Abstract
Mitochondrial dysfunction is associated with many human diseases, including cancer and neurodegeneration, that are often linked to proteins and pathways that are not well-characterized. To begin defining the functions of such poorly characterized proteins, we used mass spectrometry to map the proteomes, lipidomes and metabolomes of 174 yeast strains, each lacking a single gene related to mitochondrial biology. 144 of these genes have human homologs, 60 of which are associated with disease and 39 of which are uncharacterized. We present a multi-omic data analysis and visualization tool that we use to find covariance networks that can predict molecular functions, correlations between profiles of related gene deletions, gene-specific perturbations that reflect protein functions, and a global respiration deficiency response. Using this multi-omic approach, we link seven proteins including Hfd1p and its human homolog ALDH3A1 to mitochondrial coenzyme Q (CoQ) biosynthesis, an essential pathway disrupted in many human diseases. This Resource should provide broad molecular insights into mitochondrial protein functions.
High resolution mass spectrometry (MS) has become the primary analysis tool for many classes of biomolecules, including proteins, metabolites, and lipids. Major advancements in MS technology—particularly in the rate and depth of analysis—have enabled dozens of proteomes, metabolomes, and lipidomes to be analyzed in a single day1-3. Studies of bacteria demonstrated that parallel measurement of multiple molecule classes can synergistically enhance the biological insight afforded4, 5. Recently, proteomics has been integrated with transcriptomics and genomics in mice6, 7. However, large-scale, comprehensive (i.e., proteome-wide), multi-omic data acquisition, integration, and visualization tools remain underdeveloped, often lagging behind genomics in terms of coverage, speed, and broad accessibility for end users. Given the interdependence of proteins, lipids, and metabolites, we reasoned that coordinated analysis across all three biomolecule classes could afford new insight into eukaryotic biology. In particular, we hypothesized that this multi-omic profiling strategy, when coupled with genetic and environmental perturbations, could enable functional predictions for uncharacterized proteins.
We applied this strategy to study mitochondria, dynamic organelles whose dysfunction is associated with over 150 human diseases including cancer, diabetes, Parkinson's, and numerous genetic disorders8-10. While the yeast and mammalian mitochondrial proteomes were recently defined11-13, functional annotation of these proteins lags behind14, impeding biomedical research on the many diseases impacted by mitochondrial metabolism. Of the ~1,200 mammalian mitochondrial proteins, nearly 300 are “mitochondrial uncharacterized (x) proteins” (MXPs)15, 16 that have no well-established biochemical function within mitochondria. Here, toward defining functions for MXPs, we performed over 3,000 MS experiments in parallel to analyze the proteomes, metabolomes, and lipidomes of 174 single-gene deletion (“Δgene”) Saccharomyces cerevisiae yeast strains in biological triplicate across two metabolic conditions, fermentation and respiration (Fig. 1a). To facilitate development of biological hypotheses based on the resultant “yeast-three-thousand (Y3K)” dataset (Fig. 1b), we also developed a multi-omic data visualization approach (highlighted in Fig. 1c and online at http://y3kproject.org/). Our data establish many new connections between MXPs and proteins with well-established functions by virtue of gene-specific phenotypes or shared global biomolecular changes that result from the loss of each protein's expression. We leveraged a subset of these connections to address the incomplete mitochondrial pathway that generates ubiquinone (coenzyme Q, CoQ), an essential lipid required for oxidative phosphorylation (OxPhos) and linked to diseases ranging from severe infantile multisystemic disease to isolated myopathy and aging17, 18.
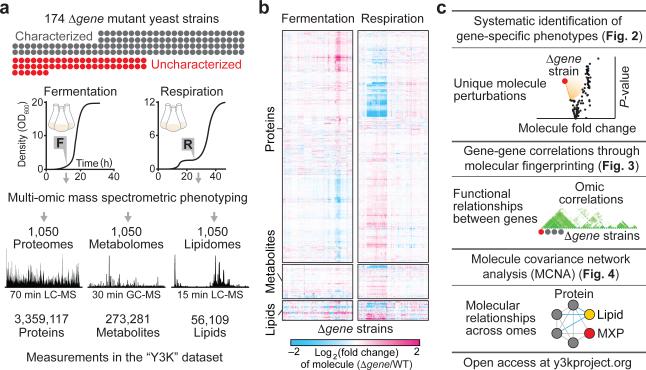
Figure 1.
Multi-omic mass spectrometry profiling and data visualization. Overviews of (a) the experimental design and high resolution quantitative MS analysis, (b) the Y3K dataset, shown as hierarchical clusters of Δgene strains and significantly perturbed molecules (relative abundances compared to WT as quantified by MS, mean, n = 3; P < 0.05, two-sided Student's t-test), and (c) the multi-omic data analysis and visualization tools developed here.
RESULTS
Multi-omic mass spectrometry profiling
The 174 Δgene yeast strains we analyzed covered 124 characterized genes that were selected to span a broad range of pathways to assist functional mapping, and 50 uncharacterized genes that encode MXPs (Fig. 1a, Supplementary Fig. 1a, and Supplementary Table 1). In selecting these targets, we prioritized genes with human homologs (144/174 genes) and those associated with disease (60/144 genes) based on primary literature analysis and online database gene annotation (e.g., omim.org). Inclusion of characterized genes, some of which could be considered as only partially characterized, also provided the ability to connect them to previously unrecognized functions. Each strain was grown in biological triplicate under two contrasting growth conditions, a standard fermentation culture condition and a carefully optimized respiration culture condition that stimulates mitochondrial function (Fig. 1a, Supplementary Fig. 1b–e, and Supplementary Note 1)—yielding six separate cultures per yeast strain.
Altogether we grew more than 1,050 yeast cultures (including WT cultures), each of which was analyzed using three separate high-resolution MS-based proteomic, metabolomic, and lipidomic techniques. These 3,000+ MS experiments yielded quantitation of 4,040 proteins, 411 metabolites, and 53 lipids (averaging 3,180 proteins, 252 metabolites, and 53 lipids per culture) (Supplementary Table 2)—over 3.5 million biomolecule measurements in total (Fig. 1a, Supplementary Fig. 2a,b, and Supplementary Table 3). Key to our approach was streamlining procedures for proteome extraction and preparation to under two hours of hands-on time (Supplementary Fig. 2c). Use of label-free quantitation negated the need for a chemical tagging step and further increased throughput. We observed a wide dynamic range across all profiled omes, with some molecule abundances spanning more than three orders of magnitude (Supplementary Fig. 2d). Additionally, we observed remarkable reproducibility between replicate cultures, with a median coefficient of variation of 12.7% considering all profiled biomolecules, and high overlap of molecules quantified across cultures (Supplementary Fig. 2e–g).
A high-level view of the Y3K dataset shows significant perturbations across all three omes, with more pronounced perturbations in respiration (Fig. 1b and Supplementary Fig. 3a). Hierarchical clustering revealed groups of functionally related molecules (along the y-axis) and groups of functionally related Δgene strains (along the x-axis). Protein clusters show significant gene ontology (GO) term enrichments for diverse processes and include both characterized and uncharacterized proteins (Supplementary Fig. 3b). For example, the uncharacterized proteins Esbp6p and Ypr010c-a cluster with proteins involved in mitochondrial ATP synthesis and electron transport chain function, respectively (Supplementary Fig. 4). Here, we leverage analyses from three different vantage points, each of which can be recapitulated with our online data visualization suite, exploiting unique biological perspectives afforded by a multi-omic dataset of diverse genetic perturbations (Fig. 1c).
Identification of gene-specific phenotypes
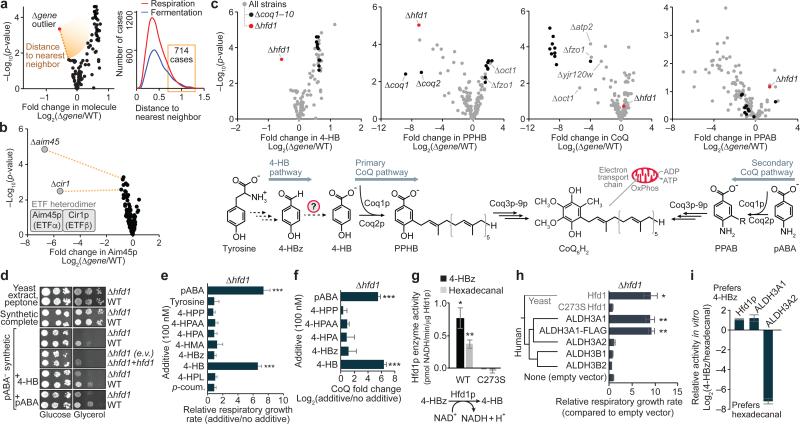
First, we systematically surveyed the Y3K dataset for significant molecule perturbations unique to just one or two of the strains in the study (Fig. 2a). This unbiased search revealed 714 Δgene-specific phenotypes (Fig. 2a, Supplementary Table 4, and Supplementary Note 2), which can reveal functional relationships. For example, the electron transfer flavoprotein (ETF) subunit Aim45p was uniquely decreased in just two Δgene strains: the Δaim45 strain, and the Δcir1 strain, which lacks the second ETF heterodimer subunit (Fig. 2b). Numerous additional Δgene-specific phenotypes were used to generate biological hypotheses (Supplementary Figs. 5 and 6). We decided to investigate one of these observations at biochemical depth: a Δhfd1-specific decrease in 4-hydroxybenzoate (4-HB), the CoQ headgroup precursor (Fig. 2c).
