The crystal structure of a phosphoribosyl anthranilate isomerase from the hyperthermophilic archaeon T. kodakaraensis was determined in space groups P1 and C2 at 1.75 and 1.85 Å resolution, respectively.
Keywords: tryptophan biosynthesis, protein stability, TIM barrel, hyperthermophilic archaea, Pyrococcus furiosus, Thermococcus kodakaraensis, phosphoribosyl anthranilate isomerase
Abstract
A phosphoribosyl anthranilate isomerase, TkTrpF, from Thermococcus kodakaraensis was expressed in Escherichia coli and purified to homogeneity. TkTrpF was crystallized and its structure was determined by molecular replacement in two different space groups (C2 and P1) using data to 1.85 and 1.75 Å resolution, respectively. TkTrpF belongs to the class of TIM-barrel proteins. Structural comparison with other phosphoribosyl anthranilate isomerases (TrpFs) showed the highest structural similarity to Pyrococcus furiosus TrpF. Similarly to P. furiosus TrpF, TkTrpF is a monomer in solution, in contrast to other thermophilic enzymes, which exist as functional dimers. Although in space group P1 TkTrpF crystallizes with two molecules in the asymmetric unit, the interface is highly improbable in solution. Potential factors for the thermostability of TkTrpF were attributed to an increase in helical structure, an increased number of charged residues and an increase in the number of salt bridges.
1. Introduction
Phosphoribosyl anthranilate isomerase (TrpF) catalyzes the third step in the committed biosynthesis of tryptophan from chorismic acid, in which N-(5-phospho-β-d-ribosyl) anthranilate (PRA) is rearranged to 1-(2-carboxyphenylamino)-1-deoxy-d-ribulose 5-phosphate (CdRP). In particular, TrpF is involved in the formation of an enolamine, which is the putative substrate for the subsequent enzyme indoleglycerol phosphate synthase (TrpC) (Taka et al., 2005 ▸).
The structural organization of TrpF differs widely among microorganisms. TrpF is found as a monofunctional enzyme in a single polypeptide chain in Saccharomyces cerevisiae (Braus, 1991 ▸), Bacillus subtilis (Hoch, 1979 ▸), Pseudomonas putida (Enatsu & Crawford, 1971 ▸) and Acinetobacter calcoaceticus (Cohn & Crawford, 1976 ▸). In contrast, in Escherichia coli (Hommel et al., 1995 ▸), Salmonella typhimurium (Bauerle et al., 1987 ▸), Aerobacter aerogenes (Egan & Gibson, 1972 ▸) and Serratia marcescens (Potts & Drapeau, 1972 ▸) TrpF is found together with TrpC in the same polypeptide chain.
TrpFs have been cloned, expressed and characterized from various organisms, but crystallization has only been reported for a few of them. The structure of TrpF has been determined from three thermophiles, Thermotoga maritima (TmTrpF; PDB entry 1lbm; Henn-Sax et al., 2002 ▸), Thermus thermophilus HB8 (TtTrpF; PDB entry 1v5x; Taka et al., 2005 ▸) and Pyrococcus furiosus (PfTrpF; PDB entry 4aaj; Repo et al., 2012 ▸), and from two mesophiles, E. coli (EcTrpF; residues 254–452 in PDB entry 1pii; Wilmanns et al., 1992 ▸) and Jonesia denitrificans DSM 20603 (JdTrpF; PDB entry 4wui; Midwest Center for Structural Genomics, unpublished work). In T. maritima and T. thermophilus TrpF functions and crystallizes as a dimer (Hennig et al., 1997 ▸; Taka et al., 2005 ▸), whereas in E. coli it exists as a monomer (Wilmanns et al., 1992 ▸). TrpF from J. denitrificans has been predicted to be monomeric (PDB entry 4wui). In most mesophilic microorganisms TrpF is monomeric and labile, whereas in most hyperthermophiles it has been reported to form a homodimer for reasons of stability (Thoma et al., 2000 ▸). A difference has been reported for PfTrpF, which is found as a monomer optimized to act at extreme temperatures (Repo et al., 2012 ▸).
T. kodakaraensis KOD1 is a hyperthemopilic archaeon that was isolated by Imanaka and coworkers from a solfatara on Kodakara Island in Kagoshima, Japan (Fukui et al., 2005 ▸). T. kodakaraensis has coccus-shaped cells and grows optimally at 358 K and pH 6.5 as an obligate heterotroph (Atomi et al., 2004 ▸). It is one of the best characterized hyperthermophiles and its whole genome (GenBank accession No. AP006878) has been sequenced and published (Fukui et al., 2005 ▸). Many novel enzymes and metabolic pathways have been identified in this archaeon, including the tryptophan-biosynthesis pathway, which plays an important role in the metabolism of nucleotides and amino acids.
Here, we report the cloning, expression, purification, crystallization and structural characterization of a TrpF from T. kodakaraensis KOD1 (TkTrpF). Studies of TkTrpF will help in understanding its mode of action and regulation in hyperthermophiles as well as the thermostability associated with TIM-barrel proteins and will provide a structural basis for enzyme engineering for biotechnological and industrial applications.
2. Materials and methods
2.1. Macromolecule production
Genomic DNA of T. kodakaraensis was used as a template to amplify tktrpF (TK0256) by polymerase chain reaction using a sequence-specific set of primers (Table 1 ▸). PCR-amplified tktrpF was purified from gel using a DNA purification kit (Fermentas Life Sciences) and ligated into cloning vector pTZ57R/T using T4 DNA ligase according to the supplier’s instructions (Thermo Fisher Scientific). The resultant recombinant plasmid was named TkTrpF-pTZ57R/T. The TkTrpF gene was liberated from TkTrpF-pTZ57R/T using NdeI (introduced in the forward primer) and HindIII (from the multiple cloning sites of pTZ57R/T). The excised TkTrpF gene product was cloned into pET-28a(+) (Novagen) utilizing the same sites and the resultant recombinant expression vector was named TkTrpF-pET28a(+). The sequence of TkTrpF was confirmed by DNA sequencing using a CEQ800 Beckman Coulter sequencing system. The recombinant protein contains 20 additional vector residues (MGSSHHHHHHSSGLVPRGSH) at the N-terminus.
