Abstract
Biomass is abundant, renewable and useful for biofuel production as well as chemical priming for plastics and composites. Deconstruction of biomass by enzymes is perceived as recalcitrant while an inclusive breakdown mechanism remains to be discovered. Fungi such as Myceliophthora thermophila M77 appear to decompose natural biomass sources quite well. This work reports on this fungus fermentation property while producing cellulolytic enzymes using natural biomass substrates. Little hydrolytic activity was detected, insufficient to explain the large amount of biomass depleted in the process. Furthermore, this work makes a comprehensive account of extracellular proteins and describes how secretomes redirect their qualitative protein content based on the nature and chemistry of the nutritional source. Fungus grown on purified cellulose or on natural biomass produced secretomes constituted by: cellobiohydrolases, cellobiose dehydrogenase, β-1,3 glucanase, β-glucosidases, aldose epimerase, glyoxal oxidase, GH74 xyloglucanase, galactosidase, aldolactonase and polysaccharide monooxygenases. Fungus grown on a mixture of purified hemicellulose fractions (xylans, arabinans and arabinoxylans) produced many enzymes, some of which are listed here: xylosidase, mixed β-1,3(4) glucanase, β-1,3 glucanases, β-glucosidases, β-mannosidase, β-glucosidases, galactosidase, chitinases, polysaccharide lyase, endo β-1,6 galactanase and aldose epimerase. Secretomes produced on natural biomass displayed a comprehensive set of enzymes involved in hydrolysis and oxidation of cellulose, hemicellulose-pectin and lignin. The participation of oxidation reactions coupled to lignin decomposition in the breakdown of natural biomass may explain the discrepancy observed for cellulose decomposition in relation to natural biomass fermentation experiments.
Electronic supplementary material
The online version of this article (doi:10.1186/s13568-016-0276-y) contains supplementary material, which is available to authorized users.
Keywords: Myceliophthora thermophila, Biomass, Cellulose degradation, Secretome composition, Cellulose hydrolysis, Cellulose oxidation
Introduction
Lignocellulosic biomass polymers are a massive and renewable source for production of biofuels and biochemicals, because they trap about 60% of all sugars produced by plants on earth. Just as it happens in nature, man-made lignocellulosic biomass such as corn stover and sugar cane bagasse that pile up alongside bio refineries and could be broken down enzymatically (Amorim et al. 2011; Lal 2005). However, currently the cost of cellulase enzyme cocktails are the bottleneck to the economical production of these second generation biofuels (Phillips et al. 2011) mainly because enzymatic conversion of lignocellulose into sugars is a slow and recalcitrant process and cellulose is an insoluble crystalline substance (Himmel and Bayer 2009) clustered within phenolic lignin (benzene ether linkages) hindering its ability to be enzymatically processed (Lacayo et al. 2013).
For cellulase aided breakdown of cellulose to take place, a single chain must be separated from the crystalline fiber and fitted into an enzyme binding site where catalytic Asp or Glu residues hydrolyze through a general acid/base mechanism the glycoside bond (Divne et al. 1994). The disconnection of the glucan chain from crystalline cellulose fibers has been proposed to be the bottleneck in enzymatic hydrolysis of cellulose (Himmel and Bayer 2009).
This recalcitrance towards the degradation of cellulose is abundantly illustrated in the repertoire of cellulose degrading enzymes produced by microorganisms that try to use this polymer as a carbon source (Segato et al. 2014). Most microorganisms produce at least three types of glycosidic bond breaking enzymes; cellobiohydrolases (also defined as exo glucanases and/or processive glucanases), endo glucanases and β-glucosidases. For comprehensive reviews of hydrolytic biomass breakdown refer to (Benz et al. 2013; Coutinho et al. 2009; Glass et al. 2013; Martens-Uzunova and Schaap 2009; Segato et al. 2014).
Recently oxidoreductase enzymes such as polysaccharide monooxygenases (PMO’s) that directly oxidize glycoside bonds generating aldones and lactones have been discovered highlighting the role of oxidation reactions in the breakdown of biomass components (Beeson et al. 2012; Horn et al. 2012; Langston et al. 2011; Quinlan et al. 2011; Vaaje-Kolstad et al. 2010).
A direct role of cellobiose dehydrogenase on cellulose depolymerization via the oxidation of glycoside bonds aided by Fenton chemistry has been suggested (Canevascini et al. 1991; Divne et al. 1994; Henriksson et al. 2000a; Mansfield et al. 1997; Mason et al. 2003; Stahlberg et al. 1996; Westermark and Eriksson 1975; Zamocky et al. 2006). Moreover, the participation of cellobiose dehydrogenase in oxidation of other biomass components such as lignin has also been considered (Henriksson et al. 2000b; Hilden et al. 2000).
Here we report on the efficiency of biomass bioconversion by Myceliophthora thermophila M77, whereas in a traditional bioreactor, the fungus completely consumes biomass sources (sugar cane bagasse) but shows little cellulase filter paper activity, leading the research to determine global secretome composition of M. thermophila growing on biomass and purified biomass components (cellulose and hemicellulose). When purified cellulose was available, the fungus produced a secretome that included hydrolytic and oxidative enzymes, almost exclusively dedicated to the breakdown of cellulose and cellulose related molecules. When natural biomass was available, the fungus produced a comprehensive collection of enzymes in addition to cellobiose dehydrogenase involved in oxidation and hydrolysis of cellulose, hemicellulose-pectin and lignin.
Materials and methods
Strains, media, solutions and biomass sources
The strain used in this work M. thermophila M77 was isolated from a sugar cane bagasse pile of the northwest region of São Paulo State, Brazil and was deposited at the Fungal Genetics Stock Center FGSC# 26436 (Moretti et al. 2012). A similar M. thermophila strain ATCC 42464 was recently sequenced by the DOE Joint Genome Institute Fungal Genomics Program (Berka et al. 2011; Kolbusz et al. 2014) and was used for DNA sequence based interpretation of LC–MS/MS data.
Myceliophthora thermophila M77 was grown on 1.8% agar petri dishes in Mandels and Sternberg salts (Mandels and Sternberg 1976) amended with 1.0% glucose and 0.1% peptone incubated at 45 °C for 7 days, or as otherwise stated. Spores were scraped off the plates with a platinum loop, suspended in 0.1% Tween 80 (Sigma-Aldrich, St Louis, MO, USA) and used to pre-inoculate (about 1 × 107 spores/mL) shaker flasks incubated at 45 °C, 250 rpm for 72 h prior to direct transfer to a bioreactor vessel or a large-scale shaker flask experiment.
In experiments using biomass substrates and derivatives, the glucose was replaced with 1.0% (w/v) of commercial microcrystalline cellulose (EC) (Celuflok 200™, Celuflok Ind. Com. São Paulo, Brazil), “in natura” milled (200-μm particle size) sugar cane bagasse (SCBIN), lignin removed (sodium hydroxide extracted) and steam exploded sugar cane bagasse (SCBDL), steam exploded sugar cane bagasse only (SCBSE), wheat bran (WB), milled soybeans (SM), and fructooligosaccharides (FOS). Sugar cane bagasse sources were prepared and chemically defined as described in (Rocha et al. 2011) and milled powders washed with water and autoclaved prior to use.
Shaken flask experiments
Twenty millilitre of pre-inoculum was added to 1 L Erlenmeyer flasks containing 200 mL of Mandels and Sternberg salts, 0.1% peptone amended with 1% (w/v) SCBSE, SCBDL, WB, EC, SM, glycerol (GLY), lactose (LAC), sucrose (SUC) and FOS alone or in combinations and proportions as indicated. Incubations were made in an orbital shaker (Innova 44R Stackable Incubator Shaker, New Brunswick, NJ, USA) for up to 120 h at 45 °C and 250 rpm and samples withdrawn daily for enzyme activity and protein quantifications.
