ABSTRACT
To achieve proper RNA transport and localization, RNA viruses exploit cellular vesicular trafficking pathways. AGFG1, a host protein essential for HIV-1 and Influenza A replication, has been shown to mediate release of intron-containing viral RNAs from the perinuclear region. It is still unknown what its precise role in this release is, or whether AGFG1 also participates in cytoplasmic transport. We report for the first time the expression patterns during oogenesis for Drongo, the fruit fly homolog of AGFG1. We find that temporally controlled Drongo expression is achieved by translational repression of drongo mRNA within P-bodies. Here we show a first link between the recycling endosome pathway and Drongo, and find that proper Drongo localization at the oocyte's cortex during mid-oogenesis requires functional Rab11.
KEYWORDS: AGFG1, cytoskeleton, Drongo, Drosophila, HIV-1, Influenza A, Rab11
Abbreviations
- AF
Alexa Fluor
- AGFG1
ADP ribosylation factor GTPase-Activating Protein (Arf-GAP) with FG repeats protein 1
- AP2
adaptor protein 2
- Armi
Armitage
- BicD
Bicaudal D
- CA
constitutively active
- capu
cappuccino
- Chc
Clathrin heavy chain
- Clc
Clathrin light chain
- DN
dominant negative
- Drongo
Drosophila neural GTS1-like ORF
- EH
EPS15 homology domain
- EPS15
epidermal growth factor receptor pathway substrate 15
- FISH
fluorescence in situ hybridization
- GEF
Guanosine Exchange Factors
- i
TRiP or Transgenic RNAi Project
- Me31B
maternal expression at 31B
- MSD
mean square displacement
- Nuf
Nuclear fallout
- osk
oskar
- P-bodies
processing bodies
- Rab
Ras-associated GTP-binding proteins
- RNAi
RNA interference
- smFISH
single molecule FISH
- Stau
Staufen
- VAMP7
vesicle-associated membrane protein 7
- WT
wild type
Introduction
RNA viruses use intracellular trafficking to achieve important steps during the viral lifecycle. For example, the HIV-1 protein Gag recruits members of cellular vesicular trafficking for long-range transport of viral components, across the cytoplasm to the plasma membrane for viral particle assembly.1 Intracellular trafficking requires one or more members of the Ras superfamily of small GTPases, such as Ras-associated GTP-binding proteins (Rab) and ADP-ribosylation factors (Arf). They use Guanosine Exchange Factors (GEF) to exchange GDP with GTP, and GTPase Activator Proteins (GAP) for the hydrolysis of bound GTP.2,3 Rabs are important for specifically docking vesicles to their target membranes, as well as F-actin remodeling, while Arf proteins are involved in vesicle coat assembly. Rab11 regulates transport of recycling endosomes and therefore is used as their marker. Rab11 associates primarily with perinuclear recycling endosomes, but also localizes at the plasma membrane.4 Rab11 and the microtubules have been shown to be essential for Influenza A virus replication.5,6
One host protein, AGFG1 (Arf-GAP with FG repeats protein 1), is required for replication of two RNA viruses, HIV-1 and Influenza A,7,8 but dispensable for cellular viability.9 AGFG1 is a 62 kDa protein that contains a C4H2 zinc finger motif also found in proteins involved in cytoskeleton modeling and vesicle transport.10,11 After its initial discovery as an HIV-1 Rev-cofactor,12,13 it has been shown that AGFG1 is essential for vesicle docking and fusion in the formation of acrosomes during spermatogenesis.9 In mammalian cells, AGFG1 localizes mainly at the perinuclear region and plasma membrane, but also throughout the cytoplasm, and participates in clathrin-mediated endocytosis of select cargo.14-16
When AGFG1's function is impaired via RNA interference (RNAi), or by expression of a dominant negative mutant, intron-containing viral RNAs are sequestered at the perinuclear region.7,16 The role of AGFG1 in HIV-1 replication is reported to be specific: downregulation of AGFG1 expression results in aberrant accumulation of viral RNAs, but does not affect the distribution of polyadenylated cellular mRNAs. However, small and discrete accumulations of cellular mRNAs were observed at the nuclear periphery of mammalian cells expressing the AGFG1 dominant negative mutant, indicating that AGFG1 may act on a small fraction of cellular mRNAs.16 To account for the dramatic effect that AGFG1 depletion has on viral RNA transport and for how AGFG1 may facilitate viral RNA release from the perinuclear region, it is likely that AGFG1s function is not limited to clathrin-mediated endocytosis of select cargo, and that it may be involved in vesicle transport and perhaps in RNA transport. To address these hypotheses, we are studying AGFG1's fruit fly homolog, Drongo (Drosophila neural GTS1-like ORF), using the Drosophila melanogaster egg chamber, an ideal model system for studying vesicular trafficking.17
The development of the fruit fly egg chamber is divided into 14 stages of oogenesis, from germarium to the mature egg. Beginning at stage 1, each egg chamber is composed of two cell types, the germline (one oocyte and 15 nurse cells) and the somatic (follicle) cells (Fig. 1A).18 Mid-oogenesis (stages 7-8) is recognized as the period when any defective egg chambers are eliminated.19 Between stages 8 and 10A a slow cytoplasmic flow takes place within the oocyte, which transitions to a fast-paced and highly ordered streaming at stage 10B, right before the contents of the nurse cell cytoplasm are dumped into the oocyte.20 Beginning at stage 8, bulk endocytosis increases within the oocyte, and it continues with high rates through stage 11.21
Figure 1.
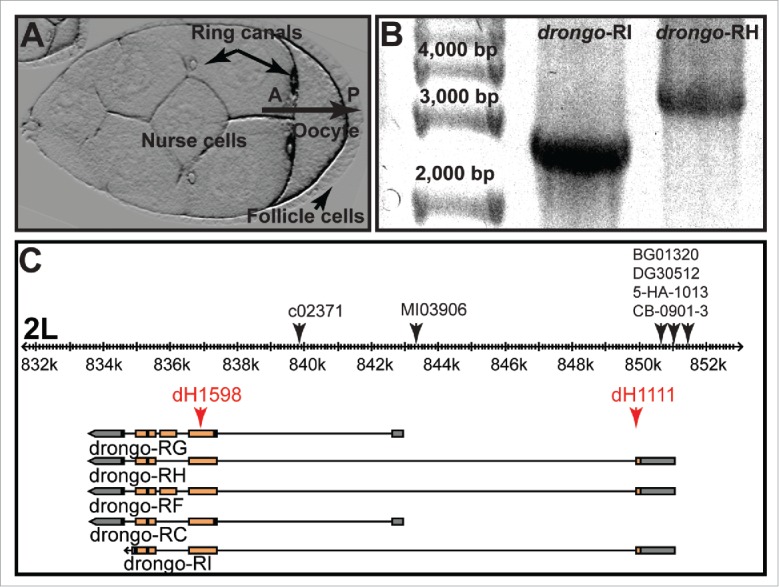
Drosophila egg chamber and drongo gene model. (A) Model system used for these studies; stage 8 Drosophila egg chamber: oocyte, nurse cells, follicle cells, and ring canals (arrows). A = anterior, P = posterior. (B) RT-PCR results for full length drongo transcripts using total RNA isolated from wild type ovaries. Expected sizes for drongo-RI and drongo-RH transcripts are 2,625 and 3,518 nucleotides, respectively. (C) drongo gene model (FlyBase: Gbrowse): boxes highlight exons, orange for coding regions and gray for UTRs. On the 2L chromosome fragment (black ruler), black arrows indicate P-element insertion location in the drongo gene. Red arrows indicate the regions targeted by drongo-specific molecular beacons.
The first studies of the drongo gene were initiated in the 1990s, but experimental difficulties (e.g. lack of efficient UAS/GAL4 system for the female germline) prevented detailed analysis of drongo's role(s) during oogenesis.22 In 2011 a large scale protein-protein interaction study revealed that Drongo directly interacts with 4 of the 7 components of the actin-related Arp2/3 complex (Arp2, Arpc1, Arpc3A and Arpc4).23,24 The actin cytoskeleton continuously undergoes temporally and spatially controlled remodeling: various proteins are used, as needed, to promote (Profilin) or inhibit (Thymosin) actin polymerization, as well as for microfilament severing (Cofilin), branching (Arp2/3) and networking (α-actinin).25 Proper F-actin assembly from actin monomers occurs by several pathways that involve nucleating factors such as the Arp2/3 complex and formins.26 Activators initiate Arp2/3-dependent actin nucleation,27 which is the limiting step in actin polymerization. Among the many cellular events that require the actin cytoskeleton, Arp2/3 is involved in endosomal sorting, recycling and Golgi-related vesicle trafficking.28
We have studied the transport and localization of drongo mRNA and Drongo protein during oogenesis using fluorescently labeled nucleic acid probes and EGFP tagging. We find that Drongo's timed expression requires translational repression via P-bodies (processing bodies) and that its localization at the oocyte's cortex requires functional Rab11. We propose that Drongo is associated with the recycling endosome trafficking pathway and may play a role in F-actin modeling during oogenesis.
Results
drongo mRNA shows enriched patterns in the oocyte
Currently, five drongo variants are predicted to be expressed in Drosophila (FlyBase).29 Previous Northern blot studies with lysates of embryos and adult female flies identified two drongo transcripts, 3.7 and 3.2 kb in length.22 Subsequent isolation and analysis of a drongo cDNA yielded the sequence for a 3,495 nucleotide-long transcript, which is similar to the drongo-RH variant (FlyBase ID: FBtr0302612). To identify which drongo mRNA variants are expressed during oogenesis, we performed RT-PCR analysis using total RNA isolated from wild type whole ovaries. We found that of the five predicted drongo variants (RC, RF, RG, RH, and RI), RH and RI are expressed during oogenesis (Fig. 1B and C). Using relative quantification RT-qPCR, we found that in wild type ovaries higher levels of drongo-RH are expressed when compared to the drongo-RI variant (∼15-fold). We sequenced and analyzed the drongo-RH coding region and 3′-UTR region using genomic DNA and drongo-RH cDNA. We identified a 33 nucleotide-long region located near the 3′ end of the drongo gene, and the 3′ end of drongo-RH cDNA, respectively, which is not included in public nucleotide databases (Materials and Methods).
