Abstract
Polycomb proteins assemble to form complexes with important roles in epigenetic regulation. The Polycomb Repressive Complex 2 (PRC2) modulates the di- and tri-methylation of lysine 27 on histone H3, each of which are associated with gene repression. Although three subunits, EZH1/2, SUZ12, and EED, form the catalytic core of PRC2, a wider group of proteins associate with low stoichiometry. This raises the question of whether dynamic variation of the PRC2 interactome results in alternative forms of the complex during differentiation. Here we compared the physical interactions of PRC2 in undifferentiated and differentiated states of NTERA2 pluripotent embryonic carcinoma cells. Label-free quantitative proteomics was used to assess endogenous immunoprecipitation of the EZH2 and SUZ12 subunits of PRC2. A high stringency data set reflecting the endogenous state of PRC2 was produced that included all previously reported core and associated PRC2 components, and several novel interacting proteins. Comparison of the interactomes obtained in undifferentiated and differentiated cells revealed candidate proteins that were enriched in complexes isolated from one of the two states. For example, SALL4 and ZNF281 associate with PRC2 in pluripotent cells, whereas PCL1 and SMAD3 preferentially associate with PRC2 in differentiating cells. Analysis of the mRNA and protein levels of these factors revealed that their association with PRC2 correlated with their cell state-specific expression. Taken together, we propose that dynamic changes to the PRC2 interactome during differentiation may contribute to directing its activity during cell fate transitions.
Proteins engage in many types of physical interaction in order to carry out the various functions of a living cell. These range from the high affinity interactions typical of protein complexes that are stable over time, to short-term interactions that occur, for example, between enzymes and substrates or between signaling proteins in regulatory pathways. Multiprotein complexes assemble and organize the individual catalytic activities required to carry out a biological process in a single place or time. Valuable insights into the organization of protein networks and the biology of the cell have been achieved by high throughput protein interaction mapping screens (1). However, dynamic changes in these networks, over time or in response to a cellular change, are more difficult to study because they require technologies such as affinity purification-quantitative mass spectrometry that are difficult to implement on a large scale. Technical developments in recent years such as high resolution high mass accuracy mass spectrometry have significantly increased the power of quantitative proteomics experiments (2). This factor, combined with software improvements, means that the researcher can have high confidence that the correct analytes (i.e. tryptic peptides accurately mapped to parent proteins) are being identified and accurately quantified.
The Polycomb proteins are a priority for such analysis because they act within protein assemblies and because they are biologically and clinically important. Polycomb genes were originally identified in screens for developmental defects in Drosophila, and the corresponding proteins form two separate families of multiprotein complex: PRC1 (Polycomb Repressor Complex 11) and PRC2 (3). Although a number of discrete PRC1 complexes have been described (all sharing a heterodimeric PCGF-RING core), the composition of PRC2 is less clearly understood. PRC2 plays a critical role in many cell phenotypes such as fate determination and stem cell identity (4). It generates and maintains repressive epigenetic mark on target genes through methylation of lysine 27 of histone H3 (H3Lys27me3). However, PRC2 has additional functions, including chromatin compaction and the reading of epigenetic marks (3, 5). The “core” PRC2 complex (i.e. the minimum showing activity in vitro) contains EZH2 (or its close ortholog EZH1), SUZ12, EED, and either RBBP4 or RBBP7 (6). The SET domain of EZH2 catalyzes the di-, and tri-methylation of H3K27, but this activity is considerably enhanced by the presence of SUZ12 and EED (7). Additional proteins (e.g. JARID2, AEBP2, PCL1, PCL2, PCL3) are often present in the complex, and although not absolutely required in vitro, they can enhance the function of the complex (8, 9).
PRC2 is therefore a classic example of a multifunctional protein complex whose activities need to be regulated over time (e.g. during differentiation or development) and at specific locations (e.g. at specific genomic domains). Such regulation may operate at the level of the protein complex itself, for example via structural changes or changes in catalytic activity induced by post-translational modification (10). Alternatively, regulation may arise from the addition or loss of interacting partners that modify the activity of the complex, or that act as recruiting factors to direct the location of the complex (11). Although protein complexes are often considered to be stable entities, there is accumulating evidence that in many cases their composition may vary dynamically (12). For example, extensive protein interactions changes take place over the course of the splicing process (13). Similarly, proteasome activity can be modulated by the presence of activating subcomplexes or tissue-specific subunit exchange (14, 15). Other workers compared interactome data with transcriptome data obtained over the yeast cell cycle and proposed that the activity of many protein complexes may be controlled through regulated expression of individual subunits (16).
Here we used a combination of immunoprecipitation and label-free quantitative mass spectrometry to study the dynamic interactome of PRC2 in a cultured cell model of differentiation. NTERA2(NT2) is a pluripotent embryonic carcinoma cell line derived from a metastatic tumor that can be induced to develop neuron-like properties upon treatment with retinoic acid (17). These cells are commonly used in studies of epigenetic change including regulation by Polycomb complexes (18–20). We favored immunoprecipitation of both SUZ12 and EZH2 subunits in order to characterize the behavior of PRC2 in a manner as close to the native state as possible, and particularly to avoid artifacts arising from exogenous expression affinity-tagged forms of PRC2 over a time course. Using this approach, we found that although all core PRC2 subunits were present in the complex in both cell states, individual interactors that associated preferentially in either the undifferentiated or differentiated state were observed. Quantitative RT-PCR and Western blot analyses showed that these associations correlated with mRNA expression of the corresponding genes. This suggests that binding of accessory proteins to the parent complex based on their differential expression is a potential regulatory mechanism controlling PRC2 activity.
EXPERIMENTAL PROCEDURES
Cell Culture and Differentiation of NTERA2 Cells
NT2/D1 cells (ATCC, CRL-1973) were cultured were cultured in 92 mm tissue culture dishes Nunclon (Fisher Scientific) in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% (v/v) Fetal Bovine Serum (Hyclone), 100U/ml penicillin and 100U/ml streptomycin (Gibco). Cells were passaged by trypsinizing with 0.25% Trypsin-EDTA (Invitrogen) and plated at a ratio of 1:6. To induce neuronal differentiation, 10 μm all-trans retinoic acid (RA)(Sigma) was added to media once cells reached a density of ∼50%. During the 8-day differentiation time course, media was changed every 2–3 days. HEK293T cells were grown in DMEM medium supplemented with 10% (v/v) FBS (Hyclone), 100 U ml-1 penicillin, and 100 U ml-1 streptomycin (Gibco).
Isolation of Nuclei
Harvested NT2 cells were washed in PBS and resuspended in Lysis buffer (25 mm Tris·HCl pH 7.6, 150 mm NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS, 2 μg/ml Aprotinin, 1 μg/ml, Leupeptin, 10 mm PMSF). The lysates were incubated for 15 min on ice and cell membranes disrupted mechanically by syringing 5 times with 21G narrow gauge needle and sonicating 3 Å∼ for 2 s at high power. Lysates were incubated on ice for another 15 min and cleared by centrifugation at 14,000 rpm 4 °C 30 min. To harvest the nuclear fraction, lysates were resuspended in an equal volume of Nuclear Buffer (20 mm HEPES pH 7.9, 0.2 mm EDTA, 1.5 mm MgCl2, 20% glycerol, 420 mm NaCl, 2 μg/ml Aprotinin, 1 μg/ml Leupeptin, 10 mm PMSF) and dounced 20 times with tight pestle type B. Lysates were incubated for 45 min rotating to dissociated chromatin-bound proteins and precleared by centrifugation at 14,000 rpm 4°C for 30 min.
