Abstract
Human metapneumovirus (hMPV) is a major cause of lower respiratory infection in young children. Repeated infections occur throughout life, but its immune evasion mechanisms are largely unknown. We recently found that hMPV M2-2 protein elicits immune evasion by targeting mitochondrial antiviral-signaling protein (MAVS), an antiviral signaling molecule. However, the molecular mechanisms underlying such inhibition are not known. Our mutagenesis studies revealed that PDZ-binding motifs, 29-DEMI-32 and 39-KEALSDGI-46, located in an immune inhibitory region of M2-2, are responsible for M2-2-mediated immune evasion. We also found both motifs prevent TRAF5 and TRAF6, the MAVS downstream adaptors, to be recruited to MAVS, while the motif 39-KEALSDGI-46 also blocks TRAF3 migrating to MAVS. In parallel, these TRAFs are important in activating transcription factors NF-kB and/or IRF-3 by hMPV. Our findings collectively demonstrate that M2-2 uses its PDZ motifs to launch the hMPV immune evasion through blocking the interaction of MAVS and its downstream TRAFs.
Keywords: hMPV, M2-2 motif, TRAF, MAVS, Innate immune response
Highlights
-
•
This manuscript describes a molecular mechanism underlying the immune evasion of hMPV.
-
•
Results create the design basis for safer and more effective hMPV vaccines/therapeutic molecules.
-
•
We demonstrate the contribution of TRAFs in antiviral responses to hMPV infection.
1. Introduction
Human metapneumovirus (hMPV), the first and only identified human pathogen belonging to the genus Metapneumovirus in the Paramyxoviridae family, is a leading cause of lower respiratory tract disease in children, the elderly, and immunocompromised patients worldwide (Williams et al., 2006, Falsey et al., 2003, Englund et al., 2006). It encodes nine proteins, among which phosphoprotein P, glycoprotein G, small hydrophobic protein SH and the M2-2 protein have been characterized with immune regulatory functions (Bao et al., 2008a, Kolli et al., 2011, Bao et al., 2008b, Goutagny et al., 2010, Ren et al., 2012, Ren et al., 2014). In this study, we focused on molecular mechanisms underlying the immune regulatory functions of M2-2 as it was identified with such a function most recently, however with regulatory mechanism largely unknown.
In response to virus infection, including infection by hMPV, host cells use several classes of pattern recognition receptors (PRRs), such as the Toll-like receptors (TLRs) and retinoic-inducible gene-I (RIG-I)-like receptors, to launch the innate immune responses in a cell type-dependent manner (reviewed in (Akira et al., 2006; Kawai and Akira, 2006)). hMPV infection of alveolar epithelial cells, the primary target of respiratory viruses, activates antiviral signaling via RIG-I and, subsequently, its adaptor MAVS, a mitochondrial protein, and further downstream kinases TRAFs/IKKs, to promote NF-κB and IRF activation (Liao et al., 2008, Seth et al., 2005, Sun et al., 2006). Recently, we have shown that the M2-2 protein contributes to the immune evasion by targeting MAVS (Ren et al., 2012). However, the molecular mechanism by which M2-2 uses to disrupt MAVS-mediated signaling has not been investigated.
M2-2 is highly conserved (>90%) between the two hMPV strains, A and B (Biacchesi et al., 2003). The M2-2 of Canadian isolate hMPV83, which belongs to A2 strain and the focus of this study, has about 70 amino acids. The domains of M2-2 responsible for the immune inhibition locate in last 45 amino acids, which also promote viral genome replication and contain a cytotoxic T-lymphocyte (CTL) epitope (Ren et al., 2012, Melendi et al., 2007, Herd et al., 2008, Herd et al., 2006). The first 25 amino acids of M2-2 are solely responsible for promoting viral gene transcription (Ren et al., 2012). These multiple functions of M2-2 highlight the need to identify the domains/motifs responsible for different M2-2 functions. In this study, we first focused on identifying M2-2 motif(s) responsible for its immune inhibition, then addressed how these motifs affect MAVS-mediated anti-hMPV signaling. This study is potentially translational as it can provide the molecular basis for the design of new, safer and more effective hMPV vaccines and therapeutic molecules. Options that can be explored include designing M2-2 mutants with reduced inhibition of innate signaling, a full CTL epitope, and proper attenuation of viral RNA synthesis. Reagents that modify M2-2-host interaction and enhance the immune capability against hMPV infection might also be explored.
2. Materials and methods
2.1. Cell lines and antibodies
LLC-MK2, A549 (human alveolar type II-like epithelial cells), and 293 (a human embryonic kidney epithelial cell line) were all from ATCC, Manassas, VA, and maintained as previously described (Ren et al., 2012, Ren et al., 2011a). BSR T7/5 cells, baby hamster kidney cells that constitutively express the T7 RNA polymerase, were a gift from Dr. Conzelmann, Munich, Germany. They were maintained in Glasgow minimal essential medium (GMEM) supplemented with 1% amino acids, 10% FBS, 12 mg/L tryptose phosphate broth, 1 mg/ml of Geneticin, 100 U/ml of Penicillin and 100 U/ml of Streptomycin. Monoclonal antibodies against Lamin b and FLAG were obtained from Sigma-Aldrich (Sigma, St. Louis, MO). The antibody against V5 was obtained from Invitrogen (Invitrogen, Carlsbad, CA). The polyclonal rabbit anti-hMPV antibodies were raised against purified hMPV by Creative Diagnostics, Shirley, NY. The polyclonal rabbit anti-MAVS antibody was a gift from Dr. Ilkka Julkunen (National Public Health Institute, Finland). The antibodies against TARF2, TRAF3, and TRAF6 were from Cell Signaling, Danvers, MA. Primary antibodies against phosphorylated IRF-3, P50 and P65 were purchased from Millipore (Millipore, Billerica, MA). FITC-conjugated goat anti-rabbit antibody was from Zymed, San Francisco, CA. Primary antibodies against TRAF5, IRF-3 and horseradish-coupled secondary antibodies were purchased from Santa Cruz (Santa Cruz, Santa Cruz, CA).