Previous RNA-FISH studies of Drosophila egg chambers using drongo digoxigenin-labeled DNA probes (150–300 nt), found that drongo is expressed in the germarium, and within follicle cells of egg chambers during stages 2 to 7.22 Up to stage 9, drongo is also localized within the oocyte, but presented low expression within the nurse cells. From stage 9 onward, its expression decreased in the oocyte and increased in the nurse cells. To determine drongo mRNA localization throughout all stages of oogenesis, we performed RNA-FISH using drongo-specific molecular beacons. In the follicle cells, drongo mRNA was present from early through late oogenesis with persistent accumulation in most follicle cells (Fig. 2A). In the nurse cells, we observed a diffuse distribution of drongo mRNA throughout oogenesis. In the oocyte, consistent with previous results, drongo was present throughout during the early stages of oogenesis, up to stage 6 (Fig. S1A), and in stage 7 oocytes an anterior-dorsal patterning was detected. After stage 7, weak posterior and lateral cortex localization was observed. At stage 9, posterior accumulation, anterior-dorsal as well as a weak lateral cortex signals were visible (Fig. 2A). The fluorescence signal was greatly reduced in egg chambers where drongo expression was downregulated using RNAi (drongoiHMJ), suggesting that our probe is drongo-specific (Fig. S1B). We also performed smFISH (single molecule FISH) using drongo-specific Stellaris® probes,30 and found that although the observed drongo mRNA localization patterns are consistent with the results we obtained when using molecular beacons (Fig. S1A and Methods), the denaturing protocol required for smFISH resulted in a very weak signal. This suggests that drongo mRNA is not well anchored and its localization is transient, which may require nondenaturing protocols and/or live cell imaging for detection.
Figure 2.
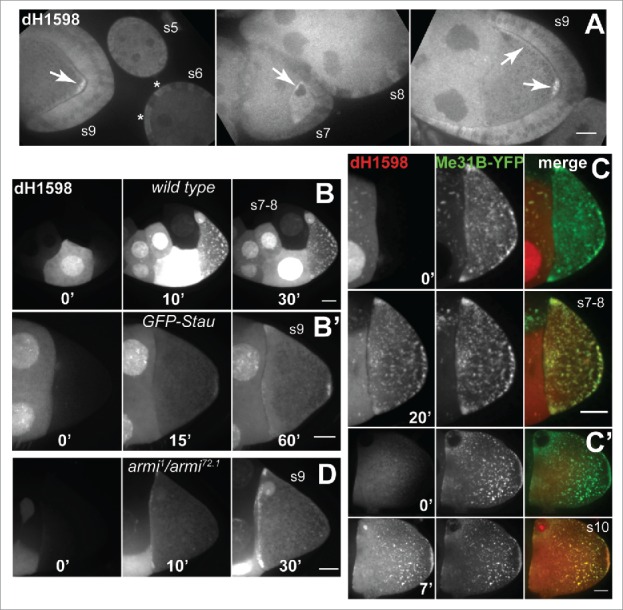
drongo mRNA localization in fixed and live oocytes using drongo-specific molecular beacons. (A) Detection of endogenous drongo transcript using dH1598-AF647 in fixed, wild type oocytes. drongo mRNA localization is indicated in the germline (arrows) and soma (asterisks). Microinjection of dH1598-AF647 in a nurse cell of a (B) wild type (XY-projection of 9 Z-slices/1 μm each) or (B’) GFP-Stau/CyO (XY-projection of 12 Z-slices/1 μm each) egg chamber at the indicated time points. Microinjection of dH1598-AF647 into a nurse cell of a (C) stage 7–8 (XY-projection of 5 Z-slices/1 μm each) or (C’) stage 10A (XY-projection of 5 Z-slices/1 μm each) Me31B-YFP egg chamber at the indicated time points. (D) Microinjection of dH1598-AF647 in a nurse cell of an armi mutant egg chamber (XY-projection of 10 Z-slices/1 μm each) at the indicated time points. s = stage. Bars, 20 μm.
We further analyzed drongo mRNA's distribution during oogenesis at higher resolution using live imaging and RNA-FISH experiments. We microinjected drongo-specific molecular beacons (Supplementary information Table S1) into live egg chambers to study the transport and localization of endogenous drongo mRNA within the oocyte at mid-oogenesis (stages 7 and 8). We found that drongo mRNA localization varied with the developmental stage. In stage 7–8 oocytes, drongo mRNA particles were distributed throughout, with a slightly preferred localization at the posterior, lateral, and anterior cortex of the oocyte (Fig. 2B, Fig. S2A, MovieS1). In stage 8–9 oocytes, drongo mRNA was concentrated at the posterior and also accumulated at the anterior (Fig. 2B’, Fig. S2B, S2B’). Moreover, at these stages (Fig. 2C, Supplementary information MovieS2) and later (Fig. 2C’), drongo mRNA associated with a marker for P-bodies, Me31B-YFP (maternal expression at 31B). P-bodies are cytoplasmic regions that are responsible for translational control of several transcripts.31 In Drosophila, Me31B is required for the translational repression of maternal mRNAs that localize in the oocyte during the early stages of development, but not for their transport.32
Polarized localization of maternal mRNAs within the oocyte is essential for embryonic development.33 One of the most studied mRNAs, oskar (osk), accumulates at the posterior end of the oocyte during mid-oogenesis, and specifies the site of assembly of abdominal and germline determinants of the future pole plasm.34 osk mRNA transport, localization and translation require precisely timed events within both the nucleus and the cytoplasm [e.g., association with the RNA-binding protein Staufen (Stau)35]. We observed partial colocalization of drongo and osk mRNAs at mid-oogenesis, using co-injections of drongo-specific and osk-specific36 molecular beacons in live egg chambers (Fig. S2B, Table S1, MovieS3). In addition, drongo mRNA colocalizes with Stau at the posterior end in live GFP-Stau egg chambers (Fig. S2B’, MovieS4).
Armitage (Armi), a protein that contains RNA helicase motifs, is required for the early onset of microtubule organization in the oocyte and is essential for the posterior localization of osk mRNA.37 Mutation of the armi gene activates the ATR/Chk2 DNA damage response pathway in the germline.38 In live stage 9 armi1/armi72.1 mutant egg chambers, where functional Armi protein is expressed in the follicle cells, but not in the germline cells,39 we observed drongo mRNA accumulation only at the anterior cortex of the oocyte (Fig. 2D, Supplementary information MovieS5). Attempts to perform quantitative analysis for drongo mRNA transport during mid-oogenesis were unsuccessful, likely due to its early localization. Taken together, these results indicate that drongo mRNA is associated with P-bodies and exhibits a cortical localization within the oocyte at mid-oogenesis and later, which is dependent on a correctly organized microtubule network.
Drongo protein is mainly localized at the oocyte's cortex
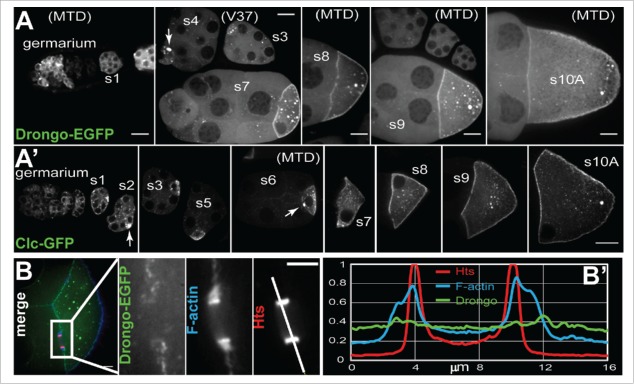
We generated a transgenic fly that overexpresses Drongo-PH with an EGFP tag at the C-terminus, driven with the UASp/GAL4 system. The Drongo-PI isoform is only 3 amino acids shorter than Drongo-PH (517 amino acids), with the first 506 amino acids being identical. Overexpression of Drongo-PH-EGFP (hereinafter referred to as Drongo-EGFP) was driven in the germline cells using 4 different drivers: V2H-GAL4, V37-GAL4, osk-GAL4, and MTD-GAL4, which allowed us to initiate expression at various stages of oogenesis and at various levels. We confirmed the size of Drongo-EGFP (∼83 kDa) using Western blot analysis of ovarian lysates and a GFP antibody (Fig. S3A). When we compared the EGFP expression driven by each driver, the observed patterns were similar for egg chambers at the same stage of oogenesis (Fig. 3A). MTD-GAL4 was the only driver that presented robust expression beginning in the germarium and persisting until late oogenesis. osk-GAL4 showed weak expression in the germarium (not shown). EGFP expression induced by V2H-GAL4 and V37-GAL4 commenced no earlier than stage 1, and the former resulted in the weakest EGFP expression when compared with the other drivers.
Figure 3.
Drongo overexpression during oogenesis. (A) Germline overexpression of Drongo-EGFP, and (A’) Clc-GFP. (B) Drongo-EGFP (MTD) localization at a nurse cell-oocyte ring canal. (B’) Normalized fluorescence intensity line plot profile along the line indicated in (B) for Drongo (green), F-actin (blue) and Hts (red). The GAL4 drivers used for Drongo-EGFP expression are indicated in parentheses. Arrows indicate (E)GFP accumulation in early stage oocytes. s = stage. Bars, 20 μm.
Up to stage 6, bright particles were observed throughout the egg chamber, mostly concentrated in the oocyte. At stages 3–6, in the oocyte, Drongo-EGFP aggregation was predominant at the anterior (Fig. 3A) while later a cortical accumulation became prominent, with particles of various sizes present in the cytoplasm. After mid-oogenesis ooplasmic aggregation was visibly reduced as the size of the oocyte increased with development, but up to stage 9 it appeared to directly correlate with the level of overexpression (i.e. more aggregates were observed under MTD control than with any other GAL4 driver). Drongo-EGFP distribution within the oocyte throughout all stages of oogenesis was very similar to the one reported for Clc-GFP overexpression (Clathrin light chain; Fig. 3A’).40 In the nurse cells, the EGFP signal was weaker, but accumulation was clearly seen around the nuclei, as well as at the nurse cell periphery (Fig. S3B) similar to AGFG1's localization in mammalian cells.14-16
Drongo-EGFP also localized closely to the filamentous actin incorporated into the ring canals that directly connect the nurse cells to the oocyte (Fig. 3B). We used higher magnification imaging to further analyze Drongo-EGFP localization at the ring canals, which were co-stained for F-actin and Hts (actin-binding protein localized at the inner rim of the ring canals). In a fluorescence line plot profile drawn across the ring canal, Drongo-EGFP appeared to surround F-actin (Fig. 3B’).
