In addition to its key role in telomerase regulation, a growing body of evidence suggests that hTERT contributes to proliferative tissue homeostasis and tumorigenesis through other activities not associated with telomere maintenance (6, 7). Despite numerous efforts to elucidate the regulatory mechanisms of hTERT, many things remain elusive (8, 9). In particular, in contrast to the long-believed view of the tight repression of hTERT in normal somatic cells, a recent finding that hTERT is expressed in cycling normal human fibroblasts at a specific cell cycle stage added further complexity to the understanding hTERT regulation (5).
Diverse approaches have been exploited to study hTERT regulation. In particular, we and one other group have successfully developed expression cloning strategies to identify the regulators of hTERT (9–12). However, this approach is based on ectopic overexpression of proteins and is often unsuccessful at revealing the genuine endogenous functions of these proteins. To explore hTERT regulatory mechanisms more efficiently, we exploited a small molecule-based chemical genetic approach (13). Our strategy here was to identify a small molecule that could derepress hTERT in normal human cells in a forward chemical genetic screen and, by finding the mode of action of this molecule, to reveal the regulatory mechanisms of hTERT.
Methods
Cell Culture, Transfection, and Luciferase Assay. IMR90 and WI38 cells were obtained from American Type Culture Collection. HFF cells were kindly provided by H. Lee (Harvard Medical School, Boston). Phoenix-ampho cells were kindly provided by G. P. Nolan (Stanford University, Stanford, CA). All cells were maintained in DMEM supplemented with 10% FBS/120 μg/ml penicillin/200 μg/ml streptomycin. Transfection was carried out with Lipofectamine (Invitrogen). Luciferase assays were performed by using the reagents from Promega.
Plasmid Constructs. The construction of p-1003, p-820, p-784, p-618, p-580, p-419, p-188, p-188-24, and p-188-51 was described in refs. 12 and 14. p24-E2F-COMPETITOR and p24-E2Fm-CONTROL were generous gifts from S. J. Weintraub (Washington University School of Medicine, St. Louis) (15). To obtain pGEM3*-188 and pGEM3*-188-E2F-m3, the hTERT promoter region was excised from p-188 with KpnI/BglII and cloned into the vector (pGEM3*) of p24-E2Fm-CONTROL after removal of its insert by SacII/BglII. pBabe-E2F-1CterSt and pBabe-puro were kindly provided by J. Campisi (16). To construct pBabe-c-Myc, pCI-puro-Myc (17) was digested with EcoRI and cloned into the EcoRI site of pBabe-puro. To generate pBabe-F-TrCP-E7 and pBabe-F-TrCP-E7(ΔDLYC), pCMV-F-TrCP-E7 and pCMV-F-TrCP-E7(ΔDLYC), kindly provided by P. M. Howley (18), were digested with EcoRI and cloned into the EcoRI site of pBabe-puro.
Screening for Small-Molecule Derepressor of hTERT. At 9 h after transfection of p-1003 into IMR90 cells, these cells were plated into 96-well plates at 15,000 cells per well and grown for 16 h before compound addition. Next, compounds in a selected set of chemical library at Korea Advanced Institute of Science and Technology were added to the plates at a final concentration of 30 μM. After 24 h, plates were assayed for luciferase activity. Twenty-five compounds increased the luciferase activity by >5-fold compared with the median value minus background for each plate. These selected compounds were evaluated by their abilities to induce hTERT mRNA in IMR90 cells by RT-PCR. Compounds were designated as hits if induction of hTERT mRNA was detected by 30 cycles of PCR amplification when used at 30 μM. Among these 12 hits, CGK1026 induced hTERT mRNA expression most significantly.
Cell Cycle Synchronization and Fluorescence-Activated Cell Sorter (FACS) Analysis. IMR90 cells were synchronized essentially as described in ref. 19. Propidium iodide staining was performed as described 19, and cells were scanned on a FACScan analyzer by using cellquest and modfit software (Becton Dickinson).
Retroviral Transduction. Infectious retroviruses were produced by transfection of retroviral plasmids into a packaging cell line, Phoenix-ampho. Supernatants from the transfected packaging cells were collected 48 h after transfection, filtered through 0.45-μm filter, and used for infection. IMR90 cells were infected with viral supernatants supplemented with 4 μg/ml polybrene. Two days later, infected cells were given puromycin (1 μg/ml) and selected for 3 days.
RNA Extraction and Semiquantitative RT-PCR. Total RNA was isolated with the Trizol reagent (Invitrogen) according to the manufacturer's instructions. The cDNA synthesis and semiquantitative PCR were performed essentially as described in ref. 14.
TRAP and X-ChIP Assays. Telomerase activity was determined by TRAP assay by using the TRAPese telomerase detection kit (Intergen, Purchase, NY) according to the manufacturer's instructions. X-ChIP assays were performed with ChIP assay kit (Upstate Biotechnology, Lake Placid, NY) as described in refs. 12 and 14.
