Summary
Understanding the population genetic consequences of declining population size is important for conserving the many species worldwide facing severe decline [1]. Thorough empirical studies on the impacts of population reduction at a genome-wide scale in the wild are scarce because they demand huge field and laboratory investments [1, 2]. Previous studies have demonstrated the importance of gene flow in introducing genetic variation to small populations [3], but few have documented both genetic and fitness consequences of decreased immigration through time in a natural population [4-6]. Here we assess temporal variation in gene flow, inbreeding, and fitness using longitudinal genomic, demographic, and phenotypic data from a long-studied population of federally Threatened Florida Scrub-Jays (Aphelocoma coerulescens; hereafter FSJ). We exhaustively sampled and genotyped the study population over two decades, providing one of the most detailed longitudinal investigations of genetics in a wild animal population to date. Immigrants were less heterozygous than residents but still introduced genetic variation into our study population. Owing to regional population declines, immigration into the study population declined from 1995-2013, resulting in increased levels of inbreeding and reduced fitness via inbreeding depression, even as the population remained demographically stable. Our results show that, contrary to conventional wisdom, small peripheral populations that already have undergone a genetic bottleneck may play a vital role in preserving genetic diversity of larger and seemingly stable populations. These findings underscore the importance of investing in the persistence of small populations and maintaining population connectivity in conservation of fragmented species.
Keywords: habitat fragmentation, gene flow, inbreeding depression, population genetics, conservation genomics
Results and Discussion
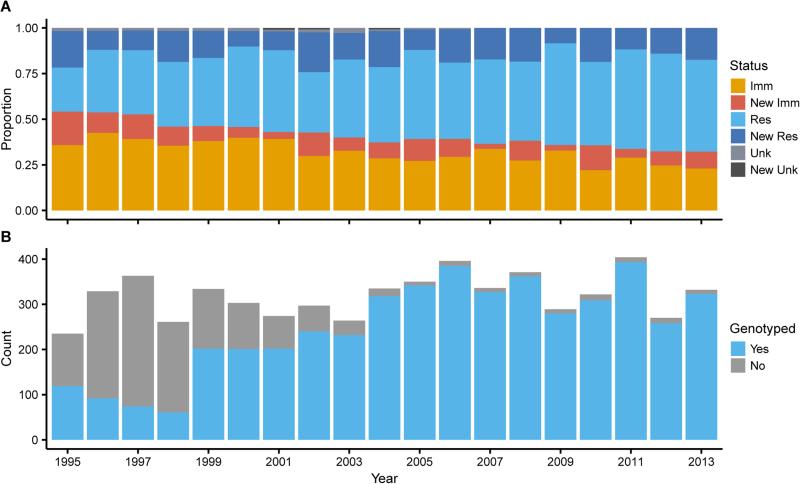
We investigated temporal changes in immigration and inbreeding from 1995-2013 in ~75 FSJ family groups. Intensive study of this population of individually-marked, non-migratory jays since 1969 provides a 14-generation pedigree and detailed lifetime reproductive histories [7]. In the past century, FSJs have undergone range-wide population declines and fragmentation caused by human-mediated habitat destruction [8]. Our study population has remained stable because of local habitat management [9]; however, the surrounding region continues to undergo declines in available habitat and numbers of jays [10]. Based on periodic surveys, we estimate that the FSJ population within 10 km surrounding the study area has dropped from ~554 families in 1985 to ~263 in 2016, largely due to habitat loss or lack of prescribed fire [10, 11]. This regional population decline corresponds with decreased immigration into our study population: the number and proportion of breeders born outside our study area declined significantly from 1995-2013 (Mann-Kendall test, p = 0.019 and 2.17 × 10−8, respectively; Figure 1A). Decreased immigration was likely due to the decline in regional jay densities, compounded by lower effective dispersal of FSJs in fragmented landscapes [12].
Figure 1. Proportion of immigrants and genotyped individuals in the study population through time.
