Abstract
Stress is a major contributor to anxiety and mood disorders. The recent discovery of epigenetic changes in the brain resulting from stress has enhanced our understanding of the mechanism by which stress is able to promote these disorders. Although epigenetics encompasses chemical modifications that occur at both DNA and histones, much attention has been focused on stress-induced DNA methylation changes on behavior. Here, we review the effect of stress-induced DNA methylation changes on physiological mechanisms that govern behavior and cognition, dysregulation of which can be harmful to mental health. A literature review was performed in the areas of DNA methylation, stress, and their impact on the brain and psychiatric illness. Key findings center on genes involved in the hypothalamic-pituitary-adrenal axis, neurotransmission and neuroplasticity. Using animal models of different stress paradigms and clinical studies, we detail how DNA methylation changes to these genes can alter physiological mechanisms that influence behavior. Appropriate levels of gene expression in the brain play an important role in mental health. This dynamic control can be disrupted by stress-induced changes to DNA methylation patterns. Advancement in other areas of epigenetics, such as histone modifications and the discovery of the novel DNA epigenetic mark, 5-hydroxymethylcytosine, could provide additional avenues to consider when determining the epigenetic effects of stress on the brain.
Keywords: DNA methylation, HPA axis, neuroplasticity, neurotransmission, genome-wide DNA methylation
INTRODUCTION
Environmental stressors are challenging events that result in a cascade of physiological changes. They have been observed to influence behavior and cognition in a variety of ways. While the focus of this review is on the negative outcomes that can result from maladaptive responses to stressors, there are some stressors that have been shown to have positive outcomes. For example, in one study, monkeys exposed to intermittent periods of moderate stress during early development–removal from their social group and exposure to unfamiliar adult monkeys–demonstrated diminished anxiety in later life in two experimental paradigms compared with controls (Parker et al., 2004). Another example from rodent studies includes one on adult rats exposed to predictable mild stressors. These animals had significantly better spatial memory performance as determined by Morris water maze and improved novel object ability as compared to control rats (Parihar et al., 2011). In addition to cognitive improvements, these stressed rats also displayed reduced depression-like behavior on the forced swim test and reduced anxiety behavior assessed by elevated plus maze. These changes were accompanied by cellular changes in their brains, such as increased neurogenesis and enhanced dendritic growth of newly born neurons. In humans, there are also examples of stressors inducing positive outcomes. For example, in a clinical study where healthy male subjects were transiently exposed to an acute stressor in the form of a cold pressure test procedure (i.e., exposure of their dominant hand to ice), stressed subjects showed a higher rate of acquisition of pavlovian conditioning in the form of trace eye blinking conditioning. In addition, these subjects also displayed improved spatial memory as determined by a virtual navigation Morris water task (Duncko et al., 2007).
In contrast to the positive outcomes that result from milder stressors, maladaptive changes result from others—often more severe and/or chronic—increase the risk of mood disorders such as major depressive disorder (MDD)(Kessler and Magee, 1994; Kendler et al., 1999; Rojo-Moreno et al., 2002). Supporting this idea, a twin study revealed that stressful life events substantially contribute to the onset of major depressive episodes over a 12-month period (Kendler et al., 1999). This was similarly observed in independent studies where chronic stressors and severity were significantly associated with the onset of major depressive episodes (Rojo-Moreno et al., 2002; Hammen et al., 2009). Likewise, early life stressors have been observed to increase the risk of MDD and its recurrence when stressors are present in later life (Kessler and Magee, 1994; Hazel et al., 2008). The effects of these stressors may be mediated by epigenetic modifications such as DNA methylation, which can dysregulate physiological processes important to behavior and cognition, including the hypothalamic-pituitary-adrenal axis, neurotransmission, and neuroplasticity. In this review, we will focus on the effects of stressors on DNA methylation change in the brain and how such changes can in some instances dysregulate physiological processes involved in behavior and cognition. To this end, we will first provide an overview of DNA methylation and its role in regulating gene expression. We will then proceed to discuss how DNA methylation changes influenced by stressors can affect these physiological mechanisms, and lead to behavioral changes.
Gene Expression Must be Appropriately Regulated for Mental Health and Cognition
Mounting evidence indicates that appropriate gene expression is required for mental health. An example that highlights this idea is the effect of altered brain derived neurotrophic factor (BDNF) expression levels on mental health. BDNF is a neurotrophin whose expression regulates behavior and cognition. In animal studies, over and under expression of BDNF in the hippocampus and prefrontal cortex results in depression-like behavior and cognitive impairment (Cunha et al., 2009; Sakata et al., 2010; Taliaz et al., 2010). Similarly, postmortem and clinical studies have revealed reduced BDNF expression in the brain and peripheral blood of suicide completers and patients with mood disorders, suggesting that reduction in BDNF expression may contribute to these phenotypes (Dwivedi et al., 2003; Cunha et al., 2006; Kim et al., 2007; Thompson Ray et al., 2011). Altered expression of other genes expressed in the brain, which include γ-aminobutyric acid and serotonin transporter, have also been observed to affect behavior (Lee et al., 2007; Luscher et al., 2011). Together, these studies underscore that altered levels of gene expression in the brain can have a negative impact on behavior. A major avenue by which stress can induce changes to gene expression levels is by altering DNA methylation patterns.
DNA Methylation Occurs in CpG and Non-CpG Contexts
DNA methylation is an epigenetic process whereby a methyl group is added to nucleotides of DNA without any alterations to DNA sequence. In mammalian cells, this process predominantly occurs on cytosine in a cytosine-guanine dinucleotide (CpG) context. Methylation of cytosine in a non-CpG context such as CH, where H can be adenosine, thymine or cytosine, has also been reported and is observed in significant proportions (~15% of methylated cytosines) in embryonic stem cells (Ramsahoye et al., 2000; Lister et al., 2011; Ziller et al., 2011) and frontal cortex of mice (~30% of methylated cytosines) (Xie et al., 2012). In contrast, a smaller proportion of methylated cytosine in the non-CpG context (~<5% of methylated cytosines) has been reported in other mammalian somatic tissues, such as fibroblasts, kidney, liver, spleen and lung (Ramsahoye et al., 2000; Ziller et al., 2011). Although the significance of methylation in the non-CpG context is yet to be fully understood, DNA methylation in the CpG context has been extensively studied and is well documented in a variety of biological paradigms.
DNA Methylation at CpG Dinucleotides Regulates Gene Expression
While much early study of CpG methylation focused on CpG islands, more recent studies have highlighted other regions with fanciful nomenclature. CpG islands are genomic regions with a high frequency of CpG dinucleotides (Gardiner-Garden and Frommer, 1987; Takai and Jones, 2002). These regions are typically present in genic regions and are commonly associated with promoters (Ioshikhes and Zhang, 2000). Indeed, CpG islands have been successfully used as a landmark to identify previously uncharacterized promoters (Illingworth and Bird, 2009). The other CpG regions have a lower CG content and have been identified by their distance from CpG islands. CpG shores are regions within 2 kb of CpG islands (Irizarry et al., 2009), while CpG shelves are regions within 2–4 kb (Rechache et al., 2012) from CpG islands. CpG open seas are regions beyond 4 kb of CpG islands (Sandoval et al., 2011). Although CpG islands are commonly targeted in DNA methylation studies, recent genome-wide DNA methylation investigations have reported a significant proportion of DNA methylation changes occurring in the non-CpG islands underscoring their functional importance (Irizarry et al., 2009; Lee et al., 2011). For example, in a genome-wide DNA methylation study by Irizarry et al. [2009] differentially methylated regions between different normal tissues and cancer tissues were observed to occur predominantly in CpG shores compared to CpG islands (Irizarry et al., 2009). This is similarly observed in other studies where differentially methylated regions in CpG shores, shelves and open-sea can occupy up to about 75% of differentially methylated regions cumulatively when compared between normal tissue and cancer tissue (Sandoval et al., 2011; Rechache et al., 2012). These studies suggest that DNA methylation in the CpG context may play an important role in establishing different cellular phenotypes. This is due to the ability of DNA methylation to regulate gene expression by regulating accessibility of transcription factors to their binding sites and influencing chromatin structure (Watt and Molloy, 1988; Iguchi-Ariga and Schaffner, 1989; Cedar and Bergman, 2009). Indeed, genome-wide DNA methylation studies using normal tissues and cancer tissues have negatively correlated DNA methylation at promoters with gene expression level (Irizarry et al., 2009; Bell et al., 2011; Dudziec et al., 2012). Methylation at CpG shores has however been observed to promote alternative transcription. For example, ~70% of differentially methylated regions, identified from different tissue types and between colon cancer and normal mucosa tissue, are present within 500 bp of alternative promoters coinciding with their alternative mRNA transcripts (Irizarry et al., 2009). This suggests that differential methylation of alternative promoters can regulate alternative transcription. These studies highlight the effect of DNA methylation pattern on gene expression.
For DNA methylation patterns to be induced and sustained, they require the function of a group of catalytic proteins called DNA methyltransferases (DNMTs). DNMTs are catalytic enzymes that transfer a methyl group from S-adenosyl methionine to DNA. Presently, DNMT1 and DNMT 3A, DNMT 3B and DNMT 3L are known to play a key role in maintaining and promoting de novo methylation of CpG dinucleotides (Okano et al., 1999; Li, 2002). In addition to DNA methylation, DNA demethylation also plays an equally important role in regulating phenotypes. DNA demethylation is a process whereby the methyl group on cytosine nucleotides is lost or removed. This process can be mediated by either a passive loss of DNA methylation signal through cell division or active DNA demethylation by enzymatic mediators (Bhutani et al., 2011; Branco et al., 2012). Importantly, reduction in DNA methylation can have an impact on phenotype by increasing gene expression level. For example, maternal separation stress of mice neonates reduced DNA methylation at the regulatory region of the arginine vasopressin (AVP) gene increasing its expression in the paraventricular nucleus. This resulted in hyperactivity of the hypothalamic-pituitary-adrenal (HPA) axis resulting in mice with a memory deficit in an inhibitory avoidance task and increased immobility in the forced swim test, which is thought to be a measurement of depression-like behavior (Murgatroyd et al., 2009). These studies demonstrate that changes to DNA methylation patterns can affect the phenotype of organisms through changes in gene expression levels.