Formaldehyde Cross-Linked Plasmid IP (Plasmid IP). At 60% confluence, IMR90 cells in a 100-mm dish were transfected with 6 μg of plasmid (pGEM3*-188 or pGEM3*-188-m3). At 30 h after transfection, cells were fixed in normal culture medium with formaldehyde at a final concentration of 1% for 10 min at 37°C. Plasmid IP was performed with ChIP assay kit (Upstate Biotechnology) essentially according to the manufacturer's instructions. An ≈250-bp fragment encompassing hTERT proximal promoter was amplified by using the primers 5′-ATACGCTCACTATAGGG-3′ and 5′-TGAGAGCTTGCATGCG-3′. PCR was carried out as follows: 1 cycle at 94°C for 3 min; 26 cycles at 94°C for 30 s, 47°C for 30 s, and 72°C for 30 s; and 1 cycle at 72°C for 1 min.
HDAC Assay. The preparation of nuclear extracts used for HDAC assay was performed essentially as described in ref. 14. HDAC assays were performed with an HDAC assay kit (Upstate Biotechnology) essentially according to the manufacturer's instructions. Briefly, 10 μg of nuclear extracts prepared from IMR90 cells were preincubated with or without CGK1026 or trichostatin A (TSA) at room temperature for 30 min. After incubation, 20,000 cpm of [3H]acetate-labeled histone peptide was added and incubated at 37°C for 5 h. Reactions were stopped by addition of acetic acid/HCl to a final concentration of 0.04/0.25 M. Samples were centrifuged, and the supernatant was counted in a scintillation counter.
Results
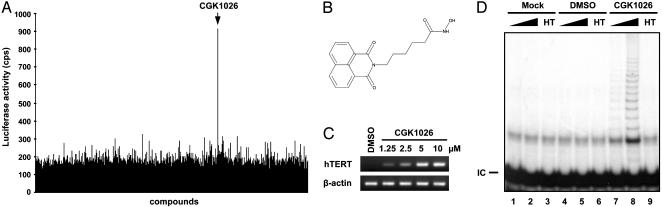
Identification of a Small-Molecule Derepressor of hTERT. To screen for small-molecule derepressors of hTERT in normal human fibroblasts, IMR90 cells were transfected with a 1,003-bp hTERT promoter cloned into a firefly luciferase reporter plasmid. Transfected cells were used in a high-throughput screen for the inducer of firefly luciferase reporter activity against a library containing ≈20,000 small synthetic organic molecules (Fig. 1A). One of the compounds identified in this screen was CGK1026 (Fig. 1 A and B). This molecule dramatically induced hTERT promoter activity in IMR90 (Fig. 1A) and other normal human fibroblasts (WI38 and HFF cells; data not shown). Consistently, CGK1026 significantly induced hTERT mRNA expression and telomerase activity in IMR90 cells (Fig. 1 C and D).
Fig. 1.
Identification of CGK1026 as a small-molecule derepressor of hTERT. (A) Screening of compounds that derepress hTERT promoter activity in IMR90 cells. (B) Chemical structure of CGK1026. (C) CGK1026 derepresses hTERT expression. Semiquantitative RT-PCR for hTERT or β-actin was performed with total RNA prepared from IMR90 cells treated with vehicle (DMSO) or CGK1026 (1.25, 2.5, 5, and 10 μM) for 24 h. (D) CGK1026 derepresses telomerase activity. TRAP assay was performed with 0.1 (lanes 1, 4, and 7) or 0.4 μg (lanes 2, 3, 5, 6, 8, and 9) of cell extract prepared from IMR90 cells incubated in the absence (mock or DMSO) or presence of 5 μM CGK1026 for 24 h. Heat-treated extract (HT) was used as a control for specificity. IC, TRAP internal control.
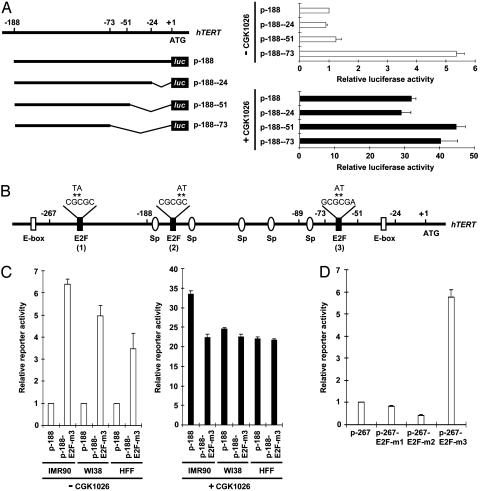
Identification of a CGK1026-Responsive Repressive Element Within the hTERT Promoter. To determine how CGK1026 derepresses hTERT, we first attempted to identify the cis-acting element within the hTERT promoter responsive to this small molecule. Serial deletion from –1,003 up to the proximal 188-bp promoter region failed to identify an element responsive to CGK1026 in IMR90 cells (Fig. 6, which is published as supporting information on the PNAS web site). Thus, we examined the reporter activities of a series of 3′-deletion reporter constructs generated from the proximal 188-bp hTERT promoter in the absence or presence of CGK1026 (Fig. 2A). Serial deletions from –1 up to –50 had almost no effect on hTERT promoter activity or its responsiveness to CGK1026. Interestingly, however, further deletion from –51 to –72 significantly increased hTERT promoter activity. Notably, CGK1026 abolished the increase in hTERT promoter activity upon deletion of this region. Sequence analysis within the region between –72 and –51 of the hTERT promoter revealed a noncanonical E2F site that matches a functional E2F-binding sequence identified in the cyclin E promoter [GCGCGA; E2F (3) in Fig. 2B] (20). Interestingly, a mutation introduced into this site (p-1003-E2F-m3) significantly derepressed hTERT promoter activity in all three types of normal fibroblasts (Fig. 2C). Moreover, in the presence of CGK1026, the derepressing effects of this mutation were not observed (Fig. 2C).