Table 1. Macromolecule-production information.
| Source organism | T. kodakaraensis |
| DNA source | Genomic DNA of T. kodakaraensis |
| Forward primer† | CATATGGTTGAGTTCGTTAAGATATGCGGCG |
| Reverse primer | TCATCCATTCCTCACCACCGCCAT |
| Cloning vector | pTZ57R/T |
| Expression vector | pET-28a(+) |
| Expression host | E. coli |
| Complete amino-acid sequence of the construct produced‡ | MGSSHHHHHHSSGLVPRGSHMVEFVKICGVKTMDELRLVERYADATGVVVNSRSKRKVPLKTAAELIEMAEIPIYLVSTMKTFPEWANAVEKTGAEYIQVHSDMHPKAVNRLKDEYGVSVMKAFMVPRESDDPAEDAERLLELIGQYEVDKILLDTGVGSGRRHDYRVSAIIAKEYPIVLAGGLTPENVGEAIRWVKPAGVDVSSGVERNGVKDRVLIEAFMAVVRNG |
The NdeI recognition site introduced in the forward primer is underlined.
The His tag at the N-terminus is underlined.
Recombinant TkTrpF-pET28a(+) plasmid was used for the production of TkTrpF in E. coli BL21 CodonPlus (DE3)-RIL cells grown in Luria–Bertani medium. In initial expression attempts at 310 K, the recombinant TkTrpF was found to be expressed in an insoluble form as inclusion bodies. To obtain soluble TkTrpF, expression was carried out at low temperature. When the OD600 of cells grown at 310 K reached 0.5–0.6, the culture was chilled on ice for 15 min and then induced with 0.5 mM IPTG. The protein was produced at 290 K in a shaking incubator overnight. Harvested cells were washed with 50 mM Tris–HCl pH 8.5, lysed by sonication in 50 mM Tris–HCl pH 8.5 buffer containing 1 mM DTT, 1 mM PMSF and 20%(w/v) glycerol (storage buffer) and centrifuged at 15 000g for 15 min at 277 K. The supernatant was heated to 338 K for 25 min and centrifuged again at 15 000g for 15 min at 277 K to remove host proteins. The resulting supernatant was loaded onto an Ni2+-charged Sepharose column, which was equilibrated with 20 mM Tris–HCl pH 8.5 containing 150 mM NaCl and 20 mM imidazole. Elution of the bound protein was performed by a stepwise increase in the imidazole concentration from 50 to 300 mM. Analysis of eluted fractions by SDS–PAGE showed that most of the protein was eluted with 150–200 mM imidazole. Purified TkTrpF was dialyzed against the storage buffer and stored at 253 K. The protein concentration was determined spectrophotometrically using the Bradford reagent (Bradford, 1976 ▸). The enzyme activity of TkTrpF was determined fluorometrically by measuring the decrease in the concentration of anthranilate in a coupled reaction (Supplementary Fig. S1) as described previously (Hommel et al., 1995 ▸; Sterner et al., 1996 ▸). A 130 molar excess of PRPP over anthranilic acid was used to ensure a constant supply of substrate owing to the thermolability of PRA. The optimum temperature for enzyme activity was found to be 328 K.
2.2. Crystallization
TkTrpF was concentrated to 2 mg ml−1 in 10 mM Tris–HCl buffer pH 8.0, 0.1 M NaCl, 0.002%(w/v) NaN3 using Amicon filters (10 000 Da cutoff). Initial crystallization screening was carried out using the sitting-drop vapour-diffusion method with the PACT screen (Hampton Research) at 289 K in 96-well Greiner CrystalQuick crystallization plates using 0.75 µl protein solution mixed with an equal amount of precipitant solution. The drops were equilibrated against 75 µl reservoir solution. Needle-shaped crystals were obtained from the initial screen. Optimization was carried out by varying the precipitant concentration using the hanging-drop vapour-diffusion method. 2 µl protein solution was mixed with an equal volume of precipitant solution and was equilibrated against 0.8 ml reservoir solution. Single crystals of suitable size for X-ray crystallographic analysis were grown using 0.1 M Tris–HCl pH 8.0, 0.2 M sodium formate, 13%(w/v) PEG 4000 (Table 2 ▸).
Table 2. Crystallization.
| Method | Hanging-drop vapour diffusion |
| Plate type | 24-well Linbro plate |
| Temperature (K) | 289 |
| Protein concentration (mg ml−1) | 2 |
| Buffer composition of protein solution | 10 mM Tris–HCl buffer pH 8.0, 0.1 M NaCl, 0.002% NaN3 |
| Composition of reservoir solution | 0.1 M Tris–HCl pH 8.0, 0.2 M sodium formate, 13%(w/v) PEG 4000 |
| Volume and ratio of drop | 4 µl (1:1) |
| Volume of reservoir (ml) | 0.8 |
2.3. Data collection and processing
Prior to data collection, crystals were flash-cooled in liquid nitrogen using mother liquor supplemented with 20%(v/v) glycerol for cryoprotection. Data were collected from single crystals at the ESRF synchrotron facility, Grenoble, France using the high-throughput fully automatic MASSIF-1 beamline (Svensson et al., 2015 ▸). Initial reference images were collected in order to calculate the best data-collection strategy with minimum radiation damage. Although all crystals were grown under the same crystallization conditions, two different space groups (C2 and P1) were identified after processing with XDS (Kabsch, 2010 ▸) followed by AIMLESS (Evans & Murshudov, 2013 ▸). The X-ray data-collection statistics are provided in Table 3 ▸.
