Abstract
The eukaryotic translation initiation factor eIF2B promotes mRNA translation as a guanine nucleotide exchange factor (GEF) for translation initiation factor 2 (eIF2). Endoplasmic reticulum (ER) stress-mediated activation of the kinase PERK and the resultant phosphorylation of eIF2’s alpha subunit (eIF2α) attenuates eIF2B GEF activity thereby inducing an integrated stress response (ISR) that defends against protein misfolding in the ER. Mutations in all five subunits of human eIF2B cause an inherited leukoencephalopathy with vanishing white matter (VWM), but the role of the ISR in its pathogenesis remains unclear. Using CRISPR-Cas9 genome editing we introduced the most severe known VWM mutation, EIF2B4A391D, into CHO cells. Compared to isogenic wildtype cells, GEF activity of cells with the VWM mutation was impaired and the mutant cells experienced modest enhancement of the ISR. However, despite their enhanced ISR, imposed by the intrinsic defect in eIF2B, disrupting the inhibitory effect of phosphorylated eIF2α on GEF by a contravening EIF2S1/eIF2αS51A mutation that functions upstream of eIF2B, selectively enfeebled both EIF2B4A391D and the related severe VWM EIF2B4R483W cells. The basis for paradoxical dependence of cells with the VWM mutations on an intact eIF2α genotype remains unclear, as both translation rates and survival from stressors that normally activate the ISR were not reproducibly affected by the VWM mutations. Nonetheless, our findings support an additional layer of complexity in the development of VWM, beyond a hyperactive ISR.
Introduction
Exchange of GDP for GTP on translation initiation factor 2 (eIF2) is an important determinant of rates of global protein synthesis in eukaryotes [1]. The activity of eIF2B, the guanine nucleotide exchange factor (GEF) charged with this task, is regulated in large part by the level of phosphorylation on the alpha subunit of its substrate (eIF2α). Phosphorylated eIF2α [eIF2(αP)] attenuates GEF activity, resulting in reduced global protein synthesis and decreased burden on the cell’s protein folding machinery [2, 3]. At the same time, eIF2(αP) and the attendant decrease in eIF2B GEF activity leads to enhanced translation of rare mRNAs that encode potent transcription factors that activate a gene expression program known as the Integrated Stress Response (ISR) in animals [4] or the General Control Response in yeast [5].
The circuitry involved in regulating the ISR, namely the kinases that phosphorylate eIF2α and the phosphatases that de-phosphorylate it, are carefully controlled. In both directions, altered function of the ISR affects fitness: abnormally low ISR activity exposes cells to the risk of protein misfolding due to unregulated protein synthesis whereas abnormally high ISR activity leads to a failure to maintain adequate levels of protein synthesis [2, 3, 6]. Balance in the ISR is especially important to fitness of the nervous system: pathological demyelination is exacerbated by abnormally low ISR activity [7] whereas cognitive dysfunction in certain neurodegenerative models is relieved by reduced ISR activity [8, 9].
The genes encoding the five subunits of the GEF eIF2B are essential. However in humans, diverse recessive missense mutations in all five are causally linked to the development of a leukoencephalopathy, known as Vanishing White Matter (VWM) or Childhood Ataxia with Cerebral Hypomyelination (CACH) [10, 11]. The recessive inheritance and the scattering of mutations widely throughout the coding sequences of the subunits are most in line with a loss-of-function phenotype [12]. This notion is supported by a trend for lower GEF activity in lysates from patient derived cells [13] and an enhanced ISR [14]; though the correlation between these biochemical features and disease severity is imperfect [15, 16].
Despite these clues, the role, if any, of altered ISR activity in the pathophysiology of VWM has remained elusive. Here we exploited the power of CRISPR-Cas9-induced recombination to create isogenic cell lines with and without a severe VWM mutation (EIF2B4A391D). Further genetic manipulation of the ISR revealed an unanticipated dependence of the VWM cells on signaling by phosphorylated eIF2α, despite their heightened ISR. We attempt to place this paradox within the context of cells that cope with fluctuating levels of unfolded protein stress.
Results
An isogenic cell culture model of a severe VWM-linked EIF2B4A391D mutation
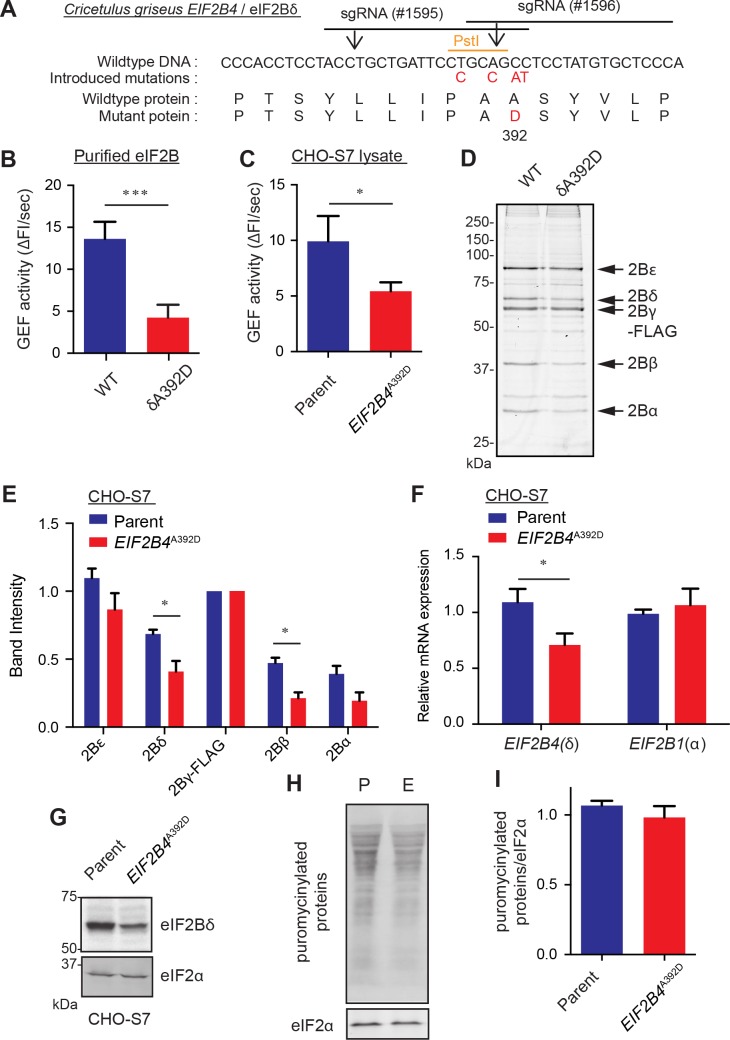
Cell biological characterization of VWM mutations has relied upon patient derived cell lines. To circumvent the confounding effects of background genotypic and phenotypic variation inherent in such studies we used CRISPR-Cas9 gene editing to create isogenic wildtype Chinese hamster ovary (CHO) cells and cells bearing a severe VWM mutation corresponding to Alanine 391 to Aspartic acid on human eIF2Bδ subunit (EIF2B4A392D in CHO) [10]. To facilitate detection of the mutant allele we introduced silent mutations that create a restriction fragment length polymorphism (RFLP), alongside the disease-associated missense mutation (Fig 1A). We generated two independent EIF2B4A392D mutant cell clones derived from different CHO parental cells; UPR-reporter containing S21 cells and S7 cells with a FLAG-tagged endogenous eIF2Bγ subunit (see below).
Fig 1. A cell model for the human VWM disease-linked mutation EIF2B4A391D.
(A) The Cricetulus griseus EIF2B4 genomic locus (NW_003613640.1, 3817:3861) targeted by CRISPR-Cas9 system and eIF2Bδ encoded protein (XP_003497100.1, 383:397). Horizontal lines and vertical arrows represent the sgRNA binding sites and Cas9 cleavage sites, respectively. Mutations, shown in red, disrupt sgRNA targeting, eliminate a PstI site and generate an A392D (corresponding to human A391D) coding sequence mutation. (B) GEF activity of purified wildtype (WT) or eIF2BδA392D mutant (δA392D) eIF2B complex. Shown are means ± S.D. of six independent experiments. *** P = 6.78 x 10−6, Unpaired t test. (C) GEF activity in the lysates of parental CHO-S7 and EIF2B4A392D mutant cells. Shown are means ± S.D. of three independent experiments. * P = 0.032, Unpaired t test. (D) Coomassie brilliant blue stained SDS-PAGE of the endogenous eIF2B complex purified from parental CHO-S7 and EIF2B4A392D cells by affinity chromatography of the FLAG-tagged endogenous eIF2Bγ subunit. The position of the individual subunits is indicated. (E) Quantification of the relative band intensity of eIF2B component in “C”, normalized to eIF2Bγ-FLAG signal. Shown are means ± SEM of three independent experiments. * P = 0.028 and 0.0103 for 2B4 and 2B2, respectively, Unpaired t test. (F) Quantitative PCR for mRNA expression of EIF2B4 and EIF2B1 in parental CHO-S7 and EIF2B4A392D cells. Shown are means ± S.D. of three independent experiments. * P = 0.013, Unpaired t test. (G) Immunoblot of eIF2Bδ and eIF2α in parental CHO-S7 and EIF2B4A392D cells. (H) Immunoblot of puromycinylated proteins following a brief pulse of puromycin, reporting on levels of translation. P and E indicate parental CHO-S7 and EIF2B4A392D mutant cells, respectively. (I) Quantification of the puromycinylated proteins in “H”, normalized to an internal control, eIF2α. Shown are means ± SEM from three experiments.
