Abstract
The invasive and metastatic properties of many human tumors have been associated with upregulation of the microRNA miR-10b, but its functional contributions in this setting have not been fully unraveled. Here we report the generation of miR-10b-deficient mice in which miR-10b is shown to be largely dispensable for normal development but critical to tumorigenesis. Loss of miR-10b delays oncogene-induced mammary tumorigenesis and suppresses epithelial-mesenchymal transition, intravasation, and metastasis in a mouse model of metastatic breast cancer. Among the target genes of miR-10b, the tumor suppressor genes Tbx5 and Pten and the metastasis suppressor gene Hoxd10 are significantly upregulated by miR-10b deletion. Mechanistically, miR-10b promotes breast cancer cell proliferation, migration, and invasion through inhibition of the expression of the transcription factor TBX5, leading to repression of the tumor suppressor genes DYRK1A and PTEN. In clinical specimens of breast cancer, the expression of TBX5, HOXD10, and DYRK1A correlates with relapse-free survival and overall survival outcomes in patients. Our results establish miR-10b as an oncomiR that drives metastasis, termed a metastamiR, and define the set of critical tumor suppressor mechanisms it overcomes to drive breast cancer progression.
Keywords: miR-10b, TBX5, breast cancer, metastasis
Introduction
MicroRNAs (miRNAs) are ~22-nucleotide non-coding RNAs that base-pair with messenger RNAs (mRNAs), leading to degradation of target mRNAs and/or inhibition of their translation (1). Although growing numbers of miRNAs have been found to be involved in breast cancer progression based on cell culture and xenograft models (2-4), their functions in the metastasis of autochthonous mammary tumors have not been demonstrated by miRNA knockout approaches. Elucidating the role of these miRNAs in normal development and in the initiation and metastasis of autochthonous tumors will provide important insights into the mechanisms of malignant progression.
It has been shown that certain miRNAs, such as miR-10b and miR-9, are upregulated in metastatic cancer cells and promote metastasis (5, 6). The first metastasis-regulating miRNA, miR-10b, was initially found to be expressed at elevated levels in metastatic cell lines and primary tumors from patients with metastatic breast cancer (5, 7). A growing body of evidence has demonstrated that miR-10b expression is also positively associated with high-grade malignancy or metastasis in other cancer types, such as pancreatic cancer (8-10), glioblastoma (11-14), bladder cancer (15), and liver cancer (16). In addition, lymph node metastases express higher levels of miR-10b than paired primary tumors in multiple cancer types (17).
When overexpressed, miR-10b can induce motility and invasiveness of various cancer cell lines (5, 15, 16, 18-21) and promote metastasis in xenograft models (5, 15, 20) through direct targeting of HOXD10, NF1, KLF4, PTEN, and likely other genes. Conversely, silencing miR-10b expression suppresses cancer cell proliferation, migration, and invasion in vitro as well as tumor growth or metastasis in vivo (14, 18, 19, 22), suggesting that miR-10b is a potential target for anti-tumor or anti-metastatic therapy. Notably, combining nanoparticle-encapsulated miR-10b inhibitors with low-dose doxorubicin achieved complete durable regression of metastases in a xenograft model of breast cancer (23). Intriguingly, miR-10b is secreted by metastatic breast cancer cells via exosomes and, upon uptake, induces invasiveness of non-malignant mammary epithelial cells (24).
In this study, we generated miR-10b-deficient mice to determine the role of miR-10b in normal development and tumor progression. Although genetic deletion of miR-10b does not cause substantial developmental defects, it impedes mammary tumor initiation, progression, and metastasis in a polyomavirus middle T (PyMT) model of breast cancer. We further identified TBX5 as a functional target of miR-10b and identified two tumor suppressor genes, DYRK1A and PTEN, as target genes of the transcription factor TBX5.
Materials and Methods
For details, see Supplementary Materials and Methods.
Generation of miR-10b knockout mice
miR-10b mutant mice were produced at Taconic Artemis. Mice with the conditional knockout allele were generated after breeding to Flp-deleter mice for the removal of the selection marker (PuroR). Mice with the constitutive knockout allele were generated after breeding to Cre-deleter mice. Both conditional and constitutive miR-10b mutant mice were maintained on a C57BL/6 background.
Tumor and metastasis studies
All animal studies were performed in accordance with a protocol approved by the Institutional Animal Care and Use Committee of MD Anderson Cancer Center. Mammary tumor-free survival was determined by daily palpation. MMTV-PyMT;miR-10b+/+ and MMTV-PyMT;miR-10b−/− female mice were euthanized at 13, 16, and 19 weeks of age and the end-point (18-26 weeks, when mice met the institutional euthanasia criteria for tumor size and overall health condition); whole mammary glands or tumors and lung tissues were collected, weighed, and prepared for histopathological analysis. The presence of metastasis was determined by histopathological review of lung sections.
Histopathology and immunostaining
Mouse tissue samples were fixed in 10% buffered formalin overnight, washed with phosphate-buffered saline (PBS), transferred to 70% ethanol, embedded in paraffin, sectioned (5 μm thick), and stained with hematoxylin and eosin (H&E). Histopathological examination was performed on all tissue sections by a pathologist (M. James You). Specific tissue areas were quantitated by counting pixels with digital imaging software (Photoshop). For immunohistochemical staining, paraffin-embedded sections were deparaffinized in xylene and rehydrated in ethanol. Antigen was retrieved using citric acid (pH 6.0, Dako). Endogenous peroxidase activity was blocked using 3% hydrogen peroxide in PBS. Sections were blocked with 5% bovine serum albumin in PBS and incubated with the primary antibody. The sections were then washed with PBS and incubated with a horseradish peroxidase-conjugated secondary antibody, followed by incubation with diaminobenzidine (DAB, Sigma).
Whole-mount staining of mammary glands
After euthanasia, the entire right #4 inguinal mammary gland was collected from wild-type and miR-10b−/− female mice, placed onto a glass slide, stretched out, and air-dried for a few minutes. The tissues were fixed overnight in Carnoy’s solution, rinsed in PBS, and stained in Carmine Alum solution overnight. The tissues were then dehydrated in ethanol and xylene and mounted.
Blood collection and blood count
Peripheral blood was drawn by retro-orbital sampling from age- and sex-matched wild-type and miR-10b−/− mice. Blood was collected in EDTA-coated tubes to prevent clotting and subjected to complete blood counting performed by Department of Veterinary Medicine & Surgery at MD Anderson Cancer Center.
Isolation and staining of circulating tumor cells (CTCs) from mice
Peripheral blood was collected via facial vein or retro-orbital bleeding and red blood cells were lysed with RBC lysing buffer (Gibco, A10492-01). Nucleated cells were spun onto the slides using Cytospin and fixed in 10% formalin. For immunofluorescent staining of the PyMT protein, cells were permeabilized with 0.25% Triton X-100. The PyMT-specific primary antibody (Abcam, ab15085, 1:50) and anti-rat secondary antibody conjugated with Fluorescein (Vector Laboratories, FI-4001, 1:100) were used. After staining, cells were mounted using mounting medium with DAPI (Vector Laboratories, H-1200). For CTC quantification, the ratio of PyMT+;DAPI+ cells to total DAPI+ cells was calculated in ×200 fields.
Quantitative PCR (qPCR)
Total RNA from mouse tissues, MEFs, and human cells was isolated using the TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. Real-time PCR and data collection were performed on a CFX96 instrument (Bio-Rad).
Plasmids and tissue culture
Human TBX5 cDNA was cloned into the pCMV-Tag2B vector using restriction enzymes BamHI and EcoRI. 293FT cells were from ThermoFisher Scientific in 2012 and the LM2 subline of MDA-MB-231 cells was from Xiang Zhang (Baylor College of Medicine) in 2014. Both cell lines were cultured in DMEM medium supplemented with 10% fetal bovine serum. Cell lines were authenticated by short tandem repeat analysis at MD Anderson’s Characterized Cell Line Core Facility.
