SUMMARY
Transcriptional gene silencing controls transposons and other repetitive elements through RNA-directed DNA methylation (RdDM) and heterochromatin formation. A key component of the Arabidopsis RdDM pathway is ARGONAUTE4 (AGO4), which associates with siRNAs to mediate DNA methylation. Here, we show that AGO4 preferentially targets transposable elements embedded within promoters of protein-coding genes. This pattern of AGO4 binding cannot be simply explained by the sequences of AGO4-bound siRNAs; instead, AGO4 binding to specific gene promoters is also mediated by long non-coding RNAs (lncRNAs) produced by RNA polymerase V. lncRNA-mediated AGO4 binding to gene promoters directs asymmetric DNA methylation to these genomic regions and is involved in regulating the expression of targeted genes. Finally, AGO4 binding overlaps sites of DNA methylation affected by the biotic stress response. Based on these findings, we propose that the targets of AGO4-directed RdDM are regulatory units responsible for controlling gene expression under specific environmental conditions.
Keywords: gene silencing, ncRNA, Argonaute, Pol V, Arabidopsis thaliana, stress
INTRODUCTION
Transcriptional gene silencing is mediated by repressive chromatin modifications directed to transposable elements and other repetitive sequences to prevent their expression, which, if uncontrolled, may have detrimental effects on the cell. In eukaryotic organisms, the primary factors driving the functional mechanism of silencing are the conserved Argonaute proteins (Hutvagner and Simard, 2008). In Arabidopsis thaliana, the RNA-mediated transcriptional gene silencing pathway (also known as RNA-mediated DNA methylation; RdDM) is mediated by ARGONAUTE4 (AGO4) (Zilberman et al., 2003). Specific genomic localization of AGO4 has been hypothesized to require the joint activity of two classes of non-coding RNAs. The first are small interfering RNAs (siRNAs), which are produced by the activities of RNA polymerase IV (Pol IV), RNA-DEPENDENT RNA POLYMERASE 2 (RDR2) and DICER-LIKE 3 (DCL3) (Law and Jacobsen, 2010). siRNAs bind AGO4 and provide sequence specificity (Qi et al., 2006) through direct base-pairing interactions with complementary loci. The other class of non-coding RNAs involved in targeting AGO4 to specific genomic loci is probably long non-coding RNA (lncRNA) produced by plant-specific RNA polymerase V (Pol V) (Wierzbicki et al., 2008), with some involvement of RNA polymerase II (Zheng et al., 2009). Pol V-produced lncRNAs have been proposed to act as binding scaffolds for AGO4–siRNA complexes (Wierzbicki et al., 2009; Wierzbicki, 2012). Upon binding to chromatin, AGO4 is believed to work with at least one more RNA-associated protein (SPT5L/KTF1) (Rowley et al., 2011), guide the de novo DNA methyltransferase DRM2 (DOMAINS REAR-RANGED METHYLASE 2), and thereby mediate DNA methylation, primarily in CHH contexts (Wierzbicki, 2012).
Little is known about the genome-wide distribution of AGO4 or other RdDM components or the mechanisms that direct them to specific loci. It is also unknown to what extent the RdDM pathway controls expression of protein-coding genes involved in specific biological processes. To answer these questions, we characterized the genome-wide distribution of AGO4 binding to chromatin. We found that AGO4 preferentially targets promoters of protein-coding genes. This specific binding pattern cannot be explained by the sequences of AGO4-associated small RNAs, and appears to be primarily mediated by Pol V-produced lncRNAs. AGO4 binding to gene promoters mediates CHH methylation, and, in some cases, affects expression levels of genes controlled by these promoters. Moreover, AGO4 binding overlaps with DNA methylation affected by the biotic stress response. This combination of results leads to the intriguing hypothesis that AGO4 binding sites are regulatory units that control gene expression under specific environmental conditions.
RESULTS
AGO4 has no preference towards TE-rich pericentromeric regions
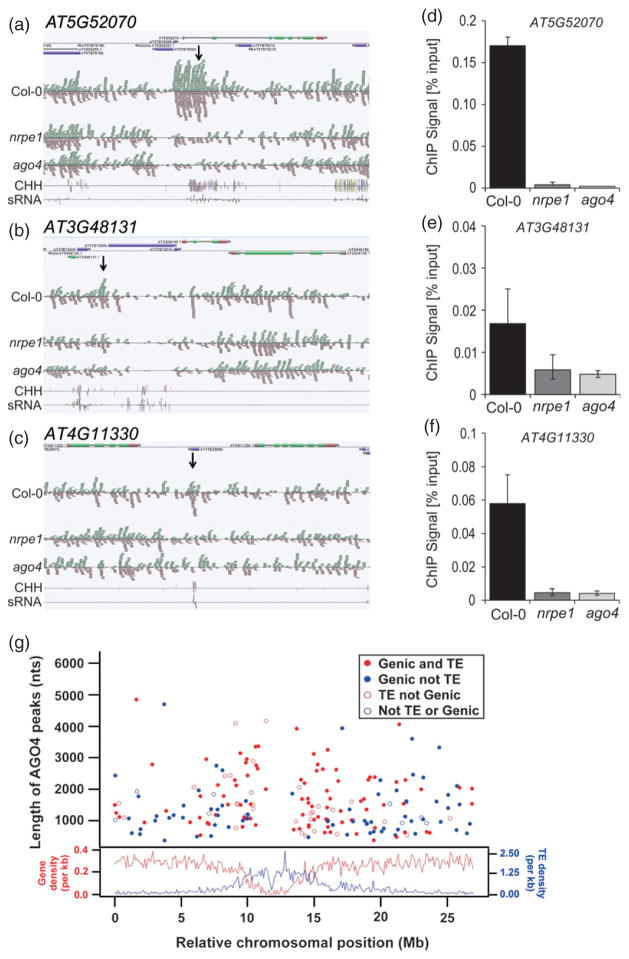
As the first step towards explaining the mechanism by which AGO4 directs RdDM-mediated silencing to specific loci, we assayed the genome-wide distribution of AGO4 binding targets using chromatin immunoprecipitation with an anti-AGO4 polyclonal antibody followed by high-throughput sequencing (ChIP-seq). Using a combinatorial comparison approach, in which ChIP-seq samples from Col-0 wild-type were compared with those from the ago4 mutant as well as with input sample controls, we identified 820 AGO4 binding regions (also referred to as peaks; Figure 1a–c, Figure S1 and Data S1). We used ChIP followed by real-time PCR (ChIP-quantitative PCR; ChIP-qPCR) to validate 24 AGO4 binding regions. Binding was confirmed at all 11 tested regions ranked in the top 20% by the peak-calling algorithm (Figure 1d and Figure S1b), all three tested regions ranked in the middle 60% (Figure 1e and Figure S1d), and 10 of 13 tested regions ranked in the bottom 20% (Figure 1f and Figure S1f). Additionally, most previously known AGO4 targets (Wierzbicki et al., 2009; Rowley et al., 2011) displayed evidence of strong AGO4 binding in our ChIP-seq; however, only IGN25 met the stringent criteria for inclusion on the list of significant AGO4 chromatin binding sites. In total, these results indicate that our analysis has a high stringency with a low proportion of false positives, even among the lowest ranking AGO4 binding sites.
