Significance
DNA sequencing has been dramatically expanding its scope in basic life science research and clinical medicine. Recently, a set of polymer-tagged nucleotides were shown to be viable substrates for replication and electronically detectable in a nanopore. Here, we describe the design and characterization of a DNA polymerase–nanopore protein construct on an integrated chip. This system incorporates all four tagged nucleotides and distinguishes single–tagged-nucleotide addition in real time. Coupling protein catalysis and nanopore-based detection to an electrode array could provide the foundation of a highly scalable, single-molecule, electronic DNA-sequencing platform.
Keywords: nanopore sequencing, protein design, polymer-tagged nucleotides, single-molecule detection, integrated electrode array
Abstract
Scalable, high-throughput DNA sequencing is a prerequisite for precision medicine and biomedical research. Recently, we presented a nanopore-based sequencing-by-synthesis (Nanopore-SBS) approach, which used a set of nucleotides with polymer tags that allow discrimination of the nucleotides in a biological nanopore. Here, we designed and covalently coupled a DNA polymerase to an α-hemolysin (αHL) heptamer using the SpyCatcher/SpyTag conjugation approach. These porin–polymerase conjugates were inserted into lipid bilayers on a complementary metal oxide semiconductor (CMOS)-based electrode array for high-throughput electrical recording of DNA synthesis. The designed nanopore construct successfully detected the capture of tagged nucleotides complementary to a DNA base on a provided template. We measured over 200 tagged-nucleotide signals for each of the four bases and developed a classification method to uniquely distinguish them from each other and background signals. The probability of falsely identifying a background event as a true capture event was less than 1.2%. In the presence of all four tagged nucleotides, we observed sequential additions in real time during polymerase-catalyzed DNA synthesis. Single-polymerase coupling to a nanopore, in combination with the Nanopore-SBS approach, can provide the foundation for a low-cost, single-molecule, electronic DNA-sequencing platform.
DNA sequencing is a fundamental technology in the biological and medical sciences (1). Advances in sequencing technology have enabled the growth of interest in individualized medicine with the hope of better treating human disease. The cost of genome sequencing has dropped by five orders of magnitude over the last decade but still remains out of reach as a conventional clinical tool (2, 3). Thus, the development of new, high-throughput, accurate, low-cost DNA-sequencing technologies is a high priority. Ensemble sequencing-by-synthesis (SBS) platforms dominate the current landscape. During SBS, a DNA polymerase binds and incorporates a nucleotide analog complementary to the template strand. Depending on the instrumentation, this nucleotide is identified either by its associated label or the appearance of a chemical by-product upon incorporation (4). These platforms take advantage of a high-fidelity polymerase reaction but require amplification and have limited read lengths (5). Recently, single-molecule strategies have been shown to have great potential to achieve long read lengths, which is critical for highly scalable and reliable genomic analysis (6–9). Pacific Biosciences’ SMRT SBS approach has been used for this purpose but has lower throughput and higher cost compared with current second-generation technology (10).
Since the first demonstration of single-molecule characterization by a biological nanopore two decades ago (11), interest has grown in using nanopores as sensors for DNA base discrimination. One approach is strand sequencing, in which each base is identified as it moves through an ion-conducting channel, ideally producing a characteristic current blockade event for each base. Progress in nanopore sequencing has been hampered by two physical limitations. First, single-base translocation can be too rapid for detection (1–3 μs per base), and second, structural similarities between bases make them difficult to identify unambiguously (12). Some attempts to address these issues have used enzymes as molecular motors to control single-stranded DNA (ssDNA) translocation speeds but still rely on identifying multiple bases simultaneously (13–15). Other approaches used exonuclease to cleave a single nucleoside-5′-monophosphate that then passes through the pore (16), or modified the pore opening with a cyclodextrin molecule to slow translocation and increase resolution for individual base detection (17, 18). All of these techniques rely on detecting similarly sized natural bases, which produce relatively similar current blockade signatures. Additionally, no strategies for covalently linking a single enzyme to a multimeric nanopore have been published.
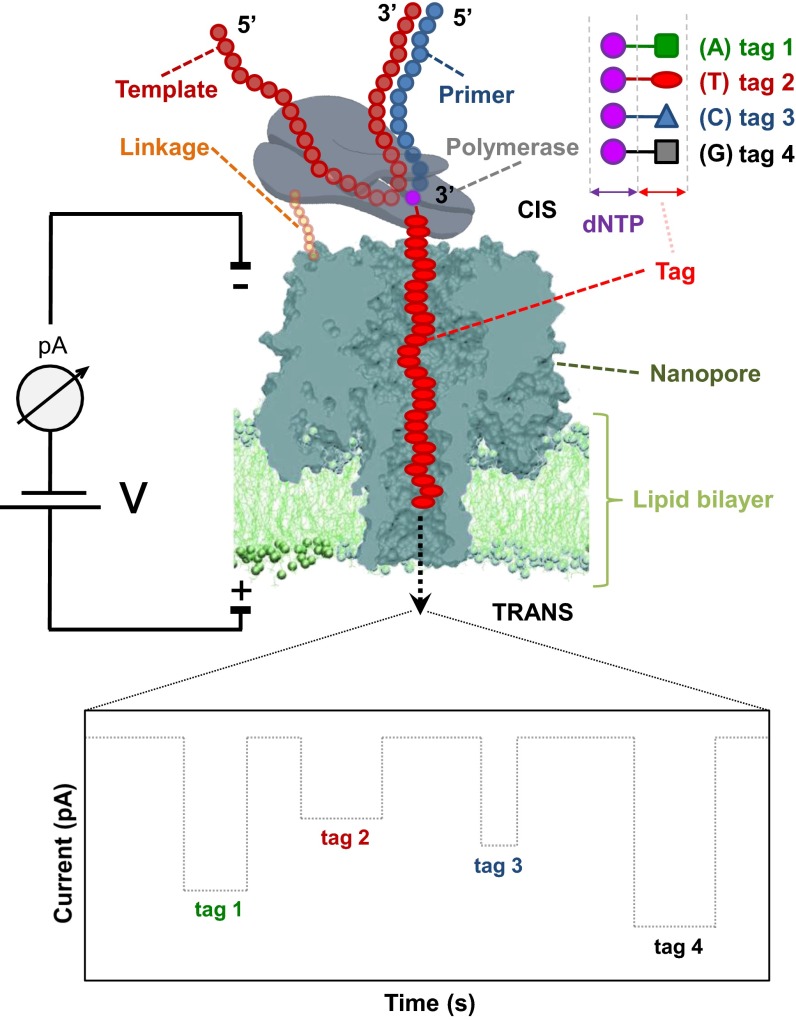
Recently, we reported a method for SBS with nanopore detection (19, 20). This approach has two distinct features: the use of nucleotides with specific tags to enhance base discrimination and a ternary DNA polymerase complex to hold the tagged nucleotides long enough for tag recognition by the nanopore. As shown in Fig. 1, a single DNA polymerase is coupled to a membrane-embedded nanopore by a short linker. Next, template and four uniquely tagged nucleotides are added to initiate DNA synthesis. During formation of the ternary complex, a polymerase binds to a complementary tagged nucleotide; the tag specific for that nucleotide is then captured in the pore. Each tag is designed to have a different size, mass, or charge, so that they generate characteristic current blockade signatures, uniquely identifying the added base. This system requires a single polymerase coupled to each nanopore to ensure any signal represents sequencing information from only one DNA template at a time. Kumar et al. (19) demonstrated that nucleotides tagged with four different polyethylene glycol (PEG) molecules at the terminal phosphate were good substrates for polymerase and that the tags could generate distinct signals as they translocate through the nanopore. These modifications enlarge the discrimination of the bases by the nanopore relative to the use of the natural nucleotides. We recently expanded upon this work by replacing the four PEG polymers with oligonucleotide-based tags and showed that a DNA polymerase coupled to the nanopore could sequentially add these tagged nucleotides to a growing DNA strand to perform Nanopore-SBS (20). Although this work showcased the promise of this technology, it did not describe in detail how to build a protein construct capable of Nanopore-SBS and did not obtain enough data to develop a statistical framework to uniquely distinguish the tagged nucleotides from each other.