Fig. 2.
Identification of a CGK1026-responsive repressive element within the hTERT promoter. (A) Progressive deletion analysis identifies a CGK1026-responsive repressive element within the hTERT promoter. The A of the translation start codon (ATG) of hTERT is designated as +1, and the name of each deletion construct was assigned according to the nucleotide number of the 5′ end/3′ end of the inserted promoter sequence. IMR90 cells were transiently transfected with each of these constructs, incubated with or without 5 μM CGK1026 for 24 h, and assayed for luciferase activity. (B) The hTERT proximal promoter showing the locations of E-boxes, Sp sites, and putative E2F sites. The sequences that were mutated are indicated by asterisks, with the substituted sequences presented above. (C) Mutation in a putative, noncanonical E2F site increases the hTERT promoter activity while decreasing CGK1026 responsiveness. IMR90, WI38, and HFF cells were transfected with the plasmids indicated, incubated with or without 5 μM CGK1026 for 24 h, and assayed for luciferase activity. In A and C, the luciferase activity of p-188 in the CGK1026-untreated cells was set to 1, and the relative luciferase activity of each construct in the absence or presence of CGK1026 treatment is presented. (D) Two other putative E2F sites are not required for the hTERT promoter repression. IMR90 cells were transfected with the plasmids indicated. Luciferase assays were performed 36 h thereafter. Results shown in A, C, and D are the average of three experiments, and bars indicate standard deviations.
In addition to the E2F site identified in our study [E2F (3) in Fig. 2B], there are two other potential E2F sites in the proximal hTERT promoter region, which were described by Crowe et al. (21) [E2F (1) and E2F (2) in Fig. 2B]. To examine whether these two putative E2F sites also contribute to the repression of hTERT in normal human fibroblasts, we mutated these sites within the hTERT promoter region of the construct p-267 (p-267-E2F-m1 and p-267-E2F-m2 in Fig. 2D). Consistently, p-267-E2F-m3 exhibited significantly enhanced promoter activity in comparison with p-267 in IMR90, WI38, and HFF cells, whereas neither p-267-E2F-m1 nor p-267-E2F-m2 affected hTERT promoter activity (Fig. 2D and data not shown).
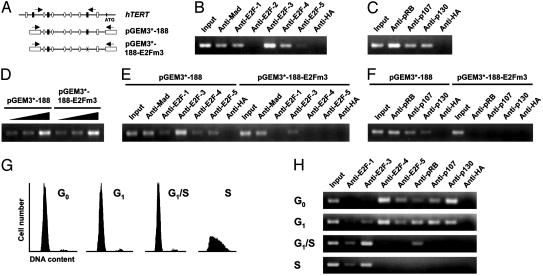
In Vivo Association of E2F and Pocket Protein Family Members with the hTERT Promoter. There are six known members of the E2F family in mammals, E2F-1 through E2F-6. E2F activity is negatively controlled mainly by interactions with members of the pocket protein family (pRB, p107, and p130) (22, 23). Thus, X-ChIP assays were performed to determine the occupancy of the endogenous hTERT promoter by E2F and pocket proteins in normal human fibroblasts (Fig. 3 A–C). Consistent with our previous findings (11, 14), endogenous hTERT promoter formed tight association with Mad in IMR90 cells (Fig. 3B). Interestingly, under these conditions, X-ChIP analysis revealed robust binding of E2F-1, E2F-3, and E2F-4 and weak binding of E2F-5 to the hTERT promoter (Fig. 3B). Consistent with the report that E2F-2 is not expressed in IMR90 cells (24), hTERT promoter was not detected from the immunoprecipitate of anti-E2F-2 antibody. All of the pocket protein family members were also found to be tightly associated with the endogenous hTERT promoter (Fig. 3C).
Fig. 3.
