ABSTRACT
Chemical modifications have been extensively exploited to circumvent shortcomings in therapeutic applications of small interfering RNAs (siRNAs). However, experimental designing and testing of these siRNAs or chemically modified siRNAs (cm-siRNAs) involves enormous resources. Therefore, in-silico intervention in designing cm-siRNAs would be of utmost importance. We developed SMEpred workbench to predict the efficacy of normal siRNAs as well as cm-siRNAs using 3031 heterogeneous cm-siRNA sequences from siRNAmod database. These include 30 frequently used chemical modifications on different positions of either siRNA strand. Support Vector Machine (SVM) was employed to develop predictive models utilizing various sequence features namely mono-, di-nucleotide composition, binary pattern and their hybrids. We achieved highest Pearson Correlation Coefficient (PCC) of 0.80 during 10-fold cross validation and similar PCC value in independent validation. We have provided the algorithm in the ‘SMEpred’ pipeline to predict the normal siRNAs from the gene or mRNA sequence. For multiple modifications, we have assembled ‘MultiModGen’ module to design multiple modifications and further process them to evaluate their predicted efficacies. SMEpred webserver will be useful to scientific community engaged in use of RNAi-based technology as well as for therapeutic development. Web server is available for public use at following URL address: http://bioinfo.imtech.res.in/manojk/smepred.
KEYWORDS: Chemically modified siRNA, cm-siRNAs, efficacy prediction, RNAi, siRNA modifications, siRNA, small interfering RNA, webserver
Introcuction
Short interfering RNA (siRNA) based mRNA knockdown technique is presently one of the standard methods in molecular biology to study gene function.1 Theoretically, siRNA can target any complementary mRNA and leads to degradation of latter employing RNA interference (RNAi) machinery.2,3 This property encouraged the idea to use siRNAs as potential next-generation class of therapeutics especially against viruses.4 Currently many siRNA-based therapeutics are already under advancement in various stages of clinical trials.5,6 However, uses of siRNA molecules as therapeutics face some challenges. These include low efficacy of mRNA inhibition, off targets, immunogenicity, low serum stability against the nucleases, cell specific delivery problems, target site recognition, accessibility and its cleavage.7-9 To overcome most of these shortcomings, siRNA molecules are engineered by introducing chemical modifications.9,10
Natural RNAs comprises of ribonucleotide-moieties, composed of sugar (Ribose), phosphate (phosphodiester bond) and nitrogenous bases Adenine (A), Guanine (G), Cytosine (C), and Uracil (U).11 With modifications on these constituent molecules, sphere of their functionality has been enhanced.8 Numerous studies on chemical modifications directed on siRNAs are reported in the literature aimed to improve their therapeutic potential.6 Testing different chemical modifications at different positions and their combinations on siRNAs are vast. For example to test 30 chemical modifications on 21-mer double-stranded siRNA sequence there would be 1260 (30 modifications × 1time × 21 per strand × 2 strands of siRNA) instances. The complexity increases exponentially by increasing permutations and combinations of multiple modifications at various positions within the same siRNA. Experimentally testing these huge number of combinations would involve loads of time and cost. Therefore, the development of an algorithm that can help in selecting the appropriate chemical modifications in a siRNA is highly desirable.
There are many bioinformatics repositories for siRNAs or related microRNA molecules available like siRecords,12 HuSiDa,13 HIVsirDB,14 VIRsiRNAdb15 miRBase,16 VIRmiRNA17 or naturally occurring modified nucleotide database, RNAMDB.11 Recently we have developed “siRNAMod” repository of chemically modified siRNAs.18 Additionally, many web servers are existing to predict the efficacy of siRNAs e.g. siPRED19 BIOPREDsi20 MysiRNA,21 desiRm,22 VIRsiRNApred23 etc. However, none of these methods predict efficacy for the chemically modified siRNAs (cm-siRNAs). We have provided assessment of various siRNA prediction algorithms and their related information in the comparison section below. For this endeavor, we have developed SMEpred workbench from experimentally validated cm-siRNAs using support vector machine (SVM) to forecast the modulation of chemical modifications on siRNAs (Fig. 1).
Figure 1.
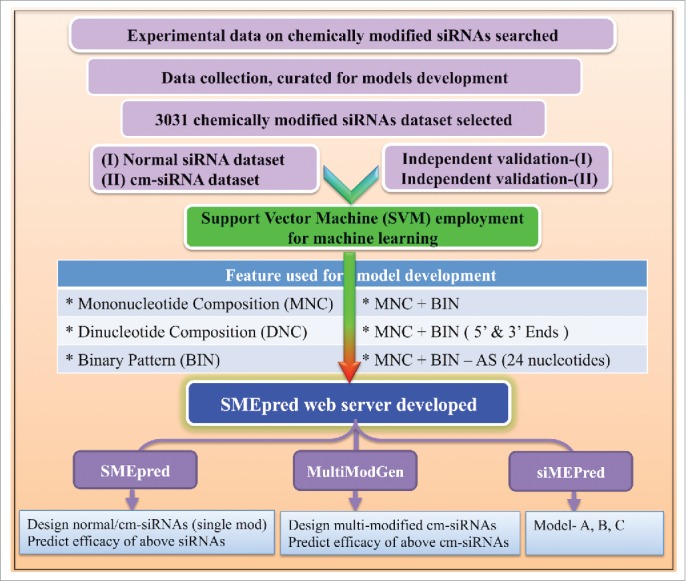
Diagramatic representation of of SMEpred workbench development and its components.
