Abstract
A fosmid library representing 10-fold coverage of the Histoplasma capsulatum G217B genome was used to construct a restriction-based physical map. The data obtained from three restriction endonuclease fingerprints, generated from each clone using BamHI, HindIII, and PstI endonucleases, were combined and used in FPC for automatic and manual contig assembly builds. Concomitantly, a whole-genome shotgun (WGS) sequencing of paired-end reads from plasmids and fosmids were assembled with PCAP, providing a predicted genome size of up to 43.5 Mbp and 17% repetitive DNA. Fosmid paired-end sequences in the WGS assembly provide anchoring information to the physical map and result in joining of existing physical map contigs into 84 clusters containing 9551 fosmid clones. Here, we detail mapping the Histoplasma capsulatum genome comprehensively in fosmids, resulting in an efficient paradigm for de novo sequencing that uses a map-assisted whole genome shotgun approach.
The dimorphic fungus, Histoplasma capsulatum, is the causative agent of histoplasmosis. In the United States, the Ohio and Mississippi river valleys are endemic regions for histoplasmosis; geographically, Histoplasma is not confined within the U.S. and can be isolated globally (Rippon 1982; Bradsher 1996). H. capsulatum grows in nitrogen-rich environments in a filamentous form (mycelia). Acquired Histoplasma infections occur when mycelia, composed of hyphae and conidia, become aerosolized by physical perturbation and are inhaled. Within the lung, hyphal fragments and conidia differentiate into yeasts that survive and proliferate within alveolar macrophages (Eissenberg et al. 1993; Newman 1999). The transition between the saprophytic mycelial phase and the parasitic yeast phase is thermally regulated and completely reversible, and blocking the phase transition prevents disease progression (Maresca and Kobayashi 1989; Woods et al 2001). Most infections remain localized in the respiratory tract and are self-limiting, and the occurrence of systemic histoplasmosis is rare and most often associated with immunocompromised patients (Sorensen et al. 1999).
Genetic elements required for morphogenesis and pathogenesis are poorly defined. CBP1, a yeast phase-specific gene that encodes a secreted calcium-binding protein essential for intracellular parasitism and pulmonary colonization, is the only genetic element defined for virulence (Kügler et al. 2000; Sebghati et al. 2000). Genomic variations between H. capsulatum environmental and clinical isolates use several molecular techniques to group isolates into classes that are unique to geographically distinct regions (Kersulyte 1992; Kasuga et al. 1999; Carter et al. 2001). Flow cytometry and reassociation kinetics estimate genome size ranges between 24 (for G186A-S) and 32 Mbp (for Downs) and repetitive DNA content ranges between 0.5% and 8.0%, respectively (Carr and Shearer Jr. 1998). Pulsed-field gel electrophoresis has resolved as many as seven chromosomes (Steele et al. 1989). Currently, sequencing of the Histoplasma genome is underway at the Washington University Genome Sequencing Center, and the resulting genome sequences of two geographically distinct strains will provide a reference for ongoing Histoplasma studies.
Our sequencing strategy requires a physical map of this relatively small eukaryotic genome. We opted to construct a fosmid library in lieu of the commonly used Bacterial Artificial Chromosome (BAC) library-based approaches (Marra et al. 1997; McPherson et al. 2001; Gregory et al. 2002). The long-range linking information provided by fosmids allows for iterative anchoring and orienting between the Histoplasma physical map and WGS assembly contigs. In this study, we detail our use of fosmids in a map-assisted whole-genome shotgun approach. This fosmid-based physical mapping approach fits readily into our physical map data generation pipeline, providing a ready source of DNA for end-sequencing analysis and using many of our established map-building informatics tools, such as Image and FPC. Taken together, the use of fosmids for physical map construction of smaller genomes (5 Mbp up to 1 Gb, for example) can provide an efficient and effective method of generating critical genomic resources.
RESULTS
Characterization of the H. capsulatum G217B Fosmid Library
Fosmid clones of the H. capsulatum G217B library were initially sized by restriction with HindIII. The average insert size was 35.9 kbp (±4.3 kbp) and ranged from 22.0 to 49.9 kbp (n = 130; data not shown). A total of 9551 fosmid clones were analyzed for the physical map, and based on the published genome size of H. capsulatum isolate G186A-S (Carr and Shearer Jr. 1998), we predict an ∼10-fold genome representation in the map.
