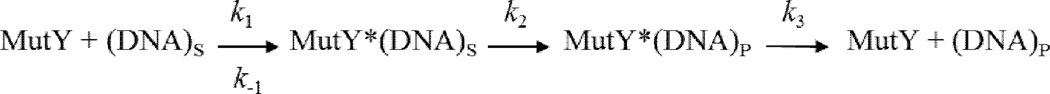Abstract
MUTYH is a base excision repair (BER) protein that prevents mutations in DNA associated with 8-oxoguanine (OG) by catalyzing the removal of adenine from inappropriately formed OG:A base-pairs. Germline mutations in the MUTYH gene are linked to colorectal polyposis and a high risk of colorectal cancer, a syndrome referred to as MUTYH-associated polyposis (MAP). There are over 300 different MUTYH mutations associated with MAP and a large fraction of these gene changes code for missense MUTYH variants. Herein, the adenine glycosylase activity, mismatch recognition properties, and interaction with relevant protein partners of human MUTYH and five MAP variants (R295C, P281L, Q324H, P502L, and R520Q) were examined. P281L MUTYH was found to be severely compromised both in DNA binding and base excision activity, consistent with the location of this variation in the iron-sulfur cluster (FCL) DNA binding motif of MUTYH. Both R295C and R520Q MUTYH were found to have low fractions of active enzyme, compromised affinity for damaged DNA, and reduced rates for adenine excision. In contrast, both Q324H and P502L MUTYH function relatively similarly to WT MUTYH in both binding and glycosylase assays. However, P502L and R520Q exhibited reduced affinity for PCNA (proliferation cell nuclear antigen), consistent with their location in the PCNA-binding motif of MUTYH. Whereas, only Q324H, and not R295C, was found to have reduced affinity for Hus1 of the Rad9-Hus1-Rad1 complex, despite both being localized to the same region implicated for interaction with Hus1. These results underscore the diversity of functional consequences due to MUTYH variants that may impact the progression of MAP.
Keywords: Base excision repair (BER), DNA glycosylase, MUTYH-Associated Polyposis (MAP), 8-oxoguanine, PCNA, Hus1, MUTYH, cancer variants, single nucleotide polymorphisms, enzyme kinetics
Graphical abstract
1. Introduction
Deleterious and mutagenic DNA damage can arise from reactions with reactive oxygen and nitrogen species (RONS)1 generated during inflammatory responses, and in response to genotoxic agents such as ionizing radiation, chemical mutagens, UV light, and chemotherapeutic drugs [1–3]. Arguably the most studied base oxidation product is 8-oxo-7,8-dihydro-2’-deoxyguanosine (OG) [1], which also serves as a key biomarker for oxidative stress [4, 5]. In the absence of repair, OG results in high levels of G:C to T:A transversion mutations due to its ability to mimic T and direct for misincorporation of A to form stable OG:A mismatches [2]. Two base excision repair glycosylases, the human OG glycosylase (hOGG1) and the human MutY homologue (MUTYH) act synergistically to prevent mutations associated with OG in duplex DNA [1, 6]. While hOGG1 initiates repair by removing the OG lesion when paired opposite C, MUTYH excises the undamaged A base when mispaired with OG [1]. Subsequent BER enzymes act sequentially at the base-less site to restore the appropriate nucleotide and preserve the originally coded G:C base pair [1, 6].
MUTYH-associated polyposis (MAP) is a colorectal cancer predisposition mechanism that was discovered based on the correlation with biallelic inheritance of two germline mutations in MUTYH encoding the missense variants Y165C and G382D MUTYH [1, 7–9]. Biallelic carriers of the these two MAP alleles have increased G to Ttransversion mutations in the tumor suppressor genes APC and K-ras in somatic tissues consistent with dysfunctional OG:A repair activity [7, 10]. Notably, adenine glycosylase activity and OG:A mismatch affinity are compromised for both variants and most dramatically for Y165C MUTYH [11–13]. Moreover, both variants have been shown to be defective in OG:A repair in a GFP-based cellular assay [12]. Structural and functional studies are consistent with the hypothesis that compromised OG:A repair activity of MUTYH variants is an initiating event leading to mutagenesis and eventual carcinogenesis in MAP [1, 14, 15].
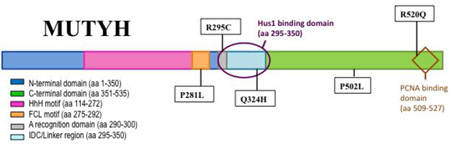
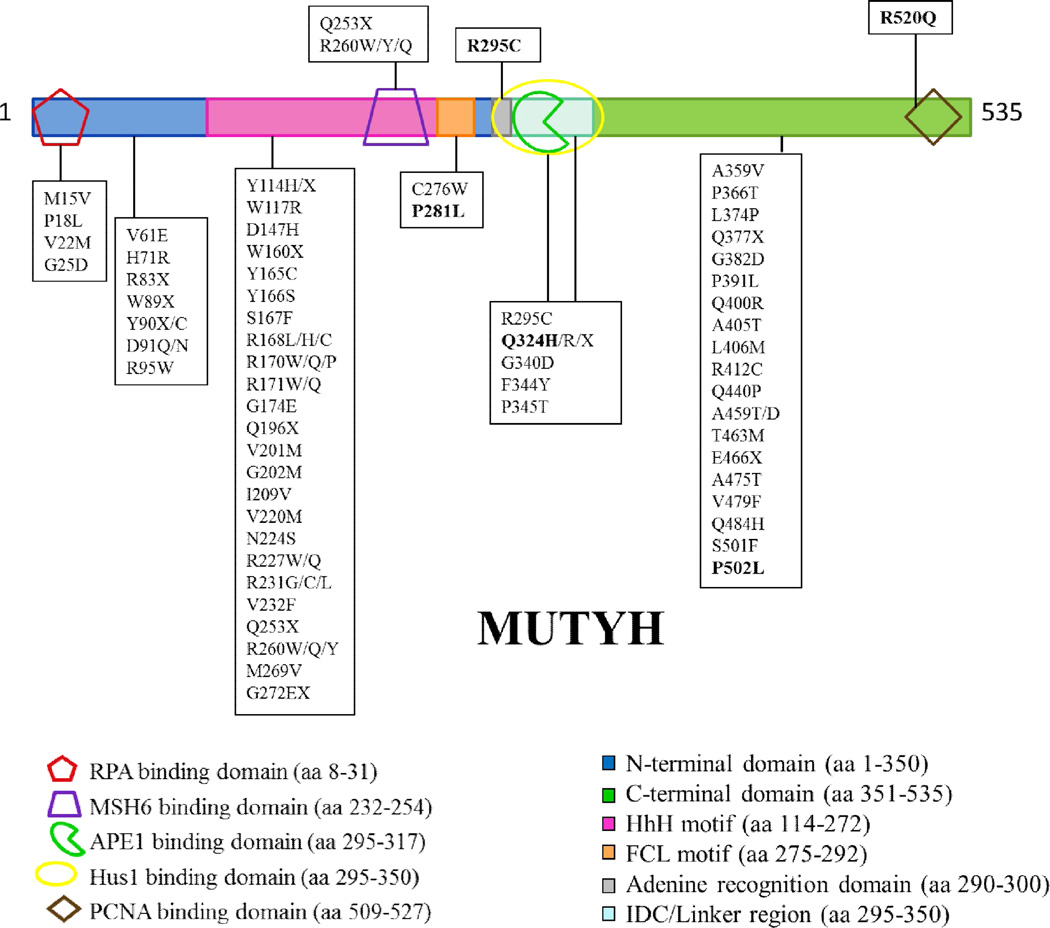
Since the initial discovery of MAP, over 300 different sequence variants in the MUTYH gene have been reported in the Leiden Open Variant Database (LOVD) [16–18]. A significant fraction of these sequence changes code for missense variants of MUTYH [16]. Many MAP-associated variants are localized in the N-terminal catalytic base excision domain (near Y165C) or the C-terminal OG recognition domain (near G382D) (Fig. 1). In these cases, based on homology to the bacterial protein, reduced OG:A recognition or adenine glycosylase activity may be anticipated for some of these variants. However, there are also many MUTYH missense variations located within regions implicated in mediating interactions of MUTYH with protein partners (Fig. 1). Indeed, MUTYH variations are found in reported binding sites for AP endonuclease 1 (APE1) [19], proliferating cell nuclear antigen (PCNA) [19–21], MutS homologue 6 (MSH6) [22], replication protein A (RPA) [19], and the Rad9-Hus1-Rad1 (9-1-1) complex [23, 24]. The correct associations between MUTYH and its various protein binding partners are necessary to ensure efficient BER of OG:A mismatches and coordination with other cellular processes.
Figure 1.
MUTYH missense and nonsense mutations located in functional domains of the protein. Shown is the α3 transcript of the human MUTYH gene which encodes a 535 amino acid protein. The various binding partners of MUTYH are represented as shapes: replication protein A (RPA), red pentagon; mutS 6 homologue (MSH6), purple trapezoid; Hus1, yellow oval; AP endonuclease 1 (APE1), green pac man symbol; and proliferating cell nuclear antigen (PCNA), brown diamond.
MUTYH variations may compromise these key interactions with protein partners, and this may result in more dramatically reduced levels of cellular OG: A repair than anticipated based on the enzyme activity analysis. Moreover, altered interactions with protein partners, such as the 9-1-1 complex, may erode proper signaling responses to DNA damage [25, 26]. To provide further insight into how MUTYH variants located in different regions of the protein impact both the intrinsic enzymatic activity and interactions with relevant protein partners, we selected a group of variants for more detailed analysis in this study.
The P502L and R520Q MUTYH variants were selected due to the location of the modified amino acids in the PCNA binding region of the protein (Fig. 1). PCNA is a ring-shaped trimeric clamp that encircles dsDNA [27, 28] and acts as an auxiliary protein for polδ and polε by enhancing the processivity of these polymerases by tethering them to DNA [29, 30]. MUTYH has been shown to directly associate with PCNA and co-localizes with PCNA to replication foci in the nucleus during G1/S phase [19, 31]. The P502L variant is a relatively rare variant [17] and has also been found in a healthy control patient of a colorectal cancer (CRC) screening cohort [32]. The R520Q varianth as been more frequently associated with CRC [17] and was also identified in a patient with lung cancer [32]. Both variants have been referred to as single nucleotide polymorphisms (SNPs) [32, 33] and as “variants of unknown significance” (VUS) [17]. Structure-based functional predictions are not possible for these variant susing the X-ray structure of the bacterial MutY bound to an OG:A-containing duplex [34] due to the location of the amino acid variation in a region of the C-terminal domain that extends beyond that of the bacterial enzyme. In addition, structural information of MUTYH is limited to an N- terminal domain fragment (amino acids 65–350) that lacks the C-terminal region completely [24].
The R295C and Q324H MUTYH variations are located in the interdomain connector (IDC) between the catalytic N-terminal and the OG-interacting C-terminal domains (Fig. 2). This region has been shown to be the binding site for the AP endonuclease APE1 [19, 35] and Hus1 of the 9-1-1 complex [23, 24]. The 9-1-1 complex encircles DNA, and it functions to facilitate the ATR-mediated phosphorylation and activation of Chk1, which in turn, elicits cellular responses such as stabilizing stalled replication forks, blocking the firing of late origins of replication, and arresting cells in G2/M phase. The Arg-to-Cys change at position 295 in MUTYH was detected in several MAP cancer patients, either in combination with the Y165C variation or in a monoallelic state [33, 36–40]. Q324H is a common MUTYH variant with an allele frequency of over 40% in some populations [41] and had previously been assumed to be a harmless polymorphism [7, 8, 42]. However, there are several clinical studies that have suggested an increased risk for CRC or lung cancer associated with this variant [43–47]. In addition, recent work from our laboratory has shown that expression of Q324H MUTYH in Mutyh-null mouse embryonic fibroblasts (MEFs) leads to significantly lower OG:A repair levels relative to WT MUTYH indicating that the Q324H allele may increase the potential risk for CRC [12]. In a similar approach, Turco et al. found that cells expressing Q324H MUTYH had a 2-fold increased amount of OG and were hypersensitive to oxidant treatment [25].