Figure 2.
ΔGene-specific phenotype detection links Hfd1p to production of 4-hydroxybenzoate for coenzyme Q biosynthesis. (a) Overview of the Δgene-specific phenotype detection approach and number of Δgene-specific phenotypes identified in the respiration and fermentation datasets (distance to nearest neighbor on a normalized scale, see Supplementary Note 2). (b) Relative abundance of Aim45p (mean, n = 3) versus statistical significance across strains. (c) Relative abundances of 4-HB, PPHB, CoQ, and PPAB (mean, n = 3) versus statistical significance across Δgene strains. (d) Serial dilutions of yeast grown on variable solid medias. E.v., empty vector; +hfd1, hfd1 plasmid transformed. (e) Relative respiratory growth rates of Δhfd1 yeast in pABA− synthetic media with the additives shown (mean ± s.d, n = 3). 4-HPP, 4-hydroxyphenylpyruvate; 4-HPAA, 4-hydroxyphenylacetaldehyde; 4-HPA, 4-hydroxyphenylacetate; 4-HMA, 4-hydroxymandelate; 4-HPL, 4-hydroxyphenyllactate; p-coum., para-coumarate. (f) Relative CoQ abundance in Δhfd1 yeast cultured in pABA− media with the additives shown (mean ± s.d., n = 3). (g) Enzyme activity of recombinant MBP-Hfd1CΔ25 in vitro against 4-HBz (200 μM) or hexadecanal (200 μM) (mean ± s.e.m., n = 3). (h) Phylogenetic relationship between yeast Hfd1p and the human ALDH3 family, and relative respiratory growth rates of Δhfd1 yeast transformed with plasmids encoding the proteins shown and cultured in pABA− synthetic media (mean ± s.d, n = 4). (i) Relative activity of the dehydrogenases shown against 4-HBz compared to hexadecanal (mean ± s.e.m., n = 3). *P < 0.05; **P < 0.01; ***P < 0.001 (two-sided Student's t-test for all panels).
Though it has been known for decades that mammals can convert tyrosine (Tyr) into 4-HB for CoQ biosynthesis19, 20, the biochemical pathway has remained undefined in mammals and yeast (Fig. 2c). The Y3K dataset reveals Δhfd1 yeast to be significantly deficient in both the metabolite 4-HB (P < 0.001) and the lipid CoQ intermediate 3-polyprenyl-4-hydroxybenzoate (PPHB) (P < 10−5) (Fig. 2c and Supplementary Fig. 7a). Despite the PPHB deficiency, Δhfd1 yeast have normal CoQ abundance (Fig. 2c), likely because of increased flux through an alternative para-amino-benzoate (pABA)- dependent CoQ pathway21, 22, as suggested by elevation of the aminated analog of PPHB (PPAB) in Δhfd1 yeast (Fig. 2c). This is in contrast to terminal CoQ biosynthesis genes (coq3–coq9), and some genes not previously linked to CoQ function (e.g. oct1 and fzo1), whose deletion causes significant (P < 0.05) CoQ deficiency and accumulation of PPHB (Fig. 2c). Because Hfd1p is predicted to be an aldehyde dehydrogenase23, we hypothesized that it catalyzes dehydrogenation of 4-hydroxybenzaldehyde (4-HBz) to form 4-HB. Consistently, 4-HBz is elevated in Δhfd1 yeast (Supplementary Fig. 7b).
We used chemical-genetics to test the proposed Hfd1p activity. Most culture media contain either 4-HB (in yeast extract) or pABA (in standard yeast nitrogen base), enabling yeast to bypass the Tyr-to-4-HB pathway, so we used a defined medium lacking pABA and 4-HB (“pABA−”). Δhfd1 yeast exhibited striking respiration deficiency on pABA− media, a phenotype rescued by pABA, 4-HB, or WT Hfd1p, but not by Hfd1p with mutations to putative catalytic residues24 (Fig. 2d and Supplementary Fig. 7c–e). Testing a panel of potential intermediates in the pathway revealed that 4-HB, but not 4-HBz, can rescue the respiratory growth and CoQ production of Δhfd1 yeast (Fig. 2e,f and Supplementary Fig. 7f,g), supporting a role for Hfd1p in dehydrogenation of 4-HBz. To directly test this activity, we purified recombinant Hfd1p for enzyme assays (Supplementary Fig. 7h). WT Hfd1p catalyzes NAD+-dependent dehydrogenation of 4-HBz, but a C273S (catalytic residue) point mutant does not (Fig. 2g). Together, these results demonstrate that Hfd1p dehydrogenates 4-HBz to produce 4-HB for CoQ biosynthesis.
Hfd1p is a member of the ancient aldehyde dehydrogenase (ALDH) superfamily, which is found across all three superkingdoms of life and includes 19 human homologs with diverse functions25. Based on phylogenetic analyses, Hfd1p is most similar to the human ALDH3 family (Supplementary Fig. 7i). ALDH3A2 (FALDH) mutations cause Sjögren–Larsson Syndrome26 due to defective fatty aldehyde metabolism. However, the endogenous functions of ALDH3A1, B1, and B2 remain obscure, and which of these human ALDH3 functions are conserved in Hfd1p has not been completely defined. Previous work showed that sphingolipid metabolism is perturbed in Δhfd1 yeast due to a defect in dehydrogenation of hexadecanal, and this defect can be rescued by ALDH3A2, but not by ALDH3A123, 27. However, a separate sphingolipid pathway defect (Δdpl1) does not disrupt the 4-HB-CoQ pathway (Supplementary Fig. 7j–m and Supplementary Note 3), suggesting that the two pathways are otherwise independent. Consistent with the idea that Hfd1p is a dual-function protein that supports both sphingolipid metabolism and CoQ biosynthesis, we observed Hfd1p activity in vitro with hexadecanal, similar to that observed with 4-HBz (Fig. 2g). However, in contrast to rescue of the sphingolipid metabolism defect, we found that ALDH3A1, but not ALDH3A2, rescues the pABA− respiratory growth phenotype of Δhfd1 yeast (Fig. 2h and Supplementary Fig. 7n). Moreover, while ALDH3A2 shows a strong substrate preference for hexadecanal over 4-HBz, Hfd1p and ALDH3A1 show a preference for 4-HBz (Fig. 2i and Supplementary Fig. 7o–q). These results suggest that the dual functions of yeast Hfd1p have diverged in human ALDH3A1 and ALDH3A2. Collectively, these results demonstrate a major cellular function for the aldehyde dehydrogenase Hfd1p in the Tyr-to-4-HB pathway and strongly suggest that ALDH3A1 plays a similar role in human CoQ biosynthesis.
Regression analysis of global perturbation profiles
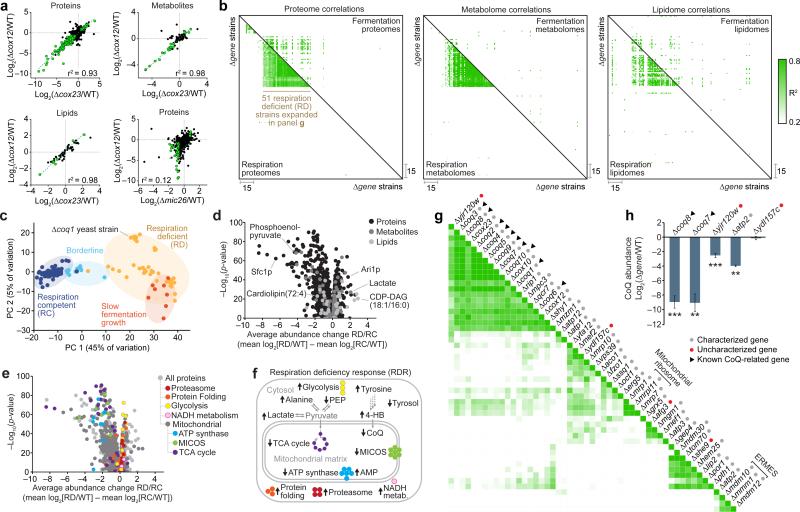
While molecular changes unique to a given Δgene strain can be functionally informative, similarities between Δgene strains can also assist characterization. In our second analysis approach, we examined Δgene–Δgene correlations through pairwise comparisons of global Δgene perturbation profiles. Deletion of functionally related genes, such as the cytochrome c oxidase genes cox12 and cox23, caused highly similar whole proteome perturbations (Fig. 3a). Notably, highly correlated phenotype changes were also observed in Δcox12 and Δcox23 metabolomes and lipidomes (Fig. 3a). However, deletion of unrelated genes, such as cox12 and mic26, generated uncorrelated phenotype changes (Fig. 3a). Examination of Δgene–Δgene correlations across the entire study indicated numerous functional relationships, with stronger correlations observed in respiration (Fig. 3b).