Environmental Pressures Induce DNA Methylation
Environmental signals play a significant role in regulating phenotypes of organisms mediated through changes in DNA methylation pattern. This was first observed in an elegant experiment where ectopic expression of the agouti gene, placed under the control of the murine retro-transposon intracisternal A particle (IAP) cryptic promoter, could be repressed by maternal diet of methyl supplements which increased DNA methylation at the IAP promoter. This resulted in mice offspring with a black fur coat and a non-obese phenotype as opposed to mice with yellow coat color and an obese phenotype (Wolff et al., 1998). Since this study, many other studies have also observed the effect of environmentally induced DNA methylation on animal phenotypes (Meaney and Szyf, 2005). Interestingly, environmentally induced DNA methylation patterns can persist over a prolonged period of time and can at least in some cases be transmitted to the next generation. Humans exposed to famine at the time of their conception showed decreased DNA methylation at the promoter of candidate imprinted genes for cardiovascular and metabolic disease, such as IGF2, GNAS and MEG3, six decades after the event (Heijmans et al., 2008; Tobi et al., 2009). Similarly, in a separate study, F1 and F2 mice whose paternal F0 ancestors had been exposed to odor potentiated startle using acetophenone, displayed enhanced behavioral sensitivity to acetophenone compared to F1 and F2 mice whose paternal F0 ancestors were not exposed to this treatment. Importantly, a reduced DNA methylation level at the M71 odorant receptor gene (Olfr151), which is a receptor activated by acetophenone, was observed from the sperm of F0 exposed to acetophenone and the sperm of its F1 progeny compared to that of F0 and F1 mice unexposed to acetophenone. This appears to be a striking transgenerational effect of the environment on DNA methylation (Dias and Ressler, 2014). There are also several examples in mice of transgenerational inheritance of these patterns. In the agouti mouse model, when the mother has the agouti phenotype, her offspring are more likely to have it (Morgan et al., 1999). Similarly, in another mouse model, axin fused mice, a kinked tail is seen when there is less methylation at a retrotransposon. Epigenetic inheritance has been reported both through maternal and paternal transmission (Rakyan et al., 2003). These studies highlight that environmental pressures might have a long-term effect on health through persistence of DNA methylation patterns.
Stress Mediates Changes to DNA Methylation Pattern of Genes Involved in the HPA Axis
Stress is a consequence of physical or psychological insults that activate physiological mechanisms to cope with the challenging event. A key mechanism involved in this process is the activation of the HPA axis. The HPA axis is a neuroendocrine system that mediates stress adaptation through metabolic and behavioral changes. In the presence of stress, the parvocellular neurons of the paraventricular nucleus (PVN) in the hypothalamus are activated, secreting hormones, such as corticotrophin releasing factor (CRF), arginine vasopressin (AVP) and oxytocin, which target the anterior pituitary. The activated anterior pituitary gland subsequently secretes adrenocorticotrophin hormone (ACTH), which targets the adrenal cortex resulting in secretion of a number of stress hormones including epinephrine, norepinephrine and glucocorticoid (cortisol in humans and corticosterone in rodents). These hormones mediate a host of physiological changes including increased heart rate, blood pressure and gluconeogenesis to prepare the body to engage in the “fight or flight” response to immediate threats. To maintain homeostatic control of the HPA axis, elevated blood levels of glucocorticoids activate the glucocorticoid receptor, which downregulates HPA axis activity by suppressing the expression of genes involved in HPA axis activation. This negative feedback mechanism is important in maintaining mental health as prolonged elevated HPA activation can increase the likelihood of psychiatric disorders such as mood disorders. Indeed, sustained HPA activation by chronic stress paradigms or chronic exposure to exogenous glucocorticoid results in anxiety and depression-like behavior in rodents (Ardayfio and Kim, 2006; Keeney et al., 2006; Murray et al., 2008). Similarly, in humans, an overactive HPA axis has also been observed in subjects with mood disorders like MDD (Watson et al., 2004, 2009). One way in which this negative feedback mechanism can be disrupted is by exposure to severe stressors.
Stressful events can negatively affect behavior and promote increased risk for psychiatric disorders. These stressors—often early-life, severe and/or chronic—have been associated with increased risk for the onset of major depression and bipolar mood symptoms in clinical studies (Ellicott et al., 1990; Daley et al., 2000; Levitan et al., 2003; Dienes et al., 2006; Johnson et al., 2008; Hammen et al., 2009). In vivo rodent models of stressors have also displayed anxiety and depression-like symptoms (Murgatroyd et al., 2009; Elliott et al., 2010). Stressors can affect behavior by altering the expression level of genes involved in the HPA axis through changes in their DNA methylation pattern. For example, mice exposed to maternal separation during their first 10 days of life had sustained reductions in DNA methylation levels in the distal promoter region of the pro-opiomelanocortin gene, which encodes the precursor of the adrenocorticotropic hormone (Wu et al., 2014). This resulted in sustained elevation of the adrenocorticotropic hormone mRNA and protein expression observed in the pituitary and plasma respectively (Wu et al., 2014). Furthermore, other studies where rodents were exposed to early life stressors in the form of maternal deprivation or chronic social defeat stress they displayed memory deficits in inhibitory avoidance, anxiety and depression-like symptoms (Murgatroyd et al., 2009; Elliott et al., 2010). These animals had reduced DNA methylation at the transcription enhancer region of the AVP gene and the promoter region of the CRF gene resulting in increased expression of AVP and CRF in the hypothalamus (Murgatroyd et al., 2009; Elliott et al., 2010; Chen et al., 2012). This resulted in elevated corticosterone levels reflecting hyperactivity of the HPA axis (Murgatroyd et al., 2009; Chen et al., 2012). In addition to heightening HPA axis activity, excessive stress can also prolong elevated HPA activity by dysregulating the expression of genes involved in the negative feedback mechanism. One such gene is FKBP5, which encodes FK506-Binding Protein-5.
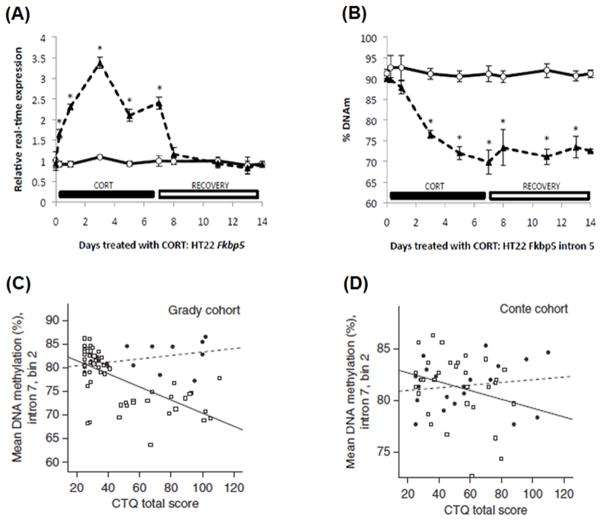
FKBP5 is a co-chaperone protein that regulates the activity of the glucocorticoid receptor (GR). It forms a complex with inactive GR to lower GR affinity for glucocorticoids and also reduce overall GR signaling by reducing GR presence in the cytoplasm through nuclear translocation (Binder, 2009). As such, altering FKBP5 expression can have physiological and behavioral consequences. A previous study has shown that single nucleotide polymorphisms in the FKBP5 locus, known to alter FKBP5 expression level and impair HPA axis negative feedback, can predict increased dorsal amygdala activity in teenagers who experienced childhood emotional neglect (White et al., 2012). Similarly, depressed subjects who express higher FKBP5 experience twice as many depressive episodes as their depressed counterparts who have lower FKBP5 expression (Binder et al., 2004). Furthermore, post-traumatic stress disorder (PTSD) subjects who possessed the risk “T” allele of rs1360780, which elevates FKBP5 expression, were at greater risk of developing lifetime PTSD than their counterparts who had the protective “C” allele, when exposed to childhood adversity (Klengel et al., 2013). Similar to the effect of genotype, glucocorticoid hormone and stress can elevate FKBP5 expression by altering DNA methylation patterns at the FKBP5 locus (Fig. 1) (Lee et al., 2010). Childhood adversity has been observed to decrease DNA methylation at a regulatory element in intron 7 of FKBP5, increasing FKBP5 expression in response to GR signaling, thus dampening the negative feedback mechanism of the HPA axis (Fig. 1)(Klengel et al., 2013).
FIG. 1.
DNA methylation of the stress-associated gene FKBP5. (A) and (B) Fkbp5 expression and DNA methylation levels following glucocorticoid exposure (Lee et al., 2010). Expression and methylation levels for Fkbp5 in the mouse hippocampal neuronal cell line HT-22. Cells were treated with corticosterone for seven days and cultured in the absence of the hormone for an additional seven days. Cells were harvested for mRNA and genomic DNA to determine the expression levels of Fkbp5 (A) and methylation differences (B). Corticosterone treated samples are depicted by black triangles with dashed lines, while vehicle-treated samples have white circles with solid lines. Asterisks indicate differences that are statistically significant (P < 0.05). (C) and (D). Correlation between DNA methylation at FKBP5 and childhood trauma questionnaire scores by FKBP5 risk genotype in two cohorts (Grady and Conte) are shown (Klengel et al., 2013). (C) Grady cohort. Risk allele carriers exhibited a strong negative correlation between DNA methylation and trauma score compared with carriers of the protective genotype. (D) The Conte cohort shows a similar correlation. Elevated corticosterone levels and increased trauma were associated with decreased DNA methylation.