Immunoprecipitation
Immunoprecipitations (IPs) were performed on nuclear protein lysates prepared in IP buffer (150 mm NaCl, 50 mm TRIS pH 8.0, 1 mm EDTA, 1% Nonidet P-40, 1 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mm PMSF). 10 μg of antibody was coupled to 50 μl packed Protein A beads (Sigma P9424) by incubation in 1 ml PBS (0.1% Tween-20) at 4 °C rotating overnight. Beads were collected by centrifugation at 1700 × g for 3 min and washed twice in 1 ml 0.2 m sodium borate pH 9.0. Antibodies were then crosslinked to beads by incubation in 1 ml 0.2 m sodium borate pH 9.0 (20 mm dimethyl pimelimidate dihydrochloride) at room temperature rotating for 30 min. The reaction was terminated by washing the beads once in 1 ml 0. 2 m ethanolamine pH 8.0 and incubating for 2 h at room temperature rotating in 1 ml 0.2 m ethanolamine pH 8.0. Beads were washed twice in Buffer C100 (20 mm HEPES pH 7.6, 0.2 mm EDTA, 1.5 mm MgCl2, 100 mm KCl, 0.5% Nonidet P-40, 20% glycerol) and blocked for 1 h min 4 °C rotating in Buffer C100 with 0.1 mg/ml insulin (Sigma, I9278), 0.2 mg/ml chicken egg albumin (Sigma A5503), 0.1% (v/v) fish skin gelatin (Sigma G7041). Antibody-crosslinked beads were incubated with nuclear lysates, in the presence of 250 U/ml Benzonase nuclease, at 4 °C rotating overnight and washed 5 × 5 min in Buffer C100 with 0.02% Nonidet P-40. After the final wash, beads destined for immunoblotting were resuspended in 50 μl 2× SDS sample buffer. Immunoprecipitated material was eluted by boiling for 5 min with shaking and associated proteins were separated by SDS-PAGE and analyzed by immunoblotting. Beads destined for mass spectrometry analysis were washed once in IP buffer containing 0.02% Nonidet P-40 followed by one wash in IP buffer with no detergent. Expression of FLAG-tagged proteins and immunoprecipitation was as described in Brien and coworkers (5). Briefly, FLAG-tagged proteins were performed using M2 anti-FLAG agarose (Sigma) overnight at 4 °C. Elution was performed at 4 °C using 250 μg/ml of 3× FLAG peptide (Sigma) in 0.05% (v/v) Nonidet P-40 with horizontal shaking.
In-solution Trypsin Digest and Mass Spectrometry
Proteins were treated with trypsin as described (21). Peptide samples were introduced into a Q Exactive mass spectrometer (Thermo Scientific), which was connected to an Ultimate 3000 RSLCnano chromatography system (Dionex) coupled to a packed C18 fused silica emitter (75 μm I.D.) Parent ion spectra (MS1) were measured at resolution 70,000, AGC target 3e6. Tandem mass spectra (MS2; up to eight scans per duty cycle) were obtained at resolution 17,500, AGC target 5e4, collision energy of 25.
Data Analysis
Data were processed using MaxQuant version 1.3.0.547 (2) using the human UniProt database (release 2013_12; 67,911 entries). The following search parameters were used: Fixed Mod: cysteine carbamidomethylation; Variable Mods: methionine oxidation; Trypsin/P digest enzyme; Missed Cleavage: 1; Precursor mass tolerances 6 ppm; Fragment ion mass tolerances 20 ppm; Peptide FDR 1%; Protein FDR 1%. “Label-Free Quantitation; LFQ,” “iBAQ,” and “Match Between Run” settings were selected. Reverse hits and contaminants that predominantly bound IgG beads (keratins, ribosomal, splicing proteins) were filtered out and not considered further. A list of removed contaminants is provided in supplemental Table S4. Features used to identify all mass spectra are available in supplemental Table S5. Raw data, including results files and software needed for viewing spectra is provided via the PRIDE database (22).
Real-time Quantitative PCR
Extracted RNA was used to generate cDNA by reverse transcriptase PCR using the TaqMan Reverse Transcription kit (Applied Biosytems). Relative mRNA expression levels were determined using the SYBR Green I detection chemistry on LightCycler 480II Real-Time PCR System (Roche). The ribosomal constituent RPO was used as normalizing gene. The primers used are listed in supplemental Table S1.
Western Blotting Analysis
Western blotting: Cells were prepared in RIPA buffer (25 mm Tris·HCl pH 7.6, 150 mm NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS, 2 μg/ml Aprotinin, 1 μg/ml Leupeptin, 10 mm PMSF). Lysates were incubated for 15 min on ice and cell membranes disrupted mechanically by syringing 5 times with 23G narrow gauge needle and sonicating 3 × 2 s at high power. Lysates were incubated on ice for another 15 min and lysates precleared by centrifugation at 14,000 rpm 4 °C 30 min to remove cellular debris. Immunoblotting was performed using the antibodies and conditions listed in supplemental Table S1.
Experimental Design and Statistical Rationale
In each of the two IP experiments (-RA, +RA), three biological replicates were compared. This was the case for both EZH2 and SUZ12 experiments, i.e. SUZ12-RA, SUZ12+RA, EZH2-RA, EZH2+RA. The Perseus program (http://coxdocs.org/doku.php?id=perseus:start) was used to carry out statistics. Briefly, the LFQ values were transformed (log2) and missing values imputed to a normal distribution (width = 0.3; shift = 1.8), and a two-tailed t test applied with correction for multiple testing (Benjamini). Volcano plots were constructed using the permutation-based approach of Tucher and coworkers (23), to implement an FDR of 0.05 as described in (24).
RESULTS
A Physical Interaction Screen for PRC2 Under Endogenous Conditions
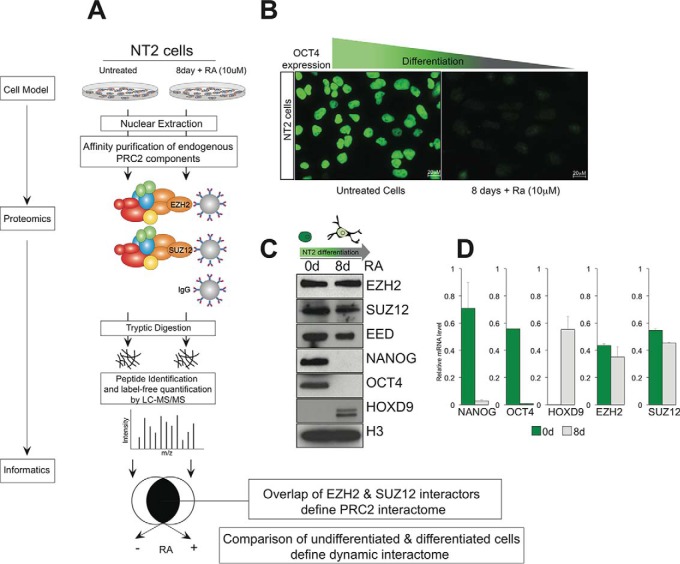
We first optimized a protocol for immunoprecipitation of the EZH2 and SUZ12 components of PRC2 from nuclear lysates. The proteins were trypsinized on the beads, and following clean-up, the resulting peptides were analyzed using Orbitrap mass spectrometry (Fig. 1A). We reasoned that the set of proteins that interact with both subunits (but not with an IgG control) would represent a high quality interactome whose composition we could compare between undifferentiated and differentiated NT2 cells. In this model of differentiation, the levels of pluripotency factors such as OCT4 and NANOG drop significantly upon addition of retinoic acid (in agreement with other reports), (20) whereas differentiation factors such as HOXD9 increased in expression (Fig. 1B, 1C). These protein level observations strongly correlate with mRNA levels measured for the corresponding genes (Fig. 1D). The levels of core PRC2 components EZH2, SUZ12 and EED were maintained throughout the 8-day experiment. Because the activity and genomic location of PRC2 varies over the NT2 differentiation cycle (18), we wondered if we could identify factors that bound PRC2 differentially in retinoic acid-treated and -untreated cells. Such dynamic interactors are of interest because they could potentially facilitate localization of PRC2 at particular genomic loci or influence its epigenetic activity.