Results
10-fold cross validation
SMEpred model evaluation done by 10-fold cross validation with Hetero-T2728 dataset and independent validation data set Hetero-V303 is presented in Table 1. The mononucleotide composition (MNC) of sequence shows the best performance via SVM technique. The composition of siRNA is one of the key criteria in defining the efficacy of siRNA. During training/testing, we achieved PCC of 0.80 for MNC (mononucleotide composition). Dinucleotide composition (DNC) and binary (BIN) sequence features did not show significant increase in PCC (Pearson correlation coefficient) value. The hybrid models also exhibited varied performance with PCC values ranging from 0.30 to 0.77 (Table 1).
Table 1.
Performance achieved by SMEpred models for 10-fold cross validation and independent validation in terms of Pearson Correlation Coefficient (PCC) using SVM for Hetero-T2728 and Hetero-V303 datasets.
| S. No. | Model Name | Feature | PCC (10-fold validation) Hetero-T2728 | PCC (Independent Validation) Hetero-V303 |
|---|---|---|---|---|
| 1A | MNC | Mononucleotide composition (SS and AS sequence) | 0.80 | 0.808 |
| 2 | DNC | Dinucleotide composition (SS and AS sequence) | 0.53 | 0.48 |
| 3 | BIN | Binary pattern (SS and AS sequence) | 0.33 | 0.37 |
| 4 | MNC + BIN | Composition + Binary (SS and AS sequence) | 0.32 | 0.38 |
| 5 | MNC + BIN (10-mer both sequences) | Composition + Binary (10ntds SS and AS sequences both 5′and 3′ end) | 0.30 | 0.38 |
| 6 | MNC + BIN (7-mer both sequences) | Composition + Binary (7 ntds SS and AS sequences both 5′and 3′ end) | 0.42 | 0.39 |
| 7 | MNC + BIN-AS (24) | Composition + AS binary (24 ntds length) | 0.75 | 0.81 |
| 8 | MNC + BIN-AS (seed-8) | Composition + AS seed Binary (8 ntds from 5′-end) | 0.78 | 0.83 |
| 9 | MNC + BIN-AS (seed-10) | Composition + AS seed Binary (10 ntds from 5′-end) | 0.78 | 0.83 |
| 10 | MNC + BIN-AS (mid-5) | Composition + AS mid Binary (9 to13 ntds from 5′-end) | 0.77 | 0.80 |
| 11B | MNC + BIN-AS (seed 13) | Composition + AS Binary (13 ntds from 5′-end) | 0.77 | 0.86 |
| 12C | MNC + BIN-AS (last 8) | Composition + AS Binary (last 8 ntds from 3′-end) | 0.76 | 0.78 |
| 13 | MNC + BIN-AS (8-5-8) | Composition + AS Binary (Initial 13+ last 8 ntds) | 0.75 | 0.81 |
| 14 | MNC-BIN-SS (24) | Composition + SS binary (only sense sequence 24 ntds) | 0.58 | 0.59 |
(S. No. = serial number; SS = sense strand; AS = Antisense strand; ntds = nucleotides; A, B, C are the models finally used on the webserver).
The hybrids models include nucleotide composition of both sequences and the BIN of sense strand as well as antisense strands, which showed PCC of 0.32. Besides, hybrid combinations like MNC with BIN of 7 nucleotides, 10 nucleotides of sense as well as antisense strand from both 5′ and 3′ends were checked. They too did not show increased performance as compared to MNC. Antisense sequence is more important as it finally bind to the cognate mRNA, so we further analyzed its component fragments. These sections include seed region (8 nucleotides from 5′-end) [model MNC + AS (seed-8)], mid region (5 nucleotides starting from 9 to 13th position) [model MNC +AS (mid-5)] and the 3′end (8 nucleotides from 3′-end) [model MNC +AS (last 8)] or combination of former 2 (13 nucleotides from 5′end). Their corresponding correlation values are provided in Table 1. We have also tested the same features on the Homo2100 dataset (Supplementary Table S1) and found the slightly higher correlation as compared to hetero data set with PCC value of 0.86 for MNC Homo-T1900. Further the hybrid models ranges in correlation with PCC value from 0.38 to 0.84.
Independent evaluation
Independent dataset (Hetero-V303) from the primary data set (Hetero-3031) was used to authorize performance of models generated. Independent dataset consists of similar sequences similar to those used in model development but are not used in the training/testing. We observed that the model based on composition features of modified siRNAs performed well in model development and validation (Table 1). For MNC independent validation, we achieved a PCC of 0.80 for Hetero-V303 data set. Other models also showed the comparable PCC values for independent validation. For hybrid model of composition and binary pattern of seed region (13 nucleotides) and binary pattern for last 8 nucleotides, we achieved PCC value of 0.86 and 0.78 respectively for independent validation dataset. The scatter plot showing the correlation between the predicted and experimental efficacies of the chemically modified siRNAs during independent validation is depicted in the Fig. 2.
Figure 2.
