Abstract
We identify a new subgroup of Type I Restriction-Modification enzymes that modify cytosine in one DNA strand and adenine in the opposite strand for host protection. Recognition specificity has been determined for ten systems using SMRT sequencing and each recognizes a novel DNA sequence motif. Previously characterized Type I systems use two identical copies of a single methyltransferase (MTase) subunit, with one bound at each half site of the specificity (S) subunit to form the MTase. The new m4C-producing Type I systems we describe have two separate yet highly similar MTase subunits that form a heterodimeric M1M2S MTase. The MTase subunits from these systems group into two families, one of which has NPPF in the highly conserved catalytic motif IV and modifies adenine to m6A, and one having an NPPY catalytic motif IV and modifying cytosine to m4C. The high degree of similarity among their cytosine-recognizing components (MTase and S) suggest they have recently evolved, most likely from the far more common m6A Type I systems. Type I enzymes that modify cytosine exclusively were formed by replacing the adenine target recognition domain (TRD) with a cytosine-recognizing TRD. These are the first examples of m4C modification in Type I RM systems.
INTRODUCTION
Restriction-Modification (R-M) systems are widespread in bacteria and archaea, where they function as a barrier to invasive DNA (1,2). R-M systems can play additional roles, such as the regulation of gene expression leading to pathogenicity through the epigenetic effect of their DNA methylation recently described (3). The first R-M system was described more than six decades ago when it was observed that bacteriophage were restricted in their ability to infect certain strains, yet phage isolated from the rare infections that did occur were subsequently able to infect the restrictive and non-restrictive strains equally, indicating they had been modified (4,5). These first systems were found to be multi-subunit molecular machines that recognize a specific DNA sequence and cleave when the sequence is unmodified, but which can protect the same site by methylation. These complex systems are classified as Type I R-M systems (6) and typically have a single specificity subunit (HsdS or S) that determines the DNA recognition sequence and which consists of two independent DNA binding domains (Target Recognition Domains, or TRDs) connected by two long alpha helices. This results in a characteristic ‘split’ DNA recognition sequence, consisting of two ‘half sites’ each containing two to five specific base pairs separated by a spacer of a fixed number of unspecified DNA bases, which is determined by the connecting helices and typically ranges from four to nine base pairs. For example, the EcoKI recognition sequence is 5′-AAC-NNNNNN-GTGC-3′. The DNA methyltransferase (MTase) complex consists of one S subunit and two methyltransferase subunits (HsdM or M), wherein each of the two TRD domains in the S subunit binds to one M subunit to form an active M2S MTase complex that methylates a base in each half site, resulting in one methylated base on each DNA strand (7). In all of the Type I R-M systems previously characterized modification was observed to occur exclusively on an adenine base in each strand (8).
DNA cleavage requires the further binding of two endonuclease subunits (HsdR or R) to a MTase complex bound at a non-modified recognition site. DNA cutting then occurs at random positions that are distant from the recognition site (see (9) for a recent review of Type I systems). Because the position of DNA cutting in Type I systems is non-specific, characterizing Type I systems has been relatively difficult and labour intensive. Due to this difficulty and their lack of use as molecular biology reagents, few Type I systems were characterized after the initial investigations into their biology. However, this situation has now changed with the advent of Single-Molecule-Real-Time (SMRT) DNA sequencing, which enables the direct detection of DNA modification. Following the bioinformatic identification of Type I system genes within a sequenced genome this often allows the unambiguous matching to their recognition specificity motifs (10,11). This has led to an exponential increase in the number of known Type I recognition specificities, from approximately 40 biochemically characterized specificities from all studies through 2011 to more than 1100 known today (8).
While sequencing the genome of Pseudomonas alcaligenes NEB 585 using SMRT sequencing to see if it contained PacI recognition sites we found an unusual Type I RM system, PacII, that contained one extra methyltransferase (hsdM or M) gene. Furthermore, the recognition sequence for this system, based on the motif deduced from the SMRT Analysis, had the expected m6A (N6-methyladenine) modification in one strand, but m4C (N4-methylcytosine) modification of a cytosine base in the other strand. In this case, one half site of the recognition sequence contained only G or C residues meaning that there was no A residue available for m6A modification. We examined other genome sequences for similar occurrences of two M genes associated with a typical Type I system specificity (hsdS or S) and endonuclease (hsdR or R) gene and found that there were many examples of this same arrangement. We proceeded to characterize in more detail the PacII system and also to check experimentally a number of additional Type I systems with this same arrangement of 2 M, 1 R and 1 S genes using SMRT sequencing. We now report the characterization of ten such systems and identify many more in other bacterial genomes.
We further report examples of Type I R-M systems that exclusively use cytosine modification, engineered by swapping out the m6A TRD and replacing it with an m4C TRD, as has previously been done to change Type I specificities (12,13).
These findings suggest Type I systems have the potential to vary their recognition specificity not only through TRD recombination events as previously observed but also through the evolution of new modification functions that allow recognition motifs modifying adenine or cytosine.
MATERIALS AND METHODS
All restriction endonucleases, Q5 DNA polymerase, S-adenosyl-methionine, T4 DNA ligase, GIBSON cloning reagents, competent cells, DNA size standards and substrate DNAs were from New England Biolabs (Ipswich, MA, USA). DNA oligonucleotides were from New England Biolabs, Organic Synthesis Division (Ipswich, MA) or Integrated DNA Technologies (Coralville, IA, USA). SMRT sequencing was performed on the Pacific Biosciences (Menlo Park, CA, USA) RSII instrument according to the manufacturer's instructions. DNA modifications were called using the SMRT Portal analysis software (Pacific Biosciences), although it should be noted that while methylation calling for m6A is usually very high even at lower coverage, calling for m4C is usually less complete at low coverage and does not necessarily reflect the true level of methylation. The detection of an m4C signal at lower coverage is usually just scored as positive or negative without making any attempt at quantification. Pseudomonas alcaligenes NEB 585 gDNA was isolated from freshly grown liquid culture by phenol-chloroform extraction and isopropanol precipitation. Genomic DNA isolated from DSM strains was purchased from the DSMZ culture collection. Pseudomonas mendocina S5-2 gDNA was kindly provided by T.M. Chong. Salmonella enterica ssp. enterica serovar Agona str. SL483 gDNA was kindly provided by David Dryden. For SMRT sequencing, gDNA samples were sheared using Covaris gTubes (Covaris, Woburn, MA, USA) before entering the Pacific Biosciences 5kb library preparation protocol according to the manufacturer's instructions. For expression in Escherichia coli, putative genes were PCR amplified from genomic DNA and joined to an expression vector using GIBSON assembly master mix (NEB). The genes were positioned downstream of the lac promoter for the pRRS plasmid (high copy), or the tet gene promoter for pACYC184 (low copy) and grown in the non-methylating E. coli strain ER2796 (14) with appropriate antibiotic selection. Following in vivo expression and modification, gDNA was isolated by enzymatic lysis (lysozyme), phenol-chloroform extraction and isopropanol precipitation from 15 ml of ER2796 cell culture expressing the systems under study. The gDNA was treated with RNaseA while undergoing shearing, and was then purified using AMPure beads (Pacific Biosciences) prior to library preparation and SMRT sequencing on the PacBio RSII sequencer to identify DNA modifications.