Constructing the H. capsulatum G217B Physical Map
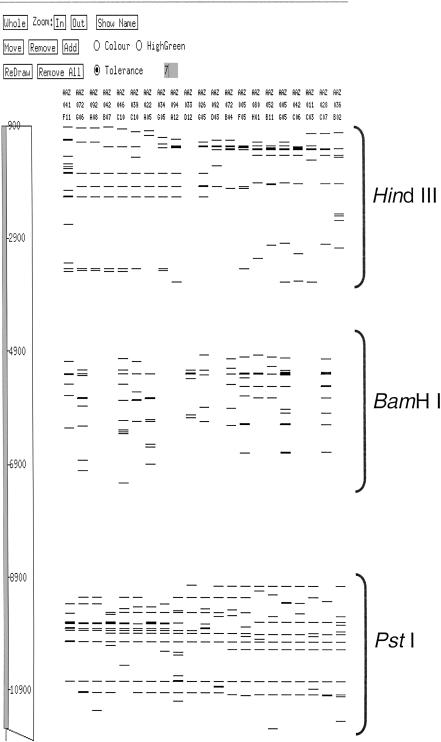
The fragment size distribution from independent HindIII, PstI, and BamHI digestion of the fosmid clones resulted in a moderate number of restriction fragments (bands) averaging 10.0 (±3.4), 12.4 (±3.9), and 8.9 (±4.3) fragments, respectively (data not shown). The restriction digest fragment pattern obtained for each clone is considered a fingerprint and is collectively used to identify clone overlaps and assemble physical maps (Marra et al. 1997). Fingerprint data from the Image software fragment mobility (or bands) files of each independent restriction digest were combined for each clone, as described in Methods. Figure 1 shows a representation of the combined fingerprints in the FPC contig assembly software. The combined fingerprints from clones with data from all three restriction digests averaged 25.6 (±9.4) fragments per fosmid clone and were used to assemble contigs at high stringency (see Methods). Several automated contig assemblies were carried out using data sets in which the order of stacking the fingerprints was varied. No differences were seen in the contigs assembled, regardless of the order in which fingerprint data were offset (data not shown).
Figure 1.
Stacked fingerprints were generated by offsetting the mobility rates within each bands file. Here, we incorporate up to three fingerprints for each clone as the combined bands file, and the mobility rates of each HindIII, BamHI, and PstI fingerprint were changed to 1–4000, 4001–8000, and 8001–12000, respectively. During manual editing, FPC builds added bands file of clones missing either one fingerprint (blank lanes in the BamHI digest) or two fingerprints (blank lanes in both HindIII and BamHI digests).
Assessment of the H. capsulatum G217B Physical Map
Starting with only clones for which three combined digests were available, an automated assembly cutoff of 3e-10 and a band tolerance of 7 produced an initial 852 contigs. We followed this initial build with an incremental addition of clones having two usable restriction digests in their derived fingerprint at cutoff of 3e-06, resulting in 1096 contigs. The manual pathfinding process, described in Methods, ultimately resulted in reducing this contig number to 113. Joining of map contigs using the linkage information of paired-fosmid end sequences present in the WGS assembly contigs results in the formation of map clusters. Therefore, by using the WGS assembly contigs to inform the map, make joins, and confirm fingerprint contigs, the final physical map is comprised of 84 contiguous clusters of ordered fosmid clones. The distribution of the 9551 clones used in generating the clusters ranged from 3 to 575, with 305 buried clones and 708 singletons (Table 1).
Table 1.
Fosmid Distribution Within Physical Map Clusters
| Clusters | Fosmids within clusters | Total fosmids | |
|---|---|---|---|
| 1 | 575 | 575 | |
| 10 | >300 | 3407 | |
| 5 | >200 | 1142 | |
| 14 | >100 | 2168 | |
| 24 | >20 | 1281 | |
| 12 | >10 | 153 | |
| 18 | >3 | 97 | |
| Singletons | 708 | ||
| Total | 84 | 9531 |
Statistics of the H. capsulatum G217B WGS Sequence Assembly
Over 1.3 million end sequence reads derived from both plasmids (2–4 kbp insert size fraction) and fosmids were assembled with the PCAP program (Huang et al. 2003). PCAP generates compatible output files that can be viewed with the graphical sequence finishing program Consed (Gordon et al. 1998). Of the total reads produced, ∼75% were included in the assembly; whereas the majority of reads excluded from the assembly contained homopolymeric and heteropolymeric runs composed of di- and trinucleotide repeats. The excluded reads were assembled using phrap (P. Green, unpubl.), generating an excluded reads assembly. This assembly and the PCAP assembly were converged by using phrap. The current WGS assembly statistics after merging of the PCAP and excluded phrap assemblies result in 1243 WGS sequence assembly contigs >2 kb (average 35 kb size) and 687 >8 kb (average 60 kb size), representing total calculated genome sizes of 43.5 and 41.4 Mbp, respectively. Presently, the composite WGS assembly is being finished.
