Abstract
Helicobacter pylori is the strongest identified risk factor for gastric cancer, the third most common cause of cancer-related death worldwide. An H. pylori constituent that augments cancer risk is the strain-specific cag pathogenicity island, which encodes a type IV secretion system (T4SS) that translocates a pro-inflammatory and oncogenic protein, CagA, into epithelial cells. However, the majority of persons colonized with CagA+ H. pylori strains do not develop cancer, suggesting that other microbial effectors also play a role in carcinogenesis. Toll-like receptor 9 (TLR9) is an endosome bound, innate immune receptor that detects and responds to hypo-methylated CpG DNA motifs that are most commonly found in microbial genomes. High expression tlr9 polymorphisms have been linked to the development of premalignant lesions in the stomach. We now demonstrate that levels of H. pylori-mediated TLR9 activation and expression are directly related to gastric cancer risk in human populations. Mechanistically, we show for the first time that the H. pylori cancer-associated cag T4SS is required for TLR9 activation and that H. pylori DNA is actively translocated by the cag T4SS to engage this host receptor. Activation of TLR9 occurs through a contact-dependent mechanism between pathogen and host, and involves transfer of microbial DNA that is both protected as well as exposed during transport. These results indicate that TLR9 activation via the cag island may modify the risk for malignancy within the context of H. pylori infection and provide an important framework for future studies investigating the microbial-epithelial interface in gastric carcinogenesis.
Introduction
Helicobacter pylori is a bacterial carcinogen that incurs the highest known risk for gastric cancer (1). Approximately 80% of the gastric cancer burden and 5.5% of all malignancies worldwide are attributable to H. pylori-induced injury (1, 2). However, only a subset of chronically colonized individuals ever develop neoplasia. Enhanced risk for gastric carcinogenesis is related to H. pylori strain differences, inflammatory responses governed by host genetic diversity, and/or specific interactions between host and microbial determinants (3).
One cancer-linked H. pylori locus is the cag pathogenicity island, which encodes a type IV secretion system (T4SS) that forms a syringe-like structure to translocate the effector oncoprotein CagA and peptidoglycan into host cells (4–19). T4SSs are common among Gram-negative bacteria due to their versatility in terms of the type and destination of secreted substrates. However, despite the ubiquity of T4SS in Gram-negative bacteria, their ability to mediate trans-kingdom DNA transfer is rare. Indeed, only a very few bacterial T4SSs have been shown to facilitate DNA transfer into yeast (20, 21) or mammalian cells (22–24). The T4SS of Agrobacterium tumefaciens is the only known example of a pathogenesis-associated T4SS that facilitates plasmid DNA transfer into a eukaryotic host (25–27). In this system, A. tumefaciens translocates the tumor-inducing Ti-plasmid into plant cells, which is then incorporated into the genome and ultimately results in malignant transformation. A. tumefaciens T4SS-mediated DNA transfer has also been shown to promote transformation of human cells under non-physiological conditions (28). Of particular interest, the A. tumefaciens archetypal T4SS retains a high level of homology to the cag type IV secretion system, suggesting that H. pylori has the ability to translocate DNA into host cells via the cag island.
In addition to the cag T4SS, host factors have also been implicated in augmenting H. pylori-induced gastric cancer risk. Toll-like receptors (TLRs) orchestrate host immune responses targeting pathogens via selective recognition of pathogen-associated molecular patterns (PAMPs) (29); however, chronic activation of TLRs in the gastric niche has been implicated in promoting carcinogenesis (30). TLR9 is an intracellular receptor that recognizes hypo-methylated CpG motifs (31), which are abundant in DNA of bacterial, viral, or synthetic origin, but are atypical within mammalian genomes (32). TLR9 expression is up-regulated in human gastric cancer specimens, and H. pylori DNA has been shown to directly promote cancer cell invasion (33–38). Moreover, polymorphisms in the tlr9 gene have also been shown to increase the risk for development of both pre-malignant and malignant gastric lesions (30, 39). Therefore, we utilized human gastric specimens and in vitro models of microbial-epithelial interactions to define the role of H. pylori DNA translocation and TLR9 activation in gastric carcinogenesis.
Results and Discussion
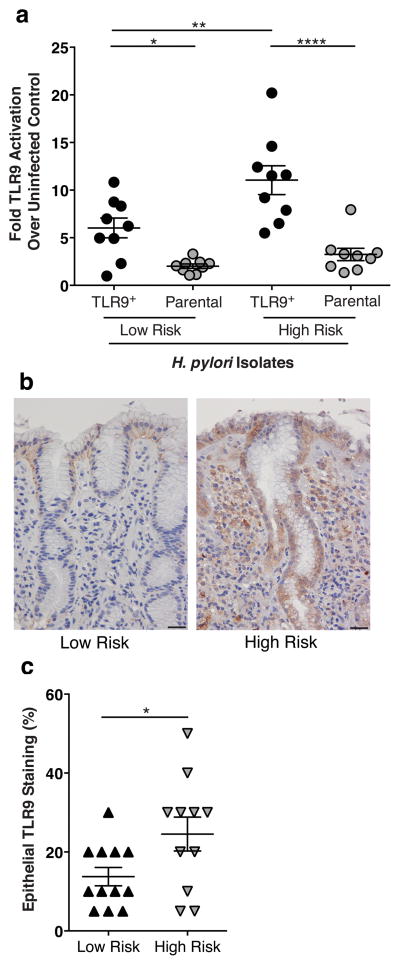
In many areas of the world, geographic differences in gastric cancer rates are present despite similarly high H. pylori prevalence rates (40). This has been well described in Colombia, where residents of the Andean Mountain region have an extraordinarily high incidence of gastric cancer (150/100 000), compared to inhabitants of the Pacific coast region (6/100 000), despite the fact that greater than 90% of the collective population is infected with H. pylori (41, 42). This disparity in gastric cancer risk but not H. pylori prevalence (15, 43–45) provides a unique opportunity to define microbial and host determinants that play a role in gastric carcinogenesis. Therefore, we first analyzed H. pylori strains isolated from the high and low gastric cancer risk regions for their ability to activate TLR9 using a specific HEK293-hTLR9 reporter assay system. In this reporter assay, HEK293 cells, which are inherently devoid of most innate immune receptors, are stably transfected with either a TLR9 expression plasmid and an NFκB/AP-1-linked SEAP reporter (TLR9+), or a control NFκB/AP-1 SEAP reporter alone (Parental). Cells were challenged with H. pylori strains isolated from patients residing in either the high-risk or low-risk region. H. pylori strains isolated from the high-risk region induced significantly higher levels of TLR9 activation compared to strains harvested from patients in the low-risk region (Figure 1a). To determine whether the ability of high-risk H. pylori isolates to activate TLR9 in vitro translated into the cognate gastric niche, levels of epithelial TLR9 expression in gastric biopsies obtained from infected patients were quantified. Gastric epithelial TLR9 expression levels were significantly increased in patients residing in the high-risk region compared to the low-risk region (Figure 1b,c), findings that mirror the in vitro data. Patients residing in the high cancer-risk region also displayed significantly increased histological scores compared to patients in the low cancer-risk region (mean ± SEM; 3.89±0.26 vs. 2.05±0.11, respectively; p≤0.0001). Additionally we observed that TLR9 expression was more extensive and involved more regions of the gastric glands in patients residing in the high-risk region, frequently extending from the base to the gastric pit. Collectively, these results indicate that H. pylori strains linked to an increased risk for gastric cancer induce more intense TLR9 activation in vitro and enhanced TLR9 expression in vivo.