Fig. 1.
Principle of single-molecule DNA sequencing by a nanopore using tagged nucleotides. Each of the four nucleotides carries a different polymer tag (green square, A; red oval, T; blue triangle, C; black square, G). During SBS, the complementary nucleotide (T shown here) forms a tight complex with primer/template DNA and the nanopore-coupled polymerase. As the tagged nucleotides are incorporated into the growing DNA template, their tags, attached via the 5′-phosphate, are captured in the pore lumen, which results in a unique current blockade signature (Bottom). At the end of the polymerase catalytic reaction, the tag is released, ending the current blockade, which returns to open-channel reading at this time. For the purpose of illustration, four distinct tag signatures are shown in the order of their sequential capture. A large array of such nanopores could lead to highly parallel, high-throughput DNA sequencing.
Here, we describe the design and characterization of a protein construct capable of carrying out Nanopore-SBS (Fig. 1). A porin attached to a single DNA polymerase molecule is inserted into a lipid bilayer formed on an electrode array. The polymerase synthesizes a new DNA strand using four uniquely tagged nucleotides. The DNA polymerase is positioned in such a way that when the ternary complex is formed with the tagged nucleotide, the tag is captured by the nanopore and identified by the resulting current blockade signature. We first describe the construction and purification of an α-hemolysin (αHL) heptamer covalently attached to a single ϕ29 DNA polymerase using the SpyTag/SpyCatcher conjugation approach (21), followed by binding of this conjugate with template DNA and its insertion into a lipid bilayer array. We confirm that this complex is stable and retains adequate pore and polymerase activities. We verify that the tagged nucleotides developed by Fuller et al. (20) can be bound by the polymerase and accurately discriminated by the nanopore. We develop an experimental approach and computational methods to uniquely and specifically distinguish true tagged-nucleotide captures from background and from other tagged nucleotides. We address ways that tagged-nucleotide captures may be misidentified and demonstrate approaches to correct for these. We further show this protein construct can capture tagged nucleotides during template-directed DNA synthesis in the presence of Mn2+, demonstrating its utility for Nanopore-SBS.
Results
Experimental Platform.
To measure current through a nanopore, we used a complementary metal oxide semiconductor (CMOS) chip containing 264 individually addressable electrodes, which was developed by Genia Technologies. In this first-generation prototype, measurements are taken every ∼1 ms, which necessitated the creation of new tagged nucleotides as described by Fuller et al. (20) To complete the development of Nanopore-SBS, we designed a porin–polymerase conjugate that could function on the Genia chip (Fig. 1). This protein assembly needs to ensure attachment of only one polymerase per pore, placement of polymerase on the cis side of the pore, and preservation of polymerase and pore function. We investigated several approaches to meet these requirements.
Construction of a Porin–Polymerase Conjugate.