Restriction assay
The Type I systems were expressed in ER2796, with the endonuclease (R gene) expressed from the high-copy plasmid pRRS and the MTase (M1, M2 and S genes) expressed from pACYC184. 100 μl of 10-fold dilutions of λvir phage (kindly supplied by Elisabeth Raleigh, NEB) was mixed with 100 μl of a fresh culture of the ER2796 constructs, incubated 15 min at 30°C, mixed with 3 ml top agar and immediately plated on LB (amp/cam), grown O/N at 37°C, and phage plaques counted. Efficiency of plating (eop) was calculated from the average of three dilutions using 2 replicates.
TRD swapping
The m6A half site TRD of S.PacII or S.DbaKI (TRD2, the carboxy-terminal TRD domain) was replaced with an m4C half site TRD (TRD1, the amino-terminal TRD domain) according to the following amino acid boundaries, leaving the connecting helices between the half sites unchanged.
TRD domains swapped: (amino acid positions delineating domains shown)
| Enzyme: | TRD1 (m4C) | TRD2 (m6A) |
| S.PacII | 1–145 | 193–342 |
| S.Mma5219I | 1–150 | (not swapped) |
| S.Asu14238II | 1–148 | (not swapped) |
| S.DbaKI | 1–145 | 198–356 |
| S.Apa12260I | 1–150 | (not swapped) |
The wt MTase expression construct, less the TRD being replaced, and the incoming TRD were PCR amplified, joined by Gibson assembly and introduced into ER2796 for in vivo expression and subsequent modification analysis.
Protein multiple sequence alignments were performed using the PROMALS 3D server (15).
SMRT sequencing methylation analyses were performed using the program RS_Modification_and_Motif_analysis.1 in the SMRTAnalysis suite of programs from Pacific Biosciences, Inc (Menlo Park, CA). ‘IPD’ (Inter-Pulse-Duration) indicates the ratio of the average time taken to incorporate the base at a given position compared to the time to incorporate that base at that given position in a DNA known to be unmodified, taking into account the surrounding sequence context of the given base (10). For m4C, an IPD ratio of 3–5 is typical, indicating that it takes on average three to five times longer to incorporate guanine opposite an m4C-modified-cytosine than an unmodified cytosine. For m6A, an IPD ratio of 5–7 is typical. Since base incorporation is stochastic, the IPD ratio becomes statistically rigorous when the coverage of sequence reads at the base under study is 25X or greater for m4C and m6A modification (11).
RESULTS
Identification of a Type I system producing m4C and m6A modification
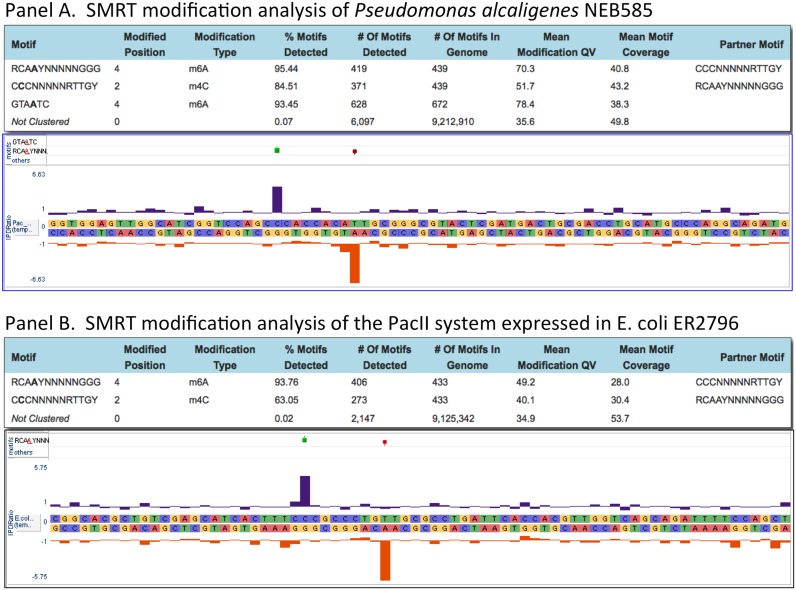
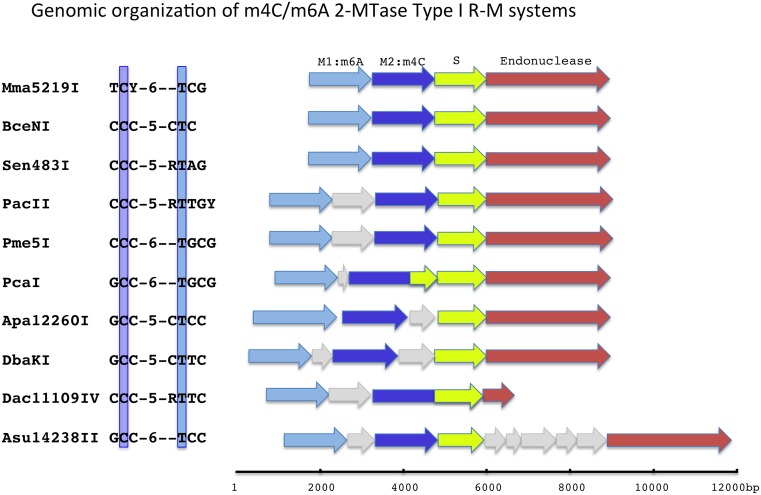
Genomic DNA from Pseudomonas alcaligenes NEB 585, the strain that produces the Type II restriction endonuclease PacI, was SMRT sequenced to determine a complete, finished genome sequence (GenBank accession CP014784) and to identify any DNA modifications and their specific DNA recognition motifs. Two methylated sequences were found, but neither corresponded to the PacI recognition sequence of TTAATTAA. Rather the genome sequence was devoid of this octanucleotide, eliminating the need for a MTase to protect against the action of the PacI REase. One modified sequence was GTA(m6A)TC with modification occurring in just one DNA strand, typical of a Type IIG-like enzyme. A second motif looked like a typical Type I specificity, 5′-CCC-N5-RTTGY-3′, but in which one half site of this split recognition sequence had a cytosine base modified at the N4 position : 5′-C(m4C)C-N5-RTTGY-3′, while the opposite strand had a modified adenine like other Type I systems: 5′-RCA(m6A)Y-N5-GGG-3′ (Figure 1, panel A). Throughout this manuscript we denote this as Cm4CCNNNNNRTTGY with the underline indicating that it is the complementary base (A in this case) that is modified. By sequence similarity a single putative Type I system was present in the P. alcaligenes genome sequence having a typical endonuclease subunit gene (R) and specificity subunit gene (S) flanked by two MTase subunit genes (M), whereas typical Type I systems have a single MTase gene (Figure 2). The presence of two MTase subunits suggested that one M subunit might modify the A residue in one half site and the second M subunit might modify the C residue in the other half site. We therefore cloned the S and both MTase genes in a non-methylating E. coli strain (ER2796) and grew the transformed strain to saturation overnight to allow in vivo modification. We then isolated total genomic DNA from the cells expressing the S and two MTase subunits and tested for methylation using SMRT sequencing. We found the E. coli genomic DNA from this clone was now modified at the same 5′-CCC-N5-RTTGY-3′ sequence motif modified in the native P. alcaligenes, demonstrating that this 2 MTase Type I system modifies both m4C and m6A (Figure 1, panel B). This system was named PacII.