Dynamic assembly of E2F–pocket protein complex on the hTERT promoter in normal human fibroblasts. (A) Schematic of the endogenous hTERT promoter region and its constructs. Notations for E-boxes, Sp sites, and putative E2F sites are identical to Fig. 2B. Arrows indicate positions of PCR primers used to detect promoter fragments. (B) In vivo occupancy of the hTERT promoter by E2F family members in IMR90 cells. (C) In vivo occupancy of the hTERT promoter by pocket protein family members in IMR90 cells. In B and C, X-ChIP assays were performed with asynchronously growing IMR90 cells. (D) Verification of transfection efficiency. Semiquantitative PCR was performed with increasing amounts of DNA from IMR90 cells transfected with pGEM3*-188 or pGEM3*-188-E2Fm3. (E) A CGK1026-responsive repressive element is required for recruitment of E2F family members onto the hTERT proximal promoter. (F) A CGK1026-responsive repressive element is required for recruitment of pocket proteins onto the hTERT proximal promoter. In E and F, plasmid IP analysis was performed with IMR90 cells transfected with pGEM3*-188 or pGEM3*-188-E2Fm3 by using the antibodies indicated. (G) FACS analysis of the synchronized IMR90 cells. IMR90 cells were rendered quiescent by serum starvation (SS), then restimulated with serum for 4, 8, or 12 h (SS + 4h, SS + 8 h, and SS + 12 h). To enrich cells at the G1/S border, serum stimulation was carried out for 24 h in aphidicolin-containing medium (Aph); after removal of aphidicolin, serum stimulation was continued for 3, 6, 9, 12, or 15 h (Aph + 3h, Aph + 6h, Aph + 9h, Aph + 12 h, and Aph + 15 h). G0, G1, G1/S, and S-phase cells shown are the cells SS, SS + 4 h combined with SS + 8 h, Aph, and Aph + 6 h, respectively. DNA content was determined by propidium iodide staining and is plotted versus cell number. (H) Changes in the binding of E2F and pocket proteins to the hTERT promoter during the cell cycle. X-ChIP assays were performed with IMR90 cells at each stage of the cell cycle. G0, G1, G1/S, and S-phase cells shown are as described for G. A portion (0.5%) of the total amount of chromatin or DNA–protein complexes used in IPs is shown (input); anti-Mad and anti-hemagglutinin (anti-HA) antibodies served as positive and negative controls, respectively, in B, C, E, F, and H.
In Vivo Assembly of E2F–Pocket Protein Complex on a CGK1026-Responsive Repressive Element Within the hTERT Promoter. We next addressed whether the CGK1026-responsive repressive element within the hTERT promoter is physically associated with E2F and pocket protein family members. To this end, we performed plasmid IP analysis by using the hTERT proximal promoter carrying the wild-type or mutated E2F (3) site in normal human fibroblasts (Fig. 3 A, D, E, and F). These hTERT promoter constructs (pGEM3*-188 and pGEM3*-188-E2Fm3) were transfected into IMR90 cells. At 30 h after transfection, the transfected cells were cross-linked, and the extracts from these cells were split into two parts. A small portion of the extracts (≈1%) was used to quantitate the amount of each transfected plasmid in these cells by PCR, and we found that similar amounts of wild-type and mutant plasmids were introduced into the cells (Fig. 3D). The remaining extracts from the transfected cells were immunoprecipitated with antibodies directed against each E2F, pocket protein family member, Mad, and hemagglutinin. Consistent with the X-ChIP results (Fig. 3B), both of the transfected hTERT promoter constructs formed a tight association with Mad in IMR90 cells (Fig. 3E). Under these conditions, we noted the association of E2F (E2F-1, E2F-3, E2F-4, and E2F-5) and all three pocket protein family members with transfected wild-type hTERT promoter construct (Fig. 3 E and F). In sharp contrast, none of the E2F or pocket proteins showed much interaction with the hTERT promoter in which the E2F (3) site was mutated (Fig. 3 E and F).
Cell Cycle-Dependent Association of E2F and Pocket Proteins with the hTERT Promoter. The E2F and pocket proteins play a pivotal role in the timely expression of an array of genes during mammalian cell cycle progression (22, 23). Based on the association of several E2F and pocket proteins with the hTERT promoter, we next examined whether complexes composed of different subunits bind at specific points during the cell cycle. IMR90 cells were synchronized by using serum starvation and aphidicolin. FACS analysis allowed us to determine the degree of enrichment of cells at each stage of the cell cycle (Fig. 3G). X-ChIP was performed with these synchronized IMR90 cells at various cell cycle time points (Fig. 3H). E2F-4 was the predominant E2F family member bound to the hTERT promoter during G0, and weaker binding of E2F-5 was also observed. E2F-4 (and E2F-5) binding persisted in cells entering G1, but binding of these factors diminished by late G1 and completely disappeared in S phase. E2F-1 and E2F-3 replaced E2F-4 and E2F-5 during late G1 and S phase. During G0, robust binding by p130 and relatively weaker binding by pRB and p107 were detected. Interestingly, concomitant with alterations of E2F members in the DNA-bound complexes, all these pocket proteins dramatically disappeared from the hTERT promoter as cells progressed through the G1/S transition. These data are consistent with the transient induction of hTERT during S phase in cycling normal human fibroblasts (5).