(A) The proportion of new and old breeders, classified as known immigrants, known residents, and unknown-origin birds. Both immigration rate (proportion of new breeders that are immigrants) and the proportion of immigrant breeders significantly decreased (Mann-Kendall test, p = 0.042 & p = 2.17 × 10−8, respectively). (B) The total number of individuals (gray) and the number of genotyped individuals (blue) each year. The study population has been exhaustively monitored since 1969, and we genotyped >60% of the population from 1999-2013.
Population genetic consequences of decreased immigration depend on the proportion of breeding pairs that included an immigrant. The proportion of resident-resident pairs increased (from 0.21 in 1995 to 0.44 in 2013; Mann-Kendall test, p = 8.75 × 10−6), and the proportion of immigrant-immigrant pairs decreased (from 0.29 in 1995 to 0.10 in 2013; Mann-Kendall test, p = 7.63 × 10−5). The proportion of immigrant-resident pairs averaged 0.49 and did not change significantly (Mann-Kendall test, p = 0.83).
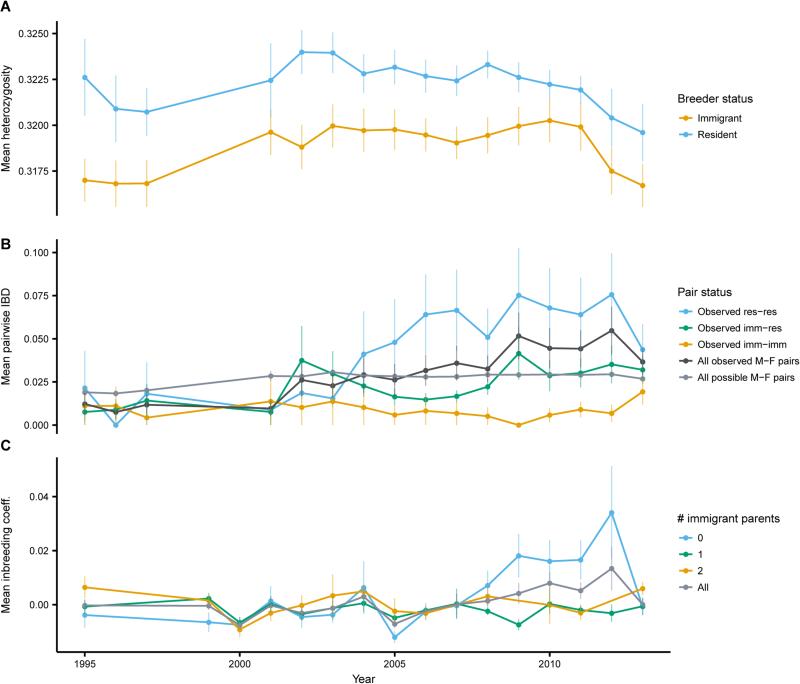
To investigate the genomic contributions of immigrants to the study population, we genotyped 3,583 individuals at 15,416 single nucleotide polymorphisms (SNPs; see Supplemental Experimental Procedures) [13]. Here, autosomal SNPs were thinned to retain 7,834 SNPs in approximate linkage equilibrium. Near-complete sampling of all nestlings and breeders (Figure 1B) allowed us to assess temporal trends in heterozygosity and relatedness, measured as the proportion of the genome shared identical-by-descent (IBD). Immigrants had significantly lower levels of observed heterozygosity compared to residents (Wilcoxon rank sum test, p = 1.80 × 10−6; Figure 2A). These data are consistent with the observation that immigrants to our study population could only have originated in smaller, more isolated, and presumably more inbred populations. We regard it as unlikely that individuals dispersing from other populations are less heterozygous than non-dispersing individuals, but we cannot rule out this possibility.
Figure 2. Changes in heterozygosity and inbreeding from 1995-2013.
(A) Mean ± SEM genome-wide observed heterozygosity for immigrant and resident breeders. Immigrants were less heterozygous compared to residents (Wilcoxon rank sum test, p < 1 × 10−12). (B) Mean ± SEM proportion of the genome shared IBD between all possible male-female (light gray), all observed male-female (dark gray), observed immigrant-immigrant (yellow), observed immigrant-resident (green), and observed resident-resident pairs (blue). Relatedness among all observed pairs significantly increased (Mann-Kendall test, p = 7.44 × 10−8). Pairs with at least one immigrant were less related than resident-resident pairs. (C) Mean ± SEM inbreeding coefficient of all Day 11 nestlings (gray), and nestlings with zero (blue), one (green), or two (yellow) immigrant parents. Mean inbreeding coefficient of the birth cohort increased (Mann-Kendall test, p = 0.0008), with elevated inbreeding in offspring of residents in 2009-2012. See also Figure S2 and Table S1.
