A novel NDH-1L-CpcG2-phycobilisome-PSI supercomplex essential to cyclic electron transport was identified in Synechocystis sp. strain PCC 6803.
Abstract
Two mutants isolated from a tagging library of Synechocystis sp. strain PCC 6803 were sensitive to high light and had a tag in sll1471 encoding CpcG2, a linker protein for photosystem I (PSI)-specific antenna. Both mutants demonstrated strongly impaired NDH-1-dependent cyclic electron transport. Blue native-polyacrylamide gel electrophoresis followed by immunoblotting and mass spectrometry analyses of the wild type and a mutant containing CpcG2 fused with yellow fluorescent protein-histidine6 indicated the presence of a novel NDH-1L-CpcG2-PSI supercomplex, which was absent in the cpcG2 deletion mutant, the PSI-less mutant, and several other strains deficient in NDH-1L and/or NDH-1M. Coimmunoprecipitation and pull-down analyses on CpcG2-yellow fluorescent protein-histidine6, using antibody against green fluorescent protein and nickel column chromatography, confirmed the association of CpcG2 with the supercomplex. Conversely, the use of antibodies against NdhH or NdhK after blue native-polyacrylamide gel electrophoresis and in coimmunoprecipitation experiments verified the necessity of CpcG2 in stabilizing the supercomplex. Furthermore, deletion of CpcG2 destabilized NDH-1L as well as its degradation product NDH-1M and significantly decreased the number of functional PSI centers, consistent with the involvement of CpcG2 in NDH-1-dependent cyclic electron transport. The CpcG2 deletion, however, had no effect on respiration. Thus, we propose that the formation of an NDH-1L-CpcG2-PSI supercomplex in cyanobacteria facilitates PSI cyclic electron transport via NDH-1L.
Cyclic electron transport (CET) around PSI is an important process for oxygenic photosynthetic organisms. In cooperation with linear electron transport, CET contributes to the formation of a proton gradient across the thylakoid membrane, which increases the production of ATP in relation to NADPH and consequently optimizes the ATP/NADPH ratio. In addition, CET plays an important role in protecting photosynthesis against various environmental stresses, such as high light (Battchikova et al., 2011a; Dai et al., 2013; Zhang et al., 2014; Zhao et al., 2014, 2015; Wang et al., 2016).
In cyanobacteria, the main route for CET involves NDH-1 complexes, which belong to the complex I family (for review, see Friedrich et al., 1995; Yagi et al., 1998; Friedrich and Scheide, 2000; Brandt et al., 2003). On the basis of sequence similarity analysis, the complex I family was suggested to originate from a common ancestor, a group 4 membrane-bound [NiFe] hydrogenase that possesses a proton-transporting hydrogen:ferredoxin (Fd) oxidoreductase activity (Böhm et al., 1990; Friedrich and Weiss, 1997; Friedrich and Scheide, 2000; Efremov and Sazanov, 2012). During evolution, however, respiratory NDH-1 and photosynthetic NDH-1 developed different catalytic activities (for review, see Peltier et al., 2016). The former has become equipped with a new NADH-oxidizing module consisting of three subunits and capable of oxidizing NADH (Efremov and Sazanov, 2012), and the latter, as was suggested recently, has retained an original electron input module that accepts electrons from Fd (Battchikova et al., 2011a; Yamamoto et al., 2011). Structurally, respiratory NDH-1 and photosynthetic NDH-1 contain a conserved L-shaped skeleton (Friedrich et al., 1995; Friedrich and Scheide, 2000; Arteni et al., 2006; Kouřil et al., 2014), but numerous oxygenic photosynthesis-specific subunits were added to the photosynthetic NDH-1 (for review, see Battchikova et al., 2011b; Ifuku et al., 2011; Peng et al., 2011a). Among them, the NdhS subunit can bind reduced Fd, which provides a functional link to the PSI complex that permits NDH-1-dependent cyclic electron transport (NDH-CET; Battchikova et al., 2011a; Yamamoto et al., 2011; Yamamoto and Shikanai, 2013; He et al., 2015).
In cyanobacteria, NDH-1 exists in multiple forms with multiple functions (for review, see Battchikova and Aro, 2007; Ogawa and Mi, 2007; Battchikova et al., 2011b). Among them, a large NDH-1 complex (NDH-1L) was shown to be involved in NDH-CET and respiration (Zhang et al., 2004; Bernát et al., 2011). Cyanobacterial NDH-1L was suggested to be an ancestor of chloroplast NDH-1 because they possess similarities in L-shaped skeleton and physiological function (for review, see Battchikova et al., 2011b; Ifuku et al., 2011; Peng et al., 2011a).
It is now well established that, in higher plants, the chloroplast NDH-1 together with PSI form an NDH-1-PSI supercomplex (Peng et al., 2008, 2009; Kouřil et al., 2014). It was demonstrated that two minor proteins of the light-harvesting complex I (LHCI), Lhca5 and Lhca6, are necessary for the supercomplex formation (Peng et al., 2009). A similar NDH-1-PSI supercomplex has been found in Physcomitrella patens that harbors the LHCI protein related to Lhca5 (Alboresi et al., 2008; Neilson and Durnford, 2010) but not in Marchantia polymorpha, which lacks analogs of Lhca5 or Lhca6 protein (Ueda et al., 2012). It was suggested earlier that the NDH-1 complex also associates with PSI in cyanobacteria (Kubota et al., 2010; Battchikova et al., 2011a). This hypothesis was supported by the results of copurification: some hydrophilic subunits of the NDH-1 complex, including NdhH to NdhK, NdhN, and NdhO, were copurified with PSI in Synechocystis sp. strain PCC 6803 (hereafter Synechocystis 6803; Kubota et al., 2010), and several PSI subunits, PsaB to PsaD and PsaF, were copurified with NDH-1 (Battchikova et al., 2011a). Despite these copurified results and the evolutionary relationship of cyanobacterial NDH-1L with chloroplast NDH-1, an NDH-1L-PSI supercomplex still remains unidentified in cyanobacteria.
In this study, we demonstrate the existence of an NDH-1L-PSI supercomplex, with a molecular mass of more than 1,000 kD, in the cyanobacterium Synechocystis 6803. Furthermore, we demonstrate that the CpcG2 protein associates with the supercomplex and is required to stabilize the supercomplex; this protein was earlier characterized as a component of the PSI-specific antenna (Kondo et al., 2007) associated with PSI (Watanabe et al., 2014). In Synechocystis 6803, the NDH-1L-PSI supercomplex is shown to be essential for NDH-CET but not for respiration.
RESULTS
Isolation of NDH-CET-Defective Mutants
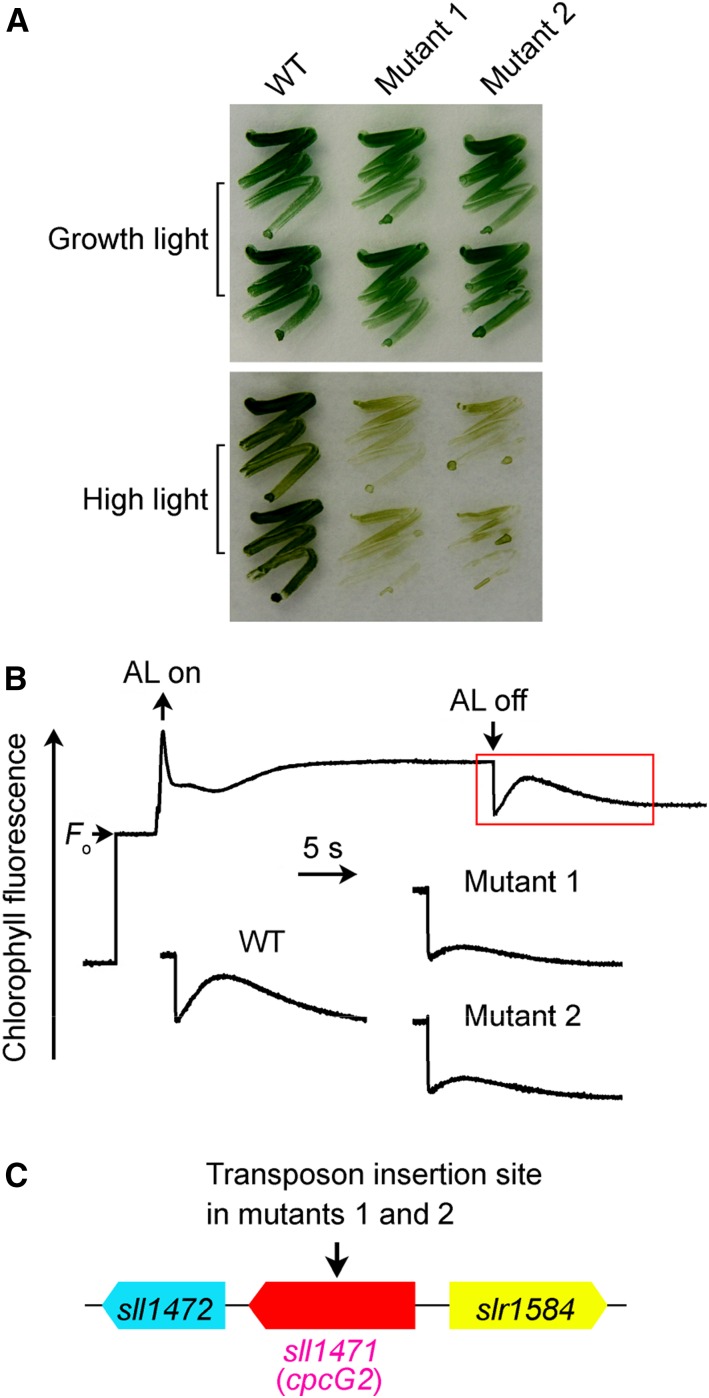
Under high-light conditions, the growth of NDH-CET-defective mutants, such as ΔndhS, is markedly slower in comparison with the wild type despite similar growth under moderate light irradiation (Battchikova et al., 2011a; Dai et al., 2013; Zhang et al., 2014; Zhao et al., 2014, 2015; Wang et al., 2016). To screen for NDH-CET-defective mutants, we transformed wild-type cells with a transposon-bearing library, thus tagging and inactivating many genes randomly, and then cultured the mutant cells under high-light conditions. We isolated two mutants, which grew slowly on plates under high light but similarly to the wild type under growth light (Fig. 1A).
Figure 1.
Growth, NDH-CET activity, and transposon insertion sites of high-light-sensitive mutants of Synechocystis 6803. A, Growth of wild-type (WT) and mutant strains under normal light (40 µmol photons m−2 s−1) and high light (300 µmol photons m−2 s−1). B, Monitoring of NDH-CET activity by Chl fluorescence analysis. The top curve shows a typical trace of Chl fluorescence in wild-type Synechocystis 6803. The Chl a concentration was adjusted to 10 µg mL−1 before measurement. Cells were exposed to AL (620 nm, 45 µmol photons m−2 s−1) for 30 s. AL was turned off, and the subsequent change in the Chl fluorescence level was monitored as an indicator of NDH-CET activity. C, The arrow schematically indicates the transposon insertion site in mutants 1 and 2 probed by PCR analysis using the primers listed in Supplemental Table S1.
