Abstract
Amyotrophic lateral sclerosis (ALS), a common motor neuron disease affecting two per 100,000 people worldwide, encompasses at least five distinct pathological subtypes, including, ALS-SOD1, ALS-C9orf72, ALS-TDP-43, ALS-FUS and Guam-ALS. The etiology of a major subset of ALS involves toxicity of the TAR DNA-binding protein-43 (TDP-43). A second RNA/DNA binding protein, fused in sarcoma/translocated in liposarcoma (FUS/TLS) has been subsequently associated with about 1% of ALS patients. While mutations in TDP-43 and FUS have been linked to ALS, the key contributing molecular mechanism(s) leading to cell death are still unclear. One unique feature of TDP-43 and FUS pathogenesis in ALS is their nuclear clearance and simultaneous cytoplasmic aggregation in affected motor neurons. Since the discoveries in the last decade implicating TDP-43 and FUS toxicity in ALS, a majority of studies have focused on their cytoplasmic aggregation and disruption of their RNA-binding functions. However, TDP-43 and FUS also bind to DNA, although the significance of their DNA binding in disease-affected neurons has been less investigated. A recent observation of accumulated genomic damage in TDP-43 and FUS-linked ALS and association of FUS with neuronal DNA damage repair pathways indicate a possible role of deregulated DNA binding function of TDP-43 and FUS in ALS. In this review, we discuss the different ALS disease subtypes, crosstalk of etiopathologies in disease progression, available animal models and their limitations, and recent advances in understanding the specific involvement of RNA/DNA binding proteins, TDP-43 and FUS, in motor neuron diseases.
Keywords: TDP-43, FUS/TLS, Amyotrophic Lateral Sclerosis, RNA processing, Genome Damage/Repair
1. Introduction
Amyotrophic lateral sclerosis (ALS), a fatal neuromuscular disease characterized by degeneration of upper and lower motor neurons of spinal cord, was first described by the French neurologist Jean-Martin Charcot in 1869, as clinical and pathological symptoms of muscular atrophy and hardening of lateral spinal cord (Chio et al, 2009b; Leblond et al, 2014). Progressive bulbar, limb, thoracic, and abdominal muscle atrophy in ALS causes death within 3-5 years after onset of symptoms in most ALS patients, normally due to respiratory failure (Chio et al, 2009a). Clinically, ALS may impact bulbar or spinal innervated muscles at onset, based on the relative loss of upper and/or lower spinal neurons, which is symptomatically consistent with bulbar or limb motor defects. While spinal neurons are predominantly affected, loss of neurons may also be observed in the brainstem and motor cortex (Al-Chalabi et al, 2012).
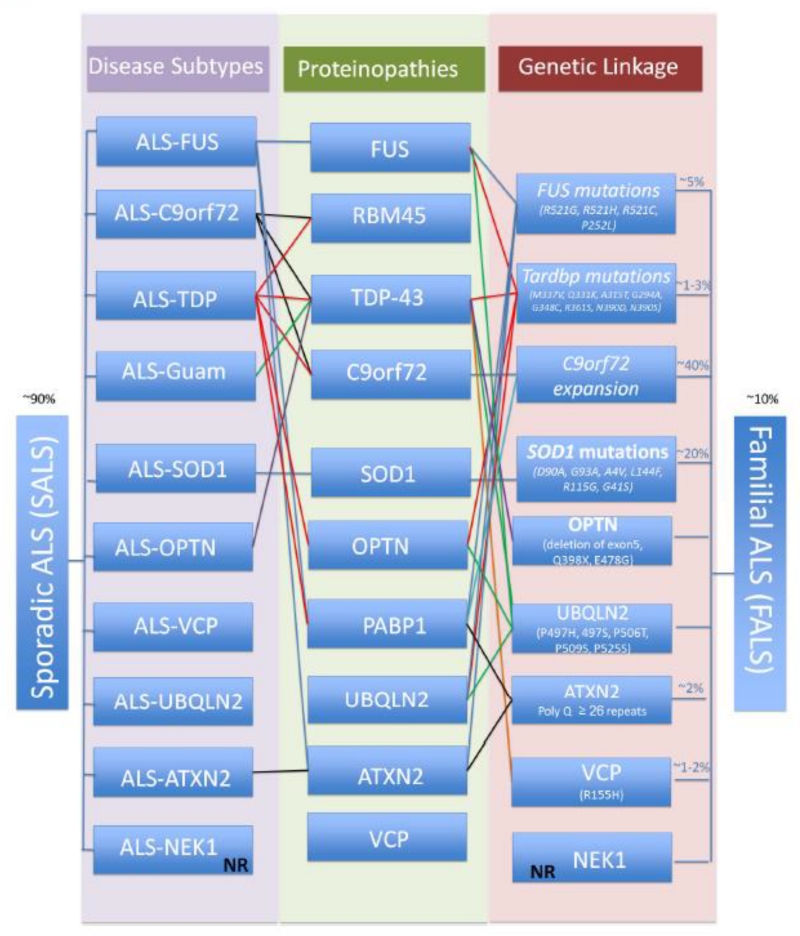
ALS is the most common degenerative disorder of motor neurons in adults, with incidence rates of 2-5 per 100,000 individuals worldwide. Men are 1.3 times more likely to develop ALS than women (Chio et al, 2013; Leblond et al, 2014). Despite its relatively low incidence compared to other neurodegenerative diseases like Alzheimer’s Disease (AD) (Hebert et al, 2013); or Parkinson’s disease (PD) (de Lau & Breteler, 2006); the devastating physiological effects and rapid lethality are the prominent features of ALS. Most ALS cases are sporadic, while about 8-10% are inherited (Figure 1).
Figure 1. Illustration of the molecular and pathological features of sporadic and familial ALS.
The sporadic disease subtypes account for ~90 % of ALS cases and which could be classified based on inclusions and the protein within. In the recent years, mutations and defects in several new ALS causing genes have been implicated in distinct subgroups of ALS patients (FALS). However, proteins encoded by these genes are also found in protein inclusions/aggregates in sporadic patients (SALS). There is a distinct pattern of co-localization or overlap of pathology among the ALS-linked protein inclusions, which are indicated. *NR: not reported.
Many ALS patients (~36-51%) also exhibit cognitive impairment, with about 20% developing frontotemporal lobar degeneration (FTLD) (Broustal et al, 2010). The reverse is also seen, with some patients with FTLD developing ALS as well (Lipton et al, 2004). This has led to the theory that ALS and FTLD are part of a clinical spectrum of disease. FTLD accounts for 10-15% of dementias, making it the second most common type of dementia for people under the age of 65, after AD. Pathologically, FTLD includes multiple subtypes, including FTLD-tau, FTLD-DPR (dipeptide repeat proteins), FTLD-UPS (ubiquitin-proteasome system), FTLD-FUS (fused in sarcoma/translocated in liposarcoma), and FTLD-TDP-43 (Tar DNA binding protein 43), although the distinction among the subtypes is not very clear. Most patients with ALS-FTLD have TDP-43 pathology, including FTLD-DPR, raising the question about the relative toxicity of DPR compared to TDP-43 pathology. Early-onset ALS-FTLD patients typically present cognitive changes, followed by muscle weakness (primarily upper body weakness), behavioral changes (such as euphoria, indifference, personality changes, and language impairments), paucity of speech, impaired comprehension, and even mutism. However, unlike AD, overall memory is relatively preserved in most patients with ALS or ALS-FTLD (Mitsuyama, 1993). The overlapping pathogenesis of ALS and FTLD suggests that these motor neuron diseases and cognitive deficits could have a common molecular basis, which is discussed later in this article.
1.1. Etiopathology and disease subtypes
The etiopathogenesis underlying degeneration of motor neurons in ALS is complex. Endogenous factors like accumulation of pro-oxidant metals and free radicals, glutamate excitoxicity, protein aggregation, mitochondrial dysfunction, and deregulation of RNA processing have been associated with dysfunctional motor neurons (Ferraiuolo et al, 2011). Various environmental factors including cigarette smoking, occupational exposure to electromagnetic radiation, heavy metals, pesticides, diesel exhaust and head traumas may increase the risk of developing ALS (Garruto et al, 1985b; Mitra et al, 2014b; Perl et al, 1982; Schmidt et al, 2010; Weisskopf et al, 2009; Yanagihara et al, 1984) and even in familial cases, an interplay of genetic and environmental factors has been linked to increased susceptibility. The first evidence of environmental triggers of sporadic ALS (SALS) was established with the indigenous populations of Guam in the Mariana Islands and in the Kii peninsula of Japan where ALS occurred with unusual incidence among these populations. Guam-ALS patients presented higher iron levels and lower zinc levels in brain and chronic nutritional deficiencies of calcium that could lead to higher intestinal absorption of toxic metals and co-deposition of calcium, aluminum and silicon in neurons (Garruto, 1991; Garruto et al, 1985a; Yasui et al, 1993) Furthermore, cyanobacteria-derived neurotoxin, beta-Methylamino-L-alanine (BMAA) is a candidate neurotoxin, like metals in the Guam ALS and Parkinsonism-dementia complex (ALS/PD), particularly prevalent in indigenous populations of Guam and Rota who consumed cyanobacteria-infested cycads (Cox & Sacks, 2002; Garruto et al, 1980; Garruto et al, 1981; Hoffman et al, 1977; Plato et al, 2003). BMAA binds to N-methyl-D-aspartate (NMDA) and (α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid) (AMPA) receptors, producing a molecule that resembles glutamate and activates glutamate receptor 5, inducing oxidative stress (OS). BMAA also inhibits the cysteine/glutamate antiporter system Xc−, leading to glutathione depletion, which further enhances OS. Moreover, BMAA may be incorporated into proteins, causing their misfolding or aggregation (Arif et al, 2014; Rao et al, 2006; Weiss et al, 1989). However, it is unclear, which of these cytotoxic properties of BMAA may be critical for Guam ALS. In addition, the Kii peninsula population was later linked to some genetic contribution/ founder effect (Arif et al, 2014).
Despite the range of pathological and clinical features among various neurodegenerative disorders, the identification of misfolded protein-rich inclusion(s) has become one of the molecular hallmarks of neurodegeneration. ALS is no exception, as its pathology involves accumulation of proteinaceous aggregates including diverse proteins like redox regulator Cu-Zn superoxide dismutase 1 (SOD1), and RNA/DNA binding proteins TDP-43 and FUS/TLS. Furthermore, the genes encoding these proteins SOD1, FUS, TARDBP and C9orf72, which encodes a yet-to-be characterized protein, have been implicated in 4 major ALS subtypes (Figure 1).
TARDBP
In 2006, the gene coding for TDP-43, an RNA/DNA binding protein, was implicated in ALS as the major component of ubiquitinated inclusions (Arai et al, 2006; Neumann et al, 2006). TDP-43 is involved in RNA processing, including splicing, transcription, and transport. The primary histopathological feature in a major subset of ALS cases is the inclusion of TDP-43 in the cytoplasm of upper and lower motor neurons and in other regions of the central nervous system (CNS), including the frontal and temporal cortex. As previously mentioned, there is evidence of an overlapping histopathology between C9orf72 genetic lesions and TDP-43 inclusions, but the mechanism is not clearly characterized (Freibaum et al, 2015). Recent studies identified co-localization of TDP-43 and RNA-binding motif-45 (RBM45) inclusions and their RNA-dependent association in motor neurons of some cases of ALS (Collins et al, 2012; Mladinic et al, 2010). Like TDP-43, RBM-45 is mainly nuclear, but migrates to cytoplasm and co-localizes in SGs to associate with Kelch-like ECH-Associated protein 1, a component of anti-oxidant machinery (Bakkar et al, 2015). Furthermore, TDP-43 co-localizes in cytoplasmic inclusions with Poly-A binding protein - 1 (PABP-1); a stress granules marker (McGurk et al, 2014).
TDP-43 is ubiquitously expressed in most tissue and cell types, including neurons and glia in the CNS. Studies of ALS, FTD, and ALS-FTD have shown that TDP-43 is cleaved, hyperphosphorylated, ubiquitinated or mis-localized in cytoplasm in the form of insoluble inclusions (Arai et al, 2006; Feiguin et al, 2009; Kwong et al, 2007; Neumann et al, 2006). ALS and ALS/PDC patients in Guam also present TDP-43 inclusions as a secondary pathology (Maekawa et al, 2009). ALS-parkinsonism disease subtypes show TDP-43 pathology in motor neurons, hippocampus, amygdala, globus pallidus, caudate, and putamen (McCluskey et al, 2009). Mutations in TARDBP gene, mostly within the glycine-rich C-terminal domain (CTD), are associated with 1-3% of cases of ALS with familial history (Daoud et al, 2009; Valdmanis et al, 2009).
FUS
The FUS/TLS protein was linked to ALS in 2009, as a component of inclusions found in ALS patients (Kwiatkowski et al, 2009; Vance et al, 2009). Although TDP-43 and FUS exhibit similar structures, functions, and pathobiology in ALS, TDP-43 pathology is notably absent in ALS-FUS cases (Kwiatkowski et al, 2009; Ticozzi et al, 2009). Like TDP-43, FUS/TLS protein is sequestered in the cytosol of ALS-affected motor neurons. Mutations in FUS cause severe loss of motor neurons in the spinal cord, moderate loss of upper motor neurons, and are associated with juvenile ALS (Kwiatkowski et al, 2009; Ticozzi et al, 2009). Patients with ALS-FUS develop distinct phenotypic patterns: early onset, with aggressive clinical progression, or late onset, with slower disease progression (Ravits et al, 2013). Pathological analysis has indicated that truncating mutations in the FUS gene can generate more aggressive phenotypes than missense mutations (Waibel et al, 2013). Interestingly, ALS-FUS appears to be clinico-pathologically distinct from FTLD-FUS. FTLD-FUS is not usually associated with mutations and the inclusions contain not only FUS, but also other FET (FUS, EWS and TAF15) proteins (Mackenzie et al, 2011). ALS-FUS present inclusions that co-localizes with stress granule marker, PABP-1 (Gal et al, 2011) and Ataxin-2 (ATXN2) (Elden et al, 2010). In addition, an unusual case has been recently reported with mutations in both FUS and TARDBP genes (King et al, 2015). This particular case exhibited a FUS P525L mutation as well as the truncating TARDBP Y374X mutation, showing moderate FUS pathology and no TDP-43 pathology, with extensive granular and p62-positive, TDP-43-negative inclusions in the spinal cord and motor/neocortex (King et al, 2015).
Other major ALS subtypes and their distinct and overlapping pathology with ALS-TDP or ALS-FUS SOD1
The primary role of antioxidant metalloenzyme, SOD1, in healthy cells is to protect cells from OS by neutralizing the toxicity of superoxide radicals to dioxygen and hydrogen peroxide molecules (Fridovich, 1978). SOD1 was the first protein to be implicated in ALS in 1993 (Rosen et al, 1993). Subsequently, more than 160 mutations in the SOD1 gene are found in ~20% of familial ALS (FALS) cases and ~2% of SALS cases (Acevedo-Arozena et al, 2011; Andersen & Al-Chalabi, 2011; Pasinelli & Brown, 2006; Saccon et al, 2013). Besides the straightforward involvement of redox imbalances, the molecular mechanism of SOD1-mediated ALS is largely unknown. The disease-linked mutations of SOD1 do not usually impair its function, while misfolding associated with mutation of proteins appears to gain toxic properties leading to neurodegeneration. Misfolded mutant SOD1 proteins accumulate in motor neurons and glial cells of the spinal cord, which mediates excitoxicity, mitochondrial dysfunction, axonopathy, and endosomal trafficking that have been implicated in ALS-SOD1 (Bosco et al, 2010). Patients with ALS-SOD1 have neuronal loss in the anterior horn region of the spinal cord and positive p62 skein-like inclusions that are negatively immunoreactive for TDP-43. P62 is a protein degradation marker related specifically to the selective autophagy system; p62-positive inclusions are widely found in TDP-43-negative-star-shaped-inclusions found in ALS cases. Furthermore, ALS-SOD1 is less frequently associated with ALS-FTD (Tan et al, 2007).
C9orf72
In 1991, a multigenerational family with a history of ALS, FTLD, and ALS-FTLD was described which was later attributed to a mutation consisting of a hexanucleotide expansion repeat (GGGGCC) in the intron between non-coding exons 1a and 1b of the gene C9orf72, which codes for a protein with unknown function (Gunnarsson et al, 1991). The pathological repeat expansion may extend to hundreds of repeats in individuals harboring the genetic lesion, presenting unusual pathological features including RNA foci and aggregates of dipeptide repeats produced from repeat associated non-ATG (RAN) translation of the repeat expansion RNA, as well as TDP-43 positive aggregates. C9orf72 is the predominant ALS gene, associated with about half of all ALS cases, including ~10% of sporadic and nearly 40% of FALS and FTD cases. Moreover, ALS-C9orf72 involves motor neuronal loss in the anterior horn, which normally overlaps with TDP-43-positive inclusions. Strikingly, these patients develop numerous p62-positive, TDP-43-negative inclusions in the dentate gyrus, neocortex, and cerebellum (Al-Sarraj et al, 2011; Boxer et al, 2011; DeJesus-Hernandez et al, 2011). Furthermore, patients with the C9orf72 hexanucleotide expansion also have RBM45 inclusions and PABP-1 (Collins et al, 2012; McGurk et al, 2014). C9orf72 mutations appear to promote mis-localization of TDP-43 to the cytoplasm, in addition to causing malformed RNA molecules (Zhang et al, 2015). C9orf72 repeat expansion partly contributes to the high prevalence of ALS cases in Kii peninsula of Japan, where 20% of ALS patients carry the hexanucleotide repeat expansion (Ishiura et al, 2012).
Optoneurin (OPTN)
OPTN was firstly linked to ALS in 2010 (Maruyama et al, 2010) and its mutations that have been identified in ALS patients, including truncation and missense mutations are thought to have a loss-of-function pathology. OPTN is localized in cytoplasmic inclusions with ubiquitin and TDP-43 in ALS affected motor neurons (Hortobagyi et al, 2011). There are conflicting studies about OPTN co-localization with SOD1 and FUS (Deng et al, 2011a; Hortobagyi et al, 2011; Keller et al, 2012). OPTN inclusions are present in several other neurodegenerative diseases (Osawa et al, 2011).
Valosin-containing protein (VCP)
In 2010, mutations in VCP gene, coding for AAA+ATPase ubiquitn-dependant segregase, were linked to 1-2% of familial cases of ALS (Johnson et al, 2010). Recent studies demonstrated that mutations in VCP cause mitochondrial dysfunction that leads to reduced ATP production. The discovery of mutations in VCP gene led to the model of multisystem proteinopathies, where multiple tissues are affected (Watts et al, 2004). Although, VCP inclusions have been reported in ALS cases, and there are no reports on its co-localization with FUS nor TDP-43 inclusions in ALS cases, TDP-43 was observed to be co-localized with VCP in the cytoplasmic inclusions of FTLD patients, and its translocation between nucleus and cytoplasm is altered by mutations in VCP gene(Gitcho et al, 2009).
Ubiquilin 2 (UBQLN2)
UBQLN2 was linked to ALS in 2011, it was shown that mutations in UBQLN2 gene caused a dominantly male-to-male inherited form of ALS, with or without dementia (Deng et al, 2011b). Given that UBQLN2 regulates degradation of ubiquitinated proteins, and dysregulation in the ubiquitin-proteasome system (UPS) has been linked to ALS; the mutations in UBQLN2 causing ALS was believed to through impaired protein degradation pathways. Besides, ALS patients with mutations in UBQLN2, present inclusions and co-localized with ubiquitin, p62, TDP-43, FUS, OPTN but there are no reports on co-localization with SOD1 aggregates (Deng et al, 2011b; Williams et al, 2012).
ATXN2
the association of extended polyQ repeats in ATXN2 gene with ALS was observed in 2010; Normally ATXN2 carries 21 to 22 polyQ repeats, whereas in ALS cases ATXN2 polyQ repeats extends to 27 to 33 (Elden et al, 2010). ATXN2 functions in mRNA polyadenylation, miRNA synthesis and stress granules formation. ATXN2 cytoplasmic inclusions have been observed in spinal cord tissue of ALS patients. Furthermore, ATXN2 and TDP-43 inclusions co-localized in FTLD patients, while ATXN2 and FUS co-localization was observed in ALS patients (Elden et al, 2010; Farg et al, 2013). In ALS patients with ATXN2 polyQ repeats, ATXN2 and PABP-1 have been observed to colocalize in inclusions (McGurk et al, 2014). ATXN2 polyQ repeats enhanced stress-induced caspase-3 activation and TDP-43 cleavage as well as its phosphorylation (Elden et al, 2010; Farg et al, 2013).
HnRNPA2B1 and hnRNPA1
Hetereogeneous ribonucleoproteins harbor prion-like domains (PLD’s) are essential to assembly into self-seeding ribonucleoprotein granules or fibrils. Mutations in hnRNPA1 and hnRNPA2B1 has been observed in cases of ALS with increased tendency of self-seeding fibrils formation and stress-granule formation (Kim et al, 2013). It is well known that hnRNPA1 and hnRNPA2B1 interact with TDP-43 via its C-terminal domain, co-localization of these proteins with TDP-43 in cytoplasmic inclusions has not been observed in ALS cases (Honda et al, 2015).
NEK1
In a recent exome-wide study, the mutation of a gene that encodes the serine/threonine kinase NIMA (never in mitosis gene-A)-related kinase, NEK1, was found in~ 3% of ALS cases in European and European-American families (Kenna et al, 2016). It is worth to note that NEK1 is an established DNA damage response factor,(Chen et al, 2011)further studies may needed to focus on NEK1 mutation mediated DNA damage repair deficiency in in ALS pathology.
In summary, although the ALS group of motor neuron diseases exhibit common phenotypes, the molecular mechanisms underlying motor neuron dysfunction appear to be distinctive among disease subtypes. This review will focus primarily on the deregulation of RNA/DNA metabolism involved in ALS-TDP-43 and ALS-FUS, as well as emerging research in our understanding of complexity associated with loss of functions versus acquired toxicity of TDP-43 and FUS.