DISCUSSION
The goal of this study was to create a fosmid-based physical map of the H. capsulatum G217B genome. An iterative process of anchoring and orienting physical map clusters to the WGS assembly contigs was employed using fosmid end-sequence information. In this project, the fosmid-based physical map supplies a genomic scaffold that both guides the genome finishing efforts and aids in resolving the differences observed between the predicted G186A-S (24 Mb) and the assembled G217B genome size. The genome size discrepancy is in conflict with preliminary flow cytometry data for H. capsulatum isolates G186A-B and G217B, which indicates that they have the same DNA mass per cell as G186A-S (Carr and Shearer Jr. 1998). In fact, a difference of >17 Mbp is predicted from the G217B WGS assembly, with an assembled genome size >41 Mbp.
To elucidate differences in the predicted and assembled genome sizes, we are in the process of anchoring physical map clusters to WGS assembly contigs. These anchors provide long-range linking information to make joins of physical map clusters and WGS assembly contigs. Graphically, Figure 2 shows map cluster 2501 from MapLink (J. Xu and J.I. Gordon, in prep.) anchored to a Consed assembly view of WGS assembly Supercontig Merge 57. After flipping the orientation of Supercontig Merge 57, these two data sets show good concordance, with two sequence gaps between contigs 26–22 and 22–19. These gaps are spanned by fosmid clones (dark lines) and can be addressed by a variety of means, including producing and sequencing shotgun libraries of the gap-spanning fosmids. Additionally, fosmid clone L_AAZ036H02 resides in contig 26 of Supercontig Merge 57 and anchors map cluster 2501 to Supercontig Merge 20 (dashed line). Using fosmid end sequences as long-range linking information, WGS assembly contigs anchored to physical map clusters allow us to evaluate and address potential joins between Supercontigs (e.g., joining Merge 20 to Merge 57)
Figure 2.
MapLink provides anchoring information between the WGS assembly contigs (represented as dotted lines) and the physical map clusters (solid bars). Physical map cluster 2501, represented by 67 overlapping fosmid clones built in FPC, anchors to WGS assembly Supercontig Merge 57. This Supercontig is composed of three contigs, that is, 26, 22, and 19 with sizes of 107.8, 136.2, and 15.0 kb, respectively. Contigs 26 and 22 are anchored by gap-spanning fosmid clones L_AAZ090H07, L_AAZ088E04, and L_AAZ059B11 (solid lines), and contigs 22 and 19 are anchored by L_AAZ081F09 and L_AAZ089B12 (solid lines). In contig 26, L_AAZ036H02 (dashed line) anchors position 80,390 and links to Supercontig Merge 20. Contig 19 contains end sequence of L_AAZ091F01 and L_AAZ080C03 (dashed lines) and anchors map cluster 5901 at positions 460 and 7700 in Supercontig Merge 57. Map cluster 5901 also anchors Supercontig Merge 81, and this anchor provides long-range linking information between WGS assembly Supercontigs Merge 57 and Merge 81. This figure is not drawn to scale, and linkage information does not indicate sequence overlap.
Reciprocally, the WGS assembly aids the physical map. In Figure 2, contig 19 contains end sequence of two fosmids (dashed lines) anchoring Supercontig Merge 57 to map cluster 5901, which in turn, is anchored to Supercontig Merge 81. In this iterative process, we can use the WGS assembly to inform the physical map of potential joins (i.e., linking physical map cluster 2501 to cluster 5901 and also joining Merge 57 and Merge 81). Using both data sets in a map-assisted whole-genome shotgun approach, we can resolve the difference between the predicted and assembled genome size of H. capsulatum G217B.