Figure 1. H. pylori activation of TLR9 in a human population.
(a) TLR9 activation by H. pylori isolates obtained from patients residing in a low or high gastric cancer risk region of Colombia. HEK-Blue-hTLR9 cells (TLR9+) and HEK-Blue-Null1 (Parental) cells (Invivogen, San Diego, CA USA) were seeded in 96 well plates (Co-Star, St. Louis, MO USA) at 80,000 cells per well in DMEM (Corning, Manassas, VA USA) without antibiotics and were challenged with either viable H. pylori (MOI 100) or media alone at 37°C with 5% CO2 for 24 hours. After 24 hours, 20μl of supernatants were added to 180μl of HEK-Blue Detection media (Invivogen). Plates were analyzed by spectrophotometer (Bitoek, Winooski, VT USA) at 650nm. All experiments were performed in duplicate and repeated at least 3 times. Data are expressed as fold over uninfected control. N=9 isolates per group, mean±SEM are shown. A one-way analysis of variance with Bonferroni correction was used to determine statistical significance between groups. (b,c) Gastric antral biopsy samples from patients residing in a high or low gastric cancer risk region in the Colombia, who were enrolled in an ongoing prospective study designed to study mechanisms of H. pylori carcinogenesis (57), were used for immunohistochemistry (histologic scores mean±SEM; low-risk, 2.05±0.11; high-risk, 3.89±0.26, p≤0.0001 Student’s t-test; histology scored as previously described (57)). Immunohistochemistry was performed on paraffin-embedded biopsy samples from de-identified, H. pylori-infected, patients with non-atrophic gastritis, intestinal metaplasia (IM), or gastric dysplasia. Tissue samples were deparaffinized and stained with a polyclonal anti-TLR9 antibody (1:1000, Imgenex San Diego, CA USA). A single pathologist (MBP) scored TLR9 IHC staining by assessing the percentage of TLR9+ epithelial cells. Magnification 40x; Scale bar, 50μm. Epithelial TLR9 staining of biopsies obtained from patients in the low (N=11) and high (N=12) gastric cancer risk regions of Colombia is quantified in (c). Student’s t-test was used to determine statistical significance between groups. (a,c) *p<0.05, **p<0.01,***p<0.001, ****p<0.0001.
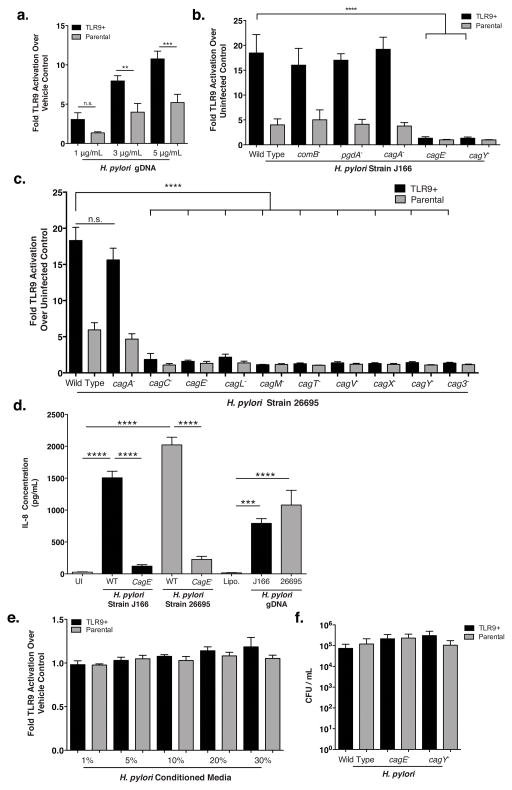
Despite the evolutionary homology of the cag T4SS to the archetypal A. tumefaciens T4SS (46), DNA translocation by the H. pylori cag system has never been demonstrated. Therefore, to define mechanisms through which H. pylori activates TLR9, we subsequently conducted experiments using the well-characterized wild-type and mutant H. pylori cag+ strains J166, 26695, and 7.13 (12, 47, 48). Previous reports have shown that H. pylori DNA induces TLR9 activation in immune cells (49–51); thus, we first confirmed that purified H. pylori genomic DNA (gDNA) could induce TLR9 activation in our HEK293 reporter assay (Figure 2a). We also demonstrated that viable wild-type cag+ H. pylori could activate TLR9 to significantly increased levels compared to controls (Figure 2b, 2c, S1). The specificity of the TLR9-dependent response was then validated in HEK293-hTLR9 reporter cells that were pre-treated with the endosomal inhibitor chloroquine, which blocks TLR9 activation. H. pylori-infected reporter cells pre-challenged with chloroquine displayed a significant decrease in TLR9 activation compared to control cells (Figure S2).
Figure 2. H. pylori activation of TLR9 requires a functional cag T4SS.
