Abstract
The differential susceptibility of skeletal muscle by myasthenia gravis (MG) is not well understood. We utilized RNA expression profiling of extraocular muscle (EOM), diaphragm (DIA), and extensor digitorum (EDL) of rats with experimental autoimmune MG (EAMG) to evaluate the hypothesis that muscles respond differentially to injury produced by EAMG. EAMG was induced in female Lewis rats by immunization with acetylcholine receptor purified from the electric organ of the Torpedo. Six weeks later after rats had developed weakness and serum antibodies directed against the AChR, animals underwent euthanasia and RNA profiling performed on DIA, EDL, and EOM. Profiling results were validated by qPCR. Across the three muscles between the experiment and control groups, 359 probes (1.16%) with greater than 2-fold changes in expression in 7 of 9 series pairwise comparisons from 31,090 probes were identified with approximately two-thirds being increased. The three muscles shared 16 genes with increased expression and 6 reduced expression. Functional annotation demonstrated that these common expression changes fell predominantly into categories of metabolism, stress response, and signaling. Evaluation of specific gene function indicated that EAMG led to a change to oxidative metabolism. Genes related to muscle regeneration and suppression of immune response were activated. Evidence of a differential immune response among muscles was not evident. Each muscle had a distinct RNA profile but with commonality in gene categories expressed that are focused on muscle repair, moderation of inflammation, and oxidative metabolism.
Keywords: myasthenia gravis, acetylcholine receptor, autoimmunity, gene expression profiles, skeletal muscle, metabolism
Introduction
Myasthenia gravis (MG) is caused by antibodies, primarily directed at skeletal muscle nicotinic acetylcholine receptor (AChR), which lead to a reduction of AChR number and damage of the muscle endplate, producing a failure of neuromuscular transmission that results in weakness (Engel et al., 1976). The pathophysiology would be expected to compromise muscles to a similar extent, but clinical investigation for over a 100 years have demonstrated a preferential involvement of certain muscles. Explanations for the differential targeting may lie in subtle aspects of the antibody-antigen engagement in vivo but are more likely to entail variations in the properties of the targeted muscles.
The differential involvement of skeletal muscles by neuromuscular disorders, including MG, is poorly understood but likely is a function of disease specific pathophysiology and properties of the individual muscles. In particular, differences in functional requirements of a muscle impact the gene expression pattern. In its role in eye movement extraocular muscle (EOM) is constantly, and this is reflected in its transcriptional profile differing from jaw and leg muscle in expression of glycogenic and gluconeogenic genes (Porter et al., 2001; Fischer et al., 2005). Further, lactate is a significant substrate for EOM, which is in stark contrast to other skeletal muscles that excess lactate produces fatigue (Andrade and McMullen, 2006). Similarly, as reflected in fiber-type distribution diaphragm also possess properties that support its high energy requirements compared to leg muscles (Polla et al., 2004). The consequences of neuromuscular disorders on whole body metabolism may then also be expected to differentially impact muscles.
Extraocular muscle (EOM) are preferentially involved by MG and several non-exclusive explanations have been proposed. A patient may develop dramatic double vision with even minimal weakness of an EOM, but a similar level of weakness of another muscle would not produce clinically evident symptoms. The extremely rapid firing rate of ocular motor neurons and the immature appearance of their neuromuscular junctions may place the EOM at particular risk for a neuromuscular transmission disorder. The RNA expression profiles of EOM, extensor digitorum longus (EDL), and diaphragm (DIA) muscle from rats with passively-transferred MG (PTMG) (Kusner et al., 2015) produced by administration of acetylcholine receptor antibody supports a greater degree of injury to EOM (Zhou et al., 2014), which supports that EOM has unique immunological characteristics that places them at specific risk for MG (Kaminski et al., 2004; Soltys et al., 2008; Pedrosa Domellof, 2013).
EAMG induced in rodents by immunization with purified AChR mimics the human disease much better than administration of AChR antibodies (Losen et al., 2015). Within 6 weeks of a single immunization, rats generate AChR antibodies and then weakness, which improves with cholinesterase inhibition. As with humans, infiltrates of inflammatory cells are not prominently observed in muscle (Nakano and Engel, 1993; Baggi et al., 2012), which is in contrast to PTMG. In order to assess, variations in intrinsic response of muscles to EAMG, we used RNA expression profiling of diaphragm (DIA), extensor digitorum longus (EDL), and EOM to assess their response.
Materials and methods
Ethics statement for animal use
Six to eight week old female Lewis rats weighing 120–150 g (Harlan, Indianapolis, IN) were maintained in the Case Western Reserve University School of Medicine animal facility. The animal facility follows IACUC, AAALAT, and AALAS standards concerning appropriate housing, cage cleaning procedure, air purity, feed, temperature, humidity, light and dark cycle. Animals were housed in isolator cages in a pathogen-free environment, and rodent chow and water were provided ad libitum. A veterinarian is on staff and will be observing the health of the animals throughout the study. All animal studies were conducted according to protocol approved by the Case Western Reserve Institutional Animal Care and Use Committee Approval Number 030185. All efforts were made to minimize animal suffering. Tissue was harvested after euthanasia by CO2 asphyxiation.
Induction and evaluation of EAMG
Torpedo AChR was purified from the electric organ of Torpedo californica by affinity chromatography as previously described (Lindstrom et al., 1983). Rats were immunized once at the base of the tail by subcutaneous injection of purified Torpedo AChR (40 μg/rat in 200 μl) emulsified in complete Freund's adjuvant supplemented with additional non-viable Mycobacterium tuberculosis H37RA (0.5 mg/rat; Difco Laboratories, Detroit, MI). Control rats were immunized with the same volume of adjuvant without AChR. Rats were monitored for evidence of weakness and their status scored based on a commonly used motor strength scale, as follows: 0 = can grip and lift lid of a cage, 1 = can grip but cannot lift the lid of a cage, 2 = unable to grip cage lid, 3 = unable to grip and has hind limb paralysis, 4 = moribund. Weight was assessed initially on a bi-weekly basis and then every other day when weakness or weight loss became evident.
Tissue preparation
After euthanasia, EOM rectus muscles, DIA, and EDL muscles were dissected from rats 6 weeks after initiation of the experiment. Muscles were pooled from 4 to 5 rats for each of three independent replicate groups. The study was then repeated twice to produce the 3 replicates for the array analysis. This procedure served to limit inter-animal and inter-experiment variability. Tissues were snap frozen in liquid nitrogen and stored at −80°C until use.
Serum AChR antibody determination
Blood was obtained at week 2 by tail vein puncture and after euthanasia from the heart at week 6. Serum was isolated and AChR antibody determination made by ELISA. Ninety-six-well immune-plates (Corning; New York, NY) were saturated with 200 μl (10 μg/ml AChR) in PBS buffer (0.1% Tween20 in PBS) per well and incubated overnight at 4°C. After washing twice with PBST buffer, the plates were incubated with 200 μl of blocking buffer (5% of bovine serum albumin in PBS) per well at 37°C for 0.5–1.5 h. The plates were washed twice with PBST buffer and then incubated for 1 h at 37°C with 100 μl of the diluted test serum (1:200). The plates were then washed twice with PBST, each well received 100 μl of peroxidase-labeled rabbit anti-rat IgG and incubated for 1 h at 37°C. One hundred microliter of substrate solution [0.05 M citrate, 0.1 M NaCl/Pi, 2,20-Azino-bis(3-ethylbenzothiazoline-6-sulfonic acid), 0.03% H2O2] was incubated for 15 min at 37°C. Color development was measured at 405 nm using a microplate reader.
Immunohistochemistry
For analysis of C9 deposition at neuromuscular junctions, cryosections of muscles were prepared. Sections were incubated with rabbit anti-ratC9 (gift of M. E. Medof) and then double-stained with FITC-labeled goat anti-rabbit Ab and Texas red–labeled αBTX (2 μg/ml; Molecular Probes Inc., Eugene, Oregon, USA) to identify neuromuscular junctions. Sections were examined with a Nikon Diaphot fluorescence microscope (Nikon Instruments Inc., Melville, New York, USA) and analyzed using ImagePro software (Media Cybernetics, Silver Spring, Maryland, USA).
