Abstract
Protein-side-chain protonation, coupled to conformational rearrangements, is one way of regulating physiological function caused by changes in protein environment. Specifically, protonation of histidine residues has been implicated in pH-dependent conformational switching in several systems, including the diphtheria toxin translocation (T) domain, which is responsible for the toxin’s cellular entry via the endosomal pathway. Our previous studies a) identified protonation of H257 as a major component of the T domain’s conformational switch and b) suggested the possibility of a neighboring H223 acting as a modulator, affecting the protonation of H257 and preventing premature conformational changes outside the endosome. To verify this “safety-latch” hypothesis, we report here the pH-dependent folding and membrane interactions of the T domain of the wild-type and that of the H223Q mutant, which lacks the latch. Thermal unfolding of the T domain, measured by circular dichroism, revealed that the reduction in the transition temperature for helical unfolding for an H223Q mutant starts at less acidic conditions (pH <7.5) relative to the wild-type protein (pH <6.5). Hydrogen-deuterium-exchange mass spectrometry demonstrates that the H223Q replacement results in a loss of stability of the amphipathic helices TH1–3 and the hydrophobic core helix TH8 at pH 6.5. That this destabilization occurs in solution correlates well with the pH-range shift for the onset of the membrane permeabilization and translocation activity of the T domain, confirming our initial hypothesis that H223 protonation guards against early refolding. Taken together, these results demonstrate that histidine protonation can fine-tune pH-dependent switching in physiologically relevant systems.
Introduction
Conformational signaling, broadly defined as structural switching in response to external stimuli, is involved in all aspects of cellular functioning. Changes in proton concentration are among the most prominent physicochemical signals capable of triggering functionally relevant structural rearrangements. pH-dependent transitions play important roles in the cellular entry of bacterial toxins (1, 2, 3, 4, 5), colicins (6), and viruses (7, 8), in the function of pH-sensors (9, 10), in the signal cascades of G-protein (11), and possibly in apoptotic regulation (12, 13, 14, 15, 16). Protonation-dependent transitions are also involved in cancer pathogenesis, because “the dysregulation of pH by cancerous cells is able to create a unique milieu that is in favor of progression, invasion, and metastasis as well as chemo-/immunoresistance of solid tumors” (17). On the other hand, novel approaches for the delivery of anticancer therapy and tumor imaging are using pH-dependent transitions for targeting diseased acidic tissues (18). Although the exact mechanism for coupling protonation and conformational changes remains largely unknown, protonation of histidine residues does cause conformational switching in several systems (7, 8, 19, 20), consistent with order-disorder transitions in intrinsically disordered proteins and their fragments (21, 22, 23). Specifically, histidines do play an important role in the functioning of the diphtheria toxin translocation domain (T domain) (24, 25, 26, 27, 28), making it an attractive model for studies of mechanisms of conformational switching by protonation (29).
Diphtheria toxin enters the cell via the endosomal pathway, which is shared by many other toxins, including botulinum, tetanus, and anthrax (1, 2, 3, 4, 5). A crucial step in cell infection is the translocation of the catalytic domain from the endosome into the cytosol, accomplished by the translocation domain. Diphtheria toxin T domain is a small, 178-residue protein fragment that forms a separate, tightly folded domain at neutral pH but changes its conformation under the acidic conditions inside the endosome. It then inserts into the lipid bilayer and translocates its own N-terminus and the attached catalytic domain across the membrane (30, 31). Although the crystallographic structure of the soluble toxin is known (32), as are the interactions of the T domain with lipid bilayers (24, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43), we report here a significantly improved understanding of the molecular mechanism of its action.
Previously, we demonstrated that the protonation of H257 is a principal element of the pH-dependent trigger, responsible for destabilizing the solution fold of the T domain in preparation for its membrane insertion (25). Our recent computational study confirmed this conclusion by estimating the free-energy penalty for protonating all six histidine residues in the T domain by thermodynamic integration along the molecular dynamics (MD) simulation trajectory (28). Protonation of H257 has the highest free-energy penalty and, hence, the most destabilizing effect on the native structure. In contrast, protonation of the neighboring H223 is favorable, suggesting that this residue is easily protonated without causing any structural rearrangements. The protonation of H223, however, has an effect on the free-energy penalty for protonation of H257, increasing it by ∼3 kcal/mole. Similar conclusions follow from an independent calculation of pKa distributions based on the continuum electrostatic model, resulting in the shift of the distribution of pKas in an MD-generated ensemble toward acidic pH by ∼1.5 units (equivalent to 2 kcal/mole in terms of free energy) (28).
Safety-latch hypothesis of conformational switching in diphtheria toxin T domain
Acidification of the endosomal environment triggers a series of conformational changes in the T domain, ultimately resulting in its refolding and bilayer insertion. The first stage occurs in solution and results in a pH-dependent conversion of the membrane-incompetent W-state into a protonated membrane-competent W+-state. The latter state is prone to aggregation, which significantly complicates its structural characterization and hampers applications of high-resolution methods. Recently, we elucidated conformational changes occurring during the W-to-W+ transition using MD simulations and fluorescence and circular dichroism spectroscopy (CD), and used various computational approaches to estimate the pKas for the T domain’s six histidine residues (28).
