Abstract
The ability to form long-lasting memories is critical to survival and thus is highly conserved across the animal kingdom. By virtue of its complexity, this same ability is vulnerable to disruption by a wide variety of neuronal traumas and pathologies. To identify effective therapies with which to treat memory disorders, it is critical to have a clear understanding of the cellular and molecular mechanisms which subserve normal learning and memory. A significant challenge to achieving this level of understanding is posed by the wide range of distinct temporal and spatial profiles of molecular signaling induced by learning-related stimuli. In this review we propose that a useful framework within which to address this challenge is to view the molecular foundation of long-lasting plasticity as composed of unique spatial and temporal molecular networks that mediate signaling both within neurons (such as via kinase signaling) as well as between neurons (such as via growth factor signaling). We propose that evaluating how cells integrate and interpret these concurrent and interacting molecular networks has the potential to significantly advance our understanding of the mechanisms underlying learning and memory formation.
Keywords: spatial network, temporal network, learning, memory, growth factors, MAPK, PKA, integration, coincidence detection
1. Introduction
The ability to form long-lasting memories is an evolutionarily conserved phenomenon that is critical to survival, and the dynamic process of acquiring and storing memories is often compromised in psychiatric and neurodegenerative disorders. It is thus important to understand how memory formation occurs in healthy individuals, and how these processes go awry in pathological states.
Memory can exist in at least three temporal domains best distinguished by the molecular mechanisms engaged in their induction and maintenance. Short-term memory (STM), which lasts on the order of minutes, is mediated by post-translational modifications [1,2]. Intermediate-term memory, which can last several hours, requires protein synthesis but not transcription [3,4], and long-term memory (LTM), which can last anywhere from days to a lifetime, requires both protein synthesis and de novo gene expression [1,5]. The study of long-lasting plasticity underlying memory formation has served as a valuable platform for the analysis of several novel molecular mechanisms, including long-term DNA modification [6] and reconsolidation [7]. However, the same complexity that yields such richness and depth in molecular signaling also poses a significant challenge to understanding the process of memory formation. One challenge lies in the temporal profile of LTM: LTMs are induced, consolidated, and expressed over a wide range of temporal scales, resulting in a virtually unlimited number of time points for assessing the molecular and cellular substrates involved. A second challenge lies in the spatial regulation required by the molecular mechanisms underlying LTMs. Specifically, LTM uniquely requires new gene expression, which raises the critical question as to how the synapse, where neural communication initially occurs, and the cell body, where transcription occurs, effectively communicate.
In this review, we will first introduce the concept of spatial and temporal molecular networks in the context of learning and memory by reviewing the spatial and temporal profiles of selected molecular signaling cascades critical for long-lasting plasticity and LTM formation. We will then focus on one specific aspect required for memory formation, growth factor signaling, which effectively captures these concepts. Finally, we will discuss how neurons might integrate and interpret these concurrent and interacting molecular networks, and how this overall framework can inform the study of LTM function and dysfunction.
2. Spatially- and Temporally-regulated Molecular Signaling in Long-lasting Plasticity and Memory Formation
It is widely appreciated that learning-related stimuli induce molecular signaling cascades (e.g. kinase activation, translation, or transcription) in discrete temporal and spatial profiles dictated by the nature of the stimulus, and these profiles are of critical importance to the resulting functional outcome. In one interesting example, Pineda and colleagues [8] demonstrated that enhancing adenylyl cyclase activity by genetically disrupting the Gi-coupled receptor that inhibits it, resulted in enhanced long-term potentiation (LTP) in the hippocampus, but this same manipulation actually impaired hippocampal-dependent memory. These data demonstrate that (i) LTP, often considered to be a cellular correlate of memory, is not always an accurate predictor of behavior, and (ii) signaling enhancement above the physiological levels induced by a learning event does not necessarily mean the memory formed will be superior. Although adenylyl cyclase activity, and cyclic adenosine monophosphate (cAMP) signaling in general, is required for memory formation (see discussion below), it is likely to be required at very specific levels and for very discrete periods of time. In the paragraphs that follow we will (i) provide a brief history of the general spatial properties of molecular signaling induced by learning-related stimuli, (ii) review the temporal and spatial profiles of two specific families of kinases critical for long-lasting plasticity and memory: MAPK and PKA, and (iii) highlight examples of the rich interactions between these kinases for molecular and cellular outcomes critical for plasticity.
2.1. Spatial properties of neural communication
Convincing evidence of the subcellular compartmentalization of distinct molecular cascades was supported by the notion that long-term synaptic strengthening induced at one synaptic site can facilitate the induction of long-term changes at another site [9,10]. Frey and Morris [9] proposed the critical idea of “synaptic tagging”, that is, a protein synthesis-independent process that sequesters proteins necessary to establish late-LTP at specific synaptic sites. For example, repeated stimulation to discrete synaptic inputs in the CA1 region of hippocampal slices has been demonstrated to induce protein synthesis and late-LTP at those inputs [9]. Subsequently, when weak stimulation, or repeated stimulation in the presence of protein synthesis inhibitors (which precludes the establishment of late-LTP), was applied to a neighboring synaptic input, late-LTP is also established at this input.
The same year, Martin and colleagues proposed in a series of experiments [11] a similar concept: “synaptic capture.” They used the model system Aplysia to investigate branch-specific protein synthesis using a bifurcated sensory-motor neuron (SN-MN) co-culture, in which two MNs are synaptically coupled to different branches of a single SN. If one set of SN-MN synapses was exposed to repeated applications of serotonin, long-term synaptic facilitation (LTF) and growth of new synaptic connections occurred in the branch which received stimulation, while the unstimulated branch remained unchanged [11]. Importantly, branch-specific facilitation required both cAMP response element binding protein (CREB)-mediated transcription and protein synthesis in the SN, despite the SN cell body being common to both MN synapses. Although CREB-dependent transcription is a uniquely somatic event, Martin and colleagues elegantly demonstrated that protein synthesis in SN neurites did not require the cell body: serotonin stimulation produced de novo protein synthesis in SN neurites regardless of a SN soma being present [11]. Moreover, when serotonin-mediated LTF was induced in one SN-MN synapse, a single exposure to serotonin, which is normally not capable of inducing LTF on its own, was able to induce LTF at the unstimulated synapse. Protein synthesis inhibitors had no effect when co-applied with a single pulse of serotonin on the second branch, but did block LTF at both branches when applied concurrently with the initial 5 pulses of serotonin on the first branch [11]. These findings highlight two critical principles: (i) there is a retrograde signal from the synapse to the soma to initiate transcription, and (ii) mRNAs can be stored locally at the synapse for rapid translation.
Taken together, these seminal experiments indicate there is an as yet undetermined contribution of the molecular signaling induced at activated synapses that also modulates the local ‘neighborhood’ of an activated synapse to facilitate more widespread activity- and location-dependent changes in neural responses. Viable candidate mechanisms that could regulate this phenomenon include the wide variety of kinases that become activated during learning and memory formation, including the MAPK and PKA families of kinases.
2.2. Protein kinases in memory formation: Mitogen-activated protein kinase (MAPK) family
The MAPK family of kinases is important for many cellular functions including proliferation, differentiation, gene expression, and mitosis, in addition to playing a crucial role in learning and memory formation [12–14]. One member of the MAPK family, ERK1/2, can be considered a molecular ‘node,’ as many upstream pathways can contribute to, and many downstream consequences result from its activity [15]. The activation of ERK1/2 itself is mediated by small G-proteins, which respond to a variety of cell stimulation, including membrane depolarization and calcium influx, both of which are fundamental to neuronal communication and plasticity, to initiate a cascade of phosphorylation events (MAPKK kinase → MAPK kinase → MAPK) [16,17].
The same upstream mediators responsible for activating ERK1/2 can additionally activate other members of the MAPK family, including c-Jun N-terminal kinases (JNKs) and p38 MAPK [12,18,19]. The JNKs are stress-activated proteins that are regulated by as many as 13 MAPKKKs, and, similarly to ERK1/2, many different stimuli may affect their activation [19,20]. JNKs phosphorylate the transcription factor c-Jun, which then binds to DNA and helps to initiate transcription [21]. c-Jun can associate with another transcription factor, c-Fos, to form the AP-1 transcriptional complex, which can in turn regulate the expression of several genes including those related to cell cycle progression and differentiation [19]. Intriguingly, AP-1 transcription factors are critical for positive feedback loops induced by brain derived neurotrophic factor (BDNF) in cortical neurons [22], a growth factor critical for long-lasting memory and plasticity (reviewed below). Moreover, AP-1 DNA binding activity is increased after LTP induction in the dentate gyrus [23], and a specific inhibitor of JNK phosphorylation, but not ERK1/2 or p38 MAPK phosphorylation, enhanced STM and blocked LTM for inhibitory avoidance in rats [24].
p38 MAPK is a particularly interesting member of the MAPK family, as its activation is required for the expression of long-term depression (long-lasting synaptic weakening; LTD) in Aplysia [25]. Upon activation by the inhibitory neuropeptide FMRFa, p38 MAPK phosphorylates CREB-2, which binds to the promoter region of the immediate early gene and transcription factor, C/EBP, preventing its transcription [25]. This is of particular consequence because C/EBP, like the MAPKs, is an evolutionarily conserved molecule required for learning and memory formation in vertebrates and invertebrates [26–28]. Indeed, inhibiting p38 MAPK activity blocks LTD and facilitates LTF, while its specific activation facilitates LTD and blocks LTF [25]. In the dentate gyrus of rats, a specific inhibitor of p38 MAPK similarly attenuated LTD, but had no effect on LTP [29]. Moreover, activation of p38 MAPK in combination with tyrosine de-phosphorylation was sufficient to induce LTD in the CA1 region of the hippocampus [30]. This is in direct opposition to the role of ERK1/2 in the induction of plasticity and memory, though as in the case of many different kinases, there may be interactions between p38 MAPK and ERK1/2 in LTD (see [31] for review). While ERK1/2 is directly activated by MEK, a MAPKK, p38 MAPK is directly activated by MKK3 and MKK6, all of which are regulated by the small G-protein Ras [32]. Because Ras activity can in theory result in the concurrent activation of p38 MAPK and ERK1/2, as is demonstrated in rat cortical neuron culture and Aplysia SNs [33], this entwined activation scheme produces the potential for different members of the MAPK family to act concurrently as integrative counterparts. This notion is important to consider when predicting the functional outcomes of parallel MAPK cascade activation, including transcriptional regulation as briefly discussed here.
Of particular interest in understanding the role of a kinase family as ubiquitous and complex as the MAPK system is its kinetic properties and temporal activation patterns in response to learning-related stimuli. ERK1/2 is activated in different temporal phases, both during and after training paradigms that induce long-term synaptic and behavioral plasticity [34–43]. Importantly, each temporal phase of ERK1/2 activity can be of critical consequence to LTM induction and maintenance. In Drosophila, Pagani and colleagues [39] reported that in response to an associative odor-shock training trial, ERK1/2 is activated at approximately the time point which corresponds to the permissive inter-trial interval (ITI) used to induce associative LTM (similar observations are reported in [36,43]). When the next training trial was delivered, ERK1/2 activation decreased to baseline, and then gradually increased again to a maximal level that coincided with when the subsequent training trial in the pattern was to be delivered [39]. Interestingly, in a model of Noonan’s syndrome in which the protein tyrosine phosphatase SHP2 is mutated, the temporal profile of ERK1/2 activation in response to a training trial is prolonged, and the pattern of repeated-trial training permissive for LTM formation in wildtype flies results in impaired LTM in SHP2 mutant flies. Pagani and colleagues [39] were able to correct this deficit simply by increasing the ITI to align with the altered kinetics of ERK1/2 activation and de-activation. Moreover, models of the temporal kinetics of kinase activation, including ERK1/2, can successfully predict non-intuitive training patterns which result in robust LTF in Aplysia [44]. These data reveal two important concepts: (i) ERK1/2 activation is important for binding multiple training trials into a unified long-lasting memory, and (ii) the temporal kinetics of kinase activation may confer ‘coincidence detection’ to the cell, a notion that we will discuss further below.
