Abstract
Selenophosphate synthetase (SPS) was initially detected in bacteria and was shown to synthesize selenophosphate, the active selenium donor. However, mammals have two SPS paralogs, which are designated SPS1 and SPS2. Although it is known that SPS2 catalyzes the synthesis of selenophosphate, the function of SPS1 remains largely unclear. To examine the role of SPS1 in mammals, we generated a Sps1 knockout mouse and found that systemic SPS1 deficiency led to embryos that were clearly underdeveloped by E8.5 and virtually resorbed by E14.5. The knockout of Sps1 in the liver preserved viability, but significantly affected the expression of a large number of mRNAs involved in cancer, embryonic development, and the glutathione system. Particularly notable was the extreme deficiency of glutaredoxin 1 (GLRX1) and glutathione-S-transferase omega 1. To assess these phenotypes at the cellular level, we targeted the removal of SPS1 in F9 cells, a mouse embryonal carcinoma cell line, which affected the glutathione system proteins and accordingly led to the accumulation of hydrogen peroxide in the cell. Further, we found that several malignant characteristics of SPS1-deficient F9 cells were reversed, suggesting that SPS1 played a role in supporting and/or sustaining cancer. In addition, the overexpression of mouse or human GLRX1 led to a reversal of observed increases in reactive oxygen species (ROS) in the F9 SPS1/GLRX1-deficient cells and resulted in levels that were similar to those in F9 SPS1-sufficient cells. The results suggested that SPS1 is an essential mammalian enzyme with roles in regulating redox homeostasis and controlling cell growth.
Keywords: cancer, reactive oxygen species (ROS), redox regulation, selenium, selenocysteine, selenophosphate synthetase 1
INTRODUCTION
Selenophosphate synthetase (SPS) was originally discovered in Escherichia coli, designated SelD [1] and was shown to synthesize monoselenophosphate from selenide and ATP [2]. Monoselenophosphate, in turn, donates selenium to an intermediate synthesized from serine attached to its tRNA to generate selenocysteine (Sec) tRNA[Ser]Sec in both bacteria [1] and eukaryotes [3]. SPS1, a protein that is highly homologous to SelD, was subsequently reported in mammals and initially thought to be SelD [4,5]. However, a second SPS protein, SPS2, was also detected in mammals, and interestingly, was determined to be a selenoprotein [6]. The fact that SPS2 is a selenoprotein suggested that it might have an autoregulatory role in selenoprotein synthesis [6,7]. Several studies have suggested that SPS2 is responsible for selenophosphate synthesis and that SPS1 plays an alternative role. As well, both in vitro and in vivo studies have subsequently demonstrated that SPS2 synthesizes monoselenophosphate for generating Sec and that SPS1 is not involved in the synthesis of Sec in mammals (see [8,9] and references therein).
However, the role of SPS1 in selenium metabolism has not yet been determined. Tamura et al. and the results from earlier studies suggested that SPS1 might have a role in recycling Sec in mammalian cells by a selenium salvage system (see [10] and references therein). SPS1 was also found to exist in a complex with selenocysteine synthase (SecS) and the transfection of selenocysteine tRNA-associated protein (Secp43) into monkey embryonic cells enhanced the formation of complexes of all three proteins and promoted their migration to the nucleus [11]. Assou et al. reported that SPS1 occurred within a group of limited genes (~500–600) that shared a common expression signature in early development and/or differentiation in both human mature oocytes and embryonic stem cells; thus, SPS1 might play an important role in development/differentiation [12]. However, the definitive role of SPS1 in liver development and function has not been determined.
SPS1 is known to be an essential protein in Drosophila, as demonstrated by the knockout of Sps1, which resulted in the loss of imaginal disc formation and was embryonic lethal [13]. Targeted removal of Sps1 mRNA in Drosophila SL2 cells resulted in mega-mitochondria formation as a result of an accumulation of glutamine [14]. As well, SPS1 was reportedly implicated in cellular defense and cell proliferation via the regulation of vitamin B6 synthesis [15]. The latter study also demonstrated an indirect involvement of SPS1 in the regulation of Sec synthesis, wherein SPS1 deficiency resulted in the down-regulation of genes involved in pyridoxal phosphate (PLP, an active form of vitamin B6), which is used as a cofactor of selenocysteine lyase (SCL), D-selenocysteine, α, β-lyase [16], and SecS [9]. It was also reported that SCL interacted with SPS1 [17]. Further, the fact that SPS1 is overexpressed in rectal carcinoma cells suggested that SPS1 levels are related to cancer development [18]. In addition to growth retardation and induction of the cellular defense system, SPS1 deficiency also led to the accumulation of reactive oxygen species (ROS) in Drosophila both in vivo and in vitro [14, 19].
Because the precise function of SPS1 is poorly understood, we undertook a study to elucidate the role of this protein in mammals using mouse models and cell culture. We generated a systemic Sps1 knockout in mice and found that the removal of Sps1 caused embryonic lethality. However, the targeted removal of Sps1 in the liver was not lethal, and transcriptome analysis revealed changes in the expression of genes that regulate cellular redox potential. The regulation of redox potential by SPS1 was confirmed using the mouse F9 embryonal carcinoma (EC) cell line, in which SPS1 deficiency resulted in the loss of some cancer characteristics.
EXPERIMENTAL
Materials
Anti-thioredoxin reductase 1 (TR1), anti-glutathione peroxidase 4 (GPx4), and anti-selenoprotein W (SelW) antibodies were purchased from Epitomics; anti-SPS1, anti-glutaredoxin 1 (GLRX1), and anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibodies, pyridoxal 5′-phosphate hydrate, semicarbazide, and NaOH were purchased from Sigma-Aldrich as well as NADPH, 5,5′-dithiobis-2-nitrobenzoic acid (DTNB), and gelatin (type A) used in the cell invasion assays. The anti-glutathione S-transferase omega (GSTO1) antibodies were obtained from Proteintech Group; anti-glutathione peroxidase 1 (GPx1) antibodies were purchased from Abcam; anti-SPS2 antibodies were obtained from Rockland; and anti-rabbit HRP conjugated secondary antibodies were purchased from Cell Signaling Technology. Dulbecco’s phosphate-buffered saline (DPBS), Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum (FBS), penicillin/streptomycin solution, puromycin, and polyacrylamide electrophoresis gels (NuPAGE®) were obtained from Life Technologies as well as the Roswell Park Memorial Institute (RPMI) 1640 medium without phenol red. Polyvinylidene fluoride (PVDF) membranes, the iScript™ cDNA synthesis kit, and SYBR® green were obtained from Bio-Rad. The TriPure isolation reagent and protease inhibitor cocktail tablets were purchased from Roche. The trypan blue stain was obtained from Lonza, and the Triton™ X-100 solution, BCA protein assay kit, and SuperSignal™ West Dura Extended Duration Substrate were purchased from ThermoScientific. LipoD293™ was obtained from SignaGen Laboratories, and the MammoCult™ Human Medium Kit was obtained from Stem Cell Technologies. The Growth Factor Reduced Matrigel™ was purchased from BD Bioscience, the Matrigel®-coated invasion chamber was obtained from Corning®, and 5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate (CM-H2DCFDA), dihydroethidium (DHE), MitoSOX™, TRIzol® reagent, and Lipofectamine® 2000 were obtained from Invitrogen. Restriction endonucleases were purchased from Enzynomics (Korea). Moloney murine leukemia virus reverse transcriptase (MoMuLV-RT) and RNase inhibitor were purchased from Promega. The CapSure® Macro LCM caps and QIAamp® DNA FFPE tissue kit were purchased from Applied Biosystems and Qiagen, respectively. The superoxide dismutase (SOD) activity assay kit was purchased from Dojindo Molecular Technologies (Japan). Glycine and perchloric acid were purchased from AMRESCO and JUNSEI, respectively. The μ-slide 8 well was purchased from Ibidi (Germany).
Construction of the conditional knockout targeting vector
The targeting vector was constructed with the neomycin resistance gene (Neo) flanked by loxP and Frt sites, Exon 2 of Sps1 flanked by loxP sites, and the regions upstream and downstream of Sps1 as shown in Figure S1. The targeting vector was linearized with NotI and electroporated into v6.4 (C57BL/6 x 129/SvJae) embryonic stem (ES) cells [20]. Candidate ES cell clones were screened for homologous recombination by 5′-end and 3′-end junction polymerase chain reaction (PCR) using the primers SPS1 gF4 + pPNT SR1 and pPNT SF2 + SPS1 gR2 (Table S1), respectively, and the resulting cells carrying the Sps1fl/Neofl allele were used to generate chimeric mice.
Generation of SPS1 knockout mice and embryo analysis
Homologous recombinant ES cell clones carrying the Sps1fl/Neofl allele were injected into C57BL/6 blastocysts and transferred to pseudopregnant females [20]. The resulting high percentage of chimeras (90% or greater based on coat color) were mated to wild type C57BL/6 mice (Jackson Labs) and the genomic DNA isolated from F1 offspring tail samples was analyzed for germline transmission. Mice carrying floxed Sps1 and containing Neo were crossed with mice expressing flippase (FLP) recombinase (C57BL/6) to remove Neo. Genomic DNA was isolated from mouse tails and screened for the loss of Neo by PCR using the SPS1 gF6 and SPS1 gR6 primers (Table S1). To obtain a standard Sps1 knockout, mice carrying Sps1fl/fl were mated with transgenic mice carrying EIIa-Cre (C57BL/6). Genomic DNA isolated from F1 offspring tail samples was analyzed for the loss of the targeted Sps1 sequence by PCR using the SPS1 gF6 and SPS1 gR6 primers (Table S1).
Heterozygous knockout Sps1 mice were mated and embryos were examined at E8.5, E10.5, E11.5, E12.5, and E14.5, where E0.5 was defined as noon on the day a mating plug was detected. For histological analysis, embryos were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) overnight at 4°C, dehydrated, and embedded in paraffin wax for sectioning. The sections (5 μm) were stained with hematoxylin and eosin, and images were acquired using an Axioimazer A1 (Zeiss). To isolate genomic DNA from the sectioned embryos, LCM was performed with a Veritas™ LCC1704 (Arcturus), using CapSure® Macro LCM caps to capture the embryo from the decidua. The capturing laser conditions were 100 mW infrared laser power and 10,000 μsec pulse time. Genomic DNA was extracted from the captured embryo fragments collected on the caps using the QIAamp® DNA FFPE tissue kit. Genotyping was performed by PCR using two primer sets: SPS1 WT-F and SPS1 WT-R for detecting the wild type (WT) allele, and SPS1 KO-F and SPS1 KO-R for detecting the knockout (KO) allele.
Liver-specific conditional knockout mice (Alb-Cre; Sps1fl/fl) were generated by initially mating Alb-Cre transgenic mice in a C57BL/6 background with Sps1fl/fl mice. The F2 generation offsprings with the Alb-Cre; Sps1fl/fl genotype were mated with Sps1fl/fl mice, and the resulting Sps1fl/fl mice, which served as controls, and Alb-Cre; Sps1fl/fl mice were used for the analyses. Mice were analyzed at 8–10 weeks of age.