Bioreactor experiments
Bioreactor assays were performed in a lab-scale Bioflo®115 (New Brunswick, NJ, USA) with a working volume of 1.5 L, using SCBSE, WB and sucrose as carbon sources. The pre-inoculum was 10% of the final volume. Cultivations were conducted in batch or pulse-fed batch mode (as indicated in Figures and Tables), the dissolved O2 concentration was >30 % of air saturation and mechanical stirring was performed with two Rushton-type impellers, in the range of 200–400 rpm. Prior to use all equipment was sterilized for 30 min at 121 °C. Automatic pH control was done using a 0.4 M HCl and NH4OH aqueous solution 3:1 (v/v) and foaming was controlled as required by manual addition of sterile antifoam polypropylene glycol 2000 (Dow Chemical, São Paulo, Brazil). Samples were withdrawn under sterile conditions daily, centrifuged at 12,000 rpm for 25 min at 4 °C and supernatants collected for cellulase (FPase), xylanase, β-glucosidase activity and total protein quantification.
Enzymatic activity assays
Cellulase activity was determined by the method of Ghose (1987) that measures the release of detectable reducing sugars removed from filter paper (FPase). Xylanase activity was determined by the method described by Bailey and Poutanen (1989). Both FPase and xylanase activities were performed measuring reducing sugars by the dinitrosalicylic acid (DNS) method (Miller 1959), using glucose and xylose standards as appropriate. β-glucosidase and cellobiohydrolase was measured using p-nitrophenol-β-d-glucopyranoside (pNPG) and p-nitrophenol-β-d-cellobioside (pNPC) (Sigma-Aldrich, USA) as substrate, respectively (Zhang et al. 2009). Total protein content was measured in micro plates using the Bio-Rad assay reagent (Bio-Rad Laboratories, Hercules, USA), using a procedure based on the Bradford method (Bradford 1976) with bovine serum albumin as standard. One enzyme unit (IU) corresponded to the amount of product (μmol) produced per minute and cellulase activity was expressed as filter paper units (FPU) calculated according to (Ghose 1987). Cellobiose dehydrogenase activity was assayed through 2,6-dichlorophenol-indophenol (DCPIP) reduction. The decrease in absorbance was measured continuously at 520 nm (ε = 6.8 × 103 M−1 cm−1) in sodium acetate buffer (50 mM; pH 5) containing DCPIP 0.3 mM, sodium lactate 30 mM and NaF 4 mM. One enzyme unit (IU) corresponded to the amount of enzyme reducing 1 μmol of DCPIP per minute (Baminger et al. 1999). Laccase activity was measured continuously by the oxidation rate of ABTS2+ to ABTS●+ at 420 nm (ε = 3.6 × 104 M−1 cm−1) in acetate buffer (50 mM; pH 3.5) containing ABTS (5 mM) in a final volume of 2 mL at 25 °C. One enzyme unit (IU) corresponded to the amount of enzyme that oxidized 1 µmol of ABTS per minute (Bourbonnais et al. 1995).
Enzymatic biomass hydrolysis
Bioconversion assays were conducted in 50 mL (125 mL Erlenmeyer flasks) final volumes, buffered with citrate 50 mM pH 5, 5% (w/v) of substrate (EC, SCBIN, SCBDL or SCBSE) and a protein load of 0.05 mg/g of glucan, incubated at 50 °C at 200 rpm. All experiments were performed in duplicates. Samples were withdrawn at 0, 6, 12, 24 and 48 h and the glucose, gluconic acid, cellobiose, cellobionic acid, xylose, arabinose, acetic, formic and levulinic acid concentrations were quantified by HPLC Dionex Ultimate 300 system equipped with a refractive index detector (HPLC-RI) using an Aminex ®HPX-87H column and eluted with 5 mM H2SO4 at 0.6 mL/min. Sugars and acids in control samples containing only the respective substrate and citrate buffer 50 mM pH 5.0 were also measured. All samples were filtered using a Millex TM 0.22 μm filter prior to further analysis.
Production of secretomes
Myceliophthora thermophila M77 was grown in Erlenmeyer flasks on Mandels & Sternberg salts, 0.1% peptone containing SCBIN (natural sugar cane bagasse, milled at 200 μm particle size) as well as modified sugar cane bagasse versions such as SCBDL (delignified with sodium hydroxide), SCBSE (steam exploded), purified celluloses containing 0.5% of avicel and 0.5% carboxymethylcellulose (Sigma Aldrich, St Louis MO), purified hemicelluloses containing 0.2% of each; birchwood-, beechwood-, oat spelt-xylan, arabinan and arabinoxylan (Megazyme International, Wicklow, Ireland) and glucose (control).
Secreted proteins were collected after a 36 h cultivation period at 45 °C, 200 rpm supernatants cleared by centrifugation (5000×g), concentrated by ultra-filtration (10,000 MWCO, PES membrane, Vivaspin, Littleton USA), rinsed twice with 5 mL of sodium acetate buffer 50 mM pH 5 and the proteins were separated by SDS-PAGE (Weber and Osborn 1969).
Secretome peptide mapping by liquid chromatography-tandem mass spectrometry (LC–MS/MS)
For secretome peptide mapping experiments two independent cultures and two protein separations through SDS-PAGE were carried out. For secretome LC–MS/MS analysis 20–30 μg of total secretome proteins were loaded onto an SDS-PAGE gel and while in Fig. 3 we show a fully resolved SDS-PAGE gel for proteomics experiments, for proteomics the SDS-PAGE was run for only about one inch into the 12% separation gel, stained with Comassie blue and the entire protein banding profile excised, processed for LC–MS/MS according to (Shevchenko et al. 1996) with modifications. Isolated gel bands were reduced with Tris (2-carboxyethyl) phosphine, alkylated by 2-Iodoacetamide, digested for 6–16 h with 8 μg/mL trypsin using ammonium bicarbonate buffer and analyzed by LC–MS/MS using LTQ-Orbitrap XL hybrid mass spectrometer (Thermo Scientific, Waltham, MA, USA). For this analysis, an Eksigent LC pump was used to separate peptide populations on analytical C18 nano-columns, with the column effluent being sprayed directly into a New Objective Picoview ion source. Using a “Top Three” MS/MS method, the Orbitrap analyzer collected accurate (5 ppm) scans of intact peptides for one second, at the same time as the LTQ ion trap simultaneously performed MS/MS fragmentation analysis of each of the three most abundant peptides eluting in that 1 s chromatographic fraction (0.8 Da mass accuracy).
Fig. 3.