Figure 1. Identification of AGO4-bound loci.
(a–c) Graphical representation of sample AGO4-bound loci identified using ChIP-seq. The genome browser screenshots show (from top) genome annotation, ChIP-seq sequencing reads from Col-0 wild-type, nrpe1 and ago4 strains, CHH DNA methylation (Lister et al., 2008) and total small RNA reads (Lister et al., 2008). More loci are shown in Figure S1.
(df) ChIP-qPCR validation of AGO4 binding to chromatin on AGO4 peaks identified using ChIP-seq in Col-0 wild-type, nrpe1 and ago4 mutants. Values are means ± SD from three independent amplifications. More loci are shown in Figure S1.
(g) AGO4 binding shows no preference towards transposon-rich pericentromeric regions. The top graph shows the distribution of AGO4 peaks along the length of chromosome 5 versus peak length in nucleotides (nt). The corresponding category for each colored dot is indicated. The lower graph shows the density of genes (red line and y axis label) and transposable elements (blue line and y axis label) along the length of chromosome 5.
The Arabidopsis RNA-mediated transcriptional gene silencing pathway mostly targets transposable elements (TEs) and other repetitive sequences (Law and Jacobsen, 2010). Because most TEs in the Arabidopsis genome cluster within pericentromeric regions (Arabidopsis Genome Initiative, 2000), silencing components may also be enriched around the centromeres. To test whether the genome-wide AGO4 chromatin binding data confirm this prediction, we mapped AGO4 peaks onto the five nuclear chromosomes of Arabidopsis. Surprisingly, we found that AGO4 peaks were distributed evenly across all five chromosomes, and their density was comparable in TE-rich pericentromeric regions and gene-rich chromosome arms (Figure 1g and Figure S2). Therefore, genome-wide identification of significant AGO4 binding sites indicates that this protein is not preferentially targeted to large heterochromatic and repetitive genomic domains.
AGO4 binds TEs within gene promoters
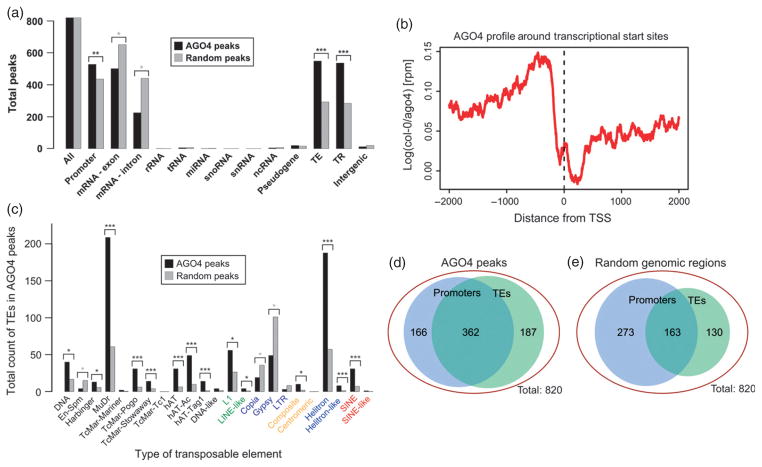
Widespread AGO4 binding within gene-rich chromosome arms often overlapped protein-coding genes (Figure 1g and Figure S2), suggesting that AGO4 binding may be enriched on genes. To test this possibility, we classified AGO4 peaks based on overlaps with annotated genomic features. AGO4 was not enriched on the transcribed regions of protein-coding genes (Figure 2a); instead we observed a significant enrichment on gene promoters defined as 1 kb regions upstream of transcription start sites (P < 0.001; Figure 2a) with 64% of all AGO4 peaks mapping to promoters of protein-coding genes. This pattern was confirmed by profiling the ChIP-seq signal around transcription start sites, which revealed preferential AGO4 binding in the region between approximately −500 and −200 bp upstream of target gene transcription start sites (Figure 2b). Moreover, AGO4 peaks were also depleted in nucleosomes (Figure S3a,b); the absence of nucleosomes being a characteristic feature of gene promoters (Chodavarapu et al., 2010). These results demonstrate that AGO4 preferentially binds promoters of protein-coding genes.
Figure 2. AGO4 is enriched on transposable elements within promoters of protein-coding genes.
(a) AGO4 binding is significantly enriched at promoters, transposable elements and tandem repeats, but deficient in gene bodies. Classification of all AGO4 peaks or a set of randomly generated peaks. TE, transposable element; TR, tandem repeats. Asterisks indicate statistically significant differences between AGO4 peaks and randomly generated peaks (*P < 0.05; **P < 0.001; ***P → 0).
(b) AGO4 binding is enriched on regions upstream of transcription start sites. Profile of AGO4 binding around transcription start sites for all annotated Arabidopsis genes, showing the proportion of ChIP-seq reads in Col-0 wild-type relative to the ago4 mutant plants. rpm, reads per million.
(c) AGO4 binding sites are significantly enriched in DNA transposons, but not in LTR transposable elements. Classification of specific transposable element levels in AGO4 peaks or a set of randomly generated peaks. Transposable elements are as specified on the x axis. Asterisks indicate statistically significant differences between AGO4 peaks and randomly generated peaks (*P < 0.05; **P < 0.001; ***P → 0).
(d,e) AGO4 preferentially binds transposable elements within gene promoters. (d) Venn diagram showing AGO4 peaks mapping to regions 1 kb upstream of transcription start sites, transposable elements or both. (e) Venn diagram showing random genomic regions 1 kb upstream of transcription start sites, transposable elements or both.
AGO4 binding was also enriched on transposable elements and tandem repeats (P → 0; Figure 2a). This enrichment was significant on most class I and class II transposable elements (Figure 2c). Interestingly, AGO4 binding was significantly depleted in En-Spm DNA transposons as well as Copia LTR retrotransposons (Figure 2c), both of which are enriched within coding sequences of protein-coding genes (Lockton and Gaut, 2009). AGO4 binding was also depleted in Gypsy LTR retrotransposons (Figure 2c). These results reveal that AGO4 has a preference towards specific families of transposable elements in the Arabidopsis genome.