Figure 3.
Functional correlations through perturbation profile regression analysis. (a) Plots comparing relative molecule abundances between pairs of Δgene strains. Strain-strain similarity assessed by linear regression analysis of Δgene perturbation profiles. Green points indicate molecules significantly perturbed in both mutants (|log2[FC]| > 0.7, P < 0.05; two-sided Student's t-test). (b) Maps of Pearson correlation coefficients (r2) for pairs of Δgene perturbation profiles across omes and metabolic conditions. Strains are clustered based on respiration proteome correlations, and this strain order is held consistent across all 6 maps. (c) Projection of respiration competent (RC) and deficient (RD) strains onto the plane defined by principal component (PC) axes 1 and 2 (full multi-omic respiration dataset). (d) Average fold change in molecule abundances (mean log2[RD strains/RC strains]) versus statistical significance (−log10[p-value, Bonferroni corrected two-sided t-test]). (e) RD versus RC proteome perturbation volcano plot (as in d) showing select functional groups (GO terms) significantly enriched in either upregulated or downregulated proteins. (f) Scheme of RDR pathways. (g) Re-clustered respiration proteome strain-strain correlation map following RDR- adjustment. (h) CoQ abundance changes in select Δgene strains (mean ± s.d., n = 3); **P < 0.01; ***P < 0.001 (two-sided Student's t-test).
A group of respiration-deficient (RD) strains showed robust correlations across all three omes (Fig. 3b), reflecting their similar broad biological functions in mitochondrial OxPhos and suggesting that they share a universal “respiration deficiency response” (RDR). Multi-omic principle component and GO term analyses revealed a coordinated RDR that provides biological insight into respiration defects—a common feature of many diseases including cancer—and suggests that a multi-omic biomarker fingerprint could afford a specific diagnostic for mitochondrial disease (Fig. 3c–f, Supplementary Fig. 8, Supplementary Note 4, and Supplementary Table 5). However, stress responses such as the RDR also pose a barrier to biochemical investigations because they can obscure functionally-informative phenotypes. To assess more specific biochemical roles for individual proteins, we normalized for the RDR across RD strains (Supplementary Fig. 9 and Supplementary Note 5). Across all of our RD strains, 776 molecules were identified as being consistently perturbed (Supplementary Table 6). The individual measurements of these RDR- associated molecules were mean normalized (“RDR-adjusted”) to reveal characteristic deviations from the general RDR and to enable visualization of Δgene-specific changes. Recalculating Δgene–Δgene correlation coefficients with RDR-adjusted plots strikingly reduces correlations between more functionally disparate genes (Supplementary Fig. 9c–e and Supplementary Table 7). Reclustering Δgene–Δgene correlations reveals new clusters of genes with similar biochemical functions (Fig. 3g). For example, known CoQ biosynthesis genes were brought into a tighter cluster that also includes the uncharacterized gene yjr120w (Fig. 3g), suggesting that yjr120w might support CoQ biosynthesis. Consistently, we observed CoQ deficiency in Δyjr120w yeast (Fig. 3h), the molecular basis of which we determined to include loss of Atp2p, an ATP synthase subunit (Supplementary Fig. 10 and Supplementary Note 6). These results show that specific ATP synthase subunits support CoQ biosynthesis and, more broadly, demonstrate how global mass spectrometry profiling can reveal functional links between genes.
Molecule covariance network analysis
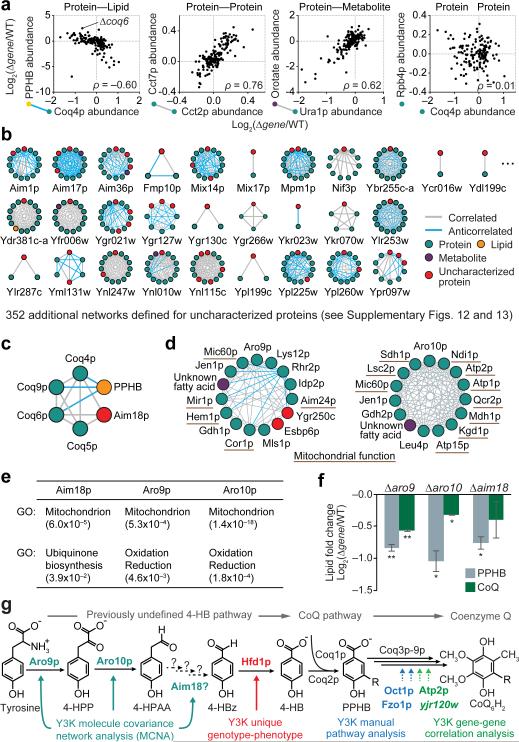
Similarly, in our third analysis approach, we leveraged the multi-omic nature of our mass spectrometry profiles to determine pairwise covariance between proteins, metabolites, and lipids. This approach is similar to mRNA coexpression profiling, which can be used to predict gene function28-30, but it integrates three complementary classes of molecules. Perturbations for functionally related molecules, such as the protein Coq4p and the lipid CoQ intermediate PPHB, show strong positive or negative correlations, while those of unrelated molecules, such as Coq4p and Rpb4p, lack correlations (Fig. 4a). Correlated molecules include proteins in complexes, such as the cytosolic TRiC/CCT chaperonin complex (Cct2p and Cct7p), and enzyme-product pairs (e.g. Ura1p and orotic acid) (Fig. 4a).
Figure 4.
Multi-omic molecule covariance network analysis assists functional characterization. (a) Relative abundances of molecule pairs across Δgene strains. Covariance assessed by Spearman's rank coefficient (ρ). (b) Nearest neighbor molecule covariance networks for a representative subset of uncharacterized proteins. (c) Network for Coq4p in the RDR-adjusted respiration dataset. (d) Networks showing the 14 molecules most strongly correlated to Aro9p or Aro10p in the RDR-adjusted respiration dataset. (e) GO term analyses of the Aim18p, Aro9p, and Aro10p networks (p-values). (f) Relative abundances of CoQ and PPHB (mean log2[Δgene/WT], n = 2) in Δaro9, Δaro10, and Δaim18 strains compared to WT yeast cultured in pABA− media; *P < 0.05; **P < 0.01 (two-sided Student's t-test). (g) Y3K-enabled characterization of proteins that support the CoQ pathway.
Examining correlations across all 4,505 molecules in the Y3K dataset through this multi-omic molecule covariance network analysis (MCNA) reveals numerous functional relationships, which can be visualized as networks of molecules (nodes) and correlations (edges) (Fig. 4b, Supplementary Fig. 11a, and Supplementary Table 8). After applying strict correlation thresholds (Bonferroni-adjusted p-value < 0.001, |ρ| ≥ 0.58), 237,342 edges remain among 2,382 nodes in the respiration dataset (Supplementary Fig. 11a–f). Many edges were observed between RDR-associated molecules (Supplementary Fig. 11g), reflecting their common relationship to mitochondrial metabolism. As described above for Δgene correlations, we deepened the molecular insight of the MCNA by RDR-adjustment, which reduced overall connectivity and increased the selectivity of functionally related molecule sub-networks (Supplementary Fig. 11g). For example, the selectivity of the mitochondrial ribosome sub-network increased 16-fold (Supplementary Fig. 11h). These RDR-adjusted networks associated the MXP Yor020w-a with the mitochondrial ribosome (Supplementary Fig. 11g). To test this association, we examined the proteome of Δyor020w-a yeast, which showed a significant decrease in the mitochondrial ribosome protein Rsm19p (Supplementary Fig. 11i), suggesting that Yor020w-a is linked to mitochondrial translation.
Hundreds of additional uncharacterized proteins were linked to characterized molecules by our MCNA, providing a foundation for generating hypotheses about their functions (Fig. 4b, Supplementary Figs. 12 and 13). For example, the MXP Aim18p was linked to a network of CoQ biosynthesis proteins, and Aro9p and Aro10p were linked to numerous mitochondrial proteins that support OxPhos (Fig. 4c–e). Based on domain homology and predicted enzymatic functions, we hypothesized that Aim18p, Aro9p, and Aro10p could function in the Tyr- to-4-HB pathway (Supplementary Fig. 14 and Supplementary Note 7). Consistently, when cultured in a pABA− media, Δaim18, Δaro9, and Δaro10 yeast are deficient in both CoQ and PPHB (Fig. 4f). This work shows how global mass spectrometry profiling can be used to generate biological hypotheses and characterize protein functions through distinct multi-omic data analysis approaches (Fig. 4g).
DISCUSSION
A constant challenge in biology is to comprehensively monitor and understand the molecular effects of a defined alteration (e.g., a disease mutation, a drug treatment, or a gene deletion). Mass spectrometry (MS) has become central to answering this challenge.