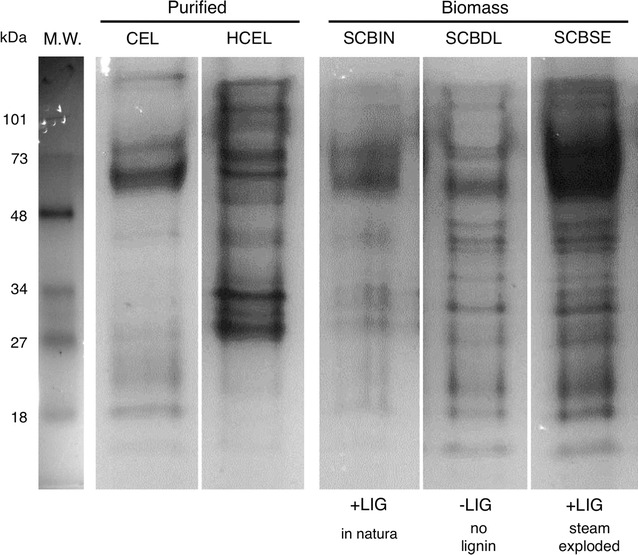
Myceliophthora thermophila M77 secretome composition. M. thermophila M77 secretomes developed on a variety of carbon sources; sugar cane bagasse (SCB) in natura (SCBIN), delignified (SCBDL) and steam-exploded (SCBSE) as well as purified cellulose (CEL) and hemicellulose (HCEL) were determined through LC–MS/MS from SDS-PAGE separated proteins. 172 proteins were identified with positive matching of 21,766 unique peptides (4019 SCBIN, 3661 SCBDL, 4269 SCBSE 3466 celluloses and 4716 hemicelluloses). CEL, purified cellulose mixture containing 0.5% of avicel and 0.5% carboxymethylcellulose; HCEL, purified hemicellulose mixture containing 0.2% of each; birchwood-, beechwood-, oat spelt-xylan, arabinan and arabinoxylan; SCBIN, milled “in natura” sugar cane bagasse; SCBDL, steam exploded and delignified with sodium hydroxide milled sugar cane bagasse and SCBSE, steam exploded milled sugar cane bagasse. +LIG, lignin present; -LIG, lignin absent; M.W., molecular weight markers shown in kDa
The LC–MS/MS raw files were used for database Mascot (version 2.2.04, Matrix Science, London UK) searches run on a NCBI M. thermophila ATCC_42464 specific subset. The DNA and amino acid sequence of M. thermophila M77 are 98.95 and 99.45% identical to M. thermophila ATCC_42464, respectively. Searches were validated using Scaffold (version 4.0.7, Proteome Software Inc. Portland, OR) with a protein threshold of 5% FDR and a peptide threshold of 99%. Further management of spectral data were performed on downloaded Excel files, total spectral counts (TSC) were normalized (against the total spectral count of each sample) and finally duplicates averaged (Additional file 1: Table S1). Thus, the quantitative value NTSC (normalized total spectrum counts) for a given protein component of a secretome reflects the amount of protein secreted as a direct response to the applied carbon source.
Results
Bioreactors and shakers producing biomass-degrading enzymes
Figure 1 reports a typical bioreactor experiment in which the carbon source was SCBSE (steam-exploded sugar cane bagasse). Extracellular protein and expected enzyme activities, cellulase (measured as activity on filter paper (FPU), xylanase and β-glucosidase accumulated in the medium over time reaching a peak at or around 96 h.
Fig. 1.
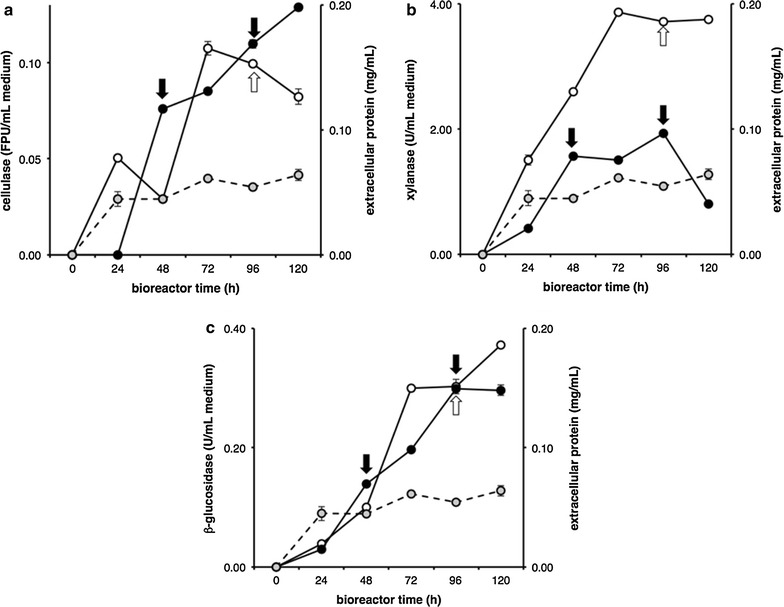
Myceliophthora thermophila M77 fed-batch bioreactor with steam exploded sugar cane bagasse (SCBSE) as the carbon source. Bioreactors containing Mandels and Sternberg salts amended with 0.1% peptone and 1% steam exploded sugar cane bagasse (SCBSE) were conducted for 120 h at 45 °C, constant pH 5.0 and feed-pulsed with SCBSE at the indicated time points (arrows). Extracellular protein accumulation (shaded symbols), cellulase (a), xylanase (b) and β-glucosidase (c) activities were followed in a single feed-pulse (open symbols) and a double feed-pulse (closed symbols) regimen
Table 1 shows a series of shaking flask experiments modifying the forms of biomass and combinations with simple carbon sources such as glycerol, lactose, sucrose and glucose designed to improve enzyme production. The highest cellulase activity, as judged by the DNS assay, was observed with steam-exploded biomass (0.23 FPU/mL) while lignin extracted biomass showed lower cellulase activity (0.10 FPU/mL) and other sources such as wheat bran and purified cellulose as well as combinations thereof did not improve cellulase activity (Table 1). Thus, none of the biomass variants produced significant improvement over cellulase activity (FPU). For xylanase activity a similar picture occurs, none of the biomass derivatives improve drastically xylanase activity, however the addition of a non-repressive carbon source such as lactose (LAC), phospho-fructo-oligosacharides (FOS) or sucrose (SUC) resulted in a slight increase in xylanase activity (Table 1).
Table 1.
Myceliophthora thermophila enzyme accumulation in shaking flask bioreactors
| Cellulase | Xylanase | β-glucosidase | Protein | |||||
|---|---|---|---|---|---|---|---|---|
| FPU/mL | p (h) | IU/mL | p (h) | IU/mL | p (h) | mg/mL | p (h) | |
| SCBSE | 0.23 ± 0.03 | 48 | 2.90 ± 0.06 | 48 | 0.43 ± 0.10 | 72 | 0.12 ± 0.01 | 120 |
| SCBDL | 0.10 ± 0.01 | 48 | 2.60 ± 0.20 | 72 | 0.33 ± 0.06 | 96 | 0.07 ± 0.01 | 96 |
| WB | 0.12 ± 0.02 | 48 | 2.00 ± 0.04 | 24 | 1.00 ± 0.04 | 120 | 0.15 ± 0.02 | 120 |
| EC | 0.10 ± 0.01 | 48 | 2.60 ± 0.14 | 72 | 0.33 ± 0.08 | 96 | 0.04 ± 0.01 | 48 |
| SCBSE + WB | 0.10 ± 0.02 | 72 | 2.80 ± 0.05 | 72 | 0.55 ± 0.05 | 72 | 0.13 ± 0.02 | 96 |
| SCBSE + SM | 0.13 ± 0.02 | 72 | 2.50 ± 0.10 | 48 | 0.53 ± 0.07 | 120 | 0.24 ± 0.03 | 96 |
| SCBSE + SM | 0.13 ± 0.03 | 72 | 2.80 ± 0.08 | 72 | 0.42 ± 0.05 | 120 | 0.10 ± 0.01 | 120 |
| 3XSCBSE + 1XFOS | – | 3.50 ± 0.05 | 48 | – | 0.11 ± 0.01 | 24 | ||
| 1XSCBSE + 1XLAC | – | 3.50 ± 0.10 | 72 | – | 0.08 ± 0.01 | 24 | ||
| 3XSCBSE + 1XLAC | 0.19 ± 0.02 | 72 | 3.10 ± 0.08 | 72 | 0.57 ± 0.05 | 120 | 0.12 ± 0.01 | 96 |
| 3XSCBSE + 1XGLY | 0.18 ± 0.02 | 96 | 2.80 ± 0.05 | 72 | 0.46 ± 0.09 | 120 | 0.10 ± 0.02 | 48 |
| 3XSCBSE + 1XSUC | 0.18 ± 0.04 | 96 | 3.30 ± 0.12 | 72 | 0.43 ± 0.08 | 120 | 0.10 ± 0.01 | 120 |
SCBSE steam-exploded sugar cane bagasse; SCBDL delignified steam-exploded sugar cane bagasse; EC celuflok 200™; WB wheat bran; SM soybean mill; GLY glycerol; LAC lactose; SUC sucrose; FOS commercial phosphofructooligosacharide. Bioreactor time (h) of peak activity (p)
Table 2 shows a series of six bioreactor runs in which we varied pH and temperature, feeding schedule as well as the combination of biomass sources, designed to overcome process side effects such as the possible interference of proteases and the onset of carbon catabolite repression. With the exception of the presence of sucrose (Table 2, run #5) that doubled the amount of cellulase, none of the other variations seemed to enhance filter paper activity.