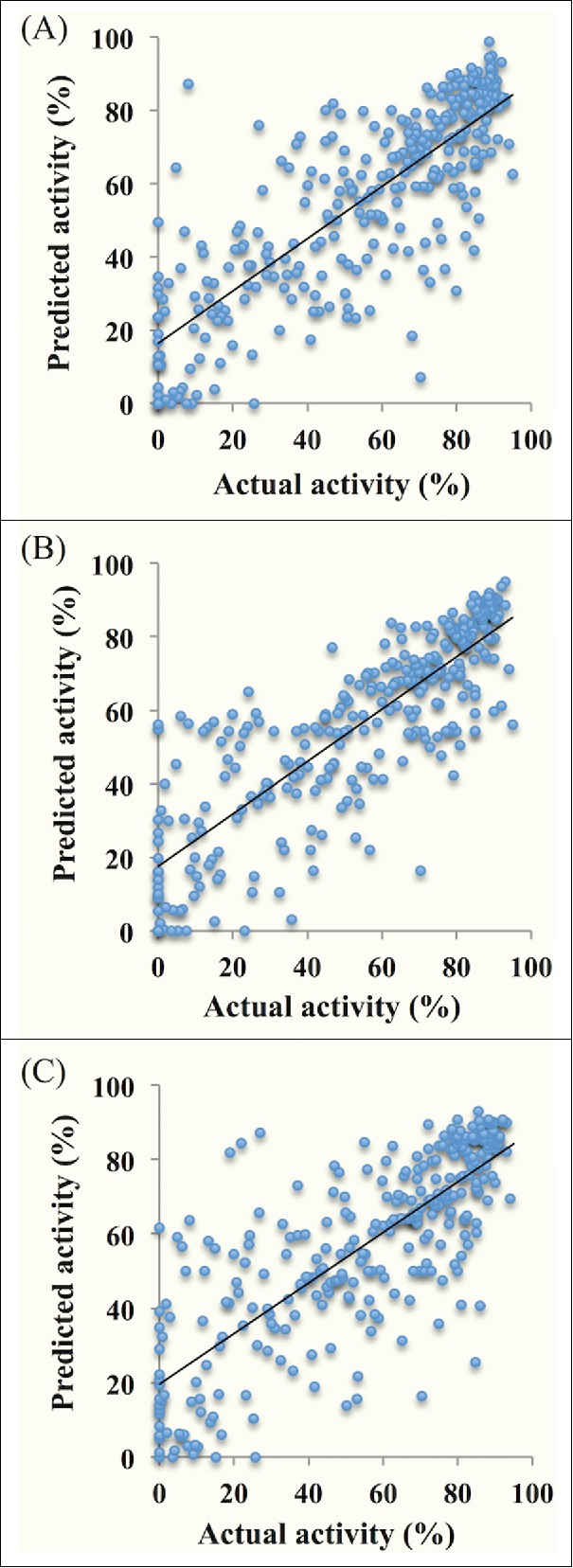
Scatter plot of percentage activities between actual and predicted chemically modified siRNA activities from independent validation data set Hetero-V303. (A) Model-A; Mononucleotide nucleotide composition (MNC); (B) Model-B; Composition (MNC) and antisense strand binary (5′-end 13 nucleotides); (C) Model-C; Composition (MNC) and antisense strand binary (3′-end 8 nucleotides).
Likewise, Homo independent validation was performed using Homo-V210 data set, which also showed the comparable PCC values as that of their training/testing models (Supplementary Table S1). Finally, we built a web server based on best performance models according to PCC value and biological significance of the cm-siRNAs.
Web server components
We developed the workbench for predicting efficacy of cm-siRNAs, with 3 components namely SMEpred, MultiModGen and SiMEPred tool.
SMEpred
SMEpred is a support vector machine based method of predicting efficacy of 21-mer long nucleotide siRNAs. This algorithm is actually the pipeline of various modules and requires mRNA or gene sequences in fasta format. In the first step, the sequence is divided in 21-mer long sequences, followed by predicting efficacy of each siRNA with a score. Score of 100, 80 to 90 and 70 to 80 represents very high efficacy, high efficacy and moderate efficacy respectively.
Further, user can select the siRNA of choice (by clicking) from the generated siRNA list, which will be subsequently chemically modified at each position computationally. Each single siRNA develops 1260 cm-siRNAs, with one modification type on each 21 positions on both siRNA strands. Next the efficacy of these modified cm-siRNAs will be predicted based on the MNC-model (Model-A). This algorithm also offers its output as efficacy values ranging form 0 to 100, with 100 as the best cm-siRNA and 0 with no effect on silencing the target gene. The pictorial representation of SMEpred is displayed in Fig. 3.
Figure 3.
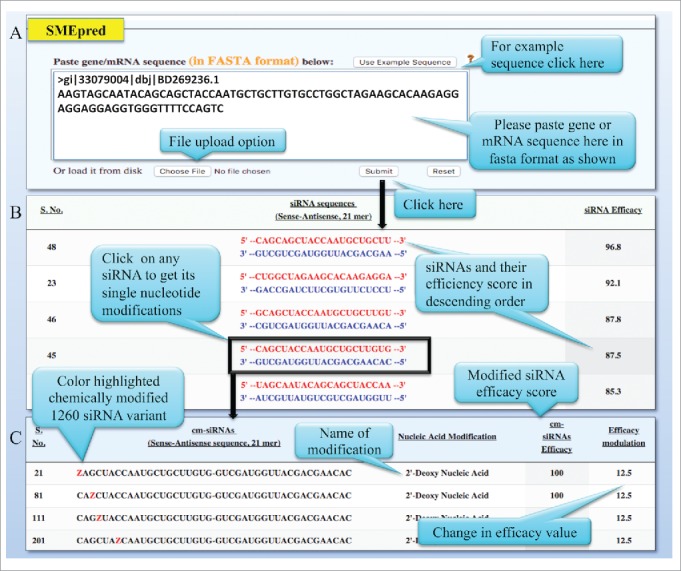
Pictorial representation of working of SMEpred pipeline. (A) Write or paste the gene or mRNA sequence in the space provided, in fasta format and click submit button. (B) Normal siRNAs that are 21-mer long nucleotides are generated from the provided sequence along with their predicted efficacy scores. (C) Further select one of the siRNA (by clicking on it) and it will redirect to the next page with 1260 cm-siRNA. This page displays cm-siRNA sequence, chemical modification highlighted in the red color predicted efficacy and value of modulated efficacy.
