Abstract
The ability of macrophages to properly migrate is crucial to their success as early responders during the innate immune response. Furthermore, improper regulation of macrophage migration is known to contribute to several pathologies. The signaling mechanisms underlying macrophage migration have been previously studied but to date the mechanical mechanism of macrophage migration has not been determined. In this study, we have created the first traction maps of motile primary human macrophages by observing their migration on compliant polyacrylamide gels. We find that the force generated by migrating macrophages is concentrated in the leading edge of the cell – so-called frontal towing - and that the magnitude of this force is dependent on the stiffness of the underlying matrix. With the aid of chemical inhibitors, we show that signaling through the RhoA kinase ROCK, myosin II, and PI3K is essential for proper macrophage force generation. Finally, we show that Rac activation by its GEF Vav1 is crucial for macrophage force generation while activation through its GEF Tiam1 is unnecessary.
Introduction
Macrophages play an important role in the innate immune response by clearing pathogens through phagocytosis and activating the adaptive immune response through cytokine production and antigen-presentation. In order to perform these functions, macrophages must be able to efficiently migrate to sites of infection. Improper regulation of macrophage function has been linked to several diseases including atherosclerosis, rheumatoid arthritis, and cancer [1]; therefore, it is crucial that we develop a better understanding of the mechanisms underlying macrophage migration. Previous work on macrophage migration has investigated the role of signaling molecules on chemokinesis and chemotaxis[2], but to our knowledge no group has studied the spatio-temporal regulation of forces during macrophage migration.
Cellular traction forces have been shown to be important for cell adhesion [3, 4], spreading [5], motility [3, 6], and extra-cellular matrix remodeling [7]. To effectively migrate on and through tissues anchorage-dependent cells must attach to their underlying substrate and generate traction against that substrate. In the towing model of cell motility the cell extends a lamellipodia and attaches to the underlying substrate through integrin binding to the extra cellular matrix. The cell then contracts which exerts traction on its underlying substrate and generates strong cellular forces at the leading edge of the cell. This contraction is ultimately followed by the release of the cell’s uropod and the forward motility of the cell [8]. The towing model has been shown to broadly apply to large contractile cells such as endothelial cells and fibroblasts [6]. The spatial distribution of forces of mesenchymal cells migrating on compliant surfaces has been studied in depth [3, 5, 6]; but despite the importance of immune cell motility relatively little work has been done to characterize the mechanical mechanisms behind the motility of immune cells.
Leukocyte motility differs from mesenchymal cell motility in several ways. Leukocytes are fast moving cells that migrate with low persistence. In order to achieve their high speed, leukocytes form weak, short-lived adhesions to their substratum. They do not form focal adhesions. This is in contrast to mesenchymal cells which form strong focal adhesions to their surface and contain stress fibers that allow for large cellular contractions [8]. Our laboratory has embarked on an effort to categorize the spatio-temporal distribution of forces exhibited by all the motile cells of the immune system. Previously, we showed that neutrophils achieve motility not through frontal towing but rather by tail-contraction or rearward-squeezing [9, 10]. In this mode of motility, the traction forces are concentrated in the cell’s uropod and the cell is pushed forward through a “squeezing” mechanism that is dependent on myosin activity. The traction stresses generated by neutrophils were found to be small compared to those generated by mesenchymal cells, which is consistent with their need to move quickly toward targets. Further work by our lab went on to show that this mode of motility is not shared by all leukocytes. Dendritic cells, which are of the monocytic lineage, display maximal stresses at the leading edge of cell indicating they use the towing model of migration. The forces displayed during dendritic cell migration were even weaker than those generated by neutrophils [11]. The contrast among the behaviors of leukocytes has thus far illustrated the importance of studying the force generation of each individual cell type individually since no one mode of motility is shared by all leukocytes.
The use of polyacrylamide gels coupled with embedded marker beads has proven a powerful method for traction force microscopy [3, 6]. Polyacrylamide hydrogels are optically clear and non-toxic which allows for effective cell culture and imaging. Furthermore, they are elastic and can be easily tuned to a variety of stiffnesses, allowing an understanding of how stiffness affects force generation. During cell migration images are taken of the cell and the embedded beads. After the experiment, cells are then removed from the gel and an image is taken of the unstressed bead locations. The tractions applied on the gel can then be calculated from the displacement of the beads from the unstressed position using elasticity theory. Our lab has used this technology to measure the traction stresses of cells undergoing adhesion and spreading as well as leukocyte migration [5, 9, 10, 12, 13].
