TWISTED SEED1 is a novel small protein located in the endomembrane system of Arabidopsis that regulates various aspects of seed and plant development.
Abstract
Small proteins have long been overlooked due to their poor annotation and the experimental challenges they pose. However, in recent years, their role in various processes has started to emerge, opening new research avenues. Here, we present the isolation and characterization of two allelic mutants, twisted seed1-1 (tws1-1) and tws1-2, which exhibit an array of developmental and biochemical phenotypes in Arabidopsis (Arabidopsis thaliana) seeds. We have identified AT5G01075 as the subtending gene encoding a small protein of 81 amino acids localized in the endoplasmic reticulum. TWS1 is strongly expressed in seeds, where it regulates both embryo development and accumulation of storage compounds. TWS1 loss-of-function seeds exhibit increased starch, sucrose, and protein accumulation at the detriment of fatty acids. TWS1 is also expressed in vegetative and reproductive tissues, where it is responsible for proper epidermal cell morphology and overall plant growth. At the cellular level, TWS1 is responsible for cuticle deposition on epidermal cells and organization of the endomembrane system. Finally, we show that TWS1 is a single-copy gene in Arabidopsis, and it is specifically conserved among angiosperms.
Developing seeds are a complex system that integrates processes implicated in the coordinated development of different tissues and the intensive activation of many metabolic pathways aimed at massive accumulation of storage compounds and secondary metabolites. As a result, young seedlings are provided with the energy and the building materials required to restarting the plant life cycle.
In oleaginous species, such as Arabidopsis (Arabidopsis thaliana), seeds typically contain 30% to 40% storage lipids and 30% to 40% proteins by weight (Baud et al., 2008). In Arabidopsis, these compounds are produced and stored by the embryo, which fills almost entirely the seed. Large amounts of starch are accumulated in the early phases of seed development, and they are later metabolized to produce proteins and lipids. Oils are accumulated as triacylglycerols (TAGs) produced in the endoplasmic reticulum (ER) and they are eventually stored in specialized ER-originated vesicles called oleosomes (Hills, 2004). Proteins, on the other hand, are synthesized on rough ER and then sorted to the protein storage vacuoles (PSVs) where they are processed to their mature form (Hills, 2004). Secondary metabolites are also accumulated in the vacuoles of the maternally derived tissues of the testa and serve complementary roles like protection against biotic and abiotic stresses and as developmental regulators (Lepiniec et al., 2006). Furthermore, during seed development, cell expansion and division are sustained by a constant supply of proteins and lipids to generate new plasma membrane (PM) and cell wall.
Therefore, a flawless secretory pathway becomes crucial to ensure the synthesis and storage of many compounds and thus to produce viable seeds. Indeed, many mutations occurring in proteins involved in the secretory machinery exhibit defects in storage compound accumulation. For example, Arabidopsis plants lacking the vacuolar sorting receptor1 missort storage proteins and secrete them from cells, resulting in the abnormal accumulation of 12S globulin and 2S albumin precursors in seeds (Shimada et al., 2003).
Moreover, plants rely on the secretory pathway to synthesize and export cell wall components. As the seed grows, cuticle is deposited on the embryo surface and on seed integuments. The cuticle provides a hydrophobic barrier that is believed to physically define organ boundaries, and thus it is fundamental for proper morphogenesis. Many mutants showing organ fusion also exhibit defects in cuticle biosynthesis and/or deposition (Lolle et al., 1998; Yang et al., 2008; Tsuwamoto et al., 2008; Pighin et al., 2004; To et al., 2012).
In the last years, many studies based on the analysis of large datasets have been carried on in order to acquire a deeper understanding of the cellular processes taking place during seed and embryo development (Belmonte et al., 2013; Wickramasuriya and Dunwell, 2015; Palovaara et al., 2013). Although these studies provide resources to support research in seed biology, much remains to be learned through classical genetic and physiology approaches, especially when 17% of Arabidopsis genes are still classified as having unknown function (The Arabidopsis Information Resource [http://www.arabidopsis.org], March 2016). Until recently, small genes and their protein products remain largely unstudied. Polypeptides of less than 100 amino acids are generally classified as small proteins (SPs). Wang and coworkers performed a statistical survey of SPs in bacteria and archaea and found that the majority of SPs in a given species is either considered a hypothetical protein or a protein of unknown function (Wang et al., 2008). Large proteins are easily annotated and more often studied, while shorter proteins tend to be ignored because they pose additional experimental challenges compared to larger proteins. However, with the recent availability of a great array of genome and transcriptome data, an increasing number of SPs have been identified and studied, demonstrating that SPs are widespread and have fundamental functions in all domains of life (Su et al., 2013; Magnani et al., 2014). SPs usually contain a single domain, tend to be nonconventional proteins, and seem to possess lineage-specific or tissue-specific functions (Zhao, 2012; Su et al., 2013). A number of chaperonins, translation initiation factors, ribosomal proteins, hormones, signaling molecules, and animal toxins, to mention a few, are all examples of SPs in multicellular organisms; thus, a great variety of biological processes involve the activity of SPs. In plants, some SPs are known to be involved in cell-to-cell communication (Jun et al., 2008; Murphy and De Smet, 2014; Meng et al., 2010) and regulatory processes (Kurata et al., 2005).
Here, we have isolated and analyzed two Arabidopsis mutants that illuminate the role of the TWISTED SEED1 (TWS1) gene, which encodes a novel SP. We demonstrate that lack of TWS1 activity causes dramatic growing abnormalities that are associated with defects in the endomembrane system, cuticle deposition, and changes in the accumulation of storage compounds in seeds.
RESULTS
Isolation of tws1-1 and tws1-2, Two Allelic Mutants Displaying Various Morphological Defects
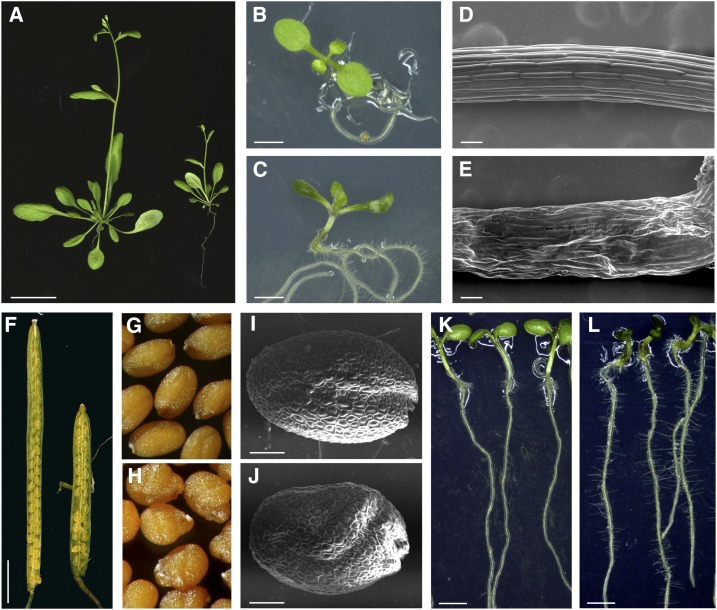
A visual screening for abnormal seed morphologies of the Versailles’ collection of T-DNA insertion lines was performed, allowing the isolation of about 250 mutants (Lepiniec et al., 2005). Two of these mutants, namely tws1-1 and tws1-2, were later found to be allelic (see below). Mutant plants exhibited a variety of growth defects at every developmental stage analyzed (Fig. 1; Supplemental Fig. S1, A–H). Bolting plants showed stunted growth when compared to wild-type plants grown in the same conditions (Fig. 1A). Seven-day-old mutant seedlings appeared darker in color, had reduced hypocotyl length, and their cotyledons were cup shaped, characteristics never observed in control ecotype Wassilewskija (Ws) seedlings (Fig. 1, B and C; Supplemental Fig. S1, B–D). Scanning electron microscopy (SEM) images of tws1-1 mutant 7-d-old seedling hypocotyls revealed how the small stature is consistent with a defect in cell morphology: Ws epidermal cells were arranged in ordered cell files, and their surface appeared smooth (Fig. 1D), whereas tws1-1 epidermal tissue organization was disrupted, and the cell surface appeared irregular (Fig. 1E). Mutant plants bore shorter siliques when compared to wild type (Fig. 1F). The short siliques phenotype was associated to a reduced seed set and to the presence of misshapen seeds (Fig. 1, G and H). Seed external morphology was analyzed through SEM. tws1-1 seeds were rounder than wild type and often presented bulges and irregularities, opposed to the smooth rugby-ball shape of a normal seed (Fig. 1, I and J; Supplemental Fig. S1, E and F). Nonetheless, the shape of the cells in the outer cell layer of the seed coat appeared normal, producing the characteristic reticulate pattern with a central columella. Furthermore, 7-d-old tws1-1 seedlings grown on vertical plates showed a higher root hair density than wild-type control plants (Fig. 1, K and L; Supplemental Fig. S1, G and H). Despite the aberrant shoot and root phenotypes, both shoot apical meristem and root apical meristem organization was not perturbed in tws1-1 (Supplemental Fig. S2).