Further analysis of AGO4 binding to gene promoters revealed that, out of 528 AGO4 binding regions identified within gene promoters, 362 (69%) overlapped with transposable elements (Figure 2d). In contrast, in a comparable set of random control regions, 436 regions mapped to gene promoters, of which 163 (37%) overlapped with transposable elements (Figure 2e, P < 7 × 10−22), indicating that AGO4 binding to gene promoters does not reflect preferential insertions of TEs into promoter regions. These results demonstrate that AGO4 binding shows a significant preference for both gene promoters and TEs. Together, our findings reveal that AGO4 preferentially binds transposons embedded within the promoters of protein-coding genes.
The AGO4 binding pattern is mediated by lncRNA
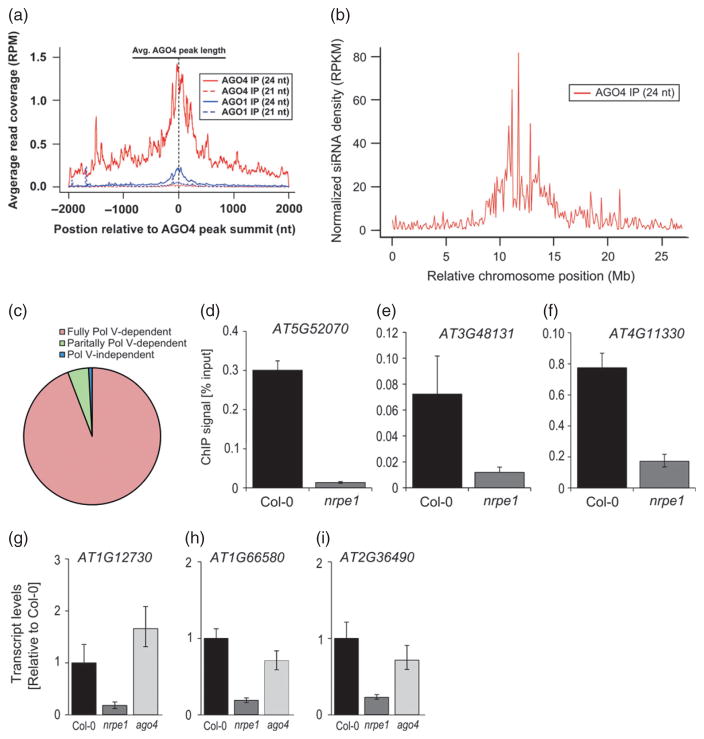
Sequence specificity of AGO4 binding to chromatin has been proposed to be directed by the sequences of incorporated 24 nt siRNAs (Qi et al., 2006). To test whether 24nt siRNAs have a function in directing AGO4 to TEs within promoters of protein-coding genes, we mapped AGO4-bound siRNAs (Wang et al., 2011) to AGO4 peaks. We found that AGO4-associated 24 nt siRNAs are enriched on AGO4 peaks (Figure 3a). As controls, similar analyses with AGO1-bound 24 nt siRNAs (Wang et al., 2011) demonstrated only minimal enrichment, and 21 nt small RNAs bound by either AGO protein revealed negligible enrichment on AGO4 peaks (Figure 3a). Consistent with these findings, we observed that the total population of 24 nt siRNAs but not 21 nt small RNAs (smRNAs), which are implicated in post-transcriptional silencing (Hutvagner and Simard, 2008), was enriched on AGO4 peaks (Figure S4a). Furthermore, only 10% of AGO4 peaks had little or no association with AGO4-bound siRNAs. These results suggest that AGO4 binding to chromatin is correlated with the presence of 24nt siRNAs, which probably have a function in guiding AGO4 to specific genomic loci.
Figure 3. AGO4 binding specificity towards TEs in gene promoters is mediated by lncRNA.
(a) AGO4 binding sites have a significant overlap with AGO4-bound 24 nt siRNAs but not 21 nt small RNAs. The plot shows the levels of previously identified (Wang et al., 2011) 24 nt and 21 nt AGO4-bound small RNAs (red lines as indicated) and AGO1-bound smRNAs (blue lines as indicated). The solid black line at the top indicates the mean AGO4 peak size. The dashed black line indicates the position of AGO4 peak summits.
(b) Density of AGO4-bound 24 nt siRNAs along the length of chromosome 5. RPKM, reads per kilobase per million.
(c) AGO4 binding is dependent on Pol V. Proportions of AGO4 binding sites identified as fully Pol V-dependent, partially Pol V-dependent and Pol V-independent are shown.
(d–f) ChIP/real-time PCR showing Pol V binding to AGO4 peaks. Values are means ± SD from three independent amplifications. More loci are shown in Figure S5b–d.
(g–i) Pol V-dependent transcripts are present on AGO4 binding sites. Accumulation of non-coding RNA accumulation was assayed using real-time RT-PCR in Col-0 wild-type, nrpe1 and ago4 mutant plants, and normalized to ACTIN 2 as a control. Values are means ± SD from three independent amplifications.
To further test whether the sequences of siRNAs are able to explain the specific pattern of AGO4 binding to chromatin, we mapped AGO4-associated 24 nt siRNAs onto the five nuclear chromosomes of Arabidopsis. Surprisingly, we found these siRNAs to be strongly enriched within TE-rich pericentromeric regions and much less abundant within gene-rich chromosome arms (Figure 3b and Figure S4b). Therefore, AGO4-associated siRNAs are not solely responsible for targeting AGO4 to its DNA interaction sites. This is consistent with a model whereby 24 nt siRNAs are necessary but not sufficient for mediating AGO4 binding to specific loci.
Another factor previously implicated in AGO4 binding to specific genomic loci is transcription by Pol V, which has been proposed to provide lncRNA scaffolds for AGO4 binding to chromatin (Wierzbicki et al., 2009). To test whether Pol V is required for genome-wide targeting of AGO4, we performed ChIP-seq using anti-AGO4 antibody on nrpe1 mutant plants, which are deficient for the largest subunit of Pol V. By comparing the ChIP-seq datasets from nrpe1 mutant plants to those of Col-0 wild-type and ago4, we tested whether AGO4 binding to specific loci requires Pol V. Surprisingly, we identified only seven Pol V-independent AGO4 peaks (0.85%) and 41 binding sites (4.96%) that demonstrated intermediate levels of AGO4 binding in nrpe1 mutant plants (Figure 3c). We also analyzed Pol V dependence by comparing normalized read counts of AGO4 binding sites in Col-0 wild-type to the nrpe1 mutant, which confirmed that the vast majority of sites have strongly reduced ChIP signals in the nrpe1 mutant (Figure S5a). These results indicate that Pol V is generally required for AGO4 binding to chromatin. The small proportion of Pol V-independent peaks and differences in AGO4 chromatin interaction strength may reflect a minor Pol V-independent mechanism of AGO4 binding, or, alternatively, may indicate that this type of targeting is not actually biologically significant. The importance of Pol V for AGO4 binding to chromatin was further supported by our ChIP-qPCR PCR validation, which demonstrated that AGO4 binding to all validated loci is dependent on Pol V (Figure 1d–f and Figure S1b,d,f). Furthermore, all 11 tested high-ranking loci, three middle-ranking loci and nine low-ranking loci show detectable Pol V binding by ChIP-qPCR with anti-NRPE1 antibody (Figure 3d–f and Figure S5b–d). Importantly, hitherto undetected AGO4 binding sites showed evidence of Pol V-dependent transcription (Figure 3g–i), indicating that Pol V produces lncRNA at these loci.