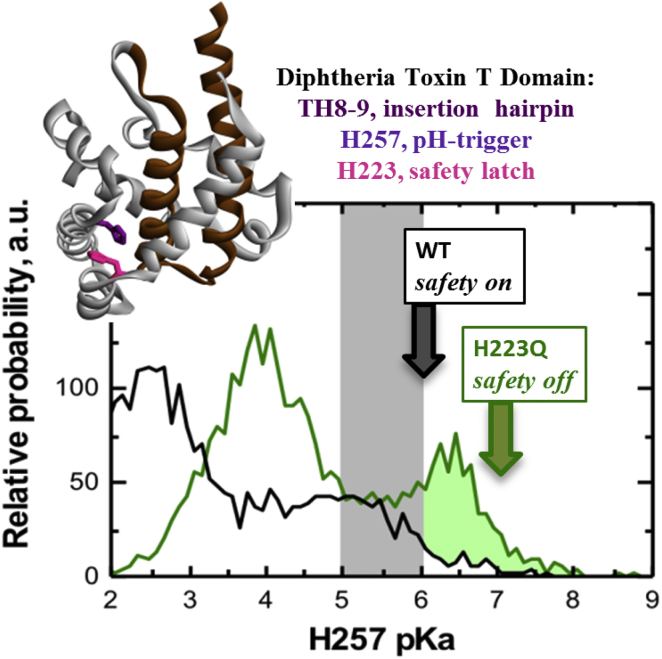
The resulting distributions of pKa probabilities for both W- and W+-states provide the thermodynamic rationale for pH-triggered conformational switching via coupling of protonation of key histidines and conformational rearrangements (see (29) and the Supporting Material for further details). These calculations also form the basis of the “safety latch” hypothesis illustrated in Fig. 1. The black and the green traces represent the pKa distribution of the main pH trigger, H257, calculated in the presence and absence, respectively, of the charge on H223 (reproduced from (28)). The protonation of H257 is coupled to a conformational transition leading to the formation of the membrane-competent state in which all histidines have pKa >7 (Fig. S1). Because protonation and conformational changes are coupled, the experimentally observed transition occurs at a pH corresponding to the rising edge of the distribution (Fig. 1, arrows) when approaching from neutral pH (as discussed in (28)). As a result, only the difference in pKa profiles above the pH of the W-to-W+ transition (Fig. 1, shaded area) affects the differences between pH-dependent folding/insertion of the wild-type (WT) and mutant of the T domain. Acidification of the endosomal environment (which is the physiological attempt by the cell to destroy the invading toxin) leads to refolding and membrane insertion of the T domain, ultimately resulting in translocation of the N-terminus with the attached catalytic domain into the cytosol. In the WT protein, the onset of the conformational change is synchronized with the pH experienced by the toxin after being internalized inside the endosome. We hypothesize that H223 plays a central role in this synchronization, acting as a “safety latch” to prohibit triggering of the refolding transition outside of the cell, where it would be nonproductive. In the absence of this safety latch, the W-to-W+ transition is expected to be dominated by the excess of pKa distribution at neutral pH, highlighted in Fig. 1 by the shaded area under the H257 pKa distribution in H223Q versus that of the WT. Here, we describe a test of this hypothesis by determining the pH-dependent stability of T domain WT and H223Q mutant in solution and their interactions with the lipid bilayers.
Figure 1.
Illustration of the “safety latch” hypothesis tested in this study. The crystallographic structure (32) of the T domain of diphtheria toxin (inset) is represented as the backbone ribbon with the consensus membrane-insertion domain, TH8–9, highlighted in brown. Histidines 223 (magenta) and 257 (purple) are shown as sticks. Solid lines represent MD-based calculations of the probability distribution that H257 has a particular pKa (reproduced from (28)) (Two cases corresponding to the neighboring H223 being neutral (green trace) or charged (black trace) are considered. Because the histidine protonation is coupled with a pH-dependent conformational transition to form a membrane-competent state (discussed in detail in (29) and illustrated in Fig. S1), the conformational transition occurs at a pH corresponding to the rising edge of the pKa distribution of the main trigger, H257 (25, 28) (vertical arrows). The protonation of H223 serves as a safety latch for the H257 trigger by moving the onset of pH-coupled refolding and insertion toward the endosomal pH range (gray band). This safety-latch hypothesis is tested here by comparing pH-dependent folding and membrane insertion of the WT T domain with that of an H223Q mutant that lacks the latch (see text for details). To see this figure in color, go online.
Materials and Methods
Materials
Diphtheria toxin T domain (segment 202–378 of the full-length toxin) was prepared as described in (44). Palmitoyloleoylphosphatidylcholine (POPC), palmitoyloleoylphosphatidylglycerol (POPG) were obtained from Avanti Polar Lipids (Alabaster, AL).
CD measurement and analysis of thermal unfolding
CD measurement and analysis of thermal unfolding were performed as described previously (26). CD spectra were measured using an upgraded Jasco-720 spectropolarimeter (Japan Spectroscopic, Tokyo, Japan) in the range 190–260 nm with samples containing 4 μM of protein in 50 mM phosphate buffer in a 1-mm optical path cuvette. 150 scans were averaged and the background was subtracted. Temperature dependency of unfolding was monitored by measuring ellipticity at 222 nm in the course of a 1°/min rate scan. The thermal unfolding was analyzed using thermodynamic equations for a two-state, N-to-U unfolding transition, where N and U are the native and unfolded states of the protein, respectively. To obtain the transition temperature (Tm) and the enthalpy changes (ΔH°), raw data were fitted applying nonlinear least-squares analysis with six fitting parameters, YN, mN, YU, mU, ΔH°, and Tm, with the following equations, which imply small heat capacity change upon unfolding (45):
| (1) |
where Y is the experimentally observed CD signal at a given temperature, YN and YU, represent the signals of the pure N and U states at 0 K, mN and mU are the temperature dependencies of these CD signals for the N and U states, respectively, and XN is the fraction of the native state at temperature T.
For measurements at different pH, appropriate volumes of 50 mM sodium phosphate buffer of pH 8 and 50 mM sodium acetate buffer pH 4 were mixed. The pH-dependence of the melting temperature Tm was fitted to the following:
| (2) |
where TN and TL are limiting values of the Tm at neutral and low pH, respectively; pKa is an apparent constant, and n is a cooperativity coefficient.