In addition to FKBP5, stress can also disrupt suppression of HPA axis activity by altering DNA methylation at the promoter of the GR gene, NR3C1. Previous studies have shown that altered gene expression levels of GR can affect behavior. Rodents with artificially reduced expression levels of GR in the cortico-limbic system displayed depression-like behavior due to impaired inhibition of the HPA axis determined by the dexamethasone suppression test (Boyle et al., 2005; Ridder et al., 2005). This test uses the synthetic glucocorticoid dexamethasone to evaluate the negative feedback response of the HPA axis to suppress endogenous glucocorticoid levels. Impairment in hippocampal repression may play a role in this phenomenon since the hippocampus is known to be involved in the negative feedback regulation of the HPA axis (Brown et al., 1999; Jankord and Herman, 2008). Indeed, acute exposure of the dentate gyrus of the hippocampus to glucocorticoid repressed HPA axis activity, while chronic exposure to glucocorticoid reduced GR expression in the hippocampus and increased HPA activity (Zhu et al., 2014). In contrast to studies that artificially reduced GR expression, overexpression of GR enhanced the HPA axis negative feedback system, reducing a depression-like phenotype (Ridder et al., 2005). Along the lines of these animal studies, excessive stress has been suggested to disrupt HPA axis suppression by increasing DNA methylation at the NR3C1 promoter to reduce GR expression. In support of this, post-mortem brain tissues from suicide subjects and blood samples from mood disorder patients showed that childhood abuse increased DNA methylation at the NR3C1 promoter corresponding with reduced GR expression (McGowan et al., 2009; Perroud et al., 2011). Together, these studies underscore how excessive stressors can dysregulate the HPA axis through changes in DNA methylation patterns. Although the HPA axis plays an important role in moderating the stress response, disruption of other physiological processes such as neurotransmission can equally be disruptive to behavior.
DNA Methylation of the Serotonin Transporter Gene Affects Behavior
Severe stressors can negatively affect behavior by altering the expression of genes involved in neurotransmission through changes to DNA methylation patterns. This is exemplified in the heavily studied serotonin transporter gene, SLC6A4. This gene is known to regulate the availability of serotonin in the synaptic cleft enabling it to moderate aspects of emotional behavior. Changes in its expression by alteration of its DNA methylation pattern have been shown to be disruptive to normal behavior. For example, childhood abuse has been shown to increase DNA methylation at the SLC6A4 promoter resulting in lowered SLC6A4 mRNA expression (Philibert et al., 2008; Beach et al., 2010; Kang et al., 2013a). The degree of DNA methylation at this promoter in depressed patients who suffered childhood abuse predicted lower quality of life, lower social and occupational functioning and a higher level of disability (Kang et al., 2013a). Likewise in animal studies, monkeys exposed to early life stress in the form of variable foraging demand, which strains the mother-infant relationship, showed enhanced reactivity to high intensity stressors with increased DNA methylation at the SLC6A4 locus (Kinnally et al., 2011). Apart from severe early life stressors, chronic stressors can also affect behavior through changes in SLC6A4 promoter DNA methylation. An investigation into the effect of chronic work stressors on a nurse cohort showed that increased DNA methylation at the promoter region of SLC6A4 was associated with a higher level of burnout (Alasaari et al., 2012). These studies are in line with clinical studies that have linked reduced expression of SLC6A4, by its promoter length polymorphism, with elevated stress reactivity (Miller et al., 2013) and with depression (Caspi et al., 2003; Karg et al., 2011; Nietzer et al., 2011; Kenna et al., 2012), though the latter point remains unsettled (Risch et al., 2009; Culverhouse et al., 2013).
Although these findings may seem at odds with the observation that reducing serotonin transporter activity with antidepressants ameliorates anxiety and depression symptoms, this contradiction could be resolved by the suggestion that reduction in serotonin transporter expression resulting from stress may result in heightened activation of the amygdala. This activation could contribute to the anxiety phenotype. Indeed, anxiety prone subjects have higher amygdala activation compared to healthy controls in emotional processing (Stein et al., 2007). Importantly, the level of serotonin transporter expression can influence amygdala activity and amygdaloid neuron morphology. Individuals who express low levels of serotonin transporter have been observed to exhibit higher amygdala activity than those who express higher levels (Hariri et al., 2002; Heinz et al., 2005). Similarly, artificially reducing serotonin transporter expression by knocking out the SLC6A4 gene in mice resulted in increased amygdaloid neuronal spine density which is presumed to be a morphological manifestation of enhanced synaptic connectivity and activity of the amygdala (Nietzer et al., 2011). These mice also display an anxiety-like phenotype (Line et al., 2011). These studies therefore suggest that stress may promote anxiety behavior by increasing amygdala activity through increased DNA methylation of the SLC6A4 gene. Further studies are required to validate this hypothesis. Besides neurotransmission, stress can also affect cognition and behavior by altering DNA methylation patterns of genes involved in neuroplasticity.
Neuroplasticity can be Impaired by Stress-Induced DNA Methylation Changes to the BDNF Locus
Neuroplasticity is a process that provides cellular adaptation to environmental signals. A gene recognized as a major player in neuroplasticity is the brain derived neurotrophic factor (BDNF). BDNF is a neurotrophin that is highly expressed in the central nervous system and mediates a number of cellular changes to facilitate brain adaptation to environmental signals. These cellular changes include synaptic plasticity (Cunha et al., 2010), arborization of neurites (Cunha et al., 2010), neurogenesis (Taliaz et al., 2010) and GABAergic neuron development and maturation (Kohara, 2003; Kohara et al., 2007; Sakata et al., 2009). The disruption of these processes have been observed to impair cognition and precipitate anxiety and depression-like behavior in rodents (Nutan and Meti, 2000; Dupret et al., 2008; Revest et al., 2009; Madroñal et al., 2010; Sakata et al., 2010).
Since BDNF regulates these neuroplasticity processes, changes to its expression by stress-induced DNA methylation patterns can have an impact on behavior. In a study where rat infants were exposed to early life stress in the form of maternal maltreatment, stressed rats exhibited anxiety behavior and showed reduced Bdnf mRNA expression in the prefrontal cortex corresponding to increased DNA methylation at promoter IV and XI compared to control rats (Roth et al., 2009). Similarly in a separate study, rats exposed to chronic stress in the form of chronic inescapable cat exposure showed reduced levels of Bdnf exon IV transcript in the dorsal dentate gyrus and dorsal CA1 corresponding to increased DNA methylation at BDNF promoter IV at these brain regions. DNA methylation changes were however absent in ventral dentate gyrus and ventral CA1 (Roth et al., 2011) (Fig. 2). This study highlights the tissue specificity of stress-induced DNA methylation changes, suggesting that tissue heterogeneity may potentially mask changes, making them difficult to detect. In line with this idea, a separate study that investigated the effect of chronic social defeat stress in whole hippocampus of mice did not observe any DNA methylation differences in Bdnf between stressed and control mice (Tsankova et al., 2006).
FIG. 2.
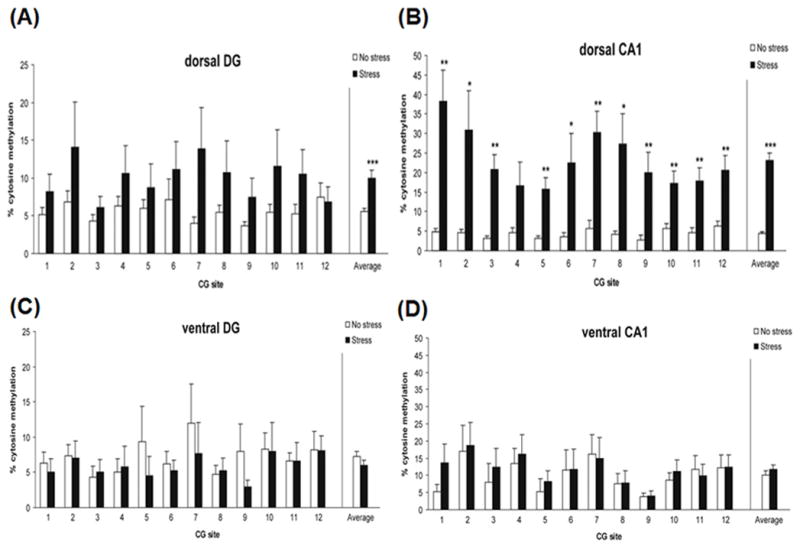
Site-specific DNA methylation of the Bdnf gene in the hippocampus of rats exposed to psychosocial stress. Rats were stressed by acute exposure to a cat followed by unstable housing. DNA methylation was increased in (A) the dorsal dentate gyrus and (B) dorsal CA1, but not in (C) the ventral dentate gyrus and (D) ventral CA1 (Roth et al., 2011).
In addition to animal studies, clinical studies have also linked the effect of BDNF DNA methylation patterns to psychiatric phenotypes. For example, increased DNA methylation in BDNF promoter IV was observed in post-mortem brain and in peripheral blood of suicidal subjects compared to non-suicidal controls (Keller et al., 2010; Kang et al., 2013b). This was also observed to be a predictor of suicidal ideation and history of previous suicide attempts (Kang et al., 2013b). Similarly, in a separate study, patients with MDD and bipolar II disorder had increased DNA methylation in BDNF promoter I in peripheral blood (Fuchikami et al., 2011; D’Addario et al., 2012). These clinical studies suggest that changes to the DNA methylation pattern at the BDNF locus may influence mood and suicidality phenotypes. Taken together, studies in rodents and patients highlight the possibility that altered BDNF DNA methylation patterns might negatively impact behavior.
Genome-Wide Approaches may Uncover Unexpected Mechanisms Through Which Epigenetic Changes can Mediate the Influence of Stress on Anxiety and Depression
Although the above-mentioned studies have successfully used locus-specific assays to identify regions where DNA methylation changes occur, this approach limits the scope of genes that can be investigated. A genome-wide approach is required to determine the extent to which stress affects DNA methylation patterns more broadly. The success of such an approach has been reported in recent studies that have not only uncovered novel genes affected by stress but have also unveiled gene networks altered by stress which may contribute to mood disorders (Fig. 3)(Labonte et al., 2012; Mehta et al., 2013; Provencal et al., 2012; Sabunciyan et al., 2012).
FIG. 3.