Figure 1.
tws1-1 phenotypic characterization. A, Wild-type (left) and tws1-1 (right) 3-week-old bolting plants. Scale bar = 2.5 cm. B and C, A wild-type (B) and a tws1-1 (C) 10-d-old seedling grown in vitro. Scale bars = 2 mm. D and E, A wild-type (D) and a tws1-1 (E) hypocotyl imaged with SEM. Scale bars = 100 µm. F, Wild-type (left) and tws1-1 (right) open silique. Scale bar = 2.5 mm. G and H, Wild-type (G) and tws1-1 (H) seeds. I and J, SEM images of a wild-type (I) and a tws1-1 (J) mature dry seed. Scale bars = 100 µm. K and L, Wild-type (K) and tws1-1 (L) 7-d-old seedlings grown in vitro on vertical plates. Scale bars = 2 mm.
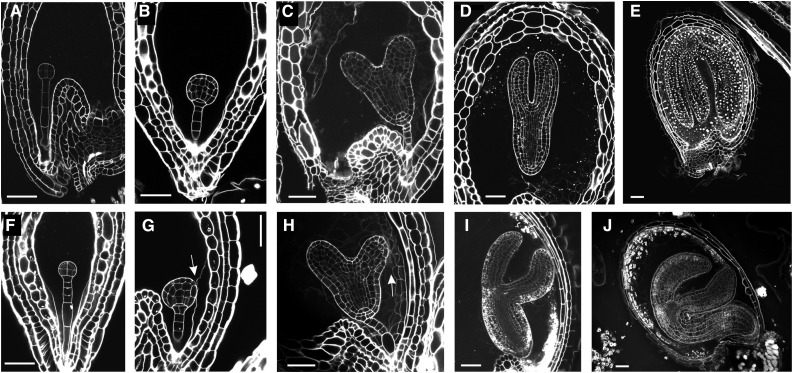
In order to precisely assess embryo morphology, we used the modified pseudo-Schiff propidium iodide imaging technique (see “Materials and Methods”). TWS1 embryos do not show any visible defect up to the late globular stage (Fig. 2, A–J). At this point in development, the embryos lost their longitudinal symmetry and appeared slightly tilted toward the side facing the testa inner wall, on the side opposite to the seed cavity (Fig. 2G, arrow). Nonetheless, embryo overall morphology was conserved, and by the heart stage it presented a thickened hypocotyl, its cotyledons were generally more separated than their wild-type counterparts, and the cotyledon facing the exterior appeared to be attached to the endothelium and/or endosperm cells in proximity (Fig. 2H, arrow). By the late torpedo stage, most of TWS1 embryos started to slightly bend toward the exterior (Fig. 2I), reaching the count of 81% (n = 196) by the mature stage (Fig. 2J), whereas this phenotype was never observed in wild-type Ws seeds (n = 329). This phenotype is reminiscent of the one caused by mutations in genes known to specify epidermal cell fate identity in Arabidopsis (Tsuwamoto et al., 2008; Yang et al., 2008).
Figure 2.
Embryo development in tws1-1. A to J, Longitudinal sections of seeds at different stages of development imaged through the modified pseudo-Schiff propidium iodide imaging technique. Wild-type (A) and tws1-1 (F) seeds at the early globular stage of embryo development. Wild-type (B) and tws1-1 (G) seeds at the globular stage of embryo development. Wild-type (C) and tws1-1 (H) seeds at the heart stage of embryo development. Wild-type (D) and tws1-1 (I) seeds at the torpedo stage of embryo development. Wild-type (E) and tws1-1 (J) seeds at the mature stage of embryo development. Scale bars = 40 µm. Arrows point to sites where tws1-1 embryo epidermal cells fuse to the surrounding tissues.
In conclusion, tws1-1 and tws1-2 pleiotropic phenotype suggests that the underlying gene is involved in a fundamental core process whose rightful execution is essential for proper seed and plant development.
Isolation and Genetic Characterization of TWS1
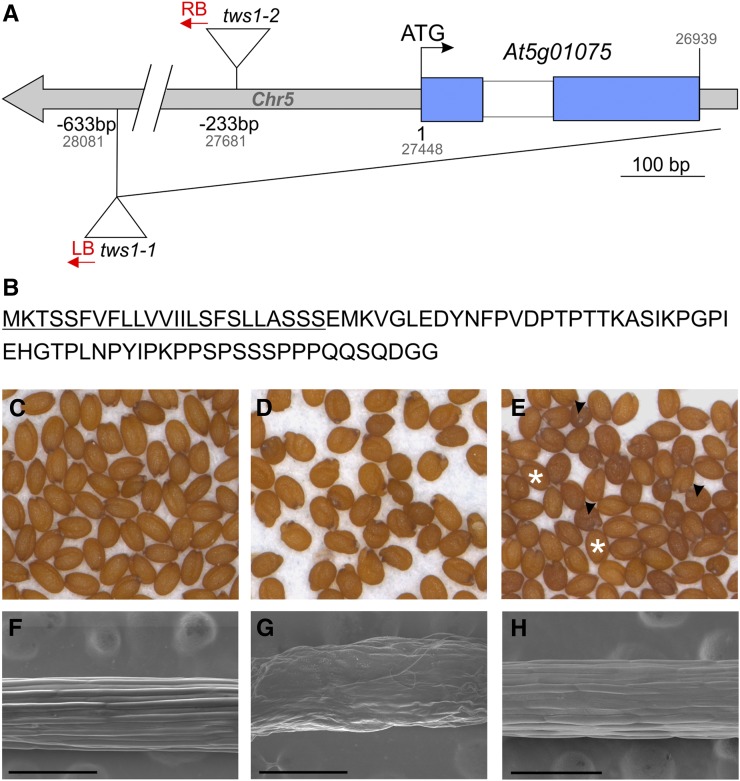
The tws1-1 and tws1-2 mutants were isolated from the Versailles T-DNA mutagenized population (http://publiclines.versailles.inra.fr/). Segregation analyses of the abnormal wrinkled seed phenotypes showed that both mutations are recessive and monogenic. Crossing the two lines did not restore the wild-type phenotype in the F1, suggesting that the two mutations are allelic. The strict cosegregation of kanamycin resistance (conferred by the T-DNA) and morphological defects showed that both mutations are genetically linked to a T-DNA insertion. Characterization of the genomic sequences flanking the T-DNA insertions confirmed that the mutations affect the same gene At5g01075 (Fig. 3A). Tws1-1 is a null allele caused by the deletion of the entire At5g01075 coding region (starting from the promoter till the downstream annotated gene), whereas tws1-2 is a weaker allele with a T-DNA insertion in its 5′ region, 233 bp before the starting codon (Fig. 3A; Supplemental Fig. S1I). The T-DNA insertion in tws1-2 allele perturbs its normal transcription: tws1-2 exhibits a decrease of mRNA accumulation and the appearance of unspliced mRNA (Supplemental Fig. S1I, star). In addition, we used genetic complementation to confirm the identity of At5g01075 as TWS1. We expressed the At5g01075 cDNA sequence under the control of the cauliflower mosaic virus 35S promoter, which restored the tws1 phenotype to the wild-type phenotype in both mutant backgrounds (Fig. 3, C–H). Figure 3, C and D, shows Ws and tws1-1 mature dry seed, respectively, whereas Figure 3E shows T2 tws1-1 transgenic plants (pro35SCaMV:TWS1) segregating wild-type-looking (stars) and tws1-looking (arrowheads) seeds. Complementation was also checked at the level of hypocotyl epidermal cell shape: complemented lines exhibited epidermal cell morphology similar to control Ws plants (Fig. 3, F–H).
Figure 3.
Molecular and genetic identification of TWS1 as At5g01075. A, Schematics of the TWS1 locus. The TWS1 gene structure is represented by blue boxes for exons, white boxes for introns, and gray boxes for other sequences. The positions of the tws1-1 and tws1-2 insertion mutations are shown both in relation to the TWS1 start codon (in black), or to the chromosome 5 (Chr5) sequence (in gray). Sequenced T-DNA borders are indicated in red (RB, right border; LB, left border), ATG indicates the first codon. B, TWS1 protein sequence. The predicted signal peptide is underlined. C to H, tws1-1 complementation analysis. C, Wild-type mature dry seeds, (D) tws1-1 mature dry seeds, and (E) mature dry seeds produced by a tws1-1 plant transformed with a construct carrying the TWS1 coding sequence under the control of a double 35S promoter sequence. The picture shows the segregating progeny of a hemizygous plant. White stars, wild-type-looking seeds; black arrowheads, tws1-1-looking seeds. F to H, Hypocotyls SEM images of 7-d-old seedlings: F, wild type; G, tws1-1; and H, complemented tws1-1. Scale bars = 300 µm.
Thus, from genetic analyses, mapping data, and the rescue of the mutant phenotypes, we conclude that At5g01075 corresponds to the TWS1 gene.
TWS1 Encodes a Novel Protein
AT5G01075 is a small gene that consists of two exons and an 83-bp intron (Fig. 3A). It encodes an 81-amino-acid peptide, which contains at its N terminus a stretch of highly hydrophobic amino acids (Fig. 3B) likely to act as a signal peptide (von Heijne, 1985). Indeed, TWS1 is predicted to enter the secretory pathway by protein subcellular localization prediction softwares such as SignalP (www.cbs.dtu.dk/services/SignalP/). The rest of the protein is predicted to be soluble and lacks any known retention signals, thus the Protein Subcellular Localization Prediction Tool (WoLF PSORT, www.genscript.com/wolf-psort.html) predicts it to be targeted to the apoplast or the vacuole.