Figure 1.
Panel (A) Methylome analysis for Pseudomonas alcaligenes NEB585, and an IPD (Inter-Pulse-Duration) plot showing modification at a representative PacII sequence motif: Cm4CC-5-RTTGY, within the genome sequence. Panel (B) methylome analysis for the genome of E. coli ER2796 expressing the PacII MTase, and an IPD plot showing modification at a representative PacII sequence motif: Cm4CC-5-RTTGY, within the ER2796 genome sequence.
Figure 2.
Genomic organization of the m4C/m6A Type I R-M system genes. M1:m6A indicates the m6A MTase subunit and M2:m4C indicates the m4C MTase subunit. The C (cytosine) that is methylated to m4C is boxed, as is the T (thymine) that pairs with the adenine that is methylated in the opposite strand. In these 10 systems the spacing between the methylated bases is conserved at 7 base pairs. Grey arrows indicate genes that are not part of the RM system.
Bioinformatic identification and characterization of additional Type I systems producing m4C and m6A modification
We performed BLAST searches using the amino acid sequence of either one of the MTase subunits, the S subunit or the R subunit of the PacII system against REBASE and the NCBI non-redundant databases. Currently, more than 300 putative Type I systems can be found that have two MTase genes in close proximity to the R and S genes (5). We obtained genomic DNA for six strains carrying such double-MTase Type I systems from the DSMZ culture collection and performed SMRT sequencing on these genomic DNAs. All six organisms exhibited methylation motifs that included a typical split Type I recognition motif in which one strand contained m4C modification and the other contained m6A modification (Table 1, Supplementary Figure S1). We cloned the putative 2-MTase Type I systems from these six strains into the non-methylating E. coli strain ER2796 and confirmed that these systems produced the Type I m4C and m6A methylation motif (Supplementary Figure S2). We also identified several additional m4C-m6A Type I systems by performing genome sequencing and SMRT sequence methylome analyses, including Pme5I (genomic DNA kindly supplied by T. M. Chong, personal communication) and Sen483I (genomic DNA kindly supplied by D.T.F. Dryden, personal communication). The Dac11109IV system was originally observed at Pacific Biosciences (T.A. Clark, personal communication). We thus obtained the recognition specificities for 10 dual-modification m4C/m6A Type I systems (Table 1, Figure 2), with the expectation that many more such systems will be described as more prokaryote methylomes are characterized through SMRT sequencing.
Table 1. Characterized m4C/m6A Type I R-M systems.
| Enzyme | Specificity | Host Organism | gDNA source |
|---|---|---|---|
| PacII | CCC-N5-RTTGY | Pseudomonas alcaligenes | NEB 585 |
| DbaKI | GCC-N5-CTTC | Desulfarculus baarsii | DSM 2075 |
| PcaI | GCC-N6–TGCG | Pelobacter carbinolicus | DSM 2380 |
| BceNI | CCC-N5-CTC | Bacillus cellulosilyticus | DSM 2522 |
| Mma5219I | TCY-N6–TCG | Methanohalophilus mahii | DSM 5219 |
| Apa12260I | GCC-N5-CTCC | Aminomonas paucivorans | DSM 12260 |
| Asu14238II | GCC-N6–TCC | Aequorivita sublithincola | DSM 14238 |
| Pme5I | CCC-N6–TGCG | Pseudomonas mendocinaS5.2 | T.M.Chong |
| Sen483I | CCC-N5-RTAG | Salmonella entericaSL483 | D.T.F.Dryden |
| Dac11109IV | CCC-N5-RTTC | Desulfobacca acetoxidans | DSM 11109 |
Restriction phenotype for m4C/m6A Type I systems expressed in E. coli
To test whether these double-MTase, m4C and m6A Type I systems are capable of restriction, we expressed several of these systems in E. coli and challenged them with lambdavir phage. We found the systems tested did restrict, though at moderate levels of roughly 30-fold, compared to the host strain carrying no restriction system (Table 2, panel A). Many of the plaques that formed in the strains expressing the Type I RM systems were significantly smaller than those in the non-restricting host, suggesting partial restriction. Phage from a single large plaque were isolated from the DbaKI and Apa12260I expressing hosts and plated against naive wild type E. coli and the RM-expressing host. For these phage passaged through the R-M expressing host, nearly the same number of plaques were obtained on the non-restricting and the restricting host, as would be expected for phage modified by the Type I MTase (Table 2, panel B). Phage from a single plaque for each system were then isolated from the non-restricting/non-modifying control host and used to challenge non-restricting and restricting host once more. For phage passaged through the restricting/modifying host, and then the non-restricting/non-modifying host, we now observed 250-fold restriction by DbaKI and 130-fold restriction by Apa12260I expressing cells (Table 2, panel C). The moderate levels of restriction are likely due to relatively poor expression in the heterologous E. coli host. These results show classic restriction and modification by these double-MTase Type I enzymes and demonstrate these are complete and active R-M systems.
Table 2. Restriction of λvir by R-M systems cloned in E. coli.
| Host | eop (efficiency of plating) | Restriction |
|---|---|---|
| A. Naive phage (1st passage: M-) | ||
| ER2796_RM- | 1 | – |
| ER2796_DbaKI_RM+ | 0.03 | 30x |
| ER2796_Apa12260I_RM+ | 0.04 | 25x |
| B. Phage isolated from restricting host (2nd passage: M+) | ||
| ER2796_RM- | 1 | – |
| ER2796_DbaKI_RM+ | 0.7 | 1.5x |
| ER2796_RM- | 1 | – |
| ER2796_Apa12260I_RM+ | 0.3 | 3x |
| C. Phage from 2nd passage isolated from non-restricting host (3rd passage: M-) | ||
| ER2796_RM- | 1 | – |
| ER2796_DbaKI_RM+ | 0.004 | 250x |
| ER2796_RM- | 1 | – |
| ER2796_Apa12260I_RM+ | 0.007 | 130x |
eop: average of 3 plates in 2 replicates.
sites: DbaKI and Apa12260I each have 8 recognition sites in Lambdavir.