To test whether the high-light-sensitive growth phenotype of the two mutants resulted from defective NDH-CET, we monitored the postillumination rise in chlorophyll (Chl) fluorescence, which has been used extensively to evaluate the NDH-CET activity in cyanobacteria (Mi et al., 1995; Deng et al., 2003a; Ma and Mi, 2005; Battchikova et al., 2011a; Dai et al., 2013; Zhang et al., 2014; Zhao et al., 2014, 2015; Wang et al., 2016) and higher plants (Burrows et al., 1998; Shikanai et al., 1998; Hashimoto et al., 2003; Wang et al., 2006; Peng et al., 2009, 2011b, 2012; Sirpiö et al., 2009; Yamamoto et al., 2011; Armbruster et al., 2013). In this method, the NDH-CET activity is manifested by a transient rise of Chl fluorescence after actinic light (AL) is turned off (the corresponding phase is shown by the red rectangle in Fig. 1B). The activity, assessed as the amplitude of the fluorescence rise, was markedly lower in both mutants than in the wild type (Fig. 1B).
To identify the genes inactivated by the transposon tagging, we analyzed the insertion sites of transposons in both mutants. Sequencing analysis of the kanamycin resistance marker (KamR) insertion region revealed that both mutants were tagged in the same gene, sll1471 (cpcG2; Fig. 1C), at position 3,403,918 of the Synechocystis 6803 genome (National Center for Biotechnology Information gi: 16332194; Kaneko et al., 1996). Therefore, it seems likely that CpcG2, a linker protein for the PSI-specific peripheral antenna CpcG2-phycobilisome (PBS; Kondo et al., 2005, 2007, 2009), is involved in NDH-CET.
Inactivation of cpcG2 Impairs NDH-CET Activity
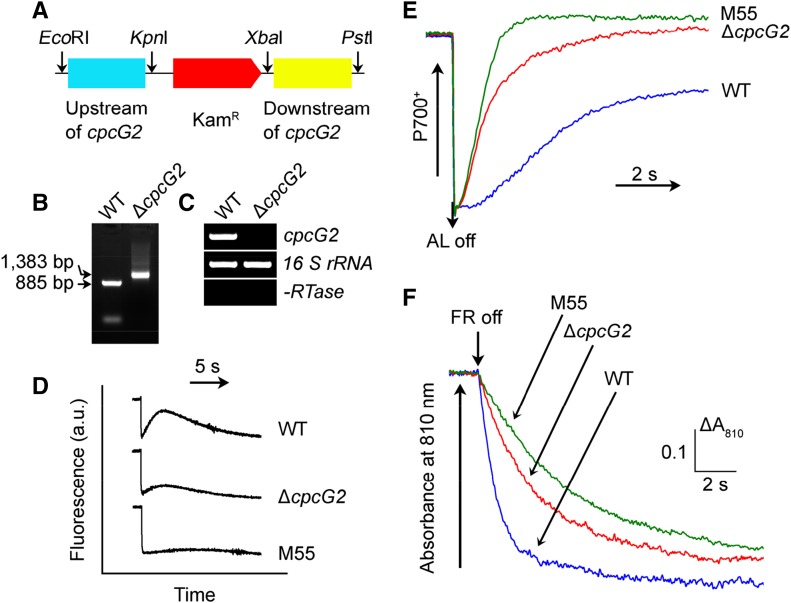
To elucidate the role of CpcG2 in NDH-CET, we constructed a cpcG2 deletion mutant (ΔcpcG2) by replacing the entire cpcG2 coding region with a KamR (Fig. 2A). PCR analysis of the cpcG2 locus confirmed complete segregation of the ΔcpcG2 mutant allele (Fig. 2B). Transcript analysis using a specific primer pair for the cpcG2-encoding gene (Supplemental Table S1) demonstrated the absence of gene product in the mutant (Fig. 2C). As expected, the activity of NDH-CET, as measured by the postillumination rise in Chl fluorescence, was lower in ∆cpcG2 than in the wild type (Fig. 2D). It is worthy of note that the NDH-CET activity was higher than that in the NdhB-deficient mutant (M55), which lacks all functional NDH-1 complexes. The significant decrease of the NDH-CET activity in the ∆cpcG2 mutant compared with the wild type was corroborated with two other methods. Cells were illuminated by AL (800 µmol photons m−2 s−1) supplemented with far-red light (FR) to store electrons in the stromal pool. After the termination of AL, P700+ was reduced transiently by electrons from the plastoquinone pool; P700 is reoxidized by the background FR. Operation of the NDH-1 complex, which transfers electrons from the reduced cytoplasmic pool to plastoquinone, hinders the reoxidation of P700 (Shikanai et al., 1998; Battchikova et al., 2011a; Dai et al., 2013; Zhang et al., 2014; Zhao et al., 2014, 2015; Wang et al., 2016). Figure 2E shows that the reoxidation of P700 was much faster in ∆cpcG2 than in the wild type. Furthermore, the rereduction rate of P700+ was monitored in darkness after the illumination of 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU)-treated cells with FR. The rereduction of P700+ was much slower in ∆cpcG2 than in the wild type (Fig. 2F), providing evidence of the scarcity of CET from reduced Fd via NDH-1 back to P700+ in darkness. Therefore, the slow growth of ∆cpcG2 under high-light conditions (Supplemental Fig. S1) could be attributed to the low NDH-CET activity.
Figure 2.
Deletion of cpcG2 and its effect on NDH-CET. A, Construction of the plasmid used to generate the cpcG2 deletion mutant (∆cpcG2). B, PCR segregation analysis of the ∆cpcG2 mutant using the cpcG2-G and cpcG2-H primer sequences (Supplemental Table S1). C, Transcript levels of cpcG2 in the wild-type (WT) and ∆cpcG2 strains. The transcript level of 16 S rRNA in each sample is shown as a control. The absence of DNA contamination was confirmed by PCR without reverse transcriptase reaction. D, Monitoring of NDH-CET activity by Chl fluorescence. For details, see the legend to Figure 1B. a.u., Arbitrary units. E, Redox kinetics of P700 after the termination of AL illumination (800 µmol photons m−2 s−1 for 30 s) under a background of FR. The cells were illuminated by AL supplemented with FR, and then AL was turned off. P700+ was reduced transiently by electrons from the plastoquinone pool and then reoxidized by background FR. The P700+ levels were standardized by their maximum levels attained by exposure to FR. F, Kinetics of the P700+ rereduction in darkness after turning off FR with a maximum at 720 nm. Wild-type, ∆cpcG2, and M55 cells were suspended in BG-11 medium containing 10 µm DCMU at the Chl a concentration of 20 µg mL−1 before measurement. Curves are normalized to the maximal signal.
A Novel Supercomplex Involving CpcG2, NDH-1L, and PSI
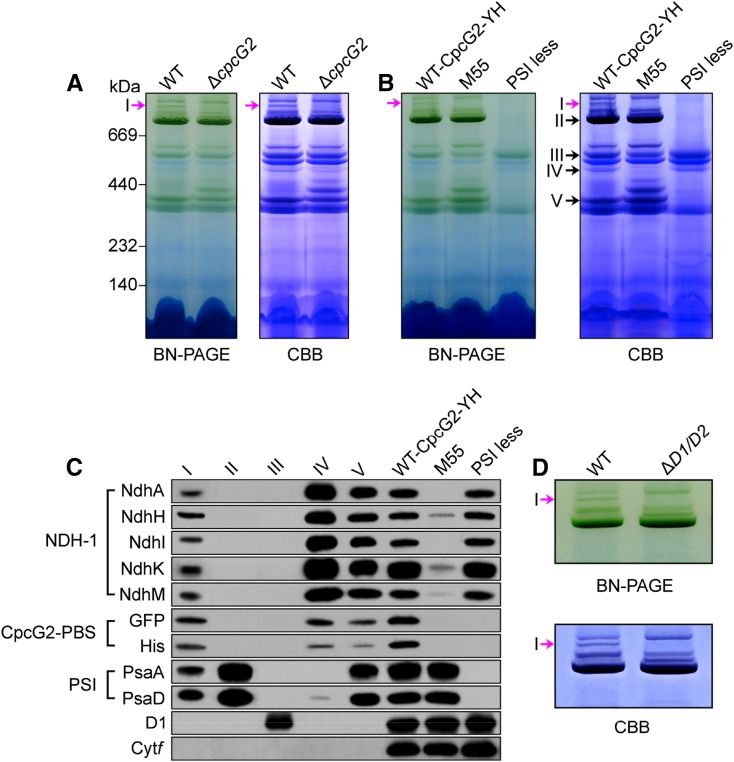
The above results suggested the presence of a supercomplex composed of NDH-1, CpcG2, and PSI (NDH-1-CpcG2-PBS-PSI; hereafter referred to as the NDH-1-PSI supercomplex). To explore this possibility, we performed blue native (BN)-PAGE of thylakoid membranes isolated from the wild-type and ∆cpcG2 strains. The results revealed that one of the Chl-containing bands with a high molecular mass (above 1,000 kD) present in the wild type was absent in the ∆cpcG2 mutant (Fig. 3A, band I shown by the pink arrows). Band I also was absent in M55 and PSI-less mutants (Fig. 3B), which lack functional NDH-1 and PSI complexes, respectively. Thus, it seemed highly likely that band I is the NDH-1-PSI supercomplex dependent on the presence of CpcG2.
Figure 3.
Identification of the NDH-1-PSI supercomplex. A, BN-PAGE and Coomassie Brilliant Blue (CBB) staining profiles of the thylakoid membranes of wild-type (WT) and ∆cpcG2 strains. Band I is shown by pink arrows. B, BN-PAGE and Coomassie Brilliant Blue staining profiles of the thylakoid membranes of WT-CpcG2-YH, M55, and PSI-less mutants. The positions of bands I to V are shown by the arrows. C, Immunodetection of subunits of NDH-1, PSI, PSII, and Cyt b6f complexes and the GFP-His tag in bands I to V excised from the gel after BN-PAGE of the thylakoid membranes of WT-CpcG2-YH (see B) and in the WT-CpcG2-YH, M55, and PSI-less strains of Synechocystis 6803. D, Analysis of band I in the wild-type and ∆ndhD1/ndhD2 (∆D1/D2) strains.
To verify the presence of the NDH-1-PSI supercomplex in Synechocystis 6803, we constructed a wild-type-CpcG2-yellow fluorescent protein (YFP)-His6 strain (hereafter referred to as WT-CpcG2-YH) by adding the YFP-His6 tag on the C terminus of CpcG2 in the wild-type background (Supplemental Fig. S2A). PCR analysis indicated complete segregation of the tagged gene (Supplemental Fig. S2B). Western-blot analysis of the thylakoid membrane from the tagged strain and confocal analysis of the tagged cells indicated the expression of CpcG2-YH (Supplemental Fig. S2, C and D). The tagging did not affect the NDH-CET activity (Supplemental Fig. S2, E–G).
Furthermore, we carried out BN-PAGE of thylakoid membranes isolated from the WT-CpcG2-YH strain. Band I and four other bands (II–V) were excised from the BN gel (Fig. 3B) and subjected to western-blot analysis using antibodies against NdhA, NdhH, NdhI, NdhK, and NdhM (NDH-1 complex), GFP and His (CpcG2-PBS antenna), PsaA and PsaD (PSI complex), D1 (PSII complex), and cytochrome (Cyt) f (Cyt b6f complex). Immunoblots demonstrated that band I contained subunits of NDH-1 and PSI complexes together with YH-tagged CpcG2 (Fig. 3C), consistent with the absence of band I in the mutants lacking its subcomponents (Fig. 3, A and B). The presence of NDH-1, CpcG2, and PSI proteins in band I was confirmed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis (Table I; Supplemental Data Set S1). It is worthy of note that components of PSII and Cyt b6f complexes, D1 and Cyt f, respectively, were not detected in the band I. Bands II to V were PSI trimer, PSII dimer, NDH-1L, and NDH-1M (overlapped by PSI monomer) complexes, respectively, and immunoblots indicated that NDH-1L and its degradation product NDH-1M retained CpcG2, as deduced from the signal of its tagging (Fig. 3C).