The mutation had conspicuous hypomorphic features, imparting an intrinsic defect in GEF activity on the purified eIF2B complex (Fig 1B and S1A Fig). Lower levels of GEF activity were also noted in lysates of mutant cells (Fig 1C), which was consistent with the combined intrinsic defect in GEF activity noted above and mild instability of the mutant eIF2B complex (Fig 1D and 1E and S1B Fig). The latter correlated with lower levels of EIF2B4 mRNA and protein in the mutant cells (Fig 1F and 1G). These were observed in independently derived mutant clones elaborated in different CHO parental lines (S1C–S1E Fig), but affected neither baseline levels of global protein synthesis (Fig 1H and 1I and S1F Fig) nor cell proliferation (S1G Fig).
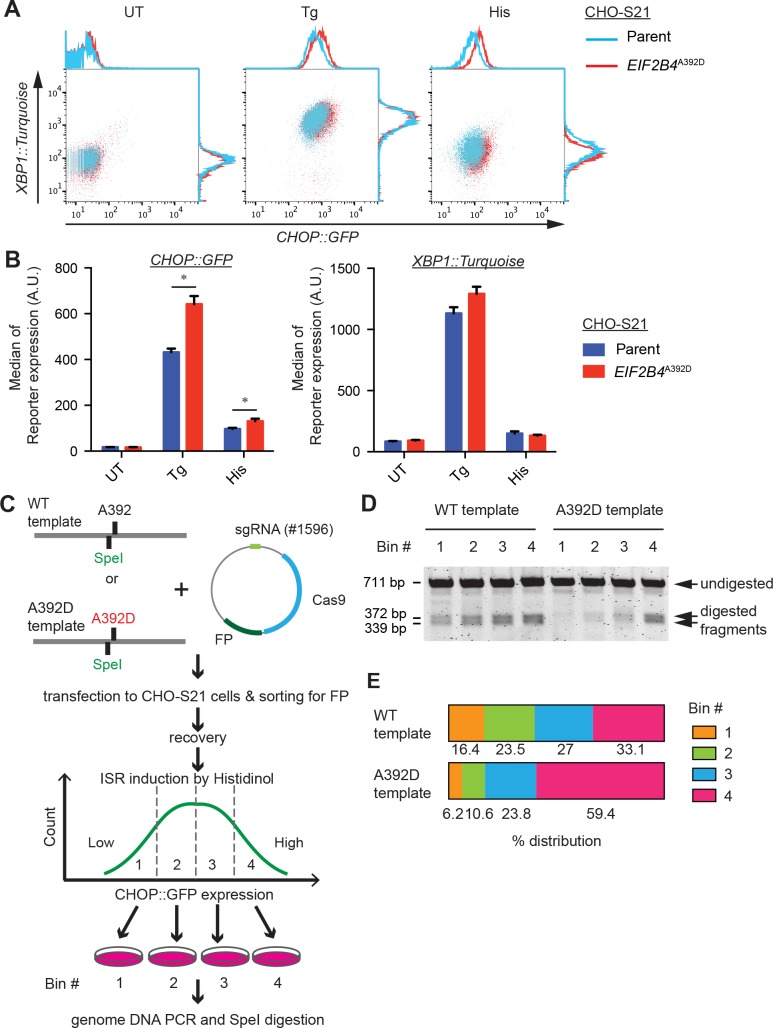
To gauge the effect of the EIF2B4A392D mutation on the activity of the ISR, we made use of a resident CHOP::GFP ISR reporter transgene [17]. The basal activity of CHOP::GFP was indistinguishable in wildtype and EIF2B4A392D mutant cells. However induction of endoplasmic reticulum (ER) stress by the ER calcium depleting agent thapsigargin (that activates the eIF2α kinase PERK) or by histidinol, an inhibitor of histidyl tRNA synthetase (that activates the eIF2α kinase GCN2), led to conspicuously more signaling in the ISR of the mutant cells (Fig 2A and 2B). Enhanced signaling in the mutant cells was specific for the ISR. Thapsigargin-mediated activity of an XBP1::Turquoise transgene (that reports on a parallel IRE1-dependent branch of the ER stress response [18]), was unaffected by the mutation. In histidinol-treated cells the induction of XBP1::Turquoise was slightly but reproducibly attenuated by the mutation, likely a reflection of feed-back emanating from the enhanced ISR activity (Fig 2A and 2B).
Fig 2. Heightened ISR in EIF2B4A392D cells.
(A) Flow cytometry analysis of CHOP::GFP and XBP1::Turquoise dual reporter-containing parental CHO-S21 and EIF2B4A392D mutant cells. The cells were untreated (UT) or stimulated with 250 nM thapsigargin (Tg) or 0.5 mM histidinol (His) for 24 hours. Note the enhanced response of the CHOP::GFP ISR reporter. (B) Bar diagram of the median ± S.D. of the reporter gene activity from experiments as shown in “A”. N = 3, *P = 0.0057 for Tg, *P = 0.037 for His, Unpaired t test. (C) Experimental design for tracking EIF2B4A392D mutations. A fluorescent protein-marked sgRNA/Cas9 plasmid targeting EIF2B4 and a wildtype or EIF2B4A392D mutant repair template marked by a silent SpeI mutation were co-transfected into CHO-S21 cells. Transfected cells (selected by FACS), were treated with histidinol and divided into four bins (Bin #1 to #4) by level of CHOP::GFP expression. After recovery, genomic DNA was isolated from cells in each bin and the targeted region of EIF2B4 was amplified by PCR and digested with SpeI to reveal frequency of targeting by either repair template. (D) PCR fragments digested with SpeI from genomic DNA of the indicated bins, visualized on an agarose gel. Shown is an image of a representative experiment reproduced twice. (E) Plot of the distribution of SpeI digested fragments in the four bins of transduced cells from the experiment in “D”. The band intensities of the digested fragments (reporting on recombination of the wildtype or mutant repair template) were normalized to total PCR product intensity and the distribution of the relative frequency of recombination in the different bins was plotted. Note the enrichment for recombination of the EIF2B4A392D mutant repair template in the ISRHigh bin.
To minimize the effect of clonal selection on the heightened ISR activity observed in EIF2B4A392D cells, we devised an unbiased assay to measure the ISR in polyclonal populations of cells that had been induced to acquire the EIF2B4A392D mutation. Parental S21 cells were co-transfected with fluorescent-tagged Cas9 and a guide targeting the EIF2B4 locus, alongside either wildtype or EIF2B4A392D repair templates that harbor an SpeI RFLP distinguishing a recombinant EIF2B4 locus from a parental one (Fig 2C and S2A Fig). The frequency of homologous recombination by this transient transfection method was low and the pool of cells offered the mutant repair template had only a minimally-elevated ISR compared to those offered a wildtype repair template (S2B Fig). The transiently transfected cells were then divided by FACS into four bins covering the range of ISR activity and the frequency of homologous recombination with either the wildtype or EIF2B4A392D repair template was measured in each bin (Fig 2C). Cells that had recombined the EIF2B4A392D repair template were enriched in the ISRHigh bin (#4), whilst the frequency of homologous recombination with the wildtype repair template was more evenly distributed amongst the four bins (Fig 2D and 2E). These observations further reveal that the EIF2B4A392D mutation disposes cells to an enhanced ISR.