(a–c) The H. pylori cag+ strains J166 and 26695 (ATCC 700392) were maintained on trypticase soy agar plates supplemented with 5% sheep blood (BD Biosciences, Sparks MD USA) as described (48, 58). Isogenic mutants were constructed as previously described (48, 55, 58–62). Flanking sequences for comB were amplified from H. pylori strain J166 DNA using primers comB8 Forward (5′-ACTAGAGCTCAAGCCTTTCAATAGCGAGCA- 3′), comB8 Reverse (5′-AGTACCGCGGAGCGATTTTCAAGCGGTTC -3’) and comB10 Forward (5’-CTGAGAATTCTTGCAATTGATGAGGCAAAG-3′) comB10 Reverse (5′-ACTAGGTACCGCGATGACTTCATTCTCTCTGG -3′). comB flanking sequences were cloned into a pBSC103 plasmid using a previously inserted kanamycin resistance cassette generated by restriction enzymes SacI and SacII (comB8) and EcoRI and KpnI (comB10). The resultant plasmid was used to transform H. pylori strain J166 and transformants were selected on Brucella agar plates supplemented with kanamycin (5 μg/mL). Correct orientation of the kanamyacin cassette with H. pylori comB was confirmed by PCR analyses. (a) H. pylori strain J166 DNA was purified by growing microbial cultures in Brucella broth supplemented with 10% neonatal calf serum (NCS) (Atlanta Biologicals, Atlanta, GA USA) overnight. Cultures were centrifuged (4000 RPM, 5 min), resuspended in 600 μL of TE buffer with 0.5% SDS and 100 μg/mL proteinase K (Qiagen Germantown MD USA), and incubated at 37°C for 1 hour. DNA was extracted using CTAB and purified by phenol-chloroform extraction. TLR9-reporter or parental cells were cultured as described in Figure 1 and challenged with purified H. pylori strain J166 DNA (1–5 μg/mL) supplemented with Lipofectamine 2000 (Life Technologies, Carlsbad CA, USA) at 37°C with 5% CO2 for 24 hours. Data are represented as fold over vehicle control. (b) TLR9 activation induced by H. pylori strain J166 or its isogenic mutants (MOI 100, T=24hrs), relative to uninfected control. (c) TLR9 activation induced by H. pylori strain 26695 or its isogenic mutants (MOI 100, T=24hrs), relative to uninfected control. (d) AGS gastric epithelial cells (ATCC) were grown in RPMI (Cell Gro, Manassas, VA, USA) supplemented with 5% fetal bovine serum (Atlanta Biologicals). Cells were seeded at 500 000 cells per well in a 12-well culture dish (Corning) and infected with either H. pylori strains J166 or 26695 (MOI 100), their respective cagE− mutants (MOI 100), or their respective purified gDNA (5 μg/mL) for 6 hours. Levels of IL-8 were quantified using Human CXCL8 ELISA (R&D Systems, Minneapolis, MN, USA) according to manufacturer’s instructions and tested in duplicate. (e) Liquid cultures of H. pylori were grown with shaking overnight in 5 mL of Brucella broth (BD Biosciences) supplemented with 10% neonatal calf serum (NCS) (Atlanta Biologicals) at 37°C and 5% CO2. Supernatants of overnight cultures were collected, filtered (0.45 μm filter) and used for TLR9 activation assays at concentrations ranging from 1–30%. Results are shown relative to vehicle (Brucella broth) control. (f) HEK293 reporter cells were co-cultured with H. pylori wild type or isogenic mutant strains of J166 at an MOI of 100 for 4 hours. Cells were washed 3 times with PBS containing gentamicin (250 μg/mL; Corning). Cells were then incubated at 37°C for an additional hour in RPMI containing gentamicin (250 μg/mL), washed 3 times with PBS, lysed in 200 μL of sterile dH2O and serial dilutions were plated on TSA plates with 5% sheep blood (BD Biosciences). Plates were incubated for 5 days at 37°C, 5% CO2 and colonies were enumerated. Experiments were repeated at least 3 times. Viable colony-forming units with mean±SEM are shown. Student’s t-test (a) or one-way analysis of variance with Bonferroni correction (b–f) was used to determine statistical significance between groups. *p<0.05,**p<0.01,***p<0.001, ****p<0.0001.
H. pylori possesses up to four potential T4SSs with the putative ability to translocate DNA to and from the bacterial cell: the comB DNA uptake (competence) system, the cag system, as well as the tfs3 and tfs4 secretion systems (52–54). Analysis of an H. pylori comB DNA uptake mutant revealed no differences in levels of TLR9 activation compared to the H. pylori wild-type strain J166 (Figure 2b). Therefore, we next investigated the role of the cag T4SS in mediating TLR9 activation. TLR9 reporter cells were challenged with either an isogenic cagA− mutant (which lacks the effector protein CagA), cagE− or cagY− mutants (which encode essential proteins for T4SS assembly), or a pgdA− mutant (which reduces peptidoglycan-mediated NOD1 activation (55)). Loss of cagA or pgdA had no effect on TLR9 activation; however, cagE− or cagY− mutants were incapable of activating TLR9, suggesting that DNA translocation to the host cell and subsequent TLR9 activation may occur via the cag T4SS (Figure 2b). The requirement for a functional cag T4SS to induce TLR9-dependent responses was also demonstrated in two independent H. pylori cag+ strains, 26695 and 7.13, as mutations in cag T4SS structural components abrogated TLR9 activation (Figure 2c, S1). Importantly, H. pylori strain 26695, which lacks intact tfs3 and tfs4 gene clusters (53), still induced robust TLR9 activation (Figure 2c). We also observed TLR9 activation following infection with H. pylori strains devoid of plasmid DNA (strain J166, 26695, Figure 2b, 2c (12, 47)), suggesting that other forms of DNA can be translocated by the cag T4SS. This observation is of particular importance because it highlights fundamental differences between H. pylori DNA translocation and DNA translocation that occurs in other bacterial species, which only translocate plasmid DNA into host cells. To extend these findings, we also infected AGS gastric epithelial cells with wild-type H. pylori cag+ strains (J166 or 26695), their respective cagE− mutants, or their purified genomic DNA and assessed the levels of IL-8 secretion as a potential downstream target of TLR9 mediated NF-κB activation (Figure 2d). We found that the cagE− mutants induced significantly decreased levels of IL-8 compared to wild-type H. pylori strains. Additionally, purified gDNA from the wild-type strains induced a significantly increased level of IL-8 production compared to vehicle control, suggesting that H. pylori gDNA can not only activate TLR9, but can also induce production of a known downstream target of this receptor. Collectively, these data indicate that a functional H. pylori cancer-linked T4SS is required for TLR9 activation, but the known effector molecules translocated by this system (CagA and peptidoglycan) are dispensable for this phenotype.
H. pylori is predominantly an extracellular pathogen and TLR9 is an endosomal receptor, implying that DNA is either directly injected into the host cell, or is taken up via a different mechanism that is enhanced by a functional T4SS. To determine whether TLR9 was activated by H. pylori DNA entering host cells via a host-induced pathway, TLR9 reporter cells were incubated with bacterial cell-free, H. pylori conditioned media at concentrations up to 30%; however, no changes in TLR9 activation were observed compared to control cells (Figure 2e). These data indicate that activating DNA is not secreted, in contrast to strategies employed by Neisseria in which DNA is translocated into the extracellular milieu (56). Although primarily extracellular, H. pylori does retain the capacity to invade host epithelial cells in a limited fashion (55). Therefore, we next quantified intracellular H. pylori viability within HEK293-hTLR9 reporter cells. Wild-type H. pylori survived equally well compared to the corresponding cag mutants (Figure 2f), despite marked differences in TLR9 activation induced by these strains (Figure 2b). These results indicate that microbial endocytosis does not affect H. pylori-induced TLR9 activation since only wild-type H. pylori were capable of activating this endosome-bound receptor.