Table I. Summary of the NDH-1, CpcG2-PBS, and PSI components identified from Q-Exactive mass analysis of the NDH-1-PSI supercomplex (band I).
The complete list of proteins identified in band I can be found in Supplemental Data Set S1.
| Component | Open Reading Frame | Protein Name | Mascot Score | Protein Match | Coverage (%) |
|---|---|---|---|---|---|
| NDH-1 complex | sll0519 | NdhA | 40.5 | 4 | 13.2 |
| sll0223 | NdhB | 24.8 | 1 | 1.9 | |
| slr0331 | NdhD1 | 43.0 | 1 | 3.4 | |
| slr0844 | NdhF1 | 41.4 | 1 | 1.6 | |
| sll0521 | NdhG | 31.0 | 1 | 6.1 | |
| slr0261 | NdhH | 64.3 | 11 | 27.9 | |
| sll0520 | NdhI | 53.0 | 6 | 29.5 | |
| slr1281 | NdhJ | 51.1 | 5 | 30.7 | |
| slr1280 | NdhK | 69.3 | 7 | 27.4 | |
| slr1623 | NdhM | 41.4 | 3 | 36.4 | |
| sll1262 | NdhN | 74.7 | 2 | 18.6 | |
| ssl1690 | NdhO | 96.8 | 3 | 52.8 | |
| CpcG2-PBS antenna | sll1471 | CpcG2 | 67.5 | 11 | 35.3 |
| sll1578 | CpcA | 47.6 | 2 | 14.8 | |
| sll1577 | CpcB | 100.5 | 6 | 41.9 | |
| sll1580 | CpcC1 | 98.0 | 9 | 40.6 | |
| sll1579 | CpcC2 | 95.3 | 15 | 52.8 | |
| PSI complex | slr1834 | PsaA | 95.6 | 20 | 21.6 |
| slr1835 | PsaB | 83.2 | 18 | 20.4 | |
| ssl0563 | PsaC | 78.7 | 8 | 85.2 | |
| slr0737 | PsaD | 100.2 | 13 | 66.0 | |
| ssr2831 | PsaE | 90.5 | 5 | 94.6 | |
| sll0819 | PsaF | 93.2 | 14 | 57.6 | |
| sml0008 | PsaJ | 20.3 | 1 | 22.5 | |
| ssr0390 | PsaK1 | 28.4 | 1 | 19.8 | |
| sll0629 | PsaK2 | 40.4 | 3 | 14.4 | |
| slr1655 | PsaL | 87.6 | 6 | 45.2 | |
| smr0005 | PsaM | 37.4 | 1 | 22.6 |
Next, we observed on the BN-PAGE gel that the NDH-1-PSI supercomplex (band I) was absent in the thylakoid membrane of the ΔndhD1/ndhD2 mutant (Fig. 3D), where the NDH-1M complex is formed instead of NDH-1L due to the absence of NdhD1 (and NdhD2) subunits (Zhang et al., 2004, 2014). In accordance with this result, NDH-1L-specific subunits, NdhD1 and NdhF1, were detected in band I that originated from the wild type (Table I; Supplemental Data Set S1). Thus, we suggest that an NDH-1L-CpcG2-PBS-PSI supercomplex is present in Synechocystis 6803. Nevertheless, the supercomplex appeared to be rather unstable and mostly decomposed during isolation and n-dodecyl β-d-maltoside (DM) treatment of the thylakoid membrane (see “Discussion”).
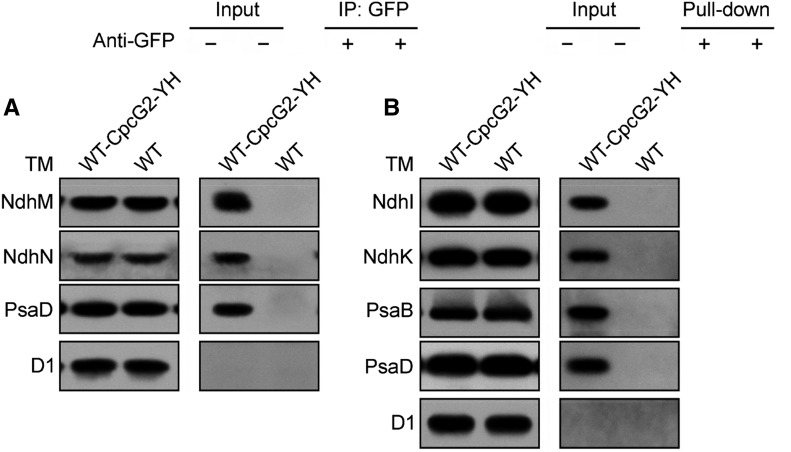
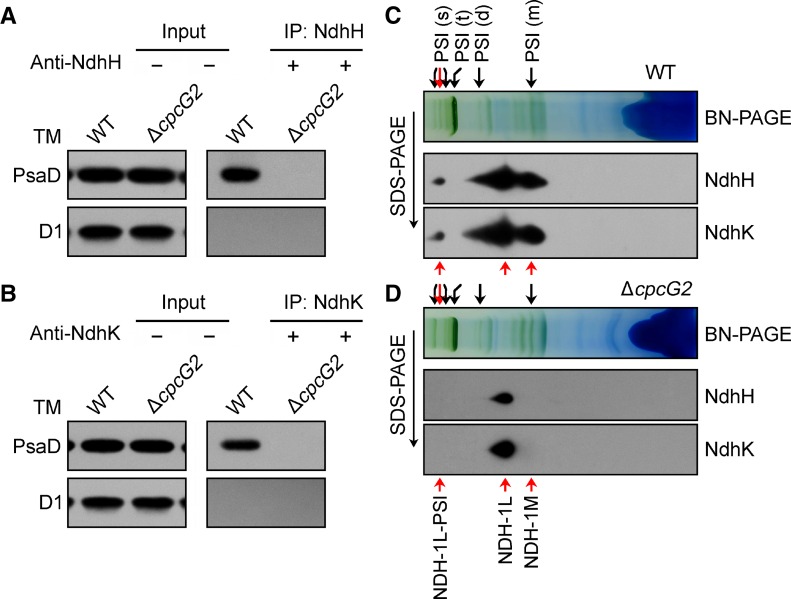
Finally, we performed a coimmunoprecipitation of CpcG2 from the WT-CpcG2-YH strain using a protein A-Sepharose resin with polyclonal anti-GFP antibodies. Wild-type cells were used as a control to monitor the specificity of the assay. Immunoblot analysis demonstrated the presence of subunits of both the NDH-1 and PSI complexes in the preparation recovered from the WT-CpcG2-YH strain but not from the wild-type strain (Fig. 4A). The D1 protein of PSII was not recovered from either strain (Fig. 4A). A pull-down analysis using Ni2+ affinity chromatography gave a similar result (Fig. 4B). Taking all these results together, we conclude that an NDH-1L-PSI supercomplex is present in Synechocystis 6803.
Figure 4.
Interaction of CpcG2 with NDH-1 and PSI complexes. A, Coimmunoprecipitation assay of the interaction of CpcG2 with NDH-1 and PSI complexes. Membrane proteins from the WT-CpcG2-YH and wild-type (WT) strains were incubated with protein A-Sepharose-coupled anti-GFP antiserum. The immunoprecipitates (IP) were probed with specific antibodies, as indicated on the left. B, Pull-down analysis of the interaction of CpcG2 with NDH-1 and PSI complexes. Membrane proteins isolated from the WT-CpcG2-YH and wild-type strains were purified using Ni2+ affinity chromatography. The purified samples were immunoblotted with antibodies against the subunits of the major photosynthetic membrane protein complexes. TM, Thylakoid membrane.
Yet another experiment was performed to corroborate the dependence of the NDH-1-PSI supercomplex on the presence of CpcG2. Coimmunoprecipitation experiments of NDH-1, using anti-NdhH and anti-NdhK polyclonal antibodies, were performed with the wild-type and ΔcpcG2 strains using a protein A-Sepharose resin. Subsequent western-blot analysis showed the presence of the PsaD subunit of PSI in the preparation recovered from the wild-type strain but not from the ΔcpcG2 strain (Fig. 5, A and B). The D1 protein of PSII was not recovered from either strain (Fig. 5, A and B). In addition, immunoblot analysis of all the PSI green bands separated by BN-PAGE indicated that, in the wild-type strain, NDH-1 associated only with PSI in band I (red arrow in the PSI supercomplex cluster; Fig. 5C), regardless of the presence of an overlap band of NDH-1M with PSI monomer in the strain (Fig. 5C). In contrast, such an association was absent in the ΔcpcG2 strain (Fig. 5D). Thus, CpcG2 is important in stabilization of the NDH-1-PSI supercomplex in Synechocystis 6803 and, thereby, is essential for the function of NDH-CET (Fig. 2).
Figure 5.
The presence of CpcG2 is necessary for the stabilization of the NDH-1-PSI supercomplex. A and B, Coimmunoprecipitation assay of the interaction of NDH-1 with PSI. Membrane proteins from the wild-type (WT) and ΔcpcG2 strains were incubated with protein A-Sepharose-coupled antiserum of anti-NdhH (A) or anti-NdhK (B). The immunoprecipitates (IP) were probed with specific antibodies, as indicated on the left. TM, Thylakoid membrane. C and D, Freshly isolated thylakoid membranes from wild-type (C) and ΔcpcG2 (D) strains were solubilized in 1.5% DM at a Chl a concentration of 0.25 µg µL−1, and the protein sample was separated by 5% to 12.5% BN-PAGE. Furthermore, thylakoid membrane complexes separated by BN-PAGE were subjected to 12% SDS-PAGE, and the proteins were immunodetected with specific antibodies against NdhH and NdhK, respectively. Red arrows in the PSI (s) cluster represents band I. PSI (s), PSI supercomplex; PSI (t), PSI trimer; PSI (d), PSI dimer; PSI (m), PSI monomer.
Deletion of CpcG2 Has Consequences for the Function of PSI
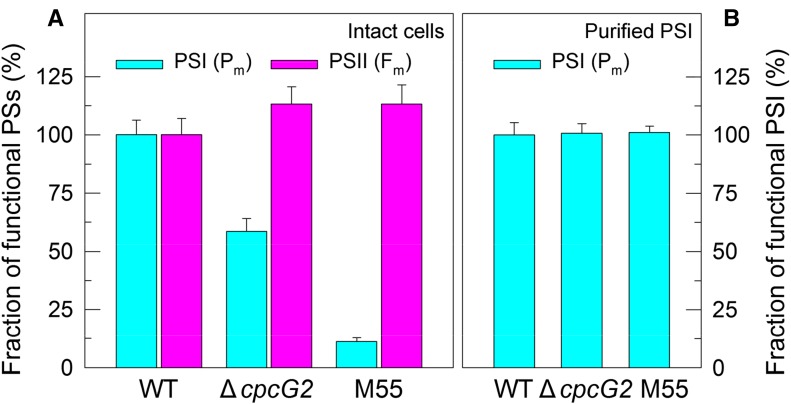
To obtain insights into the effects of CpcG2 on the function of PSI, we compared the fractions of functional PSI between the wild type and ∆cpcG2. The results indicated that deletion of CpcG2 significantly reduced the fraction of functional PSI (40%–50% reduction) under growth conditions, as deduced from the maximal P700 change (Pm) levels in DCMU-treated intact cells (Fig. 6A). The remaining functional fraction of PSI in ∆cpcG2 was still higher than that in M55, with about 90% reduction in the Pm level (Fig. 6A). In comparison, the deletion of CpcG2 did not decrease the fraction of functional PSII but rather increased it slightly under growth conditions, as deduced from the maximal fluorescence yield (Fm) values in intact cells (Fig. 6A). Taken together, these results suggest that the CpcG2 protein has a profound and direct effect on the function of PSI as an intrinsic component of the NDH-1-PSI supercomplex.