Transfection
For overexpression, the MDH control, MDH-miR-10b, pCMV-Tag2B control, and pCMV-Tag2B-TBX5 plasmids were transfected into cells using Lipofectamine 2000 (Life Technologies). For knockdown, the negative control inhibitor (AM Biotechnologies), anti-miR-10b inhibitor (AM Biotechnologies), scrambled siRNA (Sigma), and TBX5 siRNA (Sigma, SASI_Hs01_00193647 and SASI_Hs01_00193648) oligonucleotides were transfected into cells using Oligofectamine (Life Technologies). The sequence and chemical modifications of the miR-10b antisense inhibitor are: 5′-Cfs2AmCmAmAmAmUmUmCmGmGmUmUmCmUmAmCmAmGmGmGfs2Ufs2Am-3′ (m = 2′-OMe, fs = 2′-F-3′-monothioate, fs2 = 2′-F-3′-dithioate).
Luciferase reporter assay
Firefly luciferase reporter constructs containing the DYRK1A promoter (D1A-Luc) and PTEN promoter (PTEN-Luc) were from Walter Becker (RWTH Aachen University) and Jonathan Kurie (MD Anderson Cancer Center), respectively, and are described previously (25-27). In 24-well plates, pCMV-Tag2B-TBX5 (+, 50 ng; ++, 400 ng) was transfected into 293FT cells along with the pRL-SV40 Renilla luciferase construct (0.5 ng) and the firefly luciferase reporter construct (D1A-Luc or PTEN-Luc, 25 ng). Two days after transfection, luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega). Firefly luciferase activity was normalized with Renilla luciferase activity.
Cell proliferation assay
LM2 cells were transfected with the negative control inhibitor, miR-10b inhibitor, scrambled siRNA, and/or TBX5 siRNA; and in separate experiments, transfected with the pCMV-Tag2B or pCMV-Tag2B-TBX5 construct. Next day, 700 cells were plated in 96-well plates. On day 1, 2, 3, 4, and 5, cells were washed with PBS and stained with 0.05% crystal violet solution. Excess staining solution was washed with water and the stained cells were air dried. For quantitation, crystal violet was extracted with 10% acetic acid and optical density was measured using a multi-well plate reader at 595 nm.
Migration and invasion assays
Cell migration and invasion were measured using Boyden Chambers (Corning, 3422 and 354480, respectively). LM2 cells were transfected with the negative control inhibitor, miR-10b inhibitor, scrambled siRNA, and/or TBX5 siRNA; and in separate experiments, transfected with the pCMV-Tag2B or pCMV-Tag2B-TBX5 construct. Next day, cells were trypsinized, counted, and washed with PBS to remove serum and growth factors. 15,000 and 70,000 cells were re-suspended in serum-free medium and added to the upper chamber for migration and invasion assays, respectively. Serum-containing growth medium was added to the bottom chamber. 20 hours later, cells on the upper surface of the membrane were removed with a cotton swab, and cells on the lower surface of the membrane were stained and counted.
Western blot analysis
Immunoblotting was performed with precast gradient gels (Bio-Rad) using standard methods. Briefly, mouse tissues were mechanically homogenized using an electric tissue homogenizer. Homogenized mouse tissues and human cells were lysed in the RIPA buffer (Cell Signaling Technology) containing protease inhibitors (Roche) and phosphatase inhibitors (ThermoFisher Scientific). Proteins were separated by SDS-PAGE and blotted onto a PVDF membrane (Bio-Rad). Membranes were probed with the specific primary antibodies followed by horseradish peroxidase-conjugated secondary antibodies. The bands were visualized by chemiluminescence (Denville Scientific).
TCGA and computational analysis
To assess genomic alterations of TBX5, HOXD10, and DYRK1A genes, we used TCGA breast cancer somatic mutation data and copy number alteration data to determine the alteration rates. To compare the mRNA expression levels of TBX5, HOXD10, or DYRK1A between normal and tumor tissues, we used TCGA breast cancer RNA-Seq data and performed t-tests on the log2-transformed expression values (i.e., RNA-Seq by Expectation Maximization, RSEM). To assess the correlation of TBX5, HOXD10, or DYRK1A expression with clinical outcomes, we used the KM plotter (28) and performed a log-rank test to compare high and low expression groups.
Statistical analysis
We used a log-rank test for survival analysis and Wilcoxon sum rank test for tumor grade analysis. For other analyses, Student’s t test (two-tailed) was used to compare two groups for independent samples. Data are presented as mean ± s.e.m. P < 0.05 was considered statistically significant.
Results
Generation and characterization of miR-10b knockout mice
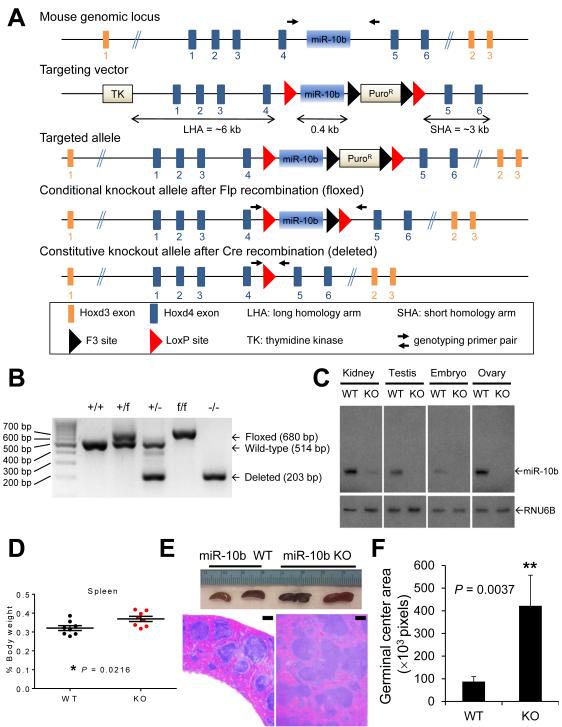
To investigate the role of miR-10b in mammalian development, we generated mice with conditional and constitutive deletion of miR-10b on a C57BL/6 background by homologous recombination and subsequent breeding to Flp- and Cre-deleter mice, which bear a ubiquitously expressed Flp or Cre recombinase transgene (Fig. 1A and 1B). Using a probe specifically detecting mature miR-10b as opposed to mature miR-10a in Northern blot analysis (Supplementary Fig. S1A and S1B), we confirmed that miR-10b expression was deficient in miR-10b-null embryos and in multiple organs of adult miR-10b−/− mice (Fig. 1C). Mouse miR-10b is embedded in intron 4 of Hoxd4 and intron 1 of Hoxd3 (Fig. 1A). Deletion of miR-10b did not alter the expression of Hoxd4 and Hoxd3 (Supplementary Fig. S1C and S1D).
Figure 1. Targeted deletion of miR-10b in mice.
(A) The targeting strategy used to generate miR-10b knockout alleles.
(B) PCR-based genotyping of wild-type (+), floxed (f), and deleted (-) alleles of miR-10b.
(C) Northern blot analysis demonstrates miR-10b deletion in the kidney, testis, ovary, and embryo of age- and sex-matched wild-type (WT) and miR-10b−/− (KO) mice.
(D) Spleen weight (relative to body weight) of age- and sex-matched wild-type (WT) and miR-10b−/− (KO) mice. n = 8 mice per group.
(E) Gross examination (top) and H&E staining (bottom) of spleens of wild-type (WT) and miR-10b−/− (KO) mice. Scale bars, 200 μm.
(F) Germinal center areas were quantitated by pixel counts. n ≥ 4 mice per group.
