Abstract
Neural crest populations along the embryonic body axis differ in developmental potential and fate, such that only cranial neural crest can contribute to craniofacial skeleton in vivo. Here, we explore the regulatory program that imbues the cranial crest with its unique features. Using axial-level specific enhancers to isolate and perform genome-wide profiling of cranial versus trunk neural crest in chick embryos, we identify and characterize regulatory relationships between a set of cranial-specific transcription factors. Introducing components of this circuit into neural crest cells of the trunk alters their identity and endows these cells with the ability to give rise to chondroblasts in vivo. Our results demonstrate that gene regulatory circuits that support formation of particular neural crest derivatives may be employed for reprogramming specific neural crest derived cell types.
Neural crest cells are characterized by multipotency and migratory ability. During embryonic development, the neural crest differentiates into multiple cell types, including chondrocytes and osteocytes, melanocytes, and neurons and glia of the peripheral nervous system (1,2). Neural crest stem cells are retained postnatally in skin and peripheral nerves, providing a potential target for replacement therapy in regenerative medicine (3–4). However, not all neural crest populations along the body axis are alike. Quail-chick grafting experiments demonstrated that the cranial and trunk neural crest differ in developmental potential: whereas the cranial neural crest forms much of the craniofacial skeleton, the trunk crest fails to contribute to skeletal lineages, even when grafted in vivo to the head (1). Approaches for engineering and replacement of specific cell types depend upon a better understanding of the molecular mechanisms that underlie establishment of specific cell types during embryonic development. Here we take advantage of differences in neural crest subpopulations (1,5–7) to identify the regulatory circuit that controls commitment of the cranial neural crest to a chondrocytic fate.
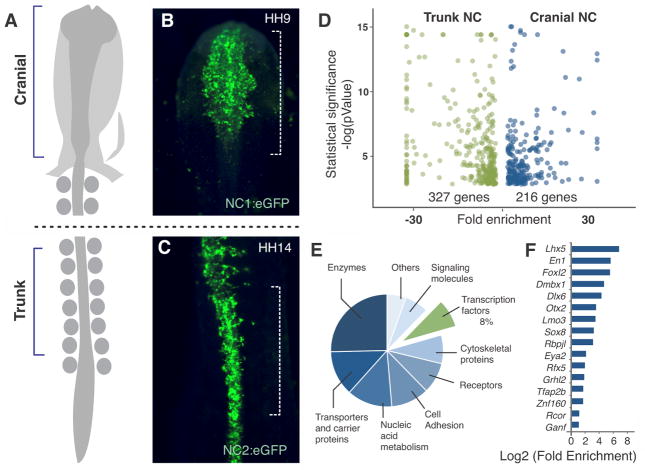
Expression of neural crest specifier genes like FoxD3 and Sox10 is controlled by enhancers specific to particular axial levels, driving onset of their transcription in either the head or trunk neural crest but not both (8,9). The enhancers are activated by different inputs, suggesting that neural crest specification is driven by distinct genetic programs in different subpopulations (2,9). In order to identify the transcriptional program that endows the cranial neural crest with its ability to give rise to ectomesenchyme (cartilage and bone of the face), we utilized the FoxD3 enhancers NC1 and NC2 (9) (Fig. 1A–C), active in the cranial and trunk neural crest, respectively, to isolate pure populations of neural crest cells for comparative transcriptional profiling. Embryos were electroporated with expression vectors containing GFP under the control of these enhancers and early-migrating cranial and trunk neural crest cells were obtained by fluorescence activated cell sorting (FACS) (10). RNA-seq analysis comparing these two populations identified 216 genes that were enriched in the cranial neural crest relative to the trunk (Fig. 1D–E, Database S1), including 16 transcription factors (listed in Fig. 1F). We confirmed the expression of these regulators in the cranial neural crest by in situ hybridization; while 6 genes were expressed throughout the cranial neural crest (Fig. S1A–F), the remainder were detected in specific subsets of cells (Fig. S1G–L). Of these, we focused on the first group which were expressed in all cranial neural crest cells, including Brain-Specific Homeobox Protein 3C (Brn3c), LIM Homeobox Protein 5 (Lhx5), Diencephalon/Mesencephalon Homeobox 1 (Dmbx1), Transcription Factor AP-2 Beta (Tfap2b), SRY Box 8 (Sox8) and the V-Ets Avian Erythroblastosis Virus E26 Oncogene Homolog 1 (Ets1).