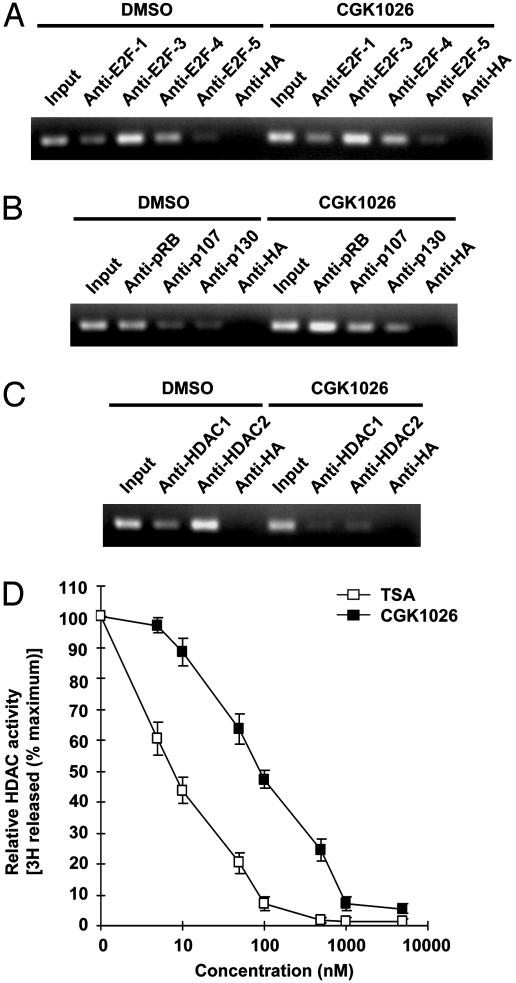
CGK1026 Inhibits the Recruitment of Functional HDAC into E2F–Pocket Protein Complex on the hTERT Promoter. As a next step to identify the direct action mechanism of CGK1026 on hTERT, we used X-ChIP analysis to examine whether CGK1026 treatment alters the in vivo hTERT promoter occupation by E2F and pocket proteins (Fig. 4 A and B). CGK1026 had no significant effects on the E2F family members; whereas, surprisingly, it increased the recruitment of pocket proteins onto the hTERT promoter. To further analyze these results, we examined the effects of the compound on the recruitment of HDAC to the E2F–pocket protein complex. It is known that one critical mechanism by which pocket proteins repress E2F target genes is the recruitment of HDAC to E2F–pocket protein complexes (22, 23). Remarkably, X-ChIP revealed a dramatic decrease in the in vivo hTERT promoter occupancy by HDAC1 and HDAC2 in response to CGK1026 treatment (Fig. 4C). TSA, a well known specific inhibitor of HDAC enzymatic activity, has been shown to displace HDAC from its associated proteins (25). Thus, we investigated whether CGK1026, like TSA, acts on hTERT by functionally suppressing HDAC activity (Fig. 4D). Interestingly, CGK1026 inhibited HDAC activity in a dose-dependent manner with an IC50 of ≈100 nM. In addition, CGK1026 induced G1 arrest of IMR90 cells as described for other HDAC inhibitors (26) (Fig. 7, which is published as supporting information on the PNAS web site), raising a possibility that the increased recruitment of pocket proteins by CGK1026 (Fig. 4B) might be an indirect cell cycle-dependent effect. Taken together, these results indicate that HDAC recruited by the pocket protein–E2F complexes are critical for hTERT repression and that CGK1026 inhibits the assembly of this repressive complex on the hTERT promoter.
Fig. 4.
The mode of action of CGK1026 on hTERT. (A) Effects of CGK1026 on the in vivo association of E2F proteins with the hTERT promoter. (B) Effects of CGK1026 on the in vivo association of pocket protein family members with the hTERT promoter. (C) Effects of CGK1026 on the in vivo association of HDACs with the hTERT promoter. In A, B, and C, IMR90 cells were incubated in the absence (DMSO) or presence of 5 μM CGK1026 for 24 h and analyzed by X-ChIP. (D) Effects of CGK1026 on the HDAC enzymatic activity. HDAC assays were performed with nuclear extracts prepared from IMR90 cells in the absence or presence of CGK1026 or TSA (5, 10, 50, 100, 500, 1,000, and 5,000 nM). Results shown are the average of three experiments, and bars indicate standard deviations.