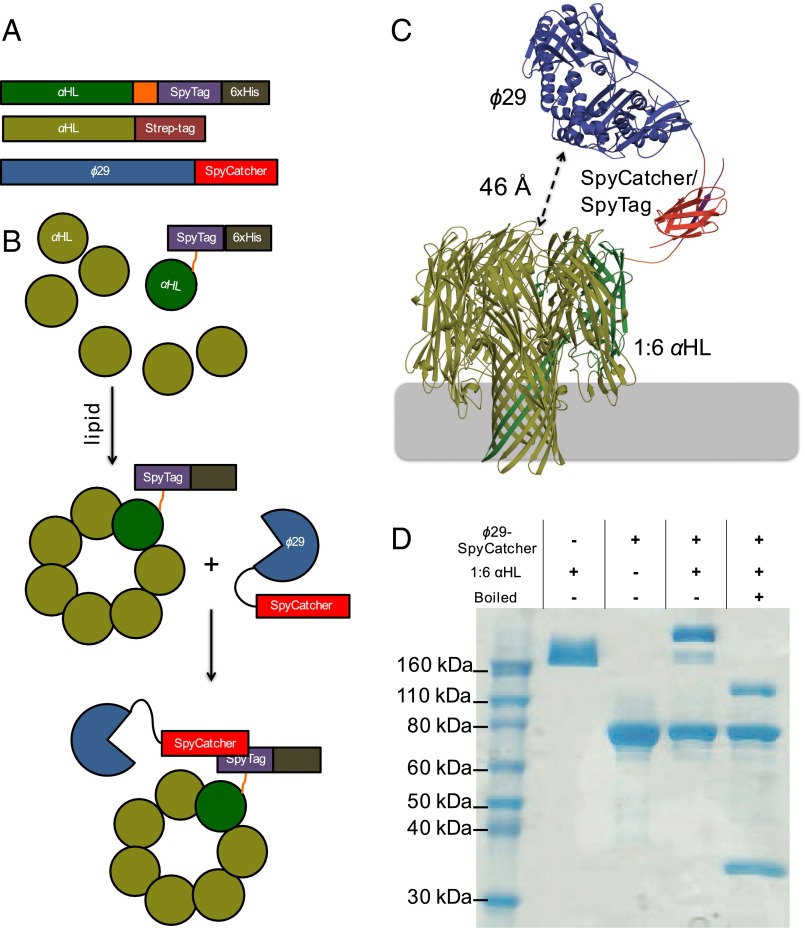
We adopted the SpyCatcher/SpyTag (21) protein conjugation system to couple a single polymerase with one αHL heptamer. Previous work had demonstrated that heteroheptameric αHL pores could be isolated by tagging some subunits with charged residues (22). We then devised a way to purify a 1:6 heptameric pore, where one subunit contains a C-terminal 6×-histidine tag (6×-His-tag) and the other six contain neutral Strep-tags (23). An αHL pore coupled to a single polymerase molecule could then be made from three proteins: αHL with a C-terminal Strep-tag, αHL with a C-terminal SpyTag peptide followed by a 6×-His-tag, and ϕ29 with a C-terminal SpyCatcher (Fig. 2A). The whole porin–polymerase conjugate can be assembled stepwise, by first forming and purifying the 1:6 (SpyTag:unmodified) αHL pore, followed by addition of ϕ29–SpyCatcher (Fig. 2B). Amino acid linker lengths between αHL–SpyTag and ϕ29–SpyCatcher were chosen based on assembling the structures of these proteins (24, 25) into a porin–polymerase conjugate in silico followed by macromolecular modeling of the linkers using Rosetta (26) (SI Appendix, Methods 1 and Figs. S1 and S2). The models demonstrated that these linkers could allow the expected tag exit site of ϕ29 to be positioned above the pore (Fig. 2C). The two αHL subunits were mixed at a ratio of one part αHL–SpyTag–6×-His-tag to six parts unmodified αHL and oligomerized by adding lipid. The αHL porins containing only one unit of SpyTag+6×-His-tag were purified by ion exchange chromatography, which allowed αHL porins containing zero, one, two, or more units of 6×-His-tag to be readily distinguished (SI Appendix, Fig. S3) (23). A single ϕ29 DNA polymerase with a C-terminal SpyCatcher was attached by incubating it with the 1:6 αHL assembly overnight (Fig. 2 A and B). Stoichiometry of the porin–polymerase conjugate was analyzed by SDS/PAGE gels stained for total protein (Fig. 2D). Polymerase function in bulk phase was determined by rolling circle amplification (SI Appendix, Methods 2 and Fig. S4).
Fig. 2.
Assembly of the porin–polymerase construct. (A) Protein constructs used to form the porin–polymerase conjugate include unmodified αHL with a Strep-tag, αHL with a C-terminal SpyTag peptide and 6×-His-tag, and ϕ29 with a C-terminal SpyCatcher domain. (B) Assembly steps. αHL–SpyTag–6×-His and unmodified αHL are oligomerized with lipid, and the 1:6 SpyTag:unmodified assembled porin is purified. Addition of ϕ29–SpyCatcher to the 1:6 pore yields one polymerase per αHL pore. (C) A molecular model generated with Rosetta using the determined structures for ϕ29 polymerase (PDB ID code 2PYJ), αHL (PDB ID code 7AHL), and SpyCatcher/SpyTag (PDB ID code 2X5P). Colors of the proteins match the cartoon representations in A and B. The expected tag exit site on the polymerase and the opening to the nanopore can be in close proximity with distances as short as 46 Å in some models. (D) The stoichiometry in solution of the porin subunits was confirmed by SDS/PAGE without boiling. To confirm the assembly, excess ϕ29–SpyCatcher was added to 1:6 pore. The combination yields only pores with one polymerase attached.
Confirmation of Nanopore Function.
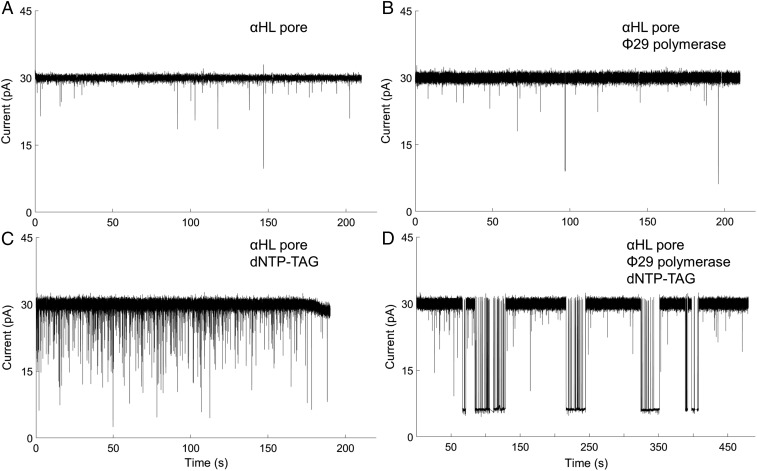
We then confirmed that our porin–polymerase construct was viable for single-molecule polymer-tag detection. First, the 1:6 αHL pore was inserted into the membrane of the Genia nanopore chip, followed by applying a 100-mV potential across the channel. The current through the pore was ∼30 pA (Fig. 3A) in a buffer containing 20 mM Hepes, pH 7.5, and 300 mM NaCl, representing a single pore insertion, thus confirming the 1:6 pore construction yielded viable, active pores. Next, the porin–polymerase conjugate was inserted into the lipid bilayer. The only change was a small increase in the fluctuation of the open channel current [root-mean-square fluctuation (RMSF) = 0.71 ± 0.24 pA; SI Appendix, Methods 3] compared with the pore alone (RMSF = 0.48 ± 0.07 pA), indicating that conjugation of the polymerase does not inhibit pore activity (Fig. 3B). To observe a detectable signal from the tagged nucleotides, the 1:6 αHL pore was inserted into the membrane, followed by addition of all four tagged nucleotides (SI Appendix, Fig. S5). There were noticeable drops in current, indicating that tagged nucleotide causes some transient blockage of the pore (Fig. 3C). Finally, we assembled the porin–polymerase conjugate, followed by addition of a self-priming DNA hairpin with a C nucleotide in the first position on the strand to be replicated (SI Appendix, Fig. S6). This complex was inserted into the membrane, and then the complementary tagged G nucleotide (SI Appendix, Fig. S5: dG6P-T30) was added in a buffer containing noncatalytic Ca2+ ions to allow capture of the tagged nucleotide but prevent base incorporation. The current versus time trace for the fully assembled nanopore–polymerase–template complex shows longer blockade events than the 1:6 αHL pore with tagged nucleotides, and produces a stable minimum current signature for the added dG6P-T30 (Fig. 3D), as well as blockade events similar to those seen in Fig. 3C. This evidence suggests that the designed pore–polymerase is a viable construct to allow single-molecule detection of captured tagged nucleotides. It also demonstrates the detection of single-molecule binding to an enzyme covalently bound to a nanopore.
