HvNramp5 is a plasma membrane-localized transporter, which is responsible for Mn and Cd uptake in barley.
Abstract
The Natural Resistance Associated Macrophage Protein (Nramp) represents a transporter family for metal ions in all organisms. Here, we functionally characterized a member of Nramp family in barley (Hordeum vulgare), HvNramp5. This member showed different expression patterns, transport substrate specificity, and cellular localization from its close homolog in rice (Oryza sativa), OsNramp5, although HvNramp5 was also localized to the plasma membrane. HvNramp5 was mainly expressed in the roots and its expression was not affected by Cd and deficiency of Zn, Cu, and Mn, but slightly up-regulated by Fe deficiency. Spatial expression analysis showed that the expression of HvNramp5 was higher in the root tips than that in the basal root regions. Furthermore, analysis with laser microdissection revealed higher expression of HvNramp5 in the outer root cell layers. HvNramp5 showed transport activity for both Mn2+ and Cd2+, but not for Fe2+ when expressed in yeast. Immunostaining with a HvNramp5 antibody showed that this protein was localized in the root epidermal cells without polarity. Knockdown of HvNramp5 in barley resulted in a significant reduction in the seedling growth at low Mn supply, but this reduction was rescued at high Mn supply. The concentration of Mn and Cd, but not other metals including Cu, Zn, and Fe, was decreased in both the roots and shoots of knockdown lines compared with the wild-type barley. These results indicate that HvNramp5 is a transporter required for uptake of Mn and Cd, but not for Fe, and that barley has a distinct uptake system from rice.
Transport of mineral elements from soil to different organs and tissues of plants requires different types of transporters (Hall and Williams, 2003; Nevo and Nelson, 2006; Yokosho et al., 2009; Olsen and Palmgren, 2014; Sasaki et al., 2016), which include the zinc-regulated transporters, iron-regulated transporter-like protein family; the natural resistance-associated macrophage protein (Nramp) family of transporters; the multidrug and toxic compound extrusion protein transporters; the heavy metal ATPase transporters; the oligopeptide transporters family; the ATP-binding cassette family of transporters; and the cation-diffusion facilitator family of transporters. Among them, Nramp represents a transporter family for metal ion in all organisms including bacteria, animals, and plants (Curie et al., 2000; Nevo and Nelson, 2006). Some members of this family in plants have been functionally characterized, especially in model plants such as Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa). In Arabidopsis, there are six members of Nramp transporter proteins. AtNramp1 is localized to the plasma membrane of root cells and functions as a high-affinity transporter for Mn uptake (Cailliatte et al., 2010). Both AtNramp3 and AtNramp4 are localized to the tonoplast and play redundant roles in Fe, exporting from the vacuole during seed germination and in Mn homeostasis at the adult stage (Thomine et al., 2000; Lanquar et al., 2005, 2010). AtNramp6 is targeted to a vesicular-shaped endomembrane compartment and is implicated in the distribution or availability of Cd within cells (Cailliatte et al., 2009). However, the function of AtNramp2 and AtNramp5 has not been characterized.
On the other hand, there are seven members of Nramp transporter family in the rice genome, of which four have been functionally characterized. They all are localized to the plasma membrane but show different roles. OsNramp1 shows transport activity for Fe and Cd in yeast and is proposed to be involved in Cd accumulation (Takahashi et al., 2011). OsNramp3 is localized at the vascular tissues of nodes and plays an important role in distribution of Mn, but not Fe and Cd (Yamaji et al., 2013). On the other hand, OsNrat1 (OsNramp4) transports trivalent Al ion (Xia et al., 2010) and is required for high Al tolerance in rice roots. Finally, OsNramp5 functions as a major transporter responsible for root Mn and Cd uptake (Ishimaru et al., 2012; Ishikawa et al., 2012; Sasaki et al., 2012). However, the function of OsNramp2, OsNramp6, and OsNramp7 is unknown.
In addition to Nramp members characterized in rice and Arabidopsis, some members in other plant species have also been characterized. For example, a soybean (Glycine max) Nramp transporter, GmDMT1 is implicated in the ferrous iron transport (Kaiser et al., 2003). Nramp1 isolated from Noccaea caerulescens, a Zn/Cd hyperaccumulator, is involved in the influx of Cd across the endodermal plasma membrane and plays a key role in Cd flux into the stele and root-to-shoot Cd transport (Milner et al., 2014). In Malus baccata, Nramp1 is capable of mediating the distribution of ions as well as transport of Fe, Mn, and Cd (Xiao et al., 2008). Besides, Nramp1 and Nramp3 in tomato (Solanum lycopersicum) have also been suggested to be involved in Mn transport (Bereczky et al., 2003). When NcNramp3 and NcNramp4 from Noccaea caerulescens were expressed in yeast, NcNramp3 transported Fe, Mn, and Cd, while NcNramp4 also transported Zn in addition to Fe, Mn, and Cd (Oomen et al., 2009). However, Nramp4 isolated from Thlaspi japonicum, a Ni hyperaccumulator, showed transport activity for Ni but not for Zn, Cd, or Mn in yeast (Mizuno et al., 2005). These findings indicate that Nramp members have a diverse role in metal transport in plants.
Barley (Hordeum vulgare) is the fourth most important cereal crop in the world; however, less progress has been made in understating of molecular mechanisms on mineral element transport in barley due to its large genome size. For example, no Nramp members in barley have been functionally characterized so far. In this study, we first isolated barley Nramp member, HvNramp5, which is a close homolog of rice OsNramp5. Detailed functional analysis revealed that HvNramp5 is involved in the uptake of both Mn and Cd, but not of Fe in barley roots. Furthermore, we found that different from OsNramp5, HvNramp5 showed a distinct pattern in the gene expression, cellular localization, and transport substrate.
RESULTS
Isolation and Phylogenetic Analysis of HvNramp5
The full-length cDNA of HvNramp5 in barley cultivar Golden Promise was amplified by PCR using primers designed based on database information (AK364374; http://www.ncbi.nlm.nih.gov/nuccore/AK364374). Blast search revealed that one copy of HvNramp5 is present in the barley genome (http://plants.ensembl.org/Hordeum_vulgare/Info/Index). Sequence analysis showed that the full-length cDNA of HvNramp5 contains 1638 bp, which encodes a peptide with 545 amino acids (Supplemental Fig. S1A). Comparison of the cDNA sequence with a genomic DNA sequence obtained from the released barley genome (Mayer et al., 2012) revealed that HvNramp5 comprises 11 introns and 12 extrons (Supplemental Fig. S1B). HvNramp5 showed 39% to 84% identity with rice Nramp members (Supplemental Fig. S1C) and the closest one is OsNramp5 (84%). HvNramp5 is predicted as a membrane protein with 11 transmembrane domains (Supplemental Fig. S1D).
