Abstract
Humans have acted as vectors for species and expanded their ranges since at least the dawn of agriculture. While relatively well characterised for macrofauna and macroflora, the extent and dynamics of human-aided microbial dispersal is poorly described. We studied the role which humans have played in manipulating the distribution of Saccharomyces cerevisiae, one of the world's most important microbes, using whole genome sequencing. We include 52 strains representative of the diversity in New Zealand to the global set of genomes for this species. Phylogenomic approaches show an exclusively European origin of the New Zealand population, with a minimum of 10 founder events mostly taking place over the last 1000 years. Our results show that humans have expanded the range of S. cerevisiae and transported it to New Zealand where it was not previously present, where it has now become established in vineyards, but radiation to native forests appears limited.
Keywords: phylogenomics, yeast, Saccharomyces cerevisiae, population genetics, genomics
Genome sequencing shows that humans have unwittingly transported wine yeast to the other side of the planet, where this species has become established in vineyards.
INTRODUCTION
Humans have transported other species beyond their natural ranges for thousands of years, both intentionally for agricultural purposes (Diamond 2002) and unintentionally as a consequence of human migration (Wichmann et al. 2009). Other than disease agents, whose effects are apparent once transposed (Mazzaglia et al. 2012), the extent to which humans have manipulated the species ranges of microbes is poorly characterised (Litchman 2010). Previous studies suggested that microbes have virtually limitless dispersal abilities (de Wit and Bouvier 2006). However, while some microbes, such as marine bacteria, appear globally distributed (Pedrós-Alió 2006), others, such as hot spring communities, are certainly not (Martiny et al. 2006; Valverde, Tuffin and Cowan 2012; Almeida et al. 2014; Talbot et al. 2014; Taylor et al. 2014; Tripathi et al. 2014), and the forces which give rise to these microbial patterns are not clear (Hanson et al. 2012; Morrison-Whittle and Goddard 2015). Microbes are key components of both natural and agricultural ecosystems, but we are generally ignorant of the means by which microbes might be dispersed, let alone the degree to which humans influence microbial species ranges (Talbot et al. 2014; Tripathi et al. 2014).
Phylogeography is the primary method used to study the distributions of organisms in relation to their genetic diversity (Avise et al. 1987), and allows inference of movements and speciation events. Phylogenomics follows this approach but utilises large portions of genomes, as opposed to a few markers (Delsuc, Brinkmann and Philippe 2005). To date, phylogenomic studies have mainly been applied to plant and animal species (del Campo et al. 2014). While a vast array of robust biogeography studies have examined the variance in microbial species distributions (reviewed in Hanson et al. 2012), there are relatively few that have employed a phylogeographic approach, and those that exist have largely used mtDNA, microsatellite or single-locus genetic markers which can be biased or lack adequate resolution (Beheregaray 2008). However, recent studies have examined the population genomics of Saccharomyces yeast species to infer their origin and signals for domestication (Almeida et al. 2014, 2015; Barbosa et al. 2016; Ludlow et al. 2016). The Saccharomyces genus is composed of seven species and originated 10–20 million years ago (Hittinger 2013). All species have complete genomes available and have been used for numerous functional (Skelly et al. 2013; Bergstrom et al. 2014), phylogenetic (Drummond et al. 2006; Scannell et al. 2011), biochemical (Piskur et al. 2006) and evolutionary (Novo et al. 2009) studies. Saccharomyces cerevisiae was the first eukaryote sequenced in its entirety due to its small 12 Mb genome. Since then, it has become the best annotated eukaryotic genome (Cherry et al. 2011) and remains a cornerstone of the genomics community with over well 200 genomes available that are being added to consistently (Cherry et al. 2011; Skelly et al. 2013; Bergstrom et al. 2014; Almeida et al. 2015; Strope et al. 2015, reviewed in Peter and Schacherer 2016), and a further 37 available for its sister species S. paradoxus (Liti et al. 2009; Bergstrom et al. 2014).
The global distribution of S. cerevisiae is becoming increasingly well characterised, as demonstrated by the recent revelation of major basal clades in China (Wang et al. 2012), discovery of an ancient European population (Almeida et al. 2015), the discovery of hybrid populations associated with coffee and coca (Ludlow et al. 2016) and novel linages in Brazil (Barbosa et al. 2016). One pattern consistently found in all studies to date is the close relatedness and short divergence time of a ‘wine/European’ group (Liti et al. 2009; Schacherer et al. 2009; Wang et al. 2012; Cromie et al. 2013). This group includes commercial winemaking strains, strains sampled in vineyards and wineries worldwide, as well as strains from European forests, and the proposed ancestral European group inhabiting Mediterranean oak (Almeida et al. 2015). Using microsatellite profiles, it has been suggested that dispersal from Europe by humans in association with the global spread of viticulture and winemaking explains this pattern (Legras et al. 2007). Together this suggests that S. cerevisiae is a species with some clades that are closely associated with and dispersed by humans, but there are other clades present in natural environments and probably dispersed only locally by other means, such as insects (Stefanini et al. 2012; Buser et al. 2014), or not at all. A very recent study described genetically distinct populations of S. cerevisiae associated with coffee and cocoa in Africa and South America (Ludlow et al. 2016); intriguingly, these do not contain novel alleles, but are inferred to have been created by the mixing of existing populations associated with European vineyards, American oak trees and the ancestral seat of this species in Far East Asia. Ludlow et al. (2016) reasonably suggest that the movement of these strains, and thus creation of these populations, was facilitated by humans. Similarly, the recently discovery of a novel lineage in Brazil shows it was formed in part by hybridisation of migrants from the European/wine group with endemic S. paradoxus, which presumably then facilitated the colonisation of native Brazilian trees (Barbosa et al. 2016); it also seems reasonable to infer that humans facilitated this radiation event.
New Zealand (NZ) is the last major landmass colonised by humans ∼1000 years ago (Hurles, Matisoo-Smith and Gray 2003) and represents a unique environment to investigate questions concerning species range expansion. Māori were the first humans to settle in NZ, and Europeans did not arrive until Captain Cook's voyage of 1769 (though Abel Tasman sighted NZ in 1642). Viticulture was introduced into NZ around 1800. Many of NZ's endemic macroscopic flora and fauna have been studied (Wallis and Trewick 2009); however, extremely limited work has been conducted on the biogeography of microbial species in NZ. Previous analyses, based on microsatellite profiles and RAD-seq, suggest that NZ harbours a diverse and globally genetically distinct metapopulation of S. cerevisiae, with some geographically distinct localised populations that are also connected by various levels of gene flow (Goddard et al. 2010; Gayevskiy and Goddard 2012; Cromie et al. 2013; Knight and Goddard 2015). Strains have been isolated from vineyard and winemaking-associated niches (Goddard et al. 2010; Knight and Goddard 2015), oak trees planted by European migrants (Zhang et al. 2010), and from native NZ forests and fruiting trees (Knight and Goddard 2015; Gayevskiy and Goddard 2016). While small in terms of global production, the NZ wine industry commands a strong position in the premium market and this sector is significant to the NZ economy. Saccharomyces yeast plays a role in the production of wine, including potentially being part of the process that geographically differentiates wines (Knight et al. 2015). Alongside the academic interest in Saccharomyces ecology and population biology (Goddard and Greig 2015), their role in winemaking adds economic interest to understand the origin of S. cerevisiae populations. One recent study suggests the presence of an ancient population of the yeast S. uvarum in Australasia, but this species is certainly not endemic to NZ, nor is there evidence for an NZ-specific population (Almeida et al. 2014). Another study only recently reported the presence of S. eubayanus and S. arboricola in NZ, but the age and origin of these species are uncertain (Gayevskiy and Goddard 2016).
Here we ask why S. cerevisiae is present in NZ, and use phylogenomic methods to evaluate its history and range expansion. Two extremes present themselves: either (1) there was an ancient S. cerevisiae population present in NZ prior to humans arriving under 1000 years ago or (2) that this species was transported to NZ by humans with winemaking who unwittingly expand this species’ range along with exotic fruit bearing plants and trees. Of course, some mix of the two is also possible.