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| Space group | C2 | P1 |
|---|---|---|
| Diffraction source | MASSIF-1, ESRF | MASSIF-1, ESRF |
| Wavelength (Å) | 0.96598 | 0.96598 |
| Temperature (K) | 100 | 100 |
| Detector | PILATUS 2M | PILATUS 2M |
| Crystal-to-detector distance (mm) | 213.57 | 234.93 |
| Rotation range per image (°) | 0.1 | 0.2 |
| Total rotation range (°) | 160 | 180 |
| Exposure time per image (s) | 0.2 | 0.2 |
| a, b, c (Å) | 88.7, 29.1, 83.9 | 29.7, 47.1, 88.5 |
| α, β, γ (°) | 90.0, 114.2, 90.0 | 102.6, 93.9, 108.3 |
| Mosaicity (°) | 0.56 | 0.32 |
| Resolution range (Å) | 25.0–1.85 (1.89–1.85) | 43.3–1.75 (1.78–1.75) |
| Total No. of reflections | 40259 | 70188 |
| No. of unique reflections | 15362 | 40054 |
| Completeness (%) | 89.8 (87.8) | 90.4 (77.8) |
| Multiplicity | 2.6 (2.5) | 1.8 (1.6) |
| 〈I/σ(I)〉 | 8.3 (2.2) | 9.6 (2.0) |
| R meas (%) | 9.7 (52.5) | 8.5 (49.4) |
| CC1/2 † | 0.992 (0.740) | 0.997 (0.760) |
| Overall B factor from Wilson plot (Å2) | 20.7 | 16.2 |
Diederichs & Karplus (2013 ▸).
2.4. Structure solution and refinement
The molecular-replacement (MR) method was employed to solve the structure using the atomic coordinates of PfTrpF (sequence identity 58%; PDB entry 4aaj) and the C2 space group in Phaser (McCoy et al., 2007 ▸). A search model was created using Sculptor (Bunkóczi & Read, 2011 ▸) and a single solution was found with a Z-score of 16.4. Refinement was carried out using simulated annealing as implemented in PHENIX (Adams et al., 2010 ▸). The progress of refinement was monitored by R free (using 5% of reflections that were excluded from refinement) and the inspection of σA-weighted 2|F o| − |F c| and |F o| − |F c| electron-density maps in Coot (Emsley & Cowtan, 2004 ▸). When the R free reached ∼30% the structure was used as a search model against the P1 data set. Molecular replacement produced a single solution. Refinement and model building were carried out as in the case of the C2 crystal data except that NCS restraints were applied in the initial stages of refinement. TLS refinement (Painter & Merritt, 2006 ▸) was carried out in the final stages of the refinement in both space groups. Ions and water molecules were assigned using PHENIX and inspected in the graphics for their chemical environment and residual positive or negative electron density in |F o| − |F c| maps. Refinement statistics for both the P1 and C2 data sets are shown in Table 4 ▸.
Table 4. Structure solution and refinement.
Values in parentheses are for the outer shell.
| Space group | C2 | P1 |
|---|---|---|
| Resolution range (Å) | 25.00–1.85 (1.89–1.85) | 43.28–1.75 (1.79–1.75) |
| Completeness (%) | 90.0 (88.0) | 90.0 (79.0) |
| σ Cutoff | 0 | 0 |
| No. of reflections, working set | 14586 | 38041 |
| No. of reflections, test set | 755 | 2008 |
| Final R cryst (%) | 21.9 (31.8) | 17.3 (25.1) |
| Final R free (%) | 26.2 (42.1) | 21.4 (33.2) |
| No. of non-H atoms | ||
| Protein | 1613 | 3266 |
| Ion | 2 [Na+] | 4 [2 Cl−, 2 Na+] |
| Water | 182 | 622 |
| Total | 1797 | 3892 |
| R.m.s. deviations | ||
| Bonds (Å) | 0.007 | 0.007 |
| Angles (°) | 1.05 | 0.82 |
| Average B factors (Å2) | ||
| Overall | 29.6 | 24.4 |
| Protein | 28.9 | 22.5 |
| Ion | 51.2 | 20.1 |
| Water | 34.6 | 34.4 |
| Ramachandran plot | ||
| Most favoured (%) | 94.1 | 96.0 |
| Allowed (%) | 5.4 | 3.8 |
| Outliers | 0.5 | 0.2 |
2.5. Structure analysis
Solvent-inaccessible charged residues, solvation energy, accessible solvent area (ASA), dimer and crystal-packing interfaces were analyzed using the Protein Interactions, Surfaces and Assemblies (PISA) server (http://www.ebi.ac.uk/msd-srv/prot_int/; Krissinel & Henrick, 2007 ▸). Secondary Structure Matching (SSM; Krissinel & Henrick, 2004 ▸) as implemented in Coot was used to superimpose homologous structures onto each other. Salt bridges were calculated by ESBRI (Costantini et al., 2008 ▸).
2.6. Size-exclusion chromatography
Size-exclusion gel chromatography (Supplementary Fig. S2) to determine the molecular mass and subunit number of TkTrpF was performed using a Superdex 200 5/150 column equilibrated with 100 mM NaCl in 10 mM Tris–HCl pH 7.2. The standard curve was obtained with carbonic dehyrogenase (29 kDa), bovine serum albumin (66 kDa), alcohol dehydrogenase (150 kDa) and β-amylase (200 kDa).