Hydrogen/deuterium exchange mass spectrometry
Hydrogen/deuterium exchange mass spectrometry (HDX) was performed as described in an earlier study (46). To perform pH-dependent HDX, the pH of various T domain solutions was adjusted to 7.5, 6.5, and 6.0 by mixing appropriately 50 mM sodium phosphate buffer (pH 7.5) with 50 mM sodium acetate buffer (pH 4). All pH values were corrected (pD = pHread + 0.4) (47) from the actual pH readings of a SevenEasy pH meter (Mettler Toledo, Columbus, OH). The HDX was initiated by mixing 1 μL of the T-domain solution (75 μM) with 19 μL of D2O exchange buffer (final D2O content was 95% and final protein concentration was 3.75 μM in labeling solution), or H2O buffer for t0 samples, and incubated for a predetermined set of times under each pH condition at 4°C. After a specific incubation time, 30 μL of ice-cold 3 M urea/1% (v/v) trifluoroacetic acid (TFA) was rapidly added to 20 μL of the incubated protein solution to quench the HDX reaction and denature the protein structure. The quenched protein mixture was immediately passed through a custom-built, freshly prepared immobilized pepsin column (2 mm × 2 cm) at a flow rate of 200 μL/min, and the resulting peptic peptides were captured on a C8 cartridge (2.1 mm × 1.5 cm, Agilent, Santa Clara, CA) for desalting; the total time for digestion and desalting was 3 min. Peptides were then separated by using a C18 high-performance liquid chromatography column (1.9 μm Hypersil Gold, Thermo Fisher, Waltham, MA) over a 5 min linear gradient (4−40% CH3CN, 0.1% (v/v) formic acid). Valves, columns, and tubing for protein digestion and peptide separation were submerged in an ice−water bath to minimize back exchange. Mass analysis was performed with an Orbitrap mass spectrometer (LTQ Orbitrap XL, Thermo Fisher, Waltham, MA) equipped with an electrospray ionization source operated at a capillary temperature of 225°C and a spray voltage of 3.5 kV. Each measurement was made in triplicate. LCMS/MS analysis to identify the peptides from digestion was performed in a data-dependent mode of an Orbitrap mass spectrometer, and the top six most abundant ions were selected for collision-induced fragmentation. MS/MS product-ion spectra were submitted to Mascot (Matrix Science, London, United Kingdom) for peptide identification. Digestion by freshly made pepsin beads yielded a set of peptides that covered 100% of the sequence of the H223Q T domain. In our earlier study, fungal XIII digestion was needed in addition to pepsin digestion to obtain nearly complete sequence coverage (46). Fungal XIII, however, was not used here because the pepsin used was more active and provided sufficient peptides with better sequence coverage.
N-terminus translocation assay
The N-terminus translocation assay was performed using thrombin-loaded large unilamellar vesicles (LUVs) made of a 3:1 molar ratio of POPC and POPG, as described previously (48). The recombinantly expressed T domain in pET15b plasmid contains an N-terminal His-tag with thrombin cleavage-site treatment. Upon insertion and translocation of the T-domain N-terminus into LUVs preloaded with thrombin, the His-tag is cleaved off by the protease, resulting in a protein of lower molecular weight; the difference between cleaved and uncleaved T domain can be detected by electrophoretic mobility. LUVs containing ∼0.02 units of bovine thrombin (Fisher Bioreagents, Fair Lawn, NJ) were prepared by the conventional extrusion method (49, 50). Nonencapsulated thrombin was removed by FPLC gel filtration on a Superose 12 1 × 30-cm column. The final concentrations were 100 nM for T domain and 2 mM for vesicles in a total volume of 40 μL. To prevent cleavage due to vesicle rupture (rather than translocation of the N-terminus), 0.02 units of the thrombin inhibitor hirudin (Sigma, St. Louis, MO) were added to the reaction mixture. After 1 h incubation at room temperature, 10 μL of 5× SDS sample buffer was added, samples were boiled for 5 min and analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (PAGE) in 4%–15% Tris-HCl gels. Other details, including control experiments demonstrating that cleavage occurs inside the vesicles and that thrombin is active at pH ≥5.8, are described in (48).
Results and Discussion
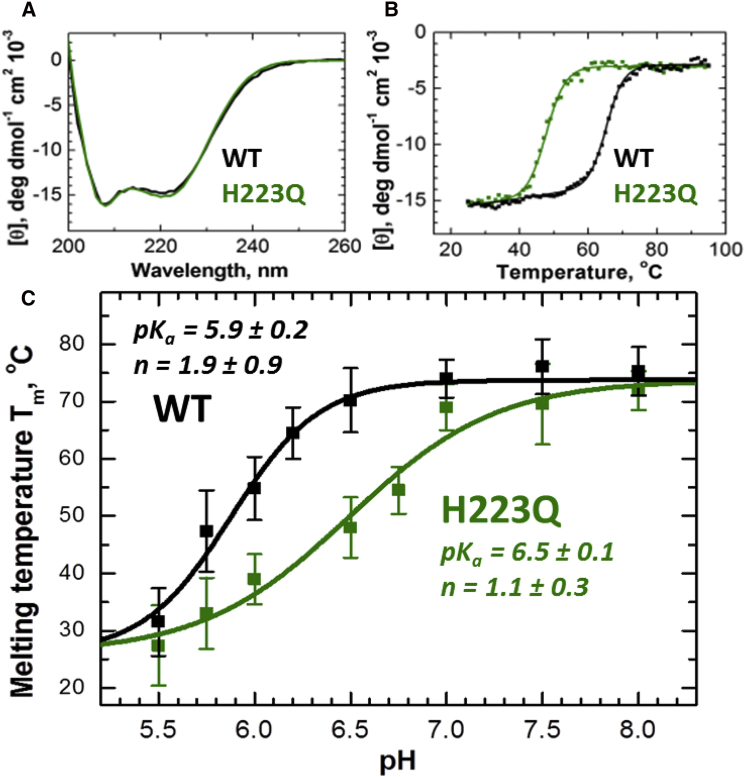
We examined the secondary structure and thermal stability of the WT and H223Q mutant T domains using CD spectroscopy at neutral and mildly acidic pH values. Histidine 223 replacement does not cause any changes of the helical content, as the CD spectra for both proteins are superimposable (Fig. 2 A). The thermal stability, however, becomes markedly different (Fig. 2 B). Whereas the enthalpy values of the transition, ΔH°, measured for mutant and WT protein coincided with each other within the precision of the experiment, the transition temperature, Tm, showed a distinctly different pattern. Consistent with our previous reports for the WT (27), the transition temperature (Fig. 2 C) decreases upon acidification below pH 6.5. In contrast, the changes in Tm for the H223Q mutant have already started at pH <7.5. The data fit to a linked model, in which both proteins share the same melting temperatures at high and low pH, yet have different apparent pKa values and cooperativity coefficients, n (Fig. 2 C, solid lines). This analysis confirms the expectations from our hypothesis that the substitution of H223 would result in the loss of conformational stability at mildly acidic pH.
