Abstract
Amplification of the epidermal growth factor receptor gene on double minutes is recurrently observed in cells of advanced gliomas, but the structure of these extrachromosomal circular DNA molecules and the mechanisms responsible for their formation are still poorly understood. By using quantitative PCR and chromosome walking, we investigated the genetic content and the organization of the repeats in the double minutes of seven gliomas. It was established that all of the amplicons of a given tumor derive from a single founding extrachromosomal DNA molecule. In each of these gliomas, the founding molecule was generated by a simple event that circularizes a chromosome fragment overlapping the epidermal growth factor receptor gene. In all cases, the fusion of the two ends of this initial amplicon resulted from microhomology-based nonhomologous end-joining. Furthermore, the corresponding chromosomal loci were not rearranged, which strongly suggests that a postreplicative event was responsible for the formation of each of these initial amplicons.
Amplification, a mutation by which a cell acquires multiple copies of part of its genome (1), is one of the mechanisms by which protooncogenes may be activated in cells of advanced tumors (2, 3). Depending on the type of tumor, metaphases mainly display chromosomes with homogeneously staining regions or small acentric circular autonomously replicating double minutes (dmins) (4). The size of dmins ranges from a few hundred kilobases to megabases.
Currently, there are few data available on the precise molecular structure of dmins, and most of these data were obtained by studying amplified mutants selected in vitro for their resistance to various cytotoxic drugs. In some cases, each circular element comprises two copies of the amplified sequence linked in inverted orientation (5, 6). In other cases, a single copy of a chromosome fragment appears to be circularized by simple ligation of its extremities (7, 8). More complex structures were found in other studies, which showed that dmins may contain multiple copies of the same amplicon or amplicons originating from various loci (9–14).
Several mechanisms have been proposed to explain the formation of dmins. One of them postulates that circular molecules are generated upon chromosome breakage across replication bubbles at stalled forks. This mechanism accounts especially well for the observation of dmin containing two palindromic amplicons but may also explain most other types of structure (15). Another model postulates that dmin may result from looping out of chromosome fragments in the G1 or G2 phase. This mechanism is supposed to give rise to dmin bearing a single amplicon and deletion of the corresponding sequence from the chromosome, without further rearrangement of that chromosome (9, 16) or gene correction of the deletion (17). A mechanism based on the circularization of the products of a chromosome fragmentation process has also been proposed (18–20). Thus, studies dealing with mutants selected in vitro suggested that different mechanisms drive extrachromosomal amplification. Their relative contribution may depend on the genetic background, human versus rodent; induced versus spontaneous formation; and/or locus and tissue specificity.
A few studies have been performed on human tumors in the form of spontaneously derived cell lines subjected to numerous in vitro passages, rather than primary tumor cells. Thus, the structure and the mechanisms of dmin formation in vivo remained to be elucidated. Cytogenetic studies have recurrently disclosed the presence of dmins in cells of gliomas, the most frequent type of tumor of the central nervous system (21). In particular, up to 50% of glioblastomas contain dmin, whereas homogeneously staining regions are rarely found (22). The epidermal growth factor receptor gene (EGFR) is the most commonly amplified gene in gliomas, representing ≈40% of the cases with amplification (reviewed in ref. 23). To improve our understanding of the mechanisms of amplification in human solid tumors, we took advantage of the recent availability of the human genome sequence. We designed primers distributed along several megabases flanking the EGFR gene and performed quantitative PCR and chromosome walking experiments to determine the genetic content and the structure of the extrachromosomal amplicons found in a series of seven gliomas. The initial event of EGFR amplification was found to be the formation of a circular DNA molecule by fusion of the two ends of a chromosome fragment through a microhomology-based nonhomologous end-joining (NHEJ) mechanism. Furthermore, it was established that the amplicons of a given tumor derive from a single founding circular DNA molecule that is most probably formed by a postreplicative event.
Materials and Methods
Biological Material. Tumors (four glioblastoma multiforme and three oligodendrogliomas) were collected at the Hôpital de la Salpêtrière (Paris). Informed consent was obtained from all patients. Five tumors were grown as xenografts in athymic mice and recovered for analysis at passage 1 or 2. With the exception of case 7, the corresponding fresh tumors were also available (Table 1). Only the fresh tumors were available in the two last cases (cases 30 and 34). Cytogenetic analysis of cases 7, 21, and 22 have been published (24).