MultiModGen
MultiModGen module complements the SMEpred component. It assists users to generate cm-siRNA sequences with multiple modifications of same or different nature on either strand of siRNA. This module requires format of customized cm-siRNA sequences that can be generated in first part. Latter, these cm-siRNAs (format) will be analyzed by the MNC-model for their predicted efficacies and will be reported in the next page. For generation of the sequence format, we need to submit siRNA sequence, position of modification from 5′-end and one-letter symbol of the chemical modification as input. Chemical modifications with one letter symbol are provided in Supplementary Table S2 as well as on the MultiModGen web page.
For input 3 boxes should be filled on the MultiModGen web page. First the siRNA sequence (GCAGCACGACUUCUUCAAGUU-CUUGAAGAAGUCGUGCUGCUU) that user wants to modify. In this sequence A, T, G, C, U stand for their universal nucleotides representing Adenine, Thymine, Guanine, Cytosine and Uracil respectively. In second and third box, one letter symbol of chemical modifications (F,T-T) and their positions (2,5,7,10,11,12,13,14,15,16,20,21-20,21) needs to be provided respectively. For more than one modification e.g., 2 different types of modifications, their one letter symbol and corresponding positions provided should be separated with double commas (,,) in between them. For further clarification please see Fig. S1. MultiModGen output can also be used as input in SiMEPred tool to predict efficacy based on different models as described below.
SiMEPred-tool
The SiMEPred algorithm predicts the efficacy of cm-siRNAs based on 3 different models. These models are based on best performance and importance of siRNA-features. Models-A is based on MNC only while model-B and C are based on MNC along with antisense strand nucleotide position. Model-B consists of information related to first 13 nucleotides from 5′end and the last one comprises of last 8 nucleotide positions from 3′end.
Cm-siRNA sequence format (e.g. GFAGFAFGAFFFFFFFAAGTT-CUUGAAGAAGUCGUGCUGCTT; Sense-Antisense) as defined above (MultiModGen) is required as input for this section. This format includes siRNA sequence, chemical modification and position of modifications. An illustrative example is displayed in Fig. S2. The output of SiMEPred displays columns in result page with serial number, siRNA sequence, length and predicted SVM model score of the corresponding sequences. The score can be sorted in ascending or descending order by clicking on the model score option to choose the best modification or their combinations.
Comparison with the existing web servers
For comparison of the various webservers designing and predicting siRNAs we have provided Table 2. As revealed in table, our webserver is the only one that can predict efficacy of cm-siRNAs as well as normal siRNA. Furthermore our pipeline is founded on the models with PCC values greater than most of these algorithms.
Table 2.
Comparison of the various siRNA prediction methods.
| S. No. | Publication PMID | Year | Technique | siRNA data set used | siRNA Prediction | cm-siRNA Prediction | Pearson correlation coefficient -R |
|---|---|---|---|---|---|---|---|
| 1 | 15201190 | 2004 | NA | 581 | Yes | No | 0.46 |
| 2 | 16025102 | 2005 | ANN | 2431 | Yes | No | 0.66 |
| 3 | 17137497 | 2006 | Linear | 2431 | Yes | No | 0.67 |
| 4 | 16870995 | 2006 | Linear | 526 | Yes | No | 0.55 |
| 5 | 16472402 | 2006 | NA | 653 | Yes | No | 0.55 |
| 6 | 17553157 | 2007 | SVM | 2431 | Yes | No | 0.78 |
| 7 | 17884914 | 2007 | Linear | 2431 | Yes | No | 0.72 |
| 8 | 17259216 | 2007 | Linear | 702 | Yes | No | 0.77 |
| 9 | 17644215 | 2007 | Rule, SVM, RFR | 3589 | Yes | No | 0.85 |
| 10 | 22102913 | 2011 | SVM | 2431 | Yes | No | 0.77 |
| 11 | 23118925 | 2012 | Linear | 2182 | Yes | No | 0.67 |
| 12 | 23241392 | 2013 | SVM | 2431 | Yes | No | 0.8 |
| 13 | 24330765 | 2013 | SVM | 1380 | Yes | No | 0.58 |
| 14 | 25888201 | 2015 | Semi supervised tensor | 2431 | Yes | No | 0.64 |
| 15 | 25725126 | 2015 | ANN | 2431 | Yes | No | 0. 74 |
| 16 | SMEpred | 2016 | SVM | 2182a;3130b | Yes | Yes | 0.72a; 0.80b |
(S. No = serial number; 2182
indicates the training set of normal siRNA sequences while 3130
represents the cm-siRNAs and 0.72a and 0.801b is their respective PCC value.)
Web server implementation
Linux platform (Apache-2.2.17), Perl are functioning at backend and PHP (5.2.14), HTML, JavaScript are employed for front-end of SMEpred web server.24,25 The following URL link provides the access to the algorithm web server for user accessibility http://bioinfo.imtech.res.in/manojk/smepred. General information for web server usage, data used etc. is provided on “HELP” section on the web server.