Overall, these in silico predictions suggest TWS1 to be a signaling peptide, although it does not share any similarity to already known plant signaling molecules (Jun et al., 2008; Murphy and De Smet, 2014).
Protein analysis softwares failed to detect any other putative conserved domain in its sequence. When TWS1 sequence is blasted against the Arabidopsis transcriptome, it only retrieves its own sequence, thus it is a single copy gene in Arabidopsis genome. TWS1 homologs are clearly detectable starting from the basal angiosperm Amborella trichopoda onwards during evolution suggesting its function has evolved to fulfill the specific adaptation needs of flowering plants (Supplemental Fig. S3).
TWS1 Is Widely Expressed during Arabidopsis Growth
Semiquantitative RT-PCR analyses performed on RNA extracted from different tissues confirmed the publicly available expression data (Supplemental Fig. S4, A and B). TWS1 mRNA strongly accumulates during seed maturation and peaks at the torpedo stage of embryo development but is also detected in flower buds and all vegetative tissues tested. RNA in situ hybridization experiments confirmed TWS1 mRNA expression in young embryos (Fig. 4, D and E).
Figure 4.
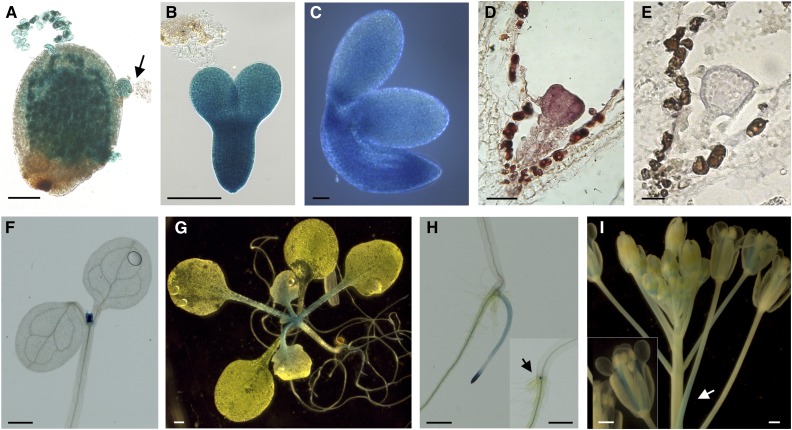
Expression analysis of TWS1. A to E, Expression analysis of TWS1 expression in developing seeds. A to C, TWS1 promoter-driven GUS activity in a seed carrying an embryo at the globular stage (arrow; A), an isolated embryo at the torpedo stage (B), and an isolated mature embryo (C). Scale bars = 50 µm. D and E, TWS1 mRNA expression pattern in a triangular stage embryo (D) and control hybridization with a TWS1 sense probe (E). Scale bars = 20 µm. F to I, Expression analysis of TWS1 expression in vegetative and reproductive tissues. TWS1 promoter-driven GUS activity in 7-d-old seedling above ground tissues (F), 2-week-old whole plant (G), 7-d-old seedling roots (H), and inflorescence (I). Scale bars = 500 µm.
Furthermore, we investigated the TWS1 transcriptional regulation by generating stably transformed Arabidopsis plants carrying the 1.1-kb putative promoter region (see “Materials and Methods” for further details) fused to the uidA-GFP reporter gene. All transgenic lines analyzed showed consistent results. We detected a strong GUS activity in developing embryos and in the testa (Fig. 4, A–C) at all developmental stages analyzed.
During vegetative development, we observed GUS staining in the shoot region and leaf primordia of 7-d-old seedlings (Fig. 4F; Supplemental Fig. S4, C and D). In 14-d-old plants, GUS staining was still present in the shoot regions and leaf primordia and extended to young leaves and petioles (Fig. 4G). In the underground organs, GUS staining was confined to the stele in the differentiation zone of primary and secondary roots, whereas it transited in the epidermal layer toward the root tip and finally appeared stronger at the root tip (Fig. 4H; Supplemental Fig. S4, E and F). We observed strong GUS signal right at the site of lateral root inception at the early stages of their development (Fig. 4H, inset). During Arabidopsis reproductive phase, GUS activity was observed at the base of the flower pedicels (Fig. 4I, arrow) and in the stamen filaments (Fig. 4I, inset) throughout flower development.
Thus, we can conclude that TWS1 is strongly expressed during seed development. Lower levels of expression in vegetative and reproductive tissues are fully consistent with the plethora of phenotypic defects observed at different developmental stages.
TWS1 Is Localized to the Endomembrane System
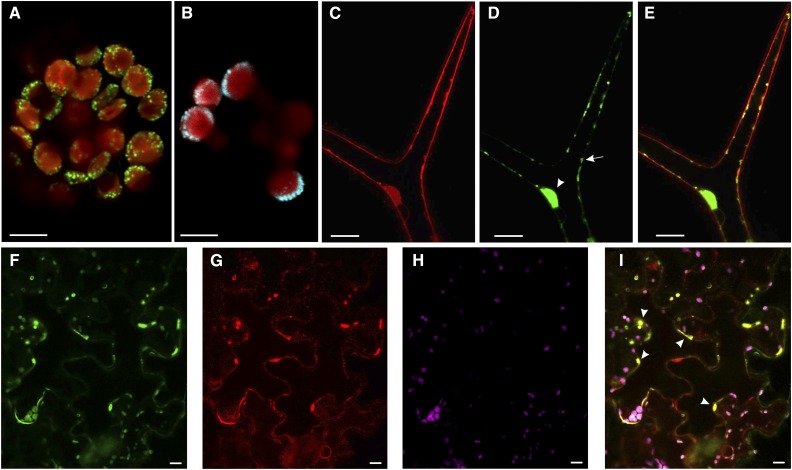
TWS1 contains a hydrophobic stretch at the 5′ portion of the protein that is likely to act as a signal peptide. To check this hypothesis, we created a TWS1 translational fusion to the GFP and transiently transformed Arabidopsis protoplasts derived from young leaves. We visualized fluorescent dots around the chloroplasts (Fig. 5A), a signal reminiscent of markers localized to the ER such as VMA21 (Fig. 5B). The visual comparison between TWS1::GFP signal and the ER marker VMA21::CFP signal suggests that TWS1 might be localized to the ER.
Figure 5.
TWS1 is localized to the ER. A, Confocal image of a mesophyll-derived protoplast transformed with a construct carrying TWS1 fused to GFP (Pro-35S:TWS1:GFP). GFP signal is shown in green, chlorophyll signal in red. Scale bar = 5 µm. B, Confocal image of a mesophyll-derived protoplast transformed with a construct carrying VMA21 fused to CFP (Pro-35S:VMA21:CFP). CFP signal is shown in cyan, chlorophyll signal in red. Scale bar = 5 µm. C to E, Confocal images of stably transformed Arabidopsis plants carrying the construct Pro-35S:TWS1:GFP. FM4-64 signal (C), TWS1::GFP signal (D), and overlay of the FM4-64 and GFP channels shown in C and D (E). Scale bars = 20 µm. F to I, Confocal images of stably transformed tobacco mesophyll cells expressing RFP fused to an ER retention signal, infiltrated with the TWS1::GFP construct. F, TWS1::GFP signal; G, ER-localized RFP signal; H, chlorophyll autofluorescence; I, superimposition of images in F to H. Arrowheads point to a few of the positions where GFP and RFP colocalize. Scale bars = 10 µm.
In order to confirm TWS1 subcellular localization, we produced stably transformed Arabidopsis plants carrying TWS1 cDNA sequence fused to GFP. This construct was able to rescue tws1-1 phenotype, demonstrating the functionality of the chimeric protein. We covisualized the internal endomembrane system with the amphiphilic styryl dye FM4-64. Confocal live imaging upon staining with FM4-64 is a valuable technique to monitor organelle morphology. After application in plants, FM4-64 immediately stains the PM and is then integrated on vesicles following endomembrane system-dependent internalization processes. Over time, FM4-64 becomes distributed throughout the full vesicular network from the PM to the vacuole, including the components of the secretory pathway (Oh-ye et al., 2011; Rigal et al., 2015).
TWS1::GFP signal was imaged in glandular trichomes, which are metabolically active cells. We could observe GFP signal in irregular spots under the cell surface (Fig. 5, C–E, arrows) and around the nucleus (Fig. 5, C–E, arrowheads), observations compatible with a localization in the cellular endomembrane system. We did not observe GFP colocalize with FM4-64 at the level of the PM or tonoplast.
Furthermore, we performed leaf infiltration experiments of transgenic Nicotiana benthamiana carrying the red fluorescent protein (RFP) fused to an ER retention signal (Martin et al., 2009) with the TWS1::GFP construct. We observed colocalization of TWS1::GFP with the RFP marker in ER subdomains (Fig. 5, F–I, arrowheads). The uneven distribution of TWS1::GFP in the ER suggests that TWS1 might play a role in a subset of ER functions, rather than as a general ER constituent.
Altogether, these results allowed us to conclude that TWS1 is localized to the endomembrane system upstream of the vacuole.