Figure 2.
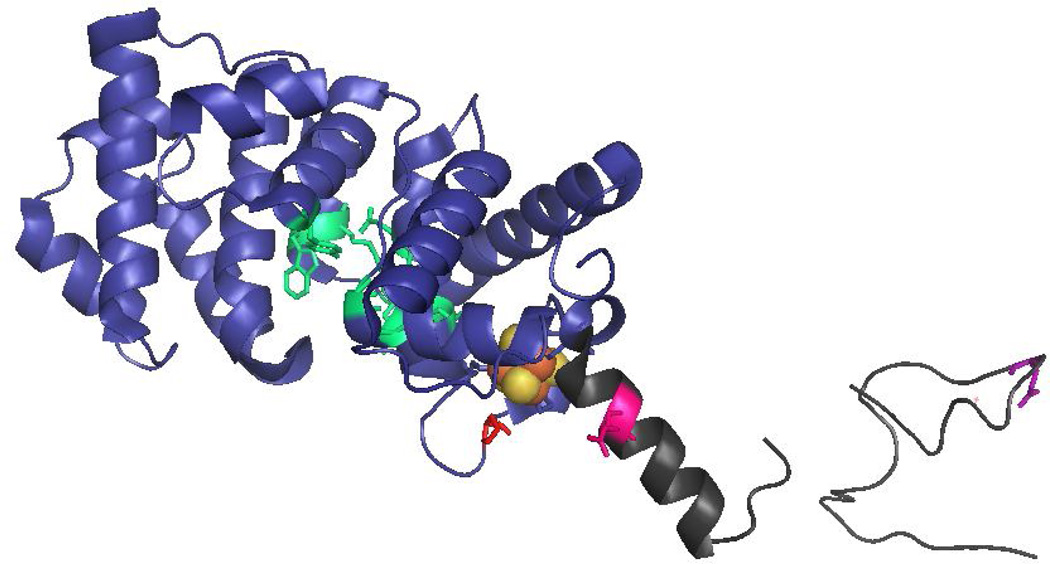
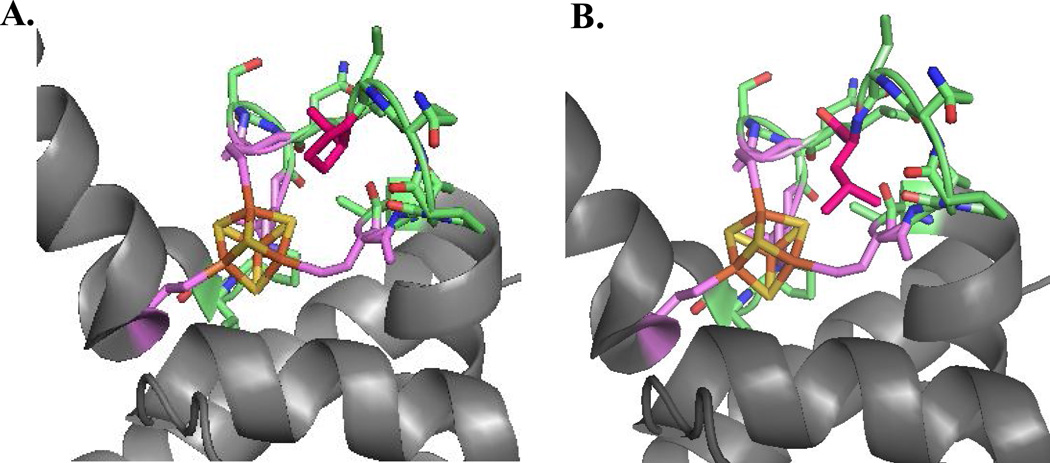
Locations of MUTYH amino acid variations. The X-ray crystal structure the N-terminal domain of MUTYH (residues 65–350) (PDB code 3N5N) illustrates the amino acid positions of P281L, R295C and Q324H MUTYH variants. The N-terminal catalytic domain is shown in blue, the adenine-specific active site pocket in green, the [4Fe-4S]2+ cluster in orange/gold spheres and the IDC/linker region in dark grey. The locations of the WT amino acids Pro 281 (red), Arg295 (pink) and Gln324 (purple) are highlighted in stick form on the ribbon backbone.
The P281L MUTYH variant has been discovered in a small subset of MAP patients [16], and is localized adjacent to the Hus1 and APE1 binding sites. Sequence alignments of MutY enzymes across species shows that Pro281 is strictly conserved and is located in the iron-sulfur cluster loop (FCL) motif of the protein (Fig. 2). The presence of a [4Fe-4S] cluster, and its associated FCL motif, has been observed in a large subset of BER glycosylases [48, 49]. The FCL motif is anchored on each end by coordination of two Cys residues to the [4Fe-4S]2+ cluster to position positively charged residues within the loop for interaction with the DNA substrate [15, 50]. Notably, replacement of the positively charged residues in E. coli MutY has been shown to compromise both damaged substrate recognition and the adenine glycosylase activity [51]. Based on the non-conservative change of a Pro-to-Leu in this key region of MUTYH, it may be anticipated that this variation would alter OG:A mismatch recognition and repair.
Herein, the adenine glycosylase activity and substrate affinity of P281L, R295C, Q324H, P502L and R520Q MUTYH were evaluated. P281L MUTYH was found to have minimal adenine glycosylase activity due to a severely compromised ability to recognize the damaged substrate. The glycosylase and binding properties of P502L and Q324H MUTYH lie at the other end of the spectrum being similar to those of the WT enzyme. However, P502L and Q324H variants exhibit reduced affinity for their respective protein partners PCNA and Hus-1 suggesting that these enzymes may be less active than WT in vivo. In contrast, the R520Q and R295C variants exhibited significant reductions in substrate DNA binding affinity, low amounts of active enzyme and reduced rates constants for the intrinsic base excision catalysis relative to the WT protein. Moreover, the affinity of R520Q MUTYH for PCNA was significantly reduced, suggesting that this feature would reduce overall repair even further in a cellular context. These data reveal the range of functional consequences of exhibited for distinct MUTYH variants. This work also provides a framework for making genetic and clinical correlations of other variants that are localized to similar regions of the protein.
2. Materials and Methods
2.1 Maintenance of Sf9 cells
A suspension culture of Spodopterafrugiperda Sf9 insect cells was passaged and split twice a week in order to maintain a healthy and >95% viable stock of host cells. The cells were supplemented with HycloneSfX-Insect media (Thermo scientific) containing gentamycin antibiotic (final concentration 25 µg/ml).
2.2 Site-directed mutagenesis of pFBDmalE-TEV-MUTYH
ApFBCmalE-TEV-MUTYH plasmid was used for expression of WT MBP-MUTYH that was modified from a previously used expression plasmid [52] by insertion of a TEV recognition site between the MBP tag and the start of the MUTYH gene [53]. Site-directed mutagenesis was used to create mutations in the WTMUTYH coding region of the pFBDmalE-TEV-MUTYH plasmid for P281L, R520Q, P502L, R295C, and Q324H MUTYH using the QuikChange mutagenesis kit (Stratagene) according to the manufacturer’s protocol. The primers are as follows, where the underlined sequence defines the codon change:
-
P281L-1: 5’-GTGTGTACCCCACAGCGCCTACTGTGCAGCCAG-3’
P281L-2: 5’-CTGGCTGCACAGTAGGCGCTGTGGGGTACACAC-3’
-
R520Q-1: 5’-CTGGATAATTTCTTTCAGTCTCACATCTCCACTG-3’
R520Q-2: 5’-CAGTGGAGATGTGAGACTGAAAGAAATTATCCAG-3’
-
P502L-1: 5’-CAGGTGTCCTCTCTGTGCAGTCGGAAAAAGCCC-3’
P502L-2: 5’-GGGCTTTTTCCGACTGCACAGAGAGGACACCTG-3’
-
R295C-1: 5’-AGCCTGTGCCGGGCATGCCAGAGAGTGGAG-3’
R295C-2: 5’-CTCCACTCTCTGGCATGCCCGGCACAGGCT-3’
-
Q324H-1: 5’-GCTCCCAACACTGGCCACTGCCACCTGTGCCTG-3’
Q324H-2: 5’-CAGGCACAGGTGGCAGTGTCCAGTGTTGGGAGC-3’
Plasmid DNA was isolated from XL-1 Blue E. coli cells using a Wizard Plus DNA purification kit (Promega) according to the manufacturer’s protocol. DNA sequencing was used to confirm the mutations, and the resulting plasmids were named pFBDmalE-TEV-P281L MUTYH, pFBDmalE-TEV-R520Q MUTYH, pFBDmalE-TEV-P502L MUTYH, pFBDmalE-TEV-R295C MUTYH, and pFBDmalE-TEV-Q324H MUTYH, respectively.
2.3 Generation of recombinant bacmid
Recombinant bacmids were generated by transformation of the above plasmids into chemically competent DH10Bac E. coli cells. Colonies were screened for the recombinant bacmid using a blue-white screening assay followed by a PCR assay according to the protocol in the Bac-to-Bac kit (Invitrogen).
2.4 Baculovirus transfections
Sf9 cells were transfected with purified recombinant bacmid DNA in a 6-well format. One × 106 cells/ml were added to each well and incubated at 28 °C for at least an hour to allow the cells to attach to the bottom. Two different transfection reactions were set up in the culture plates with 4 µl and 6 µl DNA aliquots respectively. DNA was added to 100 µl of HycloneSfX media supplemented with gentamycin and then incubated for 45 minutes at room temperature with 6 µl of Cellfectin reagent mixed in 100 µl media. Mock transfection mixtures were set up by adding no DNA. Media was removed from the attached cells and the transfection mixture was added along with 0.8 ml of additional media. The cells were incubated at 28 °C for 5 hours, followed by aspiration of the transfection mixture, addition of 2 ml of fresh media, and incubation at 28 °C for 72 hours to generate the first generation of virus particles. The supernatants were then collected and centrifuged at 500 × g for 5 minutes to remove any extraneous cellular debris. The clear supernatants were then transferred to fresh 15 ml conical tubes and stored at 4°C. Two ml of fresh growth media was added to each well and the cells were incubated for another 72 hours at 28 °C. The media containing pseudo-first generation of virus particles was processed as before and stored at 4°C.
2.5 Dot blot assays
Supernatants of transfections and amplifications of the baculovirus were assayed by dot blot using an anti-gp64 monoclonal antibody (eBioscience) to detect the presence of baculovirus as described previously [52]. A baculovirus of known high titer was used as a standard to determine the multiplicity of infection (MOI). Briefly, 2µl of each standard was dotted on a nitrocellullose membrane, in addition to 2 µl of the transfection supernatants and 2 µl of the mock transfection (negative control). The membrane was dried before it was blocked in blocking solution (5% milk in TBST, Tris-Buffered Saline + Tween 20) for one hour at room temperature with shaking. Then, the membrane was washed as follows: 1 × 15 minutes, 2 × 5 minutes in TBST. The membrane was then incubated for one hour with the mouse anti-gp64 antibody (eBiosciences) diluted 1:1000 in TBST and then washed as before. Antibody detection was performed following the procedures outlined in the GE Healthcare ECF western blotting kit.