Discussion
SMEpred is a machine learning based web server to predict the efficacy of cm-siRNAs. In this method we tried to employ bioinformatics approach to promote the siRNA technology and application one step ahead. SMEpred web server offers an advanced aspect in siRNA therapeutics research involving chemical modifications. Chemical modifications in siRNA play a cardinal role in enhancing efficiency,26-28 serum stability,29-31 reducing off-targets9,32 etc. These aspects are key to their use as therapeutic molecules.
This web server is distinct from the existing algorithms in terms of predicting efficacy of cm-siRNAs (Table 2). This characteristic adds a new domain to the collection of siRNA prediction software packages. Theoretically, this web server can predict the activity of any number and pattern of 30 modifications (along with 5 canonical nucleotides used in training/testing of models) in cm-siRNAs with high accuracy. In this study, we developed composite models and assigned complex notations for chemical modifications, which is comparatively straightforward in case of wild siRNAs.
Biologically two aspects of the siRNA (as well as cm-siRNAs) are of prime importance. One is the nucleotide composition of either strand, second is the position of the modification in general and seed region of antisense in particular.33,34 Directed at these features we have developed our models in SMEpred server. Two cm-siRNA datasets were used Hetero-T2728 (Heterogeneous data set) and Homo-T1900 (homogenous dataset) for investigation with 10-fold cross validation. Furthermore, Hetero-V303 and Homo-V210 data sets were used for their independent validations respectively. Hertero-T2728 contains dataset with varied experimental conditions and more number and combinations of chemical modifications. Heterogeneous data set gave robustness to models developed as it includes more number of chemical modification and varied experimental conditions. For Hetero-T2728 dataset MNC model performed best among all SVM models. DNC and BIN did not exhibit any increase in PCC value hence performed poorly. Next, we chose to include the MNC of both the strands and various BIN features for the development of models.23 These hybrid models using BIN with MNC23 incorporated information based on the composition as well as position of modifications (Table 1).
The hybrid model performance of MNC and BIN patterns showed lower values of PCC. For example MNC + BIN (both sequences), MNC + BIN (10-mer both sequences and MNC + BIN (7-mer both sequences) showed PCC value less than 0.50. Models with MNC and BIN-AS either entire antisense sequence as MNC + BIN-AS (24 sequence length) model 7 of Table 1 or antisense variants performed well with PCC value greater than 0.75. Furthermore, MNC and BIN of sense strand (SS) i.e. MNC + BIN-SS 24; (model 14 of Table 1) showed PCC value of 0.58. The performance of final models used in terms of PCC values of our algorithm is equivalent or better to other prediction algorithms of unmodified siRNA23 (Table 2).
With Homo-T1900 same pattern in PCC value was seen as 0.86 for MNC (Supplementary Table S1). This data set also exhibited the similar trend in PCC values on different models, as DNC and BIN individually performed poor. Whereas, increasing PCC value with hybrids as MNC + BIN-AS and its variants was seen. Nearly in all cases 2 datasets indicated a similar correlation with increased values in case of Homo-1900 data set than Hetero-3031 dataset. The reason for these decreased values of PCC in hereto data set is because of its varied experimental conditions and increased number of modifications.
The algorithm developed was further authenticated using the independent datasets. The PCC via independent validation data sets of corresponding siRNA feature was of the same order as that of the 10-fold cross validation Table 1 (and Supplementary Table S1). For instance PCC value of MNC (Hetero-3031) is 0.80 and 0.80 for 10-Fold cross validation and independent validation respectively, signifying a balanced training of the models. This provides support for the models developed for their authenticity. The correlation between the actual and predicted percentage inhibitions of independent validation dataset of Hetero-V303 is shown in Fig. 2. Each data-point represents the intersection of actual and predicted values of the cm-siRNA efficacy for final 3 webserver models (A, B, C). It suggests that the entire independent validation data set displays positive correlation with stronger agreement for higher efficacies.
For the web server execution we have chosen only MNC (Model-A) as the main model, which performed best. Furthermore, we offered 2 more models along with latter one considering the PCC value and different functional components of cm-siRNAs. These include (IV) MNC +BIN AS (seed 13) model; and (V) MNC + BIN SS (last 8) model as model B and C respectively on the web server (SiMEPred-tool). MNC shows best performance and is set as default model on the web server, based on the over all composition of the entire siRNA. Moreover if the user wants to check the effect of the modification on the first half of antisense strand he can opt for the model-B. This model involves information of MNC of entire siRNA and position of the nucleotides or modified nucleotides in the antisense strand up to 13 nucleotides from 5′-end. Likewise the model-C contains MNC for the entire siRNA and the binary pattern for the last 8 nucleotides starting from 3′-end of antisense strand.
All the 3 components (SMEpred, MultiModGen, SiMEPred-tool) of the web server can be used to complement each other. SMEpred designs and predicts normal as well as single modification siRNAs, which can be further customized using MultiModGen for more permutations and combinations.
MultiModGen generates the modified siRNA sequences as per format that act as input in SiMEPred-tool. In this tool user can select different models and explore cm-siRNA features to check the modulation in their activity.
Conclusion
The SMEpred algorithm is the first pipeline, based on experimentally validated cm-siRNA datasets to generate, design, modify and check their efficacy computationally. Prediction models were developed using SVM machine learning technique utilizing cm-siRNA data set. The compositions (including chemical modification) as well as position in siRNA sequences were applied to make prediction models more robust and comprehensive. Three models were finally integrated in the web server namely model-A based on MNC which is set as the default model. Model-B is developed on MNC with binary pattern (BIN) (13 nucleotides from 5-end) and model-C representing MNC along with 8 nucleotides from 3′-end). These models were based on their performance during model development and siRNA sequence features. SMEpred would be useful for general scientific community especially those working on the development of siRNA therapeutics via chemical modifications.