Genome organization of m4C/m6A Type I systems
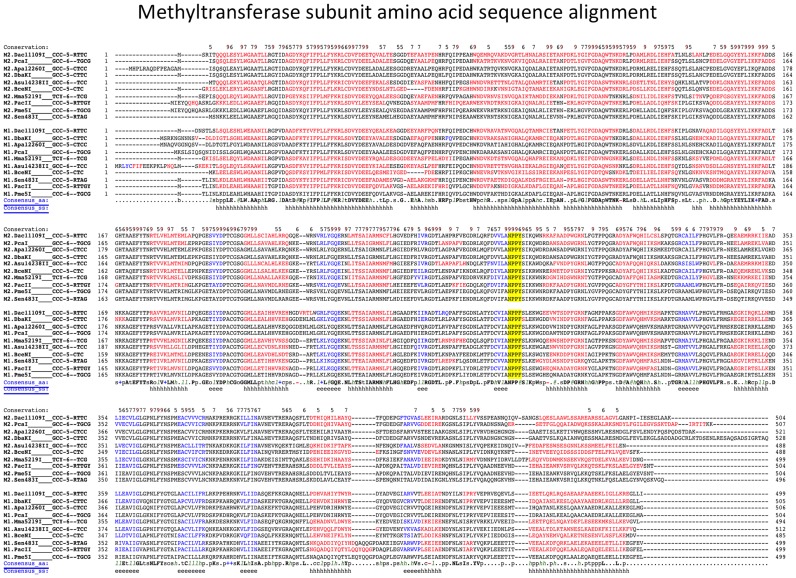
The MTase subunit amino acid sequences from the characterized systems were aligned and found to partition into two groups, with one MTase subunit of each individual Type I system sorting into each group (Figure 3). Among several highly conserved differences between the MTase subunits, at the amino-MTase catalytic motif IV, known as the ‘DPPY’ motif, one group has phenylalanine (NPPF) and the other tyrosine (NPPY). The spatial organization of the MTase subunit genes is conserved in the characterized systems and homologs, with the NPPF MTase gene located 5′ (upstream) of the NPPY MTase gene, and the NPPY MTase located 5′ (upstream), and most often directly adjacent to, the specificity gene, while the endonuclease gene is located 3′ to the specificity subunit (Figure 2). For clarity we designate the upstream MTase subunit containing the NPPF motif as ‘M1’, and the downstream MTase subunit containing the NPPY motif IV as ‘M2.’ As is common with Type I systems, some of these systems have additional open reading frames interspersed between the R-M system component genes; most commonly between the two MTase genes (6 of 10 characterized systems), but also between the M2.MTase and the S gene (two systems), or between the S gene and the R gene (one system). In one case, the M2 MTase gene is fused to the specificity subunit: M2.Dac11109IV contains both the M2 subunit and the S subunit by sequence similarity analysis, similar to the previous demonstration that the hsdM and hsdS genes of the EcoKI system could be fused into a single orf that maintained function (16). This is also consistent with the observation that the M2 (NPPY) MTase is located next to the S subunit. We also note that the M2.PcaI gene appears to encode a single TRD half-site at its carboxy-terminal end as well as the entire M2.PcaI MTase subunit, in addition to the separate, complete S subunit. The function if any of this additional TRD domain has not been investigated.
Figure 3.
Amino acid alignment of the methyltransferase proteins. The catalytic motif IV residues (NPPF or NPPY) are highlighted. Red indicates predicted helical secondary structure; blue indicates predicted beta strand secondary structure (from Promals alignment server).
The two MTase subunit protein sequences are highly similar
Overall, the protein sequences of the two MTase subunits in these systems are quite similar, suggesting that they likely share a recent common ancestor (Figure 3). For the PacII system, M1.PacII and M2.PacII share 45% identities and 63% similarities overall, while identities between the M1 and M2 MTase subunits in the 10 systems described range from 45 to 55% and similarities from 62 to 73%. That the two MTase subunits are in general more similar to each other than to Type I single MTase subunits is demonstrated by the results of BLAST searches. A BLAST search against the REBASE database with either MTase subunit returns first homologs of the same MTase subunit group from two MTase systems, with expectation values of E = 0.0 to roughly E = e-160, then mainly subunits of the other MTase subunit group from two MTase systems, with expectation values of roughly E = e-150 to E = e-115, before returning hits to MTase subunits from single MTase Type I systems at roughly E = −110 and higher (less similar) values. Because both MTase subunits must interact with the same endonuclease protein subunit to accomplish the biological function of restriction, these MTase subunits are constrained to maintain their protein-protein interface surfaces that interact with the endonuclease subunit. Structural information for Type I systems also indicates that the two MTase subunits contact each other, as well as the S subunit, to form the active MTase complex (17), so the two distinct MTase subunits in our systems are also constrained to maintain protein-protein interfaces between the C-MTase and A-MTase subunits and with their respective TRD domains.
Identification of the M subunit specificity for m4C or m6A
To determine which of the two methyltransferase proteins produces m6A and which m4C, we used two approaches. First we expressed the MTase subunit genes individually with the wild type S subunit and performed SMRT sequencing methylome analysis on the host E. coli gDNA isolated from overnight cultures. Second we mutated residues within the conserved catalytic motif IV of just one MTase subunit and performed SMRT sequencing methylome analysis on E. coli gDNA isolated from overnight cultures expressing the complete Type I MTase (M1, M2, S) carrying the mutation. The second approach was intended to leave the overall MTase complex intact while inactivating just one of the MTase subunits.
Activity of single MTase subunits with a wild type S construct
For all four systems tested, expression of just the M2_NPPY MTase with the wild type S resulted in no detectable modification (Table 3). In contrast, expression of just the M1_NPPF MTase subunit with the wild-type S resulted in partial m6A modification of the m6A half site in each of the four systems tested (Table 3). This indicates that the M1_NPPF MTase subunit is responsible for performing m6A methylation and suggested the M2_NPPY MTase is responsible for m4C modification. Furthermore, it appears that both MTase subunits are necessary to form a fully active MTase. Since the M1_MTase (NPPF) subunit alone is partially active one possibility is that these M1 subunits can bind the S subunit at both the m4C TRD and m6A TRD and are able to form the necessary intermolecular contacts between the two (now identical) MTase subunits while binding the S subunit to form an active Type I M2S MTase, though clearly not with the same efficiency as the wild type enzyme. The m4C-producing MTase subunits do not seem to have this same ability, at least in the four systems tested.
Table 3. Modification detection by SMRT sequencing.
| Enzyme | MTase construct | m6A%detected | m4C%detected | Coverage |
|---|---|---|---|---|
| PacII | RCAAY | CCC | ||
| Native organism | 95.4 | 84.5 | 41X | |
| M1 + M2 + S | 93.7 | 63.0 | 30x | |
| M1-APPA + M2 + S | – | 56.6 | 43x | |
| M1 + M2-APPA + S | 90.7 | 24.9 | 36x | |
| M1 + S | 61.7 | – | 81x | |
| M2 + S | – | – | 90x | |
| BceNI | GAG | CCC | ||
| Native organism | 97.4 | 66.4 | 38x | |
| M1 + M2 + S | 98.6 | 90.2 | 49x | |
| M1-APPA + M2 + S | – | 45.5 | 60x | |
| M1 + M2-APPA + S | 82.3 | – | 52x | |
| M1 + S | 46.6 | – | 40x | |
| M2 + S | – | – | 90x | |
| Mma5219I | GCA | TCY | ||
| Native organism | 94.0 | 72.4 | 42x | |
| M1 + M2 + S | 73.4 | 33.0 | 26x | |
| M1-APPA + M2 + S | – | – | 29x | |
| M1 + M2-APPA + S | 89.6 | – | 51x | |
| M1 + S | 80.2 | – | 40x | |
| M2 + S | – | – | 65x | |
| DbaKI | GAAG | GCC | ||
| Native organism | 91.9 | 68.8 | 29x | |
| M1 + M2 + S | 100 | 97.7 | 46x | |
| M1-APPA + M2 + S | – | 100 | 144x | |
| M1 + M2-APPA + S | 100 | 98.7 | 99x | |
| M1 + S | 35.9 | – | 60x | |
| M2 + S | – | – | 51x |
M-APPA indicates catalytic mutant (NPPF or NPPY to APPA) of the indicated MTase subunit.