P values in (D) and (F) were from a two-tailed, unpaired t-test.
From the intercrosses between miR-10b heterozygotes (miR-10b+/−, Fig. 1B), we found that mice homozygous for miR-10b deletion (miR-10b−/−) were born at the expected Mendelian ratio (Supplementary Fig. S2A). Compared with age- and sex-matched wild-type mice, miR-10b-null animals did not display significant abnormalities in total body weight (Supplementary Fig. S2B), the weight of most organs (Supplementary Fig. S2C), overall survival (Supplementary Fig. S3A), fertility (Supplementary Fig. S3B), and complete blood cell count (Supplementary Fig. S4). A comprehensive histopathological examination did not detect significant aberrations in the majority of organs in miR-10b−/− mice (Supplementary Fig. S5A). In addition, whole-mount staining of the mammary gland revealed no differences in ductal growth, ductal branching, and terminal bud formation (Supplementary Fig. S5B). However, a moderate but significant change in the weight of the spleen (P = 0.0216), ovary (P = 0.02645), and uterus (P = 0.0334) was observed in miR-10b−/− mice (Fig. 1D and Supplementary Fig. S2C). Consistent with the increase in spleen size, spleens of miR-10b-deficient animals displayed expansion of germinal centers compared with those of age- and sex-matched wild-type mice (Fig. 1E and 1F), although no lymphoma was found in a 118-week follow-up.
miR-10b deficiency impedes mammary tumor formation and progression in a PyMT mouse model of breast cancer
The mouse mammary tumor virus (MMTV)-PyMT transgenic mice on an FVB/N background develop palpable mammary tumors as early as 5 weeks of age and eventually show 100% tumor penetrance, and pulmonary metastases are observed in 94% of tumor-bearing female mice (29). In MMTV-PyMT mice on a C57BL/6 strain, tumor onset is delayed, although eventually all transgenic females develop mammary tumors (30). Because MMTV-PyMT mice recapitulate the tumor stages, pathology, metastasis, and biomarkers of patients with metastatic breast cancer (31), this model is widely used to dissect the molecular events at early and late stages of breast cancer progression.
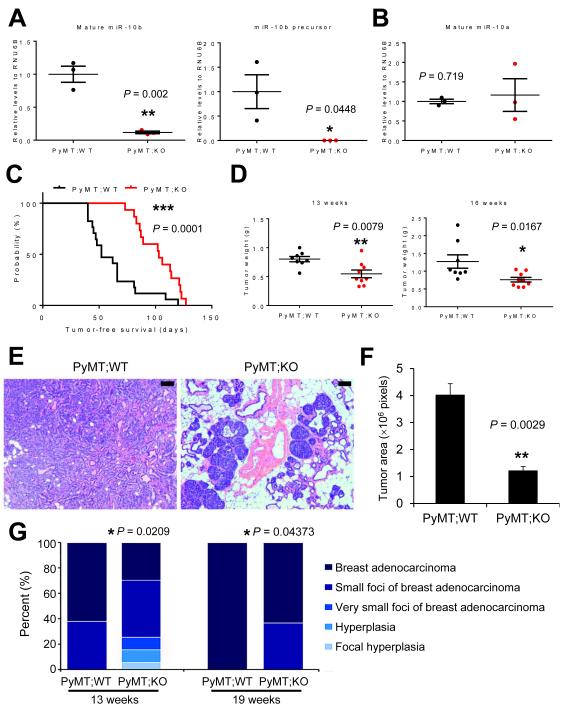
Since the miR-10b knockout mice were generated on a C57BL/6 background, we bred these animals to MMTV-PyMT mice on a C57BL/6 strain. Both mature miR-10b and its precursor were depleted in the mammary tumors of MMTV-PyMT;miR-10b−/− animals (Fig. 2A) without compensatory increase of miR-10a (Fig. 2B). In MMTV-PyMT;miR-10b+/+ mice, palpable mammary tumors appeared as early as 40 days of age, with a median onset at 52 days; by 120 days of age, all females had palpable tumors (n = 17, Fig. 2C). In contrast, MMTV-PyMT;miR-10b−/− mice developed palpable tumors starting at 73 days of age, with 50% of mice showing tumors by 103 days and 100% of mice showing tumors by 127 days of age (n = 15, Fig. 2C). Thus, deletion of miR-10b significantly delayed the onset of PyMT-induced mammary tumors (P = 0.0001).
Figure 2. Deletion of miR-10b impedes mammary tumor initiation, growth, and progression.
(A, B) qPCR of mature miR-10b (A, left panel), the miR-10b precursor (A, right panel), and mature miR-10a (B) in mammary tumors of age-matched MMTV-PyMT;miR-10b WT and MMTV-PyMT;miR-10b KO mice. n = 3 mice per group.
(C) Mammary tumor-free survival of MMTV-PyMT;miR-10b WT (n = 17) and MMTV-PyMT;miR-10b KO (n = 15) mice. The P value was from a log-rank test.
(D) Mammary tumor weight of MMTV-PyMT;miR-10b WT and MMTV-PyMT;miR-10b KO mice at 13 and 16 weeks of age. n ≥ 8 mice per group.
(E, F) H&E staining (E) and tumor area quantification (F) of mammary tissue sections from 13-week-old MMTV-PyMT;miR-10b WT and MMTV-PyMT;miR-10b KO mice. Scale bars in (E), 100 μm. n = 3 mice per group in (F).
(G) Histopathological examination of mammary glands (n = 16, 20, 10, and 11 mice per group, from left to right). P values were from Wilcoxon rank-sum test.
P values in (A), (B), (D), and (F) were from a two-tailed, unpaired t-test.
In addition to the effect on primary tumor onset, the weight of mammary tumors in MMTV-PyMT;miR-10b−/− mice was reduced by 32% (0.8052 g vs. 0.5478 g, P = 0.0079) and 40.1% (1.273 g vs. 0.7622 g, P = 0.0167) at 13 weeks and 16 weeks of age, respectively, compared with MMTV-PyMT;miR-10b+/+ mice (Fig. 2D). However, at the late stage, there was no significant difference in tumor weight (2.109 ± 0.3049 g in MMTV-PyMT;miR-10b−/− mice and 2.032g ± 0.3341 g MMTV-PyMT;miR-10b+/+ mice at 19 weeks of age; P = 0.869). We performed histopathological analysis of mammary tissues. At 13 weeks of age, 10 of 16 (62.5%) MMTV-PyMT;miR-10b+/+ mice had full-blown adenocarcinoma of the mammary gland, whereas MMTV-PyMT;miR-10b−/− mice displayed lesions ranging from focal hyperplasia to focal adenocarcinoma, with only 6 of 20 (30%, P = 0.0209) animals showing full-blown adenocarcinoma (Fig. 2E-G). At 19 weeks of age, the percentages of full-blown adenocarcinoma were 100% (10 of 10) for MMTV-PyMT;miR-10b+/+ mice and 64% (7 of 11) for MMTV-PyMT;miR-10b−/− mice, respectively (P = 0.04373, Fig. 2G). Consistent with inhibition of tumor growth, miR-10b-deficient mammary tumors exhibited a 71.3% decrease (P = 0.0205) in cell proliferation without significant changes in apoptotic cell death (P = 0.701), as gauged by phosphorylated histone H3 (Supplementary Fig. S6A and S6B) and cleaved caspase 3 (Supplementary Fig. S6C and S6D) staining, respectively.