TWS1 Shows Defects in Cuticle Deposition
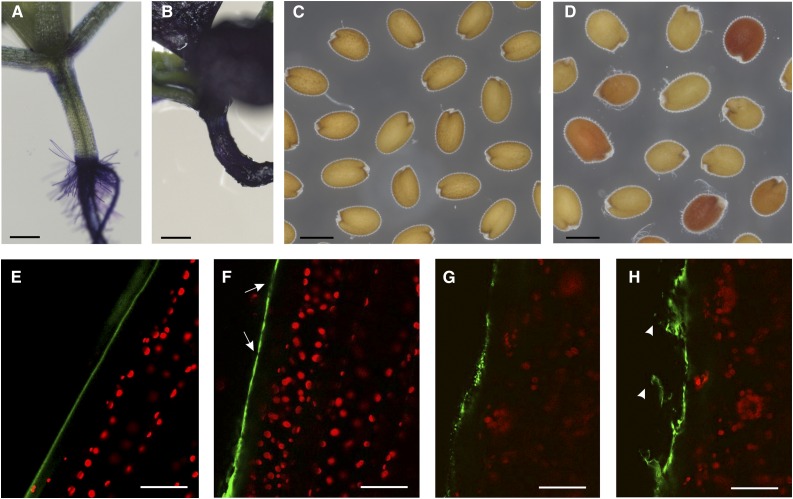
We initially characterized tws1 mutants at the level of cuticle deposition by estimating the permeability of 10-d-old seedlings by the toluidine blue test. Hypocotyls and cotyledons of wild-type seedlings incubated overnight in the staining solution repelled the toluidine blue (Fig. 6A), whereas tws1-1 seedlings stained dark blue (Fig. 6B) indicating a higher permeability of these organs in the mutant. Furthermore, staining of seeds with tetrazolium salts was performed to assess the permeability of the seed coat. When incubated in a solution of tetrazolium salts, tws1-1 seeds were found to be more sensitive to salt uptake than wild-type seeds (Fig. 6C) and, as a result, they turned from slightly red to vivid red (Fig. 6D). This suggests that tws1-1 testa integrity is impaired and some stochastic factors (such as internal pressure caused by the embryo bending in the wrong direction) might cause microscopic cracks in the outer integuments, thus resulting in seeds that uptake salts with variable efficiency.
Figure 6.
tws1 is impaired in cuticle deposition. A and B, Toluidine stained wild-type (A) and tws1-1 (B) 10-d-old seedlings. Scale bars = 2 mm. C and D, Tetrazolium salt permeability test in wild-type seeds (C) and tws1-1 seeds (D). Scale bars = 400 µm. E to H, Confocal images of Auramine O-stained cuticle in hypocotyls of wild type (E) or tws1-1 (F–H) plants. Green signal, cuticle; red signal, chlorophyll. Arrows point to sites where cuticle deposition is absent or reduced. Arrowheads point to cuticles detaching from cell surface. Scale bars = 40 µm.
Cuticle deposition has been directly observed through confocal scanning laser microscopy using the lipophilic fluorescent dye Auramine O. Auramine O has a strong affinity for regions containing acidic and unsaturated waxes, as well as cutin precursors and a moderate affinity for other lipidic cytoplasmic contents. Due to its chemical properties, Auramine O is used to visualize the cuticle in a variety of plant species (Buda et al., 2009; Lequeu et al., 2003). The hypocotyl cells of Auramine O-stained wild-type embryos showed a regular pattern of cuticle deposition: on the surface of protodermal cells, we could observe a smooth cuticle layer, free from irregularities (Fig. 6E). On the other hand, tws1-1 protodermal cells did not show a regular surface: the cuticle layer presented areas of different thickness, with points with little or no wax deposition (Fig. 6F, arrows); at times the cuticle exhibited a dotted pattern never observed in control plants (Fig. 6G). Furthermore, we could observe cuticle fragments detaching from the hypocotyl solely when imaging the mutant seedlings (Fig. 6H, arrowheads). Later in development, SEM was used to show how the density of wax crystals on stems of the tws1-1 mutant was reduced relatively to wild-type (Supplemental Fig. S5, A and B).
Overall, these data allowed us to conclude that tws1 epidermal cells are defective in cuticle deposition.
L1 Epidermal Cell Layer Is Properly Specified in tws1
In order to understand if the cuticle defects are due to a lack of epidermal cell identity specification, we analyzed the gene expression levels of known factors subtending the L1 specification: the homeodomain Leu zipper class IV proteins MERISTEM LAYER1 (AtML1) and PROTODERMAL FACTOR2 (PDF2; Abe et al., 2003; San-Bento et al., 2014). Quantitative PCR analyses performed on cDNA-derived from siliques mRNA revealed that AtML1 and PDF2 transcription levels do not change in tws1 mutant background (Supplemental Fig. S5C). In order to exclude any spatial misexpression of epidermal inducing factors, we crossed an L1 marker line carrying the GFP coding sequence under the control of the AtML1 promoter (proATML1::NLS:3xGFP; Takada and Jürgens, 2007), and we visualized GFP in both Ws and tws1 homozygous plants in a segregating F2 population. We could not appreciate any difference between Ws (Supplemental Fig. S5D) and tws1 (Supplemental Fig. S5E) embryos; GFP was observed exclusively in the outermost cell layer of developing embryos in both genetic backgrounds.
Thus, we conclude that in tws1-1 mutant the protodermal cell fate is spatially well specified.
Seed Carbon Resources Allocation Is Perturbed by TWS1 Mutations
Given the strong expression of TWS1 in seeds, we decided to analyze the content of mutant dry seeds and compared it to wild type.
First, we observed that TWS1 total loss of function in tws1-1 results in a slight increase of seed mass, whereas partial loss of TWS1 function in tws1-2 does not affect seed weight (Supplemental Fig. S1J).
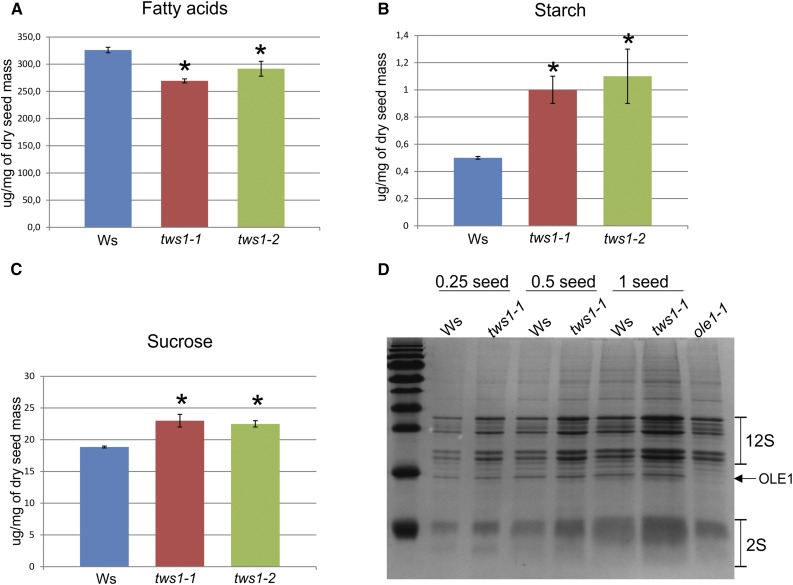
Tws1 mutant seeds showed a significant (17.4% for tws1-2 and 11% for tws1-2) reduction in total fatty acid accumulation (Fig. 7A), which might explain the cuticle defects described beforehand (Fig. 6). No particular fatty acid species was drastically affected more than the others (Supplemental Table S1). Nonetheless, the number, size, and distribution of oleosomes in tws1-1 developing embryos did not show any visible change when compared to wild type, indicating that oil biosynthesis and accumulation are not impaired (Supplemental Fig. S6, A and C), and loss of TWS1 function most likely impairs only cuticle deposition.
Figure 7.
Seed content is perturbed in tws1 mutants. A, Total fatty acid quantification in wild-type, tws1-1, and tws1-2 seeds. B, Starch quantification in wild-type, tws1-1, and tws1-2 seeds. C, Suc quantification in wild-type, tws1-1, and tws1-2 seeds. A to C, *Values are significantly different from wild type (extractions performed on 30 dry seeds per genotype, on three biological replicates, Student’s t test, P < 0.05). D, SDS-PAGE gel run with crude seed protein extracts. The volumes corresponding to a quarter of a seed, half of a seed, and one seed have been loaded on the gel.
In order to check whether the decrease in fatty acids production was compensated by changes in levels of other seed components, we analyzed starch and sugars seed content. Starch quantification revealed up to 2-fold increase in tws1 seeds compared to wild type (Fig. 7B), and a 22% increase in free Suc in tws1 mutant seeds (Fig. 7C). Moreover, a higher level of starch granules accumulation was also observed in transversal sections of 7-d-old seedling hypocotyls stained with Lugol’s solution. We never observed starch granules in Ws hypocotyl cells (Supplemental Fig. S6D), whereas epidermal as well as subepidermal cells in tws1-1 background showed starch granules accumulation (Supplemental Fig. S6E, arrowheads).