Here, we leveraged a subset of our multi-omic dataset to investigate gaps in knowledge of CoQ biosynthesis. Despite CoQ's essential function in the mitochondrial electron transport chain, role as a key cellular antioxidant, and link to numerous human diseases (e.g., ataxias, myopathies, and nephrotic syndromes), multiple steps in CoQ biosynthesis remain uncharacterized17, 31, 32. In particular, enzymes involved in the initial stage of CoQ biosynthesis— wherein the headgroup precursor 4-HB is produced—were previously undefined in mammals and yeast.
Our Δgene-specific phenotype detection approach suggested a role for the ancient aldehyde dehydrogenase superfamily member Hfd1p in 4-HB biosynthesis. Biochemical and genetic studies confirmed this role for Hfd1p in yeast and further demonstrated that the human homolog ALDH3A1 can also catalyze production of 4-HB in vivo and in vitro (Fig. 2), thereby highlighting ALDH3A1 as a candidate disease gene for primary CoQ deficiency.
Distinct Y3K dataset analyses placed additional proteins into the CoQ biosynthesis pathway. MCNA showed unexpected connections between Aro9p, Aro10p, and mitochondrial OxPhos proteins, which helped place Aro9p and Aro10p into the Tyr-to-4-HB pathway (Fig. 4). Similarly, links between Aim18p and known CoQ biosynthesis enzymes also connected Aim18p to CoQ biosynthesis. Furthermore, Y3K gene-gene correlation analyses and manual pathway analyses linked CoQ biosynthesis to other proteins whose molecular functions in this pathway are not yet fully defined (e.g. Atp2p, Fzo1p, and Oct1p). Disruption of the mammalian Fzo1p homolog, MFN2—a protein essential for mitochondrial fusion that harbors causative mutations in Charcot-Marie-Tooth disease33—was recently shown to cause CoQ deficiency through an unclear molecular mechanism34. Our results suggest that this unexpected relationship between MFN2 and CoQ biosynthesis is evolutionarily conserved, and establish yeast as a model system for further probing its mechanism.
Our Y3K dataset provides many additional leads for further biochemical studies of numerous metabolic pathways that impact human health and disease, and we expect that the open access web utility (http://y3kproject.org/) will enable others to generate their own hypotheses. With demand for multi-omic dataset analysis approaches increasing, we also hope that our multifaceted, data visualization website will serve as a useful model for future studies.
We anticipate that the multi-omic Y3K dataset will provide a resource for broader systems biology inquiries. For example, our definition of the yeast respiration deficiency response (RDR) (Fig. 3) may assist studies of how cells broadly respond to defects in OxPhos, which are observed in diverse diseases including many cancers. Our RDR work also suggests that a multi-omic fingerprint of numerous molecules could provide a highly specific biomarker panel.
ONLINE METHODS
Yeast strains and cultures
The parental (WT) Saccharomyces cerevisiae strain for this study was the haploid MATalpha BY4742. Single gene deletion (Δgene) derivatives of BY4742 were either obtained through the gene deletion consortium30 or made in-house using a KanMX deletion cassette to match those in the consortium collection. All gene deletions were confirmed by either proteomics (significant decrease in the encoded protein) or a PCR assay. Δgene strains made in-house were also confirmed by gene sequencing.
Single lots of yeast extract (‘Y’) (Research Products International, RPI), peptone (‘P’) (RPI), agar (Fisher), dextrose (‘D’) (RPI), glycerol (‘G’) (RPI), and G418 (RPI) were used for all medias. YP and YPG solutions were sterilized by automated autoclave. G418 and dextrose were sterilized by filtration (0.22 μm pore size, VWR) and added separately to sterile YP or YPG. YPD+G418 plates contained yeast extract (10 g/L), peptone (20 g/L), agar (15 g/L), dextrose (20 g/L), and G418 (200 mg/L). YPD media (fermentation cultures) contained yeast extract (10 g/L), peptone (20 g/L), and dextrose (20 g/L). YPGD media (respiration cultures) contained yeast extract (10 g/L), peptone (20 g/L), glycerol (30 g/L) and dextrose (1 g/L).
Yeast from a −80 °C glycerol stock were streaked onto YPD+G418 plates and incubated (30 °C, ~60 h). Starter cultures (3 mL YPD) were inoculated with an individual colony of yeast and incubated (30 °C, 230 rpm, 10–15 h). A WT culture was included with each set of Δgene strain cultures (usually 19 Δgene cultures and 1 WT culture). Cell density was determined by optical density at 600 nm (OD600) as described35. YPD or YPGD media (100 mL media at ambient temperature in a sterile 250 mL Erlenmeyer flask) was inoculated with 2.5×106 yeast cells and incubated (30 °C, 230 rpm). Samples of the YPD cultures were harvested 12 h after inoculation, a time point that corresponds to early fermentation (logarithmic) growth. Samples of YPGD cultures were harvested 25 h after inoculation, a time point that corresponds to early respiration growth.
Liquid chromatography tandem mass spectrometry (LC-MS/MS) proteomics
1×108 yeast cells were harvested by centrifugation (3,000 g, 3 min, 4 °C), the supernatant was removed, and the cell pellet was flash frozen in N2(l) and stored at −80 °C. Yeast pellets were resuspended in 8 M urea, 100 mM tris (pH = 8.0). Yeast cells were lysed by the addition of methanol to 90%, followed by vortexing (~30 s). Proteins were precipitated by centrifugation (12,000 g, 5 min). The supernatant was discarded, and the resultant protein pellet was resuspended in 8 M urea, 10 mM tris(2-carboxyethyl)phosphine (TCEP), 40 mM chloroacetamide (CAA) and 100 mM tris (pH = 8.0). Sample was diluted to 1.5 M urea with 50 mM tris and digested with trypsin (Promega) (overnight, ~22 °C) (1:50, enzyme:protein). Samples were desalted using Strata X columns (Phenomenex Strata-X Polymeric Reversed Phase, 10 mg/mL). Strata X columns were equilibrated with one column volume of 100% acetonitrile (ACN), followed by 0.2% formic acid. Acidified samples were loaded on column, followed by washing with three column volumes of 0.2% formic acid or 0.1% TFA. Peptides were eluted off the column by the addition of 500 μL 40% ACN with either 0.2% formic acid or 0.1% TFA and 500 μL 80% ACN with either 0.2% formic acid or 0.1% TFA. Peptide concentration was measured using a quantitative colorimetric peptide assay (Thermo). LC-MS/MS analyses were performed using previously described methodologies1, 2.
Data analysis
Raw data files were acquired in batches of 60 (3 biological replicates of 19 Δgene strains and 1 WT strain) with time between LC-MS analyses minimized to reduce run-to-run variation. Batches of raw data files were subsequently processed using MaxQuant36 (Version 1.5.0.25). Searches were performed against a target-decoy37 database of reviewed yeast proteins plus isoforms (UniProt, downloaded January 20, 2013) using the Andromeda38 search algorithm. Searches were performed using a precursor search tolerance of 4.5 ppm and a product mass tolerance of 0.35 Da. Specified search parameters included fixed modification for carbamidomethylation of cysteine residues and a variable modification for the oxidation of methionine and protein N-terminal acetylation, and a maximum of 2 missed tryptic cleavages. A 1% peptide spectrum match (PSM) false discovery rate (FDR) and a 1% protein FDR was applied according to the target-decoy method. Proteins were identified using at least one peptide (razor + unique). Proteins were quantified using MaxLFQ with an LFQ minimum ratio count of 2. LFQ intensities were calculated using the match between runs feature, and MS/MS spectra were not required for LFQ comparisons. Missing values were imputed where appropriate for proteins quantified in ≥ 50% of MS data files in a batch. Proteins not meeting this requirement were omitted from subsequent analyses. Imputation was performed on a replicate-by-replicate basis. For each replicate MS analysis a normal distribution with mean and standard deviation equivalent to that of the lowest 1% of measured LFQ intensities was generated. Missing values were filled in with values drawn from this distribution at random. Approximately 4.05% and 4.53% of quantitative measurements were imputed in the respiration and fermentation proteomic datasets, respectively. Replicate protein LFQ values from corresponding Δgene or WT strains were pooled, log2 transformed, and averaged (mean log2[strain], n = 3). Average Δgene LFQ intensities were normalized against their appropriate WT control (mean log2[Δgene/WT], n = 3) and a 2-tailed t-test (homostatic) was performed to obtain P values.
To control for batch-specific effects, proteins having unexpected and characteristic misregulation across a majority of Δgene strains processed together were identified and omitted from the dataset. For each protein quantified within a batch of Δgene strains a distribution of protein fold-changes (intra-batch) was generated. The analogous distribution of protein fold-changes from all other Δgene strains processed separately (inter-batch) was created. These two distributions were compared against each other using a Kolmogorov-Smirnov test (2-tailed) to obtain P values. If a significant difference existed at P < 0.05 (Bonferroni-adjusted) protein abundance measurements were omitted from the batch in question. This process of comparing intra-batch and inter-batch protein fold change distributions was carried iteratively and to exhaustion and resulted in the omission of an average 165 proteins/Δgene strain (~4.8% of quantified proteins) for respiration, and 188 proteins/Δgene strain (~5.9%) for fermentation.