Sample preparation for microarrays
The muscle harvested from DIA, EDL, and EOM of four rats pooled from four EAMG or control rats during RNA isolation forming three samples for subsequent array analysis. Total RNA was extracted using TRIzol reagent (GibcoBRL, Rockville, MD). RNA pellets were cleaned by RNasey kits and re-suspended at 1 mg RNA/ml DEPC-treated water and 5 μg was used in a reverse transcription reaction (SuperScript II; Life Technologies, Rockville, MD) to generate first strand cDNA. Double strand cDNA was synthesized and used in an in vitro transcription (IVT) reaction to generate biotinylated cRNA. Fragmented cRNA (15 μg) was used in a 300 μl hybridization cocktail containing herring sperm DNA and BSA as carrier molecules, spiked IVT controls, and buffering agents. A 200 μl aliquot of this cocktail was used for hybridization to Affymetrix rat REA230 (Santa Clara, CA) microarrays for 16 h at 45°C. The manufacturer's standard post-hybridization wash, double-stain, and scanning protocols used an Affymetrix GeneChip Fluidics Station 400 and a Hewlett Packard Gene Array scanner.
Microarray data analysis
Raw data from microarray scans were analyzed with Affymetrix GCOS 2.0. GCOS evaluates sets of perfect match (PM) and mismatch (MM) probe sequences to obtain both hybridization signal values and present/absent calls for each transcript. Microarrays were scaled to the same target intensity and pairwise comparisons were made between experimental and control samples. Transcripts defined as differentially regulated met the criteria of: (a) consistent increase/decrease call across 7 out of 9 replicate comparisons, based upon Wilcoxon's signed rank test (algorithm assesses probe pair saturation, calculates a p-value and determines increase, decrease, or no change calls). Any transcripts with expression intensity below 400 (5 time of background level) across all the samples were also excluded since distortion of fold difference values results when expression levels are low and may be within the level of background noise. Data were visualized as a hierarchical cluster analysis generated (Genespring software, version 7.2; Silicon Genetics, Redwood city, CA). Annotation was done according to Affymetrix NetAffyx Gene Ontology database. Data for the 18 microarray experiments used in this report can be found in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO), series accession number GSE11465.
Quantitative real-time PCR (qPCR)
Select transcripts were reanalyzed by qPCR, using the same samples as in the microarray studies. Transcript-specific primers (Supplemental Table 1) were designed using Primer Express 2.0 software [Applied Biosystems, Inc. (ABI), Foster City, CA] and specificity confirmed by NCBI BLAST. Reverse transcription was carried out on 1 μg total RNA with In vitro Reverse transcription reagent. qRT-PCR used SYBR green PCR core reagent in 24 μl volume, with an ABI PRISM 7000 Sequence Detection System. GAPDH was used as an internal positive loading control. Fold change values represent averages from triplicate measurements, using the 2−ΔΔCT method (Simon, 2003).
Results
Confirmation of EAMG induction
After the immunization, serum AChR antibody levels of the experimental group were 2.5 μg/ml ± 1.1 (2 weeks) and 8.05 μg/ml ± 3.38 (6 weeks). AChR antibody was undetectable in the control group throughout the experiment, while all rats in the experimental group developed elevations of AChR antibody levels. At weeks 4–5, weight loss was observed in EAMG rats and weakness became evident as assessed by reductions of grip strength and reduction of observed movement (data not shown). At week 6 experimental rats had a mean weight loss of 11.5% ± 3.45 compared to their peak weight, while control rats had a mean weight gain of 5.9% ±1.0 compared to the previous week weight. All rats reached the end of the experiment with no need for early euthanasia. To assess for activation of the complement system, we evaluated C9 deposition from control and experimental rats. Endplates from all control muscles had α-BTX staining and no C9 staining. Endplates from all EAMG muscles demonstrated endplates C9 deposition which overlapped with α-BTX fluorescence (not shown).
RNA profile analysis
To identify global alterations in gene expression related to EAMG in EOM, DIA, and EDL, RNA was prepared and processed for microarray hybridization from AChR immunized and control CFA immunized rats. The percentage of transcripts detected as present in each sample ranged from 48 to 63.7 with average 53.3 ± 4.2. The GAPDH probes 3–5 ratio ranged from 1.06 to 2.66 with average 1.27 ± 0.35, indicating RNA quality was appropriate and the hybridization was successful among the samples. The results are consistent with our previous expression profiling studies (Zhou et al., 2014).
We found a total of 359 transcripts (Figure 1) altered by 2-fold in 7 of 9 series pairwise comparisons between the experimental and control groups among the muscles. Figure 1 shows the degree of overlap in gene differences across the three muscles. DIA showed 147 gene changes, 85 genes were up-regulated and 62 down-regulated, EDL had 205 genes changed, 100 were up-regulated and 105 down-regulated, and EOM have 116 changed, 85 are up-regulated and 31 down-regulated. Using the 359 transcripts, hierarchical cluster analysis (Figure 2) demonstrated a distinct expression pattern of the genes found to be differentially influenced by EAMG across the three muscles. Comparison of the differentially expressed transcripts identified 16 upregulated and 6 downregulated transcripts shared among the three muscles (Table 1). Functional annotation of the differentially expressed genes identified more than a half are involved in signal transduction, metabolism and transcription regulation, suggesting the muscles experience adaptation to EAMG directly and to metabolic alterations associated with the disease (Table 2, Supplemental Table 2). Alteration of expression of 10 genes (Enc1, Nfkbia, Nfix, Errfi1, Glipr2, Dyrk2, Hbb, Hba-a1, Pnpla2, Galnt12) were identified common to the three muscles as a response to EAMG. The roles of these genes fell into two broad categories: muscle repair and suppression of inflammatory signals. The Discussion provides detailed consideration of expression alterations.
Figure 1.

Venn diagrams showing the numbers of differentially expressed transcripts in EAMG muscles compared with control rats shared by or unique to DIA, EDL, and EOM.
Figure 2.
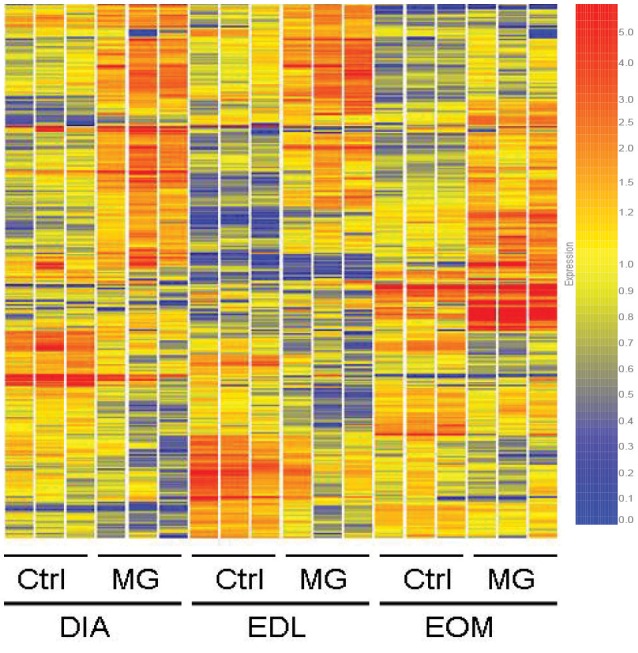
Hierarchical cluster analysis using 359 gene probes identified as differentially expressed in EAMG among DIA, EDL, and EOM. The three independent replicates of each group are represented. Expression ratios are color-coded. The scale at the right denotes normalized expression levels (red, high expression; blue, low expression).
Table 1.