Fig. 1.
Strategy for defining dynamic protein interactome of an epigenetic complex in different cell states. A, PRC2 was immunoprecipitated via the EZH2 and SUZ12 subunits from nuclear extracts of NT2 cells in undifferentiated and differentiated states. Proteins were trypsinized on beads, and the resulting peptides identified and quantified using Orbitrap mass spectrometry. The EZH2 and SUZ12 interactomes were combined to generate a high stringency PRC2 interactome, and the set of physically interacting proteins were compared across both cell states. B, Expression of the pluripotency factor OCT4 was monitored using immunofluorescence in NT2 cells before and after addition of retinoic acid. C, Western blot analysis of the same cells shows stable expression of core PRC2 components EZH2, SUZ12 and EED, and histone H3, reduced expression of pluripotency markers OCT4 and NANOG, and increased expression of the differentiation marker HOXD9, in retinoic acid treated cells. D, mRNA levels measured using qPCR correlate with the protein levels observed in panel C.
The PRC2 Interactome in Undifferentiated NT2 Cells
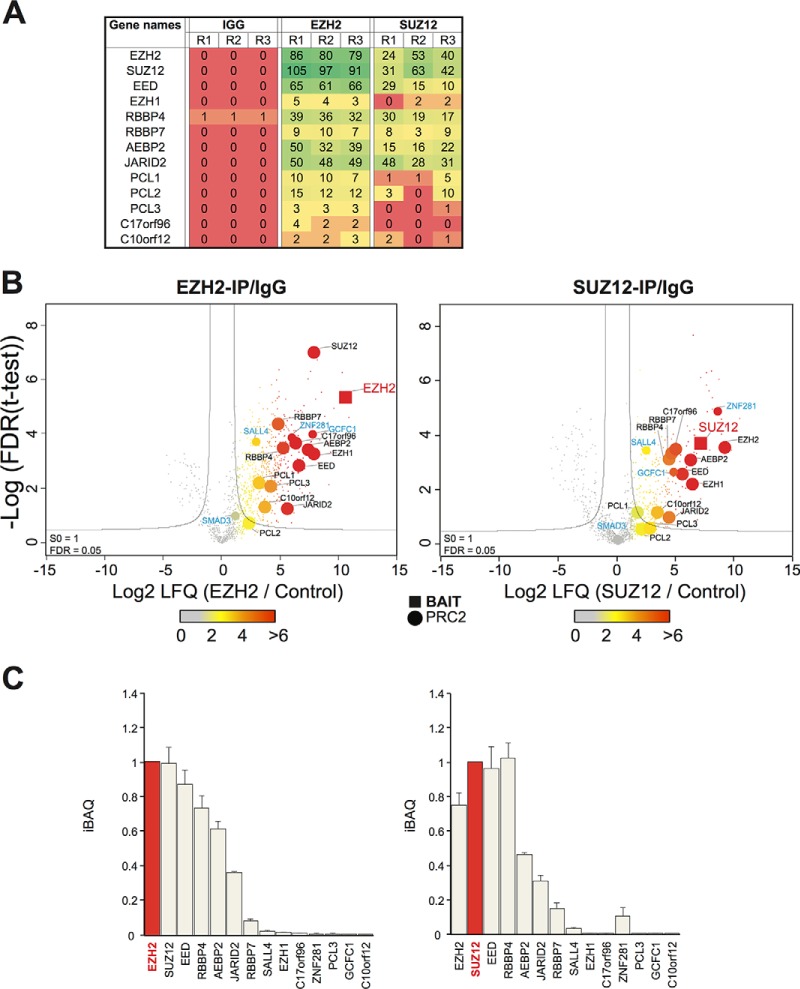
All “core” (EZH2, SUZ12, EED, RBBP4/7) and “associated” (AEBP2, JARID2, PCL1, PCL2, PCL3) PRC2 proteins, as described by Vizan and coworkers (8), were successfully immunoprecipitated and identified by LCMS with no detectable background in IgG immunoprecipitated material apart from modest levels of RBBP4 (Fig. 2A, supplemental Table S2, S3). Volcano plots were used to assess replicate IP experiments (three independent experiments) by plotting the enrichment of each detected protein (relative to an irrelevant antibody control) against the significance of that fold enrichment (t test p value). False Discovery Rate (FDR) statistics can be imposed on these results to estimate the levels of false positive assignments (here 0.05). Although there is a background of proteins enriched in the IgG control purification, in total, 366 candidate EZH2-interacting proteins, and 191 candidate SUZ12-interacting proteins, passed the FDR cut-offs. In order to increase the stringency further, we focused only on proteins that were reliably detected in both IP experiments, generating a total of 136 proteins (supplemental Table S2).
Fig. 2.
A physical interaction screen of PRC2 interactors in undifferentiated NT2 cells. A, Spectral counts (the frequency of identification quality MS2 events per 60min Orbitrap LCMS run) are plotted for control, EZH2 and SUZ12 immunoprecipitation experiments carried out in triplicate (R1, R2, R3). B, Volcano plot projects differences in t test scores (between specific and control immunoprecipitations), against log-transformed t test p values for the EZH2 and SUZ12 experiments. The data is based on the MaxQuant LFQ score. Bait proteins are labeled in red, known PRC2 interactors in black, and newly reported candidate interactors in blue. C, LFQ scores were adjusted using the iBAQ algorithm (scaled to the Bait) to plot relative stoichiometry data for core and previously-described PRC2 components, as well as for candidate PRC2 interactors identified in this study.
For both EZH2 and SUZ12 immunoprecipitations, the cognate proteins were among the most significant scoring (i.e. top right quadrant of the volcano plots; Fig. 2B), whereas all core PRC2 components and associated PRC2 interactors were within the high stringency boundary. These include AEBP2 and JARID2 and two incompletely characterized proteins (C10orf12 and C17orf96) that were reported as PRC2 interactors recently (25). Three other PRC2- associated proteins (referred to as Polycomb-like or PCL proteins) have been described: PCL1–3 (also known as PHF1, MTF2, and PHF19 respectively). These three proteins have been proposed to associate with core PRC2 in a mutually exclusive manner. All three scored as significant in the EZH2 immunoprecipitation, but only PCL3 met the high stringency FDR criteria for the SUZ12 immunoprecipitation, perhaps indicating that the PCL3-PRC2 association is stronger than that for PCL1 or PCL2, or alternatively, reflecting some steric effect of the antibody. Other factors found to interact in with both PRC2 subunits include SALL4, recently shown to recruit Polycomb complexes to the Sox2 and Sox17 loci in embryonic lineages, ZNF281, a transcription factor forming part of a pluripotency network that includes SOX2, OCT4, and NANOG, and GCFC1 (GC-rich sequence DNA-binding factor 1) (26, 27).