Figure 1. Identification of cranial-specific regulators by comparative transcriptomics.
(A) Diagram depicting dorsal view of chick embryo. (B, C) Embryos electroporated with FoxD3 axial-specific enhancers NC1 and NC2, active in cranial and trunk neural crest (NC), respectively. (D) Comparative transcriptome analysis of FACS sorted cranial and trunk neural crest populations identified 216 cranially-enriched genes. (E) Summary of gene ontology analysis for the cranial-enriched genes. (F) Enrichment levels of transcription factors expressed in the cranial neural crest. HH: Hamburger and Hamilton stages.
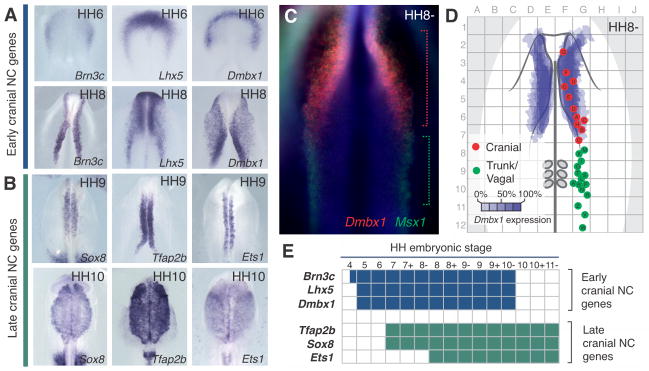
Analysis of the spatiotemporal expression of these cranial-specific regulators demonstrated that Brn3c, Lhx5 and Dmbx1 were first detected in the anterior regions of gastrula-stage embryos at Hamburger Hamilton (HH) stage 4, and persisted through stages of neural crest specification (Fig. 2A, E). These early cranial-specific genes were down-regulated after the neural crest delaminated from the neural tube. Onset of Tfap2b, Sox8 and Ets1 expression was observed later at HH7 and HH8, in neural crest progenitors residing within the cranial neural folds (Fig. 2B, E). These genes were maintained in the migratory neural crest cells during later stages of development (HH10-14; Fig. 2B, E). Co-localization of neural plate border markers Msh Homeobox 1 (Msx1) or Paired Box 7 (Pax7) with Brn3c, Lhx5 and Dmbx1 showed that the latter are expressed by an anterior subset of the neural crest progenitors (Fig. 2C). To verify that the cranial regulators mark the territory that contains cranial neural crest precursors, we analyzed the fate map of the neural plate border at the 3-somite stage using focal injections of a vital lipophilic dye (CM-DiI) to label cells along the anterior-posterior axis. The injected embryos were cultured until the 12-somite stage (HH11) when the labeled cellular progeny were scored with respect to their fate as cranial or vagal/trunk neural crest cells. The results show that the domain of expression of the early regulators (Brn3c, Lhx5 and Dmbx1) demarcates the territory that contains the progenitors of the cranial neural crest in the early neurula (HH8-) (Fig. 2D).
Figure 2. Spatial and temporal expression of cranial neural crest transcription factors (TFs).
(A, B) Dorsal views of embryos after in situ hybridization for cranial neural crest-specific TFs reveals expression at early (A) or later (B) stages. (C) Double in situ hybridization reveals that cranial regulator Dmbx1 is expressed in the anterior neural plate border, whereas Msx1 is expressed along the entire neural axis. (D) Fate map of neural crest progenitors (red and green dots) at stage HH8- confirms cranial specific expression of Dmbx1 (purple). (E) Diagram summarizing timing of expression of early and late cranial regulators.