In total, these results show that Pol V is required for AGO4 binding to most if not all of its target loci. Furthermore, our observations of (i) a strong preference for gene promoter binding by AGO4, (ii) the lack of concordance between AGO4 interaction sites and siRNA sequences bound by this protein, and (iii) Pol V transcription within AGO4 promoter-bound regions, suggest that lncRNAs produced by Pol V are also a critical factor in mediating the interaction of AGO4 with promoters of specific protein-coding genes.
lncRNA-mediated AGO4 binding controls gene activity
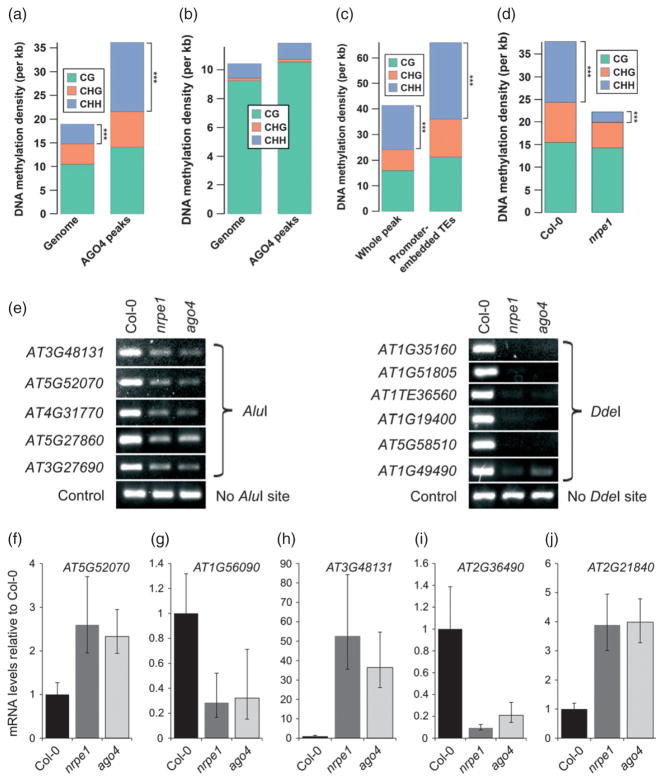
Our observation of lncRNA-mediated AGO4 binding to promoters of protein-coding genes suggests that non-coding transcription and AGO4 binding may control the expression levels of the targeted genes by mediating DNA methylation. To test this possibility, we first examined whether AGO4 binding was correlated with DNA methylation (Lister et al., 2008). AGO4 peaks showed significantly enriched (P → 0) CHH methylation relative to the genome-wide level, and also demonstrated a less pronounced enrichment in CG and CHG methylation (Figure 4a). A similar pattern of DNA methylation coincident with AGO4 binding regions was also present in ros1 dml2 dml3 triple mutant plants that are deficient in three DNA demethylases (Lister et al., 2008) (Figure S6a). Significant enrichment in CHH methylation within AGO4-bound regions was also present in met1, a mutant of the major CG methyltransferase of Arabidopsis (Lister et al., 2008) (Figure S6b). However, in drm1 drm2 cmt3 triple mutant plants (Lister et al., 2008) CHH methylation of AGO4-bound sites was strongly reduced relative to Col-0 wild-type (Figure 4a,b), suggesting that this methylation is established by the de novo methyltransferase DRM2, although involvement of CMT3 cannot be excluded. We also found that DNA methylation within AGO4 binding sites was most prominent on TEs embedded within promoters of protein-coding genes (Figure 4c). Additionally, CHH methylation within AGO4 peaks was significantly reduced (P → 0) in the nrpe1 mutant relative to wild-type (Figure 4d) (Wierzbicki et al., 2012). Together, these results demonstrate that AGO4 binding is correlated primarily with CHH methylation, and predict that AGO4 recruitment to specific genomic loci, including TEs in gene promoters, probably mediates their CHH methylation.
Figure 4. AGO4 binding mediates DNA methylation and controls gene activity.
(a) AGO4 binding sites show enriched DNA methylation, especially at CHH sites. Mean levels of CG (green), CHG (orange) and CHH (blue) methylation sites per kilobase (kb) for the Arabidopsis genome overall (left bar) or within AGO4 peaks (right bar). The methylation data have been published previously (Lister et al., 2008) for Col-0 wild-type plants. ***P → 0.
(b) CHH methylation enrichment of AGO4 binding sites requires non-CG DNA methyltransferases. Mean levels of CG (green), CHG (orange) and CHH (blue) methylation sites per kilobase (kb) for the Arabidopsis genome overall (left bar) or within AGO4 peaks (right bar). The methylation data have been published previously (Lister et al., 2008) for drm1 drm2 cmt3 triple mutant plants.
(c) DNA methylation within AGO4 binding sites is significantly higher on transposable elements embedded within gene promoters. Mean levels of CG (green), CHG (orange) and CHH (blue methylation sites) per kilobase (kb) for all promoter-associated AGO4 binding sites (left bar) or within TEs embedded in promoterassociated AGO4 binding sites (right bar). The methylation data have been published previously (Lister et al., 2008) for Col-0 wild-type plants. ***P → 0.
(d) CHH methylation within AGO4 binding sites is dependent on Pol V. Mean levels of CG (green), CHG (orange) and CHH (blue) methylation sites per kilobase (kb) for AGO4 binding sites in nrpe1 mutant (right bar) and Col-0 wild-type derived from a corresponding dataset (left bar). The methylation data have been published previously (Wierzbicki et al., 2012). ***P → 0
(e) AGO4 and Pol V are required for CHH DNA methylation on AGO4 binding sites. DNA methylation analysis using the AluI DNA methylation-sensitive restriction endonuclease. Digested genomic DNA was amplified using PCR. A sequence lacking AluI sites (IGN5) was used as a loading control. More loci are shown in Figure S6c.
(f–j) AGO4 affects the expression levels of certain protein-coding genes whose promoters contain AGO4 binding sites. mRNA accumulation was assayed using real-time RT-PCR in Col-0 wild-type, nrpe1 and ago4 mutant plants, and normalized to ACTIN 2 as a control. Values are means ± SD from three biological replicates.
