Abstract
Amplification and over expression of erbB2/neu proto-oncogene is observed in 20–30% human breast cancer and is inversely correlated with the survival of the patient. Despite this, somatic activating mutations within erbB2 in human breast cancers are rare. However, we have previously reported that a splice isoform of erbB2, containing an in-frame deletion of exon 16 (herein referred to as ErbB2ΔEx16), results in oncogenic activation of erbB2 due to constitutive dimerization of the ErbB2 receptor. Here, we demonstrate that the ErbB2ΔEx16 is a major oncogenic driver in breast cancer that constitutively signals from the cell surface. We further show that inducible expression of the ErbB2Ex16 variant in mammary gland of transgenic mice results in the rapid development of metastatic multifocal mammary tumors. Genetic and biochemical characterization of the ErbB2ΔEx16 derived mammary tumors exhibit several unique features that distinguish it from the conventional ErbB2 models expressing the erbB2 proto-oncogene in mammary epithelium. Unlike the wild-type ErbB2 derived tumors that express luminal keratins, ErbB2ΔEx16 derived tumors exhibit high degree of intra-tumoral heterogeneity co-expressing both basal and luminal keratins. Consistent with these distinct pathological features, the ErbB2ΔEx16 tumors exhibited distinct signaling and gene expression profiles that correlated with activation of number of key transcription factors implicated in breast cancer metastasis and cancer stem cell renewal.
Keywords: ErbB2, breast cancer, mouse model, splice variant
Introduction
Amplification and elevated expression of the erbB2 proto-oncogene is observed in 20 to 30% of human breast cancers and is inversely correlated with the survival of the patient 3, 39, 40. Direct evidence supporting a role for ErbB2 in mammary tumorigenesis derives from studies with transgenic mice expressing the erbB2 proto-oncogene under the transcriptional control of the mouse mammary tumor virus (MMTV) promoter/enhancer. Mammary-epithelial expression of erbB2 oncogene results in the rapid induction of multifocal mammary tumors 20, 31. By contrast, mammary epithelial expression of the proto-oncogenic form of ErbB2 (herein referred too as wild-type ErbB2) resulted in induction of focal mammary tumors after a long latency period 19. Tumor progression in these strains is associated with the activation of the ErbB2 tyrosine kinase activity due to the occurrence of somatic activating mutations in the transgene in a majority of the mammary tumors analyzed 7, 19, 36, 37. Significantly these mutations are confined to a cysteine rich region of the receptor located in a juxtatransmembrane domain and are comprised of either deletion or insertion of single cysteine residues. Further genetic and biochemical analyses has revealed that these cysteine alterations promote the formation intermolecular cysteine bridges between ErbB2 monomers, resulting in constitutive receptor dimerization and activation 37.
Although comparable somatic mutations in erbB2 have not been detected in human breast cancer, several studies have reported the expression of an alternatively spliced erbB2 isoform (ErbB2ΔEx16) that carries a 16 amino acid in-frame deletion in the juxtatransmembrane domain that closely mimics the sporadic erbB2 transgene mutations observed in the transgenic mice expressing the ErbB2 proto-oncogene 24, 38. Like sporadic transgene mutations, this erbB2 splice variant is constitutively active due to its capacity to form disulfide-bonded dimers 38. In addition to playing an important role in the oncogenic activation of ErbB2 receptor, it has been recently reported that the ErbB2ΔEx16 isoform is the principle target of the anti-ErbB2 therapeutic monoclonal Trastuzumab 1, 5.
To further validate the biological significance of the ErbB2ΔEx16 variant in mammary epithelium, we established a transgenic mouse model that expresses the ErbB2ΔEx16 variant in an inducible fashion in the mammary epithelium. In contrast to the transgenic mice expressing the wild-type ErbB2 receptor in the mammary epithelium that developed focal mammary tumors after a long latency period, the ErbB2ΔEx16 strains rapidly developed multifocal mammary tumors that frequently metastasized to the lung. Remarkably, in contrast to the ErbB2 tumors that uniformly expressed the luminal keratin 8 marker, the ErbB2ΔEx16 derived tumors exhibited a high degree intra-tumoral heterogeneity expressing both luminal and basal keratins. Consistent with basal origin of these tumors, gene expression analyses revealed that in contrast to luminal pattern for ErbB2, the ErbB2ΔEx16 derived tumors possessed many features of basal breast cancer. Molecular and genetic analyses of these tumors further showed that basal molecular phenotype was due to selective signaling of the c-Src family and p38 kinases by the ErbB2ΔEx16 variant that in-turn resulted in activation of the Smad2, HIF1α, Stat3 and YB-1 transcriptional networks. Taken together, these observations suggest that the ErbB2ΔEx16 isoform possesses distinct signaling and cellular localization processes that may impact on its role in mammary tumor progression.
Results
The ErbB2ΔEx16 isoform is expressed at different levels in a number of ErbB2 positive cell lines and confers transforming activity in vivo
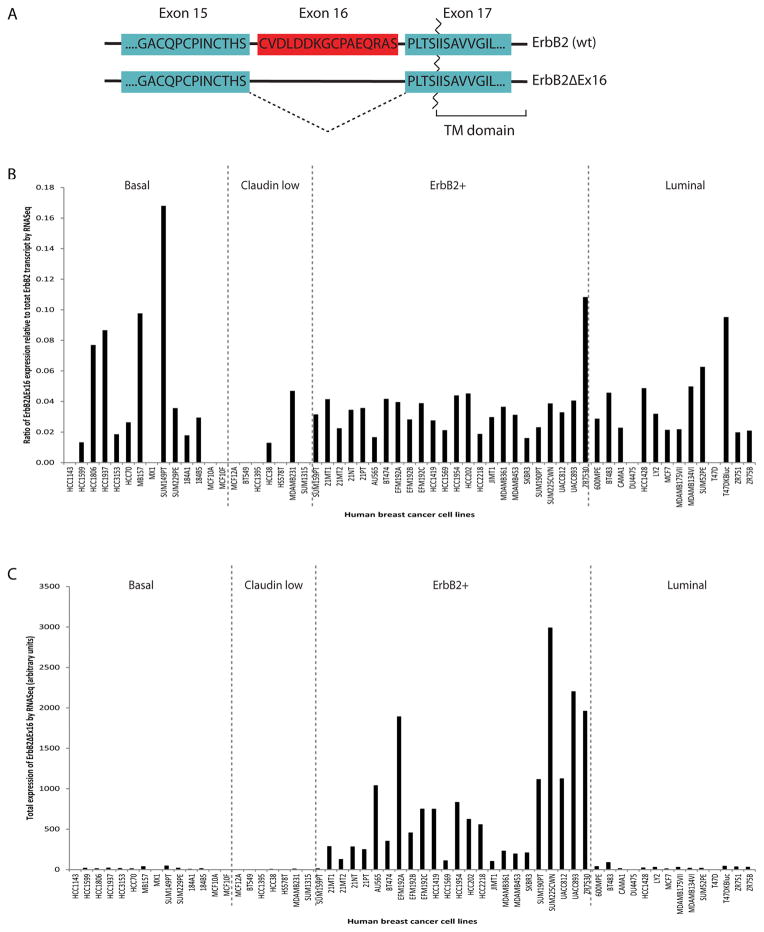
To confirm the prevalence of the ErbB2ΔEx16 isoform in ErbB2 positive category of breast cancer, we used a an RNA-seq approach to simultaneously measure the abundance of the ErbB2ΔEx16 compared to wild type transcript ErbB2 transcript in a collection of established breast cancer cell lines representing the different breast cancer subtypes 32 (Fig. 1A). The results revealed that the abundance of the ErbB2ΔEx16 isoform varied considerably with the different ErbB2 positive cells lines. Although the low levels of ErbB2 spliced form were detected in the SKBR3 (1.8% of the total ErbB2 transcript), relatively high levels of the ErbB2ΔEx16 were seen in the ZR7530 cells (12% of the total ErbB2 transcript) (Fig. 1B). Other ErbB2 cell lines maintained a modest level of the spliced variant (4–5% of total ErbB2). Although the proportion of ErbB2ΔEx16 variant varied in ErbB2 positive lines, due to amplification of ErbB2, the absolute levels of the ErbB2ΔEx16 variant were high by comparison to other breast cancer cell lines representing the other breast cancer subtypes (Fig. 1C).
Figure 1.
RNA Seq analysis of human breast cancer cell lines. (A) ErbB2ΔEx16 is an isoform of ErbB2 that lacks, by alternative splicing, the 16 amino acids encoded by exon 16. (B) Consistent with previous findings, the ratio of ErbB2ΔEx16 transcript falls between 2–10% of total ErbB2 transcript across all cell line transcriptional subtypes. (C) Total levels of ErbB2ΔEx16 are enriched in ErbB2+ cell lines.
Interestingly, the expression of the ErbB2ΔEx16 form was not restricted to tumor cells. Using RT-PCR that allowed the simultaneous measurement of wild-type ErbB2 and ErbB2ΔEx16 transcripts, we showed that the ErbB2ΔEx16 variant could be detected at variable levels (2–10% of total ErbB2 transcript) in a variety of normal human tissues (Fig. S1). Together these observations indicate that both wild-type ErbB2 and ErbB2ΔEx16 transcripts are expressed in both normal and malignant tumor samples and that breast cell lines possessing amplified ErbB2 possessed particularly elevated levels of the ErbB2ΔEx16 variant.