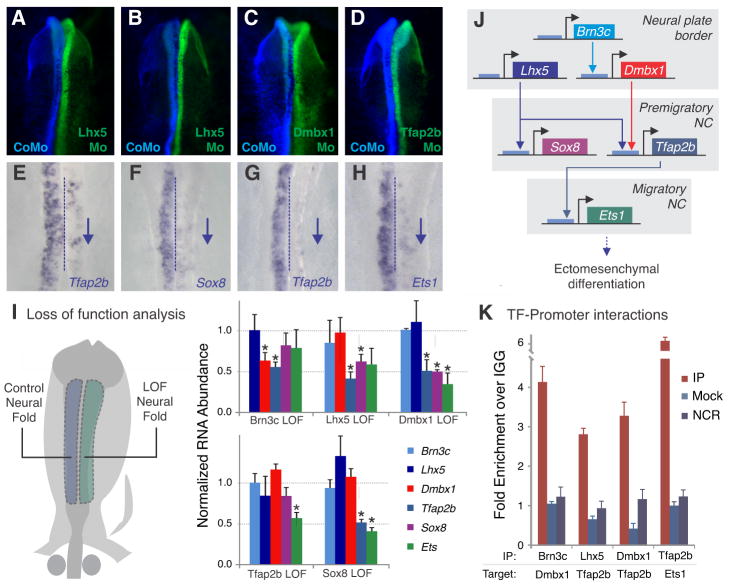
We asked whether cranial-specific regulators are part of a transcriptional circuit that underlies cranial identity by knocking down each regulator individually and assaying for changes in expression levels of the other five genes. This was done via bilateral electroporations (11), with control transfections on the left side and function-blocking morpholinos or dominant negative constructs on the right side of the same embryo (Fig. 3A–D). Transfected embryos were analyzed by in situ hybridization (Fig. 3A–H) and qPCR (Fig. 3I) for effects on putative targets; loss of the target gene in the experimental side of the embryo indicated the existence of a regulatory link between the two transcription factors. The diagram on Figure 3J contains interactions confirmed by both qPCR and in situ hybridization. Morpholino knockdown efficiency was validated by in vivo translation-blocking assays (Fig. S2).
Figure 3. Cranial-specific transcriptional circuit underlying neural crest axial identity.
(A–D) Whole mount dorsal views of embryos after morpholino (Mo) targeted to indicated transcription factor (TF) was transfected to the right side (green) and control morpholino (CoMo) to the left side (blue) of each embryo. (E–H) Same embryos as above after in situ hybridization for indicated downstream TF; blue arrows indicate transcript downregulation. (I) Comparing control to loss of function (LOF) neural folds by qPCR reveals differential regulation of downstream targets with significant changes indicated by * (Student’s t-test, P<0.05). (J) Diagram summarizing cranial specific gene regulatory circuit delineated by functional assays. (K) Chromatin immunoprecipitation (ChIP) demonstrates direct association of cranial specific TFs with promoters of their downstream targets. No enrichment was observed for intergenic negative control regions (NCR), or when the procedure was performed with mock-transfected embryos (see Materials and Methods for more information). Error bars on (I) and (K) represent standard deviation.
By testing 25 putative regulatory links, we found that the early and late cranial-specific genes constitute different hierarchical levels of a gene regulatory network (Fig. 3J). Brn3c, which is placed at the top of the circuit, is necessary for the activation of Dmbx1 in the anterior neural plate border (Fig. 3I; Fig. S3). Subsequently, Lhx5 and Dmbx1 drive expression of Tfap2b and Sox8 in the dorsal neural folds (Fig. 3A–C, E–G, I). Finally, Tfap2b activates expression of Ets1 as the neural crest becomes specified (Fig 3D, HI). Chromatin immunoprecipitation (ChIP) experiments performed in microdissected neural crest cells showed association of the cranial-specific transcription factors with promoters of predicted downstream target genes, suggesting that these regulatory links are direct (Fig. 3K; Fig. S4).
Sox9, Tfap2b and Ets1 are all retained in the migrating cranial crest cells as they move ventrally to give rise to the facial mesenchyme during stages HH10-14. To investigate if these genes play a role in the differentiation of neural crest cells into chondroblasts, we assayed the effects of disrupting the terminal module of the circuit on the expression of markers of chondrocytic differentiation. We found that Ets1 was required for the expression of ALX Homeobox 1 (Alx1, also known as Cartilage Paired-Class Homeoprotein 1) in the facial mesenchyme (Fig. S5), indicating a link between cranial identity and chondrocytic differentiation (Fig. S5). Thus, cranial-specific regulators are part of a transcriptional circuit that conveys regulatory information from the anterior neural plate border to the late migratory neural crest.