In this study we have used traction force microscopy (TFM) to determine the force generation profile of macrophages migrating on compliant surfaces. To our knowledge, this is the first measurement of force generation for motile macrophages. We sought to determine the type of motility employed by macrophages and which signaling molecules are most important for macrophage force generation. Our results indicate that macrophages use a towing mode of motility with the strongest forces concentrated at the leading edge. We have also shown that the magnitude of force generation is dependent on the stiffness of the underlying substrate. Furthermore, we have determined using a range of chemical inhibitors that like other leukocytes, macrophage force generation is dependent on signaling through PI3K, RhoA, and myosin II. Finally, we have shown that Rac signaling is critical for force generation when Rac is activated by Vav1 but not when Rac is activated by Tiam1; these results illustrate the complexity of signaling that occurs upstream of force generation.
Materials and Methods
Reagents
Bovine fibronectin and recombinant human M-CSF were obtained from Sigma (St. Louis, MO). We used the inhibitors Y27632[14] at 10μM from Millipore (Billerica, MA), Blebbistatin[15] at 20 μM from Sigma (St. Louis, MO), LY294002[16] at 50 μM from Cell Signaling (Boston, MA), NSC23766[17] at 50 μM from Millipore (San Diego, CA), 6-thio-GTP[18] at 10 μM from Jena Bioscience (Jena, Germany).
Isolation of Monocytes
Whole blood was obtained from healthy human donors by venipuncture and collected in BD Vacutainer tubes containing sodium heparin as an anticoagulant (BD Biosciences, San Jose, CA). Samples were collected with University of Pennsylvania Institutional Review Board approval from consenting adult volunteers. Blood samples were layered in a 1:1 ratio of whole blood to the density gradient 1-Step Polymorphprep (Axis-Shield, Oslo, Norway). Vials were centrifuged at 1500 rpm for 40 minutes and the mononuclear band was collected into a fresh vial.
Differentiation and Cell Culture of Macrophages
Cells were allowed to adhere to sterile non-tissue culture treated dishes in RPMI-1640 supplemented with 10% heat-inactivated FBS overnight. Non-adhered cells were removed and washed with PBS. Adherent monocytes were then differentiated for seven days in RMPI-1640 supplemented with 10% heat-inactivated FBS and 2ng/mL M-CSF (Sigma, St. Louis, MO). Cells were used for experimentation 7–12 days following the start of differentiation.
Surface Preparation
Coverslips (No 1, 45 × 50 mm, Fisher Scientific, Pittsburgh, PA) were chemically activated in preparation for covalent attachment of polyacrylamide gels using a method adapted from the protocol by Pelham and Wang. Briefly, coverslips were washed for 4 hours in 0.2 M hydrogen chloride then rinsed several times with distilled water. They were then neutralized with 0.1 M sodium hydroxide for 30 minutes and rinsed with distilled water. Coverslips were incubated on an orbital shaker in 3-aminopropyl trimethoxysilane 0.5% for 30 minutes and rinsed with distilled water. They were then activated with 0.5% glutaraldehyde for at least 1 hour. The coverslips were then air dried overnight.
Synthesis of the Bifunctional Linker
N-6-((acryloyl)amino)hexanoic acid (N-6) was synthesized using the method described by Pless et al. The N-6 copolymerizes in the acrylamide to form a reactive polyacrylamide gel. The N-6 contains an n-succinimidyl ester that is displaced by a primary amine to link the amine-containing ligand, such as fibronectin, to the polyacylamide gel.
Gel Synthesis
Acrylamide solutions were prepared containing acrylamide (40% w/v solution), n, n’-methylene-bis-acrylamide (2% w/v solution), n’-tetramethylethylene di-amine, and ammonium persulfate from Bio-Rad Laboratories (Hercules, CA). Additionally, the gels contained 0.25M HEPES, buffered to pH 8, 5.6mg of N6 dissolved in ethanol, distilled water, and carboxylate-modified fluorescent latex beads (0.5μm Fluorospheres, Molecular Probes, Eugene, OR). The concentrations of acrylamide and bis were varied to control the mechanical properties of the hydrogel.
A drop of gel solution was dispensed onto a Rainex-coated 18mm glass coverslip. A second, activated rectangular coverslip was placed on top of the gel droplet to flatten the solution; the assembly was polymerized in an inverted position to allow beads to settle to the top surface of the gel. The gels were polymerized under nitrogen for 45 minutes. The top coverslip was gently peeled away leaving a thin gel immobilized on the activated coverslip. Gels were rinsed with distilled water and incubated with 5μg/mL fibronectin in 50mM HEPES buffer overnight. Unreacted N-6 was blocked with 1:100 ethanolamine in 50mM HEPES for 30 minutes and stored in 1 x PBS at 4°C for up to two weeks.
Traction Force Microscopy of Migrating Macrophages
Traction force microscopy has been described previously [6]. Briefly, traction forces were determined based on deformations in the polyacrylamide substrate relative to the relaxed substrate as detected by movements of 0.5-μm beads embedded in the gel.