MATERIALS AND METHODS
Strain selection and sequencing
The K-means clustering algorithm used to identify maximally divergent genotypes was implemented in R (R Development Core Team 2011). Sulphite tolerance was assayed by plating onto YPD with either 10, 15 or 20 mM sodium metabisulphite in triplicate and scoring the growth of colonies as full, partial or none after 2 days at 28°C. Each strain was propagated in YPD, and high molecular weight genomic DNA was extracted using the Qiagen Blood & Cell Culture DNA Kit. Libraries were constructed using the Illumina TruSeq Nano DNA Sample Prep Kit with 550 bp insert size. Sequencing was carried out at the Beijing Genomics Institute (China) on a single 150-bp paired-end lane of an Illumina HiSeq 2000.
Genome mapping and quality control
Each sequenced genome was treated identically using a custom bioinformatics pipeline written in Perl. This pipeline is outlined below.
Quality control and trimming
FASTQC (v0.10.1; Andrews 2012) was used for quality control of each library and to determine optimal trimming parameters. Trimming was conducted with Trimmomatic (v0.25; Lohse et al. 2012) using the following parameters: ‘LEADING:3 TRAILING:3 SLIDINGWINDOW:3:20 MINLEN:30’. Following trimming, FASTQC was executed on the trimmed reads for comparison with the initial reports.
Mapping and variant calling
All trimmed reads were mapped against the Saccharomyces cerevisiae reference strain S288C using Bowtie2 (v0.12.7; Langmead and Salzberg 2012). Following mapping, samtools (v0.1.18; Li et al. 2009) was used for alignment conversion, sorting and indexing. A variant call file was produced using the mpileup command within samtools with the ‘-Bu’ parameters. The variant call file was used to create a consensus genome without the reference to allow for INDELs using the vcf2fq Perl script within samtools. Putative heterozygous positions were conservatively called as ‘N’ as the phylogenetics and population genetics methods utilised do not support ambiguous calls. Heterozygous positions were quantified with a custom Perl script which filtered out positions with a sequencing depth below 10 or above 100 and a genotype quality below 20.
Data availability
We have made our raw sequence data and consensus genomes aligned to S288C (Goffeau et al. 1996; EBI:GCA_000146045.2) publicly available at SRA: SRP042301 and BioProject: PRJNA247448.
Sequence extraction from sequenced and international genomes
In addition to the 52 genomes sequenced here, a further 72 S. cerevisiae and 37 S. paradoxus genomes were obtained from the Saccharomyces Genome Database (http://yeastgenome.org), NCBI, the Saccharomyces Genome Resequencing Project (https://sanger.ac.uk/research/projects/genomeinformatics/sgrp.html) and from Huang, Roncoroni and Gardner (2014) in the form of consensus genomes and/or raw data. To obtain an accurate estimation of the relatedness of the genomes, we extracted the well-known set of 106 orthologous loci spread through the genome of S. cerevisiae and present in all Saccharomyces species (Rokas et al. 2003). The sequences of these 106 loci were extracted by searching the S288C sequence for each locus against each consensus genome using the BLAST algorithm. Only genomes with complete sets of 106 loci were retained for phylogenetic analysis.
All sets of 106 loci were subjected to a multiple sequence alignment using clustalw (v.2.1; Larkin et al. 2007) within Geneious (v6, http://www.geneious.com; Kearse et al. 2012). Alignments were manually curated within Geneious due to the frequent homopolymer indels present in some of the genomes due to older sequencing technology. We created a second dataset comprising 13 loci sequenced from 99 S. cerevisiae strains isolated in China (Wang et al. 2012). Only these loci were sequenced from these Chinese strains with no overlap with our main dataset. The consensus sequence for each of these loci was used to search against all available genomes outlined above. Genomes with complete sets of all 13 loci were retained for phylogenetic analysis. We used five S. paradoxus genomes as an outgroup, although four of the loci include intergenic regions and the S. paradoxus genomes did not yield these loci. The remaining nine loci were sufficient for phylogenetic analysis. Multiple sequence alignments were carried out in the same way as for the 106 loci dataset.
Phylogenetics
Phylogenetic analyses were conducted using BEAST (v1.7.5; Drummond and Rambaut 2007) on the finalised sequence alignments for both datasets. A number of scenarios were run to explore the relationships between genomes and to determine the stability of inferred relationships by locus and dataset.
Substitution and clock models were unlinked for all loci in all analyses to facilitate their independent estimation. Trees were linked to obtain a consensus tree using all loci. All substitution model and rate options were left on default due to the large increase in processing time observed when any were changed. A log-normal relaxed clock (uncorrelated) was used with an exponential distribution of mean 0.3. All runs were conducted with 1 billion iterations due to the size of the datasets. We verified MCMC convergence by examining the effective sample sizes of all parameters in each analysis and with visual inspection of the traces. 10% to 40% of each run was discarded as burn-in depending on the convergence of the MCMC trace. Separate phylogenetic analyses were conducted for the two clades found housing NZ strains in the 106 loci dataset. These used S288C as the outgroup to determine high-resolution structure within these clades. Parsimony analyses including permutations of NZ and Europe terminal taxa status, and calculations of the minimum change of this state over these phylogenies, were conducted in Mesquite (Maddison and Maddison 2014).
We used the published divergence date estimates between S. cerevisiae and its sister species S. paradoxus as a calibration point for the divergences of clades within our phylogenies (Liti, Barton and Louis 2006). Divergences between clades within phylogenies are typically estimated using molecular clocks and/or by calibration time points of established species divergences using fossils. Molecular clocks for S. cerevisiae are not in wide use due to the difficulty of estimating clock-like rates of evolution in a species with unknown generation times in its natural environment and high rates of inbreeding. The time of the common ancestor of S. cerevisiae and S. paradoxus has been estimated at 0.4–3.4 mya (Liti, Barton and Louis 2006). The molecular substitution rate observed between the split of the S. cerevisiae and S. paradoxus genomes was assumed to correspond to this time period. To estimate the divergence date of a particular clade, the proportional substitution rate for the clade was calculated against the calibration point to give a date estimate.
Population metrics and structure
We utilised ANGSD (v0.588; Nielsen et al. 2012) to generate population genomic metrics. ANGSD operates on short read alignment bam files that afford statistical robustness in calculating the site frequency spectrum in comparison with traditional tools operating on a set genotype. Given this requirement, we could only use genomes with raw data available from Illumina sequencing technology. This was aided by the recent resequencing of diverse worldwide strains (Bergstrom et al. 2014). We thus created three superset subsets: the NZ strains (52), the previous and the wine/European strains (66) and the previous with all remaining strains (75). The number of sites and the number of segregating sites for each population were determined from the mean allele frequency calculations in ANGSD with the minInd parameter set to the number of strains per population and the minMaf alternatively set to 0 and 0.01. Only high-quality data were used (minQ = 20 and minMapQ = 30). Watterson's Estimator (θ) (Watterson 1975), Tajima's Pi (π) (Tajima 1989) and Tajima's D (Tajima 1989) were calculated by first calculating the site allele frequency likelihood, then the maximum likelihood estimate of the SFS, then the thetas per site and finally summarised with the thetaStat utility in ANGSD.
Tests for admixture within the S. cerevisiae genomes were conducted with Structure (Pritchard, Stephens and Donnelly 2000) due to the haploid nature of some of the genomes in our dataset. We chose to include all strains of S. cerevisiae where a consensus genome consisting of entire chromosomes was available to capture entire genomic diversity. The chromosomes of the 93 strains that met these criteria were aligned using Mauve (v2.3.1; Darling et al. 2004) within Geneious (v6, http://www.geneious.com; Kearse et al. 2012) and any nucleotide positions where either all strains showed no variation or at least one gap was present were removed. The remaining positions were run through Structure using the admixture model with 10 000 iterations of burn in followed by 20 000 iterations of analysis. K values between 2 and 20 were used with three replicate chains for each value of K to check for convergence, and the optimal number of subpopulations inferred using the Evanno method (Evanno, Regnaut and Goudet 2005). Population classifications were not used for the prior. Resulting ancestry profiles were objectively analysed using ObStruct (Gayevskiy et al. 2014) to determine the extent that geographic origin, niche of isolation and our phylogenomic analysis explains inferred population structure.