All procedures performed involving the mice were conducted in accordance with the Institutional Guidelines of the National Institutes of Health (NIH, NCI, Bethesda, MD, USA) and the Institute of Laboratory Animal Resources (Seoul National University, Seoul, Korea). All mouse experiments were approved by the Animal Ethics Committee at the National Institutes of Health and the Institutional Animal Care and Use Committee at Seoul National University.
Cell culture and cell growth rate
The mouse EC F9 cell line was obtained from the American Type Culture Collection (ATCC) and GP2-293 retroviral packaging cells were obtained from Clontech. The F9 and GP2-293 cell lines were maintained in DMEM supplemented with 10% FBS, penicillin (50 U/ml), and streptomycin (50 μg/ml) in a humidified atmosphere containing 5% CO2 at 37°C. To determine cell growth rates, the F9 cells were seeded in 24-well plates (5 × 103 cells/well) coated with 0.1% gelatin, and living cells were counted using the Trypan blue exclusion assay at the indicated time points.
Microarray analysis
Total RNA was isolated from the livers of control (Sps1fl/fl) and Sps1 liver knockout (Alb-Cre; Sps1fl/fl) mice, and from control and Sps1 knockdown F9 cells using the TriPure isolation reagent according to the manufacturer’s protocol. Microarray analysis was performed using the Affymetrix GeneChip® Mouse Genome 430 2.0 Array (n=3 per group). The results were analyzed by ANOVA and genes whose expression was significantly different from control mice (P < 0.05) were subjected to Ingenuity Pathway Analysis (IPA, v.7.5). Microarray data from the livers of control and Sps1 liver knockout mice as well as control and F9/shSPS1 cells are accessible through Gene Expression Omnibus Series accession number GSE74677 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE74677).
Preparation of SPS1 knockdown cells using retroviral transduction
The RNAi target sequences for Sps1 were placed into a pSUPER-retro vector according to the manufacturer’s instructions. The primer sequences used for the knockdown of Sps1 are provided in Table S1. The constructs, including Sps1 knockdown and its control vector, were transfected into GP2-293 cells using LipoD293™ following the manufacturer’s instructions. The medium was replaced with fresh DMEM 24 h following transfection, and the cell culture supernatants, including the virus, were harvested after an additional 24 h and incubated with F9 cells. The transfected cells were selected in the presence of 2 μg/ml puromycin.
Mouse and human Glrx1 overexpression vector construction and establishment of a stable expression cell line
To construct a mouse Glrx1 (mGLRX1) overexpression vector (pcDNA4/TO.mGLRX1), the open reading frame (ORF) of mGLRX1 was amplified from the cDNA prepared from F9 cells using the mGLRX1-KpnI-F and mGLRX1-EcoRI-R primers (Table S1). The PCR product was cloned into the KpnI/EcoRI sites of pcDNA4/TO, and pcDNA4/TO.mGLRX1 was transfected into F9/shSPS1 cells. To select for cells harboring pcDNA4/TO.mGLRX1, Zeocin™ (400 μg/ml) was added to the medium and the surviving colonies were selected. Single cell clones were isolated by diluting cells from each colony in a 96-well plate. To construct a human Glrx1 (hGLRX1) overexpression vector (pcDNA4/TO.hGLRX1), the same methods were employed except the hGLRX1 ORF was amplified from HeLa cDNAs using the BamHI-GLRX1-F and EcoRI-GLRX1-R primers (Table S1) and cloned into the BamHI/EcoRI sites of pcDNA4/TO. When the cells grew to a confluent state, each clone was transferred to a single well in a 24-well plate and allowed to expand. The cells were then analyzed for the expression of mouse Sps1, mouse Glrx1, and human Glrx1 by quantitative PCR (qPCR) as previously described [21]. Briefly, total RNA was isolated from each cell line using TRIzol®. First-strand cDNA was synthesized using Mo-MuLV reverse transcriptase and subjected to PCR using the following specific primer sets: for mouse Sps1, mSPS1-F and mSPS1-R; for mouse Glrx1, mGlrx1-F and mGlrx1-R; for human Glrx1, hGlrx1-F and hGlrx1-R; and for mouse Actb, ACTB-F and ACTB-R. The sequence of each primer is shown in Table S1.
Knockdown-resistant mSPS1 vector construction and establishment of a stable expression cell line
To construct a knockdown-resistant (rescue) SPS1 expression vector, three silent point mutations were introduced into the shRNA target sequence by two-step PCR [22]. In the first step, two DNA fragments (the 5′-half and the 3′-half) were amplified from the mouse F9 cell cDNA using two sets of primers: the SPS1 KI-F1 and the SPS1 KI-R1 for the 5′-half, and the SPS1 KI-F2 and the SPS1 KI-R2 for the 3′-half (Table S1; altered bases are under-lined). The PCR products were subjected to nested PCR using SPS1 KI-F1 and SPS1 KI-R2 to amplify the full-length SPS1 rescue construct containing the three silent mutations. The final SPS1 rescue construct was cloned into the BamHI and EcoRI restriction sites of pcDNA4/TO. The vector (pcDNA4/TO.mSPS1Rescue) was transfected into F9/shSPS1 cells and the transfected cells were selected in the presence of 400 μg/ml Zeocin™. Cells were then analyzed by qPCR for the expression of endogenous mouse Sps1, mouse Glrx1, and rescue mouse Sps1 as previously described [21]. Briefly, total RNA was isolated from each cell line, first-strand cDNA was synthesized, and the cDNA was subjected to PCR using the following specific primer sets: for endogenous mouse Sps1, mSPS1-F and mSPS1-R; for mouse Glrx1, mGlrx1-F and mGlrx1-R; for rescue mouse Sps1, mSPS1 (Rescue)-F and mSPS1 (Rescue)-R; and for mouse Actb, ACTB-F and ACTB-R. The sequence of each primer is shown in Table S1.
Western blotting
Cells were washed twice with DPBS and harvested in cold lysis buffer (DPBS with 0.5% Triton™ X-100 and protease inhibitors). The protein concentrations of the resulting cell extracts were measured using a BCA protein assay kit and 30 μg of total protein from each extract was electrophoresed on NuPAGE® polyacrylamide gels, transferred onto PVDF membranes, and incubated overnight at 4°C with antibodies against SPS1, SPS2, TRXR1, GPX1, GPX4, SELW, GLRX1, GSTO1, or GAPDH. Membranes were then washed with Tris-buffered saline containing 0.1% Tween™ 20 and incubated with secondary antibodies for 1 h. Immunolabeling was detected using the SuperSignal™ West Dura Extended Duration Substrate and exposed on x-ray films. The band intensities on Western blots were quantified using ImageJ software (NIH).
mRNA analysis from mouse liver and F9 cells using quantitative RT-PCR
Total RNA was isolated from tissues or cells using the TriPure isolation reagent according to the manufacturer’s instructions. Total RNA (500 ng) was reverse transcribed using the iScript™ cDNA synthesis kit, and qPCR was performed in triplicate using the iTaq™ Universal SYBR® Green Supermix according to the manufacturer’s instructions. The primer sequences used (primers 12–49) are shown in Table S1.
Quantification of glutathione (GSH) and glutathione disulfide (GSSG) and element concentrations in the mouse liver
Total GSH and GSSG levels in the liver and cultured cells were quantified using a GSSG/GSH quantification kit according to the manufacturer’s instructions (Dojindo Molecular Technologies). Livers from 8-week old control and Alb-Cre; Sps1fl/fl mice (n=4) were collected and Inductively Coupled Plasma-Optical Emission Spectrometry (ICP-OES) multi-element profiles were acquired by the South Dakota Agricultural Laboratories according to established procedures.
Measurement of intracellular PLP concentration
Intracellular PLP levels in F9 cells were determined using a previously described method [15] with slight modifications. Briefly, 2 × 107 cells were harvested and frozen at −70°C until analysis. Frozen cells were lysed by adding 600 μl of distilled water (DW) and vortexing thoroughly. After removing 500 μl of lysate, 40 μl of derivatization agent (250 mg/ml of both semicarbazide and glycine dissolved in DW) was added to the lysate, which was then vortexed briefly and incubated at room temperature in the dark for 30 min. Afterward, 50 μl of 60% HClO4 was added and the samples were mixed thoroughly for 1 min, centrifuged for 10 min at 13,200 rpm, and the supernatant was transferred to a new tube. The pH was adjusted to approximately 4.0 with 25% NaOH, and high performance liquid chromatography (HPLC) was performed using a Zorbax® SB-C18 column.
Measurement of SOD activity
Total SOD activity was measured using a commercial colorimetric SOD assay kit. Briefly, approximately 8 × 106 cells were harvested, washed once with PBS, and cell pellets were stored at −70°C until analysis. The cells were lysed by adding 500 μl of DW and by vortexing for 1 min. The lysate was serially diluted (1/10 scale) in dilution buffer and 20 μl of the diluted samples were used for the assay according to the manufacturer’s instructions. After the removal of cellular debris, the total protein in each sample was quantified using the Bradford assay. Manganese superoxide dismutase (MnSOD) activity was measured by adding potassium cyanide to the lysate in a final concentration of 1 mM before dilution. Cu/ZnSOD activity was calculated by subtracting MnSOD activity from total SOD activity.
Determining the type of ROS
The detection of intracellular ROS was carried out by DCFDA staining as previously described [14] with slight modifications. F9 EC cells (2 × 105) were plated onto a 0.1% gelatin-coated 35 mm dish one day before staining. The cells were incubated with 1 μM CM-H2DCFDA for 40 min at 37°C in 5% CO2, washed twice with RPMI 1640 without phenol red, and then observed under an EVOS® FL fluorescence microscope (Fisher Scientific) at an excitation wavelength of 470 nm.
The detection of superoxide was carried out by staining cells with DHE and MitoSOX™ as the cytosolic and mitochondrial superoxide probes, respectively. The staining procedures were followed as described in the manufacturer’s manual. Briefly, 2 × 105 cells were prepared as above and stained by adding DHE or MitoSOX™ to a final concentration of 1 μM and incubated at 37°C for 30 min. After washing with RPMI 1640 without phenol red, fluorescence signals were observed under an EVOS® FL fluorescence microscope at an excitation wavelength of 531 nm.
Cytosolic and mitochondrial hydrogen peroxide were detected using roGFP2-Orp1 fusion proteins as probes [24,25]. Because retroviral vectors do not replicate in F9 cells, the original probe cassettes (cytosolic roGFP2-Orp1 and mitochondrial roGPF2-Orp1) were digested with NdeI and XbaI and cloned into the pcDNA4.TO vector. Each vector was then transfected into each cell line (F9, F9/shSPS1, F9/shSP1/Rescue, and F9/shSPS1/oemGlrx1). After incubation for 24 h, 5 × 104 cells were plated in a single well of a μ-slide 8 well that had been coated with 0.1% gelatin and incubated for 12 h, washed with PBS, fixed with 3% paraformaldehyde for 7 min, and then observed under an LSM 700 confocal microscope (Zeiss). The ratio between the oxidized (405 nm) and reduced (488 nm) forms of the probes was calculated for each cell according to Morgan et al. [25]. The intensities of the 405 nm and 488 nm image from the same original field (100x magnification) were obtained separately as described [29]. The intensity of the 405 nm images was divided by the intensity of the 488 nm images to calculate the ratio. This procedure was repeated in six different fields for each cell line and the ratio images were created by dividing the 405 nm image by the 488 nm image pixel by pixel. The ImageJ ‘Fire’ Look Up Table (LUT) was used for creating false-color ratio pictures.