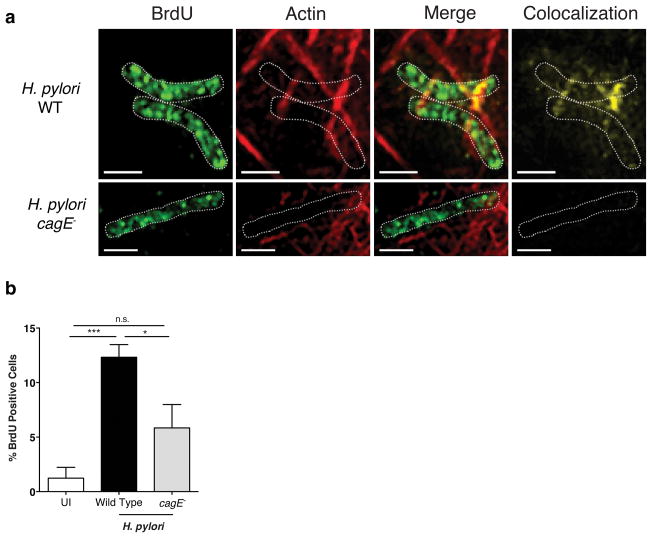
To determine whether the cag T4SS is required for direct DNA delivery into host cells, H. pylori DNA was labeled with BrdU and bacteria were subsequently co-cultured with AGS gastric epithelial cells. Using this technique, bacterial DNA can be easily distinguished from host DNA via incorporation of BrdU. Structured illumination microscopy (SIM) demonstrated that wild-type H. pylori translocated BrdU-labeled DNA into host cells (Figure 3a, Figure S3, Figure S4 movie a,b). However, intracellular BrdU-labeled DNA was not observed in host cells infected with the H. pylori cagE− mutant (Figure 3a, Fig S5 movie). To confirm and quantify these results using an independent methodology, host intracellular levels of BrdU-labeled H. pylori DNA were assessed via flow cytometry. Levels of intracellular DNA were significantly increased in H. pylori wild type-infected compared to uninfected AGS cells. A significant reduction was observed in cells infected with the cagE− mutant compared to wild-type infected AGS cells (Figure 3b), albeit not to levels observed in uninfected cells, which may be due to adherent and/or invasive H. pylori that could not be completely removed prior to analysis. Collectively, these data demonstrate that H. pylori utilizes the cag T4SS to translocate microbial DNA into eukaryotic cells, thereby activating TLR9.
Figure 3. H. pylori translocates DNA into host cells via the cag T4SS.
(a–b) H. pylori were grown overnight in Brucella broth containing 10% NCS. H. pylori were then diluted 1:10 in Brucella broth containing 5 μM BrdU (Sigma-Aldrich, St. Louis MO USA) and 10% NCS. Cultures were grown for an additional 4 hours prior to co-culture with eukaryotic cells. (a) H. pylori-mediated DNA translocation was assessed by structured illumination microscopy using AGS gastric epithelial cells co-cultured with BrdU-labeled H. pylori wild-type strain J166 or the J166 isogenic cagE− mutant. Scale bar, 1 μm. AGS cells were seeded on size 1.5 cover slips at 100 000 cells per well. BrdU-labeled H. were co-cultured at an MOI of 100 for 4 hours. Infected cells were washed 3 times in PBS, fixed with CytoFix/CytoPerm (BD Biosciences) solution for 20 minutes at 4°C and washed an additional 3 times in 1× permwash (BD Biosciences). Cells were then blocked overnight at 4°C using endogenous biotin blocking kit reagent A (Life Technologies), washed 3 times in 1× permwash and blocked again with endogenous biotin blocking kit reagent B (Life Technologies) for 30 minutes at room temperature. Cells were incubated with mouse anti-BrdU conjugated biotin (Life Technologies 1:500) overnight at 4°C diluted in 1× permwash. Cells were subsequently stained with strepdavidin-conjugated AlexaFluor488 (1:5000 for BrdU) as well as phalloidin (1:100 actin, Life Technologies) and Hoechst (1:5000 nuclei, Life Technologies) for 1 hour at room temperature. Cells were washed 3 times in 1× permwash and slides were mounted with prolong gold (Invitrogen). Slides were viewed with Delta Vision OMX confocal microscope (GE Health Care Life Sciences, Marlborough MA USA) at 63X magnification, using 1.514 immersion oil. Post-data acquisition processing was performed using SoftWorx for OMX. Images are shown as maximum intensity projections through the entire imaged area. BrdU, green; actin, red; merge, yellow. H. pylori are outlined by dotted white lines. Scale bar, 1μm. (b) BrdU-labeled H. pylori wild-type strain J166 or the J166 cagE− mutant were co-cultured with AGS cells and then subjected to flow cytometry to assess levels of host intracellular BrdU. AGS cells were co-cultured with BrdU-labeled H. pylori for 4 hours. Cells were washed and incubated for an additional hour in RPMI 1640 containing gentamicin (500 μg/mL). Adherent cells were then dissociated with 0.25% trypsin EDTA (Life Technologies) supplemented with gentamicin (500 μg/mL), fixed and permeabilized with CytoFix/CytoPerm (BD Biosciences), blocked overnight at 4°C using endogenous biotin blocking kit reagent A (Life Technologies), washed 3 times in 1× permwash (BD Biosciences) and blocked again with endogenous biotin blocking kit reagent B for 30 minutes at room temperature (Life Technologies). Cells were washed and stained with biotin conjugated anti-BrdU mouse monoclonal antibody (1:500 Life Technologies) followed by strepdavidin conjugated AlexFluor488 secondary antibody (1:5000 Life Technologies). Cells were acquired using a LSR II Flow Cytometer (BD Biosciences) and BrdU-positive cells were analyzed using FlowJo (Tree Star Inc. Ashland OR USA). Each strain was tested at least 3 times. Mean±SEM are shown. *p<0.05, ***p<0.001.
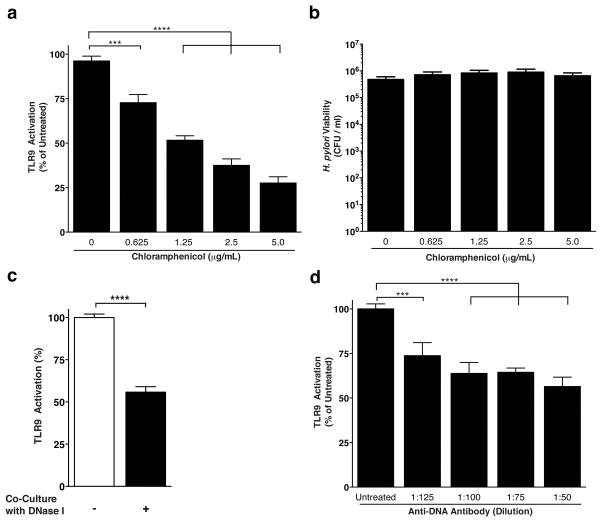
Elaboration of the cancer-associated cag T4SS develops in response to direct host cell contact, and effector translocation is dependent on assembly of the T4SS apparatus. We therefore performed orthologous mechanistic studies to investigate whether TLR9 activation required de novo synthesis of Cag proteins for apparatus biogenesis, or whether cag T4SS-dependent DNA transfer could be achieved using pre-assembled protein complexes. Treatment of H. pylori-HEK293 co-cultures with bacteriostatic, sub-lethal concentrations of chloramphenicol, an inhibitor of bacterial protein synthesis, revealed a significant reduction in levels of TLR9 activation (Figure 4a). Chloramphenicol-mediated inhibition of H. pylori-driven TLR9 activation occurred in a dose-dependent manner but did not affect H. pylori viability (Figure 4b). These data indicate that cag T4SS-mediated DNA translocation requires new protein synthesis.