2.2. Construction of M2-2 mutant antigenome and viral recovery
A plasmid encoding wild type hMPV antigenome was constructed as described (Bao et al., 2008a, Bao et al., 2008b, Biacchesi et al., 2004). Construction of M2-2 mutant cDNA was done similarly as previously illustrated (Ren et al., 2012, Bao et al., 2013). In brief, the whole hMPV antigenome was engineered by ligating three separately cloned segments in a sequential order. Herein, we used site-directed mutagenesis to individually mutate two PDZ motifs of M2-2 in the second segment, followed by the replacement of WT segment with the mutated one. The multiple-site mutagenesis does not affect M2-1 codons. Primer sequence for generating M2-2 mutant antigenome is available upon request, and the recovery of recombinant hMPV was done as previously described (Ren et al., 2012, Bao et al., 2013). Recombinant M2-2 mutant viruses were confirmed by sequencing of RT-PCR products of viral RNA. The recovered viruses were then amplified for two passages in LLC-MK2 cells and saved as stock viral preparations. Viruses with no more than 4–5 passages were used in all experiments.
2.3. Viral preparation and infection
The isolate hMPVCAN-83 and its derived recombinant viruses were propagated in LLC-MK2 cells at 35 °C in the absence of serum and in the presence of 1 µg/ml of trypsin, and were sucrose purified, as previously described (Bao et al., 2008a, Bao et al., 2008b). Viral titer was determined by immunostaining in LLC-MK2 cells, as previously described (Bao et al., 2008a, Bao et al., 2008b). To characterize the growth pattern of recombinant M2-2 mutant viruses, LLC-MK2 or Vero cell monolayers in 6-well plate were infected with rhMPV, WT or individual mutants, at multiplicity of infection (MOI) of 0.1. An equivalent amount of sucrose solution was added to uninfected LLC-MK2 or Vero cells, as control (mock infection). After initial absorption, viral inoculum was removed and cells were supplied with fresh serum-free medium with trypsin. Viruses were harvested at different times p.i. and viral titer was determined by immunostaining in LLC-MK2 cells, as previously described (Bao et al., 2008a, Bao et al., 2008b).
To investigate the role of interested M2-2 motifs in regulating innate antiviral signaling at the acute phase of infection, A549 cell monolayers were infected with rhMPV-WT or M2-2 mutant virus, at MOI of 2. Mock infection was used as negative control. Supernatants were harvested at different times p.i., and the concentrations of cytokines and chemokines were determined by ELISA or a multiplex immunoassay. Total cells were lysed to prepare mitochondrial, nuclear and cytosolic fractions, as previously described (Bao et al., 2008a, Bao et al., 2008b, Bao et al., 2013).
2.4. Plasmid construction
To investigate the role of M2-2 motifs in regulating MAVS-mediated signaling in the overexpression system, site-directed mutagenesis was used. The primers sequence will be available upon the request. The final constructs were verified by sequencing performed by the protein chemistry core laboratory at UTMB.
2.5. Reporter gene assays
To investigate the role of individual M2-2 motifs in mediating MAVS-induced anti-viral responses, logarithmically growing 293 were transfected in triplicate with luciferase reporter gene plasmids containing IFN-β promoter (designated as IFN-β-Luc) or multiple copies of NF-κB binding sites (Kb-5-Luc), together with plasmids encoding M2-2, WT or mutants, or their empty vector using FuGene 6 (Roche, Indianapolis, Indiana), as previously described (Bao et al., 2008a, Bao et al., 2008b). At 30 h post transfection, cells were lysed to measure independently luciferase and β-galactosidase reporter activity. Luciferase was normalized to the internal control β-galactosidase activity.
2.6. Quantitative RT-PCR (qRT-PCR)
The quantification of viral genome copies in rhMPV-infected cells was performed as we previously described (Ren et al., 2012). To quantify differences in the G transcription among different rhMPV infection, WT vs M2-2 mutants, a relative quantitative method was used as we previously described (Ren et al., 2012). The RT primer to measure the transcription of the hMPV G: 5′-CGTCTCAGCCAATCCCTGG TTTTTTTTTTTT CTAGTTTTGC-3′. Primers were designed to incorporate a “tag” (underlined letters) as part of the assay due to self-priming exhibited by viral RNA (Bannister et al., 2010). The tag sequence was derived from the bacterial chloramphenicol resistance (Cmr) gene. The sequence with bold letters is complementary to poly(A) tails of the transcribed hMPV G gene. The sequence in italic is G gene specific. At a 25 °C annealing temperature, the 10 nucleotides (nt) matching G-specific sequences would not be sufficient for a stable efficient priming of cDNA from an antigenome of hMPV (positive strand). On the other hand, 22 nucleotides matching transcribed N (12T’s and G-gene-specific nucleotides) are able to attain stable annealing to the transcribed G gene. The hMPV G forward primer was 5′- CATCAGTCCAGTCCGACAGC-3′, and the reverse primer against hMPV tag was 5′-CGTCTCAGCCAATCCCTGG-3′. QPCRs were run in the ABI 7500 sequence detection system under the standard default conditions: initial steps of 50 °C for 2 min and 95 °C for 10 min and PCR steps of 95 °C for 15 s and 60 °C for 1 min, for 40 cycles.
2.7. Western blot analysis
Total cellular lysates or extracts of cytosol, nuclear and mitochondria were prepared for uninfected or infected cells as previously described (Bao et al., 2008b, Tang and Wang, 2010). Proteins were then quantified with a protein quantification kit from Bio-Rad, followed by the fractionation using SDS-PAGE denaturing or native gels and protein transferring to polyvinylidene difluoride membranes as previously described (Ren et al., 2012, Ren et al., 2011b). Membranes were blocked with 5% milk in TBS-Tween 20 and incubated with the proper primary antibodies according to manufacturer’s instruction.
2.8. Statistical analysis
Statistical significance was determined using analysis of variance (ANOVA). P value of less than 0.05 was considered significant. Mean±standard error (SE) is shown.
3. Results
3.1. Identification of M2-2 motifs important for hMPV-induced immune responses
As mentioned, MAVS signaling is elicited via its downstream molecules TRAFs. The interaction between the TRAF domain of TRAFs and the TIM domain of MAVS is required for MAVS-mediated signaling (Saha et al., 2006, Kumar et al., 2006). The AA alignment revealed that M2-2 does not contain a TRAF/TIM domain, but is enriched with PDZ domains, which are common structural domains in signaling proteins for signal transduction (Ranganathan and Ross, 1997). M2-2 contains nine PDZ motifs. It is possible that M2-2 uses its PDZ domain(s) to hamper MAVS-mediated antiviral signaling (Pawson and Nash, 2003).