Measurement of ROS levels
The number of F9 cell lines that contained high amounts of ROS was measured using fluorescence-activated cell sorting (FACS) as previously described [26]. The cells were treated with CM-H2DCFDA, washed once with cold-RPMI without phenol red, and harvested. The mean fluorescent intensity was detected among a total of 50,000 cell counts from each sample, and the CM-H2DCFDA green fluorescence distributions were displayed on a histogram plot.
Soft agar assay
Anchorage-independent growth was assayed as previously described using a total of 1,000 cells from each of the stably transfected F9 cell lines, which were suspended in 0.35% agar in DMEM and spread onto 60 mm dishes masked with a basal layer of 0.7% agar in the medium [23]. The cells were incubated at 37°C and 5% CO2 for 10 days and then stained with p-iodonitrotetrazolium chloride overnight.
Invasion assay
Cell invasion assays were performed as previously described [21] using a Matrigel®-coated invasion chamber with an 8.0 μm pore size filter following the manufacturer’s protocol with minor modifications including the final concentration of cells (2 × 104 cells/mL) and the incubation time (10 h).
Statistical analysis
The values in all of the figures are presented as the standard error of the mean (SEM). Statistical analyses were performed using the GraphPad Prism 4 statistical analysis software. An unpaired Student’s t-test or one-way ANOVA test (significance level P = 0.05) followed by Tukey’s multiple comparison tests was performed to determine the statistical significance of the observed changes among the data groups.
RESULTS
Generation of Sps1 knockout mice
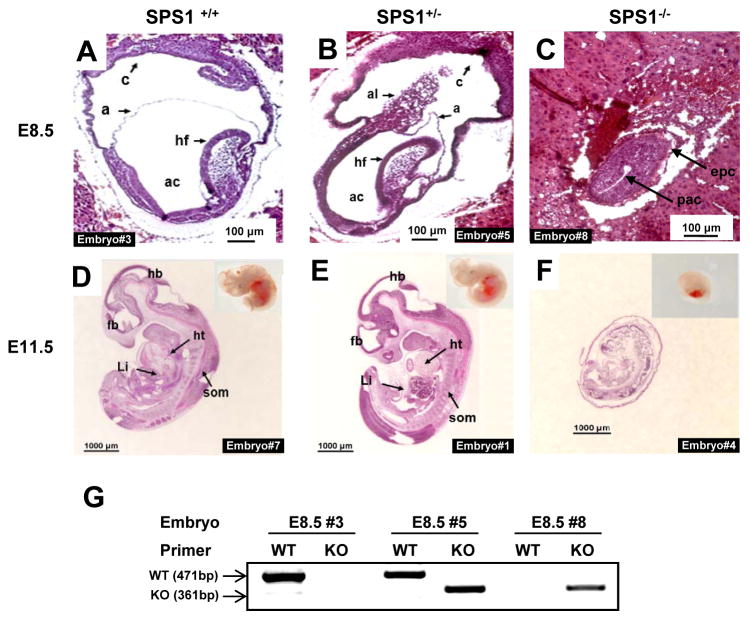
Sps1, which is located on chromosome 2 in mice, contains nine exons including eight within the coding sequence and one upstream. A knockout vector was devised wherein loxP elements flanking exon 2 were introduced (Figure S1). Sps1 was removed using EIIa-Cre, which targets gene removal in early development, and the resulting heterozygous offspring, Sps1+/− × Sps1+/− were crossed, yielding the embryonic and progeny ratios shown in Table 1. Sps1+/+and Sps1+/− embryos appeared normally developed on days 8.5 and 11.5 (Figure 1A–F), whereas Sps1−/− embryos were poorly developed on days 8.5 and 11.5 and were virtually resorbed by day 14.5 (Figure S2). Figure 1G shows an example of embryo genotyping. The genotypes of a wild type (1A), heterozygote (1B), and knockout embryo (1C) on day 8.5 were determined after capturing the embryo fragments using LCM. As shown in Figure 1C, the development of SPS1 knockout embryos was significantly retarded at E8.5 in that the embryos were smaller in size than normal embryos and the cavities were visibly much smaller. The size of the Sps1−/− embryos on day 11.5 was approximately 4-fold smaller than normal embryos (Figure 1D–F), and the organs could not be identified in intact Sps1−/− embryos, whereas normal embryos exhibited discrete organs such as the brain, eye, heart, tail bud, and somites (Figure 1D and E). Surprisingly, vestigial organs were apparent in the E11.5 Sps1−/− embryo sections, but the cell density was significantly lower compared to normal embryos, which suggested that the knockout embryos were undergoing resorption. In addition, membranes such as the amnion and yolk sac covered the embryo and could not be separated in Sps1−/− embryos at E11.5, whereas they could be removed from the wild type embryos at the same time point. Notably, no differences in development were observed between wild type and heterozygous embryos (Figures 1A and D and Figures 1B and E, respectively).
Table 1.
Targeted removal of Sps1
| Age examined | Mean number of embryos or offspring/litter | Genotypea
|
||
|---|---|---|---|---|
| +/+ | +/− | −/− | ||
| E8.5 embryo | 8.50 | 5 | 9 | 3b |
| E10.5 embryo | 9.00 | 3 | 10 | 5c |
| E11.5 embryo | 9.00 | 2 | 5 | 2c |
| E12.5 embryo | 6.00 | 3 | 5 | 4c |
| E14.5 embryo | 5.50 | 4 | 7 | 0d |
| 3 weeks (tail) | 6.15 | 46 | 77 | 0 |
= Genotypes of the total number of embryos from E8.5 to E12.5 that showed Mendelian segregation (+/+: +/−: −/−; 13:29:14);
= Development of these embryos was significantly retarded;
= Embryos were poorly developed and non-viable;
= Remnants of embryos were identified as shown in Figure S2.
Figure 1. Morphological differences in embryos generated from crosses between Sps1+/− mice.
Images from sagittal sections of E8.5 SPS1 wild type (A), heterozygous (B), and knockout mice (C), and E11.5 SPS1 wild type (D), heterozygous (E), and knockout embryos (F). Insets in (D), (E), and (F) designate whole embryos. The length of each scale bar is indicated on the bar. (G) Genotypes of E8.5 embryos were determined by PCR. As described in the Experimental section, embryo fragments were captured, genomic DNA was isolated, and PCR was performed using two sets of primers. Embryo fragments used in Figure 1G were captured from the embryos in Figures 1A, B, and C. Epc, ectoplacental cone; exc, exocoelom; pac, postamniotic cavity; a, amnion; ac, amniotic cavity; am, amniotic membrane; c, chorion; al, allantois; hf, headfold; fb, forebrain; hb, hindbrain; ht, heart; Li, liver; som, somite.
Statistical analysis provided evidence for the embryonic lethality of the Sps1−/− mice. The ratios of Sps1+/+, Sps1+/−, and Sps1−/− were approximately 13:29:14 at E8.5–12.5; however, the ratios changed after E14.5 to 1:2:0 (Table 1). Remnants of the Sps1−/− embryos were present at E14.5, which enabled genotyping, but these remnants were completely resorbed before the birth of the surviving embryos.
Next, we targeted the removal of Sps1 in the liver to obtain a population of Alb-Cre; Sps1fl/fl mice lacking SPS1 in hepatic tissue, which was confirmed by qPCR analysis (Figure 2A) and Western blotting (Figure 2B). The loss of SPS1 in the liver did not result in any apparent physical phenotype at 8–10 weeks of age. However, in preparing the SPS1 knockout construct, exon 2, which contains the translation initiation site for this protein, was deleted, but exon 3, which also encodes an ATG codon with a Kozak’s consensus sequence, was preserved. To rule out the possibility that a truncated form of SPS1 was generated from exon 3, we performed Western blotting using anti-SPS1 antibody. The truncated form of SPS1 (35.2 kD) was not detected in the wild type or heterozygous mice, suggesting that the protein was not expressed or was very rapidly degraded (Figure S3). Because we could not find any apparent differences in the physical appearances between Alb-Cre; Sps1fl/fl and control mice, we examined numerous liver functions and other metabolic functions by conducting a number of enzyme assays and assessments of other components present in the sera of SPS1 knockout and control mice (Figure S4). The only significant difference was found in aspartate aminotransferase (AST), a PLP-dependent enzyme that catalyzes the conversion of aspartate to glutamate, wherein the knockout mice exhibited lower activity than control mice. However, the biological significance of AST down-regulation in liver of Alb-Cre; Sps1fl/fl mice must await further studies.
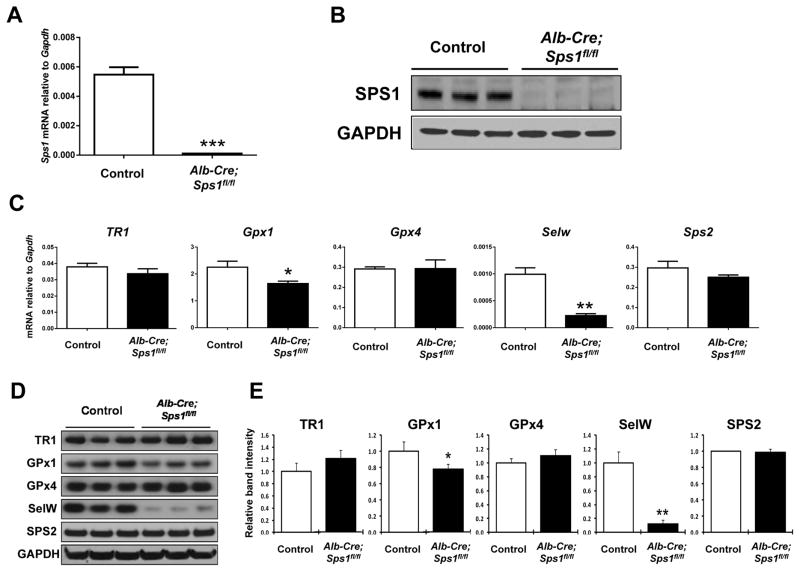
Figure 2. Expression of SPS1 and selenoproteins in control and hepatocyte-specific Sps1 knockout (Alb-Cre; Sps1fl/fl) mouse livers.