2. TDP-43 and FUS: A perspective into neurobiology and nucleic acid binding
2.1. TDP-43
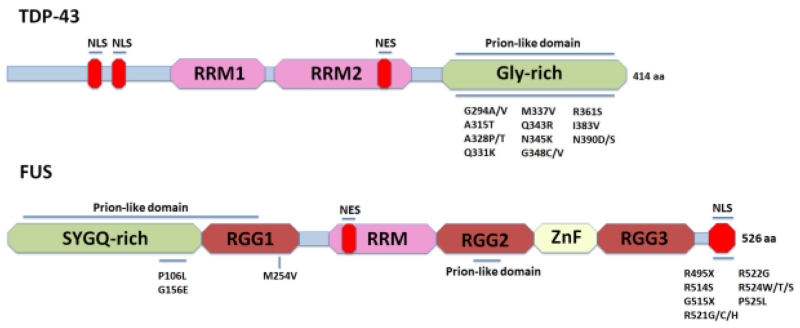
The 414 amino acid-containing TDP-43, belonging to the heterogeneous nuclear ribonucleoprotein (hnRNP) family, is encoded by the TARDBP gene and is highly conserved among human, mouse, D. melanogaster, and C. elegans genomes (Wang et al, 2004). Domain analysis revealed that TDP-43 consists of two RNA recognition motifs, RRM1 and RRM2, and a disordered glycine-rich CTD (Figure 2). Structurally, TDP-43 closely resembles other hnRNP family proteins: hnRNP A1 and hnRNP A2/B1 (Dreyfuss et al, 1993). Functionally, TDP-43 appears to be a multi-tasking protein and is essential for cell survival, due to its involvement in transcriptional repression, pre-mRNA maturation and alternative splicing, mRNA transportation, microRNA biogenesis, interaction with noncoding RNA, autoregulation, and translational regulation of a number of key proteins (Figure 3). The critical importance of TDP-43 in many cellular functions both during development and in adults is underscored by the embryonic lethal phenotype of homozygous knock-out mice and the fact that its postnatal knock-out, through conditional gene inactivation, causes rapid lethality (Shan et al, 2010). Originally discovered as a transcriptional repressor binding to TAR DNA of the human immunodeficiency virus type 1 (HIV-1) (Ou et al, 1995), TDP-43 was subsequently found to repress the transcription of mouse SP-10 gene (Abhyankar et al, 2007) and human cyclin-dependent kinase 6 (Cdk6) (Ayala et al, 2008a) by binding to the regulatory elements of the respective promoters. In this context, the association of TDP-43 with chromatin, possibly through its RRM2 motif, both in the human brain and in cultured cells, is highly significant (Ayala et al, 2008b; Casafont et al, 2009; Thorpe et al, 2008).
Figure 2.
Schematic of TDP-43 and FUS protein structure. TDP-43 and FUS, both have a Prion-like domain, nuclear localization signal, nuclear export signal and RNA recognition motif (RRM). FUS has an additional RRM as well as a Zinc finger domain. Major familial mutations are indicated. In contrast to FUS, disease-linked TDP-43 mutations are clustered in the Glycine-rich C-terminal domain; whereas FUS disease-linked mutations are mainly clustered in the nuclear localization signal domain.
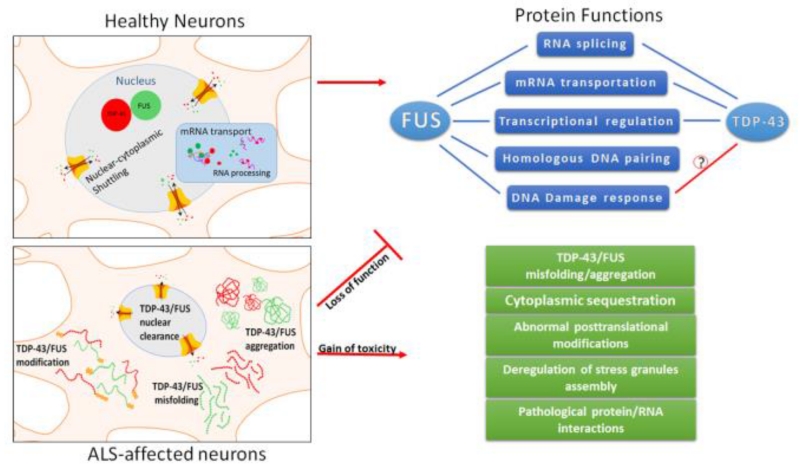
Figure 3.
TDP-43/FUS in healthy neurons bind to thousands of cellular RNA’s. They shuttle between the nucleus and cytoplasm, and play roles in miRNA biogenesis, pre-mRNA splicing, mRNA stability and transport. ALS affected motor neurons present altered cytoplasmic localization and nuclear clearance of the TDP-43 and FUS, together with deregulation in their posttranslational modification states, impacting their normal functions.
TDP-43 interacts with both UG-rich and non-UG-rich RNA sequences using its RRM domains. TDP-43 binds both single-stranded (ssDNA) and double-stranded DNA (dsDNA), with higher affinity to ssDNA at the TG repeat sequences. RRM1 may be involved in its interaction with DNA, since this region co-crystallizes with TG-rich ssDNA, forming thermostable dimeric assemblies (Kuo et al, 2014). RRMs contain two highly conserved sequences, namely RNP1 and RNP2. Residues Phe194 (conserved in RNP1) and Phe231 (conserved in RNP2) are involved in the interaction of TDP-43 with different UG- or TG-rich nucleic acid sequences (Buratti & Baralle, 2001). Mutational modification of Phe147 and Phe149 of RNP1 has been found to be sufficient to abolish the splicing regulatory functions of TDP-43 (Buratti & Baralle, 2001; Buratti et al, 2001; D’Ambrogio et al, 2009). The RRM2 domain is structurally distinct from the RRM1 domain. Interestingly, the RRM1 motif prefers to bind long (UG)6 repeats, whereas RRM2 has a stronger binding affinity for short (UG)3 repeats, suggesting that both the RRM domains are essential for normal cellular functions of TDP-43 (Kuo et al, 2009). TDP-43 also binds (UG)nUA(UG)m and poly-pyrimidine tract-containing RNA/DNA sequences (Sephton et al, 2011; Xiao et al, 2011). TDP-43 forms a homodimer with a domain arrangement similar to hnRNPA1, where the two RRM2 domains form a highly thermostable β4 strand. Thus, it is possible that in addition to its prion-like glycine-rich domain (277-414 amino acids), the RRM2 domain may also contribute to TDP-43 proteinopathy (Kuo et al, 2014).
Through its RNA-splicing functions, TDP-43 controls the expression levels and splice variants of many targets. TDP-43 autoregulates its own protein levels, as well as regulates the expression levels of other RNA-binding protein genes, including SRSF1, polypyrimidine tract-binding (PTB), and hnRNP L. (Buratti & Baralle, 2011). A UV-cross-linking immunoprecipitation-sequencing (CLIP-seq) study in mouse brain tissue showed that TDP-43 interacts with over 6,300 RNAs. The depletion of TDP-43 resulted in altered splicing of over 900 mRNAs (Polymenidou et al, 2011). The amount of TDP-43 protein in healthy cells is tightly maintained, possibly due to nonsense-mediated decay of mRNA (Lejeune & Maquat, 2005), and is likely to be critical for cellular homeostasis. TDP-43 CTD region 321-366 aa binds to the conserved 3′UTR proximal polyA1 and distal polyA4 sequences in its own TARDBP mRNA to generate its two major splice variants of 2.8kb (using intron 7 only) and 4.2kb (using both intron 6 and 7) in a ratio of ~1:3 (Ayala et al, 2011; D’Ambrogio et al, 2009). Overexpression of TDP-43 causes asymmetric interactions between TDP-43 and its nascent RNA to stall RNA polymerase II (RNA Pol II), leading to transcript degradation and maintaining the autoregulation of TDP-43 protein level (Avendano-Vazquez et al, 2012). Interestingly, TDP-35, a second splice variant of TDP-43, has been found to be expressed in the brains of ALS patients, through the use of the downstream start codon ATGMet85 (Xiao et al, 2015). Expression of TDP-35 in primary neurons causes cytoplasmic aggregation and neuronal death (Xiao et al, 2015).
Recent studies found that TDP-43, but not FUS, is a component of nuclear Drosha complex, which is involved in microRNA regulation (Kawahara & Mieda-Sato, 2012). Cytoplasmic TDP-43 associates with the Dicer complex that contains transactivation-responsive RNA binding protein (TRBP), which recruits Argonaute 2 (Ago2) for efficient processing of pre-miRNAs (Chendrimada et al, 2005; Gregory et al, 2005). These studies indicate that TDP-43 can also affect gene regulation in complex with microRNA (miRNA). For example, TDP-43 forms a complex with miR-NID1, generated from intron 5 of NRXN1 suppressing the expression of NRXN1, which is crucial for synaptic vesicle release and maintenance of synapse ultrastructure (Kang et al, 2015).
Long non-coding RNAs (lncRNAs) confer another level of gene regulation by binding to complementary mRNA sequences during post-transcriptional processing (Beltran et al, 2008). It was recently discovered that growth-arrested DNA damage-inducible gene 7 (gadd7), a 754 nucleotide (nt)-long lncRNA induced by both growth arrest and DNA damage signaling, regulates Cdk6 expression by specifically binding to and dissociating TDP-43 from Cdk6 mRNA and directing it for decay (Liu et al, 2012). This implicates the direct interaction of TDP-43 with lncRNA and its involvement in cell cycle regulation. In FTLD-TDP-43 brains, TDP-43 showed highly increased binding to metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) and nuclear enriched abundant transcript 1 (NEAT1) non-coding RNAs. In disease conditions, the binding affinity of TDP-43 increases for deep intronic regions, downstream of silenced exons (Tollervey et al, 2011).
Global attenuation of protein synthesis and induction of enhanced protein quality control are the most important and immediate cellular responses following stress. Activated protein quality control machinery removes the damaged proteins and simultaneously activates molecular chaperones in order to improve protein folding in the endoplasmic reticulum lumen (Holcik & Sonenberg, 2005; Ron & Walter, 2007). In response to stress, TDP-43 translocates from the nucleus to cytoplasm, where it binds 14-3-3 proteins. This relieves the negative inhibition of FOXO transcription factors, allowing their translocation back to the nucleus to regulate expression of genes involved in the stress response (Salih & Brunet, 2008; Zhang et al, 2014). Upon exposure to oxidative stress, cells form cytoplasmic granular particles (≤5 μm) known as stress granules (SGs). SGs are RNA-containing cytoplasmic particles composed by RNA and RNA-binding proteins involved in RNA metabolism and translation. SGs have been linked to several neurodegenerative diseases, including ALS and FTLD (Volkening et al, 2009). Although the precise components of SGs, which appear to be dynamic, are not completely understood, some of the key components that have been identified include TDP-43, T-cell intracellular antigen 1 (TIA-1) and RasGAP SH3-domain binding protein 1 (G3BP). SG formation is a rapid and transient mechanism, which starts immediately after stress exposure and dissembles within a couple of hours (Gilks et al, 2004; Kedersha & Anderson, 2002; McDonald et al, 2011; Tourriere et al, 2003). Efficient SG kinetics and dynamics are critical to neuronal cell survival to respond to both acute and chronic OS, where TDP-43 plays a major role (Aulas et al, 2012).
2.2. FUS/TLS
Following the discovery of TDP-43, mutations in a second RNA/DNA-binding protein FUS/TLS was also implicated in ALS and FTLD. Located on chromosome 16 (Aman et al, 1996), the FUS gene encodes a 526 amino acid protein that belongs to the FET/TET family. Proteins in this family are defined by the presence of an N-terminal transcription activation domain SYGQ-rich region, a C2/C2 zinc finger (ZnF) motif, and one or more arginine-glycine-glycine (RGG)-repeat sequences (Morohoshi et al, 1998). The N-terminal of FUS contains a SYGQ-rich region, followed by a RGG box (RGG1), an RRM motif, another RGG box (RGG2), a ZnF motif and an additional RGG box (RGG3). The C-terminus contains a nonclassical nuclear localization signal (NLS) with conserved proline and tyrosine residues (PY-NLS) (Burd & Dreyfuss, 1994; Dormann & Haass, 2013; Iko et al, 2004; Lanson & Pandey, 2012; Lee et al, 2006; Morohoshi et al, 1998; Prasad et al, 1994; Zinszner et al, 1994)(Figure 2). In addition, bioinformatics analysis identified two PLDs in FUS, localized in its N-terminal (residues: 1-239) and RGG2 box (residues 391-405), respectively (Cushman et al, 2010; Gitler & Shorter, 2011). PLDs are regions enriched in polar aminoacids commonly found in RNA-binding proteins; it is hypothesized that PLD drives protein aggregation in neurons. PLDs in RNA binding proteins are essential for the protein to adopt a functionally aggregated state into ribonucleoprotein granules; but the relation between physiological function and disease is not well understood (King et al, 2012).
FUS is able to bind with RNA, ssDNA, and potentially with dsDNA (Tan & Manley, 2009). The RGG-ZnF-RGG domain has been suggested as the major RNA-binding sequence, with a preference for GGUG motifs.
While FUS is ubiquitously expressed in both the nucleus and cytoplasm in many cell types, it is predominantly nuclear in glial cells and neurons (Andersson et al, 2008). FUS was originally identified as an oncogene fused with transcription repressor C/EBP homologous protein 10 (CHOP) in human malignant myxoid liposarcomas (Rabbitts et al, 1993). FUS was also identified as an activator of ETS-related gene (ERG) in acute myeloid leukemia (Ichikawa et al, 1994; Panagopoulos et al, 1994) and in Ewing’s sarcoma tumors (Shing et al, 2003) by chromosomal translocation via its N-terminus, which was later shown to possess potent transcriptional activity (Prasad et al, 1994; Zinszner et al, 1994). Subsequent studies have illuminated the transcriptional regulatory role of FUS in global or specialized components of transcriptional machinery. For example, under specific conditions, FUS may have a role in activating transcription of certain nuclear hormone receptor by interacting with their DNA-binding domain (Fay et al, 1998; Tan & Manley, 2009). Recently, FUS was also shown to affect the transcription of RNA Pol III (Tan & Manley, 2010). Additionally, FUS affects gene expression by acting as co-regulator of several transcription factors, including nuclear hormone receptors (Powers et al, 1998), Spi-1/PU.1 (Hallier et al, 1998), NF-κB (Uranishi et al, 2001), and RUNX transcription factors (Li et al, 2010). The involvement of FUS in RNA splicing by binding splicing regulator or pre-mRNA was widely investigated as well (Sama et al, 2014). FUS was identified as component of the hnRNP complex assembled on the Adeno pre-mRNA (Cinzia Calvio, 1995, RNA, 1), and subsequently as a partner of SR family splicing factors, including SRm160, SRp75, and PTB (Calvio et al, 1995). A recent study has shown that FUS is able to directly bind to thousands of pre-mRNAs, with a preference for long introns compared to exons and 3′UTRs. In addition to transcription and splicing, FUS also plays a role as an mRNA transporter between the nucleus and cytoplasm. FUS displays a bidirectional transport in neuronal dendrites to shuttle mRNA into dendritic spines, a critical step for neuronal maturation (Fujii et al, 2005).
Furthermore, due to its ssDNA/dsDNA binding properties, FUS was presumed to be involved in maintaining genomic fidelity and the DNA damage response (DDR), which was subsequently demonstrated (Wang et al, 2013b). FUS was found to be a downstream factor of ataxia-telangiectasia mutated (ATM) and Poly(ADP-ribose) Polymerase (PARP) 1, and an interacting partner of histone deacetylase 1 (HDAC1) in DDR (Baechtold et al, 1999; Gardiner et al, 2008; Mastrocola et al, 2013; Rulten et al, 2014; Wang et al, 2013b). The involvement of FUS in DDR is discussed in detail in later section of this article.
2.3. Crosstalk of TDP-43 and FUS
Both TDP-43 and FUS are involved in multiple RNA metabolic processes (Figure 3). Growing evidence suggests a functional overlap of TDP-43 and FUS (Honda et al, 2013). Tibbetts and colleagues found that TDP-43 interacts with FUS physically in vivo and in vitro, and the interaction relies on the C-terminal Gly-rich and RRM2 domains of TDP-43 (Kim et al, 2010). The interaction is required for coordinately regulating mRNA expression of their common target, histone deacetylase 6 (HDAC6) (Kim et al, 2010). Interestingly, a more recent study by Cleveland and colleagues showed enhanced interaction of ALS-associated TDP-43 mutants Q331K and M337V with FUS in HeLa cells, which was speculated to perturb normal function of FUS (Ling et al, 2010). In animal models, the TDP-43 knockdown phenotype was rescued by overexpression of WT FUS in zebrafish, which suggests a potential backup role for TDP-43 (Kabashi et al, 2011a). In D. melanogaster FUS was shown to function downstream of TDP-43 in neurons, a role required to maintain normal locomotion and regular life spans (Wang et al, 2011). Furthermore, TDP-43 or FUS-silenced primary cortical neurons obtained from mouse fetal brains exhibited significantly overlapping transcriptome profiles. Specifically, around 25% of genes with changed expression levels and around 10% of genes with altered splicing overlapped, which suggests a substantial collaboration of TDP-43 and FUS in mRNA maturation and/or transportation (Honda et al, 2013).
Potential back-up functions of TDP-43/FUS for other RNA/DNA binding proteins like SMN was also evident in several studies. In the nuclei and nuclear gemini of coiled bodies in healthy neurons, FUS together with TDP-43 and SMN have been suggested to cross-talk for spliceosome maintenance (Belly et al, 2005; Wang et al, 2008).
There is evidence showing that transport ribonucleoprotein particles (tRNP) are affected in ALS-FUS as well as in ALS-TDP-43 (Sephton & Yu, 2015). The function of TDP-43 and FUS at synapses is not known, but emerging evidence suggests that these proteins play roles in synapse integrity (Hebron et al, 2013). Furthermore, formation of granules containing TDP-43, FUS and tRNP increases upon stimuli, and genetic deletion results in altered dendritic spines and branches in cultured primary motor neurons (Sephton & Yu, 2015). Overall, although the physiological or pathological significance of the interaction of TDP-43 and FUS remains largely unknown, further investigations to understand the impact of deregulation may shed light on the common molecular mechanisms of ALS-TDP-43 and ALS-FUS.
3. Neurotoxicity of TDP-43/ FUS
3.1. Cytoplasmic aggregation and functional/nuclei-specific loss of TDP-43/FUS
Accumulation of polyubiquitinated, misfolded protein aggregates is a shared feature in most neurodegenerative disorders, such as PD, which presents α-synuclein aggregates, and AD, in which the proteins amyloid beta peptide and MAP Tau form aggregates. TDP-43 was first described in 2006 as a major component of insoluble cytoplasmic aggregates found in spinal cord and brain samples from patients with ALS and FTLD (Neumann et al, 2006). Cytosolic TDP-43 aggregates were also identified in muscle tissue of patients with inclusion body myositis and Alexander disease (Salajegheh et al, 2009; Walker et al, 2014).
In ALS, TDP-43 is fragmented at its CTD into ~35 to ~25 kDa polypeptides that form cytoplasmic insoluble inclusions. Although the molecular triggers driving TDP-43 aggregation in spinal motor neurons is not clearly understood, in vitro studies have implicated several critical factors that can induce TDP-43 cleavage and aggregation (Li et al, 2015). TDP-43 can also form dimers that are enriched in cytosolic fractions. The expression of C-terminal fragments (CTFs) in cultured cells generally reproduces the aggregation pathology found in patients. Both the CTD and N-terminal domain (NTD) of TDP-43 can form aggregates as long as they contain the C-terminal end of the RRM2 domain, a β-sheet-rich region capable of forming stable dimers (Yang et al, 2010). Moreover, aggregation of fragments can seed full-length TDP-43 aggregation, reducing nuclear TDP-43. The crystal structure of C-terminal RRM2 revealed that it folds into a structure composed of two α-helices and five β-sheets. Cell culture studies have shown that RRM2 is required for aggregation, as cleaving at these sites removes the β-strand and α-helix of RMM2, preventing native folding (Zhang et al, 2009). This observation raises the question of whether or not RRM2 plays a role in TDP-43 aggregation via misfolding. Researchers studying the domain assembly of TDP-43 found that TDP-43 without the C-terminal tail is capable of forming homodimers via its NTD. It has also been shown that truncated RRM2 in the glycine-rich region forms fibrils in vitro, similar to those found in disease models, suggesting that RRM2 plays a role in the formation of cytoplasmic inclusions of TDP-43 (Zhang et al, 2009). Inclusions within brain samples of ALS patients labeled with antibodies recognized the C-terminus of TDP-43, but not the N-terminus, suggesting that inclusions are predominantly comprised of CTFs. Inclusions in spinal cord samples of ALS patients stained positive for both C-terminal and N-terminal fragments, suggesting that inclusions contain full-length TDP-43 (Zhang et al, 2009). Furthermore, TDP-43 dimer complex was identified in human brain tissues, suggesting that dimerization is a feature of TDP-43 proteinopathies (Shiina et al, 2010).
Reactive oxygen species (ROS) and increased OS markers have been reported in patients with ALS, which are critical factors linked to TDP-43 aggregation (LoGerfo et al, 2014; Nagase et al, 2015; Niedzielska et al, 2015). A marker of OS, 4-hydroxynonenal (HNE), is found to be elevated in the spinal cord, motor cortex, cerebrospinal fluid, and serum of SALS cases (Pedersen et al, 1998; Simpson et al, 2004). HNE induces aggregation and mis-localization of TDP-43 in cultured cells (Kabuta et al, 2015). Cysteine residues are strongly involved in TDP-43 insolubility induced by HNE. Furthermore, it has been reported that cysteine residue-mediated oxidation and disulfide crosslinking alters TDP-43 solubility and impairs nuclear function. This reversible alteration of TDP-43 solubility occurs via formation of disulfide bonds through the highly conserved cysteine residues in TDP-43. These TDP-43-disulfide crosslinks are detected in control brain samples, suggesting that disulfide crosslinking occurs in normal healthy brains in response to OS (Cohen et al, 2012). In addition, OS also promotes TDP-43 acetylation, which consequently drives TDP-43 aggregation (Cohen et al, 2015). Acetylated TDP-43 (Lys-145) is found in ALS spinal cord samples (Cohen et al, 2015). Altered metal homeostasis has also been linked to TDP-43-associated neurodegeneration. Zinc, but not copper and iron salts, promotes TDP-43 nuclear depletion and formation of cytoplasmic inclusions in cultured neurons (Zinszner et al, 1994), suggesting that TDP-43 nuclear depletion is a specific consequence of increased Zn levels (Caragounis et al, 2010).
Chronic immune activation is a common feature in motor neuron diseases. It has been reported that TDP-43 mRNA and protein levels are increased in the spinal cord of ALS patients (Swarup et al, 2011b), as well as elevated levels of activated transcription factor NF-κB, which is involved in a large number of normal cellular processes, including immune and inflammatory responses. It was found that TDP-43 and NF-κB p65 interact in glial cells and neuronal cells of ALS patients, as well as in transgenic mice overexpressing human TDP-43 (Philips & Robberecht, 2011; Swarup et al, 2011b). NF-κB is a key component of the immune response and has an emerging role in ALS and other neurological disorders. It has been reported that in cell culture, exposure of microglia and astrocytes to LPS induces cytoplasmic redistribution of TDP-43 (Correia et al, 2015). Additionally, NF-κB activation increases cytoplasmic levels of TDP-43, suggesting that chronic brain inflammation can drive cytoplasmic aggregation of TDP-43 (Correia et al, 2015).