Liquid chromatography-mass spectrometry (LC-MS) lipidomics
1×108 yeast cells were harvested by centrifugation (3,000 g, 3 min, 4 °C), the supernatant was removed, and the cell pellet was flash frozen in N2(l) and stored at −80 °C. Frozen yeast pellets (1×108 cells) were thawed on ice and mixed with glass beads (0.5 mm diameter, 100 μL). CHCl3/MeOH (1:1, v/v, 4 °C) (900 μL) was added and vortexed (2 × 30 s). HCl (1 M, 200 μL, 4 °C) was added and vortexed (2 × 30 s). The samples were centrifuged (5,000 g, 2 min, 4 °C) to complete phase separation. 400 μL of the organic phase was transferred to a clean tube and dried under Ar(g). The organic residue was reconstituted in ACN/IPA/H2O (65:30:5, v/v/v) (100 μL) for LC-MS analysis.
LC-MS analysis was performed on an Ascentis Express C18 column held at 35 °C (150 mm × 2.1 mm × 2.7 μm particle size; Supelco) using an Accela LC Pump (500 μL/min flow rate; Thermo). Mobile phase A consisted of 10 mM ammonium acetate in ACN/H2O (70:30, v/v) containing 250 μL/L acetic acid. Mobile phase B consisted of 10 mM ammonium acetate in IPA/ACN (90:10, v/v) with the same additives. Initially, mobile phase B was held at 50% for 1.5 min and then increased to 95% over 6.5 min where it was held for 2 min. The column was then reequilibrated for 3.5 min before the next injection. 10 μL of sample were injected by an HTC PAL autosampler (Thermo). The LC system was coupled to a Q Exactive mass spectrometer (Build 2.3 SP2) by a HESI II heated ESI source kept at 325 °C (Thermo). The inlet capillary was kept at 350 °C, sheath gas was set to 35 units, and auxiliary gas to 15 units, and the spray voltage was set to 3,000 V. Several scan functions were used to achieve optimal data acquisition for different lipid classes. For phospholipids, MS1 data was acquired from 1–9 min at a resolving power of 35,000 with the AGC target set to 1×106, mass range to 500–900 Th, and maximum injection time to 250 ms. For fatty acids and lyso species, MS1 data was acquired from 0–3 min at a resolving power of 17,500 with the AGC target set to 5×105, mass range to 220–600 Th, and maximum injection time to 100 ms. For cardiolipins, MS1 data was acquired from 6.5–9.5 min at a resolving power of 17,500 with the AGC target set to 5×105, mass range to 1320–1500 Th, and maximum injection time to 250 ms. For cytidine diacylglycerols, MS1 data was acquired from 1–4.5 min at a resolving power of 17,500 with the AGC target set to 5×105, mass range to 920–1050 Th, and maximum injection time to 250 ms. Quantitation for all of these species was performed by integrating the MS1 peak areas of either the [M–H]− or [M+Ac]− ions. Coenzyme Q6 and demethoxycoenzyme Q6 were monitored from 4.7 to 5.8 min by positive ion tandem mass spectrometry using the 591.44 → 197.08 Th and 561.43 → 167.07 Th transitions at a normalized collision energy of 27 units, a resolving power of 17,500, a maximum injection time of 250 ms, and an isolation width of 1.5 Th. For some follow-up studies MS1 spectra were acquired from 200–1550 m/z supplemented with scheduled targeted scan modes to quantify key CoQ intermediates in their optimal polarity.
Data analysis
Peaks were automatically integrated using TraceFinder software (Thermo) and all integrations were checked manually. Missing values from undetected peaks were imputed and imputation was performed on a replicate-by-replicate basis. For each replicate MS analysis a normal distribution with mean and standard deviation equivalent to that of the lowest 5% of measured peak intensities was generated. Missing values were filled in with values drawn from this distribution at random. Approximately 1.03% and 0.85% of values were imputed in the respiration and fermentation lipidomic datasets, respectively. Total measured ion current from peaks quantified within replicate MS analyses was normalized to a corresponding WT control using a two-step procedure. First, to account for differences in cardiolipin extraction efficiency, summed cardiolipin intensities were normalized to equal the summed intensity of corresponding cardiolipin species in the WT control. All other lipid intensities were then normalized to the equal the summed intensity of non-cardiolipin species in the same control. Replicate lipid intensities from corresponding Δgene or WT strains were pooled, log2 transformed, and averaged (mean log2[strain], n = 3). Mean intensities were then normalized to WT (mean log2[Δgene/WT], n = 3) and a 2-tailed t-test (homostatic) was performed to obtain P values.
Gas chromatography-mass spectrometry (GC-MS) metabolomics
1×108 yeast cells yeast cells were isolated by rapid vacuum filtration onto a nylon filter membrane (0.45 μm pore size, Millipore) using a Glass Microanalysis Filter Holder (Millipore), briefly washed with phosphate buffered saline (1 mL), and immediately submerged into ACN/MeOH/H2O (2:2:1, v/v/v, 1.5 mL, pre-cooled to −20 °C) in a plastic tube. The time from sampling yeast from the culture to submersion in cold extraction solvent was less than 30 s. Tubes with the extraction solvent, nylon filter, and yeast were stored at −80 °C prior to analysis.
Tubes with yeast extract (also still containing insoluble yeast material and the nylon filter) were thawed at room temperature for 45 min., vortexed (~15 s), and centrifuged at room temperature (6400 rpm, 30 s) to pellet insoluble yeast material. Yeast extract (25 μL aliquot) and internal standards (25 μL aqueous mixture of isotopically labelled alanine-2,3,3,3-d4, adipic acid-d10, and xylose-13C5 acid, 5 ppm in each) were aliquoted into a 2 mL plastic tube and dried by vacuum centrifuge (~ 1 hr). The dried metabolites were resuspended in pyridine (25 μL) and vortexed. 25 μL of N-methyl-N-trimethylsilyl]trifluoroacetamide (MSTFA) with 1% trimethylchlorosilane (TMCS) was added, and the sample was vortexed and incubated (60 °C, 30 min). Samples were then transferred to a glass autosampler vials and analyzed using a GC/MS instrument comprising a Trace 1310 GC coupled to a Q Exactive Orbitrap mass spectrometer. For the yeast metabolite extracts a linear temperature gradient ranging from 50 °C to 320 °C was employed spanning a total runtime of 30 minutes. Analytes were injected onto a 30 m TraceGOLD TG-5SILMS column (Thermo) using a 1:10 split at a temperature of 275 °C and ionized using electron ionization (EI). The mass spectrometer was operated in full scan mode using a resolution of 30,000 (m/Δm) relative to 200 m/z.
Data analysis
The resulting GC-MS data were processed using an in-house developed software suite (https://github.com/coongroup/Y3K-Software). Briefly, all m/z peaks are aggregated into distinct chromatographic profiles (i.e., feature) using a 10 ppm mass tolerance. These chromatographic profiles are then grouped according to common elution apex (i.e., feature group). The collection of features (i.e., m/z peaks) sharing a common elution apex, therefore, represent an individual EI-MS spectrum of a single eluting compound. The EI-MS spectra were then compared against a matrix run and a background subtraction was performed. Remaining EI-MS spectra are then searched against the NIST 12 MS/EI library and subsequently subjected to a high resolution filtering (HRF) technique as described elsewhere. EI-MS spectra that were not identified were assigned a numeric identifier. Feature intensity, which was normalized using total metabolite signal, was used to estimate metabolite abundance. Following initial processing, raw data files were re-analyzed to extract metabolite signals which were not successfully deconvolved and registered as missing values in the dataset. This process provided measurements for ~1.87%, and 2.25% of metabolites quantified in the respiration and fermentation datasets, respectively. Remaining missing values were imputed using the same imputation strategy as described in the proteomic data processing section. Quantitative values imputed using this process account for ~0.17% and 0.13% of metabolites in the respiration and fermentation datasets, respectively.
Replicate metabolite intensities from corresponding Δgene or WT strains were pooled, log2 transformed, and averaged (mean log2[strain], n = 3). Average Δgene metabolite intensities were normalized against their appropriate WT control (mean log2[Δgene/WT], n = 3) and a 2-tailed t-test was performed to obtain P values. To account for batch-specific effects the same Kolmogorov–Smirnov testing approach as described in the proteomic data processing section was used. Distributions of inter-batch and intra-batch metabolite fold changes were compared iteratively and those that were significantly different at P < 0.05 (Bonferroni-adjusted) resulted in metabolite abundance measurements being omitted from the batch in question (~15 metabolites/Δgene strain (~5.0%) from respiration and ~21 metabolites/Δgene strain (~5.9%) from fermentation).
