Abstract
Background
Normalization is an important step for microarray data analysis to minimize biological and technical variations. Choosing a suitable approach can be critical. The default method in GeneChip expression microarray uses a constant factor, the scaling factor (SF), for every gene on an array. The SF is obtained from a trimmed average signal of the array after excluding the 2% of the probe sets with the highest and the lowest values.
Results
Among the 76 U34A GeneChip experiments, the total signals on each array showed 25.8% variations in terms of the coefficient of variation, although all microarrays were hybridized with the same amount of biotin-labeled cRNA. The 2% of the probe sets with the highest signals that were normally excluded from SF calculation accounted for 34% to 54% of the total signals (40.7% ± 4.4%, mean ± sd). In comparison with normalization factors obtained from the median signal or from the mean of the log transformed signal, SF showed the greatest variation. The normalization factors obtained from log transformed signals showed least variation.
Conclusions
Eliminating 40% of the signal data during SF calculation failed to show any benefit. Normalization factors obtained with log transformed signals performed the best. Thus, it is suggested to use the mean of the logarithm transformed data for normalization, rather than the arithmetic mean of signals in GeneChip gene expression microarrays.
Keywords: Microarray, normalization, gene expression, DNA, RNA, oligonucleotide, GeneChip, scaling
Background
The high-density oligonucleotide microarray, also known as GeneChip®, made by Affymetrix Inc (Santa Clara, CA), has been widely used in both academic institutions and industrial companies, and is considered as the "standard" of gene expression microarrays among several platforms. A single GeneChip® can hold more than 50,000 probe sets for every gene in human genome. A probe set is a collection of probe pairs that interrogates the same sequence, or set of sequences, and typically contains 11 probe pairs of 25-mer oligonucleotides [1-3]. Each pair contains the complementary sequence to the gene of interest, the so-called perfect match (PM), and a specificity control, called the Mismatch (MM) [3]. Gene expression level is obtained from the calculation of hybridization intensity to the probe pairs and is referred to as the "signal" [4-10]. The normalization method used in GeneChip software is called scaling and is defined as an adjustment of the average signal value of all arrays to a common value, the target signal value in order to make the data from multiple arrays comparable [4,11].
The purpose of data normalization is to minimize the effects of experimental and/or technical variations so that meaningful biological comparisons can be made and true biological changes can be found among multiple experiments. Several approaches have been proposed and shown to be effective and beneficial. They were mostly from studies on two-color spotted microarrays [12-19]. Some authors proposed normalization of the hybridization intensities, while others preferred to normalize the intensity ratios. Some used global, linear methods, while others used local, non-linear methods. Some suggested using the spike-in controls, or house-keeping genes, or invariant genes, while others preferred all the genes on the array. For GeneChip data, some have proposed different models to normalize signal values or normalize probe pair values [10,20-24]. Despite the presence of other alternatives, many biologists still use the default scaling method and consider that such method is satisfactory and is useful to identify biological alterations [23,25,26]. With the increasing awareness and usage of GeneChip technology and willingness to continue to use GeneChip software among many biologists, it is worth improving the performance or correcting the problems of the software. In this report, the author has demonstrated that in the scaling algorithm excluding 2% of the probe sets with the highest and the lowest values did not have much benefit. However, the logarithmic transformation of signal values prior to scaling proved to be the optimum normalization strategy and is strongly recommended.
Results
The statistical algorithm in current GeneChip software (MAS 5 and GCOS 1) for gene expression microarray data has eliminated the negative gene expression values, a problem present in earlier versions of the software [5,7]. It uses a robust averaging method based on the Tukey biweight function to calculate the gene expression level from the logarithm transformed hybridization data [3-5,11]. The reported data of a probe set is the antilog of the Tukey biweight mean multiplied by a SF and/or a normalization factor (NFaffy). When both the SF and NFaffy are equal to 1, there is no normalization or manipulation of original data. Both NFaffy and SF are computed in virtually the same way. NFaffy is calculated in comparison analysis to compare the array average of one experiment with that of a baseline experiment, while SF is obtained from the signal average of one experiment comparing with a common value, the target signal in absolute analysis [3-5,11,22]. The average value used in GeneChip is a trimmed average. It is not calculated from all probe sets, but from 96% of the probe sets after the 2% of the probe sets with the highest and the 2% of the lowest signals were removed.