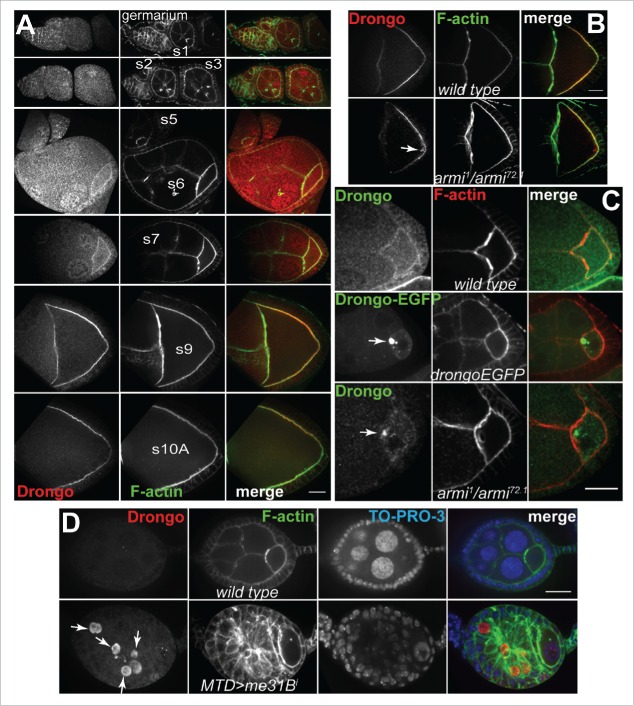
We validated the cortical localization observed in our Drongo-EGFP overexpression studies using wild type egg chambers and a peptide antibody, which was raised using a peptide (SGTGSNRQCFDCGQKGPTY) mapped to amino acids 22–40 at the N-terminus of Drongo-PH (Fig. 4A). At stage 9, in armi mutant oocytes we detected ectopic Drongo accumulation near the oocyte's cortex (Fig. 4B). However, the localization of two endosomal markers, Rab5 and Rab11, was not affected in these mutant egg chambers (Fig. S4A). Before stage 6, we detected premature Drongo accumulation at the armi mutant oocyte's anterior, which resembled the large aggregates observed at similar developmental stages in Drongo-EGFP oocytes (Fig. 4C). Moreover, we also observed premature and ectopic Drongo accumulation in the nurse cells of stages <6 me31Bi egg chambers, in which Me31B expression was downregulated in germline cells via RNAi (Fig. 4D), similar to previously reported premature Osk protein expression in me31B null egg chambers.32 Analysis of later stages was impeded by the fact that these egg chambers begin to degenerate before mid-oogenesis.
Figure 4.
Drongo distribution in fixed egg chambers. (A) Detection of endogenous Drongo using immunofluorescence studies with Drongo peptide antibody in wild type egg chambers. (B) Endogenous Drongo localization in wild type and armi mutant oocytes. Arrow indicates ectopic Drongo accumulation. (C) Premature and ectopic accumulation of Drongo in stage<6 armi mutant oocytes resembles Drongo-EGFP clumps observed at a similar stage (arrows). (D) Premature expression of Drongo when Me31B expression is reduced using RNAi (MTD-GAL4; arrows; XY-projection of 5 Z-slices/0.5 μm each). F-actin was highlighted using fluorescently labeled phalloidin. s = stage. Bars, 20 μm.
Although this antibody did recognize Drongo-EGFP in our immunofluorescence studies (Fig. S3C), in our Western blot analysis, the antibody failed to detect protein bands of the expected sizes for Drongo-EGFP and endogenous Drongo (∼56 kDa).
These results indicate that Drongo protein distribution closely resembles the localization observed for drongo mRNA, and Drongo expression reaches a peak during mid-oogenesis, while showing low levels in earlier stages. However, our results in armi mutant oocytes (Fig. 2D and 4B) suggest that mRNA localization may not be required for Drongo protein accumulation at the oocyte's cortex.
Downregulation of drongo gene expression leads to morphological defects
Our attempts to generate a deletion mutant or null fly stock through imprecise excision of P-elements inserted in the drongo gene (Fig. 1C), or to generate a drongo-knockout using CRISPR technology were not successful. When using two drongo-specific RNAi fly stocks (TRiP stocks allow tissue-specific RNAi-mediated knockdown using UASp/GAL4 system) we found efficient knockdown of drongo gene expression at the mRNA level (RT-qPCR) for the newer stock, drongoiHMJ (>65 % reduction in drongo mRNA levels vs ∼30% for drongoiHMS fly stock). In egg chambers coexpressing Drongo-EGFP and drongoiHMJ or drongoiHMS (V2H), EGFP expression was completely abolished only for the drongoiHMJ stock (data not shown).
When knockdown was initiated in the germarium (MTD), 10–15% of drongoiHMJ egg chambers presented pleiotropic defects similar to the ones reported for mutants or overexpression of genes encoding RNA-binding proteins, such as Bruno.41 Some egg chambers showed defects resulting from faulty splitting or adhesion of follicle cells and contained 32 cells, 30 nurse cells and two oocytes, which presented random positioning (Fig. 5A). We also observed tissue with a wide follicle cell band surrounding a few cells resembling nurse cells (Fig. 5A’). Other defects were due to an additional round of cystoblast division and also contained 32 cells, but with 31 nurse cells and one oocyte with five ring canals (Fig. 5B). Moreover, we observed reduced endogenous Drongo cortical signal in drongoiHMJ oocytes (Fig. 5B vs Fig. 5B’, Fig. S3D), which suggests that our antibody is selective, and likely specific for Drongo protein.
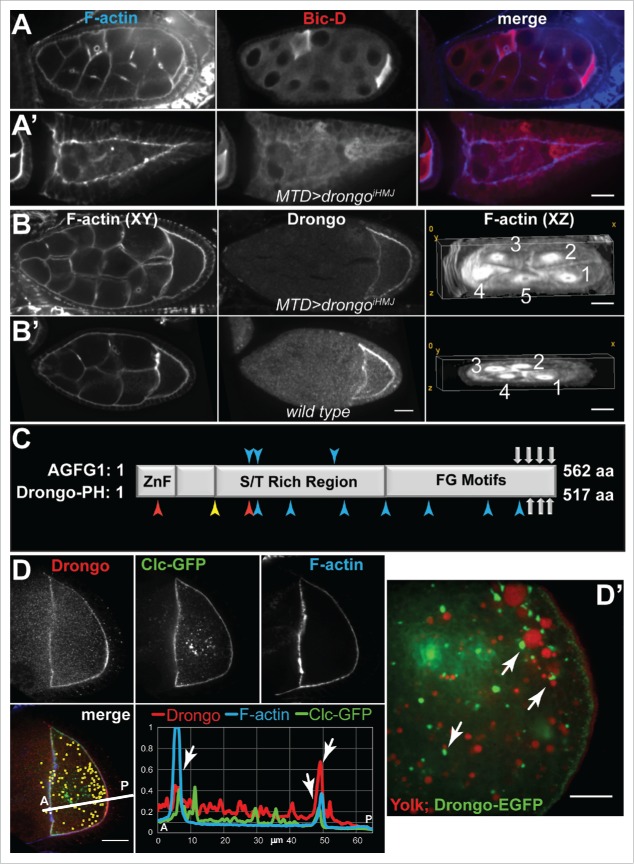
Figure 5.
drongo knockdown effects on egg morphology, and colocalization of Drongo with Clc. (A-B) Defects in drongoiHMJ as compared to (B’) wild type egg chamber. (A-A’) The oocyte position was highlighted with an oocyte marker, Bic-D (Bicaudal D). (B-B’) F-actin and Drongo distribution in one Z-slice, and rings canals in a XZ cross-section of drongoiHMJ (57 Z-slices/0.5 μm) or wild type (33 Z-slices/0.5 μm) egg chambers, respectively. (C) Comparison of AGFG1 and Drongo protein motifs (not drawn to scale). ZnF = C4H2 zinc finger, S/T = Serine/Threonine, FG = Phenylalanine and Glycine. Gray arrows indicate positions of the NPF (Asparagine-Proline-Phenylalanine) motifs. Arrow heads indicate the following: blue for AP2 (DLL, DxF or YxxΦ, Φ = hydrophobic residue), yellow for AP1 (DExxxL), and red for COPI (KKxx or WxxxW) binding motifs. (D) Colocalization between Drongo and Clc-GFP inside the oocyte (yellow: colocalization as XY-projection of 24 Z-slices/0.5 μm each, shown as an overlayer on the 9th Z-slice), and with F-actin at the anterior and posterior of a stage 8–9 oocyte. (D’) Localization of yolk granules and Drongo-EGFP particles in live egg chambers (>stage 9). (A-B’,D) F-actin was highlighted with fluorescently labeled phalloidin. A = anterior, P = posterior. Bars, (A-D) 20 μm and (D’) 10 μm.
Egg chambers with more than 16 cells were also reported when Drongo was overexpressed during oogenesis via an extended heat-shock treatment.22 Interestingly, mislocalization of Osk protein along the oocyte's cortex, but not osk mRNA, accompanied this defect, suggesting that the anchoring of Osk is compromised. However, most egg chambers appeared to develop normally and were fertilized, deposited and hatched at normal rates. In addition, we found that during mid-oogenesis most drongoiHMJ egg chambers showed wild type localization of Drongo protein at reduced levels (Fig. 5B and B’). osk mRNA localization was unaltered even in aberrant drongoiHMJ egg chambers. These results suggest that drongo knockdown in the germarium results in morphological defects that resemble those reported for mutation/overexpression of genes encoding RNA-binding proteins.
Drongo colocalizes with Clc-GFP
Upon comparing AGFG1 and Drongo protein sequences, we found that in addition to the conserved Arf-GAP motif, three of the four NPF motifs of AGFG1 are conserved in Drongo (Fig. 5C). NPF motifs are recognized by EPS15 homology (EH) domains,42 and it was reported that AGFG1 coimmunoprecipitates with EPS15 (Epidermal growth factor receptor Pathway Substrate 15), an adaptor protein that regulates intracellular trafficking at the plasma membrane and trans-Golgi network.43-45 In 2008 it was found that AGFG1 recruits VAMP7 (Vesicle-Associated Membrane Protein 7), a nonessential v-SNARE protein that participates in lysozome fusion with late endosomes and the plasma membrane, which lacks clathrin adaptor protein 2 (AP2) sorting motifs.14,15 Drongo and AGFG1 share consensus AP2 binding motifs (Fig. 5C), and it has been shown that AGFG1 directly interacts with AP2.46 Therefore, it is possible that Drongo also participates in clathrin-mediated endocytosis.
Efficient cargo uptake by endocytosis requires tightly controlled F-actin remodeling at endocytosis sites. Our immunofluorescence studies using egg chambers overexpressing Clc-GFP showed cortical colocalization for Clc-GFP, F-actin, and endogenous Drongo in the oocyte at mid-oogenesis (Fig. 5D). In the ooplasm and nurse cells adjacent to the oocyte, we observed 10 ± 2 % (s.d., n = 7) colocalization between Drongo and Clc-GFP particles. We also observed a pattern for the Drongo-EGFP distribution inside the oocyte of a >stage 9 egg chamber when we used a 63x objective with 2x additional magnification: discrete particles surrounded yolk granules, which were visualized in live egg chambers using incorporation of Trypan blue (Fig. 5D’). A similar pattern was observed for endogenous Drongo at a similar stage, but in this case the level of expression in the oocyte was greatly reduced compared to the Drongo-EGFP overexpression (Fig. S4B). This observation suggested that Drongo may play a role in endocytosis of yolk proteins and/or their transport. To determine the effect of Drongo overexpression on general endocytosis, we measured the uptake of FM 4–64x in live egg chambers, and we found that it was only mildly affected at stage 9 (Fig. S4C and Methods). In the presence of Drongo-EGFP, the posterior cortex showed a decrease in the intensity of the FM 4–64x signal when compared to the intensity in wild type oocytes. These results indicate that Drongo associates with factors essential for efficient endocytosis in the egg chamber, but bulk endocytosis is only mildly affected by Drongo overexpression.