ΔGene-Specific Phenotype Detection
For each profiled molecule (in both respiration and fermentation growth conditions) we separated potential Δgene-specific measurements into two groups: positive log2 fold change (log2[Δgene/WT]) and negative log2 fold change. These two sets were then plotted individually with log2 fold change and −log10(p-value [two-sided Student's t-test]) along the x- and y- axes, respectively. Data were normalized such that the largest log2 fold change and largest −log10(p-value) were set equal to 1. Considering the three largest fold changes where P < 0.05, we calculated the Euclidean distance to all neighboring data points and stored the smallest result. A requirement was imposed that all considered ‘neighbors’ have a smaller fold change than the data point being considered. It is anticipated that data points corresponding to Δgene-specific phenotypes will be outliers in the described plots and have large associated nearest-neighbor Euclidean distances. The described routine yielded three separate distances, the largest of which was stored for further analysis. We set a cutoff for classification as a ‘Δgene-specific phenotype’ at a Euclidean distance of 0.70.
Regression analysis of phenotype changes
Regression analysis of Δgene–Δgene perturbation profiles
For all pairwise combinations of Δgene strains from the same growth condition linear regression analysis was conducted on protein, lipid, and metabolite perturbation profiles, respectively. Fold change measurements (mean log2[Δgene/WT], n = 3) from molecules where FC > 0.7 and P < 0.05 were used and a minimum of 20 proteins, 10 metabolites, and 5 lipids, respectively, were required. These measurements were fit to a line and the associated Pearson correlation coefficient was reported. Coefficients carrying negative signs were set to 0. For pairs of Δgene strains lacking a sufficient number of molecules that met the aforementioned criteria, the Pearson coefficient was reported as 0. Hierarchical clustering of Δgene–Δgene correlations was performed as described below.
Respiration deficiency response (RDR) abundance adjustment
All Δgene strains grown under respiration conditions were classified as respiration deficient (RD) (51) or respiration competent (RC) (123) based on observation of a common perturbation profile signature. For all molecules profiled within RD Δgene strains an RDR score was calculated. This metric represents the proportion of RD Δgene strains over which the molecule was consistently perturbed, relative to all RD Δgene strains where the molecule was quantified. Considering all RD Δgene strains, 776 molecules produced an RDR score > 0.95 (consistently perturbed across more than 95% of RD Δgene strains where quantified) and were subsequently classified as RDR-associated. For each RDR-associated molecule, individual RD Δgene strain measurements were mean normalized and stored. These RDR-adjusted measurements were then used in described respiration–RDR analyses.
Regression analysis of RDR-adjusted Δgene–Δgene perturbation profiles
For all RD Δgene strains linear regression analysis was performed pairwise on RDR-adjusted protein perturbation profiles. Fold change measurements from molecules where FC > 0.7 and P < 0.05 (p-value prior to RDR adjustment) were used and a minimum of 20 proteins was required. Correlations and clustering were otherwise conducted as described above.
Hierarchical clustering
All hierarchical clustering performed in this study was done in Perseus. For all clustering operations Spearman correlation was used with average linkage, preprocessing with k-means, and the number of desired clusters set to 300 for both rows and columns.
For clustering of Δgene perturbation profiles, clustering was performed separately for fermentation and respiration datasets, and column-wise cluster order for fermentation and respiration datasets was generated using only protein fold change profiles. Column ordering was then applied to metabolite and lipid fold change datasets from the corresponding growth condition and row-wise clustering was conducted. GO term enrichment was performed in Perseus. P values were obtained from a Fisher's exact test, adjusted for multiple hypothesis testing 39 and reported where P < 0.05.
For the analysis of Δgene–Δgene correlations, clustering was performed on respiration protein perturbation profile correlation data and the resultant ordering was applied to Δgene–Δgene correlation datasets from all other omes and growth conditions for parallel visual display. The same clustering process was carried out for the analysis of Δgene–Δgene correlations of RD Δgene strains following RDR-adjustment.
Generation of Δgene strains and cloning of genes and mutants for follow-up studies
S. cerevisiae (BY4742) gene deletion strains for hfd1, atp2, ypr010c-a, and yjr120w were generated using a PCR deletion strategy in which the open reading frames were replaced by a KanMX cassette from the pFA6a-kanMX6 plasmid. Briefly, KanMX was amplified with primers containing sequence homologous to sequence just upstream of the ATG and just downstream from the terminal codon for each ORF. Amplicons were transformed into BY4742, and yeast were plated onto YEPD plates containing 100 μg/mL G418. Knockouts were confirmed by PCR and sequencing.
To generate plasmid yeast gene constructs, S. cerevisiae hfd1, atp2, and yjr120w were amplified by Accuprime Pfu polymerase (Invitrogen, USA) with primers generating a SpeI site (forward) and SalI (reverse) (BamHI forward and EcoRI reverse for yjr120w). The hfd1, atp2, and yjr120w amplicons and the yeast expression vectors p426GPD and p423GPD were digested with SpeI and SalI or BamHI and EcoRI. Hfd1 and yjr120w were ligated to p426GPD, atp2 was ligated to p423GPD, and each ligation was transformed into DH5α E. coli. Plasmid minipreps were performed and recombinants were confirmed by sequencing. Hfd1 mutants were generated via standard site-directed mutagenesis, and mutations wereconfirmed by sequencing.
To generate plasmid human gene constructs, Homo sapiens ALDH3A1 and ALDH3A2 were amplified by Accuprime Pfu polymerase with primers generating a SpeI site (forward) and SalI (reverse). The ALDH3A1 and ALDH3A2 amplicons and the yeast expression vector p426GPD were digested with SpeI and SalI. ALDH3A1 and ALDH3A2 were ligated to p426GPD and each ligation was transformed into DH5α E. coli. Plasmid minipreps were performed and recombinants were confirmed by sequencing.
Yjr120w molecular biology studies
Yeast growth assays
Δatp2 and Δyjr120w yeast were transformed with p426GPD plasmids (either encoding for Yjr120w or empty vector) and p423GPD (either encoding for Atp2p or empty vector) and grown on Ura−, His− plates containing 2% glucose. Starter cultures were inoculated with individual colonies of yeast and incubated (30 °C, ~16 h, 230 rpm). To assay Δatp2 and Δyjr120w yeast growth on agar plates, serial dilutions of yeast from a starter culture were prepared in Ura−, His− media lacking glucose. 10-fold serial dilutions of yeast cells were dropped onto Ura−, His− agar media plates containing either glucose (2%, w/v) or glycerol (3%, w/v) and incubated (30 °C, 4 d).
mRNA quantitation
BY4742 WT, Δcoq8, Δatp2, and Δyjr120w yeast were grown overnight in 3 mL YEPD. From the overnight culture, 2.5×106 cells were used to inoculate 100 mL YPGD media. 1 mL of culture was collected after 25 hours and total RNA was isolated using Masterpure Yeast RNA Purification Kit (Epicentre). 1 μg of RNA was reverse transcribed using Superscript III first strand synthesis kit (Thermo). Using the resultant cDNA as template, set up QPCR reactions: 2 μL cDNA, 12.5 μL Power Sybr Green Master Mix (Thermo), and 300 nmol/L forward and reverse primers. Primers amplifying the following targets were used: atp2, yjr120w, and ubc6 (reference gene). QPCR cycled as follows: After an initial 2 minute incubation at 50 °C, template was denatured at 95 °C for 10 minutes, cycled 40 times: 95 °C for 15 s, 60 °C for 1 minute. RNA abundance was calculated using the ΔΔCt method.
Hfd1p and ALDH3A1 biochemical studies
Media lacking pABA
A specially formulated synthetic media lacking pABA (‘pABA−’) was used for numerous follow-up studies in this project. This media consisted of CSM Mixture; Complete, 790 mg/L (# DCS0019, Formedium LTD, Hunstanton, U.K.) and yeast nitrogen base without amino acids and para-amino benzoic acid, 6.9 g/L (# CYN4102, Formedium LTD, Hunstanton, U.K.).
Yeast growth assays
Δhfd1 yeast transformed with p426GPD plasmids encoding for Hfd1p variants were grown on uracil drop-out (Ura−) synthetic media plates containing glucose (2%, w/v). Individual colonies of yeast were used to inoculate starter cultures of synthetic media lacking pABA (pABA−) but containing 20 g/L glucose. To assay WT and Δhfd1 yeast growth on agar plates, serial dilutions of yeast from a starter culture were prepared in pABA− media lacking glucose. 104, 103, or 102 yeast cells were dropped onto agar media plates containing either glucose (2%, w/v) or glycerol (3%, w/v) and incubated (30 °C, 4 d). The base medias for the agar plates consisted of either YEP (rich media), synthetic complete, pABA−, pABA− supplemented with 100 μM 4-hydroxybenzoic acid, or pABA− supplemented with 100 μM pABA.
To assay yeast growth in liquid media, yeast from a pABA− starter culture were swapped into pABA− media with glucose (0.1%, w/v) and glycerol (3%, w/v) (base medium) at an initial density of 5×106 cells/mL. To interrogate the rescue efficacy of various compounds, 100 nM (final concentrations) of pABA, tyrosine, 4-HPP, 4-HPAA, 4-HPA, 4-HMA, 4-HBz, 4-HB, 4HPL, or p-coumarate were added to the base medium. The cultures were incubated in a sterile 96 well plate with an optical, breathable coverseal (shaking at 1140 rpm). Optical density readings (OD600) were obtained every 10 min. Respiratory growth rates were determined by fitting a linear equation to the respiratory growth phase and determining the slope of the line. Relative respiratory growth rates were determined by comparing cultures with additives to those without additive.