Nonetheless, immigrants contributed genetic variation to the population. Across all years, average relatedness among residents (0.036) was higher than between residents and immigrants (0.021) and among immigrants (0.020; Wilcoxon rank sum test, p < 1 × 10−12 for both). Among observed breeding pairs, immigrant-immigrant pairs had significantly lower IBD compared to immigrant-resident pairs (0.009 and 0.025, respectively; Wilcoxon rank sum test, p = 0.015), which had significantly lower IBD compared to resident-resident pairs (0.056; Wilcoxon rank sum test, p = 0.006). These results clearly illustrate the importance of immigrants in contributing genetic variation to our population over time, even in the face of overall regional declines.
As immigration into our study population decreased from 1995-2013, relatedness of observed breeding pairs increased (Mann-Kendall test, p = 7.44 × 10−8; Figure 2B), as did mean inbreeding coefficient of the birth cohort, estimated from genome-wide SNPs [14] (Mann-Kendall test, p = 0.0008; Figure 2C). Proportion IBD sharing increased significantly for resident-resident and immigrant-resident pairs (Mann-Kendall test, p = 0.0008 and p = 0.003, respectively), but not for immigrant-immigrant pairs (Mann-Kendall test, p = 0.19; Figure 2B). Inbreeding levels decreased in 2013, primarily because of decreased inbreeding in nestlings with resident-resident parents (Figure 2C). The proportion of resident-resident offspring with at least one immigrant grandparent was higher in 2013 (0.88) compared to 2009-2012 (0.74-0.77), likely because of the unusually high influx of immigrants in 2010. Given delayed dispersal in this species, many 2010 nestlings did not successfully produce young until 2013. Temporal variation in the proportion of immigrant parents explained 34% of the variance in mean cohort inbreeding, and variation in the proportion of immigrant grandparents explained 35% of the variance in mean inbreeding of offspring of resident-resident parents (Table S1). Even though our study population had not decreased in size through time, reduced immigration clearly caused increased inbreeding in the population.
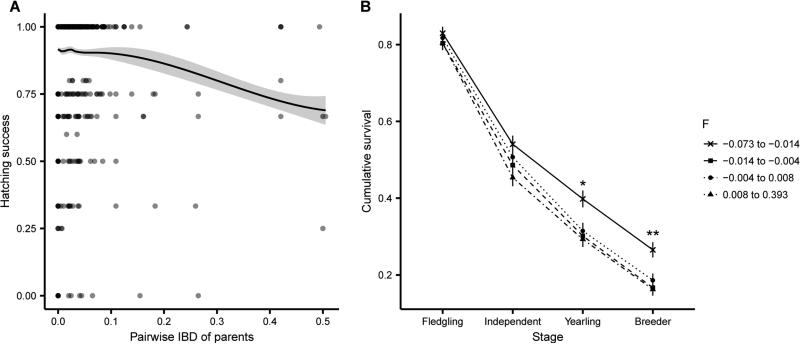
High levels of inbreeding are known to produce serious consequences on fitness [15]. We tested for inbreeding depression on several fitness-related traits: hatching success, nestling weight, juvenile survival to key life-history stages, breeder lifespan, and lifetime reproductive success (LRS, measured as the number of fledglings produced over a breeder's lifetime). Nestling weight was the strongest predictor of survival from nestling to later life stages and of survival from fledgling to independence (Table S2), which is consistent with previous findings [16]. After controlling for potential confounding factors, pairwise IBD of the parents, a proxy for expected inbreeding of the offspring, was strongly correlated with hatch failure (Figure 3A). Moreover, individual inbreeding coefficients were significantly associated with lower nestling weight and reduced survival from independence to one year (Table S3). The magnitude of the inbreeding effect increased as the number of life stages considered increased, with a strong combined effect of inbreeding on survival from Day 11 to one year and beyond (Figure 3B). The average haploid genome carries 7.48 (95% CI [1.22, 14.76]) lethal equivalents for survival from Day 11 to one year (Table S3). The fitness impact of inbreeding depression was stronger in females than males: inbreeding coefficient was significantly associated with breeder lifespan in both sexes, but with LRS in females only (Table S3). In sum, we found evidence of inbreeding depression in multiple life-history stages, from hatching success to adult fitness.