In this report, a total of 76 experiments with rat U34A GeneChip were analyzed. As shown in Table 1, the total hybridization signals varied although all arrays were hybridized with the same amount of biotin-labeled cRNA and scanned with the same scanner of identical settings. The array of the highest hybridization intensities had 2.8 times more signals than that of the lowest. The average array signals had 25.8% variation in terms of coefficient of variation. The mean signals were significantly greater than the median signals on each array, indicating a non-normal distribution. The density plot showed a long-tailed and skewed distribution (not shown) and the average of such data is known to be sensitive to the larger values in the data set.
Table 1.
Summary of signal data in 76 rat genome U34A GeneChip microarrays.
| Lowest | Highest | Mean | SD | CV (%) | |
| Total signal | 832,561.4 | 3,161,392.7 | 2,039,655.7 | 526,295.0 | 25.80% |
| Sum of signals used for SF | 524,513.7 | 1,986,236.9 | 1,212,296.5 | 336,138.0 | 27.73% |
| Trimmed total | 308,047.7 | 1,240,257.3 | 827,359.1 | 215,325.1 | 26.03% |
| Mean signal | 94.6 | 359.3 | 231.0 | 59.8 | 25.80% |
| Median of signals | 17.8 | 54.8 | 35.7 | 8.7 | 24.41% |
| Mean of log signals | 4.3 | 5.8 | 5.1 | 0.4 | 7.17% |
| Trimmed percentage | 34.4 | 54.1 | 40.7 | 4.4 | 10.70% |
"Total signal" is the sum of all the signals on each array. "Sum of signals used for SF" is the sum of signals excluding the trimmed data and used to calculate SF. "Trimmed total" is the sum of the 2% probe sets with the highest signals on the array. "Mean of log signals" is the mean of log2 transformed signals. "Trimmed percentage" = (Trimmed total/Total signal) × 100%. See also in Methods. The "lowest" and "highest" showed the lowest and highest number in the category among the 76 chips, respectively. The mean, standard deviation (SD) and coefficient of variation (CV) were also calculated.
The rat U34A GeneChip contained 8799 probe sets; hence 2% was about 176 probe sets. The sum of the 2% of the probe sets with the lowest signals accounts for less than 0.1% of the total signals (0.05% ± 0.01%, mean ± SD, n = 76) and its impact on SF calculation can be ignored. However, the sum of the 2% of the probe sets with the highest signals, the TrimTotal as used in this report, was responsible for about 40% of the total signals (from 34% to 54%, Table 1). The remaining 96% of the probe sets used for SF calculation, produced only about 60% of the signals. Excluding 4% of the probe sets did not reduce the variation, but rather slightly increased the variation, which in turn resulted in a wider range of SFs (Table 1). It was also found that the TrimTotal was highly correlated with total signal (R = 0.928), but less with medians (R = 0.536) and the mean of log signals (R = 0.643). The trimmed percentage (Tp) was found to be negatively associated with the median (R = 0.558, b = -1.116) and the mean of log signals (R = 0.495, b = -0.968), but not with the total signal of all probe sets.
Among other approaches to global linear normalization, one can also use the median signal or the mean of logarithm transformed signals to calculate the NF. NFLogMean showed a higher correlation with NFMedian than with SF. There were larger differences between NFLogMean and SF than those between NFLogMean and NFMedian (Fig. 1). To test if the larger difference was a result of removing 4% of the probe sets from the calculation, another NF, the NFTrimLogMean was obtained using the same data as for SF, but with a log transformation. There is a very significant correlation between NFTrimLogMean and NFLogMean (R = 0.9998). The 4% of the probe sets that was removed from NFTrimLogMean calculation reduced the total data by only 4% after log transformation.
Figure 1.