Quantitation of CoQ and 4-HB in pABA− Δhfd1 yeast cultures
2.5×106 Δhfd1 yeast cells from a pABA− (2% w/v glucose) starter culture were used to inoculate 100 mL of pABA− media with glucose (0.1%, w/v), glycerol (3%, w/v), and potential rescue compound (100 nM pABA, 4-HPP, 4-HPAA, 4-HPA, 4-HBz, 4-HB, or none). These 100 mL cultures were incubated (30 °C, 230 rpm). After 25 h (analogous to the primary respiration culture system used for this study), 1×108 yeast cells were harvested for lipidomic or metabolomic analyses, and CoQ and 4-HB were quantified by mass spectrometry as described above. These cultures and analyses were conducted in biological triplicate.
Hfd1p phylogenetics
The amino acid sequences of the 19 known Homo sapiens ALDH proteins25 and S. cerevisiae Hfd1p (NP_013828.1) were aligned by MUSCLE40, analyzed by ClustalW2 Phylogeny41, and visualized in iTOL42.
Mass spectrometry profiling of pABA− yeast cultures (WT, Δhfd1, Δdpl1, and Δcoq8)
2.5×106 yeast cells from a pABA− (2% w/v glucose) starter culture were used to inoculate 100 mL of pABA− media with glucose (0.1%, w/v), glycerol (3%, w/v), and rescue compound (100 μM 4-HB or none). These 100 mL cultures were incubated (30 °C, 230 rpm). After 25 h, 1×108 yeast cells were harvested for lipidomic, metabolomics, and proteomic analyses by mass spectrometry as described in the main Methods section. These cultures and analyses were conducted in biological triplicate.
Hfd1p, ALDH3A1, and ALDH3A2 expression and purification
PIPE cloning was used to generate pVP68K vectors encoding ALDH3A1, Hfd1pCΔ25, or ALDH3A2CΔ25 (Hfd1p or ALDH3A2 lacking their C-terminal 25 amino acids, which comprise putative transmembrane domains) fused to an 8His-cytoplasmically-targeted maltose-binding protein with a linker including a tobacco etch virus protease recognition site (8His-MBP-[TEV]-ALDH3A1, 8His-MBP-[TEV]-Hfd1pCΔ25, or 8His-MBP-[TEV]-ALDH3A2CΔ25). These constructs were expressed in E. coli (BL21[DE3]-RIPL strain) by autoinduction. Cells were isolated and resuspended in lysis buffer (50 mM HEPES, 300 mM NaCl, 10% glycerol, 5 mM BME, 0.25 mM PMSF, 1 mg/mL lysozyme (Sigma), pH 7.5). Cells were lysed by sonication (4 °C, 2 × 20 s), and the lysate was clarified by centrifugation (15,000 g, 30 min, 4 °C). The clarified lysate was mixed with cobalt IMAC resin (Talon resin) and incubated (4 °C, 1 h). The resin was pelleted by centrifugation (700 g, 2 min, 4 °C) and washed three times (~10 resin bed volumes each) with wash buffer (50 mM HEPES, 300 mM NaCl, 10% glycerol, 5 mM BME, 0.25 mM PMSF, 10 mM imidazole, pH 7.5). His-tagged protein was eluted with elution buffer (50 mM HEPES, 300 mM NaCl, 10% glycerol, 5 mM BME, 0.25 mM PMSF, 100 mM imidazole, pH 7.5). The eluted protein was concentrated with a 50-kDa MW-cutoff spin filter (Merck Millipore Ltd.) and exchanged into storage buffer (50 mM HEPES, 300 mM NaCl, 10% glycerol, 5 mM BME, 0.25 mM PMSF, pH 7.5). Protein concentrations were determined by absorbance at 280 nm. The MBP-fusion proteins were aliquoted, frozen in N2(l), and stored at −80 °C.
Hfd1p, ALDH3A1, and ALDH3A2 enzymology
Enzyme activity assays were conducted in groups of three replicate 100 μL reactions, each containing MBP-fusion protein (0.2–25 μg), 1 mM NAD+, and 200 μM substrate (4-HBz or hexadecanal (Avanti 857458M)) in an aqueous buffer (50 mM Tris pH 8.0, 150 mM NaCl, 0.1% Triton X-100). NADH production was observed by monitoring fluorescence (356 nm excitation, 460 nm emission) over a 30–60 minute period with a Cytation 3 Imaging Reader (BioTek). KM and kcat values were determined by measuring reaction rates in the linear range at varying substrate (4-HBz or hexadecanal) concentrations. Curve fitting to generate Michaelis-Menten parameters was performed using SigmaPlot (Systat Software, San Jose, CA). Reported activity represents the mean of three separate protein purifications.
Molecule covariance network analysis
For all pairwise combinations of molecules quantified within a particular growth condition, regression analysis was conducted using fold change measurements from all Δgene strains having a measurement for both molecules in the pair. Spearman's regression analysis was performed to obtain correlation coefficients (ρ). From these test statistics P values were calculated using a two-sided Student's t-test. All P values were corrected for multiple hypothesis testing (Bonferroni) and correlations where |ρ| ≥ 0.58 and P < 0.001 were reported. For RDR-adjusted regression analysis, the RDR adjustment procedure was carried out as described in the ‘Respiration deficiency response (RDR) abundance adjustment’ section (above). All pairs of covariant molecules are visualized as networks generated using the Gephi open graph visualization platform (version 0.9.0). Complete respiration, fermentation and RDR-adjusted respiration network layouts were generated using the Fruchterman–Reingold graph-drawing algorithm with area set to 10,000 and gravity set to 30. Gene Ontology terms were obtained from the Saccharomyces Genome Database (SGD). To calculate network selectivity the following equation was used:
Where SMCN represents the selectivity coefficient for the molecule covariance network (MCN) surrounding an individual node of interest, EObs,In is the number edges observed within a pathway of interest, ETot,In is the number of total possible edges within the pathway of interest, EObs,Out is the number of edges observed to molecules outside the pathway of interest, and ETot,Out is the number total possible edges to molecules outside the pathway of interest.
Gene ontology (GO) term enrichment analysis was performed using a Fisher's exact test with subsequent Benjamini-Hochberg FDR adjustment39 to account for multiple hypothesis testing.
Yor020w-a, Aro9p, Aro10p, and Aim18p related methods
Proteomic analysis of Δyor020w-a yeast
2.5×106 yeast cells from a pABA− (2% w/v glucose) starter culture (Δyor020w-a or WT) were used to inoculate 100 mL of pABA− media with glucose (0.1%, w/v) and glycerol (3%, w/v). These 100 mL cultures were incubated (30 °C, 230 rpm). After 25 h, 1×108 yeast cells were harvested for proteomic analyses by mass spectrometry as described in the main Methods section. These cultures and analyses were conducted in biological duplicate.
Quantitation of CoQ and PPHB in pABA− Δaro9, Δaro10, Δaim18, and WT yeast cultures
2.5×106 yeast cells from a pABA− (2% w/v glucose) starter culture were used to inoculate 100 mL of pABA− media with glucose (0.1%, w/v) and glycerol (3%, w/v). These 100 mL cultures were incubated (30 °C, 230 rpm). After 25 h, 1×108 yeast cells were harvested for lipid analysis, and CoQ and PPHB were quantified by mass spectrometry as described in the Main methods section. These cultures and analyses were conducted in biological duplicate.
Respiration deficiency response analysis
The densities of Δgene cultures were compared to those of WT cultures (2-tailed T-test). Strains with slow growth in fermentation cultures (Δgene/WT ≤ 0.2 and P < 0.05) were categorized as ‘slow fermentation growth’ strains (8 strains). Remaining strains were grouped into three categories based on their growth rates in respiration cultures. Strains with significantly decreased respiration growth (Δgene/WT < 0.6 and P < 0.05) were considered respiration deficient (RD) (41 RD strains). Strains with borderline respiration growth (0.6 ≤ Δgene/WT < 0.8) were categorized as ‘borderline respiration’ (14 strains). Strains with respiration growth rates near WT or better than WT (0.8 ≤ Δgene/WT) were categorized as respiration competent (RC) (111 RC strains).
For PCA, average log2(Δgene/WT) values for each protein, metabolite, and lipid measured in the respiration condition were analyzed using Perseus PCA software. PCA projections were exported from Perseus.
For volcano plot analyses, average log2(RD/RC) values were calculated as [mean log2(RD Δgene strains/WT)] – [mean log2(RC Δgene strains/WT)]. A t-test (2-tailed, homostatic) was performed to obtain P values. P values were corrected for multiple hypothesis testing by multiplying each P value obtained by the number of biomolecules included in this analysis (4,116) (Bonferroni correction).