Drongo colocalizes with Arpc3A, Rab11 and Arf51F
Actin is involved in uptake and transport of clathrin-coated pits via regulation of Arp2/3 activators.47 We found that Drongo-EGFP and Arpc3A (component of the Arp2/3 complex) showed colocalization at the cortex of stage 9 oocytes (Fig. 6A). A similar result was obtained when we analyzed the localization of endogenous Drongo and Arpc3A (Fig. S5A). Knockdown of another Arp2/3 component (Arpc4) using RNAi led to defective egg chambers that appeared to be the result of adhesion of more than 2 egg chambers, which displayed different morphology than the ones we observed for drongoiHMJ, but localization of Drongo was not affected (Fig. S5B).
Figure 6.
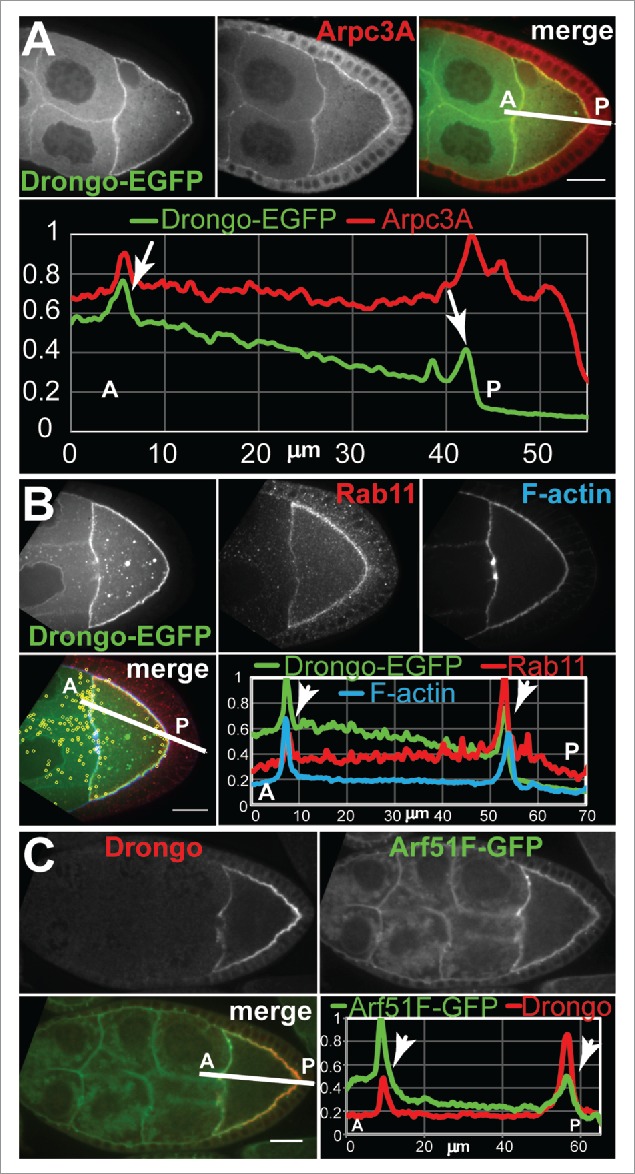
Colocalization of Drongo with actin related proteins, or small GTPases at stages 8–9 of oogenesis. (A) Drongo-EGFP and Arpc3A in a stage 8–9 egg chamber. (B) Drongo-EGFP and Rab11 (yellow: colocalization as XY-projection of 61 Z-slices/0.5 μm each, shown as an overlayer on the 27th Z-slice), in a MTD>drongoEGFP stage 8–9 oocyte. F-actin was highlighted using fluorescently labeled phalloidin. (C) Drongo and Arf51F-GFP in a stage 8–9 egg chamber expressing endogenous GFP-tagged Arf51F under native promoter control. (A-C) Colocalization at the oocyte's cortex shown in a normalized fluorescence intensity profile measured along the lines drawn as shown in the merged panels. Arrows indicate colocalization at the cortex of the oocyte. A = anterior, P = posterior. Bars, 20 μm.
In Drongo-EGFP egg chambers, we also observed colocalization at the oocyte's cortex between Drongo-EGFP and Rab11 (Fig. 6B). When we analyzed the extent of Drongo-EGFP and Rab11 association throughout the germline cells, we found 17 ± 5% colocalization in the oocyte (29 ± 1% overall, s.d., n = 4). We confirmed the cortical colocalization of endogenous Drongo and Rab11 using a monoclonal antibody of Rab11 (Fig. S6A). Rab11 localization was not affected when drongo expression was downregulated via RNAi.
Although AGFG1 has a conserved Arf-GAP motif, to date there are no reports of Arf proteins that use AGFG1. Drongo localization pattern at mid-oogenesis reflects the one observed for Arf51F-GFP48 (Fig. 6C), suggesting that Arf51F may use Drongo as GAP effector. Taken together these results indicate that Drongo is associated with the Arp2/3 complex, recycling endosomes and Arf51F.
Treatment with an Arp2/3 inhibitor leads to a discrete rearrangement of Drongo-EGFP
Drongo has been reported to directly interact with several subunits of the Arp2/3 complex.23,24 To determine if inhibition of Arp2/3-promoted actin filament branching has an effect on F-actin, and/or Drongo-EGFP localization during mid-oogenesis, we used CK-666 treatment of live wild type and Drongo-EGFP egg chambers.49 CK-666 (2-Fluoro-N-[2-(2-methyl-1H-indol-3-yl)ethyl]-benzamide) was identified in a large-scale screen of a small molecule library for inhibitors of Arp2/3-stimulated actin polymerization, and it was reported to interact with the Arp2/3 complex rather than interact with actin.50 Ectopic F-actin clumps formed only in control experiments (DMSO) in both wild type and Drongo-EGFP oocytes. In the latter, these structures also contained Drongo-EGFP, mostly at their periphery (Fig. 7A, arrows). After drug treatment, the F-actin fibers at the oocyte's posterior cortex appeared thicker and more elongated when compared to the corresponding controls (Fig. 7B vs Fig. 7A). Also, the local cortical accumulation of Drongo-EGFP appeared to undergo a rearrangement; it became punctate and more diffuse when compared with the Drongo-EGFP control, which showed a tight association with the oocyte's cortex. This was clearly visible only at higher magnification. However, Drongo-EGFP localization at the oocyte's ring canals, where Arp2/3 is required for their expansion after stage 5,51 was unaltered. These results indicate that Drongo localization becomes more diffuse when F-actin branching is impaired.
Figure 7.
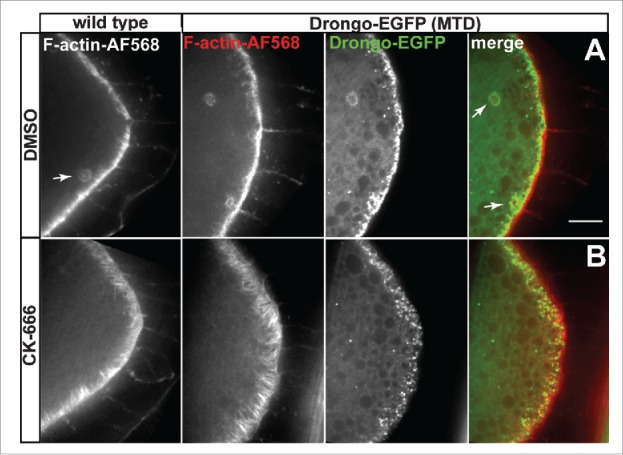
CK-666 treatment of live oocytes. F-actin and Drongo-EGFP localization for (A) control (DMSO) and (B) CK-666-treated stage 8 wild type and Drongo-EGFP oocytes. Arrows indicate ectopic F-actin clumps, and Drongo's localization at these sites. Bar, 10 μm.
Rab11 is required for Drongo localization
Rab11, a recycling endosome marker, plays an essential role in cytoskeleton organization during Drosophila oogenesis.52 Drongo distribution was not affected by expression of YFP-tagged, wild type (YFP-Rab11WT), or constitutively active (YFP-Rab11CA) Rab11 (Fig. 8A-a, 8A-b and 8B-b). However, when dominant negative Rab11 (YFP-Rab11DN) was expressed, Drongo's and Drongo-EGFP's cortical localization in the oocyte was almost completely lost (Fig. 8A-c, 8A-d and 8B-c, Supplementary information MovieS6). Early stage YFP-Rab11DN egg chambers did not present any morphological defects (Supplementary information MovieS7), but beginning at stage 7, oocyte development was delayed and many of the late stage egg chambers showed an oocyte detached from the posterior cortex (Fig. 8A-d and 8B-c). Drongo localization at the oocyte's cortex was reduced in all stage 8–9 egg chambers when low GAL4 levels (V2H) were used to express YFP-Rab11DN, but the F-actin network organization appeared intact (Fig. 8A-c). Drongo localization was less affected in stage 10 and older egg chambers (not shown). With higher GAL4 levels (MTD) Drongo's cortical localization was completely abolished in all >stage 7 oocytes and this was accompanied by morphological and F-actin defects, which increased in severity with the age of the female fly.
Figure 8.
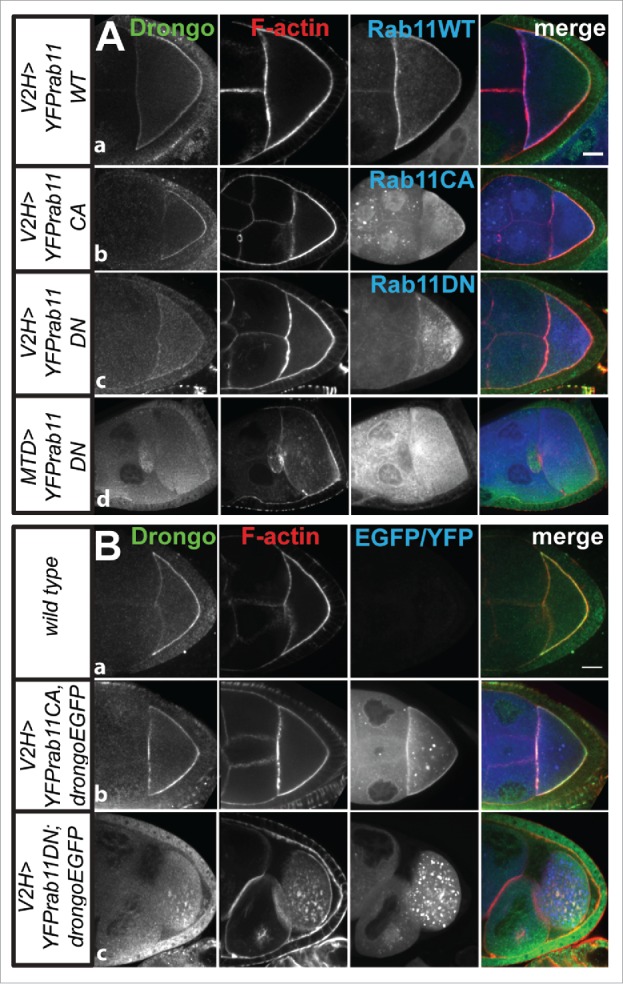
Drongo and F-actin localization in YFP-Rab11 background. Expression of YFP-Rab11 and (A) Drongo or (B) Drongo and Drongo-EGFP, in the indicated background, at stages 8–9 of oogenesis. Bars, 20 μm.