Severe VWM mutant cells are intolerant of an ISR-enfeebling EIF2S1S51A mutation
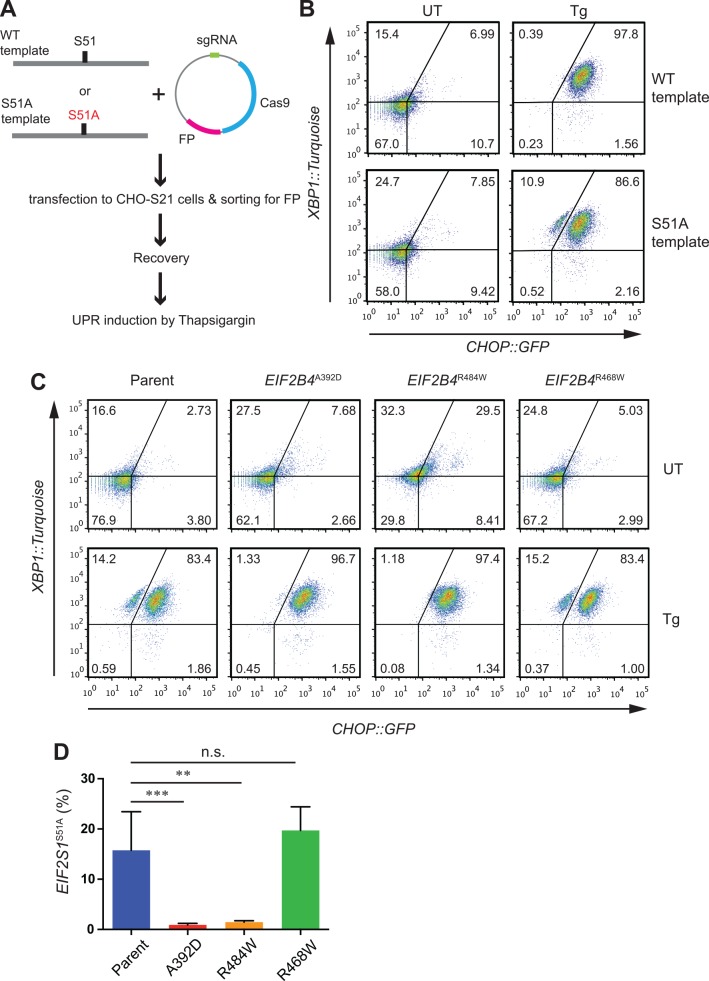
Mutations in eIF2B that enhance the yeast counterpart of the ISR (the General Control Response) buffer the effect of other mutations that lower levels of eIF2(αP) [19]. To test for similar buffering in VWM mutant cells, we sought to compare the phenotypic consequences of interfering with eIF2(αP) in wildtype and two severe VWM models: the aforementioned EIF2B4A392D and EIF2B4R484W, which corresponds to the human VWM mutation R483W [10, 11, 12] (S3A and S3B Fig).
CRISPR-Cas9 guides targeting the EIF2S1/eIF2α locus were transfected alongside a repair template with a Ser 51 to Ala mutation in eIF2α that blocks signaling to the ISR by upstream stress-activated kinases [20] (Fig 3A). In CHO cells wildtype at the EIF2B4 locus, this led to the emergence of a subpopulation of cells with impaired induction of the CHOP::GFP ISR reporter upon exposure to the ER stress-inducing agent thapsigargin. Though not studied in further detail here, the defect in the ISR likely arises both in cells that have acquired homozygous EIF2S1S51A mutation and in cells in which the EIF2S1S51A allele is present in trans to a null. Heterozygous cells with a wildtype allele of EIF2S1 are likely to be phenotypically wildtype [20]. The defect imposed by EIF2S1S51A mutation was selective for the ISR, as the parallel stress-induced pathway mediated by IRE1α and reported on by the XBP1::Turquoise reporter was unaffected (Fig 3B).
Fig 3. Severe VWM mutant cells are unable to tolerate a second EIF2S1S51A mutation.
(A) Experimental design for tracking EIF2S1S51A mutant cells. A fluorescent-tagged sgRNA/Cas9 plasmid targeting EIF2S1 was co-transfected alongside wild type (WT) or EIF2S1S51A (Mut) templates into CHO-S21 dual reporter cells. Following FACS selection for the transfected cells they were treated with 250 nM thapsigargin (Tg) for 24 hours and reporter expression was analyzed. (B) Flow cytometry analysis of reporter activity in untreated (UT) and thapsigargin-treated (Tg) CHO-S21 cells from the experiment outlined in “A”. Note the emergence of CHOP::GFP negative, XBP1::turquoise positive thapsigargin-treated cells in the pool offered an EIF2S1S51A repair template. (C) Flow cytometry analysis of reporter activity in untreated (UT) and thapsigargin-treated (Tg) parental CHO-S21 or indicated VWM mutant cells following targeting of the EIF2S1 locus with an EIF2S1S51A repair template (as described in “A”). Note the lack of CHOP::GFP negative, XBP1::turquoise positive thapsigargin-treated putative EIF2S1S51A; EIF2B4A392D or EIF2S1S51A; EIF2B4R484W double mutant cells (lower right panel). (D) Percentage of CHOP::GFP negative, XBP1::turquoise positive thapsigargin-treated putative EIF2S1S51A mutant cells in the indicated population from experiments as in “C”. Shown are means ± S.D. N = 6 (Parent), 5 (EIF2B4A392D), and 3 (EIF2B4R484W and EIF2B4R468W). *** P<0.001, ** P<0.01, n.s. not significant, One way ANOVA followed by Dunnett’s multiple comparisons test.
EIF2S1S51A mutant cells are hypersensitive to stress. This was reflected in the depletion of the ISR negative (EIF2S1S51A mutant) pool from a mixed population of wildtype and mutant cells, following exposure to histidinol (S3C and S3D Fig). Nonetheless EIF2S1S51A mutant cells gave rise to clones that could be propagated indefinitely (S3E Fig). By contrast EIF2B4A392D mutant cells targeted with the same EIF2S1S51A repair template failed to elaborate a sizeable pool of CHOP::GFP uninduced cells (Fig 3C and 3D & S3F and S3G Fig) and the few CHOP::GFP dim cells observed transiently after transduction of the EIF2S1S51A repair template failed to give rise to EIF2B4A392D; EIF2S1S51A double mutant clones. EIF2B4R484W mutant cells were similarly unable to acquire the EIF2S1S51A mutation (Fig 3C and 3D). By contrast CHO cells with a milder VWM mutation, EIF2B4R468W (a model of human EIF2B4R467W [21], S3A and S3B Fig) had no difficulty accommodating a second EIF2S1S51A mutation (Fig 3C and 3D). This feature of severe VWM mutant cells likely reflected a specific inability to tolerate the EIF2S1S51A mutation, as opposed to altered susceptibility to CRISPR-Cas9 gene editing, as targeting the ERN1/IRE1α locus with CRISPR-Cas9 guides gave rise to a similar population of XBP1::Turquoise negative daughters in wildtype or EIF2B4A392D parents (S3H and S3I Fig). Furthermore, as this analysis is conducted on pools of cells with no selection (beyond that for transient expression of the CRISPR-Cas9 encoding plasmid), the phenotypic difference between the wildtype and severe VWM mutation-bearing cells are unlikely to have arisen from random clonal selection.
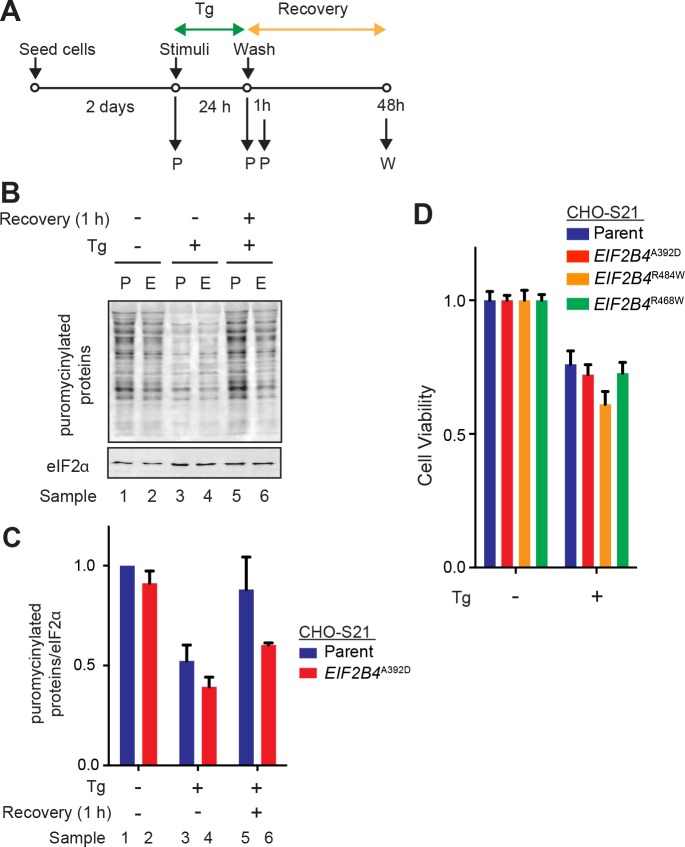
Despite the conspicuous dependence of severe VWM mutant cells on an intact ISR, we observed no consistent difference in the magnitude of the repression of protein synthesis by thapsigargin between wildtype and mutant cells (Fig 4A–4C). A modest defect in the recovery of protein synthesis in the mutant cells following washout of the thapsigargin in some experiments (Fig 4B, lanes 5 & 6), was not consistently observed in all experiments. Furthermore, the survival of cells following exposure to thapsigargin, was unaffected by the VWM mutations (Fig 4D), leaving no simple explanation for the strong genetic interaction between the VWM mutation and the EIF2S1S51A genotype.