Table 1. Characteristics of tumors and their amplicons.
| Characteristic | Case 4 | Case 7 | Case 14 | Case 21 | Case 22 | Case 30 | Case 34 |
|---|---|---|---|---|---|---|---|
| Histology* | ODA | GBM | ODA | GBM | ODA | GBM | GBM |
| Material† | N/F/X | X | N/F/X | F/X | F/X | F | F |
| Chromosomes‡ | 90 | 45 | 47 | 70 | 47 | 91 | 47 |
| Chromosomes 7§ | 6 | 3 | 3 | 3 | 3 | 6 | — |
| Amplification¶ | |||||||
| Xenograft | 9 (7-13) | 8 (5-12) | 17 (12-24) | 32 (22-45) | 55 (40-70) | — | — |
| Fresh | 10 (7-14) | — | 17 (13-25) | 30 (20-45) | 60 (55-80) | 30 (20-40) | 63 (50-80) |
| Amplicon∥ | |||||||
| 5′ Breakpoint | 54,492,001 | 54,168,756 | 47,078,805 | 53,489,375 | 54,349,769 | 53,426,589 | 54,440,318 |
| 3′ Breakpoint | 56,100,888 | 55,697,845 | 55,057,216 | 55,121,943 | 55,056,001 | 55,562,319 | 55,641,516 |
| Length, bp | 1,608,892 | 1,529,091 | 941,200 | 1,530,714 | 706,234 | 2,099,732 | 1,201,200 |
| Deletion∥ | |||||||
| 5′ Breakpoint | — | — | 47,118,693 | 54,862,131 | — | — | — |
| 3′ Breakpoint | — | — | 54,155,906 | 54,963,991 | — | — | — |
| Length, bp | — | — | 7,037,214 | 101,865 | — | — | — |
ODA, oligodendroglioma; GBM, glioblastoma multiforme; N, normal tissue; F, fresh tumor; X, xenografted tumor.
Histological types.
Available biological material.
Modal chromosome number estimated from analysis of R-banded metaphase chromosomes.
Number of chromosomes 7 determined by FISH with chromosome 7 painting probes.
Mean level of amplification. Experimental ranges are given in parentheses.
Localization of the breakpoints that generated the amplicons and their internal deletions. Breakpoints are arbitrarily located at the first base found outside of the region of microhomology (see Fig. 3). Positions are indicated according to the human genome sequence published on the University of California, Santa Cruz (UCSC) Genome Bioinformatics web site (release date July 2003).
Cytogenetic Analysis. Cell preparations were obtained after shortterm culture (1–2 days) of fresh or xenografted tumor fragments. Metaphase spreading, R-banding, and fluorescence in situ hybridization (FISH) with chromosome painting probes (Cambio, Cambridge, U.K.) and bacterial artificial chromosomes or P1-derived artificial chromosomes (BACPAC; clones RP5–1091E12 and RP11–205A3, Children's Hospital Oakland Research Institute, Oakland, CA; clone CTD-2171F16, Open Biosystems, Brussels) were performed as described (25, 26). Chromosomes stained with 4′,6-diamidino-2-phenylindole (DAPI) were identified by computer-generated reverse DAPI banding.
Amplicon Analysis. The level of amplification was measured by real-time fluorescent detection quantitative PCR. DNA from normal lymphocytes was used as a control. DNAs were extracted from lymphocytes and frozen samples of fresh or xenografted tumors by using kits obtained from Qiagen (Valencia, CA). DNAs were amplified by using the GeneAmp 5700 sequence-detection system and SYBR Green PCR kits (Applied Biosystems). PCR conditions were as described in ref. 27. All measurements were performed at least in duplicate. Primers were selected by using the primerexpress program (Applied Biosystems). Only primer pairs with efficiency of >90% were retained for further experiments. The absence of sequences identical to the PCR target in the rest of the genome was verified by using blat (28). Sequence primers are available on request. Amplification levels were calculated by using a standard curve constructed with serial dilutions of control DNA amplified in a parallel experiment. The level of amplification was measured with an experimental error of 50%. The ends of each amplicon were localized by chromosome walking using the Universal Genome Walker kit (BD Biosciences). PCR fragments corresponding to the junctions were directly sequenced by using Big Dye Terminator Sequencing kits (Applied Biosystems). Sequence data used in this work refer to the human genome sequence (released July 2003), which is available at the University of California, Santa Cruz (UCSC) Genome Bioinformatics web site (http://genome.ucsc.edu) (29, 30). blat was used for alignment searches (28).
Loss of Heterozygosity. Loss of heterozygosity was analyzed after PCR amplification of microsatellite loci in standard conditions (31). Five nanograms of template DNA were amplified with 15 μM each of 5′-(6-FAM)-labeled sense and nonlabeled antisense primers. We performed 15 PCR cycles to be at the beginning of the log phase. DNA fragments were run on an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems). Allelic size and intensities were determined by using the genescan analysis program.
Results
Tumor Selection. EGFR amplification was searched for by quantitative PCR using primers located within the gene in a series of 25 gliomas. After this first screening, only tumors displaying at least 5-fold amplification of the gene were selected. Cells from these fresh or xenografted tumors were then analyzed by FISH using a P1-derived artificial chromosome (GenBank accession no. AC006971) overlapping the EGFR gene to establish whether the amplified copies mapped onto homogeneously staining regions or dmins. Seven gliomas that contained dmins were retained for further analyses.