To test this prediction, we probed DNA methylation levels on 26 AGO4-bound promoter regions in Col-0 wild-type as well as ago4 and nrpe1 mutants. Digestion with methylation-sensitive restriction endonucleases followed by PCR revealed that tested AGO4-bound promoter regions contain CHH methylation, which was strongly reduced in both nrpe1 and ago4 mutants (Figure 4e and Figure S6c). Taken together, these results indicate that lncRNA-mediated AGO4 binding in gene promoters directs CHH methylation, and this may control transcription of these genes.
To test whether lncRNA-mediated AGO4 binding within gene promoters affects expression of proximal genes, we screened 41 genes with AGO4 peaks in their promoter regions for significant expression changes in nrpe1 and ago4 mutants. Real-time RT-PCR identified three genes that were up-regulated in nrpe1 and ago4 mutants and two genes that were down-regulated in nrpe1 and ago4 mutants (Figure 4f–j). These results demonstrate that expression of at least a subset of AGO4-bound genes is affected by AGO4 and Pol V under standard growth conditions, showing that lncRNA-mediated AGO4 binding within gene promoters is capable of affecting gene expression. One of the genes for which RNA accumulation was reduced in nrpe1 and ago4 mutants under standard growth conditions is ROS1 (AT2G36490; Figure 4i), which encodes a DNA demethylase that has previously been shown to be positively regulated by CG DNA methylation (Mathieu et al., 2007). This suggests the presence of a compensatory mechanism, whereby a reduction in CG methylation or RNA-directed CHH methylation results in a reduction of DNA demethylase production to prevent excessive loss of DNA methylation. In total, these results demonstrate that AGO4 binding within promoter regions is capable of controlling the expression of targeted genes.
AGO4 binding is correlated with DNA methylation affected by biotic stress responses
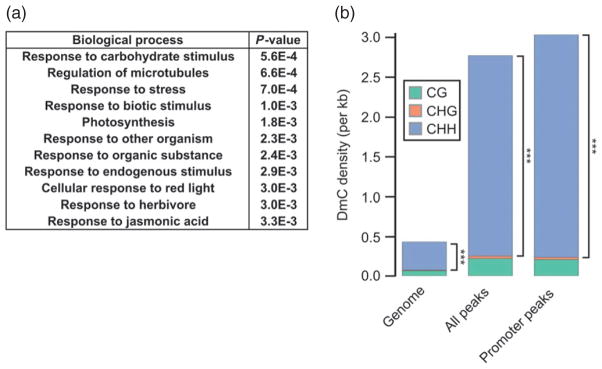
Our observation that only five of the 41 tested AGO4-associated genes are affected in ago4 and nrpe1 mutants is consistent with the lack of morphological phenotypes associated with Arabidopsis ago4 and nrpe1 mutant plants grown under optimal conditions (Zilberman et al., 2003; Kanno et al., 2005; Pontier et al., 2005). To test whether AGO4 target genes are controlled in response to stress, we performed gene ontology (GO) analysis, which revealed significant enrichment of genes that are responsive to biotic and abiotic stimuli (Figure 5a). To test whether DNA methylation levels at AGO4 binding sites are affected by stress, we calculated the mean changes in DNA methylation levels at differentially methylated regions identified in plants subjected to biotic stressors (Dowen et al., 2012). Differential methylation was significantly enriched on AGO4 binding sites relative to the genome overall (Figure 5b). In fact, stress-responsive differential CHH methylation was eight times more pronounced on AGO4 binding sites than on the genome overall (P → 0). This is much higher than the 3.5-fold enrichment of total CHH methylation on AGO4 binding sites (Figure 4a). These results suggest that enrichment of stress-induced differential methylation on AGO4 interaction regions is not merely a by-product of overall higher levels of DNA methylation at these genomic sites. This was further confirmed by the observation that AGO4 binding sites significantly overlap with salicylic acid-induced differentially methylated regions compared to 1000 random genomic permutations and vice versa (Figure S7a,b; P < 0.001 for both comparisons). These results demonstrate that a significant proportion of AGO4 binding sites contain DNA methylation that may be dynamically regulated during the plant’s response to biotic stresses. Taken together, these findings suggest that changes in DNA methylation patterns at AGO4 target genes are part of a natural gene regulatory mechanism during plant biotic stress responses.
Figure 5. AGO4 binding is enriched on genes affected by stress.
(a) AGO4 promoter binding may regulate genes encoding proteins involved in stress, environmental and hormone responses. The 11 most significantly enriched biological processes for all the genes whose promoters are bound by AGO4 are listed, together with the corresponding P values.
(b) Biotic stress-mediated differential DNA methylation is enriched on AGO4 binding sites. Mean levels of CG (green), CHG (orange) and CHH (blue) per kilobase (kb) for the Arabidopsis genome overall (left bar), within all AGO4 peaks (middle bar) or within promoter-associated AGO4 peaks (right bar). The methylation data have been published previously (Dowen et al., 2012). ***P → 0.
DISCUSSION
Argonaute proteins have been shown to recognize the sequences of specific target RNAs and genomic loci using incorporated small RNAs (Qi et al., 2006). Our findings are consistent with 24 nt siRNAs being required for AGO4 binding to chromatin, but also show that they are not sufficient. Instead, lncRNAs produced by Pol V mediate the specific binding of AGO4 to its genomic targets, many of which are transposons embedded within the promoters of protein-coding genes. Intriguingly, these results suggest that widespread AGO4-bound transposons within gene promoters may be controlling elements as proposed previously (McClintock, 1956), and identify Pol V-produced lncRNAs as the primary determinant of their status as regulatory modules.
Once the overlapping action of 24 nt siRNAs and lncRNAs guides AGO4 to specific genomic regions, chromatinmodifying enzymes are recruited, and repressive DNA and histone modifications are established. These modifications in turn affect gene expression. A possible mechanism by which RdDM controls gene expression is by affecting the binding of transcription factors or other DNA-binding proteins to cis-elements within promoters (Figure 6). This possibility is consistent with our data showing both up- and down-regulation of AGO4-controlled genes in ago4 mutant plants, reflecting the effect of DNA methylation on either repressive or activating transcription factors, respectively. However, it is also possible that AGO4 binding and RdDM affect the spread of chromatin modifications (Moshkovich et al., 2011) or RNA processing. In addition to serving as switchable regulatory elements controlled by DNA methylation status, AGO4-targeted transposable elements may also insert into novel locations, providing an additional level of transcription regulation compared with the pre-insertion promoter sequence. Our model predicts that pericentromeric silenced genomic regions that are not bound by AGO4 but give rise to siRNAs are not transcribed by Pol V. Instead, they are probably targeted by a different transcriptional silencing pathway.