RESULTS
Strain selection
We collated data from six recent studies that have surveyed for Saccharomyces cerevisiae across NZ (Table 1; Serjeant et al. 2008; Goddard et al. 2010; Zhang et al. 2010; Gayevskiy and Goddard 2012; Knight and Goddard 2015; Gayevskiy and Goddard 2016). Saccharomyces cerevisiae has been isolated from over 99% of spontaneous ferment samples, 10% of vineyard samples and only 1% of forest/tree samples. The order of magnitude difference in recovery of this species is not due to differential sampling effort as most effort was spent sampling native forests, then vineyards and least for spontaneous ferments (Table 1). Just six genotypes (characterised at nine microsatellite loci; Richards, Goddard and Gardner 2009) have been recovered from trees/forests—one of these was from an exotic oak tree (Zhang et al. 2010), but microsatellite profiling showed this to be very closely related to DBVPG1106—a strain isolated from Australian vineyards which clusters with the wine/European group for which whole genome sequence is already available, and is included in this study (Zhang et al. 2010). Population genetic analyses of the remaining five genotypes isolated from native NZ forests show these to be homogeneous with their regional vineyard and spontaneous ferment populations: there is no evidence for genetic differentiation between strains isolated from native forests and their vineyard counterparts (Knight and Goddard 2015). Thus, it is clear that S. cerevisiae is very common in NZ spontaneous ferments, and at reasonable abundance in vineyard habitats, but rare in NZ native forests. This observation, and the fact that S. cerevisiae populations in these three habitats are connected within each region of NZ (Knight and Goddard 2015), means the most likely explanation is that there is just one S. cerevisiae metapopulation in NZ closely associated with vineyards and ferments, but members of this population are transposed to native habitats at some low rate. There is no evidence that NZ harbours another genetically distinct S. cerevisiae population that is not primarily associated with ferments and vineyards. The question we ask here concerns the origin of this group. To address this question, we need data from a set of genomes that best reflects the genetic diversity in this population: this is the most pertinent parameter relevant to elucidating the origin of this species in NZ. We thus identified a set of 52 maximally divergent S. cerevisiae genotypes from those of 724 in our database using k-means clustering of microsatellite profiles; these are detailed in Table S1 (Supporting Information).
Table 1.
The collated sampling effort and recovery of Saccharomyces cerevisiae (Sc) from six recent studies in NZ (Serjeant et al. 2008; Goddard et al. 2010; Zhang et al. 2010; Gayevskiy and Goddard 2012; Knight and Goddard 2015; Gayevskiy and Goddard 2016). Native forest samples derived from soil, honeydew and fruits of native trees (Serjeant et al. 2008; Knight and Goddard 2015; Gayevskiy and Goddard 2016); vineyard samples derived from soil, fruit and flowers (Goddard et al. 2010; Gayevskiy and Goddard 2012; Knight and Goddard 2015); ferment samples derived from spontaneous ferments of Sauvignon blanc, Chardonnay and Syrah (Goddard et al. 2010; Gayevskiy and Goddard 2012; Knight and Goddard 2015).
| Colonies | Samples | Number of | Recovery | ||
|---|---|---|---|---|---|
| Habitat | Samples | analysed | yielding Sc | genotypes | rate (%) |
| Exotic oak | 190 | 1140 | 2 | 1 | 1.05 |
| Native forest | 523 | 7522 | 5 | 5 | 0.96 |
| Vineyard | 360 | 10833 | 39 | 62 | 10.83 |
| Ferment | 160 | 11590 | 159 | 656 | 99.38 |
| Total | 1233 | 31085 | 205 | 724 |
Sequencing and mapping
Sequencing of genomes derived from clonally expanded diploid populations yielded an average of 5.1 million 150-bp paired-end reads per strain for a total of 39.8 Gbp of data. An average mapping rate of 97.16% was obtained for all genomes using the S288C genome as a reference, with an average coverage of 61X, and mapping quality of 38.78 (Phred score). An average of 52 421 (SE = 532) SNPs and 4915 (SE = 36) INDELs were obtained for each genome. This number of SNPs is entirely consistent with other S. cerevisiae strains sequenced on the same platform from a diversity of international locations and niches (e.g. Bergstrom et al. 2014). The average number of heterozygous SNPs per strain was 7274 (SE = 805) which is consistent with that found for other vineyard isolates (Magwene et al. 2011), but heterozygosity levels ranged from ∼3000 to ∼31 000 (6% to 43% of all SNPs) across the 52 NZ genomes. Cursory analyses of large-scale copy number variations indicate that the genome of 6-Sol7-2 contains three copies of chromosome 4 and 27-WI_S_JASA_13 contains three copies of the first half of chromosome 7. Further, 37 of the 52 NZ genomes show large copy numbers (1.5-50X) of locus YGR201C (unknown function) on chromosome 7. We stress that the main hypothesis under test is the phylogeography of S. cerevisiae, and thus we do not concentrate on the details of fine-scale differences between genomes any further here.
Population genomic statistics
First, we compared the NZ-derived genomes with one another and then with 14 previously published genomes that either derived from Europe or are associated with winemaking, which form a tight clade, and finally to a further nine genomes derived from a diversity of locations and niches (Table S2, Supporting Information). Only genomes with high-quality data (Illumina sequencing with ∼30X or greater coverage) were included in this analysis. We only included sites that had high-quality read data for all genomes, and thus the number of comparable homologous sites reduced as more genomes were added due to the increase in missing data for some of the existing genomes. The number of segregating sites is proportionately similar between the wine/European group and the NZ population, but nearly doubles when the other genomes are added, indicating their relative divergence (Table 2). Pi is a measure of nucleotide diversity (Tajima 1989) and the NZ and wine/Europe populations appear identical in terms of nucleotide diversity, but again the inclusion of the non-wine/Europe genomes leads to a 30% increase in this statistic. This suggests that the NZ-derived genomes are more similar to the genomes deriving from Europe or associated with winemaking than to genomes derived from elsewhere. It is tempting to further investigate comparative population genetics, but since the international samples are not random representatives of a population this defies many of the assumptions underlying population genetic calculations, and so we have not pursued this here.
Table 2.
Nucleotide diversity in three superset populations of S. cerevisiae.
| Population | Sitesa | Segregating s itesb | θ (×1000) | π (×1000) | Tajima's D |
|---|---|---|---|---|---|
| NZ (52) | 11 084 457 | 124 566 (1.1%) | 3.3 | 2.3 | –1.27 |
| Wine/Europe/NZ (66) | 6 505 396 | 91 543 (1.4%) | 3.8 | 2.3 | –1.65 |
| All available (75) | 6 397 673 | 134 386 (2.1%) | 5.2 | 3 | –1.77 |
aSites where all genomes have at least one high-quality read.
bSites where all genomes have at least one high-quality read and at least one genome differs from the rest, percentage in brackets is from the total number of sites.
Phylogenomic approaches
First, we employed phylogenomic methods to evaluate long-term within-species population structure. Initially, we chose a comprehensive set of 106 orthologous loci compiled by Rokas et al. (2003) for analyses due to their distribution across the genome, presence in all Saccharomyces species and their proven capability to provide a robust phylogenetic signal. We did not use the entirety of the genomic data due to potential problems with identifying orthologues and paralogues. Of the existing 72 S. cerevisiae and 37 S. paradoxus genomes, 60 and 36 respectively contained these 106 loci; the remaining genomes had insufficient or low-quality sequencing coverage for at least some of the loci (Table S2). All 52 NZ genomes contained complete sets of these 106 loci.