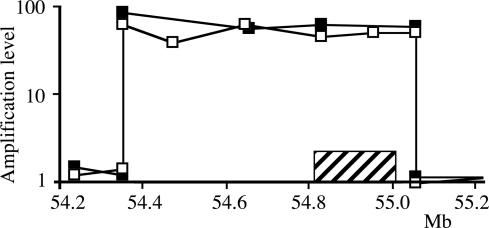
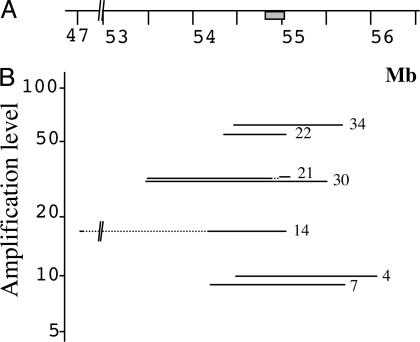
Extent of the Amplicons. Quantitative PCR was first performed by using primer pairs selected every ≈50 kb on both sides of the EGFR gene. When the regions of switch between amplified and nonamplified sequences were roughly determined, primer pairs a few kilobases apart were selected in these regions to map the transition precisely (Fig. 1). Chromosome walking was then performed toward the two ends of the amplicon to identify the junction fragment. We found that within each tumor, all of the amplicons are remarkably similar in size. Depending on the tumor, this size ranged from 0.7 to 2.1 megabases (Mb), and the level of amplification varied from 8- to 63-fold (Table 1 and Fig. 2B). No differences were found between fresh tumors, when available, and their corresponding xenografts. In case 21, a deletion of 101,865 bp in the 5′ region of the EGFR gene was observed. The copy number of that region was similar in tumor and control DNA, indicating that all amplicons harbor the deletion. This deletion removed exons 2–7, which lead to the expression of type III EGFR variant (EGFRvIII), a mutant receptor found recurrently in gliomas (32). In case 14, sequences spread along a region of ≈8 Mb appeared to be amplified. However, an internal deletion of 7 Mb was present in all amplified copies, giving an amplicon length of only 0.95 Mb, which contains a segment of 0.94 Mb, including the EGFR gene associated to a 39,888-bp segment located near position 47 Mb of chromosome 7. This latter region was not amplified in other gliomas analyzed here.
Fig. 1.
Characterization of the amplicon in case 22. The amplification levels determined by quantitative PCR are presented relative to the map (scale in megabases) of the region surrounding the EGFR gene (hatched bar). ▪, Fresh tumor; □, xenografted tumor.
Fig. 2.
Extent of the amplicons found in the studied gliomas. (A) Map of the EGFR region (scale in megabases; gray bar, EGFR gene). (B) Amplification levels, positions, and extent of the amplified regions in seven gliomas. Dotted lines indicate the localization of the internal deletions found in amplicons of cases 21 and 14.
Our results indicate that the amplification factor is constant all along the amplified sequence. However, as shown in Fig. 1, small fluctuations from one primer pair to another were observed along the amplicon, but they probably reflect the experimental limits of the approach rather than actual variations in the level of amplification. In good agreement with this interpretation, a single PCR product was obtained upon amplification of the region bearing the junction in each tumor, which reveals that all of the amplicons in a given tumor have a common ancestor. In addition, we performed long-range PCR experiments by using a single primer located near one end of the amplicons to search for inverted duplications. Amplification products were never obtained in these conditions. Together, our data suggest that, in each of these gliomas, an early event generated a single initial extra-chromosomal circular molecule by joining the two ends of a DNA segment overlapping the EGFR gene. Subsequently, tumor cells gained multiple copies of this extra-chromosomal element without noticeable rearrangements.
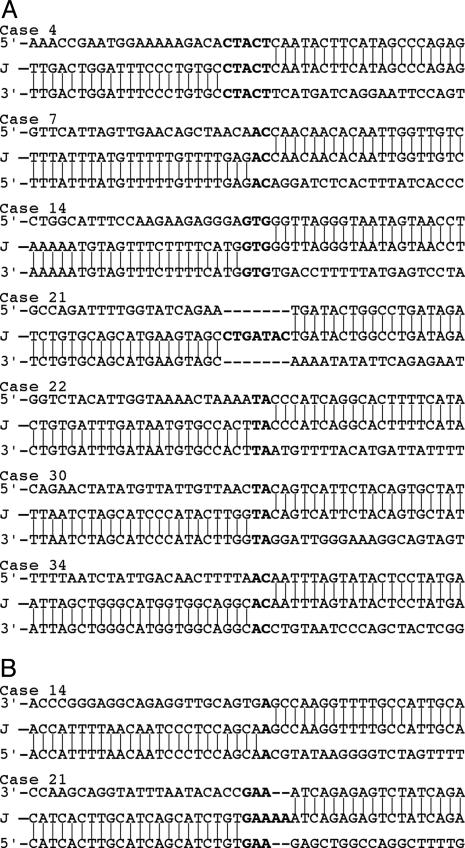
Location of the Breakpoints. The seven junction fragments were sequenced (Fig. 3). Comparison of their sequence with the human genome sequence allowed us to precisely determine the positions of the breakpoints associated with the formation of the initial extrachromosomal element in each tumor (Fig. 2B and Table 1). The seven breakpoints lying 3′ of the EGFR gene were scattered over a region of ≈1 Mb in length, with breakpoints found in cases 14 and 22 occurring within a 1.2-kb-long sequence (positions 55,056,001 and 55,057,216). At 5′ of the EGFR gene, four breakpoints (cases 4, 7, 22, and 34) were found in the region extending from a position of 54.16–54.49 Mb. Two other breakpoints were separated by 47 kb near position 53.4 Mb (cases 21 and 30). The last breakpoint (case 14) mapped near position 47 Mb (Fig. 2B).