We also studied the transport of Drongo-EGFP particles in YFP-Rab11CA and DN backgrounds. Mean squared displacement (MSD) analysis using an exponential fit of long-range transport (31 min; 94 time frames), averaged over all tracks, showed restricted diffusion for Drongo-EGFP particles in wild type (α = 0.732 ± 0 .026, s.e.m., n = 5), and in YFP-Rab11CA (α = 0.839 ± 0 .057, s.e.m., n = 3) backgrounds, similar to what we observed for Clc-GFP particles (α = 0.873 ± 0.080, s.e.m., n = 4). However, in the presence of YFP-Rab11DN, Drongo-EGFP particles moved in a more directed fashion (α = 1.009 ± 0.100, s.e.m., n = 5) (Fig. S6B). This result is in agreement with the premature streaming observed for Drongo-EGFP particles in some of the stage 9, YFP-Rab11DN oocytes (Supplementary information MovieS6). These results indicate that when Rab11 function is impaired, Drongo's cortical localization is lost at mid-oogenesis, and this is accompanied by morphological, actin-related defects.
Discussion
We report for the first time expression patterns for Drongo, fruit fly homolog of AGFG1, during oogenesis, and we find that temporally controlled Drongo expression is achieved by translational repression of drongo mRNA in P-bodies. We also show that Drongo colocalizes with F-actin, Clc, Rab11, Arpc3A and Arf51F, suggesting that Drongo is linked to endocytosis and actin modeling, as well as recycling endosomal trafficking.
drongo mRNA transport and localization depend on the cytoskeleton
drongo mRNA presents a spatially and temporally controlled distribution during oogenesis, which in fixed egg chambers is sensitive to denaturing protocols. After transcription in the nurse cells, drongo mRNA is transported and localized mainly at the oocyte's cortex as early as stage 7. These processes appear to be dependent on the proper organization of the microtubule filaments, as drongo mRNA does not localize properly in armi mutants, where microtubule network polarization is compromised. The drongo gene was identified using enhancer trapping, technique employed when functional redundancy may exist and especially when mutant phenotypes are very mild. Attempts to isolate a drongo mutant by EMS mutagenesis failed.22 By comparison with AGFG1, it is likely that drongo mutants or even a drongo-null may not exhibit severe defects during oogenesis, but it is also possible that mammalian cells express redundant factors that are not encoded in the fruit fly genome. Therefore, given the current lack of a drongo RNA-null fly stock, future studies are needed to determine whether drongo mRNA localization has functional significance during oogenesis and embryogenesis, for processes such as maternal loading. However, we propose that drongo mRNA transport and localization to the oocyte, although highly transient, is required for the correct timing of its translation at peak level.
Translational regulation directs the timed expression of Drongo protein
drongo mRNA accumulates in the oocyte early in development, but Drongo peak expression does not occur before stage 8. This is achieved by translational repression via P-bodies of the drongo transcript before mid-ogenesis and this control continues in later stages of development. Additional support for translational control of drongo gene expression comes from our observation that similar to the reported premature expression of Osk in armi mutant egg chambers,37 clumps of Drongo are formed in stage <6 oocytes. These clumps resemble the Drongo-EGFP aggregations we observed at the oocyte's anterior in early stages of oogenesis and they are likely due to increased amounts of endogenous Drongo. However, when Drongo-EGFP is expressed, the EGFP tag may also facilitate aggregation.
Although its transcript lacks UTRs, Drongo-EGFP properly localizes in the oocyte. It is possible that endogenous Drongo expression is sufficient for Drongo-EGFP's correct localization. This is supported by the increase in cortical localization of Drongo-EGFP after stage 6–7, when endogenous Drongo levels naturally begin to increase at the oocyte's cortex. However, other cellular factors could also be responsible for recruiting Drongo.
Ectopic F-actin clumps have been previously reported for oocytes of rab52 germline clones, where F-actin is not properly released from the early endosomes, following endocytosis.53 We find that the localization of Rab5 and Rab11 in the armi mutant is similar to their localization in wild type egg chambers. Therefore, the ectopic Drongo clumps observed in armi mutant are not due to Rab5-induced or Rab11-induced defects. Taken together, these results suggest that drongo mRNA localization is not required for the Drongo protein to reach the oocyte's cortex, which may be facilitated by the slow cytoplasmic flow.
Drongo protein is associated with clathrin, F-actin and Arf51F
The clathrin triskelion is composed of three Clathrin heavy chains (Chc) and three light chains (Clc). In Drosophila egg chambers, apart from its role in the formation of clathrin-coated vesicles, Chc has been reported to play a role in the polarization of the microtubule network at stage 6.54 Clc has been shown to direct endocytosis by providing direction for membrane internalization.55 After stage 7, Drongo localizes mainly at the oocyte's cortex, where it shows significant overlap with Clc-GFP, concurrent with the activation of bulk endocytosis.21 Close analysis and comparison of the Drongo and AGFG1 protein sequences reveals that they both contain AP2 binding/sorting motifs (DxF, YxxΦ, where Φ=hydrophobic residue). Drongo's tight association with Clc-GFP at the oocyte's cortex, as well as its AP2 binding motifs, suggest that it is involved in endocytosis. The fact that overexpression of Drongo-EGFP only mildly affects general endocytosis during oogenesis is not surprising, as its mammalian homolog, AGFG1, was reported to mediate endocytosis of only select cargo.14,15 However, this does not exclude a role for Drongo in endocytosis and it is possible that a lack of Drongo will have a more pronounced effect.
Drongo directly interacts with components of the Arp2/3 complex, and we observe Drongo's cortical colocalization with Arpc3A in the oocyte. Moreover, Drongo distribution near the cortex of stage 8 oocytes becomes more diffuse when Arp2/3-dependent actin branching is impaired, suggesting that F-actin is important for Drongo's tight cortical localization at mid-oogenesis. CK-666 has been reported to interfere with formation of new Arp2/3-dependent branches without affecting existing branches or the ends of microfilaments.49 Therefore, it is not surprising that following drug treatment, we observe only subtle changes in the cortical F-actin network at mid-oogenesis. It is possible that Drongo-EGFP localization is mildly affected by drug-induced local perturbations in F-actin organization. Even though Drongo distribution is not affected by downregulation of Arpc4 expression, it is likely that Arp2/3 and the microfilaments play an essential role in Drongo localization during oogenesis. It is possible that Arp2/3 still shows some functionality despite Arpc4 depletion.
Although Drongo colocalizes with Arf51F, in vitro studies are needed to determine if Drongo acts as a GAP effector for Arf51F. In our studies the decrease in FM 4–64x uptake when Drongo is overexpressed is consistent with inactivation of Arf51F by excess of GAP activity, similar to the reduction in transferrin uptake observed in HeLa cells when SMAP1 is overexpressed (GAP effector for Arf6, which is the mammalian homolog for Arf51F56). Taken together, our results indicate that Drongo may participate in endocytosis and vesicular transport, where F-actin remodeling plays an important role.
Rab11 is essential for Drongo localization
Reports that Drongo's mammalian homolog, AGFG1, and Rab11 are both essential for Influenza A virus replication led us to analyze Drongo and Rab11 localization. Rab11 is required early in egg chamber development for oocyte determination. During mid-oogenesis it is required for polarized endocytic recycling toward the posterior pole of the oocyte, and for the posterior localization of AP-2α.57 Lack of Rab11, or reduced Drongo levels in the germarium, lead to defects in oocyte specification or an additional round of cystoblast division, respectively. Therefore, it is likely that Rab11 acts upstream of Drongo during early oogenesis, and this is supported by our observation that Rab11 appears unaffected in drongoiHMJ oocytes (not shown). However, since we still observe properly localized Drongo in drongoiHMJ oocytes at mid-oogenesis, we cannot rule out that Rab11 localization and function at this stage may be affected in a drongo-null egg chamber.
Rab11 is also essential for efficient osk mRNA transport, localization and anchoring, as well as osk translation.52 The disruption of osk mRNA trafficking and anchoring is believed to be a consequence of the failure to posteriorly concentrate the plus ends of the microtubule network at mid-oogenesis in oocytes where Rab11 function is affected. Rab11 directly interacts with Nuf (Nuclear-fallout), a unique Arf effector that contains a conserved Rab11-binding motif. Rab11 and Nuf are both required for actin recruitment during metaphase furrow formation in embryogenesis.58
It was proposed that proteins associated with recycling endosome activity (e.g., Rab11 and Nuf) are also required for delivery of actin-remodeling proteins to the plasma membrane, such as Rac1 (Rho GTPase). Moreover, Arf51F regulates endosomal recycling and cortical actin remodeling, as well as trafficking of Rac1 to the plasma membrane. In addition, when coexpressed with Rab11DN, Drongo-EGFP particles show premature streaming, which is characteristic of actin-related mutations (e.g., capu and spire).59
Due to their size, vesicles cannot rely solely on diffusion to reach their destination (Chapter 3 in ref. 60); their transport is composed of an active, directed movement on the microtubules and a passive, diffusive stage. Our MSD analysis reports on the overall movement for all tracks, determined for particles of various sizes and for long-range transport, and it yields constrained diffusion for Drongo-EGFP and Clc-GFP oocytes. Given the predominantly passive nature of the long-range transport of Drongo-EGFP and Clc-GFP that we observed in a wild type background, it is impressive to observe the transition to a more directional movement that occurs in Rab11DN egg chambers.
We propose that Drongo participates in recycling endosome trafficking and we provide evidence that Drongo is associated with intracellular trafficking where F-actin remodeling plays an important role. Our results improve our understanding of AGFG1's essential role in viral RNA transport. It is possible that viruses exploit the versatility of AGFG1 to achieve efficient RNA release from the perinuclear region and to facilitate their cytoplasmic transport, thereby modulating desired changes to the cytoskeleton and to trafficking pathways. Moreover, the multicellular model system that we are using is uniquely qualified to precisely identify the steps where Drongo actively participates in the cytoplasmic transport of intron-containing viral RNAs. Elucidating Drongo's cellular role(s) will help determine why its mammalian homolog is required for transport and localization of intron-containing viral RNAs. This should open new avenues for overcoming rapid viral adaptability to drug-pressure by targeting conserved interactions between viral proteins and host cofactors, which are nonessential for cellular viability.