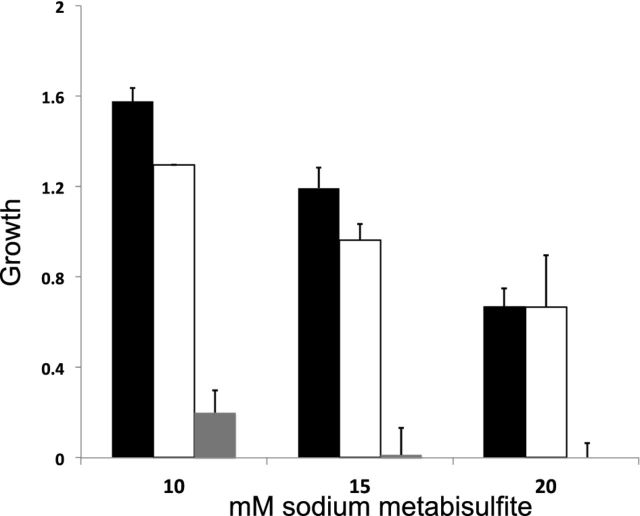
Rampant recombination among these genomes, which would be indicated by more of network-like than tree-like relationships, would significantly decrease the validity of using a phylogenetic approach for the analyses of these genomes due to its assumptions regarding bifurcating relationships. Neighbour-net (Huson and Bryant 2006) analysis (Fig. S1, Supporting Information) reveals a topology that to a first approximation shows a more tree-like than network-like structure, suggesting little recombination between major groups, and thus lends greater confidence for the use of phylogenetic approaches to evaluate some of the relationships between these genomes. To place the NZ population in a global context, we reconstructed a phylogeny using Bayesian approaches and included the 36 S. paradoxus genomes for calibration and rooting purposes (Fig. 1). The inclusion of NZ genomes reproduces an overall global topology that is comparable to earlier analyses (Liti et al. 2009; Schacherer et al. 2009). Strikingly, 85% of the NZ strains, including the strain isolated from a native forest, are interspersed within the wine/European clade (Fig. 1). The resolution within this clade is extremely poor, suggesting that this comprises a contemporaneous population experiencing gene flow and recombination, and the relatively short branch lengths show little time since divergence. This represents the first piece of evidence that the S. cerevisiae in NZ have a significant portion of ancestry, and thus derive from, and are in fact part of, the European population. Not all NZ strains fall within this wine/European group however. The remaining 15% of NZ strains form a sister clade to the wine/European group, with the inclusion of I14 and Y55, which are two soil isolates from Europe. Resistance to sulphite is a key defining phenotype of the S. cerevisiae lineage associated with viticulture and winemaking as sulphur is and has been used as an antimicrobial in both vineyards and wineries (Pretorius 2000; Aa et al. 2006). To evaluate whether this smaller group might represent a population not associated with wine, we tested the sensitivity of these to sulphite. There is no significant difference in resistance to 10, 15 and 20 mM sulphite between the NZ and European groups as determined by plate assays (F[1,105] 1.59, 0.53, 0.00 respectively and all P < 0.21), but these two groups are significantly more tolerant to 10, 15 and 20 mM sulphite than the rest of the non-wine-associated strains (F[2,129] 54.3, 20.9, 8.9 respectively and all P > 0.0002) (Fig. 2).
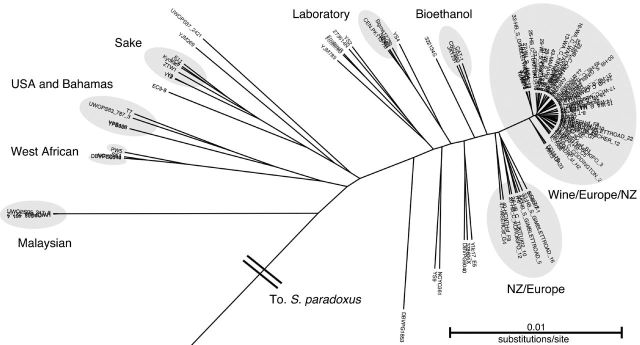
Figure 1.
Rooted phylogeny of 112 S. cerevisiae strains created using the 106 loci dataset with S. paradoxus as the outgroup reconstructed using the Bayesian methods implemented in BEAST (v1.7.5; Drummond and Rambaut 2007). The NZ strains cluster and interdigitate with the wine/European group.
Figure 2.
Sulphite tolerance of the wine/Europe/NZ group (n = 98), NZ/Europe group (n = 9) and the remaining ‘non-wine’ isolates (n = 25) shown in Fig. 1. Sulphite tolerance was assayed by plating onto YPD with either 10, 15 or 20 mM sodium metabisulphite in triplicate and scoring the growth of colonies as full (2), partial (1) or none (0) after 2 days at 28°C, and this metric is plotted on the y-axis. ANOVA reveals a significant difference in tolerance between groups at all sulphite concentrations, but Tukey's HSD tests reveal no significant difference in sulphite tolerance between wine/Europe/NZ and NZ/Europe groups at any of these three levels.
Previously identified clades (Liti et al. 2009; Schacherer et al. 2009) are reconstructed and expanded with our analyses due to the inclusion of further recently sequenced genomes. The North American clade includes additional strains sampled from Missouri (T7) and Bahamas (UWOPS83_787_3), the West African clade contains the PW5 strain sampled from Nigerian palm wine and the Sake clade contains an additional three strains (UC5, Kyokai7 and ZTW1). Several new clades are present for laboratory and bioethanol strains. Apart from the Sake clade, the ordering of the clades in relation to the S. paradoxus outgroup places strains isolated from non-agricultural niches as basal while agricultural and biotechnological associated strains are relatively derived indicating their more recent formation. Strains not residing in these clades are interspersed through the tree and tend to be positioned at the ends of longer branches and could indicate the presence of further undersampled populations or represent chimeric strains with ancestry in multiple clades.
Recently, a large novel diversity within S. cerevisiae was revealed by the sequencing of 13 loci from 99 strains isolated in China, leading to suggestions that this species originated on the Asian continent (Wang et al. 2012). We extracted these loci from all available whole genomes, resulting in 214 S. cerevisiae comprising 99 Chinese strains, 52 NZ strains, 60 strains used in the first analysis and a further three international strains containing these six loci (Table S2). We reconstructed a phylogeny with these 13 loci (Fig. S2, Supporting Information). Posterior probabilities for all labelled clades shown in Fig. S2 were >0.92 indicating adequate resolution, but the posterior probabilities for relationships of individuals within these clades, particularly within the Wine/European/NZ clade, were very poor, likely due to gene tree incongruence. Broadly, our analyses agree with earlier findings, and all eight Chinese lineages previously identified (Wang et al. 2012) were reconstructed. The tree features a large split between strains that have been sampled from non-agricultural environments regardless of sampling location, and those that are closely associated with human activity. The exception to this is the Sake clade which tends to cluster with non-agricultural strains due to a hypothesised secondary domestication event (Fay and Benavides 2005). The genetic diversity (branch lengths) within the human-associated clades is significantly lower than for the other clades, which, taken with low posterior probabilities, implies incomplete lineage sorting and/or high rates of admixture for human-associated strains.
Population genetic approaches
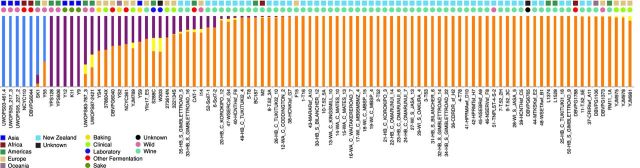
Saccharomyces cerevisiae is a sexual eukaryote, and along with the reasonable rates of heterozygosity revealed here, previous analyses show that while it tends to inbreed, there is clearly a reasonable amount of outcrossed recombination and gene flow between subpopulations occurring in the NZ population (Goddard et al. 2010; Gayevskiy and Goddard 2012; Knight and Goddard 2015), and the inference of recombination and hybridisation in global studies suggest that this may well be the case at larger scales (Liti et al. 2009; Cromie et al. 2013; Barbosa et al. 2016; Ludlow et al. 2016). The degree to which phylogenomic methods are able to recover any signal when there is diffuse population structure is not clear, i.e. when some population differentiation is present but with reasonable gene flow and recombination between subpopulations. To enable us to analyse a spectrum of possible population structures, from completely homogenised through to highly structured due to ancient divergences, we use complementary Bayesian-based population genetic methods capable of inferring finer degrees of population structure that account for recombination implemented in Structure (Pritchard, Stephens and Donnelly 2000), and the subsequent analyses of ancestry profiles by Obstruct (Gayevskiy et al. 2014). From the 11 059 143 nucleotide positions in the 93 aligned concatenated genomes, any which were uninformative or had missing data were conservatively removed leaving a total 66 316 robustly informative positions for population structure analysis. We employed Bayesian population structure approaches that account for and incorporate recombination (admixture) between strains. These population genetic approaches infer the presence of four populations using the Evanno method (Evanno, Regnaut and Goudet 2005) as implemented in Structure Harvester (Earl and vonHoldt 2011). Figure 3 shows the resulting ancestry profiles: each vertical column represents a strain and the colours show the proportion of ancestry of each strain to each of the four inferred populations. Strains that have different degrees of ancestry in different subpopulations are a result of mating and recombination between strains (or their ancestors) originating from different subpopulations. There is a progressive and gradual increase in ancestry to the orange inferred population as one moves from the assumed ‘natural’ strains on the left to the increasingly ‘human-associated’ strains on the right. Again, the NZ strains fall together and with the European strains, but with varying degrees of ancestry. It is clear that these various populations are not discrete: there are signals for some gene flow and genetic mixing among the species as a whole.