Inhibition of DNA Binding of E2F Derepresses the hTERT Promoter. To investigate the role of endogenous E2F–pocket protein–HDAC complexes in the repression of hTERT promoter, we first exploited an E2F-binding site “decoy” consisting of 24 repetitive DNA binding sites for E2F (Fig. 5A, p24-E2F-COMPETITOR) (15). Cotransfection of this decoy plasmid along with the 267-bp hTERT promoter-firefly luciferase reporter plasmid into IMR90 cells caused significant elevation of hTERT promoter activity (Fig. 5A). In sharp contrast, a similar construct containing a point mutation in the E2F binding sites (Fig. 5A, p24-E2Fm-CONTROL) exhibited almost no effect on hTERT promoter activity. Moreover, mutation in the repressive E2F (3) site of the hTERT promoter eliminated the derepression induced by the E2F decoy. Importantly, treatment with TSA or CGK1026 completely abolished the increase in the hTERT promoter activity by the E2F decoy (Fig. 5A and data not shown). Taken together, these data further support the idea that endogenous E2F complex assembled on a CGK1026-responsive E2F site of the hTERT promoter is engaged in the HDAC-mediated repression of hTERT in normal human fibroblasts.
Fig. 5.
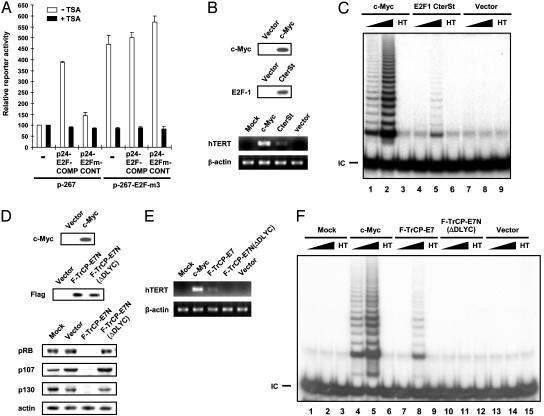
Inhibition or elimination of endogenous E2F–pocket protein complex formation derepresses hTERT and telomerase activity. (A) An E2F decoy causes derepression of hTERT promoter activity. IMR90 cells were cotransfected by using 0.5 μgof p-267 or p-267-E2F-m3 in conjunction with 2.5 μg of decoy plasmid [p24-E2F-COMPETITOR (p24-E2F-COMP) or p24-E2m-CONTROL (p24-E2m-CONT)] or empty vector (pGEM3*), incubated with or without 200 nM TSA for 24 h, and assayed for luciferase activity. For each of the two groups (± TSA), the luciferase activity of p-267 cotransfected with empty vector was set to 1, and the relative luciferase activity of each of the other transfectants is presented. TSA up-regulated the reporter activity of p-267 ≈36-fold. Results shown are the average of three experiments, and bars indicate standard deviations. (B) The hTERT mRNA expression is induced in IMR90 cells stably expressing dominant-negative E2F-1 (E2F-1 CterSt). IMR90 cells at passage 12 were infected with viruses that direct expression of c-Myc, E2F-1 CterSt, or empty vector. Stable expression of c-Myc and E2F-1 CterSt was confirmed by immunoblotting. (C) Telomerase activity is induced in IMR90 cells stably expressing E2F-1 CterSt. The amount of whole-cell lysate used was 0.04 (lanes 1, 4, and 7) or 0.4 μg (lanes 2, 3, 5, 6, 8, and 9). HT, heat treatment; IC, TRAP internal control. (D) Endogenous pRB, p107, and p130 are degraded in IMR90 cells stably expressing engineered F-TrCP-E7N ubiquitin–protein ligase. IMR90 cells at passage 12 were infected with viruses that direct expression of F-TrCP-E7N, F-TrCP-E7N(ΔDLYC) or with empty vector. Expression of c-Myc, F-TrCP-E7N, F-TrCP-E7N(ΔDLYC), pRB, p107, p130, and actin was analyzed by immunoblotting. (E) The hTERT mRNA expression is induced in IMR90 cells stably expressing F-TrCP-E7N. In B and E, semiquantitative RT-PCR for hTERT or β-actin was performed with total RNA prepared from IMR90 cells. (F) Telomerase activity is induced in IMR90 cells stably expressing F-TrCP-E7N. The amount of whole-cell lysate used for TRAP assay was 0.04 (lanes 1, 4, 7, 10, and 13) and 0.4 μg (lanes 2, 3, 5, 6, 8, 9, 11, 12, 14, and 15).
Inhibition of the Interaction of E2F with Pocket Protein Derepresses hTERT. To address whether the recruitment of pocket proteins to E2F is essential for the hTERT repression in normal human fibroblasts, we used a dominant-negative mutant of E2F-1. This mutant (E2F-1 CterSt) can bind to DNA, but lacks both the transactivation and pocket protein-binding domain (15). IMR90 cells were infected with a high-titer retrovirus that directs expression of E2F-1 CterSt. Significantly, under these conditions, we detected an induction of hTERT mRNA expression and telomerase activity by ectopic expression of E2F-1 CterSt, although to a lesser extent in comparison with c-Myc (Fig. 5 B and C). These results suggest that the association of pocket proteins with E2F may play an important role in the active repression of hTERT in normal human fibroblasts.