Primary human macrophages were plated on fibronectin-coated polyacrylamide gels at 1×104 cells/mL and incubated for 1 hour at 37°C. Unattached cells were washed away and fresh RPMI with 20ng/mL human M-CSF was added to the gel chamber to promote migration [19]. Phase contrast images of the cell were taken every 10 minutes during cell migration. Directly after a phase image was taken, a corresponding fluorescent image of the beads embedded beneath the cell was taken. Images were taken over a 4 hour period. At the end of migration the cells were removed using 0.5% SDS and an image of the beads in their unstressed state was taken. Using custom-written LIBTRC software, the bead displacements within the gel were calculated, the cell and nucleus were drawn, and a mesh that fits within the outline of the cell was created. Using the bead displacements and the material properties of the gel, the most likely surface traction vectors were calculated using the technique described by Dembo and Wang.
The overall force, |F|, exerted by the cell on its substrate, is an integral of the traction field magnitude over the area, , where T(x, y) = [Tx(x, y), Ty(x, y)] is the continuous field of traction vectors defined at any spatial position (x, y) within the cell.
Use of Pharmaceutical Inhibitors of Macrophage Motility
In experiments in which macrophages were treated with a chemical inhibitor, macrophages were seeded on 10,400 Pa gels functionalized with 5 μg/mL fibronectin and allowed to adhere for one hour. The cells were then washed and fresh media containing the correct concentration of inhibitor was applied. The macrophages were incubated for one hour to allow complete inhibition. M-CSF was then added directly prior to taking traction force measurements. All traction force measurements were taken in the continued presence of the chemical inhibitor. The forces exerted by inhibited macrophages were compared to the previously measured forces of uninhibited macrophages on 10,400Pa gels.
Results
Macrophage Force Generation is Concentrated at the Leading Edge of Migrating Cells and is Dependent on Substrate Stiffness
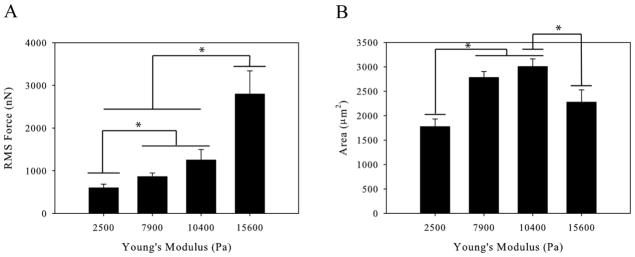
Substrate stiffness has been shown to affect a variety of cellular behaviors including differentiation, adhesion, and migration. We therefore hypothesized that changing the substrate elasticity would cause changes in macrophage force generation. Traction force microscopy was used to determine force generated by macrophages on substrates of increasing stiffness. Polyacrylamide gels were fabricated over a range of elastic moduli from 2.5 kPa to 15.6 kPa. This range of moduli encompasses the physiological range of tissue stiffnesses macrophages are exposed to in vivo including both healthy and diseased tissue [20, 21]. We used M-CSF to stimulate polarization and motility in our experiments; however macrophages require the chemokine M-CSF to differentiate and proliferate. Therefore, macrophages were differentiated and maintained in M-CSF but the M-CSF was removed for 18 hours prior to experimentation. The cells were then stimulated with 20 ng/mL M-CSF immediately before force measurements began. The force generated by macrophages was measured on polyacrylamide hydrogels of different stiffness. We found that the force exerted by macrophages increases with increasing substrate stiffness (Figure 1A). The contact area of the macrophages was also measured and found to be biphasic with respect to substrate stiffness (Figure 1B). This result indicates that the increasing force seen on substrates of increasing stiffness is directly correlated to the stiffness of the substrate and not an artifact of increased spreading of the macrophage.
Figure 1.
Primary Human Macrophages on Polyacrylamide Gels of Increasing Stiffness (A) Root-mean-squared force of primary human macrophages increases as a function of gel stiffness. (B) Spread area of primary human macrophages is biphasic with gel stiffness. (n > 46 cells per condition). Error bars are standard error, * indicates p < 0.05.