TDP-43 aggregation has been widely related to TDP-43 toxicity. However, in vitro reduction of TDP-43 aggregation by small molecules/natural compounds effectively reduced TDP-43 aggregates in ALS cell models and yeast models, but did not reduce or prevent cell death (Liu et al, 2013). This suggests that while TDP-43 aggregation is a cytotoxic effect of TDP-43 proteinopathies, its prevention alone is not sufficient to rescue neurons from degeneration and death. Jackson et al., recently showed, that expression of upframeshift protein 1 (UPF1) could rescue motor functions in ALS-like paralysis rat model based on TDP-43 overexpression. Moreover, expression of UPF1 in yeast and neuronal cell culture reduces TDP-43 cytotoxicity (Jackson et al, 2015).
Since the first report that linked a number of mutations of FUS with patients with FALS, who also developed cytoplasmic inclusions in spinal motor neurons, a pathology similar to TDP-43 (Kwiatkowski et al, 2009; Vance et al, 2009), subsequent studies identified FUS-positive cytoplasmic inclusions not only in FALS but also in a small number of SALS patients (Deng et al, 2010). Cytosolic FUS is recruited into reversible SGs (Dormann & Haass, 2011; Li et al, 2013). Furthermore, SG markers have been observed in the inclusions from patients with FUS linked ALS or FTLD (Deng et al, 2014a). In addition, ALS associated FUS mutants show increased association with SGs compared to WT FUS, mutated FUS is able to bind and sequester WT FUS into SGs, which suggests a direct pathological role of SGs in ALS. Some researchers believe that although FUS accumulation in SGs is a reversible process in healthy neurons, under chronic stress, it could lead to pathological aggregation of FUS in SGs (Ling et al, 2013). A second school of thought suggests that recruitment of FUS into SGs prevents irreversible aggregation of FUS mis-localized to the cytoplasm (Shelkovnikova et al, 2013b). Recent study in a mouse model with post-natal elimination of FUS, shows that FUS has no effect in survival of motor neurons or function, suggesting that FUS-dependent neurodegeneration is likely to be due to a gain of toxicity (Sharma et al, 2016).
3.2. Neurotoxicity of TDP-43/FUS mutations
Since the discovery of TDP-43 as the major protein found in ALS cytoplasmic aggregates, several genetic screens have been conducted to identify TARDBP gene mutations (http://alsod.iop.kcl.ac.uk/Overview/gene.aspx?gene_id=TARDBP). Three separate research groups sequenced the entire coding region of the TARDBP gene and identified mutations associated with ALS (Gitcho et al, 2008; Kabashi et al, 2008; Sreedharan et al, 2008), which were mostly substitutions in serine and threonine residues. Cell and biochemical studies of these mutations provided evidence linking TDP-43 abnormalities and neurodegeneration. In an extensive survey of patients diagnosed with ALS, (80 FALS cases and 120 SALS cases), eight heterozygous mutations were identified, with the A315T mutation found to be the most common (Kabashi et al, 2008). The M337V mutation was identified in a British family, and two other missense variations G294A and Q331K, were reported by the same investigators (Sreedharan et al, 2008).
Since then, 47 missense mutations have been identified. M337V is one of the most common mutations in patients who presented with upper-limb early onset motor neuron disease. Recent studies reported increased levels of full-length and truncated TDP-43 in differentiated neurons derived from induced pluripotent stem cells (iPSC) from ALS patient lymphoblasts carrying the M337V mutation (Rutherford et al, 2008). The same group reported additional mutations, including two previously unknown mutations (N345K, I383V). Human lymphoblastoid cells expressing N345K, I383, and M337V presented with increased caspase-cleaved ~25 kDa fragments in cytoplasmic aggregates, suggesting a novel toxic gain-of-function through protein-protein interactions or accumulation of TDP-43 fragments leading to apoptosis (Gendron et al, 2013). Other studies identified several other mutations (G348C, R361S, N390D, N390S) that also presented increased insolublê25 kDa TDP-43 fragments (Daoud et al, 2009; Del Bo et al, 2009). TDP-43 spontaneously forms aggregates; mutations within the CTD (Q331K, M337V, Q343R, N345K, R361S, N390D) increase the number of aggregates, promote toxicity in vivo, and accelerate aggregation of recombinant TDP-43 in vitro. In addition, a mutation causative of ALS and FTLD (A328T) has also been found in patients with PD, as well as in patients with FTD with Parkinsonism (Borghero et al, 2011; Cannas et al, 2013).
D. melanogaster lacking TBPH (the TDP-43 ortholog) die as pupae, and rare survivors present with synaptic dysfunction of motor neurons, reduced lifespan, and progressive motor neuron loss. These features can be rescued by expression of human TDP-43. In contrast, introducing M337V or A315T mutants fails to rescue motor neuron degeneration. (Feiguin et al, 2009; Fiesel et al, 2010; Kabashi et al, 2011b).
Studies have shown that overexpression of TDP-43 results in reduced mitochondrial density in neurites of primary motor neurons (Cozzolino & Carri, 2012). This condition is exacerbated by mutant TDP-43 Q331K/M337V overexpression. In contrast, suppression of TDP-43 results in increased mitochondrial density (Wang et al, 2013a). Also, TDP-43 co-localizes with mitochondria in motor neurons, and this co-localization is enhanced by mutant expression, suggesting that TDP-43 has a role in regulating mitochondria dynamics. TDP-43 depletion results in increased abnormal neurites and decreased cell viability. TDP-43 mutants A315T/M337V/Q331K mis-localize to the cytosol and show abnormal neurites. Cytosolic expression of TDP-43 with mutated NLS variant also showed abnormal neurite morphology and reduced cell viability (Gitcho et al, 2008; Han et al, 2013; Wang et al, 2013a).
TDP-43 mutations affect the dynamics of SG assembly by increasing the propensity for SG assembly in the presence of sodium arsenite, a potent OS inducer. This also correlates with mutant TDP-43’s decreased nuclear localization in response to sodium arsenite (McDonald et al, 2011). Mutations disrupt cytoplasmic SGs that contain translationally silenced RNA transported to target sites, enabling the cell to carry out protein synthesis. TDP-43 mutants may also be incorporated in the early stage of SG formation, resulting in larger and deregulated granules (Dewey et al, 2011; McDonald et al, 2011).
The pathogenic mechanisms of TARDBP mutations are still unclear; mutations most likely affect normal functions of TDP-43 by gain-of-toxicity or loss-of-function, mediated by enhanced aggregation and nuclear depletion. Alterations in TDP-43 function have deleterious effects, including impaired RNA metabolism and generation of toxic byproducts. The involvement of TDP-43 pathology in ALS vs. FTLD is an interesting topic, which is still not completely understood. While the location/distribution of TDP-43 aggregates differ between ALS (spinal cord) and FTLD (more widespread in the brain including frontal and temporal lobes), the familial mutations in TARDBP are unique to ALS and are not found in other neurodegenerative disorders including FTLD (Blokhuis et al, 2013; Brouwers et al, 2010; Rutherford et al, 2008; Van Deerlin et al, 2008). However, some of the common TARDBP mutations, presumably involving vulnerable residues for somatic mutations were reported in a small number of sporadic FTLD cases (Benajiba et al, 2009; Borghero et al, 2011; Borroni et al, 2009; Chiang et al, 2012; Chio et al, 2010; Corrado et al, 2009; Kovacs et al, 2009). Similar scenario may occur in other neurodegenerative diseases involving TDP-43 pathology such as inclusion body myositis and Alexander disease. Further investigations are required to understand how these sporadic diseases with TDP-43 pathology manifest in presence of somatic TARDBP mutations.
Numerous mutations of FUS have also been reported since 2009, and to date, over 50 mutations/deletions have been identified to account for ~4% FALS and rare SALS cases. Some mutations are associated with early-onset disease; for example, ALS patients exhibiting the P252L mutation are often a very young age at onset (mean: 23.7 years old) and present rapid disease progression (patients die within 12 months) (Chio et al, 2009b; Lagier-Tourenne & Cleveland, 2009). Compared to ALS, only a few FUS mutations were discovered in association with FTLD (Broustal et al, 2010). The R521H mutation was identified in a patient initially diagnosed with behavioral disorders, but rapidly developed ALS (Broustal et al, 2010). Meanwhile, R521C, G156E mutations and G174-G175 deletion were identified in a FTLD patient who was concurrently diagnosed with motor neuron disease (Blair et al, 2010; Ticozzi et al, 2009; Yan et al, 2010). M254V, P106L mutation and Gly174-Gly175 deletion were discovered in patients with FTLD (Huey et al, 2012). These cases suggest that ALS and FTLD share similar pathology and genetics.
Unlike TDP-43, in which the majority of mutations cluster in the C-terminal prion-like region, over 50% of ALS-related FUS mutations have been discovered in the C-terminal NLS domain, implying a possible involvement of its nuclear import defects. Dormann et al., showed that the last 13 amino acids (514-526) within the NLS are necessary, but not sufficient for nuclear import of FUS. In this study, FALS-associated point mutations occurred in the C-terminal region at evolutionarily conserved residues including R521 (G/H/C), R522 (G), R524 (S), and P525 (L). Immunostaining of HeLa cells expressing the above mutants showed that the P525L mutant induces dramatic nuclear import defect, whereas R522G showed a moderate phenotype. Interestingly, although the ALS-linked R521 mutation is in a highly conserved amino acid among vertebrates, is the most frequently mutated residue in FUS, the R521G/H/C mutant shows only mild cytoplasmic localization, which suggests that the mutation of R521 may result in pathological dysfunctions of FUS beyond cytosolic accumulation. The cytoplasmic accumulation of FUS P525L was subsequently observed in neurons from rat hippocampus and frontal cortex, as well as in zebrafish eggs in the same study (Dormann et al, 2010); these results were later confirmed by other groups (Niu et al, 2012; Zhou et al, 2013). As mentioned above, FUS is recruited into SGs. The FUS R495X truncation mutant (a deletion of the last 32 C-terminal amino acids of FUS), which exists in early-onset ALS, was identified by Hayward and colleagues (Bosco et al, 2010). Also, an experimental mutant of G515X (a deletion of the terminal 12 C-terminal amino acids of FUS, which includes the main cluster of ALS-linked mutations) co-localizes with SGs in HEK-293 cells extensively (in ~70-80% cells) within 4-7 min of exposure to arsenite. However, the recruitment of the R521G mutant into SGs shows significant delay and lower intensity. The same phenomenon was subsequently observed in zebrafish embryos in response to heat shock (Bosco et al, 2010). Other ALS-FUS mutants were also reported to be associated with SGs; for example, R521C co-localized with SGs in zebrafish motor neurons under the treatment of heat shock and arsenite (Acosta et al, 2014), and R514S or P525L mutants formed SGs in HeLa cells. Also, double mutations of R514S and P525L, or triple mutations of R514S, R521C, and P525L, additively enhanced SG formation (Ito et al, 2011). A detailed analysis was conducted by Suzuki and colleagues to identify functional domains that contribute to SG formation. In this study, a series of FUS deletion constructs of conserved regions, including N-terminus, SYQG-rich domain deletion (ΔNT), Gly-rich region deletion (ΔGRR), NES deletion (ΔNES), RNA recognition motif deletion (ΔRRM), first RGG box deletion (ΔRGG1), ZnF motif deletion (ΔZnF), secondary RGG box deletion (ΔRGG2) and C-terminal deletion (ΔCT), as well as WT FUS were developed and overexpressed in HeLa cells. This experiment revealed that ΔCT formed SGs extensively, whereas ΔRGG2 formed few SGs, indicating that the C-terminus plays a critical role in SG formation (Ito et al, 2011). The assembly of SGs is an emergency response for cells to survive by limiting the translation of non-essential mRNAs and focusing on producing essential proteins under stress. The observation that TDP-43 and FUS accumulate in SGs in vitro and in vivo supports its pathological significance. Several models, including a gain-of-toxicity model, loss-of-function in SGs model, and nuclear loss-of-function model, have been proposed to explain the connection between TDP-43, FUS and SGs in ALS/FTLD pathogenesis, which need to be elucidated by further investigation (Li et al, 2013).
The cytoplasmic retention of ALS-associated mutant FUS and its role in deficient RNA processing were confirmed by multiple studies. Hicks and colleagues have shown that WT FUS, but not ALS-linked FUS mutants R521G, R522G, and Δ515 (deletion of the last 12 residues in the C-terminus), are able to directly modulate the alternative splicing of exon 7 to autoregulate protein levels, and the ability to regulate splicing is tightly correlated with nuclear retention (Zhou et al, 2013). Another study showed that HEK-293T cells overexpressing FUS with the R521G or R522G mutation displays a globally-affected transcription pattern, which resembles the effect of overexpression of WT FUS but the effects of siRNAs targeting FUS (van Blitterswijk et al, 2013). Intranuclear aggregation of FUS induced by the pathogenic mutation G156E was also proposed as an alternative pathogenic mechanism of FALS. The FUS G156E mutation occurs in an evolutionarily conserved region in mammals, reptiles, and zebrafish. Clinically, a patient with the G156E mutation presented with both upper and lower motor neuron signs concurrently with dementia, and developed FTD in his fourth decade (Ticozzi et al, 2009). Nomura et al. showed that the G156E mutant increases propensity of FUS for aggregation in vitro and in vivo, which supports the model of ‘seeding reaction’, in which the aggregated protein fibrils serve as a structural template facilitating the fibrillation of unaggregated protein. Consistently, FUS G156E caused a rapid progression of ALS after its onset (Nomura et al, 2014).
3.3. Deregulation of post-translational modifications
3.3.1. Abnormal phosphorylation
Hyperphosphorylated TDP-43 has been consistently identified in the cytoplasmic inclusions of spinal motor neurons in both ALS patients and model systems. Serines 409 and 410 are the most consistently phosphorylated sites in disease, although S379, S403 and S404 are also observed in disease pathology (Hasegawa et al, 2008; Neumann et al, 2009). The functional role of TDP-43 phosphorylation is not clear, and a correlation between protein insolubility and TDP-43 phosphorylation has been reported (Neumann et al, 2006). Phosphorylated TDP-43 exhibits a longer half-life than the non-phosphorylated form. This may be due to changes in UPS-mediated degradation or increased protein stability. Mutating serine residues 409 and 410 of TDP-43 to non-phosphorylatable alanine decreased TDP-43 neurotoxicity in C. elegans models (Liachko et al, 2010; Liachko et al, 2013). There are a number of kinases identified that control TDP-43 phosphorylation, including CK1/2, CDC7, and TTBK1/2 (Hasegawa et al, 2008). TDP-43 can be directly phosphorylated by CK1/2, and expression of hyperactive CK1 promotes the accumulation of cytoplasmic insoluble phosphorylated TDP-43 (Nonaka et al, 2016). The kinases CDC7 and TTBK1/2 have also been shown to directly phosphorylate TDP-43 in vitro. Pathologically, CDC7 and phospho-TDP-43 co-localize in disease-affected neurons (Liachko et al, 2013). Furthermore, a small molecule inhibitor of CDC7 reduced phosphorylation and neurodegeneration in C. elegans model and also limited TDP-43 phosphorylation in NSC-43 cells (Liachko et al, 2013). TTBK1 and TTBK2 protein levels are elevated in motor cortex of FTLD, and the kinases co-localize with TDP-43 inclusions in ALS spinal cord (Liachko et al, 2014). TDP-43 phosphorylation is not required for its cleavage, aggregation or toxicity in cell culture systems.
Phosphorylation of FUS was recently identified. Serine 42 was identified as the phosphorylated serine in WT, but not in CHOP-fused FUS by ATM, in response to ionizing radiation (IR)-induced genomic double-strand breaks (DSBs) (Gardiner et al, 2008). In another study, DNA-dependent protein kinase (DNA-PK) mediated multiple phosphorylation on the N-terminus of FUS, and this phosphorylation facilitated its cytoplasmic accumulation upon cellular DNA damage (Deng et al, 2014b). The association of FUS phosphorylation with pathogenesis in neurodegeneration was explored by Nukina and colleagues, who found that overexpression of mouse phosphomimetic mutation of serine 505 (513 in humans) predicted potential phosphorylation sites for kinases such as CK2, DNA-PK and GSK3, S505D. This phosphorylation can enhance nuclear clearance of ALS-linked mutants including G499D, H509P, R510K, R513G, and R516S in both N2a and COS-7 cell lines, as well as impair the splicing regulatory activity of FUS mutants H509P and R510K (Kino et al, 2011). These data suggest that neurodegeneration initiated by FUS mutants could be exacerbated by phosphorylation.
3.3.2. Ubiquitination
The role of ubiquitination of TDP-43 in the pathogenesis of disease is not clearly understood. Cells dispose of soluble TDP-43 proteins via the UPS, while aggregated TDP-43 appears to be predominantly cleared through autophagy (Scotter et al, 2014). Inhibition of UPS in cell culture increases cytoplasmic accumulation of TDP-43 and formation of intracellular aggregates. Disruption of the UPS contributes to increased levels of ubiquitinated TDP-43 in ALS (Zhang et al, 2010). Ubiquitination of TDP-43 may be a late event in disease pathogenesis because most TDP-43 inclusions are not ubiquitinated in early disease stages (van Eersel et al, 2011). Autophagosome-mediated degradation is also involved in TDP-43 protein turnover. TDP-43 was proposed to interact with UBQLN2, a protein that binds ubiquitinated proteins and transports them to the proteasome for degradation. UBQLN2 also has roles in macroautophagy and chaperone-mediated autophagy. Interestingly, mutations in UBQLN2 have been found in some families with FALS (Deng et al, 2011b). UBQLN2 expression in cell culture and D. melanogaster models promotes TDP-43 autophagosomal degradation (Filimonenko et al, 2007). Recently, the E3 ubiquitin ligase Parkin was proposed to ubiquitinate TDP-43, regulating its subcellular transport (Hebron et al, 2013). In D. melanogaster models, the ubiquitin-conjugating enzyme UBE2E3 promotes ubiquitination of TDP-43; in contrast, ubiquitin isopeptidase Y (UBPY) decreased TDP-43 ubiquitination (Hans et al, 2014). Furthermore, knockdown of UBPY promotes formation of insoluble TDP-43 aggregates and induced neurotoxicity in D. melanogaster (Hans et al, 2014).
While ubiquitination and hyperphosphorylation of TDP-43 are etiologically linked to motor neuron diseases, unlike TDP-43, FUS co-localization with ubiquitin has been observed (Deng et al, 2010) in post-mortem ALS brains and spinal cord tissue. However concluding evidence of FUS ubiquitination needs to be further investigated.
3.3.3. Methylation
Arginine methylation is frequently observed in RNA-binding proteins, a process in which nuclear importation is tightly regulated (Dormann & Haass, 2013). Although it has not been reported that TDP-43 is methylated, previous work has shown that FUS is highly methylated at most of its 37 arginine residues (Du et al, 2011; Rappsilber et al, 2003), and the protein arginine transferases (PRMT) 1 and 8 have been demonstrated to interact and catalyze arginine methylation of FUS. PRMT was found to methylate both wild-type (WT) and FUS FALS mutants at a comparable overall level, even when the mutation occurred at arginine residues (Jackel et al, 2015). Methylation enhances the toxicity of ALS-associated FUS mutants by regulating their subcellular localization as well as the formation of SGs (Dormann et al, 2012; Du et al, 2011; Scaramuzzino et al, 2013; Tradewell et al, 2012), although it is controversial whether or not methyltransferase activity is involved in the incorporation of FUS into SGs (Baron et al, 2013; Sama et al, 2013). Insights into how arginine methylation affects the subcellular localization of FUS have been given by Hass and colleagues (Dormann et al, 2012), who found that PRMT1-mediated arginine methylation within the RGG3 domain (but not PY-NLS) is necessary and sufficient to restore nuclear import of mutant FUS upon inhibition of methyltransferase activity by methylation inhibitor AdOx. In addition, the unmethylated RGG3 domain tightly binds with Transportin (TRN) to compensate the weaker association between TRN and PY-NLS with mutation P525L, whereas methylation in the RGG3 domain impairs its binding with TRN significantly in FUS peptides containing P525L mutation, but only slightly in WT peptide. This binding affinity difference could play a role in PRMT-dependent methylation in ALS-FUS. Interestingly, by using an antibody specific for FUS methylation, researchers in this study also revealed that FUS is highly methylated in cytoplasmic inclusions in ALS-FUS patients, but not in FTLD-FUS patients, indicating that arginine methylation may have distinct implications in the pathogenesis of the two neurodegenerative disorders (Dormann et al, 2012). Tibshirani et al., showed that nucleo-cytoplasmic shuttling of FUS is able to affect the distribution of PRMT1 in both cultured motor neurons and transgenic mice, and in turn regulates its nuclear substrates (Tibshirani et al, 2015). In this study, PRMT1 was found to distribute with overexpressed human WT FUS in nuclei or redistribute to the cytoplasm with ectopically expressed ALS-linked FUS mutants including R521H, P525L, and R495X in neurons, in both physiological and stress conditions. Furthermore, the depletion of PRMT1 in nuclei induced by the cytoplasmic inclusion of FUS mutant R521H reduced asymmetric arginine dimethylation of histone 4 (H4), a key substrate of PRMT1, and the methylation at arginine 3, which is required for the acetylation of histone 3 (H3) at lysine residues 9 and 14 (H3K9/K14ac). Accordingly, a decreased acetylation of H3K9/K14 was also demonstrated. Additionally, decreased H3K9/K14ac was observed in either WT FUS or R521H FUS-depleted nuclei (Tibshirani et al, 2015).
3.4. Protein truncation/degradation
TDP-43 harbors caspase 3 cleavage sites, and when cleaved, TDP-43 forms ~25 kDa and ~35 kDa fragments. Researchers found that in vitro incubation of TDP-43 with caspase 3 and caspase 7 produces fragments of ~42, ~35 and ~25 kDa (Zhang et al, 2007). Fragments of ~37 kDa phosphorylated at S409 and S410 have been observed, and are presumed to be the phosphorylated forms of 35kDa fragment (Zhang et al, 2007). Research suggests that once the CTFs are generated, they are phosphorylated and ubiquitinated. The accumulation of TDP-43 CTFs may lead to neuronal dysfunction. Intermediate length poly-glutamine expansions in ATXN2 are a risk factor for ALS (Elden et al, 2010). These expansions may activate caspase 3 promoting TDP-43 cleavage and aggregate formation. Hart and colleagues show that the intermediate-length ATXN2 polyQ expansions enhance stress-induced C-terminal cleavage of TDP-43 by the activation of stress-dependent multiple caspases, including caspase 3. Accumulation of caspase 3 in motor neurons may represent a risk factor for ALS cases that harbor Ataxin2 polyQ expansions (Hart & Gitler, 2012). N-terminal sequencing of brain TDP-43 CTFs revealed that the cleavage begins at site Arg 208 or Asp 219. TDP-43 has an NLS motif that lies within the NTD. Cleavage of TDP-43 removes this NLS sequence, but not the nuclear export signal (NES) sequence, promoting cytoplasmic accumulation of TDP-43. These cytoplasmic CTFs have been proposed to serve as primary seeds for the aggregation of TDP-43 into inclusions (Hart & Gitler, 2012). Unlike TDP-43, FUS fragments have not been identified in motor neuron models.