Expression Patterns of HvNramp5
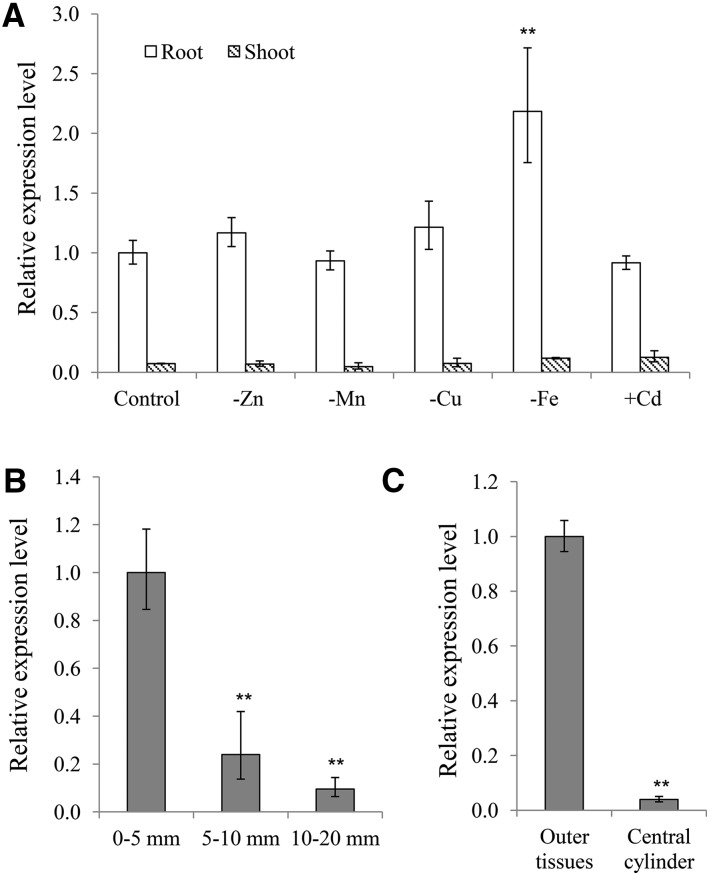
Expression analysis with quantitative RT-PCR showed that HvNramp5 was mainly expressed in the roots (Fig. 1A). The expression in the roots was not affected by deficiency of Mn, Cu, and Zn, but slightly up-regulated by Fe deficiency. The expression was also not affected by Cd exposure (Fig. 1A). Spatial expression analysis showed that the expression of HvNramp5 was markedly higher in the root tips (0 mm to 5 mm) than in the root elongation zone (5 mm to 10 mm) and root hair zone (10 mm to 20 mm; Fig. 1B). Furthermore, we investigated tissue-specificity of expression of HvNramp5 using the sample prepared before with help of laser microdissection in the root tips (2 mm; Fujii et al., 2012). The result showed that HvNramp5 was highly expressed in the outer root cell layer compared with the central cylinder tissues (Fig. 1C).
Figure 1.
Expression pattern of HvNramp5. A, Expression of HvNramp5 in response to metal deficiency or Cd. The seedlings of barley cultivar Golden Promise were cultivated in a one-fifth Hoagland’s solution with (control) or without Zn, Mn, Cu, or Fe or with 0.5 µm Cd. After 7-d treatments, the roots and shoots were sampled for RNA extraction. B, Spatial expression of HvNramp5 along the roots of Golden Promise. RNA was extracted from the root tip (0–5 mm), root elongation zone (5–10 mm), or root hair zone (10–20 mm) from 7-d-old barley roots. C, Tissue-specificity of expression of HvNramp5. Different tissues were sampled from the roots (2 mm from root tip) of barley (cv Morex) using a Veritas Laser Microdissection System as described by Fujii et al. (2012). The expression level was determined by quantitative real-time RT-PCR and expression relative to the control root (A), root tip (B), and outer tissues (C) is shown. Data are means ± sd of three biological replicates and asterisks indicate significant difference at **P < 0.01 by Student’s t test.
HvNramp5 Transports Mn and Cd, But Not Fe, in Yeast
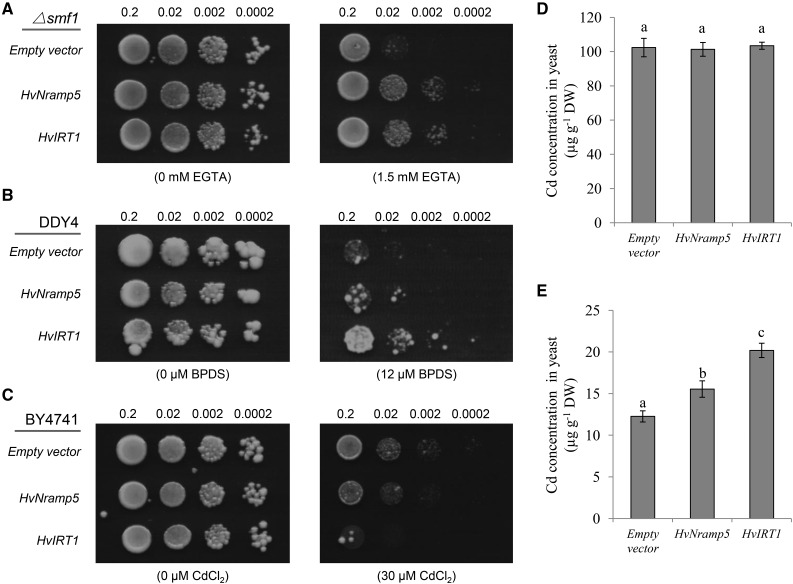
To examine transport activity of HvNramp5 for metals, we expressed an HvNramp5, HvIRT1, or pYES2 empty vector in different yeast strains, respectively. HvIRT1 as a positive control is known to transport Mn2+, Fe2+, and Cd2+ in yeast (Padas et al., 2008). In a yeast mutant defective in Mn uptake, Δsmf1, expression of HvNramp5 and HvIRT1 rescued the yeast growth under Mn-limited condition induced by Mn-chelator ethylene glycol tetraacetic acid (EGTA), but expression of the vector alone did not rescue the growth (Fig. 2A). By contrast, expression of HvNramp5 and the vector control was not able to complement the growth in a yeast mutant DDY4 (Δfet3fet4) defective in Fe uptake, whereas expression of HvIRT1 rescued the yeast growth, under Fe(II)-limited condition generated by Fe(II)-chelator4,7-biphenyl-1,10-phenanthroline-disuiphonic acid (BPDS; Fig. 2B). On the other hand, the growth of yeast strain BY4741 (a wild-type strain) expressing HvNramp5 and HvIRT1 was seriously inhibited by the presence of Cd compared with that of the vector control (Fig. 2C).
Figure 2.
Metal transport activity assay of HvNramp5 in yeast. A, Functional complementation for Mn uptake in the strain ∆smf1. B, Functional complementation for Fe uptake in the strain DDY4 (∆fet3fet4). C, Sensitivity test to Cd toxicity in the strain BY4741. Different yeast strains carrying HvNramp5, HvIRT1, or empty vector (pYES2) were cultured on the plate with or without Mn-chelator EGTA (A), Fe-chelator BPDS (B), or Cd (C). Eight microliters of cell suspension with an OD600nm of 0.2 and three serial 1:10 dilutions were spotted and incubated at 30°C for 3 to 5 d. D to E, Cd uptake in liquid medium. Yeast BY4741 carrying HvNramp5, HvIRT1, or empty vector (pYES2) was cultured in a liquid medium containing 2% Glc (D) or Gal (E) in the presence of 5 µm CdCl2 for 4 h. The Cd concentration in the yeast was determined with ICP-MS after digestion. Data are means ± sd of three biological replicates and a different small letter indicates significant difference at P < 0.05 by Tukey’s test.
The Cd accumulation in the yeast cells was also compared under the control of Gal-inducible promoter using liquid culture. The Cd accumulation in the yeast did not differ among yeast expressing HvNramp5, HvIRT1, and an empty vector in the presence of Glc (Fig. 2D), when the gene was not induced. By contrast, in the presence of Gal, yeast expressing HvNramp5 and IRT1 significantly accumulated higher Cd compared with the vector control (Fig. 2E). Furthermore, yeast expressing HvIRT1 accumulated higher Cd than that expressing HvNramp5. This result is consistent with the growth test (Fig. 2C); the growth was inhibited more in the yeast expressing HvIRT1 than that expressing HvNramp5. All these results indicate that HvNramp5 is able to transport Mn2+ and Cd2+, but not Fe2+ in yeast. Yeast cultured with Glc showed higher Cd accumulation than that cultivated with Gal (Fig. 2, D and E). This phenomenon was also reported previously by Engl and Kunz (1995), but the exact reason is unknown. Different carbon sources may affect Cd accumulation through affecting yeast growth, cell wall components, and energy metabolism.