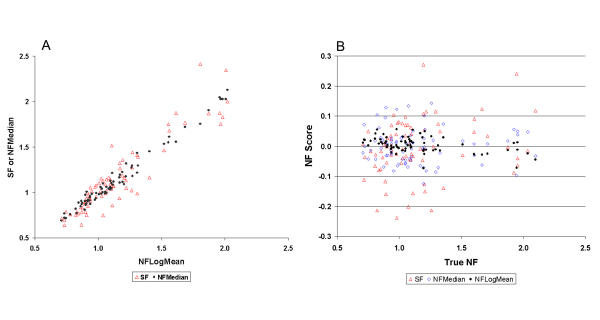
(A) Comparison among different normalization factors. NFLogMean (x-axis) is plotted against SF (red open triangle) and NFMedian (black closed circle). The correlation between NFLogMean and NFMedian is higher (R = 0.971) than that between NFLogMean and SF (R = 0.918). (B) The NF score, NFscore, for SF (red open triangle), NFMedian (blue open diamond) and NFLogMean (black closed circle) is expressed as a function of respective 'true NF'. NFTrimLogMean is not shown here to simplify the graph since it is similar to NFLogMean. See also in Methods.
Since it is impossible to obtain the true normalization factor, an average of the four global linear NFs mentioned above was used instead to estimate the 'true' NF. To compare them with the true NF, a score (NFscore) is introduced. Each NF is calculated against the respective 'true' NF to obtain its NFscore. The average NFscore (± SD) is 7.01% (± 6.24%), 4.51% (± 3.48%), 2.25%(± 2.33%) and 1.95% (± 1.61%), and the sum of NFscore is 5.33, 3.43, 1.71 and 1.48 for SF, NFMedian, NFTrimLogMean and NFLogMean, respectively (Fig. 1). The sum of NFscore indicated an accumulated variation from the true NF, and the larger the number, the larger the accumulated variation. An attempt to add a 5th NF obtained from the arithmetic mean of all probe sets of the array was also made to calculate and compare NFscore with each NFs, and the results showed the same conclusion (data not shown). It is fair to conclude that NFLogMean produced the least variation.
Discussion
Logarithmic transformation is a well-accepted approach for stabilizing variance and has become a common choice for data transformation and normalization for spotted microarrays [12,16]. Much improvement has been made in GeneChip microarray technology and accompanying software during the past few years. The current version of GeneChip software has improved its performance and is better than the earlier versions that used the Average Difference to express levels of gene expression [3,4]. However, the normalization algorithm was inherited and remains the only and default option for gene expression data processing in both MAS 5 and the newly released GeneChip Operating Software (GCOS) software. They continue to use the arithmetic mean of signals to obtain the SF in absolute analysis (single array) and the NF in comparison analysis (two arrays) [3-5,7,11,22]. It is clearly shown here that the trimmed average and the resulting SF had a larger variance than the median-based NF, or the NF based on the mean of log transformed signals. Similar results were observed in other GeneChip expression arrays, such as mouse U74A and human U133A (data not shown). Elimination of the highest and the lowest 2% of the probe set signals did not stabilize the trimmed means. When intra-array variance was reduced by 40%, this approach cannot be considered to be optimal. The logarithmic transformation of signals stabilized the variation well and made the normalization process much less dependent upon the mean and less affected by the outliers.
Although simple and popular, the global linear normalization has its drawbacks, especially when the relationship among multiple experiments or genes is not linear. To address such problems, several methods have been proposed to conduct local and non-linear normalization, [12,14-17,20,22,27]. Data normalization is a very critical and important step for microarray data mining process. The use of different approaches to normalization may have a profound impact on the selection of differentially expressed genes and conclusions about the underlying biological processes especially when subtle biological changes are investigated [12,16,28].
Conclusions
Normalization of microarray data allows direct comparison of gene expression levels among experiments. A global linear normalization, called scaling has been widely used in GeneChip microarray technology for gene expression analysis. The scaling factor (SF) is calculated from a trimmed average of gene expression level after excluding the 2% of the data points of the highest values and the lowest values. It is shown here that the 2% of the probe sets of the highest signals contained from 34% to 54% of the total signals. Elimination of the outliers did not reduce, but increased the variation among multiple arrays. Instead, normalization factors obtained from the mean of the log transformed signals had the best performance. Thus, the current scaling method, although widely used, is not optimal and needs further improvement. The mean of logarithm transformed signals is highly recommended to use for normalization factor calculation.