4. TDP-43/FUS animal models: Lessons learned and challenges
Expression of wild-type or FALS mutant TDP-43 or deletion of the endogenous TDP-43 homolog in animal models can cause motor dysfunction and neuronal death, modeling ALS and FTLD. Several loss-of-function and transgenic overexpression rodent models have been generated. TDP-43 homozygous knockout mice die between embryonic day 3.5 and 8.5; even though RNA splicing regulation function carried by TDP-43 is important, the exact cause of death is not known. (Kraemer et al, 2010; Sephton et al, 2010; Wu et al, 2010). Heterozygous mice with one functional copy of TARDBP are viable, although they develop mild motor dysfunctions and moderate pathology with age (Kraemer et al, 2010). To overcome lethality of loss of TDP-43 during development, TDP-43 was deleted specifically in motor neurons and spinal cord in post-natal mice using the Cre-lox system (Wu et al, 2012). This mice line presents muscle atrophy and motor neuron loss (Iguchi et al, 2013). A transgenic mouse line that expressed an ectopic microRNA to reduce endogenous TDP-43 ubiquitously presented muscle weakness, paralysis and neurodegeneration of cortical and spinal neurons, resulting in early lethality (Yang et al, 2014).
The transgenic rodent models vary depending on the selection of the promoter driving gene expression. Investigators have overexpressed TDP-43 and ALS-linked TDP-43 mutants using neuronal-specific promoters, inducible promoters, or ubiquitous promoters (Stallings et al, 2010; Wegorzewska et al, 2009; Xu et al, 2010; Xu et al, 2011). WT TDP-43 overexpression in mice presents accumulation of polyubiquitinated TDP-43, TDP-43 fragmentation, astrogliosis, microgliosis, axonal degeneration, neuronal loss, motor function impairments, and shortened lifespan (Sasaki et al, 2015). TDP-43 overexpressing mice also display behavioral deficits and motor neuronal loss (Tsai et al, 2010). Mice expressing human TDP-43 driven by the neuron specific Thy1 promoter (TAR4 mice) present nuclear ubiquitinated and hyperphosphorylated cytoplasmic inclusions in cortical neurons. These mice also accumulate ~25kDa and ~35kDa C-terminal fragments of TDP-43 that are characteristically found in ALS brain samples. However, the TAR4 mice do not exhibit limb paralysis (Wils et al, 2010). Mice expressing human TDP-43 driven by prion protein promoter (Prp) have decreased mouse TDP-43 mRNA levels, reactive gliosis, degenerating neurites and neurons in spinal cord. This mouse line also accumulates ~25kDa CTFs and ubiquitinated cytoplasmic inclusions. This mouse model presents motor deficits at 21 days and lethality around 2 months of age. Human TDP-43 expression affects mitochondrial distribution and integrity in mouse spinal cord neurons (Xu et al, 2010). Transgenic mice expressing human TDP-43 driven by bacterial artificial chromosome (BAC) have cognitive and motor deficits with ~25kDa and ~35kDa CTFs, reactive gliosis, and neuroinflammation (Swarup et al, 2011a; Xu et al, 2011).
Transgenic mouse models with inducible overexpression of human TDP-43 with defective NLS in forebrain, driven by a Dox-suppressible CamK2a promoter showed neuronal loss; however, phosphorylated and ubiquitinated TDP-43 pathological aggregates were less observed (Igaz et al, 2011). Interestingly, cytoplasmic expression of human TDP-43 resulted in an abnormal decrease of nuclear mouse TDP-43 and altered gene expression (Igaz et al, 2011). This mouse model did not develop ALS-like phenotype possibly due to the lack of expression in spinal cord. To overcome this, a second generation of Dox suppressible transgenic with defective NLS TDP-43 expressing line driven by neurofilament heavy chain promoter to express in brain and spinal cord was generated (Walker et al, 2015). This mouse line develops ALS-like phenotype with accumulation of phosphorylated cytoplasmic TDP-43 in brain and spinal cord with significant motor neuron loss and progressive motor impairment, eventually leading to death. Although, expression of human TDP-43 reduces endogenous mouse TDP-43 in neuronal nuclei, suppressing human TDP-43 expression after disease onset reduces the phosphorylated TDP-43 pathology, simultaneously increases nuclear mouse TDP-43 to rescue motor impairment and extension in lifespan (Walker et al, 2015).
Interestingly, while cytoplasmic mis-localization and fragmentation of TDP-43 is routinely observed in transgenic mouse models, inclusions are rarely observed. TDP-43 overexpression in D. melanogaster also results in loss of motor function and decreased dendrites and synapses (Ayala et al, 2005), while depletion of the TDP-43 ortholog results in reduced lifespan and locomotor defects due to alterations in dendrite branching and synapses (Feiguin et al, 2009). Zebrafish overexpressing TDP-43 exhibit abnormal swimming behavior and defective neuronal axon formation (Ayala et al, 2005). The fact that TDP-43 mutant expression is more toxic than WT TDP-43 is consistent with other animal models (yeast, chicken embryos, D. melanogaster, and mammalian cells). In yeast models, TDP-43 mutation accelerates aggregation of TDP-43. TDP-43 mutant expression alters SG formation, leading to increased formation of inclusions leading to toxic gain-of-function and cell death (Ash et al, 2010; Hanson et al, 2010; Kabashi et al, 2011b; Lagier-Tourenne & Cleveland, 2009; Polymenidou et al, 2011).
The first TDP-43 mouse model expressing ALS-associated TDP-43 mutant A315T driven by prion promoter presented ubiquitination with positive cortical neurons with loss of nuclear TDP-43 (Wegorzewska et al, 2009). These mice also have abnormal neuritis, decreased number of neurons in cortical layer V, and 20% motor neuron loss in ventral horn (Wegorzewska et al, 2009). In addition, TDP-43 A315T mice also develop apparent gastrointestinal dysfunction likely due to neurodegeneration of the myenteric plexus of the colon (Esmaeili et al, 2013; Guo et al, 2012). Transgenic TDP-43 M337V expression caused protein fragmentation and increased cytoplasmic expression compared to WT TDP-43 mice. Both WT and mutant TDP-43 proteins are neurotoxic upon overexpression, but mutant TDP-43 requires less overexpression than WT TDP-43 to induce neurotoxicity. TDP-43 mutant overexpression results in reactive gliosis, axonal and myelin degeneration, gait abnormalities, and early lethality (Arnold et al, 2013; Janssens et al, 2013; Swarup et al, 2011a; Xu et al, 2011).
A rat model expressing ubiquitous WT human TDP-43 displayed fragmentation, phosphorylation, and aggregation of TDP-43 and developed progressive degeneration of motor neurons without paralysis (Zhou et al, 2010). In comparison, transgenic rat models expressing M337V mutations become paralyzed and die as early as 29 days. Surprisingly, in rats expressing the M337V mutation, motor function can be rescued when overexpression is turned off, suggesting that therapeutic interventions targeting TDP-43 may be effective after disease onset (Zhou et al, 2010).
In summary, rodent models show TDP-43 toxicity is dose-dependent, few of the models have robust cytoplasmic aggregation, and TDP-43-toxicity appears to be critical in promoting disease. The major drawback of the available rodent models is that none of these models mimic the multi-faceted ALS disease phenotype and pathology, making it challenging to completely understand and address future therapeutic approaches. Most models do not demonstrate the key ALS hallmark of TDP-43 nuclear depletion and cytoplasmic sequestration and aggregation at the same time. In the case of overexpressing transgenic models, it is possible that TDP-43 autoregulation due to the presence of endogenous mouse TDP-43 could be a confounding factor. As knock-out of endogenous TDP-43 is lethal at the embryonic stage; conditional knockout model in which TDP-43 is targeted in a tissue-specific manner have been successful in recapitulating adult onset of SALS to an extent. A recently generated transgenic ΔNLS-TDP-43 mouse model did develop an ALS-like phenotype (Chiang et al, 2010).
Like ALS-TDP-43, several FUS-deficient and overexpression ALS-FUS animal models have been generated, which develop ALS-like phonotypes to varying extent. Although FUS knockout (FUS−/−) mice with different genetic backgrounds have distinct features, inbred FUS−/− mice showed defective B-lymphocyte development and activation, as well as perinatal death. Outbred FUS−/− mouse displayed defects in spermatogenesis and increased radio-sensitivity but were able to reach adulthood. Both mice lines have high genomic instability (Hicks et al, 2000; Kuroda et al, 2000). The morphological observation in neurons from embryos of inbred FUS−/− mouse was also reported, which provided the initial insight into the potential function of FUS in the CNS (Tolino et al, 2012). In this study, FUS−/− primary hippocampal neurons showed irregularly branched dendrites, and numerous long and thin processes, like immature axons, extended from cell bodies but had only a single axon. Moreover, FUS-deficient neurons isolated from mice displayed significantly reduced dendritic spine density, but increased occurrence of filopodia-like spines, with morphology similar to thin and long cytoplasmic protrusions (Fujii et al, 2005). Studies in D. melanogaster and zebrafish demonstrated that loss of FUS directly leads to neuronal cell death (Kabashi et al, 2011a; Wang et al, 2011). Together, these studies suggested a critical role of FUS in neuronal maturation.
A number of transgenic FUS models have been established by independent research teams (Robinson et al, 2015; Shelkovnikova et al, 2013a). Human FUS mutants, such as R495X, H517Q, R521G, and R521C, were ectopically expressed into D. melanogaster, zebrafish or mouse models (Lanson et al, 2011). D. melanogaster expressing human FUS mutants (R518K, R521C and R521H) developed severe neurodegeneration characterized by disorganized ommatidia and loss of mechano-sensory bristles in eyes. In zebrafish, human WT FUS, FUS H517Q and FUS R521G exhibited a predominantly nuclear localization in the spinal cord while FUS R495X and G515X truncation mutants showed cytoplasmic accumulation, and all the mutants formed SGs in response to heat shock stress. FUS lacking NLS (FUS 1-359) transgenic mouse model showed several key features of human ALS including the loss of spinal motor neurons and peripheral nerve fibers or lower motor neuron populations in the brainstem. Mice expressing FUS that lack RRM domain and carry a R522G mutation showed significant neuronal proteinopathy but no apparent neurodegeneration in brain or brainstem region. However, both mouse models displayed shorter lifespans (Robinson et al, 2015; Shelkovnikova et al, 2013a).
5. New Perspectives: Involvement of Genome Damage and Repair Defects in ALS pathology
5.1. DNA damage in ALS-affected motor neurons
To date, growing evidence supports the notion that damaged DNA accumulates in neurons as aging progresses, and neurons are particularly susceptible to accumulate damaged DNA due to their lack of self-renewal and replication. Furthermore, the brain is more susceptible to DNA damage due to high neuronal activity and higher mitochondrial respiration, a process that produces ROS that can cause the formation of DNA lesions, including single-strand breaks (SSBs) and DSBs (Acosta et al, 2014; Hegde et al, 2012; Mitra et al, 2014a). A recent study suggested that normal neuronal activity may produce endogenous DSBs, the most lethal form of DNA damage, by the activation of the NMDA glutamate receptor involved in neuroplasticity (Suberbielle et al, 2013). Increased levels of γ-H2AX, a well-known DNA DSB marker (Wang et al, 2014), was observed in postmortem brain sections of FALS patients, carrying FUS R521C or P525L mutations, in comparison to age-matched controls. This suggests that accumulated DSBs due to the loss of DNA repair function of FUS in ALS (Wang et al, 2013b), and confirmed by a transgenic mouse model harboring the FUS mutation presented elevated levels of several DNA damage markers, including γH2AX, phosphorylated p53, and activating transcription factor 3 (ATF3) in the CNS (Qiu et al, 2014).
5.2. DNA repair defects and abnormal damage response: Involvement of FUS
Defective DNA repair and its possible role in ALS was hypothesized as early as 1982 when Bradley et al., proposed that abnormal DNA in ALS may arise from a deficiency of an isozyme of one of the DNA repair enzymes (Bradley & Krasin, 1982). Currently, there is emerging evidence suggesting that defective DNA repair is present in numerous neurological disorders, raising interest in studying the role of accumulation of DNA damage in these disorders including ALS (Madabhushi et al, 2015). Oxidative damage to DNA and p53 activation have been observed in motor neurons in ALS cases (Martin et al, 2005). Abnormal activities of DNA repair components including AP endonuclease 1, DNA glycosylase OGG1, mitochondrial DNA polymerase γ and PARP1 were observed in ALS patients or mouse model, which strongly supports significantly impaired oxidative DNA damage repair in ALS. Apurinic/apyrimidinic endonuclease (APE/Ref-1) protein levels are increased in ALS cases (Coppede, 2011; Martin et al, 2007; Shaikh & Martin, 2002).
The involvement of FUS in DDR already has been indicated, in which FUS was shown to bind with and promote annealing of complementary ssDNA and D-loop formation in super helical dsDNA, an essential step for homologous recombination (HR)-mediated DNA DSB repair, while the fusion form of FUS-CHOP is unable to promote DNA pairing (Baechtold et al, 1999). Several recent studies have provided insights into the molecular mechanisms underlying how FUS is involved in DDR. Independent investigations by two research groups demonstrated that FUS is recruited to and co-localizes with γ-H2AX at DNA damage sites (induced by UVA laser and micro irradiation) in a PARP 1-dependent manner in human A549 and U2OS cell lines (Mastrocola et al, 2013; Rulten et al, 2014). Additionally, the RGG2 domain has been shown to be sufficient for the recruitment of FUS to DNA damage sites, which is significantly intensified by PLD (Mastrocola et al, 2013). Gardiner and colleagues showed that FUS, but not the FUS-CHOP fused form, is phosphorylated at Ser42 by ATM in HEK-293 cells in response to irradiation-induced DNA damage, whereas Deng et al. showed that FUS is phosphorylated by DNA-PK (Deng et al, 2014b), which indicates FUS as a novel component of the PIKK family-mediated DDR signaling pathway. Furthermore, Wang et al., showed that FUS interacts with HDAC1 in primary mouse cortical neurons, and the interaction is required for an optimal HR and non-homologous end joining (NHEJ)-mediated DSB repair. Further, increased DNA damage levels were observed in NeuN-positive neurons from FALS patients harboring R521C or P525L FUS mutations, and FALS-associated mutations FUS-R244C, R514S, or R521C overexpressed in U2OS cells showed defective HR and NHEJ-mediated DSB repair (Wang et al, 2013b). Although the phenotypes of DNA repair deficiency have been linked to the loss of FUS, the molecular mechanism of how FUS participates in DNA damage recognition and/or repair is largely unclear. The contribution and mechanisms of FUS-related unrepaired DNA damage in the initiation and progression of degenerative diseases still needs to be established.
Persistent accumulation of unrepaired genome damage due to increased damage induction coupled with their defective repair in neurons, could lead to arrest of transcription and damage-induced neuroinflammation (Shiwaku & Okazawa, 2015). Consistently, recent studies have reported accumulation of R-loop structures (presumably formed by damage induced inhibition of transcription) (Elden et al, 2010; Salvi & Mekhail, 2015), and activation of inflammatory signaling in ALS affected brain tissue (Kawamata et al, 1992; Mantovani et al, 2009; Swarup et al, 2011b; Turner et al, 2004). Moreover, transcriptional stalling together with DNA repair inhibition is a recipe for exacerbating genome instability and enhancing hypermethylation of gene promoters leading to gene silencing (Schmitz et al, 2009). Together, these recent developments open up a new avenue of research on the implications of defective DNA repair and DDR in neuronal death in ALS, and their potential as targets for therapeutic approaches.
6. Concluding remarks
The groundbreaking discoveries in 2006 implicating aggregation/fibrillation of the RNA/DNA-binding protein TDP-43 and FUS in ALS and FTLD (Kwiatkowski et al, 2009; Neumann et al, 2006; Vance et al, 2009) triggered a flurry of research activities that led to the discovery of TDP-43/FUS mutations in hereditary ALS patients and the widespread presence of TDP-43 or FUS pathology in about 40% of other neurodegenerative diseases, including, AD and PD (Arai et al, 2006; Geser et al, 2008; Hasegawa et al, 2007; Nakashima-Yasuda et al, 2007). However, how TDP-43/FUS pathology triggers neuronal apoptosis still remains unclear.
Whether neurotoxicity of RNA/DNA binding proteins in ALS is due to its ‘gain-of-toxicity’ or ‘loss-of-function’ is a key question. The ‘gain-of-toxicity’ hypothesis had initially received much attention, primarily based on the striking neurodegenerative phenotypes in multiple TDP-43/FUS-overexpression/aggregation models (Ayala et al, 2011). Yet, recent studies demonstrating widespread nuclear clearance of TDP-43 in ALS-affected neurons and significant loss of functional TDP-43/FUS pool due to its aggregation (Polymenidou et al, 2011) and strong ALS phenotype in TDP-43-deficient (partial) mice (Chiang et al, 2010; Walker et al, 2015), warrant detailed investigation of the ‘loss-of-function’ model.
Thus, despite significant increase in our understanding of the pathological and biochemical changes in ALS and other neurodegenerative disorders, there is no current cure; available treatments only temporarily slow the disease progression, but do not prevent neuronal death. This underscores the necessity of an overarching approach to unravel the fundamental mechanisms of disease initiation and progression in order to design more effective ways to prevent the onset of ALS, delay its progression, and finally develop improved treatment protocols for ALS patients.
Highlights.
Amyotrophic lateral sclerosis (ALS) is a group of motor neuron diseases involving a dozen distinct and overlapping protein inclusions.
The complex neuropathology of RNA/DNA binding proteins TDP-43 and FUS in motor neuron disease is critically discussed.
The loss of function due to nuclear clearance vs. gain of aggregating protein toxicity of TDP-43/FUS is comprehensively assessed.
New avenues of research involving the role of genome damage and repair defects in FUS/TDP-43-associated ALS.
Role of disease-linked TDP-43/FUS mutations in familial and sporadic ALS and other motor neuron diseases.
Lessons learned from TDP-43/FUS animal models.
Rationale and need for an overarching approach to unravel the fundamental mechanisms based intervention strategies.
Acknowledgements
The research in the Hegde laboratory is supported by grants from the National Institutes of Health USPHS R01 NS 088645, Muscular Dystrophy Association (MDA 294842), ALS Association (15-IIP-204), Alzheimer’s Association (NIRG-12-242135) to M.L.H. and Melo Brain Funds (M.L.H. and K.S.R.). E.N.G. is supported by a Doctoral Scholarship granted by the Institute for Training and Development of Human resources of Panama (IFARHU) and the National Secretariat for Science, Technology, and Innovation of Panama (SENACYT). K.S.R. is supported by the National System on Investigation (SNI) grant of SENACYT. B.C.K. is supported by grants from the Department of Veterans Affairs [Merit Review Grant #I01BX002619] and National Institutes of Health [R01NS064131]. N.L. is supported by a Career Development Award 2 from the Department of Veterans Affairs [#I01BX007080]. The authors thank other members of the Hegde laboratory for their assistance.