MTase subunit catalytic site mutants show M1 (NPPF) forms m6A while M2 (NPPY) forms m4C methylation
For constructs expressing the complete MTase (M1, M2 and S), mutation of the M1_MTase subunit catalytic NPPF motif to APPA abolished m6A modification in all four systems tested, but resulted in nearly wild type levels of m4C modification for three of the four systems tested, while for one system (Mma5219I) this mutation resulted in no methylation detected for either half site (Table 3). Mutation of the M2_MTase subunit catalytic NPPY motif to APPA resulted in nearly wild type levels of m6A modification, while m4C modification was abolished in three systems and greatly reduced in the fourth (Table 3). These results show that the NPPF MTase subunit is responsible for modifying adenine to form m6A, while the NPPY MTase subunit is responsible for modifying cytosine to m4C. We have designated the NPPF MTase in these systems as M1 and refer to it as the A-MTase, and the NPPY MTase as M2 and refer to it as the C-MTase.
Identification of TRD half site specificities
We were able to predict TRD half-site specificities for the various systems described based upon the sequence similarity between the individual TRD domains within the S subunits. The PacII and Pme5I systems recognize the same m4C-modified half-site motif in which the central C is modified; 5′-CCC-3′, but quite different motifs in the m6A-modified half-site; 5′-RCAAY-3′ vs 5′-CGCA-3′. Alignment of the S subunit protein sequences revealed highly significant similarity in the N-terminal TRDs (TRD1), but little sequence similarity in the C-terminal TRDs (TRD2), suggesting that TRD1 was responsible for CCC recognition and TRD2 was responsible for the m6A-modified half-site recognition. We predicted the TRD domain boundaries from a multiple sequence alignment that includes secondary structure prediction (15) (Figure 4). Here, we defined TRD1 as the amino-terminus of the proteins up to the first conserved PLPPL sequence motif that forms the turn from a globular TRD domain into the long alpha-helix connector region in Type I S subunits (17). TRD2 was defined as the sequence immediately following the predicted amino-terminal connector helix through to the second conserved PLPPL sequence motif preceding the second long helical connector. To test our bioinformatic predictions experimentally, we replaced the putative m6A TRD2 of two systems, PacII and DbaKI, with putative m4C TRD1's to create Type I systems that generate only m4C modification.
Figure 4.
Specificity subunit amino acid alignment. The conserved ‘PLPPL’ motifs connecting the globular TRDs into the two helical spacer sequences is underlined. Red indicates predicted helical secondary structure; blue indicates predicted beta strand secondary structure (from Promals alignment server).
Duplication of TRD1 in S.PacII creates a palindromic m4C specificity: 5′-Cm4CC-N5-GGG-3′
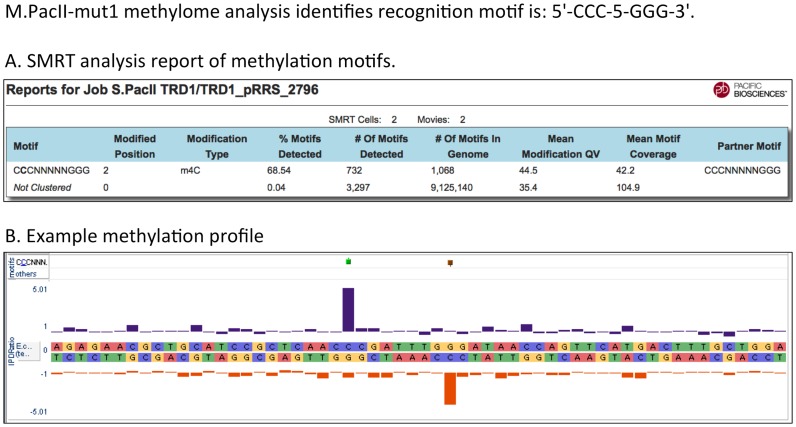
To create an all m4C Type I enzyme we replaced TRD2 of the PacII S subunit with a copy of its TRD1. S.PacII amino acids 193 through 342 (TRD2) were replaced by S.PacII amino acids 1 through 145 (TRD1). In an attempt to ensure flexibility for proper protein folding, we added a three amino acid flexible linker (Gly-Ser-Gly) between the first connecting alpha helix and the start of the duplicated TRD1 replacing TRD2. The TRD2 sequence was replaced up to the second PLPPL motif that leads into the helical connector, such that the two helical connectors between the DNA recognition domains were maintained as in the wild type protein (note that in S.PacII this TRD2 PLPPL motif is YLPPI). The altered S subunit was expressed with both MTases, M1.PacII and M2.PacII, in the non-methylating E. coli host ER2796, genomic DNA was isolated and subjected to SMRT sequencing to analyze methylation specificity. Duplication of TRD1 in the PacII system resulted in m4C modification of the palindromic recognition sequence 5′-Cm4CC-5-GGG-3′ as expected (Table 4, Figure 5). This engineered enzyme is the first example of a Type I RM system MTase that uses only m4C for host modification. This enzyme was named M.PacII-mut1. While this is a synthetic construct, there is in principle no reason such all-m4C Type I systems could not exist naturally, although we have not yet observed such a system in bacterial genome sequences.
Table 4. Hybrid all-m4C systems: modification detection by SMRT sequencing.
| Enzyme | MTase construct | Motif / m4C%detected | Coverage | |
|---|---|---|---|---|
| PacII-PacII S hybrid: CCC-5-GGG | CCC | |||
| M1 + M2 | 68.5 | 42x | ||
| M1-APPA + M2 | 45.1 | 43x | ||
| M1 + M2-APPA | – | 36x | ||
| M1 + -- | – | 81x | ||
| -- + M2 | – | 90x | ||
| PacII-Mma5219I S hybrid: CCC-5-RGA | CCC | TCY | ||
| M1 + M2 | 68.9 | 67.1 | 39x | |
| M1 + -- | – | – | 81x | |
| -- + M2 | – | – | 90x | |
| PacII-Asu14238II S hybrid: CCC-5-GGC | CCC | GCC | ||
| M1 + M2 | 48.6 | 63.8 | 43x | |
| M1 + -- | – | – | 42x | |
| -- + M2 | – | – | 46x | |
| DbaKI-DbaKI S hybrid: GCC-5-GGC | GCC | |||
| M1 + M2 | 83.3 | 100x | ||
| M1 + -- | – | 46x | ||
| -- + M2 | – | 27x | ||
| DbaKI-Apa12260I S hybrid: GCC_N5-GGC | GCC | |||
| M1 + M2 | 84.9 | 98x | ||
| M1 + -- | – | 34x | ||
| -- + M2 | – | 28x | ||
| DbaKI-Asu14238II S hybrid: GCC_N5-GGC | GCC | |||
| M1 + M2 | 86.7 | 100x | ||
| M1 + -- | – | 32x | ||
| -- + M2 | – | 27x | ||
M-APPA indicates catalytic mutant (NPPF or NPPY to APPA) of the indicated MTase subunit.