(A) mRNA levels of Sps1 were analyzed by qPCR. Data are shown as relative mRNA levels normalized to Gapdh in both control and Alb-Cre; Sps1fl/fl liver samples (n=3). ***Denotes statistically significant differences (P < 0.001). (B) Levels of SPS1 were analyzed by Western blotting in both control (Sps1fl/fl) and Alb-Cre; Sps1fl/fl liver samples (n=3). GAPDH levels are shown in the bottom panel as a protein loading control. (C) mRNA levels of TR1, Gpx1, Gpx4, SelW, and Sps2 were analyzed by qPCR. Data are shown as relative mRNA levels normalized to Gapdh in both control and Alb-Cre; Sps1fl/fl liver samples (n=3). * and ** denote statistically significant differences at P < 0.05 and P < 0.01, respectively. (D) Expression of TR1, GPx1, GPx4, SelW, and SPS2 were analyzed by Western blotting in both control and Alb-Cre; Sps1fl/fl liver samples (n=3). GAPDH levels are shown in the bottom panel as a protein loading control. (E) Quantification of the band intensities on Western blots. Relative band intensities of triplicate lanes were quantified as described in the Experimental section and Alb-Cre; Sps1fl/fl sample values were normalized to the control. * and ** denote statistically significant differences at P < 0.05 and P < 0.01, respectively.
Loss of Sps1 in the liver affects stress-related selenoproteins but not selenium status or Sec tRNA isoforms
Because SPS1 evolved from SPS and has close homology to SPS2 [6–7,14], we initially examined the possible effects that the loss of this protein might have on selenoprotein expression in mouse liver. The mRNA and protein levels of several selenoproteins known to be expressed in the mouse liver were therefore examined in SPS1-deficient (Alb-Cre; Sps1fl/fl) and control (Sps1fl/fl) hepatocytes (Figures 2C–E). The mRNA and protein levels of the stress-related selenoproteins, GPx1 and SelW, were significantly reduced, with an approximate 90% reduction in SelW in SPS1-deficient hepatocytes; however, interestingly, the corresponding levels of the housekeeping selenoproteins TR1, GPx4, and SPS2 were virtually unchanged.
The expression of stress-related selenoproteins is far more sensitive to selenium status than is that of housekeeping selenoproteins; and the synthesis of stress-related selenoproteins is dependent on the Sec-tRNA[Ser]Sec isoform that contains Um34 [27,28]. In mammals, the expression of the two Sec tRNA[Ser]Sec isoforms that are responsible for selenoprotein synthesis, 5-methoxycarbonylmethyluracil (mcm5U) and 5-methoxycarbonylmethyluracil-2′-O-methylribose (mcm5Um), are also sensitive to selenium status, as the level of the Um34 (mcm5Um) isoform is dramatically reduced under selenium-deficient conditions [27,28]. These observations prompted us to assess selenium levels and the ratios of the two Sec tRNA[Ser]Sec isoforms in SPS1-deficient livers (Figures S5A and B). However, SPS1-deficiency had no apparent effect on selenium status and little to no effect on the expression of the two Sec tRNA[Ser]Sec isoforms.
Loss of Sps1 in the liver only affects iron and manganese levels
Glutaredoxins are best known for their role in redox homeostasis. In addition, they have been shown to function in copper [29] and iron metabolism [30], and GLRX1 and GSTO1 have been reported to interact with arsenic and selenite [31]. Both of these proteins’ mRNAs, as well as some copper and zinc-related proteins’ mRNAs, were dramatically down-regulated in Sps1-deficient livers (see below). Therefore, we analyzed the levels of 13 other elements in the livers from Alb-Cre; Sps1fl/f and control mice (Figure S6) in addition to the levels of selenium (Figure S5A). Our results indicated that only the levels of iron and manganese were significantly reduced in the SPS1-deficient livers compared to controls.
Gene expression analysis suggests a link between the loss of SPS1 in liver, the GSH system, and cancer properties
To elucidate the function of SPS1, the livers from Alb-Cre; Sps1fl/fl mice were subjected to gene expression analysis (see Table S2). We subjected all genes with two-fold or greater changes in mRNA expression (P < 0.05) to IPA, which groups genes according to biological processes and displays their significance values, interacting genes, and direct or indirect association patterns. The top five networks, canonical pathways, and toxicology and hepatotoxicity lists are shown in Table S3. These networks were all related to cancer with the top network grouped as “Gene Expression, Cancer, Embryonic Development.” Interestingly, GSH-related systems appeared among the top canonical pathways and toxicology and hepatotoxicity lists, along with phase II reactions and xenobiotic metabolism pathways, which also involve GSH-related enzymes. Hence, the results motivated us to examine the GSH system in greater detail.
Loss of Sps1 in the liver alters the expression of proteins involved in GSH metabolism
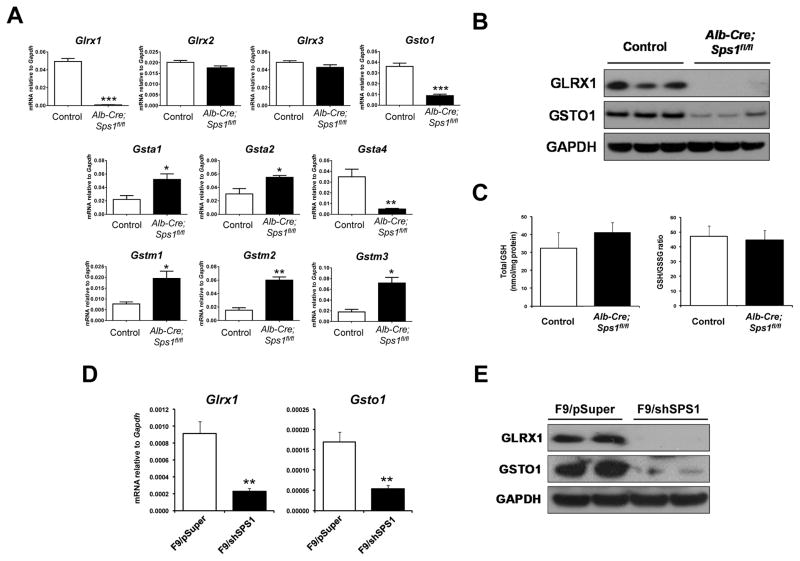
Among the GSH-related genes examined by microarray analysis, the expression of Glrx1 exhibited the largest change with an approximately 43-fold decrease in SPS1-deficient livers (Table S2B). Several other genes in the GST family of proteins were also altered in livers lacking SPS1. Gsto1 and Gsta4 exhibited greater than two-fold decreases in SPS1-deficient livers (−2.06 fold and −3.61 fold, respectively), whereas Gsta2, Gstm1, Gstm2, and Gstm3 increased more than two-fold, (2.17-fold, 2.94-fold, 2.07-fold, and 3.14-fold, respectively).
To validate the observed changes in gene expression, the mRNA levels of Glrx1, Glrx2, Glrx3, Gsto1, Gsta1, Gsta2, Gsta4, Gstm1, Gstm2, and Gstm3 were analyzed by qPCR (Figure 3A). The levels of Gsta1, Gsta2, Gstm1, Gstm2, and Gstm3 all showed a significant increase in livers lacking SPS1, whereas the levels of Gsto1 and Gsta4 showed a significant decrease and Glrx1 was virtually abolished. The levels of Glrx2 and Glrx3 mRNAs were unchanged in SPS1-deficient livers compared to control livers (Figure 3A).
Figure 3. Glutathione-S-transferase and Glrx expression in Alb-Cre; Sps1fl/fl livers and SPS1-deficient F9 cells.
(A) mRNA levels of Glrx1, Glrx2, Glrx3, Gsto1, Gsta1, Gsta2, Gsta4, Gstm1, Gstm2, and Gstm3 were analyzed by qPCR. Data are shown as relative mRNA levels normalized to Gapdh in both control and Alb-Cre; Sps1fl/fl liver samples (n=4). (B) Levels of GLRX1 and GSTO1 were analyzed by Western blotting in both control and Alb-Cre; Sps1fl/fl liver samples (n=3). GAPDH levels are shown in the bottom panel as a protein loading control. (C) Measurement of total GSH and GSSG, and the GSH/GSSG ratio in both control and Alb-Cre; Sps1fl/fl liver samples (n=3). Total GSH and GSSG were measured spectrophotometrically and the GSH/GSSG ratio was calculated as described in the Experimental section. Total GSH concentrations are the mean ± standard deviation for three independent experiments and are expressed as nmol per mg protein. (D) mRNA levels of Glrx1 and Gsto1 in both control and Sps1 knockdown F9 cells were analyzed by qPCR. The values are normalized to Gapdh and are the mean of three independent experiments. (E) Levels of GLRX1 and GSTO1 were analyzed by Western blotting in both control and Sps1 knockdown F9 cells. GAPDH levels are shown in the bottom panel as a protein loading control. *, **, and *** denote statistically significant differences at P < 0.05, P < 0.01, and P < 0.001, respectively.
The levels of GLRX1 and GSTO1 were examined by Western blotting and were found to reflect the decreases detected in mRNA levels in that GLRX1 was not detectable and GSTO1 was poorly expressed (Figure 3B). In addition, the amounts of GSH and the ratio of GSH to GSSH were assessed (Figure 3C), which revealed that the changes observed in the GSH members did not significantly affect GSH levels or the ratio of GSH to GSSH in SPS1-deficient livers (Figure 3). It should be noted that the expression of genes encoding proteins responsible for the de novo synthesis of glutathione, such as glutamate cysteine ligase catalytic subunit (GCLC), glutamate-cysteine modifier subunit (GCLM), and glutathione synthetase (GSS), were not affected by a deficiency in SPS1.
Characterization of Sps1 in F9 cells
As noted above, IPA revealed that the top network in SPS1-deficient livers was Gene Expression, Cancer, Embryonic Development (Table S3). In addition, a previous study by Assou et al. [12] demonstrated that SPS1 is highly expressed in human ES cells. Therefore, we examined the role of SPS1 in F9 cells, a mouse EC cell line.
F9 cells were transfected with an empty vector (designated F9/pSuper control cells), as well as with the same vector encoding a knockdown construct to target the removal of Sps1 mRNA (designated F9/shSPS1 cells). To confirm the knockdown of SPS1 in F9 cells, the expression levels of Sps1 and SPS1 were examined and found to be highly expressed in control cells and efficiently removed in F9/shSPS1 cells (Figure S7).
Glrx1, Gsto1, and SelW expression is significantly reduced in SPS1-deficient F9 cells
To assess the cellular effects of SPS1-deficieny in F9 cells, RNA isolated from the cells was subjected to microarray analysis. The top 25 up- and down-regulated genes with 1.5-fold or greater changes (P < 0.05) in F9/shSPS1 cells compared to those in F9/pSuper control cells are shown in Tables S4A and B, respectively. Similar to livers from Alb-Cre; Sps1fl/fl mice, Glrx1 and Gsto1 were among the most highly altered genes and were 4.5-fold and 5.2-fold down-regulated in SPS1-deficient F9 cells, respectively.
To confirm the microarray data, the mRNA levels of Glrx1 and Gsto1 were analyzed by qPCR (Figure 3D), the results of which indicated that Glrx1 and Gsto1 expression was significantly decreased. The levels of GLRX1 and GSTO1 were examined by Western blotting and GLRX1 was not detectable and GSTO1 was poorly expressed in SPS1-deficient cells (Figure 3E). The amounts of GSH and the ratio of GSH to GSSH were likewise determined, and unlike Sps1 knockout livers, SPS1-deficient F9 cell exhibited slight increases in both total GSH levels (~1.23 fold, P < 0.01) and in the ratio of GSH to GSSG (~1.28 fold, P < 0.05) compared to control cells (Figure S8).