2.6 Protein expression and purification
A 600 ml suspension culture of Sf9 insect cells at 6 × 105 cells/ml was infected with the baculovirus at a MOI of 3 pfu/cell, which was then incubated at 28°C, shaking for ~66 hours. The cells were harvested by centrifugation at 600 × g for 15–30 minutes in a hanging bucket centrifuge and washed with amylose wash buffer [20 mM Tris-HCl pH 7.5, 200 mM NaCl, 1 mM EDTA + 1X protease and phosphatase inhibitors (Roche)]. Purification was performed as previously described.
2.7 Substrate DNA preparation
The following 30-bp DNA duplex was utilized for in vitro adenine glycosylase assays and EMSAs:
-
5’-CTGTAACGGGAGCTXGTGGCTCCATGATCG-3’
3’-GACATTGCCCTCGAYCACCGAGGTACTAGC-5’,
where X = A, FA, THF; Y = OG.
The X-containing strand (2.5 pmol) was radiolabeled on the 5’ end with [32P]-ATP using T4-polynucleotide kinase (PNK). For active site titration (AST) experiments, additional unlabeled A-containing oligonucleotide strand was added to provide a final concentration of approximately 5% [32P]-phosphate-labeled DNA. The complementary OG-containing strand was then added in 10% excess to the A-containing strand in annealing buffer (20 mM Tris–HCl pH 7.6, 10 mM EDTA, 150 mM NaCl). The DNA duplex was then annealed by heating to 90 °C for 5 min followed by slow cooling to room temperature overnight. For glycosylase assays under single-turnover (STO) conditions, 10% excess of the OG-containing strand was added to 100% labeled A-containing DNA in the annealing buffer. For EMSA experiments a 20% excess of the complement OG strand was added to 100 % radiolabeled FA-containing DNA and the duplex was annealed as mentioned previously.
2.8 Adenine glycosylase assays
Kinetic assays were carried out similar to previous reports for MutY enzymes by our laboratory [52, 54]. In general, the enzyme activity was monitored by assessing the amount of strand scission at the a basic site produced by MUTYH at the A opposite OG in the 30-bp duplex. The following buffers were used for the experiments: our standard MutY assay buffer (40 mM Tris–HCl pH 7.5, 10 mM EDTA, 0.1 mg/mL BSA) or an optimized MUTYH buffer (40 mM Tris–HCl pH 7.5, 10 mM EDTA, 0.1 mg/mL BSA, 5 µM ZnCl2 and 5 mM MgCl2). Enzyme and DNA substrate were incubated at 37 °C and aliquots were removed at various times (20 s to 40 min), quenched with 0.2 M NaOH, and heated to 90 °C for 5 min. An equal volume of for mamide loading dye (0.025% bromophenol blue, 0.025% xylene cyanol, and 80% for mamide in 1XTBE) was added and the samples were heated at 90 °C for an additional 5 min. The samples were then resolved on a 15% (19:1) denaturing polyacrylamide gel in 1X TBE at 1500 V for 1.5 h and visualized by storage phosphor autoradiography.
Active site concentrations used were determined from AST experiments performed using the standard MutY assay buffer containing 60 mM NaCl. For STO experiments, the DNA substrate concentration was either 10 pM or 1 nM, and the enzyme concentration (based on AST) was at least 10-fold above the DNA concentration. Several enzyme and DNA concentrations were tested to ensure the maximum kobs and that the k2 = kobs approximation was valid. The STO experiments were performed in the optimized MUTYH buffer at either 60 or 150 mM NaCl. The analysis of the data and the determination of the rate constants were analogous to that previously reported.
2.9 Electrophoretic mobility shift assays (EMSA)
Quantitative electrophoretic mobility shift assays were performed using the 30-bp duplex substrate containing a central OG:araF Abase pair. Serial dilutions of MUTYH enzymes (based on active fraction) were made in dilution buffer (20 mM Tris-HCl pH 7.5, 10 mM EDTA, and 20% glycerol) at 4°C. Then, 10 pMOG:araF A was incubated with increasing concentrations of enzyme for 30 min. at 25 °C. Samples were electrophoresed at 4°C on an 8% non-denaturing polyacrylamide gel with 0.5X TBE at 120 V for 2 hours. Gels were dried and quantitated via storage phosphor autoradiography using ImageQuant V. 5.2. The Kd values were determined by fitting the data (percent bound substrate versus log [E]) to an equation for one-site ligand binding using Grafit 5.0. Kd values were determined from 4–6 separate experiments. In the case of P281L MUTYH protein, EMSA experiments with a 30-bp OG:THF-containing duplex were also attempted.
2.10 Flag-Hus1 protein overexpression and purification
Flag-tagged Hus1 was overexpressed in Sf9 cells and purified as described previously with a few minor modifications [55]. Sf9 insect cells were grown in HycloneSfX-Insect media supplemented with 10% FBS and 100 units of penicillin and streptomycin/ml. IPEGAL CA630 was used in place of Nonidet P-40 in the lysis buffer [50 mM Tris-HCl, pH 7.6, 0.5% IPEGAL CA630, protease inhibitors (Roche), 1 M NaCl]. After centrifugation, the supernatant was incubated with anti-FLAG agarose for 3 hours at 4 °C via batch binding. Elution was performed using the lysis buffer supplemented with 100 µg/ml Flag peptide (Sigma).
2.11 Pull-Down Assay with PCNA
Ni-NTA His-Bind Superflow resin (QIAGEN) (50 µl) was washed two times with 50 µl Buffer I (20 mM phosphate buffer, pH 7.6, 10% glycerol) and resuspended in 50 µl Buffer I. To this slurry, ~1 µg of purified recombinant human PCNA (PROSPEC) was added and incubated at 4 °C for 2 hours. After centrifugation at 8000 × g for 5 min. at 4 °C, the supernatant was saved (“unbound PCNA”). To the pellet, purified active WT, P502L, or R520Q MUTYH (~84 ng) expressed in insect Sf9 cells was added and incubated at 4 °C for 2 hours. After centrifugation at 8000 × g for 5 min. at 4 °C, the supernatant was set aside again (“unbound MUTYH”). To the pellet, 50 µl of Buffer I + 250 mM imidazole was added, and the mix was incubated for 2 hours at 4°C. After centrifugation at 8000 × g for 5 min. at 4 °C, the supernatant was saved (titled “ELUTE”). The supernatants were fractionated on a 12% SDS-polyacrylamide gel. Gels were stained with Sypro Orange. For western blot analysis, electrophoretic transfer was performed onto a PVDF membrane at 100 V for one hour. The anti-MBP antibody (NEB) was used as the primary antibody for detection of WT, P502L, and R520Q MUTYH, while the anti-His antibody (GE Healthcare) was used as the primary antibody for detection of PCNA. Immunodetection was carried out using the ECF western blotting reagent pack (GE Healthcare) for the anti-MBP and anti-His antibodies following the manufacturer’s protocol.
2.12 Pull-Down Assay with Flag-Hus1
An amylose resin (NEB) slurry (60 µl) was washed two times with 50 µl amylose wash buffer [20 mM Tris-HCl, pH 7.6, 200 mM NaCl, 1 mM EDTA, 1 mM DTT, 1X phosphatase and protease inhibitors each (Roche)]. Approximately 80 ng of active WT, Q324H, and R295C MUTYH was added to the resin with additional amylose wash buffer to a final volume of 50 µl. The MUTYH enzymes were incubated with the resin at 4 °C for 2 hours. After centrifugation at 8000 × g for 5 min. at 4 °C, the supernatant was saved (“unbound MUTYH”). To the pellet, purified Flag-Hus1 (~ 30–50 µg) was added and incubated at 4 °C for 2 hours. After centrifugation at 8000 × g for 5 min. at 4 °C, the supernatant was set aside again (titled “unbound Hus1”). To the pellet, 50 µl of amylose wash buffer + 10 mM maltose was added, and the mix was incubated for 2 hours at 4°C. After centrifugation at 8000 × g for 5 min. at 4 °C, the supernatant was saved (titled “ELUTE”). Analysis was performed as described above. The anti-MBP antibody (NEB) was used as the primary antibody for detection of WT, Q324H, and R295C MUTYH, while the anti-FLAG antibody (Sigma-Aldrich)) was used as the primary antibody for Hus1 detection. Immunodetection was carried out using the ECF western blotting reagent pack (GE Healthcare) for the anti-MBP while a HRP substrate (Thermo Scientific) was used for detection of the anti-FLAG antibody following the manufacturer’s protocol.
3. Results
3.1 Overexpression and active yields of WT MUTYH and MUTYH missense variants
WT MUTYH and the missense variants R295C, P281L, Q324H, P502L, and R520Q MUTYH were expressed as N-terminal maltose binding protein (MBP) fusion proteins. Protein overexpression and purification was carried out using an insect cell-driven baculovirus expression vector system (BEVS) [52]. The Q324H and P502L variants displayed similar expression levels as WT MUTYH (Fig. S1). In contrast, the overexpression levels of P281L, R520Q, and R295C MUTYH were 50%, 40%, and 60% of WT MUTYH, respectively (Fig. S1). The reduced expression of these three variants suggests that these particular amino acid variations may have an effecton protein stability relative to the WT protein.
The amount of total protein isolated after purification and active concentrations of MUTYH and the five variants are listed in Table 1. Measurements of active enzyme fraction are important since total protein measurements also include MUTYH that may be inappropriately folded or truncated, and therefore not catalytically competent. The active enzyme fraction of WT MUTYH and the five variants was determined using active site titration (AST) assays with a 30-bp OG:A-containing DNA substrate using methods we have previously reported [54]. The AST experiment exploits the ability to relate the amplitude of the “burst” phase of product formed under conditions of substrate excess to the amount of active enzyme. The relative amounts of active enzyme fraction in a MUTYH variant relative to the WT protein provide a useful parameter in judging the consequences of the amino acid variations on enzyme function. Notably, the active fraction measured may be influenced by the conditions of the AST experiment; therefore, we have adopted standard conditions for these assays to allow for comparison with previous studies [52, 54]. These condition are also permissive to allow for activity to be observed; notably, however, amino acid variations that significantly alter affinity for the OG:A mismatch and ability to mediate base excision will reduce the active fraction. Thus, the active fraction is a useful gauge of the critical nature of a particular region of MUTYH or amino acid in OG:A recognition and excision [56]. Owing to variation in active enzyme fraction among different preparations of the same variant, we determined the active fraction for several enzyme preparations (Table S1). Based on these analyses, the fractions of active Q324H and P502L missense variants were similar to WT MUTYH (Table 1). In contrast, the active yields of R520Q and R295C MUTYH were approximately 5% of those of WT MUTYH (Table 1). The glycosylase activity was barely detectable for the P281L MUTYH variant and indicated that the active fraction was less than 0.1% of that observed with WT MUTYH. Due to this extremely low amount of active enzyme, detailed analysis of glycosylase kinetic parameters was not feasible with P281L MUTYH.
Table 1.