Figure 2.
CD spectroscopic comparison of the secondary structure and thermal stability of the WT (black) and H223Q (green) T domains. (A) CD spectra for the two proteins coincide at all pH values, suggesting identical secondary structure (pH 6.5 shown). (B) Thermal denaturation was followed by the changes in ellipticity measured at 222 nm at pH 6.5. Solid lines represent a fit to Eq. 1 with the following parameters: Tm = 65.2 ± 0.1°C, ΔH° = −77 ± 3 kcal/mole for WT and Tm = 47.9 ± 0.1°C, ΔH° = −73 ± 3 kcal/mole for WT. (C) pH dependence of the melting temperature, Tm, calculated from CD data such as those in (B) (average of two to three measurements shown). The data were fitted to Eq. 2 to estimate the apparent pKa and n values, presented on the graph. The WT and mutant proteins were expressed in Escherichia coli and purified as described in (44). CD measurements were in cuvettes of 1 mm path length using a Jasco-720 spectropolarimeter (Japan Spectroscopic), as described previously (55). To see this figure in color, go online.
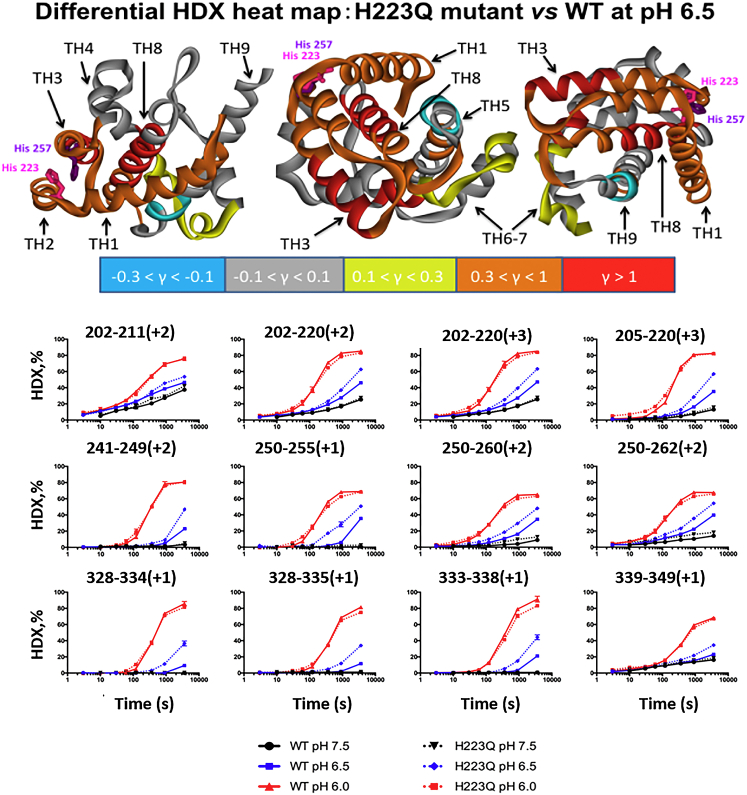
To map those elements of the H223Q secondary structure that undergo reduction in stability, we measured the kinetics of HDX and compared the results to those previously published for the WT T domain (46). The rates of protein backbone amide HDX reactions depend on hydrogen bonding, solvent accessibility, and structural flexibility of the protein. Stable secondary structures are expected to undergo slower HDX for the corresponding segments of the protein compared to unstructured parts of the protein (e.g., loops). Overall, the data for H223Q are consistent with acid-induced loosening of the structure (increase in structural flexibility), with the lowest and highest normalized exchange rates occurring at pH 7.5 and 6, respectively (Fig S2). (Note that all exchange rates are corrected for the inherent pH dependence of exchange, as described previously (46). We selected the relative extents of HDX for 55 representative peptic peptides (Fig S2) to obtain sufficient short and overlapping peptides to represent most of the protein sequence. Kinetics plots of regions with significant conformation change (shown in Fig. 3) demonstrate the following trends. a) There are virtually no differences between the WT and H223Q at pH 7.5, where little exchange is observed, and at pH 6, where both proteins show similarly fast exchange. b) Faster exchange, and hence lower helix stability, occurs for H223Q at pH 6.5. This difference in exchange is visualized with the help of a heat map demonstrating the relative difference in deuterium labeling in the two proteins at pH 6.5 (Fig. 3, top).
Figure 3.
HDX measurements of the conformational flexibility of the T domain WT and H223Q mutant. (Bottom) Kinetic HDX curves of proteolytic peptides measured at pH 7.5, 6.5, and 6.0. All time points were corrected to standard condition at pH 7.5 and 4°C, and measurements were conducted as described in (46). The title of each data subplot refers to the amino acid residue number and charge state of the peptide in the mass spectrometer. Both proteins have equally low exchange at pH 7.5 and equally high exchange at pH 6.0. At the intermediate pH value of 6.5, the exchange in H223Q is noticeably greater than that in the WT. (Top) Differential heat maps of the HDX of H223Q versus WT at pH 6.5. The ribbon diagrams are colored according to the relative differential uptake factor, γ = (HDXH223Q − HDXWT)/HDXWT, measured for the last kinetic point of 3000 s (see key below heat maps). The three versions of the map represent three different orientations of the structure. The highest differential HDX is observed for amphipathic helixes TH1–3 and for the central hydrophobic helix, TH8. To see this figure in color, go online.