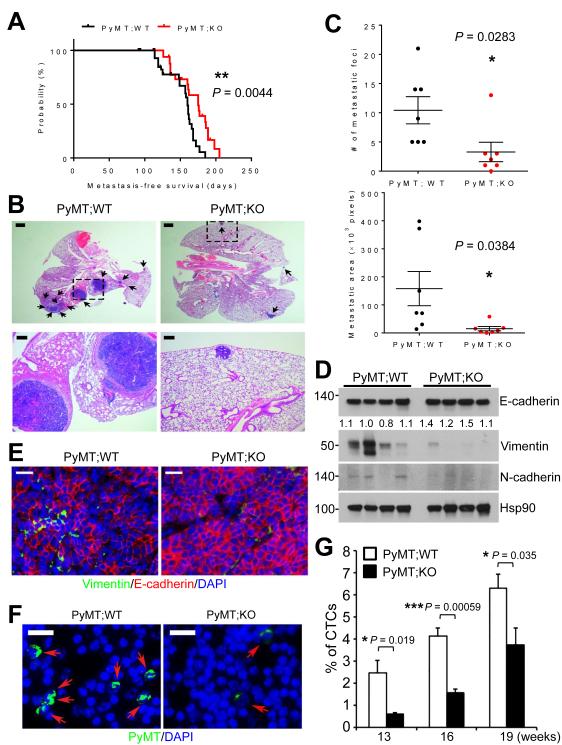
miR-10b deficiency reduces metastasis of PyMT-induced mammary tumors
In orthotopic implantation models, overexpression of miR-10b in otherwise non-metastatic breast cancer cells induced lung metastasis (5), and treatment with miR-10b antagomirs reduced metastasis formation by otherwise highly malignant mammary tumor cells (22). In the PyMT-driven mammary tumor model, miR-10b deficiency did not completely abolish metastasis; instead, it moderately but significantly prolonged metastasis-free survival (median metastasis-free survival: 160 days vs. 175 days, P = 0.0044, Fig. 3A). To quantitate the effect on metastasis, we assessed metastatic lesions in H&E-stained sections of lungs from mice at 24-26 weeks of age. Compared with MMTV-PyMT;miR-10b+/+ animals, MMTV-PyMT;miR-10b−/− mice showed 68.3% and 90.3% reduction in the number (10.4 foci vs. 3.3 foci, P = 0.0283) and area (158.0 × 103 pixels vs. 15.27 × 103 pixels, P = 0.0384) of pulmonary metastatic foci, respectively (Fig. 3B and 3C). Similar to primary tumors, metastatic tumor cells in the lungs of MMTV-PyMT;miR-10b−/− mice were also less proliferative than those in MMTV-PyMT;miR-10b+/+ mice without an increase in apoptosis (Supplementary Fig. S6E-G).
Figure 3. Deletion of miR-10b inhibits EMT, intravasation, and metastasis.
(A) Metastasis-free survival of MMTV-PyMT;miR-10b WT (n = 67) and MMTV-PyMT;miR-10b KO (n = 53) mice. The P value was from a log-rank test.
(B) H&E staining of lungs of MMTV-PyMT;miR-10b WT and MMTV-PyMT;miR-10b KO mice at 24-26 weeks of age. The bottom panels are high-magnification images of the boxed areas in the top panels. Arrows indicate metastatic foci. Scale bars, 1 mm (top) and 200 μm (bottom).
(C) Quantitation of the number (top) and area (bottom) of metastatic foci in the lungs of MMTV-PyMT;miR-10b WT and MMTV-PyMT;miR-10b KO mice at 24-26 weeks of age. n = 7 mice per group.
(D, E) Immunoblotting (D) and immunofluorescent staining (E) of EMT markers in mammary tumors of MMTV-PyMT;miR-10b WT and MMTV-PyMT;miR-10b KO mice. Scale bars in (E), 200 μm.
(F, G) Circulating tumor cells (CTCs) were immunostained with a PyMT-specific antibody (green) and nuclei were stained with DAPI (blue) (F). The percentages of CTCs were quantitated (G). Scale bars in (F), 20 μm. n ≥ 4 mice per group in (G).
P values in (C) and (G) were from a two-tailed, unpaired t-test.
miR-10b deficiency suppresses the mesenchymal phenotype and intravasation of mammary tumor cells
Previous studies demonstrated the activation of epithelial-mesenchymal transition (EMT) in the MMTV-PyMT model (32, 33). Therefore, we examined the mesenchymal markers Vimentin and N-cadherin and the epithelial marker E-cadherin in mouse mammary tumors. In MMTV-PyMT;miR-10b−/− tumors, Vimentin and N-cadherin levels were reduced and E-cadherin expression was increased (Fig. 3D and 3E). To determine the impact of miR-10b depletion on tumor cell intravasation, we compared the number of circulating tumor cells (CTCs) in the peripheral blood collected from MMTV-PyMT;miR-10b+/+ and MTV-PyMT;miR-10b−/− mice, by using an antibody that could specifically detect PyMT-positive mammary tumor cells that metastasized to the lung (Supplementary Fig. S7). In peripheral blood samples, the percentages of CTCs in MMTV-PyMT;miR-10b−/− mice were 0.59%, 1.6%, and 3.7% at 13, 16, and 19 weeks of age, respectively, which was significantly less than those in MMTV-PyMT;miR-10b+/+ mice (2.4%, 4.1%, and 6.3% at 13, 16, and 19 weeks of age, respectively, Fig. 3F and 3G). These results suggest that loss of miR-10b inhibits the ability of mammary tumor cells to disseminate, which is not simply due to the effect on the primary tumor considering similar primary tumor sizes at 19 weeks.
miR-10b regulates the expression of Tbx5, Hoxd10, and Pten in vivo
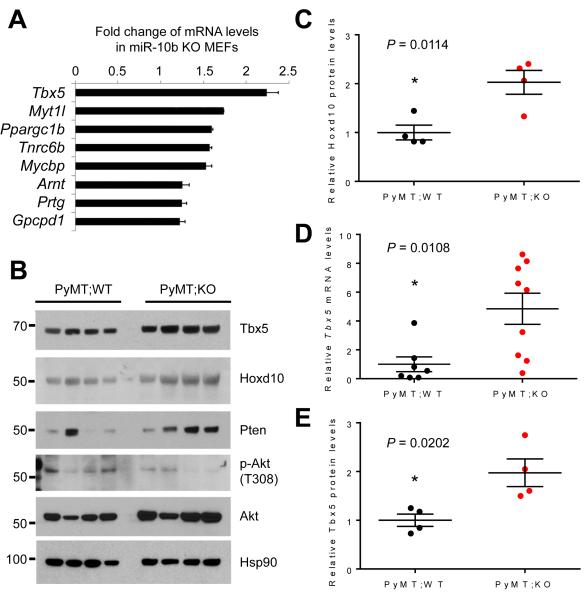
To identify miR-10b target genes whose expression is upregulated upon genetic deletion of miR-10b, we performed quantitative PCR (qPCR) analysis of approximately 100 genes which were predicted computationally to be miR-10b targets (TargetScan, mouse; version 6.0). In this initial screen, 10 genes showed a more than 1.5-fold increase in their expression levels in miR-10b-null mouse embryonic fibroblasts (MEFs), compared with wild-type MEFs (Supplementary Fig. S8). These 10 genes were tested again to confirm their upregulation by miR-10b depletion. Among the eight confirmed targets (Tbx5, Myt1l, Ppargc1b, Tnrc6b, Mycbp, Arnt, Prtg, and Gpcpd1), Tbx5 (T-box 5) stood out as the most significantly upregulated gene (Fig. 4A). In addition, Hoxd10 (homeobox D10), a metastasis suppressor gene previously reported by many groups to be targeted by miR-10b (5, 15, 16, 22-24, 34-37), was upregulated in miR-10b-deficient MEFs by 1.4 fold (Supplementary Fig. S8) and in mammary tumors from MMTV-PyMT;miR-10b−/− mice by 2 fold (Fig. 4B and 4C).
Figure 4. Tbx5, Hoxd10, and Pten are upregulated by miR-10b deletion in mice.