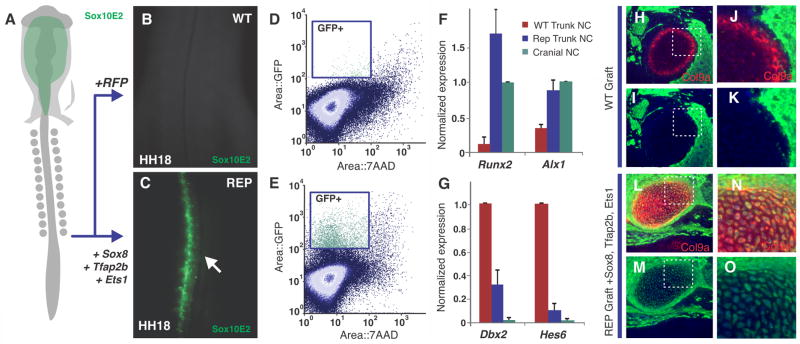
To test if this cranial-specific regulatory circuit could be used to manipulate neural crest identity, we utilized neural crest axial-specific enhancers as reporters of axial level identity. We electroporated expression constructs in the trunk neural tube of stage HH10 embryos, and found that transfection of the late cranial-specific factors (Sox8, Tfap2b and Ets1) robustly activated the cranial enhancer Sox10E2 (9, 11) in the trunk neural crest (n=15/15) (Fig. 4A–E), consistent with a shift from trunk to cranial identity. Other axial-specific enhancers were similarly affected; trunk specific enhancers NC2 and Sox10E1 were repressed after electroporation of the late factors (Fig. S6). Early cranial-specific factors (Brn3c, Lhx5 and Dmbx1), or individual late factors, were unable to activate the cranial enhancer in the trunk. To identify changes in the regulatory state of the reprogrammed trunk neural crest, we isolated transfected cells by FACS, and analyzed their expression profile by qPCR, focusing on transcription factors involved in craniofacial differentiation. The results revealed elevated expression of chondrocytic genes Runt-Related Transcription Factor 2 (Runx2) and Alx1, in the trunk Sox10E2+ cells compared with native trunk crest (Fig. 4F). The reprogrammed trunk Sox10E2+ cells also displayed loss of genes enriched in the trunk neural crest like Developing Brain Homeobox 1 (Dbx2) and Hairy And Enhancer Of Split 6 (Hes6) (Fig. 4G). This confirmed that the reprogrammed cells adopt a cranial-like expression profile, and raised the possibility that these cells might display augmented chondrocytic potential.
Figure 4. Reprogramming of neural crest axial identity and fate.
(A) Diagram of chick embryo electroporated with the Sox10E2 enhancer, expressed only in migrating cranial (Cr) neural crest (NC). (B) Transfection of trunk neural crest cells with control RFP expression vector shows no Sox10E2 expression. (C) Reprogramming of trunk neural crest cells with cranial specific regulators Sox8, Tfap2b and Ets1 results in robust expression of the cranial Sox10E2 enhancer in the trunk region (n=15/15). (D, E) Flow cytometry analysis of dissociated embryonic trunks shows a large number of Sox10E2+ trunk neural crest cells after reprogramming. (F, G) Reprogrammed (Rep) trunk neural crest display increased expression of the chondrocytic genes Runx2 and Alx1, while trunk genes Dbx2 and Hes6 are strongly downregulated. Error bars represent standard deviation. (H, I) Histological sections of E7 embryonic heads show that wild type (WT) trunk neural crest cells (GFP+; green) fail to form cartilage (Col9a+; red) after transplantation to the cranial region (n=0/5). (J, K) Inset of (H–I), showing absence of GFP+ chondrocytes. (L, M) Reprogrammed trunk neural crest cells (expressing Sox8, Tfap2b and Ets1) from GFP donor embryos transplanted to the head form ectopic cartilage nodules. (N, O) Inset of (L–M), showing chondrocytes derived from trunk neural crest cells (GFP+ and Col9a+).