3. Results and discussion
3.1. Production of soluble TkTrpF in E. coli and purification
Production of soluble recombinant TkTrpF was successful when cells harbouring TkTrpF-pET28a(+) were allowed to express at 290 K after induction with IPTG. It has been reported that expression at low temperatures leads to increased stability and correct folding patterns, which is owing to the fact that hydrophobic interactions determine inclusion-body formation (Vera et al., 2007 ▸). Increased soluble expression and activity at lower growth temperatures have also been associated with increased expression of a number of chaperones in E. coli (Ferrer et al., 2003 ▸). Moreover, growth in the temperature range 288–296 K leads to a significant reduction in degradation of the expressed protein (Hunke & Betton, 2003 ▸). Based on the thermostability of recombinant TkTrpF, the first step of purification was heat treatment at 338 K for 25 min, which resulted in the precipitation and removal of most of the heat-labile proteins from E. coli. TkTrpF was then purified to homogeneity using an Ni2+-charged Sepharose column.
3.2. Crystallization
The TkTrpF crystals in space group C2 have unit-cell parameters a = 88.7, b = 29.1, c = 83.9 Å, β = 114.2°. Assuming the presence of one molecule in the asymmetric unit, the Matthews coefficient V M (Matthews, 1968 ▸) is 2.15 Å3 Da−1, corresponding to a solvent content of ∼43%. The unit-cell parameters for the P1 crystals are a = 29.7, b = 47.1, c = 88.5 Å, α = 102.6, β = 93.9, γ = 108.3°. For two molecules in the asymmetric unit, the Matthews coefficient V M is 2.32 Å3 Da−1, corresponding to a solvent content of ∼47%.
3.3. Overall structure
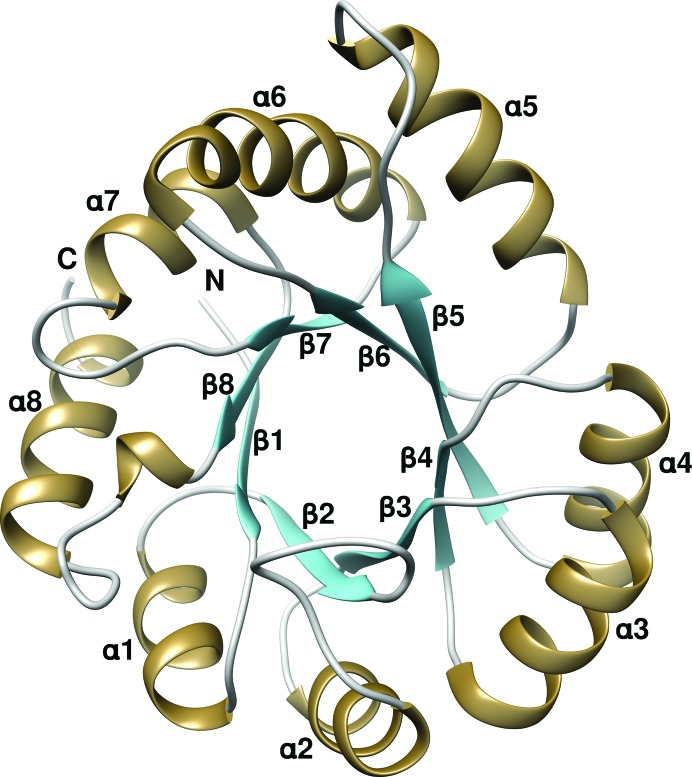
Owing to the slightly higher resolution of the P1 data set, structure analysis was carried out with the P1 crystal form, except where stated otherwise. The overall structure of TkTrpF adopts the typical (βα)8-barrel (or TIM-barrel) architecture (Wierenga, 2001 ▸) and is similar to those of other reported TrpF enzymes (Fig. 1 ▸). The refined model consists of two molecules in the asymmetric unit with a total of 417 residues. Both subunits exhibit the same structure without significant changes, as shown by the low (0.19 Å) root-mean-square deviation (r.m.s.d.) in Cα-atom positions after structural superposition. All residues in both subunits are visible in the electron density. Electron density at the N-terminus of the B subunit of the dimer allowed a His residue from the N-terminal linker region to be placed at position 0 before the starting Met residue. Alternate conformations were found at Met202 and Met49 in subunit A and Met49, Val192 and Met202 in subunit B. The refined structure of TkTrpF in space group C2 consists of one molecule of 208 residues in the asymmetric unit. Structural comparison of the TkTrpF structures in both space groups shows high similarity and no major differences (r.m.s.d. of 0.32 Å).
Figure 1.
Crystal structure of TkTrpF. The ribbon representation was created by Chimera (Pettersen et al., 2004 ▸). The alternating α-helices and β-strands of the TIM barrel are labelled.