(A) qPCR of miR-10b targets in miR-10b-null MEFs.
(B) Immunoblotting of Tbx5, Hoxd10, Pten, phospho-Akt, Akt, and Hsp90 in mammary tumors of MMTV-PyMT;miR-10b WT and MMTV-PyMT;miR-10b KO mice at 24-26 weeks of age. n = 4 mice per group.
(C) Quantification of Hoxd10 protein levels in (B).
(D) qPCR of Tbx5 in mammary tumors of MMTV-PyMT;miR-10b WT and MMTV-PyMT;miR-10b KO mice at 24-26 weeks of age. n ≥ 7 mice per group.
(E) Quantification of Tbx5 protein levels in (B).
P values in (C) – (E) were from a two-tailed, unpaired t-test.
miR-10a and miR-10b have been reported to directly target the 3′-untranslated region of TBX5 (38). Consistent with the MEF analysis, Tbx5 mRNA levels were upregulated by 4.8 fold in miR-10b-deficient PyMT tumors relative to tumors from MMTV-PyMT;miR-10b+/+ mice (Fig. 4D). This upregulation was also observed at the protein level (Fig. 4B and 4E). In addition, two recent studies reported that miR-10b directly targets PTEN (21, 39). Indeed, compared with MMTV-PyMT;miR-10b+/+ mice, MMTV-PyMT;miR-10b−/− mice displayed upregulation of Pten and downregulation of phospho-Akt in both primary mammary tumors (Fig. 4B and Supplementary Fig. S9A) and lung metastases (Supplementary Fig. S9B).
DYRK1A and PTEN are TBX5 target genes and are upregulated by miR-10b deletion
PTEN is a well-established dose-dependent tumor suppressor in breast cancer (40), and loss of PTEN expression has been found in human mammary tumors (41, 42). TBX5 expression is downregulated in colon cancer, and restored expression of TBX5 in colon tumor cells inhibited proliferation and migration (43), suggesting that TBX5 is a potential tumor suppressor. HOXD10 expression is progressively lost in mammary tumors with increasing degrees of malignancy (44, 45), and re-expression of HOXD10 in breast cancer cells suppressed migration and invasion in vitro as well as tumor progression in vivo (45), presumably by repressing genes involved in cell movement and extracellular matrix remodelling (46). However, the role of the transcription factor TBX5 in breast cancer is unknown.
To identify TBX5-regulated genes in breast cancer, we analyzed eight publicly available transcriptome profiling datasets from human breast cancer specimens and identified five genes (ADCY1, FAM134B, DYRK1A, FGFR2, and PTEN) whose expression positively correlated with TBX5. Among them, the genomic loci of DYRK1A, FGFR2, and PTEN were found to be occupied by TBX5 based on a previous ChIP-Seq study (47) (Supplementary Fig. S9C). To validate whether the expression of these three genes is indeed activated by TBX5, we overexpressed TBX5 in 293FT cells and observed upregulation of DYRK1A and PTEN mRNA levels (Supplementary Fig. S9D).
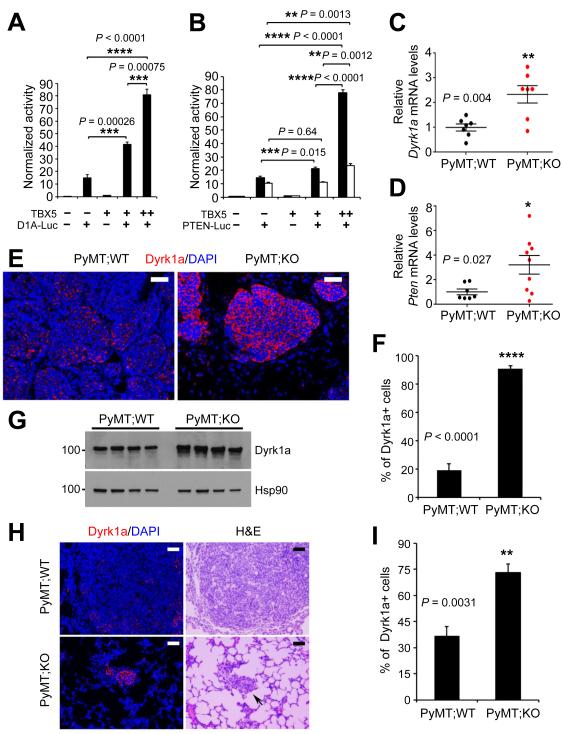
The protein kinase DYRK1A (Dual-Specificity Tyrosine-(Y)-Phosphorylation Regulated Kinase 1A) has recently been shown to promote quiescence and senescence (48), inhibit tumor growth and stemness (49), and increase chemosensitivity (50). To determine whether TBX5 activates the transcription of DYRK1A and PTEN, we performed luciferase reporter assays and found that the activity of the DYRK1A (Fig. 5A and Supplementary Fig. S9E) and PTEN (Fig. 5B and Supplementary Fig. S9E) promoters was increased by TBX5 in a dose-dependent manner. We then examined Dyrk1a and Pten mRNA levels in mouse tumors and observed upregulation of both genes in miR-10b-deficient PyMT mammary tumors (Fig. 5C and 5D). Similar to the increase in Pten protein levels (Fig. 4B; Supplementary Fig. S9A and S9B), MMTV-PyMT;miR-10b−/− mice exhibited upregulation of the percentage of Dyrk1a-positive cells and Dyrk1a protein levels in both primary mammary tumors (Fig. 5E-G) and lung metastases (Fig. 5H and 5I).
Figure 5. DYRK1A and PTEN are TBX5 target genes and are upregulated in mammary tumors and lung metastases of MMTV-PyMT;miR-10b−/− mice.
(A, B) Luciferase reporter assays show that the promoters of DYRK1A (A) and PTEN (B) are activated by TBX5 in a dose-dependent manner. D1A-Luc and PTEN-Luc are reporter constructs containing the human DYRK1A (1,497 bp) and PTEN (black bars, 1,064 bp; white bars, 1,978 bp) promoter regions, respectively.
(C, D) qPCR of Dyrk1a (C) and Pten (D) in mammary tumors of MMTV-PyMT;miR-10b WT and MMTV-PyMT;miR-10b KO mice.
(E, F) Immunofluorescent staining of Dyrk1a (E) and the percentages of Dyrk1a-positive cells (F) in mammary tumors of age-matched MMTV-PyMT;miR-10b WT and MMTV-PyMT;miR-10b KO mice. Scale bars in (E), 50 μm. n = 4 mice per group in (F).
(G) Immunoblotting of Dyrk1a and Hsp90 in mammary tumors of MMTV-PyMT;miR-10b WT and MMTV-PyMT;miR-10b KO mice 24-26 weeks of age. n = 4 mice per group.
(H, I) Immunofluorescent staining of Dyrk1a (H) and the percentages of Dyrk1a-positive cells (I) in lung metastases of age-matched MMTV-PyMT;miR-10b WT and MMTV-PyMT;miR-10b KO mice. Scale bars in (H), 50 μm. n = 4 mice per group in (I).
P values in (A) – (D), (F), and (I) were from a two-tailed, unpaired t-test.