To our knowledge neither ZNF281 nor GCFC1 have been linked to Polycomb biology before. We used the iBAQ scoring system to compare the stoichiometry of the immunoprecipitated PRC2 components. This metric generates an estimate of relative protein abundance that corrects for differences in molecular mass and for some experimental MS detection biases (28). Core members of PRC2 were found to comprise the majority of molar content (>90%) present in both SUZ12 and EZH2 immunoprecipitations (Fig. 2C), further suggesting that the data we acquired represents a reasonable biochemical description of the PRC2 complex as it exists in NT2 cells.
The PRC2 Interactome in Differentiating NT2 Cells
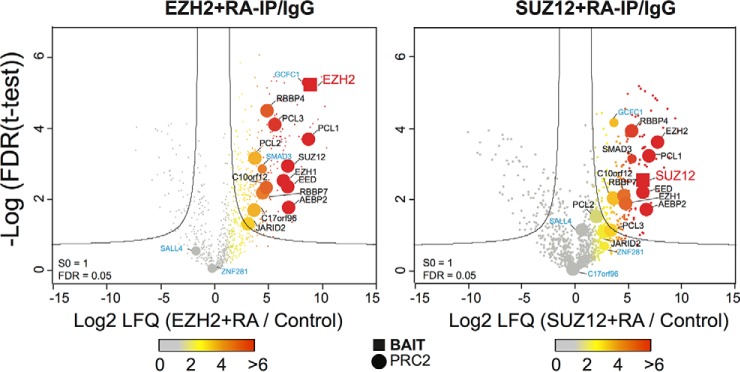
An identical experimental approach was also applied to NT2 cells that had been differentiated following treatment with retinoic acid. Again, the cognate immunoprecipitated proteins (EZH2 and SUZ12) scored highly, with nearly all core PRC2 proteins scoring above the 0.05 FDR threshold when immunoprecipitated by either subunit (Fig. 3). The exception was JARID2, which was detected in the SUZ12 immunoprecipitation with an insignificant score (but with a significant score in the EZH2 immunoprecipitation). Some other proteins that scored highly in the undifferentiated NT2 experiment, such as SALL4 and ZNF281, were detected with insignificant scores in the differentiated cell experiment (Fig. 3). Conversely, the transcription factor SMAD3, although detected with insignificant score in the undifferentiated cells, scored highly in the differentiated cells for both EZH2 and SUZ12 immunoprecipitations (Fig. 2B, 3). Intracellular signaling via the SMAD pathway has been linked in several studies to Polycomb regulation of differentiation, but to our knowledge this is the first report of a physical interaction of an Smad protein with the PRC2 complex (29, 30).
Fig. 3.
The PRC2 interactome in differentiating NT2 cells. Experiment format and Volcano plot projections were made for the PRC2 interactions in differentiating cells. A similar color scheme indicating bait proteins (red), known PRC2 interactors (black), newly reported candidate interactors (blue) was used.
By combining the overlap between the sets of EZH2- and SUZ12-interacting proteins in differentiating NT2 cells, a set of 89 proteins was produced, a slightly smaller number than the combined PRC2 “interactome” for undifferentiated cells. This suggests that in both cells states the core PRC2 complex remains relatively intact, and interacts with a similar number of accessory proteins, despite the extensive epigenetic activity and profound phenotype changes that occur during differentiation. Because the cohort of PRC2-bound genes changes substantially following differentiation of NT2 cells, such proteins could conceivably facilitate the change in activity (18). We next attempted to identify such dynamically interacting proteins.
Dynamic PRC2 Interactors Enriched in Alternative Cell States
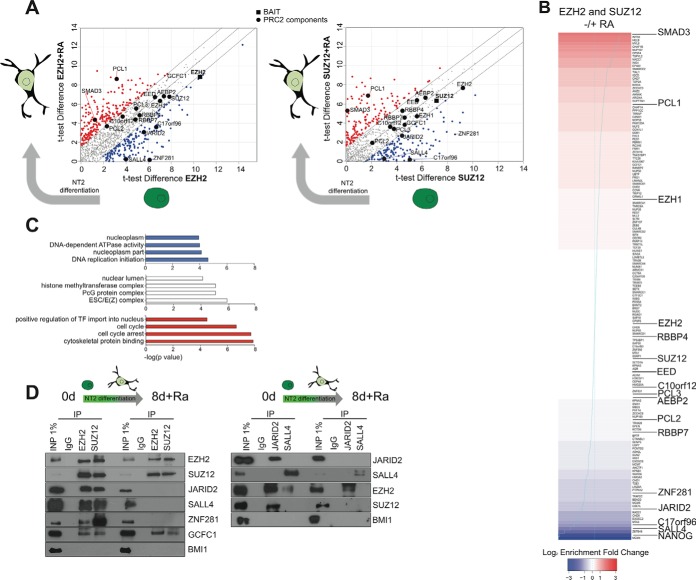
These two lists (the PRC2 interactome in undifferentiated and in differentiated cells) were compared in order to identify candidate proteins that may interact with PRC2 preferentially in either cell state. To facilitate this, we plotted on a single chart the t test scores obtained in each cell state for each protein immunoprecipitated by EZH2 or SUZ12 (Fig. 4A). Proteins mapping to the bottom of the plot are enriched in undifferentiated cells, those at the top in differentiated cells, whereas stably interacting proteins map along the diagonal. As noted above, the stable complement includes all core and most PRC2-associated components (EZH2, SUZ12, EED, RBBP7, AEBP2, PCL2, c10orf12, EZH1). This list of stable interactors also includes GCFC1, the factor newly identified as a candidate PRC2 interactor in this study.
Fig. 4.
Dynamic PRC2 interactors enriched in alternative cell states. A, t test scores obtained in the undifferentiated and differentiated cell experiments were plotted against each other for EZH2 and SUZZ12, highlighting those proteins favoring interaction with PRC2 in one or other cell state. B, The proteins were ranked according to their fold-change (calculated using the iBAQ values normalized to the immunoprecipitated protein) for combined EZH2 and SUZ12 experiments) favoring interaction in undifferentiated (blue) or differentiated (red) cells. C, Enrichment of Gene Ontology annotations for the proteins favoring static or dynamic interactions. D, Co-immunoprecipitation analysis of candidate static (EZH2, SUZ12, GCFC1) and dynamic (JARID2, SALL4) interactions in both cell states. The PRC1 component BMI1 serves as a negative control.
Proteins favoring interactions with PRC2 in undifferentiated cells included JARID2, c17orf96, SALL4 and ZNF281, whereas those favoring interactions in differentiated cells included PCL1 and SMAD3. Reassuringly, these observations were consistent across both IP experiments (i.e. were found when either EZH2 and SUZ12 antibodies were used). This prompted us to rank the PRC2 interactors according to their preference for either cell state (Fig. 4B). We used the iBAQ scores of interacting proteins, normalized to that of the immunoprecipitated protein (i.e. either EZH2 or SUZ12) to calculate the enrichments and adjust for potential differing efficiencies of either IP. Factors strongly favoring interaction with PRC2 with in undifferentiated NT2 cells (bottom of Fig. 4B, blue shading) include NANOG, SALL4, JARID2, and ZNF281, whereas factors strongly favoring interaction in differentiated cells include PCL1 and SMAD3 (top of Fig. 4B, red shading). Notably, the core PRC2 components are positioned in the mid-range of this ranking system, reflecting their stable participation within PRC2 throughout the differentiation program. When the list of PRC2-interacting proteins favoring the undifferentiated state were analyzed for Gene Ontology annotation enrichment, the terms “transcription factor import into nucleus” and “cell cycle” were found to occur most frequently (Fig. 4C); a similar analysis of those favoring the differentiated state yielded terms such as “nucleoplasm” and “DNA-dependent ATPase activity.” Meanwhile, the most enriched annotation category among proteins in the mid-range representing both cell states reflect PRC2 itself (“PcG protein complex,” “ESC/E(Z) complex”) and methyltransferase activities in general (“histone methyltransferase complex”).