3.4. Structural comparison
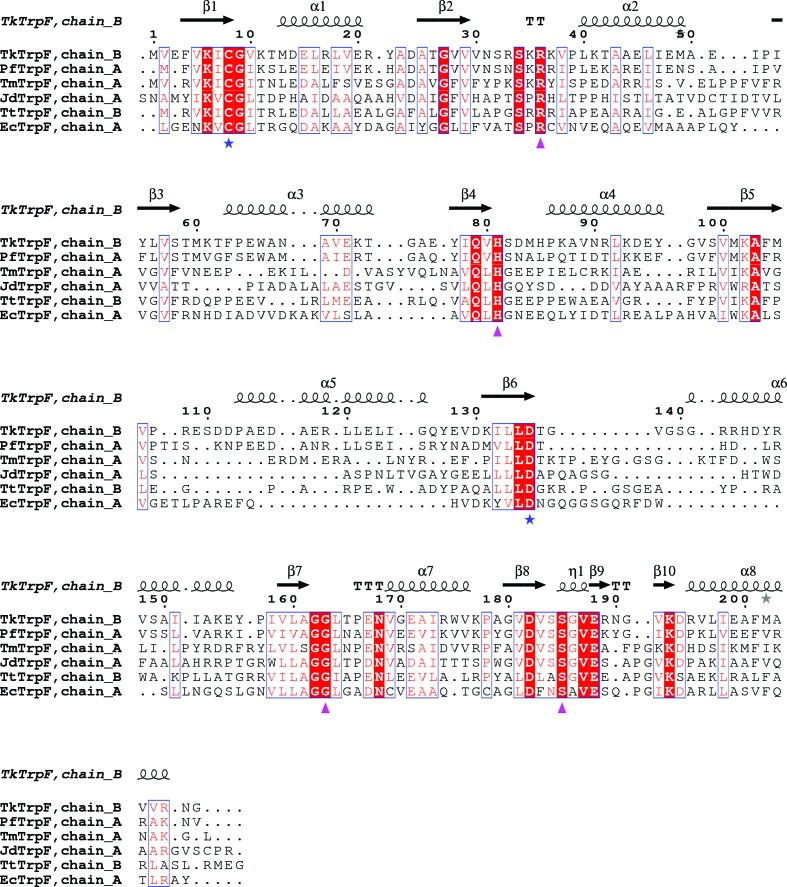
Structure-based sequence alignment of TkTrpF with other TrpF structures (Fig. 2 ▸) shows the highest sequence identity to PfTrpF (58%), followed by TmTrpF (35%), JdTrpF (32%), TtTrpF (29%) and EcTrpF (residues 254–452; 28%). JdTrpF and EcTrpF are produced by mesophiles, whilst the others are produced by thermophiles. Like all other TrpF structures deposited in the PDB to date, the β-strands are more conserved than the α-helices in the (βα)8-barrel structure.
Figure 2.
Structure-based sequence alignment of TkTrpF homologues. Conserved residues are indicated by white letters on a red background (strictly conserved) or red letters on a white background (global similarity score >0.7) and are framed in blue boxes. Residues involved in catalytic activity are indicated by blue stars and substrate-binding residues by magenta triangles. The sequences used in the alignment are those from Pyrococcus furiosus (PfTrpF; PDB entry 4aaj), Thermotoga maritima (TmTrpF; PDB entry 1nsj), Jonesia denitrificans DSM 20603 (JdTrpF; PDB entry 4wui), Thermus thermophilus HB8 (TtTrpF; PDB entry 1v5x) and Escherichia coli (EcTrpF; residues 254–452 in PDB entry 1pii). The figure was created using ESPript (v.3.0; Robert & Gouet, 2014 ▸).
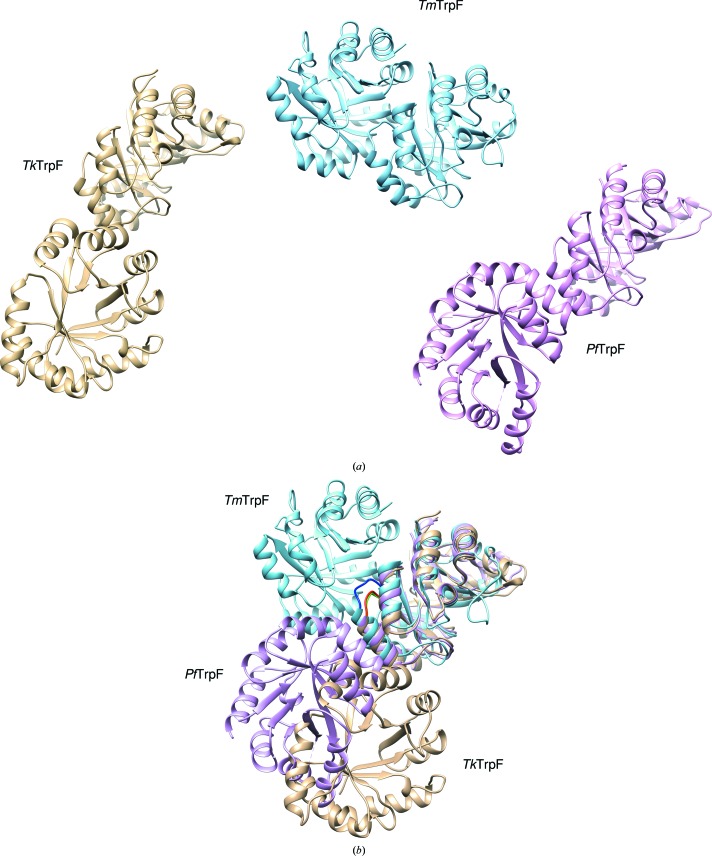
The core r.m.s.d. between TmTrpF and TkTrpF is 2.2 Å. A long loop (residues 106–112) in TkTrpF is found to be shorter in TmTrpF (107–108). Two large helices found in TkTrpF (α5, Pro113–Gln126; α6, Gly139–Glu155) show structural variations in their length compared with other dimeric TrpFs such as TmTrpF and TtTrpF. In monomeric PfTrpF the α5 helix is present but α6 (Arg145–Lys153) is short. In mesophilic EcTrpF and JdTrpF the α5 and α6 helices are missing. In EcTrpF a loop replaces helix α5, whilst helix α6 is reduced to a 310-helix (Trp391–Leu394). In JdTrpF both helices (α5 and α6) are replaced by loops. TkTrpF forms a dimer in the P1 crystal form but the interface is different from that in other reported dimeric TrpFs (TtTrpF and TmTrpF) (Fig. 3 ▸). This could be attributed to the shorter loop in TkTrpF (50–53; AEIP) compared with the corresponding loops in TmTrpF (50–54; LPPFV) and TtTrpF (49–53; LGPFV), which are implicated in normal dimer formation. In addition, the conserved residues Pro51 and Phe52, which are involved in subunit–subunit interaction in other thermophilic dimeric TrpFs, are replaced by Ala and Glu in TkTrpF. The complexation index as calculated by PDBePISA is 0 for the TkTrpF dimer found in the P1 crystal form, suggesting that the interface is unlikely to exist in solution and is probably a crystallographic artefact. This is also in agreement with the size-exclusion chromatography analysis, which suggests a TkTrpF monomer in solution. In the C2 crystal form there is one molecule in the asymmetric unit. Examination of the symmetry molecules for potential dimers revealed no dimers similar to the normal dimers found in other TrpF enzymes. However, a symmetry molecule is able to generate the dimer found in the P1 crystal form of TkTrpF, suggesting a possible rearrangement of the crystal lattice during cryocooling of the crystals (Skrzypczak-Jankun et al., 2006 ▸) that could explain the two different space groups found.