Fig. 3.
Representative current versus time traces for the various stages of the pore assembly. (A) When neither tagged nucleotide nor polymerase is present, only stable open-channel current is observable. (B) Attachment of polymerase does not change the mean open-channel current. The current root-mean-square fluctuation (RMSF) increase in B may be an indication of the polymerase coupled to the pore. (C) When no polymerase is attached to the pore and tagged nucleotide is introduced, transient events are observed. (D) When polymerase–template is attached to the pore and the complementary base dG6P-dT30 is added, there are prolonged capture events as well as transient events as observed in C.
Detection of Ternary Complex Captures.
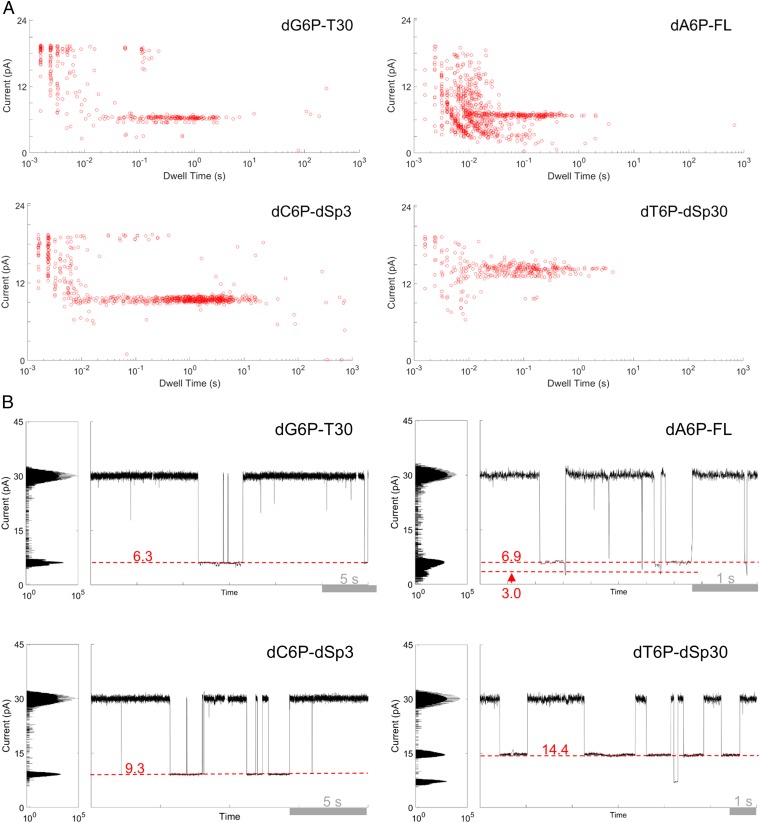
After confirming that the assembled nanopore–polymerase–template complex functioned properly, we sought to determine its efficacy for detecting all four tagged nucleotides (SI Appendix, Fig. S5). Four DNA hairpin oligonucleotides, with different bases at the first query position (SI Appendix, Fig. S6), were used as templates. Porin–polymerase conjugates loaded with these templates were then inserted into a lipid membrane, followed by addition of the complementary tagged nucleotide in a buffer containing noncatalytic divalent metal (Ca2+) ions. Whenever the current was deflected below 70% of the open channel level, the mean current of that deflection, and the duration of the deflection (dwell time) were recorded as current blockade events. The total number of recorded events (n) were as follows: n = 716 for dG6P-T30, n = 812 for dA6P-FL, n = 727 for dC6P-dSp3, and n = 717 for dT6P-dSp30. When a polymerase–template was conjugated to the porin, tagged nucleotides were captured for longer times and at distinct current levels that were not observed when polymerase was absent (Fig. 4 and SI Appendix, Methods 4 and Fig. S7). The dwell time of the tagged-nucleotide background events were <10 ms, whereas with template and polymerase present dwell times of ternary complex captures ranged from ∼10 ms to ∼5 s (SI Appendix, Figs. S8 and S9). All mean currents were outside of a SD of the next closest tag except for those between tagged A (dA6P-FL) and G (dG6P-T30) nucleotides (SI Appendix, Table S1 and Figs. S9A and S10). These two tags could be distinguished by a characteristic two-current level capture of dA6P-FL (Fig. 4 and SI Appendix, Methods 5 and Fig. S11). These results demonstrate that each of the four tagged-nucleotide signals is template specific and can be clustered into distinct current blockade groups relative to the open-channel current reading of the αHL pore (Fig. 4). We collected over 200 ternary complex capture events, which led us to develop computational approaches to accurately distinguish one tagged-nucleotide capture event from another.
Fig. 4.
Tagged-nucleotide discrimination on a semiconductor chip array. All measurements were taken on a pore–polymerase–template complex under noncatalytic conditions where the first base on the template is complementary to the added tagged nucleotide. (A) Current versus dwell time (duration of each current blockade) plots for captures of all tagged nucleotides. Capture events cluster into distinct current and dwell time regions for each tagged nucleotide. (B) Representative single-pore traces of tagged-nucleotide capture shown in A. Current blockade levels for each are marked in red. The blockades demonstrate unique, single-molecule events corresponding to the four distinct tag captures.
Discrimination Among Tagged-Nucleotide Ternary Captures.