In this study, successive modifications of the PCAP WGS assembly were used to incorporate more of the shotgun reads. The initial PCAP WGS assembly includes 75% of the end sequence data. To incorporate the missing data, we used phrap to assemble the excluded reads and combine the assembly of the excluded reads with the PCAP assembly. The current WGS assembly represents over 3100 contigs and contains up to 17% repetitive DNA. Although a comprehensive repeat analysis has not yet been performed, the theoretical repetitive nature (based on reassociation kinetics) is greater than expected with observed homopolymeric runs and small repeat units. Additionally, A:G and T:C transitions present in small repeat regions, 50–250 bp in length (data not shown) contribute to misassembled regions and may increase the repeat bias within the WGS assembly. Thus, using MapLink, discordant linking information observed between the physical map and WGS assembly targets problematic genomic regions for resolution during finishing.
Fosmid cloning is straightforward and practical for physical mapping. However, physical maps generated from BAC clones are advantageous, needing far fewer clones (when compared with fosmids) to provide the necessary depth of coverage and the fewest gaps (Schmitt et al. 1996; Marra et al. 1997; Soderlund et al. 2000). The difficulty is consistently producing BAC clones with inserts that average ≥150 kbp (desirable for the fragment pattern produced upon digestion and agarose gel separation). Routine isolation of 30–50 kbp DNA fragments produces high titer fosmid libraries (>105 CFUs/mL), and our tight insert size (35.9 ±4.3 kbp) contributes positively to whole-genome shotgun assembly algorithms, which typically perform best with end reads from insert sizes having as low a variation in length as possible (Batzoglou et al. 2002).
The fosmid-based mapping approach fits into our existing pipeline. DNA fragments from HindIII, PstI, and BamHI-digested fosmid clones were separated by agarose gel electrophoresis, stained, and scanned for interpretation by the Image software package (http://www.sanger.ac.uk/Software/Image) producing fragment mobility bands files. A combined bands file used in the initial FPC contig build (Fig. 1) was generated by offsetting the mobility values for the independent digest of each clone by a fixed amount. Advantages of combining multiple restriction patterns in the bands file, include a decreased false-negative frequency associated with the use of smaller clone sizes, as shown in Table 4 of Soderlund et al. (2000), and increased sensitivity for identifying clone overlap (Olson et al. 1986; Riles et al. 1993; Wong et al. 1997). In multiple-complete-digest restriction-fragment mapping, MCD, confirmed cosmids cloned from YACs used three independent digestions, and redundant clone coverage of from 15 to 30X was necessary for mapping accuracy (Wong et al. 1997). Our data set contained over 28,000 restriction digests encompassing the entire H. capsulatum G217B genome. In lieu of the MCD algorithm, we tested FPC and generated identical map builds when the stacking order of the fingerprints was varied. In future small mapping projects, an increase in clone coverage will most likely aid map contiguity. Future fosmid-based mapping projects will be aided by our use of libraries generated with the vector, pCC-Fos1 (EpiCentre Technologies). This inducible vector system produces sufficient fosmid DNA required for physical mapping and WGS sequencing, as well as for down-stream use in genome finishing processes as a template for custom oligonucleotide-primed sequencing reactions (to extend into gaps or improve sequence quality) from a single microtiter plate well prep.
Our results detailed mapping the Histoplasma capsulatum G217B genome exclusively using fosmid clones with existing software (Image and FPC). The map-assisted whole genome shotgun approach provides an efficient means for de novo sequencing, in which the WGS assembly and physical map are produced by different processes. The MapLink software performs automated integration between physical map clusters and WGS sequence-assembly contigs, and provides a graphical interface to view both data sets. This resulting genomic organization supplies essential linking information used in finishing, and ultimately will create an accurate genomic sequence of this unique, dimorphic fungus.
METHODS
Strains, Growth Conditions, and Vectors
Histoplasma capsulatum isolate G217B (ATCC MYA-2455) was recovered from 4°C stocks on HMM agar plates and grown as yeast at 37°C (Worsham and Goldman 1988). Colony-purified yeast isolates were confirmed to be fully virulent using a mouse model of infection (data not shown; Sebghati et al. 2000). Yeast were grown to early stationary phase.
The EpiFOS Fosmid Library Production Kit (EpiCentre Technologies) was used with vector pEpiFOS-5. Transductants were isolated on LB agar plates (Sambrook 1989) supplemented with 25 μg/mL chloramphenicol (Cm25) and S-GAL (Sigma). Clones were picked into shallow growth plates with 240 μL of TB, Cm25, supplemented with 8% glycerol and grown at 37°C overnight while shaking. Overnight growth was subcultured into 1.2 mL of LB, Cm25 and incubated overnight at 37°C. Glycerol stocks were stored at –80°C.