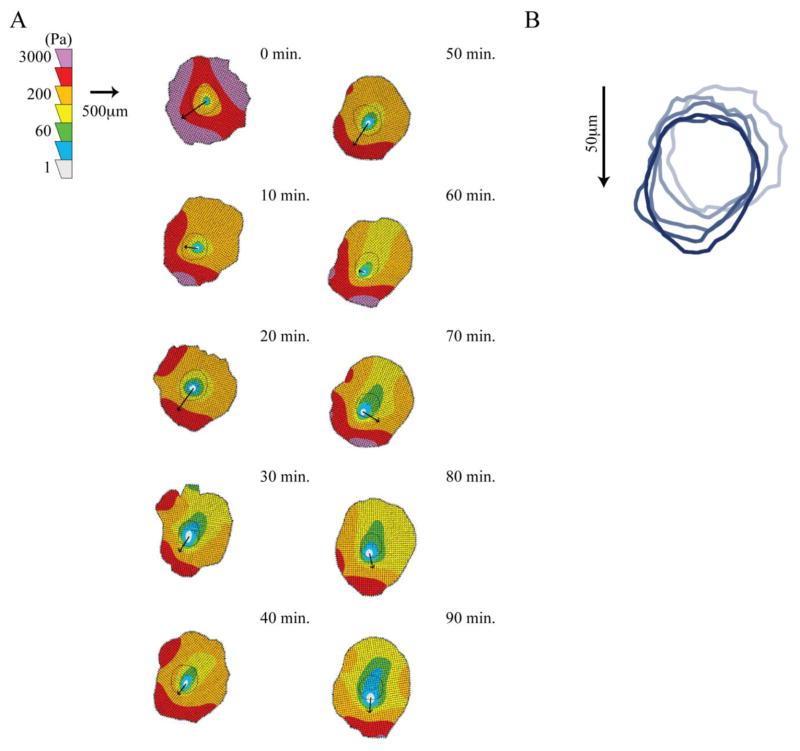
Different patterns of force organization have been seen in various motile leukocytes. Our lab has previously shows that neutrophils have high forces in the rear of the cells relative to motion indicating a rearward squeezing mode of motility [9, 10]. Conversely, dendritic cells show high forces at the front of the cells relative to motion indicating a forward towing mechanism [11]. Therefore, we measured the spatial distribution of forces in a motile macrophage to better understand the type of motility macrophages employ. A representative macrophage is shown migrating on a 10,400Pa gel in Supplementary Video 1. This video illustrates the large amount of lamellipodial ruffling associated with macrophage migration. Using traction maps of highly motile cells we found that macrophages generate the strongest forces in the front of the cell relative to motion. An illustrative cell is shown in Figure 2A and additional cell traction maps are included in Supplementary Figures 1 and 2. These figures illustrate cell tractions using heat maps to show the areas of greatest traction and arrows to indicate the direction of cell motion between the current position and the next frame. An illustration of the cell migration track for the representative cell is shown in Figure 2B. This type of force pattern suggests that macrophages use a towing mechanism of motility in which cells extend a pseudopod and attach to the substrate, generate cellular contraction through their actomyosin cytoskeleton which exerts tension on the substrate, and then release the uropod.
Figure 2.
Traction Contour Maps of Migrating Macrophage. (A) Contour plots shows traction stresses and arrows indicate the direction of motion between the indicated time-point and the next time-point of a representative macrophage on a 10,400Pa gel. (B) Outlines of cell position every 20 minutes to illustrate cell migration.
Macrophage Force Generation Requires Myosin Contraction through ROCK Signaling
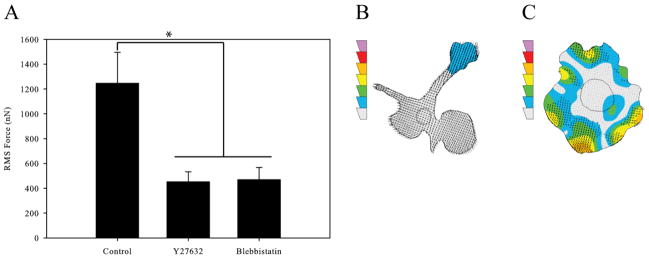
Actomyosin activity within the cell is important for cellular contraction and tail retraction during motility. This contractility depends on RhoA signaling to myosin II through its kinase ROCK. To determine the contribution of RhoA signaling on macrophage force generation we used two chemical inhibitors: Y27632 to block ROCK signaling and blebbistatin to block myosin II signaling. Both inhibitions were performed separately following the protocol described in the Materials and Methods section. We found that treatment of the cells with either Y27632 or blebbistatin lead to a significant reduction in force generation (Figure 3A). As has been previously seen, cells inhibited with Y27632 displayed long, unretracted tails and little force generation (Figure 3B) [9]. Cells treated with blebbistatin showed no polarization or significant force generation (Figure 3C).
Figure 3.