Table 2.
Substrate influence on enzyme accumulation in bioreactors
| Run | Bioreactor conditions | Cellulase | Xylanase | β-glucosidase | Protein | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Substrate | T(°C) | pH | Pulse | FPase/mL | p (h) | IU/mL | p (h) | IU/mL | p (h) | mg/mL | p(h) | |
| #1 | SCBSE | 45 | 5 | 96 | 0.10 ± 0.01 | 72 | 3.88 ± 0.12 | 72 | 0.37 ± 0.04 | 120 | 0.06 ± 0.02 | 120 |
| #2 | SCBSE | 45 | 5 | 48/96 | 0.13 ± 0.03 | 120 | 1.93 ± 0.13 | 96 | 0.30 ± 0.06 | 96 | 0.08 ± 0.02 | 96 |
| #3 | SCBSE | 45 | 6 | 48/96 | 0.10 ± 0.02 | 96 | 1.80 ± 0.03 | 48 | 0.48 ± 0.03 | 120 | 0.10 ± 0.04 | 96 |
| #4 | SCBSE | 38–29 | 6 | 96 | 0.18 ± 0.03 | 120 | 2.00 ± 0.01 | 120 | 0.44 ± 0.02 | 96 | 0.17 ± 0.01 | 120 |
| #5 | SCBSE + SUC | 45 | 6 | None | 0.21 ± 0.02 | 24 | 2.70 ± 0.07 | 48 | 0.72 ± 0.01 | 120 | 0.16 ± 0.05 | 24 |
| #6 | WB | 45 | 6 | None | 0.06 ± 0.01 | 24 | 2.88 ± 0.01 | 24 | 1.64 ± 0.10 | 96 | 0.13 ± 0.01 | 24 |
SCBSE steam-exploded sugar cane bagasse; WB wheat bran; SUC sucrose; T temperature; (p), peak activity
Figure 2 describes enzymatic biomass hydrolysis into sugars and corresponding aldonic acids of various forms of sugar cane bagasse, “in natura” (SCBIN), delignified (SCBDL), steam-exploded (SCBSE) and purified cellulose (EC) by a crude enzymatic cocktail from M. thermophila M77 produced in a bioreactor with SCBIN as the carbon source. After 24 h incubation period, 6.31 g/L of glucose and gluconic acid was produced from cellulose (EC) and 4.31, 3.16 and 1.83 g/L from SCBIN, SCBDL and SCBSE, respectively (Fig. 2a). When the conversion potential of each carbon source was considered a 19.53% conversion was determined for SCBIN and 15.89, 8.02 and 7.63% conversion for EC, SCBDL and SCBSE, respectively (Fig. 2b).
Fig. 2.
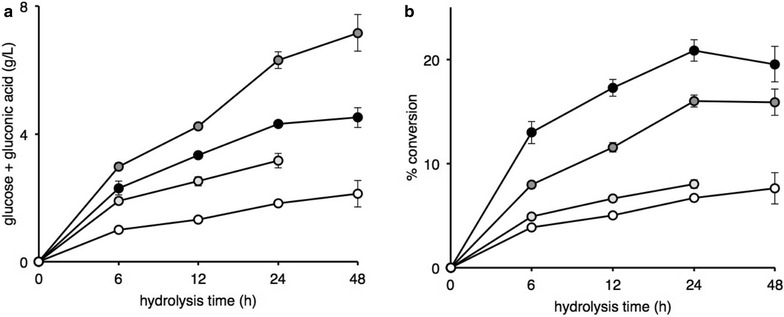
Conversion of various forms of sugar cane bagasse and crystalline cellulose into glucose, gluconic acid, cellobiose and cellobionic acid by M. thermophila M77 enzymes. M. thermophila M77 secretome produced on “in natura” sugar cane bagasse was concentrated and used to hydrolyze SCBIN (closed symbols), EC (cellufloc, dark shaded symbols), SCBDL (light shaded symbols) and SCBSE (±) at 50 °C. In a the release of glucose and gluconic acid is shown and in b the conversion percentage from total available sugars was estimated
Secretome protein composition
The secretome (all extracellular non-anchored proteins) produced by M. thermophila M77 grown in various carbon sources; SCBIN, SCBDL and SCBSE as well as purified cellulose (avicel and carboxymethylcellulose) and hemicelluloses (xylans, arabinan and arabinoxylan) were determined through LC–MS/MS (Additional file 1: Table S1; Figs. 4, 5). Total extracellular proteins (secretomes) were collected, concentrated by ultra-filtration (10 kDa cutoff), separated by SDS-PAGE, digested with trypsin, subjected to LC–MS/MS and peptides assigned through Mascot and Scaffold to M. thermophila ATCC_42464 predicted proteins. In total, 172 proteins were unambiguously identified with positive matching of 21,766 unique peptides (4019 SCBIN, 3661 SCBDL, 4269 SCBSE 3466 celluloses and 4716 hemicelluloses). The spectral counts from two independent experiments were normalized and duplicates averaged in order to enable quantitative comparisons between samples (see Additional file 1: Table S1).
Fig. 4.
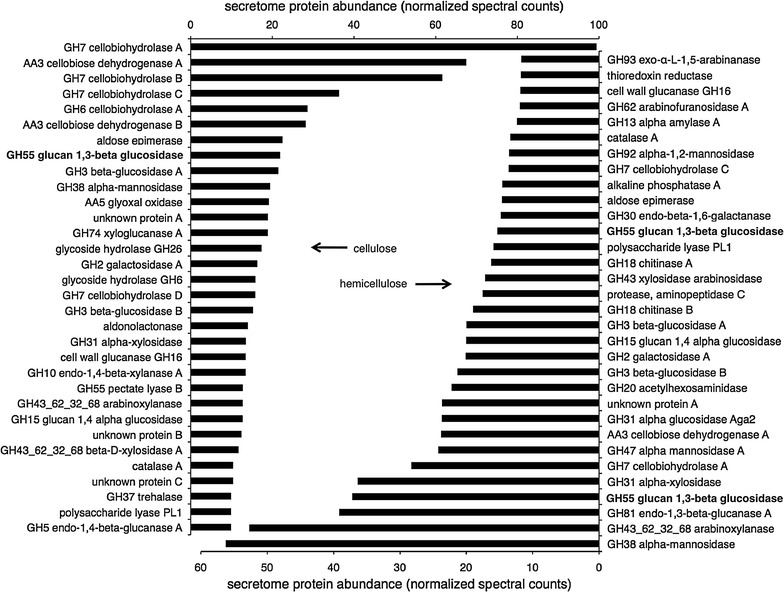
Biomass secretomes. M. thermophila M77 secretomes produced on cellulose (left panel), a mixture of 0,5% avicel and 0.5% carboxymethylcellulose, or hemicellulose (right panel), a mixture of 0.2% of each of three types of xylan, arabinan and arabinoxylan (see “Materials and methods” section) were analyzed by LC–MS/MS and protein abundance reported as normalized spectral counts. A detailed list of annotated protein names along with spectral and quantitative measurement data can be found in Additional file 1: Table S1
Fig. 5.