We quantified the accuracy of base calls among the four distinct tagged-nucleotide ternary complex captures (TCCs) probing the complementary tagged nucleotides only. First, we determined that the key signal feature to distinguish the events associated with each of the four tagged-nucleotide captures was the median residual current (SI Appendix, Fig. S10). TCCs were differentiated from background captures by requiring their dwell time to be greater than 10 ms. Then, we used a classification algorithm derived from the characteristic dwell time and residual current intervals for each set of ternary capture experiments to estimate the accuracy with which one could call a given TCC event. We found that there was a 78.8–99.2% chance of making an accurate call for each tagged-nucleotide capture by computing a confusion matrix (Table 1). We also determined that the transient captures of tagged nucleotides could be readily distinguished from polymerase-mediated ternary captures (SI Appendix, Tables S2 and S3). In addition, when all four nucleotides were added to a template where the G nucleotide was at the first position, the tagged C nucleotide, dC6P-dSp3, was captured the majority (∼69%) of the time (SI Appendix, Fig. S12 and Table S4). Longer, more distinguishable, captures of the complementary tagged nucleotide versus mismatched ones are supported by the observation that ϕ29’s Michaelis constant is 10 times lower for the correct nucleotide, versus the incorrect ones (27). This result could prove important for future polymerase-engineering steps.
Table 1.
Confusion matrix for discriminating between ternary complex captures using a capture event classification algorithm
| Actual nucleotide | ||||
| Predicted nucleotide | G | A | C | T |
| G | 96.77 | 14.38 | 0.78 | 0.00 |
| A | 2.15 | 78.77 | 0.00 | 0.00 |
| C | 1.08 | 2.05 | 99.22 | 1.61 |
| T | 0.00 | 4.80 | 0.00 | 98.39 |
Each cell represents the percent probability of classifying a particular ternary complex capture (top row labels) as any of the four variants (left column labels). The diagonal (bold text) represents the correct classification. Ternary complex captures were classified by using a custom clustering algorithm based on mean dwell time and residual current level of observed events (Methods).
Detection of Sequential Additions of Nucleotides.
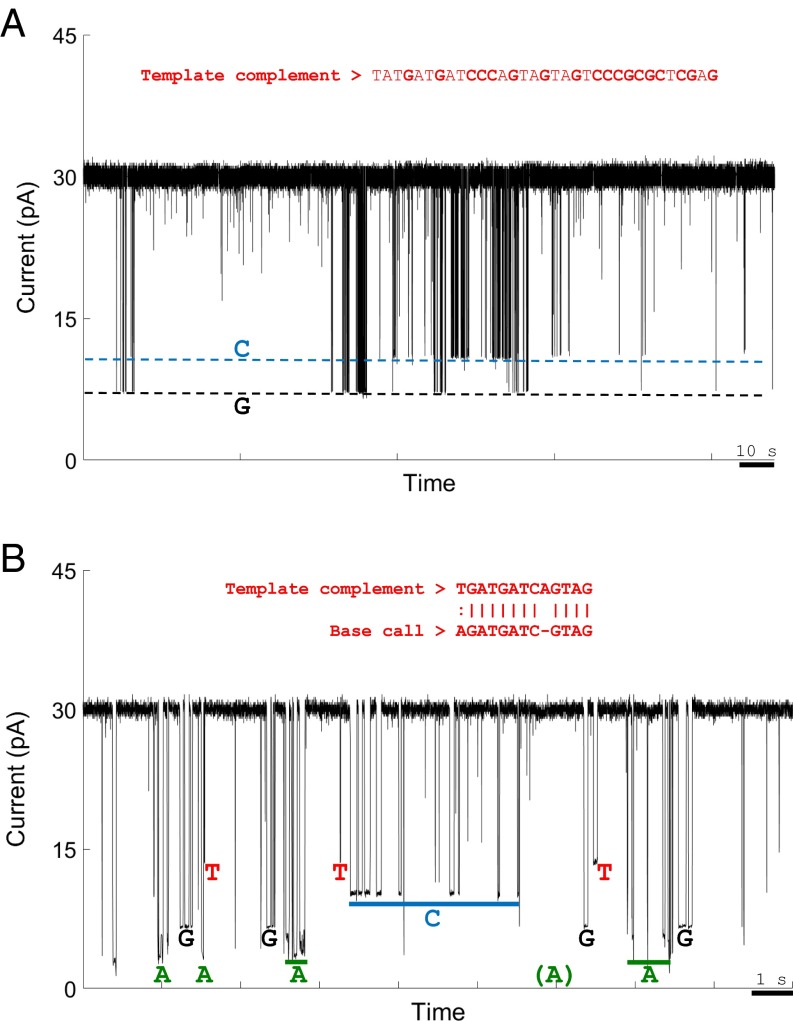
With a functioning protein construct and ability to detect single-nucleotide captures, we then tested whether sequential nucleotide additions could be detected. Tagged-nucleotides dG6P-T30 and dC6P-dSp3, along with natural dATP and dTTP, in catalytic Mn2+ ion-containing buffer were added to a nanopore–polymerase–template assembly with an A nucleotide as the first query base (SI Appendix, Fig. S6). There was clear capture of tags corresponding to the G and C nucleotides (Fig. 5A and SI Appendix, Figs. S13 and S14), and they were detected at the same frequency as predicted from the GC content of the template (SI Appendix, Table S5). The dwell times for these tagged nucleotides were shorter than in the noncatalytic condition, with average dwell times of ∼0.1 s (SI Appendix, Fig. S13C) compared with ∼1.5 s in Ca2+-containing buffer. The transient tagged-nucleotide capture profile was unaffected by the divalent metal (SI Appendix, Fig. S15), and the polymerase-mediated captures were still distinguishable from background.
Fig. 5.
Representative examples of real-time detection of numerous successive tagged-nucleotide incorporations into a self-priming DNA hairpin template catalyzed by nanopore-bound polymerase on the Genia chip. (A) Two base captures of tagged C and G nucleotides with standard A and T nucleotides. Part of the template sequence is shown in red (SI Appendix, Fig. S6). The only captures observed in the trace match the expected levels for dG6P-T30 and dC6P-dSp3. (B) Four-base sequencing. Events with dwell time >10 ms were categorized by manually assigning current blockade events to their respective tag capture boxes (Methods). Homopolymer regions in the template and raw sequencing reads were considered a single base for local sequence alignment. A 12-bp section of such an alignment is shown in red.