Figure 3.
Ancestry profiles produced by Structure showing population structure inference of 66 316 SNPs from 93 whole genomes of S. cerevisiae. Each bar represents an individual strain and colours represent different inferred populations for K = 4 (optimal). The geographic origin and niche of isolation for each strain is labelled with coloured squares and circles, respectively. Approximately the orange inferred subpopulation correlates with the European/wine group, the purple with Sake and the blue possibly Asia/Africa; there are not clear correlations with the yellow inferred subpopulation.
We went on to analyse the inferred ancestry profiles (Gayevskiy et al. 2014) to determine whether geographic origin or niche of isolation might correlate most strongly with population structure. This analysis shows that variance in genetic structure in the NZ population correlated with niche of isolation (R2 = 0.51, P < 0.0001) only marginally more than geographic origin (R2 = 0.45, P < 0.0001). Unsurprisingly, the amount of genetic variance explained (R2) is greater (R2 = 0.74, P < 0.0001) when ancestries are compared to those groups revealed from the independent phylogenetic analyses shown in Fig. 1, rather than simply geographic origin or niche of isolation. This shows that neither geographic location nor niche/‘use’ alone is sufficient to describe the observed population structure, and is exactly in line with the recent conclusion of Almeida et al. (2015) who examined the global population but included European oak population. Thus, the most accurate picture is one of the global metapopulation of connected subpopulations inhabiting various places and niches, and recapitulates the picture seen at national levels (Knight and Goddard 2015).
No evidence for hybridisation with other Saccharomyces species
Barbosa et al. (2016) recently reported novel S. cerevisiae lineages in Brazil that are related to Japanese and North American lineages, but the Brazilian populations also contain signals for mating and introgression with the wine/European group, as well as hybridisation and introgression with American S. paradoxus. This hybridisation and subsequent introgression conceivably provided novel genetic combinations better adapted to inhabit Brazilian native biomes. Ludlow et al. (2016) reported that lineages associated with coffee and cocoa were created by the hybridisation of genomes from the European/wine with North American and Chinese populations. Might the NZ S. cerevisiae have undergone a similar process—Where hybridisation with an endemic Saccharomyces, or some other S. cerevisiae population, provided an opportunity for more effective adaptive radiation in NZ? For the Saccharomyces species known to be present in NZ, S. paradoxus has been inferred to have recently migrated from Europe with oak trees (Zhang et al. 2010). The single representative of S. eubayanus is also inferred to have recently arrived from South America (Gayevskiy and Goddard 2016). However, there is evidence to suggest that S. uvarum and S. arboricola may have more ancient populations in NZ (Almeida et al. 2015; Gayevskiy and Goddard 2016). Following Gayevskiy and Goddard (2016), alignment for all 52 NZ S. cerevisiae genomes to reference genomes for the other Saccharomyces species shows that all align best to S. cerevisiae, with an average of 97.2%, and a minimum of 93.2% (Table S3, Supporting Information). The two Saccharomyces species to which the NZ S. cerevisiae align the poorest are the two candidates for potential endemic NZ species, with a maximum of just 43%. Further, there is no evidence for large blocks of NZ S. cerevisiae genomes to be more greatly related to any species other that S. cerevisiae (Table S3). Together, this provides no evidence for recent hybridisation or introgression event in the NZ S. cerevisiae group from other species. Thus, given the data available, it appears the NZ S. cerevisiae population derives exclusively from the European/wine group. Further, the earlier ancestry profile analysis shows that most of the NZ strains have the majority of their ancestry in the wine/European group—i.e. most NZ strains are ‘clean’ wine/European strains.
Number and timing of NZ incursion events
It is clear that the NZ S. cerevisiae population derived from Europe. But how many times might strains have been transferred from one side of the planet to the other, and when did this occur? We define incursion events as transfers to NZ that have become established enough for us to detect strains, or related lineages deriving from such strains, which are thus founder events. The theoretical number of incursion events ranges from just 1 to ∼2000 as this represents the best estimate for the number of different S. cerevisiae genotypes currently present in NZ (Knight and Goddard 2015). First, we evaluate whether there is any evidence to support a single founder event versus multiple incursions given these data. A single founder event would mean that all current NZ S. cerevisiae would coalesce to a single ancestor. One signal for this would be the presence of shared fixed alleles in the NZ but not European population. Of the 66 316 SNPs, none are fixed in the NZ genomes, providing no strong evidence for a single founder event.
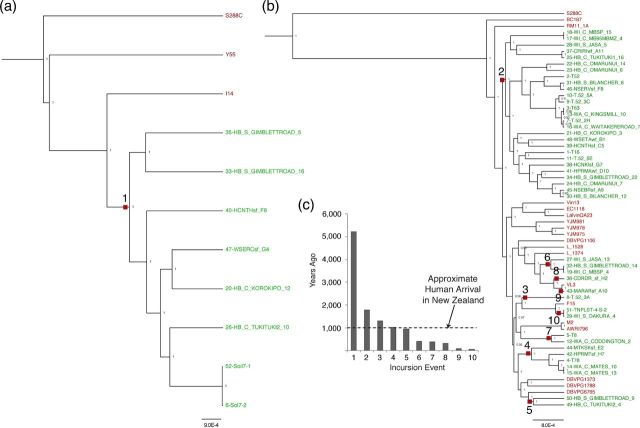
We acknowledge the tentative nature of this analyses given the relatively few strains for which we have sequences, but wished to estimate the likely minimum number of incursion events given our data to provide a lower bound to this rate, and appropriately used a maximum parsimony approach to analyse this. Under this minimal change parsimony framework, the best explanation for clades entirely comprised of NZ-derived genomes is that the ancestor of this clade was transported to NZ from Europe. Thus, we modelled the minimum possible incursion events into NZ by minimising the change from ‘wine/European’ to ‘NZ’ strain status over the phylogeny (Fig. 4). The minimum number of transfer events inferred by this analysis is 10 (one and nine in each of the two clades where NZ isolates are present). By comparisons to null distributions, this observed number of incursion events is significantly less than we would expect to see by chance (P = 0.0116 and P < 0.0001 for each clade given 10 000 permutations of terminal taxa status) given these phylogenies and proportions of NZ and European-derived genomes. As an alternate approach, strains that survived transport to and establishment in NZ might tend to sire independent lineages, and this signal might be revealed by the presence of separate subpopulations in NZ. To test this, we analysed the population structure in these genomes using Structure (Pritchard, Stephens and Donnelly 2000), and the optimal number of inferred subpopulations is 11 across the two clades that harbour NZ-derived genomes. The inferred number of subpopulations is in line with the number of incursion events suggested by parsimony analyses.
Figure 4.
Phylogenies of the (a) NZ/Europe and (b) wine/Europe/NZ clades seen in Fig. 1. NZ strains are labelled in green, while other strains are in red. Maximally parsimonious incursion events are shown as red boxes and labelled numerically corresponding to their estimated dates plotted in (c).