TBX5 is a functional target of miR-10b
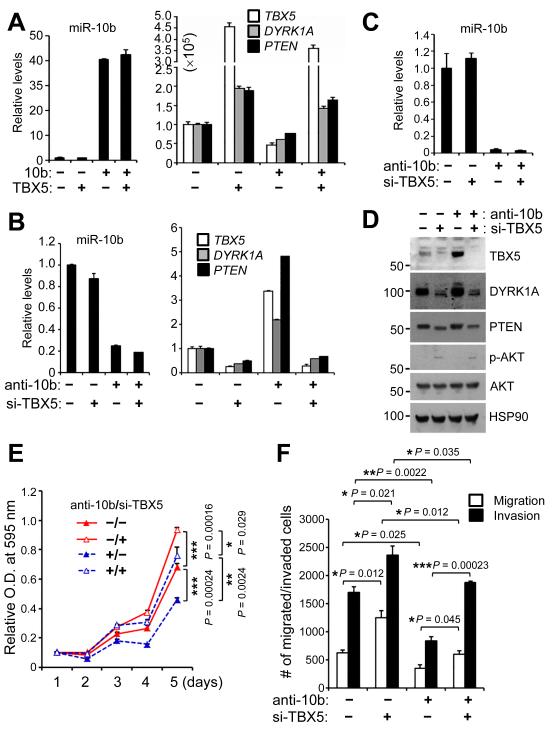
To determine whether miR-10b downregulates DYRK1A and PTEN expression through TBX5, we transfected 293FT cells with miR-10b in the absence or presence of ectopic TBX5 expression. Overexpression of miR-10b decreased mRNA levels of TBX5, DYRK1A, and PTEN, and ectopic expression TBX5 in miR-10b-overexpressing cells restored DYRK1A and PTEN expression (Fig. 6A). Conversely, we transfected 293FT cells with miR-10b antisense inhibitors in the absence or presence of TBX5 siRNA. Consistent with our observations in miR-10b knockout mice, antisense inhibitors of miR-10b upregulated TBX5, DYRK1A, and PTEN expression, which could be reversed by knockdown of TBX5 (Fig. 6B). Similarly, we observed the same effects in the LM2 lung-metastatic subline of the MDA-MB-231 human breast cancer cells (51) (Fig. 6C and 6D). Therefore, gain- and loss-of-function analyses demonstrated that miR-10b suppresses TBX5 expression, leading to downregulation of DYRK1A and PTEN.
Figure 6. TBX5 is a functional target of miR-10b.
(A) qPCR of miR-10b, TBX5, DYRK1A, and PTEN in 293FT cells transfected with miR-10b and TBX5, alone or in combination.
(B) qPCR of miR-10b, TBX5, DYRK1A, and PTEN in 293FT cells transfected with miR-10b antisense inhibitors and TBX5 siRNA, alone or in combination.
(C) qPCR of miR-10b in LM2 cells transfected with miR-10b antisense inhibitors and TBX5 siRNA, alone or in combination.
(D) Immunoblotting of TBX5, DYRK1A, PTEN, phospho-AKT, AKT, and HSP90 in LM2 cells transfected with miR-10b antisense inhibitors and TBX5 siRNA, alone or in combination.
(E, F) Growth curves (E) and migration and invasion assays (F) of LM2 cells transfected with miR-10b antisense inhibitors and TBX5 siRNA, alone or in combination. n = 4 and 3 wells per group in (E) and (F), respectively. P values were from a two-tailed, unpaired t-test.
We asked whether TBX5 mediates the functions of miR-10b in breast cancer cells. Consistent with the phenotypes of MMTV-PyMT;miR-10b−/− mice, transfecting the LM2 metastatic breast cancer cells with miR-10b antisense inhibitors suppressed cell proliferation, migration, and invasion, which could be reversed by TBX5 siRNA (Fig. 6E and 6F). Moreover, overexpression of TBX5 markedly inhibited the proliferation, migration, and invasion of LM2 cells (Supplementary Fig. S10A-C). Collectively, these results suggest that TBX5 is a functional target of miR-10b.
TBX5, HOX10, and DYRK1A are downregulated in human breast cancer and correlate with clinical outcomes
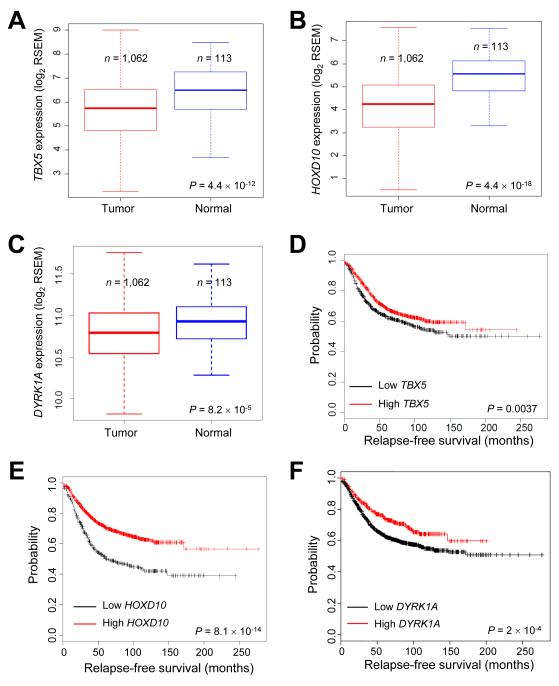
Using miR-10b knockout mice and human cell lines, we identified Tbx5, Hoxd10, and Pten as the targets of miR-10b, and DYRK1A and PTEN as the target genes of TBX5. Compared with PTEN, the relevance of TBX5, HOXD10, and DYRK1A in human breast cancer is less defined. We analyzed the breast cancer data from The Cancer Genome Atlas (TCGA) (52). Although genomic alterations of TBX5, HOXD10, and DYRK1A genes in human breast cancer were rare (TBX5: 0.83%; HOXD10: 0.93%; DYRK1A: 2.0%), their mRNA levels were significantly downregulated in breast tumors relative to normal mammary tissues (Fig. 7A-C). We then performed Kaplan-Meier (KM) plotter (28) analyses. Compared with patients with high TBX5 expression in their breast tumors, patients with low TBX5 expression had worse relapse-free survival (Fig. 7D) and overall survival (Supplementary Fig. S11A). Similarly, the expression of HOXD10 and DYRK1A also correlated with relapse-free survival (Fig. 7E and 7F) and overall survival (Supplementary Fig. S11B and S11C) of breast cancer patients. These data suggest that miR-10b-regulated genes are human breast cancer-relevant tumor suppressors or metastasis suppressors (Supplementary Fig. S11D).
Figure 7. TBX5, HOXD10, and DYRK1A are downregulated in human breast cancer and are associated with relapse-free survival.
(A-C) Box plots comparing TBX5 (A), HOXD10 (B), or DYRK1A (C) expression in normal breast tissues and breast tumors, based on the RNA-Seq data from TCGA. The boxes show the median and the interquartile range. The whiskers show the minimum and maximum. P values were from a two-tailed, unpaired t-test.
(D-F) Kaplan-Meier curves of relapse-free survival of breast cancer patients stratified by TBX5 (D), HOXD10 (E), or DYRK1A (F) expression levels. n = 1,660 patients. P values were from a log-rank test.
Discussion
In mammals, the miR-10 family consists of miR-10a and miR-10b. Their coding genes are located within two different HOX gene clusters whose protein products play important roles in development. Moreover, miR-10a and miR-10b target specific HOX mRNAs, leading to the speculation that these two miRNAs might be developmental regulators (53, 54). Surprisingly, however, extensive analyses did not reveal substantial phenotypic differences in miR-10b-deficient mice except germinal center expansion in the spleen. Similarly, miR-10a-null mice were indistinguishable from littermate controls in terms of development, growth, overall survival, and incidence of spontaneous tumorigenesis (55).