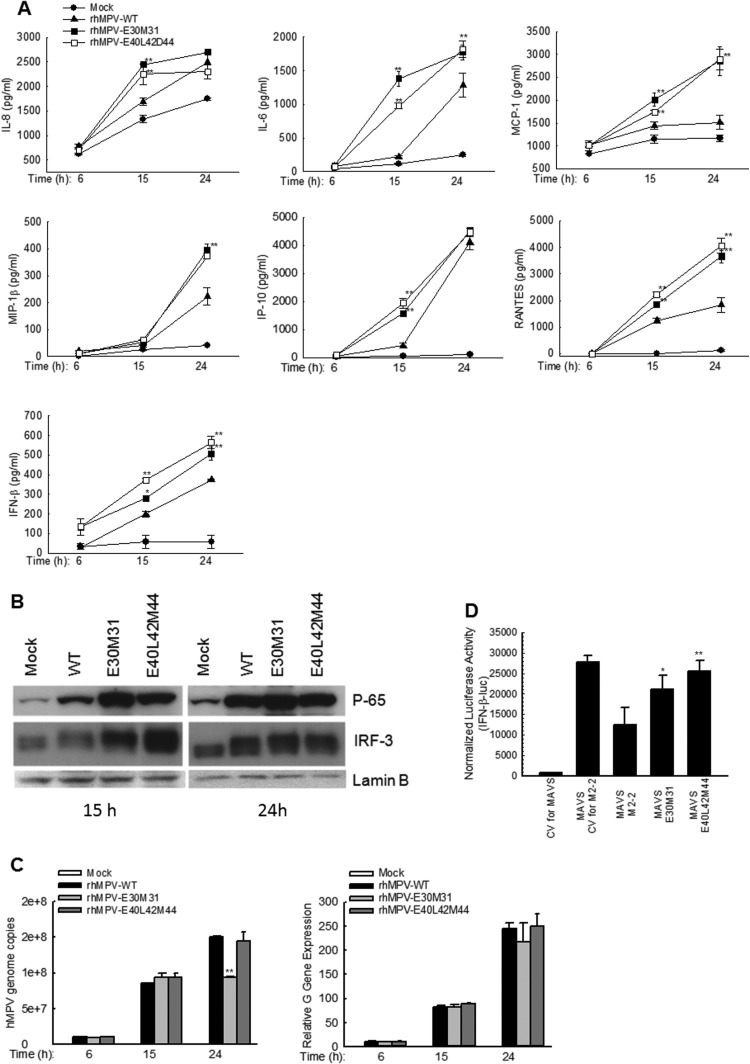
As we have previously shown, the immune inhibitory domains of M2-2 are located in the last 45 amino acids, which also regulate viral genome replication. The PDZ-binding motifs in the last 45 amino acids are 29-DEMI-32, 39-KEALSDGI-46, 58-LENI-61 and 61-IEII-64. Among them, the last two motifs contain an H-2D-restricted CTL epitope, which is important for T cells to recognize and kill virus-infected cells and contribute to immunologic control of viral replication (Melendi et al., 2007). Therefore, we first mutated motifs 29-DEMI-32 and 39-KEALSDGI-46 to initiate the immune regulatory motif identification. To do that, we generated two recombinant hMPV (rhMPV)- containing mutation in E30 and M31 of motif 29-DEMI-32 (rhMPV-E30M31) or in E40, L42 and D44 of motif 39-KEALSDGI-46 (rhMPV-E40L42D44). We found that both rhMPV-E30M31 and rhMPV-E40L42D44 induced greater secretion of several cytokines/chemokines than wild type (WT)-rhMPV at 15 and/or 24 h p.i. ( Fig. 1A).
Fig. 1.
PDZ Motifs 29-DEMI-32 and 39-KEALSDGI-46 are important for hMPV-induced innate immune responses. (A) Immune mediator induction by rhMPV. A549 cells in triplicate were mock infected or infected with rhMPV-WT, rhMPV-E30M31, or rhMPV-E40L42D44, at MOI of 2, for various time as indicated. The secretion of cytokines and chemokines in cell supernatants were measured by Bio-plex and/or ELISA. Data shown are from three independent experiments and are expressed as mean± SE. *, P<0.05, and **, P<0.01, relative to rhMPV-WT-infected A549 cells. (B) NF-κB and IRF-3 activation by rhMPV. A549 cells in flasks were mock infected or infected with rhMPV as described in A. nuclear fractions were prepared, followed by western blot to assess the nuclear translocation of p65 and IRF-3. The expression of a nuclear protein lamin B, used as an internal control, was also compared among the group. (C) Replication and gene transcription characterization of recombinant viruses. A549 cells in 6-well plates were mock infected or infected with rhMPV at MOI of 2 for various periods of time as indicated, followed by total RNA extraction using Trizol. The extracted RNAs in triplicate were then subjected to real-time PCR to assay genomic RNAs (left panel) or viral G gene transcription (right panel). The results are the representative of two independent experiments and are expressed as mean±SE of absolute copy numbers of transcribed G gene or viral genome. **,P<0.01, relative to rhMPV-WT-infected A549 cells. (D) Impact of motifs 29-DEMI-32 and 39-KEALSDGI-46 on M2-2-mediated immune evasion. A549 cells in triplicate were transfected with a luciferase reporter plasmid IFN-β-Luc (0.1 µg/well), a plasmid encoding MAVS or its control (0.1 µg/well), and a plasmid encoding hMPV M2-2 or its mutants, or a control vector. (0.1 µg/well) were transfected. After 40 h, cells were harvested for luciferase activity measurement. *P<0.05 and P<0.01 relative to MAVS+M2-2. CV: control vector for M2-2 or MAVS expression.
Among the IRF family, IRF-3 is necessary for IFN-β and RANTES gene expression in response to paramyxovirus infections (Taniguchi et al., 2001, Lin et al., 1999, Casola et al., 2002). As expected, there was a significant increase in IRF-3 nuclear translocation in M2-2 mutant-infected cells, compared to rhMPV-WT-infected cells (Fig. 1B).
P65, an importance member belonging to NF-κB superfamily, is also important in the induction of chemokines and cytokines by viruses including paramyxovirus (Garofalo et al., 1996, Tian et al., 2002). The significant role of the M2-2 motifs in modulating hMPV-induced NF-κB activation was also confirmed by enhanced p65 nuclear translocation in mutant-infected A549 cells, compared to rhMPV-WT-infected cells (Fig. 1B).
To investigate whether motif(s) are solely for immune regulation in the context of hMPV infection, the genome replication and viral gene transcription were compared among rhMPV-WT-infected cells and cells infected with two individual M2-2 mutants. As shown in Fig. 1 C, both mutants had similar genome copy numbers as WT virus following infection at the MOI of 2 at 15 h p.i. At 24 h p.i., rhMPV-E30M31- infected cells had much less virus genome copies than WT-infected cells. However, the mutations in the motif of 39-KEALSDGI-46 did not affect the genome replication, suggesting it is not important for the genome replication.