Although these observations suggest that the ErbB2ΔEx16 transcript is expressed at elevated levels in ErbB2 amplified breast cancer cell lines, the biological significance of these observations was unclear. To directly test the transforming properties of this variant, we established independent clones of immortalized NMuMG mammary cells that expressed either constitutively active rat ErbB2 (NeuNT) 45, wild-type human ErbB2 or the human ErbB2ΔEx16 variant at comparable levels (Fig. S2A). To evaluate whether cells expressing the ErbB2 variants exhibited differential tumorigenic potential, mammary cells derived from two independent clones expressing either wild-type ErbB2, activated ErbB2 (NeuNT) or ErbB2ΔEx16 were injected orthotopically into immunodeficient recipient mice and their growth monitored (Fig. S2B). In contrast to the rapid onset of tumors seen in both clones expressing NeuNT and the ErbB2ΔEx16, 2 independent clones of cells expressing the wild-type form of ErbB2 failed to form tumors (Fig. S2B). These observations argue that unlike the ErbB2ΔEx16 and activated forms of ErbB2 (Neu-NT), elevated expression of wild-type ErbB2 is not sufficient to convert these immortalized cells to the malignant phenotype.
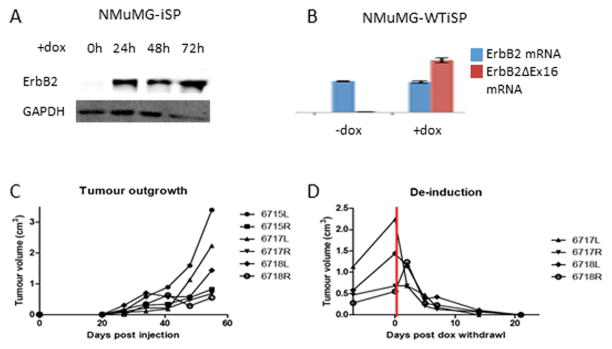
While wild-type ErbB2 appears to be incapable of transforming immortalized epithelial cells, an outstanding question is whether this isoform is capable of maintaining a tumorigenic state. In order to test this, NMuMG cells were used to generate cell lines expressing a doxycycline-inducible ErbB2ΔEx16, isolating cells with tightly regulated induction (Fig 2B). In the presence of doxycycline, robust ErbB2ΔEx16 protein expression is observed as quickly as 24 hours after in vitro doxycycline administration (Fig 2A). These cells were subsequently engineered to express wild-type ErbB2 as well, which were selected using FACS in the absence to doxycycline, in order to sort cells by robust wild-type ErbB2 expression. Cells were injected orthotopically into athymic nude mice, and the mice were treated with 2mg/ml doxycycline in the drinking water. Consistent with previous results, ErbB2ΔEx16 induction drives rapid transformation and tumor outgrowth (Fig 2C). However, when doxycycline is withdrawn, the tumors fail to express ErbB2ΔEx16 but retain robust expression of wild-type ErbB2 (Fig 2B), leading to rapid regression of the established mammary tumors (Fig. 2D). These observations demonstrate that expression of wild-type ErbB2 is insufficient to maintain a tumorigenic state in the absence of ErbB2ΔEx16 expression and indicate that the ErbB2ΔEx16 isoform and indicate that the ErbB2ΔEx16 isoform is a major oncogenic driver.
Figure 2.
ErbB2 is incapable of maintaining a tumorigenic state. (A) Doxycycline-inducible NMuMG cells (NMuMG-iSP) rapidly express ErbB2ΔEx16 in the presence of doxycycline within 24 hours of induction. (B) In cells expressing stable ErbB2 with inducible ErbB2ΔEx16 (NMuMG-WTiSP), quantitative PCR confirms robust expression of ErbB2ΔEx16 mRNA in the presence, but not the absence of doxycycline. By contrast, ErbB2 expression remains constant. (C) When NMuMG-WTiSP cells were injected orthotopically into athymic nude mice, tumors were detected 3 weeks after injection when treated with doxycycline. (D) Withdrawal of doxycycline results in rapid and complete regression of the WTiSP tumors.
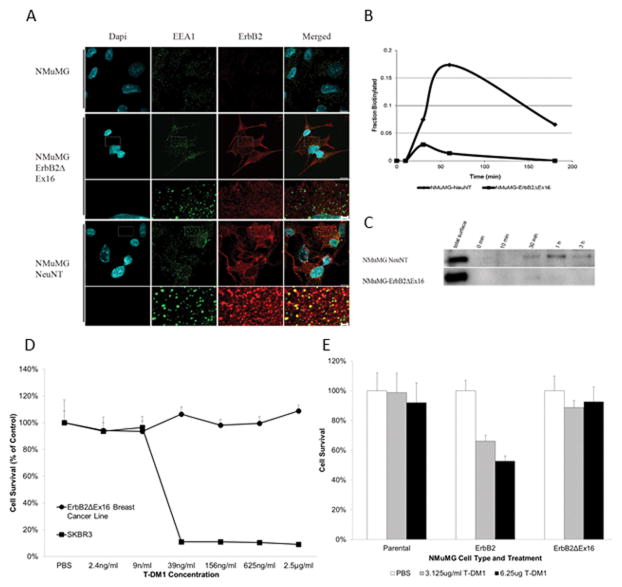
To determine whether the differential transforming activity of the ErbB2ΔEx16 variant involved altered receptor recycling, we performed immunofluorescence staining on cells expressing the different ErbB2 receptors with antibodies directed towards ErbB2 and the early endosome marker EAA1 (Fig. 3A). Cells were treated with cycloheximide in serum-free medium for one hour, followed by one hour in complete medium at 4°C. To permit receptor internalization, cells were warmed to 37°C with fresh complete medium for 15 minutes. Examination of NeuNT expressing cells revealed that ErbB2 and EAA1 were co-localized in the early endosome, indicating rapid down-regulation of ErbB2 from the cell surface (Fig. 3A). In contrast to these observations, the ErbB2ΔEx16 variant could not be detected in the endosomal compartment.
Figure 3.
ErbB2ΔEx16 displays altered internalization, which affects response to therapy. (A) Serum-induced internalization of NeuNT results in co-localization with EEA1, which is absent in ErbB2ΔEx16 cells. (B) Quantification of a biotin internalization assay (C) reveals that while NeuNT is effectively internalized, ErbB2ΔEx16 remains at the cell surface. (D) Treatment of NMuMG-ErbB2ΔEx16 cells with the anti-ErbB2 therapy T-DM1 reveals an innate resistance to treatment compared to SkBR3 cells, which express very high levels of full length ErbB2. Additionally, (E) NMuMG cells expressing full length ErbB2 are intrinsically more sensitive to T-DM1 treatment than NMuMGErbB2ΔEx16 cells.
To confirm that lack of detectable ErbB2ΔEx16 receptor in the endosomal compartment was due to defect in down-regulation from the cell surface, we performed a biotin labeling of cell surface proteins and measured the percentage of internalized ErbB2 that was protected from proteolytic degradation (see Material and Methods). Consistent with the rapid internalization of NeuNT in the endosomal compartment, a significant percentage of ErbB2 was protected 60 minutes after shifting the cells to the permissive temperature for sorting. By contrast only a small proportion of the ErbB2ΔEx16 variant was internalized (Fig. 3B and 3C).
Another indication of altered trafficking for ErbB2ΔEx16 derived variant stem from the observation that mammary tumor cell lines derived from either transgenic mice (Fig. 4) or NMuMG cells expressing the ErbB2ΔEx16 isoform are resistant to the anti-ErbB2 therapeutic T-DM1 antibody–drug conjugate whereas control cells expressing the wild-type ErbB2 were highly sensitive to T-DM1 (Fig. 3D and 3E). Given that T-DM1 drug delivery requires internalization, the inability of T-DM1 to kill these the ErbB2ΔEx16 expressing likely reflects it inability to be internalized 12. Collectively these observations demonstrate that the ErbB2ΔEx16 is primarily a cell surface receptor that is not efficiently internalized and also indicate that expression of the ErbB2ΔEx16 isoform may confer resistance to T-DM1.
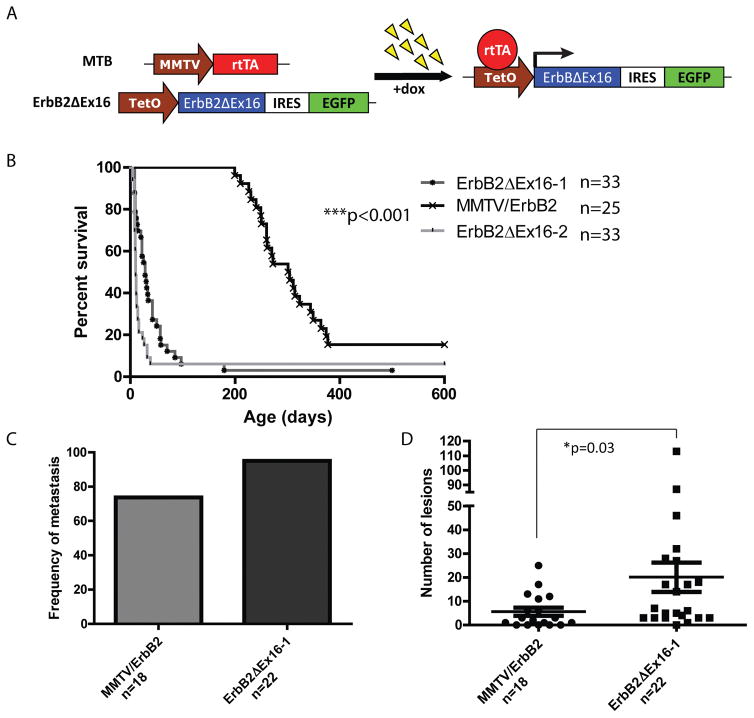
Figure 4.