Histoplasma DNA Isolation
After growth in 50 mL of HMM, H. capsulatum, G217B cells were pelleted, washed three times in 50 mL of SCE-D (1 M Sorbitol, 1 mM EDTA, 10 mM Sodium Citrate at pH 5.8, 10 mM Dithiothreitol) and suspended in 2 mL of SCE-D. DNA was isolated using the Easy DNA Kit (Invitrogen). Suspended cells were mixed with 3.5 mL of lysis solution, incubated at 65°C for 30 min, and 1.5 mL of precipitation solution was added to the lysed cells and mixed by mild inversion, to which 5 mL of chloroform was added and mixed. The suspension was separated into organic and aqueous phases by centrifugation at 1800g for 30 min at 25°C. The aqueous phase was removed, and the DNA was precipitated with the addition of 2.5 volumes of 95% ethanol. DNA was recovered by spooling on sterile Pasteur pipettes. Recovered genomic DNA was rinsed in 95% ethanol, air-dried, and incubated in 500 μL of TE buffer supplemented with 40 μg/mL RNAse A at 37°C for 1 h. Genomic DNA was column purified on Qiagen G-100 tips and precipitated with 0.7 vol of 2-propanol. The DNA was suspended in 500 μL of TE buffer.
Generating Fosmid Libraries
Approximately 20 μg of H. capsulatum G217B genomic DNA was sheared (Gene Machines Hydroshear, large orifice, speed code 22, 10 passes, retention time of 20) and end-repaired according to the manufacturer. End-repaired DNA was resolved by pulse-field gel electrophoresis using the CHEF Mapper XA Pulsed Field Electrophoresis System (Bio-Rad). The gel-run parameters were as follows: 1% GTG agarose/0.5 × TBE buffer, 6 V/cm, included angle, 120°; initial switch time, 0.5 sec.; final switch time, 1.5 sec.; run time, 11 h; ramping, linear; temperature, 14°C.
DNA fragments migrating to 30–50 kbp in length were excised and purified after a second size selection in 1% low-melting point (LMP) agarose (Bio-Rad). The second size selection gel was run at 30 V for 16 h and stained with SYBR-Green. DNA fragments were visualized with a blue light, excised, melted at 65°C, and treated with AgarAce (Promega, 1.5 U per 100 mg agarose) in 0.5× TBE buffer at 42°C for 1 h. AgarAce treatment was followed by a single phenol extraction, and the DNA solution was minimized using sec-butanol and ethanol precipitated with 0.1 M NaCl. DNA was suspended in 10 μL of molecular biology grade H20 (Sigma). DNA was desalted by drop dialysis (MF-Millipore 0.025 μm pore-size membrane filters) and ligated to pEpiFOS-5 DNA. Fosmid clones were packaged using MaxPlax λ packaging extract according to manufacturer's instructions. Packaged fosmid clones were stored at 4°C over chloroform in 1 mL of phage dilution buffer (10 mM Tris-HCl at pH 8.3, 100 mM NaCl, 10 mM MgCl2).
Fingerprinting and Physical Map Assembly of Histoplasma Fosmids
Fosmid clones were grown and fosmid DNA isolated using previously reported methods (Marra et al. 1999). Isolated fosmid DNA was digested in three separate restriction enzyme reactions using HindIII, PstI, and BamHI. Resulting fragments were loaded and separated on 1% agarose gels as previously described (Marra et al. 1997). Fragment mobility data were acquired from the images of SYBR green-stained agarose gel images using Image software (http://www.sanger.ac.uk/Software/Image), and processed for contig construction using FPC (http://www.genome.arizona.edu/software/fpc/) as follows. The fragment mobility output files from the Image software package were combined starting with the first digest and then offsetting each subsequent fragment set by a multiple of an amount (4000 and 8000 in this case) determined to avoid overlap of the fragments from any one digest with the previous one. These modified mobility files were entered into the FPC database and used to build contig assemblies at different stringencies. Initially, all clones with combined data from all three digests were assembled using the following FPC parameters: tolerance, 7; cutoff, 3e-10; Diffbury, 0.1; MinBands, 3. Next, clones with fingerprint data from only two fingerprints were added to the database and integrated into the assembly utilizing an incremental build with FPC parameters as follows: tolerance, 7; cutoff, 3e-06; Diffbury, 0.1; MinBands, 3. Finally, clones with data from only a single restriction digest were added to the database for integration, where possible, during manual editing of the assembled contigs.
