ABSTRACT
The asexual facultative aerobic haploid yeast Candida glabrata is widely used in the industrial production of various organic acids. To elucidate the physiological function of the C. glabrata transcription factor Crz1p (CgCrz1p) and its role in tolerance to acid stress, we deleted or overexpressed the corresponding gene, CgCRZ1. Deletion of CgCRZ1 resulted in a 60% decrease in the dry weight of cells (DCW) and a 50% drop in cell viability compared with those of the wild type at pH 2.0. Expression of lipid metabolism-associated genes was also significantly downregulated. Consequently, the proportion of C18:1 fatty acids, the ratio of unsaturated to saturated fatty acids, and the ergosterol content decreased by 30%, 46%, and 30%, respectively. Additionally, membrane integrity, fluidity, and H+-ATPase activity were reduced by 45%, 9%, and 50%, respectively. In contrast, overexpression of CgCrz1p increased C18:1 and ergosterol contents by 16% and 40%, respectively. Overexpression also enhanced membrane integrity, fluidity, and H+-ATPase activity by 31%, 6%, and 20%, respectively. Moreover, in the absence of pH buffering, the DCW and pyruvate titers increased by 48% and 60%, respectively, compared to that of the wild type. Together, these results suggest that CgCrz1p regulates tolerance to acidic conditions by altering membrane lipid composition in C. glabrata.
IMPORTANCE This study provides insight into the metabolism of Candida glabrata under acidic conditions, such as those encountered during the industrial production of organic acids. We found that overexpression of the transcription factor CgCrz1p improved viability, biomass, and pyruvate yields at a low pH. Analysis of plasma membrane lipid composition indicated that CgCrz1p might play an important role in its integrity and fluidity and that it enhanced the pumping of protons in acidic environments. We propose that altering the structure of the cell membrane may provide a successful strategy for increasing C. glabrata productivity at a low pH.
INTRODUCTION
The asexual facultative aerobic haploid yeast Candida glabrata is the only microorganism used for the industrial production of pyruvic (1), fumaric (2), malic (3), and α-ketoglutaric (4) acids. However, the accumulation of extracellular acid causes a significant reduction in the pH of the fermentation broth, thereby inhibiting cell growth and decreasing the synthesis of the target compound (5). To maintain the culture medium at a suitable pH, alkaline reagents, such as NaOH, Na2CO3, and CaCO3, are added, although this can result in increased osmotic stress (6). Improving the tolerance of C. glabrata to low-pH conditions should boost the economic efficiency of organic acid production. Various approaches have been attempted in this sense, including the addition of exogenous auxiliary energy substrates (7), chemical mutagenesis of microorganisms (8), rational genetic engineering (9), and adaptive evolution (10). Such approaches have achieved improvements in the titer and yield of some target organic acids (11).
Several transcription factors are known to be involved in the response to environmental stresses (12); when they are overexpressed, the resultant mutants display enhanced resistance to particular stress conditions (13–16). We previously showed that C. glabrata transcription factors Asg1p and Hal9p (CgAsg1p and CgHal9p, respectively) regulate the membrane proton pump Pma1p, which is responsible for maintaining pH homeostasis under acidic conditions (17). Manipulation of the transcription factors involved in acid stress tolerance might represent a new strategy for expanding the industrial application of C. glabrata.
The transcription factor Crz1p is a well-studied calcineurin target common to filamentous fungi and yeast and capable of responding to various external stimuli (18). In Saccharomyces cerevisiae, Crz1p (ScCrz1p) is involved in metal ion resistance (19), cell wall biogenesis (20), regulation of the transcriptional response to alkaline pH (21) or acid stress (22), and tolerance to ethanol (23). In Candida albicans, Crz1p (CaCrz1p) is implicated in antifungal susceptibility (24), virulence, lithium ion resistance, filamentation, and the regulation of pH homeostasis under alkaline and acidic conditions (24, 25). On the basis of BLAST searches, C. glabrata CRZ1 (CAGL0M06831g) shares 77% and 39.4% sequence identity with ScCRZ1 and CaCRZ1, respectively. CgCrz1p is required for virulence (26), cell wall biosynthesis, heat shock response, and calcineurin function (27). In this study, we assessed the contribution of CgCrz1p to acid stress tolerance and whether its overexpression improved cell growth at a low pH. To this end, we constructed and characterized Cgcrz1Δ deletion and Cgcrz1Δ/CgCRZ1 overexpression strains.