Methods
GeneChip experiments and data
Total RNA was isolated from rat tissues or cells in Trizol reagent and purified with Qiagen Rneasy kit. cDNA was synthesized in presence of oligo(dT)24-T4 (Genset Corp, La Jolla, CA) and biotinlated UTP and CTP were used to generate biotin labeled cRNA according to the recommended protocols [29]. Rat genome microarray, U34A GeneChip (Affymetrix Inc., Santa Clara, CA) was used and hybridized with 15 μg of gel-verified fragmented cRNA. Hybridization intensity was scanned in GeneArray 2500 scanner (Agilent, Palo Alto, CA) with Microarray Suite (MAS) 5.0 software [4]. Data from a total of 76 independent GeneChip experiments were used in this study.
Normalization factor (NF)
Gene expression data exported from MAS 5.0 were submitted to a Perl script to calculate different normalization factors. In the scaling approach, a trimmed average signal is calculated after excluding 2% probe sets with the highest signals and 2% with the lowest signal values. The scaling factor (SF) is obtained using equation (1) in comparison with a chosen fixed number, called the target signal (TS) and is verified with the results from MAS 5.0 of the same settings [3,4,11].
SFj = TS / STrimMeanj (1)
Other normalization factors for comparison were obtained by the following:
NFMedianj = TS / Smedj (2)
NFLogMeanj = 2 nfj
![]()
where i = 1..., n represents the probe sets, j = 1..., J represented the array experiments, Si is the signal of the anti-log of a robust average (Tukey biweight) of log(PM-MM) reported from MAS 5.0 [5], Smedj is the median signal on the array j, STrimMeanj is the trimmed average on array j after excluding 2% of the probe sets with the highest and the lowest signals [3,4,11,22]. NFMedianj is obtained by using the median signal on array j, and NFLogMeanj is obtained by using the mean of log transformed signals. TS was set to 150, 38 and 38 for SF, NFMedian and NFLogMean, respectively in order to have similar NFs.
In comparison with different NFs, a score, NFscore is introduced. NFscorej = (NFj - TrueNFj)/TrueNFj, and TrueNFj = (SFj + NFMedianj + NFLogMeanj + NFTrimLogMeanj)/4, where NFTrimLogMeanj, was calculated from equation (3) excluding the 2% of the probe sets with the highest and lowest signals, TrueNFj was used as a 'true' NF. Sum of 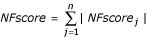 .
.
Other analysis
Unless otherwise specified, logarithm transformation is carried out with the logarithm base 2. Trimmed total signal TrimTotal is the sum of the signals from the 2% of the probe sets with the highest signal values. Total signal Total is the sum of the signals of all probe sets in the array, and trimmed percentage Tpj = (TrimTotalj / Totalj) × 100%.
Abbreviations
GeneChip® is the registered trademark owned by Affymetrix Inc.
PM: perfect Match; MM: mismatch; SF: scaling factor; NF: normalization factor; TS: target signal Short phrase: Normalization of GeneChip microarray data
Acknowledgments
Acknowledgements
I would like to acknowledge the support from Dr. H. D. Lipshitz, Dr. S. Scherer, The Centre for Applied Genomics, and The Hospital for Sick Children. The excellent technical work by Lan He is highly appreciated. I would also like to thank Drs. P. Liu, M. Post, K. Tanswell, G. Fantus and S. Keshavjee for sharing their U34A data. Review and comments from Drs. C. Greenwood, J. Beyene, C.E. M'lan and P. McLoughlin are highly appreciated. Finally, suggestions to improve this paper from the editor and referees are deeply appreciated.