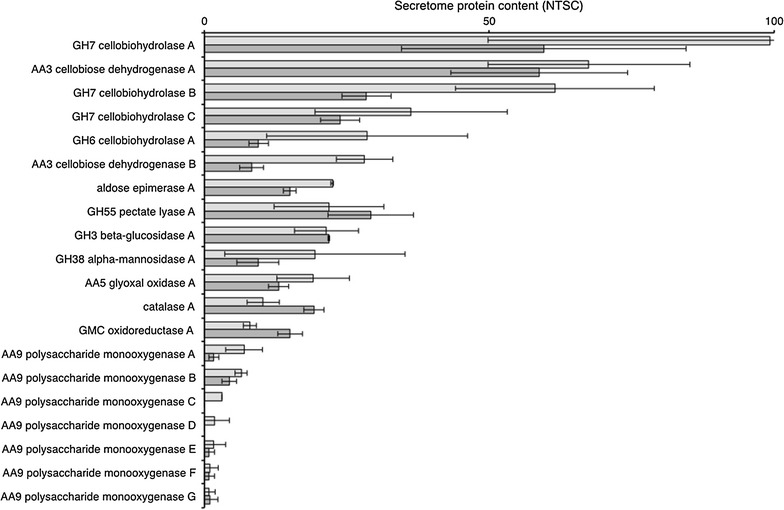
Core cellulose secretome. M. thermophila M77 secretomes produced on purified cellulose (CEL, light shaded bars) and sugar cane bagasse (SCBIN, dark shaded bars). Total extracellular protein abundance was determined by LC–MS/MS and reported as normalized spectral counts. Each bar indicates the normalized spectral counts of one enzyme and abundance ranking was done for SCBIN. Seven AA9 lytic polysaccharide monooxygenases did not rank at the top of any of the studied biomass sources however were added to the present table. A detailed list of annotated protein names along with spectral and quantitative measurement data can be found in Additional file 1: Table S1
Figure 3 shows SDS-PAGE protein profiles of enzymes secreted to the medium as a response to sugar cane bagasse, purified cellulose and hemicellulose and Fig. 4 displays secretome protein abundance profiles of M. thermophila M77 grown with purified cellulose (left panel) and a mixture of purified hemicelluloses (right panel). All major proteins in hemicellulose were associated with hemicellulose and pectin breakdown while in cellulose all major proteins were related to cellulose hydrolysis or oxidation.
In conditions where cellulose was the sole carbon source (Fig. 4, left panel) the most abundant proteins in the secretome were: GH7 cellobiohydrolase A (~10% of secretome), AA3 cellobiose dehydrogenase A (~7% of secretome), GH7 cellobiohydrolase B and C (~6 and ~4% of secretome, respectively), GH6 cellobiohydrolase A and AA3 cellobiose dehydrogenase B (~3% of each).
When M. thermophila M77 was grown in SCBIN, GH7 cellobiohydrolase A and AA3 cellobiose dehydrogenase A were the most abundant proteins both contributing with about 6% of the secretome each (Fig. 5), while other proteins such as GH55 β-1,3-glucanase, GH7 cellobiohydrolase B and C, GH81 endo-1,3-β-glucanase, GH74 xyloglucanase, GH43_62_32_68 arabinoxylanase, GH31 α-xylosidase, GH3 β-glucosidase A, catalase, hypothetical protein (MYCTH_2307339) and GH18 chitinase A contributed with less than 3% of secretome, each. Polysaccharide monooxygenases (seven in total) contributed with less than 1% each (Fig. 5). AA3 cellobiose dehydrogenase A (CdhA) on the other hand was the second most abundant protein (Fig. 4), accounting for about 6% of the secretome protein content.
We thus decided to concisely define the core cellulose secretome (Fig. 5) and further corroborate gene/protein complements. Typically, besides the classical cellobiohydrolase β-glucosidase set of proteins, cellobiose dehydrogenase, a glyoxal oxidase and an unknown GMC oxidoreductase make up the core cellulose secretome (Fig. 5).
Cellulose hydrolases such as cellobiohydrolase A, B, C and D, GH74 xyloglucanase, GH3 β-glucosidase and GH81 endo-β-1,3 glucanase did not adjust in abundance between the three types of biomass. In addition, hemicellulose hydrolases such as GH55 β-1,3-glucanase, GH43_62_32_68 arabinoxylanase, GH7 α-mannosidase and GH2 β-galactosidase also did not vary significantly in abundance among those three biomass substrates.
Figure 6 shows robust cellobiohydrolase, β-glucosidase and cellobiose dehydrogenase activity presence in the fluid of cultures grown on various forms of sugar cane bagasse (SCB). While cellobiohydrolase activity was present at similar levels in all biomass sources accumulating over a 4-day period and remained steady for up to 15 days cellobiose dehydrogenase accumulated for 9 days at differentiated levels in various forms of biomass and then sharply decreased and disappeared from the biomass cultures. β-glucosidase activity followed cellobiohydrolase with no difference in various biomass sources and steady accumulation over time. Laccase however poorly accumulated at the early stages of growth and then disappeared.
Fig. 6.
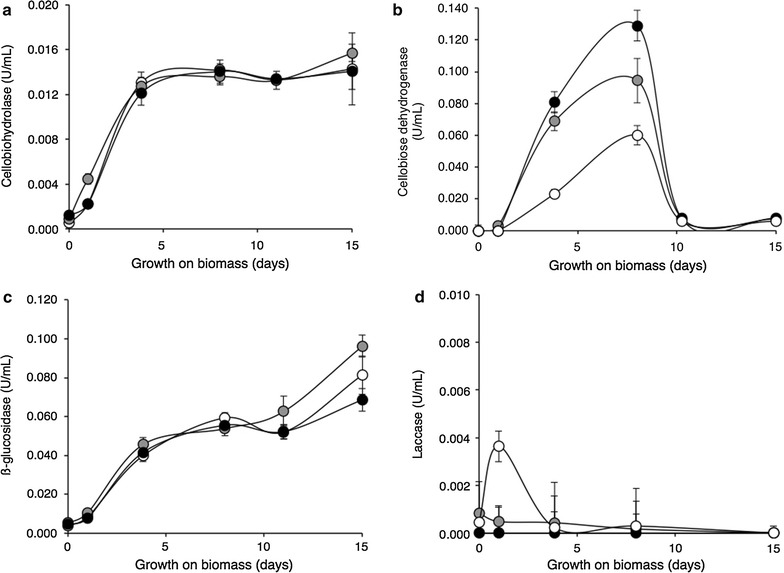
Major enzyme activities on solid biomass degradation. Cellobiohydrolase (a) and β-glucosidase (c) are the major hydrolases and cellobiose dehydrogenase (b) the major oxidase secreted by M. thermophila during growth on SCBIN (closed circles) “in natura” as well as SCBDL (shaded circles) delignified and SCBSE (open circles) “steam exploded” biomass. Other oxidative enzymes laccase (d), lignin peroxidase (not shown) and Mn-peroxidase (not shown) were detected at very low levels
Discussion
Enzymatic cocktails are typically evaluated by their biomass conversion ability, which is specified by the types of enzymes involved in cellulose breakdown producing sugars (glucose and cellobiose) and their respective aldonic acids. Experiments aimed at the production of cellulases using sugar cane bagasse as the carbon source yielded enzyme cocktails with little FPase activity even though the fungus aggressively digested the food source (Figs. 1, 2; Tables 1, 2).
We designed a series of bioreactor experiments (Table 2) to overcome process limitations. With the exception of the presence of sucrose (Table 2, run #5) that doubled the amount of cellulase, none of the other variations seemed to enhance filter paper activity. Other fungal systems such as Aspergillus nidulans and Phanerochaete chrysosporium produce similar low levels of cellulases and xylanase when growing on solid sorghum stover (Ray et al. 2012; Saykhedkar et al. 2012).