Figure 5.
Panel (A) Methylome analysis for the genome of E. coli ER2796 expressing the PacII-mut1 MTase showing recognition at the palindromic motif CCC-5-GGG. Panel (B) An IPD plot showing modification at a representative PacII-mut1 sequence motif; Cm4CC-5-GGG, within the ER2796 genome sequence.
Creation of multiple novel m4C-only Type I systems through TRD swapping
We created additional novel all-m4C Type I specificities by replacing the PacII m6A TRD with the m4C-recognizing TRD domain from two other m4C Type I systems identified in this report in which the m4C half site recognition motif differs from the PacII motif of 5′-CCC-3′. When TRD2 of PacII was replaced with TRD1 from Asu14238II, which recognizes the m4C half site motif 5′-GCC-3′, the resulting system was active and modified 5′-Cm4CC-N5-GGC-3′ as predicted (Table 4). In the same way we replaced TRD2 of PacII with TRD1 from Mma5219I, which recognizes the m4C half site motif 5′-TCY-3′, and found the hybrid was active and now recognized 5′-Cm4CC-N5-RGA-3′ as predicted (Table 4).
However, attempts to convert PacII or DbaKI to an all-m6A Type I specificity by replacing the m4C TRD with a copy of an m6A TRD, either from the same enzyme or from one of the other systems in this study, resulted in constructs that failed to produce any modification when expressed with the M1 and M2 MTase subunits.
Creation of hybrid m4C-only Type I MTases resistant to restriction by BglI
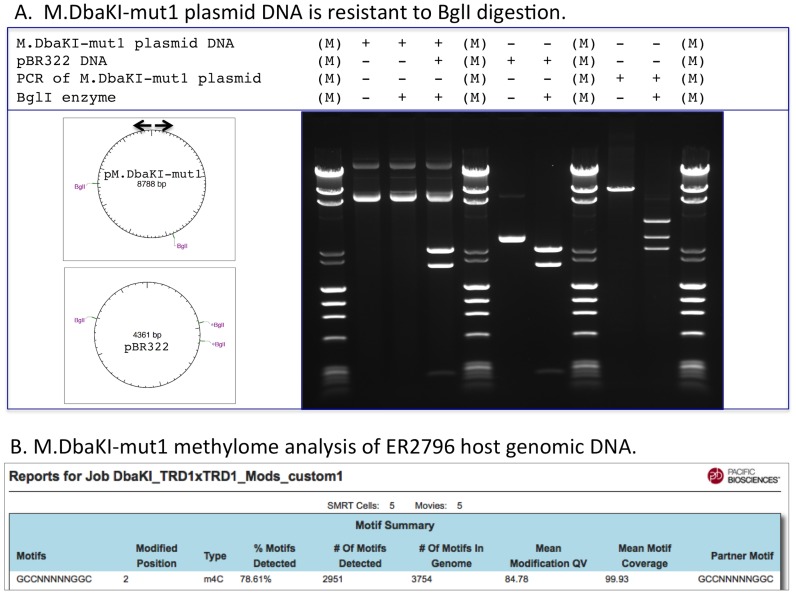
We created m4C Type I hybrids designed to recognize and methylate the palindromic motif 5′-Gm4CC-N5-GGC-3′, as this motif is methylated at exactly these positions by the Type IIP methylase M.BglI (18) and thus resistance to digestion with BglI could be used to confirm 5-Gm4CC-N5-GGC-3′ modification.
The native DbaKI MTase forms 5′-Gm4CC-N5-CTTC-3′, so we replaced the m6A-recognizing TRD2 of the DbaKI S subunit either with a duplication of DbaKI TRD1, with TRD1 from the S subunit of Apa12260I (5′-Gm4CC-N5-CTCC) or TRD1 from Asu14238II (Gm4CC-N6-TCC). All of these hybrid enzymes, if functional, are expected to recognize and modify 5′-GCC-N5-GGC-3′. ER2796 cells expressing the hybrid specificity subunit together with M1.DbaKI and M2.DbaKI were grown overnight in liquid culture to allow in vivo modification of both the plasmid carrying the MTase (M1, M2 and hybrid-S genes) and the host genomic DNA. Plasmid DNA was then prepared and digested with BglI, while gDNA was isolated and subjected to SMRT sequencing to analyze genome-wide methylation. All three hybrid constructs generated the expected modified sequence 5′-Gm4CC-N5-GGC-3′ as demonstrated by their resistance to BglI digestion of the plasmid DNAs expressing these constructs (Figure 6, panel A) and SMRT sequencing of the host E. coli gDNA (Figure 6, panel B; Table 4). This confirms the SMRT sequencing modification analyses that these hybrid MTases form 5′-Gm4CC-N5-GGC-3′.
Figure 6.
Panel (A) M.DbaKI-mut1 methylated plasmid expressing the M.DbaKI-mut1 MTase is resistant to BglI digestion, even in the same reaction in which a control unmodified DNA (pBR322) is cut. The M.DbaKI-mut1 expression plasmid has BglI recognition sites, as indicated by BglI cleavage of a PCR amplicon, which erases the M.DbaKI-mut1 modification of the plasmid. Arrows indicate the PCR primer positions. Resistance to BglI digestion confirms the SMRT sequence methylome analysis shown in Panel B that demonstrates M.DbaKI-mut1 produces Gm4CC-5-GGC modification.
MTase subunit specificity evaluated by catalytic motif mutations in all-m4C hybrid systems
We generated mutants of the PacII MTase genes by changing the conserved catalytic motif IV from NPPF or NPPY to APPA and tested these mutants for activity using the PacII hybrid all-m4C S subunit containing the duplicated TRD1 (PacII-mut1). In the full configuration containing one wild type M gene and one mutant M gene, wild type M2.PacII together with mutant M1.PacII produced 5′-Cm4CC-N5-GGG-3′, while wild-type M1.PacII together with mutant M2.PacII showed no modification as expected (Table 4). This confirmed the specificity of the individual M genes. That M1.PacII is not producing detectable m4C modification suggests that two binding events are required to modify both strands of an unmodified site, with M2.PacII modifying the first strand, then rebinding in the opposite orientation to modify the second strand. This further suggests the modifed cytosine can fit into the adenine base binding pocket of M1.PacII. Expression of either mutant M gene alone with the hybrid S subunit did not produce detectable modification (Table 4).
