Abstract
Comparative mapping and sequencing show that turnover of sex determining genes and chromosomes, and sex chromosome rearrangements, accompany speciation in many vertebrates. Here I review the evidence and propose that the evolution of therian mammals was precipitated by evolution of the male‐determining SRY gene, defining a novel XY sex chromosome pair, and interposing a reproductive barrier with the ancestral population of synapsid reptiles 190 million years ago (MYA). Divergence was reinforced by multiple translocations in monotreme sex chromosomes, the first of which supplied a novel sex determining gene. A sex chromosome‐autosome fusion may have separated eutherians (placental mammals) from marsupials 160 MYA. Another burst of sex chromosome change and speciation is occurring in rodents, precipitated by the degradation of the Y. And although primates have a more stable Y chromosome, it may be just a matter of time before the same fate overtakes our own lineage.
Also watch the video abstract.
Keywords: animal genomics, de novo sex‐determining genes, hybrid incompatibility, mammal evolution, sex chromosome differentiation and degradation, sex chromosome rearrangement
Introduction: Chromosome change and speciation
Older literature is replete with suggestions that chromosome rearrangements – particularly highly disruptive translocations between chromosomes – induce hybrid sterility, and serve to separate incipient species 1.
Many examples, particularly in grasshoppers and other insects, were put forward and discussed 2, as well as examples from reptiles 3. Many species complexes, indeed, contain species with few phenotypic differences, but chromosome exchanges that make hybrids sterile 2. The ideas were propounded that such changes occurred in an individual, were inherited by the offspring, and became fixed by mating between sibs or other relatives in small populations. Homozygotes could mate effectively only within the small group; matings between this group and the original population suffered from hybrid infertility.
However, evidence that chromosome rearrangement is important in speciation was fragmentary, and modelling has been less than convincing (reviewed 4). Views on the mechanisms of speciation, therefore, focused on the Muller‐Dobzhansky model, whereby accumulation of small mutations in populations that were physically separated promoted divergence 5, 6. Decades of work, primarily on Drosophila melanogaster, identified such ‘speciation genes’, whose mutation was associated with hybrid incompatibility, and similar genes are now found in many other organisms 7. Many alternative and complementary models have been produced to explain the processes by which one species becomes two. Also the major tenets of the hypothesis that chromosome rearrangement is critical in speciation do not receive strong support from the more detailed and incisive analysis permitted by newer molecular techniques 8. Many examples have been studied in which hybrid incompatibility is not a serious barrier to gene flow. Indeed, the importance of chromosome rearrangements has been suggested to be more in the isolation of regions of low recombination which might harbour packages of genes that work in one environment but not another 9, 10.
The role of chromosome change in speciation was also problematical because it has such drastic effects. The argument was that such changes occurred in a single individual, and could not spread through the population because of the infertility of hybrids. Because speciation was reckoned to require a long time during which changes were polymorphic, it was difficult to imagine that such drastic changes could remain in the population long enough to become fixed, and were, therefore, thought to follow, rather than to induce speciation. Yet drastic chromosome changes do get fixed, if rarely. So there must be a route from a single individual change, through the spread, at least to two individuals of the opposite sex, to enable homozygosity and fixation.
Sex chromosome turnover and rearrangement may present a special case. Sex chromosomes are known to have major effects on speciation in terms of Haldane's Rule and the large X effect, explored in detail in Drosophila but evident also in mammals 11, 12. New comparative genetic data document many vertebrates in which autosomes in one species have become sex chromosomes in another, as well as many rearrangements of sex chromosomes and autosomes. Molecular techniques have also documented the special properties of differentiated sex chromosomes in hemizygous expression and dosage compensation, and sex‐biased gene content. Hybrids between individuals with different sex pairs are much more likely to suffer from infertility because of disruptions, not only to pairing and recombination, but also to sex determination and dosage compensation and to the disruption of the activity of fertility genes on sex chromosomes 13.
Here I argue that sex chromosome change, which is both more damaging because of specialisation and degradation of sex chromosomes, but also more able to persist because incompatibility may be asymmetric (that is, has different effects in reciprocal crosses), precipitated divergence of the major mammal groups.
How are sex chromosomes rearranged or replaced?
In amniotes (mammals, birds and reptiles), the genome and the karyotype are amazingly conserved; there are few major chromosome rearrangements between an emu and a Galapagos tortoise, a snake and a chicken. Eutherian (placental) mammals are the glaring exception, showing a great variety of chromosome numbers and genome rearrangements; even the other two branches of mammals (marsupials and monotremes) have very conserved karyotypes 14.
However, even in taxa with extremely conserved karyotypes, sex chromosome turnover can take place when a novel gene on an autosome takes over the job of kick‐starting gonad differentiation.
Fish provides numerous examples of sex chromosome turnover. For instance, in medaka and its close relatives with similar karyotypes, different chromosomes show sex linkage, as a result of the rapid evolution of different sex determining genes 15, 16, 17. In sticklebacks, too, novel sex genes have rapidly arisen 18, 19, and in cichlid fish of Lake Victoria, rampant speciation is often accompanied by sex chromosome turnover 20. In frogs, too, different sex determining genes and sex chromosome systems have arisen, and one species is polymorphic for XY and ZW systems 21, 22, 23, 24.
In some reptile lineages, sex chromosome turnover is rare. For instance, all birds have a ZZ male:ZW female system and share the same Z chromosome, determining sex by means of dosage of a Z‐borne gene DMRT1 25. Snakes, with a different ZW pair, also appear to be very stable, at least in the two families (Colubridae and Viperidae) with highly degraded W chromosomes 26.
However, sex chromosome change is extremely frequent in other reptile families (e.g. lizards). One extra factor in reptiles is the ability of many species to determine sex, not by sex chromosomes and genes, but by environmental temperature. There are phylogenies of closely related agamid lizards that reveal at least four flip‐flops through genetic sex determination (GSD) and temperature sex determination (TSD), generating novel sex chromosomes 27. Some reptile species have sex chromosome systems that can be overridden by temperature 28, 29; these dual systems are extremely labile, able to change rapidly to TSD systems and even between ZW and XY 30, 31. Similar GSD‐TSD transitions may be common in some fish and may also facilitate changes in sex determination 32, 33, 34.
Much has been written about evolutionary drive to fill subtly different niches, but perhaps the triggers for speciation in some vertebrate groups include the inherent instability of the sex determining system.
Mammal sex genes and sex chromosomes are very stable
In comparison, mammals have a very stable sex determination system. Since they are homoeothermic, GSD to TSD transitions are not an option, and the XY pair of therian mammals has differentiated to extremes that make YY individuals inviable.