Gene transcript changes common to all muscles.
| Symbol | Name | DIA | EDL | EOM | Classification |
|---|---|---|---|---|---|
| Ankrd1 | Ankyrin repeat domain 1 | 5.6 | 5.5 | 3.6 | Transcription |
| Cebpd | CEBP delta | 2.9 | 8.4 | 2.5 | Transcription |
| Nfix | Nuclear factor I/X | −4.7 | −2.0 | −2.9 | Transcription |
| Nfkbia | Nuclear factor of kappa light chain | 2.3 | 2.8 | 2.3 | Transcription |
| Ddit4 | DNA-damage-inducible transcript 4 | 3.1 | 4.7 | 3.1 | Stress |
| Gadd45a | Growth arrest and DNA-damage-inducible 45 alpha | 2.3 | 2.1 | 2.5 | Stress |
| Mt1a | Metallothionein 1a | 4.7 | 8.2 | 3.3 | Stress |
| Errfi1 | ERBB receptor feedback inhibitor 1 | 2.9 | 3.0 | 2.8 | Stress |
| Mt1e | Metallothionein 1e | 3.0 | 5.5 | 2.8 | Stress |
| Gpnmb | Glycoprotein (transmembrane) nmb | 2.2 | 3.1 | 5.2 | Signal |
| Glipr2 | Golgi- associated PR-1 protein | −2.4 | −2.4 | −2.2 | Signal |
| Dyrk2 | Dual specificity tyrosine-phosphorylation-regulated kinase 2 | −2.4 | −2.3 | −2.4 | Signal |
| Enc1 | Ectodermal-neural cortex 1 | 2.4 | 2.1 | 2.1 | Signal |
| Hbb | Hemoglobin beta chain complex | −4.8 | −2.4 | −3.9 | Metabolism |
| Neu2 | Neuraminidase 2 | −2.9 | −4.0 | −2.2 | Metabolism |
| Hba-a1 | Hemoglobin alpha 2 chain | −3.6 | −2.3 | −3.5 | Metabolism |
| Pnpla2 | Patatin-like phospholipase domain containing 2 | 2.4 | 2.8 | 2.1 | Metabolism |
| pdk4 | Pyruvate dehydrogenase | 2.4 | 3.5 | 2.3 | Metabolism |
| fmo2 | Flavin containing monooxygenase 2 | 4.0 | 3.0 | 3.9 | Metabolism |
| galnt12 | GalNAc transferase 12 | 3.5 | 2.8 | 2.6 | Metabolism |
| Angptl4 | Angiopoietin-like 4 | 6.4 | 6.2 | 17.5 | Metabolism |
| Fkbp5 | FK506 binding protein 5 | 4.5 | 5.3 | 5.6 | Immune |
Table 2.
Transcript changes in percent by category.
| Shared | EOM | DIA | EDL | |
|---|---|---|---|---|
| Transcription | 18 | 5 | 7 | 19 |
| Stress | 22 | 0 | 1 | 5 |
| Metabolism | 32 | 21 | 6 | 17 |
| Signaling | 18 | 33 | 44 | 25 |
| Proteolysis | 0 | 5 | 4 | 5 |
| Muscle-related | 0 | 4 | 1 | 5 |
| Inflammatory | 5 | 8 | 7 | 2 |
| *ECM | 5 | 3 | 0 | 2 |
| EST/Unknown | 0 | 21 | 30 | 20 |
Extracellular Matrix.
In our previous study of passive transfer MG and a study of mdx mice (Porter et al., 2003a; Zhou et al., 2014), we determined a disease load index (DLI), which sums the absolute fold change values of increased and decreased transcript to provide a single transcriptional index of EAMG pathology. In the present investigation, EDL had the greatest DLI (Figure 3), although EOM had the greatest total of increased transcript levels with 351 total fold increase to 101 total fold decrease. The greater DLI of EDL suggests a greater transcriptional response to EAMG than EOM or DIA.
Figure 3.
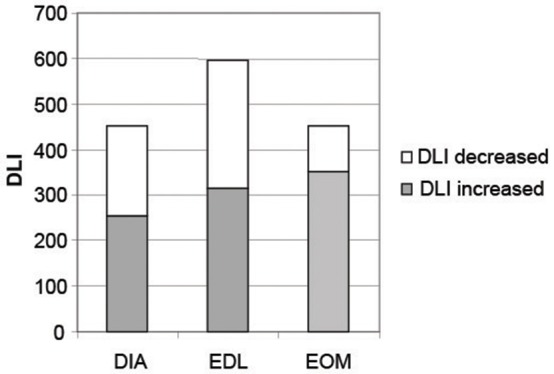
The aggregate DLI of the three muscles for EAMG illustrate disease progression by summing the absolute values of fold changes of differentially regulated transcripts in each muscle.
Validation of RNA profile
We used qPCR to validate results of the RNA profiling. We determined expression levels of 14 transcripts to provide a broad assessment of the array results. Three upregulated transcripts (Ankrd1, Mt1a, Cebpd1) and one down regulated transcript (Neu2) that by RNA profiling had been identified as altered across all three muscles. We assessed transcripts that were previously identified as altered by EAMG (Chrna1, Pde4b, Cts1, Trm63) (Mizrachi et al., 2010; Zhou et al., 2014) or involved in inflammation (NFκB, Ctse). We evaluated transcripts that were increased in the RNA profiling of EOM alone (Plunc, Pax6, Rgs2) (Porter et al., 2003b) or known to be increased in EOM (Csrp3) (Diehl et al., 2006). RNA profiling and qPCR results correlated well with correlation coefficient of 0.9 (Table 3).
Table 3.
Fold change comparison between real-time PCR and microarray.
| Gene symbol | DIA | EDL | EOM | |||
|---|---|---|---|---|---|---|
| qPCR | Array | qPCR | Array | qPCR | Array | |
| ANKRD1 | 11.7 | 6.2 | 8.1 | 5.5 | 5.6 | 3.2 |
| MT1A | 4.2 | 4.7 | 4.8 | 8.2 | 5.3 | 3.4 |
| CEBPD1 | 5.3 | 3.0 | 6.5 | 4.5 | 3.3 | 2.2 |
| PDE4B | 3.2 | 2.0 | 2.8 | 1.7 | 2.0 | 1.3 |
| CTS L | 2.8 | 1.8 | 2.7 | 2.8 | 2.2 | 1.4 |
| CHRNA1 | 2.1 | 1.9 | 2.5 | 1.7 | 3.7 | 2.3 |
| CTS E | 1.9 | 1.2 | 0.9 | 1.0 | 7.5 | 7.6 |
| CSRP3 | 5.9 | 2.4 | 5.4 | 3.3 | 3.0 | 1.4 |
| NEU2 | −3.2 | −2.7 | −3.8 | −4.0 | −1.9 | −2.2 |
| NFκB | 1.1 | 1.0 | 1.8 | 1.3 | 1.3 | 1.0 |
| PAX6 | N/A | 0.5 | N/A | 1.3 | 6.7 | 4.0 |
| PLUNC | N/A | 0.9 | N/A | 1.0 | 4.1 | 3.0 |
| RGS2 | 1.4 | 1.1 | 1.0 | 0.8 | −1.8 | −1.3 |
| TRIM63 | 6.3 | 2.4 | 4.3 | 2.5 | 1.3 | 1.0 |
Discussion
We found distinct genomic signatures for DIA, EDL, and EOM in response to EAMG with a small number of gene alterations shared among the muscles (Figure 1). The results of array analysis were validated by qPCR with a significant correlation. Across the muscles, there was wide variation in the specific gene changes, but a commonality of genes that were altered occurred in cell signaling, transcriptional factors, and metabolism categories. Genes associated with muscle injury repair were upregulated. Only, a few genes primarily associated with the immune system or inflammation were modified by EAMG, however these were focused on genes that would suppress inflammation. The observation is consistent with histology of muscle of human MG, and EAMG in the chronic stage, which does not have evidence of inflammation (Nakano and Engel, 1993; Losen et al., 2015; Tuzun et al., 2015). The expression of immunosuppressive genes provides the first insight into why EAMG, by extension human MG, lacks inflammatory infiltrates in muscle after the acute induction stage.