Fig. 3.
Sequence of the junctions resulting from circularization of the amplicons (A) and from formation of internal deletions (B). The sequence of each junction is aligned with respect to the sequences of its two normal counterparts [5′ counterpart above the junction (J) and 3′ counterpart below it]. Microhomologies and inserted sequences are shown in bold.
Characterization of the Junctions. To determine whether the initial extrachromosomal elements had been generated by illegitimate homologous recombination events, we searched for the presence of repeated sequences at the breakpoints. Of 14 breaks, 5 took place in interspersed repeated elements: 2 in an Alu, 2 in a LINE, and 1 in a MER (see Fig. 6, which is published as supporting information on the PNAS web site). One breakpoint of case 30 maps within a segmental duplication (see Fig. 7, which is published as supporting information on the PNAS web site). In no case was the same element found on both sides of the same amplicon. The remaining breaks occurred in nonrepeated sequences. One of the two internal deletions involved repeated elements (one Alu and one LINE in case 14), and the other one took place between nonrepeated regions (case 21). Thus, the rearrangements studied here were not generated by illegitimate homologous recombination events. In addition, we searched for, but failed to identify, consensus matches to sequence motifs previously associated with chromosome rearrangements. Details are given in Supporting Methods, which is published as supporting information on the PNAS web site.
We also searched for sequence features characterizing NHEJ. In six cases, microhomologies of 2 (TA or AC), 3 (GTG), or 5 bp (CTACT) in length were found in the normal counterparts of the fusion and maintained as a single copy in the junction (Fig. 3). In the last tumor (case 21), a 7-bp-long insertion (CTGATAC) was present at the junction. This insertion corresponds to the duplication of sequences flanking the breakpoints: one C from the 5′ side and the other six nucleotides from the 3′ side. The junction of the deletion identified on the extra copies of the EGFR gene in case 21 also displayed an insertion of 2 bp (AA) and a 3-bp-long (GAA) microhomology in its two counterparts. Finally, the internal deletion of the amplicon found in case 14 involved a 1-bp (A) microhomology. Together, our results strongly suggest that these junctions were formed by a microhomology-based NHEJ mechanism.
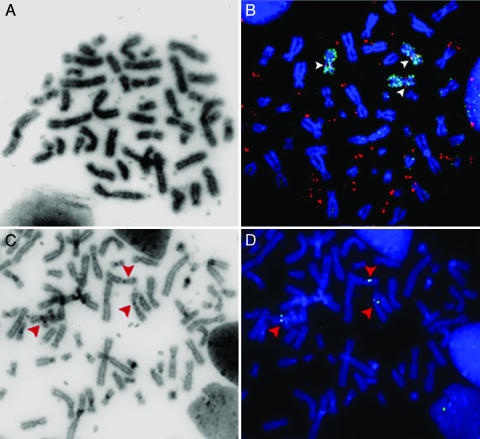
Chromosomal Status of the EGFR Locus. Analysis of R-banded chromosomes showed that the cells of four tumors were neardiploid (cases 7, 14, 22, and 34), whereas the cells of two tumors were near-tetraploid (cases 4 and 30) and the cells of one tumor (case 21) contained 70 chromosomes (Tables 1 and 2, which is published as supporting information on the PNAS web site). In all cases, dmin were present in metaphases (Fig. 4A, for example). The status of the EGFR locus in the chromosomes was analyzed by in situ hybridization with an EGFR probe (bacterial artificial chromosome RP5–1091E12) and a chromosome 7 painting probe (Fig. 4B and data not shown). We observed three copies of chromosome 7 in cells of near-diploid tumors 7, 14, and 22 (we failed to obtain data for case 34). Six copies of chromosome 7 were present in cells of near-tetraploid tumors 4 and 30, and 3 in cells of tumor 21. In most cases, chromosome 7 appeared to be normal. In cells of cases 4 and 7, a rearranged copy of chromosome 7 was observed recurrently, but neither rearrangements affected the EGFR gene region (Table 2). Hence, the EGFR locus was never deleted and was found at its expected location on each chromosome 7 present in the cells of all analyzed tumors. The chromosome regions corresponding to each breakpoint were sequenced after PCR amplification and compared with the normal sequence. In all studied cases, the sequence of the chromosomal loci was normal, which confirms at the molecular level that the chromosomal loci present in the cells are not rearranged.