Figure 4. Mechanisms of H. pylori cag-mediated DNA translocation.
(a) H. pylori strain J166 was inoculated into HEK-Blue-hTLR9 and HEK-Blue Null1 co-cultures as described in Figure 2 legend. For each biological replicate experiment, sub-lethal levels of chloramphenicol (Sigma-Aldrich) were added to triplicate wells at concentrations ranging from 0.625 μg/mL to 5 μg/mL. After 24 hours, TLR9 activation was assessed as described for TLR9 activation assays. For each concentration, OD650 values obtained for HEK-Blue-hTLR9 cells were normalized by subtracting the corresponding OD650 values obtained for HEK-Blue-Null1 cells, and TLR9 activation was calculated as a percent of untreated, antibiotic-free co-cultures. (b) After 24 hours of co-culture in chloramphenicol, H. pylori-HEK co-cultures wells were dislodged by mechanical disruption, centrifuged at 5000 x g, and were re-suspended in 1 mL sterile PBS. To calculate H. pylori viability in CFUs, 10-fold serial dilutions of resuspended co-cultures were plated, and colonies were enumerated after 72 hours of incubation. (c) H. pylori wild-type strain J166 was co-cultured with HEK-Blue-hTLR9 reporter cells at an MOI of 100. For the course of the co-culture, 20 units of Turbo DNase I (Life Technologies) were added to half of the wells, and DNase I buffer was added to the remaining wells. TLR9 activation was assessed at 24 hours. For each condition, OD650 values obtained for HEK-Blue-hTLR9 cells were normalized by subtracting the corresponding OD650 values obtained for HEK-Blue-Null1 cells, and TLR9 activation was calculated as a percent of untreated, DNase I-free co-cultures. (d) H. pylori strain J166 was inoculated into HEK-Blue-hTLR9 and HEK-Blue-Null1 co-cultures at an MOI of 100. For each biological replicate experiment, monoclonal anti-DNA antibody (Roche, Mannheim Germany) was added at varying dilutions, ranging from 1:50 – 1:200 final concentration. After 24 hours, TLR9 activation was assessed, and values were normalized as described for DNase I experiments. TLR9 activation was expressed as a percentage of untreated (no antibody) wells. Experiments were repeated at least 3 times. For all experiments, mean±SEM are shown. A one-way analysis of variance with Bonferroni correction was used to determine statistical significance between groups. *p<0.05,**p<0.01,***p<0.001, ****p<0.0001.
DNA transfer by A. tumefaciens occurs via substrate delivery both within and on the external surface of the T4SS conduit (25). To further investigate the mechanisms of cag T4SS-dependent DNA transfer into host cells in greater depth, we assessed whether exogenous DNase I treatment would compromise the integrity of transported DNA and impair TLR9 activation. Compared to untreated controls, levels of TLR9 activation decreased by approximately 50% when DNase I was added during H. pylori-HEK293 cell co-culture (Figure 4c). In parallel, we analyzed the ability of monoclonal anti-DNA antibodies to functionally block H. pylori-dependent TLR9 activation. Compared to untreated controls, H. pylori-mediated TLR9 activation was significantly reduced in the presence of pan anti-DNA antibodies, an effect that was titratible (Figure 4d). Taken together, these data indicate that cag T4SS-dependent DNA translocation occurs through a contact-dependent mechanism that involves transfer of bacterial DNA that is both protected as well as exposed during transport.
In conclusion, we have demonstrated that patients residing within a high gastric cancer risk region of Colombia express significantly higher levels of TLR9 within gastric epithelial cells compared to patients residing in the low risk region, and that the H. pylori strains isolated from these patients concomitantly induce higher levels of TLR9 activation. We further demonstrate that H. pylori utilizes the cag island, a locus known to augment cancer risk, to translocate bacterial DNA into host cells. Collectively these results define a mechanism that may explain previous epidemiologic data linking TLR9 to the development of gastric cancer.
Supplementary Material
The H. pylori cag+ strain 7.13 or its isogenic mutants were maintained on trypticase soy agar plates supplemented with 5% sheep blood (BD Biosciences) as described. TLR9-reporter or parental cells were cultured as described in Figure 2 and challenged with purified H. pylori strain 7.13 at an MOI of 100 for 24 hours. Experiments were repeated at least 3 times, mean±SEM are shown. A one-way analysis of variance with Bonferroni correction was used to determine statistical significance between groups. ****p<0.0001.
TLR9-reporter and parental cells were cultured as described in Figure 2 and pretreated for 2 hours with 50 μg/mL of chloroquine (Sigma-Aldrich). H. pylori strains J166 or 26695 or their corresponding cagE− mutants were used to infect pretreated cells at an MOI of 100 for 24 hours. The TLR9 agonist, ODN2006 (Invivogen), was used to challenge reporter cells at a concentration of 5 μM as a positive control. TLR9 activation is represented as fold over vehicle control. Experiments were repeated at least 3 times, mean±SEM are shown. A one-way analysis of variance with Bonferroni correction was used to determine statistical significance between groups. ****p<0.0001.
Supplemental Figure 3: H. pylori translocates DNA into host cells via the cag T4SS. H. pylori cag+ wild-type strain J166 was labeled with 5 μM BrdU and used to challenge AGS gastric epithelial cells. Co-cultures were stained and imaged as described in Figure 3. BrdU is labeled in green, actin in red, areas of co-localization in yellow. Scale bar equals 1 μm. (a–o) Representative z-stack images used to compile the compounded image shown in Figure 3a and supplemental movies 4a,b. Each panel represents a 0.125 μm slice. (p) Merged image of z-stacks shown in a–o.
Supplemental Movie 4: H. pylori wild-type strain J166 translocates DNA into host cells. Related to Figure 3 and supplemental Figure 3. H. pylori strain J166 was labeled with BrdU (green) and co-cultured with AGS gastric epithelial cells. Actin is stained in red, and areas of co-localization are shown in yellow. (A) X-Axis and (B) Y-Axis rotations of H. pylori adhered to an AGS cell with translocated DNA shown within the host cell. Scale bar, 1 μm.
Supplemental Movie 5: An H. pylori cag T4SS mutant fails to translocate DNA into host cells. Related to Figure 3. An H. pylori J166 cagE− mutant was labeled with BrdU (green) and co-cultured with AGS gastric epithelial cells. Animation is rotated around the Y-axis. Actin is stained in red, and areas of co-localization are shown in yellow. Scale bar, 1 μm.