DISCUSSION
Single Molecule Real Time (SMRT) sequencing permits methylome analysis of prokaryote genomes and has revolutionized the characterization of Type I RM system recognition specificity. Here we show for the first time that Type I systems can use m4C methylation for host protection in addition to m6A methylation. There is no inherent reason why Type I R-M systems should have been expected to use m6A exclusively, but the technical challenge of characterizing Type I systems by traditional approaches meant that few systems were characterized extensively until recently. While it is possible that we may find Type I systems which use 5mC (5-methylcytosine) modification for host protection, such systems are unlikely to have evolved from the canonical m6A Type I systems in view of the more extreme differences between m5C MTase structure and that of the m6A/m4C MTases. Certainly there are no bioinformatic hints of Type I systems using 5mC among the many thousand genomes so far sequenced.
One striking feature of the hundreds of m6A-Type I specificities observed in prokaryotic methylomes is their great diversity of recognition specificities. While there is likely conservation of the structural core of the TRD domains that recognize the half sites, there is great diversity in both their protein sequences and their recognition motifs (5). In contrast, the less abundant TRD domains that recognize a motif in which a cytosine is modified to m4C exhibit a much greater level of protein sequence similarity. Among the 10 recognition specificities characterized here, there are only 3 unique m4C half site motifs: 5′-CCC-3′, 5′-GCC-3′ and 5′-TCY-3′, whereas 8 out of 10 of the m6A half site motifs are different. The significant sequence conservation and limited repertoire of recognition motifs imply that m4C modification may be a relatively recent acquisition by the Type I systems.
To date there are more than 300 putative two-methyltransferase Type I systems cataloged in REBASE (8). We presume that, like the 10 systems characterized and reported here, these have split-recognition sequence motifs in which one half site is methylated at a cytosine and the other at adenine. By comparison REBASE reports almost 12,000 putative Type I systems that contain just a single M gene, which suggests that the m4C Type I systems represent about two and a half percent of the total Type I systems. This relative rarity explains why such systems were not seen before the application of SMRT sequence methylome analyses and also suggests that they are more recently evolved.
We were able to create novel Type I RM systems that use only m4C modification for host protection through swapping TRD domains, showing that these m4C Type I systems are similar to other split-recognition R-M systems where TRD swapping has been well established. We note that we were not able to generate an active all-m6A PacII system in which the m4C TRD was replaced by an m6A TRD. Further study will be necessary to see if these m4C-m6A enzymes can be engineered to m6A modifying systems. The success of our TRD swapping demonstrates both the potential to engineer new recognition specificities in the Type I R-M systems and the ability of these systems to evolve functional variation.
Such m4C systems expand the potential range of Type I recognition specificities by eliminating the need to have an adenine base in each half site, and suggests that m4C-only Type I systems may be found in Nature, although we do not find clear candidates by current bioinformatic analysis of GenBank. Putative all-m4C systems could presumably evolve to use two copies of a single (m4C) MTase subunit, just as the canonical m6A Type I systems use a single m6A MTase. While there are plenty of examples of Type I systems containing just a single MTase gene similar to the M1 MTase genes we describe, there are no putative systems containing a single MTase very highly similar to the M2-MTase gene. This is consistent with the finding that the m4C MTase subunits in this study were not functional on their own, even when paired with an all-m4C hybrid S subunits. However, we cannot rule out the possibility that there might be an alternative way of producing m4C by an MTase subunit that more closely resembles the M1-like genes.
Because an active Type I methylase uses two M subunits binding to one S subunit, which in turn binds to two separate half site targets, there are a number of protein-protein and protein-DNA interactions that need to be properly positioned. It is perhaps not surprising that when one of the M subunits or one of the TRD domains is changed that catalytic efficiency is impaired. The failure of a single M2 subunit from one of these systems to assemble correctly with its wild type S subunit and then methylate a half site probably reflects the disruption of some key protein-protein interaction in the methylase complex.
That the corresponding M1 subunit together with its wild type S subunit can assemble and at least partially modify its normal target may reflect a greater degree of tolerance in the assembly with this subunit and could perhaps be the result of memory from an earlier, ancestral time. The more extreme behaviour seen when both mutant M genes and rearranged S subunits are present are to be expected and may reflect stages in the evolution of these systems. Further insights into the assembly of these systems will likely require crystallographic structures.
The high degree of similarity between the amino acid sequences of the m4C and m6A MTase subunits in these systems suggests the m4C MTase subunit likely evolved from a duplicated m6A MTase subunit, consistent with previous phylogenetic analyses that indicated the m4C MTase M.BamHI arose from an m6A MTase ancestor (19). Although previous studies have shown that some individual amino-MTases do have the ability to modify both adenine and cytosine, the rates observed are quite different (20–22). The Type I systems we describe employ two distinct, functionally different MTases rather than a single, bi-functional MTase, suggesting the difficulty of evolving a MTase than can efficiently accommodate and methylate both cytosine and adenine. Discrimination may be due to subtle structural changes for binding and positioning the flipped cytosine or adenine bases. The similarity between the two families of m4C and m6A MTases here may provide an opportunity to gain insight into the mechanism leading to discrimination between cytosine and adenine bases among the DNA amino-MTases. We observe several highly conserved differences between the two groups of proteins that model next to the flipped base in the active site, including differences that may potentially create a smaller binding pocket in the m4C MTases for preferentially accepting cytosine. In the m4C-MTase subunits, the loop corresponding to M2.PacII Asn375-Ser376-Pro-377-Met378-Glu379 models next to the flipped base in the active site, is highly conserved and has larger amino acid side chains compared to the m6A-MTase subunits, where M1.PacII has Gly366-Thr367-Ala368-Ile369-Pro370 (model based on PDB 2Y7H, EcoKI). The M2.PacII Asn375 and Met378 residues appear well positioned to create a smaller binding pocket for preferentially accepting cytosine. While these differences require further study, it is hoped that identifying conserved crucial residues might improve the bioinformatic prediction of m4C versus m6A specificity in DNA amino-MTases.
The m4C-m6A Type I systems described present a potential evolutionary opportunity compared to the canonical Type I RM systems, in that having two distinct MTases make these systems inherently asymmetric. This asymmetry could open new paths for evolution, as one can imagine a trajectory from such asymmetric systems where an endonuclease domain fuses to just one of the now non-identical MTase subunits, leading toward the Type I-SP enzymes, which translocate and cut in just one direction (23), or the Type IIB enzymes, which recognize split sequence motifs like the Type I but cut specifically on both sides of the recognition sequence (24). These potential advantages come at the perhaps modest cost of maintaining and expressing two MTase genes. These asymmetric systems may also facilitate engineering efforts that target just one side of the Type I holoenzyme complex.