Abbreviation List
- AD
Alzheimer’s Disease
- Ago2
Argonaute 2
- ALS
Amyotrophic lateral sclerosis
- ALS/PD
Amyotrophic lateral sclerosis and Parkinsonism-dementia complex
- AMPA
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- ATF3
activating transcription factor 3
- ATM
ataxia-telangiectasia mutated
- BAC
bacterial artificial chromosome
- BMAA
beta-Methylamino-L-alanine
- C/EBP
homologous protein 10
- Cdk6
cyclin-dependent kinase 6
- CLIP-seq
UV-cross-linking immunoprecipitation-sequencing
- CNS
central nervous system
- CTD
C-terminal domain
- CTFs
C-terminal fragments
- DDR
DNA damage response
- DNA-PK
DNA-dependent protein kinase
- DSBs
doublé-strand breaks
- dsDNA
double-stranded DNA
- ERG
ETS-related gene
- FALS
familial Amyotrophic lateral sclerosis
- FET
FUS, EWS and TAF15
- FTLD
frontotemporal lobar degeneration
- FUS/TLS
fused in sarcoma/translocated in liposarcoma
- G3BP
RasGAP SH3-domain binding protein 1
- gadd7
growth-arrested DNA damage-inducible gene 7
- H3
histone 3
- H4
histone 4
- HDAC1
histone deacetylase 1
- HDAC6
histone deacetylase 6
- HIV-1
human immunodeficiency virus type 1
- HNE
4-hydroxynonenal
- hnRNP
heterogeneous nuclear ribonucleoprotein
- HR
homologous recombination
- iPSC
induced pluripotent stem cells
- IR
ionizing radiation
- LPS
lipopolysaccharides
- MALAT1
metastasis-associated lung adenocarcinoma transcript 1
- miRNA
microRNA
- NEAT1
nuclear enriched abundant transcript 1
- NES
nuclear export signal
- NHEJ
non-homologous end joining
- NLS
nuclear localization signal
- NMDA
N-methyl-D-aspartate
- NTD
N-terminal domain
- OS
OXIDATIVE STRESS
- PARP
Poly(ADP-ribose Polymerase
- PD
Parkinson’s Disease
- PLDs
Prion-like domains
- PRMT
protein arginine transferases
- Prp
Prion protein promoter
- PTB
polypyrimidine tract-binding
- PY-NLS
proline-tyrosine nuclear localization signal
- RAN
repeat associated non-ATG
- RBM45
RNA-binding motif 45
- RGG
Arginine-glycince-glycine
- RNA Pol II
RNA polymerase II
- ROS
reactive oxygen species
- RRM
RNA recognition motif
- SALS
sporadic Amyotrophic lateral sclerosis
- SGs
stress granules
- SOD1
superoxide dismutase 1
- SSBs
single-strand breaks
- ssDNA
single-stranded DNA
- TDP-43
TAR DNA-binding protein-43
- TIA-1
T-cell intracellular antigen 1
- TRBP
transactivation-responsive RNA binding protein
- tRNP
transport ribonucleoprotein particles
- UBPY
ubiquitin isopeptidase Y
- UBQLN2
ubiquilin 2
- UPF1
upframeshift protein 1
- UPS
ubiquitin-proteasome system
- WT
wild type
- ZnF
Zinc finger
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abhyankar MM, Urekar C, Reddi PP. A novel CpG-free vertebrate insulator silences the testis-specific SP-10 gene in somatic tissues: role for TDP-43 in insulator function. J Biol Chem. 2007;282:36143–36154. doi: 10.1074/jbc.M705811200. [DOI] [PubMed] [Google Scholar]
- Acevedo-Arozena A, Kalmar B, Essa S, Ricketts T, Joyce P, Kent R, Rowe C, Parker A, Gray A, Hafezparast M, Thorpe JR, Greensmith L, Fisher EM. A comprehensive assessment of the SOD1G93A low-copy transgenic mouse, which models human amyotrophic lateral sclerosis. Dis Model Mech. 2011;4:686–700. doi: 10.1242/dmm.007237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acosta JR, Goldsbury C, Winnick C, Badrock AP, Fraser ST, Laird AS, Hall TE, Don EK, Fifita JA, Blair IP, Nicholson GA, Cole NJ. Mutant human FUS Is ubiquitously mislocalized and generates persistent stress granules in primary cultured transgenic zebrafish cells. PLoS One. 2014;9:e90572. doi: 10.1371/journal.pone.0090572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Chalabi A, Jones A, Troakes C, King A, Al-Sarraj S, van den Berg LH. The genetics and neuropathology of amyotrophic lateral sclerosis. Acta Neuropathol. 2012;124:339–352. doi: 10.1007/s00401-012-1022-4. [DOI] [PubMed] [Google Scholar]
- Al-Sarraj S, King A, Troakes C, Smith B, Maekawa S, Bodi I, Rogelj B, Al-Chalabi A, Hortobagyi T, Shaw CE. p62 positive, TDP-43 negative, neuronal cytoplasmic and intranuclear inclusions in the cerebellum and hippocampus define the pathology of C9orf72-linked FTLD and MND/ALS. Acta Neuropathol. 2011;122:691–702. doi: 10.1007/s00401-011-0911-2. [DOI] [PubMed] [Google Scholar]
- Aman P, Panagopoulos I, Lassen C, Fioretos T, Mencinger M, Toresson H, Hoglund M, Forster A, Rabbitts TH, Ron D, Mandahl N, Mitelman F. Expression patterns of the human sarcoma-associated genes FUS and EWS and the genomic structure of FUS. Genomics. 1996;37:1–8. doi: 10.1006/geno.1996.0513. [DOI] [PubMed] [Google Scholar]
- Andersen PM, Al-Chalabi A. Clinical genetics of amyotrophic lateral sclerosis: what do we really know? Nat Rev Neurol. 2011;7:603–615. doi: 10.1038/nrneurol.2011.150. [DOI] [PubMed] [Google Scholar]
- Andersson MK, Stahlberg A, Arvidsson Y, Olofsson A, Semb H, Stenman G, Nilsson O, Aman P. The multifunctional FUS, EWS and TAF15 proto-oncoproteins show cell type-specific expression patterns and involvement in cell spreading and stress response. BMC Cell Biol. 2008;9:37. doi: 10.1186/1471-2121-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H, Mann D, Tsuchiya K, Yoshida M, Hashizume Y, Oda T. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- Arif M, Kazim SF, Grundke-Iqbal I, Garruto RM, Iqbal K. Tau pathology involves protein phosphatase 2A in parkinsonism-dementia of Guam. Proc Natl Acad Sci U S A. 2014;111:1144–1149. doi: 10.1073/pnas.1322614111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold ES, Ling SC, Huelga SC, Lagier-Tourenne C, Polymenidou M, Ditsworth D, Kordasiewicz HB, McAlonis-Downes M, Platoshyn O, Parone PA, Da Cruz S, Clutario KM, Swing D, Tessarollo L, Marsala M, Shaw CE, Yeo GW, Cleveland DW. ALS-linked TDP-43 mutations produce aberrant RNA splicing and adult-onset motor neuron disease without aggregation or loss of nuclear TDP-43. Proc Natl Acad Sci U S A. 2013;110:E736–745. doi: 10.1073/pnas.1222809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash PE, Zhang YJ, Roberts CM, Saldi T, Hutter H, Buratti E, Petrucelli L, Link CD. Neurotoxic effects of TDP-43 overexpression in C. elegans. Hum Mol Genet. 2010;19:3206–3218. doi: 10.1093/hmg/ddq230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulas A, Stabile S, Vande Velde C. Endogenous TDP-43, but not FUS, contributes to stress granule assembly via G3BP. Mol Neurodegener. 2012;7:54. doi: 10.1186/1750-1326-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avendano-Vazquez SE, Dhir A, Bembich S, Buratti E, Proudfoot N, Baralle FE. Autoregulation of TDP-43 mRNA levels involves interplay between transcription, splicing, and alternative polyA site selection. Genes Dev. 2012;26:1679–1684. doi: 10.1101/gad.194829.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala YM, De Conti L, Avendano-Vazquez SE, Dhir A, Romano M, D’Ambrogio A, Tollervey J, Ule J, Baralle M, Buratti E, Baralle FE. TDP-43 regulates its mRNA levels through a negative feedback loop. Embo J. 2011;30:277–288. doi: 10.1038/emboj.2010.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala YM, Misteli T, Baralle FE. TDP-43 regulates retinoblastoma protein phosphorylation through the repression of cyclin-dependent kinase 6 expression. Proc Natl Acad Sci U S A. 2008a;105:3785–3789. doi: 10.1073/pnas.0800546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala YM, Pantano S, D’Ambrogio A, Buratti E, Brindisi A, Marchetti C, Romano M, Baralle FE. Human, Drosophila, and C.elegans TDP43: nucleic acid binding properties and splicing regulatory function. J Mol Biol. 2005;348:575–588. doi: 10.1016/j.jmb.2005.02.038. [DOI] [PubMed] [Google Scholar]
- Ayala YM, Zago P, D’Ambrogio A, Xu YF, Petrucelli L, Buratti E, Baralle FE. Structural determinants of the cellular localization and shuttling of TDP-43. J Cell Sci. 2008b;121:3778–3785. doi: 10.1242/jcs.038950. [DOI] [PubMed] [Google Scholar]
- Baechtold H, Kuroda M, Sok J, Ron D, Lopez BS, Akhmedov AT. Human 75-kDa DNA-pairing protein is identical to the pro-oncoprotein TLS/FUS and is able to promote D-loop formation. J Biol Chem. 1999;274:34337–34342. doi: 10.1074/jbc.274.48.34337. [DOI] [PubMed] [Google Scholar]
- Bakkar N, Kousari A, Kovalik T, Li Y, Bowser R. RBM45 Modulates the Antioxidant Response in Amyotrophic Lateral Sclerosis through Interactions with KEAP1. Mol Cell Biol. 2015;35:2385–2399. doi: 10.1128/MCB.00087-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron DM, Kaushansky LJ, Ward CL, Sama RR, Chian RJ, Boggio KJ, Quaresma AJ, Nickerson JA, Bosco DA. Amyotrophic lateral sclerosis-linked FUS/TLS alters stress granule assembly and dynamics. Mol Neurodegener. 2013;8:30. doi: 10.1186/1750-1326-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belly A, Moreau-Gachelin F, Sadoul R, Goldberg Y. Delocalization of the multifunctional RNA splicing factor TLS/FUS in hippocampal neurones: exclusion from the nucleus and accumulation in dendritic granules and spine heads. Neurosci Lett. 2005;379:152–157. doi: 10.1016/j.neulet.2004.12.071. [DOI] [PubMed] [Google Scholar]
- Beltran M, Puig I, Pena C, Garcia JM, Alvarez AB, Pena R, Bonilla F, de Herreros AG. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22:756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benajiba L, Le Ber I, Camuzat A, Lacoste M, Thomas-Anterion C, Couratier P, Legallic S, Salachas F, Hannequin D, Decousus M, Lacomblez L, Guedj E, Golfier V, Camu W, Dubois B, Campion D, Meininger V, Brice A, French C, Genetic Research Network on Frontotemporal Lobar Degeneration/Frontotemporal Lobar Degeneration with Motoneuron, D. TARDBP mutations in motoneuron disease with frontotemporal lobar degeneration. Ann Neurol. 2009;65:470–473. doi: 10.1002/ana.21612. [DOI] [PubMed] [Google Scholar]
- Blair IP, Williams KL, Warraich ST, Durnall JC, Thoeng AD, Manavis J, Blumbergs PC, Vucic S, Kiernan MC, Nicholson GA. FUS mutations in amyotrophic lateral sclerosis: clinical, pathological, neurophysiological and genetic analysis. J Neurol Neurosurg Psychiatry. 2010;81:639–645. doi: 10.1136/jnnp.2009.194399. [DOI] [PubMed] [Google Scholar]
- Blokhuis AM, Groen EJ, Koppers M, van den Berg LH, Pasterkamp RJ. Protein aggregation in amyotrophic lateral sclerosis. Acta Neuropathol. 2013;125:777–794. doi: 10.1007/s00401-013-1125-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghero G, Floris G, Cannas A, Marrosu MG, Murru MR, Costantino E, Parish LD, Pugliatti M, Ticca A, Traynor BJ, Calvo A, Cammarosano S, Moglia C, Cistaro A, Brunetti M, Restagno G, Chio A. A patient carrying a homozygous p.A382T TARDBP missense mutation shows a syndrome including ALS, extrapyramidal symptoms, and FTD. Neurobiol Aging. 2011;32:2327, e2321–2325. doi: 10.1016/j.neurobiolaging.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borroni B, Bonvicini C, Alberici A, Buratti E, Agosti C, Archetti S, Papetti A, Stuani C, Di Luca M, Gennarelli M, Padovani A. Mutation within TARDBP leads to frontotemporal dementia without motor neuron disease. Hum Mutat. 2009;30:E974–983. doi: 10.1002/humu.21100. [DOI] [PubMed] [Google Scholar]
- Bosco DA, Lemay N, Ko HK, Zhou H, Burke C, Kwiatkowski TJ, Jr., Sapp P, McKenna-Yasek D, Brown RH, Jr., Hayward LJ. Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum Mol Genet. 2010;19:4160–4175. doi: 10.1093/hmg/ddq335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxer AL, Mackenzie IR, Boeve BF, Baker M, Seeley WW, Crook R, Feldman H, Hsiung GY, Rutherford N, Laluz V, Whitwell J, Foti D, McDade E, Molano J, Karydas A, Wojtas A, Goldman J, Mirsky J, Sengdy P, Dearmond S, Miller BL, Rademakers R. Clinical, neuroimaging and neuropathological features of a new chromosome 9p-linked FTD-ALS family. J Neurol Neurosurg Psychiatry. 2011;82:196–203. doi: 10.1136/jnnp.2009.204081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley WG, Krasin F. A new hypothesis of the etiology of amyotrophic lateral sclerosis. The DNA hypothesis. Arch Neurol. 1982;39:677–680. doi: 10.1001/archneur.1982.00510230003001. [DOI] [PubMed] [Google Scholar]
- Broustal O, Camuzat A, Guillot-Noel L, Guy N, Millecamps S, Deffond D, Lacomblez L, Golfier V, Hannequin D, Salachas F, Camu W, Didic M, Dubois B, Meininger V, Le Ber I, Brice A, French c., genetic research network on, F. F.-M. FUS mutations in frontotemporal lobar degeneration with amyotrophic lateral sclerosis. J Alzheimers Dis. 2010;22:765–769. [PubMed] [Google Scholar]
- Brouwers N, Bettens K, Gijselinck I, Engelborghs S, Pickut BA, Van Miegroet H, Montoya AG, Mattheijssens M, Peeters K, De Deyn PP, Cruts M, Sleegers K, Van Broeckhoven C. Contribution of TARDBP to Alzheimer’s disease genetic etiology. J Alzheimers Dis. 2010;21:423–430. doi: 10.3233/JAD-2010-100198. [DOI] [PubMed] [Google Scholar]
- Buratti E, Baralle FE. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem. 2001;276:36337–36343. doi: 10.1074/jbc.M104236200. [DOI] [PubMed] [Google Scholar]
- Buratti E, Baralle FE. TDP-43: new aspects of autoregulation mechanisms in RNA binding proteins and their connection with human disease. Febs j. 2011;278:3530–3538. doi: 10.1111/j.1742-4658.2011.08257.x. [DOI] [PubMed] [Google Scholar]
- Buratti E, Dork T, Zuccato E, Pagani F, Romano M, Baralle FE. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. Embo J. 2001;20:1774–1784. doi: 10.1093/emboj/20.7.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Calvio C, Neubauer G, Mann M, Lamond AI. Identification of hnRNP P2 as TLS/FUS using electrospray mass spectrometry. RNA. 1995;1:724–733. [PMC free article] [PubMed] [Google Scholar]
- Cannas A, Borghero G, Floris GL, Solla P, Chio A, Traynor BJ, Calvo A, Restagno G, Majounie E, Costantino E, Piras V, Lavra L, Pani C, Orofino G, Di Stefano F, Tacconi P, Mascia MM, Muroni A, Murru MR, Tranquilli S, Corongiu D, Rolesu M, Cuccu S, Marrosu F, Marrosu MG. The p.A382T TARDBP gene mutation in Sardinian patients affected by Parkinson’s disease and other degenerative parkinsonisms. Neurogenetics. 2013;14:161–166. doi: 10.1007/s10048-013-0360-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caragounis A, Price KA, Soon CP, Filiz G, Masters CL, Li QX, Crouch PJ, White AR. Zinc induces depletion and aggregation of endogenous TDP-43. Free Radic Biol Med. 2010;48:1152–1161. doi: 10.1016/j.freeradbiomed.2010.01.035. [DOI] [PubMed] [Google Scholar]
- Casafont I, Bengoechea R, Tapia O, Berciano MT, Lafarga M. TDP-43 localizes in mRNA transcription and processing sites in mammalian neurons. J Struct Biol. 2009;167:235–241. doi: 10.1016/j.jsb.2009.06.006. [DOI] [PubMed] [Google Scholar]
- Chen Y, Chen CF, Riley DJ, Chen PL. Nek1 kinase functions in DNA damage response and checkpoint control through a pathway independent of ATM and ATR. Cell Cycle. 2011;10:655–663. doi: 10.4161/cc.10.4.14814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang HH, Andersen PM, Tysnes OB, Gredal O, Christensen PB, Graff C. Novel TARDBP mutations in Nordic ALS patients. J Hum Genet. 2012;57:316–319. doi: 10.1038/jhg.2012.24. [DOI] [PubMed] [Google Scholar]
- Chiang PM, Ling J, Jeong YH, Price DL, Aja SM, Wong PC. Deletion of TDP-43 down-regulates Tbc1d1, a gene linked to obesity, and alters body fat metabolism. Proc Natl Acad Sci U S A. 2010;107:16320–16324. doi: 10.1073/pnas.1002176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chio A, Calvo A, Ilardi A, Cavallo E, Moglia C, Mutani R, Palmo A, Galletti R, Marinou K, Papetti L, Mora G. Lower serum lipid levels are related to respiratory impairment in patients with ALS. Neurology. 2009a;73:1681–1685. doi: 10.1212/WNL.0b013e3181c1df1e. [DOI] [PubMed] [Google Scholar]
- Chio A, Calvo A, Moglia C, Restagno G, Ossola I, Brunetti M, Montuschi A, Cistaro A, Ticca A, Traynor BJ, Schymick JC, Mutani R, Marrosu MG, Murru MR, Borghero G. Amyotrophic lateral sclerosis-frontotemporal lobar dementia in 3 families with p.Ala382Thr TARDBP mutations. Arch Neurol. 2010;67:1002–1009. doi: 10.1001/archneurol.2010.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chio A, Logroscino G, Traynor BJ, Collins J, Simeone JC, Goldstein LA, White LA. Global epidemiology of amyotrophic lateral sclerosis: a systematic review of the published literature. Neuroepidemiology. 2013;41:118–130. doi: 10.1159/000351153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chio A, Restagno G, Brunetti M, Ossola I, Calvo A, Mora G, Sabatelli M, Monsurro MR, Battistini S, Mandrioli J, Salvi F, Spataro R, Schymick J, Traynor BJ, La Bella V, Consortium, I. Two Italian kindreds with familial amyotrophic lateral sclerosis due to FUS mutation. Neurobiol Aging. 2009b;30:1272–1275. doi: 10.1016/j.neurobiolaging.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen TJ, Hwang AW, Restrepo CR, Yuan CX, Trojanowski JQ, Lee VM. An acetylation switch controls TDP-43 function and aggregation propensity. Nat Commun. 2015;6:5845. doi: 10.1038/ncomms6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen TJ, Hwang AW, Unger T, Trojanowski JQ, Lee VM. Redox signalling directly regulates TDP-43 via cysteine oxidation and disulphide cross-linking. EMBO J. 2012;31:1241–1252. doi: 10.1038/emboj.2011.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins M, Riascos D, Kovalik T, An J, Krupa K, Krupa K, Hood BL, Conrads TP, Renton AE, Traynor BJ, Bowser R. The RNA-binding motif 45 (RBM45) protein accumulates in inclusion bodies in amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration with TDP-43 inclusions (FTLD-TDP) patients. Acta Neuropathol. 2012;124:717–732. doi: 10.1007/s00401-012-1045-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppede F. An overview of DNA repair in amyotrophic lateral sclerosis. Scientific World Journal. 2011;11:1679–1691. doi: 10.1100/2011/853474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrado L, Ratti A, Gellera C, Buratti E, Castellotti B, Carlomagno Y, Ticozzi N, Mazzini L, Testa L, Taroni F, Baralle FE, Silani V, D’Alfonso S. High frequency of TARDBP gene mutations in Italian patients with amyotrophic lateral sclerosis. Hum Mutat. 2009;30:688–694. doi: 10.1002/humu.20950. [DOI] [PubMed] [Google Scholar]
- Correia AS, Patel P, Dutta K, Julien JP. Inflammation Induces TDP-43 Mislocalization and Aggregation. PLoS One. 2015;10:e0140248. doi: 10.1371/journal.pone.0140248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox PA, Sacks OW. Cycad neurotoxins, consumption of flying foxes, and ALS-PDC disease in Guam. Neurology. 2002;58:956–959. doi: 10.1212/wnl.58.6.956. [DOI] [PubMed] [Google Scholar]
- Cozzolino M, Carri MT. Mitochondrial dysfunction in ALS. Prog Neurobiol. 2012;97:54–66. doi: 10.1016/j.pneurobio.2011.06.003. [DOI] [PubMed] [Google Scholar]
- Cushman M, Johnson BS, King OD, Gitler AD, Shorter J. Prion-like disorders: blurring the divide between transmissibility and infectivity. J Cell Sci. 2010;123:1191–1201. doi: 10.1242/jcs.051672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ambrogio A, Buratti E, Stuani C, Guarnaccia C, Romano M, Ayala YM, Baralle FE. Functional mapping of the interaction between TDP-43 and hnRNP A2 in vivo. Nucleic Acids Res. 2009;37:4116–4126. doi: 10.1093/nar/gkp342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daoud H, Valdmanis PN, Kabashi E, Dion P, Dupre N, Camu W, Meininger V, Rouleau GA. Contribution of TARDBP mutations to sporadic amyotrophic lateral sclerosis. J Med Genet. 2009;46:112–114. doi: 10.1136/jmg.2008.062463. [DOI] [PubMed] [Google Scholar]
- de Lau LM, Breteler MM. Epidemiology of Parkinson’s disease. Lancet Neurol. 2006;5:525–535. doi: 10.1016/S1474-4422(06)70471-9. [DOI] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GY, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR, Rademakers R. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Bo R, Ghezzi S, Corti S, Pandolfo M, Ranieri M, Santoro D, Ghione I, Prelle A, Orsetti V, Mancuso M, Soraru G, Briani C, Angelini C, Siciliano G, Bresolin N, Comi GP. TARDBP (TDP-43) sequence analysis in patients with familial and sporadic ALS: identification of two novel mutations. Eur J Neurol. 2009;16:727–732. doi: 10.1111/j.1468-1331.2009.02574.x. [DOI] [PubMed] [Google Scholar]
- Deng H, Gao K, Jankovic J. The role of FUS gene variants in neurodegenerative diseases. Nat Rev Neurol. 2014a;10:337–348. doi: 10.1038/nrneurol.2014.78. [DOI] [PubMed] [Google Scholar]