Cellular and Subcellular Localization of HvNramp5
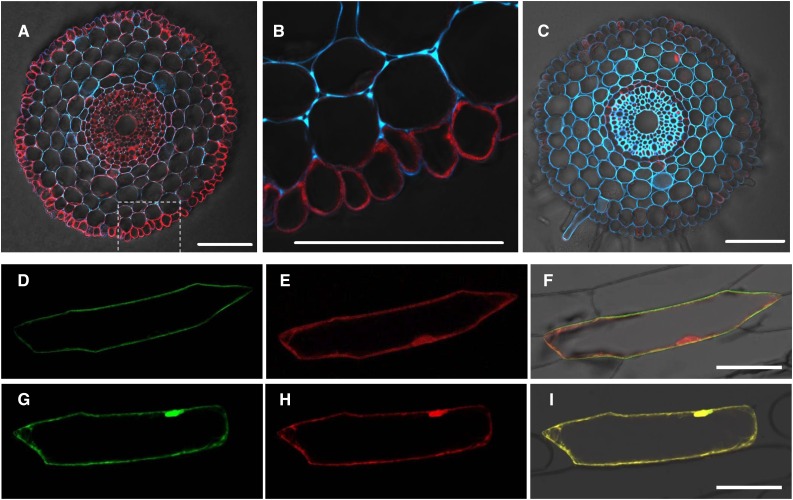
To determine the tissue and cellular localization of HvNramp5 in barley root, we conducted an immunostaining using an antibody against HvNramp5. Consistent with the expression patterns of HvNramp5 (Fig. 1B), the signal (red) was much stronger in the root tips than in the basal root region (Fig. 3, A and C). Furthermore, HvNramp5 was mainly localized at the epidermal cells of the root tips (Fig. 3A) although a weak signal was also detected in the central cylinder.
Figure 3.
Cellular and subcellular localization of HvNramp5. A to C, Cellular localization of HvNramp5 in barley roots. Immunostaining with HvNramp5 antibody was performed in root tips (A and B, 2 mm from root tips) and basal roots (C, 15 mm from root tips) of barley (cv Golden Promise). B, High-magnification image of dotted part in A. Red color shows signal from the antibody, while blue color shows autofluorescence of cell wall. Bars = 100 μm. D to I, Subcellular localization of HvNramp5. Confocal images of onion epidermis cells coexpressing HvNramp5-GFP and RFP observed with the GFP (D), RFP (E), and their merged (F) channels, and coexpressing GFP alone and RFP observed with the GFP (G), RFP (H), and their merged (I) channels. Bars = 100 μm.
Immunostaining analysis also showed that HvNramp5 is likely localized to the plasma membrane (Fig. 3B). To confirm this, we transiently expressed HvNramp5 fused with GFP or GFP alone, together with RFP (red fluorescence protein) as a cytosolic and nuclear marker in onion epidermal cells. The green fluorescence signal from GFP was observed at the plasma membrane in cells expressing HvNramp5-GFP fusion, while the red signal from RFP was localized in the cytosol and the nucleus (Fig. 3, D to F). By contrast, the signal of both green and red fluorescence was localized in the cytosol and the nucleus in the cells expressing GFP alone (Fig. 3, G–I). These results indicate that HvNramp5 is a plasma membrane-localized transporter.
Phenotypic Analysis of HvNramp5 Knockdown Lines
To understand the role of HvNramp5 in barley, we generated two independent knockdown lines of HvNramp5 using RNA interference (RNAi) methods. The expression level of HvNramp5 in two RNAi lines was 26.1% and 37.5% of the wild-type barley (Supplemental Fig. S2A). By contrast, the expression of HvIRT1 was not affected in the knockdown lines (Supplemental Fig. S2B).
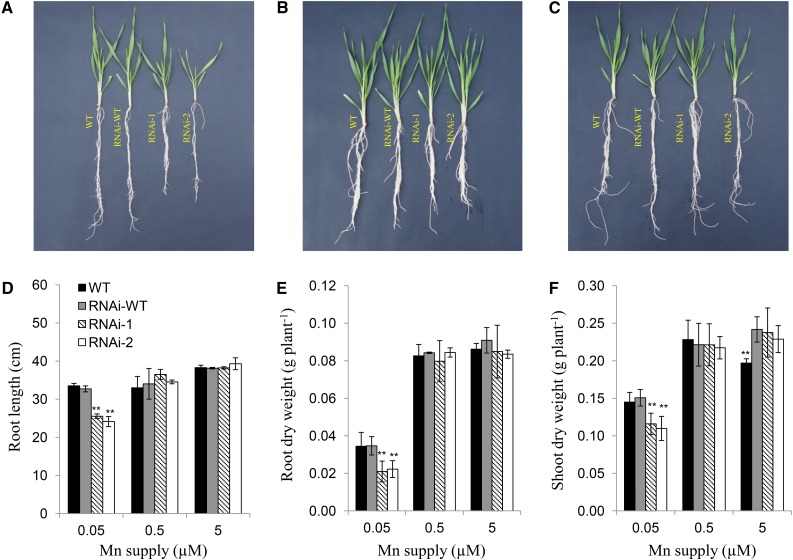
The growth was compared between the RNAi lines and the wild types (a nontransgenic line and a line showing wild-type genotype isolated from T2 population) when grown in a nutrient solution containing different Mn, Cd, and Fe concentrations. The root length and dry weight of the roots and shoots of the RNAi lines were significantly inhibited at low Mn (0.05 μm) compared with that of the wild types (Fig. 4, A and D). However, at higher Mn concentrations (0.5 μm and 5 μm), the difference in the growth was not observed (Fig. 4, B, C, E, and F). By contrast, the growth difference of the RNAi lines and wild types was not found between low and high concentrations of Cd and Fe (Supplemental Figs. S4 and S5).
Figure 4.
Effect of knockdown of HvNramp5 on plant growth at different Mn concentrations. A to C, Growth of wild-type barley (cv Golden Promise), the homozygous wild type (RNAi-wild type), and two homozygous transgenic RNAi lines (RNAi-1, RNAi-2) derived from two independent events. All lines were cultivated in a nutrient solution containing 0.05 (A), 0.5 (B), and 5 µm (C) Mn in the presence of 0.1 µm Cd for 14 d. D to F, Root length (D) and dry weight of roots (E) and shoots (F) in these lines were recorded at harvest. Data are means ± sd of three biological replicates and ** indicates significant difference at P < 0.01 by Tukey’s test.
Metal Analysis of HvNramp5 Knockdown Lines
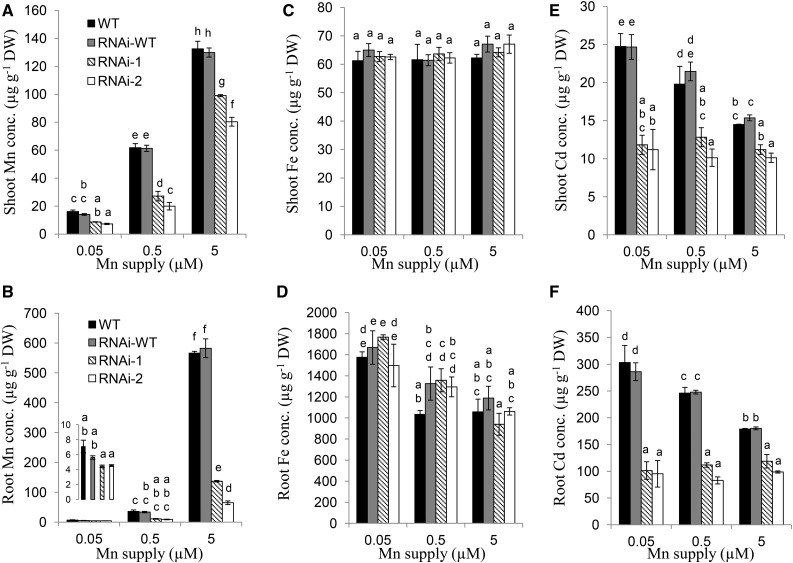
Analysis of metal concentration in the roots and shoots showed that the Mn concentration increased with increasing Mn concentrations in the nutrient solution in both RNAi lines and wild types (Fig. 5, A and B). However, the RNAi lines always showed a lower Mn concentration in both the roots and shoots than did the wild types. By contrast, there was no difference in the concentration of Fe, Cu, and Zn in both the roots and shoots between the RNAi lines and wild types (Fig. 5, C and D, and Supplemental Fig. S3). Interestingly, the Cd concentration in both the roots and shoots was decreased with increasing external Mn concentrations in the wild types, but not in the RNAi lines (Fig. 5, E and F). However, the RNAi lines showed lower Cd concentration of both the roots and shoots than did the wild types at either of the Mn concentrations (Fig. 5, E and F).
Figure 5.