There are three major groups of mammals; Eutheria (placental) and Metatheria (marsupials) together comprising subclass Theria, which diverged from the egg‐laying monotremes (Prototheria) 190 million years ago (MYA) 35 (Fig. 1). All eutherians have the same XY chromosome pair, defined by a male‐dominant testis‐determining gene identified as SRY 36. SRY activates a genetic pathway that promotes differentiation of a bipotential gonad into a testis in the embryo and suppresses ovarian development 37, 38.
Figure 1.
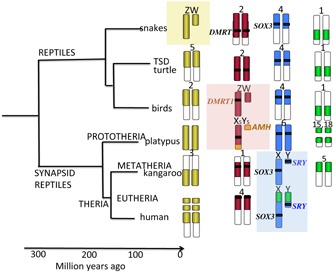
Relationships among higher vertebrate and their sex chromosomes. Placental mammals (infraclass Eutheria) diverged from marsupial mammals (Infraclass Metatheria) about 166 MYA 35. These two groups comprise mammalian Subclass Theria, which diverged from Subclass Prototheria (monotreme mammals such as the platypus) 190 MYA. Ancestral synapsid (mammal‐like) reptiles diverged from other reptile groups about 310 MYA. Comparative mapping demonstrates five orthologous segments that are sex chromosomes in one vertebrate lineage and autosomal in others. The yellow segment has become sex chromosomes in snakes (sex determining gene remains unknown). The red segment defined by DMRT1 became sex determining in birds; this segment is represented on the sex chromosomes of platypus but the sex determining gene is AMH, translocated from another block (orange). The blue region defined by SRY (which evolved from SOX3) became sex chromosomes in mammals, and the green region fused with this ancient XY in eutherian mammals.
The gene content of the X is virtually identical over even the most distantly related eutherians. The Y chromosome is a relic of the X that degraded to different extents, leaving tiny overlapping sets of active genes 38. The male‐specific region of the human Y retains only 27 active genes, though some are present in multiple copies (mostly inactive); cat and pig Y chromosomes have slightly more 39 and mice fewer 40. Only two genes on the mouse Y are critical for a male phenotype and reproduction 41.
The evolution of SRY defined de novo XY sex chromosomes in therian mammals
In line with the accepted theory of sex chromosome evolution 42, 43, the evolution of SRY is credited with initiating the differentiation of the XY pair 38 (Fig. 2). SRY is thought to have evolved from SOX3, a highly conserved gene that lies on the X in mammals 44 and is expressed in the central nervous system and germcells, but not the somatic cells of the testis, where the sex determining message must be received 45. Mis‐expression of SOX3 in the Sertoli cells causes sex reversal in humans and mice 46, suggesting that a change in tissue of expression, perhaps by fusion with a gonad‐specific promotor, created the male‐dominant SRY gene that took over sex determination from the ancestral gene 47.
Figure 2.
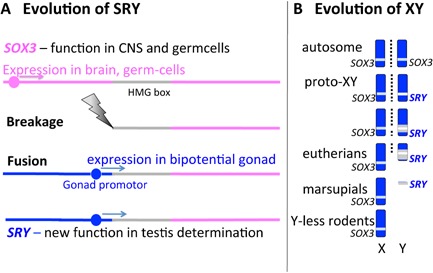
Evolution of SRY and the mammal XY sex chromosome pair. A: Evolution of SRY from an allele of autosomal SOX3. SOX3 is highly conserved throughout vertebrates, with functions in brain and germcells, but is sex reversing when mis‐expressed in the bipotential gonad. SOX3 shares with SRY a moderately conserved HMG box (grey), involved with DNA binding and bending through a specific angle. We speculate that SOX3 was truncated by breakage at the 5′ end, and rejoining with a promotor that drove its expression into the bipotential gonad. B: Differentiation of the mammal X and Y chromosomes. Originally the X and Y were an ordinary pair of autosomes bearing SOX3. Mutation of one allele to a sex‐determining SRY ensured that the chromosome that bore it became a male‐specific Y. It degenerated rapidly as nearby genes acquired male‐specific functions, and recombination (represented by dotted lines) in the region was suppressed to preserve a male‐specific gene package. Within the region of low recombination, mutation, deletion amplification and insertion of repetitive sequence was rife, hastening progressive degeneration. Genes that acquired a male‐specific function (grey) were retained. The XY of eutherian mammals retains a short pseudoautosomal segment that still recombines. However, the marsupial X and Y have lost all homology, and some rodents have completely lost the Y.
The emergence of a novel sex determining gene has been shown, in many systems, to drive the degradation of the chromosome on which it lies. In an ancient mammal, the acquisition of the dominant male determining SRY gene was, therefore, the kiss of death for the autosome that bore SOX3. Nearby genes were selected for a male‐advantage function and the region became non‐recombining to keep this male‐specific package together 38. Recruitment of more genes on the Y to a male‐specific function extended this region progressively until only the terminal few megabases remained homologous to the X as the shared pseudoautosomal region.
Not only did the Y degrade, but both the X and Y rapidly specialised. Genes on the Y acquired male‐specific functions such as in spermatogenesis. Meanwhile the X rapidly acquired sex and intelligence genes as a result of sexual antagonism and the hemizygous state in males 38. Dosage compensation evolved to silence one X in females, possibly after upregulation of genes on the X in both sexes 40.
Thus, a minor molecular accident – gene fusion and change of the transcription profile of SOX3 – was all that was required to precipitate a major turnover of sex chromosomes in ancestral mammals.
Platypus sex chromosomes rewrote mammal chromosome evolution
When did this happen? And what was the sex determining system of the ancestor of all mammals which was replaced by the novel SRY‐bearing chromosome pair in therians?
The standard approach to this question is to examine the sex chromosomes of outgroups. However, mammals and reptiles last shared a common ancestor 310 MYA, so the trail has gone a bit cold. Also, as we have seen, reptiles have an amazing array of different sex determining mechanisms, and exhibit frequent changes. It is difficult to deduce the sex determining system of a reptile ancestor, which might even have been environmental 48.
The basal group of mammals, the Prototheria, should provide some clues to the ancestral mammalian sex determining system. Monotremes (comprising two families, platypus and echidna) have multiple sex chromosomes 49. The platypus has 10 sex chromosome, five genetically distinct X and five Y. These form a chain at male meiosis which is held together by crossing over within nine pseudoautosomal regions. The five Xs segregate to one pole and the five Ys to the other 50.
The platypus orthologues of human X genes lie, not on the sex chromosomes, but on autosomes 51. Mapping the genes on the five X and five Y chromosomes confirmed that platypus sex chromosomes have no homology to the therian X. But, surprisingly, they have extensive homology to the bird ZW sex chromosome pair 52.
Other gene blocks on the platypus sex chromosomes have homology to other chicken chromosomes, consistent with the derivation of platypus chain by sequential translocations with autosomes 53. Sequence comparisons between X‐Y shared genes revealed that the large X5 and tiny Y5 that terminate the chain are the most diverged, and therefore the oldest members of the chain 54, suggesting that they represent the original mammalian sex chromosomes. X5 is largely homologous to the bird Z 52.