Acknowledgments
We acknowledge the following core laboratories and personnel at Vanderbilt University for their contributions to these studies: Tissue Acquisition and Pathology Core; Division of Animal Care; Cell Imaging Shared Resource Core (CISR) and the Digestive Disease Research Center. This work was supported by NIH R01-DK58587, R01-CA77955, P01-CA116087, and P30-DK058404 (R.M. Peek Jr.); Vanderbilt CTSA grant UL1 TR000445 from NCATS/NIH (VICTR award VR7227), APS 1-04-520-9211 and institutional funds (M. Hadjifrangiskou); R01-DK053620, R01-CA190612, and a Department of Veterans Affairs Merit Review grant I01BX001453 (K.T. Wilson); and P01-CA028842 (K.T. Wilson and P. Correa). The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- 1.Amieva M, Peek RM., Jr Pathobiology of Helicobacter pylori-induced gastric cancer. Gastroenterology. 2016;150(1):64–78. doi: 10.1053/j.gastro.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 3.Ernst PB, Peura DA, Crowe SE. The translation of Helicobacter pylori basic research to patient care. Gastroenterology. 2006;130(1):188–206. doi: 10.1053/j.gastro.2005.06.032. [DOI] [PubMed] [Google Scholar]
- 4.Segal ED, Cha J, Lo J, Falkow S, Tompkins LS. Altered states: involvement of phosphorylated CagA in the induction of host cellular growth changes by Helicobacter pylori. Proc Natl Acad Sci USA. 1999;96(25):14559–64. doi: 10.1073/pnas.96.25.14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Odenbreit S, Puls J, Sedlmaier B, Gerland E, Fischer W, Haas R. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science. 2000;287(5457):1497–500. doi: 10.1126/science.287.5457.1497. [DOI] [PubMed] [Google Scholar]
- 6.Stein M, Rappuoli R, Covacci A. Tyrosine phosphorylation of the Helicobacter pylori CagA antigen after cag-driven host cell translocation. Proc Natl Acad Sci USA. 2000;97(3):1263–8. doi: 10.1073/pnas.97.3.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Backert S, Ziska E, Brinkmann V, Zimny-Arndt U, Fauconnier A, Jungblut PR, et al. Translocation of the Helicobacter pylori CagA protein in gastric epithelial cells by a type IV secretion apparatus. Cell Microbiol. 2000;2(2):155–64. doi: 10.1046/j.1462-5822.2000.00043.x. [DOI] [PubMed] [Google Scholar]
- 8.Selbach M, Moese S, Hauck CR, Meyer TF, Backert S. Src is the kinase of the Helicobacter pylori CagA protein in vitro and in vivo. J Biol Chem. 2002;277(9):6775–8. doi: 10.1074/jbc.C100754200. [DOI] [PubMed] [Google Scholar]
- 9.Tammer I, Brandt S, Hartig R, Konig W, Backert S. Activation of Abl by Helicobacter pylori: a novel kinase for CagA and crucial mediator of host cell scattering. Gastroenterology. 2007;132(4):1309–19. doi: 10.1053/j.gastro.2007.01.050. [DOI] [PubMed] [Google Scholar]
- 10.Censini S, Lange C, Xiang Z, Crabtree JE, Ghiara P, Borodovsky M, et al. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA. 1996;93(25):14648–53. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akopyants NS, Clifton SW, Kersulyte D, Crabtree JE, Youree BE, Reece CA, et al. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol Microbiol. 1998;28(1):37–53. doi: 10.1046/j.1365-2958.1998.00770.x. [DOI] [PubMed] [Google Scholar]
- 12.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388(6642):539–47. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 13.Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397(6715):176–80. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 14.Crabtree JE, Wyatt JI, Sobala GM, Miller G, Tompkins DS, Primrose JN, et al. Systemic and mucosal humoral responses to Helicobacter pylori in gastric cancer. Gut. 1993;34(10):1339–43. doi: 10.1136/gut.34.10.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blaser MJ, Perez-Perez GI, Kleanthous H, Cover TL, Peek RM, Chyou PH, et al. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 1995;55(10):2111–5. [PubMed] [Google Scholar]
- 16.Parsonnet J, Friedman GD, Orentreich N, Vogelman H. Risk for gastric cancer in people with CagA positive or CagA negative Helicobacter pylori infection. Gut. 1997;40(3):297–301. doi: 10.1136/gut.40.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuipers EJ, Perez-Perez GI, Meuwissen SG, Blaser MJ. Helicobacter pylori and atrophic gastritis: importance of the cagA status. J Natl Cancer Inst. 1995;87(23):1777–80. doi: 10.1093/jnci/87.23.1777. [DOI] [PubMed] [Google Scholar]
- 18.Polk DB, Peek RM., Jr Helicobacter pylori: gastric cancer and beyond. Nature Rev Cancer. 2010;10(6):403–14. doi: 10.1038/nrc2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Viala J, Chaput C, Boneca IG, Cardona A, Girardin SE, Moran AP, et al. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nature Immunol. 2004;5(11):1166–74. doi: 10.1038/ni1131. [DOI] [PubMed] [Google Scholar]
- 20.Bates S, Cashmore AM, Wilkins BM. IncP plasmids are unusually effective in mediating conjugation of Escherichia coli and Saccharomyces cerevisiae: involvement of the tra2 mating system. J Bacteriol. 1998;180(24):6538–43. doi: 10.1128/jb.180.24.6538-6543.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heinemann JA, Sprague GF., Jr Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast. Nature. 1989;340(6230):205–9. doi: 10.1038/340205a0. [DOI] [PubMed] [Google Scholar]
- 22.Fernandez-Gonzalez E, de Paz HD, Alperi A, Agundez L, Faustmann M, Sangari FJ, et al. Transfer of R388 derivatives by a pathogenesis-associated type IV secretion system into both bacteria and human cells. J Bacteriol. 2011;193(22):6257–65. doi: 10.1128/JB.05905-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schroder G, Schuelein R, Quebatte M, Dehio C. Conjugative DNA transfer into human cells by the VirB/VirD4 type IV secretion system of the bacterial pathogen Bartonella henselae. Proc Natl Acad Sci USA. 2011;108(35):14643–8. doi: 10.1073/pnas.1019074108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waters VL. Conjugation between bacterial and mammalian cells. Nature Genetics. 2001;29(4):375–6. doi: 10.1038/ng779. [DOI] [PubMed] [Google Scholar]