Whereas the functional redundancy of miR-10a and miR-10b remains to be determined, a number of previous knockout studies demonstrated that many conserved miRNAs are dispensable for animal development and viability; however, some of these miRNAs play critical roles in stress responses or pathologic processes, including cardiac stress, vascular injuries, intestinal injuries, and oncogenic stress, and thus mice deficient in these miRNAs exhibit notable phenotypes in response to external or internal perturbations (56). In the present study, genetic deletion of miR-10b in a PyMT-driven mouse model of breast cancer significantly impeded mammary tumor initiation, growth, progression, and lung metastasis. The observed delay in oncogene-induced mammary tumor formation has likely been masked in previous xenograft models in which highly aggressive and late-stage tumor cells were used (5); this underscores the importance of using autochthonous tumor models to track the early changes during tumor initiation. Moreover, despite a dramatic reduction in the number and area of metastatic foci in the lung, metastasis was not completely abrogated by miR-10b deletion, suggesting that other metastasis genes and pathways are operational in this model. In addition, despite no compensatory increase of miR-10a expression, miR-10a might partially compensate for the loss of miR-10b at the functional level.
Among the target genes of miR-10b, the tumor suppressor genes Tbx5 and Pten and the metastasis suppressor gene Hoxd10 are significantly upregulated by miR-10b deletion in mice, suggesting that these genes are physiologically relevant bona fide miR-10b targets. In addition, we found that PTEN and another tumor suppressor, DYRK1A, are target genes of the transcription factor TBX5, and that both genes are downregulated by miR-10b in a TBX5-dependent manner. Furthermore, TBX5, HOXD10, and DYRK1A are underexpressed in human breast tumors and are associated with overall survival and relapse-free survival outcomes. Taken together with the well-established tumor-suppressing role of PTEN in breast cancer, these data suggest that miR-10b may downregulate multiple tumor suppressors or metastasis suppressors, ultimately contributing to malignant progression. Inhibition of miR-10b may provide therapeutic opportunities by reactivating downstream tumor-suppressing and metastasis-suppressing pathways.
Supplementary Material
Acknowledgments
We thank Department of Veterinary Medicine & Surgery and the Histology Core Laboratory at MD Anderson Cancer Center for technical assistance, and Drs. Walter Becker, Jonathan Kurie, and Xiang Zhang for providing reagents.
Grant Support
L.M. is supported by NIH grants R01CA166051 and R01CA181029, a CPRIT grant RP150319, a Stand Up To Cancer Innovative Research Grant, and a Clark Fellows Award. M.J.Y. is supported in part by NIH R01CA164346, CPRIT RP140402, and Center for Genetics and Genomics, Center for Inflammation and Cancer, Institutional Research Grant, and Sister Institution Network fund of MD Anderson Cancer Center.
Footnotes
Disclosure of Potential Conflicts of Interest
X.Y. is an employee of AM Biotechnologies LLC. The remaining authors declare no competing financial interests.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Ma L, Weinberg RA. Micromanagers of malignancy: role of microRNAs in regulating metastasis. Trends Genet. 2008;24:448–56. doi: 10.1016/j.tig.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Piao HL, Ma L. Non-coding RNAs as regulators of mammary development and breast cancer. J Mammary Gland Biol Neoplasia. 2012;17:33–42. doi: 10.1007/s10911-012-9245-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J, Ma L. MicroRNA control of epithelial-mesenchymal transition and metastasis. Cancer Metastasis Rev. 2012;31:653–62. doi: 10.1007/s10555-012-9368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–8. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 6.Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nature cell biology. 2010;12:247–56. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edmonds MD, Hurst DR, Vaidya KS, Stafford LJ, Chen D, Welch DR. Breast cancer metastasis suppressor 1 coordinately regulates metastasis-associated microRNA expression. International journal of cancer Journal international du cancer. 2009;125:1778–85. doi: 10.1002/ijc.24616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. Jama. 2007;297:1901–8. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 9.Preis M, Gardner TB, Gordon SR, Pipas JM, Mackenzie TA, Klein EE, et al. MicroRNA-10b expression correlates with response to neoadjuvant therapy and survival in pancreatic ductal adenocarcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:5812–21. doi: 10.1158/1078-0432.CCR-11-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakata K, Ohuchida K, Mizumoto K, Kayashima T, Ikenaga N, Sakai H, et al. MicroRNA-10b is overexpressed in pancreatic cancer, promotes its invasiveness, and correlates with a poor prognosis. Surgery. 2011;150:916–22. doi: 10.1016/j.surg.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 11.Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochemical and biophysical research communications. 2005;334:1351–8. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 12.Huse JT, Brennan C, Hambardzumyan D, Wee B, Pena J, Rouhanifard SH, et al. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes & development. 2009;23:1327–37. doi: 10.1101/gad.1777409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sasayama T, Nishihara M, Kondoh T, Hosoda K, Kohmura E. MicroRNA-10b is overexpressed in malignant glioma and associated with tumor invasive factors, uPAR and RhoC. International journal of cancer Journal international du cancer. 2009;125:1407–13. doi: 10.1002/ijc.24522. [DOI] [PubMed] [Google Scholar]
- 14.Gabriely G, Yi M, Narayan RS, Niers JM, Wurdinger T, Imitola J, et al. Human glioma growth is controlled by microRNA-10b. Cancer research. 2011;71:3563–72. doi: 10.1158/0008-5472.CAN-10-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao H, Li H, Yu G, Xiao W, Hu J, Tang K, et al. MicroRNA-10b promotes migration and invasion through KLF4 and HOXD10 in human bladder cancer. Oncology reports. 2014;31:1832–8. doi: 10.3892/or.2014.3048. [DOI] [PubMed] [Google Scholar]
- 16.Liao CG, Kong LM, Zhou P, Yang XL, Huang JG, Zhang HL, et al. miR-10b is overexpressed in hepatocellular carcinoma and promotes cell proliferation, migration and invasion through RhoC, uPAR and MMPs. Journal of translational medicine. 2014;12:234. doi: 10.1186/s12967-014-0234-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baffa R, Fassan M, Volinia S, O'Hara B, Liu CG, Palazzo JP, et al. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. The Journal of pathology. 2009;219:214–21. doi: 10.1002/path.2586. [DOI] [PubMed] [Google Scholar]
- 18.Chai G, Liu N, Ma J, Li H, Oblinger JL, Prahalad AK, et al. MicroRNA-10b regulates tumorigenesis in neurofibromatosis type 1. Cancer science. 2010;101:1997–2004. doi: 10.1111/j.1349-7006.2010.01616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tian Y, Luo A, Cai Y, Su Q, Ding F, Chen H, et al. MicroRNA-10b promotes migration and invasion through KLF4 in human esophageal cancer cell lines. The Journal of biological chemistry. 2010;285:7986–94. doi: 10.1074/jbc.M109.062877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li G, Wu Z, Peng Y, Liu X, Lu J, Wang L, et al. MicroRNA-10b induced by Epstein-Barr virus-encoded latent membrane protein-1 promotes the metastasis of human nasopharyngeal carcinoma cells. Cancer letters. 2010;299:29–36. doi: 10.1016/j.canlet.2010.07.021. [DOI] [PubMed] [Google Scholar]
- 21.Liu S, Sun J, Lan Q. TGF-beta-induced miR10a/b expression promotes human glioma cell migration by targeting PTEN. Molecular medicine reports. 2013;8:1741–6. doi: 10.3892/mmr.2013.1709. [DOI] [PubMed] [Google Scholar]