The value for the exchange of the WT, measured at the last time point (equivalent of 3000 s in the HDX-time plots in Fig. 3), is subtracted from that for the H223Q, and the difference is normalized to the WT value. The backbone ribbon representation of the T-domain crystallographic structure is color-coded in accordance with the differential uptake factor, γ = (HDXH223Q − HDXWT)/HDXWT. Segments with no difference in exchange (γ ∼ 0) are colored in gray, whereas those for which the exchange level is higher in the H223Q are colored in yellow (0.1 < γ < 0.3), orange (0.3 < γ < 1), and red (γ > 1). The only segment for which the levels of HDX are slightly lower in the mutant is in the middle of TH9 (light blue), probably owing to partial dimer formation occurring upon destabilization of the folded T domain. Our previous HDX study demonstrated that TH9 is part of the dimerization interface for the WT at pH <6.0 (46). We found fairly dramatic differences in HDX for TH1, TH2, and TH3, suggesting that the perturbation propagates far from the mutation site. Interestingly, our previous microsecond-range all-atom MD simulations suggest that helices TH1 and TH2 are among those undergoing the most dramatic refolding during the formation of the membrane-competent state as triggered by histidine protonation (28). We conclude that H-to-Q substitution at position 223 assists in the structural preparation necessary for membrane insertion. This is clearly also demonstrated by the dramatic increase in the accessibility of the most hydrophobic internal helix, TH8 (Fig. 3, red), for which the HDX extent in the mutant is over twice as high as that for the WT (γ > 1).
Membrane insertion and translocation of the T domain are complex, with various stages of protonation-coupled refolding occurring in solution and on a lipid bilayer (29, 44). Our previous mutagenesis studies reveal that some stages can be uncoupled by mutations of C-terminal histidines (26, 27) or of acidic residues in the TH8–9 insertion domain (51). Replacing the main trigger, H257 (25), we propagate pH dependence of the early stages to the latter stages of the pathway. To test whether the H223Q replacement leads to a shifted pH range of membrane insertion and refolding of the T domain, we compared the activity of the WT and H223Q by using two independent approaches: a) vesicle permeabilization (Fig. 4) and b) N-terminus translocation (Fig. 5).
Figure 4.
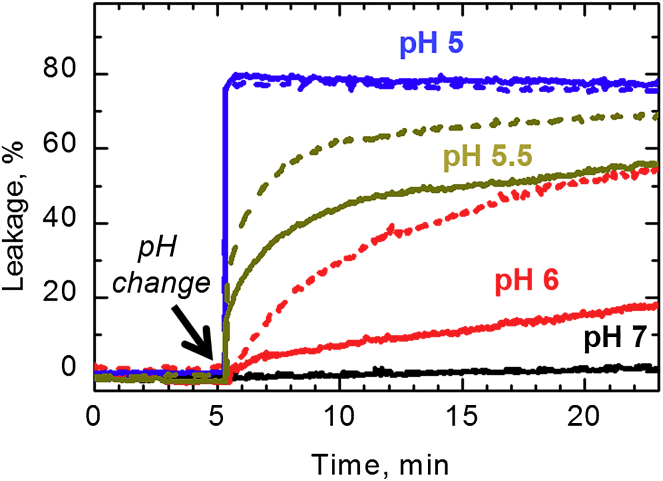
Leakage of fluorescence markers ANTS/DPX from lipid vesicles formed of a 3:1 molar ratio of POPG to POPC caused by pH-dependent vesicle permeabilization by H223Q (solid traces) and WT (dashed traces) T domains. To see this figure in color, go online.
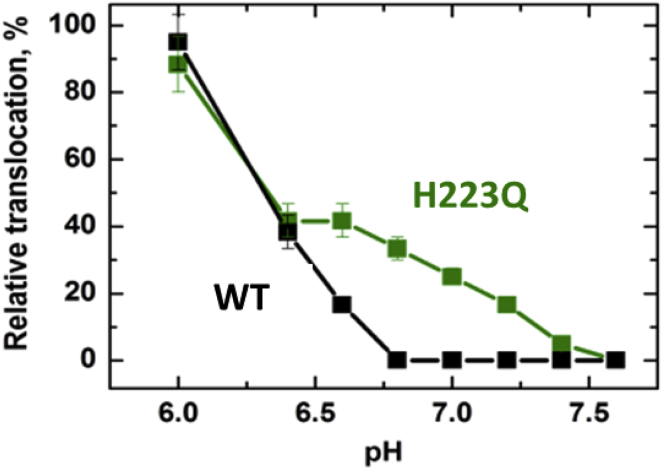
Figure 5.
pH-dependent functional activity of the WT (black) and H223Q (green) T domains, measured by the proteolysis-based translocation method of Rodnin and Ladokhin (48). The T domain was incubated with lipid vesicles preloaded with thrombin, and the translocation was assayed by proteolytic cleavage of the N-terminal 22-residue sequence of the protein. The details of experiments and controls demonstrating that cleavage occurs inside the vesicle are in (48). The higher activity of the H223Q mutant observed at 6.5 < pH < 7.5 is consistent with the difference in conformational stability (Figs. 2 and 3) predicted by the safety-latch hypothesis (cf. Fig. 1, shaded areas). To see this figure in color, go online.
We measured membrane permeabilization by using a vesicle leakage assay in which we followed changes in fluorescence intensity owing to the release of the fluorescence dye/quencher pair 8-aminonapthalene-1,3,6 trisulfonic acid (ANTS)/p-xylene-bis-pyridinium bromide (DPX) (52). When both molecules are preloaded into the vesicles, the quenching results in low fluorescence intensity, which increases upon the release of markers owing to the decrease in the concentration of DPX in the vicinity of ANTS. In our experiments, we mixed loaded vesicles with the T domain at pH 8 and triggered permeabilization by lowering the pH to a particular value (Fig. 4). Our results indicate that both H223Q (solid traces) and WT (dashed traces) T domains have equally high permeabilizing activity at strongly acidic pH (blue traces, pH 5) and no activity at neutral pH (black traces). The activity at mildly acidic pH is markedly different, with much higher activity of the mutant. Although this behavior is consistent with the predictions of the safety-latch hypothesis, we note that the difference in leakage appears to be shifted toward lower pH, as compared to changes in solution folding (compare to Figs. 2 and 3). There could be several reasons for this shift, as the membranes can affect the protonation by influencing the effective pKa and/or cause the shift in the equilibrium between the W- and W+-states by binding the latter. Both explanations are consistent with previously reported shifts of the pH-dependent binding upon the increase in the fraction of anionic lipids in the vesicles (44).