Total protein, active yield, rate constants, and dissociation constants determined for WT MUTYH and the MAP variants.a
| OG:A DNA 150 mM NaCl |
OG:A DNA 60 mM NaCl |
OG:FA DNA 100 mM NaCl |
OG:FA DNA 30 mM NaCl |
|||
|---|---|---|---|---|---|---|
| Enzyme | % Total protein relative to WTb |
% Active Fraction relative to WTb |
k2 (min−1); completion (%)c |
k2 (min−1) completion (%) |
Kd (nM) | Kd (nM) |
| WT MUTYH |
100 | 100 | 0.90 ± 0.02 100% |
0.90 ± 0.02 100% |
1.9 ± 0.6 | 0.3 ± 0.2 |
| R295C MUTYH |
60 | 5 |
kobs = 0.4 ± 0.1 30% |
0.50 ± 0.03 100% |
>9f | 0.2 ± 0.1 |
| P281L MUTYH |
50 | 0.1 | NCd | MCe | NBd | MBe |
| Q324H MUTYH |
100 | 100 |
kobs = 0.5 ± 0.1 80% |
0.80 ± 0.04 100% |
1.6 ± 0.5 | 0.4 ± 0.2 |
| P502L MUTYH |
100 | 100 | 0.8 ± 0.08 100% |
0.90 ± 0.03 100% |
2.0 ± 0.7 | 0.5 ± 0.1 |
| R520Q MUTYH |
40 | 6 |
kobs = 0.30 ± 0.02 40–60% |
0.40 ± 0.03 100% |
>8f | 0.3 ± 0.2 |
Reported k2 and Kd values are an average of at least three separate experiments; the error is one standard deviation from the average.
These are average values determined from 5–10 individual preparations for each protein. The total and active protein concentrations used to determine these average values are listed in the Supporting Information, Table S2.
Despite enzyme concentration being in excess, the substrate was not always converted completely to product at high salt, suggesting a hampered ability to properly engage the OG:A substrate for catalysis of adenine excision
No cleavage (NC) and no binding (NB) above background levels (≤1%).
Minimal cleavage (MC, ~7%) and minimal binding (MB, 6%) above background levels (≤1%).
Estimated Kd value based on the highest enzyme concentration used.
3.2 Glycosylase activity of WT MUTYH and MUTYH missense variants
Based on the similar kinetic behavior of the MUTYH variants to WT MUTYH, we used a similar minimal kinetic scheme (Scheme 1) to analyze the kinetic parameters associated with base excision catalysis [52, 54]. The rate constant k2 related to the intrinsic catalysis of adenine excision is most readily measured under conditions of single turnover (STO) in order to isolate the base excision step(s) from the product release step (k3) [54]. Since the enzyme concentration is above the Kd value in these experiments, the observed rate constant (kobs) is equal to the true rate constant (k2) for the base excision step(s).
Scheme 1.
Minimal kinetic scheme used to analyze MUTYH glycosylase activity. Substrate binding is defined by k1 and k-1; base excision is defined by k2; and product release is defined by k3 [54].
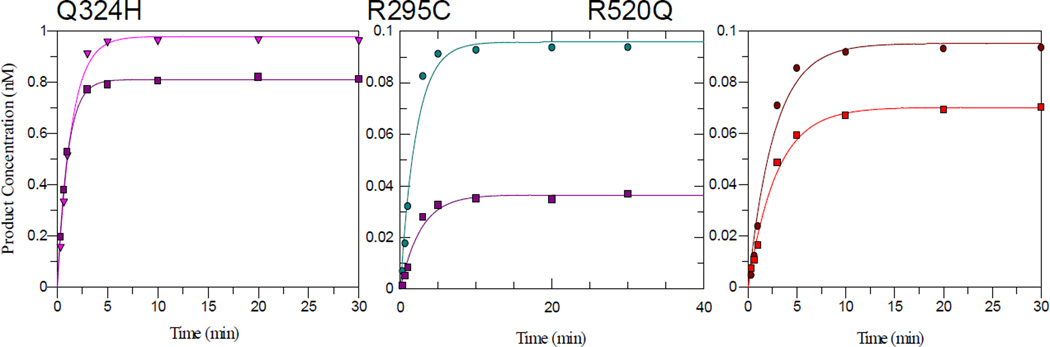
The adenine glycosylase activity of the MUTYH variants with the 30 bp duplex under STO conditions were analyzed in buffer containing 60 and 150 mM NaCl (Fig. S2, Fig. 3). The resulting k2 values determined from fitting of the data from at least three separate experiments are listed in Table 1. The k2 values for WT and P502L MUTYH were within error of each other and were the same at both NaCl concentrations. In contrast, the glycosylase reaction of Q324Husing 150 mM NaCl containing buffer (Fig. 3) reached ~70–80% completion, and fitting of the observed production curve provided a kobs of 0.5 ± 0.1 min−1 that was approximately 2-fold reduced relative to that measured for WT MUTYH. The same results were observed when the DNA concentration was lowered from 1 nM to 10 pM. However, efforts to increase the extent of completion by using a large excess of enzyme resulted in more reduced levels of reaction completion (~60%), suggesting protein aggregation at these enzyme concentrations. Notably, however, in glycosylase assays of Q324H MUTYH in buffer containing 60 mM NaCl (Fig. 3, Fig. S2), the duplex substrate was completely converted to product. Moreover, measured rate constant k2 (0.80 ± 0.04 min−1) under these conditions was within error of that measured for the WT enzyme (Table 1). Interestingly, this sensitivity to the NaCl concentration is a feature that we have previously observed with mutant MutY enzymes that have a compromised ability to bind to the damaged DNA substrate [14].
Figure 3. Adenine glycosylase activity of Q324H, R295C and R520Q MUTYH variants.
Shown are representative plots of adenine glycosylase assays under STO conditions in buffer containing 60 and 150 mM NaCl for Q324H, R295C, and R520Q MUTYH with an OG:A-containing DNA substrate at 37 °C. Complete conversion to product was only attained when the NaCl concentration was lowered to 60 mM (pink triangle for Q324H; teal circle for R295C; and brown circle for R520Q). The values for the rate constant k2 were determined from at least three separate experiments for each enzyme (from different enzyme preparations) and were averaged (Table 1). Note, WT and P502L MUTYH reactions are not altered by NaCl concentration.
The glycosylase reactions under STO conditions for both R520Q and R295C MUTYH exhibited an even greater sensitivity to the buffer salt concentration than Q324H MUTYH in terms of completion end-point (Fig. 3). The reaction of R520Q MUTYH with the OG:A duplex proceeded to levels of 40–60% completion whereas only 30% product formation was observed for the reaction of R295C MUTYH. Lowering the salt concentration to 60 mM, facilitated completion of the adenine excision reactions for both MUTYH variants. Notably, however, at both salt concentrations the rate constant k2, measured under STO conditions, for R295C and R520Q MUTYH were similar and approximately 2-fold reduced relative to WT MUTYH (Table 1). These values did not change when either the enzyme concentration or the DNA concentration was increased, thereby supporting the kobs = k2 assumption. Notably, sensitivity of the reaction completion of these two variants to the buffer NaCl concentration, and the decreased rate of intrinsic catalysis suggests that these mutations may compromise binding interactions that are necessary for base excision catalysis.
3.3 Dissociation constants Kd of WT MUTYH and MUTYH missense variants
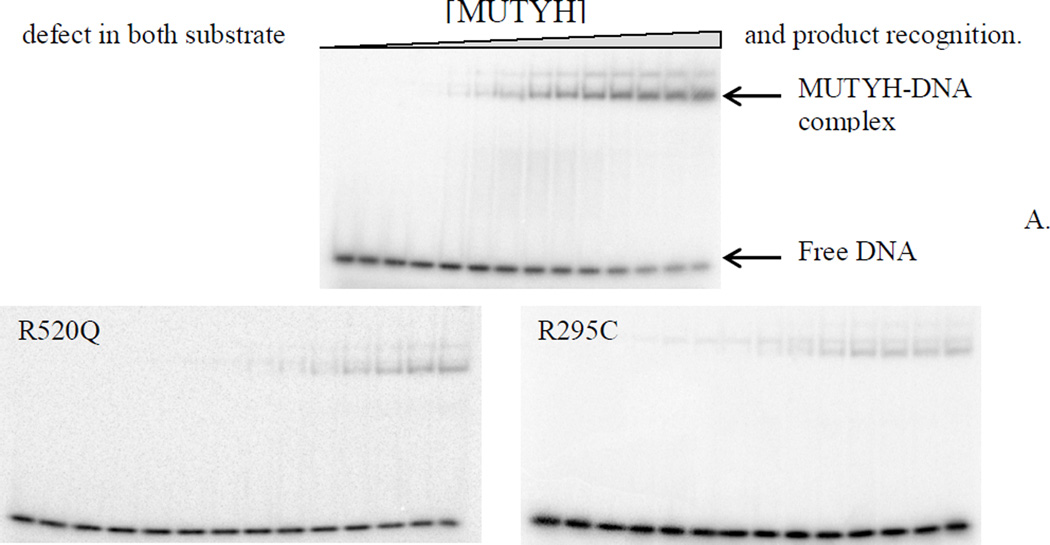
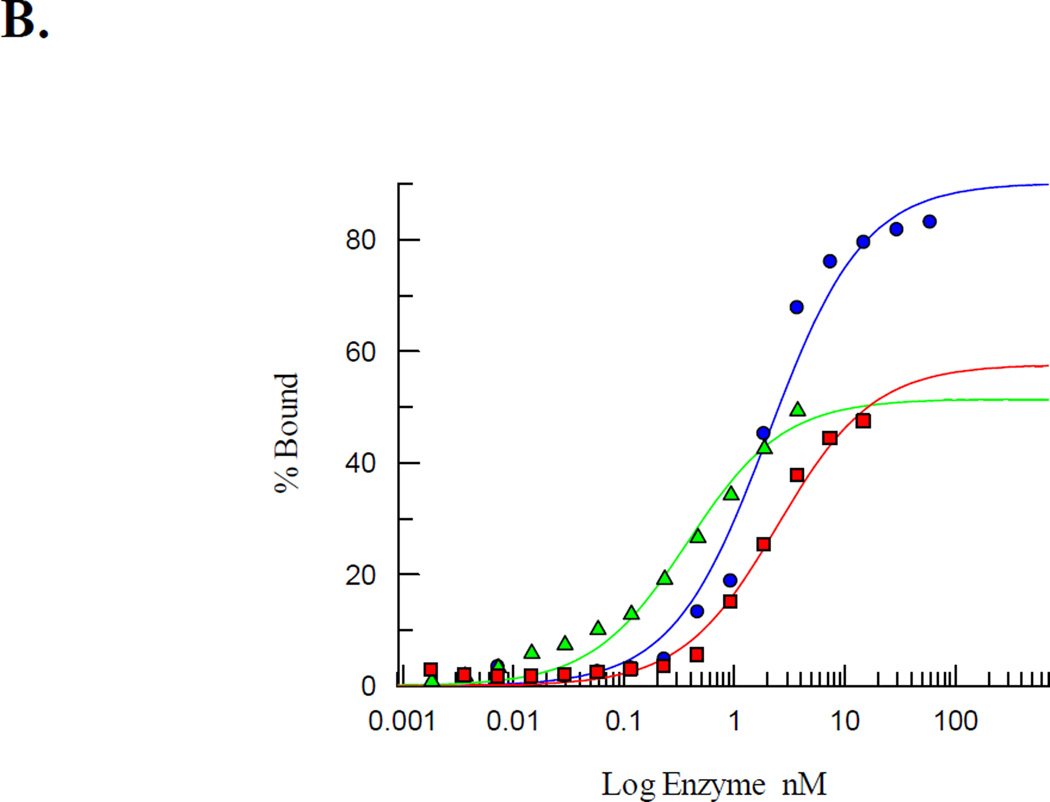
Duplexes containing the non-cleavable substrate analogue 9-(2-deoxy-2-fluoro-β-D-arabinofuranosyl) adenine (araFA) base paired opposite OG were exploited to measure the relevant dissociation constants (Kd) using electrophoretic mobility shift assays (EMSA) of WT MUTYH and the R295C, Q324H, R520Q, and P502L MUTYH variants [57]. The araFA was used in lieu of A to determine the affinity of MUTYH for a substrate-like base pair that would be resistant to enzymatic base removal [57, 58]. WT MUTYH displayed a high binding affinity for the OG:araFA-containing DNA duplex with a Kd value of 1.9 ± 0.6 nM (Table 1). Both Q324H and P502L MUTYH exhibited similar high affinities for the same DNA duplex with Kd values of 1.6 ± 0.6 nM and 2.0 ± 0.7 nM, respectively (Table 1). In contrast, both R295C and R520Q MUTYH displayed severe binding defects under the same conditions (Fig. 4). For both variants, at the highest protein concentration tested, only 30–45% of the OG:FA-containing DNA duplex formed a bound complex indicating a Kd> 8 nM. Due to the possibility that the buffer salt concentration was affecting DNA binding based on the observed sensitivity of the adenine glycosylase activity to the NaCl concentration, EMSA experiments were performed at a lower NaCl concentration of 30 mM. Suprisingly, under these conditions, the Kd values of both R520Q and R295C MUTYH were within error of the Kd determined for the WT, Q324H and P502L MUTYH (Table 1). The keen sensitivity of R520Q and R295C MUTYH to NaCl concentration suggests that Arg520 and Arg295 residues of MUTYH participate in key electrostatic interactions with the DNA substrate.