Fig. 4.
Chromosome analysis. (A and B) Metaphases from case 22. (A) Reverse DAPI staining shows the dmins. (B) Cohybridization with probe RP5–1091E12 containing the EGFR gene (red) and a chromosome 7 painting probe (green). Chromosomes are stained in blue with DAPI. Three copies of chromosome 7 are present, and each copy bears the EGFR locus at its expected location (arrowheads). Probe RP5–1091E12 reveals the dmins (extrachromosomal red signals). (C and D) Metaphase from case 21. (C) Reverse DAPI staining. (D) Hybridization with bacterial artificial chromosome CTD-2171F16 (green signal) probing for a sequence deleted from the amplicons of this tumor. FISH signals are visible on each chromosome 7 (arrowheads) but not on the dmins (see reverse DAPI in C).
FISH was also performed by using the probe CTD-2171F16 covering the deletion observed in the amplicons of case 21 (Fig. 4 C and D). Hybridization signals were observed at the expected location on every copy of chromosome 7, indicating that the deletion is specific to the extrachromosomal amplicons. A similar result was obtained for the deletion identified in case 14 by using probe RP11–205A3 (data not shown).
Loss of heterozygosity analysis was performed along chromosome 7 in cases 4 and 14 for which the corresponding normal tissue was available. In all informative loci, two alleles were present in tumor cells (see Table 3, which is published as supporting information on the PNAS web site), with a 2:1 ratio between the alleles (data not shown).
Discussion
Here, we analyzed seven gliomas, in which the cells contained dmins bearing amplified copies of the EGFR gene. Analysis of the amplified sequences at the nucleotide level showed that a single amplified junction was present in each case, indicating that all of the amplicons of a given tumor derive from a single founding circular extrachromosomal DNA molecule. Each founding element was formed upon simple joining of the ends of a chromosome fragment containing the EGFR gene. In two tumors, we identified an internal deletion present on every amplicon, confirming that all of the amplicons of a tumor have a single precursor. The size of the amplicons ranged from 0.7 to 2.1 Mb, and the amplification levels ranged from 8 to 63 Mb (Table 1). The comparison of these sizes established by the molecular approach to the size of the dmins observed on metaphases (Fig. 4) suggested that some dmins may contain several copies of the amplicons. Fusion of small extrachromosomal elements has been proposed to explain the formation of large dmins (6, 13, 14, 33, 34). Nevertheless, this phenomenon did not impede us from determining the genetic content and the organization of the initial circles. Moreover, when both types of samples were available, data were similar in fresh and xenografted tumors. Thus, xenografting does not select for particular subpopulations of cells, at least if studied after only a few passages, and so offers a valuable tool for the study of this type of tumor.
Formation of the founding amplicon and of the internal deletion observed in some of them may rely on a looping-out event driven by illegitimate homologous recombination between sequences in direct repeats or by junction of the ends of a chromosome fragment through NHEJ. Because homologous recombination requires extensive regions of sequence homology, we searched for motifs common to both counterparts of the junctions. We failed to identify adequately localized couples of repeats, which indicates that homologous recombination is not responsible for these rearrangements.
NHEJ represents the major cellular pathway activated in response to double-strand breaks in mammalian cells and gives rise to both error-free and error-prone repair events (reviewed in refs. 35–37). Error-prone NHEJ has been involved in the generation of various chromosome rearrangements, the junctions of which occur within regions presenting microhomologies one or a few base pairs in length. In addition, small insertions, duplications, or deletions are recurrently observed at these junctions. Whether error-prone NHEJ results from the operation of a specific pathway (38, 39) or the NHEJ pathway is inherently error-prone under particular circumstances is not established. Until now, the signature of the microhomology-based NHEJ has been recurrently associated with the formation of deletions and translocations found in cancers and other types of human diseases (37, 40–44). Here, we showed that the junctions present on dmin, whether they correspond to the formation of a founding circular extrachromosomal DNA molecule or to the generation of an internal deletion, involve 1- to 5-bp-long microhomologies. Moreover, short insertions were found at two of the nine junctions studied (Fig. 3). Together, these data strongly suggest that the formation of both the founding amplicons and their internal deletions also result from microhomology-based NHEJ.