Materials and methods
Fly husbandry
GFP-Stau was a gift from Daniel St Johnston (University of Cambridge). UASp-Clc-GFP was a gift from Linton M. Traub (University of Pittsburgh). osk-GAL4 was a gift from Anne Ephrussi (EMBL). Wild type (Oregon R), armi1 and armi72.1 were a gift from William E. Theurkauf (University of Massachusetts Medical School). We acquired drongoCB-0901–3, drongo5-HA-1013 and Me31B-YFP from Kyoto DGRC stock collection. We acquired the following fly stocks from Bloomington Drosophila Stock Center: drongoBG01320, CG4291DG30512, drongoMI03906, Df(2L)ast4, V37-GAL4 (BL#7063), V2H-GAL4 (BL#7062), MTD-GAL4 (BL#31777), Act5C-GAL4 (BL#25374), Arf51f-GFP (BL#60586), YFP-Rab11WT (BL#50782), YFP-Rab11CA [Rab11(Q70L), BL#9791, BL#50783], YFP-Rab11DN [Rab11(S25N), BL#9792, BL#23261], and the TRiP lines for drongo [drongoiHMS (BL#38960); drongoiHMJ (BL#60891)], me31B [me31BiGL (BL#38923)] and arpc4 [arpc4iHMS (BL#42875)]. From Exelixis Collection at Harvard we acquired drongoc02371. All stocks were raised and maintained on standard cornmeal agar food at room temperature. Female flies were fed on yeast paste for 1–3 days prior to dissection. Flies coexpressing Drongo-EGFP and an YFP-tagged protein, or an RNAi construct, were obtained from crosses of males from the V2H-GAL4; UASp:Drongo-EGFP stock with the corresponding virgin females.
Drongo-EGFP transgene
The UASp-EGFP:Drongo construct was generated by recombination of pENTR clones into the pUASp-attB-EGFP destination vector (gift from Jennifer A. Zallen, Sloan Kettering Institute) using Gateway technology (Life Technologies, Carlsbad, CA), which was inserted into attp2 sites (Genetic Services, Cambridge, MA). For PCR amplification of full length drongo-RH and drongo-RI we used LongAmp Taq DNA Polymerase (NEB, Ipswich, MA) with RNaseH-treated ovary cDNA. The cDNA coding sequence for Drongo-PH was amplified from drongo-RH cDNA using PCR. cDNA was obtained from RT reactions performed using Superscript III (Life Technologies), dT18 oligomers, and total RNA isolated from wild type ovaries. Total RNA was isolated using Trizol (Life Technologies; 15596026). The constructs were sequenced and analyzed. We amplified and sequenced the drongo-RH 3′-UTR and identified a 33 nucleotide-long region that is located just before 2L:833,663, or immediately after the 3,439th nucleotide in the drongo-RH transcript (FBtr0302612), which is not included in public databases (NCBI or Flybase; CAGATAAGAAGCAAGAATTTTTCCATTTTAGCA). These results were further confirmed with sequencing data obtained using genomic DNA isolated from wild type ovaries. DNA sequencing was performed by Genewiz, South Plainfield, NJ. Genomic DNA was isolated from 1–2 whole flies using the protocol from the Vosshall laboratory (Rockefeller University). Briefly, female flies were ground up in 50 μl of buffer (10 mM Tris-HCl pH 8, 1 mM EDTA, 25 mM NaCl, 0.2 mg/ml freshly added Proteinase K), incubated at 37˚C for 30 min, followed by 3 min at 95˚C. The tissue debris was spun down and the supernatant was saved to a new tube, from which 1 μl was used for PCR reactions.
RT-qPCR
cDNA was prepared from total RNA isolated from whole ovaries using Superscript III or GoScript enzyme (Promega, Madison, WI) as described above. Primers were designed and analyzed using PrimerQuest and OligoAnalyzer tools (Integrated DNA Technologies, Coralville, IA). Primers were acquired from Integrated DNA Technologies. qPCR was performed on white 384-well plates (E&K Scientific, Santa Clara, CA) using Lightcycler 480 with SYBR Green I Master mix (5 μl; Roche Diagnostics, Indianapolis, IN), cDNA (1 μl) from RT reactions performed with 0.25–5 μg total RNA and primer solution (4 μl) containing 5 μM of each primer. Plates were spun down to eliminate air bubbles. We used the same volume of corresponding no-RT reactions as negative control. Sequences for all primers used for this study are available upon request.
Molecular beacons
Molecular beacons probes were designed using mfold, RNAstructure and Oligowalk.61-63 The probes were synthesized, labeled and purified as previously described.64
RNA-FISH
Ovaries were dissected and fixed in 4% PFA in PBS or BRB80 (80 mM PIPES, pH 6.9; 1 mM MgCl2; 1 mM EGTA) for 10 min. Non-denaturing RNA-FISH with molecular beacon probes was performed with 200 ng of probe, in 30 μl hybridization buffer (50 mM Tris-HCl, pH 7.5; 1.5 mM MgCl2; and 100 mM NaCl) at room temperature for 2–4 h, followed by two 10 min washes with PBST (0.1 to 0.05% Triton™ X-100 in 1x PBS). Egg chambers were mounted in ProLong® Gold (Life Technologies; P36931), or glycerol based (90% glycerol, 1x PBS, 1% diazobicyclo[2.2.2]octane) media.
Immunohistochemistry
Immunofluorescence experiments were performed using egg chambers fixed for 10 min at room temperature with 4% PFA in BRB80 or PBS, permeabilized for 2 h at room temperature with 1% Triton™ X-100, 1% BSA in PBS. Egg chambers were then incubated overnight at room temperature (0.2% Triton™ X-100, 0.2% BSA in PBS) with the primary antibody and after washing, with the corresponding, fluorescently labeled, secondary antibody [Alexa Fluor (AF) or Cyanine-based (CF) fluorescent dyes (1:1,000; Life Technologies, and Biotium, Hayward, CA)]. The primary antibodies used in these studies were: rabbit anti-Drongo peptide antibody (1:300) (this study; Anaspec, Fremont, CA), mouse anti-GFP (B2; 1:300) (Santa Cruz Biotechnology, Santa Cruz, CA), guinea pig anti-Rab5 (1:5,000) and rabbit anti-Rab11 (1:7,000) (gifts from Akira Nakamura, Riken Center for Developmental Biology), mouse anti-Rab11 (1:1,000) (BD Biosciences, Franklin Lakes, NJ), rat anti-Arpc3A (1:500) (gift from Lynn Cooley, Yale University). The mouse anti-Bicaudal-D 4C2 (1:100) and mouse anti-Hts RC, developed at Rutgers University and Yale University, respectively, were obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA. Phalloidin labeled with AF488, AF568 or AF647 (Life Technologies) was used at the recommended dilution.
Trypan blue injections
Well-fed Drongo-EGFP (MTD or V2H) female flies were manually injected in the upper abdomen with a 1 mg/ml Trypan blue solution in water. After 30 min to 1 h ovaries were dissected in Halocarbon700 oil (Sigma-Aldrich, H8898) and individual egg chambers were teased directly onto a #1 cover glass (VWR, Pittsburgh, PA); Trypan blue-containing yolk granules were imaged using a 561 nm laser line.
CK-666 treatment
Whole ovaries were dissected in Schneider's Drosophila medium supplemented with 200 μM CK-666 (Sigma-Aldrich, SML0006), or DMSO for negative controls, and incubated with gentle rocking at room temperature. After 20 min ovaries were fixed with 4% PFA in PBS for 10 min, and F-actin was stained with fluorescently labeled phalloidin (Life Technologies) by incubation at room temperature in PBST for 1–2 h.
Live imaging
Egg chambers were prepared as described above. Microinjections were performed using molecular beacon solutions containing 150–300 ng/μl of probe (or of each probe for co-injections). Imaging was initiated within seconds after microinjection and performed at room temperature on a LeicaDMI-4000B inverted microscope (Leica Microsystems, Buffalo Groove, IL) mounted on a TMC isolation platform (Technical Manufacturing Corporation, Peabody, MA), with a Yokogawa CSU 10 spinning disc head and Hamamatsu C9100–13 ImagEM EMCCD camera. The microscopy set up includes diode lasers [491, 561, and 638 nm (Spectra Services, Ontario, NY)], and an Eppendorf Patchman-Femtojet microinjector (Eppendorf, Hauppauge, NY). The images were acquired as 16-bit data files, with 40x/1.25 or 63x/1.4 oil objectives (0.385 and 0.24 μm/pixel, respectively), using Volocity acquisition (PerkinElmer, Waltham, MA) and processed with ImageJ and Icy.65,66 For higher magnification analysis, we used a 2x c-mount placed in front of the camera (PerkinElmer). Z-stacks were acquired using a manual XY-stage with piezo-Z (PerkinElmer) and individual slice thicknesses ranged from 0.25 to 1 μm. For long-range transport studies, at least 13 Z-slices of 0.5 μm were acquired every 20 sec for at least 31 min, using the high-quality camera mode. Our microscope setup did not allow us to distinguish between YFP and EGFP, but we found that the EGFP signal was much stronger than the YFP signal. When using EGFP-optimal parameters for imaging egg chambers solely expressing YFP-Rab11, the YFP signal was not visible. Thus, we assumed that only the EGFP signal was acquired for egg chambers coexpressing YFP and EGFP when using the appropriate GAL4 driver and acquisition parameters.
Particle detection and tracking
(E)GFP particle tracking and analysis were performed with Icy, using “Spot Tracking.” Spots were detected using the following parameters: scale 2 (3 pixels) and 30–45 sensitivity; bright spot detection and size filtering (500 pixels or less; 240 nm/pixel). Tracking parameters were estimated for diffusive and directed motion. Tracks that spanned 10 time frames or less were removed. Tracks are displayed as tails of 10 consecutive time-frames.
MSD analysis was performed using X, Y, Z and time data for individual tracks, which were exported to MS Excel from Icy, and a custom R script (Adam Constantine and Florin Catrina, St. John's University). Results were graphed using GraphPad Prism (GraphPad Software, La Jolla, CA). Experimental data were fitted as previously described,67 using an exponential equation for volume (3D):
where “N” is 6 for 3D analysis, “D” is the diffusion coefficient, “t” is the time interval, and “α“ is a constant that depends on transport (Brownian motion/diffusion = 1, restrictive diffusive motion < 1 and superdiffusive > 1).