In addition to a complex temporal profile, ERK1/2 has a highly complex subcellular spatial activation pattern, and can be activated both at the synapse as well as at the soma to exert distinct effects unique to each compartment (e.g. receptor phosphorylation and gene expression, respectively) [45]. There is an additional level of spatial regulation that is not well appreciated in the context of learning and memory: small G-protein and scaffold localization (see [46] for review). Casar and colleagues [47] experimentally manipulated localization of the small G-protein Ras via constructs known to specifically localize to the endoplasmic reticulum (ER), lipid rafts, the disordered plasma membrane, or the Golgi complex, in HEK293T cell cultures. When ERK1/2 is activated via Ras localized to lipid rafts, but not the ER of Golgi membranes, it phosphorylates cytoplasmic/transmembrane proteins such as epidermal growth factor receptor and cytoplasmic phospholipase A2. However, when Ras was localized to the disordered plasma membrane, ERK1/2 activation resulted in phosphorylation of RSK1, a CREB kinase that shuttles between the cytoplasm and nucleus. The activation of the transcription factor Elk1, which can be phosphorylated by ERK1/2, p38 MAPK, and JNK1/2, was diminished by the inhibition of ERK1/2 regardless of the localization of Ras, though less so if ERK1/2 was activated by Ras at the Golgi membrane. Conversely, if JNK1/2 was inhibited, the most pronounced effect on Elk1 activation resulted when Ras was localized to the Golgi membrane. Casar and colleagues [47] further demonstrated that these effects resulted from the utilization of different scaffolding molecules, such as kinase suppressor of ras1 (KSR1), which is required for LTP and associative learning [48]. Interestingly, the small G-proteins Raf1 and B-Raf are widely distributed throughout the rat brain, but occupy slightly different subcellular compartments: Raf1 is primarily cytosolic surrounding the nucleus, while B-Raf is widely distributed throughout the cell [49]. These data collectively suggest that seemingly small changes in the location or mechanism by which MAPKs are activated during learning-related stimuli can in principle result in distinct routing of MAPK activation, which in turn could confer a unique message to the cell.
2.3. Protein kinases in memory formation: cAMP-dependent protein kinase (PKA)
PKA is a serine/threonine kinase that is activated by intracellular cAMP produced by adenylyl cyclase in response to cell stimulation. PKA has four subunits: two regulatory subunits that tonically inhibit two catalytic subunits. Binding of cAMP to the regulatory subunits results in the dissociation of the catalytic subunits, which are then free to phosphorylate their substrates in the cytosol and nucleus [50–52]. PKA activity has been demonstrated to be required for synaptic plasticity and memory formation in both invertebrates [53,54] and vertebrates [51,55]. Interestingly, the persistent activation of PKA was shown to be critical for 12 hours after the induction of LTF in SN-MN synapses in Aplysia[56], which is particularly remarkable given that significant PKA activation is measured in SNs within 5 minutes of a single training trial [57]. Indeed, prolonged stimulation that leads to transcription-dependent LTF and LTM in Aplysia results in a more persistent increase in cAMP levels and PKA activation, causing the catalytic subunit of PKA to translocate into the nucleus where it phosphorylates transcription factors important for inducing new gene expression required for long-term plasticity, including CREB-1 [58–60].
Surprisingly, Pu and colleagues [61] reported that chronic exposure (10 days) to opiates impairs both hippocampal LTP and LTM, which was (i) correlated with increased hippocampal PKA activity and (ii) reversed by administration of PKA inhibitors. These data suggest that over-activation of PKA can disrupt LTP induction, stressing the need for appropriate temporal regulation of PKA activity in learning-related plasticity in the hippocampus. Moreover, similar to that of ERK1/2, the temporal profile of PKA activity may encode coincident learning experiences. Mice subjected to strong contextual fear conditioning training consisting of 3 trials with a one minute ITI exhibited LTM that was sensitive to PKA disruption immediately after training, but not one or more hours after training [62]. However, mice subjected to weak contextual fear conditioning training consisting of a single trial exhibited LTM that was sensitive to PKA disruption immediately and 4 hours after training, but not 1, 6, 8, or 23.5 hours after training [62], suggesting that a single training trial induced distinct phases of PKA signaling, while multiple training trials did not.
PKA activation is also spatially regulated by scaffolding molecules, most notably A Kinase anchoring proteins (AKAPs) [63,64]. In fact, Huang and colleagues [65] suggest that AKAP anchoring of PKA may mediate synaptic tagging and capture. A competitive inhibitor of AKAP-PKA interactions, stHt31, impaired late-LTP in the CA1 region of mouse hippocampal sections induced by electrical stimulation of Schaffer collaterals without impairing the induction of chemically-induced LTP via global increases in cAMP. These data suggest that LTP induction via pathway stimulation that results in synapse-specific potentiation requires the accurate localization of PKA by AKAPs. Indeed, strong stimulation of one region of the Schaffer collaterals-CA1 pathway was no longer capable of facilitating the induction of late-LTP in a neighboring region when weak stimulation was delivered one hour later [65], suggesting that the molecular processes that induce synaptic tagging and capture (discussed above) are disrupted if PKA is inappropriately localized.
2.4. Interactions between distinct protein kinases in memory formation
Taken together, the data reflecting exquisitely precise spatial and temporal properties of molecular signaling during memory formation indicate that there is a complex and diverse network of molecular cascades induced in parallel in response to learning events. However, when each molecular event is studied in isolation, any interactions and/or synergism among these simultaneously engaged signaling cascades is difficult to determine. An example of this notion is the relationship between MAPK and PKA in transcriptional regulation in Aplysia.
Transcription can be initiated by several mechanisms, one of which is the activation of the transcription factor CREB. In Aplysia, the induction of gene expression by the isoform of CREB that activates transcription, CREB-1, can be endogenously repressed via the DNA binding capability of CREB-2 [66], suggesting that there may be two different pathways to regulate the initiation of CREB-mediated transcription: via activation and de-repression. Indeed, in Aplysia, PKA directly phosphorylates CREB-1, thereby activating it [58,59], while MAPK phosphorylates CREB-2, resulting in de-repression [67,68]. There are reports in mammalian studies of a similar mechanism by which the CREB family both activates and represses transcription. While both the mammalian homolog of CREB-2, activating transcription factor 4 (ATF4), as well as a homodimers of a related isoform, ATF3, have been reported to repress transcription (see [69] for review), ATF4 may act as a passive repressor of the transcription of certain genes while mediating differential gene expression itself [70]. Indeed, while ATF4 is upregulated in the brains of patients with and mouse models of Alzheimer’s Disease, suggesting a correlation between memory dysfunction and increased transcriptional repression, transgenic mice without ATF4 have impaired LTP, LTD, and memory [71], indicating that the relationship between CREB isoforms in mammals may be more complicated. Interestingly, ATF4 associates with Disrupted-In-Schizophrenia 1 (DISC1) and together bind to the phosphodiesterase 4D gene and repress its expression [72], suggesting that the binding partners with which ATF4 associates may in part dictate its role in transcriptional regulation.
In addition to the synergism between MAPK and PKA in regulating transcription, studies in both hippocampal and Aplysia neurons showed that PKA phosphorylation of CREB-1 requires MAPK activation [73], while nuclear translocation of MAPK requires PKA activation [74,75]. A similar inter-dependency between the consequences of MAPK and PKA signaling have been demonstrated both during LTP in the CA1 region of the hippocampus [76], LTM for inhibitory avoidance in rats [77] and for intermediate-term facilitation in Aplysia [42].
These data reviewed here collectively highlight the inter-dependent roles of distinct kinases in connecting extracellular signals to transcriptional regulation and ultimately memory formation. Furthermore, the remarkably tightly controlled temporal and spatial regulation of MAPK and PKA activity suggest that molecular signaling cascades engaged by learning-related stimuli must occur in a temporally and spatially coordinated manner to successfully support long-lasting plasticity and memory. While reviewing MAPK and PKA activity is a platform on which to begin to explore the consequences of spatial and temporal molecular networks, there are many other signaling cascades with elaborate activity profiles that are critical for learning and memory formation. These include the Ca2+/calmodulin-dependent protein kinase (CAMK) family [78,79], PKC and the persistently active atypical isoform of PKC, PKMζ [80,81], as well as mTOR-mediated protein synthesis [82,83,84,85]. Though an exhaustive evaluation of each molecular cascade is outside the scope of this review, it is important to note that several models of plasticity predict complex dynamics within and between these signaling families [44,86–88].
What remains unclear, however, is how precise spatial and temporal molecular networks, and complex interactions between intracellular signaling cascades, is ultimately initiated. Because virtually all kinases can be activated via several upstream mechanisms and second messenger systems, one hypothesis is that the extracellular signals that orchestrate intracellular signaling (i.e. neurotransmitters, hormones, neuromodulators, etc.) are themselves spatially and temporally regulated in distinct molecular networks. Growth factor (GF) signaling, which may very well mediate many of the events described above, is an excellent platform on which to begin to synthesize the data into a coherent, dynamic molecular network. We will take up this idea in the section that follows.
3. Growth Factor Signaling Networks in Learning and Memory
GF signaling cascades are tightly regulated in both space and time during developmental plasticity in order to create precise neural circuits [89–91], and these same principles are at play during GF regulation of learning and memory formation [92]. GF signaling thus affords an ideal opportunity to demonstrate the concept of temporal and spatial networks in learning and memory.
GFs are secreted molecules that bind membrane-associated extracellular receptors to initiate intracellular signaling cascades. Since the first fully characterized GF, nerve growth factor (NGF; [93,94]), it has become apparent that there are several families of GFs with unique receptors, the two major classes of which are receptor tyrosine kinases and serine-threonine kinases. Importantly, the high level of interaction between intracellular signaling cascades, including the kinases reviewed here, would predict that interactions also occur between multiple upstream signaling activators, like GFs. Indeed, a wide variety of GFs are engaged in learning and memory formation, and no GF by itself can mediate the complex molecular and cellular outcomes critical for long-lasting plasticity, (for a review on GF interactions mediating dendritic plasticity, see [92]). We thus propose that characterizing the parallel engagement of unique families of GFs supporting learning and memory formation can yield a deeper understanding of the molecular signaling networks involved in this long-lasting and dynamic plasticity.
3.1. Spatial and temporal GF signaling networks in learning and memory formation
GFs are capable of playing different roles in the different cellular and subcellular compartments in which they are engaged. In particular, mammalian neurotrophins, including NGF, BDNF, NT-3, and NT-4/5, have been extensively studied as signaling modulators critically involved in synapses undergoing learning-induced plasticity. Indeed, numerous studies have shown that the mRNAs and proteins of neurotrophins are widely distributed throughout the mammalian CNS in distinct, but largely overlapping expression patterns in regions important for learning and memory including the hippocampal formation, amygdala, and cortex [95–97]. Additionally, some studies suggest that neurotrophins can exist within different subcellular compartments within these brain regions, including axonal localization [98–103] and somatodendritic localization [98,104–107]. Furthermore, both anterograde [98,99,103] and retrograde transport [104,108] of neurotrophins has been reported, suggesting that the site of GF initiation may not be the same or only site of GF action. This can occur through endocytosis of the GF-GF receptor complex upon ligand binding, and thus may be an important means of synapse-soma communication. In addition to neurotrophins forming intracellular signaling complexes, or endosomes [109], both epidermal growth factor [110] and insulin-like growth factor II (IGF-II; [111,112]) can be regulated in a similar manner. Although not well studied in the context of learning and memory, kinase phosphorylation has been demonstrated to be a critical regulator of internalized GF receptors into either a recycled population or a degraded population [113]. Because many GF receptors are both upstream activators for kinases as well as substrates for their modulation, this sort of mechanism may represent an as yet unappreciated regulation of both spatial and temporal molecular signaling.