Materials and methods
Data collection
We have searched research articles in PubMed and mined cm-siRNAs and collected data from siRNAmod database.18 After curation, 3031 unique experimentally verified cm-siRNAs with 30 most common chemical modifications were selected based on their usage and availability of quantitative efficacy values. List of these chemical modifications along with their one letter and binary codes used in this study is provided in the Supplementary Table S2. The length of the siRNA sequences varies from 21 to 24 oligonucleotides. This dataset contains non-redundant modified siRNAs i.e., they possess different chemical modification or different positions if the modification is same. Besides, this data is obtained from diverse experimental conditions and hence termed as heterogeneous data set (Hetero-3031). To select the training/testing and the validation dataset we have used 2 approaches. In the first approach, 3031 cm-siRNAs were arranged in the decreasing order of their activities. Next every 10th sequence starting from 5th sequence (5th, 15th, 25th, and so on) was picked up for the independent validation data set, total 303 sequences represented as Dataset-1 (Supplementary Table S3). Similarly, 2 other series of cm-siRNA sequences starting from 2nd and 8th sequence were chosen with interval of 10 sequences separately as Data set-2 and Dataset-3 respectively for independent validation. While remaining 2728 sequences were used as training/testing data set in each case. This procedure insures the proper distribution of siRNA efficacy values (entire range) in the training/testing as well as in the independent validation datasets. Whereas in the second approach, cm-siRNAs were chosen randomly to make 3 independent validation data sets and remaining 2728 sequences for training/testing for each combination. Random numbers were generated using Microsoft Excel “RAND()” function. Performance of SVM models for all 6 combinations (as mentioned above) using mononucleotide composition (MNC) feature during 10-fold cross validation as well as on the respective independent validation datasets is shown in Supplementary Table S3. All the models achieved almost similar performance. It implies that the obtained correlation is independent of the data sets chosen. Of these, “Dataset-1” comprising 2728 sequences for training/testing (Hetero-T2728) and 303 sequences for independent validation (Hetero-V303) was selected for further SVM models development.
Simultaneously, we have also selected another sub data set of 2110 cm-siRNAs tested under same experimental conditions termed as Homo-2110. It was used to generate training/testing (Homo-T1900) and independent dataset (Homo-V210) as per the strategy used for Hetero-3031 data set. The outline of the SMEpred model development is shown in Fig. 1.
SMEpred normal siRNA generation and prediction
This algorithm is based on the homogenous dataset of 2182 siRNA sequences.20 This module is based on a previous work of our lab, for more details check the following link (http://imtech.res.in/raghava/sirnapred/index.html).35 The best model (Hybrid-7) is provided in its backend to design the siRNAs, which is based on binary pattern and nucleotide frequency with the PCC value of 0.72.35
siRNA sequence features
Nucleotide composition
The ratio of each type of nucleotide in the siRNA sequence is called nucleotide composition. siRNA sequences were comprised of 5 usual (A, T, G, C, U) and 30 chemically modified nucleotides in both sense and antisense strand (Supplementary Table S2). For 35 different nucleotides, the fixed length vectors of 70 for mononucleotide composition (MNC) and 2450 for dinucleotide composition (DNC) were generated for each siRNA (sense and antisense).23
Binary pattern (BIN)
To obtain descriptions reflecting the position of nucleotides, BIN is used for siRNA sequences. A BIN of size 35 is formed one for each type of nucleotide common or modified. 24 positions of siRNA for each strand resulted in 48-vector length per siRNA sequence. For list for BIN used refer Supplementary Table S2. For each nucleotide we defined 6–bit code e.g., for adenosine nucleic acid we used “000001.” Working of the BIN model is based on the position of the particular nucleotide in the siRNA sequence.23
Hybrid approach
The hybrid approach involves models developed using combinations of MNC and BIN patterns (Table 1). These were used to check the performance of the prediction method23 based on different criteria. For example, MNC and BIN were analyzed on the basis of different lengths as well as either or both siRNA strands. The BIN of antisense strand (AS) was further explored e.g. complete length of antisense or different regions as seed region (8 nucleotide), up to mid region (13 nucleotide) and remaining 3′-end region (8 nucleotides). The reason for selecting these regions is based on their importance in siRNA based interference of the cognate mRNA. See Table 1 for different combinations and their features used.
Validation
To evaluate the performance of the algorithm, we have used 10-fold cross validation technique. We divided the data set into 10 equal sized sets to carry out training and testing 10 times. In each cycle 9 data sets were used for training; besides one separate set in testing i.e. this set is not included in that particular training cycle. Performance is measured in terms of Pearson Correlation Coefficient (PCC) using the formula:
| (1) |
Where n is the size of the test set, EiPred and Eiact is its predicted and actual efficacy respectively.23
Subsequently, we performed independent validation, which includes use of independent cm-siRNA sequences excluded in model development. This data set is used only for evaluation so that our model should not be biased or over-trained toward the training dataset used during model development. Scatter plots are formed between actual (experimental) and predicted efficacy values for independent validation sets using Microsoft excel for the 3 models used in SMEpred workbench.36
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the Council of Scientific and Industrial Research (BSC0121) India and the Department of Biotechnology, Government of India (GAP001).