Although membrane leakage is a very sensitive way of detecting protein-membrane interactions, our previous studies with various mutants indicate that it provides a less specific measure of physiological activity (26), especially when compared to the N-terminus translocation assay (48). Here, we applied this protease-based method, which we previously developed to study T domain activity, to examine the translocational activity of both proteins as a function of pH. The vesicles are loaded with thrombin and incubated with externally added T domain at various pH values. The translocation is detected by cleavage of the N-terminal sequence and quantified by PAGE; appropriate control experiments confirm that the cleavage occurs only upon the translocation and not as the result of release of thrombin from the vesicles (48). The method can be used at pH >5.8, but not at a more acidic pH because the thrombin activity is pH-dependent. The examples of PAGE images used in quantitating the results are shown in Fig. S3. In Fig. 5, we present the pH dependence of translocation of the N-terminus into the vesicles, clearly demonstrating a higher activity for the H223Q mutant compared to the WT at 6.5 < pH < 7.5. These data confirm our hypothesis that histidine H223 indeed acts as a safety latch, preventing refolding and insertion into the membrane at neutral pH, corresponding to conditions before the acidification of the endosomal compartment.
Conclusions
The insertion/translocation transition in T domains is a multistep process that starts with a conformational change in solution to convert the water-soluble W-state into the membrane-competent W+-state. We established that the conformation changes by protonation of key histidine residues, specifically H257 (25, 28). On the basis of these studies, we formulated a “safety-latch” hypothesis for the role of H223 in the refolding process, positing that it prevents premature protonation of the main trigger, H257, which is confirmed here experimentally. Upon binding to membranes, the T domain goes through additional protonation at the same pH (hence, the pH-dependent transitions are staggered and not consecutive (44)), which likely involves E362, located in the middle of TH9 (51). After that, the T domain cascades through multiple conformations in the bilayer from an interfacial I-state, to an initial inserted T1-state (only the TH8–9 hairpin is inserted and the N-terminus is on the cis-side of the bilayer), and finally to a translocated T2-state (TH5 and TH8–9 are inserted and the N-terminus is translocated to the trans-side). The latter transition is modulated by C-terminal histidines (26), and mainly by H322 (27). Note that those mutations that affect the pH range of initial transitions do not necessarily lead to an overall shift in the translocation pH (51). This is not the case when we removed the safety latch; now, the H223Q T domain undergoes both solution refolding (Figs. 2 and 3) and membrane insertion/translocation (Figs. 4 and 5) at less acidic pH than does the WT T domain.
Deciphering the nature of conformational switching not only is a key to understanding the physiological functioning of many cellular systems but also is important for advancing protein engineering. On one hand, subtle changes in pH produced by tumors and other diseased tissues can target them for therapeutics (18). On the other, bacterial toxins target cancer cells as a component of immunotoxins (53, 54). Understanding the rules that govern pH-dependent conformational switching in diphtheria and other toxins has the potential to bring these two approaches together to create new tools for tumor targeting. We demonstrate here, for the first time to our knowledge, a new mechanism for histidine protonation in fine-tuning pH-dependent switching in physiologically relevant systems, advancing our ability to design protein machines that respond to external signaling via protonation.
Author Contributions
M.V.R. designed, executed, and analyzed CD, leakage, and translocation experiments and contributed to writing the manuscript. J.L. designed, executed, and analyzed HDXMS experiments and contributed to writing the manuscript. M.L.G. designed HDXMS experiments and contributed to overall project planning and writing the manuscript. A.S.L. designed the project, contributed to data analysis, and wrote the manuscript.
Acknowledgments
This research was supported by National Institutes of Health grants GM-069783 (A.S.L) and P41GM103422 (M.L.G.).
Editor: Andreas Engel.
Footnotes
Three figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30831-1.
Supporting Material
References
- 1.Hoch D.H., Romero-Mira M., Simpson L.L. Channels formed by botulinum, tetanus, and diphtheria toxins in planar lipid bilayers: relevance to translocation of proteins across membranes. Proc. Natl. Acad. Sci. USA. 1985;82:1692–1696. doi: 10.1073/pnas.82.6.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnon S.S., Schechter R., Tonat K., Working Group on Civilian Biodefense Botulinum toxin as a biological weapon: medical and public health management. JAMA. 2001;285:1059–1070. doi: 10.1001/jama.285.8.1059. [DOI] [PubMed] [Google Scholar]
- 3.Neale E.A. Moving across membranes. Nat. Struct. Biol. 2003;10:2–3. doi: 10.1038/nsb0103-2. [DOI] [PubMed] [Google Scholar]
- 4.Koriazova L.K., Montal M. Translocation of botulinum neurotoxin light chain protease through the heavy chain channel. Nat. Struct. Biol. 2003;10:13–18. doi: 10.1038/nsb879. [DOI] [PubMed] [Google Scholar]
- 5.Collier R.J., Young J.A. Anthrax toxin. Annu. Rev. Cell Dev. Biol. 2003;19:45–70. doi: 10.1146/annurev.cellbio.19.111301.140655. [DOI] [PubMed] [Google Scholar]