As for the role of motifs in viral gene transcription, we found that both motifs were not involved in viral gene transcription, consistent with our previous observation that viral gene regulatory domain is in the first 25 amino acids of M2-2 (Ren et al., 2012) (Fig. 1C). Since the expression of G, a previously described immune inhibitory viral protein (Bao et al., 2008b, Bao et al., 2013), was not altered by amino acid mutations in motifs, the gene transcription results suggested a direct immune suppression by M2-2 motifs.
In summary, these experiments revealed that both PDZ motifs, 29-DEMI-32 and 39-KEALSDGI-46, suppress immune gene expression in response to hMPV infection, with motif 29-DEMI-32 also promoting viral genome replication. Given facts that hMPV uses RIG-MAVS to launch antiviral immunity and RIG-I-MAVS pathway is known to be activated by viral RNAs, our result showing that rhMPV-E30M31 with suppressed genome replication enhanced induction of cytokines and chemokines similar to rhMPV-E40L42D44 suggested motif 29-DEMI-32 inhibits host immunity more significantly than motif 39-KEALSDGI-46 (Fig. 1A). This also raised the possibility for rhMPV-E30M31 to be a potential live attenuated vaccine candidate as it not only enhanced immune responses but also suppressed the viral genome replication. Indeed, there was about more than a log decrease in infectious particle production when rhMPV-E30M31 was seeded into MK-2 cells at MOI of 0.01, compared to rhMPV-WT. A multi-cycle growth of WT and rhMPV-E30M31 was investigated by immune staining using anti-hMPV antibody. Attenuated replication of rhMPV-E30M31 was also observed in Vero cells, a cell line deficient of type I IFN genes, following multi-cycle growth (data not shown).
The importance of motifs in disrupting MAVS-mediated signaling was also demonstrated by a luciferase reporter assay. We found that MAVS-dependent IFN-β promoter activation was inhibited by M2-2 expression, while the inhibition was attenuated by the mutations in M2-2 motifs 29-DEMI-32 or 39-KEALSDGI-46 (Fig. 1D). This experiment also suggested that M2-2-disrupted MAVS signaling is independent of virus infection.
3.2. The importance of TRAFs in hMPV-induced host responses
Our recent study has demonstrated that hMPV uses the RIG-I/MAVS pathway to initiate antiviral signaling (Liao et al., 2008). We also have demonstrated that the M2-2 protein is involved in the interference of RIG-I/MAVS pathway by binding to MAVS to disrupt MAVS-mediated antiviral signaling (Ren et al., 2012). However, how M2-2 targets MAVS signalosome is not known.
Proteins belonging to tumor necrosis factor receptor (TNFR)-Associated Factor (TRAF) family were originally identified as intracellular signaling adaptors that bind directly to the cytoplasmic regions of TNFRs, bridging upstream receptors to downstream enzymes (Inoue et al., 2000, Dempsey et al., 2003). There are six known members of the TRAF family (TRAF1 to 6) in mammalians, with variable number of RING and zinc finger domains (Wajant et al., 2001, Hacker et al., 2011, Carpenter and O’Neill, 2009, Pomerantz and Baltimore, 1999). Recently, TRAFs have been shown to be adaptors to MAVS as well (Saha et al., 2006; Nakhaei et al., 2009, Pomerantz and Baltimore, 1999, Tang and Wang, 2010). In addition, the role of TRAFs in MAVS-mediated antiviral signaling is isoform-, pathogen- and cell type-specific (Nakhaei et al., 2009, Liu et al., 2013, Feng et al., 2011, Guan et al., 2015). In hMPV-infected airway epithelial cells, the isoform(s) of TRAFs important for anti-viral signaling is currently unknown.
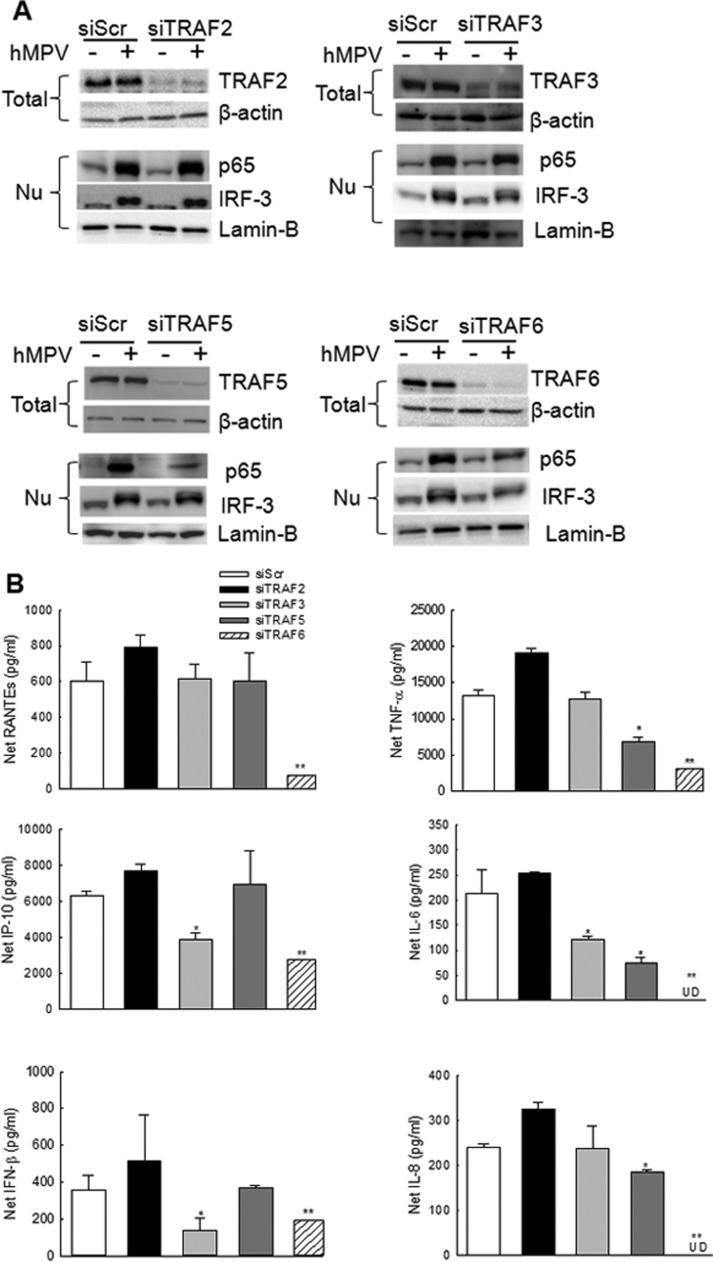
To determine the role of TRAFs in the regulation of host response to hMPV infection, we investigated cytokine, chemokine and IFN-β gene expression in A549 cells transfected with either a scramble siRNA, as control, or one specifically targeting individual TRAF and infected with hMPV. Our results showed that treatment of A549 cells with siRNA targeting TRAFs effectively blocked the protein expression (>80%) ( Fig. 2A). We also found that individual TRAF suppression by siRNA did not affect the expression of the rest of the investigated TRAFs (data not shown), suggesting that the expression of TRAFs is independent of each other. In response to hMPV, TRAF2 seemed not important in mediating the activation of p65 and IRF-3, two transcription factors critical for the induction of inflammatory/immune gene expression by hMPV (Bao et al., 2007) (Fig. 2A, upper left panel). Consequently, TRAF2 did not affect hMPV-induced cytokines/chemokines (Fig. 2B), suggesting that TRAF2 is not that critical for host anti-hMPV signaling in response to hMPV infection. In contrast with TRAF2, TRAF6 had a broad and significant effect on hMPV-induced antiviral signaling, as TRAF6 suppression by its specific siRNA led to decreased activation of p65 and IRF-3 (Fig. 2A, lower right panel) and subsequently reduced secretion of proinflammatory and antiviral molecules (Fig. 2B). We also found that TRAF3 and TRAF5 contributed to the activation of IRF-3 and p65, respectively (Fig. 2A, upper right panel for TRAF3 and lower left panel for TRAF5) confirming that the role of TRAFs in virus-mediated immune/inflammatory gene expression is isoform-specific.