Figure 4.
R520Q and R295C MUTYH exhibit a significant reduction in substrate affinity. (A) Representative storage phosphor autoradiograms of EMSA with WT, R520Q, and R295C MUTYH and OG:araFA-containing DNA substrate analog duplex at 100 mM NaCl. (B) Representative plot based on EMSA data for Kd determination. Comparison of WT (blue circles), R295C (green triangles), and R520Q (red squares) MUTYH shows that complete binding of the DNA duplex could not be achieved with the R295C or R520Q variants.
EMSA with P281L MUTYH and the OG:araFA duplex were also attempted using similar conditions. In the resulting storage phosphor autoradiograms, only a diffuse band (~6% relative to free DNA) was observed even at the highest enzyme concentration used. Notably, EMSA was also attempted using the corresponding duplex containing OG paired with product analog (THF) and gave similar results indicating that this variant has a defect in both substrate and product recognition.
3.4 Interaction of WT, P502L, and R520Q MUTYH with PCNA
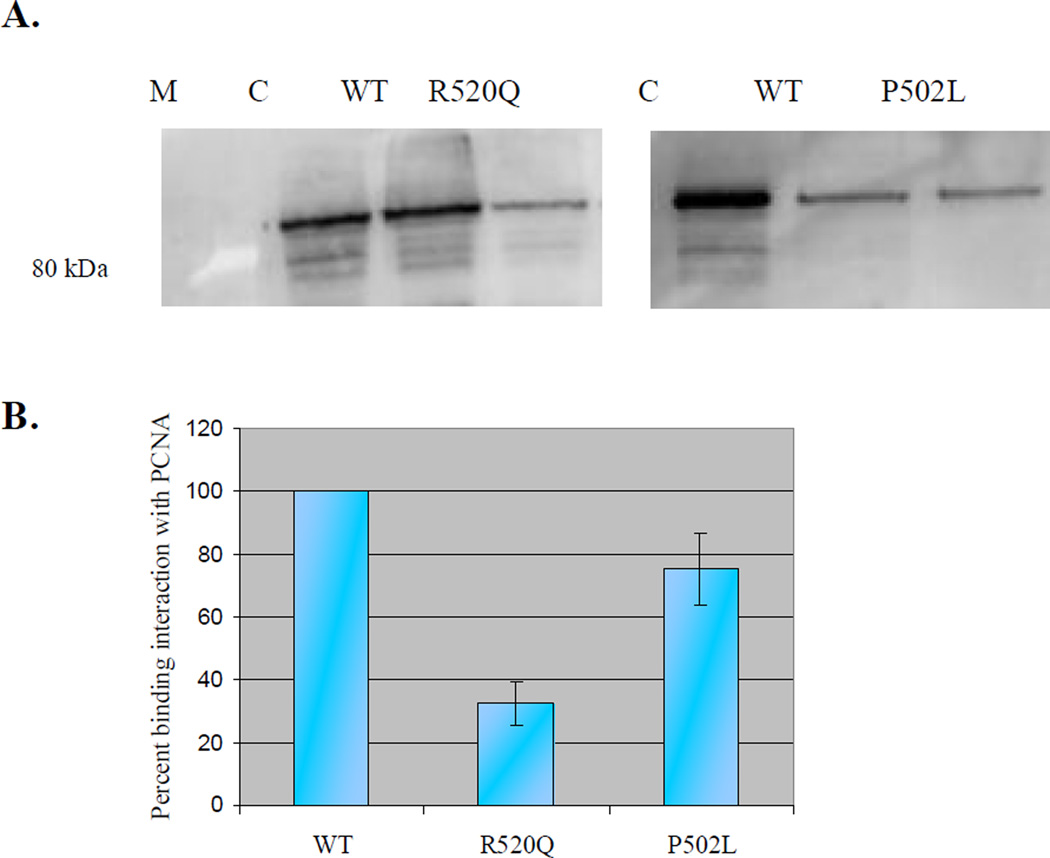
Schizosaccharomyces pombe MutY and human MUTYH have been shown to physically interact with PCNA [19, 59]. Notably, PCNA-binding proteins contain a conserved PCNA-binding motif: the PIP-box [29, 60], which has the consensus sequence Q-x-x-(h)-x-x-(a)-(a), where h represents residues with moderately hydrophobic side chains (i.e., Leu, Ile, Met), a represents residues with highly hydrophobic, aromatic side chains (i.e., Phe, Tyr), and x is any residue. The consensus sequence Q-Q-V-L-D-N-F-F in MUTYH is located at the far C-terminal end (aa 512–519) [19, 35]. To ascertain whether the P502L and R520Q missense variants alter the interaction with PCNA, pull down experiments were performed using purified MBP-tagged MUTYH enzymes and His6-tagged PCNA.
Approximately 1µg of recombinant His6-tagged human PCNA was immobilized on Ni-NTA resin and used to pull down WT MUTYH and the P502L and R520Q MUTYH variants. Neither variant bound to the Ni-NTA resin alone, but when PCNA was immobilized onto the resin prior to incubation with MUTYH, a clear signal for MBP-tagged MUTYH proteins was detected via western blot with an anti-MBP antibody (Fig. 5A). Quantification revealed that the interaction between PCNA and R520Q MUTYH was reduced approximately 75% compared to the interaction between PCNA and WT MUTYH, whereas the interaction between PCNA and P502L MUTYH was only 25% decreased (Fig. 5B). These results indicate that MUTYH mutations near or in the PCNA binding domain alter the ability of the MUTYH protein to interact with PCNA.
Figure 5.
Relative affinity of WT, P502L, and R520Q MUTYH to PCNA. A) PCNA bound to Ni-NTA resin was used to pull down P502L MUTYH (right gel), R520Q MUTYH (left gel), and WT MUTYH (equal amounts of active MUTYH proteins added to each reaction). B) Graphical representation of the binding interactions between MUTYH proteins and PCNA. Amount of WT MUTYH pulled-down was set to 100%. Averages based on at least three separate experiments; error bars represent one standard deviation from the average. C = control: purified WT MUTYH (102 kDa); M = marker (highlight at 80 kDa).
3.5 Interaction of WT, R295C, and Q324H MUTYH with Hus1
Schizosaccharomyces pombe MutY and human MUTYH have also been shown to interact with the PCNA-like protein, the Rad9-Hus1-Rad1 (9-1-1) heterotrimeric complex which plays a role in checkpoint activation in response to DNA damage [23, 24, 61]. MUTYH has been shown to specifically interact with the Hus1 subunit of the 9-1-1 complex in the absence of both the Rad9 and Rad1 subunits, and this interaction was reported to stimulate the glycosylase activity of the protein [23]. There are a handful of MUTYH missense variants located within the reported Hus1-interacting domain (aa 295–350) of the IDC linker that connects the N- and C-terminal domains (Fig. 1) [16]. To investigate whether the R295C and Q324H missense variants affect MUTYH’s ability to interact with Hus1, relative affinity was evaluated using pull-down assays.
Approximately 80 ng of active WT, Q324H, and R295C MUTYH protein was immobilized on amylose resin in amylose wash buffer and used to pull down purified Flag-tagged Hus1. Western blot analyses with both anti-FLAG and anti-MBP antibodies revealed that the larger amounts of WT MUTYH bound to the resin compared to the MUTYH variants. Consequently, all data was normalized to the amount of WT MUTYH immobilized on the amylose resin. The interaction of Q324H MUTYH with Hus1 was decreased ~20% relative to the interaction between WT MUTYH and Hus1, whereas the R295C mutation did not affect the binding of MUTYH with Hus1 (Fig. 6).
Figure 6.
Relative binding of WT, Q324H, and R295C MUTYH enzymes with the Hus1 subunit of the 9-1-1 complex. All data was normalized to the amount of WT MUTYH protein immobilized on resin. Averages based on at least three separate experiments and the error bars represent one standard deviation from the average.
4. Discussion
The discovery of MAP provided the first example of an inherited form of cancer originating as a consequence of defective BER [62, 63]. As part of establishing the MUTYH variant-CRC connection, the variants corresponding to the founder missense variants Y165C and G382D in the bacterial enzyme were analyzed, and were found to be catalytically compromised but not completely inactive [64]. Subsequent work on the mouse homolog provided similar results on the functional consequences for both variants [65]. Analysis of MUTYH activity has been hampered by the low levels of active protein obtained from expression in E. coli; notably, however, the activity analysis is aided by correction of the active fraction to allow for more accurate comparisons between enzymes and preparations [66]. We previously reported improved levels and quality of MUTYH isolated using a baculovirus-infected insect cell expression system (BEVS) [67]. In BEVS expressed Y165C and G382D MUTYH, we observed trends that mirrored faithfully those previously reported for bacterial MutY [68]. These results are consistent with the strict conservation of Tyr 165 and Gly 382 in all MutY enzymes, and their location in key functional regions of the protein [15]. In contrast, in many other MUTYH missense variants, the WT MUTYH amino acid is not strictly conserved across species or lies in a region that is not present in the bacterial enzyme. In order to further illuminate the potential spectrum of functional consequences of MUTYH variants, the adenine glycosylase activity and the mismatch binding affinity of five variants (R295C, P281L, Q324H, P502L, and R520Q MUTYH) in distinct locations were analyzed herein. Additionally, the effects of the relevant alterations on affinity for PCNA and Hus1 were analyzed using pull-down assays and western blotting experiments.
The original classification of Q324H MUTYH as a polymorphism was based in part on the report of glycosylase activity similar to the WT enzyme [69]. Several subsequent studies have indicated similar or small differences in activity [12, 25, 70]. In this study, we found the glycosylase activity to be similar to WT MUTYH under conditions of low buffer salt. A subtle decrease in adenine glycosylase activity of Q324H relative to WT is only observed when using a higher salt containing buffer (150 mM NaCl). Notably, the well-established MAP variant G382D MUTYH also exhibits modest alterations in adenine glycosylase activity similar to Q324H MUTYH [12]. One distinction between G382D and Q324H is that the former showed a reduced affinity for an OG:FA duplex relative to WT [71], while Q324H exhibits WT-levels of affinity with the substrate analog.