Last, we observed the effect of tws1 deficiency on the quality and quantity of the storage proteins in dry seeds running crude seed protein extracts on SDS-polyacrylamide (SDS-PAGE) gels. Figure 7D shows a comparison of the seed storage protein profiles of Ws, tws1-1 and oleosin1 (ole1) seeds (Siloto et al., 2006) on an SDS-PAGE gel stained with Coomassie Blue. Interestingly, tws1-1 seeds accumulated on average 1.7 times more 12S globulins and 1.9 times more 2S albumins than wild type (n = 3 biological replicates, Z test, P < 0.01). This increase in protein accumulation was specific to storage proteins targeted to the PSVs, as other proteins known to be accumulated in oleosomes, such as OLE1, did not show any quantitative change (the ole1 mutant protein seed extract is present as a control to pinpoint to the OLE1 protein). Furthermore, no seed storage protein precursors were accumulated, indicating they are properly sorted to the PSVs, where they are processed to their mature form.
TWS1 Mutation Results in Altered Vacuole Morphology
Because of the increase in seed protein accumulation in tws1-1 mutant and TWS1 localization in the endomembrane system, we decided to investigate vacuole morphology in the mutant background.
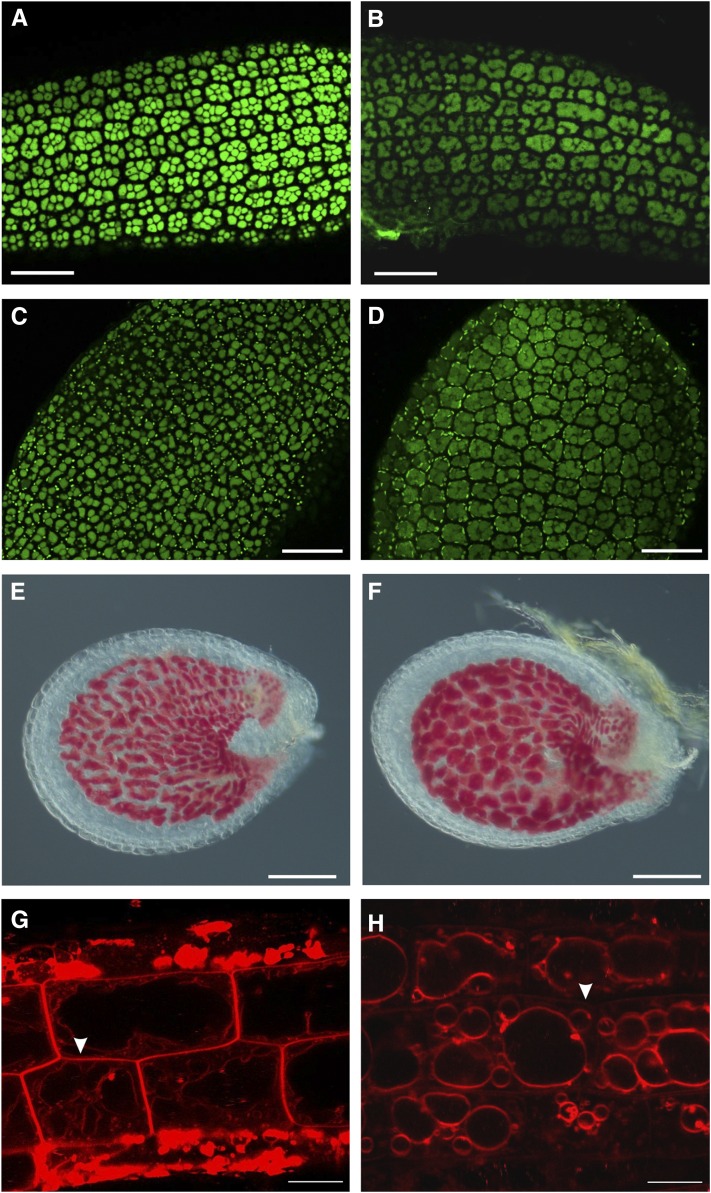
Initially, we investigated whether tws1 mutation affected the PSV morphology in mature embryos, where they can be easily visualized by autofluorescence. PSVs are a special type of vacuoles that accumulate proteins in dry seed cells. Figure 8 shows the morphology of PSVs in embryo hypocotyl (Fig. 8, A and B) and cotyledon protodermal cells (Fig. 8, C and D). Wild-type hypocotyl protodermal cells imaged with a confocal microscope showed eight to 10 PSVs in each cell, depending on the focal plan observed (Fig. 8A). On the other hand, tws1-1 hypocotyl protodermal cells showed less fluorescent vacuoles, which appeared enlarged or fused together (Fig. 8B). The same defect was observed in cotyledon protodermal cells (Fig. 8, C and D).
Figure 8.
Altered vacuole morphology in tws1. A to D, Confocal images of protein autofluorescence in embryo hypocotyls (A and B), and cotyledons (C and D). A and C, Wild-type embryos; B and D, tws1-1 embryos. Scale bars = 40 µm. E and F, Wild-type (E) and tws1-1 (F) vanillin-stained seed. Both seeds contain embryos at the heart stage. Scale bars = 100 µm. G and H, Confocal images of FM4-64-stained root epidermal cells of wild-type (G) and tws1-1 (H) seedlings. Scale bars = 10 µm.
Furthermore, Arabidopsis seeds accumulate large amounts of flavonoid polymers (proanthocyanidins) and leucoanthocyanidin precursors in the seed coat (Debeaujon et al., 2003). Vanillin staining of developing seeds allowed us to visualize vacuoles of the innermost testa integument as proanthocyanidins are mainly sequestrated into the vacuole (Lepiniec et al., 2006). Figure 8F clearly shows how vanillin stained tws1 endothelium exhibited larger vacuoles than control wild type (Fig. 8E).
In order to check if lack of TWS1 function only perturbs PSV morphology during seed development, we observed root lytic vacuoles in both mutant and wild-type plants. Root epidermal cells were imaged after being incubated in a FM4-64 solution for few hours. A wild-type epidermal cell located at the transition zone appeared elongated and filled almost entirely by one vacuole. The cytoplasm and all organelles were squeezed around the cell periphery by the vacuole turgor (Fig. 8G). However, tws1-1 epidermal cells exhibited multiple and rounder vacuoles (Fig. 8H). Furthermore, compared to the control, the PM of mutant cells was lightly stained (Fig. 8, G and H, arrowheads), suggesting an accelerated rate of membrane internalization.
Taken together, these results provide evidence that TWS1 function is required for proper vacuolar morphology in plants.
DISCUSSION
In this study, we present the isolation and characterization of TWS1, a single copy gene in the Arabidopsis genome. Database searches revealed homologs in all flowering plants and in the basal angiosperm Amborella trichopoda. This observation suggests that TWS1 fulfills a function proper to seed plants, although we cannot exclude a rapid and divergent evolution from its putative ancestral form that impairs its identification in more basal plants. Indeed, SP evolutionary studies suggest that they tend to evolve more rapidly and therefore are less conserved than long proteins (Guigo et al., 2003; Wei et al., 2005; Windsor and Mitchell-Olds, 2006).
Storage compound accumulation in tws1 mutant seeds is affected with a 17% reduction in fatty acid content followed by an increase in starch, Suc, and storage protein accumulation (Fig. 7). However, the speculated reduction in TAGs accumulation is too small to be visually appreciated as a change in oleosomes size and/or distribution (Supplemental Fig. S5, A and B). Wild-type embryos are filled with protein and lipid bodies but contain relatively small amounts of starch at maturity (Baud et al., 2008). When the accumulation of one storage compound is compromised by specific genetic mutations, a compensatory effect in the accumulation of the other storage product is often observed. This is the case for the mutants wrinkled1 and trialcylglycerol1, whose seeds contain considerably fewer oils and increased Suc levels (Focks and Benning, 1998; Lu et al., 2002), or for leafy cotyledon1 seeds, where the primary storage product is starch at the detriment of oil and protein accumulation (Meinke et al., 1994). We hypothesize that changes in carbon resource allocation are part of the pleiotropic effects of a mutation in a protein implicated in intracellular trafficking. We speculate that TWS1 mutation might negatively affect the functionality of parts of the ER involved in the biosynthesis of storage oils and cutin precursors, thus slowing down the flux of Suc to oils and other cellular components. As a consequence starch and Suc accumulate and part of these carbon resources might be diverted toward the production of extra storage proteins.