We than designed a biomass hydrolysis experiment using the M. thermophila M77 enzymatic cocktail and determined the release of glucose and corresponding gluconic acid. Nevertheless, when the conversion potential of each carbon source was considered, 19.53% for SCBIN and 15.89, 8.02 and 7.63% conversion for EC, SCBDL and SCBSE was observed, respectively (Fig. 2b). However, considering our experiment, the highest cellulase activity detected (0.23 FPU/mL) was applied on SCBIN 5% (w/v) representing an enzyme loading of only ~5 FPU/g of glucan in the hydrolysis experiment, making the observed 20% conversion rate unjustifiable, since other authors have loaded higher filter paper units (FPU) to get similar conversion rates (Adsul et al. 2005; da Silva et al. 2010; Ishihama et al. 2005; Pietrobon et al. 2011; Visser et al. 2015).
Measuring cellulose degradation by methods that only detect hydrolysis mechanisms (for instance FPase activity) may not be sufficient to evaluate the cellulose breakdown power of these enzyme mixtures because the fungus may secrete enzymes that instead of hydrolyzing cellulose, oxidize glycosidic linkages instead.
Secretome protein abundance profiles of M. thermophila M77 grown with purified cellulose (Fig. 4, left panel) and a mixture of purified hemicelluloses (Fig. 4, right panel) were constructed. Secretome protein profiles were substrate specific reflecting the nature of the substrate. All major proteins in hemicellulose were associated with hemicellulose and pectin breakdown while in cellulose all major proteins were related to cellulose hydrolysis or oxidation.
GH7 cellobiohydrolases (CbhA) act at the reducing end of a single cellulose chain and CbhA is the only major GH7 cellobiohydrolase that contains a cellulose-binding domain (CBM1). The presence of cellobiohydrolases devoid of cellulose binding domains, CbhB and CbhC in M. thermophila M77 secretomes followed similar observations made in other fungi suggesting that these CBM-devoid enzymes collaborate with CBM-bearing exo enzymes on cellulosic chains that have already been pulled apart from the crystalline fiber (Segato et al. 2012).
When grown on SCBIN, GH7 cellobiohydrolase (CbhA) and AA3 cellobiose dehydrogenase (CdhA) were the most abundant proteins both contributing with about 6% of the secretome each (Fig. 5), while other proteins contributed with less than 3% of secretome, each.
The M. thermophila M77 CdhA is a complete cellobiose dehydrogenase, a flavin-dependent dehydrogenase connected through a flexible linker to a heme-binding cytochrome and a true cellulose-binding domain (CBM1) (Tan et al. 2015). CdhA generates electrons by oxidation of cellobiose (perhaps generated by CbhA) and longer cellodextrins to 1-5-δ-lactones (Westermark and Eriksson 1975). Lactones hydrolyze spontaneously in solution, or enzymatically by lactonases (also present in the secretome), to generate aldonic acids (Beeson et al. 2011). Electrons generated by the flavin-dependent dehydrogenase are shuttled via heme-binding cytochrome to the recently discovered copper dependent polysaccharide monooxygenases (PMO’s) that in turn oxidize glycoside bonds in crystalline cellulose, hemicellulose and pectin (Beeson et al. 2011; Canevascini et al. 1991).
Remarkably, Fig. 5 shows that when the fungus grew in biomass (SCBIN) or purified cellulose (CEL) AA3 cellobiose dehydrogenase A was present in about the same concentration (about 50%) while GH7 and GH6 cellobiohydrolases were present at higher levels in CEL. Thus, the CdhA mediated electrons could be transferred to a wide range of oxygenases (Hemsworth et al. 2013; Westermark and Eriksson 1975; Zamocky et al. 2006). Other enzymes such as GH31 α-xylosidase, GH18 chitinase A (but not chitinase B), GMC oxidoreductase (an unknown oxidoreductase), GH43 xylosidase arabinosidase and AA5 glyoxal oxidase, followed a similar pattern, low abundance in delignified biomass and abundant in whole forms of biomass. The function of GMC oxidoreductase (GloA), glyoxal oxidase (GoxA) and GH18 chitinase even though clearly annotated by bioinformatics remain unclear and undefined.
Thus, based on the protein profile of secreted proteins and enzymatic activity detected it appears that the fungus accesses all available polymers, cellulose hemicellulose-pectin and lignin but does not produce hydrolysis products exclusively.
The participation of oxidation reactions coupled to lignin decomposition in the breakdown of cellulose chains (Beeson et al. 2012; Phillips et al. 2011), may explain the discrepancy observed between absolute FPase activity values in bioreactor experiments and the real (total) power of cellulose breakdown observed in biomass hydrolysis experiments. Furthermore, it is possible that CdhA, GloA and GoxA fail to interact and transfer electrons to cellulose, lignin and hemicellulose acceptor proteins, they generate an excess of hydrogen peroxide, which may directly oxidize glycosidic and phenolic bonds by a currently unknown mechanism.
Myceliophthora thermophila M77 produces specific secretomes that mirror the cell wall composition, formulate a mixed set of enzymes that in addition to hydrolyze glycoside bonds also promote coupled oxidation of cellulose and other biomass components enhancing the overall biomass degradation process. The secretome protein signature of M. thermophila M77 revealed cellobiose dehydrogenase (21% of the total secretome) as the major player in cellulose oxidation partnering perhaps with oxidation proteins such as glyoxal oxidase (4% of the secretome content) or glucose oxidase (not very abundant). The exact function of these enzymes remains uncertain.
Authors’ contributions
JGCP, EG and RAP conceived and designed the study. HBS, TMSB SDH JR conducted protein analysis experiments; JGCP, PD, DL and EG conducted bioreactor experiments, BC acquired data, performed the bioinformatics analysis and RAP drafted the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We value Raymond L. Huhnke’s biofuels leadership with funding from the Oklahoma Bioenergy Center.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Strains used in this study are available from the Fungal Genetics Stock Center (Kansas City, MO). Ancillary data are submitted as Additional file 1: Table S1.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Funding
Department of Energy, awards 06103-OKL, ZDJ-7-77608-01 and CNPq (Brazil) Science Without Borders Program awards 403090/2012-1 to RAP and 201319/2012-8 to HBS.
Abbreviations
- AA3
auxiliary activities 3
- ABTS
2,2′-Azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt
- CBM
carbohydrate binding domain
- DCPIP
2,6-dichlorophenol-indophenol
- DNS
dinitrosalicylic acid
- EC
commercial microcrystalline cellulose
- FDR
false discovery rate
- FOS
fructoligosaccharides
- FPase
filter paper cellulase activity
- FPU
filter paper unit
- HPLC
high performance liquid chromatography
- IU
enzyme unit
- GH3 7, 18, 74, 81
glycoside hydrolase family 3, 7,18, 74 and 81
- GLY
glycerol
- LAC
lactose
- LC–MS/MS
liquid chromatography–tandem mass spectrometry
- NCBI
National Center for Biotechnology Information (NIH, US)
- NTSC
normalized total spectral counts
- PMO
polysaccharide monoxygenase
- pNPC
p-nitrophenol-β-d-cellobioside
- pNPG
p-nitrophenol-β-d-glucopyranoside
- SCB
sugar cane bagasse
- SCBDL
lignin removed (sodium hydroxide extracted) and steam exploded sugar cane bagasse
- SCBIN
in natura” milled (200-μm particle size) sugar cane bagasse
- SCBSE
steam exploded sugar cane bagasse
- SDS-PAGE
SDS polyacrylamide gel electrophoresis
- SM
milled soybeans
- SUC
sucrose
- TSC
total spectral counts
- WB
wheat bran
Additional file
Additional file 1. Comprehensive LC–MS/MS secretome analysis.
Contributor Information
Hévila Brognaro dos Santos, Email: hbrognaro@yahoo.com.br.