Metal concentration in HvNramp5 RNAi lines at different Mn concentrations. Wild-type barley (cv Golden Promise), the homozygous wild type (RNAi-wild type), and two homozygous transgenic RNAi lines (RNAi-1, RNAi-2) derived from two independent events were cultivated in a nutrient solution containing 0.05, 0.5, and 5 µm Mn in the presence of 0.1 µm Cd for 14 d. The concentration of Mn (A and B), Fe (C and D), and Cd (E and F) in the shoots (A, C, and E) and roots (B, D, and F) was determined by ICP-MS. Data are means ± sd of three biological replicates and a different small letter indicates significant difference at P < 0.05 by Tukey’s test.
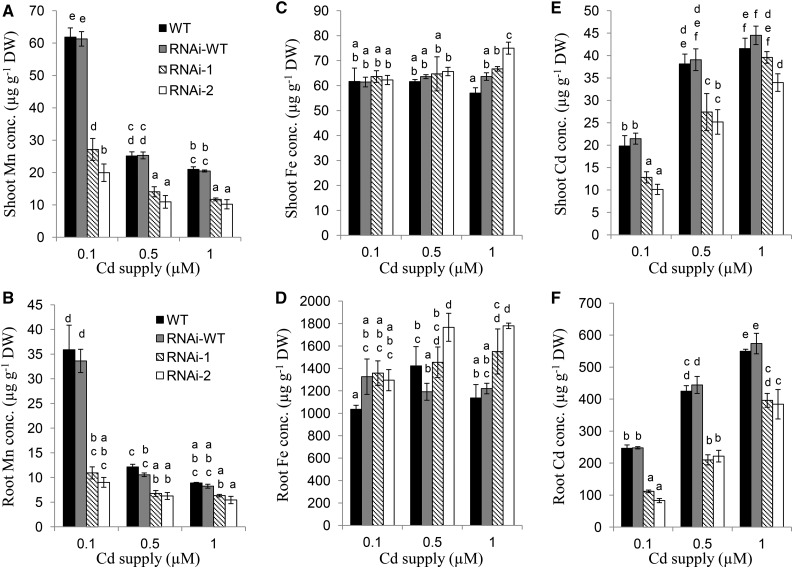
When the seedlings of the RNAi lines and the wild types were exposed to different Cd concentrations (0.1, 0.5, and 1 μm), the concentration of Mn in the roots and shoots was decreased with increasing Cd concentrations in the external solution in both the RNAi lines and the wild types (Fig. 6, A and B). However, compared with the wild type, the Mn concentration in the RNAi lines was lower at either of the Cd concentrations (Fig. 6, A and B). There was almost no difference in the concentration of Cu and Zn in both the roots and shoots between the knockdown lines and wild types (Supplemental Fig. S4), but Fe concentration in the roots and shoots was slightly higher in RNAi lines than in wild types at 1 μm Cd (Fig. 6, C and D). The reason for this increased Fe accumulation is unknown. By contrast, the concentration of Cd in the roots and the shoots was lower in knockdown lines than in wild types, although the Cd concentration of the roots and shoots increased with increasing external Cd concentrations in both the RNAi lines and wild types (Fig. 6, E and F). When the seedlings of the RNAi lines and the wild types were exposed to different Fe concentrations (0.1, 2, and 10 μm), the Fe concentration in the roots and shoots did not differ between different lines although the Fe concentration in all lines increased with increasing Fe concentrations in the external solution (Supplemental Fig. S5). These results indicate that knockdown of HvNramp5 expression caused decreased accumulation of Mn and Cd, but not Fe—and that Mn and Cd competed with each other in the uptake.
Figure 6.
Metal concentration in HvNramp5 RNAi lines at different Cd concentrations. Wild-type barley (cv Golden Promise), the homozygous wild type (RNAi-wild type), and two homozygous transgenic RNAi lines (RNAi-1, RNAi-2) derived from two independent events were cultivated in a nutrient solution containing 0.1, 0.5, or 1 µm Cd for 14 d. The concentration of Mn (A and B), Fe (C and D), and Cd (E and F) in the shoots (A, C, and E) and roots (B, D, and F) was determined by ICP-MS. Data are means ± sd of three biological replicates and a different small letter indicates significant difference at P < 0.05 by Tukey’s test.
Comparison of Mn Accumulation and Nramp5 Expression between Rice and Barley
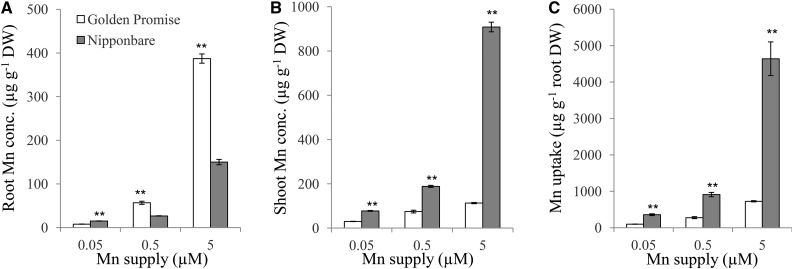
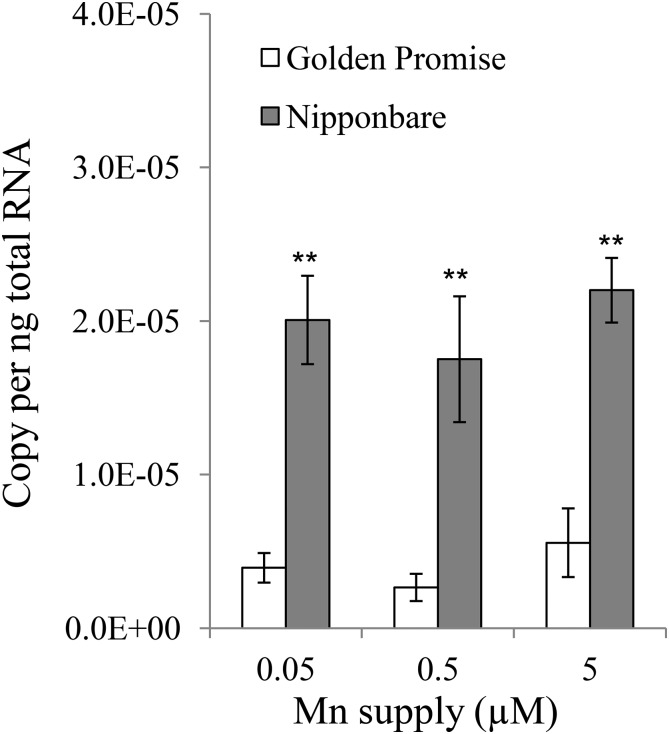
To compare differences in Mn accumulation and expression of HvNramp5 and OsNramp5 between barley (cv Golden Promise) and rice (cv Nipponbare), we grew both plants in a different Mn supply for one week. Barley roots accumulated higher Mn than rice roots, but rice shoots accumulated much higher Mn than did barley shoots, irrespectively of external Mn concentrations (Fig. 7, A and B). The total uptake of Mn of rice was 3 times to 6 times higher than that of barley under different Mn concentrations (Fig. 7C). The expression of HvNramp5 and OsNramp5 in the roots was compared between rice and barley by absolute quantitative RT-PCR. The expression of Nramp5 was 4 times to 7 times higher in rice than in barley at all Mn supply conditions tested (Fig. 8).
Figure 7.
Accumulation of Mn in barley and rice. Concentration of Mn in the roots (A) and shoots (B) and total uptake of Mn (C) in barley (cv Golden Promise) and rice (cv Nipponbare). Rice and barley were grown in nutrient solutions containing 0.05, 0.5, or 5 µm Mn for 7 d. The concentration of Mn in the roots and shoots was determined by ICP-MS. Total uptake of Mn was calculated as total Mn content in the whole plant divided by the root dry weight. Data are means ± sd of three biological replicates and ** indicates significant difference at P < 0.01 by Student’s t test.
Figure 8.
Comparison of HvNramp5 and OsNramp5 expression level in the root between barley and rice. The expression level of HvNramp5 and OsNramp5 in the roots was compared between barley (cv Golden Promise) and rice (cv Nipponbare) by absolute quantitative real-time PCR using common primers. Rice and barley were grown in nutrient solutions containing 0.05, 0.5, or 5 µm Mn for 7 d and then the roots were sampled for RNA extraction and gene expression analysis. Data are means ± sd of three biological replicates and ** indicates significant difference at P < 0.01 by Student’s t test.