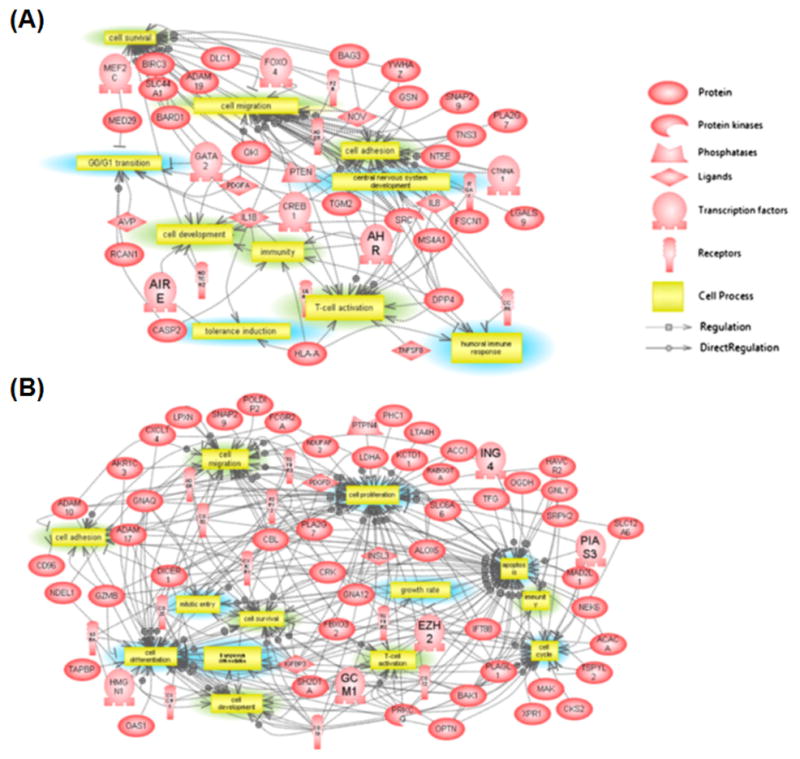
Differences in gene networks between post-traumatic stressed patients with childhood abuse versus no childhood abuse. (A) Cellular processes overrepresented in PTSD patients (A) with or (B) without history of child abuse (Mehta at al., 2013).
Currently, a few different approaches are available to study genome-wide DNA methylation. These approaches rely on either protein-affinity enrichment of DNA methylated regions, bisulfite conversion or methylated cytosine-sensitive restriction enzymes. They are typically used in conjunction with microarray or next-generation sequencing platforms. Although widely used, these methods suffer from several limitations. For example, protein affinity-based enrichment of methylated cytosine has been reported to enrich for low CpG density regions and does not provide base-pair resolution of methylated cytosines (Harris et al., 2010; Nair et al., 2011). In contrast, methods using methylated cytosine-sensitive restriction enzymes can provide base-pair resolution, but enrich for CpG dense regions (Harris et al., 2010). Although these two approaches can be used complementarily, limited starting material and increased cost of sequencing and microarray may be prohibitive. An alternative unbiased method to detect methylated CpG at a base-pair resolution is whole-genome bisulfite sequencing. However, this approach requires increased sequencing costs to provide adequate sequencing depth. Due to these limitations, a method that provides cost-effective detection of CpG methylation at base-pair resolution without bias of CpG dense and poor regions would be advantageous to the study of the genome-wide DNA methylation pattern. This can be satisfied using genome-wide targeted capture technology.
The genome-wide targeted capture approach relies on DNA or RNA sequence baits to enrich for candidate regions which can be interrogated by next-generation sequencing. This approach has been widely used in exome sequencing studies (Girotto et al., 2013; Madhavan et al., 2013) and has more recently been adapted to investigate DNA methylation at base-pair resolution through the use of bisulfite conversion (Wang et al., 2011; Ivanov et al., 2013). As baits can be designed against regions of interest, this eliminates any bias for CpG dense and poor regions while enabling sequencing reactions to be focused on candidate regions of interest improving yield per sequencing cost. Due to these benefits, we are currently using this approach in an ongoing study to investigate the effects of stress on DNA methylation in mice that have been exposed to chronic social defeat stress.
Some Mood Disorder Medications can Mediate DNA Methylation Changes in the Brain
As we have discussed in preceding section, some forms of stressors, such as childhood abuse, can induce maladaptive epigenetic changes. In this section, we explore whether it is possible that therapeutic treatments which are able to reverse these changes might alleviate symptoms of mood disorders. Supporting this possibility, a recent study where rats were treated with the DNMT inhibitor 5-aza-2′-deoxycytidine showed reduced depression-like behavior determined by reduced immobility in the forced swim test (Sales et al., 2011). This corresponded to reduced global DNA methylation level and increased BDNF expression in the hippocampus (Sales et al., 2011). Psychiatric medications such as valproate, used to treat mood disorders, as well as the antidepressants, such as escitalopram and imipramine, have also been shown to induce DNA methylation changes in rodent brain (Dong et al., 2010; Elliott et al., 2010; Melas et al., 2012). Furthermore, the most effective treatment for depression, electroconvulsive treatment, was shown to cause demethylation of DNA in the dentate gyrus of the hippocampus in growth factors that have been implicated in regulating adult neurogenesis (Ma et al., 2009). It remains a possibility that the epigenetic changes observed in these treatment studies are epiphenomena. However, these studies do raise the question of whether novel therapeutics might be developed that would be aimed at reversing stress-induced epigenetic changes associated with anxiety and mood disorders. Such interventions have recently been suggested in epilepsy (Williams-Karnesky et al., 2013). Currently, four medications are FDA-approved that target epigenetic changes to treat cancer (Ho et al., 2013).
DISCUSSION
In this review, we have highlighted how changes to DNA methylation patterns induced by stress can affect behavior by disrupting key physiological mechanisms that govern brain function. Although these mechanisms have been discussed separately, they do not function independently, but rather interact to influence each other’s function. For instance, elevated CRF and corticosterone levels in mice, which reflect heightened HPA axis activity, have been observed to reduce BDNF expression in the hippocampus, which can impair cellular adaptation by neuroplasticity (Table I) (Murakami et al., 2005; Flandreau et al., 2012). Conversely, stress-mediated increase of BDNF in the PVN may promote elevated HPA axis activity (Givalois et al., 2004; Flandreau et al., 2012). In addition to the cross-talk between BDNF and the HPA axis, studies have reported that BDNF and serotonin can regulate each other’s expression and function. For example, brain exposure to BDNF has been shown to increase sprouting of serotonergic axons (Mamounas et al., 1995), increase rate of 5HT synthesis by increasing expression of tryptophan hydroxylase (Mamounas et al., 1995), and increase activity-mediated release of 5HT (Goggi et al., 2002). Similarly, inhibiting 5HT transporter function by the use of selective serotonin reuptake inhibitors has been observed to increase BDNF expression (De Foubert et al., 2004; Wolkowitz et al., 2011).
TABLE I.
Primary Macro and Micro Effects of Chronic Stress on the Brain (McEwen, 2007)
| Regions affected | Neuroanatomic changes produced | Cell types affected | Genes affected |
|---|---|---|---|
| 1. Hippocampus | 1. Decreased branching of dendrites in hippocampus and prefrontal cortex, and corresponding decreased synaptic function. | 1. Neurons (in particular, dentate gyrus and CA3 pyramidal cells in hippocampus) | 1. BDNF (Brain-derived neurotrophic factor) - decreased expression. |
| 2. Prefrontal cortex | 2. Reduced survival and decreased proliferation of neurons in hippocampus (dentate gyrus and CA3 regions in particular) and prefrontal cortex. | 2. FKBP5 (FK506 Binding Protein 5) - increased expression. | |
| 3. Amygdala | 3. Increased survival and proliferation of neurons in amygdala. |
In addition to DNA methylation, other epigenetic modifications are equally important in regulating expression of genes involved in brain function. Indeed, a chronic inescapable stress study showed that changes to BDNF exon IV expression in the ventral CA1 of the hippocampus was not accompanied by increased DNA methylation at BDNF promoter IV, highlighting the involvement of additional epigenetic mechanisms involved in regulating this promoter in chronic stress (Roth et al., 2011). One such mechanism known to be regulated by stress is histone modification. This mechanism mediates transfer of chemical groups, such as acetyl and methyl groups, onto amino acid residues on histones to regulate the permisssiveness of chromatin structure to transcription. Stress has been observed to induce histone modifications that influence behavior. In a chronic social defeat stress study, increased methylation of lysine residue 27 of histone 3, a transcription repressive histone mark, at the Bdnf promoter reduced Bdnf expression in mouse hippocampus resulting in anxiety behavior. No changes to DNA methylation patterns were observed in this study (Tsankova et al., 2006).
There are several new directions that might prove fruitful for the field going forward. First, very recently, stress has been found to induce changes to a novel epigenetic mark, 5-hydroxymethylcytosine (5hmC), that affect behavior. 5hmC is thought to be an intermediate metabolic product of oxidized methylated cytosine and has been observed to alleviate transcriptional repression mediated by 5-methylcytosines (Branco et al., 2012). 5hmC is currently known to be involved in regulating mammalian brain development and physiological mechanisms such as neurogenesis (Hahn et al., 2013; Lister et al., 2013). Alteration to 5hmC patterns has been suggested to contribute to diseases such as cancer (Pfeifer et al., 2013). Although the involvement of 5hmC in stress in currently unknown, this epigenetic mark is known to regulate gene expression (Lister et al., 2013). Future studies should incorporate methods to distinguish 5mC from 5hmC, such as Tet-assisted bisulfite sequencing (TAB-seq) or oxidative bisulfite-sequencing (oxo-seq)(Yu et al., 2012; Booth et al., 2013). Description of these techniques has been extensively discussed elsewhere (Nestor et al., 2014).
A second future direction involves the study of non-CpG methylation in the stressed brain. Previous studies have provided correlative evidence to suggest that non-CpG methylation may regulate gene expression. For example, depletion of non-CpG methylation was previously observed in the embryonic stem cell line H1, but not in the fibroblast cell line IMR90 at enhancers specific for H1 (Lister et al., 2009). Non-CpG methylation at promoters of imprinted genes have also been negatively correlated with gene expression in mouse prefrontal cortex (Xie et al., 2012). This was similarly observed in a separate study where mouse and human brain showed depleted non-CpG methylation in expressed genes with genic non-CpG methylation inversely proportional to the abundance of the associated transcripts (Lister et al., 2013). In line with these studies, a recent investigation by Gou et al. provided direct molecular evidence to show that non-CpG methylation affects gene expression. Using a reporter gene assay, cells transfected with plasmids containing non-CpG methylation were less likely to express the reporter gene (Guo et al., 2014). Furthermore, knockdown of DNMT3A significantly reduced non-CpG methylation, but left CpG methylation unaffected at particular loci resulting in increased mRNA expression of those non-CpG methylated genes (Guo et al., 2014). Importantly, non-CpG methylation in adult mouse neurons was observed to be conserved in the adult human brain (Guo et al., 2014). Given that these studies collectively show that non-CpG methylation is able to regulate gene expression, future studies of mouse models of stress could shed light on whether stress may mediate its effect by altering non-CpG methylation in the brain.