It appears the movement of S. cerevisiae from Europe to NZ is thus not only detectable but also constrained. The question under scrutiny here is the extent to which humans have expanded microbial species ranges. Just because we infer at least 10 incursion events from Europe, this does not necessarily prove that humans were the agents of transfer; S. cerevisiae might have been moved by other means and been present before humans arrived. Humans only arrived in NZ about 1000 years ago, and winemaking only in the last ∼200 years (Hurles, Matisoo-Smith and Gray 2003), so next we attempted to estimate the ages of the NZ clades. Again, under a parsimony framework, these potentially represent the ages of lineages and populations that have expanded since their ancestors arrived in NZ. Given our data, the substitution rate between S. paradoxus and S. cerevisiae is 0.3366. Dating microbial phylogenies is difficult, and the time to the common ancestor of S. cerevisiae and S. paradoxus has been estimated between 0.4 and 3.4 mya (Liti, Barton and Louis 2006). If the molecular substitution rate is assumed to be constant across this time period, then the potential ages of these 10 clades may be simply estimated by calculating the proportional distance of the relevant nodes compared to the node defining the S. paradoxus/S. cerevisiae split. With this approach, the lower bound timing estimates of S. cerevisiae incursions into NZ from Europe spans from ∼60 to 5000 years ago (Fig. 4). However, we note that large confidence limits around the timing of the S. paradoxus and S. cerevisiae split (Liti, Barton and Louis 2006) clearly translate into large limits around the estimates for incursions into NZ. Without wanting to overly extrapolate these tentative timings, it is interesting to note that most inferred incursion events are just above or well below the 1000 year cut-off, and just one is substantially older. This one ‘older’ event is the inferred incursion event from the smaller sister clade to the wine/European group, where eight NZ-derived genomes cluster with soil isolates from Europe (Fig. 4). Given the uncertain nature of the dating of this phylogeny combined with the assumptions of constant substitution rates, apart from one possible exception, there is no compelling evidence to suggest that S. cerevisiae has been in NZ significantly longer than humans have. Thus, human introduction appears the most likely explanation for S. cerevisiae's presence in NZ.
Lastly, we estimated the node containing the entire wine/European/NZ group to be between 4635 and 39 394 years old. This estimate overlaps with the earliest evidence for humans producing fermented drinks some 9000 years ago in China (McGovern et al. 2004), and this places all the S. cerevisiae found in NZ in the group that expanded along with the human passion for viticulture and winemaking.
DISCUSSION
These data reveal that the Saccharomyces cerevisiae inhabiting NZ originated from Europe. We estimate a minimum of 10 incursion events founded the NZ population. It appears that this species has been transported to NZ and has become successfully established in vineyards, but that radiation to native forest habitats is rare, but detectable. This may be due to low rates of movement or that this S. cerevisiae population is poorly adapted to NZ native forest niches, or both. Saccharomyces arboricola inhabits NZ native forests, but unlike S. cerevisiae populations in habiting Brazilian native forests, which hybridised with endemic S. paradoxus, there is no evidence that the NZ S. cerevisiae have hybridised with endemic S. arboricola. Due to the very recent arrival of S. cerevisiae to NZ, perhaps this is occurring currently, or will do soon.
Permutation analyses suggest that the rate of incursion into NZ is not rampant. However, analyses estimating the number of incursions might produce an erroneous result not necessarily due to analytical reasons, but mostly due to the unequal number of samples deriving from NZ and European populations. Recall the NZ strains were deliberately chosen to represent the genetic diversity in NZ based on surveys from both vineyard and native forest habitats, but it might well be that with increased sampling in the European wine group one finds strains that interdigitate among NZ strains in various clades. This would elevate the number of estimated transitions from Europe. However, additional data may not show this pattern; either way, here we estimate the minimum number of incursion events given the data available. Under the assumption that the largest NZ clade was founded by a single strain, which has then radiated in NZ, one can compare metrics that might provide insights into the evolution of S. cerevisiae since arriving in NZ. The largest NZ clade (defined by node 2 in Fig. 4) has values of π = 1909, θw = 2114 and Tajima's D = –0.41. This compares to π = 1155, θw = 1305 and Tajima's D = –0.43 for a set of 10 wine/European Genomes, and implies possibly greater genetic diversity, but no more compelling signal for selection, in this NZ clade compared to the wine/European group generally.
The estimates concerning timings of these incursion events are less certain. This is due to the problems associated with dating microbial phylogenies in general, owing to the lack of fossils, and then extrapolating the uncertain estimates we have to relatively recent divergence events. While the mutation rate of S. cerevisiae has been estimated (Lang and Murray 2008), this has been deduced using a few strains under laboratory conditions. A further complication is that we have very little idea of absolute mitotic and meiotic generation times in nature, making calibrations of absolute timings using mutation rates a fruitless way forward (Goddard and Greig 2015). Here we make assumptions about the constancy of the rates of molecular evolution. Given these caveats, we estimate the likely timings of these incursions, and their lower bounds are not greatly above, and indeed are mostly below 1000 years ago (Fig. 4). Another possible reason for an inaccurate inference in terms of transfer timings also stems from a lack of sampling: strains might have migrated only very recently to NZ even though their last common ancestor with a European strain occurred thousands of years ago.
Previous analyses, using repeat regions, showed the NZ S. cerevisiae population as internationally genetically distinct (Goddard et al. 2010). The analysis of whole genomes here does not agree with this. This discrepancy might be explained by the fact that repeat loci evolve rapidly and thus are capable of resolving finer levels of population differentiation than average signals from whole genomes (or many loci) can. Significant signals for differentiation revealed by analyses using repeat regions would occur if rates of gene flow (incursion events) between Europe and NZ are relatively low, and less than the rates of evolution at these repeat regions: analysis here suggests the number of incursion events into NZ have not been that great, and thus correlates with this idea. The sequencing of diploid genomes here allows rates of heterozygosity to be calculated, and these are on average similar to those previously estimated (Magwene et al. 2011); however, the variance in rates of heterozygosity in the NZ genomes is substantial (from 6% to 43% of SNPs). Such rates of heterozygosity may be explained, at least in the NZ group, by the inference that ∼20% of mating events are outbred combined with reasonable levels of gene flow between subpopulations (Knight and Goddard 2015), and that the European/ wine group more generally have elevated outcrossing rates (Peter and Schacherer 2016).
Together, these estimates of origins and timings strongly suggest that humans introduced S. cerevisiae into NZ recently, and thus expanded the range of this species. This pattern correlates with the previous observation of S. cerevisiae presence in new oak barrels from Europe once arrived in NZ (Goddard et al. 2010). The signals provided by these phylogenomic analyses are also in line with work showing trends in S. cerevisiae population division that correlates with the expansion of viticulture globally (Legras et al. 2007). This extent of human-mediated movement is also consistent with the analyses of cocoa and coffee populations in Africa and Europe, where approximately three significant movements from Europe, North America and Asia to Africa and South America were inferred, and migration of wine strains to Brazil which hybridised with S. paradoxus. However, there were no estimations regarding the timing of either the Brazil or cocoa and coffee strain movements (Barbosa et al. 2016; Ludlow et al. 2016). In addition, analysis of a handful of S. paradoxus isolates from NZ also infers transfer from Europe to NZ associated with the movement of another plant species by humans: Quercus (oak trees) (Zhang et al. 2010). It is interesting to note that S. uvarum is inferred to have been present in Australasia, and possibly S. arboricola in NZ, well before humans might have been, and so it seems that the ranges, modes and ages of dispersal of these sister taxa differ.
The earliest evidence for the human use of S. cerevisiae for fermentation has been dated to ∼9000 years ago and comes from pottery jars in China (McGovern et al. 2004). The earliest evidence for wine production comes from Iran ∼7400 years ago and seeds of domesticated grapes have been found in Georgia and Turkey and dated to ∼8000 years ago (This, Lacombe and Thomas 2006). Winemaking then spread to adjacent areas and around the Mediterranean ∼5000–5500 years ago (This, Lacombe and Thomas 2006). Our results show that strains associated with winemaking are closely related to one another regardless of geographic location. The archaeological dates allow another way to calibrate the dating on this phylogeny and indicate that the split between S. cerevisiae and S. paradoxus is closer to the lower bound of 0.4 mya than the upper of 3.4 mya.