DISCUSSION
HvNramp5 Is a Transporter for Uptake of Mn and Cd, But Not Fe, in Barley
In this study, we isolated and functionally characterized the first Nramp member, HvNramp5 in barley. HvNramp5 encodes a plasma membrane-localized protein and transports Mn2+ and Cd2+, but not Fe2+ in yeast (Figs. 2 and 3). HvNramp5 is mainly localized at the epidermal cells of the root tips (Fig. 3A). Knockdown of HvNramp5 resulted in decreased accumulation of Mn and Cd, but not Fe, Cu, and Zn in both roots and shoots (Fig. 5 and Supplemental Fig. S3). Furthermore, the growth of knockdown lines was inhibited at low Mn concentration (Fig. 4A). These results indicate that HvNramp5 is involved in the uptake of Mn and Cd in barley.
Mn uptake was previously reported to be mediated by HvIRT1 (Pedas et al., 2008). HvIRT1 is also a plasma membrane-localized protein and is able to transport Mn, Fe, Zn, and Cd in yeast. The expression of HvIRT1 was induced by Mn and Fe-deficiency (Pedas et al., 2008). Furthermore, different HvIRT1 expression level was associated with genotypic difference in Mn-uptake efficiency of barley (Pedas et al., 2008). However, the exact role of HvIRT1 in Mn uptake in planta has not been examined. In this study, we found that knockdown of HvNramp5 did not affect the expression of HvIRT1 (Supplemental Fig. S2), but significantly reduced Mn uptake at all Mn supply conditions tested (Fig. 5, A and B). The growth of RNAi lines was inhibited at limited Mn supply (Fig. 4A). This strongly supports that HvNramp5 is involved in Mn uptake in barley. One possibility is that HvNramp5 and HvIRT1 may play different roles in Mn uptake in barley. HvIRT1 was up-regulated by Mn-deficiency (Pedas et al., 2008), while HvNramp5 was constitutively expressed in the roots (Fig. 1A). Therefore, HvNramp5 may constitutively contribute to Mn uptake, while HvIRT1 may play an additional role in Mn uptake under a Mn-deficiency condition, although further works are required to make this conclusion. HvNramp5 was also detected in the central cylinder in addition to the epidermal cells (Fig. 3A). This implicates that HvNramp5 may also be involved in the root-to-shoot translocation of Mn, although further evidence is required.
There is worldwide concern regarding Cd accumulation in cereal grains because the intake of Cd from grains through the food chain will affect our health. The threshold value of the Cd concentration in barley grain is 0.1 mg kg−1, which is the most critical among major cereal crops (0.4 mg kg−1 for rice and 0.2 mg kg−1 for wheat (Triticum spp.; CODEX STAN 193-1995, 2013). However, little is known about the molecular mechanisms underlying the Cd uptake in barley. There is wide genotypic variation in Cd accumulation in barley grain, and a recent study with genome-wide association mapping detected several quantitative trait loci controlling Cd accumulation in different organs (Wu et al., 2015). However, the genes responsible for Cd accumulation have not been identified in barley. In this study, we found that Cd uptake is also mediated by HvNramp5 because knockdown of HvNramp5 decreased Cd accumulation in barley (Fig. 5, E and F). This means that Mn and Cd share the same transporter for the uptake. This is supported by competitive interaction between Mn and Cd for the uptake; increasing Mn in the external solution decreased Cd accumulation and vice versa (Figs. 5 and 6). In rice, Mn and Cd also share the same transporter, OsNramp5, for the uptake (Ishimaru et al., 2012; Ishikawa et al., 2012; Sasaki et al., 2012), although HvNramp5 differs from OsNramp5 in terms of expression patterns, tissue localization, and specificity of transport substrate as discussed below.
Different from Cd and Mn, which are taken up in an ionic form, Fe is taken up in the form of Fe(III)-phytosiderophore complex, which is transported by HvYS1 localized at the epidermal cells of barley roots (Murata et al., 2006). This may explain why HvNramp5 is not involved in Fe uptake (Fig. 5, C and D).
Different Uptake System for Mn and Cd between Rice and Barley
Rice is usually cultivated under anaerobic conditions, where Mn concentration in soil solution is very high due to soil reduction (Sasaki et al., 2011), while barley is an upland plant, which is cultivated in low Mn conditions (Pedas et al., 2005). Although both OsNramp5 in rice and HvNramp5 in barley mediate Mn uptake by the roots (Sasaki et al., 2012, Fig. 5), rice takes up and accumulates much more Mn than barley (Fig. 7). This difference could be attributed to their different expression pattern, tissue, and cellular localization. OsNramp5 showed higher expression than HvNramp5 at all Mn supply conditions tested (Fig. 8). Furthermore, in contrast to OsNramp5, which is highly expressed in the basal region of the rice roots (Sasaki et al., 2012), HvNramp5 is highly expressed in the barley root tips (Fig. 1B). In the barley root tips, HvNramp5 is localized to the epidermal cells without polarity (Fig. 3A), while OsNramp5 in rice is polarly localized at the distal side of the exodermises and endodermis of the basal root region (Sasaki et al., 2012). Recently, a Mn efflux transporter (OsMTP9) has been identified in rice, which is localized at the proximal side of both the exodermis and endodermis (Ueno et al., 2015). Rice basal roots are characterized by two Casparian strips: one at the exodermis and one at the endodermis, and by formation of aerenchyma (Ma et al., 2006; Yamaji and Ma, 2007). A recent study showed that both Casparian strips and polar localization of influx and efflux transporters are required for efficient Si uptake in rice (Sakurai et al., 2015). Rice has a similar uptake system for Mn to that for Si, constructing an efficient uptake system by cooperative transporters OsNramp5-OsMPT9 (Ueno et al., 2015). By contrast, in barley root tips, the Casparian strip is usually weakly developed at the endodermis and there is no formation of aerenchyma. Therefore, Mn taken up through HvNramp5 at the epidermal cells is radially transported to the stele symplastically with less efficiency compared with rice uptake system. In barley roots, an efflux Si transporter (HvLsi2) localized at the endodermis was found to be required for further releasing Si to the xylem (Mitani et al., 2009), but it is unknown whether there is a similar efflux transporter for Mn like OsMTP9 in barley. According to the barley genome database (http://webblast.ipk-gatersleben.de/barley/), a close homolog with 85% identity of OsMTP9 was found in barley. Its function and role in Mn uptake needs to be examined and compared with OsMTP9 in future.
Although HvNramp5 shares 84% identity with OsNramp5, there is some difference in the specificity of transport substrates between OsNramp5 and HvNramp5; OsNramp5 transports Mn2+, Fe2+, and Cd2+ in yeast (Ishimaru et al., 2012; Ishikawa et al., 2012), while HvNramp5 transports Mn2+ and Cd2+, but not Fe2+ (Fig. 2). Knockout of OsNramp5 resulted in decreased accumulation of Mn, Fe, and Cd when the plants were grown in nutrient solution (Sasaki et al., 2012). By contrast, knockdown of HvNramp5 resulted in decreased Mn and Cd concentration in the roots and shoots, but the Fe concentration was unaffected (Fig. 5, C and D). In addition to HvNramp5 and OsNramp5, other Nramp members also show different specificity. For example, OsNramp3 only transports Mn, but not Cd and Fe (Yamaji et al., 2013), while OsNrat1 (OsNramp4) transports trivalent Al ion (Xia et al., 2010). The mechanism underlying this specificity of transport substrate remains to be examined in future. Recently, three residues of AtNramp4 have been identified to be involved in metal selectivity in Arabidopsis (Pottier et al., 2015).
In conclusion, HvNramp5 is a plasma membrane-localized transporter for Mn and Cd. It is constitutively expressed in the root epidermal cells and responsible for Mn and Cd uptake in barley.