Manual Contig Editing
The optimized FPC assembly for H. capsulatum was used as a starting point to merge contigs in a process called pathfinding. The pathfinding process is as follows: (1) Each contig is visually examined by an experienced technician who verifies proper clone order and searches for possible misassemblies, (2) clone order is established and misassemblies are addressed, (3) merges are made by extending contig ends with singletons and by comparing the ends of all contigs with all others in the assembly at a lower stringency than the original build, (4) after all contigs have been edited and merged at the established parameters, fosmid end sequences are aligned to the H. capsulatum G217B sequence assembly using BLAST (Altschul et al. 1990) and grouped in clusters and, (5) then edited to achieve contiguity and the ends again searched at a lower stringency to make all possible merges.
Clusters of fingerprint contigs and individual clones were created on the basis of the order of the 15,655 H. capsulatum G217B fosmid-end sequences that are contained within the assembly. The boundaries on the assembly of fingerprint contigs and clones with two end sequences were determined. Overlapping contigs and clones were positioned in clusters, separated by gaps for easy viewing and editing in the FPC graphical interface. The orientations of contigs were flipped as needed. The fingerprints within each cluster were manually reviewed and merges made on the basis of fingerprint matches. Of the 294 merges suggested by the sequence assembly, 94% were confirmed by fingerprints. This process was repeated using a later sequence assembly, resulting in the incorporation of an additional 134 single clones and 38 contig merges.
Fosmid End Sequencing
The fosmid DNA utilized for fingerprinting was of sufficient quality for DNA sequencing. The protocol used to end sequence fosmid clones was as described (http://www.tigr.org/tdb/bac_ends/mouse/bac_end_intro.html), with minor modifications. The forward sequencing primer sequence used was 5′-GGTGTGTGCAA GGCGATTAAGTTGG-3′, and the reverse sequencing primer sequence was 5′-CTCGTATGTTGTGTGGAATTGTGAGC-3′. Trace files generated on our ABI sequencing instruments were processed using the phred program (Ewing and Green 1998; Ewing et al. 1998). Once traces are generated, they are checked for quality and trimmed before being deposited into our internal end-sequence database. Quality-trimmed fosmid end sequences (reads >50 bp of sequence and phred score of >20) were coassembled with our whole-genome shotgun sequence assembly using the PCAP algorithm (Huang et al. 2003) and phrap. Embedded fosmid end reads were used to orient and order FPC contig assemblies. In addition, our analysis of the initial sequence assemblies enabled us to identify putative sequence gaps that could be spanned by fosmids whose ends were found within assembled sequence contigs. A set of ∼400 such fosmids were then prepared by conventional methods and used to make plasmid subclone libraries. These libraries were sequenced at a light coverage (384 reactions per fosmid) and added to the assembly to pull in small contigs and gap-filling sequence reads from the initial sequence data.
Genome Sequencing Strategy
Our sequencing strategy involved a whole-genome shotgun approach to generate initial 8× coverage of the H. capsulatum G217B genome. Specifically, DNA was sheared, end-repaired, linker-ligated, and size-selected. Gel-purified DNA was ligated to plasmid pOTW13 (Cliften et al. 2003). The resulting subclones were processed through our production-sequencing pipeline, including magnetic bead-based DNA purification, dual-end sequencing with Big Dye version 3.1 terminator chemistry (Applied Biosystems), and analysis on ABI 3700 and 3730xl DNA sequencers. Each fosmid fingerprinted generated paired-end read sequence data, as described.
Data Availability
H. capsulatum G217B fingerprint contigs can be viewed in FPC format (http://genome.wustl.edu/projects/hcapsulatum/index.php?fpc=1). The PCAP and phrap WGS assembly contigs are available for BLAST (http://www.genome.wustl.edu/blast/histo_client.cgi). Fosmid end sequences have been submitted to the NCBI trace archive database.