Fig. 2.
Impact of TRAFs on hMPV-induced innate immunity. A549 cells were transfected with 100 nM siRNA targeting TRAF2 (siTRAF2), TRAF3 (siTRAF3), TRAF5 (siTRAF5) or TRAF6 (siTRAF6). A scrambled siRNA (siScr) was used as a negative control. At 48 h post transfection, cells were mock infected or infected with hMPV for 15 h at a multiplicity of infection (MOI) of 2. (A). Cell were harvested to prepare total cell lysates or nuclear fractions. Total cell lysates were subjected to 8% SDS-PAGE, followed by Western blot analysis of TRAFs expression. Nuclear fractions were used to assess the nuclear translocation of p65 and IRF-3. Membrane was stripped and reprobed with anti-β-actin or lamin-B antibody to control for equal loading of the total cell lysates or nuclear fractions respectively. Results are representative of three separate experiments. (B) Supernatants were harvested to measure the induction of cytokines/chemokines by Bio-plex and/or ELISA. Data shown are from three independent experiments and are expressed as mean± SE. *, P<0.05, and **, P<0.01, relative to siScr –treated A549 cells.
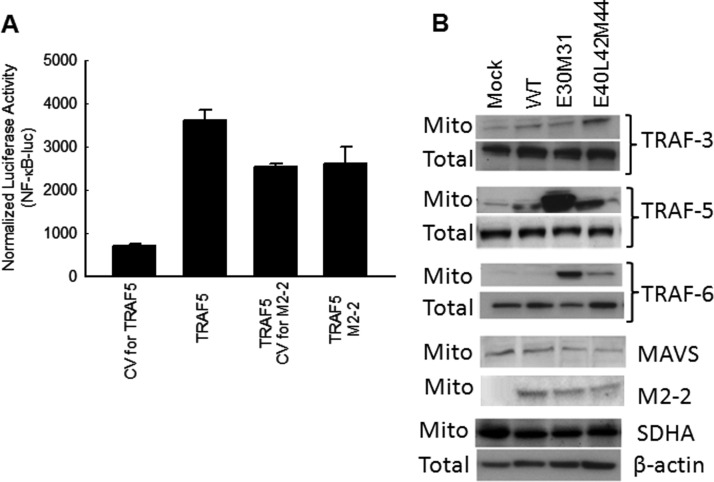
3.3. The effects of M2-2 PDZ motifs on MAVS/TRAFs signalosome
As mentioned, hMPV M2-2 protein interacts with MAVS and subsequently blocks MAVS-mediated immune signaling (Ren et al., 2012). However, M2-2 does not block the signaling induced by MAVS's downstream signaling molecules, including TRAF6 and IKKs (Ren et al., 2012). In this study, we also found that M2-2 did not block TRAF-5-induced signaling, confirming MAVS, but not its downstream signaling factors, as the target of M2-2 ( Fig. 3A). One of the possible consequences of M2-2-mediated targeting of MAVS is that M2-2 disrupts MAVS-mediated signaling via blocking the interaction between MAVS and its downstream signaling molecules TRAF(s). To test whether the M2-2 protein blocks MAVS-TRAF interaction in airway epithelial cells, we investigated the TARF recruitment to mitochondria in response to rhMPV-WT and M2-2 mutants infection. Three TRAFs, i.e. TRAF 3, 5, and 6, were investigated, as they played a role in the activation of transcription factors NF-kB and/or IRF-3 and secretion of cytokines/chemokines in response to hMPV infection (Fig. 2). Uninfected and infected A549 cells were harvested at 15 h p.i. to prepare the mitochondrial fraction. The mitochondrion purity was well controlled, as demonstrated by the expression of specific marker for cellular fractions during the preparation (Supplementary Fig1). We found that rhMPV-E40L42D44-infected cells, but not rhMPV-E30M31-infected cells, had a slightly higher TRAF3 recruitment to mitochondria than WT-infected cells (Fig. 3B). We also found that there was a significant increase in the abundance of TRAF5 in both rhMPV-E40L42D44- and rhMPV-E30M31-infected cells compared to WT-infected cells. Compared to rhMPV-E30M31-infected cells, the increase of TRAF5 in rhMPV-E40L42D44-infected was much less. In terms of TRAF6 recruitment, both M2-2 mutants were able to recruit more TRAF6, with more recruitment in rhMPV-E30M31 -infected cells than in rhMPV-E40L42D44-infected cells (Fig. 3B). To exclude the possibility that mutant-enhanced TRAF recruitment resulted from more MAVS in mitochondria, we investigated the abundance of MAVS and the mitochondrial protein SDHA (succinate dehydrogenase complex, subunit A, used as loading control). Slightly decreased MAVS in the mitochondria and stable levels of SDHA on exposure to mutant viruses suggested this is not the case. In parallel to the slight decreased MAVS expression, mitochondrial M2-2 expression was also slightly declined in mutant-infected cells, suggesting that the binding of M2-2 protein to MAVS is proportionally even between WT- and mutant-infected cells. All of these results supported that the PDZ motifs of M2-2 inhibit MAVS-TRAF/3/5/6 interaction in infected cells.