MATERIALS AND METHODS
Cloning of Full-Length cDNA of HvNramp5
To isolate the full-length cDNA sequence of HvNramp5, total RNA was extracted from barley (Hordeum vulgare cv Golden Promise) roots using the RNeasy Plant Mini Kit (Qiagen). The total RNA was then converted to cDNA according to the protocol supplied by the manufacturers of SuperScript II (Invitrogen). The full-length cDNA was amplified by the PCR using primers 5′-TACGTTCACGCAAAGGGACA-3′ and 5′-ACGTATACACGGTCGCACAG-3′, which were designed based on a reference sequence of AK364374 by blasting the mRNA sequence of OsNramp5 (Sasaki et al., 2012), found in the barley genome database (http://webblast.ipk-gatersleben.de/barley/). The fragment amplified was then introduced into the pGEM vector by pGEM-T (Easy) Vector Systems (Promega) for sequencing. The sequence of the amplified fragments was confirmed by a sequence analyzer (ABI Prism 310 genetic analyzer; Applied Biosystems) using the Big Dye Terminators V3.1 cycle sequencing kit.
Phylogenetic Analysis
Amino acid sequence of HvNramp5 from barley cultivar Golden Promise was translated by software DNASTAR (http://www.dnastar.com/), based on the full-length cDNA sequence. Transmembrane domains of HvNramp5 were predicted by SOSUI, ver. 1.11, software (http://bp.nuap.nagoya-u.ac.jp/sosui/). For phylogenetic analysis, peptide sequence alignment was analyzed by ClustalW software (http://clustalw.ddbj.nig.ac.jp/), comparing HvNramp5 with seven Nramp proteins in rice (Oryza sativa), including OsNramp1 (LOC_Os07g15460), OsNramp2 (LOC_Os03g11010), OsNramp3 (LOC_Os06g46310), OsNramp4 (LOC_Os02g03900), OsNramp5 (LOC_Os07g15370), OsNramp6 (LOC_Os01g31870), and OsNramp7 (LOC_Os12g39180). The phylogenetic tree was constructed with MEGA 5 software (http://www.megasoftware.net/) after ClustalW alignment.
Yeast Experiments
The cDNA fragment containing an entire ORF of HvNramp5 was amplified by RT-PCR using the primers 5′-TTTAAGCTTAAAATGGAGATCGAGAGGGAGGC-3′ and 5′-TTTCTCGAGCTACTGCATATCTCTAGTGC-3′, containing the HindIII and XhoI restriction sites, respectively. The cDNA fragment of HvIRT1 ORF was also amplified by RT-PCR using the primers 5′-TTTAAGCTTAAAATGTCGTCGTCGTCGTCG-3′ and 5′-TTTGCGGCCGCTCACGCCCATTTGGCCATG-3′, containing HindIII and NotI restriction site, respectively. These fragments were cloned into a pYES2 vector (Invitrogen). After sequence confirmation, both the constructed plasmids and the empty vector were introduced into different yeast strains according to the manufacturer’s protocols (S.C. Easy Comp Transformation Kit; Invitrogen). The yeast strains used in this study were BY4741, the Mn uptake-deficient mutant ∆smf1, and the Fe uptake-deficient double-mutant DDY4 (∆fet3fet4), as described by Yamaji et al. (2013).
Complementation of the ∆smf1 and DDY4 phenotypes and evaluation of Cd tolerance in yeast (BY4741, a wild-type strain) were performed according to Yamaji et al. (2013) with some modifications. We supplemented with 0 or 1.5 mm EGTA in the growth medium for ∆smf1, 0 or 12 mm BPDS in the growth medium for DDY4, and 0 or 30 µm CdCl2 in the growth medium for BY4741. After spotting at different yeast-cell dilutions (optical densities at 600 nm of 0.2, 0.02, 0.002, and 0.0002), plates were incubated for 3 to 5 d at 30°C and then the plates were photographed.
For liquid experiment of Cd uptake in yeast (BY4741), each transformant was precultured overnight in liquid medium containing 2% Glc, 0.67% yeast nitrogen base without amino acids, and 0.2% appropriate amino acids. Yeast cells were pelleted by centrifugation and washed three times with Milli-Q water (Millipore), and the resulting pellets were resuspended in the same liquid medium (control) in the presence of 5 µm CdCl2 or in another liquid medium containing 2% Gal, 0.67% yeast nitrogen base without amino acids, 0.2% appropriate amino acids, and 5 µm CdCl2. The uptake experiment was done at 30°C with shaking horizontally at 200 rpm for 4 h and cells were washed three times by Milli-Q water (Millipore) at the end of the experiment. The dry weight of yeast cells was recorded after drying 24 h at 70°C in an oven and the yeast was extracted with a 5% HNO3 for Cd measurement. All yeast experiments were conducted at least two times independently and yielded the similar results.
Expression Pattern of HvNramp5
For spatial expression of HvNramp5, the roots of barley (cv Golden Promise) grown in a 0.5 mm CaCl2 solution for 4 d, were separated into different segments (0–5, 5–10, 10–20 mm) with a razor. To investigate tissue specific expression of HvNramp5, cDNA samples of central cylinder and outer tissues, which were prepared before (Fujii et al., 2012), were used. The samples were collected from the root tip section (2 mm) of barley (cv Morex) using a Veritas Laser Microdissection System LCC1704 (Molecular Devices). To determine the effect of metal deficiency on HvNramp5 expression, seedlings (2-week-old) were transferred to one-fifth Hoagland’s solution with or without each essential metal including Fe, Zn, Cu, and Mn. To determine the effect of Cd addition on HvNramp5 expression, a final concentration of 0.5 μΜ CdSO4 was added to the nutrient solution as described below. Culture solution was renewed every 2 d. After 7 d of metal depletion or Cd treatment, root and shoot samples were harvested separately for RNA extraction and gene expression analysis. Extraction of total RNA was as same as described above. Specific cDNAs were amplified by SYBR premix EX Taq (TAKARA) and quantitative real-time RT-PCR (ABI Prism 7500; Applied Biosystems) with the following primer sets: 5′-TCCCTCGCCTACCTGGAT-3′ and 5′-GCTTCGGATACTCGCTCTT-3′ for HvNramp5; and 5′-GACTCTGGTCATGGTGTCAGC-3′ and 5′-GGCTGGAAGAGGACCTCAGG-3′ for Actin as an internal standard. The relative expression was normalized based on these two genes by the 2∆∆Ct method using the CFX Manager software (Bio-Rad).
Subcellular and Cellular Localization of HvNramp5
For observation of the subcellular localization of HvNramp5, HvNramp5 cDNA containing SalI and NcoI restriction sites, but not the stop codon, was amplified by RT- PCR using the primers 5′-TTGTCGACATGGAGATCGAGAGGGAGGC-3′ and 5′-TTTCCATGGGCTGCATATCTCTAGTGCTGT-3′. After sequence confirmation, the amplified cDNA fragment was then subcloned in-frame in front of the GFP-coding region in a pBluescript vector, producing the HvNramp5-GFP construct under the control of the 35S promoter. RFP was used as a marker of cytosol and nucleus (Matz et al., 1999). Gold particles with a diameter of 1 µm coated with HvNramp5-GFP or GFP alone and RFP were introduced into onion (Allium cepa L.) epidermal cells via particle bombardment (PDS1000/He particle delivery system; Bio-Rad) using 1100 psi rupture disks. After incubation in the dark at room temperature for 16 h, the fluorescence was observed by confocal laser scanning microscopy (LSM700; Carl Zeiss).
To examine the cellular localization, the synthetic peptide MEIEREAPGSERGRSWRAN-C (positions 1–19 of HvNramp5 amino acids) was used to immunize rabbits to obtain antibodies against HvNramp5. The obtained antiserum was purified through a peptide affinity column before use. Different root cross sections (2 mm and 15 mm from the root tip) from 10-d-old seedlings of cultivar Golden Promise were used for immunostaining of HvNramp5 according to Sasaki et al. (2012). Fluorescence of the secondary antibody (Alexa Fluor 555 goat anti-rabbit IgG; Molecular Probes) was observed with a confocal laser scanning microscope (LSM700; Carl Zeiss).