The other monotreme family, the echidnas, also have a sex chromosome chain but it consists of only nine elements, with the tiny terminal Y5 fused onto another Y 55. Chromosome painting and gene mapping shows that four of the five XY pairs in the echidna chain are the same, but the fifth is represented by an autosome in platypus. Likewise, one of the platypus XY elements is represented by an autosome in echidnas. This implies that the three translocations in common occurred before platypus and echidna diverged, probably about 50 MYA 56, 57, but one further translocation occurred independently in each of the two lineages at or after divergence, and involved different autosomes.
The identity of the monotreme sex determining gene has been controversial. Not surprisingly, monotremes lack SRY, and SOX3 is autosomal 58, 59, lying with orthologues of other human X genes on chromosome 6. The bird sex determining gene DMRT1 lies on an X chromosome in monotremes, so is unlikely to promote dosage‐dependent determination in which two copies promote male development. There is no trace of a Y‐borne DMRT1 variant that could act as a male‐determining gene. The most convincing candidate is an AMH orthologue that lies with syntenic markers on the tiny Y5 chromosome 54. This suggests that translocation of the ancestral sex pair with an autosome provided a novel sex gene in an ancestral monotreme. The other four Y chromosomes are very degenerate; it will be interesting to discover whether they retain gene variants that have acquired functions in fertility.
We can date the evolution of SRY and the XY system of therian mammals from the known divergence dates of reptiles and mammal lineages (Fig. 1). Since birds and reptiles, and even monotremes do not share the therian XY, it must have evolved at or after the Prototheria‐Theria divergence 190 MYA. And since marsupials and eutherians, which diverged 166 MYA, share SRY and the XY chromosome pair, SRY and the therian XY must have evolved in the 24 million year window between 190 and 166 million years ago.
Sex chromosome‐autosome fusion accompanied evolution of eutherian mammals
Although the XY pair of marsupials and eutherians is monophyletic, there is an important difference. Comparative mapping of the marsupial orthologues of genes on the long arm and the pericentric region of the human X lie on the marsupial X, but more distal genes on the short arm of the human X map together on an autosome in marsupials 60.
This defines two ancient evolutionary blocks, the X conserved region XCR, which is shared by all therian mammals, and the X added region XAR, which is autosomal in marsupials.
Conserved and added regions YCR and YAR can be identified also on the human Y by mapping the X homologues of Y genes. The ancestor of SRY (SOX3) is one of the few Y genes that survive from the ancient XY; most human Y genes derive from the added region 61.
Regions orthologous to XCR and XAR are separate also in birds and other vertebrates 62, implying that they represent two separate ancient gene blocks that fused in eutherians. The addition of an autosomal region to the Y as well as to the X chromosome implies that fusion occurred within a large pseudoautosomal region when the XY pair was only partially differentiated.
Again we can date this fusion by comparing distantly related eutherians, humans and the basal afrotherians (Fig. 1). The finding that genes on the human X all lie also on the elephant X chromosome implies that the fusion occurred before the eutherian radiation about 105 MYA 63. And the finding that in elephants, XCR and XAR are separated by the centromere, suggests that the two ancestral blocks underwent centromere‐centromere (Robertsonian) fusion 64.
Sex chromosome changes define the three major mammal groups
These data imply that three major sex chromosome changes separate the three major mammal lineages:
de novo evolution of SRY and degeneration of the neo Y chromosome in a therian ancestor 190–166 MYA,
three X‐autosome translocations in monotremes 190–55 MYA, and a further independent translocation in platypus and echidna lineages,
Robertsonian fusion of XY and autosome in eutherians 166–105 MYA.
We do not know how sex determination operated in the ancestor of all mammals, but it is clear that it did not involve SRY, whose ancestor SOX3 is autosomal in monotremes as well as other vertebrates. The homology between monotreme and bird sex chromosomes initially suggested that the mammalian ancestor had a bird‐like ZW system, driven by DMRT1 dosage. However, this need not represent identity by descent, for a flatfish also has a ZW sex pair with homology to the bird ZW, and uses DMRT1 dosage to determine sex 32. DMRT1 has also been pressed into service independently by spawning copies that became a dominant male‐determiner in medaka 16, 65 or female determiner in Xenopus 66; clearly this gene is simply good at sex determination and has been independently recruited in several lineages 67.
In order to explore specific consequences of a take‐over by a de novo SRY‐driven XY system, I shall posit that an ancient ZW system, similar to that in birds, prevailed in the ancestor of all mammals. I propose that SRY arose on a ZZ background after the mis‐expression of SOX3 in the bipotential gonad, and became fixed in a small population, which could no longer effectively mate with the original population. This population gave rise to therian mammals.
Each of these changes would have been highly disruptive to hybrids. Matings between animals with a primitive bird‐like ZW system and the de novo XY system could produce offspring with mixtures of sex chromosomes that are likely to be infertile or even inviable (Fig. 3). The degree of disruption would depend on the rapidity of sex chromosome differentiation and degradation. If Y chromosome change and dosage compensation were rapidly established, offspring of heterogametic XY males and ZW females would suffer from haploinsufficiency, exacerbated by specialisation of the X and the evolution of dosage compensation (Fig. 3b). If the de novo XY system maintained homomorphy for longer, the two systems might co‐exist for many generations. Interestingly, the results of reciprocal crosses might differ; a cross between homogametic XX females and ZZ males might deliver some viable and fertile individuals (Fig. 3c), and thus might maintain a polymorphism for many generations.
Figure 3.
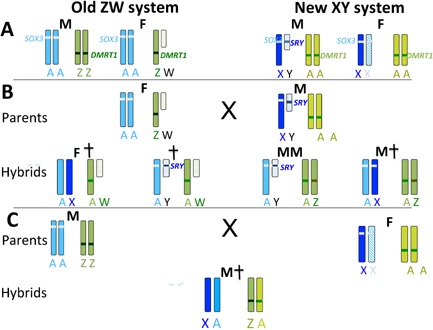
Interaction of ancient ZW and de novo XY sex determining systems. A: In the old ZW system, the male (M) has two Z chromosomes (green), bearing DMRT1. Females (F) have a single Z and a degenerate W. Dosage of DMRT1 determines sex; two copies are required for male development (dark green). Some upregulation of Z genes occurs in females. An autosome (blue) bearing SOX3 (light blue) is present in two copies in both sexes. In the new XY system, with the evolution of SRY (dark blue) from SOX3, the blue autosome has become a sex pair. Males have a single X and a degenerate Y bearing the male‐dominant testis determining gene SRY; genes on single X are upregulated (darker blue). Females have two X chromosomes; one upregulated (darker blue) and one inactive (blue hatching). Gene content of the X has become specialised. The Z chromosome has become an autosome (A, lighter green), losing its dosage compensation and DMRT1 is no longer sex determining (lighter green). B: Matings between heterogametic female (ZW) and heterogametic male (XY) produce hybrid offspring with four combinations of ZW and XY chromosomes, with predicted phenotypes as shown (male M, female F, dead †). Hybrids with two green chromosomes may be male with or without SRY. Only offspring with two blue chromosomes and a single green chromosome are likely to be female. Offspring with both a degenerate W and Y are likely to be inviable, and offspring with two blue chromosomes (an X and autosome) could suffer from dosage problems (too much product if the X is upregulated; too little if it is inactivated). C: Matings between homogametic male (ZZ) and homogametic female (XX) produces offspring with only one combination of sex chromosomes. With two DMRT1 copies these are likely to be male; however, the combination of an X and the original autosome may not be viable if the X is upreguated or inactivated.