- 22.Ma L, Reinhardt F, Pan E, Soutschek J, Bhat B, Marcusson EG, et al. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol. 2010;28:341–7. doi: 10.1038/nbt.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoo B, Kavishwar A, Ross A, Wang P, Tabassum DP, Polyak K, et al. Combining miR-10b-Targeted Nanotherapy with Low-Dose Doxorubicin Elicits Durable Regressions of Metastatic Breast Cancer. Cancer research. 2015;75:4407–15. doi: 10.1158/0008-5472.CAN-15-0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh R, Pochampally R, Watabe K, Lu Z, Mo YY. Exosome-mediated transfer of miR-10b promotes cell invasion in breast cancer. Molecular cancer. 2014;13:256. doi: 10.1186/1476-4598-13-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Virolle T, Adamson ED, Baron V, Birle D, Mercola D, Mustelin T, et al. The Egr-1 transcription factor directly activates PTEN during irradiation-induced signalling. Nature cell biology. 2001;3:1124–8. doi: 10.1038/ncb1201-1124. [DOI] [PubMed] [Google Scholar]
- 26.Sheng X, Koul D, Liu JL, Liu TJ, Yung WK. Promoter analysis of tumor suppressor gene PTEN: identification of minimum promoter region. Biochemical and biophysical research communications. 2002;292:422–6. doi: 10.1006/bbrc.2002.6662. [DOI] [PubMed] [Google Scholar]
- 27.Maenz B, Hekerman P, Vela EM, Galceran J, Becker W. Characterization of the human DYRK1A promoter and its regulation by the transcription factor E2F1. BMC molecular biology. 2008;9:30. doi: 10.1186/1471-2199-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast cancer research and treatment. 2010;123:725–31. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 29.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Molecular and cellular biology. 1992;12:954–61. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davie SA, Maglione JE, Manner CK, Young D, Cardiff RD, MacLeod CL, et al. Effects of FVB/NJ and C57Bl/6J strain backgrounds on mammary tumor phenotype in inducible nitric oxide synthase deficient mice. Transgenic research. 2007;16:193–201. doi: 10.1007/s11248-006-9056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin EY, Jones JG, Li P, Zhu L, Whitney KD, Muller WJ, et al. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. The American journal of pathology. 2003;163:2113–26. doi: 10.1016/S0002-9440(10)63568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trimboli AJ, Fukino K, de Bruin A, Wei G, Shen L, Tanner SM, et al. Direct evidence for epithelial-mesenchymal transitions in breast cancer. Cancer research. 2008;68:937–45. doi: 10.1158/0008-5472.CAN-07-2148. [DOI] [PubMed] [Google Scholar]
- 33.Ye X, Tam WL, Shibue T, Kaygusuz Y, Reinhardt F, Ng Eaton E, et al. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature. 2015;525:256–60. doi: 10.1038/nature14897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Z, Zhu J, Cao H, Ren H, Fang X. miR-10b promotes cell invasion through RhoC-AKT signaling pathway by targeting HOXD10 in gastric cancer. International journal of oncology. 2012;40:1553–60. doi: 10.3892/ijo.2012.1342. [DOI] [PubMed] [Google Scholar]
- 35.Nakayama I, Shibazaki M, Yashima-Abo A, Miura F, Sugiyama T, Masuda T, et al. Loss of HOXD10 expression induced by upregulation of miR-10b accelerates the migration and invasion activities of ovarian cancer cells. International journal of oncology. 2013;43:63–71. doi: 10.3892/ijo.2013.1935. [DOI] [PubMed] [Google Scholar]
- 36.Sun L, Yan W, Wang Y, Sun G, Luo H, Zhang J, et al. MicroRNA-10b induces glioma cell invasion by modulating MMP-14 and uPAR expression via HOXD10. Brain research. 2011;1389:9–18. doi: 10.1016/j.brainres.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 37.Yu X, Li Z, Shen J, Wu WK, Liang J, Weng X, et al. MicroRNA-10b promotes nucleus pulposus cell proliferation through RhoC-Akt pathway by targeting HOXD10 in intervetebral disc degeneration. PloS one. 2013;8:e83080. doi: 10.1371/journal.pone.0083080. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Wang F, Yang XY, Zhao JY, Yu LW, Zhang P, Duan WY, et al. miR-10a and miR-10b target the 3'-untranslated region of TBX5 to repress its expression. Pediatric cardiology. 2014;35:1072–9. doi: 10.1007/s00246-014-0901-y. [DOI] [PubMed] [Google Scholar]
- 39.Mussnich P, D'Angelo D, Leone V, Croce CM, Fusco A. The High Mobility Group A proteins contribute to thyroid cell transformation by regulating miR-603 and miR-10b expression. Molecular oncology. 2013;7:531–42. doi: 10.1016/j.molonc.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alimonti A, Carracedo A, Clohessy JG, Trotman LC, Nardella C, Egia A, et al. Subtle variations in Pten dose determine cancer susceptibility. Nat Genet. 2010;42:454–8. doi: 10.1038/ng.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perez-Tenorio G, Alkhori L, Olsson B, Waltersson MA, Nordenskjold B, Rutqvist LE, et al. PIK3CA mutations and PTEN loss correlate with similar prognostic factors and are not mutually exclusive in breast cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007;13:3577–84. doi: 10.1158/1078-0432.CCR-06-1609. [DOI] [PubMed] [Google Scholar]
- 42.Zhang J, Zhang P, Wei Y, Piao HL, Wang W, Maddika S, et al. Deubiquitylation and stabilization of PTEN by USP13. Nature cell biology. 2013;15:1486–94. doi: 10.1038/ncb2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu J, Ma X, Cheung KF, Li X, Tian L, Wang S, et al. Epigenetic inactivation of T-box transcription factor 5, a novel tumor suppressor gene, is associated with colon cancer. Oncogene. 2010;29:6464–74. doi: 10.1038/onc.2010.370. [DOI] [PubMed] [Google Scholar]
- 44.Makiyama K, Hamada J, Takada M, Murakawa K, Takahashi Y, Tada M, et al. Aberrant expression of HOX genes in human invasive breast carcinoma. Oncology reports. 2005;13:673–9. [PubMed] [Google Scholar]
- 45.Carrio M, Arderiu G, Myers C, Boudreau NJ. Homeobox D10 induces phenotypic reversion of breast tumor cells in a three-dimensional culture model. Cancer research. 2005;65:7177–85. doi: 10.1158/0008-5472.CAN-04-1717. [DOI] [PubMed] [Google Scholar]
- 46.Myers C, Charboneau A, Cheung I, Hanks D, Boudreau N. Sustained expression of homeobox D10 inhibits angiogenesis. The American journal of pathology. 2002;161:2099–109. doi: 10.1016/S0002-9440(10)64488-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He A, Kong SW, Ma Q, Pu WT. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:5632–7. doi: 10.1073/pnas.1016959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Litovchick L, Florens LA, Swanson SK, Washburn MP, DeCaprio JA. DYRK1A protein kinase promotes quiescence and senescence through DREAM complex assembly. Genes Dev. 2011;25:801–13. doi: 10.1101/gad.2034211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee SB, Frattini V, Bansal M, Castano AM, Sherman D, Hutchinson K, et al. An ID2-dependent mechanism for VHL inactivation in cancer. Nature. 2016;529:172–7. doi: 10.1038/nature16475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Q, Liu N, Zang S, Liu H, Wang P, Ji C, et al. Tumor suppressor DYRK1A effects on proliferation and chemoresistance of AML cells by downregulating c-Myc. PLoS One. 2014;9:e98853. doi: 10.1371/journal.pone.0098853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–24. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cancer Genome Atlas N Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tehler D, Hoyland-Kroghsbo NM, Lund AH. The miR-10 microRNA precursor family. RNA biology. 2011;8:728–34. doi: 10.4161/rna.8.5.16324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lund AH. miR-10 in development and cancer. Cell death and differentiation. 2010;17:209–14. doi: 10.1038/cdd.2009.58. [DOI] [PubMed] [Google Scholar]
- 55.Stadthagen G, Tehler D, Hoyland-Kroghsbo NM, Wen J, Krogh A, Jensen KT, et al. Loss of miR-10a activates lpo and collaborates with activated Wnt signaling in inducing intestinal neoplasia in female mice. PLoS genetics. 2013;9:e1003913. doi: 10.1371/journal.pgen.1003913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vidigal JA, Ventura A. The biological functions of miRNAs: lessons from in vivo studies. Trends in cell biology. 2015;25:137–47. doi: 10.1016/j.tcb.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.