We further examined the expression of mRNA and protein levels to assess whether the expression of the stress-related selenoproteins SelW and GPx1, which were significantly reduced in Sps1 knockout livers (see Figure 2), were also affected in SPS1-deficient F9 cells (Figure S9). Only SelW mRNA and protein expression levels were significantly down-regulated as measured by qPCR and Western blotting, respectively. Further, a likely explanation as to why the mRNA of this gene was not detected by microarray analysis was that it was present in very low levels in F9 cells.
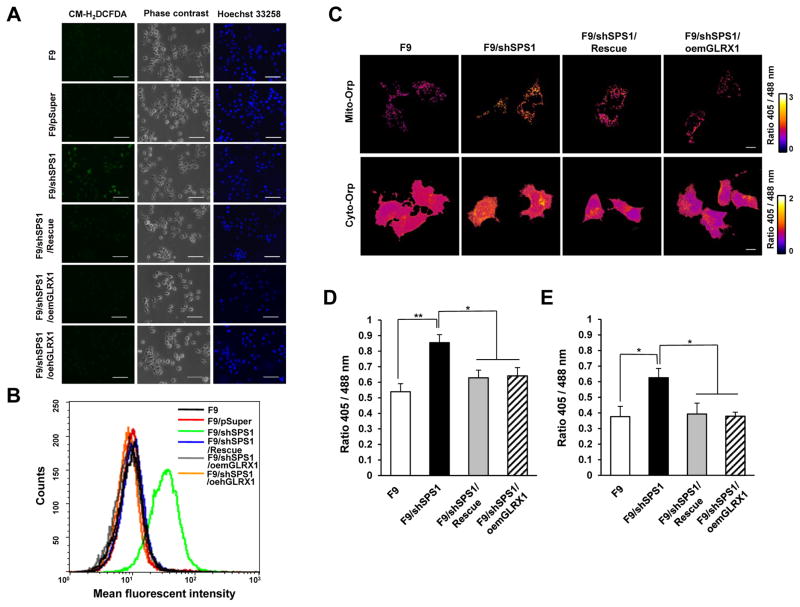
Hydrogen peroxide accumulates due to the down-regulation of GLRX1 in SPS1-deficient F9 cells
GLRX1 and GSTO1, which have been reported to participate in regulating redox homeostasis [32,33], were significantly decreased in SPS1-deficient mouse livers and F9 cells. These results suggested that SPS1 deficiency led to ROS accumulation. To examine the role of GLRX in ROS accumulation, mGLRX1 and hGLRX1 were overexpressed separately in SPS1-deficient F9 cells, and the cells were assayed for ROS levels. Each cell line was treated with CM-H2DCFDA and the cells were examined by fluorescence microscopy (Figure 4A). ROS levels were found to be significantly increased in the F9/shSPS1 cells compared to F9 or F9/pSuper cells. However, interestingly, the overexpression of mGLRX1 and hGLRX1 in SPS1-deficient F9 cells (designated F9/shSPS1/oemGLRX1 and F9/shSPS1/oehGLRX1, respectively) significantly diminished ROS levels. Phase contrast images and Hoechst staining of each cell line are shown to demonstrate that the ROS signals were derived from live cells (Figure 4A). To exclude a possible off-target effect, we introduced a knockdown-resistant rescue construct (designated F9/shSPS1/Rescue) into SPS1 knockdown cells. As shown in Figure 4A, ROS levels decreased to levels that were comparable to negative controls (F9 or F9/pSuper), suggesting that ROS generation was dependent on the deficiency of SPS1. When cells were sorted by FACS to monitor the amount of ROS that accumulated in each cell line, the mean fluorescence of SPS1-deficient cells was significantly increased compared to the fluorescence in F9, F9/pSuper control, F9/shSPS1/oemGLRX1, F9/shSPS1/oehGLRX1, and in F9/shSPS1/Rescue cells (Figure 4B).
Figure 4. Accumulation of intracellular hydrogen peroxide in SPS1-deficient F9 cells.
(A) Cells were stained with CM-H2DCFDA to detect intracellular ROS and then counterstained with Hoechst 33258 as described in the Experimental section. The cells were photographed under a fluorescence microscope. Scale bars represent 100 μm. (B) Measurement of intracellular ROS levels. After the cells were stained with CM-H2DCFDA, ROS levels were measured using FACS. Representative FACS plot showing the amount of intracellular ROS. (C–E) Detection of intracellular hydrogen peroxide using roGFP2-Orp1 probes. (C) Vectors encoding mitochondrial roGFP2-Orp1 or cytosolic roGFP2-Orp1 were transfected into SPS1-deficient F9 and control cells and the 405/488 nm ratio images were obtained as described in the Experimental section. Scale bars represent 10 μm. Quantification of microscopy data of mitochondrial (D) and cytosolic (E) hydrogen peroxide. Emission ratios from six randomly selected fields (100× image) for each sample were averaged. Error bars represent standard deviation. * and ** denote statistically significant differences at P < 0.05 and P < 0.01, respectively.
It has been established that DCFDA staining does not distinguish between the type of ROS present [34], and that specific probes should be used to identify ROS types. To determine the cytosolic and mitochondrial superoxide levels, the cells were stained with hydroethidine (HE) and MitoSOX™, respectively. The results indicated that the levels of superoxide in the cytosol and mitochondria were not affected by the SPS1 deficiency (Figure S10). Positive signals were not detected in all cells except the positive controls, which were treated with antimycin A. Conversely, as shown in Figure 4C–E, both intracellular and mitochondrial hydrogen peroxide levels were increased in SPS1-deficient cells when measured using cytosolic roGFP2-Orp1 and mitochondrial roGFP2-Orp1 probes [24]. The introduction of the knockdown-resistant rescue vector or Glrx1 overexpressing vector decreased the hydrogen peroxide concentration to levels that were similar to those of control cells. These results strongly suggested that the increased DCF signals in SPS-deficient cells reflected the increase in hydrogen peroxide levels and that the accumulation of hydrogen peroxide in SPS1-deficient F9 cells was mainly due to the lack of intracellular GLRX1 activity.
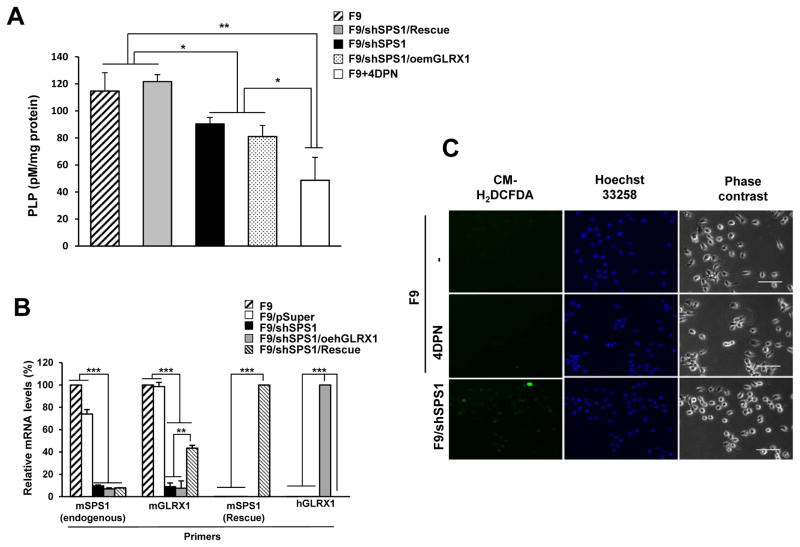
SPS1 deficiency decreases PLP levels
In our previous study involving Drosophila cells, the biosynthesis of PLP (the active form of vitamin B6) was decreased by Sps1 knockdown. Hence, we also measured the intracellular PLP levels in SPS1-deficient F9 cells. Although the expression pattern of genes involved in PLP biosynthesis was not changed by the knockdown of Sps1, PLP levels were decreased by approximately 30% (P < 0.05, Fig 5A). The decreased PLP levels were recovered by the transfection of a rescue vector (F9/shSPS1/Rescue), indicating that intracellular PLP levels were regulated by SPS1. Interestingly, PLP levels were not recovered by the overexpression of mouse GLRX1 (F9/shSPS1/oemGLRX1). However, the levels of Glrx1 mRNAs were significantly increased in F9/shSPS1/Rescue cells (Figure 5B). The levels of PLP and mouse Glrx1 mRNA in F9/shSPS1/oemGLRX1 cells suggested that the PLP levels in the mouse F9 cells were not responsible for ROS generation. To confirm this possibility, we inhibited PLP biosynthesis and examined the cells for ROS accumulation. When F9 cells were treated with 4-deoxypyridoxine (4-DPN), an inhibitor of PLP biosynthesis, ROS was not detected; however, SPS1-deficient cells exhibited strong ROS signals (Figure 5C). These results suggested that SPS1 was required for maintaining PLP levels by a mechanism other than the regulation of the transcription of genes involved in PLP biosynthesis and that the intracellular PLPs were not involved in ROS scavenging in F9 cells.
Figure 5. Measurement of intracellular PLP levels and their effect on ROS generation.
(A) After cells were cultured, PLP levels were measured as described in the Experimental section. Treatment with 4-DPN was used as a control. Data shown are representative of three independent experiments. (B) mRNA levels were measured by qPCR. cDNAs were amplified using specific primer sets as described in the Experimental section. β-actin was used as an internal control. Statistical significance was tested by one-way ANOVA followed by Tukey’s multiple comparison tests. Error bars represent standard deviation. *, **, and *** denote statistically significant differences at P < 0.05, P < 0.01, and P < 0.001, respectively. (C) The effect of inhibition of PLP biosynthesis on ROS generation. F9 cells were incubated with 4-DPN overnight and stained with DCFDA. F9/shSPS1 was used as a positive control. Scale bars represent 50 μm.
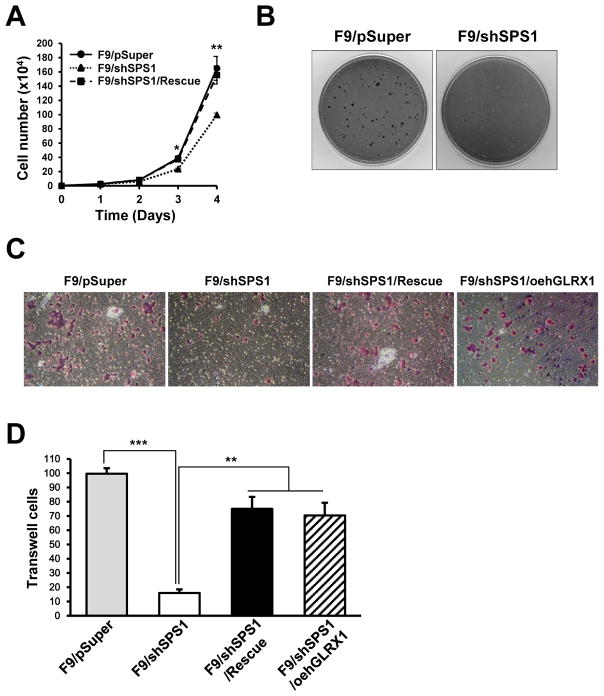
SPS1-deficiency in F9 cells reverses cancer characteristics
An examination of the growth rates of F9/shSPS1 and control F9 cells revealed significant reductions in SPS1-deficient cells compared to control cells at 72 and 96 h of growth (Figure 6A). Interestingly, the growth of F9/shSPS1 cells in soft agar was dramatically inhibited compared to control cells (Figure 6B), suggesting that some of the malignant properties of SPS1-deficient cells had changed to become more like those of normal cells [23,35], prompting us to explore additional cancer properties in SPS1-deficient F9 cells.