The effects of translocations and fusions between sex chromosomes and autosomes are well known in mice and humans 68, 69. Individuals heterozygous for translocation produce half balanced and half unbalanced gametes, a severe reproductive barrier between old and new systems. Interference with the dosage compensation system appears to contribute to infertility in mouse translocation hybrids 70.
It is also interesting to explore what might happen when a sex chromosome autosome translocation takes place, as was the case in ancestral monotremes. Again, for argument's sake, I will propose that monotremes originally had a bird‐like ZW system driven by dosage of DMRT1. The first translocation to this ancestral Z (probably in a ZZ male) added a region that contained the AMH gene, an important player in the sex determining network in reptiles, and the sex determining gene in some fish 16, 71. I propose that AMH took over as a male‐dominant sex determining gene, defining a neo‐Y. A system of alternate segregation evolved to segregate multiple X and Y chromosomes into X‐ and Y‐bearing sperm. This change could have promoted or reinforced monotreme divergence, since translocation heterozygotes would produce unbalanced gametes, as well as having two, possibly antagonistic, sex determining genes. Subsequent translocations would have been less disruptive, since alternate anaphase segregation had already evolved; however, a fourth translocation, different in platypus and echidna, may have promoted divergence of families Ornothorhynchidae (platypus) and Tachyglossidae (echidnas).
Either change could have been first; de novo XY evolution may have precipitated divergence of Theria, leaving the ancient ZW system to be later salvaged by sex chromosome‐autosome translocation in Prototheria. Alternatively an initial translocation may have been sufficient to drive apart the ancestors of Theria and Prototheria, with one lineage evolving alternate segregation, and the other soldiering on until it evolved a novel sex chromosome system.
In the therian lineage, a further major change, fusion of the XY with an autosome, may have promoted divergence of eutherian mammals from marsupials.
Thus the differences in sex chromosomes of the three major mammal groups would all be highly disruptive to hybrid matings. I propose here that these changes, rather than merely following the isolation of mammal populations, had a major role in precipitating divergence.
How does sex chromosome turnover occur?
This scenario for the sex chromosome‐driven divergence of major mammal groups calls into question how sex chromosome turnover occurs, how it can be fixed in a population, and the selective forces promoting and opposing fixation. There is a catch‐22 here; divergence depends on a reproductive barrier being posed by hybrid infertility, and the barrier is highest after the most drastic changes. However, the more drastic the change, the less likely it is to be fixed.
The problems of accounting for chromosome change have been debated over decades. A chromosome rearrangement must occur, or a novel sex determining gene must arise, in a single individual. How can this spread in a population when it leads to infertility? Turnover of mammal sex chromosomes is equally difficult to explain, whether or not it promotes divergence.
However, sex chromosome turnover has occurred in mammals, though rarely. What needs to be explored, therefore, is how and why does it get fixed? Is there selection for the new system, and on what characteristic?
In examining how a novel sex chromosome can take over a population, it is useful to distinguish between young sex chromosome systems and highly differentiated sex chromosomes.
Young sex chromosomes, such as those in many fish and some reptiles, show little differentiation. Thus the Y (or W) chromosome has not yet degraded to the extent that vital genes have been lost, nor has dosage compensation evolved to any great extent. This means that YY (or WW) individuals are likely to be viable, and a new sex gene on another chromosome may exist as a polymorphism for a considerable time. This seems to be the case for the Japanese wrinkled frog Rana rugosa, in which XY and ZW systems co‐exist in a hybrid zone where many different sex chromosome combinations occur 23.
In such systems, the original sex chromosomes can readily revert to their previous status as autosomes. This appears to have occurred in medaka and related fish, in which an ancestral sex determining gene on one chromosome was replaced with a copy of the DMRT1 gene on a different chromosome 65. This can occur more easily if the sex chromosomes have differentiated to only a minor extent; however, highly diverged sex chromosomes may survive as autosomes if the de novo system arises on either an XX female or a ZZ male background.
The extent to which replacement of highly differentiated sex chromosomes with a novel system poses a reproductive barrier between old and new sex systems will depend on the rapidity with which the novel system differentiates. Hybrids will be subfertile, producing unbalanced gametes and duplication‐deficient offspring (Fig. 3). In addition, there is likely to be derangement of expression of genes on the X (or Z) that are dosage compensated.
How do new sex chromosomes become fixed?
However, it is more difficult to explain how such a drastic alteration in a single individual can become fixed in a population. The classic explanation is that random mutations or rearrangements occur by chance in small populations and can achieve homozygosity by inbreeding 1. There is ample evidence that chromosome rearrangements may occur and be randomly fixed in small populations without being adaptive. For instance, many different chromosome fusions have become fixed in isolated populations of mice introduced to Mauritius only 700 years ago; these populations are now unable to hybridise 72. This is far too short a time to envisage gradual accumulation of mutants in isolated populations that ultimately pose a reproductive barrier, and the most sensible conclusion is that non‐adaptive chromosome change and drift has promoted speciation.
Although many authors have discounted the possibility that drastic chromosome change or sex chromosome turnover can occur and be fixed, we have two examples of recent turnover in groups of rodents that have replaced SRY and lost the entire Y chromosome.
The better characterised of these are species of the spiny rat Tokudaia, which live a precarious existence on islands of south Japan. Two species have XO males and females; they lack SRY, and other fertility genes on the mouse Y have been moved or replaced. There are mutations in the target gene SOX9 73, and a novel gene CBX2 (Chromobox protein homologue 2) appears to have been elevated from its subservient role in gene regulation via chromatin remodelling to the head of the sex determining pathway 74.
Independently, loss of the Y and SRY has occurred in two species of the mole vole Ellobius 75, which presumably now determine sex by an as yet undiscovered gene on the X or autosomes. The loss of the SRY sex determining mechanism seems to have been accompanied by changes in its target gene SOX9 that may have led to its upregulation in the absence of SRY 76.