Targeted Degradation of Endogenous Pocket Proteins Derepresses hTERT. We sought to address more directly the endogenous function of pocket proteins in the regulation of hTERT by eliminating pocket proteins in normal human fibroblasts. Because all three pocket protein family members were associated with the endogenous hTERT promoter in IMR90 cells (Fig. 3C), we chose to eliminate all these proteins together by using a protein-knockout strategy (18). The E7 protein encoded by human papillomavirus type 16 can complex with pRB, p107, and p130, inhibiting their normal cellular functions (27–29). For selective degradation of cellular pocket proteins, we used a chimeric protein with full-length FLAG-tagged βTrCP fused to the N-terminal 35 residues of human papillomavirus type 16 E7 (E7N), which contains a conserved LXCXE pocket protein-binding motif (Fig. 5 D, E, and F; F-TrCP-E7N); thus, this motif of E7 recruits cellular pocket proteins to ubiquitin ligase βTrCP for ubiquitin-dependent proteolysis (18). As a negative control, the same N-terminal domain of E7 with a deletion of the 4-aa segment required for pocket protein binding, E7N (ΔDLYC), was fused to FLAG-tagged βTrCP [Fig. 5 D, E, and F; F-TrCP-E7N(ΔDLYC)]. IMR90 cells were transduced with high-titer retroviruses that direct expression of these proteins. As expected, pRB, p107, and p130 were all degraded in cells expressing F-TrCP-E7N, whereas no degradation was observed in the cells expressing F-TrCP-E7(ΔDLYC) (Fig. 5D). Interestingly, hTERT mRNA was induced in IMR90 cells in which all of the pocket proteins were efficiently degraded (Fig. 5E). Correspondingly, telomerase activity was also induced in these cells (Fig. 5F). These results further support the physiological role of pocket proteins in the active repression of hTERT in normal human fibroblasts.
Discussion
Recently, an interesting but embarrassing finding was reported that hTERT is expressed in cycling normal human fibroblasts, previously believed to lack hTERT expression, during their transit through S phase (5). By exploiting a chemical genetic approach, we have shown that assembly and disassembly of complexes consisting of E2F, pocket protein, and HDAC provide a mechanistic basis for the repression and activation of hTERT in cycling normal human fibroblasts (Fig. 8A, which is published as supporting information on the PNAS web site). Notably, X-ChIP analysis showed dynamic alterations in hTERT promoter occupancy by E2F and pocket proteins during the cell cycle (Fig. 3 G and H). A small-molecule CGK1026 was found to inhibit the recruitment of functional HDAC into these E2F–pocket protein complexes assembled on the hTERT promoter. Moreover, our studies show that artificial accumulation of S phase-specific E2F form, freed from pocket protein either by ectopic expression of dominant-negative E2F (Fig. 5 B and C) or by targeted degradation of cellular pocket proteins (Fig. 5 D, E, and F), can induce hTERT and telomerase activity in normal fibroblasts.
In contrast to cycling fibroblasts, those that have entered senescence do not express hTERT (5). This phenomenon is also consistent with the regulatory model based on the E2F–pocket protein–HDAC complex (Fig. 8A). It is well known that expression of cyclin-dependent kinase inhibitors (e.g., p16INK4a and p21Waf1/Cip1) progressively increases with cumulative population doublings and telomere shortening (30), resulting in the accumulation of hypophosphorylated pocket proteins in senescent cells. These activated pocket proteins stably suppress E2F target genes by facilitating heterochromatin formation during cellular senescence (31). Our results suggest that this potentiated pRB pathway may cause the previously observed senescence-associated constitutive repression of hTERT in normal human cells, which suggests an action of the pRB pathway as a tumor suppressing mechanism; by more stringently repressing hTERT in the course of aging, the pRB pathway may suppress spontaneous immortalization or tumorigenesis. Taken together, our findings revise a well known unidirectional link from telomerase (telomere) to the E2F–pRB pathway in normal human cells (4, 30) as a bidirectional feedback link (Fig. 8B).
Supplementary Material
Acknowledgments
We thank H. Lee, G. P. Nolan, S. J. Weintraub, J. Campisi, and P. M. Howley for gifts of reagents. This work was supported by the Brain Korea 21 Project of the Korean Ministry of Education, the Molecular and Cellular BioDiscovery Research Program, the Center for Biological Modulators of the 21st Century Frontier R&D Program, and the R&D Program for Fusion Strategy of Advanced Technologies of the Korean Ministry of Science and Technology.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: hTERT, human telomerase reverse transcriptase; HDAC, histone deacetylase; IP, immunoprecipitation; X-ChIP, formaldehyde cross-linked chromatin IP; plasmid IP, formaldehyde cross-linked plasmid IP; TSA, trichostatin A; E7N, human papillomavirus type 16 E7.