Adding to the complexity, neurotrophins can be secreted from either postsynaptic dendritic spines or from presynaptic terminals, and both their synthesis and release in the adult central nervous system seems to be activity-dependent [95,97,114,115]. Indeed, membrane depolarization in hippocampal cultures via high potassium solutions or glutamate treatment increases mRNA expression of BDNF and NGF in addition to increasing the release of BDNF, NGF, and NT-3 [116–120]. Moreover, the induction of GF mRNAs in hippocampal slices appears to be spatially restricted to the area of stimulation in response to LTP-inducing stimuli: Patterson and colleagues [121] reported that stimulation of the CA1 region leads to an increase in mRNA levels of both BDNF and NT-3 in CA1, but not in neurons in the CA3 or dentate gyrus regions. Interestingly, in addition to activity-dependent release, there is also evidence for neurotrophin-induced neurotrophin secretion [122,123], which raises the interesting question regarding the functional significance of activity-dependent synthesis and release of multiple neurotrophins at the same synapse. Autocrine or paracrine actions of released neurotrophins have been reported [95,96,124], however the simultaneous actions of multiple growth factors released at the same synapse have yet to be explored. Moreover, it will be important to examine and dissect the parallel or convergent signaling cascades downstream of GF receptor activation within the same cell.
Once released at a synapse, neurotrophins as well as other GFs bind to their putative receptors and induce intracellular signaling leading to changes in synaptic transmission and connectivity. Multiple studies in hippocampal and cortical cultures and slices [125,126] have reported that treatment with each of the neurotrophins (BDNF, NGF, NT-3, and NT-4/5) induces potentiation of excitatory glutamatergic synaptic transmission through either presynaptic [127,128] and/or postsynaptic mechanisms [129,130]. Interestingly, in addition to facilitation of excitatory synaptic transmission, there have also been reports of neurotrophin-mediated depression of inhibitory synaptic transmission [125,131], which would result in a similar functional outcome (increased excitability) albeit through a very different molecular cascade (decreased inhibition).
Activity-dependent release of GFs, which is common during a learning event, often occurs at the synapse [97]; however, GFs can mediate both synaptic and somatic events. For example, BDNF signaling induced by IA training in rats regulates the activity of several proteins important for LTM formation at the synapse, such as CAMKIIα, ERK1/2, and AKT, as well as CREB phosphorylation, which is a specifically nuclear, and thus somatic, protein [132]. Moreover, Kanhema and colleagues [133] demonstrated that an infusion of BDNF into the dentate gyrus of anesthetized rats induced LTP, and increased phosphorylation of translational machinery proteins in a spatially segregated manner. Specifically, BDNF increased eIF4E phosphorylation in synaptically-enriched samples, and increased eEF2 phosphorylation in global, but not synaptically-enriched samples. Interestingly, both effects were MEK-dependent, suggesting that BDNF signaling can make use of the same signaling broker, perhaps in different cellular compartments, for different outcomes.
GF signaling is also engaged in temporally discrete profiles during learning and memory formation. BDNF signaling, as discussed above, is often engaged very early in response to neuronal activity [97]. Interestingly however, in general GFs are recruited in a delayed fashion well after a learning event or learning-related stimulus. For example, IGF-II mRNA and protein is increased in the dorsal hippocampus 20 hours, but not 6 or 9 hours after inhibitory avoidance (IA) training in rats, and its signaling via the IGF-II receptor is required for more than 1, but less than 4 days, after IA training for memory consolidation [134]. Moreover, NGF levels in the hippocampal CA1 region were increased one week after contextual fear conditioning in rats, but not earlier or later [135], and inhibiting NGF signaling 1 week, but not 4 weeks, after training impaired memory [135]. Activin, a member of the TGFβ superfamily, was required for specifically late-LTP several hours after high frequency stimulation [136], and LTP could be enhanced by exogenous activin application or blocked by activin inhibitors 1 hour, but not 3 hours after induction [136]. There is even evidence that BDNF is additionally required long after training to support memory persistence. Blocking protein synthesis in the hippocampus 12 hours after IA training disrupts the expression of memory 7 days, but not 2 days, after training [137,138], which can be rescued by co-injection of BDNF [138]. Moreover, Bambah-Mukku and colleagues [124] have recently demonstrated a BDNF-induced positive feedback loop that supports long-term IA memory in rats. IA training induces BDNF-dependent CREB activation and C/EBPβ expression; C/EBPβ then mediates an increase in BDNF gene and protein expression, which constitutes an additional phase of BDNF signaling required for LTM formation.
Despite extensive data reporting very precise spatial and temporal regulation of both intracellular signaling (i.e. kinase activation) as well as their upstream activators (i.e. GFs), a critical issue remains: what is the meaning of this signaling for the cell? Specifically, how are these disparate (both temporally and spatially) molecular networks integrated into a coherent message? And, what is the value of this complexity? One possible answer is that the integration of individual spatiotemporal profiles is capable of relaying vastly more information than each signal considered in isolation. It is thus possible that individual temporal and spatial molecular networks interact to serve “coincidence detection” functions. In the section that follows we will examine possible mechanisms of spatial and temporal integration, specifically highlighting mechanisms of synapse—soma communication as this trans-compartmental communication contains both a temporal and spatial component. We will then review in detail a specific example of the notion of coincidence detection in GF signaling networks: Two-Trial LTM formation in Aplysia.
4. Cellular Interpretation of Spatial and Temporal Molecular Networks
4.1. Integration of distinct molecular networks: How do synapses and cell bodies communicate?
Long-term forms of cellular and behavioral plasticity are distinguished from other forms of memory (e.g. working, short-term, and intermediate-term memory) by their requirement of de novo gene expression [26,53]. However, there is abundant evidence that synapses are the initial site of plasticity, making the mechanisms by which large numbers of synapses on any one neuron communicate with a single cell body, and vice versa, of particular interest.
Microtubules are dynamic elements of the cell structure that serve as ‘tracks’ upon which molecular motors can transport cargo. Microtubules typically run throughout the cell and are able to integrate indirectly with the actin framework through cross-linking cytoskeletal proteins such as the CLASP family of proteins [139–141]. There are diverse families of microtubule motors, such as kinesins, for transporting molecular cargo in the anterograde direction (i.e. from soma to synapse), but not nearly the same diversity in retrograde (i.e. from synapse to soma) motor proteins, such as dyneins [142,143]. Additionally, the actin cytoskeleton is traversed by myosin proteins [142]. Both kinesins and myosins move across microtubules and actin, respectively, via phosphorylation-induced changes in protein conformation, whereas dyneins move in a more complex manner, with their head regions making contact with microtubules [142]. Moreover, while kinesins and myosins directly attach to vesicles via anchoring proteins [142], dyneins complex with cargo via sets of scaffold proteins, such as the dynactin protein complex, where vesicular cargo must be attached indirectly to the dynein protein [144]. The kinesins and dyneins also yield different temporal kinetics of transport. Kinesins have a high duty ratio, meaning that they can quickly transport vesicular cargo long distances (several micrometers) without falling off of microtubules, while dyneins have a low duty ratio [144], collectively resulting in much faster transport from soma to synapse than vice versa.
Microtubules tend not to extend into dendritic spines, the primary sites of synaptic input, where instead there is a high concentration of actin filaments [139,140]. Myosin-mediated transport on actin filaments is largely responsible for organelle translocation and relatively slow cellular movement [139], in contrast to kinesin-mediated transport on microtubules which is responsible for fast, axonal transport of small vesicular cargo [144]. Interestingly, microtubules have been reported to ‘invade’ dendritic spines in mouse hippocampus in response to NMDA receptor-dependent calcium influx, a phenomenon dependent on actin polymerization and the expression of Drebrin, a protein that facilitates the interaction between actin filaments and microtubules [145]. Importantly, microtubule content in dendritic spines is primarily observed when dendritic spines are active [145], suggesting that microtubule invasion into active dendritic spines could serve as an activity-dependent and transient trafficking route for vesicular cargo traveling between the soma and synapse. Indeed, increased vesicular trafficking as well as increased kinesin heavy chain mRNA have been reported in response to learning-related stimuli in Aplysia [146]. Together, these data raise the intriguing possibility that somatic gene and protein expression must be spatially and temporally coordinated in such a way as to take advantage of the transient availability of transport to activated synapses. Moreover, the molecular mechanisms which regulate these windows of transport may be of critical importance to the successful induction of long-lasting plasticity, and to our knowledge have not yet been addressed.
4.2. Integration of distinct molecular networks: A case study of coincidence detection in Aplysia
We have briefly discussed how the soma and synapse can communicate, and some aspects of the temporal regulation of this communication. However, because molecular cascades are often studied in isolation, it is experimentally difficult to determine if there are instances in which distinct molecular networks are integrated to mediate functional outcomes that neither network, on its own, can support. We will now review a study using the marine mollusk, Aplysia, as a model, which demonstrates the spatial and temporal integration of distinct GF signaling networks to synergistically support LTM formation.
The simple central nervous system of Aplysia is well-suited for interrogating the molecular mechanisms of learning and memory formation [53]. The defensive withdrawal reflexes are largely governed by monosynaptic sensory neuron (SN) communication with motor neurons (MNs) [147–149]. Upon training with electrical stimulation or the neuromodulator serotonin, the withdrawal reflexes become enhanced, or sensitized, and similar to vertebrates, LTM for sensitization in Aplysia lasts days to weeks and requires both translation and transcription [150].
Memory formation in Aplysia, like other species, is responsive to the pattern of training. For instance, four or five repeated, spaced training trials [151] yields superior LTM than the same amount of training given with little time in between training trials [43,152], which is a well-conserved phenomenon known as the spacing effect [153]. MAPK activation (homologous to ERK1/2) at SN cell bodies occurs in two different phases in response to sensitizing stimuli in Aplysia: (i) during the training interval and (ii) after training. A single training trial induces an early phase of MAPK activation in SN somata 45 minutes after Trial 1 [37,43]. This early phase is transient, and in the absence of additional training trials it decays to baseline by 60 minutes [37,43]. However, following repeated trial training that is permissive for the induction of LTM, MAPK is persistently activated in the SN somata 1–3 hours after training [35,42,154]. Importantly, both the early and late phases of MAPK activation are required for LTM formation [35,37].