Overall, the analyses conducted here add further support to the concept that humans have facilitated the global transfer of this microbial species through our agricultural activities, and thus have significantly expanded this species’ range. In doing so, it appears that humans have provided an opportunity for one lineage of S. cerevisiae to radiate to and become established in areas well beyond the ancestral range for this species. Not only has the transfer of this species provided an opportunity for it to become established in NZ's agricultural ecosystems, but it is now also found in natural forest ecosystems. Whether S. cerevisiae has or may become established in NZ native forest ecosystems is debatable as the low rate of recovery may simply reflect rare transposition events, by humans or insects perhaps (Buser et al. 2014), that will perhaps ultimately fail to seed successful lineages in that inhabit. Indeed, we do not have a good understanding of the niches to which S. cerevisiae is adapted, if any at all (Goddard and Greig 2015). Alternatively, it is possible that S. cerevisiae may become established in native habitats. NZ has a list of human introduced invasive species that has decimated portions of endemic ecosystems—stoats and rats destroying native NZ birds as a prime example (Norton 2009). The interesting question is whether S. cerevisiae is classed as an invasive species in NZ: while this species has been introduced by humans, at the moment its invasion is primarily restricted to agricultural ecosystems, where it arguably adds value.
Supplementary Material
Acknowledgments
The completion of this research would not have been possible without the support and assistance of the many collaborating companies who allowed access to their land and donated juice: Amisfield, Ata Rangi, Coal Pit, Constellation, Delegats, Domain Road, Frey Vineyard, Misha's Vineyard, Mt Difficulty, Mt Riley, Neudorf, Palliser, Pernod Ricard, Rippon, Seifried, Te Kairanga, Tohu, Trinity Hill, Villa Maria and Vita Brevis.
SUPPLEMENTARY DATA
FUNDING
This work was supported by Faculty Research Development Fund (FRDF) (grant ), , the , and grants to MRG.
Conflict of interest. None declared.
REFERENCES
- Aa E, Townsend JP, Adams RI, et al. Population structure and gene evolution in Saccharomyces cerevisiae. FEMS Yeast Res. 2006;6:702–15. doi: 10.1111/j.1567-1364.2006.00059.x. [DOI] [PubMed] [Google Scholar]
- Almeida P, Barbosa R, Zalar P, et al. A population genomics insight into the Mediterranean origins of wine yeast domestication. Mol Ecol. 2015;24:5412–27. doi: 10.1111/mec.13341. [DOI] [PubMed] [Google Scholar]
- Almeida P, Gonçalves C, Teixeira S, et al. A Gondwanan imprint on global diversity and domestication of wine and cider yeast Saccharomyces uvarum. Nat Commun. 2014;5:4044. doi: 10.1038/ncomms5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2012. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (15 October 2016, date last accessed) [Google Scholar]
- Avise J, Arnold J, Ball R, et al. Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Annu Rev Ecol Syst. 1987;18:489–522. [Google Scholar]
- Barbosa R, Almeida P, Safar SVB, et al. Evidence of natural hybridization in Brazilian wild lineages of Saccharomyces cerevisiae. Genome Biol Evol. 2016;8:317–29. doi: 10.1093/gbe/evv263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beheregaray LB. Twenty years of phylogeography: the state of the field and the challenges for the Southern Hemisphere. Mol Ecol. 2008;17:3754–74. doi: 10.1111/j.1365-294X.2008.03857.x. [DOI] [PubMed] [Google Scholar]
- Bergstrom A, Simpson JT, Salinas F, et al. A high-definition view of functional genetic variation from natural yeast genomes. Mol Biol Evol. 2014;31:872–88. doi: 10.1093/molbev/msu037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buser CC, Newcomb RD, Gaskett AC, et al. Niche construction initiates the evolution of mutualistic interactions. Ecol Lett. 2014;17:1257–64. doi: 10.1111/ele.12331. [DOI] [PubMed] [Google Scholar]
- Cherry JM, Hong EL, Amundsen C, et al. Saccharomyces genome database: the genomics resource of budding yeast. Nucleic Acids Res. 2011;40:D700–5. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie GA, Hyma KE, Ludlow CL, et al. Genomic sequence diversity and population structure of Saccharomyces cerevisiae assessed by RAD-seq. G3. 2013;3:2163–71. doi: 10.1534/g3.113.007492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling ACE, Mau B, Blattner FR, et al. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit R, Bouvier T. “Everything is everywhere, but, the environment selects;” what did Baas Becking and Beijerinck really say? Environ Microbiol. 2006;8:755–8. doi: 10.1111/j.1462-2920.2006.01017.x. [DOI] [PubMed] [Google Scholar]
- del Campo J, Sieracki ME, Molestina R, et al. The others: our biased perspective of eukaryotic genomes. Trends Ecol Evol. 2014;29:252–9. doi: 10.1016/j.tree.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Philippe H. Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet. 2005;6:361–75. doi: 10.1038/nrg1603. [DOI] [PubMed] [Google Scholar]
- Diamond J. Evolution, consequences and future of plant and animal domestication. Nature. 2002;418:700–7. doi: 10.1038/nature01019. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Ho SYW, Phillips MJ, et al. Relaxed phylogenetics and dating with confidence Penny D, ed. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214–8. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl DA, vonHoldt BM. Structure harvester: a website and program for visualizing structure output and implementing the Evanno method. Conserv Genet Resour. 2011;4:359–61. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol. 2005;14:2611–20. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Fay JC, Benavides JA. Evidence for domesticated and wild populations of Saccharomyces cerevisiae. PLoS Genet. 2005;1:66–71. doi: 10.1371/journal.pgen.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayevskiy V, Goddard MR. Geographic delineations of yeast communities and populations associated with vines and wines in New Zealand. ISME J. 2012;6:1281–90. doi: 10.1038/ismej.2011.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayevskiy V, Goddard MR. Saccharomyces eubayanus and Saccharomyces arboricola reside in North Island native New Zealand forests. Environ Microbiol. 2016;18:1137–47. doi: 10.1111/1462-2920.13107. [DOI] [PubMed] [Google Scholar]
- Gayevskiy V, Klaere S, Knight S, et al. ObStruct: a method to objectively analyse factors driving population structure using Bayesian ancestry profiles Pajewski NM, ed. PLoS One. 2014;9:e85196. doi: 10.1371/journal.pone.0085196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard MR, Anfang N, Tang R, et al. A distinct population of Saccharomyces cerevisiae in New Zealand: evidence for local dispersal by insects and human-aided global dispersal in oak barrels. Environ Microbiol. 2010;12:63–73. doi: 10.1111/j.1462-2920.2009.02035.x. [DOI] [PubMed] [Google Scholar]
- Goddard MR, Greig D. Saccharomyces cerevisiae: a nomadic yeast with no niche? FEMS Yeast Res. 2015;15:fov009. doi: 10.1093/femsyr/fov009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, et al. Life with 6000 Genes. Science. 1996;274:546–67. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- Hanson CA, Fuhrman JA, Horner-Devine MC, et al. Beyond biogeographic patterns: processes shaping the microbial landscape. Nat Rev Microbiol. 2012;10:497–506. doi: 10.1038/nrmicro2795. [DOI] [PubMed] [Google Scholar]
- Hittinger CT. Saccharomyces diversity and evolution: a budding model genus. Trends Genet. 2013;29:309–17. doi: 10.1016/j.tig.2013.01.002. [DOI] [PubMed] [Google Scholar]
- Huang C, Roncoroni M, Gardner RC. MET2 affects production of hydrogen sulfide during wine fermentation. Appl Microbiol Biot. 2014;98:7125–35. doi: 10.1007/s00253-014-5789-1. [DOI] [PubMed] [Google Scholar]