Fig 4. Stress-resistance of wildtype and VWM cells.
(A) Schema of experiments to compare the effect of thapsigargin in parental CHO-S21 and VWM mutant cells. Cells were treated with thapsigargin (Tg; 250 nM) for the indicated time, washed free of compounds and allowed to recover before assay. W = WST-1 assay, P = Puromycin labeling. (B) Immunoblot of puromycinylated proteins following a brief pulse of puromycin, reporting on levels of translation under the indicated experimental conditions. Shown is a representative experiment reproduced four times. P and E indicate parental CHO-S21 and EIF2B4A392D mutant cells, respectively. (C) Quantification of “B”. Signal intensities of puromycinylated proteins were normalized by eIF2α. Shown are means ± SEM of four independent experiments. (D) Cell viability measured by the WST-1 assay in the experiment described in “A”. Shown are the mean ± SEM of four replicates of a representative experiment repeated three (R484W, R468W) to six (A392D) times.
Discussion
Stress, imposed by infection or head trauma is frequently cited as a precipitating event in VWM [22]. But hypersensitivity to stress has not been evident in cell culture models of the disease. Both theoretical considerations (built on the hypomorphic features of disease-associated mutations in eIF2B) and empirical observations suggest, if anything, that VWM cells have heightened activity of the ISR–a major stress resistance pathway that counteracts proteotoxicity. To help address this conundrum and detect potentially subtle disease-associated phenotypes we chose to introduce severe, early onset VWM mutations into otherwise isogenic CHO cell lines with built-in reporters for the UPR and ISR.
Whilst CHO cells are a limited model for the complex neurobiology of VWM, they do provide a useful tool to elucidate general aspects of the effect of the mutation. Further important advantages of CHO cells are their relative clonal stability, the availability of integrated ISR reporters and the ease of CRISPR-Cas9 gene editing in these cells (this is an important improvement over lymphoblast cell lines isolated from patients used in previous studies of VWM, as the latter lack isogenic wildtype counterparts and are subject to clonal variation during immortalization). Finally, severe VWM mutations also compromise organs such as the female ovary [23] and CHO cells, derived from hamster ovaries, might capture some of the relevant biology. Thus, whilst one may not safely extrapolate from CHO cells to the astrocytes and oligodendrocytes that are the target cells implicated in VWM, it seems reasonable to imagine that the fundamental biology is shared.
In an isogenic context, where gene dosage affects have been eliminated, we confirmed that a severe VWM model in CHO cells is associated with lowered eIF2B GEF activity. The basis for this is likely multifactorial as the mutation studied in detail, EIF2B4A392D, negatively affected both the abundance of the eIF2B complex and its intrinsic GEF activity. This feature likely accounts for the enhanced ISR activity of the VWM cells; a subtle phenotype, but one that is observed both in single mutant clones (where clonal variation is always a concern) and in unselected pools of mutant cells.
The inability of cells with either of two severe VWM mutations (EIF2B4A392D and EIF2B4R484W) to tolerate a second mutation in eIF2α that prevents its phosphorylation on serine 51 and thus eliminates ISR activity (EIF2S1S51A), occurred against this backdrop of diminished eIF2B activity and a heightened ISR activity of the VWM cells. This is a robust phenotype, observed in multiple EIF2B4A392D clones and in the related severe EIF2B4R484W mutation. By contrast CHO cells with a weaker EIF2B4R468W VWM mutation, readily tolerated the ISR-blocking EIF2S1S51A mutation, suggesting a correlation between the severity of the VWM mutation and the dependence of the cells on a serine residue in position 51 of eIF2α.
The mechanism by which the severe VWM mutations sensitize cells to the loss of ISR signaling through an EIF2S1S51A mutation remains unclear. It is formally possible that an alanine in position 51 of eIF2α has hypomorphic features that are unrelated to the loss of the ISR-initiating phosphorylation event at that site; features that become critical in presence of a second, severe VWM mutation. However, in mice, the EIF2S1S51A mutation is largely mimicked by mutations in the upstream kinases [20] leading us to disfavor this possibility. An alternative explanation might be that severe VWM mutations are impaired in tolerating severe swings in unfolded protein stress that occur when the ISR is dysregulated, but this possibility will need to be explored in further detail.
Materials and Methods
Cell culture and Reagents
CHO-K1 (ATCC, CCL-61) based cell lines were maintained in regular media consisting of Nutrient Mixture F12 Ham (SIGMA), 10% Fetal calf serum (FetalClone II, ThermoFisher), 2 mM L-glutamine (SIGMA), and 1x Penicillin/Streptomycin at 37°C with 5% CO2.
CHO-S21 cells containing the CHOP::GFP and XBP1::Turquoise reporters were derived from G418r CHOP::GFP CHO-C30 cells [17] by introduction of a stable XBP1::Turquoise reporter derived by replacing the Venus fluorescent protein in pCAX-F-XBP1ΔDBD-venus [18] with Turquoise alongside a puromycin resistance marker and selection for clones that activate both reporters in response to ER stress.
Reagents were sourced as indicated: thapsigargin (Calbiochem), L-histidinol dihydrochloride (ACROS Organics), puromycin (Calbiochem), and tunicamycin (MELFORD).
Generation of the genome edited cells by CRISPR-Cas9
CHO-S7 cells
A 3xFLAG tag was incorporated into the EIF2B3/eIF2Bγ subunit of CHO-C30 cells [17] by a CRISPR-Cas9 mediated homology-directed repair (HDR). A single guide (sg) RNA sequence for targeting C-terminal CDS of EIF2B3 was selected from the CRISPy database [URL: http://staff.biosustain.dtu.dk/laeb/crispy/ [24]] and a duplex DNA of the sequence (made of oligo DNA No. 1 and No. 2 in S1 Table) was inserted into the pCas9(BB)-2A-GFP plasmid (Addgene plasmid #48138) following published procedures [25], making the sgRNA/Cas9 plasmid (lab ID; UK1491). To construct a repair template containing the 3xFLAG sequence at the C-terminal end of EIF2B3 CDS without a stop codon, 5’ and 3’ homology arms were amplified from CHO genomic DNA by PCR using the primer sets; oligo DNA No. 3 & 4 and oligo DNA No. 5 & 6, respectively. A 3xFLAG sequence was recovered from the pCEFL_mCherry_3xFLAG_C plasmid (lab ID; UK1314) digested with EcoRI and ApaI. The 5’ homology arm (digested with SacI/EcoRI), 3’ homology arm (digested with ApaI/KpnI), and 3xFLAG sequence were sequentially ligated into a correspondingly digested pBS KS(+) plasmid (lab ID; UK1), making a repair template plasmid (lab ID; UK1500).
CHO-C30 cells were plated in 6 well plates at a density of 2 x 105 per well. 24 hours later the cells were transfected with the repair template plasmid (1 μg) and an sgRNA/Cas9 plasmid (1 μg) using Lipofectamine LTX (Invitrogen) according to manufacturer’s instruction. The next day, GFP positive cells, which indicate plasmid transfected cells, were collected using a Beckman Coulter MoFlo Cell sorter, and expanded as single clones in 96 well plates. Genomic DNAs isolated from the clones were screened by PCR using primers (oligo DNA No. 7 & 8) and EcoRI digestion (3xFLAG inserted cell should have exogenous EcoRI site at C-terminus of EIF2B3 CDS derived from the repair template). Finally, the insertion of 3xFLAG was confirmed by sequencing. The clone (S7) we used in this study has an insertion of 3xFLAG in one allele while another allele has intact EIF2B3.