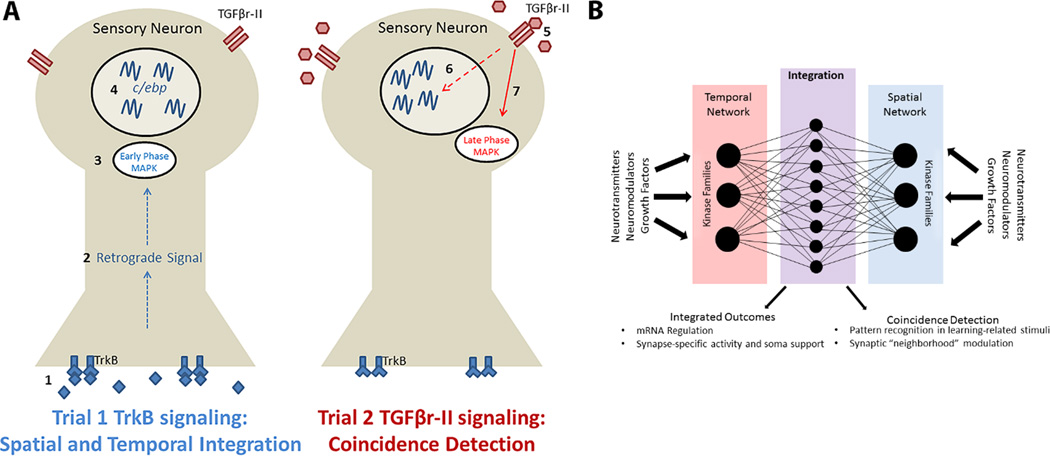
Philips and colleagues [37,43,149] developed a novel LTM training paradigm, consisting of only two training trials spaced by 45 minutes (Two-Trial training), which is sufficient to induce LTM in Aplysia. Interestingly, Trial 2 must be delivered within a restricted temporal window: if occurring at 15 minutes or 60 minutes after Trial 1, no LTM is induced [37,43]. Collectively, data from Carew and colleagues suggest that Trial 1 induces a “molecular context” at 45 minutes that includes MAPK activation, its translocation into the nucleus of SNs, and increased gene expression of the immediate early gene c/ebp [37]. Trial 2 then interacts with this molecular context and causes changes which make the circuit permissive for long-lasting information storage. With this minimalistic training pattern, Two-Trial training affords the unique opportunity to assay molecular and cellular changes induced by each training trial individually, as well as to explore the interaction between the molecular context established by Trial 1 and the changes induced by Trial 2. Two GF families have been reported to activate MAPK and be required for long-term plasticity in Aplysia: neurotrophins signaling via the receptor TrkB [155,156] and the TGFβ superfamily signaling via the receptor TGFβr-II [154,157–160]. Thus, as a first step in characterizing the role of different GF families during LTM formation, Kopec and colleagues [161] analyzed the necessity for TrkB and TGFβr-II signaling upstream of the early and late phases of MAPK activation induced by Two-Trial training.
The data demonstrated a striking double-dissociation in the temporal and spatial regulation of these GFs during learning-related stimuli. Trial 1 initiated TrkB signaling at the SN-MN synapse, which was required for early-phase MAPK activation in SN cell bodies. Importantly, these data necessitate an intracellularly transported retrograde signaling to exert the somatic effects of synaptic TrkB signaling. Trial 2 initiated TGFβr-II signaling at SN cell bodies, which was required for late-phase MAPK activation in the same compartment. Surprisingly, the later, TGFβr-II-dependent phase of MAPK was not disrupted if Trial 1 TrkB signaling was inhibited, suggesting that these two GF families were signaling independently despite being in temporal sequence.
Kopec and colleagues [161] next sought to determine if these spatiotemporally regulated GF signaling cascades regulated a molecular event uniquely required for long-lasting plasticity and memory: transcription. C/EBP, briefly mentioned above, is an immediate early gene and transcription factor that is evolutionarily conserved and required for LTM formation [26]. Moreover, both Trial 1 and Trial 2 result in increased c/ebp mRNA expression in Aplysia [37,161]. A unique syngergistic interaction emerged when GF signaling upstream of c/ebp expression was examined. In agreement with the MAPK data, Trial 1 TrkB signaling was required for Trial 1 c/ebp expression, and Trial 2 TGFβr-II signaling was required for Trial 2 c/ebp expression. However, unlike the MAPK data, when TrkB signaling was inhibited during Trial 1, Trial 2 c/ebp expression was also disrupted. These data suggest that Trial 1 TrkB signaling increases the expression of c/ebp mRNAs (presumably through transcriptional activation, though this has not yet been tested), and Trial 2 TGFβr-II signaling, rather than engaging a new wave of transcription, prolongs the expression of Trial 1 mRNAs. Finally, in a set of experiments to determine if these molecular observations were behaviorally relevant, Kopec and colleagues [161] demonstrated that Trial 1 TrkB signaling and Trial 2 TGFβr-II signaling were, indeed, required for Two-Trial LTM for sensitization in Aplysia.
This report exemplifies many of the principles discussed in the present review (see Figure 1A). First, there are spatially and temporally distinct molecular networks, specifically GF signaling networks: TrkB signaling was initiated synaptically to mediate early, Trial-1 dependent molecular outcomes (i.e. MAPK activation and c/ebp expression). Conversely, TGFβr-II signaling was initiated somatically to mediate later, Trial-2 dependent molecular outcomes. Moreover, TrkB signaling resulted in synapse-soma communication, while TGFβr-II signaling prolonged the expression of existing mRNAs. Importantly, Trial 1, despite activating TrkB signaling, synapse-soma communication, MAPK, and transcription, cannot by itself produce LTM [43]. Transcription is a complex, highly integrative cellular function, requiring the collaborative activity of several different families of proteins and protein complexes. The fact that de novo gene expression readily occurs in response to stimuli that alone cannot induce LTM is of critical importance: transcription is necessary, but not sufficient, for the formation long-lasting plasticity. Instead, the coincident arrival of another learning-related signaling cascade, at the appropriate time and in the appropriate subcellular compartment, must occur. Thus, the molecular events induced by Trial 2 serve a coincidence detection role in the cell such that the circuit initiates long-lasting information storage.
Figure 1.
Neuronal interpretation of spatial and temporal molecular networks in long-lasting plasticity and memory
A.Distinct GF signaling networks (TrkB and TGFβr-II) synergistically interact to mediate LTM formation in Aplysia. (1) Trial 1 induces release of TrkB ligands at the SN-MN synapse, which results in (2) an intracellularly transported retrograde signaling that is responsible for (3) a transient and delayed phase of MAPK activation in SN somata. Concurrently, TrkB signaling mediates (4) an increase in expression of the immediate early gene, c/ebp. The delivery of Trial 2 during this molecular context induces (5) TGFβr-II ligand release at SN somata. TGFβr-II signaling then interacts with the TrkB signaling cascade to (6) prolong the expression of c/ebp mRNA established by Trial 1, and (7) independently mediates a persistent phase of MAPK activation. These molecular observations demonstrate both the spatial and temporal integration of distinct molecular networks as well as one possible role this complexity serves: coincidence detection. B. Reports reviewed here indicate that there are exceedingly complex spatial and temporal profiles of both extracellular (e.g. GFs) and intracellular (e.g. kinases) molecular signaling cascades. We propose that the integration of these distinct molecular networks serves to provide the neuron with more information than any one network can provide alone (e.g. coincidence detection). Thus, the initiation of complex molecular programs, including those underlying long-lasting cellular and behavioral plasticity, is best understood as synergistic interactions between multiple signaling networks.
Precisely what signal induced by Trial 2 is most salient at conferring coincidence is at present unclear, and it is likely that an interaction of several events contribute to this process. However, because TGFβr-II signaling during Trial 2 is required for Two-Trial LTM formation, consequences of the signaling via this GF are likely to be involved. These include, but are not limited to (i) an independent phase of MAPK activity, which could have numerous downstream outcomes, and (ii) prolonged c/ebp expression, which could result in the increased likelihood of its translation, and thus increased activity of its functions as a transcription factor (i.e. more new gene expression). Interestingly, c/ebp mRNAs contain an AU-rich element in their 3’ untranslated regions, which is a motif targeted by the Hu/ELAV family of RNA binding proteins that can stabilize mRNAs [162–164]. Post-transcriptional regulation by RNA binding proteins has been proposed to serve as a method by which the cell filters out transcriptional noise and ensures the degradation of contextually irrelevant transcripts [165], and thus RNA binding proteins are one way in which newly transcribed mRNAs may be regulated. The phosphorylation state of Hu, which has been reported to be modulated by PKCδ, p38 MAPK, and AMP kinase dictates its ability to bind to target mRNAs and shuttle them between the nucleus and the cytoplasm, which in turn regulates the probability that mRNAs associate with ribosomes for translation [166–168]. Thus post-transcriptional regulation of mRNAs could be a site of integration for the signaling of several different kinases and potentially GF signaling cascades. If TGFβr-II signaling does, indeed, recruit RNA binding proteins, other mRNAs not yet reported to be upregulated by Trial 1 may additionally be stabilized.
5. Consequences of Spatial and Temporal Molecular Networks: Experimental and Therapeutic Implications
Collectively, the studies reviewed here support the notion depicted in Fig. 1B: neurons integrate spatially- and temporally-regulated molecular signaling networks, and in response, initiate complex molecular programs required for long-lasting cellular and behavioral plasticity. Moreover, this integration provides more information than any one signaling network alone, such as coincidence detection. Importantly, two recent publications utilizing transcriptomic profiling in development support this notion and demonstrate unexpected instances of interaction and integration [169,170]. The task of characterizing the temporal and spatial profiles of a molecular network is itself a difficult undertaking. A scenario involving two or more GFs (or other signaling brokers) would significantly increase the number of possible combinatorial effects, making these studies inherently difficult. Monosynaptic connections such as those underlying the defensive withdrawal reflexes in Aplysia offer the spatial resolution needed for these mechanistically detailed studies, as exemplified in the case study above. As it stands, five different GFs have been reported to have a role in plasticity in Aplysia [156,160,171–173], and there are many more GFs implicated in mammalian plasticity [92].
The notion of spatial and temporal molecular networks is particularly important from the perspective of translational research and therapeutic intervention. Many GFs, including BDNF-TrkB and TGFβ-TGFβr signaling, have been suggested to be dysregulated during cognitive dysfunction of various etiologies in humans, including Alzheimer’s disease [174–176]. GF receptors are ideal candidates for therapeutic interventions because their ligand binding domain is extracellular. However, the results reported by Kopec and colleagues [161] reviewed above emphasize three reasons why a therapy that targets a single GF family may be insufficient to ameliorate cognitive deficits. First, the targeted GF family may be only one of several GFs in a molecular network disrupted by pathology. Thus, restoring signaling from one GF family may restore only a single component of the signaling required for behavior. Furthermore, the targeted GF family may require the interaction with other GFs to fully achieve the functional outcomes of its molecular signaling. For instance, in the case of Two-Trial LTM in Aplysia, augmented or restored TrkB signaling may not be sufficient to restore LTM if TGFβr-II signaling is not efficiently interacting with TrkB-dependent signaling. Second, GF signaling is highly spatially regulated and this specificity may well be important for the neural circuit to correctly and effectively interpret the molecular signaling. Utilizing a systemic drug, despite having restored GF signaling, may disrupt the spatial signature of a GF family and thus disrupt behavior. Taking the data from Kopec and colleagues [161] as an illustrative example, if TrkB signaling was blocked at the synapse during Trial 1, but at the same time augmented at the soma, would this result in successful Two-Trial LTM formation? The delayed nature of the synaptic TrkB-dependent signal positions it to effectively interact with TGFβr-II signaling during Trial 2, and this may not be the result if TrkB signaling is instead engaged at the soma during Trial 1. And third, GF signaling also follows a strict temporal profile during LTM formation in Aplysia, which may well be the case for other forms of plasticity underlying various forms of learning. Without an understanding of when a GF family is recruited to support behavior, therapeutic interventions may miss the time frame in which it could be most effective. For instance, if TGFβr-II signaling was augmented one hour after Trial 1, apc/ebp expression would have returned to baseline levels and TGFβr-II signaling would not have the opportunity to interact with the TrkB-dependent signaling cascade, and thus may not result in the same behavioral outcome.