Given this result, we used the same template to see whether all tagged nucleotides could be detected under catalytic conditions. Equimolar quantities of the four tagged nucleotides were added in the presence of Mn2+ to perform Nanopore-SBS. Out of 70 single pores obtained, 25 captured two or more tags, whereas six of those showed detectable captures of all four tagged nucleotides. The pore with the most transitions between tag capture levels is shown in Fig. 5B. The other five are displayed in SI Appendix, Fig. S16. All four characteristic current levels for the tags and transitions between them can be readily distinguished. The ability to observe all four tagged nucleotides without the presence of noncatalytic divalent cations, which slows tag release, demonstrates greater potential for sequencing speed then previously shown (20). Homopolymer sequences in the template, and repeated, high-frequency tag capture events of the same nucleotide in the raw sequencing reads were considered a single base for sequence alignment. We recognized 12 clear sequence transitions in a 20-s period. Out of the 12 base transitions observed in the data, ∼85% match the template strand, showing that this method can produce results that closely align to the template sequence. Improved methods that use the time between tag capture events could allow discrimination between high-frequency captures of the same tag and captures due to new complementary tagged-nucleotide binding (SI Appendix, Methods 6 and Fig. S17), which may further enhance the observed sequencing accuracy. These methods could allow more confident sequencing of homopolymer regions in a template.
Discussion
Our results demonstrate that the binding and incorporation of tagged nucleotides by DNA polymerase can be detected on a nanopore array to perform Nanopore-SBS. By constructing a protein conjugate with one polymerase per porin, we ensure the observed activity comes from only one polymerase. The nanopore-attached polymerase retains its ability to bind and incorporate the complementary nucleotide for detection of real-time DNA synthesis. We improved upon our previous work by demonstrating the polymerase can capture tagged nucleotides over a long enough time period to be detected in the pore without the need for noncatalytic divalent cations, which slow the overall DNA synthesis rate. This represents a comprehensive characterization study of a single enzyme conjugated to a protein nanopore.
Previous uses of polymerases to guide DNA through a nanopore (14, 15) did not couple the polymerase directly to the protein pore, instead relying on voltage to initiate the entry of ssDNA into the pore. In contrast, our Nanopore-SBS approach allows clear, template-dependent single-molecule binding observations for a DNA polymerase replicating a target strand of DNA. The use of tags with distinct electrostatic properties enhances the difference between bases and provides a way to perform accurate single-molecule SBS using nanopore detection.
Improvement in the Nanopore-SBS platform allowed the generation of hundreds of tagged-nucleotide captures, an order of magnitude more data than our previous publication, necessitating the implementation of a series of methods to capture, analyze, and interpret this large amount of data. We developed an experimental approach and computational algorithms to uniquely and specifically distinguish true tagged-nucleotide captures from background and from other tagged nucleotides. We also addressed ways that tagged-nucleotide captures can be misidentified and demonstrate approaches to correct for these.
Ongoing work centers on overcoming several challenges such as homopolymer sequencing, improving the yield of functional pores, increasing pore lifetime, and demonstrating chip reusability (SI Appendix, Methods 6–8 and Fig. S18). To increase accuracy, we will continue to improve tag design to achieve better discrimination. Future efforts include optimizing the linker length (28) and composition (29, 30) between αHL and SpyTag, as well as between ϕ29 and SpyCatcher. A better linker should allow more reliable capture and ensure detection of all incorporated tagged nucleotides.
The method of isolating a multimeric nanopore with one unique subunit, followed by covalently attaching a single polymerase, could be extended to other methods of single-molecule detection via a nanopore. Single-molecule enzyme activity, or protein–protein interactions, could be observed by coupling the desired molecular event to the alteration of current through the pore. This technology could serve as the basis for the design of a host of high-throughput molecular sensors. It is likely that other applications of using molecular motors, such as a polymerase (13, 14), helicase (31), or unfoldase (32), to observe DNA or protein in a nanopore could benefit from this work.
The nanopore measurements described here were obtained on a first-generation CMOS-based electrode array chip developed by Genia Technologies, which can potentially scale to billions of sensors (33). Our progress in protein engineering for Nanopore-SBS is currently being carried forward to inform development of the next-generation device and protein constructs. Future work will focus on the development of new polymerases that have more desirable kinetics, new porin–polymerase conjugation strategies, and new tags that produce more distinguishable current blockade signatures. These improvements are being implemented on Genia’s state-of-the-art, massively parallelized nanopore arrays, which can serve as a high-throughput single-molecule sequencing system.
Methods
Protein Expression and Purification.
The ϕ29 DNA polymerase–SpyCatcher construct with an N-terminal Strep-tag was expressed in BL21 DE3 Star cells by growing them in Magic Media (Invitrogen) at 37 °C until OD ∼0.6, followed by overnight growth at 25 °C. Cells were resuspended and lysed by sonication in Polymerase Buffer (PolBuff): 50 mM Tris, pH 7.5, 150 mM NaCl, 0.1 mM EDTA, 0.05% (vol/vol) Tween 20, and 5 mM 2-mercaptoethanol. Benzolase nuclease was added after cell lysis to remove excess bound DNA. The protein was purified using Streptactin columns per the manufacturer’s instructions (IBA). Purified protein was eluted with PolBuff with added desthiobiotin. Both αHL–Strep-tag and αHL–SpyTag-6×-His were expressed in BL21 DE3 Star pLys-S cells grown in Magic Media for 8 h at 37 °C. Each was lysed by sonication in 50 mM Tris, pH 8.0, 200 mM NaCl. Strep-tagged αHL was purified on Streptactin columns and eluted in the same buffer with desthiobiotin. His-tagged αHL was purified with a cobalt column and eluted with 300 mM imidazole.
1:6 Porin Assembly Formation and Isolation.