For GO term analyses, proteins were separated as increasing in RD strains (positive log2[RD/RC]) or decreasing in RD strains (negative log2[RD/RC]). Proteins with Bonferroni-corrected P < 1×10−20 were collected from each group and subjected to GO term enrichment analysis (http://geneontology.org/page/go-enrichment-analysis). Select GO terms were highlighted because they were significantly enriched (Bonferroni corrected P < 0.05) in proteins that were reduced (−) or increased (+) in RD strains. Boxplots of select molecules were generated using matplotlib in python to compare particular molecules across all RD and RC strains.
For ROC analysis, RD strains were considered positive examples whereas RC cells were considered negative examples. Using the log2(Δgene/WT) values for individual biomolecules as a discriminator, ROCs were generated by calculating false positive rate (FPR) and true positive rate (TPR) for values that fall above a particular cutoff for molecules that are increased in RD strains relative to WT and below that cutoff for molecules that are decreased in RD strains relative to WT. A + sign indicates that an increase in that molecule is predictive of RD whereas a – sign indicates that a reduction in that molecule is predictive of RD.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Pagliarini and Coon labs for helpful discussions. This work was supported by a Searle Scholars Award and NIH grants R01DK098672, R01GM112057, and R01GM115591 (to D.J.P.); NIH grant R35GM118110 (to J.J.C.); NIH Ruth L. Kirschstein NRSA F30AG043282 (to J.A.S.); DOE Great Lakes Bioenergy Research Center (DOE Office of Science BER DE-FC02-07ER64494 to N.W.K. and A.U.); ACS Analytical Chemistry and Society of Analytical Chemists of Pittsburgh awards (to A.L.R.); NSF Graduate Research Fellowship and NIH T32GM007215 (to M.T.V.); NIH T32DK007665 (to Z.A.K.); and NIH T32HG002760 (to E.A.T).
Footnotes
Accession codes. Raw MS data files from all proteome, metabolome, and lipidome analyses are available through the CHORUS project data repository (Project ID 1119). All software used for GC/MS quantitation and data analysis is available online at Github (https://github.com/coongroup/Y3K-Software) along with corresponding usage documentation.
Supplementary Information includes Supplementary Notes 1–7, Supplementary Figures 1–14, and Supplementary Tables 1–8 (as Excel files).
AUTHOR CONTRIBUTIONS
J.A.S., N.W.K., D.J.P., and J.J.C. conceived of the project and its design and wrote the manuscript. J.A.S., A.J., K.P.R., X.G., Z.A.K., and J.S. prepared samples and performed biochemical experiments. E.C.F., A.L.R., M.J.P.R., A.U., P.D.H., X.G., K.J.C., E.A.T., and A.S.H. acquired MS data. J.A.S., N.W.K., M.T.V., H.M., M.S.W., D.J.P., and J.J.C. analyzed data.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Hebert AS, et al. The one hour yeast proteome. Mol Cell Proteomics. 2014;13:339–347. doi: 10.1074/mcp.M113.034769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richards AL, et al. One-hour proteome analysis in yeast. Nat Protoc. 2015;10:701–714. doi: 10.1038/nprot.2015.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peterson AC, et al. Development of a GC/Quadrupole-Orbitrap mass spectrometer, part I: design and characterization. Anal Chem. 2014;86:10036–10043. doi: 10.1021/ac5014767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishii N, et al. Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science. 2007;316:593–597. doi: 10.1126/science.1132067. [DOI] [PubMed] [Google Scholar]
- 5.Buescher JM, et al. Global network reorganization during dynamic adaptations of Bacillus subtilis metabolism. Science. 2012;335:1099–1103. doi: 10.1126/science.1206871. [DOI] [PubMed] [Google Scholar]
- 6.Williams EG, et al. Systems proteomics of liver mitochondria function. Science. 2016;352:aad0189. doi: 10.1126/science.aad0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chick JM, et al. Defining the consequences of genetic variation on a proteome-wide scale. Nature. 2016;534:500–505. doi: 10.1038/nature18270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koopman WJ, Willems PH, Smeitink JA. Monogenic mitochondrial disorders. N Engl J Med. 2012;366:1132–1141. doi: 10.1056/NEJMra1012478. [DOI] [PubMed] [Google Scholar]
- 10.Vafai SB, Mootha VK. Mitochondrial disorders as windows into an ancient organelle. Nature. 2012;491:374–383. doi: 10.1038/nature11707. [DOI] [PubMed] [Google Scholar]
- 11.Pagliarini DJ, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calvo SE, Clauser KR, Mootha VK. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016;44:D1251–1257. doi: 10.1093/nar/gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sickmann A, et al. The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci U S A. 2003;100:13207–13212. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green ED, Guyer MS, National Human Genome Research, I Charting a course for genomic medicine from base pairs to bedside. Nature. 2011;470:204–213. doi: 10.1038/nature09764. [DOI] [PubMed] [Google Scholar]
- 15.Pagliarini DJ, Rutter J. Hallmarks of a new era in mitochondrial biochemistry. Genes Dev. 2013;27:2615–2627. doi: 10.1101/gad.229724.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Floyd BJ, et al. Mitochondrial protein interaction mapping identifies new regulators of respiratory chain function. Mol Cell In Press. 2016 doi: 10.1016/j.molcel.2016.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quinzii CM, Hirano M. Coenzyme Q and mitochondrial disease. Dev Disabil Res Rev. 2010;16:183–188. doi: 10.1002/ddrr.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalen A, Appelkvist EL, Dallner G. Age-related changes in the lipid compositions of rat and human tissues. Lipids. 1989;24:579–584. doi: 10.1007/BF02535072. [DOI] [PubMed] [Google Scholar]
- 19.Bentley RR, V. G., Springer CM, Dialameh GH, Olson RE. The Origin of the Benzoquinone Ring of Coenzyme Q9 in the Rat. Biochem Biophys Res Commun. 1961;5:443–446. [Google Scholar]
- 20.Booth ANM, M. S., Robbins DJ, Emerson OH, Jones FT, Deeds F. Urinary Phenolic Acid Metabolites of Tyrosine. J Biol Chem. 1960;235:2649–2652. [Google Scholar]
- 21.Pierrel F, et al. Involvement of mitochondrial ferredoxin and para-aminobenzoic acid in yeast coenzyme Q biosynthesis. Chem Biol. 2010;17:449–459. doi: 10.1016/j.chembiol.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 22.Marbois B, et al. para-Aminobenzoic acid is a precursor in coenzyme Q6 biosynthesis in Saccharomyces cerevisiae. J Biol Chem. 2010;285:27827–27838. doi: 10.1074/jbc.M110.151894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakahara K, et al. The Sjogren-Larsson syndrome gene encodes a hexadecenal dehydrogenase of the sphingosine 1-phosphate degradation pathway. Mol Cell. 2012;46:461–471. doi: 10.1016/j.molcel.2012.04.033. [DOI] [PubMed] [Google Scholar]
- 24.Liu ZJ, et al. The first structure of an aldehyde dehydrogenase reveals novel interactions between NAD and the Rossmann fold. Nat Struct Biol. 1997;4:317–326. doi: 10.1038/nsb0497-317. [DOI] [PubMed] [Google Scholar]
- 25.Jackson B, et al. Update on the aldehyde dehydrogenase gene (ALDH) superfamily. Hum Genomics. 2011;5:283–303. doi: 10.1186/1479-7364-5-4-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Laurenzi V, et al. Sjogren-Larsson syndrome is caused by mutations in the fatty aldehyde dehydrogenase gene. Nat Genet. 1996;12:52–57. doi: 10.1038/ng0196-52. [DOI] [PubMed] [Google Scholar]
- 27.Kitamura T, et al. Substrate specificity, plasma membrane localization, and lipid modification of the aldehyde dehydrogenase ALDH3B1. Biochim Biophys Acta. 2013;1831:1395–1401. doi: 10.1016/j.bbalip.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 28.Hughes TR, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 29.Kemmeren P, et al. Large-scale genetic perturbations reveal regulatory networks and an abundance of gene-specific repressors. Cell. 2014;157:740–752. doi: 10.1016/j.cell.2014.02.054. [DOI] [PubMed] [Google Scholar]
- 30.Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 31.Laredj LN, Licitra F, Puccio HM. The molecular genetics of coenzyme Q biosynthesis in health and disease. Biochimie. 2014;100:78–87. doi: 10.1016/j.biochi.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 32.Tran UC, Clarke CF. Endogenous synthesis of coenzyme Q in eukaryotes. Mitochondrion. 2007;7(Suppl):S62–71. doi: 10.1016/j.mito.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuchner S, et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet. 2004;36:449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]
- 34.Mourier A, et al. Mitofusin 2 is required to maintain mitochondrial coenzyme Q levels. J Cell Biol. 2015;208:429–442. doi: 10.1083/jcb.201411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hebert AS, et al. Amine-reactive neutron-encoded labels for highly plexed proteomic quantitation. Mol Cell Proteomics. 2013;12:3360–3369. doi: 10.1074/mcp.M113.032011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 37.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 38.Cox J, et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 39.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Statist. Soc. B. 1995;57:289–300. [Google Scholar]
- 40.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 42.Letunic I, Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011;39:W475–478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.