Fig. 3.
PDZ Motifs 29-DEMI-32 and 39-KEALSDGI-46 of M2-2 protein suppress IRF-3 and NF-κB activation by inhibiting mitochondrial signalosome formation. (A). The effect of M2-2 on TRAF-activated signaling. A549 cells were transfected a luciferase reporter plasmid NF-κB-Luc (0.5 µg/well), a plasmid encoding TRAF5 or its control (0.5 µg/well), and a plasmid encoding hMPV M2-2 or its mutants, or a control vector (0.1 µg/well). After 30 h, cells were harvested for luciferase activity measurement. (B). The effect of motifs on the recruitment of TRAFs to mitochondrial compartment. A549 cells were infected with rhMPV-WT, rhMPV-E30M31, or rhMPV-E40L42D44, at MOI of 2, for 15 h and harvested to purify mitochondria. The abundance of mitochondria-associated MAVS, M2-2 and TRAF proteins was investigated by Western blot. Membranes were stripped and reprobed with anti-SDHA, as control for comparable loading of samples. Data shown are representative of two independent experiments.
4. Discussion
In this study, we identified the molecular mechanisms by which M2-2 interferes with MAVS-mediated antiviral signaling. This is an important study as it will help elucidate the physiopathology of hMPV infection. Therefore, the results should provide important insights into the development of therapeutic strategies. As discussed, the dissection of functional motifs also provides the conceptual framework for developing safer, more effective live attenuated vaccine candidates to reduce the morbidity and mortality of hMPV infection.
Compared to the M2-2 protein of respiratory syncytial virus (RSV), a family member of hMPV and also a leading cause of LTRI in infants, the M2-2 of hMPV shares common and distinctive features with that of RSV. Both viruses with the deletion of M2-2 are attenuated. Both M2-2 proteins promote viral genome replication. However, they have opposite roles in viral mRNA synthesis. The M2-2 protein of RSV suppresses viral mRNA synthesis, while our publication demonstrated a stimulus role of hMPV M2-2 in mRNA transcription (Bermingham and Collins, 1999, Jin et al., 2000, Teng et al., 2000, Karron et al., 2015). By comparing the sequences of these two proteins, they are quite different from each other. Whether the sequence difference accounts for the distinctive role of M2-2 proteins needs to the investigated in the future.
The innate immune response functions as a first line of host defense against invading pathogens, as well as a critical component in regulating adaptive immune responses. The effectiveness of innate immune response against viral infection depends on the interactive nature of virus components with the host innate antiviral immune systems, including the MAVS centered signalosome (Akira et al., 2006). TRAFs, as important signaling adaptors of MAVS, are critical for antiviral function in a cell-type and pathogen-specific manner (Saha et al., 2006, Nakhaei et al., 2009, Pomerantz and Baltimore, 1999, Tang and Wang, 2010). Among the six TRAF members, except TRAF-1, other members have an N-terminal RING finger domain, followed by a variable number of zinc fingers (Wajant et al., 2001). As shown in Fig. 2, TRAF-activated NF-kB and IRF-3 in response to hMPV is also isotype-specific. TRAF2 seems not critical in the antiviral signaling at all. TRAF3 and TRAF5 contribute to the activation of IRF-3 and NF-kB, respectively, while TRAF-6 controls the activation of both transcription factors. There are still many questions that need to be addressed for a better understanding of the molecular mechanisms underlying the regulation of antiviral signaling by TRAFs. Do TRAFs use their structural motif(s), such as RING and zinc fingers, to control the activation the NF-kB and/or IRF-3? If so, which ones are responsible? This information is important as it should provide insights into developing reagents to modify the interaction of TRAFs with their up- and down-stream signaling molecules to control inflammation and enhance antiviral immunity during hMPV infection.
Viruses inhibit MAVS-mediated antiviral signaling via several mechanisms. Hepatitis C virus uses its NS3-4A to cleave MAVS to block MAVS-dependent antiviral signaling (Welsch et al., 2015). Rotavirus NS1 protein inhibits MAVS expression to attenuate its signaling (Nandi et al., 2014). Coronavirus targets MAVS/TRAF3/TRAF6 signalosome to suppress MAVS-mediated signaling (Shi et al., 2014). These studies also demonstrate that the molecular mechanisms used by viruses to block MAVS-mediated antiviral signaling are virus- specific. Here we demonstrated that hMPV M2-2 protein uses its PDZ motifs 29-DEMI-32 and 39-KEALSDGI-46 to suppress MAVS signaling by preventing the migration of adaptors TRAF3/5/6 to the mitochondrial MAVS. As mentioned, the interaction between the TRAF domain of TRAFs and the TIM of MAVS is required for MAVS-mediated signaling (Saha et al., 2006, Kumar et al., 2006). However, M2-2 does not contain a TRAF/TIM domain, but is enriched with PDZ domains. Ranganathan and Ross (1997) Among four PDZ-binding motifs in innate immune inhibitory domains of M2-2, motifs 29-DEMI-32 and 39-KEALSDGI-46 were investigated since the other two PDZ motifs, 58-LENI-61 and 61-IEII-64, have H-2d-restricted CTL epitopes (Melendi et al., 2007). We found that both motif 29-DEMI-32 and motif 39-KEALSDGI-46 are important for the activation of NF-kB- and IRF-3, and, subsequently, the induction of antiviral and proinflammatory mediators. The molecular mechanisms associated with 29-DEMI-32 -inhibited MAVS signaling is likely via preventing the recruitment of TRAF5 and TRAF6, two significant molecules controlling NF-kB and/or IRF-3 activation in hMPV infection. In addition, motif 39-KEALSDGI-46 also hampered the migration of TRAF3 to the mitochondria. Our findings are consistent with what we have previously reported—the domains where the motifs belong are not critical to viral gene transcription because mutations in these two motifs did not affect the gene transcription of viral proteins. However, the motif 29-DEMI-32 seemed also important for viral genome replication consistent with the fact that rhMPV-E30M31 was much more attenuated than rhMPV-E40L42D44. Collectively, results from both overexpression and recombinant virus systems complimentarily revealed novel motifs of M2-2 as innate immunity regulators and provided additional insights into the attenuation mechanism of rhMPV with complete M2-2 deletion (ΔM2-2). Currently, we are investigating whether rhMPV-E30M31 and rhMPV-E40L42D44 can be used to as more effective live attenuated vaccines against hMPV by comparing their immunogenicity, attenuation, and Th1/Th2 balance with those of ΔM2-2 (manuscript in preparation). This study reveals novel motifs of hMPV M2-2 protein responsible for regulating host antiviral response, a critical starting point for the accomplishment of preventive and therapeutic objectives.