Macrophage Force Generation with Rear Contraction Inhibition. (A) Root-mean-squared forces of uninhibited macrophages, macrophages inhibited with 10μM Y27632 to reduce ROCK signaling, and macrophages inhibited with 20μM Blebbistatin to reduce myosin II activity. (B) Traction contour plot of a representative cell inhibited with Y27632 imaged at 10 minutes. (C) Traction contour plot of a representative cell inhibited with Blebbistatin imaged at 340 minutes. The traction contours are plotted using the same force scale as in Figure 2A. All results from macrophages plated on 10,400Pa gels with 5μg/mL fibronectin. (n > 44 for each condition). Error bars are standard error. * indicates p < 0.05.
Macrophage Force Generation is Dependent on PI3K Signaling and Rac Signaling Downstream of Vav1 but not Tiam1
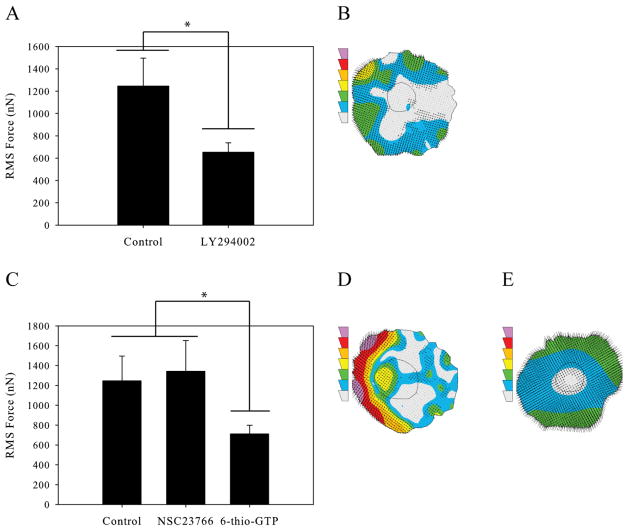
Phosphoinostitide 3-kinase (PI3K) is a membrane bound signaling protein that interacts with both integrin receptors as well as the M-CSF receptor [22]. Upon stimulation, PI3K signals downstream to several pathways inducing membrane ruffling, cell polarization, and motility [23–25]. We investigated the role of PI3K activity in macrophage force generation by inhibiting cells with 50μM LY294002. Cells were inhibited using the same protocol outlined in Materials and Methods section. We found that inhibition of PI3K caused a significant decrease in macrophage force generation (Figure 4A); a representative traction map of an LY294002 inhibited cell is shown in Figure 4B.
Figure 4.
Macrophage force generation with leading edge inhibition. (A) Root-mean-squared force of uninhibited macrophages and macrophages inhibited with 50μM LY294002 to reduce PI3K signaling. (B) Traction contour plot of a representative cell inhibited with LY294002 imaged at 230 minutes. (C) Root-mean-squared force of uninhibited macrophages, macrophages inhibited with 50μM NSC23766 to reduce Rac-Tiam1 binding, and macrophages inhibited with 10μM 6-thio-GTP to block Rac-Vav1 binding. (D) Traction contour plot of a representative cell inhibited with NSC23766 imaged at 320 minutes. (E) Traction contour plot of a representative cell inhibited with 6-thio-GTP imaged at 240 minutes. The traction contours are plotted using the same force scale as in Figure 2A. All results from macrophages plated on 10,400Pa gels with 5μg/mL fibronectin. (n > 58 for each condition). Error bars are standard error. * indicates p < 0.05.
Rac, a GTPase downstream of PI3K signaling, is known to be involved in lamellipodial protrusion at the leading edge of migrating macrophages [26]. We hypothesized that Rac would be important for macrophage force generation because of its role in motility at the leading edge of cells and our previous finding that macrophage forces during motility are the strongest at the front of the cell. We first inhibited cells with 50μM NSC23766, a chemical inhibitor that primarily prevents activation of Rac by blocking the interaction of Rac and Tiam1, a Rac GEF. We found that this inhibition caused no significant change in the ability of macrophages to generate force (Figure 4C and D). However, others have previously found that the specific GEF involved in Rac activation can determine Rac’s downstream function [27]. We therefore sought to inhibit Rac through a GEF known to be important in cell motility. Macrophages were treated with 10μM 6-thio-GTP which prevents Rac binding to its GEF Vav1. Contrary to our results with NSC23766, this inhibition of Rac lead to a significant reduction in the force produced (Figure 4C and E). This result suggests that the activation of Rac for force generation is, at least to some degree, GEF-specific.
Discussion
Previous work has shown that matrix stiffness has a significant effect on force-mediated cell behaviors including cell adhesion [3, 4], spreading [5], and migration [3, 6]. We have shown that in primary human macrophages force generation is a stiffness-dependent process with increasing stiffness of the underlying matrix resulting in increased force generation. This trend has been seen in other cell types including leukocytes [9]. This result is important physiologically because macrophages must migrate through tissues of different densities in the body. In addition, many of the disease associated with macrophage migration are accompanied by changes in the stiffness of native tissues such as hardening of the arteries in atherosclerosis and development of solid tumors in cancer [1, 21, 28, 29]. We have shown that macrophages are able to sense the stiffness of the underlying tissue and modulate their mechanobehavior accordingly. They are able to generate very large traction stresses in response to stiff substrates which may be necessary for migration through tissues in the body.