In conclusion, plasticity underlying memory formation is an exquisitely complex interplay between differential temporal and spatial molecular networks. Different signaling cascades, including kinases and GF signaling reviewed here, could be initiated in (i) an autocrine or paracrine fashion, (ii) simultaneously or in different temporal phases, resulting in (iii) local or long-distance (e.g. synaptic vs. somatic) molecular consequences, all of which increase the likelihood of interactions and synergism between cascades. Moreover, there can be salient information in these interactions and instances of synergy, such that cells can interpret ‘coincidence,’ pattern, and statistical probability in a series of learning-related stimuli. Discovering how cells integrate and interpret these dynamic networks is an extraordinary challenge, but this challenge is essential to tackle, as it is critical not only for understanding how the plasticity underlying learning and memory formation normally occurs, but also for designing effective therapies to intervene when this plasticity goes awry.
Acknowledgments
This work was supported by NIMH RO1 MH 041083 to TJC.
Footnotes
Conflict of Interest
The authors declare no competing financial interests.
References
- 1.Castellucci VF, Blumenfeld H, Goelet P, et al. Inhibitor of protein synthesis blocks long-term behavioral sensitization in the isolated gill-withdrawal reflex of Aplysia. J Neurobiol. 1989;20:1–9. doi: 10.1002/neu.480200102. [DOI] [PubMed] [Google Scholar]
- 2.Xia SZ, Feng CH, Guo AK. Multiple-phase model of memory consolidation confirmed by behavioral and pharmacological analyses of operant conditioning in Drosophila. Pharmacol Biochem Behav. 1998;60:809–816. doi: 10.1016/s0091-3057(98)00060-4. [DOI] [PubMed] [Google Scholar]
- 3.Sutton MA, Masters SE, Bagnall MW, et al. Molecular mechanisms underlying a unique intermediate phase of memory in aplysia. Neuron. 2001;31:143–154. doi: 10.1016/s0896-6273(01)00342-7. [DOI] [PubMed] [Google Scholar]
- 4.Stough S, Shobe JL, Carew TJ. Intermediate-term processes in memory formation. Curr Opin Neurobiol. 2006;16:672–678. doi: 10.1016/j.conb.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 5.Bailey CH, Bartsch D, Kandel ER. Toward a molecular definition of long-term memory storage. Proc Natl Acad Sci U S A. 1996;93:13445–13452. doi: 10.1073/pnas.93.24.13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heyward FD, Sweatt JD. DNA Methylation in Memory Formation: Emerging Insights. Neuroscientist. 2015;21:475–489. doi: 10.1177/1073858415579635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alberini CM. Mechanisms of memory stabilization: are consolidation and reconsolidation similar or distinct processes? Trends Neurosci. 2005;28:51–56. doi: 10.1016/j.tins.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 8.Pineda VV, Athos JI, Wang H, et al. Removal of G(ialpha1) constraints on adenylyl cyclase in the hippocampus enhances LTP and impairs memory formation. Neuron. 2004;41:153–163. doi: 10.1016/s0896-6273(03)00813-4. [DOI] [PubMed] [Google Scholar]
- 9.Frey U, Morris RG. Synaptic tagging and long-term potentiation. Nature. 1997;385:533–536. doi: 10.1038/385533a0. [DOI] [PubMed] [Google Scholar]
- 10.Sherff CM, Carew TJ. Coincident induction of long-term facilitation in Aplysia: cooperativity between cell bodies and remote synapses. Science. 1999;285:1911–1914. doi: 10.1126/science.285.5435.1911. [DOI] [PubMed] [Google Scholar]
- 11.Martin KC, Casadio A, Zhu H, et al. Synapse-specific, long-term facilitation of aplysia sensory to motor synapses: a function for local protein synthesis in memory storage. Cell. 1997;91:927–938. doi: 10.1016/s0092-8674(00)80484-5. [DOI] [PubMed] [Google Scholar]
- 12.Giese KP, Mizuno K. The roles of protein kinases in learning and memory. Learn Mem. 2013;20:540–552. doi: 10.1101/lm.028449.112. [DOI] [PubMed] [Google Scholar]
- 13.Graves JD, Campbell JS, Krebs EG. Protein serine/threonine kinases of the MAPK cascade. Ann N Y Acad Sci. 1995;766:320–343. doi: 10.1111/j.1749-6632.1995.tb26684.x. [DOI] [PubMed] [Google Scholar]
- 14.Sweatt JD. Mitogen-activated protein kinases in synaptic plasticity and memory. Curr Opin Neurobiol. 2004;14:311–317. doi: 10.1016/j.conb.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 15.Reissner KJ, Shobe JL, Carew TJ. Molecular nodes in memory processing: Insights from Aplysia. Cell Moll Life Sci. 2006;63 doi: 10.1007/s00018-006-6022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosen LB, Ginty DD, Weber MJ, et al. Membrane depolarization and calcium influx stimulate MEK and MAP kinase via activation of Ras. Neuron. 1994;12:1207–1221. doi: 10.1016/0896-6273(94)90438-3. [DOI] [PubMed] [Google Scholar]
- 17.Farnsworth CL, Freshney NW, Rosen LB, et al. Calcium activation of Ras mediated by neuronal exchange factor Ras-GRF. Nature. 1995;376:524–527. doi: 10.1038/376524a0. [DOI] [PubMed] [Google Scholar]
- 18.Sherrin T, Blank T, Todorovic C. c-Jun N-terminal kinases in memory and synaptic plasticity. Rev Neurosci. 2011;22:403–410. doi: 10.1515/RNS.2011.032. [DOI] [PubMed] [Google Scholar]
- 19.Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002;298:1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- 20.Tournier C, Hess P, Yang DD, et al. Requirement of JNK for stress-induced activation of the cytochrome c-mediated death pathway. Science. 2000;288:870–874. doi: 10.1126/science.288.5467.870. [DOI] [PubMed] [Google Scholar]
- 21.Kyriakis JM, Banerjee P, Nikolakaki E, et al. The stress-activated protein kinase subfamily of c-Jun kinases. Nature. 1994;369:156–160. doi: 10.1038/369156a0. [DOI] [PubMed] [Google Scholar]
- 22.Tuvikene J, Pruunsild P, Orav E, et al. AP-1 Transcription Factors Mediate BDNF-Positive Feedback Loop in Cortical Neurons. J Neurosci. 2016;36:1290–1305. doi: 10.1523/JNEUROSCI.3360-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams JM, Beckmann AM, Mason-Parker SE, et al. Sequential increase in Egr-1 and AP-1 DNA binding activity in the dentate gyrus following the induction of long-term potentiation. Brain Res Mol Brain Res. 2000;77:258–266. doi: 10.1016/s0169-328x(00)00061-9. [DOI] [PubMed] [Google Scholar]
- 24.Bevilaqua LR, Kerr DS, Medina JH, et al. Inhibition of hippocampal Jun N-terminal kinase enhances short-term memory but blocks long-term memory formation and retrieval of an inhibitory avoidance task. Eur J Neurosci. 2003;17:897–902. doi: 10.1046/j.1460-9568.2003.02524.x. [DOI] [PubMed] [Google Scholar]
- 25.Guan Z, Kim JH, Lomvardas S, et al. p38 MAP kinase mediates both short-term and long-term synaptic depression in aplysia. J Neurosci. 2003;23:7317–7325. doi: 10.1523/JNEUROSCI.23-19-07317.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alberini CM. Transcription factors in long-term memory and synaptic plasticity. Physiol Rev. 2009;89:121–145. doi: 10.1152/physrev.00017.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alberini CM, Ghirardi M, Metz R, et al. C/EBP is an immediate-early gene required for the consolidation of long-term facilitation in Aplysia. Cell. 1994;76:1099–1114. doi: 10.1016/0092-8674(94)90386-7. [DOI] [PubMed] [Google Scholar]
- 28.Taubenfeld SM, Milekic MH, Monti B, et al. The consolidation of new but not reactivated memory requires hippocampal C/EBPbeta. Nat Neurosci. 2001;4:813–818. doi: 10.1038/90520. [DOI] [PubMed] [Google Scholar]
- 29.Murray HJ, O'Connor JJ. A role for COX-2 and p38 mitogen activated protein kinase in long-term depression in the rat dentate gyrus in vitro. Neuropharmacology. 2003;44:374–380. doi: 10.1016/s0028-3908(02)00375-1. [DOI] [PubMed] [Google Scholar]
- 30.Moult PR, Correa SA, Collingridge GL, et al. Co-activation of p38 mitogen-activated protein kinase and protein tyrosine phosphatase underlies metabotropic glutamate receptor-dependent long-term depression. J Physiol. 2008;586:2499–2510. doi: 10.1113/jphysiol.2008.153122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanderson TM, Hogg EL, Collingridge GL, et al. Hippocampal mGluR-LTD in health and disease: focus on the p38 MAPK and ERK1/2 pathways. J Neurochem. 2016 doi: 10.1111/jnc.13592. [DOI] [PubMed] [Google Scholar]
- 32.Zarubin T, Han J. Activation and signaling of the p38 MAP kinase pathway. Cell Res. 2005;15:11–18. doi: 10.1038/sj.cr.7290257. [DOI] [PubMed] [Google Scholar]
- 33.Liu RY, Zhang Y, Coughlin BL, et al. Doxorubicin attenuates serotonin-induced long-term synaptic facilitation by phosphorylation of p38 mitogen-activated protein kinase. J Neurosci. 2014;34:13289–13300. doi: 10.1523/JNEUROSCI.0538-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Atkins CM, Selcher JC, Petraitis JJ, et al. The MAPK cascade is required for mammalian associative learning. Nat Neurosci. 1998;1:602–609. doi: 10.1038/2836. [DOI] [PubMed] [Google Scholar]
- 35.Sharma SK, Sherff CM, Shobe J, et al. Differential role of mitogen-activated protein kinase in three distinct phases of memory for sensitization in Aplysia. J Neurosci. 2003;23:3899–3907. doi: 10.1523/JNEUROSCI.23-09-03899.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ajay SM, Bhalla US. A role for ERKII in synaptic selectivity on the time-scale of minutes. Eur J Neurosci. 2004;20:2671–2680. doi: 10.1111/j.1460-9568.2004.03725.x. [DOI] [PubMed] [Google Scholar]
- 37.Philips GT, Ye X, Kopec AM, et al. MAPK establishes a molecular context that defines effective training patterns for long-term memory formation. J Neurosci. 2013;33:7565–7573. doi: 10.1523/JNEUROSCI.5561-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ye X, Shobe JL, Sharma SK, et al. Small G proteins exhibit pattern sensitivity in MAPK activation during the induction of memory and synaptic facilitation in Aplysia. Proc Natl Acad Sci U S A. 2008;105:20511–20516. doi: 10.1073/pnas.0808110105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pagani MR, Oishi K, Gelb BD, et al. The phosphatase SHP2 regulates the spacing effect for long-term memory induction. Cell. 2009;139:186–198. doi: 10.1016/j.cell.2009.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shobe JL, Zhao Y, Stough S, et al. Temporal phases of activity-dependent plasticity and memory are mediated by compartmentalized routing of MAPK signaling in aplysia sensory neurons. Neuron. 2009;61:113–125. doi: 10.1016/j.neuron.2008.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michel M, Green CL, Eskin A, et al. PKG-mediated MAPK signaling is necessary for long-term operant memory in Aplysia. Learn Mem. 2011;18:108–117. doi: 10.1101/lm.2063611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye X, Marina A, Carew TJ. Local synaptic integration of mitogen-activated protein kinase and protein kinase A signaling mediates intermediate-term synaptic facilitation in Aplysia. Proc Natl Acad Sci U S A. 2012;109:18162–18167. doi: 10.1073/pnas.1209956109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Philips GT, Tzvetkova EI, Carew TJ. Transient mitogen-activated protein kinase activation is confined to a narrow temporal window required for the induction of two-trial long-term memory in Aplysia. J Neurosci. 2007;27:13701–13705. doi: 10.1523/JNEUROSCI.4262-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Liu RY, Heberton GA, et al. Computational design of enhanced learning protocols. Nat Neurosci. 2012;15:294–297. doi: 10.1038/nn.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sweatt JD. The neuronal MAP kinase cascade: a biochemical signal integration system subserving synaptic plasticity and memory. J Neurochem. 2001;76:1–10. doi: 10.1046/j.1471-4159.2001.00054.x. [DOI] [PubMed] [Google Scholar]