- Hurles ME, Matisoo-Smith E, Gray RD. Untangling Oceanic settlement: the edge of the knowable. Trends Ecol Evol. 2003;18:531–40. [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–67. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight S, Goddard MR. Quantifying separation and similarity in a Saccharomyces cerevisiae metapopulation. ISME J. 2015;9:361–70. doi: 10.1038/ismej.2014.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight S, Klaere S, Fedrizzi B, et al. Regional microbial signatures positively correlate with differential wine phenotypes: evidence for a microbial aspect to terroir. Sci Rep. 2015;5:14233. doi: 10.1038/srep14233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang GI, Murray AW. Estimating the per-base-pair mutation rate in the yeast Saccharomyces cerevisiae. Genetics. 2008;178:67–82. doi: 10.1534/genetics.107.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Legras J-L, Merdinoglu D, Cornuet J-M, et al. Bread, beer and wine: Saccharomyces cerevisiae diversity reflects human history. Mol Ecol. 2007;16:2091–102. doi: 10.1111/j.1365-294X.2007.03266.x. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, et al. Subgroup 1GPDP The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litchman E. Invisible invaders: non-pathogenic invasive microbes in aquatic and terrestrial ecosystems. Ecol Lett. 2010;13:1560–72. doi: 10.1111/j.1461-0248.2010.01544.x. [DOI] [PubMed] [Google Scholar]
- Liti G, Barton DBH, Louis EJ. Sequence diversity, reproductive isolation and species concepts in Saccharomyces. Genetics. 2006;174:839–50. doi: 10.1534/genetics.106.062166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, Carter DM, Moses AM, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–41. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Bolger AM, Nagel A, et al. RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res. 2012;40:W622–7. doi: 10.1093/nar/gks540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludlow CL, Cromie GA, Garmendia-Torres C, et al. Independent origins of yeast associated with coffee and cacao fermentation. Curr Biol. 2016;26:965–71. doi: 10.1016/j.cub.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern PE, Zhang J, Tang J, et al. Fermented beverages of pre-and proto-historic China. P Natl Acad Sci USA. 2004;101:17593–8. doi: 10.1073/pnas.0407921102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. Mesquite: A Modular System for Evolutionary Analysis. 2014. http://mesquiteproject.org (15 October 2016, date last accessed) [Google Scholar]
- Magwene PM, Kayikci O, Granek JA, et al. Outcrossing, mitotic recombination, and life-history trade-offs shape genome evolution in Saccharomyces cerevisiae. P Natl Acad Sci USA. 2011;108:1987–92. doi: 10.1073/pnas.1012544108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny JBH, Bohannan BJM, Brown JH, et al. Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol. 2006;4:102–12. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- Mazzaglia A, Studholme DJ, Taratufolo MC, et al. Pseudomonas syringae pv. actinidiae (PSA) isolates from recent bacterial canker of kiwifruit outbreaks belong to the same genetic lineage. PLoS One. 2012;7:e36518. doi: 10.1371/journal.pone.0036518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison-Whittle P, Goddard MR. Quantifying the relative roles of selective and neutral processes in defining eukaryotic microbial communities. ISME J. 2015;9:1–9. doi: 10.1038/ismej.2015.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Korneliussen T, Albrechtsen A, et al. SNP calling, genotype calling, and sample allele frequency estimation from new-generation sequencing data. PLoS One. 2012;7:e37558. doi: 10.1371/journal.pone.0037558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton D. Species invasions and the limits to restoration: learning from the New Zealand experience. Science. 2009;325:569–71. doi: 10.1126/science.1172978. [DOI] [PubMed] [Google Scholar]
- Novo M, Bigey F, Beyne E, et al. Eukaryote-to-eukaryote gene transfer events revealed by the genome sequence of the wine yeast Saccharomyces cerevisiae EC1118. P Natl Acad Sci USA. 2009;106:16333–8. doi: 10.1073/pnas.0904673106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrós-Alió C. Marine microbial diversity: can it be determined? Trends Microbiol 200614257–63. [DOI] [PubMed] [Google Scholar]
- Peter J, Schacherer J. Population genomics of yeasts: towards a comprehensive view across a broad evolutionary scale. Yeast. 2016;33:73–81. doi: 10.1002/yea.3142. [DOI] [PubMed] [Google Scholar]
- Piskur J, Rozpedowska E, Polakova S, et al. How did Saccharomyces evolve to become a good brewer? Trends Genet. 2006;22:183–6. doi: 10.1016/j.tig.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Pretorius IS. Tailoring wine yeast for the new millennium: novel approaches to the ancient art of winemaking. Yeast. 2000;16:675–729. doi: 10.1002/1097-0061(20000615)16:8<675::AID-YEA585>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–59. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2011. http://www.R-project.org/ (15 October 2016, date last accessed) [Google Scholar]
- Richards KD, Goddard MR, Gardner RC. A database of microsatellite genotypes for Saccharomyces cerevisiae. Anton Leeuw. 2009;96:355–9. doi: 10.1007/s10482-009-9346-3. [DOI] [PubMed] [Google Scholar]
- Rokas A, Williams BL, King N, et al. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature. 2003;425:798–804. doi: 10.1038/nature02053. [DOI] [PubMed] [Google Scholar]
- Scannell DR, Zill OA, Rokas A, et al. The awesome power of yeast evolutionary genetics: new genome sequences and strain resources for the Saccharomyces sensu stricto genus. G3. 2011;1:11–25. doi: 10.1534/g3.111.000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schacherer J, Shapiro JA, Ruderfer DM, et al. Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature. 2009;458:342–5. doi: 10.1038/nature07670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serjeant K, Tang R, Anfang N, et al. Yeasts associated with the New Zealand Nothofagus honeydew system. New Zeal J Ecol. 2008;32:209–13. [Google Scholar]
- Skelly DA, Merrihew GE, Riffle M, et al. Integrative phenomics reveals insight into the structure of phenotypic diversity in budding yeast. Genome Res. 2013;23:1496–504. doi: 10.1101/gr.155762.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanini I, Dapporto L, Legras J-L, et al. Role of social wasps in Saccharomyces cerevisiae ecology and evolution. P Nat Acad Sci USA. 2012;109:13398–403. doi: 10.1073/pnas.1208362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strope PK, Skelly DA, Kozmin SG, et al. The 100-genomes strains, an S. cerevisiae resource that illuminates its natural phenotypic and genotypic variation and emergence as an opportunistic pathogen. Genome Res. 2015;25:762–74. doi: 10.1101/gr.185538.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–95. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot JM, Bruns TD, Taylor JW, et al. Endemism and functional convergence across the North American soil mycobiome. P Natl Acad Sci USA. 2014;111:6341–6. doi: 10.1073/pnas.1402584111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MW, Tsai P, Anfang N, et al. Pyrosequencing reveals regional differences in fruit-associated fungal communities. Environ Microbiol. 2014;16:2848–58. doi: 10.1111/1462-2920.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- This P, Lacombe T, Thomas MR. Historical origins and genetic diversity of wine grapes. Trends Genet. 2006;22:511–9. doi: 10.1016/j.tig.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Tripathi BM, Lee-Cruz L, Kim M, et al. Spatial scaling effects on soil bacterial communities in Malaysian tropical forests. Microb Ecol. 2014;68:247–58. doi: 10.1007/s00248-014-0404-7. [DOI] [PubMed] [Google Scholar]
- Valverde A, Tuffin M, Cowan DA. Biogeography of bacterial communities in hot springs: a focus on the actinobacteria. Extremophiles. 2012;16:669–79. doi: 10.1007/s00792-012-0465-9. [DOI] [PubMed] [Google Scholar]
- Wallis GP, Trewick SA. New Zealand phylogeography: evolution on a small continent. Mol Ecol. 2009;18:3548–80. doi: 10.1111/j.1365-294X.2009.04294.x. [DOI] [PubMed] [Google Scholar]
- Wang Q-M, Liu W-Q, Liti G, et al. Surprisingly diverged populations of Saccharomyces cerevisiae in natural environments remote from human activity. Mol Ecol. 2012;21:5404–17. doi: 10.1111/j.1365-294X.2012.05732.x. [DOI] [PubMed] [Google Scholar]
- Watterson GA. On the number of segregating sites in genetical models without recombination. Theor Popul Biol. 1975;7:256–76. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Wichmann MC, Alexander MJ, Soons MB, et al. Human-mediated dispersal of seeds over long distances. P Roy Soc B: Biol Sci. 2009;276:523–32. doi: 10.1098/rspb.2008.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Skelton A, Gardner RC, et al. Saccharomyces paradoxus and Saccharomyces cerevisiae reside on oak trees in New Zealand: evidence for migration from Europe and interspecies hybrids. FEMS Yeast Res. 2010;10:941–7. doi: 10.1111/j.1567-1364.2010.00681.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We have made our raw sequence data and consensus genomes aligned to S288C (Goffeau et al. 1996; EBI:GCA_000146045.2) publicly available at SRA: SRP042301 and BioProject: PRJNA247448.