We sought to validate some of these results by coimmunoprecipitation (Fig. 4D). As expected, the core PRC2 members EZH2 and SUZ12 could be reciprocally coimmunoprecipitated in both cell states. The apparent loss of JARID2 from the complex in differentiated NT2 cells was confirmed, as was the association with SALL4 and ZNF281. The association with GCFC1 was maintained through the differentiation cycle. Importantly, reciprocal experiments using antibodies to JARID2 and SALL4 broadly confirmed these results. The exception was that some residual EZH2 signal was observed in JARID2 immunoprecipitations from differentiated cells. However, no such staining was observed when SUZ12 was used to probe the JARID2 precipitate (Fig. 4D). This might indicate that EZH2 can bind JARID2 independently of the PRC2 complex, although additional biophysical experiments would be needed to confirm this. Interestingly, this “exception” was also observed in the label-free LCMS experiments (Fig. 4A), arguing that proteomics technologies, if applied carefully, are a reliable and sensitive measure of protein abundance that accurately reflects traditional biochemical approaches. Several additional experiments were attempted but unfortunately had to be abandoned due to poor antibody performance in coimmunoprecipitation experiments (e.g. SMAD3, GCFC1, ZNF281). The ranked set of dynamic PRC2 interacting proteins is therefore a valuable resource, but individual candidate interactors will require further study to understand their precise role in Polycomb biology.
Dynamic PRC2 Interactions in NT2 Cells Can be Explained by Differential Gene Expression
The simplest explanation for why a protein might interact with another protein in a specific cell state is that it is only present in the cell in one or other state. We therefore looked for evidence that proteins scored as dynamically interacting with PRC2 in our NT2 model were in fact differentially expressed in retinoic acid treated or untreated cells. We compared our data with transcriptome data obtained from retinoic acid treated NT2 cells (31).
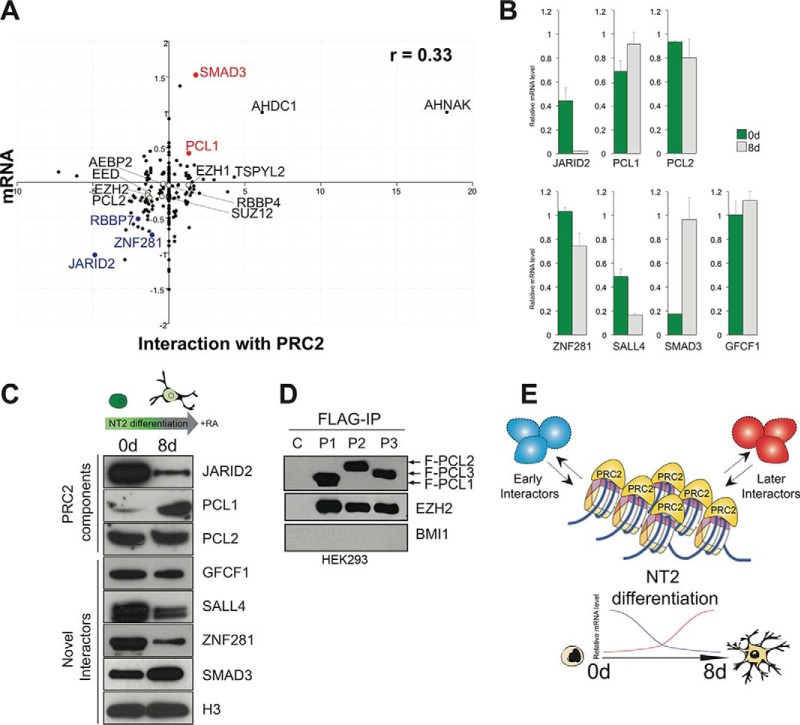
This analysis found broad correlation between the fold-change observed for mRNAs and their corresponding proteins following RA treatment (Spearman's Rank Correlation, r = 0.33; Fig. 5A). Specifically, the core PRC2 members were present in PRC2 in both cell states and displayed good mRNA-protein correlation. In fact, most proteins that our interactome analysis found to favor either of the cell states also showed mRNA-protein correlation for the relevant state. In particular, JARID2 and ZNF281 show high expression of both mRNA and protein for undifferentiated cells, whereas PCL1 and SMAD3 show similar patterns for differentiated cells. In contrast, a number of candidate PRC2 interactors found in our screen to favor interaction in differentiating cells did show large mRNA increases in differentiated cells compared with the levels of the cognate protein. These include AHNAK (a neuroblast differentiation-associated protein), AHDC1 (a protein linked to mental retardation), and TSPYL2 (a transcription protein linked to neuronal synapse function), and MYL6 (a myosin light chain protein). Although they passed the high stringency statistical analysis, these proteins will require independent confirmation as PRC2 interactions in order to rule out nonspecific binding because of high level expression. No mRNA data was available for GCFC1, PCL3, AEBP2, SALL4, NANOG, or c17orf96.
Fig. 5.
Dynamic PRC2 interactions in NT2 cells can be explained by differential gene expression. A, Enrichment scores for proteins interacting with PRC2 in either cell state were plotted against mRNA expression for the corresponding gene in differentiated NT2 cells (-Log10; 55). B, mRNA expression in untreated and retinoic acid-treated NT2 cells were measured using qPCR. C, Protein levels for PRC2 interactors were compared in untreated and treated cells by Western blot. D, Coimmunoprecipitation analysis of FLAG-tagged PCL1, PCL2 and PCL3 expressed in HEK293 cells shows that all three co-purify with EZH2. The PRC1 component BMI1 serves as a negative control. E, In differentiating cells, the core PRC2 complex appears to be stable, whereas the subset of interactions that interact dynamically in general correspond to their expression in the relevant cell state.
In order to confirm our observations for a selection of the candidate dynamically interacting proteins, we measured mRNA levels in undifferentiated and differentiated NT2 cells using RT-PCR. This analysis found that mRNAs for Jarid2, Sall4, and Znf281 were significantly decreased upon differentiation, whereas mRNAs for Pcl1 and Smad3 were significantly increased (Fig 5B). These observations directly correlate to the differential PRC2 interactions of the corresponding proteins in the same cell states.
The presence of mRNA does not prove that the cognate protein is expressed, nor that it is stable in the cell even if it is successfully translated. We therefore assessed the levels of JARID2, SALL4, ZNF281, PCL1, SMAD3, PCL2, GCFC1, and histone H3 protein over the NT2 differentiation cycle using Western blot (Fig. 5C). This experiment found that the levels of mRNA and protein were indeed correlated. The data suggest that these proteins associate with PRC2 based on their expression in NT2 cells in a particular cell state, rather than alternative potential regulatory mechanisms such as differential localization, altered affinity for PRC2 based on PTMs, or the presence of co-factors such as ncRNAs (32, 33). In support of this, we exogenously expressed FLAG-tagged PCL3 (as well as PCL1 and PCL2) in HEK293 cells, where it is not endogenously expressed (5). Coimmunoprecipitation experiments confirmed that the PCL3 copurified with EZH2 (but not the PRC1 component BMI1) if FLAG tag was used for IP of PCL3. This suggests that the presence of the differentially interacting protein is sufficient to trigger the interaction with PRC2, at least for the case of PCL3 (Fig. 5D).