We have presented several lines of evidence that indicate TWS1 as required for proper cuticle deposition throughout Arabidopsis development (Figs. 2 and 6). The plant cuticle is a complex hydrophobic layer covering the areal surfaces which has many critical functions, not only at the physiological level through limiting water loss and pathogen resistance, but also at the developmental level acting as a morphogenic signal and preventing organ fusion during organogenesis (Nawrath, 2006; Riederer, 2006). Cuticle deposition defects have been previously described in either mutants failing to differentiate a proper epidermis or mutants impaired in waxes biosynthesis and/or transport, thus TWS1 might well be implicated in one of these processes. The best-characterized regulators of protodermal cell fate in Arabidopsis are the functionally redundant AtML1 and PDF2 (Abe et al., 2003): AtML1 and PDF2 double mutants lack the L1 layer and are not viable. Among other factors involved in specifying protodermal cell fate, the bHLH transcription factor ZHOUPI (ZOU; Yang et al., 2008) plays a central role. ZOU has been suggested to act through molecules implicated in signaling, such as the subtilisin protease ABNORMAL LEAF SHAPE1, the redundant Leu-rich repeat receptor-like kinases GASSHO1 (GSO1) and GSO2 (Tsuwamoto et al., 2008), and the receptor-like kinases ARABIDOPSIS CRINKLY4 and ABNORMAL LEAF SHAPE2 to activate the expression of AtML1 and PDF2 in developing embryos, thus maintaining epidermal cell fate (Javelle et al., 2011). ZOU mutants form a defective cuticle and show embryos abnormal adhesion to the endosperm. As a consequence, they produce shriveled seeds that germinate into seedlings that survive only if grown in conditions of high humidity (Yang et al., 2008). zou seed phenotype is not only due to excess water loss but also to the adhesion of the embryos to the surrounding endosperm cells, thus impairing its elongation and eventually promoting its bending toward the micropylar pole rather than the chalazal pole as observed in tws1 mutants. In a remarkable parallel with tws1, gso1/gso2 double-mutant embryos are indistinguishable from wild-type embryos until the heart-torpedo transition stage, when they start to expand laterally. At the late torpedo stage, gso1/gso2 embryos adhere to the peripheral endosperm cells and bend in reverse in the final stages of seed development. As for zou mutant, the pleiotropic gso1/gso2 phenotype has been pinned down to a defective epidermis and more specifically to an impaired cuticle deposition. Given the fact that L1 layer determinants such as AtML1 and PDF2 are properly expressed (Supplemental Fig. S5, C–E), we think that the epidermal layer is well specified in the mutant background. Overall, we can conclude that TWS1 likely acts as a downstream effector of epidermal cell differentiation.
The cuticle defects exhibited by tws1 mutants could be directly responsible for their root phenotype (Fig. 1, K and L; Supplemental Fig. S1, G and H). Root hairs become visible on seedling roots shortly after germination and are produced by epidermal cells. In wild-type plants, root hair cells are interspersed with nonhair cells following a distinct position-dependent pattern whose formation has been extensively studied (Grierson et al., 2014). Root hair cell differentiation requires polarized cell expansion at the growing tip, a process that relies on many other integrated processes such as secretion, endomembrane trafficking, cytoskeletal organization, and cell wall modifications. The result of this differentiation is a long tubular-shaped cell that lacks a cuticle layer to allow nutrient intake, anchorage, and microbe interactions (Hofer, 1991). The absence of the cuticle layer in root hair cells is generally viewed as a final result of differentiation, rather than as an early inducing factor of root hair cell fate specification. However, it is well known from experiments conducted in Fucus embryos that cell wall components are important to maintain and direct cell fate in plant development (Berger et al., 1994). In this light, we can interpret tws1 ectopic root hairs phenotype as a result of an imperfect cuticle deposition which exposes more root epidermal cells to the external environment, a condition that is thus perceived at the cellular level as a lack of cuticle, which ultimately causes induction of root hair cell differentiation. Indeed, a number of cuticle-deficient mutants are known to exhibit defects in trichome density, stomatal density, and show altered planes of epidermal cell division, suggesting that cuticular components and/or their precursors are required for the patterning of epidermis (Yephremov et al., 1999; Gray, 2005; Sieber et al., 2000).
Given its cuticle defects, TWS1 could have a role in wax trafficking. Cuticular waxes are produced in the ER of epidermal cells before being exported to the cell surface facing the environment through mechanisms that are still largely unknown. Among the hypotheses that have been proposed to explain intracellular trafficking of wax compounds, the physical contact between ER and the PM is supported by preliminary data based on the observation of wax compounds inside ER-derived membrane inclusions in ATP binding cassette transporter mutant cells (McFarlane et al., 2010). In this scenario, TWS1 could play a role in specialized ER sites that would come in contact with specialized PM site to ensure waxes transport to the cell surface. The irregular and reduced cuticle deposition exhibited by tws1 epidermal cells (Fig. 6, E–H; Supplemental Fig. S5 A and B) is in line with this hypothesis.
Nevertheless, TWS1 could play a wider role being implicated in some general aspects of ER functioning, thus affecting other processes mediated by the secretory pathway. This hypothesis is supported by the observation that PSV and lytic vacuole morphology is also altered in tws1 mutants (Fig. 8), as well as protein content in mature seeds (Fig. 7D). TWS1 localization in the upstream compartments of the secretory pathway is consistent with this hypothesis, as vacuoles are downstream compartments of the secretory pathway; thus, mutations affecting genes involved in upstream trafficking can result in vacuole morphology defects. For example, the trans-Golgi network/early endosome-localized protein KEG affects plant vacuole morphology and transport to the vacuole (Gu and Innes, 2012). Furthermore, loss-of-function mutants of the subunits of the adaptor protein complex 3 show defects in vacuole formation although adaptor protein complex 3 has been shown to function in the post-Golgi trafficking machinery (Viotti et al., 2013). Regardless of its precise biochemical role, TWS1 function is likely dependent on its interaction with other ER or Golgi residents, as it is lacking any known ER retention signal or Golgi targeting signal. In this light, we are currently working on identifying its partners in order to unveil its precise biochemical function.
The isolation and characterization of the TWS1 gene have not only provided insights into how a cellular process can widely influence overall plant development and metabolism but has also revealed the activity of a to our knowledge previously uncharacterized SP in Arabidopsis. TWS1 does not have any known protein domain, a characteristic that is shared with many other SPs (Su et al., 2013). The set of SPs studied thus far in different organisms suggests they are unlikely to possess enzymatic functions. Instead, they appear to act in a mechanical fashion, blocking a domain of a target protein, favoring the interactions between proteins and other molecules, bringing conformational changes to larger proteins and as membrane anchors (Storz et al., 2014). As TWS1::GFP colocalizes in some domains of the ER with an RFP carrying an ER retention signal (Fig. 5), it could influence the biochemical activity of specific enzymatic complexes such as the ER-bound multienzymatic fatty acid elongase complex, resulting in waxes with altered chemistry. Indeed, cutin composition could well be compromised, as we observed Auramine O-stained cuticle detaching from the cell surface (Fig. 6, E–H), a phenomenon never observed in wild-type samples. Evidences suggest that cuticle permeability is not necessarily correlated to physical thickness, and both chemical composition and structural assembly are likely to play important roles (Burghardt and Riederer, 2006). Given its small size, TWS1 could act as a small bridge to bring together enzymes implicated in the very long-chain fatty acids biosynthesis pathway or as a cofactor for one of the enzymes. In order to gain more clues on TWS1 biochemical role, we are currently analyzing wax deposition in the mutant background. Furthermore, in stably transformed plants, we have sometimes observed TWS1::GFP signal in the nucleus (Fig. 5, C–E), and we are currently exploring the possibility that TWS1 might be able to move between cellular compartments by means of interaction with protein partners.
Overall, our results pave the way to further molecular and biochemical characterizations of the TWS1 protein and open new routes for elucidating some aspects of wax biosynthesis and/or transport and storage compound allocation in seeds.
MATERIALS AND METHODS
Plant Material, Transformation, and Selection of Transgenic Lines
All Arabidopsis (Arabidopsis thaliana) genotypes were in the Ws accession. Mutant seeds can be obtained from http://publiclines.versailles.inra.fr/tdna/index: tws1-1 is T-DNA line CON41; tws1-2 is T-DNA line DXX33. Flanking sequence tags are publicly available and have been confirmed following the instructions on http://publiclines.versailles.inra.fr/. Plants were grown either on Murashige and Skoog medium or on soil, under long day conditions at 22°C. Seeds were stratified at 4°C for 4 d prior to light exposure. Prior to in vitro culture, seeds were surface-sterilized for 40 min with chlorine gas. Control seeds were collected and grown side by side with experimental seeds for each experiment. Agrobacterium tumefaciens strain C58C1 was used to stably transform Arabidopsis plants by the floral dip method (Clough and Bent, 1998). For each plant transformation, more than 30 resistant lines were recovered, and at least 10 lines were used to carry on further experiments. For the complementation experiment, we isolated 32 transgenic lines and 10 showed complementation (complementation has been scored based on wild-type seed phenotype recovery).
RNA in Situ Hybridization and Cytological Analyses
RNA in situ hybridization analyses were conducted as described in Carles et al. (2010), using the TWS1 full-length coding sequence as the antisense probe.
Whole-mount vanillin assays for proanthocyanidins detection were performed as previously described (Debeaujon et al., 2000).
Seedling hypocotyls for histological analysis were fixed overnight at room temperature in a solution of 45% ethanol, 2.5% glacial acetic acid, and 2.5% formaldehyde (v/v). The samples were then dehydrated by sequential 30-min incubations in 50%, 70%, 85%, 95%, and 99.5% (v/v) ethanol, followed by two incubations of 1 h each in 100% (v/v) ethanol. The dehydrated samples were set in Technovit 7100 resin (Heraeus Kulzer) at room temperature, once in 50% (v/v) resin/ethanol and twice in 100% resin. Five-micrometer sections of the plant tissues were cut with a rotary microtome and stained with Lugol’s solution (Sigma-Aldrich).