Barley Transformation and Knockdown Mutant Identification
To generate the hairpin RNAi construct, we cloned a 244 bp fragment (−5 to 239 bp from ATG) of HvNramp5 cDNA as inverted repeats into the pANDA vector (Miki and Shimamoto, 2004) under control of the maize (Zea mays) ubiquitin 1 promoter and subsequently transformed the vector into Agrobacterium tumefaciens (strain EHA101). The primer sequences used for amplifying a 244-bp fragment of HvNramp5 cDNA were 5′-AAAAAGCAGGCTCGGCAATGGAGATCGAGAGG-3′ and 5′-AGAAAGCTGGGTTATCTGTGATTGGCTCCGGC-3′. Immature embryos of barley cultivar Golden Promise were used for Agrobacterium-mediated transformation. Transformation was basically performed using the protocols of Hensel et al. (2008), Hiei et al. (2006), and Hiei and Komari (2006).
After PCR confirmation using the primers 5′-GTCGTCGGTGAACAGGTATGG-3′ and 5′-AGAAAGCTGGGTTATCTGTGATTGGCTCCGGC-3′, we obtained several independent transgenic lines, and two independent transgenic homozygous lines (T2 generation) of them were used for further analysis. The expression levels of HvNramp5 and HvIRT1 were determined in the roots of the RNAi lines, the homozygous wild type from transgenic plants (RNAi-wild type), and Golden Promise (wild type), using real-time RT-PCR. The primers used for HvIRT1 were 5′-CCAGATGTTTGAGGGGATGG-3′ and 5′-GATAGACACAAGACACACCC-3′, and the primers for HvNramp5 and Actin were the same as described above.
Plant Materials and Growth Condition
Seeds of wild-type barley (cv Golden Promise) and RNAi lines were soaked in deionized water for 2 h, and then incubated overnight on a moist paper in the dark at 23°C. Germinated seeds were transferred to a nylon mesh floating on a continuously aerated solution in a 1.5-l plastic container containing 0.5 mm CaCl2 (pH 5.6) in the dark. After a 4-d culture, the seedlings were then transferred into an aerated one-fifth Hoagland’s solution (pH 6.0). The solution contained 1 mm KNO3, 1 mm Ca(NO3)2, 0.4 mm MgSO4, and 0.2 mm NH4H2PO4, and micronutrients comprising 20 µm Fe-EDTA, 3 µm H3BO3, 1.0 µm (NH4)6Mo7O24, 0.5 µm MnCl2, 0.4 µm ZnSO4, and 0.2 µm CuSO4. All plants were grown in a growth chamber at 23°C of 14 h d/18°C of 10 h night. Culture solution was renewed every 2 d.
For phenotypic analysis of the HvNramp5 RNAi lines, the seedlings of two transgenic homozygous RNAi lines, the homozygous wild type, and the Golden Promise were grown in a one-fifth Hoagland’s solution with different concentrations of Mn (0.05, 0.5, and 5 μm) as MnCl2; or Cd (0.1, 0.5, and 1 μm) as CdSO4; or Fe (0.1, 2, and 10 μm) as FeSO4. The Cd concentration in the solution for different Mn and Fe treatments was 0.1 μm. After 14 d, the roots were washed three times with 5 mm CaCl2 and separated from the shoots. The samples were dried at 70°C for 2 d and used for metal concentration determination as described below.
For comparing Mn accumulation and Nramp5 expression level between barley and rice, seedlings of barley (cv Golden Promise) and rice (cv Nipponbare) were grown in one-fifth Hoagland’s solution as mentioned above for barley and one-half-strength Kimura B solution for rice containing 0.05, 0.5 or 5 μm Mn, respectively. After exposure for 7 d, the roots and shoots were sampled for Mn concentration determination. The fresh roots were also used for RNA extraction and absolute expression analysis as described below.
Absolute Expression Analysis of Nramp5 in Barley and Rice
After alignment of the cDNA sequences of Nramp5 genes in barley and rice, a 362 bp identical fragment was amplified from cv Golden Promise using common primers (5′-CTCTGGGTGATTCTGATTG-3′ and 5′-CTCAGCTCCCCGAAGAAG-3′). The fragment amplified was then introduced into the pGEM vector by pGEM-T (Easy) Vector Systems (Promega). To generate standard curves for absolute quantification, a series of dilutions (from 1 × 10−1 to 10−6 ng) of the plasmids prepared were made and then assayed by real-time PCR. CT values for each sample were converted into absolute copy numbers using the standard curves. The expression levels of Nramp5 in the roots of barley and rice were quantified by absolute quantitative real-time PCR using the common primers.
Metal Concentration Determination
After being dried in an oven at 70°C for 2 d, samples were digested in concentrated nitric acid at 140°C. The concentration of mineral elements including Mn, Fe, Cu, Zn, and Cd in the digest solution was determined by ICP-MS (7700X; Agilent Technologies) after dilution as described by Wu et al. (2015).
Statistical Analysis
Significance analysis was performed by Student’s t test or Tukey’s test using SPSS software (IBM SPSS Statistics Ver. 16). The difference at P < 0.05 and P < 0.01 was considered as significant and highly significant, respectively.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number LC184278.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Sequence, gene, and protein structure, and phylogenetic analysis of HvNramp5.
Supplemental Figure S2. Gene expression level in the roots of HvNramp5 RNAi lines and wild types.
Supplemental Figure S3. Concentration of Cu and Zn in the HvNramp5 RNAi lines at different Mn concentrations.
Supplemental Figure S4. Phenotypic analysis of HvNramp5 RNAi lines at different Cd concentrations.
Supplemental Figure S5. Phenotypic analysis of HvNramp5 RNAi lines at different Fe concentrations.
Supplementary Material
Footnotes
This work was supported by a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Genomics-based Technology for Agricultural Improvement grant no. TRS-1006 and the Scientific Technique Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry grant no. 25013A to J.F.M.) and by a Grant-in-Aid for Specially Promoted Research (Japan Society for the Promotion of Science KAKENHI grant no. 16H06296 to J.F.M.).
Articles can be viewed without a subscription.