Interestingly, the X chromosome in both these groups of Y‐less rodents has not reverted to an autosomal state 77, perhaps because X‐borne genes are upregulated so that two copies are deleterious.
Clues to how loss of the Y could have occurred in spiny rats are provided by a third species (Fig. 4b) which has a bizarre Y, containing more than 100 amplified copies of a mutationally damaged SRY. This suggests that the SRY gene of a common ancestor underwent sequence changes that weakened the binding of SRY protein to DNA, so amplification of this mutant gene was strongly selected to compensate for its poor DNA binding. A change of function of CBX2 to activate the autosomal SOX9 could then have occurred, and this more robust system could have rapidly replaced the compromised SRY sex determination system.
Figure 4.
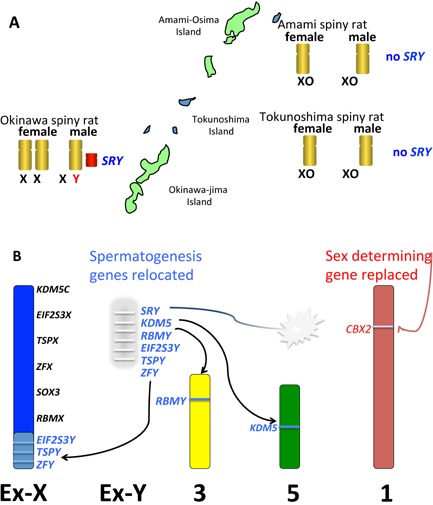
Turnover of sex chromosomes and speciation in the spiny rat. A: Three species of spiny rat (genus Tokudaia) live on islands in south Japan. The Amami spiny rat retains a Y chromosome but the Amami and Tokunoshima spiny rat have no Y and no SRY 80. B: The process of loss of the Y. Fertility genes have been relocated to autosomes (chromosomes 3, 5) or the old X as shown, and the rest of the Y, bearing SRY (amplified and possibly inactive), was lost. A novel sex determining function was acquired by CBX2 on chromosome 1.
More generally, I propose that a terminally differentiated and mutated Y chromosome may specify a compromised sex determination system, so its replacement by a de novo gene on an autosome has a selective advantage. Specifically for mammals I suggest that an old system was nearing the end of its useful life, with terminally differentiated sex chromosomes that bore a mutated sex determining gene, and were full of repetitive sequences and highly prone to rearrangement or deletion. Thus novel sex determining systems that arose independently in monotremes and therians could have been positively selected.
Can sex chromosome turnover promote divergence?
I have discussed how sex chromosome turnover, even drastic chromosome rearrangement and change from one differentiated sex pair to a novel system, can and does occur, and can be fixed in a population, even in mammals, and can contribute to reproductive isolation.
But it is extremely difficult to show that chromosome change causes reproductive isolation, rather than simply following or being coincident with an accumulation of mutations that traditionally is credited with reproductive isolation. Obviously there are many divergence events that are decidedly not caused by chromosome change or sex chromosome turnover; witness the near identical gene content of X chromosomes across all eutherian lineages, and the completely stable karyotypes of 60 species of dasyurid marsupials.
However, it is remarkable that the major mammal groups are characterised by sex chromosome rearrangement and turnover (Fig. 5), which are uncommon in mammals.
Figure 5.
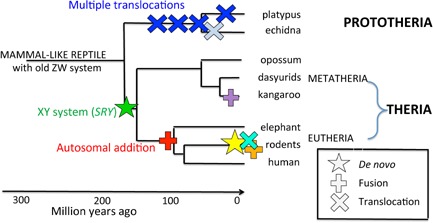
Major sex chromosome changes associated with divergence of mammal lineages. De novo evolution of SRY and the XY sex chromosome system (green star) occurred early in therian evolution and may have triggered divergence of Theria from Prototheria. At the same time, the ancient ZW system was bolstered by translocation with an autosome (blue cross), which may also have promoted divergence of Prototheria from Theria. Two further translocations occurred before divergence of platypus and echidna; different final translocations may have triggered divergence of Families Ornithorhynchidae (platypus) and Tachyglossidae (echidna). In the Theria, fusion of the XY with an autosome (denoted by +) occurred before eutherian radiation, and may have triggered divergence of Metatheria and Eutheria. Fusions are evident in some marsupial lineages, and many changes in sex determination (symbols in other colours) are observed in different rodent lineages which may signify a recent burst of sex chromosome turnover and speciation.
Remarkable though this coincidence is, it does not tell us that divergence was caused, or even promoted by, chromosome change. How might we distinguish the two hypotheses? Information about populations at deep divergence points such as those that separated the major three groups of mammals is inaccessible. Even where change has occurred over a brief evolutionary time, as in spiny rats, it remains possible that small island populations accumulated mutations that interposed reproductive barriers independently of sex chromosome turnover.
The most advantageous systems to analyse might be rodent groups that have undergone rapid divergence and seem to be undergoing sex chromosome turnover.
Is there a new round of mammalian sex chromosome turnover and speciation?
The mammal Y chromosome shows all the signs of extreme differentiation. The mouse Y, particularly, seems to be near the end of its useful life. It contains only two genes essential for male phenotype, and these can be readily replaced; SRY by upregulating SOX9, and EIF2S3Y a sexually dimorphic transcription factor, by upregulating its X‐borne partner EIF2S3X 78.
Many rodent species seem now to be experimenting with novel systems 79. There are several examples of closely related rodent species with variant sex chromosomes. As well as the two genera that contain Y‐less species discussed above, there are three species of Patagonian akodont mice that have evolved an inactive SRY gene, leading to SRY+ female mice 80. Since other species in the same clade do not have SRY+ females, these changes were thought to be independent; however, they may have occurred after SRY was mutationally damaged, or relocated to part of the Y subject to epigenetic silencing in an ancestral animal. Wood lemmings and a pygmy mouse species have independently evolved variant X chromosomes that suppress SRY action, again leading to XY SRY+ females 79.
Pygmy mice are famous for rearrangements of sex chromosomes and autosomes 81. There are X‐ and Y‐autosome fusions that differentiate closely related species, and some species have both, requiring segregation from a chain of four or even six chromosomes at male meiosis. This is reminiscent of the monotreme translocation complex – perhaps both were selected because they rescue an ancestral system that has become error‐prone.
Nor are rodents the only taxon that is prone to sexual experimentation. Bats and shrews, too, show many sex chromosome‐autosome fusions. Marsupials have XXY and XYY systems 82, and one fusion of a NOR‐carrying autosomal regions 83 may have separated the kangaroo family from other marsupials 11 MYA.