- 6.Davidson V.L., Brunden K.R., Cramer W.A. Acidic pH requirement for insertion of colicin E1 into artificial membrane vesicles: relevance to the mechanism of action of colicins and certain toxins. Proc. Natl. Acad. Sci. USA. 1985;82:1386–1390. doi: 10.1073/pnas.82.5.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mair C.M., Meyer T., Herrmann A. A histidine residue of the influenza virus hemagglutinin controls the pH dependence of the conformational change mediating membrane fusion. J. Virol. 2014;88:13189–13200. doi: 10.1128/JVI.01704-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalani M.R., Moradi A., Tajkhorshid E. Characterizing a histidine switch controlling pH-dependent conformational changes of the influenza virus hemagglutinin. Biophys. J. 2013;105:993–1003. doi: 10.1016/j.bpj.2013.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benčina M. Illumination of the spatial order of intracellular pH by genetically encoded pH-sensitive sensors. Sensors (Basel) 2013;13:16736–16758. doi: 10.3390/s131216736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casey J.R., Grinstein S., Orlowski J. Sensors and regulators of intracellular pH. Nat. Rev. Mol. Cell Biol. 2010;11:50–61. doi: 10.1038/nrm2820. [DOI] [PubMed] [Google Scholar]
- 11.Isom D.G., Sridharan V., Dohlman H.G. Protons as second messenger regulators of G protein signaling. Mol. Cell. 2013;51:531–538. doi: 10.1016/j.molcel.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minn A.J., Vélez P., Thompson C.B. Bcl-x(L) forms an ion channel in synthetic lipid membranes. Nature. 1997;385:353–357. doi: 10.1038/385353a0. [DOI] [PubMed] [Google Scholar]
- 13.Thuduppathy G.R., Terrones O., Hill R.B. The N-terminal domain of Bcl-xL reversibly binds membranes in a pH-dependent manner. Biochemistry. 2006;45:14533–14542. doi: 10.1021/bi0616652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thuduppathy G.R., Craig J.W., Hill R.B. Evidence that membrane insertion of the cytosolic domain of Bcl-xL is governed by an electrostatic mechanism. J. Mol. Biol. 2006;359:1045–1058. doi: 10.1016/j.jmb.2006.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García-Sáez A.J., Ries J., Schwille P. Membrane promotes tBID interaction with BCL(XL) Nat. Struct. Mol. Biol. 2009;16:1178–1185. doi: 10.1038/nsmb.1671. [DOI] [PubMed] [Google Scholar]
- 16.Vargas-Uribe M., Rodnin M.V., Ladokhin A.S. Comparison of membrane insertion pathways of the apoptotic regulator Bcl-xL and the diphtheria toxin translocation domain. Biochemistry. 2013;52:7901–7909. doi: 10.1021/bi400926k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barar J., Omidi Y. Dysregulated pH in tumor microenvironment checkmates cancer therapy. Bioimpacts. 2013;3:149–162. doi: 10.5681/bi.2013.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andreev O.A., Engelman D.M., Reshetnyak Y.K. Targeting diseased tissues by pHLIP insertion at low cell surface pH. Front. Physiol. 2014;5:97. doi: 10.3389/fphys.2014.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buckley J.T., Wilmsen H.U., van der Goot F.G. Protonation of histidine-132 promotes oligomerization of the channel-forming toxin aerolysin. Biochemistry. 1995;34:16450–16455. doi: 10.1021/bi00050a028. [DOI] [PubMed] [Google Scholar]
- 20.Wimalasena D.S., Cramer J.C., Bann J.G. Effect of 2-fluorohistidine labeling of the anthrax protective antigen on stability, pore formation, and translocation. Biochemistry. 2007;46:14928–14936. doi: 10.1021/bi701763z. [DOI] [PubMed] [Google Scholar]
- 21.Goh G.K., Dunker A.K., Uversky V.N. Protein intrinsic disorder and influenza virulence: the 1918 H1N1 and H5N1 viruses. Virol. J. 2009;6:69. doi: 10.1186/1743-422X-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uversky V.N. Intrinsically disordered proteins and their environment: effects of strong denaturants, temperature, pH, counter ions, membranes, binding partners, osmolytes, and macromolecular crowding. Protein J. 2009;28:305–325. doi: 10.1007/s10930-009-9201-4. [DOI] [PubMed] [Google Scholar]
- 23.Xue B., Li L., Dunker A.K. Analysis of structured and intrinsically disordered regions of transmembrane proteins. Mol. Biosyst. 2009;5:1688–1702. doi: 10.1039/B905913J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perier A., Chassaing A., Gillet D. Concerted protonation of key histidines triggers membrane interaction of the diphtheria toxin T domain. J. Biol. Chem. 2007;282:24239–24245. doi: 10.1074/jbc.M703392200. [DOI] [PubMed] [Google Scholar]
- 25.Rodnin M.V., Kyrychenko A., Ladokhin A.S. Conformational switching of the diphtheria toxin T domain. J. Mol. Biol. 2010;402:1–7. doi: 10.1016/j.jmb.2010.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodnin M.V., Kyrychenko A., Ladokhin A.S. Replacement of C-terminal histidines uncouples membrane insertion and translocation in diphtheria toxin T-domain. Biophys. J. 2011;101:L41–L43. doi: 10.1016/j.bpj.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vargas-Uribe M., Rodnin M.V., Ladokhin A.S. Crucial role of H322 in folding of the diphtheria toxin T-domain into the open-channel state. Biochemistry. 2013;52:3457–3463. doi: 10.1021/bi400249f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurnikov I.V., Kyrychenko A., Ladokhin A.S. pH-triggered conformational switching of the diphtheria toxin T-domain: the roles of N-terminal histidines. J. Mol. Biol. 2013;425:2752–2764. doi: 10.1016/j.jmb.2013.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ladokhin A.S. pH-triggered conformational switching along the membrane insertion pathway of the diphtheria toxin T-domain. Toxins (Basel) 2013;5:1362–1380. doi: 10.3390/toxins5081362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Senzel L., Huynh P.D., Finkelstein A. The diphtheria toxin channel-forming T domain translocates its own NH2-terminal region across planar bilayers. J. Gen. Physiol. 1998;112:317–324. doi: 10.1085/jgp.112.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oh K.J., Senzel L., Finkelstein A. Translocation of the catalytic domain of diphtheria toxin across planar phospholipid bilayers by its own T domain. Proc. Natl. Acad. Sci. USA. 1999;96:8467–8470. doi: 10.1073/pnas.96.15.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bennett M.J., Eisenberg D. Refined structure of monomeric diphtheria toxin at 2.3 Å resolution. Protein Sci. 1994;3:1464–1475. doi: 10.1002/pro.5560030912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oh K.J., Zhan H., Hubbell W.L. Organization of diphtheria toxin T domain in bilayers: a site-directed spin labeling study. Science. 1996;273:810–812. doi: 10.1126/science.273.5276.810. [DOI] [PubMed] [Google Scholar]