Figure 3. Inbreeding depression in hatching success and juvenile survival.
(A) Proportion of the genome shared IBD between the parents was negatively correlated with hatching success (n = 769 nests). The line shows predicted values from the fitted model, and the shaded area shows the 95% CI. (B) Survival of Day 11 nestlings in each inbreeding coefficient quartile to different life-stages (n = 2019 individuals). Error bars ± SEM. Asterisks indicate stages significantly associated with inbreeding (*p < 0.05, **p < 0.01). Compared to individuals in the bottom quartile, individuals in the top three quartiles (more inbred) are 24% less likely to survive one year and 35% less likely to survive to breed. See also Figure S3, Table S2, and Table S3.
Decreasing immigration rates, associated with increased inbreeding and consequent inbreeding depression in a number of fitness-related traits, appeared to affect key parameters of fitness in our population. Along with increasing inbreeding, hatching success decreased significantly over time (Mann-Kendall test, p = 0.006), as did survival from fledgling to independence (Mann-Kendall test, p = 9.04 × 10−6).
To our knowledge, this study is the first to demonstrate negative genetic and fitness consequences of decreased gene flow through time within a large natural population that seemingly remains demographically stable. Our study population is one of the largest and best-managed remaining populations of this endangered species [10] and is historically considered to be of sufficient size and protection to ensure long-term population viability [17]. The steady decrease in immigration rate currently underway is presumably a direct consequence of ongoing population declines and local extirpations in the surrounding region. Even though our study population is in one of the most contiguous landscapes remaining in the extant distribution of FSJs, regional declines of >50% over the past two decades led to elevated local inbreeding. The population has remained demographically stable because FSJs are long-lived and breeder mortality is negatively correlated with density, but we anticipate that population size will decline soon if immigration rate continues to decrease. Demographic effects are likely to be more rapid and severe in smaller populations that are not saturated with jays and have even lower immigration rates. This suggests that range-wide, genetic consequences of habitat fragmentation are likely to be much higher than previously anticipated. Because immigrants into our study population likely originated from multiple populations, we suspect that the greater average fitness we found among offspring of immigrants was driven by heterosis, rather than by a handful of specific beneficial alleles carried by immigrants. Regardless, our results underscore that fighting off the “extinction vortex” [18, 19] needs to begin early and involve active conservation efforts even while some populations outwardly appear healthy and stable. When prioritizing investments in landscape protection for fragmented endangered species, attention should be paid to preserving small and even inbred populations, as they can play a vital role in preserving and enhancing genetic diversity among larger and seemingly stable populations.
Experimental Procedures
Study population
The FSJ is a non-migratory, federally Threatened bird restricted to xeric oak scrub habitat unique to Florida. A population of FSJs at Archbold Biological Station (Highlands County, FL) has been intensively studied since 1969 [20]. Every individual is uniquely banded, allowing continuous documentation of immigrants. All nests of all family-groups are monitored, producing fully documented annual fecundity and fitness measures for all breeding birds. Individuals born in the study site are measured, banded, and blood-sampled as 11-day-old nestlings and again as juveniles (50-100 days post-hatch). Unbanded adults are captured for sampling and banding as soon as possible. We have blood samples for every nestling and immigrant recruited into the study population in 1989-1991, 1995, and 1999 to the present. All work was approved by the Cornell University Institutional Animal Care and Use Committee (IACUC 2010-0015) and authorized by permits from the U.S. Fish and Wildlife Service (TE824723-8), the U.S. Geological Survey (banding permit 07732), and the Florida Fish and Wildlife Conservation Commission (LSSC-10-00205).