A critical issue that cannot be addressed by our investigation is the degree to which individual mechanisms alter transcriptional profiles. For example, reduced muscle activity produced by EAMG, or for that matter, passive transfer MG in our previous investigation influences the gene expression pattern. The EOM in particular have unique neuromuscular transmission properties with synapses subject to extremely high stimulation rates by their motor neurons and a reduction in muscle stimulus would likely lead to transcriptional alterations (Spencer and Porter, 2005). Knowing that post-synaptic damage is likely to influence presynaptic properties, we cannot dissociate our results to one specific alteration produced by EAMG (Ouanounou et al., 2016). As we discuss the influence of alterations in whole body metabolism produced by weight loss further will alter gene expression of each muscle in a differential pattern given their unique characteristics.
Gene expression alterations unique to EAMG and common to all muscles
The enhanced expression of five genes (Nfix, Enc, Errfi1, Glipr2, Dyrk2, Galnt12) modified by EAMG appear to enhance muscle repair. Nfix, is a transcription factor known to be involved in myogenesis and muscle regeneration (Déak et al., 2014). Enc1 is an actin-binding protein, which is involved in neuronal process formation (García-Calero and Puelles, 2005; Kim et al., 2009). Its expression has not been observed previously in muscle, and presumably, Enc1 would function to aid recovery of damaged neuromuscular junctions. Errfi1 (also known as Mig6) is an inhibitor of epidermal growth factor signaling and is increased with cell stress and positively regulates cell growth. Elevation of Errfi1 expression would be expected as a response to muscle injury, either from increased oxidative metabolism or neuromuscular junction injury (Pante et al., 2005). Our previous PTMG profiling study identified a homolog of Mig6 to be upregulated across all three muscles. Glipr2 is a signaling molecule, which has not been well-characterized, but is upregulated in sciatic nerve injury from experimental diabetes (Zhang et al., 2015). Glipr2 is involved in transition of epithelial to mesenchymal cells by way of epidermal growth factor signaling pathways (Huang et al., 2013), which are also known to be involved in recovery from muscle injury. Dyrk2 encodes a tyrosine kinase, which negatively regulates growth of cardiac myocytes (Weiss et al., 2013), and the reduced expression of Dyrk2 in EAMG would be expected to promote recovery from injury. Galnt12 encodes an acetylgalactosaminyl transferase and polymorphisms in the gene are associated colonic cancer (Clarke et al., 2012; Wang et al., 2014). The increased expression of Galnt12 likely enhances protein modifications that may be involved in the regenerative process.
Nfkbia (nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha) is a transcriptional factor with increased expression among all three muscles exclusive to EAMG. Nfkbia inhibits cell apoptosis by inhibition of caspase. In rats, Nfkbia is decreased in diet-induced obesity and hyperlipidemia. Nfkbia can inhibit NFκB by direct binding and regulation of transcriptional responses to NFκB, including cell adhesion, immune and proinflammatory responses, apoptosis, differentiation and growth (Hotamisligil, 2006). In addition to its effects on contractility, Fkbp5 modulates NFκB activity and in tandem with Nfkbia would reduce inflammatory signaling in muscle (Erlejman et al., 2014).
In addition, to the primary categories of muscle repair and immunosuppression, there were three gene changes that would influence metabolism. The Dyrk2 gene product phosphorylates glycogen synthase (Skurat and Dietrich, 2004) and therefore, the gene's down-regulation would be a response to a shift to fatty acid oxidation (see discussion below). Pnpla2 encodes a lipase that hydrolyzes fatty acids from triacylglycerol and mutations of the gene produce a myopathy. The upregulation of the gene's expression is also in keeping with the overall shift to fatty acid oxidation (Henriksson, 1995). This change to oxidation is a reflection of decreased availability of glucose and would further be reflected in a reduction of muscle force generation. The reason for the reduced expression of the two hemoglobin gene transcripts (Hbb, Hba-a1) in muscle is not clear.
Gene expression changes specific to EAMG
Consistent with overall profile, alterations of genes involved in metabolism were the most common category for EOM. The gene encoding the alpha subunit of the acetylcholine receptor was increased in response to EAMG in skeletal muscle an EOM previously (Asher et al., 1990; Léger et al., 2006). We evaluated expression of four genes by qPCR expressed at high levels in EOM, three of which appear to be responsive to injury. Pax6 is a transcriptional regulator involved in eye development and during development influences muscle formation (Davis-Silberman et al., 2005). Its increased expression in EOM suggests that it may be responding to injury, perhaps through the elevated activity of muscle satellite cells in EOM (McLoon et al., 2004). Rgs2, a regulator of G-protein signaling, also appears to be involved quiescent stem cell renewal of muscle (Subramaniam et al., 2014). Cspr1, a member of the LIM protein family, has many functions in skeletal and cardiac muscle including myocyte differentiation (Vafiadaki et al., 2015). Plunc is expressed in nasal epithelial tissue and has a bactericidal effects suggesting a role in innate immunity (Liu et al., 2016). Its upregulation in EOM in response to EAMG suggests a response to antibody-mediated injury but otherwise is unclear.
Gene expression alterations common to all muscles and PTMG
PTMG is produced by administration of either mono- or poly-clonal antibody specifically directed toward the autoantigen, in the present study the skeletal muscle AChR (Kusner et al., 2015). The model mimics the final effector pathway of autoantibody destruction observed in humans with MG. The onset of muscle injury is rapid and is accompanied by muscle inflammation not seen in the active model or the human disease. In our previous study of PTMG of EDL, DIA, and EOM (Zhou et al., 2014), RNA profiling demonstrated a greater number of gene alterations, a preponderance of immune-related gene expression alterations, and EOM had the greatest DLI. There were specific gene alterations common among the three muscles and interestingly among these, several were shared with our present RNA profile of EAMG (Zhou et al., 2014). These ten genes were Ankrd1, Gpnmb, Cebpd, Pdk4, Angptl4, Ddit4, Gadd45a, Mt1a, Fmo2, and Fkbp5.
Evidence of immunosuppressive response across muscles
Ankrd1 has been found to be increased in response in several forms of muscle injury, including denervation, motor neuron disease, stretch, and starvation (Miller et al., 2003; Wu et al., 2011; Calvo et al., 2012). Ankrd1 also down regulates NFκB as an anti-inflammatory signal (Liu et al., 2015), which would contribute to the lack of local muscle inflammation in EAMG, while its increase in PTMG may be a response to acute inflammation observed. Osteoactivin (Gpnmb) is a type I transmembrane glycoprotein that is expressed in numerous tissues including skeletal muscle. Osteoactivin has immunosuppressive effects (Schwarzbich et al., 2012) and promotes maintenance of innervation (Furochi et al., 2007), these properties would aid recovery from injury by EAMG. In contrast to the PTMG study in which an inflammatory infiltrate was present, we can be confident that the source of Gpnmb transcripts is from the skeletal muscle. The increase of Cebpd, a transcriptional factor known to reduce pro-inflammatory cytokines (Pedersen and Febbraio, 2008; Scheler et al., 2013), would also serve to limit muscle inflammation in response to EAMG (Moore et al., 2012). Increased expression of cortisone-regulated target genes also would indicate a general anti-inflammatory state, which would also be reflected in metabolic alterations.