Figure 3.
Structural comparison of TkTrpF (brown) with PfTrpF (magenta; PDB entry 4aaj) and TmTrpF (cyan; PDB entry 1nsj). (a) Dimers of TkTrpF, TmTrpF and PfTrpF are shown separately using the same orientation of subunit A. The PfTrpF dimer was generated from a symmetry molecule (Repo et al., 2012 ▸). (b) Structural superposition of subunit A. The different orientation of subunit B is shown. TkTrpF and PfTrpF have different dimeric packing to the normal dimer found in TmTrpF. The loop which is essential for dimer formation in TmTrpF is shown in blue and the corresponding loops in TkTrpF and PfTrpF are depicted in green and red, respectively.
Structural comparison by superposition of the TkTrpF dimer found in the crystals with the TmTrpF dimer and the crystallographically generated dimeric form of PfTrpF shows different dimer interfaces from the normal functional dimeric TrpFs (Fig. 3 ▸), where TmTrpF is a dimer whereas TkTrpF and TmTrpF are monomers in solution. The PfTrpF subunit interface is different from that of TmTrpF and TkTrpF, with the second barrel subunit occupying an intermediate position when the structures are superimposed. The dimer crystal-packing interface in TkTrpF and PfTrpF most likely plays no functional role. However, the use of the same region in each barrel molecule to make contacts with a second molecule and the different orientations may provide some insights into dimer interfaces and their evolution to transform nonfunctional dimers into functional dimers.
3.5. Active site
TkTrpF performs an Amadori rearrangement involving general acid–base catalysis. In TmTrpF, Cys7 and Asp126 were found to be essential active-site residues, with Cys7 acting as the general base and Asp126 as the general acid (Henn-Sax et al., 2002 ▸). The corresponding residues in TkTrpF are Cys8 and Asp135 at the C-terminal end of the β1 strand and at the end of the β6 strand, respectively. Arg36 and His83 in TmTrpF are known to be involved in binding CdRP, where Arg36 forms a salt bridge with the carboxylic group of the anthranilate moiety of CdRP and His83 forms a hydrogen bond to CdRP (Henn-Sax et al., 2002 ▸). The equivalent residues in TkTrpF are Arg36 and His81 found in the loop following α2 and at the C-terminus of β4, respectively.
In TmTrpF, there are eight hydrogen bonds between residues (Gly158, Gly159, Ser180, Ser181 and Gly182) and the phosphate group of PRA, while in EcTrpF Gly407 and Ser428 were involved in the formation of four hydrogen bonds to the phosphate of PRA (Hennig et al., 1997 ▸). The corresponding residues in TkTrpF are Ser185 and Gly163. Ser185 is found in a small helical area before helix α8 and Gly163 is found in a loop between α7 and β7. All of these residues involved in catalysis and substrate binding (Cys8, Asp135, Arg36 and His81, Ser185 and Gly163; residue numbering refers to TkTrpF) are found to be conserved in all PDB-deposited TrpF structures. In general, residues involved in catalysis are found at the C-termini of strands β1 and β6 and those involved in phosphate binding between helices α7 and α8.
3.6. Structural basis of TkTrpF thermostability
Protein thermostability has been attributed to various factors (Berezovsky & Shakhnovich, 2005 ▸; Kumar et al., 2000 ▸; Szilágyi & Závodszky, 2000 ▸; Suhre & Claverie, 2003 ▸; Unsworth et al., 2007 ▸). An analysis applied to TkTrpF is presented in Table 5 ▸. The percentages of various amino acids known to affect thermostability was investigated. Proline residues, for example, have been found to increase protein thermostability by decreasing the entropy of the unfolded state and conferring rigidity (Vieille & Zeikus, 2001 ▸). Glycines, on the other hand, can adopt various conformations and therefore their presence increases flexibility. The Pro:Gly ratio was calculated following similar calculations as used for PfTrpF (Repo et al., 2012 ▸). The number of Pro residues is less than that of Gly residues (a ratio of <1) in TkTrpF and PfTrpF, whilst a ratio of 1 was identified in thermophilic TrpFs and mesophilic JdTrpF and a ratio of 0.24 was identified in EcTrpF. Hence, no role of the ratio of Pro and Gly residues in TrpF thermostability can be deduced. The aliphatic amino acid Val is found to be preferred in hyperthermophiles, whereas the small uncharged nonpolar amino acid alanine is found in a higher proportion in mesophiles and is avoided in hyperthermophiles (Suhre & Claverie, 2003 ▸). A similar trend has also been found in TrpFs. No preference was found amongst the different TrpFs in the total number of hydrophobic residues, suggesting that hydrophobicity plays no role in TrpF thermostability. In fact, the number of hydrophobic residues in hyperthermophilic TrpFs is lower than that in thermophilic and mesophilic TrpFs. A strong preference for the use of charged residues (Asp, Glu, Lys and Arg) has been shown in hyperthermophilic proteins. Acidic residues (Asp + Glu) were found in the highest proportion in TkTrpF among all of the TrpF structures currently deposited in the PDB. The amount of Lys + Arg residues is also elevated in TkTrpF, as in other thermophiles. In contrast, mesophilic TrpFs lag behind thermophiles in the number of these positively charged residues. Salt bridges have also been suggested to contribute to the elevated stability of proteins. In TkTrpF the number of intra-chain salt bridges within a 3.5 Å cutoff limit is greater than in other TrpFs, presumably as a result of the increased number of charged residues. Finally, an additional mechanism of stabilization compared with other TrpFs is found in TkTrpF. Helical content, which is known to be associated with thermostability of proteins (Kumar et al., 2000 ▸), is higher in TkTrpF and PfTrpF compared with all other reported TrpFs. Both the α5 and α6 helices are longer in TkTrpF compared with thermophilic TrpFs and both of these helices are absent in mesophiles. Thus, the higher helical content in hyperthermophilic TrpFs may also contribute to their enhanced thermostability.