We were also able to show that the increase in force seen on substrates of increasing stiffness was not solely due to an increase in cell area because cell spread area was found to be biphasic with increasing matrix stiffness. Although many cells increase their cell area as a function of matrix stiffness [5] others have found a similar biphasic relationship of area with increasing stiffness [30]. This could be explained by the recent result indicating that an increase in substrate stiffness leads to an increase in integrin clustering [31]. This clustering could prevent the cells from spreading over a large area on stiff substrates.
We have created the first traction maps of migrating macrophages and have shown that the highest areas of traction stress at the leading edge of a migrating macrophage. This result indicates that macrophages utilize a forward towing mechanism of motility. Our lab has previously shown that this type of force distribution is seen in dendritic cells, another monocytic cell lineage [11]. However, even though they share a frontal towing mechanism, there are substantial differences. Although a direct comparison of the magnitude of the forces is impossible because these forces were measured using a micropost array, the forces exerted by dendritic cells are much smaller than those exerted by macrophages and associated with the tips of filopodia that extend from the front of the cell. In contrast, macrophages have a clear lamellipodia. However, the frontal towing mechanism exhibited among leukocytes of monocytic lineage is in contrast to the rearward squeezing seen in neutrophils [9], suggesting that leukocyte motility is diverse and the mechanisms used by cells undergoing ameboid motility are not uniform.
We next sought to investigate the signaling involved in the generation of traction force by macrophages. We have previously shown that inhibition of the RhoA kinase ROCK in a macrophage cell line does not significantly decrease the cell’s motility but it has a strong effect on cell morphology. Others have also shown that ROCK is important for the generation of cellular traction forces [9]. Therefore, we used the chemical inhibitor Y27632 to investigate the effect of ROCK signaling on macrophage force generation. We found that cells inhibited with Y27632 show long unretracted tails due to a defect in myosin contraction. These cells also exhibited little to no force (Figure 3A and B). Therefore, RhoA signaling through ROCK is clearly necessary for macrophage force generation. These results support previous findings that ROCK activity is strongly correlated with the stabilization of the actin cytoskeleton and integrin activation in monocytic cells [32, 33]. We also found that the myosin II inhibitor, blebbistatin, lead to a significant reduction in traction force (Figure 3A and C). As expected, this result indicated that myosin II is necessary for proper cytoskeletal contraction and force generation.
Several signaling pathways localized in the front of migrating cells have been shown to be important for macrophage migration. Phosphoinostitide 3-kinase (PI3K) is activated at the cell membrane and has been shown to be upstream of many signaling pathways involved in macrophage migration [23–25]. We found that in addition to macrophage migration, PI3K signaling is also important for macrophage force generation. Inhibition of macrophages with the chemical inhibitor LY294002 lead to a significant decrease in force generation (Figure 4A and B).
One GTPase known to be activated downstream of PI3K is Rac. Rac has been previously shown to be important for macrophage ruffling and motility. Like other GTPases, Rac is activated by several guanine nucleotide exchange factors or GEFs. These GEFs exchange a GDP bound to the GTPase for a GTP, thereby activating the GTPase. We have shown that inhibition of Rac through NSC23766, which blocks Rac binding to the GEF Tiam1, leads to no change in force generation but inhibition of Rac through 6-thio-GTP, which blocking binding between Rac and Vav1, leads to a significant reduction in force. This result indicates that Rac’s downstream activity is affected by which GEF activated it. Others have previously shown that in chronic lymphocytic leukemia cells Rac activated through Tiam1 was not necessary for cell motility but was important for proliferation [34]. Furthermore, it has been shown that in neutrophils Vav1 is essential for motility and the mechanosensing under flow[35]. Vav1 has also been shown to be important in F-actin reorganization in macrophages [36]. These previous results along with the results presented in this study indicate that Rac activation by Vav1 is crucial for macrophage force generation, potentially due to Vav1’s role in mediating signals from integrins to Rac and its ability to reorganize the cytoskeleton. Rac activation by Tiam1, however, leads to signaling events that are not necessary for macrophage force generation.