Figure 6. Model for AGO4 function on gene promoters.
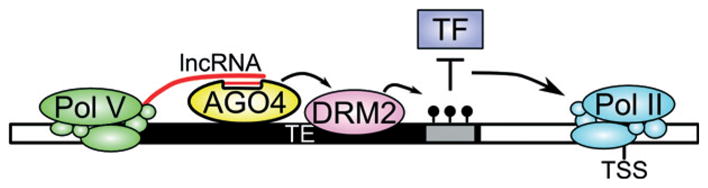
Pol V produces long non-coding RNA, which is a scaffold for AGO4-siRNA binding. AGO4 recruits the de novo DNA methyltransferase DRM2. CHH methylation affects transcription factor binding within gene promoters, which in turn positively or negatively affects Pol II transcription and gene expression.
Our work provides direct evidence of preferential binding of an RdDM component to promoters of protein-coding genes. The results are consistent with immunostaining data showing the presence of AGO4 outside chromocenters (Li et al., 2006; Pontes et al., 2006), with preferential up-regulation of euchromatic genes in the drm1 drm2 cmt3 triple mutant (Zhang et al., 2006), and the presence of some well-characterized RdDM targets in euchromatin (Huettel et al., 2006; Henderson and Jacobsen, 2008). They are also consistent with recently published genome-wide localization of Pol V (Wierzbicki et al., 2012; Zhong et al., 2012). Targeting of AGO4 towards promoters of protein-coding genes also reveals an additional level of gene expression control that is probably conserved between plants and animals (Cernilogar et al., 2011; Moshkovich et al., 2011). It is interesting that only minimal morphological phenotypes are observed in Arabidopsis RdDM mutant plants grown under standard conditions (Zilberman et al., 2003; Pontier et al., 2005), which suggests that this mechanism may be more prevalent in organisms with higher transposon content, such as maize, in which disruption of RdDM results in more dramatic phenotypes (Alleman et al., 2006; Erhard et al., 2009). This mechanism may also a have much greater impact in plants such as tomato, where the majority of 24 nt siRNAs map to gene-rich chromosomal regions (Tomato Genome Consortium, 2012). AGO4-mediated control of gene expression may also operate in certain developmental stages, as suggested for early embryonic development (Mosher et al., 2008; Autran et al., 2011), or provide a common response to environmental stimuli (Figure 5) (Agorio and Vera, 2007; Pecinka et al., 2010; Tittel-Elmer et al., 2010).
A potential involvement of RdDM-targeted TEs in response to environmental stimuli is supported by our observations that AGO4 binding sites significantly overlap genomic regions, at which biotic stresses have been shown to affect DNA methylation levels (Figure 5b) (Dowen et al., 2012). Thus, our findings probably provide an explanation for previous reports showing the involvement of AGO4 and Pol V in pathogen responses (Agorio and Vera, 2007; López et al., 2011). We propose that pathogen infection affects siRNA production and/or Pol V transcription, which in turn causes changes in promoter DNA methylation and affects gene expression levels. In conclusion, our findings establish that determination of the regulatory functions of AGO4 and the entire RdDM pathway in normal plant development and stress responses is an important goal for future research.
EXPERIMENTAL PROCEDURES
Plant material
Arabidopsis thaliana nrpe1 (nrpd1b-11) and ago4 [ago4-1 (Zilberman et al., 2003), introgressed into the Col-0 background] have been described previously (Onodera et al., 2005; Wierzbicki et al., 2009). Plants were cultivated at 22°C under long-day conditions (16 h day/8 h night).
RNA analysis
For assays of mRNA accumulation, total RNA was extracted from 2 to 3-week-old plants using an RNeasy plant mini kit (Qiagen; www.qiagen.com), and three biological replicates were amplified using a SuperScript III Platinum SYBR Green one-step quantitative RT-PCR kit (Invitrogen; www.invitrogen.com) in an Applied Biosystems (www.appliedbiosystems.com) 7500 real-time PCR machine. For assays of Pol V transcript accumulation, total RNA was extracted from 2 to 3-week-old plants using an RNeasy plant mini kit (Qiagen) and assayed as described previously (Wierzbicki et al., 2008), except that random primers were used and cDNA was amplified in a Bio-Rad (www.bio-rad.com) CFX Connect real-time PCR machine. Two independent biological repetitions were performed. Oligonucleotides used in these and other PCR assays can be found in Table S1.
DNA methylation analysis
Genomic DNA was extracted from above-ground tissue of 2-week-old plants using an DNeasy Plant Mini kit (Qiagen). Genomic DNA (100 ng) was digested using 10 units of AluI, DdeI or Sau3AI restriction enzymes (NEB; www-neb.com) for 20 min. After heat inactivation of the enzyme, DNA was amplified using 0.75 units of platinum Taq (Invitrogen).
Antibodies
The affinity-purified rabbit polyclonal anti-AGO4 and anti-NRPE1 antibodies have been described previously (Ream et al., 2009; Wierzbicki et al., 2009).
Chromatin immunoprecipitation
ChIP was performed as described previously (Rowley et al., 2011) with slight modifications. A detailed protocol is provided in Methods S1.
ChIP-seq library preparation and sequencing
All ChIP-seq and input libraries were prepared according to the Illumina (www.illumina.com) ChIP-seq library preparation protocol, and subjected to sequencing on a Genome Analyzer IIx according to the manufacturer’s instructions.
Sequence processing
Raw reads were pre-processed and mapped to the Arabidopsis genome using a pipeline as previously described (Zheng et al., 2010) with slight modifications. Specifically, we used the Bowtie program (Langmead et al., 2009) instead of the original cross_match aligner. All valid alignments were reported in order to tolerate non-uniquely mapping reads, as AGO4 is thought to target heterochromatin and repetitive elements in Arabidopsis. A detailed procedure is provided in Methods S1.
AGO4 binding site identification
AGO4-bound peaks (AGO4 binding regions) were called using the CSAR R package (Muino et al., 2011). To do this, all mapped reads were extended to 250 nucleotides and merged from both strands. Peaks were required to reach a significant fold-enrichment between test and control with a false discovery rate <0.05. To identify high-quality peaks with minimum false positives, five sets of peaks were called either between ChIP and input samples (‘traditional calls’) or between Col-0 and ago4 or nrpe1 mutants (‘direct comparison’) as our basis for defining substantial peaks. Then Pol V-dependent and Pol V-independent peaks were determined by ‘peak arithmetic’ manipulations, which reliably identify peaks that are enriched for both ChIP versus input and wild-type versus mutant comparisons. Descriptions of these manipulations are provided in Methods S1.