RNA Extraction, and Semiquantitative and Quantitative RT-PCR
Total RNA was isolated using the RNeasy Plant Kit (Qiagen) and treated with RNase-free DNase on column, as specified by the manufacturer manual. The first-strand cDNA synthesis was performed on 1 mg of total RNA using Superscript III RNase H reverse transcriptase (Invitrogen), according to the manufacturer’s instructions. The annealing temperature was 58°C to 60°C for all primer pairs. Quantification of transcripts by real-time PCR was performed using the SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) and a CFX Connect Real-Time PCR Detection System (Bio-Rad). Three technical replicates were run for each sample. The specificity of the amplification was determined by performing a dissociation curve analysis. Relative quantification values were calculated using the 2−ΔCt method (Livak and Schmittgen, 2001).
DNA Extraction and Cloning
Genomic DNA was extracted using the DNeasy Plant Kit (Qiagen). PCR-amplified products were cloned into the pENTR/D-TOPO vector (Invitrogen) and then moved into the appropriate destination vector from the pMDC series (Curtis and Grossniklaus, 2003) or pBGWFS7 (Karimi et al., 2002) through an LR reaction (Invitrogen), according to the manufacturer’s instructions. The TWS1 promoter sequence, consisting of 1108 bp from the stop codon of the nearest upstream gene to the TWS1 start codon, was PCR-amplified and cloned into the binary vector pBGWFS7 for expression analysis. The TWS1 coding sequence was PCR-amplified and cloned into the binary vector pMDC32 under the control of a double cauliflower mosaic virus 35S promoter for tws1-1 and tws1-2 complementation analysis and in pMDC83 for TWS1 subcellular localization (TWS1:GFP fusion). TWS1 mRNA transcripts were PCR-amplified and cloned into the pGEM-T Easy vector (Promega) for in vitro transcription of an in situ hybridization probe. Primer sequences are listed in Supplemental Table S2.
SEM
Uncoated samples were analyzed and imaged using the tabletop Scanning Electron Microscope SH 1500 (Hirox).
GUS Assays
Histochemical detection of GUS activity was performed as previously described (Jefferson, 1989), with the modification that 2 mm potassium ferrocyanide and 2 mm potassium ferricyanide were used. Whole-mount incubation times ranged from 8 to 12 h after vacuum infiltration.
Prior to the performance of the GUS assay, seeds at early stages of development were squeezed between a slide and a coverslip in order to pop out the embryos and improve image quality.
Protoplast Preparation
Arabidopsis protoplasts were isolated from leaves and transformed as previously described (Wu et al., 2009).
Transient Infiltration of Tobacco Leaves
Nicotiana benthamiana leaf infiltration has been performed as previously described (Sparkes et al., 2006) using the A. tumefaciens strain GV3101.
Confocal Microscopy
All samples were imaged with a Leica TCS-SP5 spectral confocal laser scanning microscope (Leica Microsystems) using a 40× or 63× magnification lens.
For Auramine O staining, 7-d-old seedlings were imaged upon immersion in a 0.01% w/v Auramine O (Sigma; 0.01% w/v in 0.05 m Tris/HCl, pH 7.2) aqueous solution for 10 min and subsequently rinsed with water.
For pseudo-Schiff propidium iodide staining, developing seeds were harvested and treated as previously described (Xu et al., 2016). Upon staining, the samples were transferred onto microscope slides for confocal laser scanning microscopy analysis.
Protein autofluorescence in PSVs was imaged using the filter set for GFP.
For FM4-64 staining, samples were incubated in a 5 μL/mL dye solution for 2 h, subsequently rinsed in water, and transferred to a microscope slides for imaging.
Oleosomes have been visualized upon staining with Nile Red.
Biochemical Analyses
Total fatty acids from dry seeds were extracted and quantified as previously described (Li et al., 2006). Starch and Suc measurements in dry seeds were performed as previously described (Caspar et al., 1991). Protein extracts were prepared from 20 dry seeds per sample and suspended in Leammli buffer (2×) with dithiothreitol. The crude seed protein extracts were incubated at 95°C for 5 min, followed by a centrifugation at max speed for 3 min, and the supernatants were pipetted into a clean tube. The volumes corresponding to a quarter of a seed, half of a seed, and 1 seed have been loaded on a 15% SDS-PAGE gel followed by Coomassie Blue staining.
Sequence Analysis and Image Processing
Multiple sequence alignments were performed using the software MUSCLE (Edgar, 2004). Phylogenetic analyses were conducted using MEGA7 software (Kumar et al., 2016). SDS-PAGE gel band intensity has been measured using the National Center for Biotechnology Information Image J software for image processing and analysis (https://imagej.nih.gov/ij/).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: TWS1, At5g01075; AtML1, At4g21750; PDF2, At4g04890; ZOU, AT1G49770; GSO1, AT4G20140; GSO2, AT5G44700; ABNORMAL LEAF SHAPE1, AT1G62340; ABNORMAL LEAF SHAPE2, AT2G20300; ARABIDOPSIS CRINKLY4, AT1G69040; WRINKLED1, AT3G54320; TRIALCYLGLYCEROL1, AT2G19450; LEAFY COTYLEDON1, AT1G21970.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phenotypic and molecular characterization of tws1-1 and tws1-2 mutant alleles.
Supplemental Figure S2. tws1-1 shoot and root apical meristem morphology.
Supplemental Figure S3. TWS1 homologs in plants and their evolutionary relationships.
Supplemental Figure S4. TWS1 expression.
Supplemental Figure S5. Stem cuticle deposition defects and L1 marker analysis in tws1-1.
Supplemental Figure S6. Oleosomes and starch granule visualization in tws1-1.
Supplemental Table S1. Fatty acids composition of tws1-1 and tws1-2 mutant seeds.
Supplemental Table S2. Primers used in the experimental procedures.
Supplementary Material
Acknowledgments
We thank Dr. Karl-Josef Dietz for the Vma21::CFP marker line, and V. Tanty, J. Porret, and J. Kronenberger for their technical help in the initial characterization of the mutants supported by Genoplante programme, Gerd Jürgens’ laboratory for proATML1::NLS:3xGFP marker line. We thank the “Observatoire du Végétal” for plant culture and access to imaging facility and assistance.
Glossary
- PSV
protein storage vacuole
- PM
plasma membrane
- SP
small protein
Footnotes
This work was supported by the LabEx Saclay Plant Sciences (ANR-10-LABX-0040-SPS). E.F. is supported by fellowships from EMBO (no. 243-2012) and the LabEx Saclay Plant Sciences (project GESEM).
Articles can be viewed without a subscription.
References
- Abe M, Katsumata H, Komeda Y, Takahashi T (2003) Regulation of shoot epidermal cell differentiation by a pair of homeodomain proteins in Arabidopsis. Development 130: 635–643 [DOI] [PubMed] [Google Scholar]
- Baud S, Dubreucq B, Miquel M, Rochat C, Lepiniec L (2008) Storage reserve accumulation in Arabidopsis: metabolic and developmental control of seed filling. Arabidopsis Book 6: e0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmonte MF, Kirkbride RC, Stone SL, Pelletier JM, Bui AQ, Yeung EC, Hashimoto M, Fei J, Harada CM, Munoz MD, et al. (2013) Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc Natl Acad Sci USA 110: E435–E444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F, Taylor A, Brownlee C (1994) Cell fate determination by the cell wall in early fucus development. Science 263: 1421–1423 [DOI] [PubMed] [Google Scholar]
- Buda GJ, Isaacson T, Matas AJ, Paolillo DJ Rose JK (2009) Three-dimensional imaging of plant cuticle architecture using confocal scanning laser microscopy. Plant J,60: 378–385. [DOI] [PubMed] [Google Scholar]
- Burghardt M, Riederer M (2006) Cuticular transpiration. In Riederer M, Müller C, eds, Annual Plant Reviews, Vol 23 Blackwell Publishing, Oxford, pp 292–311 [Google Scholar]
- Carles CC, Ha CM, Jun JH, Fiume E, Fletcher JC (2010) Analyzing shoot apical meristem development. Methods Mol Biol 655: 105–129 [DOI] [PubMed] [Google Scholar]
- Caspar T, Lin TP, Kakefuda G, Benbow L, Preiss J, Somerville C (1991) Mutants of Arabidopsis with altered regulation of starch degradation. Plant Physiol 95: 1181–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Léon-Kloosterziel KM, Koornneef M (2000) Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol 122: 403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Nesi N, Perez P, Devic M, Grandjean O, Caboche M, Lepiniec L (2003) Proanthocyanidin-accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development. Plant Cell 15: 2514–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focks N, Benning C (1998) wrinkled1: a novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol 118: 91–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J. (2005) Guard cells: transcription factors regulate stomatal movements. Curr Biol 15: R593–R595 [DOI] [PubMed] [Google Scholar]