We also studied the status of chromosome 7, from which the amplicons originate. We observed three chromosome 7 per near-diploid genome or six per near-tetraploid genome. In tumors, the presence of three copies of a chromosome in a near-diploid cell is generally explained by a mitotic nondisjunction followed by selection of the cells that acquire a growth advantage during tumor development (45). Three copies of chromosome 7 are recurrently observed in astrocytomas of grade II and III and in glioblastomas, whereas EGFR amplification is relatively specific to glioblastomas (reviewed in ref. 46). In addition, three chromosomes 7 were also observed in nonneoplastic brain cells after short time in culture, an indication that trisomy for chromosome 7 is selected for in this tissue (47). Hence, the gain of one chromosome 7 most probably represents an early event in glioma development, independent of the formation of the extrachromosomal DNA molecules, and near-tetraploid cells containing six chromosomes 7 were probably generated by secondary endoreduplication events. This hypothesis is supported by loss of heterozygosity analyses showing that the two alleles are present in a ratio of 2:1 in two studied cases.
The presence or the absence of chromosomal scars resulting from the events that gave rise to the founding circular molecule may help us to determine the stage of the cell cycle at which these events took place. Excision of a sequence out of the chromosome before replication is expected to generate a rearranged copy of chromosome 7. Indeed, the broken parts of the chromosome may fuse together or with another broken chromosome, giving rise to a chromosome 7 or to marker chromosomes from which the EGFR locus is deleted, respectively (48). In contrast, postreplicative events may lead to cells containing nonrearranged chromosome 7, either because the founding extrachromosomal DNA molecule does not originate from the copies of chromosome 7 retained in tumor cells or because the deletion created upon excision of the founding sequence has been precisely repaired (see below). Six of the seven gliomas gave informative results upon study by chromosome banding and FISH. We found that all chromosomes 7 present in the cells bear the EGFR locus at its normal location. Only two abnormal chromosomes 7 were detected with the EGFR region not being involved in the rearrangement. Thus, our results favor models postulating that the formation of the extrachromosomal DNA molecules result from postreplicative events.
A first type of model supposes that excision of a sequence out of a duplicated chromosome gives rise to one deleted sister chromatid, the second one remaining unaffected. After mitosis, the deleted and the normal sister chromatid each segregate in a daughter cell (Fig. 5A). A cell receiving the normal chromatid and the extrachromosomal DNA molecule may acquire a growth advantage because it has one more copy of the gene than the cells that did not enter the amplification process. In contrast, a cell that receives a deleted chromosome and the founding extrachromosomal DNA molecule does not gain an EGFR copy at that stage and may be counterselected. However, any cell bearing an extrachromosomal DNA molecule acquires the potential to evolve rapidly through uneven segregation of that molecule at mitosis. Hence, the transient advantage of cells having three normal copies of chromosome 7 rather than two may be insufficient to explain the observation that all gliomas studied here derive from the former type of cells.
Fig. 5.
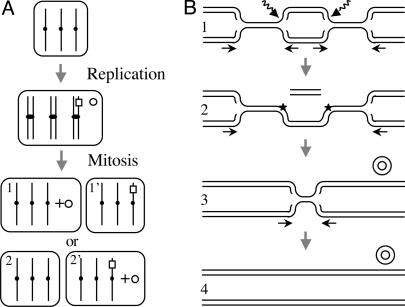
Postreplicative models for the formation of extrachromosomal molecules without a chromosome scar. (A) Segregation-based model. The excision of a chromosome fragment gives rise to a deleted chromosome 7 (□) and an extrachromosomal circular element (○). At mitosis, the element may segregate together with three normal copies of chromosome 7 (daughter cell 1) or with two normal copies and one rearranged copy of that chromosome (daughter cell 2′). The model postulates that cell 1, which has four copies of the EGFR gene, is selected. (B) Rereplication-based model. In lane 1, two breaks occur in a replication eye on the same strand (broken arrows). Arrows indicate the directions of fork progression. In lane 2, a chromosome fragment is excised, and two single-strand breaks remain in the chromosome (stars). They may be repaired at this stage or bypassed at stage 3 by fork regression. In lane 3, the linear fragment is circularized and the corresponding chromosomal region rereplicated by forks emanating from flanking regions. In lane 4, an extrachromosomal element has been formed without a chromosome scar.
A second model postulates that the deletion may be precisely filled by recombination-dependent DNA replication by using the normal sister chromatid as the template (reviewed in refs. 49 and 50). Alternatively, as previously proposed (reviewed in ref. 15), the initial extrachromosomal DNA molecules may be formed upon breakage of a replication eye. This hypothesis is now supported by recent works suggesting that the replication process is a major source of double-strand breaks in cells growing in the absence of extrinsic insults. Indeed, replication forks can collide when encountering nonrepaired lesions or DNA-bound proteins or when the replication machinery is perturbed (reviewed in 49). Hence, the postreplicative rearrangements described here may rely on double-strand breaks generated upstream and downstream of the EGFR gene after replication of the EGFR region (Fig. 5B). The excision of the intervening fragment may be corrected through rereplication of the remaining sequence by forks emanating from the flanking regions.