- 46.Casar B, Pinto A, Crespo P. ERK dimers and scaffold proteins: unexpected partners for a forgotten (cytoplasmic) task. Cell Cycle. 2009;8:1007–1013. doi: 10.4161/cc.8.7.8078. [DOI] [PubMed] [Google Scholar]
- 47.Casar B, Arozarena I, Sanz-Moreno V, et al. Ras subcellular localization defines extracellular signal-regulated kinase 1 and 2 substrate specificity through distinct utilization of scaffold proteins. Mol Cell Biol. 2009;29:1338–1353. doi: 10.1128/MCB.01359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shalin SC, Hernandez CM, Dougherty MK, et al. Kinase suppressor of Ras1 compartmentalizes hippocampal signal transduction and subserves synaptic plasticity and memory formation. Neuron. 2006;50:765–779. doi: 10.1016/j.neuron.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 49.Morice C, Nothias F, Konig S, et al. Raf-1 and B-Raf proteins have similar regional distributions but differential subcellular localization in adult rat brain. Eur J Neurosci. 1999;11:1995–2006. doi: 10.1046/j.1460-9568.1999.00609.x. [DOI] [PubMed] [Google Scholar]
- 50.Micheau J, Riedel G. Protein kinases: which one is the memory molecule? Cell Mol Life Sci. 1999;55:534–548. doi: 10.1007/s000180050312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nguyen PV, Woo NH. Regulation of hippocampal synaptic plasticity by cyclic AMP-dependent protein kinases. Prog Neurobiol. 2003;71:401–437. doi: 10.1016/j.pneurobio.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 52.Baudry M, Zhu G, Liu Y, et al. Multiple cellular cascades participate in long-term potentiation and in hippocampus-dependent learning. Brain Res. 2015;1621:73–81. doi: 10.1016/j.brainres.2014.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kandel ER. The molecular biology of memory storage: a dialog between genes and synapses. Biosci Rep. 2001;21:565–611. doi: 10.1023/a:1014775008533. [DOI] [PubMed] [Google Scholar]
- 54.Kandel ER. The molecular biology of memory: cAMP, PKA, CRE, CREB-1, CREB-2, and CPEB. Mol Brain. 2012;5:14. doi: 10.1186/1756-6606-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abel T, Nguyen PV, Barad M, et al. Genetic demonstration of a role for PKA in the late phase of LTP and in hippocampus-based long-term memory. Cell. 1997;88:615–626. doi: 10.1016/s0092-8674(00)81904-2. [DOI] [PubMed] [Google Scholar]
- 56.Hegde AN, Inokuchi K, Pei W, et al. Ubiquitin C-terminal hydrolase is an immediate-early gene essential for long-term facilitation in Aplysia. Cell. 1997;89:115–126. doi: 10.1016/s0092-8674(00)80188-9. [DOI] [PubMed] [Google Scholar]
- 57.Muller U, Carew TJ. Serotonin induces temporally and mechanistically distinct phases of persistent PKA activity in Aplysia sensory neurons. Neuron. 1998;21:1423–1434. doi: 10.1016/s0896-6273(00)80660-1. [DOI] [PubMed] [Google Scholar]
- 58.Bacskai BJ, Hochner B, Mahaut-Smith M, et al. Spatially resolved dynamics of cAMP and protein kinase A subunits in Aplysia sensory neurons. Science. 1993;260:222–226. doi: 10.1126/science.7682336. [DOI] [PubMed] [Google Scholar]
- 59.Kaang BK, Kandel ER, Grant SG. Activation of cAMP-responsive genes by stimuli that produce long-term facilitation in Aplysia sensory neurons. Neuron. 1993;10:427–435. doi: 10.1016/0896-6273(93)90331-k. [DOI] [PubMed] [Google Scholar]
- 60.Dash PK, Hochner B, Kandel ER. Injection of the cAMP-responsive element into the nucleus of Aplysia sensory neurons blocks long-term facilitation. Nature. 1990;345:718–721. doi: 10.1038/345718a0. [DOI] [PubMed] [Google Scholar]
- 61.Pu L, Bao GB, Xu NJ, et al. Hippocampal long-term potentiation is reduced by chronic opiate treatment and can be restored by re-exposure to opiates. J Neurosci. 2002;22:1914–1921. doi: 10.1523/JNEUROSCI.22-05-01914.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bourtchuladze R, Frenguelli B, Blendy J, et al. Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell. 1994;79:59–68. doi: 10.1016/0092-8674(94)90400-6. [DOI] [PubMed] [Google Scholar]
- 63.Bauman AL, Goehring AS, Scott JD. Orchestration of synaptic plasticity through AKAP signaling complexes. Neuropharmacology. 2004;46:299–310. doi: 10.1016/j.neuropharm.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 64.Sanderson JL, Dell'Acqua ML. AKAP signaling complexes in regulation of excitatory synaptic plasticity. Neuroscientist. 2011;17:321–336. doi: 10.1177/1073858410384740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang T, McDonough CB, Abel T. Compartmentalized PKA signaling events are required for synaptic tagging and capture during hippocampal late-phase long-term potentiation. Eur J Cell Biol. 2006;85:635–642. doi: 10.1016/j.ejcb.2006.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guan Z, Giustetto M, Lomvardas S, et al. Integration of long-term-memory-related synaptic plasticity involves bidirectional regulation of gene expression and chromatin structure. Cell. 2002;111:483–493. doi: 10.1016/s0092-8674(02)01074-7. [DOI] [PubMed] [Google Scholar]
- 67.Bartsch D, Ghirardi M, Skehel PA, et al. Aplysia CREB2 represses long-term facilitation: relief of repression converts transient facilitation into long-term functional and structural change. Cell. 1995;83:979–992. doi: 10.1016/0092-8674(95)90213-9. [DOI] [PubMed] [Google Scholar]
- 68.Abel T, Martin KC, Bartsch D, et al. Memory suppressor genes: inhibitory constraints on the storage of long-term memory. Science. 1998;279:338–341. doi: 10.1126/science.279.5349.338. [DOI] [PubMed] [Google Scholar]
- 69.Hai T, Hartman MG. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene. 2001;273:1–11. doi: 10.1016/s0378-1119(01)00551-0. [DOI] [PubMed] [Google Scholar]
- 70.Schoch S, Cibelli G, Magin A, et al. Modular structure of cAMP response element binding protein 2 (CREB2) Neurochem Int. 2001;38:601–608. doi: 10.1016/s0197-0186(00)00127-3. [DOI] [PubMed] [Google Scholar]
- 71.Pasini S, Corona C, Liu J, et al. Specific downregulation of hippocampal ATF4 reveals a necessary role in synaptic plasticity and memory. Cell Rep. 2015;11:183–191. doi: 10.1016/j.celrep.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Soda T, Frank C, Ishizuka K, et al. DISC1-ATF4 transcriptional repression complex: dual regulation of the cAMP-PDE4 cascade by DISC1. Mol Psychiatry. 2013;18:898–908. doi: 10.1038/mp.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roberson ED, English JD, Adams JP, et al. The mitogen-activated protein kinase cascade couples PKA and PKC to cAMP response element binding protein phosphorylation in area CA1 of hippocampus. J Neurosci. 1999;19:4337–4348. doi: 10.1523/JNEUROSCI.19-11-04337.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Martin KC, Michael D, Rose JC, et al. MAP kinase translocates into the nucleus of the presynaptic cell and is required for long-term facilitation in Aplysia. Neuron. 1997;18:899–912. doi: 10.1016/s0896-6273(00)80330-x. [DOI] [PubMed] [Google Scholar]
- 75.Impey S, Obrietan K, Wong ST, et al. Cross talk between ERK and PKA is required for Ca2+ stimulation of CREB-dependent transcription and ERK nuclear translocation. Neuron. 1998;21:869–883. doi: 10.1016/s0896-6273(00)80602-9. [DOI] [PubMed] [Google Scholar]
- 76.Rosenkranz JA, Frick A, Johnston D. Kinase-dependent modification of dendritic excitability after long-term potentiation. J Physiol. 2009;587:115–125. doi: 10.1113/jphysiol.2008.158816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Quevedo J, Vianna MR, Roesler R, et al. Pretraining but not preexposure to the task apparatus prevents the memory impairment induced by blockade of protein synthesis, PKA or MAP kinase in rats. Neurochem Res. 2005;30:61–67. doi: 10.1007/s11064-004-9686-3. [DOI] [PubMed] [Google Scholar]
- 78.Wu GY, Deisseroth K, Tsien RW. Activity-dependent CREB phosphorylation: convergence of a fast, sensitive calmodulin kinase pathway and a slow, less sensitive mitogen-activated protein kinase pathway. Proc Natl Acad Sci U S A. 2001;98:2808–2813. doi: 10.1073/pnas.051634198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wayman GA, Lee YS, Tokumitsu H, et al. Calmodulin-kinases: modulators of neuronal development and plasticity. Neuron. 2008;59:914–931. doi: 10.1016/j.neuron.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kelly MT, Crary JF, Sacktor TC. Regulation of protein kinase Mzeta synthesis by multiple kinases in long-term potentiation. J Neurosci. 2007;27:3439–3444. doi: 10.1523/JNEUROSCI.5612-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Newton AC. Protein kinase C: structure, function, and regulation. J Biol Chem. 1995;270:28495–28498. doi: 10.1074/jbc.270.48.28495. [DOI] [PubMed] [Google Scholar]
- 82.Tsokas P, Ma T, Iyengar R, et al. Mitogen-activated protein kinase upregulates the dendritic translation machinery in long-term potentiation by controlling the mammalian target of rapamycin pathway. J Neurosci. 2007;27:5885–5894. doi: 10.1523/JNEUROSCI.4548-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cammalleri M, Lutjens R, Berton F, et al. Time-restricted role for dendritic activation of the mTOR-p70S6K pathway in the induction of late-phase long-term potentiation in the CA1. Proc Natl Acad Sci U S A. 2003;100:14368–14373. doi: 10.1073/pnas.2336098100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kelleher RJ, 3rd, Govindarajan A, Tonegawa S. Translational regulatory mechanisms in persistent forms of synaptic plasticity. Neuron. 2004;44:59–73. doi: 10.1016/j.neuron.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 85.Hoeffer CA, Klann E. mTOR signaling: at the crossroads of plasticity, memory and disease. Trends Neurosci. 2010;33:67–75. doi: 10.1016/j.tins.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hayer A, Bhalla US. Molecular switches at the synapse emerge from receptor and kinase traffic. PLoS Comput Biol. 2005;1:137–154. doi: 10.1371/journal.pcbi.0010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Smolen P, Baxter DA, Byrne JH. A model of the roles of essential kinases in the induction and expression of late long-term potentiation. Biophys J. 2006;90:2760–2775. doi: 10.1529/biophysj.105.072470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jalil SJ, Sacktor TC, Shouval HZ. Atypical PKCs in memory maintenance: the roles of feedback and redundancy. Learn Mem. 2015;22:344–353. doi: 10.1101/lm.038844.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Heerssen HM, Segal RA. Location, location, location: a spatial view of neurotrophin signal transduction. Trends Neurosci. 2002;25:160–165. doi: 10.1016/s0166-2236(02)02144-6. [DOI] [PubMed] [Google Scholar]