CHO EIF2B4A392D, EIF2B4R484W and EIF2B4R468W cells
The EIF2B4A392D mutation was incorporated into the CHO-S21 cells (see Cell culture section) or CHO-S7 (see above) cells by a similar CRISPR-Cas9 mediated HDR as above. The duplex DNAs [made of oligo DNAs (No. 9 and No. 10) or (No. 11 and No. 12) in S1 Table] were inserted into the pCas9(BB)-2A-GFP plasmid, making the sgRNA/Cas9 plasmids (lab ID; UK1595, or UK1596, respectively). For a repair template, 5’ and 3’ homology arms amplified by oligo DNAs [(No. 13 and No. 14) and (No. 15 and No. 16), respectively] were PCR knitted by primers No. 13 and No. 16. PCR products were purified from agarose gel using GeneJET Gel Extraction Kit (ThermoFisher) and used as a repair template. Transfection, cell sorting and cell cloning were similarly done as described above. For genotyping screen at EIF2B4 A392 locus, genomic DNA was amplified by nested PCR using oligo DNAs No. 17 and No. 18 as a first primer set and No. 13 and No. 16 as second. The PCR products were digested by PstI because the repair template contains silent mutations to disrupt the endogenous PstI site for a marker of HDR as shown in Fig 1A. The incorporated mutations were confirmed by sequencing for genomic DNA and cDNA derived from mutant clones. We obtained clones harboring homozygous EIF2B4A392D mutation in CHO-S21 cells (c6) from the UK1595 transfected clones, and CHO-S7 cell line (c3) from the UK1596 transfected clones. The EIF2B4R484W or EIF2B4R468W mutation in CHO-S21 cells was generated using similar methods as EIF2B4A392D. For EIF2B4R484W, the duplex DNAs [made of oligo DNAs (No. 33 and No. 34) in S1 Table] were inserted into the pCas9(BB)-2A-mCherry plasmid, making the sgRNA/Cas9 plasmids (lab ID; UK1633). For a repair template, 5’ and 3’ homology arms amplified by oligo DNAs [(No. 35 and No. 36) and (No. 37 and No. 38), respectively] were PCR knitted by primers No. 35 and No. 38. For EIF2B4R468W, the duplex DNAs [made of oligo DNAs (No. 39 and No. 40) in S1 Table] were inserted into the pCas9(BB)-2A-GFP plasmid, making the sgRNA/Cas9 plasmids (lab ID; UK1597). For a repair template, 5’ and 3’ homology arms amplified by oligo DNAs [(No. 35 and No. 41) and (No. 42 and No. 38), respectively] were PCR knitted by primers No. 35 and No. 38.
EIF2B4A392D mutation pool analysis in CHO-S21 cells
For the EIF2B4A392D mutation repair template, 5’ and 3’ homology arms amplified by oligo DNAs [(No. 13 & No. 31) and (No. 15 & No. 16), respectively] were PCR knitted by primers No. 13 and No. 16. For A392 wildtype control, oligo DNA No. 32 was used instead of No. 31. PCR products were purified using GeneJET Gel Extraction Kit (ThermoFisher) and used as a repair template.
CHO-S21 cells were plated in 6 well plates at a density of 2 x 105 per well. 24 hours later the cells were transfected with the repair template plasmid (0.5 μg) and an sgRNA/Cas9 plasmid (UK1596, 0.5 μg) using Lipofectamine LTX (Invitrogen). Two days later, GFP positive cells, which indicate plasmid transfected cells, were collected by using a Beckman Coulter MoFlo Cell sorter, and recovered in the medium for five to six days till the fluorescent marker expression was eliminated. The cells were re-plated in 6 well plates at a density of 4 x 104 cells per well and two days later, the cells were treated with 0.5 mM histidinol. After 18 hours, the cells were subjected to cell sorting using a Beckman Coulter MoFlo Cell sorter. Sorting gates were set according CHOP::GFP reporter expression as bin#1; top 10%, bin#2; upper middle 35%, bin#3 lower middle 35%, and bin#4 bottom 10% GFP expression, respectively (See also Fig 2C). Cells located between the gate boundaries (10% of total) were not collected. Collected cells were recovered and expanded to obtain enough cells for genomic DNA isolation.
Genomic DNA was isolated using standard phenol-chloroform extraction and isopropanol precipitation protocol. EIF2B4 A392 genomic locus was amplified by nested PCR using oligo DNAs No. 17 and No. 18 as a first primer set and No. 13 and No. 16 as second. The PCR products were digested by SpeI (BcuI, ThermoFisher) and run on 2% agarose gel containing SYTO60 red fluorescent nucleic acid stain (ThermoFisher). The gel was scanned on an Odyssey near infrared imager (LI-COR).
EIF2S1/eIF2α S51A mutation or ERN1/IRE1α deletion pool analysis in CHO-S21 cells
The duplex DNA for targeting EIF2S1 S51A [made of oligo DNAs (No. 25 and No. 26) in S1 Table] or ERN1 [made of oligo DNAs (No. 27 and No. 28)] were inserted into the pCas9-2A-mCherry plasmid [lab ID; UK1610, which was made by replacing the eGFP portion of pCas9(BB)-2A-GFP plasmid to mCherry using the Gibson assembly (NEB) according the manufacturer’s instruction], making the sgRNA/Cas9 plasmids (lab ID; UK1696, or UK1615, respectively). For a repair template of EIF2S1 S51A or control S51 wildtype, single strand oligo DNAs (ssODN; No. 29 or No. 30 in S1 Table, respectively) were synthesized (Integrated DNA Technologies).
CHO-S21 cells were plated in 6 well plates at a density of 2 x 105 per well. 24 hours later the cells were transfected with UK1696 plasmid (0.5 μg) and an ssODN (0.5 μg) for EIF2S1 or UK1615 plasmid (0.5 μg) alone for ERN1 using Lipofectamine LTX (Invitrogen). Two days later, mCherry positive cells, which indicate plasmid-transfected cells, were collected using a Beckman Coulter MoFlo Cell sorter, and recovered in the medium for five to six days. The cells were re-plated in 6 well plates at a density of 4 x 104 cells per well, and two days later, the cells were treated with 250 nM thapsigargin for 24 hours. The reporter fluorescent protein expression in the cells was analyzed by Flow cytometry as described below.
Quantitative PCR
Total RNA was isolated from CHO derived cells by acid guanidinium thiocyanate-phenol-chloroform extraction using RNA STAT 60 (Amsbio) and isopropanol precipitation. 1.5 μg of RNA was reverse transcribed by a reverse transcriptase RevertAid (ThermoFisher) with oligo(dT)18 primer. Quantitative PCR analysis was performed using Power SYBR Green PCR Master Mix (Applied Biosystems) according the manufacturer’s instruction on a 7900HT Fast Real-Time PCR system (Applied Biosystems). Oligo DNAs in Tabel I were used for PCR reaction; No. 19 and No. 20 for EIF2B4, No. 21 and No. 22 for EIF2B1, No. 23 and No. 24 for RPL27. Relative quantities of amplified PCR products were determined using SDS 2.4.1 software (Applied Biosystems) and normalized to RPL27 values.
Puromycin labeling and immunoblot analysis
For puromycin labeling experiments, 3 x 105 cells were plated in 60 mm dishes. Two days later, culture media was changed to fresh media. For monitoring basal translation, the cells were treated with 10 μg/ml puromycin for 10 min and harvested. For monitoring translation in stimulated cells, cells were treated with 250 nM thapsigargin for 24 hours and 10 μg/ml puromycin was added during the last 10 min before harvest. For monitoring translation in recovering cells, the stimulated cells were washed by 3 ml of culture media and incubated for 1 hour, followed by treatment of 10 μg/ml puromycin during the last 10 min before harvest. Cells were lysed in a lysis buffer containing 1% Triton X-100, 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 10% Glycerol, 2 mM PMSF, 10 μg/ml aprotinin, 4 μg/μl pepstatin and 4 μM leupeptin. After centrifugation at 21130 x g for 15 min, supernatants were mixed with a standard SDS-PAGE sample buffer. 40 μg of total protein was subjected to 12.5% SDS–polyacrylamide gels electrophoresis and transferred onto Immobilin-P PVDF membrane (EMD Millipore). Immunoblot detection was conducted using primary antibodies for puromycinylated protein [26], total eIF2α [27], and IRDye 800 conjugated secondary antisera (LI-COR) followed by scanning on a Odyssey near infrared imager (LI-COR). Scanned images were quantified using imageJ software.
For detection of eIF2Bδ, a rabbit anti-eIF2Bδ primary antibody (Proteintech), HRP-linked rabbit secondary antibody and SuperSignal West Pico Chemiluminescent Substrate (ThermoFisher) were used.