ORCID
Showkat Ahmad Dar http://orcid.org/0000-0002-5077-1925
Manoj Kumar http://orcid.org/0000-0003-3769-052X
References
- 1.Gaglione M, Messere A. Recent progress in chemically modified siRNAs. Mini Rev Med Chem 2010; 7:578-95; PMID:20500149; http://dx.doi.org/ 10.2174/138955710791384036 [DOI] [PubMed] [Google Scholar]
- 2.Kurreck J. RNA interference: from basic research to therapeutic applications. Angewandte Chemie (International ed in English) 2009; 48:1378-98; PMID:19153977; http://dx.doi.org/ 10.1002/anie.200802092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tafer H, Ameres SL, Obernosterer G, Gebeshuber CA, Schroeder R, Martinez J, Hofacker IL. The impact of target site accessibility on the design of effective siRNAs. Nat Biotechnol 2008; 26:578-83; PMID:18438400; http://dx.doi.org/ 10.1038/nbt1404 [DOI] [PubMed] [Google Scholar]
- 4.ElHefnawi M, Hassan N, Kamar M, Siam R, Remoli AL, El-Azab I, AlAidy O, Marsili G, Sgarbanti M. The design of optimal therapeutic small interfering RNA molecules targeting diverse strains of influenza A virus. Bioinformatics 2011; 27:3364-70; PMID:21994230; http://dx.doi.org/ 10.1093/bioinformatics/btr555 [DOI] [PubMed] [Google Scholar]
- 5.Behlke MA. Progress towards in vivo use of siRNAs. Mol Ther 2006; 13:644-70; PMID:16481219; http://dx.doi.org/ 10.1016/j.ymthe.2006.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bramsen JB, Laursen MB, Nielsen AF, Hansen TB, Bus C, Langkjaer N, Babu BR, Højland T, Abramov M, Van Aerschot A, et al.. A large-scale chemical modification screen identifies design rules to generate siRNAs with high activity, high stability and low toxicity. Nucl Acids Res 2009; 37:2867-81; PMID:19282453; http://dx.doi.org/ 10.1093/nar/gkp106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ameres SL, Martinez J, Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell 2007; 130:101-12; PMID:17632058; http://dx.doi.org/ 10.1016/j.cell.2007.04.037 [DOI] [PubMed] [Google Scholar]
- 8.Shukla S, Sumaria CS, Pradeepkumar PI. Exploring chemical modifications for siRNA therapeutics: a structural and functional outlook. Chem Med Chem 2010; 5:328-49; PMID:20043313; http://dx.doi.org/ 10.1002/cmdc.200900444 [DOI] [PubMed] [Google Scholar]
- 9.Bramsen JB, Kjems J. Development of therapeutic-grade small interfering RNAs by chemical engineering. Front Gen 2012; 3:154; PMID:22934103; http://dx.doi.org/24152142 10.3389/fgene.2012.00154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Onizuka K, Harrison JG, Ball-Jones AA, Ibarra-Soza JM, Zheng Y, Ly D, Lam W, Mac S, Tantillo DJ, Beal PA. Short interfering RNA guide strand modifiers from computational screening. J Am Chem Soc 2013; 135:17069-77; PMID:24152142; http://dx.doi.org/ 10.1021/ja4079754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FA, Fabris D, Agris PF. The RNA modification database, RNAMDB: 2011 update. Nucl Acids Res 2011; 39:D195-201; PMID:21071406; http://dx.doi.org/ 10.1093/nar/gkq1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren Y, Gong W, Zhou H, Wang Y, Xiao F, Li T. siRecords: a database of mammalian RNAi experiments and efficacies. Nucl Acids Res 2009; 37:D146-9; PMID:18996894; http://dx.doi.org/ 10.1093/nar/gkn817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Truss M, Swat M, Kielbasa SM, Schafer R, Herzel H, Hagemeier C. HuSiDa–the human siRNA database: an open-access database for published functional siRNA sequences and technical details of efficient transfer into recipient cells. Nucl Acids Res 2005; 33:D108-11; PMID:15608157; http://dx.doi.org/ 10.1093/nar/gki131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tyagi A, Ahmed F, Thakur N, Sharma A, Raghava GP, Kumar M. HIVsirDB: a database of HIV inhibiting siRNAs. PloS One 2011; 6:e25917; PMID:22022467; http://dx.doi.org/ 10.1371/journal.pone.0025917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thakur N, Qureshi A, Kumar M. VIRsiRNAdb: a curated database of experimentally validated viral siRNA/shRNA. Nucl Acids Res 2012; 40:D230-6; PMID:22139916; http://dx.doi.org/ 10.1093/nar/gkr1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucl Acids Res 2014; 42:D68-73; PMID:24275495; http://dx.doi.org/ 10.1093/nar/gkt1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qureshi A, Thakur N, Monga I, Thakur A, Kumar M. VIRmiRNA: a comprehensive resource for experimentally validated viral miRNAs and their targets. Database : the journal of biological databases and curation 2014; 2014:1-10; PMID: 25380780; http://dx.doi.org/22102913 10.1093/database/bau103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dar SA, Thakur A, Qureshi A, Kumar M. siRNAmod: A database of experimentally validated chemically modified siRNAs. Scient Rep 2016; 6:20031; PMID:26818131; http://dx.doi.org/22102913 10.1038/srep20031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan WJ, Chen CW, Chu YW. siPRED: predicting siRNA efficacy using various characteristic methods. PloS one 2011; 6:e27602; PMID:22102913; http://dx.doi.org/ 10.1371/journal.pone.0027602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huesken D, Lange J, Mickanin C, Weiler J, Asselbergs F, Warner J, Meloon B, Engel S, Rosenberg A, Cohen D. Design of a genome-wide siRNA library using an artificial neural network. Nat Biotechnol 2005; 23:995-1001; PMID:16025102; http://dx.doi.org/ 10.1038/nbt1118 [DOI] [PubMed] [Google Scholar]