Data analysis
Fluorescence intensity line plot profile analyses were performed using ImageJ. Absolute values for each channel were normalized using the maximum fluorescence intensity from all channels and data was graphed using MS Excel. All projections are maximum intensity projections for the corresponding Z-stacks. Colocalization studies were performed for the whole image (512×512, pixels), or for the region of interest (nurse cells, oocyte) using Icy colocalization protocols. All statistical analyses to determine standard deviations were performed with GraphPad Prism.
Supplementary Material
Disclosure of potential conflicts of interest
S.A.E.M. is among a group of inventors who earn royalties for molecular beacon usage.
Acknowledgments
We thank William E. Theurkauf, Linton M. Traub, Anne Ephrussi, Daniel St Johnston, as well as to the BDSC Indiana, Exelixis Harvard and Kyoto DGRC for kindly sharing and providing the fly stocks for our study. We thank the TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947) for providing transgenic RNAi fly stocks used in this study. We thank Akira Nakamura and Lynn Cooley for kindly sharing antibodies. We thank Jennifer A. Zallen for the generous sharing of pUASp vectors and the Gateway protocol. We thank Paul Feinstein for allowing us to use the Roche Lightcycler instrument. We are grateful to Sherry L. Spinelli, Laura Owen, Omar S. Omar and Fred R. Kramer for their critical comments and editorial advice on the manuscript. We thank Bridget S. Koppetsch for her advice on maintenance of fly stocks. We thank Adam Constantine and Florin Catrina for writing the R script for MSD analyses. We are grateful to Ellen L. Kittler and Maria L. Zapp for their invaluable advice and for lending their expertise throughout these studies.
Funding
This work was supported in part by a National Science Foundation CAREER Award 1149738 and by several internal awards: three Professional Staff Congress-CUNY Awards, Innovation Seed Funding and Presidential Fund for Faculty Advancement Award, to DPB.
References
- 1.Dong X, Li H, Derdowski A, Ding L, Burnett A, Chen X, Peters TR, Dermody TS, Woodruff E, Wang JJ, et al.. AP-3 directs the intracellular trafficking of HIV-1 Gag and plays a key role in particle assembly. Cell 2005; 120:663-74; PMID:15766529; http://dx.doi.org/ 10.1016/j.cell.2004.12.023 [DOI] [PubMed] [Google Scholar]
- 2.Mizuno-Yamasaki E, Rivera-Molina F, Novick P. GTPase networks in membrane traffic. Annu Rev Biochem 2012; 81:637-59; PMID:22463690; http://dx.doi.org/ 10.1146/annurev-biochem-052810-093700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang J, Schulze KL, Hiesinger PR, Suyama K, Wang S, Fish M, Acar M, Hoskins RA, Bellen HJ, Scott MP. Thirty-one flavors of Drosophila Rab proteins. Genetics 2007; 176:1307-22; PMID:17409086; http://dx.doi.org/ 10.1534/genetics.106.066761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi S, Kubo K, Waguri S, Yabashi A, Shin HW, Katoh Y, Nakayama K. Rab11 regulates exocytosis of recycling vesicles at the plasma membrane. J Cell Sci 2012; 125:4049-57; PMID:22685325; http://dx.doi.org/ 10.1242/jcs.102913 [DOI] [PubMed] [Google Scholar]
- 5.Amorim MJ, Bruce EA, Read EK, Foeglein A, Mahen R, Stuart AD, Digard P. A Rab11- and microtubule-dependent mechanism for cytoplasmic transport of influenza A virus viral RNA. J Virol 2011; 85:4143-56; PMID:21307188; http://dx.doi.org/ 10.1128/JVI.02606-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eisfeld AJ, Kawakami E, Watanabe T, Neumann G, Kawaoka Y. RAB11A is essential for transport of the influenza virus genome to the plasma membrane. J Virol 2011; 85:6117-26; PMID:21525351; http://dx.doi.org/ 10.1128/JVI.00378-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eisfeld AJ, Neumann G, Kawaoka Y. Human immunodeficiency virus Rev-binding protein is essential for influenza a virus replication and promotes genome trafficking in late-stage infection. J Virol 2011; 85:9588-98; PMID:21752912; http://dx.doi.org/ 10.1128/JVI.05064-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu Z, Sanchez-Velar N, Catrina IE, Kittler EL, Udofia EB, Zapp ML. The cellular HIV-1 Rev cofactor hRIP is required for viral replication. Proc Natl Acad Sci U S A 2005; 102:4027-32; PMID:15749819; http://dx.doi.org/ 10.1073/pnas.0408889102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kang-Decker N, Mantchev GT, Juneja SC, McNiven MA, van Deursen JM. Lack of acrosome formation in Hrb-deficient mice. Science 2001; 294:1531-3; PMID:11711676; http://dx.doi.org/ 10.1126/science.1063665 [DOI] [PubMed] [Google Scholar]
- 10.Cukierman E, Huber I, Rotman M, Cassel D. The ARF1 GTPase-activating protein: zinc finger motif and Golgi complex localization. Science 1995; 270:1999-2002; PMID:8533093; http://dx.doi.org/ 10.1126/science.270.5244.1999 [DOI] [PubMed] [Google Scholar]
- 11.Kirchhausen T. Three ways to make a vesicle. Nat Rev Mol Cell Biol 2000; 1:187-98; PMID:11252894; http://dx.doi.org/ 10.1038/35043117 [DOI] [PubMed] [Google Scholar]
- 12.Bogerd HP, Fridell RA, Madore S, Cullen BR. Identification of a novel cellular cofactor for the Rev/Rex class of retroviral regulatory proteins. Cell 1995; 82:485-94; PMID:7634337; http://dx.doi.org/ 10.1016/0092-8674(95)90437-9 [DOI] [PubMed] [Google Scholar]
- 13.Fritz CC, Zapp ML, Green MR. A human nucleoporin-like protein that specifically interacts with HIV Rev. Nature 1995; 376:530-3; PMID:7637788; http://dx.doi.org/ 10.1038/376530a0 [DOI] [PubMed] [Google Scholar]
- 14.Chaineau M, Danglot L, Proux-Gillardeaux V, Galli T. Role of HRB in clathrin-dependent endocytosis. J Biol Chem 2008; 283:34365-73; PMID:18819912; http://dx.doi.org/ 10.1074/jbc.M804587200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pryor PR, Jackson L, Gray SR, Edeling MA, Thompson A, Sanderson CM, Evans PR, Owen DJ, Luzio JP. Molecular basis for the sorting of the SNARE VAMP7 into endocytic clathrin-coated vesicles by the ArfGAP Hrb. Cell 2008; 134:817-27; PMID:18775314; http://dx.doi.org/ 10.1016/j.cell.2008.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanchez-Velar N, Udofia EB, Yu Z, Zapp ML. hRIP, a cellular cofactor for Rev function, promotes release of HIV RNAs from the perinuclear region. Genes Dev 2004; 18:23-34; PMID:14701878; http://dx.doi.org/ 10.1101/gad.1149704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kondylis V, Rabouille C. The Golgi apparatus: lessons from Drosophila. FEBS Letters 2009; 583:3827-38; PMID:19800333; http://dx.doi.org/ 10.1016/j.febslet.2009.09.048 [DOI] [PubMed] [Google Scholar]
- 18.McLaughlin JM, Bratu DP. Drosophila melanogaster Oogenesis: An overview. Methods Mol Biol 2015; 1328:1-20; PMID:26324426; http://dx.doi.org/ 10.1007/978-1-4939-2851-4_1 [DOI] [PubMed] [Google Scholar]
- 19.Buszczak M, Cooley L. Eggs to die for: cell death during Drosophila oogenesis. Cell Death Differ 2000; 7:1071-4; PMID:11139280; http://dx.doi.org/ 10.1038/sj.cdd.4400755 [DOI] [PubMed] [Google Scholar]
- 20.Serbus LR, Cha BJ, Theurkauf WE, Saxton WM. Dynein and the actin cytoskeleton control kinesin-driven cytoplasmic streaming in Drosophila oocytes. Development 2005; 132:3743-52; PMID:16077093; http://dx.doi.org/ 10.1242/dev.01956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schonbaum CP, Perrino JJ, Mahowald AP. Regulation of the vitellogenin receptor during Drosophila melanogaster oogenesis. Mol Biol Cell 2000; 11:511-21; PMID:10679010; http://dx.doi.org/ 10.1091/mbc.11.2.511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pritchard J. Analysis of drongo, a new Drosophila zinc finger gene expressed during oogenesis and neurogenesis. University of Warwick, 1999:Ph.D. Dissertation; (UMI)AAIU133506 [Google Scholar]
- 23.Guruharsha KG, Rual JF, Zhai B, Mintseris J, Vaidya P, Vaidya N, Beekman C, Wong C, Rhee DY, Cenaj O. A protein complex network of Drosophila melanogaster. Cell 2011; 147:690-703; PMID:22036573; http://dx.doi.org/ 10.1016/j.cell.2011.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murali T, Pacifico S, Yu J, Guest S, Roberts GG 3rd, Finley RL Jr. DroID 2011: a comprehensive, integrated resource for protein, transcription factor, RNA and gene interactions for Drosophila. Nucleic Acids Res 2011; 39:D736-43; PMID:21036869; http://dx.doi.org/ 10.1093/nar/gkq1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alberts B, Johnson A, Lewis J, Morgan D, Raff M, Roberts K, Walter P. Molecular Biology of the Cell Garland Sciences, Taylor & Francis Group, LLC, New York, 2015; ISBN-13: 978-0815344322 [Google Scholar]
- 26.Rotty JD, Wu C, Haynes EM, Suarez C, Winkelman JD, Johnson HE, Haugh JM, Kovar DR, Bear JE. Profilin-1 serves as a gatekeeper for actin assembly by Arp2/3-dependent and -independent pathways. Dev Cell 2015; 32:54-67; PMID:25543281; http://dx.doi.org/ 10.1016/j.devcel.2014.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zallen JA, Cohen Y, Hudson AM, Cooley L, Wieschaus E, Schejter ED. SCAR is a primary regulator of Arp2/3-dependent morphological events in Drosophila. J Cell Biol 2002; 156:689-701; PMID:11854309; http://dx.doi.org/ 10.1083/jcb.200109057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anitei M, Hoflack B. Bridging membrane and cytoskeleton dynamics in the secretory and endocytic pathways. Nat Cell Biol 2012; 14:11-9; http://dx.doi.org/ 10.1038/ncb2409 [DOI] [PubMed] [Google Scholar]
- 29.St Pierre SE, Ponting L, Stefancsik R, McQuilton P, FlyBase C. FlyBase 102–advanced approaches to interrogating FlyBase. Nucleic Acids Res 2014; 42:D780-8; PMID:24234449; http://dx.doi.org/ 10.1093/nar/gkt1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bayer LV, Batish M, Formel SK, Bratu DP. Single-Molecule RNA In Situ Hybridization (smFISH) and Immunofluorescence (IF) in the Drosophila Egg Chamber. Methods Mol Biol 2015; 1328:125-36; PMID:26324434; http://dx.doi.org/ 10.1007/978-1-4939-2851-4_9 [DOI] [PubMed] [Google Scholar]