Figure 6. Detection and measurement of additional cancer characteristics in F9 cells.
(A) Effect of Sps1 knockdown on the cell growth rate of F9 cells. Living cells were stained with Trypan blue and counted every 24 h over a 96 h period. * and ** denote statistically significant differences at P < 0.05 and P < 0.01, respectively. (B) Effect of Sps1 knockdown on colony formation in soft agar assay. Cells were grown in soft agar for 10 days and stained with p-iodonitrotetrazolium as described in the Experimental section. (C) Invasion assay in F9 cells. Invaded cells were stained as described in the Experimental section and photographed (100×). (D) Invaded cells per 3.3 mm2 were counted. Error bars represent standard deviation. ** and *** denote statistically significant differences at P < 0.01 and P < 0.001, respectively. Data shown are representative of three independent experiments.
The results indicating reduced growth rates (Figures 6A, B) and ROS accumulation (Figures 5A–C) in SPS1-deficient cells suggested that the SPS1-deficient F9 cells underwent changes in their malignant properties such as cell invasion. The number of F9/shSPS1 cells that migrated through the Matrigel®-coated membrane was significantly decreased compared to control F9/pSuper cells (Figure 6C). Interestingly, the invasiveness of both F9/shSPS1/Rescue and F9/shSPS1/oehGLRX1 cells appeared to be significantly restored (Figure 6C). The number of cells in each cell line were counted and the number of F9/shSPS1 cells was reduced six-fold compared to the F9/pSuper cells; however, the number of F9/shSPS1/Rescue and F9/shSPS1/oehGLRX1 cells increased 4.5- and 4.2-fold, respectively (Figure 6D). The reason we overexpressed human Glrx1 in F9/shSPS1 cells was that the resulting gene product has greater than 90% homology with mouse GLRX1; thus, we did not anticipate that its expression and function would be affected by the F9/shSPS1 knockdown vector, which was the resulting observation. In addition, the expression of the knockdown-resistant Sps1 in F9 cells (F9/shSPS1/Rescue) resulted in similar effects to the overexpression of human Glrx1 in SPS1-deficient cells.
The results of the studies demonstrating a reduced growth rate, altered growth in soft agar, higher ROS levels, and decreased motility in F9/shSPS1 cells, as well as the subsequent restoration of the invasive properties in GLRX1 knock-in F9/shSPS1/oehGLRX1 cells (Figures 4 and 6), suggested that the accumulation of ROS might inhibit the migratory properties of embryonic cancer cells and that GLRX1 could restore the invasiveness in SPS1-deficient cells.
DISCUSSION
SPS1 has been an enigmatic protein. Being a paralog of SPS2, a selenoprotein critical for the biological use of selenium, SPS1 does not play such a prominent role as SPS2 in biosynthesis of this protein class. What then is its function? In fruit flies, SPS1 is an essential protein (as discussed in the Introduction and further discussed below), but the consequences of its deficiency in mammals are poorly understood. We found that Sps1 knockout was embryonic lethal in early development in mice (Table 1). On the other hand, there were no obvious phenotypes when the knockout was specifically targeted to liver. We selected liver as a target tissue to examine the effects of SPS1 deficiency on cellular metabolism as this organ is known to tolerate the loss of different proteins, and in particular, proteins involved in selenium metabolism and redox homeostasis, which permitted us to assess the effects of SPS1 loss on cellular metabolism. In this regard, Sps1 is similar to other genes including TR1 [36–38], Secisbp2 [39], and Secp43 [40], which are lethal in early development but do not result in any apparent physical phenotypic differences following targeted knockout in the liver.
While SPS1 does not appear to be directly involved in selenoprotein biosynthesis, it affects selenoprotein expression. Of the selenoproteins examined in liver-specific Sps1 knockout mice, only SelW and GPx1, both stress-related selenoproteins, were significantly down-regulated in liver at the level of gene expression suggesting a role of SPS1, either directly or indirectly, in their expression. GPx1 is a well-known redox regulator that reduces cellular hydroperoxides at the expense of glutathione oxidation. SelW was suggested to participate in cellular redox regulation and is a thioredoxin-like fold that is covalently modified with glutathione [41]. SelW has also been reported to regulate cell proliferation and muscle differentiation [42], but its exact function remains unknown. It is interesting that the expression of these two stress-related selenoproteins is not associated with the levels of selenium or the expression of the Sec tRNA[Ser]Sec mcm5Um isoform considering these proteins and this tRNA are extremely sensitive to selenium status [43–45]. In contrast to SelW and GPx1, we found a slight elevation in TR1 levels. Such a pattern is often observed under conditions of oxidative stress. Therefore, the most likely possibility is that SPS1 deficiency leads to oxidative stress, which in turn alters expression of certain selenoproteins.
Oxidative stress may also affect the levels of transition metals and other redox-related elements. Indeed, among the 13 elements (other than selenium) measured in the livers from Alb-Cre; Sps1fl/fl and control mice, iron and manganese levels were significantly reduced in Sps1 knockout livers. Interestingly, several cytochrome P450 (Cyp) genes, which encode iron-containing monooxygenases involved in the metabolism of a wide variety of compounds, were altered in Alb-Cre; Sps1fl/fl mice. The relationship of the P450 (Cyp) genes to SPS1 was not apparent. On the other hand, the activity of total SOD and MnSOD were not affected by the SPS1 deficiency (Figure S11) suggesting that the reduced levels of Mn did not affect MnSOD activity.
The most striking observation was the dramatic change in the expression of Glrx1 as a result of SPS1 deficiency. GLRX1 is a thioredoxin-fold oxidoreductase and has glutathione-dependent oxidoreductase functions and is responsible for reducing ribonucleotide reductase. It also can support glutathione-dependent reduction of the oxidized deiodinase 3, reduction of dehydroascorbate and regeneration of oxidatively damaged proteins ([46,47] and references therein). After reducing its target molecule, GLRX1 is oxidized to form an intramolecular disulfide, which is in turn reduced by GSH. There was a possibility that the decreased levels of GLRX1 would lead to an increase of GSH levels and/or GSH/GSSG ratio. Slight increases (~1.2 fold) in these two parameters were observed in SPS1-deficient F9 cells. However, GSH levels and GSH/GSSG ratios were not significantly changed in the livers from Alb-Cre; Sps1fl/fl mice compared to control mice. It can be speculated that the reason why these two parameters were not changed in the SPS1 knockout liver was because they are maintained by other enzymes in the GSH system in the liver.
Consistent with the observations discussed above, significantly higher levels of ROS were detected in F9/shSPS1 cells than in control cells, and this phenotype of SPS1-deficient F9 cells was also reversed by overexpressing either mouse or human GLRX1. This finding suggested that SPS1 could play a role in maintaining low ROS levels by regulating expression of GSH metabolic enzymes involved in regulating redox homeostasis, including GLRX1 (see also [44]). Thus, SPS1 appears to function in the protection of cells from oxidative stress. Notably, the expression of knockdown-resistant Sps1 in F9 cells (F9/shSPS1/Rescue) reduced ROS levels to those of control cells, even though it recovered Glrx1 mRNA levels to only 45% of the parent F9 cells. These results further suggested that the down-regulation of Glrx1 expression by SPS1 knockdown was not entirely responsible for the accumulation of ROS. It appears that other regulatory redox proteins such as GSTO1 and GSTA4 may also be involved in ROS metabolism linked to SPS1 status. It is interesting that hydrogen peroxide was altered by SPS1 deficiency, while SOD and MnSOD activities were not affected suggesting that superoxide was likely not affected. In contrast, knockout of Sps1 in Drosophila led to the accumulation of superoxide [20]. It is not clear why mouse and Drosophila exhibited these differences, but it is not unusual to see this at such great evolutionary distances.
We think that low levels of GLRX1 in response to SPS1 deficiency contribute to the accumulation of ROS in F9 cells (Figure 4), because the overexpression of GLRX1 reduced ROS up to the level of control cells, whereas GPx1 levels were not changed significantly. The increased ROS levels in SPS1-deficient F9 cells would result in changes in the redox state of GLRX1. When the redox state of GLRX1 was measured using GFP-fused GLRX1 probes that can detect oxidized and reduced forms of GLRX1 in the cytosol or mitochondria, we found that the ratio of oxidized/reduced GLRX1 was not changed in either compartment in SPS1-deficient F9 cells (Figure S12).
Another consequence of SPS1 deficiency was related to cell proliferation and cancer phenotypes. The growth rates of F9/shSPS1 cells in normal culture medium and in soft agar were significantly decreased. In addition, the invasiveness of the SPS1-deficient cells was dramatically reduced. Hence, our studies demonstrated that SPS1 is an essential protein in early development that has critical roles in regulating the expression of enzymes involved in redox homeostasis and in promoting and/or sustaining cancer. These findings raise additional questions regarding the function of SPS1. As noted above, SPS1 has been shown to regulate redox homeostasis in insects [14,19], and its deficiency resulted in megamitochondria formation [14]. It also participates in cellular defense and proliferation [15]. In mammals, SPS1 appears to function similarly in redox homeostasis, but it also has additional roles. Although both Drosophila and mouse SPS1 regulate the expression of genes controlling cellular redox homeostasis, the specific pathways are somewhat different between these two organisms. Among the genes that participate in cellular redox homeostasis, no common genes or homologs were observed whose expression is regulated by SPS1 in both Drosophila and mouse cells (compare GEO accession number GSE17685 [8] with GEO accession number GSE74677 in this study).
In summary, this study revealed critical roles of SPS1 in redox homeostasis, wherein this protein regulates the expression of a subset of oxidoreductases and detoxification proteins. The expression of GLRX1, an important oxidoreductase, is almost completely dependent on SPS1. As expected, these changes disrupt redox homeostasis of SPS1 deficient cells, whereas overexpression of GLRX1 reverses some of these phenotypes. Whether the altered redox homeostasis is fully responsible for the observed embryonic lethality of SPS1 deficiency is unclear, but our study provides clear avenues for further investigation of this remarkable protein.
Supplementary Material
Figure S1. Targeted removal of SPS1 in mice.
Figure S2. Remnants of Sps1 knockout mice at E14.5.
Figure S3. SPS1 levels in control and hepatocyte-specific Sps1 knockout (Alb-Cre; Sps1fl/fl) mouse liver.
Figure S4. Clinical chemistry analysis in plasma from control and Alb-Cre; Sps1fl/fl mice.
Figure S5. Selenium levels and Sec tRNA isoform distribution in livers from control and Alb-Cre; Sps1fl/fl mice.
Figure S6. Element analysis in livers from control and Alb-Cre; Sps1fl/fl mice.
Figure S7. Knockdown of Sps1 in F9 cells.