References
- 1.Hahn, W. C. & Meyerson, M. (2001) Ann. Med. 33, 123–129. [DOI] [PubMed] [Google Scholar]
- 2.Shay, J. W. & Bacchetti, S. (1997) Eur. J. Cancer 33, 787–791. [DOI] [PubMed] [Google Scholar]
- 3.Mason, P. J. (2003) BioEssays 25, 126–133. [DOI] [PubMed] [Google Scholar]
- 4.Wright, W. E. & Shay, J. W. (2002) Nat. Biotechnol. 20, 682–688. [DOI] [PubMed] [Google Scholar]
- 5.Masutomi, K., Yu, E. Y., Khurts, S., Ben-Porath, I., Currier, J. L., Metz, G. B., Brooks, M. W., Kaneko, S., Murakami, S., DeCaprio, J. A., et al. (2003) Cell 114, 241–253. [DOI] [PubMed] [Google Scholar]
- 6.Chang, S. & DePinho, R. A. (2002) Proc. Natl. Acad. Sci. USA 99, 12520–12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith, L. L., Coller, H. A. & Roberts, J. M. (2003) Nat. Cell Biol. 5, 474–479. [DOI] [PubMed] [Google Scholar]
- 8.Kyo, S. & Inoue, M. (2002) Oncogene 21, 688–697. [DOI] [PubMed] [Google Scholar]
- 9.Lin, S. Y. & Elledge, S. J. (2003) Cell 113, 881–889. [DOI] [PubMed] [Google Scholar]
- 10.Oh, S., Song, Y., Yim, J. & Kim, T. K. (1999) J. Biol. Chem. 274, 37473–37478. [DOI] [PubMed] [Google Scholar]
- 11.Oh, S., Song, Y. H., Yim, J. & Kim, T. K. (2000) Oncogene 19, 1485–1490. [DOI] [PubMed] [Google Scholar]
- 12.Won, J., Yim, J. & Kim, T. K. (2002) FASEB J. 16, 1943–1945. [DOI] [PubMed] [Google Scholar]
- 13.Stockwell, B. R. (2000) Nat. Rev. Genet. 1, 116–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Won, J., Yim, J. & Kim, T. K. (2002) J. Biol. Chem. 277, 38230–38238. [DOI] [PubMed] [Google Scholar]
- 15.He, S., Cook, B. L., Deverman, B. E., Weihe, U., Zhang, F., Prachand, V., Zheng, J. & Weintraub, S. J. (2002) Mol. Cell. Biol. 20, 363–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dimri, G. P., Itahana, K., Acosta, M. & Campisi, J. (2000) Mol. Cell. Biol. 20, 273–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oh, S., Song, Y. H., Kim, U. J., Yim, J. & Kim, T. K. (1999) Biochem. Biophys. Res. Commun. 263, 361–365. [DOI] [PubMed] [Google Scholar]
- 18.Zhou, P., Bogacki, R., McReynolds, L. & Howley, P. M. (2000) Mol. Cell 6, 751–756. [DOI] [PubMed] [Google Scholar]
- 19.Abdurashidova, G., Riva, S., Biamonti, G., Giacca, M. & Falaschi, A. (1998) EMBO J. 17, 2961–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Botz, J., Zerfass-Thome, K., Spitkovsky, D., Delius, H., Vogt, B., Eilers, M., Hatzigeorgiou, A. & Jansen-Durr, P. (1996) Mol. Cell. Biol. 16, 3401–3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crowe, D. L., Nguyen, D. C., Tsang, K. J. & Kyo, S. (2001) Nucleic Acids Res. 29, 2789–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dyson, N. (1998) Genes Dev. 12, 2245–2262. [DOI] [PubMed] [Google Scholar]
- 23.Classon, M. & Dyson, N. (2001) Exp. Cell Res., 264, 135–147. [DOI] [PubMed] [Google Scholar]
- 24.Good, L., Dimri, G. P., Campisi, J. & Chen, K. Y. (1996) J. Cell Physiol. 168, 580–588. [DOI] [PubMed] [Google Scholar]
- 25.Choi, H. S., Lee, J. H., Park, J. G. & Lee, Y. I. (2002) Biochem. Biophys. Res. Commun. 296, 1005–1012. [DOI] [PubMed] [Google Scholar]
- 26.Sambucetti, L. C., Fischer, D. D., Zabludoff, S., Kwon, P. O., Chamberlin, H., Trogani, N., Xu, H. & Cohen, D. (1999) J. Biol. Chem. 274, 34940–34947. [DOI] [PubMed] [Google Scholar]
- 27.Dyson, N., Howley, P. M., Munger, K. & Harlow, E. (1989) Science 243, 934–937. [DOI] [PubMed] [Google Scholar]
- 28.Munger, K., Werness, B. A., Dyson, N., Phelps, W. C., Harlow, E. & Howley, P. M. (1989) EMBO J. 8, 4099–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dyson, N., Guida, P., Munger, K. & Harlow, E. (1992) J. Virol. 66, 6893–6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campisi, J. (2001) Trends Cell Biol. 11, S27–S31. [DOI] [PubMed] [Google Scholar]
- 31.Narita, M., Nunez, S., Heard, E., Narita, M., Lin, A. W., Hearn, S. A., Spector, D. L., Hannon, G. J. & Lowe, S.W. (2003) Cell 113, 703–716. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.