An additional filtering step was also implemented to exclude peaks with a potential ecotype bias, because the ago4 mutant plants used in this study were originally identified (Zilberman et al., 2003) in the Landsberg (Ler-1) ecotype of Arabidopsis and subsequently back-crossed to Col-0 plants three times. To do this, any peak that either (i) cannot be mapped to the Ler-1 draft genome (‘Ler-1 unmappable’) or (ii) can be better mapped to the Ler-1 draft genome (‘Ler-1 better mapped’) were discarded from further analysis.
To distinguish the AGO4 peaks that are completely dependent from those that are partially dependent on Pol V activity, we determined whether the clone abundance of AGO4 binding sites was comparable (less than twofold difference) between nrpe1 ChIP and ago4 ChIP samples (Pol V-dependent) or not (Pol V partially dependent). The vast majority of Pol V-dependent peaks were completely dependent, and therefore we did not separate these peaks in further analyses.
AGO4 binding site classification
To classify and annotate AGO4 peaks, their genomic coordinates were compared to various classes of known genetic elements annotated by the Arabidopsis Information Resource (TAIR9 release) on the Arabidopsis genome, including protein-coding genes (exons and introns), rRNAs, tRNAs, miRNAs, snoRNAs, snRNAs, ncRNAs, pseudogenes and TEs. To supplement this analysis, additional repetitive elements were defined using the Repeat-Masker program (http://www.repeatmasker.org/). We defined gene promoters as regions 1 kb upstream from the transcription start sites of protein-coding genes. As a negative control, 1000 sets of random peaks (NC peaks) were sampled from the genome, classified and annotated similarly, and the P values for enrichment or depletion in specific categories were estimated using a boot-strapping method based on these NC peaks. To comprehensively characterize the classes and families of transposable elements in AGO4 peaks, we used the TEs identified by RepeatMasker and their corresponding annotation information.
To characterize small RNA profiles near AGO4 peaks, small RNA immunoprecipitation datasets and total small RNA datasets (Wang et al., 2011) for both AGO4 and AGO1 from Arabidopsis seedlings were used; the small RNA immunoprecipitation datasets or total small RNA reads were searched within the AGO4 peaks as well as their flanking regions (2 kb upstream and downstream).
To characterize the cytosine methylation (mC) in AGO4 peaks, we used published single-nucleotide mC datasets, including genome-wide mC profiles from Col-0, met1, ddc and rdd mutant plants (Lister et al., 2008), kindly provided by Dr Ryan Lister. The mC sites were searched within all AGO4 peaks as well as NC peaks, and the mC density was calculated and compared between AGO4 peaks and NC peaks for CG, CHG and CHH methylation or as a whole. The mC density was also directly compared between Col-0 and nrpe1 mutant plants using recently published mC datasets (Wierzbicki et al., 2012).
To characterize AGO4 binding profiles around transcription start sites, a log fold change profile of ChIP-seq reads between Col-0 and ago4 samples relative to positions in the transcription start site of all protein-coding genes was generated using the CEAS program (Shin et al., 2009). Similarly, to characterize the nucleosome profile around AGO4 peaks, we used published MNase-seq datasets (Chodavarapu et al., 2010), and calculated the log fold change of MNase-seq reads between Col-0 and ago4 samples relative to positions in the transcription start site using the CEAS program (Shin et al., 2009). We also called the well-positioned nucleosomes as previously described (Kaplan et al., 2009), and determined nucleosome density profiles for all or promoter-overlapping AGO4 peaks.
GO analysis
To identify significantly enriched biological processes for the genes corresponding to AGO4-bound promoters, the gene IDs of these loci were analyzed using the GOEAST online analysis tool (Zheng and Wang, 2008) with a false discovery rate < 0.05.
Accession number and genome browser link
All six ChIP-seq library datasets were deposited into the National Center for Biotechnology Information Gene Expression Omnibus under accession number GSE35381. The AnnoJ genome browser for all ChIP-seq libraries and external datasets (smRNAs and DNA methylation) presented in this paper is http://gregorylab.bio.upenn.edu/annoj_atAGO4/.
Supplementary Material
Identification of AGO4-bound loci.
AGO4 binding shows no preference towards transposon-rich pericentromeric regions.
The summit of AGO4 peaks lacks nucleosomes.
The pattern of AGO4 binding cannot be explained by AGO4-bound siRNAs.
AGO4 binding specificity towards TEs in gene promoters is mediated by lncRNA.
AGO4 binding mediates DNA methylation.
AGO4 binding overlaps genomic regions showing stress-induced differential CHH methylation.
Oligonucleotides used in this study.
AGO4-bound genomic regions identified in this study.
Detailed description of various methods.
Acknowledgments
We thank Eran Pichersky, David Engelke (University of Michigan, Department of Biological Chemistry) and Eric Richards (Boyce Thompson Institute) for critical reading of the manuscript. This work was supported by US National Science Foundation grants MCB 1120271 to A.T.W and CAREER Award MCB 1053846 to B.D. G., and Austrian Science Fund (FWF) fellowship J3199-B09 to G. B., and National Institutes of Health National Research Service Award 5-T32-GM07544.