- 90.Dailey L, Ambrosetti D, Mansukhani A, et al. Mechanisms underlying differential responses to FGF signaling. Cytokine Growth Factor Rev. 2005;16:233–247. doi: 10.1016/j.cytogfr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 91.Ramel MC, Hill CS. Spatial regulation of BMP activity. FEBS Lett. 2012;586:1929–1941. doi: 10.1016/j.febslet.2012.02.035. [DOI] [PubMed] [Google Scholar]
- 92.Kopec AM, Carew TJ. Growth factor signaling and memory formation: temporal and spatial integration of a molecular network. Learn Mem. 2013;20:531–539. doi: 10.1101/lm.031377.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cohen S, Levi-Montalcini R, Hamburger V. A nerve growth-stimulating factor isolated from sarcomas 37 and 180. Proc Natl Acad Sci U S A. 1954;40:1014–1018. doi: 10.1073/pnas.40.10.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Levi-Montalcini R, Skaper SD, Dal Toso R, et al. Nerve growth factor: from neurotrophin to neurokine. Trends Neurosci. 1996;19:514–520. doi: 10.1016/S0166-2236(96)10058-8. [DOI] [PubMed] [Google Scholar]
- 95.Edelmann E, Lessmann V, Brigadski T. Pre- and postsynaptic twists in BDNF secretion and action in synaptic plasticity. Neuropharmacology. 2014;76(Pt C):610–627. doi: 10.1016/j.neuropharm.2013.05.043. [DOI] [PubMed] [Google Scholar]
- 96.Lessmann V, Gottmann K, Malcangio M. Neurotrophin secretion: current facts and future prospects. Prog Neurobiol. 2003;69:341–374. doi: 10.1016/s0301-0082(03)00019-4. [DOI] [PubMed] [Google Scholar]
- 97.Tongiorgi E. Activity-dependent expression of brain-derived neurotrophic factor in dendrites: facts and open questions. Neurosci Res. 2008;61:335–346. doi: 10.1016/j.neures.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 98.Conner JM, Lauterborn JC, Yan Q, et al. Distribution of brain-derived neurotrophic factor (BDNF) protein and mRNA in the normal adult rat CNS: evidence for anterograde axonal transport. J Neurosci. 1997;17:2295–2313. doi: 10.1523/JNEUROSCI.17-07-02295.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Smith MA, Zhang LX, Lyons WE, et al. Anterograde transport of endogenous brain-derived neurotrophic factor in hippocampal mossy fibers. Neuroreport. 1997;8:1829–1834. doi: 10.1097/00001756-199705260-00008. [DOI] [PubMed] [Google Scholar]
- 100.Yan Q, Rosenfeld RD, Matheson CR, et al. Expression of brain-derived neurotrophic factor protein in the adult rat central nervous system. Neuroscience. 1997;78:431–448. doi: 10.1016/s0306-4522(96)00613-6. [DOI] [PubMed] [Google Scholar]
- 101.Salio C, Averill S, Priestley JV, et al. Costorage of BDNF and neuropeptides within individual dense-core vesicles in central and peripheral neurons. Dev Neurobiol. 2007;67:326–338. doi: 10.1002/dneu.20358. [DOI] [PubMed] [Google Scholar]
- 102.Dieni S, Matsumoto T, Dekkers M, et al. BDNF and its pro-peptide are stored in presynaptic dense core vesicles in brain neurons. J Cell Biol. 2012;196:775–788. doi: 10.1083/jcb.201201038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Altar CA, Cai N, Bliven T, et al. Anterograde transport of brain-derived neurotrophic factor and its role in the brain. Nature. 1997;389:856–860. doi: 10.1038/39885. [DOI] [PubMed] [Google Scholar]
- 104.Wetmore C, Cao YH, Pettersson RF, et al. Brain-derived neurotrophic factor: subcellular compartmentalization and interneuronal transfer as visualized with anti-peptide antibodies. Proc Natl Acad Sci U S A. 1991;88:9843–9847. doi: 10.1073/pnas.88.21.9843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Aoki C, Wu K, Elste A, et al. Localization of brain-derived neurotrophic factor and TrkB receptors to postsynaptic densities of adult rat cerebral cortex. J Neurosci Res. 2000;59:454–463. doi: 10.1002/(SICI)1097-4547(20000201)59:3<454::AID-JNR21>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 106.Tongiorgi E, Armellin M, Giulianini PG, et al. Brain-derived neurotrophic factor mRNA and protein are targeted to discrete dendritic laminas by events that trigger epileptogenesis. J Neurosci. 2004;24:6842–6852. doi: 10.1523/JNEUROSCI.5471-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.An JJ, Gharami K, Liao GY, et al. Distinct role of long 3' UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134:175–187. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.DiStefano PS, Friedman B, Radziejewski C, et al. The neurotrophins BDNF, NT-3, and NGF display distinct patterns of retrograde axonal transport in peripheral and central neurons. Neuron. 1992;8:983–993. doi: 10.1016/0896-6273(92)90213-w. [DOI] [PubMed] [Google Scholar]
- 109.Zweifel LS, Kuruvilla R, Ginty DD. Functions and mechanisms of retrograde neurotrophin signalling. Nat Rev Neurosci. 2005;6:615–625. doi: 10.1038/nrn1727. [DOI] [PubMed] [Google Scholar]
- 110.Baulida J, Kraus MH, Alimandi M, et al. All ErbB receptors other than the epidermal growth factor receptor are endocytosis impaired. Journal of Biological Chemistry. 1996;271:5251–5257. doi: 10.1074/jbc.271.9.5251. [DOI] [PubMed] [Google Scholar]
- 111.Lau MM, Stewart CE, Liu Z, et al. Loss of the imprinted IGF2/cation-independent mannose 6-phosphate receptor results in fetal overgrowth and perinatal lethality. Genes Dev. 1994;8:2953–2963. doi: 10.1101/gad.8.24.2953. [DOI] [PubMed] [Google Scholar]
- 112.Hawkes C, Kar S. The insulin-like growth factor-II/mannose-6-phosphate receptor: structure, distribution and function in the central nervous system. Brain Res Brain Res Rev. 2004;44:117–140. doi: 10.1016/j.brainresrev.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 113.Bao J, Alroy I, Waterman H, et al. Threonine phosphorylation diverts internalized epidermal growth factor receptors from a degradative pathway to the recycling endosome. Journal of Biological Chemistry. 2000;275:26178–26186. doi: 10.1074/jbc.M002367200. [DOI] [PubMed] [Google Scholar]
- 114.Lu B. BDNF and activity-dependent synaptic modulation. Learn Mem. 2003;10:86–98. doi: 10.1101/lm.54603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lessmann V, Brigadski T. Mechanisms, locations, and kinetics of synaptic BDNF secretion: an update. Neurosci Res. 2009;65:11–22. doi: 10.1016/j.neures.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 116.Zafra F, Hengerer B, Leibrock J, et al. Activity dependent regulation of BDNF and NGF mRNAs in the rat hippocampus is mediated by non-NMDA glutamate receptors. Embo j. 1990;9:3545–3550. doi: 10.1002/j.1460-2075.1990.tb07564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Blochl A, Thoenen H. Characterization of nerve growth factor (NGF) release from hippocampal neurons: evidence for a constitutive and an unconventional sodium-dependent regulated pathway. Eur J Neurosci. 1995;7:1220–1228. doi: 10.1111/j.1460-9568.1995.tb01112.x. [DOI] [PubMed] [Google Scholar]
- 118.Blochl A, Thoenen H. Localization of cellular storage compartments and sites of constitutive and activity-dependent release of nerve growth factor (NGF) in primary cultures of hippocampal neurons. Mol Cell Neurosci. 1996;7:173–190. doi: 10.1006/mcne.1996.0014. [DOI] [PubMed] [Google Scholar]
- 119.Goodman LJ, Valverde J, Lim F, et al. Regulated release and polarized localization of brain-derived neurotrophic factor in hippocampal neurons. Mol Cell Neurosci. 1996;7:222–238. doi: 10.1006/mcne.1996.0017. [DOI] [PubMed] [Google Scholar]
- 120.Boukhaddaoui H, Sieso V, Scamps F, et al. An activity-dependent neurotrophin-3 autocrine loop regulates the phenotype of developing hippocampal pyramidal neurons before target contact. J Neurosci. 2001;21:8789–8797. doi: 10.1523/JNEUROSCI.21-22-08789.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Patterson SL, Grover LM, Schwartzkroin PA, et al. Neurotrophin expression in rat hippocampal slices: a stimulus paradigm inducing LTP in CA1 evokes increases in BDNF and NT-3 mRNAs. Neuron. 1992;9:1081–1088. doi: 10.1016/0896-6273(92)90067-n. [DOI] [PubMed] [Google Scholar]
- 122.Canossa M, Griesbeck O, Berninger B, et al. Neurotrophin release by neurotrophins: implications for activity-dependent neuronal plasticity. Proc Natl Acad Sci U S A. 1997;94:13279–13286. doi: 10.1073/pnas.94.24.13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kruttgen A, Moller JC, Heymach JV, Jr, et al. Neurotrophins induce release of neurotrophins by the regulated secretory pathway. Proc Natl Acad Sci U S A. 1998;95:9614–9619. doi: 10.1073/pnas.95.16.9614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bambah-Mukku D, Travaglia A, Chen DY, et al. A Positive Autoregulatory BDNF Feedback Loop via C/EBPbeta Mediates Hippocampal Memory Consolidation. J Neurosci. 2014;34:12547–12559. doi: 10.1523/JNEUROSCI.0324-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kim HG, Wang T, Olafsson P, et al. Neurotrophin 3 potentiates neuronal activity and inhibits gamma-aminobutyratergic synaptic transmission in cortical neurons. Proc Natl Acad Sci U S A. 1994;91:12341–12345. doi: 10.1073/pnas.91.25.12341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Carmignoto G, Pizzorusso T, Tia S, et al. Brain-derived neurotrophic factor and nerve growth factor potentiate excitatory synaptic transmission in the rat visual cortex. J Physiol. 1997;498(Pt 1):153–164. doi: 10.1113/jphysiol.1997.sp021848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lessmann V, Gottmann K, Heumann R. BDNF and NT-4/5 enhance glutamatergic synaptic transmission in cultured hippocampal neurones. Neuroreport. 1994;6:21–25. doi: 10.1097/00001756-199412300-00007. [DOI] [PubMed] [Google Scholar]
- 128.Kang H, Schuman EM. Long-lasting neurotrophin-induced enhancement of synaptic transmission in the adult hippocampus. Science. 1995;267:1658–1662. doi: 10.1126/science.7886457. [DOI] [PubMed] [Google Scholar]