A transgenic mouse model of ErbB2ΔEx16-driven tumorigenesis. (A) A reverse tetracycline-dependent transactivator (rtTA) is expressed in the mammary epithelium of transgenic mice via the MMTV promoter. In the presence of doxycycline, ErbB2ΔEx16 is expressed in mammary epithelial cells through rtTA binding to the tetracycline-dependent promoter (TetO). (B) Expression of ErbB2ΔEx16 drives rapid tumor onset with a latency of 28 and 10 days for two independent founder lines, compared to 280 days for full length ErbB2. (C) ErbB2 and ErbB2ΔEx16 models metastasize to the lungs with a frequency of 74% and 95%, respectively, with (D) a significantly increased number of lung lesions in ErbB2ΔEx16 mice (p=0.03).
Mammary specific expression of the ErbB2ΔEx16 isoform results in the rapid induction of metastatic breast cancers
To directly evaluate the oncogenic potential of the ErbB2ΔEx16 isoform in the mammary epithelium, we placed the ErbB2ΔEx16 under the transcriptional control of the tetracycline response element and crossed these strains of FVB mice to a separate FVB strain expressing the Reverse Tetracycline Trans activator (rtTA) under the transcriptional control of the mouse mammary tumor virus promoter (MMTV) 29. To facilitate detection of transgene expressers, we placed an IRES EGFP expression cassette downstream of the ErbB2ΔEx16 cDNA (Fig. 4A). In this manner, we can induce expression of the ErbB2ΔEx16 in the mammary epithelium by the addition of doxycycline to the drinking water. Of the initial 7 independent founder lines, 2 independent the founder strains could be induced to express elevated levels of the ErbB2ΔEx16 isoform upon doxycycline administration (Fig. S3A and S3B). Using RT-PCR analyses, we show that ErbB2ΔEx16 expression is primarily restricted to the mammary and salivary glands (Fig S4).
To further characterize the kinetics of tumor onset in the inducible ErbB2ΔEx16, we generated cohorts of female mice from two of these independent strains and monitored tumor induction following addition of doxycycline to drinking water. Groups were unblinded throughout the study, and a target sample size of 20 per group was chosen to ensure sufficient statistical power. The results revealed that in contrast to MMTV/ErbB2 strain that developed focal mammary tumors after long latency period (T50=280 days) 15, both inducible ErbB2ΔEx16 strains rapidly induced multifocal mammary tumors with an extremely short latency period (T50=10 and 28 days) (Fig. 4B). Additionally, ErbB2ΔEx16 tumors frequently gave rise to metastatic lesions in the lung in a majority of tumor bearing animals (Fig 4C–D).
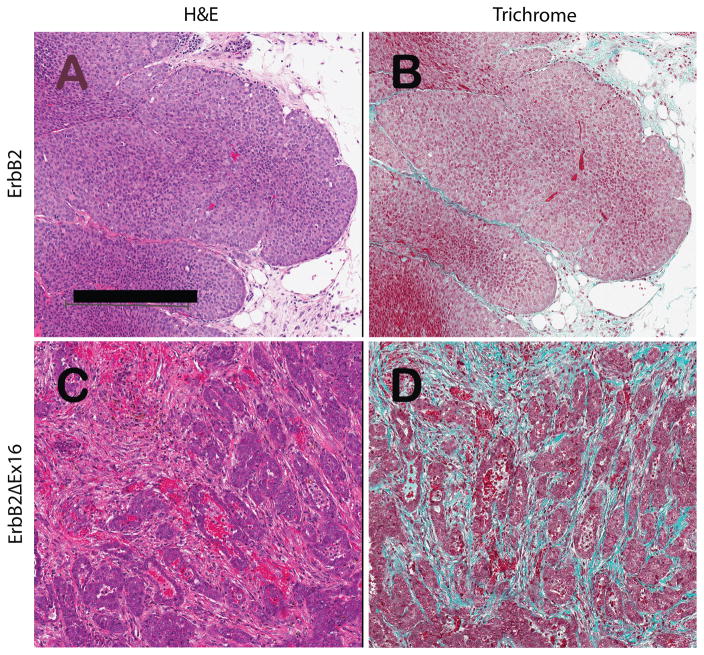
Histological analyses of these tumors revealed that that the ErbB2ΔEx16 possessed tumor pathology that is distinct from those expressing the erbB2 proto-oncogene (Fig. 5A and 5C). Using trichrome staining, which stains for collagen deposition, we confirmed that ErbB2ΔEx16 derived tumors possess much higher levels of matrix deposition than the comparable ErbB2 tumors (Fig. 5B and 5D).
Figure 5.
Histopathology of ErbB2-driven tumor phenotypes described by hematoxylin and eosin stained slides (A and C) and comparing the relative amounts of ECM deposition using green trichrome staining for connective tissue (B and D). Figures 1A and 1B illustrate a signature solid nodular tumor from an ErbB2 bearing mouse. In contrast, Panels C and D illustrates the more glandular phenotype induced by the ErbB2ΔEx16 protein, which has abundant connective tissue separating the nests and cords of tumor cells. These tumors infiltrate through the adipose tissue and do not have pushing margins seen in the signature phenotype (Panels A and B). The scale bar indicates 250 microns (Panel A).
To further evaluate whether the observed pathological differences between the two ErbB2 tumor models was due to different cellular origin, we next stained both full length and the ErbB2ΔEx16 derived tumors with either luminal (cytokeratin 8) or basal keratin (cytokeratin 14, 6 and 5) markers. In contrast to tumors expressing full length ErbB2 that were uniformly positive for Keratin 8 marker and negative for any of the basal keratin markers, the ErbB2ΔEx16 derived tumors stained positive for both luminal and basal keratins (Fig. S5A). Indeed, co-staining with different keratin markers revealed a diversity of cells that co-expressed both basal and luminal keratin markers. (Fig. S5B). Collectively these observations argue that in contrast to ErbB2 derived tumors, mammary epithelial expression of ErbB2ΔEx16 results in rapid induction metastatic mammary tumors that possess a high degree of intra-tumoral heterogeneity.
To further establish the critical importance of the ErbB2ΔEx16 as major oncogenic driver in mammary tumorigenesis, we next determined whether withdrawal of doxycycline from the drinking water in ErbB2ΔEx16 tumor bearing mice would result in tumor regression. The results showed that removal of doxycycline from tumor-bearing animals resulted in rapid regression of the ErbB2ΔEx16 derived tumors. However after a variable tumor-free period, focal mammary tumors reappeared (Fig. S6A). Molecular and pathological analyses of these recurrent tumors revealed that exhibited EMT phenotype (Fig. S6C) that was further correlated with reduced expression of the epithelial marker E-Cadherin and complete loss of the ErbB2ΔEx16 transgene expression (Fig. S6B). Thus like our previous cell culture data (Fig. 2), these observations indicate that sustained expression of the ErbB2ΔEx16 is required to maintain the transformed state.
The ErbB2ΔEx16 derived tumors exhibit unique signaling and transcriptional profiles due to activation of transcription factor network
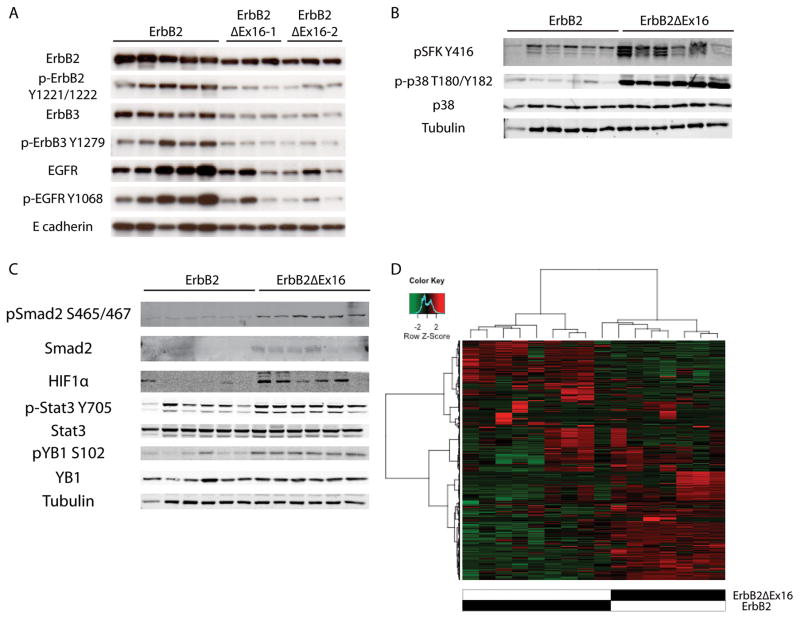
To further elucidate the molecular basis for the potent transforming ability of the ErbB2ΔEx16 isoform we performed both RPPA and immunoblot analyses on a number of downstream signaling pathways. Despite the constitutive dimerization of the ErbB2ΔEx16 receptor 38, the levels of tyrosine phosphorylated ErbB2, EGFR and ErbB3 were impaired by comparison ErbB2 tumors (Fig. 6A). Conversely, the ErbB2ΔEx16 derived tumors exhibited a profound activation of p38 and c-Src family kinases compared to the tumors expressing the ErbB2 receptor (Fig. 6B). Another important set of phosphorylated proteins that were clearly activated by RPPA as measured by the phosphorylation status in the ErbB2ΔEx16 derived tumors were a number of transcription factors, including Stat-3, YB-1, Smad-2 and HIF1α (Fig. S7). To confirm that activation status of this transcription factor network in the ErbB2ΔEx16 class of mammary tumors, we performed immunoblot analyses with phospho-specific antibodies directed against each of these transcription factors. Consistent with RPPA analyses, the results showed that although there was no absolute increase in the levels in these transcription factors, the ErbB2ΔEx16 derived tumors exhibited a dramatic elevation in activation status of these key transcription factors (Fig. 6C).
Figure 6.