Are rodents showing us the future for our own lineage? Human, indeed primate, sex chromosomes seem to be unusually stable 84, so the human Y may last far longer than the 4.6 MY predicted on the basis of linear decay, or even the more generous lifetime afforded by exponential decay and positive selection 38, 85. But if primates survive for long enough, change will happen. Will new rounds of fusions rescue the dying Y as in monotremes? Or will novel genes evolve and define new sex chromosomes as in medaka fish and ancient therians? If it happens, will the novel sex chromosomes and sex determining systems promote hominid speciation?
Conclusion and outlook
New sex determining genes, and novel and rearranged sex chromosomes, characterise all three major branches of mammals. These types of sex chromosome turnover leads to infertility of hybrid offspring, because of unbalanced gametes and disruption of dosage compensation mechanisms.
How are such radical changes perpetuated and fixed in a population? Spread may have been facilitated by the unequal effects of reciprocal crosses between homogametic and heterogametic males and females. They may even have been selected for if the system they replaced was in a terminal stage of degradation.
Did these changes drive, or follow, divergence of mammal lineages? Each would provide a barrier to mating between individuals with old and new sex chromosome systems. Chromosome speciation was discounted for decades in favour of hypotheses that populations first become isolated (physically or behaviorally), and diverged gradually by the accumulation of small mutations.
To distinguish between these two hypotheses, mammals with variant sex chromosome systems are particularly valuable. Several rodent lineages show recent chromosome rearrangement or turnover, with novel genes that substitute for – or repress – SRY. These novel systems could provide new insight about the role of sex chromosome turnover in speciation.
The other source of information might be species that recently entered a new niche, such as the mice in Mauritius in which chromosome change and speciation occurred too rapidly to allow for slow accumulation of mutants. Particularly revealing may be captive colonies of zebrafish, which independently lost their W and now rely on different genes in the sex determining pathway 86.
Complete genome sequencing of non‐model vertebrates promoted by Genome 10 K 87 may reveal many more systems that are in the process of sex chromosome change, and which we can exploit to determine under what conditions sex chromosome turnover leads to reproductive isolation, and how this translates into speciation.
The author has declared no conflict of interest.
References
- 1. White MJD. 1973. Animal Cytology and Evolution. 3d Ed Cambridge, England: University Press; p 961. [Google Scholar]
- 2. King M. 1993. Species Evolution: The Role of Chromosome Change. Cambridge, England: Cambridge University Press. [Google Scholar]
- 3. Olmo E. 2005. Rate of chromosome changes and speciation in reptiles. Genetica 125: 185–203. [DOI] [PubMed] [Google Scholar]
- 4. Faria R, Navarro A. 2010. Chromosomal speciation revisited: rearranging theory with pieces of evidence. Trends Ecol Evol 25: 660–9. [DOI] [PubMed] [Google Scholar]
- 5. Mayr E. 1963. Animal Species and Evolution. Cambridge, MA: Harvard University Press. [Google Scholar]
- 6. Via S. 2009. Natural selection in action during speciation. Proc Natl Acad Sci USA 106: 9939–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Presgraves DC. 2010. The molecular evolutionary basis of species formation. Nat Rev Genet 11: 175–80. [DOI] [PubMed] [Google Scholar]
- 8. Navarro A, Faria R. 2014. Pool and conquer: new tricks for (c)old problems. Mol Ecol 23: 1653–5. [DOI] [PubMed] [Google Scholar]
- 9. Navarro A, Barton NH. 2003. Chromosomal speciation and molecular divergence‐accelerated evolution in rearranged chromosomes. Science 300: 321–4. [DOI] [PubMed] [Google Scholar]
- 10. Kirkpatrick M, Barton N. 2006. Chromosome inversions, local adaptation and speciation. Genetics 173: 419–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Civetta A. 2016. Misregulation of gene expression and sterility in interspecies hybrids: causal links and alternative hypotheses. J Mol Evol 82: 176–82. [DOI] [PubMed] [Google Scholar]
- 12. Forejt J. 1996. Hybrid sterility in the mouse. Trends Genet 12: 412–7. [DOI] [PubMed] [Google Scholar]
- 13. Graves JAM, O'Neill RJ. 1997. Sex chromosome evolution and Haldane's rule. J Hered 88: 358–60. [DOI] [PubMed] [Google Scholar]
- 14. Deakin JE, Graves JAM, Rens W. 2012. The evolution of marsupial and monotreme chromosomes. Cytogenet Genome Res 137: 113–29. [DOI] [PubMed] [Google Scholar]
- 15. Bender HS, Graves JAM, Deakin JE. 2014. Pathogenesis and molecular biology of a transmissible tumor in the Tasmanian devil. Annu Rev Anim Biosci 2: 165–87. [DOI] [PubMed] [Google Scholar]
- 16. Kikuchi K, Hamaguchi S. 2013. Novel sex‐determining genes in fish and sex chromosome evolution. Dev Dyn 242: 339–53. [DOI] [PubMed] [Google Scholar]
- 17. Myosho T, Takehana Y, Hamaguchi S, Sakaizumi M. 2015. Turnover of sex chromosomes in celebensis group medaka fishes. G3 (Bethesda) 5: 2685–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Natri HM, Shikano T, Merila J. 2013. Progressive recombination suppression and differentiation in recently evolved neo‐sex chromosomes. Mol Biol Evol 30: 1131–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kitano J, Peichel CL. 2012. Turnover of sex chromosomes and speciation in fishes. Environ Biol Fishes 94: 549–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ser JR, Roberts RB, Kocher TD. 2010. Multiple interacting loci control sex determination in lake Malawi cichlid fish. Evolution 64: 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Malcom JW, Kudra RS, Malone JH. 2014. The sex chromosomes of frogs: variability and tolerance offer clues to genome evolution and function. J Genomics 2: 68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Uno Y, Nishida C, Takagi C, Igawa T, et al. 2015. Extraordinary diversity in the origins of sex chromosomes in anurans inferred from comparative gene mapping. Cytogenet Genome Res 145: 218–29. [DOI] [PubMed] [Google Scholar]
- 23. Miura I, Ohtani H, Ogata M. 2012. Independent degeneration of W and Y sex chromosomes in frog Rana rugosa . Chromosome Res 20: 47–55. [DOI] [PubMed] [Google Scholar]