Acknowledgments
We thank the members within Mapping, Finishing and support, and Informatics groups of the GSC directly involved in this endeavor. Special thanks go to Jacquelyn Engle and Linda Eissenberg of the Goldman laboratory for their assistance in strain handling and virulence assays. This work was supported by NIAID and Public Health Service grant AI25584 (to E.R.M.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2361404.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Batzoglou, S., Jaffe, D.B., Stanley, K., Butler, J., Gnerre, S., Mauceli, E., Berger, B., Mesirov, J.P., and Lander, E.S. 2002. ARACHNE: A whole-genome shotgun assembler. Genome Res. 12: 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradsher, R.W. 1996. Histoplasmosis and blastomycosis. Clin. Infect. Dis. 22 Suppl 2: S102–S11. [DOI] [PubMed] [Google Scholar]
- Carr, J. and Shearer Jr., G. 1998. Genome size, complexity, and ploidy of the pathogenic fungus Histoplasma capsulatum. J. Bacteriol. 180: 6697–6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter, D.A., Taylor, J.W., Dechairo, B., Burt, A., Koenig, G.L., and White, T.J. 2001. Amplified single-nucleotide polymorphisms and a (GA)(n) microsatellite marker reveal genetic differentiation between populations of Histoplasma capsulatum from the Americas. Fungal Genet. Biol. 34: 37–48. [DOI] [PubMed] [Google Scholar]
- Cliften, P., Sudarsanam, P., Desikan, A., Fulton, L., Fulton, B., Majors, J., Waterston, R., Cohen, B.A., and Johnston, M. 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 301: 71–76. [DOI] [PubMed] [Google Scholar]
- Eissenberg, L.G., Goldman, W.E., and Schlesinger, P.H. 1993. Histoplasma capsulatum modulates the acidification of phagolysosomes. J. Exp. Med. 177: 1605–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing, B. and Green, P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8: 186–194. [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175–185. [DOI] [PubMed] [Google Scholar]
- Gordon, D., Abajian, C, and Green, P. 1998. Consed: A graphical tool for sequence finishing. Genome Res. 8: 195–202. [DOI] [PubMed] [Google Scholar]
- Gregory, S.G., Sekhon, M., Schein, J., Zhao, S., Osoegawa, K., Scott, C.E., Evans, R.S., Burridge, P.W., Cox, T.V., Fox, C.A., et al. 2002. A physical map of the mouse genome. Nature 418: 743–750. [DOI] [PubMed] [Google Scholar]
- Huang, X., Wang, J., Aluru, S., Yang, S-P., and Hillier, L. 2003. PCAP: A whole-genome assembly program. Genome Res. 13: 2164–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga, T., Taylor, J.W., and White, T.J. 1999. Phylogenetic relationships of varieties and geographical groups of the human pathogenic fungus Histoplasma capsulatum Darling. J. Clin. Microbiol. 37: 653–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersulyte, D., Woods, J.P., Keath, E.J., Goldman, W.E., and Berg, D.E. 1992. Diversity among clinical isolates of Histoplasma capsulatum detected by polymerase chain reaction with arbitrary primers. J. Bacteriol. 174: 7075–7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kügler, S., Schurtz-Sebghati, T., Groppe-Eissenberg, L., and Goldman, W.E. 2000. Phenotypic variation and intracellular parasitism by Histoplasma capsulatum. Proc. Natl. Acad. Sci. 97: 8794–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca, B. and Kobayashi, G.S. 1989. Dimorphism in Histoplasma capsulatum: A model for the study of cell differentiation in pathogenic fungi. Microbiol. Rev. 53: 186–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra, M.A., Kucaba, T.A., Dietrich, N.L., Green, E.D., Brownstein, B., Wilson, R.K., McDonald, K.M., Hillier, L.W., McPherson, J.D., and Waterston, R.H. 1997. High throughput fingerprint analysis of large-insert clones. Genome Res. 7: 1072–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra, M.A., Kucaba, T.A., Hillier, L.W., and Waterston, R.H. 1999. High-throughput plasmid DNA purification for 3 cents per sample. Nucleic Acids Res. 27: e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson, J.D., Marra, M., Hillier, L., Waterston, R.H., Chinwalla, A., Wallis, J., Sekhon, M., Wylie, K., Mardis, E.R., Wilson, R.K., et al. 2001. A physical map of the human genome. Nature 409: 934–941. [DOI] [PubMed] [Google Scholar]
- Newman, S.L. 1999. Macrophages in host defense against Histoplasma capsulatum. Trends. Microbiol. 7: 67–71. [DOI] [PubMed] [Google Scholar]
- Olson, M.V., Dutchik, J.E., Graham, M.Y., Brodeur, G.M., Helms, C., Frank, M., MacCollin, M., Scheinman, R., and Frank, T. 1986. Random-clone strategy for genomic restriction mapping in yeast. Proc. Natl. Acad. Sci. 83: 7826–7830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riles, L., Dutchik, J.E., Baktha, A., McCauley, B.K., Thayer, E.C., Leckie, M.P., Braden, V.V., Depke, J.E., and Olson, M.V. 1993. Physical maps of the six smallest chromosomes of Saccharomyces cerevisiae at a resolution of 2.6 kilobase pairs. Genetics 134: 81–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippon, J.W. 1982. Histoplasmosis (Histoplasmosis capsulati). In: Medical mycology. The Pathogenic fungi and the pathogenic actinomycetes (ed. M.L.W.R. Wonsiewicz), pp. 201–205. W.B. Saunders & Co., Philadelphia, PA.