Although these factors certainly do play a role in PcG biology, our work here suggests that differential expression of Polycomb-interacting factors is a potentially important mode of regulation for this key epigenetic complex.
DISCUSSION
It is unsurprising that the complicated task of regulating the expression of hundreds of genes during development might require a suite of related protein complexes, each specializing or refining a general biochemical function. For PRC1 in mammals, this appears to be at least partly explained by the presence of multiple paralogs of each subunit. For PRC2, the situation is less clear, with generally only a single gene present for core PRC2 subunits. The exceptions to this are EZH2, where a nonessential close paralog (EZH1) is mainly expressed in adult tissue, and EED, where alternate splice variants can display different phenotypes (11, 34).
The presence of multiple PRC2-associated proteins has therefore led to the proposal that different PRC2-related complexes exist, and that these alternate forms modulate the central catalytic activity by: (a) localizing it to particular genomic loci via targeting factors; and/or (b) recruiting additional epigenetic activities to the complex (e.g. readers of histone marks or chromatin remodelling activities) (35). Although pioneering work on the composition of the PRC2 complex has been carried out in Drosophila and mouse embryonic stem cells (36–38), few studies have attempted to directly compare the physical interactions of PRC2 subunits in different states. Here we carried out such a study and found that, although the majority of reported PRC2 proteins appear to form stable associations with the complex throughout the NT2 differentiation cycle, a smaller number of proteins were found to associate with the complex preferentially in one cell state or the other (Fig. 5E). These dynamically interacting proteins include two that are generally considered to be bone fide PRC2 components (JARID2, PCL1), as well as several whose functional relationship to PRC2 is less clear (e.g. ZNF281, SALL4).
We found that JARID2 favored interaction with PRC2 in undifferentiated NT2 cells. JARID2 contains a JmjC domain that normally encodes a demethylase activity, but that appears to be inactive in this case (39). Different outcomes arising from loss of Jarid2 have been reported suggesting that the specific biochemical activity of JARID2 might be altered depending on cell context (40, 41). Furthermore, in mice, the phenotype expressed by disruption of Jarid2 is milder and more pleiotropic than that observed for the genes of core PRC2 subunits, suggesting that JARID2 is at least partly dispensable for PRC2 activity (42). Another protein found to favor interaction with PRC2 in undifferentiated NT2 cells is SALL4, a zinc finger transcription factor that is essential for ES cell formation (43). It is a regulator of the PRC1 protein BMI1 in hematopoietic cells (44, 45). To our knowledge, our data is the first report of a physical interaction between SALL4 and a PRC2 protein. However, SALL4 was one of a group of neuronal stem cell regulators identified in a network topology analysis of transcriptome data that link it to PRC2. Other genes identified within this network included Ezh2, Suz12, Jarid2, pcl2, and Sox2 (46).
Among the proteins found to favor interaction with PRC2 in undifferentiated NT2 cells, c17orf96 has been reported to co-purify with JARID2 and PCL2 in embryonic stem cells but not in HEK293 cells (47, 48). C17orf96 contains no recognizable domains and appears to be unstructured. It was recently shown to have affinity for CpG islands, and loss of the gene affects H3K27me3 levels, SUZ12 binding, and gene repression (but also, interestingly, RNA polymerase II activity) in mouse ES cells (49). Thus, this incompletely characterized gene appears to play both PRC2-dependent and PRC2-independent roles in stem cell biology. ZNF281 is another relatively unstudied zinc finger transcription factor involved in the regulation of the embryonic stem cell state, and was recently found to be linked to the DNA damage response (50). In contrast, several proteins favor interaction with PRC2 in differentiating NT2 cells. Interestingly, these included PCL1, one of the three PCL paralogs in mammalian cells. Notably, a role for PCL1 was recently demonstrated in quiescent cells (9). This contrasted with PCL2 and PCL3, which favor proliferating cells. In general, no obvious pattern can be discerned among the sets of proteins favoring interaction with PRC2 in either cell state. In some cases, the set of “undifferentiated interactors” were linked to ES cell biology (SALL4, JARID2), whereas some of the 'differentiated interactors' have well known functions in differentiation (SMAD3). Furthermore, many dynamic interactors have DNA-binding function or are confirmed transcription factors, consistent with possible roles as recruiters of PRC2 to specific loci.
The study of dynamic protein interactions is technically challenging and relatively few systematic studies have been reported in any system (51). Very recently, a study of the dynamic interactions of PRC2 and PRC1 was carried out in mouse embryonic stem cells that were differentiated into neural progenitor cells (NPCs) (52). Although our study employs direct immunoprecipitation and Kloet and coworkers used a Bacterial Artificial Chromosome insertion strategy, in general the identities and stoichiometry of the PRC2 interacting proteins reported are very similar. In fact, many of the proteins found to be dynamically interacting overlapped in both studies (PHF3, MTF2, C17orf96, JAIRID2). The major difference between the studies was that Kloet and coworkers found that the overall levels of PRC2 core components (EZH2, SUZ12, EED) dropped significantly upon differentiation into NPCs, whereas we did not observe a similar reduction upon differentiation of human NT2 cells. The two experimental models are very different however, and the different observation presumably highlights the diverse behavior of Polycomb regulation in different cellular contexts.
In this work, we identified context-dependent interactors of PRC2 and show that they strongly correlate with expression of the interacting proteins in the relevant cell context. This observation broadly agrees with other workers who have proposed that protein complexes may be at least partly regulated by expression of component parts (16). All generally accepted members of PRC2 were detected, but some at low levels (e.g. PCL3 was only detected by a single peptide on one replicate of the SUZ12 IP experiment). Additional upstream separation of protein or peptide, for example in gel digest or online cation exchange, might improve sensitivity. Alternatively, the stringency of the washing conditions could be adjusted by varying salt of detergent concentrations, or a cross-linking approach employed.
Our work also highlights the limitations of such studies. First, by interrogating individual subunits, information about the composition of variant complexes is lost. Here, we attempted to partly overcome this by investigating two independent subunits of PRC2 and using quantitative mass spectrometry, but it is not clear how many different forms of PRC2 are present in the nucleus at a given time. For example, relatively small amounts of EZH1 were found by us in replicate EZH2 and SUZ12 immunoprecipitation experiments, even though it is unclear why EZH1 would be present in a complex that already contains EZH2. Other authors have similarly reported a direct interaction between EZH1 and EZH2 (53), whereas others have presented evidence against this (54). Use of techniques such as gel filtration failed to resolve these issues (data not shown). In future, a combination of physical interaction maps and biophysical methods (e.g. thermal shift assay) may be needed to obtain a more fine-grained view of the PRC2 interactome. Second, current studies generally do not link the composition of the complexes directly to their function. Large-scale chromatin immunoprecipitation sequencing experiments (ChIPseq: to determine which loci the PRC2 interactors are binding to) and methyltransferase assays (to measure the functional consequence of loss of individual subunits) can be used to address this. These are very challenging experiments however, the former requiring high quality antibody reagents that are not always available, whereas the latter require purification of PRC2 variants whose composition is known with certainly, again very difficult to achieve in practice. Lastly, the contribution of other regulatory mechanisms needs to be assessed, in particular the role of PTM-induced structural change to dynamic PRC2 activity, or the presence of ncRNAs. Nevertheless, our work extends our understanding of PRC2 biology by showing that a core complex is maintained throughout an experimentally induced cell state change, whereas individual proteins favor interaction during one or other state.