While m4C is the least common of the three methylated bases that provide host protection in R-M systems, it had previously been observed in the Type II restriction systems—both regular ones such as BamHI and in Type IIG-like systems, and it is now showing up in Type I systems as well as in some Type III systems (Murray, I.A. and Roberts, R.J. unpublished). In all cases the numbers are low indicating it is probably a recent evolutionary discovery, but it is not uncommon. One wonders whether other kinds of methylation or more extreme modification might also be associated with restriction systems. Certainly, phages such as bacteriophage Mu and the T-even phages use more complicated base decorations to overcome restriction enzymes and it would be surprising if the bacteria themselves have not found a use for additional exotic modifications. If they have, then the usual bioinformatic methods of finding RM systems might require a more open approach. Once again Nature has offered some surprises and reminded us that we are still at the beginning of understanding the lifestyle of the bacteria.
Supplementary Material
Acknowledgments
We thank David Dryden (University of Edinburgh) and T.M. Chong (University of Malaya) for providing DNA samples. R.D.M., Y.A.L. and R.J.R. work for New England Biolabs, a company that sells research reagents, including restriction enzymes and DNA methylases to the scientific community. T.A.C. works for Pacific Biosciences, a company developing single molecule, real-time sequencing technologies, including the sequencing platform used in this project.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
New England Biolabs, Inc. Funding for open access charge: New England Biolabs.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wilson G.G., Murray N.E. Restriction and Modification Systems. Annu. Rev. Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- 2.Oliveira P.H., Touchon M., Rocha E.P. The interplay of restriction-modification systems with mobile genetic elements and their prokaryotic hosts. Nucleic Acids Res. 2014;42:10618–10631. doi: 10.1093/nar/gku734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seib K.L., Jen F.E., Tan A., Scott A.L., Kumar R., Power P.M., Chen L.T., Wu H.J., Wang A.H., Hill D.M., et al. Specificity of the ModA11, ModA12 and ModD1 epigenetic regulator N(6)-adenine DNA methyltransferases of Neisseria meningitidis. Nucleic Acids Res. 2015;43:4150–4162. doi: 10.1093/nar/gkv219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arber W., Linn S. DNA modification and restriction. Annu. Rev. Biochem. 1969;38:467–500. doi: 10.1146/annurev.bi.38.070169.002343. [DOI] [PubMed] [Google Scholar]
- 5.Bickle T., Arber W. Host-controlled restriction and modification of filamentous 1- and F-specific bacteriophages. Virology. 1969;39:605–607. doi: 10.1016/0042-6822(69)90112-3. [DOI] [PubMed] [Google Scholar]
- 6.Roberts R.J., et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–1812. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper L.P., Dryden D.T.F. The domains of a type I DNA methyltransferase: interactions and role in recognition of DNA methylation. J. Mol. Biol. 1994;236:1011–1021. doi: 10.1016/0022-2836(94)90008-6. [DOI] [PubMed] [Google Scholar]
- 8.Roberts R.J., Vincze T., Posfai J., Macelis D. REBASE–a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2015;43:D298–D299. doi: 10.1093/nar/gku1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loenen W.A., Dryden D.T., Raleigh E.A., Wilson G.G. Type I restriction enzymes and their relatives. Nucleic Acids Res. 2014;42:20–44. doi: 10.1093/nar/gkt847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flusberg B.A., Webster D.R., Lee J.H., Travers K.J., Olivares E.C., Clark T.A., Korlach J., Turner S.W. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nature Methods. 2010;7:461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark T.A., Murray I.A., Morgan R.D., Kislyuk A.O., Spittle K.E., Boitano M., Fomenkov A., Roberts R.J., Korlach J. Characterization of DNA methyltransferase specificities using single-molecule, real-time DNA sequencing. Nucleic Acids Res. 2012;40:e29. doi: 10.1093/nar/gkr1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Sullivan D., Twomey D.P., Coffey A., Hill C., Fitzgerald G.F., Ross R.P. Novel type I restriction specificities through domain shuffling of HsdS subunits in lactococcus lactis. Mol. Microbiol. 2000;36:866–875. doi: 10.1046/j.1365-2958.2000.01901.x. [DOI] [PubMed] [Google Scholar]
- 13.Gann A.A.F., Campbell A.J.B., Collins J.F., Coulson A.F.W., Murray N.E. Reassortment of DNA recognition domains and the evolution of new specificities. Mol. Microbiol. 1987;1:13–22. doi: 10.1111/j.1365-2958.1987.tb00521.x. [DOI] [PubMed] [Google Scholar]
- 14.Anton B.P., Mongodin E.F., Agrawal S., Fomenkov A., Byrd D.R., Roberts R.J., Raleigh E.A. Complete Genome Sequence of ER2796, a DNA Methyltransferase-Deficient Strain of Escherichia coli K-12. PLoS One. 2015;10:e0127446. doi: 10.1371/journal.pone.0127446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pei J., Kim B.H., Grishin N.V. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roberts G.A., Chen K., Cooper L.P., White J.H., Blakely G.W., Dryden D.T. Removal of a frameshift between the hsdM and hsdS genes of the EcoKI Type IA DNA restriction and modification system produces a new type of system and links the different families of Type I systems. Nucleic Acids Res. 2012;40:10916–10924. doi: 10.1093/nar/gks876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kennaway C.K., Obarska-Kosinska A., White J.H., Tuszynska I., Cooper L.P., Bujnicki J.M., Trinick J., Dryden D.T. The structure of M.EcoKI Type I DNA methyltransferase with a DNA mimic antirestriction protein. Nucleic Acids Res. 2009;37:762–770. doi: 10.1093/nar/gkn988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morgan R.D. Complete Genome Sequence and Methylome Analysis of Bacillus globigii ATCC 49760. Genome Announc. 2016;4 doi: 10.1128/genomeA.00427-16. doi:10.1128/genomeA.00427-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bujnicki J.M., Radlinska M. Molecular evolution of DNA-(cytosine-N4) methyltransferases: evidence for their polyphyletic origin. Nucleic Acids Res. 1999;27:4501–4509. doi: 10.1093/nar/27.22.4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeltsch A., Christ F., Fatemi M., Roth M. On the substrate specificity of DNA methyltransferases. J. Biol. Chem. 1999;274:19538–19544. doi: 10.1074/jbc.274.28.19538. [DOI] [PubMed] [Google Scholar]
- 21.Jeltsch A. The cytosine N4-methyltransferase M.PvuII also modifies adenine residues. Biol. Chem. 2001;382:707–710. doi: 10.1515/BC.2001.084. [DOI] [PubMed] [Google Scholar]
- 22.Roth M., Jeltsch A. Changing the target base specificity of the EcoRV DNA methyltransferase by rational de novo protein-design. Nucleic Acids Res. 2001;29:3137–3144. doi: 10.1093/nar/29.15.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith R.M., Josephsen J., Szczelkun M.D. The single polypeptide restriction-modification enzyme LlaGI is a self-contained molecular motor that translocates DNA loops. Nucleic Acids Res. 2009;37:7219–7230. doi: 10.1093/nar/gkp794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marshall J.J., Halford S.E. The type IIB restriction endonucleases. Biochemical Soc. Trans. 2010;38:410–416. doi: 10.1042/BST0380410. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.