References
- Lipshutz RJ, Fodor SP, Gingeras TR, Lockhart DJ. High density synthetic oligonucleotide arrays. Nat Genet. 1999;21:20–24. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
- Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H, Brown EL. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- Affymetrix GeneChip Expression Analysis: Data Analysis Fundamentals. http://wwwaffymetrixcom/ http://www.affymetrix.com/
- Affymetrix . In: Microarray Suite 5.0 User's Guide. 2002. Affymetrix, editor. Santa Clara, CA, USA, Affymetrix Inc; 2001. [Google Scholar]
- Hubbell E, Liu WM, Mei R. Robust estimators for expression analysis. Bioinformatics. 2002;18:1585–1592. doi: 10.1093/bioinformatics/18.12.1585. [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WM, Mei R, Di X, Ryder TB, Hubbell E, Dee S, Webster TA, Harrington CA, Ho MH, Baid J, Smeekens SP. Analysis of high density expression microarrays with signed-rank call algorithms. Bioinformatics. 2002;18:1593–1599. doi: 10.1093/bioinformatics/18.12.1593. [DOI] [PubMed] [Google Scholar]
- Sasik R., Calvo, E., and Corbeil, J. Statistical analysis of high-density oligonucleotide arrays: a multiplicative noise model. Bioinformatics. 2002;18:1633–1640. doi: 10.1093/bioinformatics/18.12.1633. [DOI] [PubMed] [Google Scholar]
- Naef F, Hacker CR, Patil N, Magnasco M. Empirical characterization of the expression ratio noise structure in high-density oligonucleotide arrays. Genome Biol. 2002;3:RESEARCH0018. doi: 10.1186/gb-2002-3-4-research0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Affymetrix . http://wwwaffymetrixcom/ 1.0 GeneChip Operating Software: User's Guide.http://www.affymetrix.com/index.affx [Google Scholar]
- Quackenbush J. Microarray data normalization and transformation. Nat Genet. 2002;32 Suppl:496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
- Culhane AC, Perriere G, Considine EC, Cotter TG, Higgins DG. Between-group analysis of microarray data. Bioinformatics. 2002;18:1600–1608. doi: 10.1093/bioinformatics/18.12.1600. [DOI] [PubMed] [Google Scholar]
- Durbin BP, Hardin JS, Hawkins DM, Rocke DM. A variance-stabilizing transformation for gene-expression microarray data. Bioinformatics. 2002;18 Suppl 1:S105–10. doi: 10.1093/bioinformatics/18.suppl_1.s105. [DOI] [PubMed] [Google Scholar]
- Kepler TB, Crosby L, Morgan KT. Normalization and analysis of DNA microarray data by self-consistency and local regression. Genome Biol. 2002;3:RESEARCH0037. doi: 10.1186/gb-2002-3-7-research0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Li C, Ellis B, Wong WH. Feature extraction and normalization algorithms for high-density oligonucleotide gene expression array data. J Cell Biochem Suppl. 2001;Suppl 37:120–125. doi: 10.1002/jcb.10073. [DOI] [PubMed] [Google Scholar]
- Hill AA, Brown EL, Whitley MZ, Tucker-Kellogg G, Hunter CP, Slonim DK. Evaluation of normalization procedures for oligonucleotide array data based on spiked cRNA controls. Genome Biol. 2001;2:RESEARCH0055. doi: 10.1186/gb-2001-2-12-research0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang IV, Chen E, Hasseman JP, Liang W, Frank BC, Wang S, Sharov V, Saeed AI, White J, Li J, Lee NH, Yeatman TJ, Quackenbush J. Within the fold: assessing differential expression measures and reproducibility in microarray assays. Genome Biol. 2002;3:research0062. doi: 10.1186/gb-2002-3-11-research0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Li C, Hung Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2:RESEARCH0032. doi: 10.1186/gb-2001-2-8-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Geller SC, Gregg JP, Hagerman P, Rocke DM. Transformation and normalization of oligonucleotide microarray data. Bioinformatics. 2003;19:1817–1823. doi: 10.1093/bioinformatics/btg245. [DOI] [PubMed] [Google Scholar]
- Stuart RO, Bush KT, Nigam SK. Changes in global gene expression patterns during development and maturation of the rat kidney. Proc Natl Acad Sci U S A. 2001;98:5649–5654. doi: 10.1073/pnas.091110798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudtson KL, Griffin C, Iacobas DA, Johnson K, Khitrov G, Levy S, Massimi A, Nowak N, Viale A, Grill G, Brooks AI. A current profile of microarray laboratories: the 2002-2003 ABRF microarray research group survey of laboratories using microarray technologies. http://wwwabrforg.
- Tseng GC, Oh MK, Rohlin L, Liao JC, Wong WH. Issues in cDNA microarray analysis: quality filtering, channel normalization, models of variations and assessment of gene effects. Nucleic Acids Res. 2001;29:2549–2557. doi: 10.1093/nar/29.12.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann R, Seidl T, Dugas M. Profound effect of normalization on detection of differentially expressed genes in oligonucleotide microarray data analysis. Genome Biol. 2002;3:RESEARCH0033. doi: 10.1186/gb-2002-3-7-research0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Affymetrix GeneChip Expression Analysis: Technical Manual. http://wwwaffymetrixcom/ http://www.affymetrix.com/