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- Agorio A, Vera P. ARGONAUTE4 is required for resistance to Pseudomonas syringae in Arabidopsis. Plant Cell. 2007;19:3778–3790. doi: 10.1105/tpc.107.054494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alleman M, Sidorenko L, McGinnis K, Seshadri V, Dorweiler JE, White J, Sikkink K, Chandler VL. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature. 2006;442:295–298. doi: 10.1038/nature04884. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Autran D, Baroux C, Raissig MT, et al. Maternal epigenetic pathways control parental contributions to Arabidopsis early embryogenesis. Cell. 2011;145:707–719. doi: 10.1016/j.cell.2011.04.014. [DOI] [PubMed] [Google Scholar]
- Cernilogar FM, Onorati MC, Kothe GO, et al. Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature. 2011;480:391–395. doi: 10.1038/nature10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Bernatavichute YV, et al. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466:388–392. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen RH, Pelizzola M, Schmitz RJ, Lister R, Dowen JM, Nery JR, Dixon JE, Ecker JR. Widespread dynamic DNA methylation in response to biotic stress. Proc Natl Acad Sci USA. 2012;109:E2183–E2191. doi: 10.1073/pnas.1209329109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhard KF, Jr, Stonaker JL, Parkinson SE, Lim JP, Hale CJ, Hollick JB. RNA polymerase IV functions in paramutation in Zea mays. Science. 2009;323:1201–1205. doi: 10.1126/science.1164508. [DOI] [PubMed] [Google Scholar]
- Henderson IR, Jacobsen SE. Tandem repeats upstream of the Arabidopsis endogene SDC recruit non-CG DNA methylation and initiate siRNA spreading. Genes Dev. 2008;22:1597–1606. doi: 10.1101/gad.1667808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huettel B, Kanno T, Daxinger L, Aufsatz W, Matzke AJM, Matzke M. Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J. 2006;25:2828–2836. doi: 10.1038/sj.emboj.7601150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, Daxinger L, Kreil DP, Matzke M, Matzke AJM. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet. 2005;37:761–765. doi: 10.1038/ng1580. [DOI] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. Establishing, maintaining and modifying DNAmethylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CF, Pontes O, El-Shami M, Henderson IR, Bernatavichute YV, Chan SW-L, Lagrange T, Pikaard CS, Jacobsen SE. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell. 2006;126:93–106. doi: 10.1016/j.cell.2006.05.032. [DOI] [PubMed] [Google Scholar]
- Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockton S, Gaut BS. The contribution of transposable elements to expressed coding sequence in Arabidopsis thaliana. J Mol Evol. 2009;68:80–89. doi: 10.1007/s00239-008-9190-5. [DOI] [PubMed] [Google Scholar]
- López A, Ramírez V, García-Andrade J, Flors V, Vera P. The RNA silencing enzyme RNA polymerase V is required for plant immunity. PLoS Genet. 2011;7:e1002434. doi: 10.1371/journal.pgen.1002434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu O, Reinders J, Caikovski M, Smathajitt C, Paszkowski J. Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell. 2007;130:851–862. doi: 10.1016/j.cell.2007.07.007. [DOI] [PubMed] [Google Scholar]
- McClintock B. Controlling elements and the gene. Cold Spring Harbor Symp Quant Biol. 1956;21:197–216. doi: 10.1101/sqb.1956.021.01.017. [DOI] [PubMed] [Google Scholar]
- Mosher RA, Schwach F, Studholme D, Baulcombe DC. PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc Natl Acad Sci USA. 2008;105:3145–3150. doi: 10.1073/pnas.0709632105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshkovich N, Nisha P, Boyle PJ, Thompson BA, Dale RK, Lei EP. RNAi-independent role for Argonaute2 in CTCF/CP190 chromatin insulator function. Genes Dev. 2011;25:1686–1701. doi: 10.1101/gad.16651211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muino JM, Kaufmann K, Ham RC, van Angenent GC, Krajewski P. ChIP-seq analysis in R (CSAR): an R package for the statistical detection of protein-bound genomic regions. Plant Methods. 2011;7:11. doi: 10.1186/1746-4811-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y, Haag JR, Ream T, Costa Nunes P, Pontes O, Pikaard CS. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Pecinka A, Dinh HQ, Baubec T, Rosa M, Lettner N, Mittelsten Scheid O. Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell. 2010;22:3118–3129. doi: 10.1105/tpc.110.078493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O, Li CF, Costa Nunes P, Haag J, Ream T, Vitins A, Jacobsen SE, Pikaard CS. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell. 2006;126:79–92. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
- Pontier D, Yahubyan G, Vega D, Bulski A, Saez-Vasquez J, Hakimi M-A, Lerbs-Mache S, Colot V, Lagrange T. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 2005;19:2030–2040. doi: 10.1101/gad.348405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, He X, Wang XJ, Kohany O, Jurka J, Hannon GJ. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature. 2006;443:1008–1012. doi: 10.1038/nature05198. [DOI] [PubMed] [Google Scholar]
- Ream TS, Haag JR, Wierzbicki AT, et al. Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol Cell. 2009;33:192–203. doi: 10.1016/j.molcel.2008.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley MJ, Avrutsky MI, Sifuentes CJ, Pereira L, Wierzbicki AT. Independent chromatin binding of ARGONAUTE4 and SPT5L/KTF1 mediates transcriptional gene silencing. PLoS Genet. 2011;7:e1002120. doi: 10.1371/journal.pgen.1002120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Liu T, Manrai AK, Liu XS. CEAS: cis-regulatory element annotation system. Bioinformatics. 2009;25:2605–2606. doi: 10.1093/bioinformatics/btp479. [DOI] [PubMed] [Google Scholar]
- Tittel-Elmer M, Bucher E, Broger L, Mathieu O, Paszkowski J, Vaillant I. Stress-induced activation of heterochromatic transcription. PLoS Genet. 2010;6:e1001175. doi: 10.1371/journal.pgen.1001175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635–641. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhang, Xiuren, Liu J, et al. Deep sequencing of small RNAs specifically associated with Arabidopsis AGO1 and AGO4 uncovers new AGO functions. Plant J. 2011;67:292–304. doi: 10.1111/j.1365-313X.2011.04594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT. The role of long non-coding RNA in transcriptional gene silencing. Curr Opin Plant Biol. 2012 doi: 10.1016/j.pbi.2012.08.008. [DOI] [PubMed] [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Ream TS, Haag JR, Pikaard CS. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet. 2009;41:630–634. doi: 10.1038/ng.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Cocklin R, Mayampurath A, Lister R, Rowley MJ, Gregory BD, Ecker JR, Tang H, Pikaard CS. Spatial and functional relationships among Pol V-associated loci, Pol IV-dependent siRNAs, and cytosine methylation in the Arabidopsis epigenome. Genes Dev. 2012;26:1825–1836. doi: 10.1101/gad.197772.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Zheng Q, Wang X-J. GOEAST: a web-based software toolkit for Gene Ontology enrichment analysis. Nucleic Acids Res. 2008;36:W358–W363. doi: 10.1093/nar/gkn276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Wang Z, Li S, Yu B, Liu J-Y, Chen X. Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev. 2009;23:2850–2860. doi: 10.1101/gad.1868009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Ryvkin P, Li F, Dragomir I, Valladares O, Yang J, Cao K, Wang L-S, Gregory BD. Genome-wide double-stranded RNA sequencing reveals the functional significance of base-paired RNAs in Arabidopsis. PLoS Genet. 2010;6:e1001141. doi: 10.1371/journal.pgen.1001141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Hale CJ, Law JA, Johnson LM, Feng S, Tu A, Jacobsen SE. DDR complex facilitates global association of RNA polymerase V to promoters and evolutionarily young transposons. Nat Struct Mol Biol. 2012;19:870–875. doi: 10.1038/nsmb.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Identification of AGO4-bound loci.
AGO4 binding shows no preference towards transposon-rich pericentromeric regions.
The summit of AGO4 peaks lacks nucleosomes.
The pattern of AGO4 binding cannot be explained by AGO4-bound siRNAs.
AGO4 binding specificity towards TEs in gene promoters is mediated by lncRNA.
AGO4 binding mediates DNA methylation.
AGO4 binding overlaps genomic regions showing stress-induced differential CHH methylation.
Oligonucleotides used in this study.
AGO4-bound genomic regions identified in this study.
Detailed description of various methods.