- Deng HX, Bigio EH, Zhai H, Fecto F, Ajroud K, Shi Y, Yan J, Mishra M, Ajroud-Driss S, Heller S, Sufit R, Siddique N, Mugnaini E, Siddique T. Differential involvement of optineurin in amyotrophic lateral sclerosis with or without SOD1 mutations. Arch Neurol. 2011a;68:1057–1061. doi: 10.1001/archneurol.2011.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng HX, Chen W, Hong ST, Boycott KM, Gorrie GH, Siddique N, Yang Y, Fecto F, Shi Y, Zhai H, Jiang H, Hirano M, Rampersaud E, Jansen GH, Donkervoort S, Bigio EH, Brooks BR, Ajroud K, Sufit RL, Haines JL, Mugnaini E, Pericak-Vance MA, Siddique T. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011b;477:211–215. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng HX, Zhai H, Bigio EH, Yan J, Fecto F, Ajroud K, Mishra M, Ajroud-Driss S, Heller S, Sufit R, Siddique N, Mugnaini E, Siddique T. FUS-immunoreactive inclusions are a common feature in sporadic and non-SOD1 familial amyotrophic lateral sclerosis. Ann Neurol. 2010;67:739–748. doi: 10.1002/ana.22051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Q, Holler CJ, Taylor G, Hudson KF, Watkins W, Gearing M, Ito D, Murray ME, Dickson DW, Seyfried NT, Kukar T. FUS is phosphorylated by DNA-PK and accumulates in the cytoplasm after DNA damage. J Neurosci. 2014b;34:7802–7813. doi: 10.1523/JNEUROSCI.0172-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey CM, Cenik B, Sephton CF, Dries DR, Mayer P, 3rd, Good SK, Johnson BA, Herz J, Yu G. TDP-43 is directed to stress granules by sorbitol, a novel physiological osmotic and oxidative stressor. Mol Cell Biol. 2011;31:1098–1108. doi: 10.1128/MCB.01279-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann D, Haass C. TDP-43 and FUS: a nuclear affair. Trends Neurosci. 2011;34:339–348. doi: 10.1016/j.tins.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Dormann D, Haass C. Fused in sarcoma (FUS): an oncogene goes awry in neurodegeneration. Mol Cell Neurosci. 2013;56:475–486. doi: 10.1016/j.mcn.2013.03.006. [DOI] [PubMed] [Google Scholar]
- Dormann D, Madl T, Valori CF, Bentmann E, Tahirovic S, Abou-Ajram C, Kremmer E, Ansorge O, Mackenzie IR, Neumann M, Haass C. Arginine methylation next to the PY-NLS modulates Transportin binding and nuclear import of FUS. EMBO J. 2012;31:4258–4275. doi: 10.1038/emboj.2012.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann D, Rodde R, Edbauer D, Bentmann E, Fischer I, Hruscha A, Than ME, Mackenzie IR, Capell A, Schmid B, Neumann M, Haass C. ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J. 2010;29:2841–2857. doi: 10.1038/emboj.2010.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G, Matunis MJ, Pinol-Roma S, Burd CG. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- Du K, Arai S, Kawamura T, Matsushita A, Kurokawa R. TLS and PRMT1 synergistically coactivate transcription at the survivin promoter through TLS arginine methylation. Biochem Biophys Res Commun. 2011;404:991–996. doi: 10.1016/j.bbrc.2010.12.097. [DOI] [PubMed] [Google Scholar]
- Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, Armakola M, Geser F, Greene R, Lu MM, Padmanabhan A, Clay-Falcone D, McCluskey L, Elman L, Juhr D, Gruber PJ, Rub U, Auburger G, Trojanowski JQ, Lee VM, Van Deerlin VM, Bonini NM, Gitler AD. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmaeili MA, Panahi M, Yadav S, Hennings L, Kiaei M. Premature death of TDP-43 (A315T) transgenic mice due to gastrointestinal complications prior to development of full neurological symptoms of amyotrophic lateral sclerosis. Int J Exp Pathol. 2013;94:56–64. doi: 10.1111/iep.12006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farg MA, Soo KY, Warraich ST, Sundaramoorthy V, Blair IP, Atkin JD. Ataxin-2 interacts with FUS and intermediate-length polyglutamine expansions enhance FUS-related pathology in amyotrophic lateral sclerosis. Hum Mol Genet. 2013;22:717–728. doi: 10.1093/hmg/dds479. [DOI] [PubMed] [Google Scholar]
- Fay RM, Walker CS, Powers JM. Discoloration of a compomer by stains. J Gt Houst Dent Soc. 1998;69:12–13. [PubMed] [Google Scholar]
- Feiguin F, Godena VK, Romano G, D’Ambrogio A, Klima R, Baralle FE. Depletion of TDP-43 affects Drosophila motoneurons terminal synapsis and locomotive behavior. FEBS Lett. 2009;583:1586–1592. doi: 10.1016/j.febslet.2009.04.019. [DOI] [PubMed] [Google Scholar]
- Ferraiuolo L, Kirby J, Grierson AJ, Sendtner M, Shaw PJ. Molecular pathways of motor neuron injury in amyotrophic lateral sclerosis. Nat Rev Neurol. 2011;7:616–630. doi: 10.1038/nrneurol.2011.152. [DOI] [PubMed] [Google Scholar]
- Fiesel FC, Voigt A, Weber SS, Van den Haute C, Waldenmaier A, Gorner K, Walter M, Anderson ML, Kern JV, Rasse TM, Schmidt T, Springer W, Kirchner R, Bonin M, Neumann M, Baekelandt V, Alunni-Fabbroni M, Schulz JB, Kahle PJ. Knockdown of transactive response DNA-binding protein (TDP-43) downregulates histone deacetylase 6. EMBO J. 2010;29:209–221. doi: 10.1038/emboj.2009.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filimonenko M, Stuffers S, Raiborg C, Yamamoto A, Malerod L, Fisher EM, Isaacs A, Brech A, Stenmark H, Simonsen A. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, Lu Y, Lopez-Gonzalez R, Kim NC, Almeida S, Lee KH, Badders N, Valentine M, Miller BL, Wong PC, Petrucelli L, Kim HJ, Gao FB, Taylor JP. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature. 2015;525:129–133. doi: 10.1038/nature14974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridovich I. The biology of oxygen radicals. Science. 1978;201:875–880. doi: 10.1126/science.210504. [DOI] [PubMed] [Google Scholar]
- Fujii R, Okabe S, Urushido T, Inoue K, Yoshimura A, Tachibana T, Nishikawa T, Hicks GG, Takumi T. The RNA binding protein TLS is translocated to dendritic spines by mGluR5 activation and regulates spine morphology. Curr Biol. 2005;15:587–593. doi: 10.1016/j.cub.2005.01.058. [DOI] [PubMed] [Google Scholar]
- Gal J, Zhang J, Kwinter DM, Zhai J, Jia H, Jia J, Zhu H. Nuclear localization sequence of FUS and induction of stress granules by ALS mutants. Neurobiol Aging. 2011;32:2323, e2327–2340. doi: 10.1016/j.neurobiolaging.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner M, Toth R, Vandermoere F, Morrice NA, Rouse J. Identification and characterization of FUS/TLS as a new target of ATM. Biochem J. 2008;415:297–307. doi: 10.1042/BJ20081135. [DOI] [PubMed] [Google Scholar]
- Garruto RM. Pacific paradigms of environmentally-induced neurological disorders: clinical, epidemiological and molecular perspectives. Neurotoxicology. 1991;12:347–377. [PubMed] [Google Scholar]
- Garruto RM, Gajdusek C, Chen KM. Amyotrophic lateral sclerosis among Chamorro migrants from Guam. Ann Neurol. 1980;8:612–619. doi: 10.1002/ana.410080612. [DOI] [PubMed] [Google Scholar]
- Garruto RM, Gajdusek DC, Chen KM. Amyotrophic lateral sclerosis and parkinsonism-dementia among Filipino migrants to Guam. Ann Neurol. 1981;10:341–350. doi: 10.1002/ana.410100405. [DOI] [PubMed] [Google Scholar]
- Garruto RM, Swyt C, Fiori CE, Yanagihara R, Gajdusek DC. Intraneuronal deposition of calcium and aluminium in amyotropic lateral sclerosis of Guam. Lancet. 1985a;2:1353. doi: 10.1016/s0140-6736(85)92642-x. [DOI] [PubMed] [Google Scholar]
- Garruto RM, Yanagihara R, Gajdusek DC. Disappearance of high-incidence amyotrophic lateral sclerosis and parkinsonism-dementia on Guam. Neurology. 1985b;35:193–198. doi: 10.1212/wnl.35.2.193. [DOI] [PubMed] [Google Scholar]
- Gendron TF, Rademakers R, Petrucelli L. TARDBP mutation analysis in TDP-43 proteinopathies and deciphering the toxicity of mutant TDP-43. J Alzheimers Dis. 2013;33(Suppl 1):S35–45. doi: 10.3233/JAD-2012-129036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geser F, Winton MJ, Kwong LK, Xu Y, Xie SX, Igaz LM, Garruto RM, Perl DP, Galasko D, Lee VM, Trojanowski JQ. Pathological TDP-43 in parkinsonism-dementia complex and amyotrophic lateral sclerosis of Guam. Acta Neuropathol. 2008;115:133–145. doi: 10.1007/s00401-007-0257-y. [DOI] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitcho MA, Baloh RH, Chakraverty S, Mayo K, Norton JB, Levitch D, Hatanpaa KJ, White CL, 3rd, Bigio EH, Caselli R, Baker M, Al-Lozi MT, Morris JC, Pestronk A, Rademakers R, Goate AM, Cairns NJ. TDP-43 A315T mutation in familial motor neuron disease. Ann Neurol. 2008;63:535–538. doi: 10.1002/ana.21344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitcho MA, Strider J, Carter D, Taylor-Reinwald L, Forman MS, Goate AM, Cairns NJ. VCP mutations causing frontotemporal lobar degeneration disrupt localization of TDP-43 and induce cell death. J Biol Chem. 2009;284:12384–12398. doi: 10.1074/jbc.M900992200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitler AD, Shorter J. RNA-binding proteins with prion-like domains in ALS and FTLD-U. Prion. 2011;5:179–187. doi: 10.4161/pri.5.3.17230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Gunnarsson LG, Dahlbom K, Strandman E. Motor neuron disease and dementia reported among 13 members of a single family. Acta Neurol Scand. 1991;84:429–433. doi: 10.1111/j.1600-0404.1991.tb04983.x. [DOI] [PubMed] [Google Scholar]
- Guo Y, Wang Q, Zhang K, An T, Shi P, Li Z, Duan W, Li C. HO-1 induction in motor cortex and intestinal dysfunction in TDP-43 A315T transgenic mice. Brain Res. 2012;1460:88–95. doi: 10.1016/j.brainres.2012.04.003. [DOI] [PubMed] [Google Scholar]
- Hallier M, Lerga A, Barnache S, Tavitian A, Moreau-Gachelin F. The transcription factor Spi-1/PU.1 interacts with the potential splicing factor TLS. J Biol Chem. 1998;273:4838–4842. doi: 10.1074/jbc.273.9.4838. [DOI] [PubMed] [Google Scholar]
- Han JH, Yu TH, Ryu HH, Jun MH, Ban BK, Jang DJ, Lee JA. ALS/FTLD-linked TDP-43 regulates neurite morphology and cell survival in differentiated neurons. Exp Cell Res. 2013;319:1998–2005. doi: 10.1016/j.yexcr.2013.05.025. [DOI] [PubMed] [Google Scholar]
- Hans F, Fiesel FC, Strong JC, Jackel S, Rasse TM, Geisler S, Springer W, Schulz JB, Voigt A, Kahle PJ. UBE2E ubiquitin-conjugating enzymes and ubiquitin isopeptidase Y regulate TDP-43 protein ubiquitination. J Biol Chem. 2014;289:19164–19179. doi: 10.1074/jbc.M114.561704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson KA, Kim SH, Wassarman DA, Tibbetts RS. Ubiquilin modifies TDP-43 toxicity in a Drosophila model of amyotrophic lateral sclerosis (ALS) J Biol Chem. 2010;285:11068–11072. doi: 10.1074/jbc.C109.078527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart MP, Gitler AD. ALS-associated ataxin 2 polyQ expansions enhance stress-induced caspase 3 activation and increase TDP-43 pathological modifications. J Neurosci. 2012;32:9133–9142. doi: 10.1523/JNEUROSCI.0996-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Arai T, Akiyama H, Nonaka T, Mori H, Hashimoto T, Yamazaki M, Oyanagi K. TDP-43 is deposited in the Guam parkinsonism-dementia complex brains. Brain. 2007;130:1386–1394. doi: 10.1093/brain/awm065. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Arai T, Nonaka T, Kametani F, Yoshida M, Hashizume Y, Beach TG, Buratti E, Baralle F, Morita M, Nakano I, Oda T, Tsuchiya K, Akiyama H. Phosphorylated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Ann Neurol. 2008;64:60–70. doi: 10.1002/ana.21425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert LE, Weuve J, Scherr PA, Evans DA. Alzheimer disease in the United States (2010-2050) estimated using the 2010 census. Neurology. 2013;80:1778–1783. doi: 10.1212/WNL.0b013e31828726f5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebron ML, Lonskaya I, Sharpe K, Weerasinghe PP, Algarzae NK, Shekoyan AR, Moussa CE. Parkin ubiquitinates Tar-DNA binding protein-43 (TDP-43) and promotes its cytosolic accumulation via interaction with histone deacetylase 6 (HDAC6) J Biol Chem. 2013;288:4103–4115. doi: 10.1074/jbc.M112.419945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde ML, Mantha AK, Hazra TK, Bhakat KK, Mitra S, Szczesny B. Oxidative genome damage and its repair: implications in aging and neurodegenerative diseases. Mech Ageing Dev. 2012;133:157–168. doi: 10.1016/j.mad.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks GG, Singh N, Nashabi A, Mai S, Bozek G, Klewes L, Arapovic D, White EK, Koury MJ, Oltz EM, Van Kaer L, Ruley HE. Fus deficiency in mice results in defective B-lymphocyte development and activation, high levels of chromosomal instability and perinatal death. Nat Genet. 2000;24:175–179. doi: 10.1038/72842. [DOI] [PubMed] [Google Scholar]
- Hoffman PM, Robbins DS, Gibbs CJ, Jr., Gajdusek DC, Garruto RM, Terasaki OI. Histocompatibility antigens in amyotrophic lateral sclerosis and parkinsonism-dementia on Guam. Lancet. 1977;2:717. doi: 10.1016/s0140-6736(77)90525-6. [DOI] [PubMed] [Google Scholar]
- Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- Honda D, Ishigaki S, Iguchi Y, Fujioka Y, Udagawa T, Masuda A, Ohno K, Katsuno M, Sobue G. The ALS/FTLD-related RNA-binding proteins TDP-43 and FUS have common downstream RNA targets in cortical neurons. FEBS Open Bio. 2013;4:1–10. doi: 10.1016/j.fob.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda H, Hamasaki H, Wakamiya T, Koyama S, Suzuki SO, Fujii N, Iwaki T. Loss of hnRNPA1 in ALS spinal cord motor neurons with TDP-43-positive inclusions. Neuropathology. 2015;35:37–43. doi: 10.1111/neup.12153. [DOI] [PubMed] [Google Scholar]
- Hortobagyi T, Troakes C, Nishimura AL, Vance C, van Swieten JC, Seelaar H, King A, Al-Sarraj S, Rogelj B, Shaw CE. Optineurin inclusions occur in a minority of TDP-43 positive ALS and FTLD-TDP cases and are rarely observed in other neurodegenerative disorders. Acta Neuropathol. 2011;121:519–527. doi: 10.1007/s00401-011-0813-3. [DOI] [PubMed] [Google Scholar]
- Huey ED, Ferrari R, Moreno JH, Jensen C, Morris CM, Potocnik F, Kalaria RN, Tierney M, Wassermann EM, Hardy J, Grafman J, Momeni P. FUS and TDP43 genetic variability in FTD and CBS. Neurobiol Aging. 2012;33:1016, e1019–1017. doi: 10.1016/j.neurobiolaging.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa H, Shimizu K, Hayashi Y, Ohki M. An RNA-binding protein gene, TLS/FUS, is fused to ERG in human myeloid leukemia with t(16;21) chromosomal translocation. Cancer Res. 1994;54:2865–2868. [PubMed] [Google Scholar]
- Igaz LM, Kwong LK, Lee EB, Chen-Plotkin A, Swanson E, Unger T, Malunda J, Xu Y, Winton MJ, Trojanowski JQ, Lee VM. Dysregulation of the ALS-associated gene TDP-43 leads to neuronal death and degeneration in mice. J Clin Invest. 2011;121:726–738. doi: 10.1172/JCI44867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iguchi Y, Katsuno M, Niwa J, Takagi S, Ishigaki S, Ikenaka K, Kawai K, Watanabe H, Yamanaka K, Takahashi R, Misawa H, Sasaki S, Tanaka F, Sobue G. Loss of TDP-43 causes age-dependent progressive motor neuron degeneration. Brain. 2013;136:1371–1382. doi: 10.1093/brain/awt029. [DOI] [PubMed] [Google Scholar]
- Iko Y, Kodama TS, Kasai N, Oyama T, Morita EH, Muto T, Okumura M, Fujii R, Takumi T, Tate S, Morikawa K. Domain architectures and characterization of an RNA-binding protein, TLS. J Biol Chem. 2004;279:44834–44840. doi: 10.1074/jbc.M408552200. [DOI] [PubMed] [Google Scholar]
- Ishiura H, Takahashi Y, Mitsui J, Yoshida S, Kihira T, Kokubo Y, Kuzuhara S, Ranum LP, Tamaoki T, Ichikawa Y, Date H, Goto J, Tsuji S. C9ORF72 repeat expansion in amyotrophic lateral sclerosis in the Kii peninsula of Japan. Arch Neurol. 2012;69:1154–1158. doi: 10.1001/archneurol.2012.1219. [DOI] [PubMed] [Google Scholar]
- Ito D, Seki M, Tsunoda Y, Uchiyama H, Suzuki N. Nuclear transport impairment of amyotrophic lateral sclerosis-linked mutations in FUS/TLS. Ann Neurol. 2011;69:152–162. doi: 10.1002/ana.22246. [DOI] [PubMed] [Google Scholar]
- Jackel S, Summerer AK, Thommes CM, Pan X, Voigt A, Schulz JB, Rasse TM, Dormann D, Haass C, Kahle PJ. Nuclear import factor transportin and arginine methyltransferase 1 modify FUS neurotoxicity in Drosophila. Neurobiol Dis. 2015;74:76–88. doi: 10.1016/j.nbd.2014.11.003. [DOI] [PubMed] [Google Scholar]
- Jackson KL, Dayton RD, Orchard EA, Ju S, Ringe D, Petsko GA, Maquat LE, Klein RL. Preservation of forelimb function by UPF1 gene therapy in a rat model of TDP-43-induced motor paralysis. Gene Ther. 2015;22:20–28. doi: 10.1038/gt.2014.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens J, Wils H, Kleinberger G, Joris G, Cuijt I, Ceuterick-de Groote C, Van Broeckhoven C, Kumar-Singh S. Overexpression of ALS-associated p.M337V human TDP-43 in mice worsens disease features compared to wild-type human TDP-43 mice. Mol Neurobiol. 2013;48:22–35. doi: 10.1007/s12035-013-8427-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, Gibbs JR, Brunetti M, Gronka S, Wuu J, Ding J, McCluskey L, Martinez-Lage M, Falcone D, Hernandez DG, Arepalli S, Chong S, Schymick JC, Rothstein J, Landi F, Wang YD, Calvo A, Mora G, Sabatelli M, Monsurro MR, Battistini S, Salvi F, Spataro R, Sola P, Borghero G, Consortium I, Galassi G, Scholz SW, Taylor JP, Restagno G, Chio A, Traynor BJ. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E, Bercier V, Lissouba A, Liao M, Brustein E, Rouleau GA, Drapeau P. FUS and TARDBP but not SOD1 interact in genetic models of amyotrophic lateral sclerosis. PLoS Genet. 2011a;7:e1002214. doi: 10.1371/journal.pgen.1002214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E, Brustein E, Champagne N, Drapeau P. Zebrafish models for the functional genomics of neurogenetic disorders. Biochim Biophys Acta. 2011b;1812:335–345. doi: 10.1016/j.bbadis.2010.09.011. [DOI] [PubMed] [Google Scholar]
- Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, Vande Velde C, Bouchard JP, Lacomblez L, Pochigaeva K, Salachas F, Pradat PF, Camu W, Meininger V, Dupre N, Rouleau GA. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40:572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- Kabuta C, Kono K, Wada K, Kabuta T. 4-Hydroxynonenal induces persistent insolubilization of TDP-43 and alters its intracellular localization. Biochem Biophys Res Commun. 2015;463:82–87. doi: 10.1016/j.bbrc.2015.05.027. [DOI] [PubMed] [Google Scholar]
- Kang L, Liu X, Gong Z, Zheng H, Wang J, Li Y, Yang H, Hardwick J, Dai H, Poon RT, Lee NP, Mao M, Peng Z, Chen R. Genome-wide identification of RNA editing in hepatocellular carcinoma. Genomics. 2015;105:76–82. doi: 10.1016/j.ygeno.2014.11.005. [DOI] [PubMed] [Google Scholar]
- Kawahara Y, Mieda-Sato A. TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc Natl Acad Sci U S A. 2012;109:3347–3352. doi: 10.1073/pnas.1112427109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T, Akiyama H, Yamada T, McGeer PL. Immunologic reactions in amyotrophic lateral sclerosis brain and spinal cord tissue. Am J Pathol. 1992;140:691–707. [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Anderson P. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem Soc Trans. 2002;30:963–969. doi: 10.1042/bst0300963. [DOI] [PubMed] [Google Scholar]
- Keller BA, Volkening K, Droppelmann CA, Ang LC, Rademakers R, Strong MJ. Co-aggregation of RNA binding proteins in ALS spinal motor neurons: evidence of a common pathogenic mechanism. Acta Neuropathol. 2012;124:733–747. doi: 10.1007/s00401-012-1035-z. [DOI] [PubMed] [Google Scholar]