Metabolic alterations influenced by EAMG
The other genes in common across MG models and the three muscles under study were primarily involved in metabolism. At the time of euthanasia, rats were demonstrating weight loss, however, in comparison an RNA profile of rat gastrocnemius after fasting, the gene alterations observed with EAMG are distinct (de Lange et al., 2004). The pyruvate dehydrogenase kinase gene is a key regulatory enzyme in skeletal muscle involved in switching the primary energy source from glucose to fatty acids in response to physiological conditions. Transcription of the PDK4 gene is activated by fasting in a tissue-specific manner (Wende et al., 2005; Yumuk, 2006) and after both short-term high-intensity and prolonged low-intensity exercise. Angptl4 (an inhibitor of lipoprotein lipase) expression was increased, and its protein product is directly involved in reduction of glucose utilization, enhanced lipid metabolism, and increased insulin sensitivity during fasting (Dutton and Trayhurn, 2008; Staiger et al., 2009). Metallothioneins are low molecular weight, cysteine–rich zinc binding proteins and serve to protect cells after oxidative stress injury (Reinecke et al., 2006). Mt1a is a metallothionein, which is induced in skeletal muscle of animals in catabolic states and physiological stress situations (Lecker et al., 2004), which also result in elevated levels of glucocorticoids and reactive oxygen species. Another metallothionein, Mt1e, was elevated among all muscles in the present study, but not the PTMG investigation. The increase in flavin-containing mono-oxygenase 2 (Fmo2) also would serve as a response to an increase in reactive oxygen species in the switch to fatty acid oxidation (Krueger and Williams, 2005). These observations suggest that EAMG directly or indirectly may increase intracellular reactive oxygen species with muscle responding with a potential adaptive response to systemic metabolic alterations. The uniform response to stress is further confirmed by increased expression of Ddit4 and Gadd45a. Ddit4 is a DNA-damage-inducible transcript, inhibits mTOR (downstream kinase of IGF pathway) functional control of cell growth in response to energy stress (Sofer et al., 2005; Wang, 2006). Gadd45a is activated by physiological stress and DNA damage serving to modulate cell cycle arrest and apoptosis (Prosperi, 1997). Increased Cebpd expression occurs with denervation and food deprivation (Allen et al., 2010; Zhang et al., 2013). In skeletal muscle Cebpd expression would moderate myostatin expression and potentially produce muscle atrophy (Yang et al., 2005).
Muscle contractility gene influences
The increase of Fkbp5 (FK506 binding protein) may serve to moderate calcium activation through the ryanodine receptor (Krueger and Williams, 2005). In PTMG, muscle contractility is compromised to a greater extent than expected from the neuromuscular transmission defect alone. An increase in Fkbp5 would reduce influx of calcium and negatively impact muscle contractility. Muscle force generation is also reduced in isolated muscle preparations treated with sera from patients with MG (Imai et al., 2011, 2012). In EDL, Zfn28 was found to be increased. Zfn28 is a RING zinc finger protein family member, which localizes to the Z-line and M-line lattices of myofibrils. In vitro binding studies indicate that Zfn28 binds to titin near the region responsible for kinase activity. Since these family members can form heterodimers, this suggests that these proteins may serve as a link between titin kinase and microtubule-dependent signal pathways in muscle (Ng et al., 2003). Alterations in the signaling complex could also moderate contractility through alterations in elasticity.
Neu2 was the only gene reduced in expression among all three muscles and common to the RNA profile of PTMG. Neu2 encodes a glycohydrolytic enzyme that is primarily expressed in mature muscle (Miyagi and Yamaguchi, 2012). The function of Neu2 has not been well characterized but is considered to enhance muscle regeneration and development, and its reduced expression in EAMG may compromise muscle repair.
RNA profile across muscles
Numerous clinical observations demonstrate a differential effect of neuromuscular disorders on specific muscle groups. Our previous RNA expression analysis of PTMG showed that EOM had the greatest DLI, which we had considered consistent with the greater susceptibility of EOM to disease observed in human MG. In the PTMG, transcript alterations were largely related to immune activity. In EAMG, EDL had the greatest transcriptional response to EAMG but this was related to preponderance of metabolic alterations. EOM given their specialized function in moving the globe and small size would not contribute to whole body metabolic control in contrast to EDL and other large muscles, which are critical to regulation of glucose, fatty acid, and amino acid synthesis and utilization.
Regarding specific gene expression alterations of note, the AChR subunit α-subunit was increased consistent with previous EAMG investigations. The expression difference was likely detectable in EOM because of its high innervation ratio and the increased expression reserved to subsynaptic nuclei (Hippenmeyer et al., 2007). Spp1 (osteopontin) was upregulated in EOM. OPN is a glycosylated phosphoprotein originally identified in bone matrix, but now found to be produced in many cell types. OPN is considered a pro-inflammatory cytokine, and increased levels have been associated with inflammatory muscle disease (Urganus et al., 2009; Niewold et al., 2010; Kim et al., 2012) and muscular dystrophy (Kyriakides et al., 2011). Its elevated expression is in distinction from the upregulation of an anti-inflammatory state of RNA profile, which would support the contention that EOM has a unique immune environment, which in certain conditions enhances susceptibility to autoimmune and inflammatory disorders (Soltys et al., 2008).
Metallothionein family members are highly induced by catabolic states where found to be elevated across all muscle groups (Sacheck et al., 2007). Foxo1a, a transcription factor, was increased in EDL and diaphragm by EAMG and is also increased in states of catabolism (Sacheck et al., 2007). Cathepsin L was elevated in EDL with EAMG which is also the case for muscle atrophy.
Clinical relevance
As with any animal study there are limits in application of results to human disease. The rat and human immune systems possess unique characteristics, and the exogenous administration of autoantigen with adjuvant does not mimic the spontaneous development of the breakdown in tolerance of the human disease (Losen et al., 2015). However, the ultimate common pathway of antibody attack with complement activation is shared between EAMG and human MG. The subsequent influence of generalized weakness on the animal and human also are likely to have commonalities.
The EAMG rats developed moderate weight loss, which is likely driving the gene expression changes related to metabolism. The three muscles shared similar responses in shifting to from glycolytic to oxidative metabolism. In keeping with this alteration pathway the same response in terms of stress and free radical metabolism pathways, and a number of nuclear receptors target genes involved in lipid and glucose metabolism were altered by EAMG. Cortisone or dexamethasone target genes were induced and insulin-moderated pathways were inhibited. This was true for EOM, which relies on glucose and lactate for generation of energy (Porter et al., 2001; Andrade and McMullen, 2006). These observations demonstrate that RNA profiles related to metabolism undergo significant alterations by EAMG.
In muscle, there are four pathways for protein degradation: lysosomal proteases including the cathepsins; calcium-dependent proteases; cytosolic ATP-dependent (proteasome); and cytosolic ATP-independent proteolytic pathways. In EDL and DIA FBXO32(atrogin-1) and Trim63(MuRF-1) were upregulated, these two genes are E3 ubiquitin-ligase, which are involved in ubiquitin-proteasome proteolysis pathway and are markers of muscle atrophy (Clavel et al., 2006; Edström et al., 2006). Their common transcription factor, Foxo1a was also increased in expression (Léger et al., 2006; Nakashima et al., 2006). Proteasome pathway related genes were more prominently expressed in EDL compared with DIA and EOM. EDL can be considered a “standard” skeletal muscle, which participates in regulation of whole body metabolism. These data suggest the EAMG leads to accelerated proteolysis, which is also likely to occur among patients with significant weakness. Since the active model of EAMG more closely mimics MG in humans, it is likely that metabolic alterations should be a focus of investigation in clinical studies.
Conclusion
Our study demonstrates the complex alterations occurring on a transcriptional level in response to the direct effects of acetylcholine receptor antibody attack on the neuromuscular junction as well as secondary influences on nerve-muscle communication and development of weight loss. In our investigation we cannot uncouple these effects. We show that alterations in metabolism related genes occurs, an anti-inflammatory response develops, and muscle repair programs develop. Despite an expectation that EOM would have a greater DLI, this was not the case indicating that the reasons for the greater susceptibility of EOM to MG are not reflected in the transcriptional profile.
Author contributions
The experimental work was performed in the laboratory of HK. The study was designed and supervised by HK. LK, GC, and BG performed animal experiments. BG and GC performed bioinformatics analysis. KH and JA assisted in data analysis. All authors participated in writing the initial drafts of the manuscript. All authors read and approved the final manuscript.