- Sambrook, J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schmitt, H., Kim, U.J., Slepak, T., Blin, N., Simon, M.I., and Shizuya, H. 1996. Framework for a physical map of the human 22q13 region using bacterial artificial chromosomes (BACs). Genomics 33: 9–20. [DOI] [PubMed] [Google Scholar]
- Sebghati, T.S., Engle, J.T., and Goldman, W.E. 2000. Intracellular parasitism by Histoplasma capsulatum: Fungal virulence and calcium dependence. Science 290: 1368–1372. [DOI] [PubMed] [Google Scholar]
- Soderlund, C., Humphray, S., Dunham, A., and French, L. 2000. Contigs built with fingerprints, markers, and FPC V4.7. Genome Res. 10: 1772–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen, K.N., Clemons, K.V., and Stevens, D.A. 1999. Murine models of blastomycosis, coccidioidomycosis, and histoplasmosis. Mycopathologia 146: 53–65. [DOI] [PubMed] [Google Scholar]
- Steele, P.E., Carle, G.F., Kobayashi, G.S., and Medoff, G. 1989. Electrophoretic analysis of Histoplasma capsulatum chromosomal DNA. Mol. Cell. Biol. 9: 983–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, G.K., Yu, J., Thayer, E.C., and Olson, M.V. 1997. Multiple-complete-digest restriction fragment mapping: Generating sequence-ready maps for large-scale DNA sequencing. Proc. Natl. Acad. Sci. 94: 5225–5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods, J.P., Heinecke, E.L., Luecke, J.W., Maldonado, E., Ng, J.Z., Retallack, D.M., and Timmerman, M.M. 2001. Pathogenesis of Histoplasma capsulatum. Semin. Respir. Infect. 16: 91–101. [DOI] [PubMed] [Google Scholar]
- Worsham, P.L. and Goldman, W.E. 1988. Quantitative plating of Histoplasma capsulatum without addition of conditioned medium or siderophores. J. Med. Vet. Mycol. 26: 137–143. [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.genome.arizona.edu/software/fpc/; The FPC and Friends Web site is associated with the Arizona Genomics Institute (AGI) and Arizona Genomics Computational Laboratory (AGCoL). This site provides FPC software download and documentation links.
- http://www.genome.wustl.edu/blast/histo_client.cgi; This link is part of the Histoplasma capsulatum project maintained on the Genome Sequencing Center Web site. The Histoplasma capsulatum G217B and G186A-R Whole Genome Shotgun (WGS) PCAP and ARACHNE assemblies are available for BLASTIN searches.
- http://genome.wustl.edu/projects/hcapsulatum/index.php?fpc=1; An additional link where the Histoplasma capsulatum FPC database can be downloaded.
- http://www.sanger.ac.uk/Software/Image; A link to IMAGE, the fingerprint image analysis system, at the Sanger Institute.
- http://www.tigr.org/tdb/bac_ends/mouse/bac_end_intro.html; This site provides links to BAC End Sequencing Protocols and BAC library resources.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
H. capsulatum G217B fingerprint contigs can be viewed in FPC format (http://genome.wustl.edu/projects/hcapsulatum/index.php?fpc=1). The PCAP and phrap WGS assembly contigs are available for BLAST (http://www.genome.wustl.edu/blast/histo_client.cgi). Fosmid end sequences have been submitted to the NCBI trace archive database.