Immunoblot analysis of ErbB2-expressing tumors. (A) Full length ErbB2-driven tumors co-express heterodimer partners ErbB3 and EGFR, whereas ErbB2ΔEx16 tumors arise in the absence of heterodimerization partners. (B) ErbB2ΔEx16 displays heightened activation of Src-family kinases (SFK) and p38 MAPK. (C) Additionally, ErbB2ΔEx16-expressing tumors preferentially activate a distinct subset of transcription factors, primarily Smad2, HIF1α, Stat3, and YB1. (D) Unsupervised hierarchical clustering of tumors results in grouping of tumors by genotype.
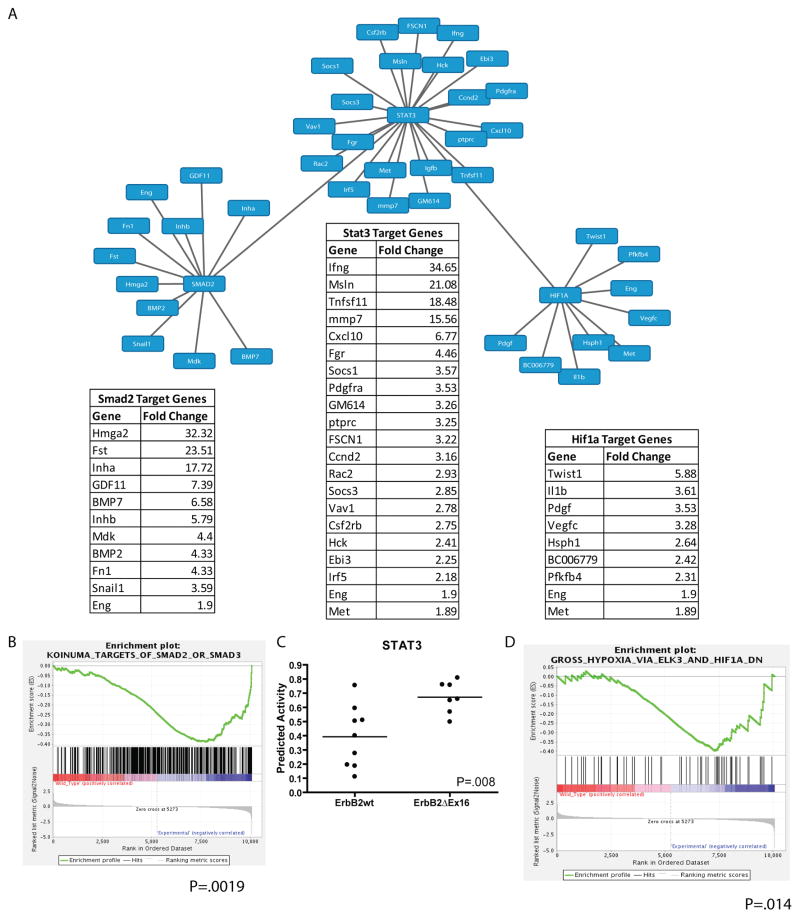
The observations outlined above indicate that the ErbB2ΔEx16 derived tumors have pathological and molecular features that are distinct from tumors induced by the ErbB2 receptor. To further explore the molecular basis for the striking differences between these closely related ErbB2 receptors, we performed gene expression profiling on 8 independent tumors from either the wild-type ErbB2 or the ErbB2ΔEx16 strains using the Agilent platform. Consistent with the molecular and pathological analyses, unsupervised hierarchical clustering of gene expression profiles revealed that the ErbB2Δ16 tumors co-clustered and could be readily distinguished from their ErbB2 counterparts (Fig. 6D). Given that our RPPA data indicated that ErbB2ΔEx16 selectively activated Stat3, Smad2 and HIF-1α transcription factors, we next evaluated whether differential gene expression profiles between two ErbB2 tumor subtypes could be due to selective activation of this transcriptional factor network. To accomplish this, we determined whether known target genes of these transcription factors were selectively up-regulated in the ErbB2ΔEx16 derived tumors. The results of these analyses confirmed that differential gene expression profile exhibited by the ErbB2ΔEx16 derived tumors can in part, be accounted for by activation of this transcriptional network (Fig. 7A). Target genes downstream of Smad-2 (p=0.0019), HIF1α (p=0.014) and Stat3 (p=0.008) were significantly enriched in ErbB2ΔEx16 samples (Fig. 7B, 7C, and 7D). Notably, expression of Stat3 target genes such interferon γ and Cxcl10 that have implicated in regulation of tumor immune microenvironment were dramatically up-regulated in the ErbB2ΔEx16 derived tumors. Similarly, selective up-regulation of Smad-2 targets such TGF β superfamily was also observed in the ErbB2ΔEx16 derived tumor setting (Fig. 7A). In addition, transcription factors Snail1 and Twist1 that are also targets of Smad-2 and HIF1α in ErbB2ΔEx16 tumors. Interestingly, these latter transcription factors are thought to play a critical role in EMT process 14. Collectively, these observations indicate that the ErbB2ΔEx16 derived tumors possess transcriptional signature associated with invasive breast cancers.
Figure 7.
Analysis of transcription factor activation from gene expression profiling. (A) Downstream of ErbB2ΔEx16, the transcription factors Smad2, Stat3, and Hif1α are preferentially activated, as evidenced by enrichment in known target gene expression obtained from gene expression profiling. Enrichment plots by GSEA (B and D) suggest strong association of Smad2 (p=0.0019) and HIF1α (p=0.014) with ErbB2ΔEx16 expression. (C) Activity of the Stat3 transcriptional networks is also is enriched in ErbB2ΔEx16-expressing tumors (p=0.008).
Discussion
In this study, we demonstrate that the oncogenic ErbB2ΔEx16 variant is expressed at appreciable levels in a number of ErbB2 expressing breast cancer cell lines. Additionally, we show that the ErbB2ΔEx16 expression is both sufficient and necessary for the maintenance of the transformed phenotype (Fig. 2 and Fig. S6). Specifically, we demonstrate that in contrast to another oncogenic ErbB2 mutant (NeuNT), the ErbB2ΔEx16 isoform is constitutively localized to the plasma membrane compartment (Fig. 3). Consistent with the unique properties of the ErbB2ΔEx16, mammary specific expression of the ErbB2ΔEx16 resulted in rapid induction of multifocal metastatic mammary tumors that possessed molecular and pathological features that were distinct from the conventional MMTV ErbB2 models. These features include high levels of intra-tumoral heterogeneity that is characterized by co-expression of both luminal and basal keratins and is reflected at the molecular level by a distinct gene expression profile (Figs. 4–7).
Although a number of studies have detected the presence of the ErbB2ΔEx16 variant in both established breast cancer cell lines and primary breast cancers (Fig. 1) 1, 5, 24, 38, the precise role of this variant in ErbB2-induced tumor progression was unclear. Using NMuMG immortalized mammary epithelial cell model, we demonstrate that elevated expression of wild-type ErbB2 is not sufficient to drive morphological transformation of this immortalized mammary epithelial cell line (Fig. S2). By contrast, expression of the activated ErbB2 (NT mutation) or the ErbB2ΔEx16 form was sufficient to drive the tumorigenic conversion of NMuMG cells (Fig. S2). Furthermore, expression of wild-type ErbB2 in the context of an ErbB2ΔEx16-driven tumor is incapable of maintaining transformation following loss of ErbB2ΔEx16 expression (Fig. 2). These observations indicate that at least in this immortalized cell system, the ErbB2ΔEx16 is the major oncogenic driver.
The potent transforming property of the ErbB2ΔEx16 isoform were further correlated with constitutive localization to the plasma membrane. By contrast, other activated versions of ErbB2 (NeuNT) are rapidly down-regulated from the cell surface (Fig. 3A and 3B). The difference in receptor trafficking in ErbB2ΔEx16 tumors may account for the distinct signaling output from the ErbB2ΔEx16 receptor. In this regard, it has been reported that the ErbB2ΔEx16 variant is specifically associated within a membrane compartment harboring c-Src 28. Another important consequence of the differential trafficking properties of the ErbB2ΔEx16 variant, is that tumor cells expressing this variant are highly resistant to T-DM1 (Fig. 3C and 3D). Because the biological activity of Trastuzumab-drug conjugate is dependent on its ability to be internalized to release the drug toxin 12, the inability T-DM1 to kill the ErbB2ΔEx16 variant expressing tumor cells reflects its inability to be efficient internalized. These data contrast with a recent report indicating that Trastuzumab can effectively eliminate ErbB2ΔEx16 expressing tumors in vivo 5. The difference between the in vitro resistance phenotype we observed (Fig. 3) and the in vivo sensitivity may reflect the importance of the Antibody Directed Cellular Cytotoxicity (ADCC) in T-DM1 anti-tumor response 11.
Consistent with the unique signaling properties of the ErbB2ΔEx16 variant, mammary epithelial expression of this isoform resulted in rapid induction of multifocal mammary tumors that frequently metastasized to the lung, whereas comparable strains expressing ErbB2 in the mammary gland develop focal mammary tumors only after a long onset period (Fig. 4). In agreement with these observations, a transgenic line expressing the ErbB2ΔEx16 variant with constitutive MMTV promoter rapidly developed mammary tumors 6. However, detailed molecular and pathological analyses of mammary tumors were not reported.