In vitro measurement of GEF activity on GDP-bound eIF2
The fluorescent intensity-based assay of GDP release from eIF2 (GEF assay) was performed as previously described [28]. Substrate eIF2 was prepared from 10 confluent 10 cm dishes of CHO cells harboring eIF2αS51A-3xFLAG. Cells were washed with ice cold PBS and collected with PBS containing 1 mM EDTA. After centrifugation at 376 x g for 5 min, the cell pellets were lysed with a 4 x volume of the lysis buffer described in the immunoblot analysis section. The lysates were centrifuged at 21130 x g for 15 min. The supernatant was pre-cleared with Protein A sepharose (ZYMED) at 4°C for 30 min. After removal of Protein A sepharose, 30 μl of FLAG-M2 sepharose (SIGMA) was added into the supernatant and rotated for 30 min at 4°C. The beads were washed with 1 ml of high salt (500 mM NaCl) lysis buffer three times and with 50 μl of GDP-mounting buffer [50 mM Tris-HCl pH7.4, 150 mM NaCl, 1 mM DTT] twice.
3xFLAG-eIF2 complex was eluted with 125 μg/ml 3xFLAG peptide (SIGMA) in 40 μl of GDP-mounting buffer at 4°C for 30 min mixing at 1000 rpm. The 40 μl of eluate, which contains approx. 30 nM eIF2 complex, was mixed with 20 μl of 150 nM Bodipy-FL-GDP (Invitrogen) and incubated at 25°C for 20 min to reach equilibrium binding of Bodipy-FL-GDP in eIF2. Then, the reactant was mixed with 10 μl of 12 mM MgCl2 and was passed through a G-50 sephadex column (GE healthcare) equilibrated with GEF assay buffer [50 mM Tris-HCl (pH7.4), 150 mM NaCl, 1 mM DTT, 2 mM MgCl2, 0.01% Triton X-100] and used as a substrate in the GEF assay.
CHO cells from 80–90% confluent 10 cm dishes were collected as described above and mixed with homogenization buffer [50 mM Tris-HCl (pH7.4), 375 μM magnesium acetate, 75 μM EDTA, 95 mM potassium acetate, 2.5 mg/ml digitonin, 10% Glycerol, 1 mM DTT] supplemented with protease and phosphatase inhibitors as the lysis buffer. The mixture was set on ice for 15 min and passed through a 0.5 ml syringe with 29G needle five times. After centrifugation at 21130 x g for 15 min, the supernatants were subjected to the GEF assay.
For the GEF assay, 2 μl of eIF2 substrate and 3 μl of GEF assay buffer containing 1.5 mM non-labeled GDP were mixed with 5 μl of 4.5 μg/μl cell lysate in 384 well round bottom black polystyrene assay plates (Corning, Cat #3667). Fluorescence intensity (excitation wavelength: 485 nm, bandwidth 20 nm, emission wavelength: 535 nm, band width 25 nm) was measured using a TECAN F500 plate reader every 20 seconds till the fluorescence intensity plateaued. GEF activity was calculated as a decrease of the fluorescent intensity (ΔFI) per second at the initial linear phase of the reaction.
Immuno-affinity Purification of endogenous eIF2B complex
Fourteen 10 cm dishes of confluent CHO-S7 parental or EIF2B4A392D cells were washed with ice cold PBS and collected with PBS containing 1 mM EDTA. After centrifugation at 376 x g for 5 min, the cell pellets were lysed with a 4 x volume of the modified homogenization (MH) buffer [50 mM Tris-HCl (pH7.4), 375 μM magnesium acetate, 75 μM EDTA, 95 mM potassium acetate, 10% Glycerol, 0.5% Triton X-100, 1 mM DTT] supplemented with protease and phosphatase inhibitors as the lysis buffer. The lysates were centrifuged at 21130 x g for 15 min. The supernatant was pre-cleared with Protein A sepharose at 4°C for 30 min. After removal of protein A sepharose (ZYMED), 30 μl of FLAG-M2 sepharose (SIGMA) was added into the supernatant and rotated for 30 min at 4°C. The beads were washed in 1 ml of MH buffer three times. FLAG-eIF2B complex on the beads was eluted with 250 μg/ml 3xFLAG peptide (SIGMA) in 20 μl of MH buffer at 4°C for 30 min mixing at 1200 rpm. After spinning down the beads, 18 μl of the supernatant was mixed with 6 μl of 4x SDS-sample buffer. The whole sample was subjected to 12.5% SDS-PAGE and the gel was stained by InstantBlue (Expedeon). The identity of eIF2B complex was confirmed by Mass spectrometry.
For a large scale purification of eIF2B complex, which was subjected to in vitro GEF assay as shown in Fig 1A and S1A and S1B Fig, CHO-S7 parental or EIF2B4A392D cells were adapted to suspension culture. 3.5 litter of cultured cells (1 x106 cells/mL) were collected by spin down and washed with 2 x 25 ml of ice cold PBS. Cells were lysed with 2x pellet volume of lysis buffer [50mM Tris pH7.5, 150mM NaCl, 5mM MgCl2, 0.5% Triton, 10% Glycerol, 1mM DTT, 1x protease inhibitors]. Lysate were clarified by centrifuge at 20,000 rpm for 30 min and ~ 35 ml of supernatant were obtained. 600 μl (300 μl bed volume) of FLAG-M2 sepharose (SIGMA) was added into the supernatant and rotated for 60 min at 4°C. The beads were washed with high salt (500 mM NaCl) lysis buffer four times and with GEF assay buffer four times. FLAG-eIF2B complex on the beads was eluted with 125 μg/ml 3xFLAG peptide (SIGMA) in 400 μl of GEF assay buffer twice. The eluted wildtype and mutant eIF2B complexes, which were adjusted to equal amount of eIF2Bε catalytic subunit (3 nM), were subjected to GEF assay as described above.
Flow Cytometry analysis
CHO-S21 cells were plated at a density of 4 x 104 cells per well on a 6-well tissue culture plate. Two days later the culture medium was replaced with 2 mL of fresh medium and cells were treated with indicated compounds for 24 hours. Immediately before analysis, the cells were washed with PBS and collected in PBS containing 4 mM EDTA. Single cell fluorescent signals (10,000 cells/sample) were measured by a dual-channel flow cytometry with LSRFortessa cell analyzer (Beckton Dickinson). GFP (excitation laser 488 nm, filter 530/30), Turquoise (modified CFP) (excitation laser 405 nm, filter 450/50) signals were detected. FlowJo software was used to analyze the data.
Cell viability analysis (WST-1 assay)
CHO-S21 parental or VWM cells were plated at a density of 2,000 cells per well in a 24-well tissue culture plate. Two days later the culture medium was replaced with the medium containing the indicated compounds as shown in Fig 4B. 24 hours later, the cells were washed once in regular medium and then maintained in regular medium for 48 hours. Then, the medium was replaced with fresh medium containing 50 μM WST-1 (Dojindo) and 20 μM 1-methoxy phenazine methosulfate (Sigma), and the cells were incubated for 2 hours in the cell culture incubator before absorbance at 440 nm in the culture media was measured.
Statistical analysis
Statistical significances were determined by unpaired t test or one way ANOVA followed by Dunnett’s multiple comparisons test as indicated in the Figure legends, using Prism software (GraphPad Software, Inc.).
Supporting Information
(A) Time dependent decline of fluorescent intensity (FI) of eIF2-Bodipy-GDP substrate by purified wildtype (WT) or eIF2BδA392D mutant (δA392D) eIF2B complex or control buffer (-). FI was measured every 20 seconds. (B) Coomassie brilliant blue stained SDS-PAGE of purified WT or δA392D eIF2B complexes used for measuring GEF activity in Fig 1B and S1A Fig. Note that the δA392D eIF2B complex possessed less eIF2Bε catalytic subunit when the same amounts of eIF2Bγ-FLAG were loaded (compare lane 2 and 3). The amount of purified eIF2B introduced into the GEF assay was adjusted to equalize the content of the eIF2Bε catalytic subunit. (C) GEF activity in the lysates of parental CHO-S21 and EIF2B4A392D cells. Shown are means ± S.D. of three independent experiments. *P = 0.0066, Unpaired t test. (D) Quantitative PCR for mRNA expression of EIF2B4 and EIF2B1 in CHO-S21 parental and EIF2B4A392D cells. Shown are means ± S.D. of three independent experiments. * P = 0.001, Unpaired t test. (E) Immunoblot of total eIF2Bδ and eIF2α in parental CHO-S21 and EIF2B4A392D cell lysate. (F) Translation monitored by immunoblot for puromycinylated proteins in parental CHO-S21 and EIF2B4A392D cells. Quantification of these measurements from four independent experiments is presented in Fig 4C. (G) Cell growth of parental CHO-S21 and EIF2B4A392D cells under standard culture condition. Shown are the means ± S.D. of three independent experiments. Cells reached full confluence at day 5.