Table 5. Comparison of the TkTrpF structure with other TrpF structures.
Amino-acid calculations were based on sequence data. Surface-exposed residues were calculated using PDBePISA (Krissinel & Henrick, 2007 ▸) and charged residues using ProtParam at the ExPASy portal (Gasteiger et al., 2005 ▸). Helical content was calculated by PDBSUM (Laskowski et al., 2005 ▸).
| Protein | TkTrpF | PfTrpF | TmTrpF | TtTrpF | EcTrpF | JdTrpF |
|---|---|---|---|---|---|---|
| Oligomerization state in solution | Monomer | Monomer | Dimer | Dimer | Monomer | Monomer |
| Amino-acid residues | 208 | 200 | 205 | 200 | 199 | 205 |
| Surface-exposed residues | 185 (88.9%) | 181 (90.5%) | 179 (87.3%) | 184 (92.0%) | 174 (87.4%) | 180 (87.8%) |
| Pro:Gly | 0.6 | 0.72 | 1.0 | 1.0 | 0.24 | 1.0 |
| Asp + Glu | 32 (15.3%) | 27 (13.5%) | 29 (14.1%) | 26 (13.0%) | 22 (11.1%) | 19 (9.3%) |
| Lys + Arg | 28 (13.4%) | 29 (14.5%) | 28 (13.7%) | 26 (13.0%) | 15 (7.5%) | 12 (5.8%) |
| Val | 28 (13.4%) | 29 (14.5%) | 23 (11.2%) | 14 (7.0%) | 19 (9.5%) | 19 (9.3%) |
| Ala | 19 (9.1%) | 16 (8.0%) | 13 (6.3%) | 34 (17.0%) | 28 (14.1%) | 35 (17.0%) |
| Hydrophobic residues (Ala + Pro + Met + Val + Leu + Trp + Phe) | 56 (26.8%) | 56 (26.5%) | 58 (28.4%) | 92 (45.6%) | 65 (32.7%) | 77 (37.2%) |
| ASA (Å2) | 10260 | 9720 | 9980 | 9230 | 9100 | 9240 |
| Active dimer interface (Å2) | NA | NA | 990 | 1240 | NA | NA |
| Complexation index | NA | NA | 0.49 | 1 | NA | NA |
| Intra-chain salt bridges | 14 | 7 | 13 | 5 | 9 | 2 |
| Helical content (%) | 42.3 | 42.0 | 32.0 | 31.0 | 33.1 | 28.8 |
4. Conclusions
The structure of TkTrpF was solved by X-ray crystallography in two different space groups (C2 and P1), with one and two molecules in the crystallographic asymmetric unit, using data to 1.85 and 1.75 Å resolution, respectively. TkTrpF belongs to the family of TIM-barrel proteins. Crystallographic analysis suggested that TkTrpF is unable to form a dimer similar to that found in dimeric TrpF enzymes owing to a shorter loop. TkTrpF shows the highest structural similarity to monomeric PfTrpF. TkTrpF is the second example of a TrpF enzyme from a hyperthermophilic archaeon after PfTrpF. TkTrpF, similar to PfTrpF, also exists in a monomeric form, suggesting that dimer formation is not required for improved thermostability in hyperthermophilic archaea. An elevated number of charged residues and salt bridges, and a higher helical content are suggested to increase the thermostabiliy of TkTrpF.