Another distinct feature of these the ErbB2ΔEx16 expressing tumors is that they have several unique pathological features, including major deposition of extracellular matrix (ECM) (Fig. 5) and co-expression of both luminal and basal keratin markers (Fig. S5). The dense ECM depositions observed in the ErbB2ΔEx16 derived tumors was further correlated with the metastatic dissemination to the lung (Fig. 5C–5D). In many respects, this ECM deposition phenotype resembles high breast density that is a known risk factor for development of breast cancer 27. Other studies have linked matrix density to directly impact on metastatic fate of tumor cells 10, 30
Although both of the ErbB2-driven mouse models expressed the luminal marker cytokeratin 8 (Krt8; Fig. S5) only the ErbB2ΔEx16 derived tumors exhibited expression of basal cytokeratin markers such as Krt5, Krt6, and Krt14 (Fig. S5). These findings indicate that the ErbB2ΔEx16 derived mammary tumors exhibit a high degree of intra-tumoral heterogeneity comprising of mixed cell lineages, whereas MMTV wild-type ErbB2 tumors are comprised of a uniform luminal cell type.
The ErbB2ΔEx16 derived tumors also exhibited a profound difference in signaling pathways activated compared to their full-length counterparts. Using RPPA and immunoblot analyses (Fig. 6, Fig. S7), we demonstrate that the ErbB2ΔEx16 derived tumors exhibit reduced tyrosine phosphorylation of ErbB2 and its heterodimer partners EGFR and ErbB3 (Fig. 6A) and elevated activation c-Src family of tyrosine kinases and activation of p38 kinase (Fig. 6B) compared to ErbB2 derived tumors. These analyses also revealed that ErbB2ΔEx16 dependent signaling was responsible for activation of number of key transcription factors including Stat3, Smad2, YB-1 and HIF1α (Fig. 6C). Activation of these transcription factor networks has been implicated in regulating EMT process that is involved in breast cancer stem cell renewal 23, 46. Consistent with these biochemical analyses, analyses of the gene expression profiles of ErbB2ΔEx16 derived tumors revealed that many of the known target genes of this transcriptional network are selectively up-regulated in ErbB2ΔEx16 derived tumors (Fig. 7). Critically, comparison of the ErbB2ΔEx16 transcriptional signature to the human data sets revealed that they align with basal category of human breast cancers that have particularly bad outcome (Fig. S9).
One important set of Stat3 target genes that are up-regulated in the ErbB2ΔEx16 tumors, such as interferon γ and Cxcl10, are important modulators of immune micro-environment (Fig 7). Given that activation of Stat3 is primarily in the tumor epithelium, these Stat3 dependent cytokines are acting in a paracrine fashion to promote immune infiltration (Fig. S8). In this regard, it is noteworthy that mammary tumor specific disruption of Stat3 in an ErbB2 model results in a decrease in infiltrating macrophages due to down-regulation of a number of Stat3 dependent inflammatory cytokines 34. Interestingly, the loss of an inflammatory tumor microenvironment is associated with a dramatic decrease in ErbB2 dependent metastasis 34. Indeed, the increase in spontaneous metastasis observed in ErbB2ΔEx16 derived tumors (Fig. 4) may be due to enhanced inflammatory tumor immune microenvironment. In addition to modulating macrophage infiltration, our recent studies have indicated that epithelial activation Stat3 plays a critical role in maintaining an immune-suppressive tumor microenvironment. For example, mammary epithelial deletion of Stat3 in PyV mT model results in rapid immune-mediated clearance of emerging PyV mT tumors 22. Collectively, these observations argue that the potent transforming properties of ErbB2Δ16 variant is due to its impact on the tumor immune microenvironment.
Recent studies have suggested that the ErbB2ΔEx16 variant may play an important role in biological effects of ErBB2 targeted therapies, such as the anti-ErbB2 antibody Trastuzumab. In one report, mammary cell lines expressing ErbB2ΔEx16 were resistant to Trastuzumab treatment 28 whereas several recent publications assert that Trastuzumab treatment can eliminate ErbB2ΔEx16-expressing tumors. Indeed, the authors of the latter set of studies have argued that the ErbB2ΔEx16 variant is the primary target of Trastuzumab 1, 5. In light of our observations that the ErbB2ΔEx16 variant confers resistance to Trastuzumab based T-DM1 raise the possibility that its expression may be important determinant of resistance to these agents. Another interesting finding from our studies is that the ErbB2ΔEx16 variant is also expressed to some level in a number of normal tissue sites (Fig. S1) indicating a potential role in normal tissue homeostasis. In this regard, we have shown that mice expressing ErbB2 cDNA (hence lacking the ErbB2ΔEx16 variant) develop both spindle and cardiac cell dysfunction indicating that the ErbB2ΔEx16 variant may have important physiological functions 2, 33. More recently, several RNA splicing factors have been identified that facilitate exon 16 skipping 17. Future studies directed towards understanding the role of the ErbB2ΔEx16 form in development and tumorigenesis will shed important insight into targeting this key oncogenic driver.
Materials and Methods
RNA-seq analysis of breast cancer cell lines
Whole transcriptome shotgun sequencing (RNAseq) was completed on breast cancer cell lines and expression analysis was performed with the ALEXA-seq software package as previously described 18. Briefly, this approach comprises (i) creation of a database of expression and alternative expression sequence ‘features’ (genes, transcripts, exons, junctions, boundaries, introns, and intergenic sequences) based on Ensembl gene models, (ii) mapping of short paired-end sequence reads to these features, (iii) identification of features that are expressed above background noise while taking into account locus-by-locus noise, (iv) identification of features that are differentially expressed in samples and (v) identification of the subset of differentially expressed features that correspond to alternative expression of mRNA isoforms. RNAseq data were available for 59 lines. An average of 71.0 million (76bp paired-end) reads passed quality control per sample. Of these, 54.0 million reads mapped to the transcriptome on average, resulting in an average coverage of 48.5X across all known genes. Log2 transformed estimates of ErbB2 exon-level and junction-level expression were extracted for analysis. The abundance of ErbB2ΔEx16 was determined using the estimated expression of the E15:E17 junction. Full length expression was determined by taking the mean of E15, E16 and E17 exons and the E15:E16 and E16:E17 junctions. Error bars represent standard deviation.
Immunoblot analysis
Flash frozen tumor tissue was disrupted by mortar and pestle in liquid nitrogen, and lysed in a modified Hepes lysis buffer (50mM Hepes pH 7.5 (Bioshop HEP001), 150mM NaCl (Biobasic SB0476), 10% Glycerol (GB0232), 0.5% NP-40 (Biobasic NDB0385), 1mM EDTA pH 8.0 (Biobasic EB0185), 10mM NaF (Bioshop SFL001), 1mM PMSF (Bioshop PMS123), 10mM β Glycerophosphate (Bioshop GYP001), 1mM Na3VO4 (Bioshop SOV664), 10μg/ml Aprotinin (Biobasic AD0153), 10μg/ml Leupeptin (Biobasic LDJ691). Lysates were quantified by Bradford assay, and reduced in SDS loading buffer with β-mercaptoethanol (Sigma M6250). Immunoblots are representative of 3 independent experiments.
Antibodies
The following primary antibodies were used for immunoblotting: (Santa Cruz Biotechnology) ErbB3 C17 (sc-285), (Cell Signaling Technology) phospho-ErbB2 Y1221/1222 (2249), EGFR (2232), phospho-Src Family Kinase Y416 (2101), phospho-ErbB3 Y1289 (4791), phospho-p38 (9215), p38 T180/Y182 (9212), tubulin (2148), phospho-Smad2 S245/250/255 (3104), Smad2/3 (3102), HIF1α (3716), phospho-Stat3 Y705 (9145), Stat3 (9139), phospho-YB1 S102 (2900), (Millipore) c-ErbB2 Ab-3 (OP-15), (BD Transduction) E-cadherin (610182), (Abcam) YB1 (12148).
Cell surface biotinylation
Surface receptors were biotinylated with Sulfo NHS ss Biotin (Fisher-Pierce 21331) to monitor their rates of internalization. Control cells were either not incubated at 37°C (0 minutes control) or incubated for 3 hours at 37°C without subsequent incubation in the biotin stripping buffer (total surface receptor control). Cells were lysed in PLCγ lysis buffer. Biotinlyated protein was precipitated from 500 g of total lysate with streptavidin-coated agarose beads (Pierce 20347) overnight at 4°C. The lysate was then washed and prepared for immunoprecipitation as described previously, and analyzed by immunoblot using ImageQuant TL software (GE Healthcare Biosciences). The experiment was repeated 3 times, and representative results are shown. Error bars represent standard deviation.
RPPA of transgenic mammary tumors
Flash frozen tumor pieces were disrupted using a mortar and pestle while immersed in liquid nitrogen, and transferred to a 50ml conical tube. Once nitrogen has evaporated, tissue was lysed in NP40 Lysis buffer (50mM HEPES pH 7.5, 150mM NaCl, 10% glycerol, 1% NP-40, 1mM EDTA, 10mM NaF, 1mM PMSF, 10mM β Glycerophosphate, 1mm Na3VO4, 10μg/ml aprotinin, 10μg/ml leupeptin) on ice for 10 minutes. For RPPA analysis, cellular proteins were denatured by 1% SDS (with β-mercaptoethanol) and diluted in five 2-fold serial dilutions in dilution buffer (lysis buffer containing 1% SDS). Serial diluted lysates were arrayed on nitrocellulose-coated slides (Grace Biolab) by Aushon 2470 Arrayer (Aushon BioSystems) and subjected to RPPA as described previously 13. Analysis of the RPPA data was performed by pooling the sample data by genotype (n=5 for MMTV/ErbB2, n=10 for ErbB2ΔEx16), and statistical significance was determined by subjecting the means to an unpaired Student’s T-test, with a p value cut off of < 0.05. Error bars represent standard deviation.