To form a 1:6 SpyTag:unmodified αHL pore, purified αHL proteins were mixed in a ratio of 1:6 SpyTag construct:unmodified. The lipid 1,2-diphytanoyl-sn-glycero-3-phosphocholine (DPhPC) was added to a final concentration of 5 mg/mL, followed by incubation at 40 °C for 30 min. Lipid vesicles were subsequently popped by adding n-octyl-β-d-glucoside (βOG) to 5% (vol/vol). Fully formed oligomers were separated from vesicles and monomers by size exclusion chromatography (SEC) in 20 mM Hepes, pH 7.5, 75 mM KCl, and 30 mM βOG. Oligomeric protein obtained from the SEC was then run on a MonoS column in 20 mM MES buffer, pH 5.0, 0.1% Tween 20, and eluted with a linear gradient of 0 M to 2 M NaCl. The desired 1:6 assembly eluted after the 0:7 porin because the 1:6 assembly contains a 6×-His-tag. The 1:6 composition was confirmed by adding SpyCatcher protein and observing a size shift of the conjugate on an SDS polyacrylamide gel indicative of only one SpyCatcher molecule per assembled pore.
Polymerase and Template Attachment.
Purified ϕ29 and the desired template were bound to the pore by incubating two molar equivalents of polymerase and four equivalents of DNA template per 1:6 pore overnight at 4 °C. The full tertiary complex was isolated by SEC in 20 mM Hepes, pH 7.5, 150 mM KCl, 0.01% Tween 20, and 5 mM tris(2-carboxyethyl)phosphine. Isolated fractions were characterized by SDS/PAGE to confirm the presence of ϕ29 and αHL conjugate. Formed complexes were tested for polymerase function by rolling circle amplification.
Lipid Bilayer Formation.
Synthetic lipid 1,2-di-O-phytanyl-sn-glycero-3-phosphocholine (Avanti Polar Lipids) was diluted in tridecane (Sigma-Aldrich) to a final concentration of 15 mg/mL. A single lipid bilayer was formed on a silanized CMOS chip surface containing an array of 264 Ag/AgCl electrodes. The automated lipid spreading protocol used an iterative buffer and air bubble flow to mechanically thin the membrane. During this step, voltage was applied across the lipid bilayer to detect its capacitance, which directly correlates to structural integrity of the membrane. An empirically determined capacitance threshold value of 5 fF/μm2 was used to classify the properly formed, single lipid bilayer to conclude the thinning protocols.
Pore Insertion.
The automated pore insertion method consisted of two voltage protocols: (i) initially constant DC voltage was applied at 160 mV for 1 min, immediately followed by (ii) a linearly increasing voltage ramp from 50 to 600 mV with a 1 mV/s incremental step. The smoothly increasing voltage gradient amplified the electrical driving force guiding the nanopores into the lipid bilayer. If a cell became active, that is, had a measured current between 10 and 50 pA, we considered this event a pore insertion, due to the measured increase in conductance across the bilayer. Immediately after this event, this cell was turned off to prevent additional pore insertions. In this way, the probability of multiple pore insertions above the same electrode array element was minimized.
Nanopore Experiments.
All TCC experiments were performed in a buffer containing 300 mM NaCl, 3 mM CaCl2—providing the noncatalytic divalent cations to probe nucleotide binding/unbinding events—and 20 mM Hepes, pH 7.5. For sequencing experiments, this buffer was modified by replacing CaCl2 with 0.1 mM MnCl2 as a catalytic cation source during the polymerase extension reaction to initiate and sustain sequential nucleotide additions along the template DNA. Purified porin–polymerase–template conjugates were diluted in buffer to a final concentration of 2 nM. After pumping a 5-μL aliquot to the cis compartment, single pores were embedded in the planar lipid bilayer that separates two compartments (denoted cis and trans), each containing ∼3 μL of buffer solution. Experiments were conducted at 27 °C with 5 μM tagged nucleotides added to the cis well.
Data Acquisition.
The ionic current through the nanopore was measured between individually addressable Ag/AgCl electrodes coupled to a silicon substrate integrated electrical circuit. This consisted of an integrating patch-clamp amplifier (Genia Technologies), which provided a constant 100-mV potential across the lipid bilayer in voltage-clamp mode. Data were recorded at a 1-kHz bandwidth in an asynchronous configuration at each cell using circuit-based analog-to-digital conversion and noise filtering (Genia Technologies), which allows independent sequence reads at each pore complex. During the various experimental steps, a precision syringe pump (Tecan) was used in an automated fashion to deliver reagents into the microfluidic chamber of the CMOS chip at a flow rate of 1 μL/s. Software control was implemented in Python, which interfaced with the pump via an RS 232 communication protocol.
Event Detection and Data Analysis.
Ionic current blockade events were identified using a custom event detection algorithm implemented in MATLAB (2014b; MathWorks). Briefly, an event was identified by selecting segments that deflected from open-channel current (IO = ∼30 pA at 100 mV in 300 mM NaCl, 3 mM CaCl2, and 20 mM Hepes, pH 7.5) below a cutoff value of 70% of IO (21 pA) to a stable current level (IB) with a minimum dwell time of >10 ms. For each nanopore experiment, event searches were performed to obtain the average residual current level (with respect to open channel) for each capture event (IRES). Statistical analysis was performed to determine the mean, median, and SD of each capture event by fitting a Gaussian to a histogram of IB values. The residual current blockade was defined as follows: IRES% = IRES/IO, whereas the duration of the event in the deflected segment corresponds to the dwell time. Mean dwell time and residual current of each event in an experimental set was accumulatively quantified using scatter plots and box-and-whisker plots. On each box plot, the central red mark represents the median, whereas the bottom and top blue edges of the box are the first and third quartile median values, respectively. The whiskers extend to the lowest and highest values within 1.5 interquartile range of the first and third quartile medians. Alternatively, average dwell time/residual current probability histograms were generated by plotting each bin normalized by the total number of observed events.
Classification of Capture Events.
As a conservative classification method, we have identified the TCC events as all events clustered inside the tag capture box defined by a mean dwell time interval of 10−2 to 10+1 s and a normalized current blockade (or residual current) region bounded by the first and third quartile median values (lower/upper bounds) of the normalized current blockage boxplots—for a particular tagged nucleotide—respectively (Fig. 4). The lower bound of the dwell time interval (10 ms) corresponds to the background cutoff (SI Appendix, Fig. S9), whereas the upper bound was selected to filter out clogged pores from the TCC event set. Mean and median residual currents and SDs are determined after Gaussian fitting of the TCC event histograms.
Supplementary Material
Acknowledgments
This work was supported by NIH Grant R01 HG007415 and Genia Technologies, Inc.