To avoid biases caused by study tract expansion during the late 1980s, we restricted our analyses to a set of 54-76 territories that have been consistently monitored since 1990 and span a geographic area of approximately constant size. We classified a breeder as an immigrant if it was known to be born outside the core study tract. Unbanded individuals that appeared before 1990 could have been born in the geographic area considered in our analysis before population monitoring expanded, and were classified as unknown-origin. The proportion of unknown-origin breeders dropped to below 4% by 1995; therefore we started our analyses in 1995. Data are available at figshare (10.6084/m9.figshare.3593088).
SNP discovery and genotyping
Genomic DNA was extracted from blood samples in lysis buffer. We used genotypingby-sequencing of immigrants and residents from 1978–2008 to discover SNPs [13], then designed custom Illumina iSelect BeadChips for 15,416 genome-wide SNPs (Figure S1). We genotyped 4,032 samples at Geneseek, Inc. (Lincoln, NE). SNP quality control (Gentrain score > 0.7, SNP and individual call rate > 95%) and pedigree validation were performed in GenomeStudio (Illumina, San Diego, CA), PLINK [21], and PedCheck [22]. To obtain unbiased estimates of genetic diversity and relatedness, we removed Z-linked SNPs and pruned SNPs in high linkage disequilibrium. Our final dataset consisted of 7,834 autosomal SNPs in approximate linkage equilibrium in 3,583 individuals. See Supplemental Experimental Procedures for details.
Heterozygosity and inbreeding
To investigate the genetic contribution of immigrants to the population, we used PLINK (options --het, --ibc, and --genome) to estimate individual inbreeding coefficients and mean observed heterozygosity as well as pairwise IBD. We only included years with >50 genotyped breeders or nestlings. The genomic estimator of inbreeding coefficient used here reflects the correlation between uniting gametes (FIII from [14]; see Supplemental Experimental Procedures and Figure S2). Mean site-based observed heterozygosity was calculated as the number of heterozygous loci divided by the number of loci genotyped in that individual. We used Mann-Kendall tests adjusted for autocorrelation to test for temporal trends using the R package fume [23].
We modeled temporal variation in expected mean offspring inbreeding as a function of the proportion of resident-resident, resident-immigrant, and immigrant-immigrant parents and mean IBD between these pairs. We fit models using both constant and time-varying values for the two variable sets and calculated the coefficient of determination to determine the impact of immigrants on temporal changes in offspring inbreeding. Similar models were fit for grandparents of children of two residents. See Supplemental Experimental Procedures and Table S1. All statistical analyses were performed in R [24].
Inbreeding depression
We tested for inbreeding depression in hatching success, several juvenile traits, breeder lifespan (number of years an individual bred in the study population), and LRS (measured as the total number of fledglings). Juvenile traits included nestling weight (at Day 11) and survival to key life stages: Day 11 post-hatch, fledgling (~Day 18), nutritional independence (~Day 85), yearling (~Day 300, when birds are physiologically capable of breeding), and recruited breeder. We obtained hatching success from 769 nests with genotyped parents from 1987-2013. An egg was considered a hatching failure if it remained unpipped more than 5 days after the other eggs had hatched. To only include eggs that failed to hatch due to infertility or early embryo mortality in this analysis, nests that were depredated or abandoned fewer than three days after the eggs hatched were excluded. Our estimate of survival to breed is an underestimate because it fails to account for birds who emigrate and breed in other populations. Adult analyses only included breeders that died before 2014. Inbreeding depression analyses used data from the core territories in 1980-2013; sample sizes are listed in Table S3.