- 21.Mysara M, Elhefnawi M, Garibaldi JM. MysiRNA: improving siRNA efficacy prediction using a machine-learning model combining multi-tools and whole stacking energy (DeltaG). J Biomed Informat 2012; 45:528-34.; PMID:22388012; http://dx.doi.org/21853133 10.1016/j.jbi.2012.02.005 [DOI] [PubMed] [Google Scholar]
- 22.Ahmed F, Raghava GP. Designing of highly effective complementary and mismatch siRNAs for silencing a gene. PloS One 2011; 6:e23443; PMID:21853133; http://dx.doi.org/ 10.1371/journal.pone.0023443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qureshi A, Thakur N, Kumar M. VIRsiRNApred: a web server for predicting inhibition efficacy of siRNAs targeting human viruses. J Translat Med 2013; 11:305; PMID:24330765; http://dx.doi.org/25781990 10.1186/1479-5876-11-305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajput A, Gupta AK, Kumar M. Prediction and analysis of quorum sensing peptides based on sequence features. PloS One 2015; 10:e0120066; PMID:25781990; http://dx.doi.org/ 10.1371/journal.pone.0120066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar Gupta A, Kumar M. HPVbase–a knowledgebase of viral integrations, methylation patterns and microRNAs aberrant expression: As potential biomarkers for Human papillomaviruses mediated carcinomas. Scient Rep 2015; 5:12522; PMID:26205472; http://dx.doi.org/21899349 10.1038/srep12522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abe N, Abe H, Nagai C, Harada M, Hatakeyama H, Harashima H, Ohshiro T, Nishihara M, Furukawa K, Maeda M, et al.. Synthesis, structure, and biological activity of dumbbell-shaped nanocircular RNAs for RNA interference. Bioconjugate Chem 2011; 22:2082-92; PMID:21899349; http://dx.doi.org/ 10.1021/bc2003154 [DOI] [PubMed] [Google Scholar]
- 27.Strapps WR, Pickering V, Muiru GT, Rice J, Orsborn S, Polisky BA, Sachs A, Bartz SR. The siRNA sequence and guide strand overhangs are determinants of in vivo duration of silencing. Nucl Acids Res 2010; 38:4788-97; PMID:20360048; http://dx.doi.org/ 10.1093/nar/gkq206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi M, Nagai C, Hatakeyama H, Minakawa N, Harashima H, Matsuda A. Intracellular stability of 2′-OMe-4′-thioribonucleoside modified siRNA leads to long-term RNAi effect. Nucl Acids Res 2012; 40:5787-93; PMID:22411910; http://dx.doi.org/ 10.1093/nar/gks204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Czauderna F, Fechtner M, Dames S, Aygun H, Klippel A, Pronk GJ, Giese K, Kaufmann J. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucl Acids Res 2003; 31:2705-16; PMID:12771196; http://dx.doi.org/ 10.1093/nar/gkg393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dowler T, Bergeron D, Tedeschi AL, Paquet L, Ferrari N, Damha MJ. Improvements in siRNA properties mediated by 2′-deoxy-2′-fluoro-beta-D-arabinonucleic acid (FANA). Nucl Acids Res 2006; 34:1669-75; PMID:16554553; http://dx.doi.org/ 10.1093/nar/gkl033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall AH, Wan J, Shaughnessy EE, Ramsay Shaw B, Alexander KA. RNA interference using boranophosphate siRNAs: structure-activity relationships. Nucl Acids Res 2004; 32:5991-6000; PMID:15545637; http://dx.doi.org/ 10.1093/nar/gkh936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ui-Tei K, Naito Y, Nishi K, Juni A, Saigo K. Thermodynamic stability and Watson-Crick base pairing in the seed duplex are major determinants of the efficiency of the siRNA-based off-target effect. Nucl Acids Res 2008; 36:7100-9; PMID:18988625; http://dx.doi.org/ 10.1093/nar/gkn902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan CY, Carmack CS, Long DD, Maliyekkel A, Shao Y, Roninson IB, Ding Y. A structural interpretation of the effect of GC-content on efficiency of RNA interference. BMC Bioinformat 2009; 10(Suppl 1):S33; PMID:19208134; http://dx.doi.org/12804036 10.1186/1471-2105-10-S1-S33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harborth J, Elbashir SM, Vandenburgh K, Manninga H, Scaringe SA, Weber K, Tuschl T. Sequence, chemical, and structural variation of small interfering RNAs and short hairpin RNAs and the effect on mammalian gene silencing. Antisense Nucleic Acid Drug Dev 2003; 13:83-105; PMID:12804036; http://dx.doi.org/ 10.1089/108729003321629638 [DOI] [PubMed] [Google Scholar]
- 35.Kumar M, Lata S, Raghava G. siRNApred: SVM based method for predicting efficacy value of siRNA. Proceedings of the first international conference on Open Source for Computer Aided Drug Discovery (OSCADD), 2009. [Google Scholar]
- 36.Qureshi A, Kaur G, Kumar M. AVCpred: An integrated web server for prediction and design of antiviral compounds. Chem Biol Drug Design 2016; Aug 4:1-10; PMID: 27490990; http://dx.doi.org/ 10.1111/cbdd.12834 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