In conclusion, we demonstrate here that extrachromosomal amplicons bearing the EGFR gene in gliomas derive from a single precursor in each tumor. This initial circular extrachromosomal DNA most probably results from the postreplicative excision of a chromosomal fragment, the two ends of which are ligated by using the microhomology-based error-prone NHEJ pathway. These data extend the known roles of this double-strand break repair pathway to the formation of dmin in tumors.
Supplementary Material
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: dmin, double minute; EGFR, epidermal growth factor receptor; NHEJ, nonhomologous end-joining; FISH, fluorescence in situ hybridization; DAPI, 4′,6-diamidino-2-phenylindole.
References
- 1.Alt, F. W., Kellems, R. E., Bertino, J. R. & Schimke, R. T. (1978) J. Biol. Chem. 253, 1357–1370. [PubMed] [Google Scholar]
- 2.Schwab, M. (1999) Semin. Cancer Biol. 9, 319–325. [DOI] [PubMed] [Google Scholar]
- 3.Knudson, A. G. (2000) Annu. Rev. Genet. 34, 1–19. [DOI] [PubMed] [Google Scholar]
- 4.Hahn, P. J. (1993) BioEssays 15, 477–484. [DOI] [PubMed] [Google Scholar]
- 5.Fakharzadeh, S. S., Rosenblum-Vos, L., Murphy, M., Hoffman, E. K. & George, D. L. (1993) Genomics 15, 283–290. [DOI] [PubMed] [Google Scholar]
- 6.Nonet, G. H., Carroll, S. M., DeRose, M. L. & Wahl, G. M. (1993) Genomics 15, 543–558. [DOI] [PubMed] [Google Scholar]
- 7.Schneider, S. S., Hiemstra, J. L., Zehnbauer, B. A., Taillon-Miller, P., Le Paslier, D. L., Vogelstein, B. & Brodeur, G. M. (1992) Mol. Cell. Biol. 12, 5563–5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stahl, F., Wettergren, Y. & Levan, G. (1992) Mol. Cell. Biol. 12, 1179–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coquelle, A., Toledo, F., Stern, S., Bieth, A. & Debatisse, M. (1998) Mol. Cell 2, 259–265. [DOI] [PubMed] [Google Scholar]
- 10.Rizwana, R. & Hahn, P. J. (1998) Genomics 51, 207–215. [DOI] [PubMed] [Google Scholar]
- 11.Schoenlein, P. V., Shen, D. W., Barrett, J. T., Pastan, I. & Gottesman, M. M. (1992) Mol. Biol. Cell 3, 507–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hahn, P. J., Nevaldine, B. & Longo, J. A. (1992) Mol. Cell. Biol. 12, 2911–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wahl, G. M. (1989) Cancer Res. 49, 1333–1340. [PubMed] [Google Scholar]
- 14.Carroll, S. M., DeRose, M. L., Gaudray, P., Moore, C. M., Needham-Vandevanter, D. R., Von Hoff, D. D. & Wahl, G. M. (1988) Mol. Cell. Biol. 8, 1525–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Windle, B. E. & Wahl, G. M. (1992) Mutat. Res. 276, 199–224. [DOI] [PubMed] [Google Scholar]
- 16.Toledo, F., Buttin, G., Debatisse, M. (1993) Curr. Biol. 3, 255–264. [DOI] [PubMed] [Google Scholar]
- 17.Roelofs, H., Tasseron-de Jong, J. G., van der Wal-Aker, J., Rodenburg, R. J., van Houten, G. B., van de Putte, P. & Giphart-Gassler, M. (1992) Mutat. Res. 276, 241–260. [DOI] [PubMed] [Google Scholar]
- 18.Sen, S., Hittelman, W. N., Teeter, L. D. & Kuo, M. T. (1989) Cancer Res. 49, 6731–6737. [PubMed] [Google Scholar]
- 19.Hahn, H., Kaap, L. N., Morgan, W. F., Painter, R. B. (1986) Cancer Res. 46, 4607–4612. [PubMed] [Google Scholar]
- 20.Hahn, H., Morgan, W. F., Painter, R. B. (1987) Somatic Cell Mol. Genetic 13, 597–608. [DOI] [PubMed] [Google Scholar]
- 21.Mahaley, M. S. Jr., Mettlin, C., Natarajan, N., Laws, E. R. Jr. & Peace, B. B. (1989) J. Neurosurg. 71, 826–836. [DOI] [PubMed] [Google Scholar]
- 22.Bigner, S. H., Mark, J. & Bigner, D. D. (1990) Cancer Genet. Cytogenet. 47, 141–154. [DOI] [PubMed] [Google Scholar]
- 23.Collins, V. P. (1995) Glia 15, 289–296. [DOI] [PubMed] [Google Scholar]
- 24.Muleris, M., Almeida, A., Dutrillaux, A. M., Pruchon, E., Vega, F., Delattre, J. Y., Poisson, M., Malfoy, B. & Dutrillaux, B. (1994) Oncogene 9, 2717–2722. [PubMed] [Google Scholar]
- 25.Bernardino, J., Bourgeois, C. A., Muleris, M., Dutrillaux, A. M., Malfoy, B. & Dutrillaux, B. (1997) Cancer Genet. Cytogenet. 96, 123–128. [DOI] [PubMed] [Google Scholar]
- 26.Dutrillaux, B. & Couturier, J. (1981) La Pratique de l'Analyse Chromosomique (Masson, Paris).