Genotyping
Transgenic mice were genotyped using the following primers for MTB (Fwd ACCGTACTCGTCAATTCCAAGGG Rev TGCCGCCATTATTACGACAAGC), EGFP for ErbB2ΔE16 (Fwd CACAAGTTCAGCGTGTCC Rev TGTACAGCTCGTCCATGC), and MMTV-ErbB2 (Fwd CCTCCTAAAGGACCTAGAGGAAGGC, Rev CAAGGCCAGGAGAGGCACTGGGGAG)
Generation of transgenic mice
A DNA fragment containing the Tet-responsive promoter and erbB2ΔEx16 cDNA was generated by PvuII restriction digest, and sent for pronuclear injection by the McGill LSC Transgenic Core Facility. Putative founder lines were interbred with transgenic mice harboring the MMTV/rtTA transgene (MTB, a kind gift from Dr. Lewis Chodosh). Bigenic female mice were induced for 5 days with 2mg/ml doxycycline in their drinking water. Transgene expression was assessed by immunoblot analysis using the thoracic mammary gland, and by whole-mount EGFP and hematoxylin staining of the abdominal mammary gland. MMTV/ErbB2 mice were a generous gift from Genentech (Roche) 15. Female transgenic mice carrying the MTB and ErbB2ΔEx16 transgenes were induced at 8–10 weeks of age with 2mg/ml doxycycline (Wisent 450-185-EG) in the drinking water, with tumor onset monitored by weekly palpation. Statistical significance was determined using a Rank Log (Mandel-Cox) test (n=25 for MMTV/ErbB2, n=33 for ErbB2ΔEx16).
Histological staining
Tumor pieces were fixed in 10% neutral buffered formalin for 24 hours, processed and embedded in paraffin at the McGill LSC Histology core facility. Sections were cut and stained with Harris’ Haematoxylin and Eosin, or Gomori’s trichrome Stain kit (Polysciences 24205-1) according to manufacturer’s protocol. Images are representative of at least 10 independent samples. For immunohistochemistry, cells were treated as described previously 25, using Signalstain ABC kit (Vector Laboratories PK-4000) secondary antibody, and Liquid DAB+ substrate chromogen system (Dako 8059S) to develop, as per manufacturer’s instructions. Primary antibodies were anti-keratin 14 (1:500, Covance, Cat#PRB-155P), anti-keratin 5 (1:500 Covance, Cat#PRB-160P), anti-keratin 6 (1:500, Covance, Cat#PRB-169P) anti-keratin 8/18 (1:200, Fitzgerald, Cat# RDI-PROGP11), CD3ε (1:200, Abcam, Ab16669), phospho-Stat3 Y705 (1:200, Cell Signaling, #9145). Stained sections were quantified using a nuclear staining algorithm on the Aperio Imagescope software. All processed samples were single-blinded for genotype during experiment and analysis. Statistical significance was determined by two-tailed, unpaired, Student’s T test (CD3ε; n=10, pStat3; n=5). Error bars represent standard deviation. For immunohistofluorescence, fluorescent secondary antibodies Alexa488, Alexa555, and Alexa647 (1:1000, Molecular Probes), and counterstained using DAPI (Sigma, 32670).
Lung metastasis
At necropsy, lung tissue was fixed and processed similarly to tumor pieces. FFPE lung samples were step sectioned to obtain 5 sections at 50μm intervals, and stained with Harris’ Haematoxylin and Eosin. Samples were blinded for genotype, and metastases were counted by microscopic inspection (n=18 for MMTV/ErbB2, and n=22 for ErbB2ΔEx16). Metastatic burden analysis was performed by first determining whether the populations presented with different variances using an F-test. As the variances of the two groups were found to be significantly different, evaluation of significance of the means was determined using an unpaired Welch’s T-test. Error bars represent standard deviation.
RNA extraction and microarray analysis
Total RNA from transgenic tumor samples (n=9 for MMTV/ErbB2, n=7 for ErbB2ΔEx16) was harvested using the RNEasy mini kit (Qiagen 74106) according to manufacturer’s instructions. Total RNA was quantified using a NanoDrop Spectrophotometer ND-1000 (NanoDrop Technologies, Inc.) and its integrity was assessed using a 2100 Bioanalyzer (Agilent Technologies). Cyanine 3-labeled CTP cRNA was produced with 1ug of total RNA using the Low Input Quick Amp Labeling Kit, according to manufacturer’s instructions (Agilent Technologies, Inc).
The labeled cRNA was then normalized at 600ng, fragmented and hybridized on SurePrint G3 Mouse GE 8×60K. The arrays were incubated in an Agilent Hybridization oven at 65°C for 17 hours at 10 rpm. They were washed and scanned on an Agilent DNA Microarray Scanner C. All these steps were done according to Agilent One-Color Microarray-Based Gene Expression Analysis protocol (Agilent Technologies, Inc). Data was obtained using the Agilent Feature Extraction software version 11.0.1.1. Using the LIMMA R package 42, normalization was performed through the application of background correction with the half setting 35, and subsequent quantile between-array normalization 41. Probe data was collapsed to RefSeq features by taking the median value. Unsupervised hierarchical clustering was accomplished through the use of Ward’s algorithm using the Pearson correlation distance metric applied on the features having the top 2% highest variance across samples. Array data from ErbB2 samples (n=9) and ErbB2ΔEx16 (n=7) samples were analyzed using the SAM 44 algorithm to identify significant fold changes up and down regulated in the ErbB2ΔEx16 samples as compared to the ErbB2 positive samples. The top 1000 differentially regulated genes were placed into iRegulon 21 with Smad2, STAT3 and Hif1α to view gene interactions between differentially regulated genes and Smad2, STAT3, and Hif1α. Gene Set Enrichment Analysis 43 was performed to identify Smad2 and Hif1α gene set enrichment in the ErbB2ΔEx16 samples using genepattern. Stat3 activity was predicted using a gene signature approach according to previous studies4, 8, 16. Immune gene signature was determined by subjecting the processed gene signatures to Enrichr Gene Enrichment Analysis 9.
Cell culture
NMuMG (ATCC CRL1636) and SKBR3 (ATCC HTB30) cells were cultured in DMEM (Wisent 319-005-CL) supplemented with 10% FBS (Wisent 095-150), 10mM HEPES pH 7.5 (Wisent 330-050-EL), 10μg/ml insulin (Wisent 511-016-UG), 100IU/ml Penicillin and 100μg/ml Streptomycin (Wisent 450-201-EL). NMuMGErbB2wt and NMuMG-ErbB2ΔEx16 cell lines were transfected with either pMSCVErbB2wt or pMSCV-ErbB2ΔEx16 plasmids using Genejuice (Novagen 70967), and selected with 2μg/ml puromycin (Clontech 631305). NMuMG-NeuNT cells were generated as reported previously45. For doxycycline-inducible cell lines, prior to pMSCV transfection, NMuMG cells were first infected with lentivirus carrying pTIBZ-ErbB2wt or pTIBZ-ErbB2ΔEx16, selected in 8μg/ml Blasticidin (Thermo Fisher A1113903), induced with 10μg/ml doxycycline, and sorted by FACS analysis for robust ErbB2 expression. All cell lines were negative for mycoplasma.
RT-PCR analysis
1μg of total RNA is converted to cDNA using the NEB ProtoScript II First Strand cDNA Synthesis Kit (New England Biolabs E6560S). Q-PCR reactions were performed in triplicate using 1/10 of the RT product with the Roche LightCycler 480 SYBR Green I Master kit (Roche 04707516001). ErbB2ΔEx16 mRNA was detected using specific primers (Fwd CAGCGGTGTGAAACCTGACC, Rev TGGACGTCAGAGGGGAGTGG), normalized to either wild-type ErbB2 (Fwd GTGGACCTGGATGACAAGGG, Rev TGCTGCCGTCGCTTGATGAG) or GAPDH (Fwd GTGGTCTCCTCTGACTTCAAC Rev GTTGCTGTAGCCAAATTCGTTG). All samples were analyzed in triplicate with error bars representing standard deviation.
Orthotopic injections
Experiments were conducted according to an approved institutional Animal Use Protocol. 1×105 cells were injected into the left inguinal fat pad of athymic nude mice (Charles River Laboratories International), with mice fed 2mg/ml doxycycline in the drinking water, changed weekly. Tumors were monitored by weekly measurements. When tumors reached an approximate size of 1 cm3, doxycycline treatment was withdrawn, and tumors were monitored twice weekly for outgrowth. The results are representative of 3 repetitions, with n=5 injections per cell line.
Immunofluorescence
2.5×104 cells were seeded on coverslips and allowed to grow for 48 hours. Cells were incubated with 100μg/ml of cycloheximide (Sigma-Aldrich C1988-1G) in serum-free media for one hour. Immunofluorescence was performed as described in Marcotte et al.26 with the primary antibodies c-ErbB2 (1:500, DAKO A048529), and EEA1 (1:100, Santa Cruz sc-6415). Cells were counterstained with DAPI (Invitrogen D1306). Confocal images were obtained using an Axiovert 200M microscope (Carl Zeiss MicroImaging, Inc.) and analyzed using LSM5 Image browser (Empix Imaging). Images are representative of 3 replicates.
MTT assay
A total of 3000 cells (SKBR3) or 5000 cells (NMuMG and ErbB2ΔEx16 transgenic breast cancer cell lines) were plated in a final volume of 0.2 ml in 96-well flat bottom plates with indicated concentrations of Trastuzumab-DM1. After 3 days, 20 μl of a 5 mg/ml MTT solution in PBS were added to each well for 4 hours. After removal of the medium, 100 μl of DMSO were added to each well. The absorbance at 540 nm was determined using a Bio-Rad Model 680 microplate reader. At minimum, triplicate wells were assayed for each condition.
Study approval
All animal studies were carried out in accordance with the guidelines of the approved Animal Use Protocol by the McGill University Animal Care Committee (UACC) and the Canadian Council on Animal Care (CCAC).
Supplementary Material
Figure S1: ErbB2ΔEx16 mRNA expression in normal human tissue. (A) Quantitative PCR, differentiating wild type ErbB2 mRNA from ErbB2ΔEx16, reveals a relative expression level of 2–10% of total ErbB2 transcript across a panel of normal human tissues.