Figure 6.
Functional fractions of PSI and PSII in the wild type (WT), ΔcpcG2, and M55. Cells were grown under 2% CO2 at 40 µmol photons m−2 s−1 and were collected during their logarithmic growth phase. The Chl a concentration of intact cells (A) or purified PSI samples (B) in the medium was adjusted to 20 µg mL−1. Prior to the Pm measurements, DCMU (10 µm final concentration; the same below) was added to the medium containing intact cells or to the medium containing purified PSI, 200 µm 2,6-dichlorophenol-indophenol, 5 mm sodium ascorbate, and 1 mm methyl viologen. The functionality of PSI and PSII reaction centers was determined by the Pm and Fm parameters, respectively, expressed as a percentage of the wild type (100%). Values are means ± sd (n = 5). PSs, Photosystems.
Either impaired activity of PSI CET and/or an impairment of the functionality of PSI itself would explain the reduced fraction of functional PSI in DCMU-treated ΔcpcG2 intact cells. Deletion of CpcG2 impaired PSI CET (Fig. 2, D–F) and, consequently, in the presence of DCMU decreased the production of ATP and suppressed the Calvin-Benson cycle. Thus, it appears that the reduced fraction of functional PSI in ∆cpcG2 probably resulted from the charge recombination because of overreduction of PSI acceptor side. To test alternative possibilities, we purified the PSI complexes from the wild type and ΔcpcG2 and determined their fractions of functional PSI in the presence of exogenous PSI electron donor and acceptor (see “Materials and Methods”). In these circumstances, the Pm value reflects the functionality of PSI itself. As shown in Figure 6B, the deletion of CpcG2 did not alter the Pm value of isolated PSI complexes, consistent with the fact that the deletion did not affect the accumulation of the PSI complex in the thylakoid membrane (Fig. 3). Thus, it appears plausible that, in the absence of CpcG2, the impaired PSI CET but not the functionality of PSI as such decreases the fraction of functional PSI under growth conditions.
CpcG2 Is Essential to Stabilize NDH-1L and NDH-1M Complexes
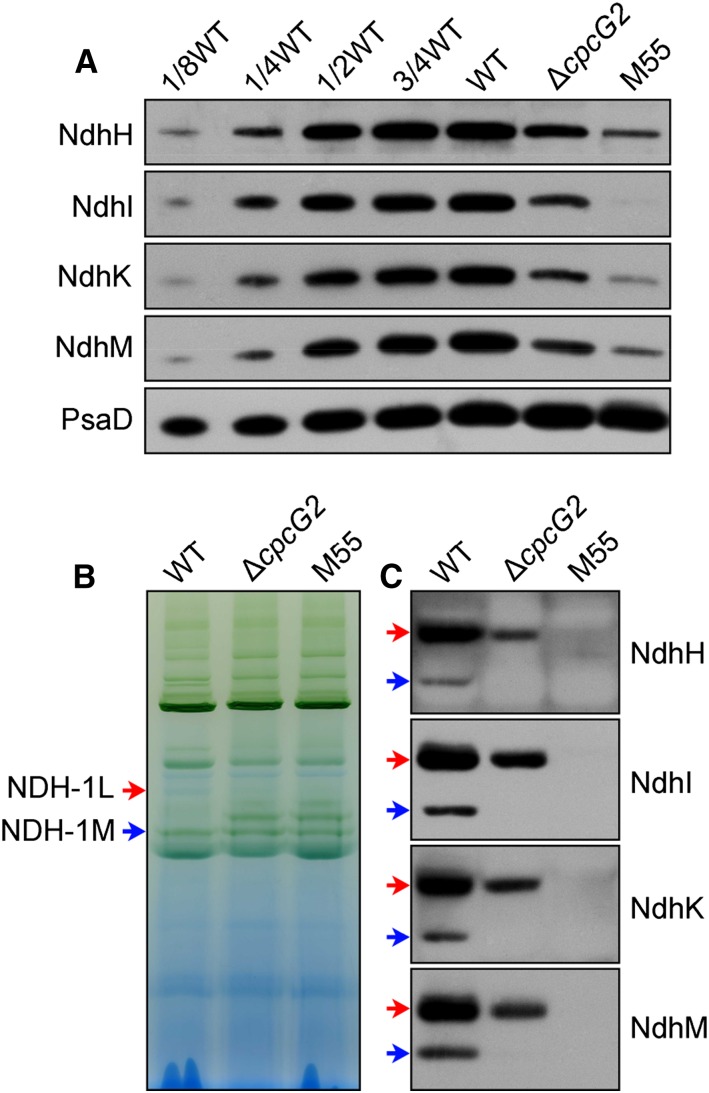
To investigate the effects of CpcG2 on the stabilization of NDH-1, we compared the accumulation and assembly of NDH-1L and NDH-1M complexes in wild-type, ΔcpcG2, and M55 strains. Deletion of cpcG2 significantly decreased the amount of NDH-1 subunits in the thylakoid membrane (Fig. 7A). Immunoblotting with antibodies specific to the NdhH, NdhK, NdhI, and NdhM subunits showed that the abundance of NDH-1L in the ΔcpcG2 mutant was about one-third of that in the wild type and that its degradation product NDH-1M had disappeared (Fig. 7, B and C). This indicated that the CpcG2 protein is essential to stabilize the majority of the NDH-1L complex, another component of the NDH-1-PSI supercomplex, corroborating its effect on NDH-CET (Fig. 2). Based on the above results, we conclude that the formation of the NDH-1L-PSI supercomplex is very important for the performance of NDH-1L and PSI in NDH-CET.
Figure 7.
Accumulation and assembly of NDH-1 complexes. A, Immunodetection of Ndh subunits in the thylakoid membranes from the wild type (WT; including the indicated serial dilutions) and ∆cpcG2 and M55 mutants. Protein blotting was performed with antibodies against several hydrophilic Ndh subunits (NdhH, NdhI, NdhK, and NdhM). Lanes were loaded with thylakoid membrane proteins corresponding to 1 µg of Chl a (100%). PsaD was used as a loading control. B, BN-PAGE profiles of the wild type and mutants used in A. Thylakoid membrane extract corresponding to 9 µg of Chl a was loaded on each lane. Red and blue arrows indicate the positions of NDH-1L and NDH-1M complexes, respectively. C, Western-blot analysis of the BN gel in B with antibodies against specific Ndh subunits, showing a cross-reaction with the NDH-1L and NDH-1M complexes.
Respiration Was Unaffected in the ∆cpcG2 Mutant
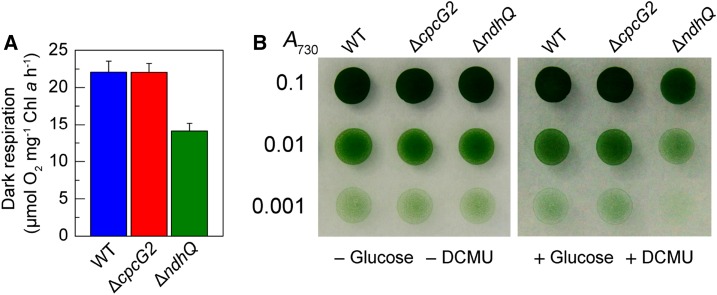
In addition to NDH-CET, NDH-1L also is involved in respiration (Zhang et al., 2004; Bernát et al., 2011). Hence, we examined the effect of cpcG2 deletion on respiration. The results indicated that deletion of cpcG2 influenced neither the respiration activity (Fig. 8A) nor the growth on plates with or without Glc and DCMU (Fig. 8B). This indicated that the NDH-1L in the NDH-1-PSI supercomplex is involved mainly in NDH-CET and suggests that the NDH-1L unconnected to the supercomplex may be responsible for respiration.
Figure 8.
Respiration of wild-type (WT) and mutant cells. A, Rate of oxygen uptake in the dark at 30°C of wild-type, ∆cpcG2, and ∆ndhQ cells grown under 2% (v/v) CO2. The Chl a concentration was adjusted to 10 µg mL−1 before measurement. Error bars indicate se (n = 5). B, Growth of wild-type, ∆cpcG2, and ∆ndhQ cells on agar plates in the absence (left) and presence (right) of Glc (5 mm) and DCMU (10 µm). Three microliters of cell suspensions with densities corresponding to A730 values of 0.1 (top rows), 0.01 (middle rows), and 0.001 (bottom rows) was spotted on agar plates and incubated under 2% (v/v) CO2 in air for 6 d at 40 µmol photons m−2 s−1.
DISCUSSION
The NDH-1L-PSI Supercomplex Is Formed in Synechocystis 6803
Recently, the NDH-1-PSI supercomplex was identified and characterized in angiosperms (Peng et al., 2008, 2009; Kouřil et al., 2014). Stable formation of the supercomplex additionally needs two LHCI proteins, Lhca5 and Lhca6, which link the PSI complex to NDH-1 (Peng et al., 2009). Although cyanobacteria lack the homologs of Lhca5 and Lhca6 (Peng et al., 2009), we discovered an NDH-1L-PSI supercomplex in the thylakoid membrane of Synechocystis 6803 as a high-molecular-mass protein complex (more than 1,000 kD) by BN-PAGE (see band I in Fig. 3). Instead of the Lhca5 and Lhca6 proteins, the Synechocystis 6803 NDH-1L-PSI is shown to require the presence of the CpcG2 protein, which belongs to PBS, the main light-harvesting antenna in cyanobacteria. Synechocystis 6803 has two distinct types of PBS that are assembled with two different CpcG copies (Kondo et al., 2005). The conventional PBS consists of peripheral rods and a central tricylindrical core with CpcG1. This antenna is not involved in the formation of the NDH-1L-PSI supercomplex, since deletion of CpcG1 and ApcD, its specific components, did not affect the intensity of band I (Supplemental Figs. S3–S5). Another type of PBS, comprising CpcG2, retains phycocyanin rods but is devoid of the typical allophycocyanin-containing central core (Kondo et al., 2005; Watanabe and Ikeuchi, 2013). CpcG2-PBS was characterized recently as the PSI-specific antenna (Kondo et al., 2007) capable of forming a CpcG2-PBS-PSI complex in cyanobacteria (Watanabe et al., 2014). Here, we demonstrate that CpcG2-PBS-PSI forms a supercomplex with NDH-1L in Synechocystis 6803.
The identity of the NDH-1-PSI supercomplex, represented by band I in Figure 3, was established by BN-PAGE of thylakoid membrane preparations obtained from wild-type and several mutant strains, followed by western blot (Fig. 3C) and LC-MS/MS (Table I; Supplemental Data Set S1). The NDH-1-PSI supercomplex was present in the wild type and WT-CpcG2-YH, while it was absent from the M55, ∆cpcG2, and PSI-less mutants (Fig. 3, A and B), confirming the necessity of all three components, NDH-1, CpcG2, and PSI. Furthermore, NdhD1 and NdhF1, the specific subunits of NDH-1L, were present in the NDH-1-PSI supercomplex of the wild-type strain (Table I; Supplemental Data Set S1). In line with this result, the ∆D1/D2 mutant lacked the band I corresponding to the NDH-1-PSI supercomplex (Fig. 3D), providing evidence that the NDH-1L complex is capable of forming the supercomplex in Synechocystis 6803.