We have shown that macrophages are mechanoresponsive cells capable of exerting large forces on their underlying substrate. This ability may offer an advantage to macrophages which spend large amounts of time navigating through tissues of different densities. We have also shown that macrophages concentrate their forces at the leading edge of migrating cells and that signaling events that occur at the front of migrating cells are critical for macrophage force generation. PI3K is known to translocate to the leading edge of polarized cells [25] and it has been shown that Vav1 is a PI3K-dependent activator for Rac1 in macrophages stimulated with CSF-1 [37]. Therefore, it is significant that inhibition of either PI3K activity or the Vav1-Rac1 interaction leads to a significant reduction in force generation by macrophages. It is plausible that the signaling activity through PI3K and Rac1 has a significant influence on the frontal-towing mechanism of migrating macrophages.
Conclusions
We have been able to show that macrophages produce large forces during migration on compliant surfaces. These traction maps indicate that macrophages use a pulling mechanism of motility with large forces in the front of migrating cells. We have found some of the molecules responsible for this force and have shown that the activation path for GTPases is important when considering their downstream effecter functions. To our knowledge, this is the first demonstration of force generation during macrophage migration. In the future, studies like this will be crucial in understanding the role of mechanosensing in macrophage migration and the signaling events involved in motility.
Supplementary Material
Supplemental Figure 1. Traction Contour Maps of Migrating Macrophage on a 10,400Pa gel. Contour plots shows traction stresses and arrows indicate the direction of motion between the indicated timepoint and the next timepoint.
Supplemental Figure 2. Traction Contour Maps of Migrating Macrophage on a 15,600Pa gel. Contour plots shows traction stresses and arrows indicate the direction of motion between the indicated timepoint and the next timepoint.
References
- 1.Pollard JW. Trophic macrophages in development and disease. Nat Rev Immunol. 2009;9(4):259–70. doi: 10.1038/nri2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ridley AJ. Rho proteins, PI 3-kinases, and monocyte/macrophage motility. FEBS Lett. 2001;498(2–3):168–71. doi: 10.1016/s0014-5793(01)02481-4. [DOI] [PubMed] [Google Scholar]
- 3.Pelham RJ, Jr, Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci U S A. 1997;94(25):13661–5. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reinhart-King CA. Endothelial cell adhesion and migration. Methods Enzymol. 2008;443:45–64. doi: 10.1016/S0076-6879(08)02003-X. [DOI] [PubMed] [Google Scholar]
- 5.Reinhart-King CA, Dembo M, Hammer DA. The dynamics and mechanics of endothelial cell spreading. Biophys J. 2005;89(1):676–89. doi: 10.1529/biophysj.104.054320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dembo M, Wang YL. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys J. 1999;76(4):2307–16. doi: 10.1016/S0006-3495(99)77386-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lemmon CA, Chen CS, Romer LH. Cell traction forces direct fibronectin matrix assembly. Biophys J. 2009;96(2):729–38. doi: 10.1016/j.bpj.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lauffenberger DA, Linderman JJ. Receptors: Models for Binging, Trafficking, and Signaling. Oxford University Press; 1993. [Google Scholar]
- 9.Jannat RA, Dembo M, Hammer DA. Traction forces of neutrophils migrating on compliant substrates. Biophys J. 2011;101(3):575–84. doi: 10.1016/j.bpj.2011.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith LA, et al. Neutrophil traction stresses are concentrated in the uropod during migration. Biophys J. 2007;92(7):L58–60. doi: 10.1529/biophysj.106.102822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ricart BG, et al. Measuring traction forces of motile dendritic cells on micropost arrays. Biophys J. 2011;101(11):2620–8. doi: 10.1016/j.bpj.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jannat RA, et al. Neutrophil adhesion and chemotaxis depend on substrate mechanics. J Phys Condens Matter. 2010;22(19):194117. doi: 10.1088/0953-8984/22/19/194117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reinhart-King CA, Dembo M, Hammer DA. Cell-cell mechanical communication through compliant substrates. Biophys J. 2008;95(12):6044–51. doi: 10.1529/biophysj.107.127662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishizaki T, et al. Pharmacological properties of Y-27632, a specific inhibitor of rho-associated kinases. Mol Pharmacol. 2000;57(5):976–83. [PubMed] [Google Scholar]
- 15.Kovacs M, et al. Mechanism of blebbistatin inhibition of myosin II. J Biol Chem. 2004;279(34):35557–63. doi: 10.1074/jbc.M405319200. [DOI] [PubMed] [Google Scholar]
- 16.Vlahos CJ, et al. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002) J Biol Chem. 1994;269(7):5241–8. [PubMed] [Google Scholar]