Thaís Milena Souza Bezerra, Email: milenatsb@gmail.com.
José G. C. Pradella, Email: jpradella@bioetanol.org.br
Priscila Delabona, Email: priscila.delabona@bioetanol.org.br.
Deise Lima, Email: deise.lima@bioetanol.org.br.
Eleni Gomes, Email: eleni@ibilce.unesp.br.
Steve D. Hartson, Email: hartson.steve@gmail.com
Janet Rogers, Email: janet.rogers@okstate.edu.
Brian Couger, Email: mbcouger@gmail.com.
Rolf Prade, Phone: 1-405-744-7522, Email: prade@okstate.edu.
References
- Adsul MG, Ghule JE, Shaikh H, Singh R, Bastawde KB, Gokhale DV, Varma AJ. Enzymatic hydrolysis of delignified bagasse polysaccharides. Carbohydr Polym. 2005;62(1):6–10. doi: 10.1016/j.carbpol.2005.07.010. [DOI] [Google Scholar]
- Amorim HV, Lopes ML, de Castro Oliveira JV, Buckeridge MS, Goldman GH. Scientific challenges of bioethanol production in Brazil. Appl Microbiol Biotechnol. 2011;91(5):1267–1275. doi: 10.1007/s00253-011-3437-6. [DOI] [PubMed] [Google Scholar]
- Bailey M, Poutanen K. Production of xylanolytic enzymes by strains of Aspergillus. Appl Microbiol Biotechnol. 1989;30(1):5–10. doi: 10.1007/BF00255989. [DOI] [Google Scholar]
- Baminger U, Nidetzky B, Kulbe KD, Haltrich D. A simple assay for measuring cellobiose dehydrogenase activity in the presence of laccase. J Microbiol Methods. 1999;35(3):253–259. doi: 10.1016/S0167-7012(99)00022-6. [DOI] [PubMed] [Google Scholar]
- Beeson WT, Iavarone AT, Hausmann CD, Cate JH, Marletta MA. Extracellular aldonolactonase from Myceliophthora thermophila. Appl Environ Microbiol. 2011;77(2):650–656. doi: 10.1128/AEM.01922-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeson WT, Phillips CM, Cate JH, Marletta MA. Oxidative cleavage of cellulose by fungal copper-dependent polysaccharide monooxygenases. J Am Chem Soc. 2012;134(2):890–892. doi: 10.1021/ja210657t. [DOI] [PubMed] [Google Scholar]
- Benz JP, Chau BH, Zheng D, Bauer S, Glass NL, Somerville CR. A comparative systems analysis of polysaccharide-elicited responses in Neurospora crassa reveals carbon source-specific cellular adaptations. Mol Microbiol. 2013;91(2):275–299. doi: 10.1111/mmi.12459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berka RM, Grigoriev IV, Otillar R, Salamov A, Grimwood J, Reid I, Ishmael N, John T, Darmond C, Moisan MC, Henrissat B, Coutinho PM, Lombard V, Natvig DO, Lindquist E, Schmutz J, Lucas S, Harris P, Powlowski J, Bellemare A, Taylor D, Butler G, de Vries RP, Allijn IE, van den Brink J, Ushinsky S, Storms R, Powell AJ, Paulsen IT, Elbourne LD, Baker SE, Magnuson J, Laboissiere S, Clutterbuck AJ, Martinez D, Wogulis M, de Leon AL, Rey MW, Tsang A. Comparative genomic analysis of the thermophilic biomass-degrading fungi Myceliophthora thermophila and Thielavia terrestris. Nat Biotechnol. 2011;29(10):922–927. doi: 10.1038/nbt.1976. [DOI] [PubMed] [Google Scholar]
- Bourbonnais R, Paice MG, Reid ID, Lanthier P, Yaguchi M. Lignin oxidation by laccase isozymes from Trametes versicolor and role of the mediator 2,2′- azinobis (3-ethylbenzthiazoline-6-sulfonate) in kraft lignin depolymerization. Appl Environ Microbiol. 1995;61(5):1876–1880. doi: 10.1128/aem.61.5.1876-1880.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72(1–2):248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Canevascini G, Borer P, Dreyer JL. Cellobiose dehydrogenases of Sporotrichum (Chrysosporium) thermophile. Eur J Biochem. 1991;198(1):43–52. doi: 10.1111/j.1432-1033.1991.tb15984.x. [DOI] [PubMed] [Google Scholar]
- Coutinho PM, Andersen MR, Kolenova K, vanKuyk PA, Benoit I, Gruben BS, Trejo- Aguilar B, Visser H, van Solingen P, Pakula T, Seiboth B, Battaglia E, Aguilar- Osorio G, de Jong JF, Ohm RA, Aguilar M, Henrissat B, Nielsen J, Stalbrand H, de Vries RP. Post-genomic insights into the plant polysaccharide degradation potential of Aspergillus nidulans and comparison to Aspergillus niger and Aspergillus oryzae. Fungal Genet Biol. 2009;46(Suppl 1):S161–S169. doi: 10.1016/j.fgb.2008.07.020. [DOI] [PubMed] [Google Scholar]
- da Silva AS, Inoue H, Endo T, Yano S, Bon EP. Milling pretreatment of sugarcane bagasse and straw for enzymatic hydrolysis and ethanol fermentation. Bioresour Technol. 2010;101(19):7402–7409. doi: 10.1016/j.biortech.2010.05.008. [DOI] [PubMed] [Google Scholar]
- Divne C, Stahlberg J, Reinikainen T, Ruohonen L, Pettersson G, Knowles JK, Teeri TT, Jones TA. The three-dimensional crystal structure of the catalytic core of cellobiohydrolase I from Trichoderma reesei. Science. 1994;265(5171):524–528. doi: 10.1126/science.8036495. [DOI] [PubMed] [Google Scholar]
- Ghose TK. Measurement of cellulase activities. Pure Appl Chem. 1987;59(2):257–268. doi: 10.1351/pac198759020257. [DOI] [Google Scholar]
- Glass NL, Schmoll M, Cate JH, Coradetti S. Plant cell wall deconstruction by ascomycete fungi. Annu Rev Microbiol. 2013;67:477–498. doi: 10.1146/annurev-micro-092611-150044. [DOI] [PubMed] [Google Scholar]
- Hemsworth GR, Davies GJ, Walton PH. Recent insights into copper-containing lytic polysaccharide mono-oxygenases. Curr Opin Struct Biol. 2013;23(5):660–668. doi: 10.1016/j.sbi.2013.05.006. [DOI] [PubMed] [Google Scholar]
- Henriksson G, Johansson G, Pettersson G. A critical review of cellobiose dehydrogenases. J Biotechnol. 2000;78(2):93–113. doi: 10.1016/S0168-1656(00)00206-6. [DOI] [PubMed] [Google Scholar]
- Henriksson G, Zhang L, Li J, Ljungquist P, Reitberger T, Pettersson G, Johansson G. Is cellobiose dehydrogenase from Phanerochaete chrysosporium a lignin degrading enzyme? Biochim Biophys Acta. 2000;1480(1–2):83–91. doi: 10.1016/S0167-4838(00)00096-0. [DOI] [PubMed] [Google Scholar]
- Hilden L, Johansson G, Pettersson G, Li J, Ljungquist P, Henriksson G. Do the extracellular enzymes cellobiose dehydrogenase and manganese peroxidase form a pathway in lignin biodegradation? FEBS Lett. 2000;477(1–2):79–83. doi: 10.1016/S0014-5793(00)01757-9. [DOI] [PubMed] [Google Scholar]
- Himmel ME, Bayer EA. Lignocellulose conversion to biofuels: current challenges, global perspectives. Curr Opin Biotechnol. 2009;20(3):316–317. doi: 10.1016/j.copbio.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Horn SJ, Vaaje-Kolstad G, Westereng B, Eijsink VG. Novel enzymes for the degradation of cellulose. Biotechnol Biofuels. 2012;5(45):1–12. doi: 10.1186/1754-6834-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteom. 2005;4(9):1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- Kolbusz MA, Di Falco M, Ishmael N, Marqueteau S, Moisan M-C, Baptista CdS, Powlowski J, Tsang A. Transcriptome and exoproteome analysis of utilization of plant-derived biomass by Myceliophthora thermophila. Fungal Genet Biol. 2014;72:10–20. doi: 10.1016/j.fgb.2014.05.006. [DOI] [PubMed] [Google Scholar]