References
- Bereczky Z, Wang HY, Schubert V, Ganal M, Bauer P (2003) Differential regulation of nramp and irt metal transporter genes in wild type and iron uptake mutants of tomato. J Biol Chem 278: 24697–24704 [DOI] [PubMed] [Google Scholar]
- Cailliatte R, Lapeyre B, Briat JF, Mari S, Curie C (2009) The NRAMP6 metal transporter contributes to cadmium toxicity. Biochem J 422: 217–228 [DOI] [PubMed] [Google Scholar]
- Cailliatte R, Schikora A, Briat JF, Mari S, Curie C (2010) High-affinity manganese uptake by the metal transporter NRAMP1 is essential for Arabidopsis growth in low manganese conditions. Plant Cell 22: 904–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CODEX STAN 193-1995 (2013) General standard for contaminants and toxins in foods and feed. www.fao.org/input/download/standards/17/CXS_193e_2015.pdf (accessed March 10, 2015)
- Curie C, Alonso JM, Le Jean M, Ecker JR, Briat JF (2000) Involvement of NRAMP1 from Arabidopsis thaliana in iron transport. Biochem J 347: 749–755 [PMC free article] [PubMed] [Google Scholar]
- Engl A, Kunz B (1995) Biosorption of heavy metals by Saccharomyces cerevisiae: effects of nutrient conditions. J Chem Technol Biotechnol 63: 257–261 [Google Scholar]
- Fujii M, Yokosho K, Yamaji N, Saisho D, Yamane M, Takahashi H, Sato K, Nakazono M, Ma JF (2012) Acquisition of aluminium tolerance by modification of a single gene in barley. Nat Commun 3: 713–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JL, Williams LE (2003) Transition metal transporters in plants. J Exp Bot 54: 2601–2613 [DOI] [PubMed] [Google Scholar]
- Hensel G, Valkov V, Middlefell-Williams J, Kumlehn J (2008) Efficient generation of transgenic barley: the way forward to modulate plant-microbe interactions. J Plant Physiol 165: 71–82 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ishida Y, Kasaoka K, Komari T (2006) Improved frequency of transformation in rice and maize by treatment of immature embryos with centrifugation and heat prior to infection with Agrobacterium tumefaciens. Plant Cell Tiss Org 87: 233–243 [Google Scholar]
- Hiei Y, Komari T (2006) Improved protocols for transformation of indica rice mediated by Agrobacterium tumefaciens. Plant Cell Tiss Org 85: 271–283 [Google Scholar]
- Mayer KF, Waugh R, Brown JW, Schulman A, Langridge P, Platzer M, Fincher GB, Muehlbauer GJ, Sato K, Close TJ, Wise RP, Stein N (2012) A physical, genetic and functional sequence assembly of the barley genome. Nature 491: 711–716 [DOI] [PubMed] [Google Scholar]
- Ishikawa S, Ishimaru Y, Igura M, Kuramata M, Abe T, Senoura T, Hase Y, Arao T, Nishizawa NK, Nakanishi H (2012) Ion-beam irradiation, gene identification, and marker-assisted breeding in the development of low-cadmium rice. Proc Natl Acad Sci USA 109: 19166–19171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru Y, Takahashi R, Bashir K, Shimo H, Senoura T, Sugimoto K, Ono K, Yano M, Ishikawa S, Arao T, Nakanishi H, Nishizawa NK (2012) Characterizing the role of rice NRAMP5 in manganese, iron and cadmium transport. Sci Rep 2: 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser BN, Moreau S, Castelli J, Thomson R, Lambert A, Bogliolo S, Puppo A, Day DA (2003) The soybean NRAMP homologue, GmDMT1, is a symbiotic divalent metal transporter capable of ferrous iron transport. Plant J 35: 295–304 [DOI] [PubMed] [Google Scholar]
- Lanquar V, Lelièvre F, Bolte S, Hamès C, Alcon C, Neumann D, Vansuyt G, Curie C, Schröder A, Krämer U, Barbier-Brygoo H, Thomine S (2005) Mobilization of vacuolar iron by AtNRAMP3 and AtNRAMP4 is essential for seed germination on low iron. EMBO J 24: 4041–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanquar V, Ramos MS, Lelièvre F, Barbier-Brygoo H, Krieger-Liszkay A, Krämer U, Thomine S (2010) Export of vacuolar manganese by AtNRAMP3 and AtNRAMP4 is required for optimal photosynthesis and growth under manganese deficiency. Plant Physiol 152: 1986–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JF, Tamai K, Yamaji N, Mitani N, Konishi S, Katsuhara M, Ishiguro M, Murata Y, Yano M (2006) A silicon transporter in rice. Nature 440: 688–691 [DOI] [PubMed] [Google Scholar]
- Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA (1999) Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol 17: 969–973 [DOI] [PubMed] [Google Scholar]
- Miki D, Shimamoto K (2004) Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol 45: 490–495 [DOI] [PubMed] [Google Scholar]
- Milner MJ, Mitani-Ueno N, Yamaji N, Yokosho K, Craft E, Fei Z, Ebbs S, Clemencia Zambrano M, Ma JF, Kochian LV (2014) Root and shoot transcriptome analysis of two ecotypes of Noccaea caerulescens uncovers the role of NcNramp1 in Cd hyperaccumulation. Plant J 78: 398–410 [DOI] [PubMed] [Google Scholar]
- Mitani N, Chiba Y, Yamaji N, Ma JF (2009) Identification and characterization of maize and barley Lsi2-like silicon efflux transporters reveals a distinct silicon uptake system from that in rice. Plant Cell 21: 2133–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T, Usui K, Horie K, Nosaka S, Mizuno N, Obata H (2005) Cloning of three ZIP/Nramp transporter genes from a Ni hyperaccumulator plant Thlaspi japonicum and their Ni2+-transport abilities. Plant Physiol Biochem 43: 793–801 [DOI] [PubMed] [Google Scholar]
- Murata Y, Ma JF, Yamaji N, Ueno D, Nomoto K, Iwashita T (2006) A specific transporter for iron(III)-phytosiderophore in barley roots. Plant J 46: 563–572 [DOI] [PubMed] [Google Scholar]
- Nevo Y, Nelson N (2006) The NRAMP family of metal-ion transporters. Biochim Biophys Acta 1763: 609–620 [DOI] [PubMed] [Google Scholar]
- Olsen LI, Palmgren MG (2014) Many rivers to cross: the journey of zinc from soil to seed. Front Plant Sci 5: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oomen RJ, Wu J, Lelièvre F, Blanchet S, Richaud P, Barbier-Brygoo H, Aarts MG, Thomine S (2009) Functional characterization of NRAMP3 and NRAMP4 from the metal hyperaccumulator Thlaspi caerulescens. New Phytol 181: 637–650 [DOI] [PubMed] [Google Scholar]
- Pedas P, Hebbern CA, Schjoerring JK, Holm PE, Husted S (2005) Differential capacity for high-affinity manganese uptake contributes to differences between barley genotypes in tolerance to low manganese availability. Plant Physiol 139: 1411–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedas P, Ytting CK, Fuglsang AT, Jahn TP, Schjoerring JK, Husted S (2008) Manganese efficiency in barley: identification and characterization of the metal ion transporter HvIRT1. Plant Physiol 148: 455–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pottier M, Oomen R, Picco C, Giraudat J, Scholz-Starke J, Richaud P, Carpaneto A, Thomine S (2015) Identification of mutations allowing Natural Resistance Associated Macrophage Proteins (NRAMP) to discriminate against cadmium. Plant J 83: 625–637 [DOI] [PubMed] [Google Scholar]
- Sakurai G, Satake A, Yamaji N, Mitani-Ueno N, Yokozawa M, Feugier FG, Ma JF (2015) In silico simulation modeling reveals the importance of the Casparian strip for efficient silicon uptake in rice roots. Plant Cell Physiol 56: 631–639 [DOI] [PubMed] [Google Scholar]
- Sasaki A, Yamaji N, Ma JF (2016) Transporters involved in mineral nutrient uptake in rice. J Exp Bot 67: 3645–3653 [DOI] [PubMed] [Google Scholar]
- Sasaki A, Yamaji N, Xia J, Ma JF (2011) OsYSL6 is involved in the detoxification of excess manganese in rice. Plant Physiol 157: 1832–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A, Yamaji N, Yokosho K, Ma JF (2012) Nramp5 is a major transporter responsible for manganese and cadmium uptake in rice. Plant Cell 24: 2155–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi R, Ishimaru Y, Senoura T, Shimo H, Ishikawa S, Arao T, Nakanishi H, Nishizawa NK (2011) The OsNRAMP1 iron transporter is involved in Cd accumulation in rice. J Exp Bot 62: 4843–4850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomine S, Wang R, Ward JM, Crawford NM, Schroeder JI (2000) Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to Nramp genes. Proc Natl Acad Sci USA 97: 4991–4996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno D, Sasaki A, Yamaji N, Miyaji T, Fujii Y, Takemoto Y, Moriyama S, Che J, Moriyama Y, Iwasaki K, Ma JF (2015) A polarly localized transporter for efficient manganese uptake in rice. Nat Plants 1: 15170. [DOI] [PubMed] [Google Scholar]
- Wu D, Sato K, Ma JF (2015) Genome-wide association mapping of cadmium accumulation in different organs of barley. New Phytol 208: 817–829 [DOI] [PubMed] [Google Scholar]
- Xia J, Yamaji N, Kasai T, Ma JF (2010) Plasma membrane-localized transporter for aluminum in rice. Proc Natl Acad Sci USA 107: 18381–18385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Yin L, Xu X, Li T, Han Z (2008) The iron-regulated transporter, MbNRAMP1, isolated from Malus baccata is involved in Fe, Mn and Cd trafficking. Ann Bot (Lond) 102: 881–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji N, Ma JF (2007) Spatial distribution and temporal variation of the rice silicon transporter Lsi1. Plant Physiol 143: 1306–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji N, Sasaki A, Xia JX, Yokosho K, Ma JF (2013) A node-based switch for preferential distribution of manganese in rice. Nat Commun 4: 2442. [DOI] [PubMed] [Google Scholar]
- Yokosho K, Yamaji N, Ueno D, Mitani N, Ma JF (2009) OsFRDL1 is a citrate transporter required for efficient translocation of iron in rice. Plant Physiol 149: 297–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.