Acknowledegments
All authors concur there are no conflicts of interest associated in this published work. This work was supported by grants from the National Institutes of Health-National Institute of Allergy and Infectious Diseases 1R01AI107033-01 and R21AI113771-01A1, the American Lung Association RG232529N, and American Heart Association 12BGIA12060008 to X.B. Authors thank Dr. Animesh Chandra for assistance with manuscript editing and submission, supported by the UTMB CTSA funded by NCATS grant UL1TR001439.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2016.09.026.
Appendix A. Supplementary material
Supplementary Material Supplementary Fig 1. Purity of isolated cellular compartments. A549 cells were infected with rhMPV-WT at MOI of 2. Mock infection was used as a control. At 15 h p.i., mitochondrial, cytosol and nuclear extracts were prepared. The abundance of mitochondria-associated MAVS and TRAF6 proteins was investigated by Western blot. The purity was demonstrated by the mitochondrial marker SDHA, nuclear marker lamin-b and cytosolic marker β-actin. Data shown are representative of two independent experiments.
.
References
- Akira S., Uematsu S., Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Bannister R. Use of a highly sensitive strand-specific quantitative PCR to identify abortive replication in the mouse model of respiratory syncytial virus disease. Virol. J. 2010;7:250. doi: 10.1186/1743-422X-7-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X. Airway epithelial cell response to human metapneumovirus infection. Virology. 2007;368:91–101. doi: 10.1016/j.virol.2007.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X. Human metapneumovirus small hydrophobic protein inhibits NF-kappaB transcriptional activity. J. Virol. 2008;82:8224–8229. doi: 10.1128/JVI.02584-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X. Human metapneumovirus glycoprotein G inhibits innate immune responses. PLoS Pathog. 2008;4:e1000077. doi: 10.1371/journal.ppat.1000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X. Human metapneumovirus glycoprotein G disrupts mitochondrial signaling in airway epithelial cells. PLoS One. 2013;8:e62568. doi: 10.1371/journal.pone.0062568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham A., Collins P.L. The M2-2 protein of human respiratory syncytial virus is a regulatory factor involved in the balance between RNA replication and transcription. Proc. Natl. Acad. Sci. USA. 1999;96:11259–11264. doi: 10.1073/pnas.96.20.11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biacchesi S. Genetic diversity between human metapneumovirus subgroups. Virology. 2003;315:1–9. doi: 10.1016/s0042-6822(03)00528-2. [DOI] [PubMed] [Google Scholar]
- Biacchesi S. Recovery of human metapneumovirus from cDNA: optimization of growth in vitro and expression of additional genes. Virology. 2004;321:247–259. doi: 10.1016/j.virol.2003.12.020. [DOI] [PubMed] [Google Scholar]
- Carpenter S., O’Neill L.A. Recent insights into the structure of Toll-like receptors and post-translational modifications of their associated signalling proteins. Biochem. J. 2009;422:1–10. doi: 10.1042/BJ20090616. [DOI] [PubMed] [Google Scholar]
- Casola A., Henderson A., Liu T., Garofalo R.P., Brasier A.R. Regulation of RANTES promoter activation in alveolar epithelial cells after cytokine stimulation. Am. J. Physiol. Lung Cell Mol. Physiol. 2002;283:L1280–L1290. doi: 10.1152/ajplung.00162.2002. [DOI] [PubMed] [Google Scholar]
- Dempsey P.W., Doyle S.E., He J.Q., Cheng G. The signaling adaptors and pathways activated by TNF superfamily. Cytokine Growth Factor Rev. 2003;14:193–209. doi: 10.1016/s1359-6101(03)00021-2. [DOI] [PubMed] [Google Scholar]
- Englund J.A. Brief communication: fatal human metapneumovirus infection in stem-cell transplant recipients. Ann. Intern. Med. 2006;144:344–349. doi: 10.7326/0003-4819-144-5-200603070-00010. [DOI] [PubMed] [Google Scholar]
- Falsey A.R., Erdman D., Anderson L.J., Walsh E.E. Human metapneumovirus infections in young and elderly adults. J. Infect. Dis. 2003;187:785–790. doi: 10.1086/367901. [DOI] [PubMed] [Google Scholar]
- Feng H. Expression profiles of carp IRF-3/-7 correlate with the up-regulation of RIG-I/MAVS/TRAF3/TBK1, four pivotal molecules in RIG-I signaling pathway. Fish Shellfish Immunol. 2011;30:1159–1169. doi: 10.1016/j.fsi.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Garofalo R.P. Transcriptional activation of the interleukin-8 gene by respiratory syncytial virus infection in alveolar epithelial cells: nuclear translocation of the RelA transcription factor as a mechanism producing airway mucosal inflammation. J. Virol. 1996;70:8773–8781. doi: 10.1128/jvi.70.12.8773-8781.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goutagny N. Cell type-specific recognition of human metapneumoviruses (HMPVs) by retinoic acid-inducible gene I (RIG-I) and TLR7 and viral interference of RIG-I ligand recognition by HMPV-B1 phosphoprotein. J. Immunol. 2010;184:1168–1179. doi: 10.4049/jimmunol.0902750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan K. MAVS promotes inflammasome activation by targeting ASC for K63-linked ubiquitination via the E3 ligase TRAF3. J. Immunol. 2015;194:4880–4890. doi: 10.4049/jimmunol.1402851. [DOI] [PubMed] [Google Scholar]
- Hacker H., Tseng P.H., Karin M. Expanding TRAF function: traf3 as a tri-faced immune regulator. Nat. Rev. Immunol. 2011;11:457–468. doi: 10.1038/nri2998. [DOI] [PubMed] [Google Scholar]
- Herd K.A. Cytotoxic T-lymphocyte epitope vaccination protects against human metapneumovirus infection and disease in mice. J. Virol. 2006;80:2034–2044. doi: 10.1128/JVI.80.4.2034-2044.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herd K.A., Nissen M.D., Hopkins P.M., Sloots T.P., Tindle R.W. Major histocompatibility complex class I cytotoxic T lymphocyte immunity to human metapneumovirus (hMPV) in individuals with previous hMPV infection and respiratory disease. J. Infect. Dis. 2008;197:584–592. doi: 10.1086/526536. [DOI] [PubMed] [Google Scholar]
- Inoue J. Tumor necrosis factor receptor-associated factor (TRAF) family: adapter proteins that mediate cytokine signaling. Exp. Cell Res. 2000;254:14–24. doi: 10.1006/excr.1999.4733. [DOI] [PubMed] [Google Scholar]