A third future direction involves a shift in focus from the promoter regions to a broader array of regulatory regions. Complementing epigenetic modifications, the location where these modifications occur is equally important for influencing appropriate gene expression. Presently, numerous studies have been promoter-centric in reports on efforts to understand the epigenetics of stress. Although epigenetic changes at promoters are easily associated with genes they regulate, genome-wide DNA methylation studies have shown that differentially methylated regions are also commonly observed in regions outside of promoters (Lee et al., 2011). In addition, these non-promoter regions can contain regulatory elements, such as enhancers and suppressors, which are critical regulators of appropriate gene expression. In support of this, the ENCODE project has identified numerous regulatory elements flanking promoter regions (Consortium, 2012). Reporter gene studies have also shown that regulatory elements can direct tissue and cell type activity of promoters and regulate promoter response to environmental signals (Davidson et al., 2011; Hing et al., 2012; Klengel et al., 2013). In addition, early life stress studies have shown that stress can induce DNA methylation changes in non-promoter regions and thereby affect behavior (Murgatroyd et al., 2009; Klengel et al., 2013). Promoter-centric efforts may prevent stress-induced epigenetic changes at important regulatory elements from being noticed. Although challenges exist for identifying and studying the function of regulatory elements, experimental techniques have been described to overcome this problem (Mackenzie et al., 2013). As the cost of DNA sequencing continues to drop, it will soon be possible to cover all positions in the genome with bisulfite sequencing.
A fourth direction for the future involves accounting for variation in cell types in DNA methylation studies of the stressed brain. Although important epigenetic variation might exist in disease-relevant genes, they may be confined to one cell type. For example, a CpG island in CACNA1C, a gene strongly implicated in bipolar disorder, was found to display less methylation in neurons than in glial cells (Nishioka et al., 2013). To account for this variation, different approaches have been developed to study DNA methylation signatures for a specific cell type such as neurons or glial cells in general, or for particular subsets of these. One such approach is the use of fluorescence-activated cell sorting. This approach takes advantage of fluorescence tagged antibodies to mark cells that express a cell type-specific marker. These immuno-tagged cells can subsequently be separated by flow cytometry. Studies using this approach have successfully identified cell type-specific DNA methylation signatures from neuronal-enriched cell populations determined by NeuN-positive staining and non-neuronal cell populations determined by NeuN-negative staining (Iwamoto et al., 2011; Lister et al., 2013).
In addition to experimental approaches, bioinformatic approaches have been developed to distinguish between celltype-specific DNA methylation patterns. For instance, in a study by Guintivano et al., the investigators used a stringent selection of neuronal and non-neuronal DNA methylation signatures across different loci to generate a model that could accurately quantify the proportion of neuronal and glial cell populations from genome-wide DNA methylation data (Guintivano et al., 2013). Using this approach, they analyzed a data set to investigate DNA methylation signatures from post-mortem frontal cortex of major depressive disorder patients (Sabunciyan et al., 2012). Although no regions were observed to be significantly differentially methylated in the initial analysis, adjustment for cell heterogeneity using their model revealed three differentially methylated regions that retained nominal significance after correction for multiple testing (Guintivano et al., 2013). Together, these experimental and bioinformatic approaches provide an opportunity to investigate how stress might alter DNA methylation patterns in different cell types in the brain.
A fifth future direction that may hold promise is the use of induced pluripotent stem (iPS) cells from patients with mood and anxiety disorders. Such an approach has been reported in schizophrenia research where neurons created from fibroblasts derived from schizophrenic patients displayed a cellular and molecular phenotype that may reflect the disease trait (Brennand et al., 2011). For example, these neurons showed reduced neural connectivity determined by reduced expression of the neural cell adhesion molecule, reduced dendritic arborization, increased expression of neuregulin 1, and reduced expression of genes involved in activity dependent refinement of synaptic connections and long term potentiation (Brennand et al., 2011). Since iPS cells carry the genetic vulnerability of the patients from whom they are derived and produce a cellular phenotype that may reflect the disease trait, they can be used to compare with neurons created from control iPS cells to look for differential methylation in response to glucocorticoids or other HPA axis factors. Similarly, one could assess whether differential methylation predicts differential rescue by antidepressants of glucocorticoid-induced changes in the cells.
In conclusion, the brain relies on the appropriate level of gene expression to regulate behavior and cognition. Disruption to this control by stress-induced DNA methylation changes may lead to anxiety and mood disorders. Advancement in other areas of epigenetics such as the study of histone modifications, and of the novel DNA epigenetic marks 5-hydroxymethylcytosine and non-CpG methylation, could provide additional avenues to consider when studying the epigenetic effects of stress on the brain. It is hoped that by combining the study of various epigenetic marks, investigators can provide a global view of the effects of stress on the epigenome, which may aid in the development of new therapies.
References
- Alasaari JS, Lagus M, Ollila HM, Toivola A, Kivimaki M, Vahtera J, Kronholm E, et al. Environmental stress affects DNA methylation of a CpG rich promoter region of serotonin transporter gene in a nurse cohort. PLoS One. 2012;7:e45813. doi: 10.1371/journal.pone.0045813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardayfio P, Kim KS. Anxiogenic-like effect of chronic corticosterone in the light-dark emergence task in mice. Behav Neurosci. 2006;120:249–256. doi: 10.1037/0735-7044.120.2.249. [DOI] [PubMed] [Google Scholar]
- Beach SRH, Brody GH, Todorov AA, Gunter TD, Philibert RA. Methylation at SLC6A4 is linked to family history of child abuse: An examination of the Iowa Adoptee sample. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:710–713. doi: 10.1002/ajmg.b.31028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JT, Pai AA, Pickrell JK, Gaffney DJ, Pique-Regi R, Degner JF, Gilad Y, Pritchard JK. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011;12:R10. doi: 10.1186/gb-2011-12-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutani N, Burns DM, Blau HM. DNA demethylation dynamics. Cell. 2011;146:866–872. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder EB. The role of FKBP5, a co-chaperone of the glucocorticoid receptor in the pathogenesis and therapy of affective and anxiety disorders. Psychoneuroendocrinology. 2009;34(Suppl 1):S186–195. doi: 10.1016/j.psyneuen.2009.05.021. [DOI] [PubMed] [Google Scholar]
- Binder EB, Salyakina D, Lichtner P, Wochnik GM, Ising M, Putz B, Papiol S, et al. Polymorphisms in FKBP5 are associated with increased recurrence of depressive episodes and rapid response to antidepressant treatment. Nat Genet. 2004;36:1319–1325. doi: 10.1038/ng1479. [DOI] [PubMed] [Google Scholar]
- Booth MJ, Ost TWB, Beraldi D, Bell NM, Branco MR, Reik W, Balasubramanian S. Oxidative bisulfite sequencing of 5-methylcytosine and 5-hydroxymethylcytosine. Nat Protoc. 2013;8:1841–1851. doi: 10.1038/nprot.2013.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle MP, Brewer JA, Funatsu M, Wozniak DF, Tsien JZ, Izumi Y, Muglia LJ. Acquired deficit of forebrain glucocorticoid receptor produces depression-like changes in adrenal axis regulation and behavior. Proc Natl Acad Sci U S A. 2005;102:473–478. doi: 10.1073/pnas.0406458102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet. 2012;13:7–13. doi: 10.1038/nrg3080. [DOI] [PubMed] [Google Scholar]
- Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, Li Y, et al. Modelling schizophrenia using human induced pluripotent stem cells (vol 473, pg 221, 2011) Nature. 2011;479:556–556. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown ES, Rush AJ, Mcewen BS. Hippocampal remodeling and damage by corticosteroids: Implications for mood disorders. Neuropsychopharmacology. 1999;21:474–484. doi: 10.1016/S0893-133X(99)00054-8. [DOI] [PubMed] [Google Scholar]
- Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, Mcclay J, et al. Influence of life stress on depression: Moderation by a polymorphism in the 5-HTT gene. Science. 2003;301:386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Linking DNA methylation and histone modification: Patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- Chen J, Evans AN, Liu Y, Honda M, Saavedra JM, Aguilera G. Maternal deprivation in rats is associated with corticotrophin-releasing hormone (CRH) promoter hypomethylation and enhances CRH transcriptional responses to stress in adulthood. J Neuroendocrinol. 2012;24:1055–1064. doi: 10.1111/j.1365-2826.2012.02306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium T.E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culverhouse RC, Bowes L, Breslau N, Nurnberger JI, Jr, Burmeister M, Fergusson DM, Munafo M, et al. Protocol for a collaborative meta-analysis of 5-HTTLPR, stress, and depression. BMC Psychiatry. 2013;13:304. doi: 10.1186/1471-244X-13-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha ABM, Frey BN, Andreazza AC, Goi JD, Rosa AR, Gonçalves CA, Santin A, Kapczinski F. Serum brain-derived neurotrophic factor is decreased in bipolar disorder during depressive and manic episodes. Neurosci Lett. 2006;398:215–219. doi: 10.1016/j.neulet.2005.12.085. [DOI] [PubMed] [Google Scholar]