- 31.Weil TT, Parton RM, Herpers B, Soetaert J, Veenendaal T, Xanthakis D, Dobbie IM, Halstead JM, Hayashi R, Rabouille C, et al.. Drosophila patterning is established by differential association of mRNAs with P bodies. Nat Cell Biol 2012; 14:1305-13; PMID:23178881; http://dx.doi.org/ 10.1038/ncb2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamura A, Amikura R, Hanyu K, Kobayashi S. Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development 2001; 128:3233-42; PMID:11546740 [DOI] [PubMed] [Google Scholar]
- 33.Jansen RP. mRNA localization: message on the move. Nat Rev Mol Cell Biol 2001; 2:247-56; PMID:11283722; http://dx.doi.org/ 10.1038/35067016 [DOI] [PubMed] [Google Scholar]
- 34.Ephrussi A, Dickinson LK, Lehmann R. Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell 1991; 66:37-50; PMID:2070417; http://dx.doi.org/ 10.1016/0092-8674(91)90137-N [DOI] [PubMed] [Google Scholar]
- 35.St Johnston D, Beuchle D, Nusslein-Volhard C. Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell 1991; 66:51-63; PMID:1712672; http://dx.doi.org/ 10.1016/0092-8674(91)90138-O [DOI] [PubMed] [Google Scholar]
- 36.Bratu DP. Molecular beacons light the way: Imaging native mRNAs in living cells. Discov Med 2003; 3:44-7; PMID:20705038 [PubMed] [Google Scholar]
- 37.Cook HA, Koppetsch BS, Wu J, Theurkauf WE. The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell 2004; 116:817-29; PMID:15035984; http://dx.doi.org/ 10.1016/S0092-8674(04)00250-8 [DOI] [PubMed] [Google Scholar]
- 38.Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA, Theurkauf WE. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev Cell 2007; 12:45-55; PMID:17199040; http://dx.doi.org/ 10.1016/j.devcel.2006.12.001 [DOI] [PubMed] [Google Scholar]
- 39.Olivieri D, Sykora MM, Sachidanandam R, Mechtler K, Brennecke J. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J 2010; 29:3301-17; PMID:20818334; http://dx.doi.org/ 10.1038/emboj.2010.212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jha A, Watkins SC, Traub LM. The apoptotic engulfment protein Ced-6 participates in clathrin-mediated yolk uptake in Drosophila egg chambers. Mol Biol Cell 2012; 23:1742-64; PMID:22398720; http://dx.doi.org/ 10.1091/mbc.E11-11-0939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Snee M, Benz D, Jen J, Macdonald PM. Two distinct domains of Bruno bind specifically to the oskar mRNA. RNA Biol 2008; 5:1-9; PMID:18388491; http://dx.doi.org/ 10.4161/rna.5.1.5735 [DOI] [PubMed] [Google Scholar]
- 42.Salcini AE, Confalonieri S, Doria M, Santolini E, Tassi E, Minenkova O, Cesareni G, Pelicci PG, Di Fiore PP. Binding specificity and in vivo targets of the EH domain, a novel protein-protein interaction module. Genes dev 1997; 11:2239-49; PMID:9303539; http://dx.doi.org/ 10.1101/gad.11.17.2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chi S, Cao H, Chen J, McNiven MA. Eps15 mediates vesicle trafficking from the trans-Golgi network via an interaction with the clathrin adaptor AP-1. Mol Biol Cell 2008; 19:3564-75; PMID:18524853; http://dx.doi.org/ 10.1091/mbc.E07-10-0997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doria M, Salcini AE, Colombo E, Parslow TG, Pelicci PG, Di Fiore PP. The eps15 homology (EH) domain-based interaction between eps15 and hrb connects the molecular machinery of endocytosis to that of nucleocytosolic transport. J Cell Biol 1999; 147:1379-84; PMID:10613896; http://dx.doi.org/ 10.1083/jcb.147.7.1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Bergen En , Henegouwen PM. Eps15: a multifunctional adaptor protein regulating intracellular trafficking. Cell Commun Signal 2009; 7:24; PMID:19814798; http://dx.doi.org/ 10.1186/1478-811X-7-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Umasankar PK, Sanker S, Thieman JR, Chakraborty S, Wendland B, Tsang M, Traub LM. Distinct and separable activities of the endocytic clathrin-coat components Fcho1/2 and AP-2 in developmental patterning. Nat Cell Biol 2012; 14:488-501; PMID:22484487; http://dx.doi.org/ 10.1038/ncb2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mooren OL, Galletta BJ, Cooper JA. Roles for actin assembly in endocytosis. Ann Rev Biochem 2012; 81:661-86; PMID:22663081; http://dx.doi.org/ 10.1146/annurev-biochem-060910-094416 [DOI] [PubMed] [Google Scholar]
- 48.Huang J, Zhou W, Dong W, Watson AM, Hong Y. Directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering. Proc Natl Acad Sci USA 2009; 106:8284-9; PMID:19429710; http://dx.doi.org/ 10.1073/pnas.0900641106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hetrick B, Han MS, Helgeson LA, Nolen BJ. Small molecules CK-666 and CK-869 inhibit actin-related protein 2/3 complex by blocking an activating conformational change. Chem Biol 2013; 20:701-12; PMID:23623350; http://dx.doi.org/ 10.1016/j.chembiol.2013.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nolen BJ, Tomasevic N, Russell A, Pierce DW, Jia Z, McCormick CD, Hartman J, Sakowicz R, Pollard TD. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature 2009; 460:1031-4; PMID:19648907; http://dx.doi.org/ 10.1038/nature08231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hudson AM, Cooley L. A subset of dynamic actin rearrangements in Drosophila requires the Arp2/3 complex. J Cell Biol 2002; 156:677-87; PMID:11854308; http://dx.doi.org/ 10.1083/jcb.200109065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jankovics F, Sinka R, Erdelyi M. An interaction type of genetic screen reveals a role of the Rab11 gene in oskar mRNA localization in the developing Drosophila melanogaster oocyte. Genetics 2001; 158:1177-88; PMID:11454766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Compagnon J, Gervais L, Roman MS, Chamot-Boeuf S, Guichet A. Interplay between Rab5 and PtdIns(4,5)P2 controls early endocytosis in the Drosophila germline. J Cell Sci 2009; 122:25-35; PMID:19050045; http://dx.doi.org/ 10.1242/jcs.033027 [DOI] [PubMed] [Google Scholar]
- 54.Vazquez-Pianzola P, Adam J, Haldemann D, Hain D, Urlaub H, Suter B. Clathrin heavy chain plays multiple roles in polarizing the Drosophila oocyte downstream of Bic-D. Development 2014; 141:1915-26; PMID:24718986; http://dx.doi.org/ 10.1242/dev.099432 [DOI] [PubMed] [Google Scholar]
- 55.Boettner DR, Friesen H, Andrews B, Lemmon SK. Clathrin light chain directs endocytosis by influencing the binding of the yeast Hip1R homologue, Sla2, to F-actin. Mol Biol Cell 2011; 22:3699-714; PMID:21849475; http://dx.doi.org/ 10.1091/mbc.E11-07-0628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tanabe K, Torii T, Natsume W, Braesch-Andersen S, Watanabe T, Satake M. A novel GTPase-activating protein for ARF6 directly interacts with clathrin and regulates clathrin-dependent endocytosis. Mol Biol Cell 2005; 16:1617-28; PMID:15659652; http://dx.doi.org/ 10.1091/mbc.E04-08-0683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dollar G, Struckhoff E, Michaud J, Cohen RS. Rab11 polarization of the Drosophila oocyte: a novel link between membrane trafficking, microtubule organization, and oskar mRNA localization and translation. Development 2002; 129:517-26; PMID:11807042 [DOI] [PubMed] [Google Scholar]
- 58.Riggs B, Rothwell W, Mische S, Hickson GR, Matheson J, Hays TS, Gould GW, Sullivan W. Actin cytoskeleton remodeling during early Drosophila furrow formation requires recycling endosomal components Nuclear-fallout and Rab11. J Cell Biol 2003; 163:143-54; PMID:14530382; http://dx.doi.org/ 10.1083/jcb.200305115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dahlgaard K, Raposo AA, Niccoli T, St Johnston D. Capu and Spire assemble a cytoplasmic actin mesh that maintains microtubule organization in the Drosophila oocyte. Dev Cell 2007; 13:539-53; PMID:17925229; http://dx.doi.org/ 10.1016/j.devcel.2007.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Friedman MH. Principles and Models of Biological Transport Springer Science+Business Media, LLC, New York, 2008; ISBN 978-0-387-79240-8; http://dx.doi.org/ 10.1007/978-0-387-79240-8 [DOI] [Google Scholar]
- 61.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 2003; 31:3406-15; PMID:12824337; http://dx.doi.org/ 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mathews DH. RNA secondary structure analysis using RNAstructure. Curr ProtocolsBioinformatics / editoral board, Andreas D Baxevanis [et al] 2006; Chapter 12:Unit 12 6; 13:12.6:12.6.1–12.6.14; http://dx.doi.org/ 10.1002/0471250953.bi1206s13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mathews DH. Using OligoWalk to identify efficient siRNA sequences. Methods Mol Biol 2010; 629:109-21; PMID:20387146; http://dx.doi.org/ 10.1007/978-1-60761-657-3_8 [DOI] [PubMed] [Google Scholar]
- 64.Bratu DP, Catrina IE, Marras SA. Tiny molecular beacons for in vivo mRNA detection. Methods Mol Biol 2011; 714:141-57; PMID:21431739; http://dx.doi.org/ 10.1007/978-1-61779-005-8_9 [DOI] [PubMed] [Google Scholar]
- 65.de Chaumont F, Dallongeville S, Chenouard N, Herve N, Pop S, Provoost T, Meas-Yedid V, Pankajakshan P, Lecomte T, Le Montagner Y, et al.. Icy: an open bioimage informatics platform for extended reproducible research. Nat Methods 2012; 9:690-6; PMID:22743774; http://dx.doi.org/ 10.1038/nmeth.2075 [DOI] [PubMed] [Google Scholar]
- 66.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al.. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012; 9:676-82; PMID:22743772; http://dx.doi.org/ 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mhlanga MM, Bratu DP, Genovesio A, Rybarska A, Chenouard N, Nehrbass U, Olivo-Marin JC. In vivo colocalisation of oskar mRNA and trans-acting proteins revealed by quantitative imaging of the Drosophila oocyte. PloS One 2009; 4:e6241; PMID:19597554; http://dx.doi.org/ 10.1371/journal.pone.0006241 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.