- Jin H. Recombinant respiratory syncytial viruses with deletions in the NS1, NS2, SH, and M2-2 genes are attenuated in vitro and in vivo. Virology. 2000;273:210–218. doi: 10.1006/viro.2000.0393. [DOI] [PubMed] [Google Scholar]
- Karron R.A. A gene deletion that up-regulates viral gene expression yields an attenuated RSV vaccine with improved antibody responses in children. Sci. Transl. Med. 2015;7:312ra175. doi: 10.1126/scitranslmed.aac8463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T., Akira S. Antiviral signaling through pattern recognition receptors. J. Biochem. 2006 doi: 10.1093/jb/mvm032. [DOI] [PubMed] [Google Scholar]
- Kolli D. Human metapneumovirus glycoprotein G inhibits TLR4-dependent signaling in monocyte-derived dendritic cells. J. Immunol. 2011;187:47–54. doi: 10.4049/jimmunol.1002589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar H. Essential role of IPS-1 in innate immune responses against RNA viruses. J. Exp. Med. 2006;203:1795–1803. doi: 10.1084/jem.20060792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S. Role of retinoic acid inducible gene-I in human metapneumovirus-induced cellular signalling. J. Gen. Virol. 2008;89:1978–1986. doi: 10.1099/vir.0.2008/000778-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Heylbroeck C., Genin P., Pitha P.M., Hiscott J. Essential role of interferon regulatory factor 3 in direct activation of RANTES chemokine transcription. Mol. Cell. Biol. 1999;19:959–966. doi: 10.1128/mcb.19.2.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. Elife. 2013;2:e00785. doi: 10.7554/eLife.00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melendi G.A. Mapping and characterization of the primary and anamnestic H-2d-restricted cytotoxic T-lymphocyte response in mice against human metapneumovirus. J. Virol. 2007;81:11461–11467. doi: 10.1128/JVI.02423-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakhaei P. The E3 ubiquitin ligase Triad3A negatively regulates the RIG-I/MAVS signaling pathway by targeting TRAF3 for degradation. PLoS Pathog. 2009;5:e1000650. doi: 10.1371/journal.ppat.1000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi S. MAVS protein is attenuated by rotavirus nonstructural protein 1. PLoS One. 2014;9:e92126. doi: 10.1371/journal.pone.0092126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T., Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- Pomerantz J.L., Baltimore D. NF-kappaB activation by a signaling complex containing TRAF2, TANK and TBK1, a novel IKK-related kinase. EMBO J. 1999;18:6694–6704. doi: 10.1093/emboj/18.23.6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan R., Ross E.M. PDZ domain proteins: scaffolds for signaling complexes. Curr. Biol. 1997;7:R770–R773. doi: 10.1016/s0960-9822(06)00401-5. [DOI] [PubMed] [Google Scholar]
- Ren J. Human metapneumovirus inhibits IFN-beta signaling by downregulating Jak1 and Tyk2 cellular levels. PLoS One. 2011;6:e24496. doi: 10.1371/journal.pone.0024496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J. A novel mechanism for the inhibition of interferon regulatory factor-3-dependent gene expression by human respiratory syncytial virus NS1 protein. J. Gen. Virol. 2011;92:2153–2159. doi: 10.1099/vir.0.032987-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J. Human metapneumovirus M2-2 protein inhibits innate cellular signaling by targeting MAVS. J. Virol. 2012;86:13049–13061. doi: 10.1128/JVI.01248-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J. Human metapneumovirus m2-2 protein inhibits innate immune response in monocyte-derived dendritic cells. PLoS One. 2014;9:e91865. doi: 10.1371/journal.pone.0091865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S.K. Regulation of antiviral responses by a direct and specific interaction between TRAF3 and Cardif. EMBO J. 2006;25:3257–3263. doi: 10.1038/sj.emboj.7601220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth R.B., Sun L., Ea C.K., Chen Z.J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Shi C.S. SARS-coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J. Immunol. 2014;193:3080–3089. doi: 10.4049/jimmunol.1303196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006;24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Tang E.D., Wang C.Y. TRAF5 is a downstream target of MAVS in antiviral innate immune signaling. PLoS One. 2010;5:e9172. doi: 10.1371/journal.pone.0009172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T., Ogasawara K., Takaoka A., Tanaka N. IRF family of transcription factors as regulators of host defense. Annu Rev. Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- Teng M.N. Recombinant respiratory syncytial virus that does not express the NS1 or M2-2 protein is highly attenuated and immunogenic in chimpanzees. J. Virol. 2000;74:9317–9321. doi: 10.1128/jvi.74.19.9317-9321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B. Identification of NF-kB dependent gene networks in respiratory syncytial virus-infected cells. J. Virol. 2002;76:6800–6814. doi: 10.1128/JVI.76.13.6800-6814.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajant H., Henkler F., Scheurich P. The TNF-receptor-associated factor family: scaffold molecules for cytokine receptors, kinases and their regulators. Cell Signal. 2001;13:389–400. doi: 10.1016/s0898-6568(01)00160-7. [DOI] [PubMed] [Google Scholar]
- Welsch C. Hepatitis C virus variants resistant to macrocyclic NS3-4A inhibitors subvert IFN-beta induction by efficient MAVS cleavage. J. Hepatol. 2015;62:779–784. doi: 10.1016/j.jhep.2014.11.009. [DOI] [PubMed] [Google Scholar]
- Williams J.V. The role of human metapneumovirus in upper respiratory tract infections in children: a 20-year experience. J. Infect. Dis. 2006;193:387–395. doi: 10.1086/499274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material Supplementary Fig 1. Purity of isolated cellular compartments. A549 cells were infected with rhMPV-WT at MOI of 2. Mock infection was used as a control. At 15 h p.i., mitochondrial, cytosol and nuclear extracts were prepared. The abundance of mitochondria-associated MAVS and TRAF6 proteins was investigated by Western blot. The purity was demonstrated by the mitochondrial marker SDHA, nuclear marker lamin-b and cytosolic marker β-actin. Data shown are representative of two independent experiments.