We recently demonstrated using a GFP-based repair assay that cells expressing Q324H exhibited reduced levels of OG:A mismatch repair that approach the levels observed for the two known cancer variants Y165C and G382D MUTYH [12]. The subtle defect in the glycosylase activity of the Q324H variant suggested that other factors may reduce OG:A repair efficiency in vivo. Several studies have demonstrated that human MUTYH physically interacts with the Hus1 subunit of the 9-1-1 complex via the IDC region of the protein [23, 24]. In this work, pull-down experiments using the WT, Q324H, and R295C MUTYH enzymes and BEVS-expressed and purified Hus1 protein revealed that the Q324H amino acid variation caused a 20% reduction in the binding interaction between MUTYH and Hus1. This result is consistent with a previous study that showed the Q324H MUTYH had a reduced ability relative to WT MUTYH to pull-down Hus1 and Rad9 from nuclear cell extracts [25]. The interaction of MUTYH with 9-1-1 has also been previously shown to stimulate the glycosylase activity [23]. Moreover, Turco et al. [25] showed that cells expressing Q324H were hypersensitive to oxidant treatment and improperly accumulated in the S-phase of the cell cycle.
Gln 324 is localized within the IDC that links the catalytic N- and OG recognition C-terminal domains. In the structure of an N-terminal fragment of MUTYH lacking the C-terminal domain, the IDC region forms a “docking scaffold” for interactions with other proteins, such as the 9-1-1 complex [24]. We recently discovered a coordinated Zn(II) ion within the IDC and have shown that MUTYH lacking the Zn(II) ion has low levels of active enzyme [56]. We have dubbed this region a “Zinc-Linchpin Motif” and suggest that Zn(II) coordination in the IDC facilitates the ability of the OG-recognition and catalytic domains to effectively engage the DNA substrate to effect adenine excision. The Zn(II) ion is absent in the truncated MUTYH structure presumably due to loss during purification; in addition, parts of the IDC in the X-ray structure are disordered, suggesting that this region may be flexible and adopt alternative conformations [24]. Interestingly, structural studies of hNEIL1 [72, 73] and hTDG [74] showed the predicted 9-1-1 interacting domains to be unstructured, suggesting that the flexibility of these regions enables transient interactions with multiple protein partners. The alteration of Gln with His in the middle of the IDC may alter specific contacts with Hus1 or indirectly affect the ability of this region to adopt the correct conformation necessary for binding to Hus1. Indeed, the replacement of Gln with a His residue in close proximity to the Zn(II) Cys ligands may also alter the Zn(II) site coordination and structure.
P502L, like Q324H, has been proposed to be a single nucleotide polymorphism. Herein, the overexpression levels and the amount of active protein were similar to the WT protein, suggesting that the Pro to Leu change does not alter the folding properties or the stability of MUTYH. Moreover, the glycosylase activity and the binding affinity for an OG:araFA-containing DNA duplex were within error of WT MUTYH. The lack of an effect on the glycosylase activity or mismatch affinity suggests that P502L MUTYH may indeed be a harmless polymorph; however, the observed reduction in the interaction with PCNA suggests that P502L MUTYH may not be efficiently delivered to newly formed OG:A mismatches during DNA replication. Pro 502 is located just outside of the conserved PCNA binding motif (QXXLXXFF) of MUTYH near the end of the C-terminal domain (aa 512–519). The location outside of the PCNA binding motif suggests that the Pro-to-Leu change may indirectly alter the conformation of this region to reduce the interaction with PCNA rather than directly alter specific protein-protein contacts. The reduced affinity for PCNA suggests that the ability to repair OG:A mismatches may be reduced in vivo, and that an increased risk for CRC may be associated with the P502L variation. In this case, additional functional and clinical data is needed to provide a clearer picture of the magnitude of risk associated with this variant.
It has also been suggested that the R520Q missense variation constitutes a SNP [32, 33]; nevertheless, the results on this variant herein strongly suggest that this variant is functionally defective. The levels of overexpressed protein were 40% relative to WT MUTYH; moreover, the levels of active protein were only 6% of that for WT MUTYH preparations. Thus, even if the intrinsic activity is similar to the WT protein, the reduced fraction of active enzyme would lead to low levels of OG:A repair. In addition, the adenine glycosylase assays also showed that catalysis is hampered with this variant under standard assays conditions. In glycosylase reactions, the R520Q variant failed to reach completion and exhibited a 3-fold reduction in rate. Moreover, even using less stringent buffer conditions where reactions reached completion, the rate of adenine excision was found to be 2-fold reduced compared to WT MUTYH. The sensitivity of R520Q to the buffer salt concentration suggests that Arg520 is involved in electrostatic interactions with the DNA phosphodiester backbone that are important for the catalysis of adenine excision [14]. Consistent with the importance of this residue in binding interactions with the substrate, R520Q MUTYH exhibited a significant binding defect compared to WT MUTYH at 100 mM NaCl. Indeed, the lower limit estimated Kd for the OG:araFA duplex with R520Q is at least 4-fold greater than the measured Kd for the WT enzyme. Interestingly, we previously showed that the nearby residue Ser 524 of MUTYH is a phosphorylation site in vivo [52]. Notably, S524A and S524D MUTYH also exhibited compromised affinity for the DNA substrate. These results suggest that this C-terminal tail region participates in engagement of DNA substrate and phosphorylation may serve as a mechanism to fine-tune the interaction with DNA.
The localization of Arg 520 in the middle of the PCNA binding domain of MUTYH and adjacent to the PIP box is consistent with significant reduction (75%) in affinity of R520Q MUTYH for PCNA. Of note, PCNA has also been reported to interact with hMSH6 and to facilitate its localization to base-base mismatches in the newly replicated strand of DNA during MMR [75, 76]. Moreover, MUTYH has been also been shown to physically interact with hMSH6 [77]. These findings suggest that MUTYH may exist in a multi-protein complex with PCNA and hMSH6 to facilitate post-replicative repair of mismatched DNA by directing MUTYH to misincorporated adenine bases opposite OG lesions in the parental DNA strand. Distinct from the other variants, the Arg-to-Gln change appears to alter both specific interactions and proper folding of the C-terminal end of MUTYH needed for efficient base excision and interaction with PCNA. Based on the many functional defects, along with the reduced fraction of catalytically competent enzyme, we would anticipate that R520Q MUTYH is a deleterious allele.
The R295C variation is located in the IDC region of MUTYH near Q324H. The R295C MUTYH protein exhibited low levels of active enzyme (5%) compared to WT MUTYH. In addition, our studies revealed that the adenine glycosylase reactions of the R295C variant only went to completion when the buffer NaCl concentration was lowered to 60 mM. Under these conditions, the rate constant k2 for R295C was reduced 2-fold compared to WT MUTYH. In addition, R295C displayed a significant decrease in substrate binding affinity at 100 mM NaCl that is at least 10-fold reduced relative to the WT enzyme. These data show that the damage recognition and base excision properties are compromised by the Arg-to-Cys change at position 295. This conclusion based on our results is in stark contrast to a previous study of the bacterially expressed R295C MUTYH type 2 nuclear protein that indicated activity similar to the corresponding WT protein [13]. The differences between the two studies may be due in part to our use of the mitochondrial form of the protein; however, those differences are likely to be small in vitro [13]. Indeed, the detailed analysis of the adenine glycosylase kinetics, including active site titration and rate constant measurements, coupled with measurements of substrate affinity using the OG:FA substrate analog described herein was needed to reveal the functional defects of R295C MUTYH. In fact, in activity assays of R295C MUTYH in excess over DNA (STO) at low NaCl concentrations, we also observed complete conversion of the OG:A substrate to product with only a 2-fold slower rate. Since many MUTYH variants are not completely devoid of activity, qualitative analyses of the enzyme activity may be misleading and therefore prompts a more thorough analysis as we have described herein to fully expose functional defects of MUTYH variants, like R295C.
Overall, the enzymatic properties of R295C resemble those of R520Q MUTYH. The consequences of these two variations suggest that both Arg 295 and Arg 520 participate in electrostatic interactions between MUTYH and the DNA phosphodiester backbone that are important for catalysis. Due to the extensive MUTYH-DNA binding interface, the combined effects of numerous electrostatic interactions facilitate formation of the catalytically competent enzyme-OG:A mismatch DNA complex. However, at high NaCl concentration, the sodium cations compete for the phosphate-binding sites more effectively, thereby preventing proper substrate DNA-MUTYH interaction. Thus, loss of important interactions due to the mutated residue in the variant enzyme are revealed more keenly at higher salt concentrations where bolstering non-specific contacts have been removed. The results observed herein are reminiscent of our previous studies with the mutations of Tyr 82 in Ec MutY that corresponds to Tyr 165 in MUTYH [14]. Replacement of the intercalating Tyr residue with large, bulky amino acids such as Phe and Leu did not have nearly as negative an effect on catalysis as substitutions with small side chains like Cys, suggesting that the bulky residue helped stabilize the distorted DNA conformation required for base excision catalysis. Moreover, only Y82C MutY exhibited the high sensitivity to the NaCl concentration suggesting that in the absence of the stabilizing intercalating interaction, the enzyme was more reliant on nonspecific electrostatic interactions to support catalysis.
Despite the location of Arg 295 within the 9-1-1 binding region of the IDC, the Arg to Cys mutation at position 295 did not affect the interactions between MUTYH and Hus1. Arg 295 is located at the beginning to the alpha helix in the IDC that projects away from the globular N-terminal domain (Fig. 2). Several residues including Val 315 and Glu 316 have been shown to be mediators of the interaction with the 9-1-1 complex and are localized after the IDC alpha helix in a region that would be away from the DNA duplex and accessible [24]. Notably, the MUTYH structure reveals the involvement of Arg 295 in a hydrogen-bond network with residues in the N-terminal domain that likely aid in fixing the orientation of the alpha helical portion of IDC [24]. Thus, the role of Arg 295 appears to be two-fold in stabilizing the structure of the IDC and mediating interaction with the DNA duplex. These roles manifest themselves in a reduction in active fraction and reduced affinity for DNA. This is consistent with a role of the IDC not only in mediating protein-protein interactions but in coordinating the proper engagement of the N- and C-terminal domains on OG:A mispairs. The low active enzyme concentration, compromised DNA affinity and reduced catalysis, similar to R520Q and the known cancer variant Y165C MUTYH, argue that R295C is also a dysfunctional allele.
SIFT (Sorting Intolerant From Tolerant), a computational predictor based on the degree of conservation of amino acid residues in alignments of closely related sequences, classified the P281L variation as not tolerated. Herein, we find protein overexpression levels of the P281L MAP variant were approximately half of WT, however, the fraction of the enzyme capable of engaging the OG:A substrate and mediating adenine excision was exceedingly low. The inability to engage the DNA substrate was also reflected by the lack of a distinct P281L-DNA complex using EMSA. These results are also consistent with a previous report using P281L MUTYH expressed in E. coli [78]. The minimal activity and binding observed for P281L MUTYH suggests that this variant will be unable to mediate effective repair of OG:A mismatches in vivo.