Finally, we tested whether this cranial neural crest circuit could reprogram not only enhancer activity and expression of axial-specific neural crest genes, but also cell fate such that reprogrammed trunk neural crest cells could differentiate into craniofacial cartilage. We co-transfected three constructs encoding late transcription factors Sox8, Tfap2b and Ets1 into the posterior epiblast of HH5 chicken embryos transgenic for GFP by electroporating DNA in the region posterior to the Hensen’s node. The GFP+ trunk neural folds then were microdissected at HH11 and immediately transplanted to the cranial regions of HH9 wild-type chick embryos (Fig. S7). The grafted embryos were incubated until host embryonic day (E)7, by which time endogenous cartilage cells have differentiated. The fate of donor tissue was assayed using markers for neuronal, melanocytic and chondrocytic differentiation. By E7, wild type (host) and reprogrammed (donor) trunk neural crest migrated to the proximal part of the jaw. As observed with chick-quail chimeras (5–7), we found that wild type or mock-transfected trunk neural crest cells grafted into the cephalic region gave rise to neurons and melanocytes, but were unable to differentiate into chondroblasts (n=0/5) (Fig. 4H–K). The same was observed with trunk neural crest transfected with the early cranial-specific factors (n=0/6). Reprogrammed trunk neural crest, however, acquired chondrogenic potential and formed ectopic cartilage nodules (n=4/7) (Fig. 4L–O) in the proximal jaw. Thus, introducing components of the cranial-specific transcriptional circuit is sufficient to reprogram trunk neural crest and to drive them to adopt an additional cartilaginous fate. These results definitively show that the cranial-specific regulatory circuit (Fig. 3J) we have defined confers chondrocytic potential to the trunk neural crest.
The development and differentiation of neural crest cells is controlled by a complex gene regulatory network, composed of transcription factors, signaling molecules and epigenetic modifiers (12–13). Here, we have expanded the cranial neural crest gene regulatory network by identifying transcriptional interactions specific to the cranial crest and absent from other subpopulations. By linking anterior identity in the gastrula to the expression of drivers of chondrocytic differentiation, we have identified a cranial-specific circuit (Fig. 3J) that endows the neural crest with its unique potential to differentiate into the craniofacial skeleton of vertebrates. Our results highlight how transcriptional circuits can be rewired to alter progenitor cell identity and fate during embryonic development.
Supplementary Material
Acknowledgments
We thank Joanna Tan-Cabugao, Michael Stone, Brian Jun and Daniel S. E. Koo for technical assistance. The Caltech Millard and Muriel Jacobs Genetics and Genomics Laboratory provided sequencing and bioinformatics support. We are indebted to Diana Perez, Keith Beadle, and Rochelle Diamond for cell-sorting assistance at the Caltech Flow Cytometry Cell Sorting Facility. We also thank Meyer Barembaum for the Sox8 and Tfap2b expression constructs. This work was supported by NIH grants DE024157 and HD037105 to MEB. MS-C was funded by a fellowship from the Pew Fellows Program in Biomedical Sciences and by NIH grant K99DE024232. Supplement contains additional data.
Footnotes
Materials and Methods
References and Notes
- 1.Le Douarin N. The neural crest. Cambridge University Press; 1982. [Google Scholar]
- 2.Simoes-Costa M, Bronner ME. Insights into neural crest development and evolution from genomic analysis. Genome research. 2013;23:1069–1080. doi: 10.1101/gr.157586.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uesaka T, Nagashimada M, Enomoto H. Neuronal Differentiation in Schwann Cell Lineage Underlies Postnatal Neurogenesis in the Enteric Nervous System. J Neurosci. 2015;35:9879–9888. doi: 10.1523/JNEUROSCI.1239-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dyachuk V, et al. Parasympathetic neurons originate from nerve associated peripheral glial progenitors. Science. 2014;345:82–87. doi: 10.1126/science.1253281. [DOI] [PubMed] [Google Scholar]
- 5.Le Lievre CS, Le Douarin NM. Mesenchymal derivatives of the neural crest: analysis of chimaeric quail and chick embryos. Journal of embryology and experimental morphology. 1975;34:125–154. [PubMed] [Google Scholar]
- 6.Le Lievre CS, Schweizer GG, Ziller CM, Le Douarin NM. Restrictions of developmental capabilities in neural crest cell derivatives as tested by in vivo transplantation experiments. Developmental biology. 1980;77:362–378. doi: 10.1016/0012-1606(80)90481-9. [DOI] [PubMed] [Google Scholar]