- Kenna KP, van Doormaal PT, Dekker AM, Ticozzi N, Kenna BJ, Diekstra FP, van Rheenen W, van Eijk KR, Jones AR, Keagle P, Shatunov A, Sproviero W, Smith BN, van Es MA, Topp SD, Kenna A, Miller JW, Fallini C, Tiloca C, McLaughlin RL, Vance C, Troakes C, Colombrita C, Mora G, Calvo A, Verde F, Al-Sarraj S, King A, Calini D, de Belleroche J, Baas F, van der Kooi AJ, de Visser M, Ten Asbroek AL, Sapp PC, McKenna-Yasek D, Polak M, Asress S, Munoz-Blanco JL, Strom TM, Meitinger T, Morrison KE, Consortium S, Lauria G, Williams KL, Leigh PN, Nicholson GA, Blair IP, Leblond CS, Dion PA, Rouleau GA, Pall H, Shaw PJ, Turner MR, Talbot K, Taroni F, Boylan KB, Van Blitterswijk M, Rademakers R, Esteban-Perez J, Garcia-Redondo A, Van Damme P, Robberecht W, Chio A, Gellera C, Drepper C, Sendtner M, Ratti A, Glass JD, Mora JS, Basak NA, Hardiman O, Ludolph AC, Andersen PM, Weishaupt JH, Brown RH, Jr., Al-Chalabi A, Silani V, Shaw CE, van den Berg LH, Veldink JH, Landers JE. NEK1 variants confer susceptibility to amyotrophic lateral sclerosis. Nat Genet. 2016 doi: 10.1038/ng.3626. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, Kanagaraj AP, Carter R, Boylan KB, Wojtas AM, Rademakers R, Pinkus JL, Greenberg SA, Trojanowski JQ, Traynor BJ, Smith BN, Topp S, Gkazi AS, Miller J, Shaw CE, Kottlors M, Kirschner J, Pestronk A, Li YR, Ford AF, Gitler AD, Benatar M, King OD, Kimonis VE, Ross ED, Weihl CC, Shorter J, Taylor JP. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Shanware NP, Bowler MJ, Tibbetts RS. Amyotrophic lateral sclerosis-associated proteins TDP-43 and FUS/TLS function in a common biochemical complex to co-regulate HDAC6 mRNA. J Biol Chem. 2010;285:34097–34105. doi: 10.1074/jbc.M110.154831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A, Troakes C, Smith B, Nolan M, Curran O, Vance C, Shaw CE, Al-Sarraj S. ALS-FUS pathology revisited: singleton FUS mutations and an unusual case with both a FUS and TARDBP mutation. Acta Neuropathol Commun. 2015;3:62. doi: 10.1186/s40478-015-0235-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King OD, Gitler AD, Shorter J. The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res. 2012;1462:61–80. doi: 10.1016/j.brainres.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino Y, Washizu C, Aquilanti E, Okuno M, Kurosawa M, Yamada M, Doi H, Nukina N. Intracellular localization and splicing regulation of FUS/TLS are variably affected by amyotrophic lateral sclerosis-linked mutations. Nucleic Acids Res. 2011;39:2781–2798. doi: 10.1093/nar/gkq1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs GG, Murrell JR, Horvath S, Haraszti L, Majtenyi K, Molnar MJ, Budka H, Ghetti B, Spina S. TARDBP variation associated with frontotemporal dementia, supranuclear gaze palsy, and chorea. Mov Disord. 2009;24:1843–1847. doi: 10.1002/mds.22697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer BC, Schuck T, Wheeler JM, Robinson LC, Trojanowski JQ, Lee VM, Schellenberg GD. Loss of murine TDP-43 disrupts motor function and plays an essential role in embryogenesis. Acta Neuropathol. 2010;119:409–419. doi: 10.1007/s00401-010-0659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo PH, Chiang CH, Wang YT, Doudeva LG, Yuan HS. The crystal structure of TDP-43 RRM1-DNA complex reveals the specific recognition for UG- and TG-rich nucleic acids. Nucleic Acids Res. 2014;42:4712–4722. doi: 10.1093/nar/gkt1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo PH, Doudeva LG, Wang YT, Shen CK, Yuan HS. Structural insights into TDP-43 in nucleic-acid binding and domain interactions. Nucleic Acids Res. 2009;37:1799–1808. doi: 10.1093/nar/gkp013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda M, Sok J, Webb L, Baechtold H, Urano F, Yin Y, Chung P, de Rooij DG, Akhmedov A, Ashley T, Ron D. Male sterility and enhanced radiation sensitivity in TLS(-/-) mice. EMBO J. 2000;19:453–462. doi: 10.1093/emboj/19.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski TJ, Jr., Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, Valdmanis P, Rouleau GA, Hosler BA, Cortelli P, de Jong PJ, Yoshinaga Y, Haines JL, Pericak-Vance MA, Yan J, Ticozzi N, Siddique T, McKenna-Yasek D, Sapp PC, Horvitz HR, Landers JE, Brown RH., Jr. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- Kwong LK, Neumann M, Sampathu DM, Lee VM, Trojanowski JQ. TDP-43 proteinopathy: the neuropathology underlying major forms of sporadic and familial frontotemporal lobar degeneration and motor neuron disease. Acta Neuropathol. 2007;114:63–70. doi: 10.1007/s00401-007-0226-5. [DOI] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Cleveland DW. Rethinking ALS: the FUS about TDP-43. Cell. 2009;136:1001–1004. doi: 10.1016/j.cell.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanson NA, Jr., Maltare A, King H, Smith R, Kim JH, Taylor JP, Lloyd TE, Pandey UB. A Drosophila model of FUS-related neurodegeneration reveals genetic interaction between FUS and TDP-43. Hum Mol Genet. 2011;20:2510–2523. doi: 10.1093/hmg/ddr150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanson NA, Jr., Pandey UB. FUS-related proteinopathies: lessons from animal models. Brain Res. 2012;1462:44–60. doi: 10.1016/j.brainres.2012.01.039. [DOI] [PubMed] [Google Scholar]
- Leblond CS, Kaneb HM, Dion PA, Rouleau GA. Dissection of genetic factors associated with amyotrophic lateral sclerosis. Exp Neurol. 2014;262(Pt B):91–101. doi: 10.1016/j.expneurol.2014.04.013. [DOI] [PubMed] [Google Scholar]
- Lee BJ, Cansizoglu AE, Suel KE, Louis TH, Zhang Z, Chook YM. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell. 2006;126:543–558. doi: 10.1016/j.cell.2006.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F, Maquat LE. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr Opin Cell Biol. 2005;17:309–315. doi: 10.1016/j.ceb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Li Q, Yokoshi M, Okada H, Kawahara Y. The cleavage pattern of TDP-43 determines its rate of clearance and cytotoxicity. Nat Commun. 2015;6:6183. doi: 10.1038/ncomms7183. [DOI] [PubMed] [Google Scholar]
- Li X, Decker M, Westendorf JJ. TEThered to Runx: novel binding partners for runx factors. Blood Cells Mol Dis. 2010;45:82–85. doi: 10.1016/j.bcmd.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YR, King OD, Shorter J, Gitler AD. Stress granules as crucibles of ALS pathogenesis. J Cell Biol. 2013;201:361–372. doi: 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liachko NF, Guthrie CR, Kraemer BC. Phosphorylation promotes neurotoxicity in a Caenorhabditis elegans model of TDP-43 proteinopathy. J Neurosci. 2010;30:16208–16219. doi: 10.1523/JNEUROSCI.2911-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liachko NF, McMillan PJ, Guthrie CR, Bird TD, Leverenz JB, Kraemer BC. CDC7 inhibition blocks pathological TDP-43 phosphorylation and neurodegeneration. Ann Neurol. 2013;74:39–52. doi: 10.1002/ana.23870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liachko NF, McMillan PJ, Strovas TJ, Loomis E, Greenup L, Murrell JR, Ghetti B, Raskind MA, Montine TJ, Bird TD, Leverenz JB, Kraemer BC. The tau tubulin kinases TTBK1/2 promote accumulation of pathological TDP-43. PLoS Genet. 2014;10:e1004803. doi: 10.1371/journal.pgen.1004803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling SC, Albuquerque CP, Han JS, Lagier-Tourenne C, Tokunaga S, Zhou H, Cleveland DW. ALS-associated mutations in TDP-43 increase its stability and promote TDP-43 complexes with FUS/TLS. Proc Natl Acad Sci U S A. 2010;107:13318–13323. doi: 10.1073/pnas.1008227107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79:416–438. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipton AM, White CL, 3rd, Bigio EH. Frontotemporal lobar degeneration with motor neuron disease-type inclusions predominates in 76 cases of frontotemporal degeneration. Acta Neuropathol. 2004;108:379–385. doi: 10.1007/s00401-004-0900-9. [DOI] [PubMed] [Google Scholar]
- Liu R, Yang G, Nonaka T, Arai T, Jia W, Cynader MS. Reducing TDP-43 aggregation does not prevent its cytotoxicity. Acta Neuropathol Commun. 2013;1:49. doi: 10.1186/2051-5960-1-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Li D, Zhang W, Guo M, Zhan Q. Long non-coding RNA gadd7 interacts with TDP-43 and regulates Cdk6 mRNA decay. Embo j. 2012;31:4415–4427. doi: 10.1038/emboj.2012.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoGerfo A, Chico L, Borgia L, Petrozzi L, Rocchi A, D’Amelio A, Carlesi C, Caldarazzo Ienco E, Mancuso M, Siciliano G. Lack of association between nuclear factor erythroid-derived 2-like 2 promoter gene polymorphisms and oxidative stress biomarkers in amyotrophic lateral sclerosis patients. Oxid Med Cell Longev. 2014;2014:432626. doi: 10.1155/2014/432626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, Munoz DG, Kusaka H, Yokota O, Ishihara K, Roeber S, Kretzschmar HA, Cairns NJ, Neumann M. Distinct pathological subtypes of FTLD-FUS. Acta Neuropathol. 2011;121:207–218. doi: 10.1007/s00401-010-0764-0. [DOI] [PubMed] [Google Scholar]
- Madabhushi R, Gao F, Pfenning AR, Pan L, Yamakawa S, Seo J, Rueda R, Phan TX, Yamakawa H, Pao PC, Stott RT, Gjoneska E, Nott A, Cho S, Kellis M, Tsai LH. Activity-Induced DNA Breaks Govern the Expression of Neuronal Early-Response Genes. Cell. 2015;161:1592–1605. doi: 10.1016/j.cell.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa S, Leigh PN, King A, Jones E, Steele JC, Bodi I, Shaw CE, Hortobagyi T, Al-Sarraj S. TDP-43 is consistently co-localized with ubiquitinated inclusions in sporadic and Guam amyotrophic lateral sclerosis but not in familial amyotrophic lateral sclerosis with and without SOD1 mutations. Neuropathology. 2009;29:672–683. doi: 10.1111/j.1440-1789.2009.01029.x. [DOI] [PubMed] [Google Scholar]
- Mantovani S, Garbelli S, Pasini A, Alimonti D, Perotti C, Melazzini M, Bendotti C, Mora G. Immune system alterations in sporadic amyotrophic lateral sclerosis patients suggest an ongoing neuroinflammatory process. J Neuroimmunol. 2009;210:73–79. doi: 10.1016/j.jneuroim.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Martin LJ, Chen K, Liu Z. Adult motor neuron apoptosis is mediated by nitric oxide and Fas death receptor linked by DNA damage and p53 activation. J Neurosci. 2005;25:6449–6459. doi: 10.1523/JNEUROSCI.0911-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin LJ, Liu Z, Chen K, Price AC, Pan Y, Swaby JA, Golden WC. Motor neuron degeneration in amyotrophic lateral sclerosis mutant superoxide dismutase-1 transgenic mice: mechanisms of mitochondriopathy and cell death. J Comp Neurol. 2007;500:20–46. doi: 10.1002/cne.21160. [DOI] [PubMed] [Google Scholar]
- Maruyama H, Morino H, Ito H, Izumi Y, Kato H, Watanabe Y, Kinoshita Y, Kamada M, Nodera H, Suzuki H, Komure O, Matsuura S, Kobatake K, Morimoto N, Abe K, Suzuki N, Aoki M, Kawata A, Hirai T, Kato T, Ogasawara K, Hirano A, Takumi T, Kusaka H, Hagiwara K, Kaji R, Kawakami H. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465:223–226. doi: 10.1038/nature08971. [DOI] [PubMed] [Google Scholar]
- Mastrocola AS, Kim SH, Trinh AT, Rodenkirch LA, Tibbetts RS. The RNA-binding protein fused in sarcoma (FUS) functions downstream of poly(ADP-ribose) polymerase (PARP) in response to DNA damage. J Biol Chem. 2013;288:24731–24741. doi: 10.1074/jbc.M113.497974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCluskey LF, Elman LB, Martinez-Lage M, Van Deerlin V, Yuan W, Clay D, Siderowf A, Trojanowski JQ. Amyotrophic lateral sclerosis-plus syndrome with TAR DNA-binding protein-43 pathology. Arch Neurol. 2009;66:121–124. doi: 10.1001/archneur.66.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald KK, Aulas A, Destroismaisons L, Pickles S, Beleac E, Camu W, Rouleau GA, Vande Velde C. TAR DNA-binding protein 43 (TDP-43) regulates stress granule dynamics via differential regulation of G3BP and TIA-1. Hum Mol Genet. 2011;20:1400–1410. doi: 10.1093/hmg/ddr021. [DOI] [PubMed] [Google Scholar]
- McGurk L, Lee VM, Trojanowksi JQ, Van Deerlin VM, Lee EB, Bonini NM. Poly-A binding protein-1 localization to a subset of TDP-43 inclusions in amyotrophic lateral sclerosis occurs more frequently in patients harboring an expansion in C9orf72. J Neuropathol Exp Neurol. 2014;73:837–845. doi: 10.1097/NEN.0000000000000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra J, Guerrero EN, Hegde PM, Wang H, Boldogh I, Rao KS, Mitra S, Hegde ML. New perspectives on oxidized genome damage and repair inhibition by pro-oxidant metals in neurological diseases. Biomolecules. 2014a;4:678–703. doi: 10.3390/biom4030678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra J, Vasquez V, Hegde PM, Boldogh I, Mitra S, Kent TA, Rao KS, Hegde ML. Revisiting Metal Toxicity in Neurodegenerative Diseases and Stroke: Therapeutic Potential. Neurol Res Ther. 2014b;1 doi: 10.14437/NRTOA-1-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuyama Y. Presenile dementia with motor neuron disease. Dementia. 1993;4:137–142. doi: 10.1159/000107312. [DOI] [PubMed] [Google Scholar]
- Mladinic M, Lefevre C, Del Bel E, Nicholls J, Digby M. Developmental changes of gene expression after spinal cord injury in neonatal opossums. Brain Res. 2010;1363:20–39. doi: 10.1016/j.brainres.2010.09.024. [DOI] [PubMed] [Google Scholar]
- Morohoshi F, Ootsuka Y, Arai K, Ichikawa H, Mitani S, Munakata N, Ohki M. Genomic structure of the human RBP56/hTAFII68 and FUS/TLS genes. Gene. 1998;221:191–198. doi: 10.1016/s0378-1119(98)00463-6. [DOI] [PubMed] [Google Scholar]
- Nagase M, Yamamoto Y, Miyazaki Y, Yoshino H. Increased oxidative stress in patients with amyotrophic lateral sclerosis and the effect of edaravone administration. Redox Rep. 2015 doi: 10.1179/1351000215Y.0000000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima-Yasuda H, Uryu K, Robinson J, Xie SX, Hurtig H, Duda JE, Arnold SE, Siderowf A, Grossman M, Leverenz JB, Woltjer R, Lopez OL, Hamilton R, Tsuang DW, Galasko D, Masliah E, Kaye J, Clark CM, Montine TJ, Lee VM, Trojanowski JQ. Co-morbidity of TDP-43 proteinopathy in Lewy body related diseases. Acta Neuropathol. 2007;114:221–229. doi: 10.1007/s00401-007-0261-2. [DOI] [PubMed] [Google Scholar]
- Neumann M, Kwong LK, Lee EB, Kremmer E, Flatley A, Xu Y, Forman MS, Troost D, Kretzschmar HA, Trojanowski JQ, Lee VM. Phosphorylation of S409/410 of TDP-43 is a consistent feature in all sporadic and familial forms of TDP-43 proteinopathies. Acta Neuropathol. 2009;117:137–149. doi: 10.1007/s00401-008-0477-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Niedzielska E, Smaga I, Gawlik M, Moniczewski A, Stankowicz P, Pera J, Filip M. Oxidative Stress in Neurodegenerative Diseases. Mol Neurobiol. 2015 doi: 10.1007/s12035-015-9337-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu C, Zhang J, Gao F, Yang L, Jia M, Zhu H, Gong W. FUS-NLS/Transportin 1 complex structure provides insights into the nuclear targeting mechanism of FUS and the implications in ALS. PLoS One. 2012;7:e47056. doi: 10.1371/journal.pone.0047056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Watanabe S, Kaneko K, Yamanaka K, Nukina N, Furukawa Y. Intranuclear aggregation of mutant FUS/TLS as a molecular pathomechanism of amyotrophic lateral sclerosis. J Biol Chem. 2014;289:1192–1202. doi: 10.1074/jbc.M113.516492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka T, Suzuki G, Tanaka Y, Kametani F, Hirai S, Okado H, Miyashita T, Saitoe M, Akiyama H, Masai H, Hasegawa M. Phosphorylation of TAR DNA-binding Protein of 43 kDa (TDP-43) by Truncated Casein Kinase 1delta Triggers Mislocalization and Accumulation of TDP-43. J Biol Chem. 2016;291:5473–5483. doi: 10.1074/jbc.M115.695379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa T, Mizuno Y, Fujita Y, Takatama M, Nakazato Y, Okamoto K. Optineurin in neurodegenerative diseases. Neuropathology. 2011;31:569–574. doi: 10.1111/j.1440-1789.2011.01199.x. [DOI] [PubMed] [Google Scholar]
- Ou SH, Wu F, Harrich D, Garcia-Martinez LF, Gaynor RB. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J Virol. 1995;69:3584–3596. doi: 10.1128/jvi.69.6.3584-3596.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panagopoulos I, Aman P, Fioretos T, Hoglund M, Johansson B, Mandahl N, Heim S, Behrendtz M, Mitelman F. Fusion of the FUS gene with ERG in acute myeloid leukemia with t(16;21)(p11;q22) Genes Chromosomes Cancer. 1994;11:256–262. doi: 10.1002/gcc.2870110408. [DOI] [PubMed] [Google Scholar]
- Pasinelli P, Brown RH. Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat Rev Neurosci. 2006;7:710–723. doi: 10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- Pedersen WA, Fu W, Keller JN, Markesbery WR, Appel S, Smith RG, Kasarskis E, Mattson MP. Protein modification by the lipid peroxidation product 4-hydroxynonenal in the spinal cords of amyotrophic lateral sclerosis patients. Ann Neurol. 1998;44:819–824. doi: 10.1002/ana.410440518. [DOI] [PubMed] [Google Scholar]
- Perl DP, Gajdusek DC, Garruto RM, Yanagihara RT, Gibbs CJ. Intraneuronal aluminum accumulation in amyotrophic lateral sclerosis and Parkinsonism-dementia of Guam. Science. 1982;217:1053–1055. doi: 10.1126/science.7112111. [DOI] [PubMed] [Google Scholar]
- Philips T, Robberecht W. Neuroinflammation in amyotrophic lateral sclerosis: role of glial activation in motor neuron disease. Lancet Neurol. 2011;10:253–263. doi: 10.1016/S1474-4422(11)70015-1. [DOI] [PubMed] [Google Scholar]
- Plato CC, Garruto RM, Galasko D, Craig UK, Plato M, Gamst A, Torres JM, Wiederholt W. Amyotrophic lateral sclerosis and parkinsonism-dementia complex of Guam: changing incidence rates during the past 60 years. Am J Epidemiol. 2003;157:149–157. doi: 10.1093/aje/kwf175. [DOI] [PubMed] [Google Scholar]
- Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, Ling SC, Sun E, Wancewicz E, Mazur C, Kordasiewicz H, Sedaghat Y, Donohue JP, Shiue L, Bennett CF, Yeo GW, Cleveland DW. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers CA, Mathur M, Raaka BM, Ron D, Samuels HH. TLS (translocated-in-liposarcoma) is a high-affinity interactor for steroid, thyroid hormone, and retinoid receptors. Mol Endocrinol. 1998;12:4–18. doi: 10.1210/mend.12.1.0043. [DOI] [PubMed] [Google Scholar]
- Prasad DD, Ouchida M, Lee L, Rao VN, Reddy ES. TLS/FUS fusion domain of TLS/FUS-erg chimeric protein resulting from the t(16;21) chromosomal translocation in human myeloid leukemia functions as a transcriptional activation domain. Oncogene. 1994;9:3717–3729. [PubMed] [Google Scholar]
- Qiu H, Lee S, Shang Y, Wang WY, Au KF, Kamiya S, Barmada SJ, Finkbeiner S, Lui H, Carlton CE, Tang AA, Oldham MC, Wang H, Shorter J, Filiano AJ, Roberson ED, Tourtellotte WG, Chen B, Tsai LH, Huang EJ. ALS-associated mutation FUS-R521C causes DNA damage and RNA splicing defects. J Clin Invest. 2014;124:981–999. doi: 10.1172/JCI72723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbitts TH, Forster A, Larson R, Nathan P. Fusion of the dominant negative transcription regulator CHOP with a novel gene FUS by translocation t(12;16) in malignant liposarcoma. Nat Genet. 1993;4:175–180. doi: 10.1038/ng0693-175. [DOI] [PubMed] [Google Scholar]
- Rao SD, Banack SA, Cox PA, Weiss JH. BMAA selectively injures motor neurons via AMPA/kainate receptor activation. Exp Neurol. 2006;201:244–252. doi: 10.1016/j.expneurol.2006.04.017. [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Friesen WJ, Paushkin S, Dreyfuss G, Mann M. Detection of arginine dimethylated peptides by parallel precursor ion scanning mass spectrometry in positive ion mode. Anal Chem. 2003;75:3107–3114. doi: 10.1021/ac026283q. [DOI] [PubMed] [Google Scholar]
- Ravits J, Appel S, Baloh RH, Barohn R, Brooks BR, Elman L, Floeter MK, Henderson C, Lomen-Hoerth C, Macklis JD, McCluskey L, Mitsumoto H, Przedborski S, Rothstein J, Trojanowski JQ, van den Berg LH, Ringel S. Deciphering amyotrophic lateral sclerosis: what phenotype, neuropathology and genetics are telling us about pathogenesis. Amyotroph Lateral Scler Frontotemporal Degener. 2013;14(Suppl 1):5–18. doi: 10.3109/21678421.2013.778548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson HK, Deykin AV, Bronovitsky EV, Ovchinnikov RK, Ustyugov AA, Shelkovnikova TA, Kukharsky MS, Ermolkevich TG, Goldman IL, Sadchikova ER, Kovrazhkina EA, Bachurin SO, Buchman VL, Ninkina NN. Early lethality and neuronal proteinopathy in mice expressing cytoplasm-targeted FUS that lacks the RNA recognition motif. Amyotroph Lateral Scler Frontotemporal Degener. 2015;16:402–409. doi: 10.3109/21678421.2015.1040994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O’Regan JP, Deng HX, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- Rulten SL, Rotheray A, Green RL, Grundy GJ, Moore DA, Gomez-Herreros F, Hafezparast M, Caldecott KW. PARP-1 dependent recruitment of the amyotrophic lateral sclerosis-associated protein FUS/TLS to sites of oxidative DNA damage. Nucleic Acids Res. 2014;42:307–314. doi: 10.1093/nar/gkt835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford NJ, Zhang YJ, Baker M, Gass JM, Finch NA, Xu YF, Stewart H, Kelley BJ, Kuntz K, Crook RJ, Sreedharan J, Vance C, Sorenson E, Lippa C, Bigio EH, Geschwind DH, Knopman DS, Mitsumoto H, Petersen RC, Cashman NR, Hutton M, Shaw CE, Boylan KB, Boeve B, Graff-Radford NR, Wszolek ZK, Caselli RJ, Dickson DW, Mackenzie IR, Petrucelli L, Rademakers R. Novel mutations in TARDBP (TDP-43) in patients with familial amyotrophic lateral sclerosis. PLoS Genet. 2008;4:e1000193. doi: 10.1371/journal.pgen.1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccon RA, Bunton-Stasyshyn RK, Fisher EM, Fratta P. Is SOD1 loss of function involved in amyotrophic lateral sclerosis? Brain. 2013;136:2342–2358. doi: 10.1093/brain/awt097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salajegheh M, Pinkus JL, Taylor JP, Amato AA, Nazareno R, Baloh RH, Greenberg SA. Sarcoplasmic redistribution of nuclear TDP-43 in inclusion body myositis. Muscle Nerve. 2009;40:19–31. doi: 10.1002/mus.21386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salih DA, Brunet A. FoxO transcription factors in the maintenance of cellular homeostasis during aging. Curr Opin Cell Biol. 2008;20:126–136. doi: 10.1016/j.ceb.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvi JS, Mekhail K. R-loops highlight the nucleus in ALS. Nucleus. 2015;6:23–29. doi: 10.1080/19491034.2015.1004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sama RR, Ward CL, Bosco DA. Functions of FUS/TLS from DNA repair to stress response: implications for ALS. ASN Neuro. 2014;6 doi: 10.1177/1759091414544472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sama RR, Ward CL, Kaushansky LJ, Lemay N, Ishigaki S, Urano F, Bosco DA. FUS/TLS assembles into stress granules and is a prosurvival factor during hyperosmolar stress. J Cell Physiol. 2013;228:2222–2231. doi: 10.1002/jcp.24395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki S, Iguchi Y, Katsuno M, Sobue G. Alterations in the blood-spinal cord barrier in TDP-43 conditional knockout mice. Neurosci Lett. 2015;598:1–5. doi: 10.1016/j.neulet.2015.05.005. [DOI] [PubMed] [Google Scholar]