(PDF)
(A) The Cricetulus griseus EIF2B4 genomic locus as in Fig 1A, showing the position of the silent SpeI site introduced by recombination of the repair template encoding a wildtype protein and the one encoding the A392D mutation. Note that whilst the parental and repaired chromosomes could also be distinguished by loss of the PstI site from the repaired version, a variable background of undigested PCR product eroded the discriminatory value of the PstI RFLP. (B) Histogram of the CHOP::GFP reporter expression in untreated and histidinol-treated cells described in Fig 2C.
(PDF)
(A) Allele structure of the Cricetulus griseus EIF2B4 genomic locus (NW_003613640.1, 5027:5071 for R468W, 5084:5146 for R484W) targeted by CRISPR-Cas9 system and eIF2Bδ encoded protein (XP_003497100.1, 461:475 for R468W, 480:500 for R484W). Horizontal lines and vertical arrows represent the sgRNA binding sites and Cas9 cleavage sites, respectively. Mutations, shown in red, disrupt sgRNA targeting, eliminate or generate restriction enzyme site (a PstI site for R468W or a SalI site for R484W, respectively), and generate R468W or R484W mutation. The EIF2B4R484W mutant clone possesses R484W mutation on one allele and 7 nucleotides deletion on another, which causes three missense mutations and subsequent premature stop codon. (B) Immunoblot of eIF2Bδ and eIF2α in parental and indicated VWM mutant CHO-S21 cells. (C) Schema of the experiment to measure the sensitivity for EIF2S1S51A mutant cells to histidinol. After targeting the EIF2S1 locus with an EIF2S1S51A repair template, CHO-S21 cells were either left untreated (“Control”) or exposed to 0.5 mM histidinol for 2 days and allowed to recover for additional 2 days before treatment with 250 nM thapsigargin (Tg) for 1 day and flow cytometry to quantify the fraction of ISR negative (putative EIF2S1S51A mutant) cells in the population. (D) Flow cytometry analysis of cells subjected to the experiment described in “C”. Note the depletion of CHOP::GFP negative, XBP1::turquoise positive putative EIF2S1S51A mutant cells from the population of cells exposed to histidinol. (E) Flow cytometry analysis of reporter activity in parental CHO-S21 cells or a representative stable EIF2S1S51A mutant clone (C8) isolated from the EIF2S1S51A template-transfected CHO-S21 cell pool. Cells were treated with 2 μg/ml tunicamycin (Tm) for 20 hours before analysis. (F) Flow cytometry analysis of reporter activity in untreated (UT) and thapsigargin-treated (Tg) parental CHO-S7 or EIF2B4A392D mutant cells following targeting of the EIF2S1 locus with an EIF2S1S51A repair template (as in Fig 3A). Note the lack of CHOP::GFP negative thapsigargin-treated putative EIF2S1S51A; EIF2B4A392D double mutant cells (lower right panel) and the absence of a signal in the turquoise channel of these CHO-S7 cells lacking XBP1::turquoise reporter. (G) Percentage of CHOP::GFP negative thapsigargin-treated putative EIF2S1S51A cells in the parental or EIF2B4A392D population from experiments as in “F”. Shown are means ± S.D. of three independent experiments. ** P = 0.0021, Unpaired t test. (H) As in Fig 3A, but cells were transfected with an sgRNA/Cas9 plasmid targeting ERN1/IRE1α. Note the emergence of CHOP::GFP positive, XBP1::turquoise negative ERN1/IRE1α mutant cells in both the parental and EIF2B4A392D pools. (I) Percentage of CHOP::GFP positive, XBP1::turquoise negative putative ERN1/IRE1α deleted cells in the parental or EIF2B4A392D population from experiments shown in “H”. Shown are means ± S.D. of three independent experiments. No statistical significance in unpaired t test.
(PDF)
(PDF)
Acknowledgments
We thank the CIMR flow cytometry core facility team (R. Schulte, M. Maj and C. Cossetti) for assistance with FACS and the CIMR proteomics core facility for protein identification and T. Iwawaki (Gunma University) for the gift of the XBP1::Venus reporter gene.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Supported by grants from the Wellcome Trust (Wellcome 200848/Z/16/Z) and the European Commission (EU FP7 Beta-Bat No: 277713) and, a Wellcome Trust Strategic Award for core facilities to the Cambridge Institute for Medical Research (Wellcome 100140). YS is a Japanese Society for the Promotion of Science Postdoctoral Fellow for Research Abroad. NAW is a Medical Research Council supported PhD student. DR is a Wellcome Trust Principal Research Fellow. Wellcome Trust URL; http://www.wellcome.ac.uk, JSPS URL; https://www.jsps.go.jp, MRC URL; http://www.mrc.ac.uk. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Mohammad-Qureshi SS, Jennings MD, Pavitt GD. Clues to the mechanism of action of eIF2B, the guanine-nucleotide-exchange factor for translation initiation. Biochem Soc Trans. 2008;36(Pt 4):658–64. 10.1042/BST0360658 . [DOI] [PubMed] [Google Scholar]
- 2.Proud CG. eIF2 and the control of cell physiology. Semin Cell Dev Biol. 2005;16(1):3–12. 10.1016/j.semcdb.2004.11.004 [DOI] [PubMed] [Google Scholar]
- 3.Baird TD, Wek RC. Eukaryotic initiation factor 2 phosphorylation and translational control in metabolism. Advances in nutrition. 2012;3(3):307–21. Epub 2012/05/16. 10.3945/an.112.002113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11(3):619–33. Epub 2003/04/02. . [DOI] [PubMed] [Google Scholar]
- 5.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–50. Epub 2005/09/13. 10.1146/annurev.micro.59.031805.133833 . [DOI] [PubMed] [Google Scholar]
- 6.Ron D, Harding H. eIF2α phosphorylation in cellular stress responses and disease In: Sonenberg N, Hershey J, Mathews M, editors. Translational Control. Cold Spring Harbor Monograph Series. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2007. p. 345–68. [Google Scholar]
- 7.Lin W, Popko B. Endoplasmic reticulum stress in disorders of myelinating cells. Nat Neurosci. 2009;12(4):379–85. 10.1038/nn.2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma T, Trinh MA, Wexler AJ, Bourbon C, Gatti E, Pierre P, et al. Suppression of eIF2 alpha kinases alleviates Alzheimer's disease-related plasticity and memory deficits. Nat Neurosci. 2013;16(9):1299–305. 10.1038/nn.3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moreno JA, Radford H, Peretti D, Steinert JR, Verity N, Martin MG, et al. Sustained translational repression by eIF2 alpha-P mediates prion neurodegeneration. Nature. 2012;485(7399):507–11. 10.1038/nature11058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Knaap MS, van Berkel CG, Herms J, van Coster R, Baethmann M, Naidu S, et al. eIF2B-related disorders: antenatal onset and involvement of multiple organs. Am J Hum Genet. 2003;73(5):1199–207. 10.1086/379524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fogli A, Boespflug-Tanguy O. The large spectrum of eIF2B-related diseases. Biochem Soc Trans. 2006;34(Pt 1):22–9. Epub 2005/10/26. BST20060022 [pii] 10.1042/BST20060022 . [DOI] [PubMed] [Google Scholar]
- 12.Pavitt GD, Proud CG. Protein synthesis and its control in neuronal cells with a focus on vanishing white matter disease. Biochem Soc Trans. 2009;37(Pt 6):1298–310. 10.1042/BST0371298 . [DOI] [PubMed] [Google Scholar]
- 13.Li W, Wang X, Van Der Knaap MS, Proud CG. Mutations linked to leukoencephalopathy with vanishing white matter impair the function of the eukaryotic initiation factor 2B complex in diverse ways. Mol Cell Biol. 2004;24(8):3295–306. 10.1128/MCB.24.8.3295-3306.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kantor L, Harding HP, Ron D, Schiffmann R, Kaneski CR, Kimball SR, et al. Heightened stress response in primary fibroblasts expressing mutant eIF2B genes from CACH/VWM leukodystrophy patients Hum Genet. 2005;118(1):99–106. [DOI] [PubMed] [Google Scholar]
- 15.van Kollenburg B, Thomas AA, Vermeulen G, Bertrand GA, van Berkel CG, Pronk JC, et al. Regulation of protein synthesis in lymphoblasts from vanishing white matter patients. Neurobiol Dis. 2006;21(3):496–504. 10.1016/j.nbd.2005.08.009 . [DOI] [PubMed] [Google Scholar]