We fitted mixed models with the appropriate error structure for each trait. Specifically, we used logistic for hatching success and juvenile survival, Gaussian for nestling weight, negative binomial for breeder lifespan, and zero-inflated negative binomial for LRS. Since nestlings are banded at Day 11, we used the proportion of IBD sharing between the parents as a proxy for inbreeding for hatching success and survival from hatching to day 11. For each trait, we first tested for significant predictors other than inbreeding by fitting models for each independent variable separately as a fixed effect. Fixed effects considered in the inbreeding depression models included characteristics of the individual (sex, nestling and juvenile weight), the natal nest (clutch size, brood size, incubation date, hatch date, age at fledgling, number of helpers, pair experience, ages of mom and dad), the natal territory (territory size, time since fire), and the natal year (acorn abundance, breeding density, drought index, precipitation, temperature). Random effects included natal year and either the identity of the pair for the nest-based models (hatching success and survival from hatch to Day 11) or the natal nest for the individual-based models (all other traits). Then, we constructed models for all combinations of significant predictors and performed model selection using the Akaike information criterion. To test for the importance of inbreeding while controlling for other potential confounding effects, we fitted all fixed and random effects from the best model in addition to the inbreeding coefficient or relatedness of the parents. Fixed effects were standardized to ensure model convergence. We fitted models using the R packages lme4 [25], lmerTest [26], pscl [27] and glmmADMB [28]. We estimated the average number of lethal equivalents using methods from [29]. See Supplemental Experimental Procedures for details, including a discussion on temporal autocorrelation.
Supplementary Material
Highlights.
Inbreeding increased over 19 years in a pedigreed population of an endangered bird
Genomic sampling reveals that the increase was caused by reduced immigration
Inbreeding affects many life-history stages, from hatching success to adult fitness
Gene flow from small populations is vital for conservation of large populations
Acknowledgments
We thank J. Grenier, C. Acharya, and L. Stenzler for help with lab work. G. Coop, W. Hochachka, and V. Buffalo provided statistical advice. We thank the students, interns, and staff at Archbold Biological Station who collected the demographic data. We thank S. Pruett for assistance in demographic data quality control. Thanks to R. Harrison, the Clark and Harrison labs, D. Irwin, and an anonymous reviewer for comments. This work was supported by NSF (DEB0855879 and DEB1257628) and the Cornell Lab of Ornithology Athena Fund. N.C. was supported by a NSF Graduate Research Fellowship, a Cornell Center for Comparative and Population Genomics Fellowship, and NIH grant RO1GM107374 to G. Coop.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
N.C., J.W.F., and A.G.C. designed the study. N.C. collected genotype data, and N.C., E.J.C., J.W.F., and R.B. organized FSJ demographic data. N.C. analyzed data and wrote the paper, with comments from all authors.
References
- 1.Ouborg NJ, Vergeer P, Mix C. The rough edges of the conservation genetics paradigm for plants. J Ecol. 2006;94:1233–1248. [Google Scholar]
- 2.Kohn MH, Murphy WJ, Ostrander EA, Wayne RK. Genomics and conservation genetics. Trends Ecol Evol. 2006;21:629–637. doi: 10.1016/j.tree.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 3.Keller LF, Jeffery KJ, Arcese P, Beaumont MA, Hochachka WM, Smith JNM, Bruford MW. Immigration and the ephemerality of a natural population bottleneck: evidence from molecular markers. Proc Biol Sci. 2001;268:1387–1394. doi: 10.1098/rspb.2001.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hogg JT, Forbes SH, Steele BM, Luikart G. Genetic rescue of an insular population of large mammals. Proc Biol Sci. 2006;273:1491–1499. doi: 10.1098/rspb.2006.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Westemeier RL, Brawn JD, Simpson SA, Esker TL, Jansen RW, Walk JW, Kershner EL, Bouzat JL, Paige KN. Tracking the long-term decline and recovery of an isolated population. Science. 1998;282:1695–1698. doi: 10.1126/science.282.5394.1695. [DOI] [PubMed] [Google Scholar]