The Pro281 residue is located in the solvent-exposed and highly conserved FCL motif of MUTYH. These results underscore the critical role of the FCL motif in efficient recognition of OG:A mismatches. Indeed, the effects on catalysis with this variant are much more dramatic than those observed for the known cancer variant Y165C MUTYH. Tyr 165 is intimately involved in OG:A mismatch detection and adenine excision by intercalating 5’ to the OG [1, 14, 15]. We modeled replacement of Pro 281 with Leu in the crystal structure of the human MUTYH fragment to provide insight into the low activity of this variant (Fig. 7). The bulkiness of a Leu residue and the uniqueness of Pro in facilitating turns, suggests that the Leu residue will not be accommodated as shown in Fig. 7 and will require a substantial conformational change of the FCL. Alterations in FCL conformation will likely restrict the ability of the FCL to effectively recognize and engage the DNA substrate.
Figure 7.
Impact of replacement of Pro 281 in FCL motif of MUTYH. (A) A view of the FCL motif in the structure of MUTYH (PDB code 3N5N) is shown with [4Fe-4S]2+ cluster in orange/gold spheres, the FCL motif in green with side chains shown, and Pro 281 colored pink. (B) In the same orientation, Pro 281 is replaced with a Leu in silico. The snug fit and potential steric clashes suggest that this replacement would significantly alter the conformation of the FCL motif and disrupt ability to bind properly to the DNA substrate.
In summary, this work describes the functional characterization of a handful of MUTYH missense variants that are located within distinct domains of the protein. It highlights the importance of the FCL motif in DNA damage recognition and base removal as the P281L variant displayed the most dramatic deficiencies in overall active yield, glycosylase activity, and DNA binding affinity. Both the R295C and R520Q missense variants exhibited significant reductions in substrate binding affinity and active enzyme fraction, which suggested that these two Arg residues may be involved in electrostatic and hydrogen-bonding interactions that are critical for the formation of the MUYTH-OG:A mismatch DNA complex. The Q324H and P502L MUTYH variants behaved similar to the WT protein with respect to glycosylase activity and mismatch recognition; however, P502L caused a 25% decrease in the interaction with PCNA while Q324H caused a 20% reduction in the MUTYH-Hus1 binding interaction. The reduced interaction of these two variants with their protein partners highlights additional factors that may lead to reduced repair in vivo.
MAP-associated variants provide insight into important features of MUTYH that are needed for MUTYH function in preventing OG-associated mutations and how dysfunction of MUTYH sets the stage for carcinogenesis. In addition, understanding the detailed functional properties of MAP variants provides critical information for clinicians counseling MAP patients. For a given variant, relative dysfunction can be gauged by the intrinsic adenine glycosylase activity, mismatch binding affinity, total and active protein expression levels and altered interactions with cellular protein partners. Other parameters may also come into play such as altered post-translational modification due to the variation. All these factors together should be considered when assessing MUTYH variants as they all have the potential to play a role in MAP. Indeed, confidence in the clinical classification of a particular variant as pathogenic, mildly pathogenic or harmless is engendered when several lines of evidence point in the same direction. In addition, such information on relative CRC risk depending on the harbored variant, will likely become increasingly important as DNA sequencing of known cancer susceptibility genes, like MUTYH, becomes more commonplace.
Supplementary Material
Research Highlights.
We provide a detailed analysis of the glycosylase activity and binding affinity of five MUTYH variants.
A variant in the FCL motif of MUTYH (P281L) has minimal glycosylase activity due to an inability to bind to substrate DNA.
R295C and R520Q MUTYH have low levels of active enzyme, exhibit compromised affinity for damaged DNA and reduced base excision catalysis.
Q324H and P502L have nearly WT glycosylase activity, but exhibit reduced affinity for protein partners Hus1 (of the 9-1-1 complex) and PCNA, respectively.
R520Q MUTYH also exhibits compromised affinity for PCNA suggesting a dual role for Arg 520 in OG:A mismatch recognition and interaction with PCNA.
Acknowledgements
We thank Dr. Lindsey-Boltz and Dr. Aziz Sancar for providing the Hus1 expression plasmid. We also thank Dr. Sheng Cao for synthesis of the araFA-containing oligonucleotide used in this work. The help of Dr. Marc Greenblatt (UVM) in the SIFT analysis of MUTYH is also greatly appreciated. This work is supported by a grant from the National Cancer Institute of the National Institutes of Health (CA067985).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations: 9-1-1, Rad9-Hus1-Rad1; araFA, 9-(2-deoxy-2-fluoro-β-D-arabinofuranosyl) adenine; AST, active site titration; bp, base-pair; BER, base excision repair; ds, CRC, colorectal cancer; double-stranded; Ec, Escherichia coli, EMSA, electrophoretic mobility shift assay; FCL, iron-sulfur cluster loop; GFP, green fluorescent protein; IDC, interdomain connector; MAP, MUTYH-associated polyposis; MBP, maltose-binding protein; MOI, multiplicity of infection; PCNA, proliferation cell nuclear antigen; SIFT, sorting intolerant from tolerant; STO, single turnover; TBE, tris-borate-EDTA; TBST, Tris-buffered saline + Tween; TEV, THF, tetrahydrofuranabasic site analog; tobacco etch virus.
Conflict of Interest statement
The authors declare that there is no conflict of interest
References
- 1.David SS, O’Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neeley WL, Essigmann JM. Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem Res. Toxicol. 2006;19:491–505. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- 3.Dizdaroglu M. Oxidatively induced DNA damage: mechanisms, repair and disease. Cancer Lett. 2012;327:26–47. doi: 10.1016/j.canlet.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 4.Klaunig JE, Kamendulis LM. The role of oxidative stress in carcinogenesis. Annu. Rev. Pharmacol. Toxicol. 2004;44:239–267. doi: 10.1146/annurev.pharmtox.44.101802.121851. [DOI] [PubMed] [Google Scholar]
- 5.Valavanidis A, Vlachogianni T, Fiotakis C. 8-hydroxy-2’ -deoxyguanosine (8-OHdG): A critical biomarker of oxidative stress and carcinogenesis. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2009;27:120–139. doi: 10.1080/10590500902885684. [DOI] [PubMed] [Google Scholar]
- 6.Van Loon B, Markkanen E, Hubscher U. Oxygen as a friend and enemy: How to combat the mutational potential of 8-oxo-guanine. DNA Repair. 2010;9:604–616. doi: 10.1016/j.dnarep.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Al-Tassan N, et al. Inherited variants of MYH associated with somatic G:C to T:A mutations in colorectal tumors. Nature Gen. 2002;30:227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 8.Cheadle JP, Sampson JR. MUTYH-associated polyposis-from defect in base excision repair to clinical genetic testing. DNA Repair. 2006;6:274–279. doi: 10.1016/j.dnarep.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Mazzei F, Viel A, Bignami M. Role of MUTYH in human cancer. Mutat. Research. 2013;743–744:33–43. doi: 10.1016/j.mrfmmm.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 10.Lipton L, et al. Carcinogenesis in MYH-Associated Polyposis Follows a Distinct Genetic Pathway. Cancer Res. 2003;63:7595–7599. [PubMed] [Google Scholar]
- 11.Kundu S, et al. Adenine removal activity and bacterial complementation with the human MutY homologue (MUTYH) and Y165C, G382D, P391L and Q324R variants associated with colorectal cancer. DNA Repair (Amst) 2009;8:1400–1410. doi: 10.1016/j.dnarep.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raetz AG, et al. Cancer-associated variants and a common polymorphism of MUTYH exhibit reduced repair of oxidative DNA damage using a GFP-based assay in mammalian cells. Carcinogenesis. 2012;33:2301–2309. doi: 10.1093/carcin/bgs270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goto M, et al. Adenine DNA glycosylase activity of 14 human MutY homolog (MUTYH) variant proteins found in patients with colorectal polyposis and cancer. Hum Mutat. 2010;31:E1861–E1874. doi: 10.1002/humu.21363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livingston AL, et al. Insight into the roles of tyrosine 82 and glycine 253 in the Escherichia coli adenine glycosylase MutY. Biochemistry. 2005;44:14179–14190. doi: 10.1021/bi050976u. [DOI] [PubMed] [Google Scholar]
- 15.Fromme JC, et al. Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature. 2004;427:652–656. doi: 10.1038/nature02306. [DOI] [PubMed] [Google Scholar]
- 16.Out AA, et al. Leiden Open Variation Database of the MUTYH gene. Hum Mutat. 2010;31:1205–1215. doi: 10.1002/humu.21343. [DOI] [PubMed] [Google Scholar]
- 17.Out AA, Tops CM. 2015 [Google Scholar]
- 18.Yamaguchi S, et al. MUTYH-associated colorectal cancer and adenomatous polyposis. Surg Today. 2014;44:593–600. doi: 10.1007/s00595-013-0592-7. [DOI] [PubMed] [Google Scholar]
- 19.Parker A, et al. Human homolog of the MutY repair protein (hMYH) physically interacts with proteins involved in long patch DNA base excision repair. J. Biol. Chem. 2001;276:5547–5555. doi: 10.1074/jbc.M008463200. [DOI] [PubMed] [Google Scholar]
- 20.Matsumoto Y. Molecular mechanism of PCNA-dependent base excision repair. Prog Nucleic Acid Res Mol Biol. 2001;68:129–138. doi: 10.1016/s0079-6603(01)68095-4. [DOI] [PubMed] [Google Scholar]
- 21.Hayashi H, et al. Replication-associated repair of adenine:8-oxoguanine mispairs by MYH. Curr. Biol. 2002;12:335–339. doi: 10.1016/s0960-9822(02)00686-3. [DOI] [PubMed] [Google Scholar]
- 22.Gu Y, et al. Human MutY homolog, a DNA glycosylase involved in base excision repair, physically and functionally interacts with mismatch repair proteins human MutS homolog 2/human MutS homolog 6. J. Biol. Chem. 2002;277:11135–11142. doi: 10.1074/jbc.M108618200. [DOI] [PubMed] [Google Scholar]
- 23.Shi G, et al. Physical and functional interactions bwtween MutY glycosylase homologue (MYH) and checkpoint proteins Rad9-Rad1-Hus1. Biochem. J. 2006;400:53–62. doi: 10.1042/BJ20060774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luncsford PJ, et al. A Structural Hinge in Eukaryotic MutY Homologues Meidates Catalytic Activity and Rad9-Hus1-Hus1 Checkpoint Complex Interactions. J. Mol. Biol. 2010;403:351–370. doi: 10.1016/j.jmb.2010.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turco E, et al. Understanding the role of the Q338H MUTYH variant in oxidative damage repair. Nucleic Acids Res. 2013;41:4093–4103. doi: 10.1093/nar/gkt130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oka S, Nakabeppu Y. DNA glycosylase encoded by MUTYH functions as a molecular switch for programmed cell death under oxidative stress to suppress tumorigenesis. Cancer Sci. 2011;102:677–682. doi: 10.1111/j.1349-7006.2011.01869.x. [DOI] [PubMed] [Google Scholar]
- 27.Krishna TS, et al. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 28.Gulbis JM, et al. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- 29.Jonsson ZO, Hubscher U. Proliferating cell nuclear antigen: more than a clamp for DNA polymerases. Bioessays. 1997;19:967–975. doi: 10.1002/bies.950191106. [DOI] [PubMed] [Google Scholar]