Figure S2: Ectopic expression of ErbB2 isoforms in NMuMG cells. (A) Immunoblot analysis confirms ectopic ErbB2 overexpression in NMuMG cells. (B) When NMuMG cells were injected orthotopically into athymic nude mice, only cells expressing ErbB2ΔEx16 or the oncogenic mutant NeuNT produced tumours in mice, whereas full length ErbB2 (ErbB2wt) expression did not.
Figure S3: Characterization of a doxycycline-inducible ErbB2ΔEx16 transgenic mouse. (A) Mice from two separate founder lines were interbred with the MMTV/rtTA (MTB) transgenic strain. Female bigenic mice were treated with 2mg/ml doxycycline for 5 days, and mammary glands subjected to wholemount analysis. After 5 days of induction, EGFP fluorescence is clearly visible in the presence, but not the absence, of doxycycline. Evidence of transformation is visible in haematoxylin-stained mammary glands. (B) Immunoblot analysis of flash-frozen tissue confirms ectopic ErbB2ΔEx16 expression in a doxycycline-dependent manner.
Figure S4: Organ survey of ErbB2ΔEx16 mRNA levels in ErbB2ΔEx16 transgenic mice. ErbB2ΔEx16 transcripts in major organs of ErbB2ΔEx16 transgenic animals without (− Dox) or with (+Dox) a 5-day doxycycline induction are measured with RT-qPCR, which demonstrates drastic elevations of ErbB2ΔEx16 mRNA level in mammary and salivary tissues in the presence of doxycycline.
Figure S5: (A) Mouse mammary tumors driven by full length ErbB2 stain positive for the epithelial cytokeratin CK8. By contrast, tumors driven by ErbB2ΔEx16 also express myoepithelial cytokeratins CK5 and CK14, in addition to CK8. (B) ErbB2ΔEx16-driven tumors contain cells positive for CK6 (green) and CK14 (red), as well as double-positive cells (yellow), which are absent in ErbB2 tumors.
Figure S6: Doxycycline withdrawal of ErbB2ΔEx16-driven tumors. (A) Upon withdrawal of doxycycline, tumors rapidly regress and disappear. After a variable latency, approximately 60% of mice display recurrent tumors in the absence of doxycycline. (B) In contrast to the primary lesions, the recurrent tumors lack ErbB2 and E-cadherin expression. (C) Recurrent tumors arising in the absence of doxycycline present with a pathology displaying spindle-like EMT characteristics. Scale bars represent 50μm.
Figure S7: RPPA analysis of transgenic tumors indicates ErbB2ΔEx16 activates a distinct set of pathways from full length ErbB2 tumors. Unsupervised hierarchical clustering of independent tumors separates tumors by genotype, indicating inherent similarities within genotype, and differences between groups.
Figure S8: Validation of microarray data suggests an immune infiltration of ErbB2ΔEx16 tumors. (A) Enrichr gene enrichment analysis revealed a strong gene signature corresponding to immune system infiltration in the tumors. (B) Immunohistochemical staining indicated increased Stat3 phosphorylation in ErbB2ΔEx16 tumors, (C) which was significant by quantification (p-0.002). (D) Immunohistochemical staining also revealed an increase in CD3-positive cells, which was significant by quantification (p=0.003). Scale bars represent 50μm.
Figure S9: ErbB2ΔEx16-driven tumors transcriptionally resemble basal and claudin-low tumors. When subjecting ErbB2 (WildTypeHer2) and ErbB2ΔEx16 (Delta16) tumors to cross-species comparisons, the gene expression profile of ErbB2 tumors most resemble Her2/ErbB2-amplified human breast cancer, whereas ErbB2ΔEx16 tumors align more closely with basal or claudin-low subtypes.
Acknowledgments
This study was supported by grants awarded to W.J.M from the Terry Fox Foundation (#020002), the Canadian Institutes of Health Research (MOP 93525 and MOP 89751), and the National Institutes of Health PO1 (2PO1CA099031-06A1). W.J. M is supported by CRC Chair in Molecular Oncology. J.T was supported by the Department of Defense Breast Cancer Predoctoral Traineeship award #W81XWH 10-1-0114. Z.H was supported by Susan G. Komen Breast Cancer Foundation (CCR14299200) and NIH-NCI (T32-CA009111). E.R.A is supported by NIH grant R01CA160514. R.D.C is supported by a US-NCI grant U01 CA141582. MMTV/ErbB2 transgenic mice were a generous donation from Genentech.
Footnotes
Conflicts of Interest
There are no conflicts of interest to declare.
References
- 1.Alajati A, Sausgruber N, Aceto N, Duss S, Sarret S, Voshol H, et al. Mammary tumor formation and metastasis evoked by a HER2 splice variant. Cancer Res. 2014;73:5320–5327. doi: 10.1158/0008-5472.CAN-12-3186. [DOI] [PubMed] [Google Scholar]
- 2.Andrechek ER, Hardy WR, Girgis-Gabardo AA, Perry RL, Butler R, Graham FL, et al. ErbB2 is required for muscle spindle and myoblast cell survival. Mol Cell Biol. 2002;22:4714–4722. doi: 10.1128/MCB.22.13.4714-4722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrulis IL, Bull SB, Blackstein ME, Sutherland D, Mak C, Sidlofsky S, et al. neu/erbB-2 amplification identifies a poor-prognosis group of women with node-negative breast cancer. Toronto Breast Cancer Study Group. J Clin Oncol. 1998;16:1340–1349. doi: 10.1200/JCO.1998.16.4.1340. [DOI] [PubMed] [Google Scholar]
- 4.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 5.Castagnoli L, Iezzi M, Ghedini GC, Ciravolo V, Marzano G, Lamolinara A, et al. Activated d16HER2 homodimers and Src kinase mediate optimal efficacy for trastuzumab. Cancer Res. 2014 doi: 10.1158/0008-5472.CAN-14-0983. [DOI] [PubMed] [Google Scholar]
- 6.Castiglioni F, Tagliabue E, Campiglio M, Pupa SM, Balsari A, Menard S. Role of exon-16-deleted HER2 in breast carcinomas. Endocrine-related cancer. 2006;13:221–232. doi: 10.1677/erc.1.01047. [DOI] [PubMed] [Google Scholar]
- 7.Chan R, Muller WJ, Siegel PM. Oncogenic activating mutations in the neu/erbB-2 oncogene are involved in the induction of mammary tumors. Annals of the New York Academy of Sciences. 1999;889:45–51. doi: 10.1111/j.1749-6632.1999.tb08722.x. [DOI] [PubMed] [Google Scholar]
- 8.Chang JT, Carvalho C, Mori S, Bild AH, Gatza ML, Wang Q, et al. A genomic strategy to elucidate modules of oncogenic pathway signaling networks. Molecular cell. 2009;34:104–114. doi: 10.1016/j.molcel.2009.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y, Terajima M, Yang Y, Sun L, Ahn YH, Pankova D, et al. Lysyl hydroxylase 2 induces a collagen cross-link switch in tumor stroma. J Clin Invest. 2015;125:1147–1162. doi: 10.1172/JCI74725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clynes RA, Towers TL, Presta LG, Ravetch JV. Inhibitory Fc receptors modulate in vivo cytotoxicity against tumor targets. Nat Med. 2000;6:443–446. doi: 10.1038/74704. [DOI] [PubMed] [Google Scholar]
- 12.Diessner J, Bruttel V, Stein RG, Horn E, Hausler SF, Dietl J, et al. Targeting of preexisting and induced breast cancer stem cells with trastuzumab and trastuzumab emtansine (T-DM1) Cell death & disease. 2014;5:e1149. doi: 10.1038/cddis.2014.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dillon RL, Marcotte R, Hennessy BT, Woodgett JR, Mills GB, Muller WJ. Akt1 and akt2 play distinct roles in the initiation and metastatic phases of mammary tumor progression. Cancer Res. 2009;69:5057–5064. doi: 10.1158/0008-5472.CAN-08-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiMeo TA, Anderson K, Phadke P, Fan C, Perou CM, Naber S, et al. A novel lung metastasis signature links Wnt signaling with cancer cell self-renewal and epithelial-mesenchymal transition in basal-like breast cancer. Cancer Res. 2009;69:5364–5373. doi: 10.1158/0008-5472.CAN-08-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finkle D, Quan ZR, Asghari V, Kloss J, Ghaboosi N, Mai E, et al. HER2-targeted therapy reduces incidence and progression of midlife mammary tumors in female murine mammary tumor virus huHER2-transgenic mice. Clin Cancer Res. 2004;10:2499–2511. doi: 10.1158/1078-0432.ccr-03-0448. [DOI] [PubMed] [Google Scholar]
- 16.Gatza ML, Lucas JE, Barry WT, Kim JW, Wang Q, Crawford MD, et al. A pathway-based classification of human breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:6994–6999. doi: 10.1073/pnas.0912708107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gautrey H, Jackson C, Dittrich AL, Browell D, Lennard T, Tyson-Capper A. SRSF3 and hnRNP H1 regulate a splicing hotspot of HER2 in breast cancer cells. RNA Biol. 2015;12:1139–1151. doi: 10.1080/15476286.2015.1076610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffith M, Griffith OL, Mwenifumbo J, Goya R, Morrissy AS, Morin RD, et al. Alternative expression analysis by RNA sequencing. Nat Methods. 2010;7:843–847. doi: 10.1038/nmeth.1503. [DOI] [PubMed] [Google Scholar]
- 19.Guy CT, Webster MA, Schaller M, Parsons TJ, Cardiff RD, Muller WJ. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:10578–10582. doi: 10.1073/pnas.89.22.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guy CT, Cardiff RD, Muller WJ. Activated neu induces rapid tumor progression. J Biol Chem. 1996;271:7673–7678. doi: 10.1074/jbc.271.13.7673. [DOI] [PubMed] [Google Scholar]