- 129.Levine ES, Crozier RA, Black IB, et al. Brain-derived neurotrophic factor modulates hippocampal synaptic transmission by increasing N-methyl-D-aspartic acid receptor activity. Proc Natl Acad Sci U S A. 1998;95:10235–10239. doi: 10.1073/pnas.95.17.10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Levine ES, Dreyfus CF, Black IB, et al. Brain-derived neurotrophic factor rapidly enhances synaptic transmission in hippocampal neurons via postsynaptic tyrosine kinase receptors. Proc Natl Acad Sci U S A. 1995;92:8074–8077. doi: 10.1073/pnas.92.17.8074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tanaka T, Saito H, Matsuki N. Inhibition of GABAA synaptic responses by brain-derived neurotrophic factor (BDNF) in rat hippocampus. J Neurosci. 1997;17:2959–2966. doi: 10.1523/JNEUROSCI.17-09-02959.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chen DY, Bambah-Mukku D, Pollonini G, et al. Glucocorticoid receptors recruit the CaMKIIalpha-BDNF-CREB pathways to mediate memory consolidation. Nat Neurosci. 2012;15:1707–1714. doi: 10.1038/nn.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kanhema T, Dagestad G, Panja D, et al. Dual regulation of translation initiation and peptide chain elongation during BDNF-induced LTP in vivo: evidence for compartment-specific translation control. J Neurochem. 2006;99:1328–1337. doi: 10.1111/j.1471-4159.2006.04158.x. [DOI] [PubMed] [Google Scholar]
- 134.Chen DY, Stern SA, Garcia-Osta A, et al. A critical role for IGF-II in memory consolidation and enhancement. Nature. 2011;469:491–497. doi: 10.1038/nature09667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Woolf NJ, Milov AM, Schweitzer ES, et al. Elevation of nerve growth factor and antisense knockdown of TrkA receptor during contextual memory consolidation. J Neurosci. 2001;21:1047–1055. doi: 10.1523/JNEUROSCI.21-03-01047.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ageta H, Ikegami S, Miura M, et al. Activin plays a key role in the maintenance of long-term memory and late-LTP. Learning & Memory. 2010;17:176–185. doi: 10.1101/lm.16659010. [DOI] [PubMed] [Google Scholar]
- 137.Bekinschtein P, Cammarota M, Igaz LM, et al. Persistence of long-term memory storage requires a late protein synthesis- and BDNF- dependent phase in the hippocampus. Neuron. 2007;53:261–277. doi: 10.1016/j.neuron.2006.11.025. [DOI] [PubMed] [Google Scholar]
- 138.Bekinschtein P, Cammarota M, Katche C, et al. BDNF is essential to promote persistence of long-term memory storage. Proc Natl Acad Sci U S A. 2008;105:2711–2716. doi: 10.1073/pnas.0711863105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Matus A. Actin-based plasticity in dendritic spines. Science. 2000;290:754–758. doi: 10.1126/science.290.5492.754. [DOI] [PubMed] [Google Scholar]
- 140.Jaworski J, Kapitein LC, Gouveia SM, et al. Dynamic microtubules regulate dendritic spine morphology and synaptic plasticity. Neuron. 2009;61:85–100. doi: 10.1016/j.neuron.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 141.Tsvetkov A, Popov S. Analysis of tubulin transport in nerve processes. Methods Mol Med. 2007;137:161–173. doi: 10.1007/978-1-59745-442-1_11. [DOI] [PubMed] [Google Scholar]
- 142.Hirokawa N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science. 1998;279:519–526. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- 143.Goldstein LS, Philp AV. The road less traveled: emerging principles of kinesin motor utilization. Annu Rev Cell Dev Biol. 1999;15:141–183. doi: 10.1146/annurev.cellbio.15.1.141. [DOI] [PubMed] [Google Scholar]
- 144.Woehlke G, Schliwa M. Walking on two heads: the many talents of kinesin. Nat Rev Mol Cell Biol. 2000;1:50–58. doi: 10.1038/35036069. [DOI] [PubMed] [Google Scholar]
- 145.Merriam EB, Millette M, Lumbard DC, et al. Synaptic regulation of microtubule dynamics in dendritic spines by calcium, F-actin, and drebrin. J Neurosci. 2013;33:16471–16482. doi: 10.1523/JNEUROSCI.0661-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Puthanveettil SV, Monje FJ, Miniaci MC, et al. A new component in synaptic plasticity: upregulation of kinesin in the neurons of the gill-withdrawal reflex. Cell. 2008;135:960–973. doi: 10.1016/j.cell.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Pinsker HM, Hening WA, Carew TJ, et al. Long-term sensitization of a defensive withdrawal reflex in Aplysia. Science. 1973;182:1039–1042. doi: 10.1126/science.182.4116.1039. [DOI] [PubMed] [Google Scholar]
- 148.Marinesco S, Carew TJ. Serotonin release evoked by tail nerve stimulation in the CNS of aplysia: characterization and relationship to heterosynaptic plasticity. J Neurosci. 2002;22:2299–2312. doi: 10.1523/JNEUROSCI.22-06-02299.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Philips GT, Sherff CM, Menges SA, et al. The tail-elicited tail withdrawal reflex of Aplysia is mediated centrally at tail sensory-motor synapses and exhibits sensitization across multiple temporal domains. Learn Mem. 2011;18:272–282. doi: 10.1101/lm.2125311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hawkins RD, Kandel ER, Bailey CH. Molecular mechanisms of memory storage in Aplysia. Biol Bull. 2006;210:174–191. doi: 10.2307/4134556. [DOI] [PubMed] [Google Scholar]
- 151.Sutton MA, Ide J, Masters SE, et al. Interaction between amount and pattern of training in the induction of intermediate- and long-term memory for sensitization in aplysia. Learn Mem. 2002;9:29–40. doi: 10.1101/lm.44802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mauelshagen J, Sherff CM, Carew TJ. Differential induction of long-term synaptic facilitation by spaced and massed applications of serotonin at sensory neuron synapses of Aplysia californica. Learn Mem. 1998;5:246–256. [PMC free article] [PubMed] [Google Scholar]
- 153.Philips GT, Kopec AM, Carew TJ. Pattern and predictability in memory formation: From molecular mechanisms to clinical relevance. Neurobiol Learn Mem. 2013 doi: 10.1016/j.nlm.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Shobe J, Philips GT, Carew TJ. Transforming growth factor beta recruits persistent MAPK signaling to regulate long-term memory consolidation in Aplysia californica. Learn Mem. 2016;23:182–188. doi: 10.1101/lm.040915.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sharma SK, Sherff CM, Stough S, et al. A tropomyosin-related kinase B ligand is required for ERK activation, long-term synaptic facilitation, and long-term memory in aplysia. Proc Natl Acad Sci U S A. 2006;103:14206–14210. doi: 10.1073/pnas.0603412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kassabov SR, Choi YB, Karl KA, et al. A Single Aplysia Neurotrophin Mediates Synaptic Facilitation via Differentially Processed Isoforms. Cell Rep. 2013;3:1213–1227. doi: 10.1016/j.celrep.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chin J, Angers A, Cleary LJ, et al. TGF-beta 1 in Aplysia: Role in long-term changes in the excitability of sensory neurons and distribution of T beta R-II-like immunoreactivity. Learning & Memory. 1999;6:317–330. [PMC free article] [PubMed] [Google Scholar]
- 158.Chin J, Angers A, Cleary LJ, et al. Transforming growth factor beta1 alters synapsin distribution and modulates synaptic depression in Aplysia. J Neurosci. 2002;22:RC220. doi: 10.1523/JNEUROSCI.22-09-j0004.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chin J, Liu RY, Cleary LJ, et al. TGF-beta1-induced long-term changes in neuronal excitability in aplysia sensory neurons depend on MAPK. J Neurophysiol. 2006;95:3286–3290. doi: 10.1152/jn.00770.2005. [DOI] [PubMed] [Google Scholar]
- 160.Zhang F, Endo S, Cleary LJ, et al. Role of transforming growth factor-beta in long-term synaptic facilitation in Aplysia. Science. 1997;275:1318–1320. doi: 10.1126/science.275.5304.1318. [DOI] [PubMed] [Google Scholar]
- 161.Kopec AM, Philips GT, Carew TJ. Distinct Growth Factor Families Are Recruited in Unique Spatiotemporal Domains during Long-Term Memory Formation in Aplysia californica. Neuron. 2015;86:1228–1239. doi: 10.1016/j.neuron.2015.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yim SJ, Lee YS, Lee JA, et al. Regulation of ApC/EBP mRNA by the Aplysia AU-rich element-binding protein, ApELAV, and its effects on 5-hydroxytryptamine-induced long-term facilitation. J Neurochem. 2006;98:420–429. doi: 10.1111/j.1471-4159.2006.03887.x. [DOI] [PubMed] [Google Scholar]
- 163.Glisovic T, Bachorik JL, Yong J, et al. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sanchez-Diaz P, Penalva LO. Post-transcription meets post-genomic: the saga of RNA binding proteins in a new era. RNA Biol. 2006;3:101–109. doi: 10.4161/rna.3.3.3373. [DOI] [PubMed] [Google Scholar]
- 165.Wyers F, Rougemaille M, Badis G, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 166.Doller A, Schlepckow K, Schwalbe H, et al. Tandem phosphorylation of serines 221 and 318 by protein kinase Cdelta coordinates mRNA binding and nucleocytoplasmic shuttling of HuR. Mol Cell Biol. 2010;30:1397–1410. doi: 10.1128/MCB.01373-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lafarga V, Cuadrado A, Lopez de Silanes I, et al. p38 Mitogen-activated protein kinase- and HuR-dependent stabilization of p21(Cip1) mRNA mediates the G(1)/S checkpoint. Mol Cell Biol. 2009;29:4341–4351. doi: 10.1128/MCB.00210-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wang W, Fan J, Yang X, et al. AMP-activated kinase regulates cytoplasmic HuR. Mol Cell Biol. 2002;22:3425–3436. doi: 10.1128/MCB.22.10.3425-3436.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Bakken TE, Miller JA, Ding SL, et al. A comprehensive transcriptional map of primate brain development. Nature. 2016;535:367–375. doi: 10.1038/nature18637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yuzwa SA, Yang G, Borrett MJ, et al. Proneurogenic Ligands Defined by Modeling Developing Cortex Growth Factor Communication Networks. Neuron. 2016 doi: 10.1016/j.neuron.2016.07.037. [DOI] [PubMed] [Google Scholar]
- 171.Pollak DD, Minh BQ, Cicvaric A, et al. A novel fibroblast growth factor receptor family member promotes neuronal outgrowth and synaptic plasticity in aplysia. Amino Acids. 2014;46:2477–2488. doi: 10.1007/s00726-014-1803-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Pu L, Kopec AM, Boyle HD, et al. A novel cysteine-rich neurotrophic factor in Aplysia facilitates growth, MAPK activation, and long-term synaptic facilitation. Learn Mem. 2014;21:215–222. doi: 10.1101/lm.033662.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Brunet JF, Shapiro E, Foster SA, et al. Identification of a peptide specific for Aplysia sensory neurons by PCR-based differential screening. Science. 1991;252:856–859. doi: 10.1126/science.1840700. [DOI] [PubMed] [Google Scholar]
- 174.Phillips HS, Hains JM, Armanini M, et al. BDNF mRNA is decreased in the hippocampus of individuals with Alzheimer's disease. Neuron. 1991;7:695–702. doi: 10.1016/0896-6273(91)90273-3. [DOI] [PubMed] [Google Scholar]
- 175.Peress NS, Perillo E. Differential expression of TGF-beta 1, 2 and 3 isotypes in Alzheimer's disease: a comparative immunohistochemical study with cerebral infarction, aged human and mouse control brains. J Neuropathol Exp Neurol. 1995;54:802–811. doi: 10.1097/00005072-199511000-00007. [DOI] [PubMed] [Google Scholar]
- 176.Lim YY, Villemagne VL, Laws SM, et al. APOE and BDNF polymorphisms moderate amyloid beta-related cognitive decline in preclinical Alzheimer's disease. Mol Psychiatry. 2014 doi: 10.1038/mp.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]