- Cunha C, Angelucci A, D’antoni A, Dobrossy MD, Dunnett SB, Berardi N, Brambilla R. Brain-derived neurotrophic factor (BDNF) overexpression in the forebrain results in learning and memory impairments. Neurobiol Dis. 2009;33:358–368. doi: 10.1016/j.nbd.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Cunha C, Brambilla R, Thomas KL. A simple role for BDNF in learning and memory? Front Mol Neurosci. 2010;3:1. doi: 10.3389/neuro.02.001.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’addario C, Dell’osso B, Palazzo MC, Benatti B, Lietti L, Cattaneo E, Galimberti D, et al. Selective DNA methylation of BDNF promoter in bipolar disorder: Differences among patients with BDI and BDII. Neuropsychopharmacology. 2012;37:1647–1655. doi: 10.1038/npp.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley SE, Hammen C, Rao U. Predictors of first onset and recurrence of major depression in young women during the 5 years following high school graduation. J Abnorm Psychol. 2000;109:525–533. [PubMed] [Google Scholar]
- Davidson S, Lear M, Shanley L, Hing B, Baizan-Edge A, Herwig A, Quinn JP, et al. Differential activity by polymorphic variants of a remote enhancer that supports galanin expression in the hypothalamus and amygdala: Implications for obesity, depression and alcoholism. Neuropsychopharmacology. 2011;36:2211–2221. doi: 10.1038/npp.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Foubert G, Carney SL, Robinson CS, Destexhe EJ, Tomlinson R, Hicks CA, Murray TK, et al. Fluoxetine-induced change in rat brain expression of brain-derived neurotrophic factor varies depending on length of treatment. Neuroscience. 2004;128:597–604. doi: 10.1016/j.neuroscience.2004.06.054. [DOI] [PubMed] [Google Scholar]
- Dias BG, Ressler KJ. Parental olfactory experience influences behavior and neural structure in subsequent generations. Nat Neurosci. 2014;17:89–96. doi: 10.1038/nn.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dienes KA, Hammen C, Henry RM, Cohen AN, Daley SE. The stress sensitization hypothesis: Understanding the course of bipolar disorder. J Affect Disord. 2006;95:43–49. doi: 10.1016/j.jad.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Dong E, Chen Y, Gavin DP, Grayson DR, Guidotti A. Valproate induces DNA demethylation in nuclear extracts from adult mouse brain. Epigenetics. 2010;5:730–735. doi: 10.4161/epi.5.8.13053. [DOI] [PubMed] [Google Scholar]
- Dudziec E, Gogol-Doring A, Cookson V, Chen W, Catto J. Integrated epigenome profiling of repressive histone modifications, DNA methylation and gene expression in normal and malignant urothelial cells. PLoS One. 2012;7:e32750. doi: 10.1371/journal.pone.0032750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncko R, Cornwell B, Cui L, Merikangas KR, Grillon C. Acute exposure to stress improves performance in trace eyeblink conditioning and spatial learning tasks in healthy men. Learn Mem. 2007;14:329–335. doi: 10.1101/lm.483807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupret D, Revest JM, Koehl M, Ichas F, De Giorgi F, Costet P, Abrous DN, Piazza PV. Spatial relational memory requires hippocampal adult neurogenesis. PLoS One. 2008;3:e1959. doi: 10.1371/journal.pone.0001959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi Y, Rizavi HS, Conley RR, Roberts RC, Tamminga CA, Pandey GN. Altered gene expression of brain-derived neurotrophic factor and receptor tyrosine kinase B in postmortem brain of suicide subjects. Arch Gen Psychiatry. 2003;60:804–815. doi: 10.1001/archpsyc.60.8.804. [DOI] [PubMed] [Google Scholar]
- Ellicott A, Hammen C, Gitlin M, Brown G, Jamison K. Life events and the course of bipolar disorder. Am J Psychiatry. 1990;147:1194–1198. doi: 10.1176/ajp.147.9.1194. [DOI] [PubMed] [Google Scholar]
- Elliott E, Ezra-Nevo G, Regev L, Neufeld-Cohen A, Chen A. Resilience to social stress coincides with functional DNA methylation of the Crf gene in adult mice. Nat Neurosci. 2010;13:1351–1353. doi: 10.1038/nn.2642. [DOI] [PubMed] [Google Scholar]
- Flandreau EI, Ressler KJ, Owens MJ, Nemeroff CB. Chronic overexpression of corticotropin-releasing factor from the central amygdala produces HPA axis hyperactivity and behavioral anxiety associated with gene-expression changes in the hippocampus and paraventricular nucleus of the hypothalamus. Psychoneuroendocrinology. 2012;37:27–38. doi: 10.1016/j.psyneuen.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchikami M, Morinobu S, Segawa M, Okamoto Y, Yamawaki S, Ozaki N, Inoue T, et al. DNA methylation profiles of the brain-derived neurotrophic factor (BDNF) gene as a potent diagnostic biomarker in major depression. PLoS One. 2011:6. doi: 10.1371/journal.pone.0023881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Girotto G, Abdulhadi K, Buniello A, Vozzi D, Licastro D, D’eustacchio A, Vuckovic D, et al. Linkage study and exome sequencing identify a BDP1 mutation associated with hereditary hearing loss. PLoS One. 2013;8:e80323. doi: 10.1371/journal.pone.0080323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givalois L, Naert G, Rage F, Ixart G, Arancibia S, Tapia-Arancibia L. A single brain-derived neurotrophic factor injection modifies hypothalamo–pituitary–adrenocortical axis activity in adult male rats. Mol Cell Neurosci. 2004;27:280–295. doi: 10.1016/j.mcn.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Goggi J, Pullar IA, Carney SL, Bradford HF. Modulation of neurotransmitter release induced by brain-derived neurotrophic factor in rat brain striatal slices in vitro. Brain Res. 2002;941:34–42. doi: 10.1016/s0006-8993(02)02505-2. [DOI] [PubMed] [Google Scholar]
- Guintivano J, Aryee MJ, Kaminsky ZA. A cell epigenotype specific model for the correction of brain cellular heterogeneity bias and its application to age, brain region and major depression. Epigenetics. 2013;8:290–302. doi: 10.4161/epi.23924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Shin JH, Shin J, Li H, Xie B, Zhong C, et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat Neurosci. 2014;17:215–222. doi: 10.1038/nn.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MA, Qiu R, Wu X, Li AX, Zhang H, Wang J, Jui J, et al. Dynamics of 5-hydroxymethylcytosine and chromatin marks in mammalian neurogenesis. Cell Rep. 2013;3:291–300. doi: 10.1016/j.celrep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammen C, Kim EY, Eberhart NK, Brennan PA. Chronic and acute stress and the prediction of major depression in women. Depress Anxiety. 2009;26:718–723. doi: 10.1002/da.20571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariri AR, Mattay VS, Tessitore A, Kolachana B, Fera F, Goldman D, Egan MF, Weinberger DR. Serotonin transporter genetic variation and the response of the human amygdala. Science. 2002;297:400–403. doi: 10.1126/science.1071829. [DOI] [PubMed] [Google Scholar]
- Harris E, Ponts N, Levchuk A, Roch K, Lonardi S. BRAT: bisulfite-treated reads analysis tool. Bioinformatics. 2010;26:572–573. doi: 10.1093/bioinformatics/btp706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazel NA, Hammen C, Brennan PA, Najman J. Early childhood adversity and adolescent depression: The mediating role of continued stress. Psychol Med. 2008;38:581–589. doi: 10.1017/S0033291708002857. [DOI] [PubMed] [Google Scholar]
- Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A. 2008;105:17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz A, Braus DF, Smolka MN, Wrase J, Puls I, Hermann D, Klein S, et al. Amygdala-prefrontal coupling depends on a genetic variation of the serotonin transporter. Nat Neurosci. 2005;8:20–21. doi: 10.1038/nn1366. [DOI] [PubMed] [Google Scholar]
- Hing B, Davidson S, Lear M, Breen G, Quinn J, Mcguffin P, Mackenzie A. A polymorphism associated with depressive disorders differentially regulates brain derived neurotrophic factor promoter IV activity. Biol Psychiatry. 2012;71:618–626. doi: 10.1016/j.biopsych.2011.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho AS, Turcan S, Chan TA. Epigenetic therapy: Use of agents targeting deacetylation and methylation in cancer management. Onco Targets Ther. 2013;6:223–232. doi: 10.2147/OTT.S34680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iguchi-Ariga SM, Schaffner W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989;3:612–619. doi: 10.1101/gad.3.5.612. [DOI] [PubMed] [Google Scholar]
- Illingworth RS, Bird AP. CpG islands–‘a rough guide’. FEBS Lett. 2009;583:1713–1720. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- Ioshikhes IP, Zhang MQ. Large-scale human promoter mapping using CpG islands. Nat Genet. 2000;26:61–63. doi: 10.1038/79189. [DOI] [PubMed] [Google Scholar]
- Irizarry R, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov M, Kals M, Kacevska M, Metspalu A, Ingelman-Sundberg M, Milani L. In-solution hybrid capture of bisulfite-converted DNA for targeted bisulfite sequencing of 174 ADME genes. Nucleic Acids Res. 2013;41:e72. doi: 10.1093/nar/gks1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto K, Bundo M, Ueda J, Oldham MC, Ukai W, Hashimoto E, Saito T, et al. Neurons show distinctive DNA methylation profile and higher interindividual variations compared with non-neurons. Genome Res. 2011;21:688–696. doi: 10.1101/gr.112755.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankord R, Herman JP. Limbic regulation of hypothalamo-pituitary-adrenocortical function during acute and chronic stress. Ann N Y Acad Sci. 2008;1148:64–73. doi: 10.1196/annals.1410.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SL, Cuellar AK, Ruggero C, Winett-Perlman C, Goodnick P, White R, Miller I. Life events as predictors of mania and depression in bipolar I disorder. J Abnorm Psychol. 2008;117:268–277. doi: 10.1037/0021-843X.117.2.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Kim JM, Stewart R, Kim SY, Bae KY, Kim SW, Shin IS, Shin MG, Yoon JS. Association of SLC6A4 methylation with early adversity, characteristics and outcomes in depression. Prog Neuropsychopharmacol Biol Psychiatry. 2013a;44:23–28. doi: 10.1016/j.pnpbp.2013.01.006. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Kim JM, Lee JY, Kim SY, Bae KY, Kim SW, Shin IS, et al. BDNF promoter methylation and suicidal behavior in depressive patients. J Affect Disord. 2013b doi: 10.1016/j.jad.2013.08.001. [DOI] [PubMed] [Google Scholar]