- 7.Lwigale PY, Conrad GW, Bronner-Fraser M. Graded potential of neural crest to form cornea, sensory neurons and cartilage along the rostrocaudal axis. Development. 2004;131:1979–1991. doi: 10.1242/dev.01106. [DOI] [PubMed] [Google Scholar]
- 8.Betancur P, Bronner-Fraser M, Sauka-Spengler T. Genomic code for Sox10 activation reveals a key regulatory enhancer for cranial neural crest. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3570–3575. doi: 10.1073/pnas.0906596107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simoes-Costa MS, McKeown SJ, Tan-Cabugao J, Sauka-Spengler T, Bronner ME. Dynamic and differential regulation of stem cell factor FoxD3 in the neural crest is Encrypted in the genome. PLoS genetics. 2012;8:e1003142. doi: 10.1371/journal.pgen.1003142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simoes-Costa M, Tan-Cabugao J, Antoshechkin I, Sauka-Spengler T, Bronner ME. Transcriptome analysis reveals novel players in the cranial neural crest gene regulatory network. Genome research. 2014;24:281–290. doi: 10.1101/gr.161182.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simoes-Costa M, Stone M, Bronner ME. Axud1 Integrates Wnt Signaling and Transcriptional Inputs to Drive Neural Crest Formation. Developmental cell. 2015;35(5):644–554. doi: 10.1016/j.devcel.2015.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simoes-Costa M, Bronner ME. Establishing neural crest identity: a gene regulatory recipe. Development. 2015;142:242–257. doi: 10.1242/dev.105445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green SA, Simoes-Costa M, Bronner ME. Evolution of vertebrates as viewed from the crest. Nature. 2015;520:474–482. doi: 10.1038/nature14436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sauka-Spengler T, Barembaum M. Gain- and loss-of-function approaches in the chick embryo. Methods in cell biology. 2008;87:237–256. doi: 10.1016/S0091-679X(08)00212-4. [DOI] [PubMed] [Google Scholar]
- 15.Chapman SC, Collignon J, Schoenwolf GC, Lumsden A. Improved method for chick whole-embryo culture using a filter paper carrier. Developmental dynamics. 2001;220:284–289. doi: 10.1002/1097-0177(20010301)220:3<284::AID-DVDY1102>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 16.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28:511–U174. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Theveneau E, Duband JL, Altabef M. Ets-1 confers cranial features on neural crest delamination. PloS one. 2007;2:e1142. doi: 10.1371/journal.pone.0001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilkinson DG. In situ hybridization : a practical approach. IRL Press at Oxford University Press; 1992. [Google Scholar]
- 20.Denkers N, Garcia-Villalba P, Rodesch CK, Nielson KR, Mauch TJ. FISHing for chick genes: Triple-label whole-mount fluorescence in situ hybridization detects simultaneous and overlapping gene expression in avian embryos. Developmental dynamics. 2004;229:651–657. doi: 10.1002/dvdy.20005. [DOI] [PubMed] [Google Scholar]
- 21.Ezin AM, Fraser SE, Bronner-Fraser M. Fate map and morphogenesis of presumptive neural crest and dorsal neural tube. Developmental biology. 2009;330:221–236. doi: 10.1016/j.ydbio.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim DS, Matsuda T, Cepko CL. A core paired-type and POU homeodomain containing transcription factor program drives retinal bipolar cell gene expression. Journal of Neuroscience. 2008;28:7748–7764. doi: 10.1523/JNEUROSCI.0397-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim J, Cantor AB, Orkin SH, Wang J. Use of in vivo biotinylation to study protein–protein and protein–DNA interactions in mouse embryonic stem cells. Nature Protocols. 2009;4:506–517. doi: 10.1038/nprot.2009.23. [DOI] [PubMed] [Google Scholar]
- 24.Kolodziej KE, Pourfarzad F, de Boer E, Krpic S, Grosveld F, Strouboulis J. Optimal use of tandem biotin and V5 tags in ChIP assay. BMC Molecular Biology. 2009;10:6. doi: 10.1186/1471-2199-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakamura H, Funahashi J. Introduction of DNA into chick embryos by in ovo electroporation. Methods. 2001;24:43–48. doi: 10.1006/meth.2001.1155. [DOI] [PubMed] [Google Scholar]
- 26.Barembaum M, Bronner ME. Identification and dissection of a key enhancer mediating cranial neural crest specific expression of transcription factor, Ets-1. Developmental Biology. 2013;382:567–575. doi: 10.1016/j.ydbio.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.