- 17.Gao Y, et al. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc Natl Acad Sci U S A. 2004;101(20):7618–23. doi: 10.1073/pnas.0307512101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poppe D, et al. Azathioprine suppresses ezrin-radixin-moesin-dependent T cell-APC conjugation through inhibition of Vav guanosine exchange activity on Rac proteins. J Immunol. 2006;176(1):640–51. doi: 10.4049/jimmunol.176.1.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abou-Kheir W, et al. A WAVE2-Abi1 complex mediates CSF-1-induced F-actin-rich membrane protrusions and migration in macrophages. J Cell Sci. 2005;118(Pt 22):5369–79. doi: 10.1242/jcs.02638. [DOI] [PubMed] [Google Scholar]
- 20.Engler AJ, et al. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–89. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 21.Paszek MJ, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8(3):241–54. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 22.Jones GE, et al. Requirement for PI 3-kinase gamma in macrophage migration to MCP-1 and CSF-1. Exp Cell Res. 2003;290(1):120–31. doi: 10.1016/s0014-4827(03)00318-5. [DOI] [PubMed] [Google Scholar]
- 23.Munugalavadla V, et al. p85alpha subunit of class IA PI-3 kinase is crucial for macrophage growth and migration. Blood. 2005;106(1):103–9. doi: 10.1182/blood-2004-10-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papakonstanti EA, et al. Distinct roles of class IA PI3K isoforms in primary and immortalised macrophages. J Cell Sci. 2008;121(Pt 24):4124–33. doi: 10.1242/jcs.032763. [DOI] [PubMed] [Google Scholar]
- 25.Vanhaesebroeck B, et al. Distinct PI(3)Ks mediate mitogenic signalling and cell migration in macrophages. Nat Cell Biol. 1999;1(1):69–71. doi: 10.1038/9045. [DOI] [PubMed] [Google Scholar]
- 26.Pixley FJ. Macrophage Migration and Its Regulation by CSF-1. Int J Cell Biol. 2012;2012:501962. doi: 10.1155/2012/501962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park YM, et al. Oxidized LDL/CD36 interaction induces loss of cell polarity and inhibits macrophage locomotion. Mol Biol Cell. 2012;23(16):3057–68. doi: 10.1091/mbc.E11-12-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bussy C, et al. Intrinsic stiffness of the carotid arterial wall material in essential hypertensives. Hypertension. 2000;35(5):1049–54. doi: 10.1161/01.hyp.35.5.1049. [DOI] [PubMed] [Google Scholar]
- 29.Kothapalli D, et al. Cardiovascular protection by ApoE and ApoE-HDL linked to suppression of ECM gene expression and arterial stiffening. Cell Rep. 2012;2(5):1259–71. doi: 10.1016/j.celrep.2012.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kraning-Rush CM, Califano JP, Reinhart-King CA. Cellular traction stresses increase with increasing metastatic potential. PLoS One. 2012;7(2):e32572. doi: 10.1371/journal.pone.0032572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paszek MJ, et al. Integrin clustering is driven by mechanical resistance from the glycocalyx and the substrate. PLoS Comput Biol. 2009;5(12):e1000604. doi: 10.1371/journal.pcbi.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Worthylake RA, Burridge K. RhoA and ROCK promote migration by limiting membrane protrusions. J Biol Chem. 2003;278(15):13578–84. doi: 10.1074/jbc.M211584200. [DOI] [PubMed] [Google Scholar]
- 33.Worthylake RA, et al. RhoA is required for monocyte tail retraction during transendothelial migration. J Cell Biol. 2001;154(1):147–60. doi: 10.1083/jcb.200103048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hofbauer SW, et al. Tiam1/Rac1 signals contribute to the proliferation and chemoresistance, but not motility, of chronic lymphocytic leukemia cells. Blood. 2014;123(14):2181–8. doi: 10.1182/blood-2013-08-523563. [DOI] [PubMed] [Google Scholar]
- 35.Phillipson M, et al. Vav1 is essential for mechanotactic crawling and migration of neutrophils out of the inflamed microvasculature. J Immunol. 2009;182(11):6870–8. doi: 10.4049/jimmunol.0803414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhavsar PJ, et al. Vav GEFs regulate macrophage morphology and adhesion-induced Rac and Rho activation. Exp Cell Res. 2009;315(19):3345–58. doi: 10.1016/j.yexcr.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 37.Vedham V, Phee H, Coggeshall KM. Vav activation and function as a rac guanine nucleotide exchange factor in macrophage colony-stimulating factor-induced macrophage chemotaxis. Mol Cell Biol. 2005;25(10):4211–20. doi: 10.1128/MCB.25.10.4211-4220.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Traction Contour Maps of Migrating Macrophage on a 10,400Pa gel. Contour plots shows traction stresses and arrows indicate the direction of motion between the indicated timepoint and the next timepoint.
Supplemental Figure 2. Traction Contour Maps of Migrating Macrophage on a 15,600Pa gel. Contour plots shows traction stresses and arrows indicate the direction of motion between the indicated timepoint and the next timepoint.