Footnotes
Conflict of interest statement: The Nanopore SBS technology has been exclusively licensed by Genia. In accordance with the policy of Columbia University, the coinventors (S. Kumar, M.C., C.T., Z.L., S. Kalachikov, J.J.R., and J.J.) are entitled to royalties through this license. G.M.C. is a member of the Scientific Advisory Board of Genia, other potential conflicts are described here: arep.med.harvard.edu/gmc/tech.html.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1608271113/-/DCSupplemental.
References
- 1.Shendure J, Lieberman Aiden E. The expanding scope of DNA sequencing. Nat Biotechnol. 2012;30(11):1084–1094. doi: 10.1038/nbt.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470(7333):187–197. doi: 10.1038/nature09792. [DOI] [PubMed] [Google Scholar]
- 3.Soon WW, Hariharan M, Snyder MP. High-throughput sequencing for biology and medicine. Mol Syst Biol. 2013;9(1):640. doi: 10.1038/msb.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen CY. DNA polymerases drive DNA sequencing-by-synthesis technologies: Both past and present. Front Microbiol. 2014;5(JUN):305. doi: 10.3389/fmicb.2014.00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuller CW, et al. The challenges of sequencing by synthesis. Nat Biotechnol. 2009;27(11):1013–1023. doi: 10.1038/nbt.1585. [DOI] [PubMed] [Google Scholar]
- 6.Perkins TT, Quake SR, Smith DE, Chu S. Relaxation of a single DNA molecule observed by optical microscopy. Science. 1994;264(5160):822–826. doi: 10.1126/science.8171336. [DOI] [PubMed] [Google Scholar]
- 7.Smith SB, Cui Y, Bustamante C. Overstretching B-DNA: The elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271(5250):795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 8.Rief M, Clausen-Schaumann H, Gaub HE. Sequence-dependent mechanics of single DNA molecules. Nat Struct Biol. 1999;6(4):346–349. doi: 10.1038/7582. [DOI] [PubMed] [Google Scholar]
- 9.Harris TD, et al. Single-molecule DNA sequencing of a viral genome. Science. 2008;320(5872):106–109. doi: 10.1126/science.1150427. [DOI] [PubMed] [Google Scholar]
- 10.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323(5910):133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 11.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci USA. 1996;93(24):13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng Y, Zhang Y, Ying C, Wang D, Du C. Nanopore-based fourth-generation DNA sequencing technology. Genomics Proteomics Bioinformatics. 2015;13(1):4–16. doi: 10.1016/j.gpb.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cherf GM, et al. Automated forward and reverse ratcheting of DNA in a nanopore at 5-Å precision. Nat Biotechnol. 2012;30(4):344–348. doi: 10.1038/nbt.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manrao EA, et al. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat Biotechnol. 2012;30(4):349–353. doi: 10.1038/nbt.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laszlo AH, et al. Decoding long nanopore sequencing reads of natural DNA. Nat Biotechnol. 2014;32(8):829–833. doi: 10.1038/nbt.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clarke J, et al. Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nanotechnol. 2009;4(4):265–270. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- 17.Astier Y, Braha O, Bayley H. Toward single molecule DNA sequencing: Direct identification of ribonucleoside and deoxyribonucleoside 5′-monophosphates by using an engineered protein nanopore equipped with a molecular adapter. J Am Chem Soc. 2006;128(5):1705–1710. doi: 10.1021/ja057123+. [DOI] [PubMed] [Google Scholar]
- 18.Ayub M, Hardwick SW, Luisi BF, Bayley H. Nanopore-based identification of individual nucleotides for direct RNA sequencing. Nano Lett. 2013;13(12):6144–6150. doi: 10.1021/nl403469r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar S, et al. PEG-labeled nucleotides and nanopore detection for single molecule DNA sequencing by synthesis. Sci Rep. 2012;2:684. doi: 10.1038/srep00684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuller CW, et al. Real-time single-molecule electronic DNA sequencing by synthesis using polymer-tagged nucleotides on a nanopore array. Proc Natl Acad Sci USA. 2016;113(19):5233–5238. doi: 10.1073/pnas.1601782113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zakeri B, et al. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc Natl Acad Sci USA. 2012;109(12):E690–E697. doi: 10.1073/pnas.1115485109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howorka S, Cheley S, Bayley H. Sequence-specific detection of individual DNA strands using engineered nanopores. Nat Biotechnol. 2001;19(7):636–639. doi: 10.1038/90236. [DOI] [PubMed] [Google Scholar]
- 23.Davis R, Chen R, Bibillo A, Korenblum D, Dorwart M. 2014. Nucleic acid sequencing using tags. US Patent Application 14/073,445.
- 24.Song L, et al. Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science. 1996;274(5294):1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 25.Berman AJ, et al. Structures of phi29 DNA polymerase complexed with substrate: The mechanism of translocation in B-family polymerases. EMBO J. 2007;26(14):3494–3505. doi: 10.1038/sj.emboj.7601780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leaver-Fay A, et al. ROSETTA3: An object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santos E, Lázaro JM, Pérez-Arnaiz P, Salas M, de Vega M. Role of the LEXE motif of protein-primed DNA polymerases in the interaction with the incoming nucleotide. J Biol Chem. 2014;289(5):2888–2898. doi: 10.1074/jbc.M113.530980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson-Mosher A, Shinar T, Silver PA, Way J. Dynamics simulations for engineering macromolecular interactions. Chaos. 2013;23(2):025110. doi: 10.1063/1.4810915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reddy Chichili VP, Kumar V, Sivaraman J. Linkers in the structural biology of protein-protein interactions. Protein Sci. 2013;22(2):153–167. doi: 10.1002/pro.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein JS, Jiang S, Galimidi RP, Keeffe JR, Bjorkman PJ. Design and characterization of structured protein linkers with differing flexibilities. Protein Eng Des Sel. 2014;27(10):325–330. doi: 10.1093/protein/gzu043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Derrington IM, et al. Subangstrom single-molecule measurements of motor proteins using a nanopore. Nat Biotechnol. 2015;33(10):1073–1075. doi: 10.1038/nbt.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nivala J, Marks DB, Akeson M. Unfoldase-mediated protein translocation through an α-hemolysin nanopore. Nat Biotechnol. 2013;31(3):247–250. doi: 10.1038/nbt.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merolla PA, et al. Artificial brains. A million spiking-neuron integrated circuit with a scalable communication network and interface. Science. 2014;345(6197):668–673. doi: 10.1126/science.1254642. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.