Supplementary Material
Acknowledgments
We thank the UCD Conway Proteomics Facility and Genome Facility for help in this study. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD004462.
Footnotes
Author contributions: G.O. carried out the majority of experimental work, with assistance from G.B., E.J., D.A., and G.S. (immunoprecipitation experiments), A.W., N.M., B.D., and K.W. (mass spectrometry). A.B. and G.C. designed the study with assistance from J.C. G.O., A.B., and G.C. wrote the paper.
* This work was supported by a Science Foundation Ireland Principal Investigator Award (SFI 10/1N.1/B3.19).
 This article contains supplemental material.
This article contains supplemental material.
Competing Financial Interests statement: The authors declare no competing financial interests.
1 The abbreviations used are:
- PRC
- polycomb repressor complex
- FDR
- false discovery rate.
REFERENCES
- 1. Phizicky E., Bastiaens P. I. H., Zhu H., Snyder M., and Fields S. (2003) Protein analysis on a proteomic scale. Nature 422, 208–215 [DOI] [PubMed] [Google Scholar]
- 2. Cox J., and Mann M. (2011) Quantitative, High-Resolution Proteomics for Data-Driven Systems Biology. Annu. Rev. Biochem. 80, 273–299 [DOI] [PubMed] [Google Scholar]
- 3. Di Croce L., and Helin K. (2013) Transcriptional regulation by Polycomb group proteins. Nat. Structural Mol. Biol. 20, 1147–1155 [DOI] [PubMed] [Google Scholar]
- 4. Schuettengruber B., Chourrout D., Vervoort M., Leblanc B., and Cavalli G. (2007) Genome regulation by polycomb and trithorax proteins. Cell 128, 735–745 [DOI] [PubMed] [Google Scholar]
- 5. Brien G. L., Gambero G., O'Connell D. J., Jerman E., Turner S. A., Egan C. M., Dunne E. J., Jurgens M. C., Wynne K., Piao L., Lohan A. J., Ferguson N., Shi X., Sinha K. M., Loftus B. J., Cagney G., and Bracken A. P. (2012) Polycomb PHF19 binds H3K36me3 and recruits PRC2 and demethylase NO66 to embryonic stem cell genes during differentiation. Nat. Structural Mol. Biol. 19, 1273–1281 [DOI] [PubMed] [Google Scholar]
- 6. Margueron R., and Reinberg D. (2011) The Polycomb complex PRC2 and its mark in life. Nature 469, 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Conway E., Healy E., and Bracken A. P. (2015) PRC2 mediated H3K27 methylations in cellular identity and cancer. Curr. Opin. Cell Biol. 37, 42–48 [DOI] [PubMed] [Google Scholar]
- 8. Vizán P., Beringer M., Ballaré C., and Di Croce L. (2015) Role of PRC2-associated factors in stem cells and disease. FEBS J. 282, 1723–1735 [DOI] [PubMed] [Google Scholar]
- 9. Brien G. L., Healy E., Jerman E., Conway E., Fadda E., O'Donovan D., Krivtsov A. V., Rice A. M., Kearney C. J., Flaus A., McDade S. S., Martin S. J., McLysaght A., O'Connell D. J., Armstrong S. A., and Bracken A. P. (2015) A chromatin-independent role of Polycomb-like 1 to stabilize p53 and promote cellular quiescence. Genes Dev. 29, 2231–2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kikuchi J., Koyama D., Wada T., Izumi T., Hofgaard P. O., Bogen B., and Furukawa Y. (2015) Phosphorylation-mediated EZH2 inactivation promotes drug resistance in multiple myeloma. J. Clin. Investig. 125, 4375–4390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schwartz Y. B., and Pirrotta V. (2013) A new world of Polycombs: unexpected partnerships and emerging functions. Nat. Rev. Genetics 14, 853–864 [DOI] [PubMed] [Google Scholar]
- 12. Yeger-Lotem E., and Sharan R. (2015) Human protein interaction networks across tissues and diseases. Front. Genetics 6, 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hegele A., Kamburov A., Grossmann A., Sourlis C., Wowro S., Weimann M., Will C. L., Pena V., Lührmann R., and Stelzl U. (2012) Dynamic protein-protein interaction wiring of the human spliceosome. Mol. Cell 45, 567–580 [DOI] [PubMed] [Google Scholar]
- 14. Ruschak A. M., and Kay L. E. (2012) Proteasome allostery as a population shift between interchanging conformers. Proc. Natl. Acad. Sci. U.S.A. 109, E3454–E62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mishto M., Liepe J., Textoris-Taube K., Keller C., Henklein P., Weberruβ M., Dahlmann B., Enenkel C., Voigt A., Kuckelkorn U., Stumpf M. P. H., and Kloetzel P. M. (2014) Proteasome isoforms exhibit only quantitative differences in cleavage and epitope generation. Eur. J. Immunol. 44, 3508–3521 [DOI] [PubMed] [Google Scholar]
- 16. de Lichtenberg U., Jensen L. J., Brunak S., and Bork P. (2005) Dynamic complex formation during the yeast cell cycle. Science 307, 724–727 [DOI] [PubMed] [Google Scholar]
- 17. Lee V. M., and Andrews P. W. (1986) Differentiation of NTERA-2 clonal human embryonal carcinoma cells into neurons involves the induction of all three neurofilament proteins. J. Neurosci. 6, 514–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bracken A. P., Dietrich N., Pasini D., Hansen K. H., and Helin K. (2006) Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Develop. 20, 1123–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Richly H., Rocha-Viegas L., Ribeiro J. D., Demajo S., Gundem G., Lopez-Bigas N., Nakagawa T., Rospert S., Ito T., and Di Croce L. (2010) Transcriptional activation of polycomb-repressed genes by ZRF1. Nature 468, 1124–1128 [DOI] [PubMed] [Google Scholar]
- 20. Dietrich N., Lerdrup M., Landt E., Agrawal-Singh S., Bak M., Tommerup N., Rappsilber J., Södersten E., and Hansen K. (2012) REST-mediated recruitment of polycomb repressor complexes in mammalian cells. PLoS Genetics 8, e1002494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Turriziani B., Garcia-Munoz A., Pilkington R., Raso C., Kolch W., and von Kriegsheim A. (2014) On-Beads Digestion in Conjunction with Data-Dependent Mass Spectrometry: A Shortcut to Quantitative and Dynamic Interaction Proteomics. Biology 3, 320–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vizcaíno J. A., Csordas A., del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q-W, Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tusher V. G., Tibshirani R., and Chu G. (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U.S.A. 98, 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M. Y., Geiger T., Mann M., and Cox J. (2016) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods doi: 10.1038/nmeth.3901 [DOI] [PubMed] [Google Scholar]
- 25. Smits A. H., Jansen P. W. T. C., Poser I., Hyman A. A., and Vermeulen M. (2012) Stoichiometry of chromatin-associated protein complexes revealed by label-free quantitative mass spectrometry-based proteomics. Nucleic Acids Res. 41, e28–e28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Abboud N., Moore-Morris T., Hiriart E., Yang H., Bezerra H., Gualazzi M-G, Stefanovic S., Guénantin A-C, Evans S. M., and Pucéat M. (2015) A cohesin-OCT4 complex mediates Sox enhancers to prime an early embryonic lineage. Nat. Commun. 6, 6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang Z-X, Teh CH-L, Chan CM-Y, Chu C., Rossbach M., Kunarso G., Allapitchay T. B., Wong K. Y., and Stanton L. W. (2008) The transcription factor Zfp281 controls embryonic stem cell pluripotency by direct activation and repression of target genes. Stem Cells 26, 2791–2799 [DOI] [PubMed] [Google Scholar]