MATERIALS AND METHODS
Strains, media, and growth conditions.
Strains used in this study are listed in Table S1 in the supplemental material. Unless stated otherwise, strains were grown in synthetically defined yeast nitrogen base (YNB; 0.67% yeast nitrogen base, 2% glucose, pH 6.0) medium supplemented with essential nutrients, whereas yeast peptone dextrose (1% yeast nitrogen base, 2% tryptone, 2% glucose) medium was used to determine cell viability. To achieve the desired pH, the medium was buffered with 2-(4-[2-hydroxyethyl] piperazin-1-yl) ethanesulfonic acid to a final concentration of 100 mM with HCl or NaOH as required. All strains were grown at 30°C with shaking at 200 rpm.
The Cgcrz1Δ mutant was obtained by genomic integration of a HIS3 marker in the CgCRZ1 locus. The CgHIS3 marker gene was amplified from the C. glabrata strain ATCC 2001 (a gift from Karl Kuchler) genome by PCR with 5′ and 3′ flanking regions corresponding to CgCRZ1. PCR products were transformed in C. glabrata ATCC 55 as described previously (28). CgCRZ1 deletion by homologous recombination of CgHIS3 was confirmed by genomic PCR and DNA sequencing.
CgCRZ1 was amplified from the genome of C. glabrata ATCC 2001 and was flanked by BamHI and StuI restriction sites. The fragment was inserted in the pY26 vector under the control of the glycerol-3-phosphate dehydrogenase promoter following digestion with BamHI and StuI and ligation with T4 ligase. Primer sequences and DNA sequencing are shown in Tables S2 to S4 and Fig. S1 in the supplemental material. The pY26-CgCRZI construct was then transformed into the Cgcrz1Δ mutant to generate the Cgcrz1Δ/CgCRZ1 overexpression strain according to the steps mentioned above.
Spot assays.
A C. glabrata culture in logarithmic phase was diluted to an absorbance at 600 nm (A600) of 1.0 in phosphate-buffered saline. Aliquots (4 μl) of 10-fold serial dilutions were spotted onto YNB agar plates at different pH values. Growth was assessed after incubation for 4 days.
Growth and CFU counts.
To analyze the growth of C. glabrata strains at a low pH, cells were grown to logarithmic phase in YNB and then diluted in fresh medium at pH 6.0 or pH 2.0 to an initial A600 of 0.1. A600 was recorded at regular intervals. To analyze cell viability, appropriate dilutions of YNB pH 2.0 cultures were plated onto yeast peptone dextrose agar at various time points. Total CFU values were calculated by counting the number of viable colonies that had grown after incubation for 2 days. Data are presented as histograms to illustrate cell survival over time.
Pyruvate production.
The wild-type, Cgcrz1Δ, and Cgcrz1Δ/CgCRZ1 strains were cultivated in 250-ml flasks containing 25 ml medium A (30 g/liter glucose, 10 g/liter peptone, 1 g/liter KH2PO4, 0.5 g/liter MgSO4·7H2O) for 24 h. Following centrifugation, pellets were resuspended in distilled water and cell suspensions were inoculated into flasks containing 50 ml medium B (100 g/liter glucose, 3 g/liter KH2PO4, 0.8 g/liter MgSO4·7H2O, 3 g/liter sodium acetate, 18 μg/liter thiamine-HCl, 4 mg/liter biotin, 0.04 g/liter pyridoxine-HCl, 0.8 g/liter nicotinic acid) at an initial biomass dry weight of 0.5 g/liter. Fermentation was performed over 48 h in a 500-ml flask containing 50 ml medium B alone or medium B buffered with 40 g/liter CaCO3.
Analytical methods.
A660 was converted to dry weight of cells (DCW) according to the standard curve reported in reference 29 and the following formula: A660/DCW = 1/0.23 g/liter.
The concentration of pyruvate was determined by high-performance liquid chromatography (Dionex, Shanghai, China) as described previously (30) and extracellular pH was measured with a pH meter (Dionex).
Genome-wide transcription analysis.