Figure S8. Measurement of total GSH and GSSG, and the GSH/GSSG ratio in the control (F9/pSuper) and Sps1-deficient cells.
Figure S9. Expression of selenoproteins in SPS1-deficient mouse F9 embryonal carcinoma cells.
Figure S10. Detection of intracellular superoxide in SPS1-deficient F9 cells.
Figure S11. Measurement of superoxide dismutase activity.
Figure S12. Measurement of the ratio of disulfide/dithiol GLRX1 in the mitochondria and cytosol of F9 cells using Grx1-roGFP2 probes.
SM Text. Materials and Methods and References
Table S1. Primer sequences.
Table S2. Microarray analysis of the 25 most highly (A) up- and (B) down-regulated genes in Alb-Cre; Sps1fl/fl mouse liver
Table S3. Ingenuity Pathway Analysis of liver from control and Alb-Cre; Sps1fl/fl mice
Table S4. Microarray analysis of the 25 most highly (A) up- and (B) down-regulated genes in SPS1 knockdown F9 mouse embryonic carcinoma cells.
SUMMARY STATEMENT.
Systemic knockout of Sps1 caused embryonic lethality in mice, but no apparent physical phenotype when targeted to liver. SPS1 deficiency in mouse liver and embryonic cancer cells permitted us to elucidate roles of this protein in redox homeostasis.
Acknowledgments
FUNDING
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2015R1D1A1A01059170), and the Bio & Medical Technology Development Program of the National Research Foundation (NRF), awarded by the Ministry of Science (2012M3A9D1054622) to BJL, as well as the Intramural Research Program of the National Institutes of Health (NIH), National Cancer Institute Center for Cancer Research to DLH, DSS, and LT, NIH grants CA080946, GM061603, and GM065204 to VNG, DFG SCHW914/2-1 to US, and Towson University’s Jess and Mildred Fisher College of Science and Mathematics Endowed Chair to PAT.
Abbreviations List
- SPS
selenophosphate synthetase
- Sec
selenocysteine
- SCL
selenocysteine lyase
- PLP
pyridoxal phosphate
- ROS
reactive oxygen species
- EC
embryonic carcinoma
- TR1
thioredoxin reductase 1
- GPx
glutathione peroxidase
- SelW
selenoprotein W
- GLRX1
glutaredoxin 1
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GSTO1
glutathione S-transferase omega 1
- DPBS
Dulbecco’s phosphate buffered saline
- DMEM
Dulbecco’s modified Eagle’s medium
- FBS
fetal bovine serum
- DTNB
5,5′-dithiobis-2-nitrobenzoic acid
- PVDF
polyvinylidene fluoride
- RPMI
Roswell Park Memorial Institute
- CM-H2DCFDA
5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate
- DHE
dihydroethidium
- MoMuLV-RT
Moloney murine leukemia virus reverse transcriptase
- LCM
laser capture microdissection
- SOD
superoxide dismutase
- Neo
neomycin resistance gene
- ES
embryonic stem cell
- PCR
polymerase chain reaction
- FLP
flippase
- PBS
phosphate-buffered saline
- IPA
Ingenuity Pathway Analysis
- ORF
open reading frame
- qPCR
quantitative PCR
- GSH
glutathione
- GSSG
glutathione disulfide
- ICP-OES
Inductively Coupled Plasma-Optical Emission Spectrometry
- DW
distilled water
- FACS
fluorescence-activated cell sorting
- AST
aspartate aminotransferase
- mcm5U
5-methoxycarbonylmethyluracil
- mcm5Um
5-methoxycarbonylmethyluracil-2′-O-methylribose
- GST
glutathione S-transferase
- GCLC
glutamate cysteine ligase catalytic subunit
- GCLM
glutamate-cysteine modifier subunit
- GSS
glutathione synthetase
- HE
hydroethidine
- 4-DPN
4-deoxypyridoxine
- Cyp
cytochrome P450
Footnotes
AUTHOR CONTRIBUTIONS
Byeong Jae Lee and Dolph L. Hatfield contributed to the study design and concept. Ryuta Tobe, Bradley A. Carlson, Jang Hoe Huh, Nadia P. Castro, Xue-Ming Xu, Petra A. Tsuji, Sang Goo Lee, Jeyoung Bang, Ji-Woon Na, Daniel Beaglehole, Eileen Southon, Harold Seifried, and Lino Tessarollo performed the experiments. David S. Salomon, Ulrich Schweizer, Vadim N. Gladyshev, Dolph L. Hatfield, and Byeong Jae Lee analyzed the data. Young-Yun Kong provided technical assistance. Byeong Jae Lee and Dolph L. Hatfield wrote the manuscript and directed the project.
References
- 1.Böck A, Forchhammer K, Heider J, Baron C. Selenoprotein synthesis: an expansion of the genetic code. Trends Biochem Sci. 1991;16(12):463–7. doi: 10.1016/0968-0004(91)90180-4. [DOI] [PubMed] [Google Scholar]
- 2.Glass RS, Singh WP, Jung W, Veres Z, Scholz TD, Stadtman TC. Monoselenophosphate: synthesis, characterization, and identity with the prokaryotic biological selenium donor, compound SePX. Biochemistry. 1999;32(47):12555–9. doi: 10.1021/bi00210a001. [DOI] [PubMed] [Google Scholar]
- 3.Carlson BA, Tobe R, Tsuji PA, Yoo MH, Feigenbaum L, Tessarollo L, Lee BJ, Schweizer U, Gladyshev VN, Hatfield DL. Selenocysteine tRNA[Ser]Sec: The central component of selenoprotein biosynthesis. In: Brigelius-Flohé R, Sies H, editors. Selenium Functions in Health and Disease. CRC Press; Taylor & Francis Group; Boca Raton, FL: 2015. pp. 55–82. [Google Scholar]
- 4.Kim IY, Stadtman TC. Selenophosphate synthetase: detection in extracts of rat tissues by immunoblot assay and partial purification of the enzyme from the archaean Methanococcus vannielii. Proc Nat’l Acad Sci U S A. 1995;92(17):7710–3. doi: 10.1073/pnas.92.17.7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Low SC, Harney JW, Berry MJ. Cloning and functional characterization of human selenophosphate synthetase, an essential component of selenoprotein synthesis. J Biol Chem. 1995;270(37):21659–64. doi: 10.1074/jbc.270.37.21659. [DOI] [PubMed] [Google Scholar]
- 6.Guimarães MJ, Peterson D, Vicari A, Cocks BG, Copeland NG, Gilbert DJ, Jenkins NA, Ferrick DA, Kastelein RA, Bazan JF, Zlotnik A. Identification of a novel selD homolog from eukaryotes, bacteria, and archaea: is there an autoregulatory mechanism in selenocysteine metabolism? Proc Nat’l Acad Sci U S A. 1996;93(26):15086–91. doi: 10.1073/pnas.93.26.15086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim IY, Guimarães MJ, Zlotnik A, Bazan JF, Stadtman TC. Fetal mouse selenophosphate synthetase 2 (SPS2): characterization of the cysteine mutant form overproduced in a baculovirus-insect cell system. Proc Nat’l Acad Sci U S A. 1997;94(2):418–21. doi: 10.1073/pnas.94.2.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu XM, Carlson BA, Mix H, Zhang Y, Saira K, Glass RS, Berry MJ, Gladyshev VN, Hatfield DL. Biosynthesis of selenocysteine on its tRNA in eukaryotes. PLoS Biol. 2007;5(1):e4. doi: 10.1371/journal.pbio.0050004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu XM, Carlson BA, Irons R, Mix H, Zhong N, Gladyshev VN, Hatfield DL. Selenophosphate synthetase 2 is essential for selenoprotein biosynthesis. Biochem J. 2007;404(1):115–20. doi: 10.1042/BJ20070165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamura T, Yamamoto S, Takahata M, Sakaguchi H, Tanaka H, Stadtman TC, Inagaki K. Selenophosphate synthetase genes from lung adenocarcinoma cells: Sps1 for recycling L-selenocysteine and Sps2 for selenite assimilation. Proc Na’tl Acad Sci U S A. 2004;101(46):16162–7. doi: 10.1073/pnas.0406313101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Small-Howard A, Morozova N, Stoytcheva Z, Forry EP, Mansell JB, Harney JW, Carlson BA, Xu XM, Hatfield DL, Berry MJ. Supramolecular complexes mediate selenocysteine incorporation in vivo. Mol Cell Biol. 2006;26(6):2337–46. doi: 10.1128/MCB.26.6.2337-2346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Assou S, Cerecedo D, Tondeur S, Pantesco V, Hovatta O, Klein B, Hamamah S, De Vos J. A gene expression signature shared by human mature oocytes and embryonic stem cells. BMC Genomics. 2009;10:10. doi: 10.1186/1471-2164-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alsina B, Corominas M, Berry MJ, Baguna J, Serras F. Disruption of selenoprotein biosynthesis affects cell proliferation in the imaginal discs and brain of Drosophila melanogaster. J Cell Sci. 1999;112( Pt 17):2875–84. doi: 10.1242/jcs.112.17.2875. [DOI] [PubMed] [Google Scholar]
- 14.Shim MS, Kim JY, Jung HK, Lee KH, Xu XM, Carlson BA, Kim KW, Kim IY, Hatfield DL, Lee BJ. Elevation of glutamine level by selenophosphate synthetase 1 knockdown induces megamitochondrial formation in Drosophila cells. J Biol Chem. 2009;284(47):32881–94. doi: 10.1074/jbc.M109.026492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee KH, Shim MS, Kim JY, Jung HK, Lee E, Carlson BA, Xu XM, Park JM, Hatfield DL, Park T, Lee BJ. Drosophila selenophosphate synthetase 1 regulates vitamin B6 metabolism: prediction and confirmation. BMC Genomics. 2011;12:426. doi: 10.1186/1471-2164-12-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soda K, Oikawa T, Esaki N. Vitamin B6 enzymes participating in selenium amino acid metabolism. Biofactors. 1990;10(2–3):257–62. doi: 10.1002/biof.5520100225. [DOI] [PubMed] [Google Scholar]
- 17.Tobe R, Mihara H, Kurihara T, Esaki N. Identification of proteins interacting with selenocysteine lyase. Biosci Biotechnol Biochem. 2009;73(5):1230–2. doi: 10.1271/bbb.90065. [DOI] [PubMed] [Google Scholar]
- 18.Choi SY, Jang JH, Kim KR. Analysis of differentially expressed genes in human rectal carcinoma using suppression subtractive hybridization. Clin Exp Med. 2011;11(4):219–26. doi: 10.1007/s10238-010-0130-5. [DOI] [PubMed] [Google Scholar]
- 19.Morey M, Corominas M, Serras F. DIAP1 suppresses ROS-induced apoptosis caused by impairment of the selD/sps1 homolog in Drosophila. J Cell Sci. 2003;116(22):4597–604. doi: 10.1242/jcs.00783. [DOI] [PubMed] [Google Scholar]
- 20.Reid SW, Tessarollo L. Isolation, microinjection and transfer of mouse blastocysts. Methods Mol Biol. 2009;530:269–85. doi: 10.1007/978-1-59745-471-1_14. [DOI] [PubMed] [Google Scholar]