- 16.Liu R, van der Lei HD, Wang X, Wortham NC, Tang H, van Berkel CG, et al. Severity of vanishing white matter disease does not correlate with deficits in eIF2B activity or the integrity of eIF2B complexes. Hum Mutat. 2011;32(9):1036–45. 10.1002/humu.21535 . [DOI] [PubMed] [Google Scholar]
- 17.Novoa I, Zeng H, Harding H, Ron D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2α. J Cell Biol. 2001;153(5):1011–22. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwawaki T, Akai R, Kohno K, Miura M. A transgenic mouse model for monitoring endoplasmic reticulum stress. Nat Med. 2004;10(1):98–102. 10.1038/nm970 [DOI] [PubMed] [Google Scholar]
- 19.Harashima S, Hinnebusch AG. Multiple GCD genes required for repression of GCN4, a transcriptional activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1986;6(11):3990–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheuner D, Song B, McEwen E, Gillespie P, Saunders T, Bonner-Weir S, et al. Translational control is required for the unfolded protein response and in-vivo glucose homeostasis. Mol Cell. 2001;7(6):1165–76. Epub 2001/06/30. . [DOI] [PubMed] [Google Scholar]
- 21.Maletkovic J, Schiffmann R, Gorospe JR, Gordon ES, Mintz M, Hoffman EP, et al. Genetic and clinical heterogeneity in eIF2B-related disorder. J Child Neurol. 2008;23(2):205–15. 10.1177/0883073807308705 . [DOI] [PubMed] [Google Scholar]
- 22.van der Knaap MS, Pronk JC, Scheper GC. Vanishing white matter disease. Lancet Neurol. 2006;5(5):413–23. 10.1016/S1474-4422(06)70440-9 . [DOI] [PubMed] [Google Scholar]
- 23.Fogli A, Rodriguez D, Eymard-Pierre E, Bouhour F, Labauge P, Meaney BF, et al. Ovarian Failure Related to Eukaryotic Initiation Factor 2B Mutations. Am J Hum Genet. 2003;72(6):1544–50. 10.1086/375404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ronda C, Pedersen LE, Hansen HG, Kallehauge TB, Betenbaugh MJ, Nielsen AT, et al. Accelerating genome editing in CHO cells using CRISPR Cas9 and CRISPy, a web-based target finding tool. Biotechnol Bioeng. 2014;111(8):1604–16. Epub 2014/05/16. 10.1002/bit.25233 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–308. Epub 2013/10/26. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nature methods. 2009;6(4):275–7. Epub 2009/03/24. nmeth.1314 [pii] 10.1038/nmeth.1314 . [DOI] [PubMed] [Google Scholar]
- 27.Scorsone KA, Panniers R, Rowlands AG, Henshaw EC. Phosphorylation of eukaryotic initiation factor 2 during physiological stresses which affect protein synthesis. J Biol Chem. 1987;262(30):14538–43. . [PubMed] [Google Scholar]
- 28.Sekine Y, Zyryanova A, Crespillo-Casado A, Fischer PM, Harding HP, Ron D. Mutations in a translation initiation factor identify the target of a memory-enhancing compound. Science. 2015;348(6238):1027–30. Epub 2015/04/11. 10.1126/science.aaa6986 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Time dependent decline of fluorescent intensity (FI) of eIF2-Bodipy-GDP substrate by purified wildtype (WT) or eIF2BδA392D mutant (δA392D) eIF2B complex or control buffer (-). FI was measured every 20 seconds. (B) Coomassie brilliant blue stained SDS-PAGE of purified WT or δA392D eIF2B complexes used for measuring GEF activity in Fig 1B and S1A Fig. Note that the δA392D eIF2B complex possessed less eIF2Bε catalytic subunit when the same amounts of eIF2Bγ-FLAG were loaded (compare lane 2 and 3). The amount of purified eIF2B introduced into the GEF assay was adjusted to equalize the content of the eIF2Bε catalytic subunit. (C) GEF activity in the lysates of parental CHO-S21 and EIF2B4A392D cells. Shown are means ± S.D. of three independent experiments. *P = 0.0066, Unpaired t test. (D) Quantitative PCR for mRNA expression of EIF2B4 and EIF2B1 in CHO-S21 parental and EIF2B4A392D cells. Shown are means ± S.D. of three independent experiments. * P = 0.001, Unpaired t test. (E) Immunoblot of total eIF2Bδ and eIF2α in parental CHO-S21 and EIF2B4A392D cell lysate. (F) Translation monitored by immunoblot for puromycinylated proteins in parental CHO-S21 and EIF2B4A392D cells. Quantification of these measurements from four independent experiments is presented in Fig 4C. (G) Cell growth of parental CHO-S21 and EIF2B4A392D cells under standard culture condition. Shown are the means ± S.D. of three independent experiments. Cells reached full confluence at day 5.
(PDF)
(A) The Cricetulus griseus EIF2B4 genomic locus as in Fig 1A, showing the position of the silent SpeI site introduced by recombination of the repair template encoding a wildtype protein and the one encoding the A392D mutation. Note that whilst the parental and repaired chromosomes could also be distinguished by loss of the PstI site from the repaired version, a variable background of undigested PCR product eroded the discriminatory value of the PstI RFLP. (B) Histogram of the CHOP::GFP reporter expression in untreated and histidinol-treated cells described in Fig 2C.
(PDF)
(A) Allele structure of the Cricetulus griseus EIF2B4 genomic locus (NW_003613640.1, 5027:5071 for R468W, 5084:5146 for R484W) targeted by CRISPR-Cas9 system and eIF2Bδ encoded protein (XP_003497100.1, 461:475 for R468W, 480:500 for R484W). Horizontal lines and vertical arrows represent the sgRNA binding sites and Cas9 cleavage sites, respectively. Mutations, shown in red, disrupt sgRNA targeting, eliminate or generate restriction enzyme site (a PstI site for R468W or a SalI site for R484W, respectively), and generate R468W or R484W mutation. The EIF2B4R484W mutant clone possesses R484W mutation on one allele and 7 nucleotides deletion on another, which causes three missense mutations and subsequent premature stop codon. (B) Immunoblot of eIF2Bδ and eIF2α in parental and indicated VWM mutant CHO-S21 cells. (C) Schema of the experiment to measure the sensitivity for EIF2S1S51A mutant cells to histidinol. After targeting the EIF2S1 locus with an EIF2S1S51A repair template, CHO-S21 cells were either left untreated (“Control”) or exposed to 0.5 mM histidinol for 2 days and allowed to recover for additional 2 days before treatment with 250 nM thapsigargin (Tg) for 1 day and flow cytometry to quantify the fraction of ISR negative (putative EIF2S1S51A mutant) cells in the population. (D) Flow cytometry analysis of cells subjected to the experiment described in “C”. Note the depletion of CHOP::GFP negative, XBP1::turquoise positive putative EIF2S1S51A mutant cells from the population of cells exposed to histidinol. (E) Flow cytometry analysis of reporter activity in parental CHO-S21 cells or a representative stable EIF2S1S51A mutant clone (C8) isolated from the EIF2S1S51A template-transfected CHO-S21 cell pool. Cells were treated with 2 μg/ml tunicamycin (Tm) for 20 hours before analysis. (F) Flow cytometry analysis of reporter activity in untreated (UT) and thapsigargin-treated (Tg) parental CHO-S7 or EIF2B4A392D mutant cells following targeting of the EIF2S1 locus with an EIF2S1S51A repair template (as in Fig 3A). Note the lack of CHOP::GFP negative thapsigargin-treated putative EIF2S1S51A; EIF2B4A392D double mutant cells (lower right panel) and the absence of a signal in the turquoise channel of these CHO-S7 cells lacking XBP1::turquoise reporter. (G) Percentage of CHOP::GFP negative thapsigargin-treated putative EIF2S1S51A cells in the parental or EIF2B4A392D population from experiments as in “F”. Shown are means ± S.D. of three independent experiments. ** P = 0.0021, Unpaired t test. (H) As in Fig 3A, but cells were transfected with an sgRNA/Cas9 plasmid targeting ERN1/IRE1α. Note the emergence of CHOP::GFP positive, XBP1::turquoise negative ERN1/IRE1α mutant cells in both the parental and EIF2B4A392D pools. (I) Percentage of CHOP::GFP positive, XBP1::turquoise negative putative ERN1/IRE1α deleted cells in the parental or EIF2B4A392D population from experiments shown in “H”. Shown are means ± S.D. of three independent experiments. No statistical significance in unpaired t test.
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.