Funding
Supported by National Institutes of Health R24EY014837 (HK).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fphys.2016.00524/full#supplementary-material
References
- Allen D. L., Cleary A. S., Hanson A. M., Lindsay S. F., Reed J. M. (2010). CCAAT/enhancer binding protein-delta expression is increased in fast skeletal muscle by food deprivation and regulates myostatin transcription in vitro. Am. J. Physiol. Regul. Integr. Comp. Physiol. 299, R1592–R1601. 10.1152/ajpregu.00247.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade F. H., McMullen C. A. (2006). Lactate is a metabolic substrate that sustains extraocular muscle function. Pflugers Arch. 452, 102–108. 10.1007/s00424-005-0010-0 [DOI] [PubMed] [Google Scholar]
- Asher O., Neumann D., Witzemann V., Fuchs S. (1990). Acetylcholine receptor gene expression in experimental autoimmune myasthenia gravis. FEBS Lett. 261, 231–235. 10.1016/0014-5793(90)80932-9 [DOI] [PubMed] [Google Scholar]
- Baggi F., Antozzi C., Toscani C., Cordiglieri C. (2012). Acetylcholine receptor-induced experimental myasthenia gravis: what have we learned from animal models after three decades? Arch. Immunol. Ther. Exp. (Warsz). 60, 19–30. 10.1007/s00005-011-0158-6 [DOI] [PubMed] [Google Scholar]
- Calvo A. C., Manzano R., Atencia-Cibreiro G., Oliván S., Muñoz M. J., Zaragoza P., et al. (2012). Genetic biomarkers for ALS disease in transgenic SOD1(G93A) mice. PLoS ONE 7:e32632. 10.1371/journal.pone.0032632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke E., Green R. C., Green J. S., Mahoney K., Parfrey P. S., Younghusband H. B., et al. (2012). Inherited deleterious variants in GALNT12 are associated with CRC susceptibility. Hum. Mutat. 33, 1056–1058. 10.1002/humu.22088 [DOI] [PubMed] [Google Scholar]
- Clavel S., Coldefy A. S., Kurkdjian E., Salles J., Margaritis I., Derijard B. (2006). Atrophy-related ubiquitin ligases, atrogin-1 and MuRF1 are up-regulated in aged rat Tibialis Anterior muscle. Mech. Ageing Dev. 127, 794–801. 10.1016/j.mad.2006.07.005 [DOI] [PubMed] [Google Scholar]
- Davis-Silberman N., Kalich T., Oron-Karni V., Marquardt T., Kroeber M., Tamm E. R., et al. (2005). Genetic dissection of Pax6 dosage requirements in the developing mouse eye. Hum. Mol. Genet. 14, 2265–2276. 10.1093/hmg/ddi231 [DOI] [PubMed] [Google Scholar]
- Déak F., Mátés L., Korpos E., Zvara A., Szenasi T., Kiricsi M., et al. (2014). Extracellular deposition of matrilin-2 controls the timing of the myogenic program during muscle regeneration. J. Cell Sci. 127, 3240–3256. 10.1242/jcs.141556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange P., Ragni M., Silvestri E., Moreno M., Schiavo L., Lombardi A., et al. (2004). Combined cDNA array/RT-PCR analysis of gene expression profile in rat gastrocnemius muscle: relation to its adaptive function in energy metabolism during fasting. FASEB J. 18, 350–352. 10.1096/fj.03-0342fje [DOI] [PubMed] [Google Scholar]
- Diehl A. G., Zareparsi S., Qian M., Khanna R., Angeles R., Gage P. J. (2006). Extraocular muscle morphogenesis and gene expression are regulated by Pitx2 gene dose. Invest. Ophthalmol. Vis. Sci. 47, 1785–1793. 10.1167/iovs.05-1424 [DOI] [PubMed] [Google Scholar]
- Dutton S., Trayhurn P. (2008). Regulation of angiopoietin-like protein 4/fasting-induced adipose factor (Angptl4/FIAF) expression in mouse white adipose tissue and 3T3-L1 adipocytes. Br. J. Nutr. 100, 18–26. 10.1017/S0007114507882961 [DOI] [PubMed] [Google Scholar]
- Edström E., Altun M., Hägglund M., Ulfhake B. (2006). Atrogin-1/MAFbx and MuRF1 are downregulated in aging-related loss of skeletal muscle. J. Gerontol. A Biol. Sci. Med. Sci. 61, 663–674. 10.1093/gerona/61.7.663 [DOI] [PubMed] [Google Scholar]
- Engel A. G., Tsujihata M., Lambert E. H., Lindstrom J. M., Lennon V. A. (1976). Experimental autoimmune myasthenia gravis: a sequential and quantitative study of the neuromuscular junction ultrastructure and electrophysiologic correlations. J. Neuropathol. Exp. Neurol. 35, 569–587. 10.1097/00005072-197609000-00008 [DOI] [PubMed] [Google Scholar]
- Erlejman A. G., De Leo S. A., Mazaira G. I., Molinari A. M., Camisay M. F., Fontana V., et al. (2014). NFκB transcriptional activity is modulated by FK506-binding proteins FKBP51 and FKBP52: a role for peptidyl-prolyl isomerase activity. J. Biol. Chem. 289, 26263–26276. 10.1074/jbc.M114.582882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M. D., Budak M. T., Bakay M., Gorospe J. R., Kjellgren D., Pedrosa-Domellof F., et al. (2005). Definition of the unique human extraocular muscle allotype by expression profiling. Physiol. Genomics 22, 283–291. 10.1152/physiolgenomics.00158.2004 [DOI] [PubMed] [Google Scholar]
- Furochi H., Tamura S., Takeshima K., Hirasaka K., Nakao R., Kishi K., et al. (2007). Overexpression of osteoactivin protects skeletal muscle from severe degeneration caused by long-term denervation in mice. J. Med. Invest. 54, 248–254. 10.2152/jmi.54.248 [DOI] [PubMed] [Google Scholar]
- García-Calero E., Puelles L. (2005). Pallial expression of Enc1 RNA in postnatal mouse telencephalon. Brain Res. Bull. 66, 445–448. 10.1016/j.brainresbull.2005.05.003 [DOI] [PubMed] [Google Scholar]
- Henriksson J. (1995). Muscle fuel selection: effect of exercise and training. Proc. Nutr. Soc. 54, 125–138. 10.1079/PNS19950042 [DOI] [PubMed] [Google Scholar]
- Hippenmeyer S., Huber R. M., Ladle D. R., Murphy K., Arber S. (2007). ETS transcription factor Erm controls subsynaptic gene expression in skeletal muscles. Neuron 55, 726–740. 10.1016/j.neuron.2007.07.028 [DOI] [PubMed] [Google Scholar]
- Hotamisligil G. S. (2006). Inflammation and metabolic disorders. Nature 444, 860–867. 10.1038/nature05485 [DOI] [PubMed] [Google Scholar]
- Huang S., Liu F., Niu Q., Li Y., Liu C., Zhang L., et al. (2013). GLIPR-2 overexpression in HK-2 cells promotes cell EMT and migration through ERK1/2 activation. PLoS ONE 8:e58574. 10.1371/journal.pone.0058574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T., Tsuda E., Hozuki T., Yoshikawa H., Yamauchi R., Saitoh M., et al. (2012). Contribution of anti-ryanodine receptor antibody to impairment of excitation-contraction coupling in myasthenia gravis. Clin Neurophysiol. 123, 1242–1247. 10.1016/j.clinph.2011.10.038 [DOI] [PubMed] [Google Scholar]
- Imai T., Tsuda E., Toyoshima T., Yoshikawa H., Motomura M., Shimohama S. (2011). Anti-ryanodine receptor-positive acetylcholine receptor-negative myasthenia gravis: evidence of impaired excitation-contraction coupling. Muscle Nerve 43, 294–295. 10.1002/mus.21887 [DOI] [PubMed] [Google Scholar]
- Kaminski H. J., Li Z., Richmonds C., Lin F., Medof M. E. (2004). Complement regulators in extraocular muscle and experimental autoimmune myasthenia gravis. Exp. Neurol. 189, 333–342. 10.1016/j.expneurol.2004.06.005 [DOI] [PubMed] [Google Scholar]