- 27.Vilain, A., Vogt, N., Dutrillaux, B. & Malfoy, B. (1999) FEBS Lett. 460, 231–234. [DOI] [PubMed] [Google Scholar]
- 28.Kent, W. J. (2002) Genome Res. 12, 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kent, W. J., Sugnet, C. W., Furey, T. S., Roskin, K. M., Pringle, T. H., Zahler, A. M. & Haussler, D. (2002) Genome Res. 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karolchik, D., Baertsch, R., Diekhans, M., Furey, T. S., Hinrichs, A., Lu, Y. T., Roskin, K. M., Schwartz, M., Sugnet, C. W., Thomas, D. J., et al. (2003) Nucleic Acids Res. 31, 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lefèvre, S. H., Vogt, N., Dutrillaux, A. M., Chauveinc, L., Stoppa-Lyonnet, D., Doz, F., Desjardins, L., Dutrillaux, B., Chevillard, S. & Malfoy, B. (2001) Oncogene 20, 8092–8099. [DOI] [PubMed] [Google Scholar]
- 32.Frederick, L., Wang, X. Y., Eley, G. & James, C. D. (2000) Cancer Res. 60, 1383–1387. [PubMed] [Google Scholar]
- 33.Ruiz, J. C., Choi, K. H., von Hoff, D. D., Roninson, I. B. & Wahl, G. M. (1989) Mol. Cell. Biol. 9, 109–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Von Hoff, D. D., Needham-VanDevanter, D. R., Yucel, J., Windle, B. E. & Wahl, G. M. (1988) Proc. Natl. Acad. Sci. USA 85, 4804–4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfeiffer, P., Goedecke, W. & Obe, G. (2000) Mutagenesis 15, 289–302. [DOI] [PubMed] [Google Scholar]
- 36.Mills, K. D., Ferguson, D. O. & Alt, F. W. (2003) Immunol. Rev. 194, 77–95. [DOI] [PubMed] [Google Scholar]
- 37.Rassool, F. V. (2003) Cancer Lett. 193, 1–9. [DOI] [PubMed] [Google Scholar]
- 38.Zhu, C., Mills, K. D., Ferguson, D. O., Lee, C., Manis, J., Fleming, J., Gao, Y., Morton, C. C. & Alt, F. W. (2002) Cell 109, 811–821. [DOI] [PubMed] [Google Scholar]
- 39.Difilippantonio, M. J., Petersen, S., Chen, H. T., Johnson, R., Jasin, M., Kanaar, R., Ried, T. & Nussenzweig, A. (2002) J. Exp. Med. 196, 469–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Florl, A. R. & Schulz, W. A. (2003) Genes Chromosomes Cancer 37, 141–148. [DOI] [PubMed] [Google Scholar]
- 41.Sasaki, S., Kitagawa, Y., Sekido, Y., Minna, J. D., Kuwano, H., Yokota, J. & Kohno, T. (2003) Oncogene 22, 3792–3798. [DOI] [PubMed] [Google Scholar]
- 42.Sironi, M., Pozzoli, U., Cagliani, R., Giorda, R., Comi, G. P., Bardoni, A., Menozzi, G. & Bresolin, N. (2003) Hum. Genet 112, 272–288. [DOI] [PubMed] [Google Scholar]
- 43.Klugbauer, S., Pfeiffer, P., Gassenhuber, H., Beimfohr, C. & Rabes, H. M. (2001) Genomics 73, 149–160. [DOI] [PubMed] [Google Scholar]
- 44.Okuno, Y., Hahn, P. J. & Gilbert, D. M. (2004) Nucleic Acids Res. 32, 749–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dutrillaux, B. (1995) Adv. Cancer Res. 67, 59–82. [DOI] [PubMed] [Google Scholar]
- 46.Shapiro, J. R. (2002) Am. J. Med. Genet. 115, 194–201. [DOI] [PubMed] [Google Scholar]
- 47.Heim, S., Mandahl, N., Jin, Y., Stromblad, S., Lindstrom, E., Salford, L. G. & Mitelman, F. (1989) Cytogenet. Cell Genet. 52, 136–138. [DOI] [PubMed] [Google Scholar]
- 48.Coquelle, A., Rozier, L., Dutrillaux, B. & Debatisse, M. (2002) Oncogene 21, 7671–7679. [DOI] [PubMed] [Google Scholar]
- 49.Klein, H. L. & Kreuzer, K. N. (2002) Mol. Cell 9, 471–480. [DOI] [PubMed] [Google Scholar]
- 50.Helleday, T. (2003) Mutat. Res. 532, 103–115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.