- 21.Janky R, Verfaillie A, Imrichova H, Van de Sande B, Standaert L, Christiaens V, et al. iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS computational biology. 2014;10:e1003731. doi: 10.1371/journal.pcbi.1003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones LM, Broz ML, Ranger JJ, Ozcelik J, Ahn R, Zuo D-M, Ursini-Siegel J, Hallett M, Krummel M, Muller WJ. Stat3 establishes an immunosuppressive microenvironment during the early stages of breast carcinogenesis to promote tumor growth and metastasis. Cancer Research. 2015 doi: 10.1158/0008-5472.CAN-15-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korsching E, Packeisen J, Liedtke C, Hungermann D, Wulfing P, van Diest PJ, et al. The origin of vimentin expression in invasive breast cancer: epithelial-mesenchymal transition, myoepithelial histogenesis or histogenesis from progenitor cells with bilinear differentiation potential? J Pathol. 2005;206:451–457. doi: 10.1002/path.1797. [DOI] [PubMed] [Google Scholar]
- 24.Kwong KY, Hung MC. A novel splice variant of HER2 with increased transformation activity. Mol Carcinog. 1998;23:62–68. doi: 10.1002/(sici)1098-2744(199810)23:2<62::aid-mc2>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 25.Lahlou H, Sanguin-Gendreau V, Zuo D, Cardiff RD, McLean GW, Frame MC, et al. Mammary epithelial-specific disruption of the focal adhesion kinase blocks mammary tumor progression. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:20302–20307. doi: 10.1073/pnas.0710091104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marcotte R, Zhou L, Kim H, Roskelly CD, Muller WJ. c-Src associates with ErbB2 through an interaction between catalytic domains and confers enhanced transforming potential. Mol Cell Biol. 2009;29:5858–5871. doi: 10.1128/MCB.01731-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCormack VA, Burton A, Dos-Santos-Silva I, Hipwell JH, Dickens C, Salem D, et al. International Consortium on Mammographic Density: Methodology and population diversity captured across 22 countries. Cancer epidemiology. 2015;40:141–151. doi: 10.1016/j.canep.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitra D, Brumlik MJ, Okamgba SU, Zhu Y, Duplessis TT, Parvani JG, et al. An oncogenic isoform of HER2 associated with locally disseminated breast cancer and trastuzumab resistance. Molecular cancer therapeutics. 2009;8:2152–2162. doi: 10.1158/1535-7163.MCT-09-0295. [DOI] [PubMed] [Google Scholar]
- 29.Moody SE, Sarkisian CJ, Hahn KT, Gunther EJ, Pickup S, Dugan KD, et al. Conditional activation of Neu in the mammary epithelium of transgenic mice results in reversible pulmonary metastasis. Cancer Cell. 2002;2:451–461. doi: 10.1016/s1535-6108(02)00212-x. [DOI] [PubMed] [Google Scholar]
- 30.Mouw JK, Yui Y, Damiano L, Bainer RO, Lakins JN, Acerbi I, et al. Tissue mechanics modulate microRNA-dependent PTEN expression to regulate malignant progression. Nat Med. 2014;20:360–367. doi: 10.1038/nm.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muller WJ, Sinn E, Pattengale PK, Wallace R, Leder P. Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell. 1988;54:105–115. doi: 10.1016/0092-8674(88)90184-5. [DOI] [PubMed] [Google Scholar]
- 32.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perry MC, Dufour CR, Eichner LJ, Tsang DW, Deblois G, Muller WJ, et al. ERBB2 deficiency alters an E2F-1-dependent adaptive stress response and leads to cardiac dysfunction. Mol Cell Biol. 2014;34:4232–4243. doi: 10.1128/MCB.00895-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ranger JJ, Levy DE, Shahalizadeh S, Hallet M, Muller WJ. Identification of a Stat3-dependent Transcription regulatory Network involved in metastatic progression. Cancer Res. 2009;69 doi: 10.1158/0008-5472.CAN-09-1684. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A, et al. A comparison of background correction methods for two-colour microarrays. Bioinformatics. 2007;23:2700–2707. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- 36.Siegel PM, Dankort DL, Hardy WR, Muller WJ. Novel activating mutations in the neu proto-oncogene involved in induction of mammary tumors. Mol Cell Biol. 1994;14:7068–7077. doi: 10.1128/mcb.14.11.7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siegel PM, Muller WJ. Mutations affecting conserved cysteine residues within the extracellular domain of Neu promote receptor dimerization and activation. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:8878–8883. doi: 10.1073/pnas.93.17.8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siegel PM, Ryan ED, Cardiff RD, Muller WJ. Elevated expression of activated forms of Neu/ErbB-2 and ErbB-3 are involved in the induction of mammary tumors in transgenic mice: implications for human breast cancer [In Process Citation] Embo J. 1999;18:2149–2164. doi: 10.1093/emboj/18.8.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 40.Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A, Press MF. Studies of the HER-2/neu proto-oncogene in human breast and ovaian cancer. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 41.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 42.Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics (Oxford, England) 2005;21:2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- 43.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ursini-Siegel J, Rajput AB, Lu H, Sanguin-Gendreau V, Zuo D, Papavasiliou V, et al. Elevated expression of DecR1 impairs ErbB2/Neu-induced mammary tumor development. Mol Cell Biol. 2007;27:6361–6371. doi: 10.1128/MCB.00686-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu M, Smolen GA, Zhang J, Wittner B, Schott BJ, Brachtel E, et al. A developmentally regulated inducer of EMT, LBX1, contributes to breast cancer progression. Genes Dev. 2009;23:1737–1742. doi: 10.1101/gad.1809309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: ErbB2ΔEx16 mRNA expression in normal human tissue. (A) Quantitative PCR, differentiating wild type ErbB2 mRNA from ErbB2ΔEx16, reveals a relative expression level of 2–10% of total ErbB2 transcript across a panel of normal human tissues.
Figure S2: Ectopic expression of ErbB2 isoforms in NMuMG cells. (A) Immunoblot analysis confirms ectopic ErbB2 overexpression in NMuMG cells. (B) When NMuMG cells were injected orthotopically into athymic nude mice, only cells expressing ErbB2ΔEx16 or the oncogenic mutant NeuNT produced tumours in mice, whereas full length ErbB2 (ErbB2wt) expression did not.
Figure S3: Characterization of a doxycycline-inducible ErbB2ΔEx16 transgenic mouse. (A) Mice from two separate founder lines were interbred with the MMTV/rtTA (MTB) transgenic strain. Female bigenic mice were treated with 2mg/ml doxycycline for 5 days, and mammary glands subjected to wholemount analysis. After 5 days of induction, EGFP fluorescence is clearly visible in the presence, but not the absence, of doxycycline. Evidence of transformation is visible in haematoxylin-stained mammary glands. (B) Immunoblot analysis of flash-frozen tissue confirms ectopic ErbB2ΔEx16 expression in a doxycycline-dependent manner.
Figure S4: Organ survey of ErbB2ΔEx16 mRNA levels in ErbB2ΔEx16 transgenic mice. ErbB2ΔEx16 transcripts in major organs of ErbB2ΔEx16 transgenic animals without (− Dox) or with (+Dox) a 5-day doxycycline induction are measured with RT-qPCR, which demonstrates drastic elevations of ErbB2ΔEx16 mRNA level in mammary and salivary tissues in the presence of doxycycline.
Figure S5: (A) Mouse mammary tumors driven by full length ErbB2 stain positive for the epithelial cytokeratin CK8. By contrast, tumors driven by ErbB2ΔEx16 also express myoepithelial cytokeratins CK5 and CK14, in addition to CK8. (B) ErbB2ΔEx16-driven tumors contain cells positive for CK6 (green) and CK14 (red), as well as double-positive cells (yellow), which are absent in ErbB2 tumors.
Figure S6: Doxycycline withdrawal of ErbB2ΔEx16-driven tumors. (A) Upon withdrawal of doxycycline, tumors rapidly regress and disappear. After a variable latency, approximately 60% of mice display recurrent tumors in the absence of doxycycline. (B) In contrast to the primary lesions, the recurrent tumors lack ErbB2 and E-cadherin expression. (C) Recurrent tumors arising in the absence of doxycycline present with a pathology displaying spindle-like EMT characteristics. Scale bars represent 50μm.
Figure S7: RPPA analysis of transgenic tumors indicates ErbB2ΔEx16 activates a distinct set of pathways from full length ErbB2 tumors. Unsupervised hierarchical clustering of independent tumors separates tumors by genotype, indicating inherent similarities within genotype, and differences between groups.
Figure S8: Validation of microarray data suggests an immune infiltration of ErbB2ΔEx16 tumors. (A) Enrichr gene enrichment analysis revealed a strong gene signature corresponding to immune system infiltration in the tumors. (B) Immunohistochemical staining indicated increased Stat3 phosphorylation in ErbB2ΔEx16 tumors, (C) which was significant by quantification (p-0.002). (D) Immunohistochemical staining also revealed an increase in CD3-positive cells, which was significant by quantification (p=0.003). Scale bars represent 50μm.
Figure S9: ErbB2ΔEx16-driven tumors transcriptionally resemble basal and claudin-low tumors. When subjecting ErbB2 (WildTypeHer2) and ErbB2ΔEx16 (Delta16) tumors to cross-species comparisons, the gene expression profile of ErbB2 tumors most resemble Her2/ErbB2-amplified human breast cancer, whereas ErbB2ΔEx16 tumors align more closely with basal or claudin-low subtypes.