- Karg K, Burmeister M, Shedden K, Sen S. The serotonin transporter promoter variant (5-HTTLPR), stress, and depression meta-analysis revisited: Evidence of genetic moderation. Arch Gen Psychiatry. 2011;68:444–454. doi: 10.1001/archgenpsychiatry.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney A, Jessop DS, Harbuz MS, Marsden CA, Hogg S, Blackburn-Munro RE. Differential effects of acute and chronic social defeat stress on hypothalamic-pituitary-adrenal axis function and hippocampal serotonin release in mice. J Neuroendocrinol. 2006;18:330–338. doi: 10.1111/j.1365-2826.2006.01422.x. [DOI] [PubMed] [Google Scholar]
- Keller S, Sarchiapone M, Zarrilli F, et al. Increased BDNF promoter methylation in the wernicke area of suicide subjects. Arch Gen Psychiatry. 2010;67:258–267. doi: 10.1001/archgenpsychiatry.2010.9. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Karkowski LM, Prescott CA. Causal relationship between stressful life events and the onset of major depression. Am J Psychiatry. 1999;156:837–841. doi: 10.1176/ajp.156.6.837. [DOI] [PubMed] [Google Scholar]
- Kenna GA, Roder-Hanna N, Leggio L, Zywiak WH, Clifford J, Edwards S, Kenna JA, et al. Association of the 5-HTT gene-linked promoter region (5-HTTLPR) polymorphism with psychiatric disorders: review of psychopathology and pharmacotherapy. Pharmgenomics Pers Med. 2012;5:19–35. doi: 10.2147/PGPM.S23462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler RC, Magee WJ. Childhood family violence and adult recurrent depression. J Health Soc Behav. 1994;35:13–27. [PubMed] [Google Scholar]
- Kim YK, Lee HP, Won SD, Park EY, Lee HY, Lee BH, Lee SW, et al. Low plasma BDNF is associated with suicidal behavior in major depression. Prog Neuropsychopharmacol Biol Psychiatry. 2007;31:78–85. doi: 10.1016/j.pnpbp.2006.06.024. [DOI] [PubMed] [Google Scholar]
- Kinnally EL, Feinberg C, Kim D, Ferguson K, Leibel R, Coplan JD, John Mann J. DNA methylation as a risk factor in the effects of early life stress. Brain Behav Immun. 2011;25:1548–1553. doi: 10.1016/j.bbi.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klengel T, Mehta D, Anacker C, Rex-Haffner M, Pruessner JC, Pariante CM, Pace TW, et al. Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat Neurosci. 2013;16:33–41. doi: 10.1038/nn.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara K, Yasuda H, Huang Y, Adachi N, Sohya K, Tsumoto T. A local reduction in cortical GABAergic synapses after a loss of endogenous brain-derived neurotrophic factor, as revealed by single-cell gene knockout method. J Neurosci. 2007;27:7234–7244. doi: 10.1523/JNEUROSCI.1943-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara KKA, Adachi N, Nishida M, Itami C, Nakamura S, Tsumoto T. Inhibitory but not excitatory cortical neurons require presynaptic brain-derived neurotrophic factor for dendritic development. J Neurosci. 2003;23:6123–6131. doi: 10.1523/JNEUROSCI.23-14-06123.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonte B, Suderman M, Maussion G, Navaro L, Yerko V, Mahar I, Bureau A, et al. Genome-wide epigenetic regulation by early-life trauma. Arch Gen Psychiatry. 2012;69:722–731. doi: 10.1001/archgenpsychiatry.2011.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Kim HJ, Kim JG, Ryu V, Kim BT, Kang DW, Jahng JW. Depressive behaviors and decreased expression of serotonin reuptake transporter in rats that experienced neonatal maternal separation. Neurosci Res. 2007;58:32–39. doi: 10.1016/j.neures.2007.01.008. [DOI] [PubMed] [Google Scholar]
- Lee RS, Tamashiro KL, Aryee MJ, Murakami P, Seifuddin F, Herb B, Huo Y, et al. Adaptation of the CHARM DNA methylation platform for the rat genome reveals novel brain region-specific differences. Epigenetics. 2011;6:1378–1390. doi: 10.4161/epi.6.11.18072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RS, Tamashiro KL, Yang X, Purcell RH, Harvey A, Willour VL, Huo Y, et al. Chronic corticosterone exposure increases expression and decreases deoxyribonucleic acid methylation of Fkbp5 in mice. Endocrinology. 2010;151:4332–4343. doi: 10.1210/en.2010-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitan RD, Rector NA, Sheldon T, Goering P. Childhood adversities associated with major depression and/or anxiety disorders in a community sample of Ontario: Issues of co-morbidity and specificity. Depress Anxiety. 2003;17:34–42. doi: 10.1002/da.10077. [DOI] [PubMed] [Google Scholar]
- Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- Line SJ, Barkus C, Coyle C, Jennings KA, Deacon RM, Lesch KP, Sharp T, Bannerman DM. Opposing alterations in anxiety and species-typical behaviours in serotonin transporter overexpressor and knockout mice. Eur Neuropsychopharmacol. 2011;21:108–116. doi: 10.1016/j.euroneuro.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013:341. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen R, Hawkins R, Hon G, Tonti-Filippini J, Nery J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Kida Y, Hawkins R, Nery J, Hon G, Antosiewicz-Bourget J, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscher B, Shen Q, Sahir N. The GABAergic deficit hypothesis of major depressive disorder. Mol Psychiatry. 2011;16:383–406. doi: 10.1038/mp.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, Flavell RA, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie A, Hing B, Davidson S. Exploring the effects of polymorphisms on cis-regulatory signal transduction response. Trends Mol Med. 2013;19:99–107. doi: 10.1016/j.molmed.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhavan S, Gusev Y, Natarajan TG, Song L, Bhuvaneshwar K, Gauba R, Pandey A, et al. Genome-wide multi-omics profiling of colorectal cancer identifies immune determinants strongly associated with relapse. Front Genet. 2013:4. doi: 10.3389/fgene.2013.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madroñal N, Gruart A, Sacktor TC, Delgado-García JM. PKMζ inhibition reverses learning-induced increases in hippocampal synaptic strength and memory during trace eyeblink conditioning. PLoS One. 2010;5:e10400. doi: 10.1371/journal.pone.0010400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamounas LA, Blue ME, Siuciak JA, Altar CA. Brain-derived neurotrophic factor promotes the survival and sprouting of serotonergic axons in rat brain. J Neurosci. 1995;15:7929–7939. doi: 10.1523/JNEUROSCI.15-12-07929.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcewen BS. Physiology and neurobiology of stress and adaptation: Central role of the brain. Physiol Rev. 2007;87:873–904. doi: 10.1152/physrev.00041.2006. [DOI] [PubMed] [Google Scholar]
- Mcgowan PO, Sasaki A, D’alessio AC, Dymov S, Labonte B, Szyf M, Turecki G, Meaney MJ. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaney MJ, Szyf M. Environmental programming of stress responses through DNA methylation: Life at the interface between a dynamic environment and a fixed genome. Dialogues Clin Neurosci. 2005;7:103–123. doi: 10.31887/DCNS.2005.7.2/mmeaney. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta D, Klengel T, Conneely KN, Smith AK, Altmann A, Pace TW, Rex-Haffner M, et al. Childhood maltreatment is associated with distinct genomic and epigenetic profiles in posttraumatic stress disorder. Proc Natl Acad Sci U S A. 2013;110:8302–8307. doi: 10.1073/pnas.1217750110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melas PA, Rogdaki M, Lennartsson A, Bjork K, Qi H, Witasp A, Werme M, et al. Antidepressant treatment is associated with epigenetic alterations in the promoter of P11 in a genetic model of depression. Int J Neuropsychopharmacol. 2012;15:669–679. doi: 10.1017/S1461145711000940. [DOI] [PubMed] [Google Scholar]
- Miller R, Wankerl M, Stalder T, Kirschbaum C, Alexander N. The serotonin transporter gene-linked polymorphic region (5-HTTLPR) and cortisol stress reactivity: a meta-analysis. Mol Psychiatry. 2013;18:1018–1024. doi: 10.1038/mp.2012.124. [DOI] [PubMed] [Google Scholar]
- Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- Murakami S, Imbe H, Morikawa Y, Kubo C, Senba E. Chronic stress, as well as acute stress, reduces BDNF mRNA expression in the rat hippocampus but less robustly. Neurosci Res. 2005;53:129–139. doi: 10.1016/j.neures.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Murgatroyd C, Patchev AV, Wu Y, Micale V, Bockmuhl Y, Fischer D, Holsboer F, et al. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat Neurosci. 2009;12:1559–1566. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- Murray F, Smith DW, Hutson PH. Chronic low dose corticosterone exposure decreased hippocampal cell proliferation, volume and induced anxiety and depression like behaviours in mice. Eur J Pharmacol. 2008;583:115–127. doi: 10.1016/j.ejphar.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Nair SS, Coolen MW, Stirzaker C, Song JZ, Statham AL, Strbenac D, Robinson MD, Clark SJ. Comparison of methyl-DNA immunoprecipitation (MeDIP) and methyl-CpG binding domain (MBD) protein capture for genome-wide DNA methylation analysis reveal CpG sequence coverage bias. Epigenetics. 2011;6:34–44. doi: 10.4161/epi.6.1.13313. [DOI] [PubMed] [Google Scholar]
- Nestor CE, Reddington JP, Benson M, Meehan RR. Investigating 5-hydroxymethylcytosine (5hmC): The state of the art. Methods Mol Biol. 2014;1094:243–258. doi: 10.1007/978-1-62703-706-8_19. [DOI] [PubMed] [Google Scholar]
- Nietzer SL, Bonn M, Jansen F, Heiming RS, Lewejohann L, Sachser N, Asan ES, et al. Serotonin transporter knockout and repeated social defeat stress: Impact on neuronal morphology and plasticity in limbic brain areas. Behav Brain Res. 2011;220:42–54. doi: 10.1016/j.bbr.2011.01.011. [DOI] [PubMed] [Google Scholar]