- 24. Roco AS, Olmstead AW, Degitz SJ, Amano T, et al. 2015. Coexistence of Y, W, and Z sex chromosomes in Xenopus tropicalis . Proc Natl Acad Sci USA 112: E4752–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Smith CA, Roeszler KN, Ohnesorg T, Cummins DM, et al. 2009. The avian Z‐linked gene DMRT1 is required for male sex determination in the chicken. Nature 461: 267–71. [DOI] [PubMed] [Google Scholar]
- 26. Vicoso B, Emerson JJ, Zektser Y, Mahajan S, et al. 2013. Comparative sex chromosome genomics in snakes: differentiation, evolutionary strata, and lack of global dosage compensation. PLoS Biol 11: e1001643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ezaz T, Sarre SD, O'Meally D, Graves JAM, et al. 2009. Sex chromosome evolution in lizards: independent origins and rapid transitions. Cytogenet Genome Res 127: 249–60. [DOI] [PubMed] [Google Scholar]
- 28. Ezaz T, Quinn AE, Miura I, Sarre SD, et al. 2005. The dragon lizard Pogona vitticeps has ZZ/ZW micro‐sex chromosomes. Chromosome Res 13: 763–76. [DOI] [PubMed] [Google Scholar]
- 29. Quinn AE, Georges A, Sarre S, Guarino F, et al. 2007. Temperature sex reversal implies sex gene dosage in a reptile. Science 316: 411. [DOI] [PubMed] [Google Scholar]
- 30. Quinn AE, Sarre SD, Ezaz T, Graves JAM, et al. 2011. Evolutionary transitions between mechanisms of sex determination in vertebrates. Biol Lett 7: 443–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Holleley CE, O'Meally D, Sarre SD, JAM Graves, et al. 2015. Sex reversal triggers the rapid transition from genetic to temperature‐dependent sex. Nature 523: 79–82. [DOI] [PubMed] [Google Scholar]
- 32. Chen S, Zhang G, Shao C, Huang Q, et al. 2014. Whole‐genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat Genet 46: 253–60. [DOI] [PubMed] [Google Scholar]
- 33. Baroiller JF, D'Cotta H, Saillant E. 2009. Environmental effects on fish sex determination and differentiation. Sex Dev 3: 118–35. [DOI] [PubMed] [Google Scholar]
- 34. Piferrer F, Blazquez M, Navarro L, Gonzalez A. 2005. Genetic, endocrine, and environmental components of sex determination and differentiation in the European sea bass (Dicentrarchus labrax L.). Gen Comp Endocrinol 142: 102–10. [DOI] [PubMed] [Google Scholar]
- 35. Luo ZX, Yuan CX, Meng QJ, Ji Q. 2011. A Jurassic eutherian mammal and divergence of marsupials and placentals. Nature 476: 442–5. [DOI] [PubMed] [Google Scholar]
- 36. Sinclair AH, Berta P, Palmer MS, Hawkins JR, et al. 1990. A gene from the human sex‐determining region encodes a protein with homology to a conserved DNA‐binding motif. Nature 346: 240–4. [DOI] [PubMed] [Google Scholar]
- 37. Eggers S, Ohnesorg T, Sinclair AH. 2014. Genetic regulation of mammalian gonad development. Nat Rev Endocrinol 10: 673–83. [DOI] [PubMed] [Google Scholar]
- 38. Graves JAM. 2006. Sex chromosome specialization and degeneration in mammals. Cell 124: 901–14. [DOI] [PubMed] [Google Scholar]
- 39. Skinner BM, Sargent CA, Churcher C, Hunt T, et al. 2016. The pig X and Y Chromosomes: structure, sequence, and evolution. Genome Res 26: 130–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Graves JAM. 2016. Evolution of vertebrate sex chromosomes and dosage compensation. Nat Rev Genet 17: 33–46. [DOI] [PubMed] [Google Scholar]
- 41. Yamauchi Y, Riel JM, Stoytcheva Z, Ward MA. 2014. Two Y genes can replace the entire Y chromosome for assisted reproduction in the mouse. Science 343: 69–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ohno S. 1967. Sex Chromosomes and Sex‐Linked Genes. Berlin Heidelberg: Springer‐Verlag. [Google Scholar]
- 43. Charlesworth B. 1991. The evolution of sex chromosomes. Science 251: 1030–3. [DOI] [PubMed] [Google Scholar]
- 44. Foster JW, Graves JAM. 1994. An SRY‐related sequence on the marsupial X chromosome: implications for the evolution of the mammalian testis‐determining gene. Proc Natl Acad Sci USA 91: 1927–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Burgoyne PS, Buehr M, Koopman P, Rossant J, et al. 1988. Cell‐autonomous action of the testis‐determining gene: sertoli cells are exclusively XY in XX‐XY chimaeric mouse testes. Development 102: 443–50. [DOI] [PubMed] [Google Scholar]
- 46. Sutton E, Hughes J, White S, Sekido R, et al. 2011. Identification of SOX3 as an XX male sex reversal gene in mice and humans. J Clin Invest 121: 328–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Graves JAM. 2013. How to evolve new vertebrate sex determining genes. Dev Dyn 242: 354–59. [DOI] [PubMed] [Google Scholar]
- 48. Johnson Pokorna MJ, Kratochvil L. 2014. What was the ancestral sex‐determining mechanism in amniote vertebrates? Biol Rev Camb Philos Soc 91: 1–2. [DOI] [PubMed] [Google Scholar]
- 49. Murtagh CEA. 1977. A unique cytogenetic system in monotremes. Chromosoma 65: 37–57. [Google Scholar]
- 50. Grutzner F, Rens W, Tsend‐Ayush W, El‐Mogharbel N, et al. 2004. In the platypus a meiotic chain of ten sex chromosomes shares genes with the bird Z and mammal X chromosomes. Nature 432: 913–7. [DOI] [PubMed] [Google Scholar]
- 51. Waters PD, Delbridge ML, Deakin JE, El‐Mogharbel, et al. 2005. Autosomal location of genes from the conserved mammalian X in the platypus (Ornithorhynchus anatinus) implications for mammalian sex chromosome evolution. Chromosome Res 13: 401–10. [DOI] [PubMed] [Google Scholar]
- 52. Veyrunes F, Waters PD, Miethke P, Rens W, et al. 2008. Bird‐like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res 18: 965–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gruetzner F, Ashley T, Rowell DM, Graves JAM. 2006. How did the platypus get its sex chromosome chain? A comparison of meiotic multiples and sex chromosomes in plants and animals. Chromosoma 115: 75–88. [DOI] [PubMed] [Google Scholar]
- 54. Cortez D, Marin R, Toledo‐Flores D, Froidevaux D, et al. 2014. Origins and functional evolution of Y chromosomes across mammals. Nature 508: 488–93. [DOI] [PubMed] [Google Scholar]