- 25.Cascales E, Christie PJ. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science. 2004;304(5674):1170–3. doi: 10.1126/science.1095211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chilton MD, Drummond MH, Merio DJ, Sciaky D, Montoya AL, Gordon MP, et al. Stable incorporation of plasmid DNA into higher plant cells: the molecular basis of crown gall tumorigenesis. Cell. 1977;11(2):263–71. doi: 10.1016/0092-8674(77)90043-5. [DOI] [PubMed] [Google Scholar]
- 27.Christie PJ. Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J Bacteriol. 1997;179(10):3085–94. doi: 10.1128/jb.179.10.3085-3094.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kunik T, Tzfira T, Kapulnik Y, Gafni Y, Dingwall C, Citovsky V. Genetic transformation of HeLa cells by Agrobacterium. Proc Natl Acad Sci USA. 2001;98(4):1871–6. doi: 10.1073/pnas.041327598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barton GM. A calculated response: control of inflammation by the innate immune system. J Clin Invest. 2008;118(2):413–20. doi: 10.1172/JCI34431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castano-Rodriguez N, Kaakoush NO, Mitchell HM. Pattern-recognition receptors and gastric cancer. Front Immunol. 2014;5:336. doi: 10.3389/fimmu.2014.00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408(6813):740–5. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 32.Pachathundikandi SK, Lind J, Tegtmeyer N, El-Omar EM, Backert S. Interplay of the gastric pathogen Helicobacter pylori with toll-like receptors. Biomed Res Int. 2015;2015:192420. doi: 10.1155/2015/192420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang YJ, Wu MS, Lin JT, Chen CC. Helicobacter pylori-Induced invasion and angiogenesis of gastric cells is mediated by cyclooxygenase-2 induction through TLR2/TLR9 and promoter regulation. J Immunol. 2005;175(12):8242–52. doi: 10.4049/jimmunol.175.12.8242. [DOI] [PubMed] [Google Scholar]
- 34.Chang YJ, Wu MS, Lin JT, Sheu BS, Muta T, Inoue H, et al. Induction of cyclooxygenase-2 overexpression in human gastric epithelial cells by Helicobacter pylori involves TLR2/TLR9 and c-Src-dependent nuclear factor-kappaB activation. Mol Pharmacol. 2004;66(6):1465–77. doi: 10.1124/mol.104.005199. [DOI] [PubMed] [Google Scholar]
- 35.Ilvesaro JM, Merrell MA, Li L, Wakchoure S, Graves D, Brooks S, et al. Toll-like receptor 9 mediates CpG oligonucleotide-induced cellular invasion. Mol Cancer Res. 2008;6(10):1534–43. doi: 10.1158/1541-7786.MCR-07-2005. [DOI] [PubMed] [Google Scholar]
- 36.Kauppila JH, Karttunen TJ, Saarnio J, Nyberg P, Salo T, Graves DE, et al. Short DNA sequences and bacterial DNA induce esophageal, gastric, and colorectal cancer cell invasion. APMIS. 2013;121(6):511–22. doi: 10.1111/apm.12016. [DOI] [PubMed] [Google Scholar]
- 37.Schmausser B, Andrulis M, Endrich S, Lee SK, Josenhans C, Muller-Hermelink HK, et al. Expression and subcellular distribution of toll-like receptors TLR4, TLR5 and TLR9 on the gastric epithelium in Helicobacter pylori infection. Clin Exp Immunol. 2004;136(3):521–6. doi: 10.1111/j.1365-2249.2004.02464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang TR, Peng JC, Qiao YQ, Zhu MM, Zhao D, Shen J, et al. Helicobacter pylori regulates TLR4 and TLR9 during gastric carcinogenesis. Int J Clin Exp Pathol. 2014;7(10):6950–5. [PMC free article] [PubMed] [Google Scholar]
- 39.Castano-Rodriguez N, Kaakoush NO, Pardo AL, Goh KL, Fock KM, Mitchell HM. Genetic polymorphisms in the Toll-like receptor signalling pathway in Helicobacter pylori infection and related gastric cancer. Human Immunol. 2014;75(8):808–15. doi: 10.1016/j.humimm.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 40.Torres J, Lopez L, Lazcano E, Camorlinga M, Flores L, Munoz O. Trends in Helicobacter pylori infection and gastric cancer in Mexico. Cancer Epidemiol Biomarkers Prev. 2005;14(8):1874–7. doi: 10.1158/1055-9965.EPI-05-0113. [DOI] [PubMed] [Google Scholar]
- 41.Correa P, Cuello C, Duque E, Burbano LC, Garcia FT, Bolanos O, et al. Gastric cancer in Colombia. III. Natural history of precursor lesions. J Natl Cancer Inst. 1976;57(5):1027–35. doi: 10.1093/jnci/57.5.1027. [DOI] [PubMed] [Google Scholar]
- 42.Cuello C, Correa P, Haenszel W, Gordillo G, Brown C, Archer M, et al. Gastric cancer in Colombia. I. Cancer risk and suspect environmental agents. J Natl Cancer Inst. 1976;57(5):1015–20. doi: 10.1093/jnci/57.5.1015. [DOI] [PubMed] [Google Scholar]
- 43.Basso D, Zambon CF, Letley DP, Stranges A, Marchet A, Rhead JL, et al. Clinical relevance of Helicobacter pylori cagA and vacA gene polymorphisms. Gastroenterology. 2008;135(1):91–9. doi: 10.1053/j.gastro.2008.03.041. [DOI] [PubMed] [Google Scholar]
- 44.Figueiredo C, Machado JC, Pharoah P, Seruca R, Sousa S, Carvalho R, et al. Helicobacter pylori and interleukin 1 genotyping: an opportunity to identify high-risk individuals for gastric carcinoma. J Natl Cancer Inst. 2002;94(22):1680–7. doi: 10.1093/jnci/94.22.1680. [DOI] [PubMed] [Google Scholar]
- 45.Bravo LE, van Doom LJ, Realpe JL, Correa P. Virulence-associated genotypes of Helicobacter pylori: do they explain the African enigma? Am J Gastroenterol. 2002;97(11):2839–42. doi: 10.1111/j.1572-0241.2002.07031.x. [DOI] [PubMed] [Google Scholar]
- 46.Christie PJ, Whitaker N, Gonzalez-Rivera C. Mechanism and structure of the bacterial type IV secretion systems. Biochim Biophys Acta. 2014;1843(8):1578–91. doi: 10.1016/j.bbamcr.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Linz B, Windsor HM, McGraw JJ, Hansen LM, Gajewski JP, Tomsho LP, et al. A mutation burst during the acute phase of Helicobacter pylori infection in humans and rhesus macaques. Nat Commun. 2014;5:4165. doi: 10.1038/ncomms5165. [DOI] [PubMed] [Google Scholar]
- 48.Franco AT, Israel DA, Washington MK, Krishna U, Fox JG, Rogers AB, et al. Activation of beta-catenin by carcinogenic Helicobacter pylori. Proc Natl Acad Sci USA. 2005;102(30):10646–51. doi: 10.1073/pnas.0504927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alvarez-Arellano L, Cortes-Reynosa P, Sanchez-Zauco N, Salazar E, Torres J, Maldonado-Bernal C. TLR9 and NF-kappaB are partially involved in activation of human neutrophils by Helicobacter pylori and its purified DNA. PLoS One. 2014;9(7):e101342. doi: 10.1371/journal.pone.0101342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Otani K, Tanigawa T, Watanabe T, Nadatani Y, Sogawa M, Yamagami H, et al. Toll-like receptor 9 signaling has anti-inflammatory effects on the early phase of Helicobacter pylori-induced gastritis. Biochem Biophys Res Commun. 2012;426(3):342–9. doi: 10.1016/j.bbrc.2012.08.080. [DOI] [PubMed] [Google Scholar]