Attempts to purify an intact NDH-1L-PSI supercomplex by Suc density gradient centrifugation were not successful. The supercomplex could not be separated from PSI by this method (Supplemental Figs. S6 and S7). Furthermore, amounts of the diagnostic NdhH or NdhK subunit in the PSI-containing heavy fractions were at most 6% of total NDH-1 in the cells, indicating that the supercomplex was not stable and mostly decomposed during isolation and DM treatment of thylakoid membranes. Nonetheless, the existence of NDH-1-PSI in Synechocystis 6803 was confirmed by coimmunoprecipitation (Fig. 4A) and pull-down assay (Fig. 4B). Regardless of the different parts of the CpcG2-YH protein employed as anchors, these two approaches produced similar results: subunits of both NDH-1 and PSI complexes were recovered from the WT-CpcG2-YH strain but not from the wild-type strain. It is important to note that the D1 subunit of PSII was not recovered from either strain, in agreement with the earlier observation that CpcG2 is a component of the PSI-specific antenna (Kondo et al., 2007; Watanabe and Ikeuchi, 2013). The high specificity of these two methods corroborates the fact that the NDH-1-PSI supercomplex is formed in Synechocystis 6803.
CpcG2-PBS Affects the Stabilization of NDH-1L and NDH-1M
The fragility of the NDH-1L-PSI supercomplex was evidently influenced by the instability of NDH-1L in the supercomplex. Indeed, the majority of NDH-1L was detected in its free form (Fig. 5C). The results of this study further showed that deletion of the CpcG2 protein significantly reduced the amounts of both NDH-1L and NDH-1M compared with the wild type (Fig. 7). The amount of NDH-1L was diminished to about one-third of the wild-type level, and NDH-1M disappeared (Fig. 7, B and C). In the PSI-less strain, however, the amount of NDH-1 subunits was similar to that in the WT-CpcG2-tagged strain (Fig. 3C), which is in a good agreement with previously published data reporting that NDH-1L remains stable in the PSI-less strain, generating only a small amount of NDH-1M (Zhang et al., 2004). CpcG2-PBS, in turn, has been characterized as the PSI-specific antenna (Kondo et al., 2007), localized mostly at the surface of the PSI complex and being capable of forming a CpcG2-PBS-PSI complex in cyanobacteria (Watanabe et al., 2014). Thus, the drastic effect of CpcG2 deletion on NDH-1L and NDH-1M appears to be indirect (Fig. 9).
Figure 9.
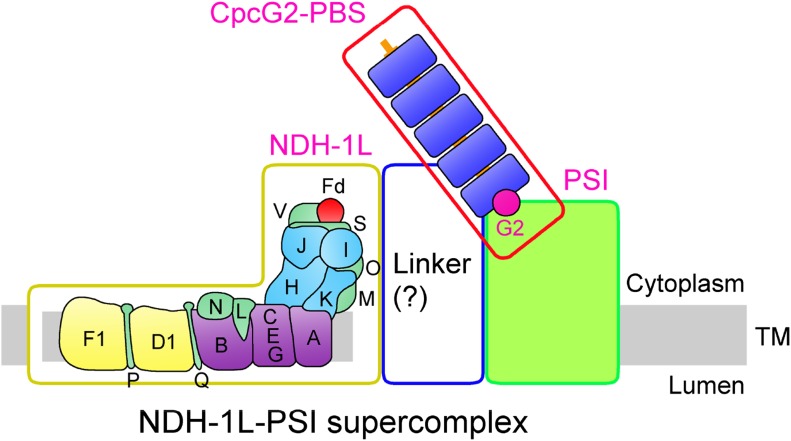
Schematic model of the NDH-1L-PSI supercomplex in cyanobacteria. CpcG2-PBS-PSI interacts with NDH-1L to form the NDH-1L-PSI supercomplex via an unidentified linker between NDH-1L and PSI. TM, Thylakoid membrane.
The reason for the instability of NDH-1L and NDH-1M in the absence of CpcG2 still remains unclear. It appears plausible that the CpcG2-PBS antenna interacts directly with a still unidentified linker between NDH-1L and PSI (question mark in Fig. 9), which is assumed to be necessary to stabilize the NDH-1L complex. Thus, we anticipate that, in the absence of CpcG2-PBS, an unknown linker would become exposed to an abnormal environment, resulting in the breakdown of the NDH-1L complex.
Model of the Cyanobacterial NDH-1L-PSI Supercomplex
The results described above, including patterns of NDH-1L-PSI degradation observed in various mutants, give some insights into the arrangement of components in the cyanobacterial NDH-1L-PSI supercomplex. At least three modules, NDH-1L, PSI, and the CpcG2-PBS antenna, form the supercomplex; the presence of proteins forming phycocyanin rods in the supercomplex was confirmed by mass spectrometry analysis (Table I; Supplemental Table S2; Supplemental Data Set S1), immunodetection (Supplemental Fig. S8A), and absorption spectrum (Supplemental Fig. S8B). Elimination of any of these three components abolished the supercomplex. Furthermore, the absence of the NDH-1L complex did not affect the stability of the PSI complex, and vice versa (Fig. 3, B and D). In contrast, the elimination of CpcG2 resulted in major losses of NDH-1L and NDH-1M complexes (Fig. 7, B and C). Taking all these results together, it appears that the NDH-1M domain interacts with the CpcG2-PBS antenna but not with PSI. However, the interaction may be mostly indirect, since the CpcG2-PBS antenna was recently assumed to be localized at the surface of the PSI complex to form a CpcG2-PBS-PSI complex (Watanabe et al., 2014). Based on the above analyses, we suggest that the CpcG2-PBS-PSI complex associates with NDH-1L to form an NDH-1L-PSI supercomplex, possibly via an unidentified linker between NDH-1L and PSI, as presented schematically in Figure 9.
Role of the NDH-1L-PSI Supercomplex in NDH-CET
In higher plants, the formation of the NDH-1-PSI supercomplex is not connected directly to NDH-1 activity, although its formation stabilizes the chloroplast NDH-1 complex, in particular under high-light conditions (Peng and Shikanai, 2011). We show here that, in Synechocystis 6803, in contrast to higher plant chloroplasts, the formation of the NDH-1L-PSI supercomplex is likely connected directly to the activity of NDH-CET (Figs. 1 and 2). This possibility is supported by the fact that the deletion of ndhL (M9 mutant) that impairs NDH-CET activity (Mi et al., 1992) abolishes the NDH-1L-PSI supercomplex but does not affect the assembly of NDH-1L and NDH-1M complexes even under high-light conditions (Supplemental Fig. S9).
The chloroplast NDH-1 complex was proposed to accept electrons from Fd, and NdhS was suggested to be the Fd-binding subunit based on the overall spatial homology of NdhS and the PsaE subunit of the PSI complex (Yamamoto et al., 2011; Yamamoto and Shikanai, 2013). Cyanobacterial NdhS is homologous to its counterpart in chloroplasts, and the interaction of NdhS with Fd has been demonstrated in the cyanobacterium Thermosynechococcus elongatus (He et al., 2015). Thus, Fd is likely to be an electron donor to cyanobacterial NDH-1 complexes, at least to NDH-1L. We propose that CpcG2-PBS has multiple roles in the process of electron donation from Fd to NDH-1L: first, it stabilizes the NDH-1M part of the NDH-1L complex, possibly via an unidentified linker (question mark in Fig. 9); second, it helps to position PSI in proximity to NDH-1L so that Fd reduced at PSI can be captured efficiently by NdhS, strongly facilitating NDH-CET; and third, it optimizes the PSI function, which is essential to NDH-CET.
Besides NdhS, NdhV was suggested recently to be a member of the Fd-binding domain structure (Gao et al., 2016). Deletion of NdhS or NdhV significantly impairs NDH-CET but has only little effect on respiration and the growth of cells under air at acidic pH (Battchikova et al., 2011a; Ma and Ogawa, 2015; Gao et al., 2016), similar to the results obtained with the ΔcpcG2 mutant (Figs. 1, 2, and 8; Supplemental Fig. S10). Similar properties of these three mutants lacking CpcG2 or components of the Fd-binding domain structure strongly suggest that formation of the NDH-1L-PSI supercomplex is important to stabilize the structure of the Fd-binding domain in the Synechocystis 6803 thylakoid membrane.
In addition to Fd, NADPH has been suggested as an alternative electron donor to NDH-1 in cyanobacteria (Mi et al., 1995; Matsuo et al., 1998; Deng et al., 2003a, 2003b; Ma et al., 2006; Ma and Mi, 2008; Hu et al., 2013; Ma and Ogawa, 2015). Electron donation from reduced Fd to NDH-1 may contribute principally to NDH-CET, while NADPH may contribute mainly to CO2 uptake and respiration, since deletion of NdhS or NdhV greatly impaired NDH-CET but did not affect CO2 uptake and respiration (Battchikova et al., 2011a; Ma and Ogawa, 2015; Gao et al., 2016). However, this hypothesis requires further investigation.
MATERIALS AND METHODS
Culture Conditions
A Glc-tolerant strain of wild-type Synechocystis sp. strain PCC 6803 and its mutants, ∆cpcG1, ∆cpcG2, ∆apcD, ∆ndhB (M55; Ogawa, 1991a), ∆ndhD1/ndhD2 (∆D1/D2; Ohkawa et al., 2000), ∆ndhD3/ndhD4 (∆D3/D4; Ohkawa et al., 2000), ∆ndhL (M9; Ogawa, 1991b, 1992), and WT-CpcG2-YH, were cultured at 30°C in BG-11 medium (Allen, 1968) buffered with Tris-HCl (5 mm; pH 8) by bubbling with 2% (v/v) CO2 in air. Solid medium was BG-11 supplemented with 1.5% agar. Continuous illumination was provided by fluorescent lamps at 40 µmol photons m−2 s−1. The PSI-less mutant (Shen et al., 1993) was grown in BG-11 medium supplemented with 5 mm Glc at 30°C at 5 µmol photons m−2 s−1. The mutant strains were grown in the presence of appropriate antibiotics.
Isolation and Construction of Mutants
A cosmid library of the Synechocystis 6803 genome was constructed. The library, which contained 105 clones with inserts of 35 to 38.5 kb, was subjected to in vitro transposon mutagenesis using the EZ-Tn5 < KAN-2 > Insertion Kit (Epicentre Biotechnologies) and then used to transform wild-type cells of Synechocystis 6803. Following transformation, cells were spread on 1.5% BG-11 agar plates supplemented with 5 µg mL−1 kanamycin, and KamR mutants that grew slowly under high light but normally under growth light were isolated. Genomic DNA isolated from each mutant was digested with HhaI and, after self-ligation, was used as a template for inverse PCR with primers (Supplemental Table S1) complementary to the N- and C-terminal regions of the KamR cassette. The exact position of the cassette in the mutant genome was determined by sequencing the PCR product.
The ΔcpcG2 mutant was constructed as follows. The upstream and downstream regions of sll1471 (cpcG2) were amplified by PCR, creating appropriate restriction sites. A DNA fragment encoding a KamR cassette also was amplified by PCR, creating KpnI and XbaI sites using appropriate PCR primers, cpcG2-C and cpcG2-D (Supplemental Table S1). These three PCR products were ligated into the multiple cloning site of pUC19 (Fig. 2A) and used to transform the wild-type cells of Synechocystis 6803 to generate the ΔcpcG2 mutant. The transformants were spread on agar plates containing BG-11 medium and kanamycin (10 µg mL−1) buffered at pH 8, and the plates were incubated in 2% (v/v) CO2 in air under illumination by fluorescent lamps at 40 µmol photons m−2 s−1. The mutated cpcG2 in the transformants was segregated to homogeneity (by successive streak purification) as determined by PCR amplification and reverse transcription (RT)-PCR analysis (Fig. 2, B and C).