- Kim J. S., Bashir M. M., Werth V. P. (2012). Gottron's papules exhibit dermal accumulation of CD44 variant 7 (CD44v7) and its binding partner osteopontin: a unique molecular signature. J. Invest. Dermatol. 132, 1825–1832. 10.1038/jid.2012.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. G., Jang S. J., Soh J., Lee K., Park J. K., Chang W. K., et al. (2009). Expression of ectodermal neural cortex 1 and its association with actin during the ovulatory process in the rat. Endocrinology 150, 3800–3806. 10.1210/en.2008-1587 [DOI] [PubMed] [Google Scholar]
- Krueger S. K., Williams D. E. (2005). Mammalian flavin-containing monooxygenases: structure/function, genetic polymorphisms and role in drug metabolism. Pharmacol. Ther. 106, 357–387. 10.1016/j.pharmthera.2005.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusner L. L., Losen M., Vincent A., Lindstrom J., Tzartos S., Lazaridis K., et al. (2015). Guidelines for pre-clinical assessment of the acetylcholine receptor-specific passive transfer myasthenia gravis model-Recommendations for methods and experimental designs. Exp. Neurol. 270, 3–10. 10.1016/j.expneurol.2015.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakides T., Pegoraro E., Hoffman E. P., Piva L., Cagnin S., Lanfranchi G., et al. (2011). SPP1 genotype is a determinant of disease severity in Duchenne muscular dystrophy: predicting the severity of Duchenne muscular dystrophy: implications for treatment. Neurology 77, 1858; author reply 1858–1859. 10.1212/wnl.0b013e318239b9ae [DOI] [PubMed] [Google Scholar]
- Lecker S. H., Jagoe R. T., Gilbert A., Gomes M., Baracos V., Bailey J., et al. (2004). Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 18, 39–51. 10.1096/fj.03-0610com [DOI] [PubMed] [Google Scholar]
- Léger B., Cartoni R., Praz M., Lamon S., Dériaz O., Crettenand A., et al. (2006). Akt signalling through GSK-3beta, mTOR and Foxo1 is involved in human skeletal muscle hypertrophy and atrophy. J. Physiol. 576, 923–933. 10.1113/jphysiol.2006.116715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom J. M., Cooper J. F., Swanson L. W. (1983). Purification of acetylcholine receptors from the muscle of Electrophorus electricus. Biochemistry 22, 3796–3800. 10.1021/bi00285a013 [DOI] [PubMed] [Google Scholar]
- Liu H., Zhang X., Wu J., French S. W., He Z. (2016). New insights on the palate, lung, and nasal epithelium clone (PLUNC) proteins: based on molecular and functional analysis of its homolog of YH1/SPLUNC1. Exp. Mol. Pathol. 100, 363–369. 10.1016/j.yexmp.2015.12.002 [DOI] [PubMed] [Google Scholar]
- Liu X. H., Bauman W. A., Cardozo C. (2015). ANKRD1 modulates inflammatory responses in C2C12 myoblasts through feedback inhibition of NF-kappaB signaling activity. Biochem. Biophys. Res. Commun. 464, 208–213. 10.1016/j.bbrc.2015.06.118 [DOI] [PubMed] [Google Scholar]
- Losen M., Martinez-Martinez P., Molenaar P. C., Lazaridis K., Tzartos S., Brenner T., et al. (2015). Standardization of the experimental autoimmune myasthenia gravis (EAMG) model by immunization of rats with Torpedo californica acetylcholine receptors - Recommendations for methods and experimental designs. Exp. Neurol. 270, 18–28. 10.1016/j.expneurol.2015.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLoon L. K., Rowe J., Wirtschafter J., McCormick K. M. (2004). Continuous myofiber remodeling in uninjured extraocular myofibers: myonuclear turnover and evidence for apoptosis. Muscle Nerve 29, 707–715. 10.1002/mus.20012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. K., Bang M. L., Witt C. C., Labeit D., Trombitas C., Watanabe K., et al. (2003). The muscle ankyrin repeat proteins: CARP, ankrd2/Arpp and DARP as a family of titin filament-based stress response molecules. J. Mol. Biol. 333, 951–964. 10.1016/j.jmb.2003.09.012 [DOI] [PubMed] [Google Scholar]
- Miyagi T., Yamaguchi K. (2012). Mammalian sialidases: physiological and pathological roles in cellular functions. Glycobiology 22, 880–896. 10.1093/glycob/cws057 [DOI] [PubMed] [Google Scholar]
- Mizrachi K., Aricha R., Feferman T., Kela-Madar N., Mandel I., Paperna T., et al. (2010). Involvement of phosphodiesterases in autoimmune diseases. J. Neuroimmunol. 220, 43–51. 10.1016/j.jneuroim.2009.12.012 [DOI] [PubMed] [Google Scholar]
- Moore F., Santin I., Nogueira T. C., Gurzov E. N., Marselli L., Marchetti P., et al. (2012). The transcription factor C/EBP delta has anti-apoptotic and anti-inflammatory roles in pancreatic beta cells. PLoS ONE 7:e31062. 10.1371/journal.pone.0031062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano S., Engel A. G. (1993). Myasthenia gravis: quantitative immunocytochemical analysis of inflammatory cells and detection of complement membrane attack complex at the end-plate in 30 patients. Neurology 43, 1167–1172. 10.1212/WNL.43.6.1167 [DOI] [PubMed] [Google Scholar]
- Nakashima K., Yakabe Y., Yamazaki M., Abe H. (2006). Effects of fasting and refeeding on expression of atrogin-1 and Akt/FOXO signaling pathway in skeletal muscle of chicks. Biosci. Biotechnol. Biochem. 70, 2775–2778. 10.1271/bbb.60274 [DOI] [PubMed] [Google Scholar]
- Ng C. C., Arakawa H., Fukuda S., Kondoh H., Nakamura Y. (2003). p53RFP, a p53-inducible RING-finger protein, regulates the stability of p21WAF1. Oncogene 22, 4449–4458. 10.1038/sj.onc.1206586 [DOI] [PubMed] [Google Scholar]
- Niewold T. B., Kariuki S. N., Morgan G. A., Shrestha S., Pachman L. M. (2010). Gene-gene-sex interaction in cytokine gene polymorphisms revealed by serum interferon alpha phenotype in juvenile dermatomyositis. J. Pediatr. 157, 653–657. 10.1016/j.jpeds.2010.04.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouanounou G., Baux G., Bal T. (2016). A novel synaptic plasticity rule explains homeostasis of neuromuscular transmission. eLife 5:e12190. 10.7554/eLife.12190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pante G., Thompson J., Lamballe F., Iwata T., Ferby I., Barr F. A., et al. (2005). Mitogen-inducible gene 6 is an endogenous inhibitor of HGF/Met-induced cell migration and neurite growth. J. Cell Biol. 171, 337–348. 10.1083/jcb.200502013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen B. K., Febbraio M. A. (2008). Muscle as an endocrine organ: focus on muscle-derived interleukin-6. Physiol. Rev. 88, 1379–1406. 10.1152/physrev.90100.2007 [DOI] [PubMed] [Google Scholar]
- Pedrosa Domellof F. (2013). Extraocular muscles: response to neuromuscular diseases and specific pathologies, in Craniofacial Muscles: A New Framework for Understanding the Effector Side of Craniofacial Muscle Control, eds McLoon L. K., Andrade F. (New York, NY: Springer-Verlad; ), 75–90. [Google Scholar]
- Polla B., D'Antona G., Bottinelli R., Reggiani C. (2004). Respiratory muscle fibres: specialisation and plasticity. Thorax 59, 808–817. 10.1136/thx.2003.009894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter J. D., Khanna S., Kaminski H. J., Rao J. S., Merriam A. P., Richmonds C. R., et al. (2001). Extraocular muscle is defined by a fundamentally distinct gene expression profile. Proc. Natl. Acad. Sci. U.S.A. 98, 12062–12067. 10.1073/pnas.211257298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter J. D., Merriam A. P., Khanna S., Andrade F. H., Richmonds C. R., Leahy P., et al. (2003b). Constitutive properties, not molecular adaptations, mediate extraocular muscle sparing in dystrophic mdx mice. FASEB J. 17, 893–895. 10.1096/fj.02-0810fje [DOI] [PubMed] [Google Scholar]