- 34.Oh K.J., Zhan H., Collier R.J. Conformation of the diphtheria toxin T domain in membranes: a site-directed spin-labeling study of the TH8 helix and TL5 loop. Biochemistry. 1999;38:10336–10343. doi: 10.1021/bi990520a. [DOI] [PubMed] [Google Scholar]
- 35.Kachel K., Ren J., London E. Identifying transmembrane states and defining the membrane insertion boundaries of hydrophobic helices in membrane-inserted diphtheria toxin T domain. J. Biol. Chem. 1998;273:22950–22956. doi: 10.1074/jbc.273.36.22950. [DOI] [PubMed] [Google Scholar]
- 36.Senzel L., Gordon M., Finkelstein A. Topography of diphtheria Toxin’s T domain in the open channel state. J. Gen. Physiol. 2000;115:421–434. doi: 10.1085/jgp.115.4.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao G., London E. Behavior of diphtheria toxin T domain containing substitutions that block normal membrane insertion at Pro345 and Leu307: control of deep membrane insertion and coupling between deep insertion of hydrophobic subdomains. Biochemistry. 2005;44:4488–4498. doi: 10.1021/bi047705o. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y., Malenbaum S.E., London E. Identification of shallow and deep membrane-penetrating forms of diphtheria toxin T domain that are regulated by protein concentration and bilayer width. J. Biol. Chem. 1997;272:25091–25098. doi: 10.1074/jbc.272.40.25091. [DOI] [PubMed] [Google Scholar]
- 39.Chenal A., Savarin P., Forge V. Membrane protein insertion regulated by bringing electrostatic and hydrophobic interactions into play. A case study with the translocation domain of diphtheria toxin. J. Biol. Chem. 2002;277:43425–43432. doi: 10.1074/jbc.M204148200. [DOI] [PubMed] [Google Scholar]
- 40.Ladokhin A.S., Legmann R., White S.H. Reversible refolding of the diphtheria toxin T-domain on lipid membranes. Biochemistry. 2004;43:7451–7458. doi: 10.1021/bi036157w. [DOI] [PubMed] [Google Scholar]
- 41.Palchevskyy S.S., Posokhov Y.O., Ladokhin A.S. Chaperoning of insertion of membrane proteins into lipid bilayers by hemifluorinated surfactants: application to diphtheria toxin. Biochemistry. 2006;45:2629–2635. doi: 10.1021/bi052257l. [DOI] [PubMed] [Google Scholar]
- 42.Montagner C., Perier A., Chenal A. Behavior of the N-terminal helices of the diphtheria toxin T domain during the successive steps of membrane interaction. Biochemistry. 2007;46:1878–1887. doi: 10.1021/bi602381z. [DOI] [PubMed] [Google Scholar]
- 43.Posokhov Y.O., Rodnin M.V., Ladokhin A.S. FCS study of the thermodynamics of membrane protein insertion into the lipid bilayer chaperoned by fluorinated surfactants. Biophys. J. 2008;95:L54–L56. doi: 10.1529/biophysj.108.141002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kyrychenko A., Posokhov Y.O., Ladokhin A.S. Kinetic intermediate reveals staggered pH-dependent transitions along the membrane insertion pathway of the diphtheria toxin T-domain. Biochemistry. 2009;48:7584–7594. doi: 10.1021/bi9009264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eftink M.R. The use of fluorescence methods to monitor unfolding transitions in proteins. Biophys. J. 1994;66:482–501. doi: 10.1016/s0006-3495(94)80799-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J., Rodnin M.V., Gross M.L. Hydrogen-deuterium exchange and mass spectrometry reveal the pH-dependent conformational changes of diphtheria toxin T domain. Biochemistry. 2014;53:6849–6856. doi: 10.1021/bi500893y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Glasoe P.K., Long F.A. Use of glass electrodes to measure acidities in deuterium oxide1,2. J. Phys. Chem. 1960;64:188–190. [Google Scholar]
- 48.Rodnin M.V., Ladokhin A.S. Membrane translocation assay based on proteolytic cleavage: application to diphtheria toxin T domain. Biochim. Biophys. Acta. 2014;1848:35–40. doi: 10.1016/j.bbamem.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hope M.J., Bally M.B., Cullis P.R. Generation of multilamellar and unilamellar phospholipid vesicles. Chem. Phys. Lipids. 1986;40:89–107. [Google Scholar]
- 50.Mayer L.D., Hope M.J., Cullis P.R. Vesicles of variable sizes produced by a rapid extrusion procedure. Biochim. Biophys. Acta. 1986;858:161–168. doi: 10.1016/0005-2736(86)90302-0. [DOI] [PubMed] [Google Scholar]
- 51.Ghatak C., Rodnin M.V., Ladokhin A.S. Role of acidic residues in helices TH8-TH9 in membrane interactions of the diphtheria toxin T domain. Toxins (Basel) 2015;7:1303–1323. doi: 10.3390/toxins7041303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ladokhin A.S., Wimley W.C., White S.H. Mechanism of leakage of contents of membrane vesicles determined by fluorescence requenching. Methods Enzymol. 1997;278:474–486. doi: 10.1016/s0076-6879(97)78025-x. [DOI] [PubMed] [Google Scholar]
- 53.Mathew M., Verma R.S. Humanized immunotoxins: a new generation of immunotoxins for targeted cancer therapy. Cancer Sci. 2009;100:1359–1365. doi: 10.1111/j.1349-7006.2009.01192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Antignani A., Fitzgerald D. Immunotoxins: the role of the toxin. Toxins (Basel) 2013;5:1486–1502. doi: 10.3390/toxins5081486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rodnin M.V., Posokhov Y.O., Ladokhin A.S. Interactions of fluorinated surfactants with diphtheria toxin T-domain: testing new media for studies of membrane proteins. Biophys. J. 2008;94:4348–4357. doi: 10.1529/biophysj.107.126235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.