- 28. Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., and Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473, 337–342 [DOI] [PubMed] [Google Scholar]
- 29. Kim S. W., Yoon S-J, Chuong E., Oyolu C., Wills A. E., Gupta R., and Baker J. (2011) Chromatin and transcriptional signatures for Nodal signaling during endoderm formation in hESCs. Develop. Biol. 357, 492–504 [DOI] [PubMed] [Google Scholar]
- 30. Dahle Ø., and and Kuehn M. R. (2013) Polycomb Determines Responses to Smad2/3 Signaling in Embryonic Stem Cell Differentiation and in Reprogramming. Stem Cells 31, 1488–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Houldsworth J., Heath S. C., Bosl G. J., Studer L., and Chaganti R. S. K. (2002) Expression profiling of lineage differentiation in pluripotential human embryonal carcinoma cells. Cell Growth Differentiation 13, 257–264 [PubMed] [Google Scholar]
- 32. Yang L., Lin C., Liu W., Zhang J., Ohgi K. A., Grinstein J. D., Dorrestein P. C., and Rosenfeld M. G. (2011) ncRNA- and Pc2 Methylation-Dependent Gene Relocation between Nuclear Structures Mediates Gene Activation Programs. Cell 147, 773–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jenuwein T., and Allis C. D. (2001) Translating the histone code. Science 293, 1074–1080 [DOI] [PubMed] [Google Scholar]
- 34. Kuzmichev A., Jenuwein T., Tempst P., and Reinberg D. (2004) Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol. Cell 14, 183–193 [DOI] [PubMed] [Google Scholar]
- 35. Simon J. A., and Kingston R. E. (2013) Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell 49, 808–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kennison J. A. (1995) The Polycomb and trithorax group proteins of Drosophila: trans-regulators of homeotic gene function. Annu. Rev. Gen. 29, 289–303 [DOI] [PubMed] [Google Scholar]
- 37. Daubresse G., Deuring R., Moore L., Papoulas O., Zakrajsek I., Waldrip W. R., Scott M. P., Kennison J. A., and Tamkun J. W. (1999) The Drosophila kismet gene is related to chromatin-remodeling factors and is required for both segmentation and segment identity. Development 126, 1175–1187 [DOI] [PubMed] [Google Scholar]
- 38. Coulson M., Robert S., Eyre H. J., and Saint R. (1998) The identification and localization of a human gene with sequence similarity to Polycomblike of Drosophila melanogaster. Genomics 48, 381–383 [DOI] [PubMed] [Google Scholar]
- 39. Klose R. J., Kallin E. M., and Zhang Y. (2006) JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 7, 715–727 [DOI] [PubMed] [Google Scholar]
- 40. Shen X., Kim W., Fujiwara Y., Simon M. D., Liu Y., Mysliwiec M. R., Yuan G-C, Lee Y., and Orkin S. H. (2009) Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell 139, 1303–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pasini D., Cloos P. A. C., Walfridsson J., Olsson L., Bukowski J-P, Johansen J. V., Bak M., Tommerup N., Rappsilber J., and Helin K. (2010) JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature 464, 306–310 [DOI] [PubMed] [Google Scholar]
- 42. Takeuchi T., Yamazaki Y., Katoh-Fukui Y., Tsuchiya R., Kondo S., Motoyama J., and Higashinakagawa T. (1995) Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes Develop. 9, 1211–1222 [DOI] [PubMed] [Google Scholar]
- 43. Yang J., Gao C., Chai L., and Ma Y. (2010) A novel SALL4/OCT4 transcriptional feedback network for pluripotency of embryonic stem cells. PloS one 5, e10766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang J., Chai L., Liu F., Fink L. M., Lin P., Silberstein L. E., Amin H. M., Ward D. C., and Ma Y. (2007) Bmi-1 is a target gene for SALL4 in hematopoietic and leukemic cells. Proc. Natl. Acad. Sci. U.S.A. 104, 10494–10499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Milanovich S., Peterson J., Allred J., Stelloh C., Rajasekaran K., Fisher J., Duncan S. A., Malarkannan S., and Rao S. (2015) Sall4 overexpression blocks murine hematopoiesis in a dose-dependent manner. Exp. Hematol. 43, 53–64.e1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yaqubi M., Mohammadnia A., and Fallahi H. (2015) Predicting involvement of polycomb repressive complex 2 in direct conversion of mouse fibroblasts into induced neural stem cells. Stem Cell Res. Therapy 6, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang Z., Jones A., Sun C-W, Li C., Chang C-W, Joo H-Y, Dai Q., Mysliwiec M. R., Wu L-C, Guo Y., Yang W., Liu K., Pawlik K. M., Erdjument-Bromage H., Tempst P., Lee Y., Min J., Townes T. M., and Wang H. (2011) PRC2 complexes with JARID2, MTF2, and esPRC2p48 in ES cells to modulate ES cell pluripotency and somatic cell reprogramming. Stem Cells 29, 229–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Alekseyenko A. A., Gorchakov A. A., Kharchenko P. V., and Kuroda M. I. (2014) Reciprocal interactions of human C10orf12 and C17orf96 with PRC2 revealed by BioTAP-XL cross-linking and affinity purification. Proc. Natl. Acad. Sci. U.S.A. 111, 2488–2493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liefke R., and Shi Y. (2015) The PRC2-associated factor C17orf96 is a novel CpG island regulator in mouse ES cells. Cell Discovery 1, 15008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pieraccioli M., Nicolai S., Antonov A., Somers J., Malewicz M., Melino G., and Raschellà G. (2015) ZNF281 contributes to the DNA damage response by controlling the expression of XRCC2 and XRCC4. Oncogene 35, 2592–2601 [DOI] [PubMed] [Google Scholar]
- 51. Przytycka T. M., Singh M., and Slonim D. K. (2010) Toward the dynamic interactome: it's about time. Briefings Bioinformatics 11, 15–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kloet S. L., Makowski M. M., Baymaz H. I., van Voorthuijsen L., Karemaker I. D., Santanach A., Jansen P. W. T. C., Di Croce L., and Vermeulen M. (2016) The dynamic interactome and genomic targets of Polycomb complexes during stem-cell differentiation. Nat. Structural Mol. Biol. 23, 682–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shen X., Liu Y., Hsu Y-J, Fujiwara Y., Kim J., Mao X., Yuan G-C, and Orkin S. H. (2008) EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell 32, 491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Margueron R., Li G., Sarma K., Blais A., Zavadil J., Woodcock C. L., Dynlacht B. D., and Reinberg D. (2008) Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol. Cell 32, 503–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Houldsworth J., Heath S. C., Bosl G. J., Studer L., and Chaganti R. S. (2002) Expression profiling of lineage differentiation in pluripotential human embryonal carcinoma cells. Cell Growth Differ. 13, 257–264 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.