Supplementary Material
PDB reference: TkTrpF, space group C2, 5lhe
PDB reference: space group P1, 5lhf
Supporting Information: Supplementary Figures S1 and S2.. DOI: 10.1107/S2053230X16015223/hv5338sup1.pdf
Acknowledgments
We thank Juha Määttä for size-exclusion chromatography measurements. Financial support to SP from the Erasmus–Mundus student exchange program is acknowledged. Biocenter Finland is thanked for infrastructure support.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Atomi, H., Fukui, T., Kanai, T., Morikawa, M. & Imanaka, T. (2004). Archaea, 1, 263–267. [DOI] [PMC free article] [PubMed]
- Bauerle, R., Hess, J. & French, S. (1987). Methods Enzymol. 142, 366–386. [DOI] [PubMed]
- Berezovsky, I. N. & Shakhnovich, E. I. (2005). Proc. Natl Acad. Sci. USA, 102, 12742–12747. [DOI] [PMC free article] [PubMed]
- Bradford, M. M. (1976). Anal. Biochem. 72, 248–254. [DOI] [PubMed]
- Braus, G. H. (1991). Microbiol. Rev. 55, 349–370. [DOI] [PMC free article] [PubMed]
- Bunkóczi, G. & Read, R. J. (2011). Acta Cryst. D67, 303–312. [DOI] [PMC free article] [PubMed]
- Cohn, W. & Crawford, I. P. (1976). J. Bacteriol. 127, 367–379. [DOI] [PMC free article] [PubMed]
- Costantini, S., Colonna, G. & Facchiano, A. M. (2008). Bioinformation, 3, 137–138. [DOI] [PMC free article] [PubMed]
- Diederichs, K. & Karplus, P. A. (2013). Acta Cryst. D69, 1215–1222. [DOI] [PMC free article] [PubMed]
- Egan, A. F. & Gibson, F. (1972). Biochem. J. 130, 847–859. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Enatsu, T. & Crawford, I. P. (1971). J. Bacteriol. 108, 431–438. [DOI] [PMC free article] [PubMed]
- Evans, P. R. & Murshudov, G. N. (2013). Acta Cryst. D69, 1204–1214. [DOI] [PMC free article] [PubMed]
- Ferrer, M., Chernikova, T. N., Yakimov, M. M., Golyshin, P. N. & Timmis, K. N. (2003). Nature Biotechnol. 21, 1266–1267. [DOI] [PubMed]
- Fukui, T., Atomi, H., Kanai, T., Matsumi, R., Fujiwara, S. & Imanaka, T. (2005). Genome Res. 15, 352–363. [DOI] [PMC free article] [PubMed]
- Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D. & Bairoch, A. (2005). The Proteomics Protocols Handbook, edited by J. M. Walker, pp. 571–607. Totowa: Humana Press.
- Hennig, M., Sterner, R., Kirschner, K. & Jansonius, J. N. (1997). Biochemistry, 36, 6009–6016. [DOI] [PubMed]
- Henn-Sax, M., Thoma, R., Schmidt, S., Hennig, M., Kirschner, K. & Sterner, R. (2002). Biochemistry, 41, 12032–12042. [DOI] [PubMed]
- Hoch, S. O. (1979). J. Bacteriol. 139, 362–368. [DOI] [PMC free article] [PubMed]
- Hommel, U., Eberhard, M. & Kirschner, K. (1995). Biochemistry, 34, 5429–5439. [DOI] [PubMed]
- Hunke, S. & Betton, J.-M. (2003). Mol. Microbiol. 50, 1579–1589. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Krissinel, E. & Henrick, K. (2004). Acta Cryst. D60, 2256–2268. [DOI] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Kumar, S., Tsai, C.-J. & Nussinov, R. (2000). Protein Eng. Des. Sel. 13, 179–191. [DOI] [PubMed]
- Laskowski, R. A., Chistyakov, V. V. & Thornton, J. M. (2005). Nucleic Acids Res. 33, D266–D268. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Painter, J. & Merritt, E. A. (2006). J. Appl. Cryst. 39, 109–111.
- Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C. & Ferrin, T. E. (2004). J. Comput. Chem. 25, 1605–1612. [DOI] [PubMed]
- Potts, J. M. & Drapeau, G. R. (1972). J. Bacteriol. 111, 334–339. [DOI] [PMC free article] [PubMed]
- Repo, H., Oeemig, J. S., Djupsjöbacka, J., Iwaï, H. & Heikinheimo, P. (2012). Acta Cryst. D68, 1479–1487. [DOI] [PubMed]
- Robert, X. & Gouet, P. (2014). Nucleic Acids Res. 42, W320–W324. [DOI] [PMC free article] [PubMed]
- Skrzypczak-Jankun, E., Borbulevych, O. Y., Zavodszky, M. I., Baranski, M. R., Padmanabhan, K., Petricek, V. & Jankun, J. (2006). Acta Cryst. D62, 766–775. [DOI] [PubMed]
- Sterner, R., Kleemann, G. R., Szadkowski, H., Lustig, A., Hennig, M. & Kirschner, K. (1996). Protein Sci. 5, 2000–2008. [DOI] [PMC free article] [PubMed]
- Suhre, K. & Claverie, J.-M. (2003). J. Biol. Chem. 278, 17198–17202. [DOI] [PubMed]
- Svensson, O., Malbet-Monaco, S., Popov, A., Nurizzo, D. & Bowler, M. W. (2015). Acta Cryst. D71, 1757–1767. [DOI] [PMC free article] [PubMed]
- Szilágyi, A. & Závodszky, P. (2000). Structure, 8, 493–504. [DOI] [PubMed]
- Taka, J., Ogasahara, K., Jeyakanthan, J., Kunishima, N., Kuroishi, C., Sugahara, M., Yokoyama, S. & Yutani, K. (2005). J. Biochem. 137, 569–578. [DOI] [PubMed]
- Thoma, R., Hennig, M., Sterner, R. & Kirschner, K. (2000). Structure, 8, 265–276. [DOI] [PubMed]
- Unsworth, L. D., van der Oost, J. & Koutsopoulos, S. (2007). FEBS J. 274, 4044–4056. [DOI] [PubMed]
- Vera, A., González-Montalbán, N., Arís, A. & Villaverde, A. (2007). Biotechnol. Bioeng. 96, 1101–1106. [DOI] [PubMed]
- Vieille, C. & Zeikus, G. J. (2001). Microbiol. Mol. Biol. Rev. 65, 1–43. [DOI] [PMC free article] [PubMed]
- Wierenga, R. K. (2001). FEBS Lett. 492, 193–198. [DOI] [PubMed]
- Wilmanns, M., Priestle, J. P., Niermann, T. & Jansonius, J. N. (1992). J. Mol. Biol. 223, 477–507. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: TkTrpF, space group C2, 5lhe
PDB reference: space group P1, 5lhf
Supporting Information: Supplementary Figures S1 and S2.. DOI: 10.1107/S2053230X16015223/hv5338sup1.pdf