- 55. Rens W, O'Brien PC, Grutzner F, Clarke O, et al. 2007. The multiple sex chromosomes of platypus and echidna are not completely identical and several share homology with the avian Z. Genome Biol 8: R243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rowe T, Rich TH, Vickers‐Rich P, Springer M, et al. 2008. The oldest platypus and its bearing on divergence timing of the platypus and echidna clades. Proc Natl Acad Sci USA 105: 1238–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Phillips MJ, Bennett TH, Lee MS. 2009. Molecules, morphology, and ecology indicate a recent, amphibious ancestry for echidnas. Proc Natl Acad Sci USA 106: 17089–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wallis MC, Waters PD, Delbridge ML, Kirby PJ, et al. 2007. Sex determination in platypus and echidna: autosomal location of SOX3 confirms the absence of SRY from monotremes. Chromosome Res 15: 949–59. [DOI] [PubMed] [Google Scholar]
- 59. Warren WC, Hillier LW, Graves JAM, Birney E, et al. 2008. Genome analysis of the platypus reveals unique signatures of evolution. Nature 453: 175–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Graves JAM. 1995. The origin and function of the mammalian Y chromosome and Y‐borne genes‐an evolving understanding. Bioessays 17: 311–20. [DOI] [PubMed] [Google Scholar]
- 61. Waters PD, Duffy B, Frost CJ, Delbridge ML, et al. 2001. The human Y chromosome derives largely from a single autosomal region added to the sex chromosomes 80‐130 million years ago. Cytogenet Cell Genet 92: 74–9. [DOI] [PubMed] [Google Scholar]
- 62.International Chicken Genome Sequencing Consortium. 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432: 695–716. [DOI] [PubMed] [Google Scholar]
- 63. Benton MJ, Donoghue PC. 2007. Paleontological evidence to date the tree of life. Mol Biol Evol 24: 26–53. [DOI] [PubMed] [Google Scholar]
- 64. Delgado CL, Waters PD, Gilbert C, Robinson TJ, et al. 2009. Physical mapping of the elephant X chromosome: conservation of gene order over 105 million years. Chromosome Res 17: 917–26. [DOI] [PubMed] [Google Scholar]
- 65. Kondo M, Nanda I, Schmid M, Schartl M. 2009. Sex determination and sex chromosome evolution: insights from medaka. Sex Dev 3: 88–98. [DOI] [PubMed] [Google Scholar]
- 66. Yoshimoto S, Okada E, Umemoto H, Tamura K, et al. 2008. A W‐linked DM‐domain gene, DM‐W, participates in primary ovary development in Xenopus laevis. Proc Natl Acad Sci USA 105: 2469–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. O'Meally D, Ezaz T, Georges A, Sarre SD, et al. 2012. Are some chromosomes particularly good at sex? Insights from amniotes. Chromosome Res 20: 7–19. [DOI] [PubMed] [Google Scholar]
- 68. Russell LB, Montgomery CS. 1969. Comparative studies on X‐autosome translocations in the mouse. I. Origin, viability, fertility, and weight of five T(X;1)'S. Genetics 63: 103–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chandley AC, Christie S, Fletcher J, Frackiewicz A, et al. 1972. Translocation heterozygosity and associated subfertility in man. Cytogenetics 11: 516–33. [DOI] [PubMed] [Google Scholar]
- 70. Homolka D, Ivanek R, Capkova J, Jansa P, et al. 2007. Chromosomal rearrangement interferes with meiotic X chromosome inactivation. Genome Res 17: 1431–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wessels S, Sharifi RA, Luehmann LM, Rueangsri S, et al. 2014. Allelic variant in the anti‐Mullerian hormone gene leads to autosomal and temperature‐dependent sex reversal in a selected Nile tilapia line. PLoS ONE 9: e104795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Britton‐Davidian J, Catalan J, da Graca Ramalhinho M, Ganem G, et al. 2000. Rapid chromosomal evolution in island mice. Nature 403: 158. [DOI] [PubMed] [Google Scholar]
- 73. Kimura R, Murata C, Kuroki Y, Kuroiwa A. 2014. Mutations in the testis‐specific enhancer of SOX9 in the SRY independent sex‐determining mechanism in the genus Tokudaia. PLoS ONE 9: e108779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kuroiwa A, Handa S, Nishiyama C, Chiba E, et al. 2011. Additional copies of CBX2 in the genomes of males of mammals lacking SRY, the Amami spiny rat (Tokudaia osimensis) and the Tokunoshima spiny rat (Tokudaia tokunoshimensis). Chromosome Res 19: 635–44. [DOI] [PubMed] [Google Scholar]
- 75. Just W, Rau W, Vogel W, Akhverdian M, et al. 1995. Absence of Sry in species of the vole Ellobius. Nat Genet 11: 117–8. [DOI] [PubMed] [Google Scholar]
- 76. Bagheri‐Fam S, Sreenivasan R, Bernard P, Knower KC, et al. 2012. Sox9 gene regulation and the loss of the XY/XX sex‐determining mechanism in the mole vole Ellobius lutescens. Chromosome Res 20: 191–9. [DOI] [PubMed] [Google Scholar]
- 77. Just W, Baumstark A, Suss A, Graphodatsky A, et al. 2007. Ellobius lutescens: sex determination and sex chromosome. Sex Dev 1: 211–21. [DOI] [PubMed] [Google Scholar]
- 78. Yamauchi Y, Riel JM, Ruthig VA, Ortega EA, et al. 2016. Two genes substitute for the mouse Y chromosome for spermatogenesis and reproduction. Science 351: 514–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Fredga K. 1988. Aberrant chromosomal sex‐determining mechanisms in mammals, with special reference to species with XY females. Philos Trans R Soc Lond B Biol Sci B 322: 83–95. [DOI] [PubMed] [Google Scholar]
- 80. Hoekstra HE, Edwards SV. 2000. Multiple origins of XY female mice (genus Akodon): phylogenetic and chromosomal evidence. Proc Biol Sci 267: 1825–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Veyrunes F, Chevret P, Catalan J, Castiglia R, et al. 2010. A novel sex determination system in a close relative of the house mouse. Proc Biol Sci 277: 1049–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Martin PG, Hayman DL. 1967. Quantitative comparisons between the karyotypes of Australian marsupials from three different superfamilies. Chromosoma 20: 290–310. [DOI] [PubMed] [Google Scholar]
- 83. Toder R, Wienberg J, Voullaire L, O'Brien PC, et al. 1997. Shared DNA sequences between the X and Y chromosomes in the tammar wallaby? Evidence for independent additions to eutherian and marsupial sex chromosomes. Chromosoma 106: 94–8. [DOI] [PubMed] [Google Scholar]
- 84. Hughes JF, Rozen S. 2012. Genomics and genetics of human and primate Y chromosomes. Annu Rev Genomics Hum Genet 13: 83–108. [DOI] [PubMed] [Google Scholar]
- 85. Aitken RJ, Graves JAM. 2002. The future of sex. Nature 415: 963. [DOI] [PubMed] [Google Scholar]
- 86. Wilson CA, High SK, McCluskey BM, Amores A, et al. 2014. Wild sex in zebrafish: loss of the natural sex determinant in domesticated strains. Genetics 198: 1291–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Koepfli KP, Paten B, Genome CoS K, O'Brien SJ. 2015. The Genome 10 K Project: a way forward. Annu Rev Anim Biosci 3: 57–111. [DOI] [PMC free article] [PubMed] [Google Scholar]