- 30.Kelman Z. PCNA: structure, functions and interactions. Oncogene. 1997;14:629–640. doi: 10.1038/sj.onc.1200886. [DOI] [PubMed] [Google Scholar]
- 31.Boldogh I, et al. hMYH cell cycle-dependent expression, subcellular localization and association with replication foci: evidence suggesting replication-coupled repair of adenine:8-oxoguanine mispairs. Nucleic Acids Res. 2001;29:2802–2809. doi: 10.1093/nar/29.13.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fleischmann C, et al. Comprehensive analysis of the contribution of germline MYH variation to early-onset colorectal cancer. Int. J. Cancer. 2004;109:554–558. doi: 10.1002/ijc.20020. [DOI] [PubMed] [Google Scholar]
- 33.Olschwang S, et al. Similar colorectal cancer risk in patients with monoallelic and biallelic mutations in the MYH gene identified in a population with adenomatous polyposis. Genet Test. 2007;11:315–320. doi: 10.1089/gte.2007.9995. [DOI] [PubMed] [Google Scholar]
- 34.Lee S, Verdine GL. Atomic substitution reveals the structural basis for substrate adenine recognition and removal by adenine DNA glycosylase. Proc. Natl. Acad. Sci. U.S.A. 2009;106:18497–18502. doi: 10.1073/pnas.0902908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parker AR, Eschleman JR. Human MutY: gene structure, protein functions and interactions, and role in carcinogenesis. Cell. Mol. Life Sci. 2003;60:2064–2083. doi: 10.1007/s00018-003-3053-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sieber OM, et al. Multiple colorectal adenomas, classic adenomatous polyposis, and germ-line mutations in MYH. N. Engl. J. Med. 2003;348:791–799. doi: 10.1056/NEJMoa025283. [DOI] [PubMed] [Google Scholar]
- 37.Aretz S, et al. MUTYH-associated Polyposis: 70 of 71 patients with biallelic mutations present with an attenuated or atypical phenotype. Int. J. Cancer. 2006;119:807–814. doi: 10.1002/ijc.21905. [DOI] [PubMed] [Google Scholar]
- 38.Niessen RC, et al. MUTYH and the mismatch repair system: partners in crime? Hum Genet. 2006;119:206–211. doi: 10.1007/s00439-005-0118-5. [DOI] [PubMed] [Google Scholar]
- 39.Jones N, et al. Increased Colorectal Cancer Incidence in Obligate Carriers of Heterozygous Mutations in MUTYH. Gastroenterology. 2009 doi: 10.1053/j.gastro.2009.04.047. [DOI] [PubMed] [Google Scholar]
- 40.Nielsen MJ, et al. Analysis of MUTYH genotypes and colorectal phenotypes in patients with MUTYH-Associated Polyposis. Gastroenterology. 2009;136:471–476. doi: 10.1053/j.gastro.2008.10.056. [DOI] [PubMed] [Google Scholar]
- 41.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shinmura K, et al. Somatic mutations and single nucleotide polymorphisms of base excision repair genes involved in the repair of 8-hydroxyguanine in damaged DNA. Cancer Lett. 2001;166:65–69. doi: 10.1016/s0304-3835(01)00435-9. [DOI] [PubMed] [Google Scholar]
- 43.Picelli S, et al. Common variants in human CRC genes as low-risk alleles. Eur J Cancer. 2010;46:1041–1048. doi: 10.1016/j.ejca.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 44.Tao H, et al. Association between genetic polymorphisms of the base excision repair gene MUTYH and increased colorectal cancer risk in a Japanese population. Cancer Sci. 2008;99:355–360. doi: 10.1111/j.1349-7006.2007.00694.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kasahara M, et al. Association of MUTYH Gln324His and APEX1 Asp148Glu with colorectal cancer and smoking in a Japanese population. J. Exp. Clin. Canc. Res. 2008;27 doi: 10.1186/1756-9966-27-49. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miyaishi A, et al. MUTYH Gln324His gene polymorphism and genetic susceptibility for lung cancer in a Japanese population. J Exp Clin Cancer Res. 2009;28:10. doi: 10.1186/1756-9966-28-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan Z, Shin J, Fordyce K. Q324H, A germline variant of the MUTY homolog (MYH) gene is strongly associated with early onset adenomatous colorectal polyp formation among African Americans. The 2nd biennial scientific meeting of international society for gastrointestinal hereditary tumours; 2007; Pacifico-Yokohama. [Google Scholar]
- 48.Lukianova OL, David SS. A role for iron-sulfur clusters in DNA repair. Curr. Opin. Chem. Biol. 2005;9:145–151. doi: 10.1016/j.cbpa.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 49.Fuss JO, et al. Emerging critical roles of Fe-S clusters in DNA replication and repair. Biochim Biophys Acta. 2015;1853:1253–1271. doi: 10.1016/j.bbamcr.2015.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thayer MM, et al. Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO. 1995;14:4108–4120. doi: 10.1002/j.1460-2075.1995.tb00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chepanoske CL, et al. Positively charged residues within the Iron-Sulfur Cluster Loop of E. coli MutY participate in damage recogniton and removal. Arch. Biochem. Biophys. 2000;380:11–19. doi: 10.1006/abbi.2000.1890. [DOI] [PubMed] [Google Scholar]
- 52.Kundu S, et al. Ser 524 is a phosphorylation site in MUTYH and Ser 524 mutations alter 8-oxoguanine (OG): A mismatch recognition. DNA Repair. 2010;9:1026–1037. doi: 10.1016/j.dnarep.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brinkmeyer MK. MutY and MUTYH, in Chemistry. Davis CA: University of California, Davis; 2012. Mismatch Recognition and Base Excision Repair of Natural and Unnatural Amino Acid Variations of the Adenine DNA Glycosylases. [Google Scholar]
- 54.Porello SL, Leyes AE, David SS. Single-turnover and pre-steady-state kinetics of the reaction of the adenine glycosylase MutY with mismatch-containing DNA substrates. Biochemistry. 1998;37:14756–14764. doi: 10.1021/bi981594+. [DOI] [PubMed] [Google Scholar]
- 55.Lindsey-Boltz LA, et al. Purification and characterization of human DNA damage checkpoint Rad complexes. Proc Natl Acad Sci U S A. 2001;98:11236–11241. doi: 10.1073/pnas.201373498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Engstrom LM, et al. A zinc linchpin motif in the MUTYH glycosylase interdomain connector is required for efficient repair of DNA damage. J Am Chem Soc. 2014;136:7829–7832. doi: 10.1021/ja502942d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chepanoske CL, Porello SP, Fujiwara T, Sugiyama H, David SS. Investigation of substrate recognition by E. Coli MutY using substrate analogs. Nucleic Acids Res. 1999;27:3197–3204. doi: 10.1093/nar/27.15.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cao S. Department of Chemistry. Salt Lake City, UT: University of Utah; 2010. Synthesis of Fluorinated Analogs of Oxidative DNA Lesions and Their Use to Probe Features of Recognition and Repair by Base Excision Repair Glycosylases. [Google Scholar]
- 59.Chang DY, Lu AL. Functional interaction of MutY homolog with proliferating cell nuclear antigen in fission yeast, Schizosaccharomyces pombe. J Biol Chem. 2002;277:11853–11858. doi: 10.1074/jbc.M111739200. [DOI] [PubMed] [Google Scholar]
- 60.Warbrick E. The puzzle of PCNA’s many partners. Bioessays. 2000;22:997–1006. doi: 10.1002/1521-1878(200011)22:11<997::AID-BIES6>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 61.Chang DY, Lu AL. Interaction of checkpoint proteins Hus1/Rad1/Rad9 with DNA base excision repair enzyme MutY homolog in fission yeast, Schizosaccharomyces pombe. J Biol Chem. 2005;280:408–417. doi: 10.1074/jbc.M406800200. [DOI] [PubMed] [Google Scholar]
- 62.Cheadle JP, Sampson JR. MUTYH-associated polyposis--from defect in base excision repair to clinical genetic testing. DNA Repair (Amst) 2007;6:274–279. doi: 10.1016/j.dnarep.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 63.Nemec AA, Wallace SS, Sweasy JB. Variant base excision repair proteins: contributors to genomic instability. Semin Cancer Biol. 2010;20:320–328. doi: 10.1016/j.semcancer.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Al-Tassan N, et al. Inherited variants of MYH associated with somatic G:C-->T:A mutations in colorectal tumors. Nat Genet. 2002;30:227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 65.Pope MA, Chmiel NH, David SS. Insight into the functional consequences of hMYH variants associated with colorectal cancer: distinct differences in the adenine glycosylase activity and the response to AP endonucleases of Y150C and G365D murine MYH. DNA Repair (Amst) 2005;4:315–325. doi: 10.1016/j.dnarep.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 66.Kundu S, et al. Adenine removal activity and bacterial complementation with the human MutY homologue (MUTYH) and Y165C, G382D, P391L and Q324R variants associated with colorectal cancer. DNA Repair (Amst) 2009;8:1400–1410. doi: 10.1016/j.dnarep.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kundu S, et al. Ser 524 is a phosphorylation site in MUTYH and Ser 524 mutations alter 8-oxoguanine (OG): a mismatch recognition. DNA Repair (Amst) 2010;9:1026–1037. doi: 10.1016/j.dnarep.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Raetz AG, et al. Cancer-associated variants and a common polymorphism of MUTYH exhibit reduced repair of oxidative DNA damage using a GFP-based assay in mammalian cells. Carcinogensis. 2012 doi: 10.1093/carcin/bgs270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shinmura K, et al. Adenine excisional repair function of MYH protein on the adenine:8-hydroxyguanine base pair in double-stranded DNA. Nucleic Acids Res. 2000;28:4912–4918. doi: 10.1093/nar/28.24.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ali M, et al. Characterization of mutant MUTYH proteins associated with familial colorectal cancer. Gastroenterology. 2008;135:499–507. doi: 10.1053/j.gastro.2008.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kundu S. Department of Chemistry. Salt Lake City: University of Utah; 2009. MUTYH-mediated repair of oxidative DNA Damage: Functional charcterization and investigation into post-translation modifications of MUTYH and Variants implicated in colon cancer. [Google Scholar]
- 72.Doublie S, et al. The crystal structure of human endonuclease VIII-like 1 (NEIL1) reveals a zincless finger motif required for glycosylase activity. Proc. Natl. Acad. Sci. USA. 2004;101:10284–10289. doi: 10.1073/pnas.0402051101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hegde ML, et al. The Disordered C-Terminal Domain of Human DNA Glycosylase NEIL1 Contributes to Its Stability via Intramolecular Interactions. J. Mol. Biol. 2013;425:2359–2371. doi: 10.1016/j.jmb.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smet-Nocca C, et al. The thymine-DNA glycosylase regulatory domain: residual structure and DNA binding. Biochemistry. 2008;47:6519–6530. doi: 10.1021/bi7022283. [DOI] [PubMed] [Google Scholar]
- 75.Lau PJ, Kolodner RD. Transfer of the MSH2.MSH6 complex from proliferating cell nuclear antigen to mispaired bases in DNA. J Biol Chem. 2003;278:14–17. doi: 10.1074/jbc.C200627200. [DOI] [PubMed] [Google Scholar]
- 76.Shell SS, Putnam CD, Kolodner RD. The N terminus of Saccharomyces cerevisiae Msh6 is an unstructured tether to PCNA. Mol Cell. 2007;26:565–578. doi: 10.1016/j.molcel.2007.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gu Y, et al. Human MutY homolog, a DNA glycosylase involved in base excision repair, physically and functionally interacts with mismatch repair proteins human MutS homolog 2/human MutS homolog 6. J Biol Chem. 2002;277:11135–11142. doi: 10.1074/jbc.M108618200. [DOI] [PubMed] [Google Scholar]
- 78.Ali M, et al. Characterization of mutant MUTYH proteins associated with familial colorectal cancer. Gastroenterology. 2008;135:499–507. doi: 10.1053/j.gastro.2008.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.