- 6.Johnson WE, Onorato DP, Roelke ME, Land ED, Cunningham M, Belden RC, McBride R, Jansen D, Lotz M, Shindle D, et al. Genetic Restoration of the Florida Panther. Science. 2010;329:1641–1645. doi: 10.1126/science.1192891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fitzpatrick JW, Bowman R. Florida scrub-jays: oversized territories and group defense in a fire-maintained habitat. In: Koenig WD, Dickinson JL, editors. Cooperative Breeding in Vertebrates. Cambridge University Press; Cambridge: 2016. pp. 77–96. [Google Scholar]
- 8.Woolfenden GE, Fitzpatrick JW. Florida Scrub-Jay (Aphelocoma coerulescens). In: Poole A, editor. The Birds of North America online. Vol. 228. Cornell Lab of Ornithology; Ithaca: 1996. [Google Scholar]
- 9.Turner WR, Wilcove DS, Swain HM. Assessing the effectiveness of reserve acquisition programs in protecting rare and threatened species. Conserv Biol. 2006;20:1657–1669. doi: 10.1111/j.1523-1739.2006.00536.x. [DOI] [PubMed] [Google Scholar]
- 10.Boughton RK, Bowman R. State wide assessment of Florida Scrub-Jays on managed areas: A comparison of current populations to the results of the 1992-1993 survey, (Report submitted to the USFWS) 2011 [Google Scholar]
- 11.Fitzpatrick J, Pranty B, Stith B. Florida Scrub Jay statewide map, 1992-1993. Archbold Biological Station; Lake Placid, FL, USA: 1994. p. 16. [Google Scholar]
- 12.Coulon A, Fitzpatrick JW, Bowman R, Lovette IJ. Effects of habitat fragmentation on effective dispersal of Florida Scrub-Jays. Conserv Biol. 2010;24:1080–1088. doi: 10.1111/j.1523-1739.2009.01438.x. [DOI] [PubMed] [Google Scholar]
- 13.Chen N, Van Hout CV, Gottipati S, Clark AG. Using Mendelian Inheritance To Improve High-Throughput SNP Discovery. Genetics. 2014;198:847–857. doi: 10.1534/genetics.114.169052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A Tool for Genome- wide Complex Trait Analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keller LF, Waller DM. Inbreeding effects in wild populations. Trends Ecol Evol. 2002;17 [Google Scholar]
- 16.Mumme RL, Bowman R, Pruett MS, Fitzpatrick JW. Natal territory size, group size, and body mass affect lifetime fitness in the cooperatively breeding Florida Scrub-Jay. The Auk. 2015;132:634–646. [Google Scholar]
- 17.Fitzpatrick JW, Woolfenden GE, Kopeny MT. Ecology and development- related habitat requirements of the Florida Scrub Jay (Aphelocoma coerulescens coerulescens) Office of Environmental Services, Florida Game and Fresh Water Fish Commission; 1991. [Google Scholar]
- 18.Gilpin ME, Soulé ME. Minimum viable populations: processes of extinction. In: Soulé ME, editor. Conservation Biology: The Science of Scarcity and Diversity. Sinauer Associates; Sunderland, MA: 1986. pp. 19–34. [Google Scholar]
- 19.Lynch M, Conery J, Burger R. Mutational Meltdowns in Sexual Populations. Evolution. 1995;49:1067–1080. doi: 10.1111/j.1558-5646.1995.tb04434.x. [DOI] [PubMed] [Google Scholar]
- 20.Woolfenden GE, Fitzpatrick JW. The Florida Scrub Jay - Demography of a cooperative-breeding bird. Princeton University Press; Princeton: 1984. [Google Scholar]
- 21.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santander Meteorology Group fume: FUME package. (R package version 1.0) 2012 [Google Scholar]
- 24.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2015. [Google Scholar]
- 25.Bates D, Maechler M, Bolker B, Walker S. Fitting Linear Mixed-Effects Models Using {lme4}. J Stat Softw. 2015;67:1–48. [Google Scholar]
- 26.Kuznetsova A, Bruun Brockhoff P, Haubo Bojesen Christensen R. lmerTest: Tests in Linear Mixed Effects Models. (R package version 2.0-30) 2016 [Google Scholar]
- 27.Zeileis A, Kleiber C, Jackman S. Regression Models for Count Data in R. J Stat Softw. 2008;27 [Google Scholar]
- 28.Fournier DA, Skaug HJ, Ancheta J, Ianelli J, Magnusson A, Maunder MN, Nielsen A, Sibert J. AD Model Builder: using automatic differentiation for statistical inference of highly parameterized complex nonlinear models. Optim Methods Softw. 2012;27:233–249. [Google Scholar]
- 29.Grueber CE, Nakagawa S, Laws RJ, Jamieson IG. Multimodel inference in ecology and evolution: challenges and solutions. J Evol Biol. 2011;24:699–711. doi: 10.1111/j.1420-9101.2010.02210.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.