- 21.Bang J, Jang M, Huh JH, Na JW, Shim M, Carlson BA, Tobe R, Tsuji PA, Gladyshev VN, Hatfield DL, Lee BJ. Deficiency of the 15-kDa selenoprotein led to cytoskeleton remodeling and non-apoptotic membrane blebbing through a RhoA/ROCK pathway. Biochem Biophys Res Commun. 2014;456(4):884–90. doi: 10.1016/j.bbrc.2014.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bang J, Huh JH, Na JW, Lu Q, Carlson BA, Tobe R, Tsuji PA, Gladyshev VN, Hatfield DL, Lee BJ. Cell Proliferation and Motility Are Inhibited by G1 Phase Arrest in 15-kDa Selenoprotein-Deficient Chang Liver Cells. Mol Cells. 2015;38(5):457–65. doi: 10.14348/molcells.2015.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irons R, Tsuji PA, Carlson BA, Ouyang P, Yoo MH, Xu XM, Hatfield DL, Gladyshev VN, Davis CD. Deficiency in the 15-kDa selenoprotein inhibits tumorigenicity and metastasis of colon cancer cells. Cancer Prev Res (Phila) 2010;3(5):630–9. doi: 10.1158/1940-6207.CAPR-10-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morgan B, Sobotta MC, Dick TP. Measuring E(GSH) and H2O2 with roGFP2-based redox probes. Free Radic Biol Med. 2011;51(11):1943–51. doi: 10.1016/j.freeradbiomed.2011.08.035. [DOI] [PubMed] [Google Scholar]
- 25.Gutscher M, Pauleau AL, Marty L, Brach T, Wabnitz GH, Samstag Y, Meyer AJ, Dick TP. Real-time imaging of the intracellular glutathione redox potential. Nat Methods. 2008;5(6):553–9. doi: 10.1038/nmeth.1212. [DOI] [PubMed] [Google Scholar]
- 26.Yoo MH, Gu X, Xu XM, Kim JY, Carlson BA, Patterson AD, Cai H, Gladyshev VN, Hatfield DL. Delineating the role of glutathione peroxidase 4 in protecting cells against lipid hydroperoxide damage and in Alzheimer’s disease. Antioxid Redox Signal. 2010;12(7):819–27. doi: 10.1089/ars.2009.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carlson BA, Moustafa ME, Sengupta A, Schweizer U, Shrimali R, Rao M, Zhong N, Wang S, Feigenbaum L, Lee BJ, Gladyshev VN, Hatfield DL. Selective restoration of the selenoprotein population in a mouse hepatocyte selenoproteinless background with different mutant selenocysteine tRNAs lacking Um34. J Biol Chem. 2007;282(45):32591–602. doi: 10.1074/jbc.M707036200. [DOI] [PubMed] [Google Scholar]
- 28.Carlson BA, Xu XM, Gladyshev VN, Hatfield DL. Selective rescue of selenoprotein expression in mice lacking a highly specialized methyl group in selenocysteine tRNA. J Biol Chem. 2005;280(7):5542–8. doi: 10.1074/jbc.M411725200. [DOI] [PubMed] [Google Scholar]
- 29.De Benedetto ML, Capo CR, Ferri A, Valle C, Polimanti R, Carrì MT, Rossi L. Glutaredoxin 1 is a major player in copper metabolism in neuroblastoma cells. Biochim Biophys Acta. 2013;1840(1):255–61. doi: 10.1016/j.bbagen.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 30.Couturier J, Przybyla-Toscano J, Roret T, Didierjean C, Rouhier N. The roles of glutaredoxins ligating Fe-S clusters: Sensing transfer or repair functions? Biochim Biophys Acta. 2015;1853(6):1513–27. doi: 10.1016/j.bbamcr.2014.09.018. [DOI] [PubMed] [Google Scholar]
- 31.Zakharyan RA, Tsaprailis G, Chowdhury UK, Hernandez A, Aposhian HV. Interactions of sodium selenite, glutathione, arsenic species, and omega class human glutathione transferase. Chem Res Toxicol. 2005;18(8):1287–95. doi: 10.1021/tx0500530. [DOI] [PubMed] [Google Scholar]
- 32.Fernandes AP, Holmgren A. Glutaredoxins: glutathione-dependent redox enzymes with functions far beyond a simple thioredoxin backup system. Antioxid Redox Signal. 2004;6(1):63–74. doi: 10.1089/152308604771978354. [DOI] [PubMed] [Google Scholar]
- 33.Menon D, Board PG. A role for glutathione transferase Omega 1 (GSTO1-1) in the glutathionylation cycle. J Biol Chem. 2013;288(36):25769–79. doi: 10.1074/jbc.M113.487785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalyanaraman B, Darley-Usmar V, Davies KJ, Dennery PA, Forman HJ, Grisham MB, Mann GE, Moore K, Roberts LJ, 2nd, Ischiropoulos H. Measuring reactive oxygen and nitrogen species with fluorescent probes: challenges and limitations. Free Radic Biol Med. 2012;52(1):1–6. doi: 10.1016/j.freeradbiomed.2011.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoo MH, Xu XM, Carlson BA, Gladyshev VN, Hatfield DL. Thioredoxin reductase 1 deficiency reverses tumor phenotype and tumorigenicity of lung carcinoma cells. J Biol Chem. 2006;281(19):13005–8. doi: 10.1074/jbc.C600012200. [DOI] [PubMed] [Google Scholar]
- 36.Jakupoglu C, Przemeck GK, Schneider M, Moreno SG, Mayr N, Hatzopoulos AK, de Angelis MH, Wurst W, Bornkamm GW, Brielmeier M, Conrad M. Cytoplasmic thioredoxin reductase is essential for embryogenesis but dispensable for cardiac development. Mol Cell Biol. 2005;25(5):1980–8. doi: 10.1128/MCB.25.5.1980-1988.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suvorova ES, Lucas O, Weisend CM, Rollins MF, Merrill GF, Capecchi MR, Schmidt EE. Cytoprotective Nrf2 pathway is induced in chronically txnrd 1-deficient hepatocytes. PLoS One. 2009;4(7):e6158. doi: 10.1371/journal.pone.0006158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carlson BA, Yoo MH, Tobe R, Mueller C, Naranjo-Suarez S, Hoffmann VJ, Gladyshev VN, Hatfield DL. Thioredoxin reductase 1 protects against chemically induced hepatocarcinogenesis via control of cellular redox homeostasis. Carcinogenesis. 2012;33(9):1806–13. doi: 10.1093/carcin/bgs230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seeher S, Atassi T, Mahdi Y, Carlson BA, Braun D, Wirth EK, Klein MO, Reix N, Miniard AC, Schomburg L, Hatfield DL, Driscoll DM, Schweizer U. Secisbp2 is essential for embryonic development and enhances selenoprotein expression. Antioxid Redox Signal. 2014;21(6):835–49. doi: 10.1089/ars.2013.5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahdi Xu XM, Carlson BA, Fradejas N, Günter P, Braun D, Southon E, Tessarollo L, Hatfield DL, Schweizer U. Expression of Selenoproteins Is Maintained in Mice Carrying Mutations in SECp43, the tRNA Selenocysteine 1 Associated Protein (Trnau1ap) PLoS One. 2015;10(6):e0127349. doi: 10.1371/journal.pone.0127349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dikiy A, Novoselov SV, Fomenko DE, Sengupta A, Carlson BA, Cerny RL, Ginalski K, Grishin NV, Hatfield DL, Gladyshev VN. SelT, SelW, SelH, and Rdx12: Genomics and Molecular Insights into the Functions of Selenoproteins of a Novel Thioredoxin-like Family. Biochemistry. 2007;46(23):6871–82. doi: 10.1021/bi602462q. [DOI] [PubMed] [Google Scholar]
- 42.Jeon YH, Ko KY, Lee JH, Park KJ, Jang JK, Kim IY. Identification of a redox-modulatory interaction between selenoprotein W and 14-3-3 protein. Biochim Biophys Acta. 2015;1863:10–8. doi: 10.1016/j.bbamcr.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 43.Carlson BA, Moustafa ME, Sengupta A, Schweizer U, Shrimali R, Rao M, Zhong N, Wang S, Feigenbaum L, Lee BJ, Gladyshev VN, Hatfield DL. Selective restoration of the selenoprotein population in a mouse hepatocyte selenoproteinless background with different mutant selenocysteine tRNAs lacking Um34. J Biol Chem. 2007;282:32591–602. doi: 10.1074/jbc.M707036200. [DOI] [PubMed] [Google Scholar]
- 44.Hatfield DL, Gladyshev VN. How selenium has altered our understanding of the genetic code. Mol Cell Biol. 2002;22:3565–76. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sundae RA, Raines AM, Barnes KM, Evenson JK. Selenium status highly regulates selenoprotein mRNA levels for only a subset of the selenoproteins in the selenoproteome. 2009;29:329–38. doi: 10.1042/BSR20080146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lillig CH, Berndt, Holmgren A. Glutaredoxin systems. Biochim Biophys Acta. 2008;1780:1304–17. doi: 10.1016/j.bbagen.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 47.Schweizer U, Schlicker C, Brauna D, Köhrle J, Steegborn C. Crystal structure of mammalian selenocysteinedependent iodothyronine deiodinase suggests a peroxiredoxin-like catalytic mechanism. Proc Natl Acad Sci USA. 2014;111:10526–31. doi: 10.1073/pnas.1323873111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Targeted removal of SPS1 in mice.
Figure S2. Remnants of Sps1 knockout mice at E14.5.
Figure S3. SPS1 levels in control and hepatocyte-specific Sps1 knockout (Alb-Cre; Sps1fl/fl) mouse liver.
Figure S4. Clinical chemistry analysis in plasma from control and Alb-Cre; Sps1fl/fl mice.
Figure S5. Selenium levels and Sec tRNA isoform distribution in livers from control and Alb-Cre; Sps1fl/fl mice.
Figure S6. Element analysis in livers from control and Alb-Cre; Sps1fl/fl mice.
Figure S7. Knockdown of Sps1 in F9 cells.
Figure S8. Measurement of total GSH and GSSG, and the GSH/GSSG ratio in the control (F9/pSuper) and Sps1-deficient cells.
Figure S9. Expression of selenoproteins in SPS1-deficient mouse F9 embryonal carcinoma cells.
Figure S10. Detection of intracellular superoxide in SPS1-deficient F9 cells.
Figure S11. Measurement of superoxide dismutase activity.
Figure S12. Measurement of the ratio of disulfide/dithiol GLRX1 in the mitochondria and cytosol of F9 cells using Grx1-roGFP2 probes.
SM Text. Materials and Methods and References
Table S1. Primer sequences.
Table S2. Microarray analysis of the 25 most highly (A) up- and (B) down-regulated genes in Alb-Cre; Sps1fl/fl mouse liver
Table S3. Ingenuity Pathway Analysis of liver from control and Alb-Cre; Sps1fl/fl mice
Table S4. Microarray analysis of the 25 most highly (A) up- and (B) down-regulated genes in SPS1 knockdown F9 mouse embryonic carcinoma cells.