- Lacayo CI, Hwang MS, Ding SY, Thelen MP. Lignin depletion enhances the digestibility of cellulose in cultured xylem cells. Plos ONE. 2013;8(7):e68266. doi: 10.1371/journal.pone.0068266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal R. World crop residues production and implications of its use as a biofuel. Environ Int. 2005;31(4):575–584. doi: 10.1016/j.envint.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Langston JA, Shaghasi T, Abbate E, Xu F, Vlasenko E, Sweeney MD. Oxidoreductive cellulose depolymerization by the enzymes cellobiose dehydrogenase and glycoside hydrolase 61. Appl Environ Microbiol. 2011;77(19):7007–7015. doi: 10.1128/AEM.05815-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandels M, Sternberg D. Recent advances in cellulose technology. Ferment Technol. 1976;54:267–286. [Google Scholar]
- Mansfield SD, De Jong E, Saddler JN. Cellobiose dehydrogenase, an active agent in cellulose depolymerization. Appl Environ Microbiol. 1997;63(10):3804–3809. doi: 10.1128/aem.63.10.3804-3809.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens-Uzunova ES, Schaap PJ. Assessment of the pectin degrading enzyme network of Aspergillus niger by functional genomics. Fungal Genet Biol. 2009;46(Suppl 1):S170–S179. doi: 10.1016/j.fgb.2008.07.021. [DOI] [PubMed] [Google Scholar]
- Mason MG, Nicholls P, Wilson MT. Rotting by radicals—the role of cellobiose oxidoreductase? Biochem Soc Trans. 2003;31(Pt 6):1335–1336. doi: 10.1042/bst0311335. [DOI] [PubMed] [Google Scholar]
- Miller GL. Use of dintirosalicilic acid reagent for determination of reducing sugar. Anal Chem. 1959;31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- Moretti MM, Bocchini-Martins DA, Silva RD, Rodrigues A, Sette LD, Gomes E. Selection of thermophilic and thermotolerant fungi for the production of cellulases and xylanases under solid-state fermentation. Braz J Microbiol. 2012;43(3):1062–1071. doi: 10.1590/S1517-83822012000300032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips CM, Beeson WT, Cate JH, Marletta MA. Cellobiose dehydrogenase and a copper-dependent polysaccharide monooxygenase potentiate cellulose degradation by Neurospora crassa. ACS Chem Biol. 2011;6(12):1399–1406. doi: 10.1021/cb200351y. [DOI] [PubMed] [Google Scholar]
- Pietrobon VC, Monteiro RTR, Pompeu GB, Borges EPE, Lopes ML, de Amorim HV, da Cruz SH, Viegas EKD. Enzymatic hydrolysis of sugarcane bagasse pretreated with acid or alkali. Braz Arch Biol Technol. 2011;54(2):229–233. doi: 10.1590/S1516-89132011000200002. [DOI] [Google Scholar]
- Quinlan RJ, Sweeney MD, Lo Leggio L, Otten H, Poulsen JC, Johansen KS, Krogh KB, Jorgensen CI, Tovborg M, Anthonsen A, Tryfona T, Walter CP, Dupree P, Xu F, Davies GJ, Walton PH. Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc Natl Acad Sci USA. 2011;108(37):15079–15084. doi: 10.1073/pnas.1105776108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, Saykhedkar S, Ayoubi-Canaan P, Hartson SD, Prade R, Mort AJ. Phanerochaete chrysosporium produces a diverse array of extracellular enzymes when grown on sorghum. Appl Microbiol Biotechnol. 2012;93(5):2075–2089. doi: 10.1007/s00253-012-3907-5. [DOI] [PubMed] [Google Scholar]
- Rocha GJM, Gonçalves AR, Oliveira BR, Olivares EG, Rossell CEV. Steam explosion pretreatment reproduction and alkaline delignification reactions performed on a pilot scale with sugarcane bagasse for bioethanol production. Ind Crops Prod. 2011;35(1):274–279. doi: 10.1016/j.indcrop.2011.07.010. [DOI] [Google Scholar]
- Saykhedkar S, Ray A, Ayoubi-Canaan P, Hartson SD, Prade R, Mort AJ. A time course analysis of the extracellular proteome of Aspergillus nidulans growing on sorghum stover. Biotechnol Biofuels. 2012;5(1):52. doi: 10.1186/1754-6834-5-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segato F, Damasio AR, Goncalves TA, Murakami MT, Squina FM, Polizeli M, Mort AJ, Prade RA. Two structurally discrete GH7-cellobiohydrolases compete for the same cellulosic substrate fiber. Biotechnol Biofuels. 2012;5:21. doi: 10.1186/1754-6834-5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segato F, Damasio AR, de Lucas RC, Squina FM, Prade RA. Genomics review of holocellulose deconstruction by aspergilli. Microbiol Mol Biol Rev. 2014;78(4):588–613. doi: 10.1128/MMBR.00019-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68(5):850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Stahlberg J, Divne C, Koivula A, Piens K, Claeyssens M, Teeri TT, Jones TA. Activity studies and crystal structures of catalytically deficient mutants of cellobiohydrolase I from Trichoderma reesei. J Mol Biol. 1996;264(2):337–349. doi: 10.1006/jmbi.1996.0644. [DOI] [PubMed] [Google Scholar]
- Tan TC, Kracher D, Gandini R, Sygmund C, Kittl R, Haltrich D, Hallberg BM, Ludwig R, Divne C. Structural basis for cellobiose dehydrogenase action during oxidative cellulose degradation. Nat Commun. 2015;6:7542. doi: 10.1038/ncomms8542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaaje-Kolstad G, Westereng B, Horn SJ, Liu Z, Zhai H, Sorlie M, Eijsink VG. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science. 2010;330(6001):219–222. doi: 10.1126/science.1192231. [DOI] [PubMed] [Google Scholar]
- Visser EM, Leal TF, de Almeida MN, Guimaraes VM. Increased enzymatic hydrolysis of sugarcane bagasse from enzyme recycling. Biotechnol Biofuels. 2015;8(1):5. doi: 10.1186/s13068-014-0185-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber K, Osborn M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem. 1969;244(16):4406–4412. [PubMed] [Google Scholar]
- Westermark U, Eriksson KE. Purification and properties of cellobiose: quinone oxidoreductase from Sporotrichum pulverulentum. Acta Chem Scand B. 1975;29(4):419–424. doi: 10.3891/acta.chem.scand.29b-0419. [DOI] [PubMed] [Google Scholar]
- Zamocky M, Ludwig R, Peterbauer C, Hallberg BM, Divne C, Nicholls P, Haltrich D. Cellobiose dehydrogenase—a flavocytochrome from wood-degrading, phytopathogenic and saprotrophic fungi. Curr Protein Pept Sci. 2006;7(3):255–280. doi: 10.2174/138920306777452367. [DOI] [PubMed] [Google Scholar]
- Zhang YHP, Hong J, Ye X. Cellulase assays. Biofuels. In: Mielenz JR, editor. Methods in molecular biology. New York: Humana Press; 2009. pp. 213–231. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains used in this study are available from the Fungal Genetics Stock Center (Kansas City, MO). Ancillary data are submitted as Additional file 1: Table S1.