- Nishioka M, Shimada T, Bundo M, Ukai W, Hashimoto E, Saito T, Kano Y, et al. Neuronal cell-type specific DNA methylation patterns of the Cacna1c gene. Int J Dev Neurosci. 2013;31:89–95. doi: 10.1016/j.ijdevneu.2012.11.007. [DOI] [PubMed] [Google Scholar]
- Nutan KS, Meti BL. Deficits in operant behavior and alteration of CA1, CA3 hippocampal dendritic arborization due to subicular lesions. J Neurosci Res. 2000;59:806–812. doi: 10.1002/(SICI)1097-4547(20000315)59:6<806::AID-JNR13>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Parker KJ, Buckmaster CL, Schatzberg AF, Lyons DM. Prospective investigation of stress inoculation in young monkeys. Arch Gen Psychiatry. 2004;61:933–941. doi: 10.1001/archpsyc.61.9.933. [DOI] [PubMed] [Google Scholar]
- Parihar VK, Hattiangady B, Kuruba R, Shuai B, Shetty AK. Predictable chronic mild stress improves mood, hippocampal neurogenesis and memory. Mol Psychiatry. 2011;16:171–183. doi: 10.1038/mp.2009.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perroud N, Paoloni-Giacobino A, Prada P, Olie E, Salzmann A, Nicastro R, Guillaume S, et al. Increased methylation of glucocorticoid receptor gene (NR3C1) in adults with a history of childhood maltreatment: A link with the severity and type of trauma. Transl Psychiatry. 2011;1:e59. doi: 10.1038/tp.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer GP, Kadam S, Jin SG. 5-hydroxymethylcytosine and its potential roles in development and cancer. Epigenetics Chromatin. 2013;6:10. doi: 10.1186/1756-8935-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philibert RA, Sandhu H, Hollenbeck N, Gunter T, Adams W, Madan A. The relationship of 5HTT (SLC6A4) methylation and genotype on mRNA expression and liability to major depression and alcohol dependence in subjects from the Iowa Adoption Studies. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:543–549. doi: 10.1002/ajmg.b.30657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencal N, Suderman MJ, Guillemin C, Massart R, Ruggiero A, Wang D, Bennett AJ, et al. The signature of maternal rearing in the methylome in rhesus macaque prefrontal cortex and T cells. J Neurosci. 2012;32:15626–15642. doi: 10.1523/JNEUROSCI.1470-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan VK, Chong S, Champ ME, Cuthbert PC, Morgan HD, Luu KV, Whitelaw E. Transgenerational inheritance of epigenetic states at the murine Axin(Fu) allele occurs after maternal and paternal transmission. Proc Natl Acad Sci U S A. 2003;100:2538–2543. doi: 10.1073/pnas.0436776100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci U S A. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechache NS, Wang Y, Stevenson HS, Killian JK, Edelman DC, Merino M, Zhang L, et al. DNA methylation profiling identifies global methylation differences and markers of adrenocortical tumors. J Clin Endocrinol Metab. 2012;97:E1004–1013. doi: 10.1210/jc.2011-3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revest JM, Dupret D, Koehl M, Funk-Reiter C, Grosjean N, Piazza PV, Abrous DN. Adult hippocampal neurogenesis is involved in anxiety-related behaviors. Mol Psychiatry. 2009;14:959–967. doi: 10.1038/mp.2009.15. [DOI] [PubMed] [Google Scholar]
- Ridder S, Chourbaji S, Hellweg R, Urani A, Zacher C, Schmid W, Zink M, et al. Mice with genetically altered glucocorticoid receptor expression show altered sensitivity for stress-induced depressive reactions. J Neurosci. 2005;25:6243–6250. doi: 10.1523/JNEUROSCI.0736-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Herrell R, Lehner T, Liang KY, Eaves L, Hoh J, Griem A, et al. Interaction between the serotonin transporter gene (5-HTTLPR), stressful life events, and risk of depression: a meta-analysis. JAMA. 2009;301:2462–2471. doi: 10.1001/jama.2009.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo-Moreno L, Livianos-Aldana L, Cervera-Martínez G, Dominguez-Carabantes JA, Reig-Cebrian MJ. The role of stress in the onset of depressive disorders. Soc Psychiatry Psychiatr Epidemiol. 2002;37:592–598. doi: 10.1007/s00127-002-0595-y. [DOI] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol Psychiatry. 2009;65:760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, Zoladz PR, Sweatt JD, Diamond DM. Epigenetic modification of hippocampal Bdnf DNA in adult rats in an animal model of post-traumatic stress disorder. J Psychiatr Res. 2011;45:919–926. doi: 10.1016/j.jpsychires.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabunciyan S, Aryee MJ, Irizarry RA, Rongione M, Webster MJ, Kaufman WE, Murakami P, et al. Genome-wide DNA methylation scan in major depressive disorder. PLoS One. 2012;7:e34451. doi: 10.1371/journal.pone.0034451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata K, Jin L, Jha S. Lack of promoter IV-driven BDNF transcription results in depression-like behavior. Genes Brain Behav. 2010;9:712–721. doi: 10.1111/j.1601-183X.2010.00605.x. [DOI] [PubMed] [Google Scholar]
- Sakata K, Woo NH, Martinowich K, Greene JS, Schloesser RJ, Shen L, Lu B. Critical role of promoter IV-driven BDNF transcription in GABAergic transmission and synaptic plasticity in the prefrontal cortex. Proc Natl Acad Sci. 2009;106:5942–5947. doi: 10.1073/pnas.0811431106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sales AJ, Biojone C, Terceti MS, Guimaraes FS, Gomes MV, Joca SR. Antidepressant-like effect induced by systemic and intra-hippocampal administration of DNA methylation inhibitors. Br J Pharmacol. 2011;164:1711–1721. doi: 10.1111/j.1476-5381.2011.01489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval J, Heyn H, Moran S, Serra-Musach J, Pujana MA, Bibikova M, Esteller M. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6:692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- Stein MB, Simmons AN, Feinstein JS, Paulus MP. Increased amygdala and insula activation during emotion processing in anxiety-prone subjects. Am J Psychiatry. 2007;164:318–327. doi: 10.1176/ajp.2007.164.2.318. [DOI] [PubMed] [Google Scholar]
- Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taliaz D, Stall N, Dar DE, Zangen A. Knockdown of brain-derived neurotrophic factor in specific brain sites precipitates behaviors associated with depression and reduces neurogenesis. Mol Psychiatry. 2010;15:80–92. doi: 10.1038/mp.2009.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson Ray M, Weickert CS, Wyatt E, Webster MJ. Decreased BDNF, trkB-TK+ and GAD67 mRNA expression in the hippocampus of individuals with schizophrenia and mood disorders. J Psychiatry Neurosci. 2011;36:195–203. doi: 10.1503/jpn.100048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobi EW, Lumey LH, Talens RP, Kremer D, Putter H, Stein AD, Slagboom PE, Heijmans BT. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum Mol Genet. 2009;18:4046–4053. doi: 10.1093/hmg/ddp353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9:519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- Vreeburg Sa HWGVPJ, et al. Major depressive disorder and hypothalamic-pituitary-adrenal axis activity: Results from a large cohort study. Arch Gen Psychiatry. 2009;66:617–626. doi: 10.1001/archgenpsychiatry.2009.50. [DOI] [PubMed] [Google Scholar]
- Wang J, Jiang H, Ji G, Gao F, Wu M, Sun J, Luo H, et al. High resolution profiling of human exon methylation by liquid hybridization capture-based bisulfite sequencing. BMC Genomics. 2011;12:597. doi: 10.1186/1471-2164-12-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson S, Gallagher P, Ritchie JC, Ferrier IN, Young AH. Hypothalamic-pituitary-adrenal axis function in patients with bipolar disorder. Br J Psychiatry. 2004;184:496–502. doi: 10.1192/bjp.184.6.496. [DOI] [PubMed] [Google Scholar]
- Watt F, Molloy PL. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 1988;2:1136–1143. doi: 10.1101/gad.2.9.1136. [DOI] [PubMed] [Google Scholar]
- White MG, Bogdan R, Fisher PM, Munoz KE, Williamson DE, Hariri AR. FKBP5 and emotional neglect interact to predict individual differences in amygdala reactivity. Genes Brain Behav. 2012;11:869–878. doi: 10.1111/j.1601-183X.2012.00837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams-Karnesky RL, Sandau US, Lusardi TA, Lytle NK, Farrell JM, Pritchard EM, Kaplan DL, Boison D. Epigenetic changes induced by adenosine augmentation therapy prevent epileptogenesis. J Clin Invest. 2013;123:3552–3563. doi: 10.1172/JCI65636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff GL, Kodell RL, Moore SR, Cooney CA. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 1998;12:949–957. [PubMed] [Google Scholar]
- Wolkowitz OM, Wolf J, Shelly W, Rosser R, Burke HM, Lerner GK, Reus VI, et al. Serum BDNF levels before treatment predict SSRI response in depression. Prog Neuro-Psychopharmacol Biol Psychiatry. 2011;35:1623–1630. doi: 10.1016/j.pnpbp.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Patchev AV, Daniel G, Almeida OF, Spengler D. Early-life stress reduces DNA methylation of the Pomc gene in male mice. Endocrinology. 2014 doi: 10.1210/en.2013-1868. en20131868. [DOI] [PubMed] [Google Scholar]
- Xie W, Barr CL, Kim A, Yue F, Lee AY, Eubanks J, Dempster EL, Ren B. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell. 2012;148:816–831. doi: 10.1016/j.cell.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Hon GC, Szulwach KE, Song CX, Jin P, Ren B, He C. Tet-assisted bisulfite sequencing of 5-hydroxymethylcytosine. Nat Protoc. 2012;7:2159–2170. doi: 10.1038/nprot.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu LJ, Liu MY, Li H, Liu X, Chen C, Han Z, Wu HY, et al. The different roles of glucocorticoids in the hippocampus and hypothalamus in chronic stress-induced HPA axis hyperactivity. PLoS One. 2014;9:e97689. doi: 10.1371/journal.pone.0097689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Müller F, Liao J, Zhang Y, Gu H, Bock C, Boyle P, et al. Genomic distribution and inter-sample variation of non-CpG methylation across human cell types. PLoS Genet. 2011;7:e1002389. doi: 10.1371/journal.pgen.1002389. [DOI] [PMC free article] [PubMed] [Google Scholar]