The WT-CpcG2-YH mutant was constructed as follows. A DNA fragment containing cpcG2 and its upstream region was amplified by PCR, creating SalI and KpnI sites on both ends, and was ligated to the SalI and KpnI sites in the multiple cloning site of the pEYFP-His6-SpR plasmid (Birungi et al., 2010). A fragment containing the downstream region of cpcG2 also was amplified by PCR, creating EcoRI and SpeI sites, and was ligated to the downstream region of the SpR gene (Supplemental Fig. S2A). The vector thus constructed was used to transform the wild-type cells of Synechocystis 6803 to generate the WT-CpcG2-YH mutant strain. The transformation was performed as described previously (Williams and Szalay, 1983; Long et al., 2011). The yfp and his6 region in the transformants was segregated to homogeneity (by successive streak purification) as determined by PCR amplification (Supplemental Fig. S2B).
RNA Extraction and RT-PCR Analysis
Total RNA was isolated and analyzed as described previously (McGinn et al., 2003). RT-PCR was performed using the Access RT-PCR system (Promega) to generate products corresponding to cpcG1, cpcG2, apcD, and 16 S rRNA, with 0.5 µg of DNase-treated total RNA as starting material. RT-PCR conditions were 95°C for 5 min followed by cycles of 95°C, 62°C, and 72°C for 30 s each. The reactions were stopped after 25 cycles for 16 S rRNA and after 35 cycles for cpcG1, cpcG2, and apcD. The primers used are summarized in Supplemental Table S1.
Chl Fluorescence and P700 Analysis
The transient rise in Chl fluorescence after AL had been turned off was monitored as described (Ma and Mi, 2005). The redox kinetics of P700 was measured according to previously described methods (Battchikova et al., 2011a; Dai et al., 2013; Zhang et al., 2014; Zhao et al., 2014, 2015). The rereduction of P700+ in darkness was measured with a Dual-PAM-100 (Walz) with an emitter-detector unit (ED-101US/MD) by monitoring absorbance changes at 830 nm and using 875 nm as a reference. Cells were kept in the dark for 2 min, and 10 µm DCMU was added to the cultures prior to the measurement. The P700 was oxidized by far-red light with a maximum at 720 nm from a light-emitting diode lamp for 30 s, and the subsequent rereduction of P700+ in the dark was monitored.
The amounts of functional PSI and PSII reaction centers were estimated by the Pm and Fm parameters, respectively. The Chl a concentration of intact cells or PSI isolated by Suc density gradient (Supplemental Figs. S6 and S7) in the medium was adjusted to 20 µg mL−1. Prior to the Pm measurements, DCMU (10 µm final concentration; the same below) was added to the medium containing intact cells or to the medium containing PSI, 200 µm 2,6-dichlorophenol-indophenol, 5 mm sodium ascorbate, and 1 mm methyl viologen. The Pm values were determined using a saturation pulse (100 ms; 10,000 µmol photons m−2 s−1) under an FR background (720 nm; about 0.3 µmol photons m−2 s−1) as described previously (Klüghammer and Schreiber, 2008).
Isolation of Crude Thylakoid Membranes
The cell cultures (800 mL) were harvested at the logarithmic phase and washed twice by suspending in 50 mL of fresh BG-11 medium, and the thylakoid membranes were isolated according to Gombos et al. (1994) with some modifications as follows. The cells suspended in 5 mL of disruption buffer (10 mm HEPES-NaOH, 5 mm sodium phosphate, pH 7.5, 10 mm MgCl2, 10 mm NaCl, and 25% [v/v] glycerol) were supplemented by zirconia/silica beads and broken by vortexing 20 times at the highest speed for 30 s at 4°C with 5 min of cooling on ice between the runs. The crude extract was centrifuged at 5,000g for 5 min to remove the glass beads and unbroken cells. By further centrifugation at 20,000g for 30 min, we obtained crude thylakoid membranes from the precipitation.
Electrophoresis and Immunoblotting
BN-PAGE of Synechocystis 6803 membranes was performed as described previously (Kügler et al., 1997) with slight modifications (Battchikova et al., 2011a; Dai et al., 2013; Zhang et al., 2014; Zhao et al., 2014, 2015). Isolated membranes were prepared for BN-PAGE as follows. Membranes were washed with 330 mm sorbitol, 50 mm Bis-Tris, pH 7, and 0.5 mm phenylmethylsulfonyl fluoride (PMSF; Sigma) and resuspended in 20% (w/v) glycerol, 25 mm Bis-Tris, pH 7, 10 mm MgCl2, 0.1 units of RNase-free DNase RQ1 (Promega) at a Chl a concentration of 0.25 mg mL−1, and 0.5 mm PMSF. The samples were incubated on ice for 10 min, and an equal volume of 3% DM was added. Solubilization was performed for 40 min on ice. Insoluble components were removed by centrifugation at 18,000g for 15 min. The collected supernatant was mixed with one-tenth volume of sample buffer, 5% Serva Blue G, 100 mm Bis-Tris, pH 7, 30% (w/v) Suc, 500 mm ε-amino-n-caproic acid, and 10 mm EDTA. Solubilized membranes were then applied to a 0.75-mm-thick, 5% to 12.5% acrylamide gradient gel (Hoefer Mighty Small mini-vertical unit). Samples were loaded on an equal Chl a basis per lane. Electrophoresis was performed at 4°C by increasing the voltage gradually from 50 up to 200 V during the 5.5-h run.
For electrophoresis in the second dimension, several lanes of the BN gel were cut out and incubated in Laemmli SDS sample buffer containing 5% β-mercaptoethanol and 6 m urea for 1 h at 25°C. The lanes were then laid onto a 1-mm-thick 12% polyacrylamide gel with 6 m urea (Laemmli, 1970). After electrophoresis, the proteins were visualized by Coomassie Brilliant Blue staining.
For immunoblotting, the proteins were electrotransferred to a polyvinylidene difluoride membrane (Immobilon-P; Millipore) and detected by protein-specific antibodies using an ECL assay kit (Amersham) according to the manufacturer’s protocol. Antibodies against NdhA and NdhN proteins of Synechocystis 6803 were raised in our laboratory. Primer sequences used to amplify the nucleotide sequences encoding amino acids 54 to 93 of NdhA and the entire NdhN are listed in Supplemental Table S1. The PCR products were ligated into the pGEX-5X-1 and pET32a vectors, respectively, and the constructs were amplified in Escherichia coli DH-5α. The plasmids were used to transform E. coli strain BL21 (DE3) pLysS for protein expression. The expression products from E. coli were purified and used as antigens to immunize rabbits to produce polyclonal antibodies. Antibodies against NdhH, NdhI, NdhK, and NdhM were raised previously in our laboratory (Ma and Mi, 2005; Zhao et al., 2014). Antibody against His was purchased from Shanghai Immune Biotech, and antibodies against GFP, PSI subunits (PsaA, PsaB, and PsaD), a PSII subunit (D1), and a Cyt b6f subunit (Cyt f) were purchased from Agrisera.
Peptide Preparation for Tandem Mass Spectrometry Analysis
Thylakoid membrane complexes isolated from the wild-type strain were solubilized and subjected to BN-PAGE. Coomassie Brilliant Blue-stained band I from the wild type (pink arrows in Fig. 3A) was excised from the BN gel. Peptide preparation and liquid chromatography-electrospray ionization-tandem mass spectrometry analyses were performed as described previously (Battchikova et al., 2011a). The excised bands were treated twice with 50 mm ammonium bicarbonate in 30% (v/v) acetonitrile for 10 min and 100% (v/v) acetonitrile for 15 min and then dried in a vacuum concentrator. The dried gel pieces were treated with 0.01 mg mL−1 trypsin (sequence grade; Promega) and 50 mm ammonium bicarbonate and incubated at 37°C for 20 h. The digested peptides in the gel pieces were recovered twice with 20 µL of 5% (v/v) formic acid and 50% (v/v) acetonitrile. The extracted peptides were combined and then dried in a vacuum concentrator.
Mass Spectrometry Analysis and Database Searching
LC-MS/MS analyses were performed on a Q-Exactive mass spectrometer (Thermo Finnigan) coupled with an Easy-nLC1000 HPLC system (Thermo Fisher Scientific). Trypsin-digested peptides were dissolved in 12 µL of 2% formic acid, loaded onto a C18 reverse-phase column (10 cm × 75 μm i.d.) in buffer A (2% acetonitrile and 0.1% formic acid), and separated by a linear gradient of buffer B (80% acetonitrile and 0.1% formic acid) at a flow rate of 250 nL min−1 controlled by IntelliFlow technology over 90 min. Mass spectrometry data were acquired using a data-dependent top-10 method dynamically choosing the most abundant precursor ions from the survey scan (300–1,800 mass-to-charge ratio [m/z]) for high-energy collisional dissociation fragmentation. Survey scans were acquired at a resolution of 70,000 at m/z 200, and the resolution for high-energy collisional dissociation spectra was set to 17,500 at m/z 200. The raw data were processed with Protein Discoverer software version 1.1 (Thermo Fisher Scientific) using default parameters to generate concatenated Mascot generic files. Database searches were performed using an in-house Mascot server (version 2.2) against a UniProt database of Synechocystis 6803 proteins. The search criteria allowed for one missed cleavage of trypsin, oxidation of Met, and 6-ppm and 0.1-D mass accuracies for mass spectrometry and tandem mass spectrometry modes, respectively.
Suc Density Gradient Analysis
Freshly isolated thylakoid membranes were washed twice with buffer containing 5 mm sodium phosphate, 10 mm MgCl2, 10 mm NaCl, 0.15 m Suc, 10 mm HEPES-NaOH (pH 7.5), and 1 mm PMSF and then solubilized in the same buffer containing 1% DM at a Chl concentration of 1 mg mL−1 and 1 mm PMSF for 20 min on ice. After centrifugation at 20,000g for 10 min at 4°C, the cleared supernatant (200 µL) was loaded on top of an 11-mL linear Suc gradient (5%–40%) prepared with buffer containing 5 mm sodium phosphate, 10 mm MgCl2, 10 mm NaCl, 0.02% DM, and 10 mm HEPES-NaOH (pH 7.5). Thylakoid membrane protein complexes were separated by ultracentrifuge for 8 h at 110,000g at 4°C in a P40ST rotor (Hitachi). The gradients were then fractionated from top to bottom into 36 fractions by a microliter syringe, and equal amounts of fractions were used for further immunoblot analysis.
Coimmunoprecipitation
The coimmunoprecipitation assay was performed as described previously (Komenda et al., 2005) with some modifications. Two milligrams of thylakoid membrane protein isolated from WT-CpcG2-YH or the wild-type strain in 500 µL of TNE buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 2 mm EDTA, and 1 mm PMSF) was solubilized with 0.05% (w/w) DM for 30 min at 4°C. After centrifugation at 20,000g for 10 min at 4°C, the supernatant was incubated with 20 µL of anti-GFP serum overnight at 4°C with gentle rotation. The solution was then incubated with an equal amount of protein A-Sepharose (GE Healthcare) for 2 h at 4°C. The protein A-Sepharose 4B with bound protein immunoglobulin complexes was subsequently sedimented and washed twice with 1 mL of TNE buffer containing 0.05% (w/w) DM and twice with TNE buffer. The beads were then suspended in 30 µL of 1× SDS (2%) loading buffer and then boiled for 5 min. After centrifugation, the supernatant was collected and subjected to immunoblot analysis.