- Grierson C, Nielsen E, Ketelaarc T, Schiefelbein J (2014) Root hairs. Arabidopsis Book 12: e0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Innes RW (2012) The KEEP ON GOING protein of Arabidopsis regulates intracellular protein trafficking and is degraded during fungal infection. Plant Cell 11: 4717–4730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guigo R, Dermitzakis ET, Agarwal P, Ponting CP, Parra G, Reymond A, Abril JF, Keibler E, Lyle R, Ucla C, et al. (2003) Comparison of mouse and human genomes followed by experimental verification yields an estimated 1,019 additional genes. Proc Natl Acad Sci USA 100: 1140–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hills MJ. (2004) Control of storage-product synthesis in seeds. Curr Opin Plant Biol 7: 302–308 [DOI] [PubMed] [Google Scholar]
- Hofer AM. (1991) Root hairs. In Waisel Y, Eshel A, Kafkafi U, eds, Plant Roots—The Hidden Half. Marcel Dekker, New York, pp 111–126 [Google Scholar]
- Javelle M, Vernoud V, Rogowsky PM, Ingram GC (2011) Epidermis: the formation and functions of a fundamental plant tissue. New Phytol 189: 17–39 [DOI] [PubMed] [Google Scholar]
- Jefferson RA. (1989) The GUS reporter gene system. Nature 342: 837–838 [DOI] [PubMed] [Google Scholar]
- Jun JH, Fiume E, Fletcher JC (2008) The CLE family of plant polypeptide signaling molecules. Cell Mol Life Sci 65: 743–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata T, Ishida T, Kawabata-Awai C, Noguchi M, Hattori S, Sano R, Nagasaka R, Tominaga R, Koshino-Kimura Y, Kato T, et al. (2005) Cell-to-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development 132: 5387–5398 [DOI] [PubMed] [Google Scholar]
- Lepiniec L, Debeaujon I, Routaboul JM, Baudry A, Pourcel L, Nesi N, Caboche M (2006) Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol 57: 405–430 [DOI] [PubMed] [Google Scholar]
- Lepiniec L, Devic M, Berger F (2005) Genetic and molecular control of seed development in Arabidopsis. In Leister D, ed, Plant Functional Genomics. Food Products Press, Binghamton, NY, pp 511–564. [Google Scholar]
- Lequeu J, Fauconnier ML, Chammaï A, Bronner R, Blee E (2003) Formation of plant cuticle: evidence for the occurrence of the peroxygenase pathway. Plant J 36: 155–164 [DOI] [PubMed] [Google Scholar]
- Li Y, Beisson F, Pollard M, Ohlrogge J (2006) Oil content of Arabidopsis seeds: the influence of seed anatomy, light and plant-to-plant variation. Phytochemistry 67: 904–915 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lolle SJ, Hsu W, Pruitt RE (1998) Genetic analysis of organ fusion in Arabidopsis thaliana. Genetics 149: 607–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CA, Ho TH, Ho SL, Yu SM (2002) Three novel MYB proteins with one DNA binding repeat mediate sugar and hormone regulation of alpha-amylase gene expression. Plant Cell 14: 1963–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani E, de Klein N, Nam HI, Kim JG, Pham K, Fiume E, Mudgett MB, Rhee SY (2014) A comprehensive analysis of microProteins reveals their potentially widespread mechanism of transcriptional regulation. Plant Physiol 165: 149–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K, Kopperud K, Chakrabarty R, Banerjee R, Brooks R, Goodin MM (2009) Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J 59: 150–162 [DOI] [PubMed] [Google Scholar]
- McFarlane HE, Shin JJ, Bird DA, Samuels AL (2010) Arabidopsis ABCG transporters, which are required for export of diverse cuticular lipids, dimerize in different combinations. Plant Cell 22: 3066–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke DW, Franzmann LH, Nickle TC, Yeung EC (1994) Leafy cotyledon mutants of Arabidopsis. Plant Cell 6: 1049–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Wong JH, Feldman LJ, Lemaux PG, Buchanan BB (2010) A membrane-associated thioredoxin required for plant growth moves from cell to cell, suggestive of a role in intercellular communication. Proc Natl Acad Sci USA 107: 3900–3905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E, De Smet I (2014) Understanding the RALF family: a tale of many species. Trends Plant Sci 19: 664–671 [DOI] [PubMed] [Google Scholar]
- Nawrath C. (2006) Unraveling the complex network of cuticular structure and function. Curr Opin Plant Biol 9: 281–287 [DOI] [PubMed] [Google Scholar]
- Oh-ye Y, Inoue Y, Moriyasu Y (2011) Detecting autophagy in Arabidopsis roots by membrane-permeable cysteine protease inhibitor E-64d and endocytosis tracer FM4-64. Plant Signal Behav 6: 1946–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palovaara J, Saiga S, Weijers D (2013) Transcriptomics approaches in the early Arabidopsis embryo. Trends Plant Sci 18: 514–521 [DOI] [PubMed] [Google Scholar]
- Pighin JA, Zheng H, Balakshin LJ, Goodman IP, Western TL, Jetter R, Kunst L, Samuels AL (2004) Plant cuticular lipid export requires an ABC transporter. Science 306: 702–704 [DOI] [PubMed] [Google Scholar]
- Riederer M. (2006) Thermodynamics of the water permeability of plant cuticles: characterization of the polar pathway. J Exp Bot 57: 2937–2942 [DOI] [PubMed] [Google Scholar]
- Rigal A, Doyle SM, Robert S (2015) Live cell imaging of FM4-64, a tool for tracing the endocytic pathways in Arabidopsis root cells. Methods Mol Biol 1242: 93–103 [DOI] [PubMed] [Google Scholar]
- San-Bento R, Farcot E, Galletti R, Creff A, Ingram G (2014) Epidermal identity is maintained by cell-cell communication via a universally active feedback loop in Arabidopsis thaliana. Plant J 77: 46–58 [DOI] [PubMed] [Google Scholar]
- Shimada T, Fuji K, Tamura K, Kondo M, Nishimura M, Hara-Nishimura I (2003) Vacuolar sorting receptor for seed storage proteins in Arabidopsis thaliana. Proc Natl Acad Sci USA 100: 16095–16100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber P, Schorderet M, Ryser U, Buchala A, Kolattukudy P, Métraux JP, Nawrath C (2000) Transgenic Arabidopsis plants expressing a fungal cutinase show alterations in the structure and properties of the cuticle and postgenital organ fusions. Plant Cell 12: 721–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siloto RM, Findlay K, Lopez-Villalobos A, Yeung EC, Nykiforuk CL, Moloney MM (2006) The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell 18: 1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc 1: 2019–2025 [DOI] [PubMed] [Google Scholar]
- Storz G, Wolf YI, Ramamurthi KS (2014) Small proteins can no longer be ignored. Annu Rev Biochem 83: 753–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su M, Ling Y, Yu J, Wu J, Xiao J (2013) Small proteins: untapped area of potential biological importance. Front Genet 4: 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S, Jürgens G (2007) Transcriptional regulation of epidermal cell fate in the Arabidopsis embryo. Development 134: 1141–1150 [DOI] [PubMed] [Google Scholar]
- To A, Joubès J, Barthole G, Lécureuil A, Scagnelli A, Jasinski S, Lepiniec L, Baud S (2012) WRINKLED transcription factors orchestrate tissue-specific regulation of fatty acid biosynthesis in Arabidopsis. Plant Cell 24: 5007–5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuwamoto R, Fukuoka H, Takahata Y (2008) GASSHO1 and GASSHO2 encoding a putative leucine-rich repeat transmembrane-type receptor kinase are essential for the normal development of the epidermal surface in Arabidopsis embryos. Plant J 54: 30–42 [DOI] [PubMed] [Google Scholar]
- Viotti C, Krüger F, Krebs M, Neubert C, Fink F, Lupanga U, Scheuring D, Boutté Y, Frescatada-Rosa M, Wolfenstetter S, et al. (2013) The endoplasmic reticulum is the main membrane source for biogenesis of the lytic vacuole in Arabidopsis. Plant Cell 25: 3434–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. (1985) Signal sequences. The limits of variation. J Mol Biol 184: 99–105 [DOI] [PubMed] [Google Scholar]
- Wang F, Xiao J, Pan L, Yang M, Zhang G, Jin S, Yu J (2008) A systematic survey of mini-proteins in bacteria and archaea. PLoS One 3: e4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C, Lamesch P, Arumugam M, Rosenberg J, Hu P, Vidal M, Brent MR (2005) Closing in on the C. elegans ORFeome by cloning TWINSCAN predictions. Genome Res 15: 577–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickramasuriya AM, Dunwell JM (2015) Global scale transcriptome analysis of Arabidopsis embryogenesis in vitro. BMC Genomics 16: 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windsor AJ, Mitchell-Olds T (2006) Comparative genomics as a tool for gene discovery. Curr Opin Biotechnol 17: 161–167 [DOI] [PubMed] [Google Scholar]
- Wu FH, Shen SC, Lee LY, Lee SH, Chan MT, Lin CS (2009) Tape-Arabidopsis Sandwich—a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Fiume E, Coen O, Longin C, Lepiniec L, Magnani E (2016) The antagonistic development of endosperm and nucellus in Arabidopsis seeds. Plant Cell 28: 1343–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Johnston N, Talideh E, Mitchell S, Jeffree C, Goodrich J, Ingram G (2008) The endosperm-specific ZHOUPI gene of Arabidopsis thaliana regulates endosperm breakdown and embryonic epidermal development. Development 135: 3501–3509 [DOI] [PubMed] [Google Scholar]
- Yephremov A, Wisman E, Huijser P, Huijser C, Wellesen K, Saedler H (1999) Characterization of the FIDDLEHEAD gene of Arabidopsis reveals a link between adhesion response and cell differentiation in the epidermis. Plant Cell 11: 2187–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Xiao J-F, Yu J (2012) An integrated analysis of lineage-specific small proteins across eight eukaryotes reveals functional and evolutionary significance. Prog Biochem Biophys 39: 359–367 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.