- Porter J. D., Merriam A. P., Leahy P., Gong B., Khanna S. (2003a). Dissection of temporal gene expression signatures of affected and spared muscle groups in dystrophin-deficient (mdx) mice. Hum. Mol. Genet. 12, 1813–1821. 10.1093/hmg/ddg197 [DOI] [PubMed] [Google Scholar]
- Prosperi E. (1997). Multiple roles of the proliferating cell nuclear antigen: DNA replication, repair and cell cycle control. Prog. Cell Cycle Res. 3, 193–210. 10.1007/978-1-4615-5371-7_15 [DOI] [PubMed] [Google Scholar]
- Reinecke F., Levanets O., Olivier Y., Louw R., Semete B., Grobler A., et al. (2006). Metallothionein isoform 2A expression is inducible and protects against ROS-mediated cell death in rotenone-treated HeLa cells. Biochem. J. 395, 405–415. 10.1042/BJ20051253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacheck J. M., Hyatt J. P., Raffaello A., Jagoe R. T., Roy R. R., Edgerton V. R., et al. (2007). Rapid disuse and denervation atrophy involve transcriptional changes similar to those of muscle wasting during systemic diseases. FASEB J. 21, 140–155. 10.1096/fj.06-6604com [DOI] [PubMed] [Google Scholar]
- Scheler M., Irmler M., Lehr S., Hartwig S., Staiger H., Al-Hasani H., et al. (2013). Cytokine response of primary human myotubes in an in vitro exercise model. Am. J. Physiol. Cell Physiol. 305, C877–C886. 10.1152/ajpcell.00043.2013 [DOI] [PubMed] [Google Scholar]
- Schwarzbich M. A., Gutknecht M., Salih J., Salih H. R., Brossart P., Rittig S. M., et al. (2012). The immune inhibitory receptor osteoactivin is upregulated in monocyte-derived dendritic cells by BCR-ABL tyrosine kinase inhibitors. Cancer Immunol. Immunother. 61, 193–202. 10.1007/s00262-011-1096-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon P. (2003). Q-Gene: processing quantitative real-time RT-PCR data. Bioinformatics 19, 1439–1440. 10.1093/bioinformatics/btg157 [DOI] [PubMed] [Google Scholar]
- Skurat A. V., Dietrich A. D. (2004). Phosphorylation of Ser640 in muscle glycogen synthase by DYRK family protein kinases. J. Biol. Chem. 279, 2490–2498. 10.1074/jbc.M301769200 [DOI] [PubMed] [Google Scholar]
- Sofer A., Lei K., Johannessen C. M., Ellisen L. W. (2005). Regulation of mTOR and cell growth in response to energy stress by REDD1. Mol. Cell. Biol. 25, 5834–5845. 10.1128/MCB.25.14.5834-5845.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltys J., Gong B., Kaminski H. J., Zhou Y., Kusner L. L. (2008). Extraocular muscle susceptibility to myasthenia gravis: unique immunological environment? Ann. N. Y. Acad. Sci. 1132, 220–224. 10.1196/annals.1405.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer R. F., Porter J. D. (2005). Biological organization of the extraocular muscles. Prog. Brain Res. 151, 43–80. 10.1016/S0079-6123(05)51002-1 [DOI] [PubMed] [Google Scholar]
- Staiger H., Haas C., Machann J., Werner R., Weisser M., Schick F., et al. (2009). Muscle-derived angiopoietin-like protein 4 is induced by fatty acids via peroxisome proliferator-activated receptor (PPAR)-delta and is of metabolic relevance in humans. Diabetes 58, 579–589. 10.2337/db07-1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam S., Sreenivas P., Cheedipudi S., Reddy V. R., Shashidhara L. S., Chilukoti R. K., et al. (2014). Distinct transcriptional networks in quiescent myoblasts: a role for Wnt signaling in reversible vs. irreversible arrest. PLoS ONE 8:e65097. 10.1371/journal.pone.0065097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzun E., Berrih-Aknin S., Brenner T., Kusner L. L., Le Panse R., Yang H., et al. (2015). Guidelines for standard preclinical experiments in the mouse model of myasthenia gravis induced by acetylcholine receptor immunization. Exp. Neurol. 270, 11–17. 10.1016/j.expneurol.2015.02.009 [DOI] [PubMed] [Google Scholar]
- Urganus A. L., Zhao Y. D., Pachman L. M. (2009). Juvenile dermatomyositis calcifications selectively displayed markers of bone formation. Arthritis Rheum. 61, 501–508. 10.1002/art.24391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafiadaki E., Arvanitis D. A., Sanoudou D. (2015). Muscle LIM Protein: master regulator of cardiac and skeletal muscle functions. Gene 566, 1–7. 10.1016/j.gene.2015.04.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Ju L., Fan J., Zhu Y., Liu X., Zhu K., et al. (2014). Histone H3K4 methyltransferase Mll1 regulates protein glycosylation and tunicamycin-induced apoptosis through transcriptional regulation. Biochim. Biophys. Acta 1843, 2592–2602. 10.1016/j.bbamcr.2014.06.013 [DOI] [PubMed] [Google Scholar]
- Wang Y. (2006). Complementary therapies for inflammation. Nat. Biotechnol. 24, 1224–1226. 10.1038/nbt1006-1224 [DOI] [PubMed] [Google Scholar]
- Weiss C. S., Ochs M. M., Hagenmueller M., Streit M. R., Malekar P., Riffel J. H., et al. (2013). DYRK2 negatively regulates cardiomyocyte growth by mediating repressor function of GSK-3beta on eIF2Bepsilon. PLoS ONE 8:e70848. 10.1371/journal.pone.0070848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wende A. R., Huss J. M., Schaeffer P. J., Giguère V., Kelly D. P. (2005). PGC-1alpha coactivates PDK4 gene expression via the orphan nuclear receptor ERRalpha: a mechanism for transcriptional control of muscle glucose metabolism. Mol. Cell. Biol. 25, 10684–10694. 10.1128/MCB.25.24.10684-10694.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. L., Kandarian S. C., Jackman R. W. (2011). Identification of genes that elicit disuse muscle atrophy via the transcription factors p50 and Bcl-3. PLoS ONE 6:e16171. 10.1371/journal.pone.0016171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Mammen J., Wei W., Menconi M., Evenson A., Fareed M., et al. (2005). Expression and activity of C/EBPbeta and delta are upregulated by dexamethasone in skeletal muscle. J. Cell. Physiol. 204, 219–226. 10.1002/jcp.20278 [DOI] [PubMed] [Google Scholar]
- Yumuk V. D. (2006). Targeting components of the stress system as potential therapies for the metabolic syndrome: the peroxisome-proliferator-activated receptors. Ann. N. Y. Acad. Sci. 1083, 306–318. 10.1196/annals.1367.019 [DOI] [PubMed] [Google Scholar]
- Zhang L., Pan J., Dong Y., Tweardy D. J., Dong Y., Garibotto G., et al. (2013). Stat3 activation links a C/EBPδ to myostatin pathway to stimulate loss of muscle mass. Cell Metab. 18, 368–379. 10.1016/j.cmet.2013.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Qu S., Liang A., Jiang H., Wang H. (2015). Gene expression microarray analysis of the sciatic nerve of mice with diabetic neuropathy. Int. J. Mol. Med. 35, 333–339. 10.3892/ijmm.2014.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Kaminski H. J., Gong B., Cheng G., Feuerman J. M., Kusner L. (2014). RNA expression analysis of passive transfer myasthenia supports extraocular muscle as a unique immunological environment. Invest. Ophthalmol. Vis. Sci. 55, 4348–4359. 10.1167/iovs.14-14422 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