- Scaramuzzino C, Monaghan J, Milioto C, Lanson NA, Jr., Maltare A, Aggarwal T, Casci I, Fackelmayer FO, Pennuto M, Pandey UB. Protein arginine methyltransferase 1 and 8 interact with FUS to modify its sub-cellular distribution and toxicity in vitro and in vivo. PLoS One. 2013;8:e61576. doi: 10.1371/journal.pone.0061576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S, Kwee LC, Allen KD, Oddone EZ. Association of ALS with head injury, cigarette smoking and APOE genotypes. J Neurol Sci. 2010;291:22–29. doi: 10.1016/j.jns.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz KM, Schmitt N, Hoffmann-Rohrer U, Schafer A, Grummt I, Mayer C. TAF12 recruits Gadd45a and the nucleotide excision repair complex to the promoter of rRNA genes leading to active DNA demethylation. Mol Cell. 2009;33:344–353. doi: 10.1016/j.molcel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- Scotter EL, Vance C, Nishimura AL, Lee YB, Chen HJ, Urwin H, Sardone V, Mitchell JC, Rogelj B, Rubinsztein DC, Shaw CE. Differential roles of the ubiquitin proteasome system and autophagy in the clearance of soluble and aggregated TDP-43 species. J Cell Sci. 2014;127:1263–1278. doi: 10.1242/jcs.140087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sephton CF, Cenik C, Kucukural A, Dammer EB, Cenik B, Han Y, Dewey CM, Roth FP, Herz J, Peng J, Moore MJ, Yu G. Identification of neuronal RNA targets of TDP-43-containing ribonucleoprotein complexes. J Biol Chem. 2011;286:1204–1215. doi: 10.1074/jbc.M110.190884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sephton CF, Good SK, Atkin S, Dewey CM, Mayer P, 3rd, Herz J, Yu G. TDP-43 is a developmentally regulated protein essential for early embryonic development. J Biol Chem. 2010;285:6826–6834. doi: 10.1074/jbc.M109.061846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sephton CF, Yu G. The function of RNA-binding proteins at the synapse: implications for neurodegeneration. Cell Mol Life Sci. 2015;72:3621–3635. doi: 10.1007/s00018-015-1943-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh AY, Martin LJ. DNA base-excision repair enzyme apurinic/apyrimidinic endonuclease/redox factor-1 is increased and competent in the brain and spinal cord of individuals with amyotrophic lateral sclerosis. Neuromolecular Med. 2002;2:47–60. doi: 10.1007/s12017-002-0038-7. [DOI] [PubMed] [Google Scholar]
- Shan X, Chiang PM, Price DL, Wong PC. Altered distributions of Gemini of coiled bodies and mitochondria in motor neurons of TDP-43 transgenic mice. Proc Natl Acad Sci U S A. 2010;107:16325–16330. doi: 10.1073/pnas.1003459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Lyashchenko AK, Lu L, Nasrabady SE, Elmaleh M, Mendelsohn M, Nemes A, Tapia JC, Mentis GZ, Shneider NA. ALS-associated mutant FUS induces selective motor neuron degeneration through toxic gain of function. Nat Commun. 2016;7:10465. doi: 10.1038/ncomms10465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelkovnikova TA, Peters OM, Deykin AV, Connor-Robson N, Robinson H, Ustyugov AA, Bachurin SO, Ermolkevich TG, Goldman IL, Sadchikova ER, Kovrazhkina EA, Skvortsova VI, Ling SC, Da Cruz S, Parone PA, Buchman VL, Ninkina NN. Fused in sarcoma (FUS) protein lacking nuclear localization signal (NLS) and major RNA binding motifs triggers proteinopathy and severe motor phenotype in transgenic mice. J Biol Chem. 2013a;288:25266–25274. doi: 10.1074/jbc.M113.492017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelkovnikova TA, Robinson HK, Connor-Robson N, Buchman VL. Recruitment into stress granules prevents irreversible aggregation of FUS protein mislocalized to the cytoplasm. Cell Cycle. 2013b;12:3194–3202. doi: 10.4161/cc.26241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina Y, Arima K, Tabunoki H, Satoh J. TDP-43 dimerizes in human cells in culture. Cell Mol Neurobiol. 2010;30:641–652. doi: 10.1007/s10571-009-9489-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shing DC, McMullan DJ, Roberts P, Smith K, Chin SF, Nicholson J, Tillman RM, Ramani P, Cullinane C, Coleman N. FUS/ERG gene fusions in Ewing’s tumors. Cancer Res. 2003;63:4568–4576. [PubMed] [Google Scholar]
- Shiwaku H, Okazawa H. Impaired DNA damage repair as a common feature of neurodegenerative diseases and psychiatric disorders. Curr Mol Med. 2015;15:119–128. doi: 10.2174/1566524015666150303002556. [DOI] [PubMed] [Google Scholar]
- Simpson EP, Henry YK, Henkel JS, Smith RG, Appel SH. Increased lipid peroxidation in sera of ALS patients: a potential biomarker of disease burden. Neurology. 2004;62:1758–1765. doi: 10.1212/wnl.62.10.1758. [DOI] [PubMed] [Google Scholar]
- Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, Baralle F, de Belleroche J, Mitchell JD, Leigh PN, Al-Chalabi A, Miller CC, Nicholson G, Shaw CE. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallings NR, Puttaparthi K, Luther CM, Burns DK, Elliott JL. Progressive motor weakness in transgenic mice expressing human TDP-43. Neurobiol Dis. 2010;40:404–414. doi: 10.1016/j.nbd.2010.06.017. [DOI] [PubMed] [Google Scholar]
- Suberbielle E, Sanchez PE, Kravitz AV, Wang X, Ho K, Eilertson K, Devidze N, Kreitzer AC, Mucke L. Physiologic brain activity causes DNA double-strand breaks in neurons, with exacerbation by amyloid-beta. Nat Neurosci. 2013;16:613–621. doi: 10.1038/nn.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup V, Phaneuf D, Bareil C, Robertson J, Rouleau GA, Kriz J, Julien JP. Pathological hallmarks of amyotrophic lateral sclerosis/frontotemporal lobar degeneration in transgenic mice produced with TDP-43 genomic fragments. Brain. 2011a;134:2610–2626. doi: 10.1093/brain/awr159. [DOI] [PubMed] [Google Scholar]
- Swarup V, Phaneuf D, Dupre N, Petri S, Strong M, Kriz J, Julien JP. Deregulation of TDP-43 in amyotrophic lateral sclerosis triggers nuclear factor kappaB-mediated pathogenic pathways. J Exp Med. 2011b;208:2429–2447. doi: 10.1084/jem.20111313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan AY, Manley JL. The TET family of proteins: functions and roles in disease. J Mol Cell Biol. 2009;1:82–92. doi: 10.1093/jmcb/mjp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan AY, Manley JL. TLS inhibits RNA polymerase III transcription. Mol Cell Biol. 2010;30:186–196. doi: 10.1128/MCB.00884-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan CF, Eguchi H, Tagawa A, Onodera O, Iwasaki T, Tsujino A, Nishizawa M, Kakita A, Takahashi H. TDP-43 immunoreactivity in neuronal inclusions in familial amyotrophic lateral sclerosis with or without SOD1 gene mutation. Acta Neuropathol. 2007;113:535–542. doi: 10.1007/s00401-007-0206-9. [DOI] [PubMed] [Google Scholar]
- Thorpe JR, Tang H, Atherton J, Cairns NJ. Fine structural analysis of the neuronal inclusions of frontotemporal lobar degeneration with TDP-43 proteinopathy. J Neural Transm. 2008;115:1661–1671. doi: 10.1007/s00702-008-0137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani M, Tradewell ML, Mattina KR, Minotti S, Yang W, Zhou H, Strong MJ, Hayward LJ, Durham HD. Cytoplasmic sequestration of FUS/TLS associated with ALS alters histone marks through loss of nuclear protein arginine methyltransferase 1. Hum Mol Genet. 2015;24:773–786. doi: 10.1093/hmg/ddu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticozzi N, Silani V, LeClerc AL, Keagle P, Gellera C, Ratti A, Taroni F, Kwiatkowski TJ, Jr., McKenna-Yasek DM, Sapp PC, Brown RH, Jr., Landers JE. Analysis of FUS gene mutation in familial amyotrophic lateral sclerosis within an Italian cohort. Neurology. 2009;73:1180–1185. doi: 10.1212/WNL.0b013e3181bbff05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolino M, Kohrmann M, Kiebler MA. RNA-binding proteins involved in RNA localization and their implications in neuronal diseases. Eur J Neurosci. 2012;35:1818–1836. doi: 10.1111/j.1460-9568.2012.08160.x. [DOI] [PubMed] [Google Scholar]
- Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, Konig J, Hortobagyi T, Nishimura AL, Zupunski V, Patani R, Chandran S, Rot G, Zupan B, Shaw CE, Ule J. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourriere H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol. 2003;160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tradewell ML, Yu Z, Tibshirani M, Boulanger MC, Durham HD, Richard S. Arginine methylation by PRMT1 regulates nuclear-cytoplasmic localization and toxicity of FUS/TLS harbouring ALS-linked mutations. Hum Mol Genet. 2012;21:136–149. doi: 10.1093/hmg/ddr448. [DOI] [PubMed] [Google Scholar]
- Tsai KJ, Yang CH, Fang YH, Cho KH, Chien WL, Wang WT, Wu TW, Lin CP, Fu WM, Shen CK. Elevated expression of TDP-43 in the forebrain of mice is sufficient to cause neurological and pathological phenotypes mimicking FTLD-U. J Exp Med. 2010;207:1661–1673. doi: 10.1084/jem.20092164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner MR, Cagnin A, Turkheimer FE, Miller CC, Shaw CE, Brooks DJ, Leigh PN, Banati RB. Evidence of widespread cerebral microglial activation in amyotrophic lateral sclerosis: an [11C](R)-PK11195 positron emission tomography study. Neurobiol Dis. 2004;15:601–609. doi: 10.1016/j.nbd.2003.12.012. [DOI] [PubMed] [Google Scholar]
- Uranishi H, Tetsuka T, Yamashita M, Asamitsu K, Shimizu M, Itoh M, Okamoto T. Involvement of the pro-oncoprotein TLS (translocated in liposarcoma) in nuclear factor-kappa B p65-mediated transcription as a coactivator. J Biol Chem. 2001;276:13395–13401. doi: 10.1074/jbc.M011176200. [DOI] [PubMed] [Google Scholar]
- Valdmanis PN, Daoud H, Dion PA, Rouleau GA. Recent advances in the genetics of amyotrophic lateral sclerosis. Curr Neurol Neurosci Rep. 2009;9:198–205. doi: 10.1007/s11910-009-0030-9. [DOI] [PubMed] [Google Scholar]
- van Blitterswijk M, Wang ET, Friedman BA, Keagle PJ, Lowe P, Leclerc AL, van den Berg LH, Housman DE, Veldink JH, Landers JE. Characterization of FUS mutations in amyotrophic lateral sclerosis using RNA-Seq. PLoS One. 2013;8:e60788. doi: 10.1371/journal.pone.0060788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Deerlin VM, Leverenz JB, Bekris LM, Bird TD, Yuan W, Elman LB, Clay D, Wood EM, Chen-Plotkin AS, Martinez-Lage M, Steinbart E, McCluskey L, Grossman M, Neumann M, Wu IL, Yang WS, Kalb R, Galasko DR, Montine TJ, Trojanowski JQ, Lee VM, Schellenberg GD, Yu CE. TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis. Lancet Neurol. 2008;7:409–416. doi: 10.1016/S1474-4422(08)70071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eersel J, Ke YD, Gladbach A, Bi M, Gotz J, Kril JJ, Ittner LM. Cytoplasmic accumulation and aggregation of TDP-43 upon proteasome inhibition in cultured neurons. PLoS One. 2011;6:e22850. doi: 10.1371/journal.pone.0022850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, Ganesalingam J, Williams KL, Tripathi V, Al-Saraj S, Al-Chalabi A, Leigh PN, Blair IP, Nicholson G, de Belleroche J, Gallo JM, Miller CC, Shaw CE. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkening K, Leystra-Lantz C, Yang W, Jaffee H, Strong MJ. Tar DNA binding protein of 43 kDa (TDP-43), 14-3-3 proteins and copper/zinc superoxide dismutase (SOD1) interact to modulate NFL mRNA stability. Implications for altered RNA processing in amyotrophic lateral sclerosis (ALS) Brain Res. 2009;1305:168–182. doi: 10.1016/j.brainres.2009.09.105. [DOI] [PubMed] [Google Scholar]
- Waibel S, Neumann M, Rosenbohm A, Birve A, Volk AE, Weishaupt JH, Meyer T, Muller U, Andersen PM, Ludolph AC. Truncating mutations in FUS/TLS give rise to a more aggressive ALS-phenotype than missense mutations: a clinico-genetic study in Germany. Eur J Neurol. 2013;20:540–546. doi: 10.1111/ene.12031. [DOI] [PubMed] [Google Scholar]
- Walker AK, Daniels CM, Goldman JE, Trojanowski JQ, Lee VM, Messing A. Astrocytic TDP-43 pathology in Alexander disease. J Neurosci. 2014;34:6448–6458. doi: 10.1523/JNEUROSCI.0248-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AK, Spiller KJ, Ge G, Zheng A, Xu Y, Zhou M, Tripathy K, Kwong LK, Trojanowski JQ, Lee VM. Functional recovery in new mouse models of ALS/FTLD after clearance of pathological cytoplasmic TDP-43. Acta Neuropathol. 2015;130:643–660. doi: 10.1007/s00401-015-1460-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Adhikari S, Butler BE, Pandita TK, Mitra S, Hegde ML. A Perspective on Chromosomal Double Strand Break Markers in Mammalian Cells. Jacobs J Radiat Oncol. 2014;1 [PMC free article] [PubMed] [Google Scholar]
- Wang HY, Wang IF, Bose J, Shen CK. Structural diversity and functional implications of the eukaryotic TDP gene family. Genomics. 2004;83:130–139. doi: 10.1016/s0888-7543(03)00214-3. [DOI] [PubMed] [Google Scholar]
- Wang IF, Wu LS, Chang HY, Shen CK. TDP-43, the signature protein of FTLD-U, is a neuronal activity-responsive factor. J Neurochem. 2008;105:797–806. doi: 10.1111/j.1471-4159.2007.05190.x. [DOI] [PubMed] [Google Scholar]
- Wang JW, Brent JR, Tomlinson A, Shneider NA, McCabe BD. The ALS-associated proteins FUS and TDP-43 function together to affect Drosophila locomotion and life span. J Clin Invest. 2011;121:4118–4126. doi: 10.1172/JCI57883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Li L, Lin WL, Dickson DW, Petrucelli L, Zhang T, Wang X. The ALS disease-associated mutant TDP-43 impairs mitochondrial dynamics and function in motor neurons. Hum Mol Genet. 2013a;22:4706–4719. doi: 10.1093/hmg/ddt319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WY, Pan L, Su SC, Quinn EJ, Sasaki M, Jimenez JC, Mackenzie IR, Huang EJ, Tsai LH. Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. Nat Neurosci. 2013b;16:1383–1391. doi: 10.1038/nn.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts GD, Wymer J, Kovach MJ, Mehta SG, Mumm S, Darvish D, Pestronk A, Whyte MP, Kimonis VE. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet. 2004;36:377–381. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- Wegorzewska I, Bell S, Cairns NJ, Miller TM, Baloh RH. TDP-43 mutant transgenic mice develop features of ALS and frontotemporal lobar degeneration. Proc Natl Acad Sci U S A. 2009;106:18809–18814. doi: 10.1073/pnas.0908767106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss JH, Koh JY, Choi DW. Neurotoxicity of beta-N-methylamino-L-alanine (BMAA) and beta-N-oxalylamino-L-alanine (BOAA) on cultured cortical neurons. Brain Res. 1989;497:64–71. doi: 10.1016/0006-8993(89)90970-0. [DOI] [PubMed] [Google Scholar]
- Weisskopf MG, Morozova N, O’Reilly EJ, McCullough ML, Calle EE, Thun MJ, Ascherio A. Prospective study of chemical exposures and amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 2009;80:558–561. doi: 10.1136/jnnp.2008.156976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KL, Warraich ST, Yang S, Solski JA, Fernando R, Rouleau GA, Nicholson GA, Blair IP. UBQLN2/ubiquilin 2 mutation and pathology in familial amyotrophic lateral sclerosis. Neurobiol Aging. 2012;33:2527, e2523–2510. doi: 10.1016/j.neurobiolaging.2012.05.008. [DOI] [PubMed] [Google Scholar]
- Wils H, Kleinberger G, Janssens J, Pereson S, Joris G, Cuijt I, Smits V, Ceuterick-de Groote C, Van Broeckhoven C, Kumar-Singh S. TDP-43 transgenic mice develop spastic paralysis and neuronal inclusions characteristic of ALS and frontotemporal lobar degeneration. Proc Natl Acad Sci U S A. 2010;107:3858–3863. doi: 10.1073/pnas.0912417107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LS, Cheng WC, Hou SC, Yan YT, Jiang ST, Shen CK. TDP-43, a neuro-pathosignature factor, is essential for early mouse embryogenesis. Genesis. 2010;48:56–62. doi: 10.1002/dvg.20584. [DOI] [PubMed] [Google Scholar]
- Wu LS, Cheng WC, Shen CK. Targeted depletion of TDP-43 expression in the spinal cord motor neurons leads to the development of amyotrophic lateral sclerosis-like phenotypes in mice. J Biol Chem. 2012;287:27335–27344. doi: 10.1074/jbc.M112.359000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Sanelli T, Chiang H, Sun Y, Chakrabartty A, Keith J, Rogaeva E, Zinman L, Robertson J. Low molecular weight species of TDP-43 generated by abnormal splicing form inclusions in amyotrophic lateral sclerosis and result in motor neuron death. Acta Neuropathol. 2015;130:49–61. doi: 10.1007/s00401-015-1412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Sanelli T, Dib S, Sheps D, Findlater J, Bilbao J, Keith J, Zinman L, Rogaeva E, Robertson J. RNA targets of TDP-43 identified by UV-CLIP are deregulated in ALS. Mol Cell Neurosci. 2011;47:167–180. doi: 10.1016/j.mcn.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Xu YF, Gendron TF, Zhang YJ, Lin WL, D’Alton S, Sheng H, Casey MC, Tong J, Knight J, Yu X, Rademakers R, Boylan K, Hutton M, McGowan E, Dickson DW, Lewis J, Petrucelli L. Wild-type human TDP-43 expression causes TDP-43 phosphorylation, mitochondrial aggregation, motor deficits, and early mortality in transgenic mice. J Neurosci. 2010;30:10851–10859. doi: 10.1523/JNEUROSCI.1630-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YF, Zhang YJ, Lin WL, Cao X, Stetler C, Dickson DW, Lewis J, Petrucelli L. Expression of mutant TDP-43 induces neuronal dysfunction in transgenic mice. Mol Neurodegener. 2011;6:73. doi: 10.1186/1750-1326-6-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Deng HX, Siddique N, Fecto F, Chen W, Yang Y, Liu E, Donkervoort S, Zheng JG, Shi Y, Ahmeti KB, Brooks B, Engel WK, Siddique T. Frameshift and novel mutations in FUS in familial amyotrophic lateral sclerosis and ALS/dementia. Neurology. 2010;75:807–814. doi: 10.1212/WNL.0b013e3181f07e0c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagihara R, Garruto RM, Gajdusek DC, Tomita A, Uchikawa T, Konagaya Y, Chen KM, Sobue I, Plato CC, Gibbs CJ., Jr. Calcium and vitamin D metabolism in Guamanian Chamorros with amyotrophic lateral sclerosis and parkinsonism-dementia. Ann Neurol. 1984;15:42–48. doi: 10.1002/ana.410150108. [DOI] [PubMed] [Google Scholar]
- Yang C, Tan W, Whittle C, Qiu L, Cao L, Akbarian S, Xu Z. The C-terminal TDP-43 fragments have a high aggregation propensity and harm neurons by a dominant-negative mechanism. PLoS One. 2010;5:e15878. doi: 10.1371/journal.pone.0015878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Wang H, Qiao T, Yang B, Aliaga L, Qiu L, Tan W, Salameh J, McKenna-Yasek DM, Smith T, Peng L, Moore MJ, Brown RH, Jr., Cai H, Xu Z. Partial loss of TDP-43 function causes phenotypes of amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2014;111:E1121–1129. doi: 10.1073/pnas.1322641111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui M, Ota K, Garruto RM. Concentrations of zinc and iron in the brains of Guamanian patients with amyotrophic lateral sclerosis and parkinsonism-dementia. Neurotoxicology. 1993;14:445–450. [PubMed] [Google Scholar]
- Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, Daley EL, Miller SJ, Cunningham KM, Vidensky S, Gupta S, Thomas MA, Hong I, Chiu SL, Huganir RL, Ostrow LW, Matunis MJ, Wang J, Sattler R, Lloyd TE, Rothstein JD. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525:56–61. doi: 10.1038/nature14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Baldie G, Periz G, Wang J. RNA-processing protein TDP-43 regulates FOXO-dependent protein quality control in stress response. PLoS Genet. 2014;10:e1004693. doi: 10.1371/journal.pgen.1004693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Gendron TF, Xu YF, Ko LW, Yen SH, Petrucelli L. Phosphorylation regulates proteasomal-mediated degradation and solubility of TAR DNA binding protein-43 C-terminal fragments. Mol Neurodegener. 2010;5:33. doi: 10.1186/1750-1326-5-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Xu YF, Cook C, Gendron TF, Roettges P, Link CD, Lin WL, Tong J, Castanedes-Casey M, Ash P, Gass J, Rangachari V, Buratti E, Baralle F, Golde TE, Dickson DW, Petrucelli L. Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc Natl Acad Sci U S A. 2009;106:7607–7612. doi: 10.1073/pnas.0900688106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Xu YF, Dickey CA, Buratti E, Baralle F, Bailey R, Pickering-Brown S, Dickson D, Petrucelli L. Progranulin mediates caspase-dependent cleavage of TAR DNA binding protein-43. J Neurosci. 2007;27:10530–10534. doi: 10.1523/JNEUROSCI.3421-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Huang C, Chen H, Wang D, Landel CP, Xia PY, Bowser R, Liu YJ, Xia XG. Transgenic rat model of neurodegeneration caused by mutation in the TDP gene. PLoS Genet. 2010;6:e1000887. doi: 10.1371/journal.pgen.1000887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Liu S, Liu G, Ozturk A, Hicks GG. ALS-associated FUS mutations result in compromised FUS alternative splicing and autoregulation. PLoS Genet. 2013;9:e1003895. doi: 10.1371/journal.pgen.1003895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinszner H, Albalat R, Ron D. A novel effector domain from the RNA-binding protein TLS or EWS is required for oncogenic transformation by CHOP. Genes Dev. 1994;8:2513–2526. doi: 10.1101/gad.8.21.2513. [DOI] [PubMed] [Google Scholar]