Affinity Chromatography
Purification of protein complexes containing the His6 tag was performed using a nickel-nitrilotriacetic His-Bind resin column (Novagen). Chromatography buffers contained 20 mm HEPES-KOH, pH 7.5, 10% (w/v) glycerol, and 100 mm NaCl supplemented with 10, 20, and 250 mm imidazole for binding, washing, and elution of the protein complexes, respectively.
Growth Curve
The cell density of wild-type and ∆cpcG2 cells cultured at growth light and high light was determined every 12 and 6 h, respectively, and that of wild-type, ∆cpcG2, and ∆D3/D4 cells cultured under pH 6.5 and 2% (v/v) CO2 in air or air was determined every 12 h, by measuring the A730 using a spectrophotometer (UV3000; Shimadzu).
Wild-type, ∆cpcG2, and ∆D3/D4 cells (1.5 mL) were collected by centrifugation, and pigments in the pellet were extracted by 1.5 mL of methyl alcohol. Chl a in the extract was determined using a spectrophotometer (UV3000; Shimadzu) according to the formula Chl a (µg mL−1) = 13.9 × A665 (Arnon, 1949).
Dark Respiration
Oxygen uptake was measured at 30°C (Hansatech) with a Clark-type oxygen electrode according to the method of Ma and Mi (2005). Cells were suspended in fresh BG-11 medium at a Chl a concentration of 10 µg mL−1.
Confocal Laser Scanning Microscopy
Cell imaging was performed using a laser scanning confocal microscope (FV1000; Olympus) with a 100×/1.4 plan apochromat oil-immersion objective and with an 80-µm confocal pinhole. The argon laser line, 515 nm, and the helium laser line, 543 nm, were used for the excitation of YFP and autofluorescence, respectively. YFP fluorescence emission was selected with a 515-nm dichroic mirror and a band-pass filter of 535 to 590 nm. Autofluorescence was collected through a 545-nm dichroic mirror and a long-pass filter of 560 nm.
Accession Numbers
Sequence data from this article can be found in the International Nucleotide Sequence Database Collaboration under the following accession numbers: CpcG1, NP_440438.2; CpcG2, NP_442922.1; NdhB, NP_441103.1; PsaA, NP_440757.1; and PsaB, NP_440758.1.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Growth curve of wild-type and ∆cpcG2 cells under different light intensities.
Supplemental Figure S2. Construction and characterization of the WT-CpcG2-YH strain.
Supplemental Figure S3. Construction and analysis of the cpcG1 deletion mutant.
Supplemental Figure S4. Construction and analysis of the apcD deletion mutant.
Supplemental Figure S5. Analysis of band I in the wild-type, ∆cpcG1, and ∆apcD strains.
Supplemental Figure S6. Suc density gradient analysis of NDH-1 complexes isolated from the WT-CpcG2-YH strain.
Supplemental Figure S7. Suc density gradient analysis of thylakoid membrane protein complexes isolated from the WT-CpcG2-YH and ∆cpcG2 strains.
Supplemental Figure S8. Identification of phycobilisome components in the NDH-1L-PSI supercomplex.
Supplemental Figure S9. Analysis of NDH-1L-PSI, NDH-1L, and NDH-1M in wild-type and M9 strains.
Supplemental Figure S10. Growth curve of wild-type, ∆cpcG2, and ∆D3/D4 cells under different CO2 concentrations at pH 6.5.
Supplemental Table S1. Primers used in this study.
Supplemental Table S2. Summary of the CpcG2-PBS antenna components identified from Q-Exactive mass analysis of the NDH-1-PSI supercomplex (band I).
Supplemental Data Set S1. Total proteins identified from band I.
Supplementary Material
Acknowledgments
We thank Toshiharu Shikanai (Kyoto University) for reading the article with critical comments and Wim Vermaas (Arizona State University) for the PSI-less mutant; mass spectrometry was carried out at Shanghai Applied Protein Technology; confocal microscopy was performed at the Shanghai Institute of Plant Physiology and Ecology, and Dr. Xiaoshu Gao of the Institute is thanked for help in cell imaging.
Glossary
- CET
cyclic electron transport
- NDH-CET
NDH-1-dependent cyclic electron transport
- Fd
ferredoxin
- Chl
chlorophyll
- AL
actinic light
- FR
far-red light
- DCMU
3-(3,4-dichlorophenyl)-1,1-dimethylurea
- BN
blue native
- Cyt
cytochrome
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- DM
n-dodecyl β-d-maltoside
- RT
reverse transcription
- PMSF
phenylmethylsulfonyl fluoride
- m/z
mass-to-charge ratio
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31370270 and 31570235 to W.M.), the Shanghai Natural Science Foundation (grant no. 14ZR1430000 to W.M.), the Academy of Finland (project no. 273870 and Center of Excellence project no. 271832 to E.-M.A.), and the People Program (Marie Curie Actions) of the European Union’s Seventh Framework Program FP7/2007–2013/ (Research Executive Agency grant no. 317184, PHOTO.COMM, to E.-M.A.).
References
- Alboresi A, Caffarri S, Nogue F, Bassi R, Morosinotto T (2008) In silico and biochemical analysis of Physcomitrella patens photosynthetic antenna: identification of subunits which evolved upon land adaptation. PLoS ONE 3: e2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen MM. (1968) Simple conditions for growth of unicellular blue-green algae on plates. J Phycol 4: 1–4 [DOI] [PubMed] [Google Scholar]
- Armbruster U, Rühle T, Kreller R, Strotbek C, Zühlke J, Tadini L, Blunder T, Hertle AP, Qi Y, Rengstl B, et al. (2013) The photosynthesis affected mutant68-like protein evolved from a PSII assembly factor to mediate assembly of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis. Plant Cell 25: 3926–3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon DI. (1949) Copper enzymes in isolated chloroplasts: polyphenoloxidase in Beta vulgaris. Plant Physiol 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arteni AA, Zhang P, Battchikova N, Ogawa T, Aro EM, Boekema EJ (2006) Structural characterization of NDH-1 complexes of Thermosynechococcus elongatus by single particle electron microscopy. Biochim Biophys Acta 1757: 1469–1475 [DOI] [PubMed] [Google Scholar]
- Battchikova N, Aro EM (2007) Cyanobacterial NDH-1 complexes: multiplicity in function and subunit composition. Physiol Plant 131: 22–32 [DOI] [PubMed] [Google Scholar]
- Battchikova N, Eisenhut M, Aro EM (2011b) Cyanobacterial NDH-1 complexes: novel insights and remaining puzzles. Biochim Biophys Acta 1807: 935–944 [DOI] [PubMed] [Google Scholar]
- Battchikova N, Wei L, Du L, Bersanini L, Aro EM, Ma W (2011a) Identification of novel Ssl0352 protein (NdhS), essential for efficient operation of cyclic electron transport around photosystem I, in NADPH:plastoquinone oxidoreductase (NDH-1) complexes of Synechocystis sp. PCC 6803. J Biol Chem 286: 36992–37001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernát G, Appel J, Ogawa T, Rögner M (2011) Distinct roles of multiple NDH-1 complexes in the cyanobacterial electron transport network as revealed by kinetic analysis of P700+ reduction in various Ndh-deficient mutants of Synechocystis sp. strain PCC6803. J Bacteriol 193: 292–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birungi M, Folea M, Battchikova N, Xu M, Mi H, Ogawa T, Aro EM, Boekema EJ (2010) Possibilities of subunit localization with fluorescent protein tags and electron microscopy exemplified by a cyanobacterial NDH-1 study. Biochim Biophys Acta 1797: 1681–1686 [DOI] [PubMed] [Google Scholar]
- Böhm R, Sauter M, Böck A (1990) Nucleotide sequence and expression of an operon in Escherichia coli coding for formate hydrogenlyase components. Mol Microbiol 4: 231–243 [DOI] [PubMed] [Google Scholar]
- Brandt U, Kerscher S, Dröse S, Zwicker K, Zickermann V (2003) Proton pumping by NADH:ubiquinone oxidoreductase: a redox driven conformational change mechanism? FEBS Lett 545: 9–17 [DOI] [PubMed] [Google Scholar]
- Burrows PA, Sazanov LA, Svab Z, Maliga P, Nixon PJ (1998) Identification of a functional respiratory complex in chloroplasts through analysis of tobacco mutants containing disrupted plastid ndh genes. EMBO J 17: 868–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai H, Zhang L, Zhang J, Mi H, Ogawa T, Ma W (2013) Identification of a cyanobacterial CRR6 protein, Slr1097, required for efficient assembly of NDH-1 complexes in Synechocystis sp. PCC 6803. Plant J 75: 858–866 [DOI] [PubMed] [Google Scholar]
- Deng Y, Ye J, Mi H (2003a) Effects of low CO2 on NAD(P)H dehydrogenase, a mediator of cyclic electron transport around photosystem I in the cyanobacterium Synechocystis PCC6803. Plant Cell Physiol 44: 534–540 [DOI] [PubMed] [Google Scholar]
- Deng Y, Ye JY, Mi HL, Shen YG (2003b) [Separation of hydrophobic NAD(P)H dehydrogenase subcomplexes from cyanobacterium Synechocystis PCC6803]. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai) 35: 723–727 [PubMed] [Google Scholar]
- Efremov RG, Sazanov LA (2012) The coupling mechanism of respiratory complex I: a structural and evolutionary perspective. Biochim Biophys Acta 1817: 1785–1795 [DOI] [PubMed] [Google Scholar]
- Friedrich T, Scheide D (2000) The respiratory complex I of bacteria, archaea and eukarya and its module common with membrane-bound multisubunit hydrogenases. FEBS Lett 479: 1–5 [DOI] [PubMed] [Google Scholar]
- Friedrich T, Steinmüller K, Weiss H (1995) The proton-pumping respiratory complex I of bacteria and mitochondria and its homologue in chloroplasts. FEBS Lett 367: 107–111 [DOI] [PubMed] [Google Scholar]
- Friedrich T, Weiss H (1997) Modular evolution of the respiratory NADH:ubiquinone oxidoreductase and the origin of its modules. J Theor Biol 187: 529–540 [DOI] [PubMed] [Google Scholar]
- Gao F, Zhao J, Wang X, Qin S, Wei L, Ma W (2016) NdhV is a subunit of NADPH dehydrogenase essential for cyclic electron transport in Synechocystis sp. strain PCC 6803. Plant Physiol 170: 752–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gombos Z, Wada H, Murata N (1994) The recovery of photosynthesis from low-temperature photoinhibition is accelerated by the unsaturation of membrane lipids: a mechanism of chilling tolerance. Proc Natl Acad Sci USA 91: 8787–8791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Endo T, Peltier G, Tasaka M, Shikanai T (2003) A nucleus-encoded factor, CRR2, is essential for the expression of chloroplast ndhB in Arabidopsis. Plant J 36: 541–549 [DOI] [PubMed] [Google Scholar]
- He Z, Zheng F, Wu Y, Li Q, Lv J, Fu P, Mi H (2015) NDH-1L interacts with ferredoxin via the subunit NdhS in Thermosynechococcus elongatus. Photosynth Res 126: 341–349 [DOI] [PubMed] [Google Scholar]
- Hu P, Lv J, Fu P, Hualing M (2013) Enzymatic characterization of an active NDH complex from Thermosynechococcus elongatus. FEBS Lett 587: 2340–2345 [DOI] [PubMed] [Google Scholar]
- Ifuku K, Endo T, Shikanai T, Aro EM (2011) Structure of the chloroplast NADH dehydrogenase-like complex: nomenclature for nuclear-encoded subunits. Plant Cell Physiol 52: 1560–1568 [DOI] [PubMed] [Google Scholar]
- Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, et al. (1996) Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res 3: 109–136 [DOI] [PubMed] [Google Scholar]
- Klüghammer C, Schreiber U (2008) Saturation pulse method for assessment of energy conversion in PS I. PAM Application Notes 1: 11–14 [Google Scholar]
- Komenda J, Tichý M, Eichacker LA (2005) The PsbH protein is associated with the inner antenna CP47 and facilitates D1 processing and incorporation into PSII in the cyanobacterium Synechocystis PCC 6803. Plant Cell Physiol 46: 1477–1483 [DOI] [PubMed] [Google Scholar]
- Kondo K, Geng XX, Katayama M, Ikeuchi M (2005) Distinct roles of CpcG1 and CpcG2 in phycobilisome assembly in the cyanobacterium Synechocystis sp. PCC 6803. Photosynth Res 84: 269–273 [DOI] [PubMed] [Google Scholar]
- Kondo K, Mullineaux CW, Ikeuchi M (2009) Distinct roles of CpcG1-phycobilisome and CpcG2-phycobilisome in state transitions in a cyanobacterium Synechocystis sp. PCC 6803. Photosynth Res 99: 217–225 [DOI] [PubMed] [Google Scholar]
- Kondo K, Ochiai Y, Katayama M, Ikeuchi M (2007) The membrane-associated CpcG2-phycobilisome in Synechocystis: a new photosystem I antenna. Plant Physiol 144: 1200–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouřil R, Strouhal O, Nosek L, Lenobel R, Chamrád I, Boekema EJ, Šebela M, Ilík P (2014) Structural characterization of a plant photosystem I and NAD(P)H dehydrogenase supercomplex. Plant J 77: 568–576 [DOI] [PubMed] [Google Scholar]
- Kubota H, Sakurai I, Katayama K, Mizusawa N, Ohashi S, Kobayashi M, Zhang P, Aro EM, Wada H (2010) Purification and characterization of photosystem I complex from Synechocystis sp. PCC 6803 by expressing histidine-tagged subunits. Biochim Biophys Acta 1797: 98–105 [DOI] [PubMed] [Google Scholar]
- Kügler M, Jänsch L, Kruft V, Schmitz UK, Braun HP (1997) Analysis of the chloroplast protein complexes by blue-native polyacrylamide gel electrophoresis (BN-PAGE). Photosynth Res 53: 35–44 [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Long Z, Zhao J, Zhang J, Wei L, Wang Q, Ma W (2011) Effects of different light treatments on the natural transformation of Synechocystis sp. strain PCC 6803. Afr J Microbiol Res 5: 3603–3610 [Google Scholar]
- Ma W, Deng Y, Ogawa T, Mi H (2006) Active NDH-1 complexes from the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Cell Physiol 47: 1432–1436 [DOI] [PubMed] [Google Scholar]
- Ma W, Mi H (2005) Expression and activity of type 1 NAD(P)H dehydrogenase at different growth phases of the cyanobacterium, Synechocystis PCC6803. Physiol Plant 125: 135–140 [Google Scholar]
- Ma W, Mi H (2008) Effect of exogenous glucose on the expression and activity of NADPH dehydrogenase complexes in the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Physiol Biochem 46: 775–779 [DOI] [PubMed] [Google Scholar]
- Ma W, Ogawa T (2015) Oxygenic photosynthesis-specific subunits of cyanobacterial NADPH dehydrogenases. IUBMB Life 67: 3–8 [DOI] [PubMed] [Google Scholar]
- Matsuo M, Endo T, Asada K (1998) Properties of the respiratory NAD(P)H dehydrogenase isolated from the cyanobacterium Synechocystis PCC6803. Plant Cell Physiol 39: 263–267 [DOI] [PubMed] [Google Scholar]
- McGinn PJ, Price GD, Maleszka R, Badger MR (2003) Inorganic carbon limitation and light control the expression of transcripts related to the CO2-concentrating mechanism in the cyanobacterium Synechocystis sp. strain PCC6803. Plant Physiol 132: 218–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Endo T, Ogawa T, Asada K (1995) Thylakoid membrane-bound, NADPH-specific pyridine nucleotide dehydrogenase complex mediated cyclic electron transport in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol 36: 661–668 [Google Scholar]
- Mi H, Endo T, Schreiber U, Ogawa T, Asada K (1992) Electron donation from cyclic and respiratory flows to the photosynthetic intersystem chain is mediated by pyridine nucleotide dehydrogenase in the cyanobacterium Synechocystis PCC 6803. Plant Cell Physiol 33: 1233–1237 [Google Scholar]
- Neilson JAD, Durnford DG (2010) Structural and functional diversification of the light-harvesting complexes in photosynthetic eukaryotes. Photosynth Res 106: 57–71 [DOI] [PubMed] [Google Scholar]
- Ogawa T. (1991a) A gene homologous to the subunit-2 gene of NADH dehydrogenase is essential to inorganic carbon transport of Synechocystis PCC6803. Proc Natl Acad Sci USA 88: 4275–4279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T. (1991b) Cloning and inactivation of a gene essential to inorganic carbon transport of Synechocystis PCC6803. Plant Physiol 96: 280–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T. (1992) Identification and characterization of the ictA/ndhL gene product essential to inorganic carbon transport of Synechocystis PCC6803. Plant Physiol 99: 1604–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T, Mi H (2007) Cyanobacterial NADPH dehydrogenase complexes. Photosynth Res 93: 69–77 [DOI] [PubMed] [Google Scholar]
- Ohkawa H, Pakrasi HB, Ogawa T (2000) Two types of functionally distinct NAD(P)H dehydrogenases in Synechocystis sp. strain PCC6803. J Biol Chem 275: 31630–31634 [DOI] [PubMed] [Google Scholar]
- Peltier G, Aro EM, Shikanai T (2016) NDH-1 and NDH-2 plastoquinone reductases in oxygenic photosynthesis. Annu Rev Plant Biol 67: 55–80 [DOI] [PubMed] [Google Scholar]
- Peng L, Fukao Y, Fujiwara M, Shikanai T (2012) Multistep assembly of chloroplast NADH dehydrogenase-like subcomplex A requires several nucleus-encoded proteins, including CRR41 and CRR42, in Arabidopsis. Plant Cell 24: 202–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Fukao Y, Fujiwara M, Takami T, Shikanai T (2009) Efficient operation of NAD(P)H dehydrogenase requires supercomplex formation with photosystem I via minor LHCI in Arabidopsis. Plant Cell 21: 3623–3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Fukao Y, Myouga F, Motohashi R, Shinozaki K, Shikanai T (2011b) A chaperonin subunit with unique structures is essential for folding of a specific substrate. PLoS Biol 9: e1001040. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Peng L, Shikanai T (2011) Supercomplex formation with photosystem I is required for the stabilization of the chloroplast NADH dehydrogenase-like complex in Arabidopsis. Plant Physiol 155: 1629–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Shimizu H, Shikanai T (2008) The chloroplast NAD(P)H dehydrogenase complex interacts with photosystem I in Arabidopsis. J Biol Chem 283: 34873–34879 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Peng L, Yamamoto H, Shikanai T (2011a) Structure and biogenesis of the chloroplast NAD(P)H dehydrogenase complex. Biochim Biophys Acta 1807: 945–953 [DOI] [PubMed] [Google Scholar]
- Shen G, Boussiba S, Vermaas WF (1993) Synechocystis sp PCC 6803 strains lacking photosystem I and phycobilisome function. Plant Cell 5: 1853–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T, Endo T, Hashimoto T, Yamada Y, Asada K, Yokota A (1998) Directed disruption of the tobacco ndhB gene impairs cyclic electron flow around photosystem I. Proc Natl Acad Sci USA 95: 9705–9709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirpiö S, Allahverdiyeva Y, Holmström M, Khrouchtchova A, Haldrup A, Battchikova N, Aro EM (2009) Novel nuclear-encoded subunits of the chloroplast NAD(P)H dehydrogenase complex. J Biol Chem 284: 905–912 [DOI] [PubMed] [Google Scholar]
- Ueda M, Kuniyoshi T, Yamamoto H, Sugimoto K, Ishizaki K, Kohchi T, Nishimura Y, Shikanai T (2012) Composition and physiological function of the chloroplast NADH dehydrogenase-like complex in Marchantia polymorpha. Plant J 72: 683–693 [DOI] [PubMed] [Google Scholar]
- Wang P, Duan W, Takabayashi A, Endo T, Shikanai T, Ye JY, Mi H (2006) Chloroplastic NAD(P)H dehydrogenase in tobacco leaves functions in alleviation of oxidative damage caused by temperature stress. Plant Physiol 141: 465–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Gao F, Zhang J, Zhao J, Ogawa T, Ma W (2016) A cytoplasmic protein Ssl3829 is important for NDH-1 hydrophilic arm assembly in Synechocystis sp. strain PCC 6803. Plant Physiol 171: 864–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Ikeuchi M (2013) Phycobilisome: architecture of a light-harvesting supercomplex. Photosynth Res 116: 265–276 [DOI] [PubMed] [Google Scholar]
- Watanabe M, Semchonok DA, Webber-Birungi MT, Ehira S, Kondo K, Narikawa R, Ohmori M, Boekema EJ, Ikeuchi M (2014) Attachment of phycobilisomes in an antenna-photosystem I supercomplex of cyanobacteria. Proc Natl Acad Sci USA 111: 2512–2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JGK, Szalay AA (1983) Stable integration of foreign DNA into the chromosome of the cyanobacterium Synechococcus R2. Gene 24: 37–51 [DOI] [PubMed] [Google Scholar]
- Yagi T, Yano T, Di Bernardo S, Matsuno-Yagi A (1998) Procaryotic complex I (NDH-1), an overview. Biochim Biophys Acta 1364: 125–133 [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Peng L, Fukao Y, Shikanai T (2011) An Src homology 3 domain-like fold protein forms a ferredoxin binding site for the chloroplast NADH dehydrogenase-like complex in Arabidopsis. Plant Cell 23: 1480–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Shikanai T (2013) In planta mutagenesis of Src homology 3 domain-like fold of NdhS, a ferredoxin-binding subunit of the chloroplast NADH dehydrogenase-like complex in Arabidopsis: a conserved Arg-193 plays a critical role in ferredoxin binding. J Biol Chem 288: 36328–36337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Gao F, Zhao J, Ogawa T, Wang Q, Ma W (2014) NdhP is an exclusive subunit of large complex of NADPH dehydrogenase essential to stabilize the complex in Synechocystis sp. strain PCC 6803. J Biol Chem 289: 18770–18781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Battchikova N, Jansen T, Appel J, Ogawa T, Aro EM (2004) Expression and functional roles of the two distinct NDH-1 complexes and the carbon acquisition complex NdhD3/NdhF3/CupA/Sll1735 in Synechocystis sp PCC 6803. Plant Cell 16: 3326–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Gao F, Zhang J, Ogawa T, Ma W (2014) NdhO, a subunit of NADPH dehydrogenase, destabilizes medium size complex of the enzyme in Synechocystis sp. strain PCC 6803. J Biol Chem 289: 26669–26676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Rong W, Gao F, Ogawa T, Ma W (2015) Subunit Q is required to stabilize the large complex of NADPH dehydrogenase in Synechocystis sp. strain PCC 6803. Plant Physiol 168: 443–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.