- 51.Rad R, Ballhorn W, Voland P, Eisenacher K, Mages J, Rad L, et al. Extracellular and intracellular pattern recognition receptors cooperate in the recognition of Helicobacter pylori. Gastroenterology. 2009;136(7):2247–57. doi: 10.1053/j.gastro.2009.02.066. [DOI] [PubMed] [Google Scholar]
- 52.Fernandez-Gonzalez E, Backert S. DNA transfer in the gastric pathogen Helicobacter pylori. J Gastroenterol. 2014;49(4):594–604. doi: 10.1007/s00535-014-0938-y. [DOI] [PubMed] [Google Scholar]
- 53.Fischer W, Windhager L, Rohrer S, Zeiller M, Karnholz A, Hoffmann R, et al. Strain-specific genes of Helicobacter pylori: genome evolution driven by a novel type IV secretion system and genomic island transfer. Nucleic Acids Res. 2010;38(18):6089–101. doi: 10.1093/nar/gkq378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kersulyte D, Velapatino B, Mukhopadhyay AK, Cahuayme L, Bussalleu A, Combe J, et al. Cluster of type IV secretion genes in Helicobacter pylori’s plasticity zone. J Bacteriol. 2003;185(13):3764–72. doi: 10.1128/JB.185.13.3764-3772.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suarez G, Romero-Gallo J, Piazuelo MB, Wang G, Maier R, Forsberg LS, et al. Modification of Helicobacter pylori peptidoglycan enhances NOD1 activation and promotes cancer of the stomach. Cancer Res. 2015;75:1749–59. doi: 10.1158/0008-5472.CAN-14-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hamilton HL, Dominguez NM, Schwartz KJ, Hackett KT, Dillard JP. Neisseria gonorrhoeae secretes chromosomal DNA via a novel type IV secretion system. Mol Microbiol. 2005;55(6):1704–21. doi: 10.1111/j.1365-2958.2005.04521.x. [DOI] [PubMed] [Google Scholar]
- 57.Chaturvedi R, de Sablet T, Asim M, Piazuelo MB, Barry DP, Verriere TG, et al. Increased Helicobacter pylori-associated gastric cancer risk in the Andean region of Colombia is mediated by spermine oxidase. Oncogene. 2015;34(26):3429–40. doi: 10.1038/onc.2014.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sierra JC, Hobbs S, Chaturvedi R, Yan F, Wilson KT, Peek RM, Jr, et al. Induction of COX-2 expression by Helicobacter pylori is mediated by activation of epidermal growth factor receptor in gastric epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2013;305(2):G196–203. doi: 10.1152/ajpgi.00495.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barrozo RM, Cooke CL, Hansen LM, Lam AM, Gaddy JA, Johnson EM, et al. Functional plasticity in the type IV secretion system of Helicobacter pylori. PLoS Pathog. 2013;9(2):e1003189. doi: 10.1371/journal.ppat.1003189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Crawford HC, Krishna US, Israel DA, Matrisian LM, Washington MK, Peek RM., Jr Helicobacter pylori strain-selective induction of matrix metalloproteinase-7 in vitro and within gastric mucosa. Gastroenterology. 2003;125(4):1125–36. doi: 10.1016/s0016-5085(03)01206-x. [DOI] [PubMed] [Google Scholar]
- 61.Johnson EM, Gaddy JA, Voss BJ, Hennig EE, Cover TL. Genes required for assembly of pili associated with the Helicobacter pylori cag type IV secretion system. Infect Immun. 2014;82(8):3457–70. doi: 10.1128/IAI.01640-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shaffer CL, Gaddy JA, Loh JT, Johnson EM, Hill S, Hennig EE, et al. Helicobacter pylori exploits a unique repertoire of type IV secretion system components for pilus assembly at the bacteria-host cell interface. PLoS Pathog. 2011;7(9):e1002237. doi: 10.1371/journal.ppat.1002237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The H. pylori cag+ strain 7.13 or its isogenic mutants were maintained on trypticase soy agar plates supplemented with 5% sheep blood (BD Biosciences) as described. TLR9-reporter or parental cells were cultured as described in Figure 2 and challenged with purified H. pylori strain 7.13 at an MOI of 100 for 24 hours. Experiments were repeated at least 3 times, mean±SEM are shown. A one-way analysis of variance with Bonferroni correction was used to determine statistical significance between groups. ****p<0.0001.
TLR9-reporter and parental cells were cultured as described in Figure 2 and pretreated for 2 hours with 50 μg/mL of chloroquine (Sigma-Aldrich). H. pylori strains J166 or 26695 or their corresponding cagE− mutants were used to infect pretreated cells at an MOI of 100 for 24 hours. The TLR9 agonist, ODN2006 (Invivogen), was used to challenge reporter cells at a concentration of 5 μM as a positive control. TLR9 activation is represented as fold over vehicle control. Experiments were repeated at least 3 times, mean±SEM are shown. A one-way analysis of variance with Bonferroni correction was used to determine statistical significance between groups. ****p<0.0001.
Supplemental Figure 3: H. pylori translocates DNA into host cells via the cag T4SS. H. pylori cag+ wild-type strain J166 was labeled with 5 μM BrdU and used to challenge AGS gastric epithelial cells. Co-cultures were stained and imaged as described in Figure 3. BrdU is labeled in green, actin in red, areas of co-localization in yellow. Scale bar equals 1 μm. (a–o) Representative z-stack images used to compile the compounded image shown in Figure 3a and supplemental movies 4a,b. Each panel represents a 0.125 μm slice. (p) Merged image of z-stacks shown in a–o.
Supplemental Movie 4: H. pylori wild-type strain J166 translocates DNA into host cells. Related to Figure 3 and supplemental Figure 3. H. pylori strain J166 was labeled with BrdU (green) and co-cultured with AGS gastric epithelial cells. Actin is stained in red, and areas of co-localization are shown in yellow. (A) X-Axis and (B) Y-Axis rotations of H. pylori adhered to an AGS cell with translocated DNA shown within the host cell. Scale bar, 1 μm.
Supplemental Movie 5: An H. pylori cag T4SS mutant fails to translocate DNA into host cells. Related to Figure 3. An H. pylori J166 cagE− mutant was labeled with BrdU (green) and co-cultured with AGS gastric epithelial cells. Animation is rotated around the Y-axis. Actin is stained in red, and areas of co-localization are shown in yellow. Scale bar, 1 μm.