The wild-type and Cgcrz1Δ strains were cultured to logarithmic phase and then reinoculated into YNB medium at pH 6.0 or pH 2.0 at an initial A600 of 1.0. After incubation for 6 h, cells were harvested and washed twice with phosphate-buffered saline by resuspension and centrifugation at 5,000 × g for 4 min at 4°C. Total RNA was isolated using the MiniBEST universal RNA extraction kit (TaKaRa Bio, Shiga, Japan). The concentration and quality of total RNA were determined by microspectrophotometry using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Frozen samples were sent to the Beijing Genomics Institute (http://www.genomics.cn/index; Beijing, China) for global gene analysis. The NEBNext poly(A) mRNA magnetic isolation module (E7490; New England BioLabs, Ipswich, MA, USA) was used to remove rRNA and enrich for mRNA. The NEBNext mRNA library prep master mix set for Illumina (E6110) and NEBNext multiplex oligonucleotides for Illumina (E7500) were used to construct the libraries. The qualified libraries generated clusters on an Illumina cBot (Illumina, San Diego, CA, USA), and sequencing was performed on an Illumina HiSeq 2500. C. glabrata genes were annotated on the basis of their sequence similarity to putative orthologs in other yeast species.
qRT-PCR.
C. glabrata cells in logarithmic phase were inoculated into YNB at pH 6.0 or pH 2.0 at an initial A600 of 1.0 and cultured for 6 h. Total RNA was extracted using the MiniBEST universal RNA extraction kit, and 1 μg was used to synthesize cDNA with the PrimeScript II 1st-strand cDNA synthesis kit (TaKaRa Bio). The cDNA mixture was diluted to ∼100 ng/μl and used for quantitative reverse transcription-PCR (qRT-PCR) with SYBR Premix ExTaq (Takara Bio) on an iQ5 continuous fluorescence detector system (Bio-Rad, Hercules, CA). Data were normalized to the actin gene. The primers used for qRT-PCR verification are given in Table S8 in the supplemental material.
Membrane fatty acid measurement.
C. glabrata cells were cultured in YNB at pH 6.0 or pH 2.0 for 6 to 8 h, harvested, and washed twice with distilled water by resuspension and centrifugation at 5,000 × g for 4 min at 4°C. Pellets of similar weights (50 mg) were freeze-dried and samples were used for extraction and methylation of total lipids as previously described (31). For gas chromatography, components were separated by passage through a polyethylene glycol capillary column as previously described (32).
Sterol measurement.
Freeze-dried samples, collected as described above, were saponified by adding 40 μl cholesterol (0.5 mg/ml), which was used as an internal standard sterol. Saponified samples were resuspended in 1.0 ml hexane and mixed by vortexing them for 5 min. After low-speed centrifugation at 1,000 × g for 5 min, the organic phase was collected and hexane was removed by evaporation under a flow of nitrogen gas. The resulting mixture of sterol esters was dissolved in hexamethyldisilazane and trimethylchlorosilane in anhydrous pyridine and then derivatized at 30°C for 30 min prior to gas chromatography. Finally, the organic reagent was evaporated under a flow of nitrogen gas, and the resulting material was suspended in hexane.
Samples were analyzed with a gas chromatograph-mass spectrometer (Waters, Santa Clara, MA) as previously described (33). Column dimensions were 30 m by 0.25 mm, and film thickness was 0.25 μm (DB-5MS stationary phase; J&W Scientific, Folsom, CA, USA).
Plasma membrane H+-ATPase activity assay.
C. glabrata cells in logarithmic phase were inoculated into YNB at pH 6.0 or pH 2.0 at an initial A600 of 1.0 and incubated for 6 to 8 h. Total membrane fractions were prepared as described previously (34). Plasma membrane ATPase activity was determined on the basis of the amount of phosphate released by hydrolysis of ATP in the reaction mixture as described previously (35). The membrane fraction (5 μg) was incubated at 30°C for 30 min in 120 μl of assay mixture [5 mM ATP, 10 mM MgSO4, 50 mM KCl in 50 mM 2-(N-morpholino)ethanesulfonic acid, pH 6.0]. To eliminate any non-plasma membrane ATPase activity, KNO3, NaN3, and ammonium molybdate were added to final concentrations of 50 mM, 5 mM, and 0.2 mM, respectively. After incubation at 30°C for 30 min, the reaction was stopped by adding 130 μl of 1% (wt/vol) SDS, 0.6 M H2SO4, 1.2% (wt/vol) ammonium molybdate, and 1.6% (wt/vol) ascorbic acid. The amount of inorganic phosphate released was calculated from the A750 value after incubation at room temperature for 10 min. Plasma membrane ATPase activity was expressed in micromoles of inorganic phosphate released per minute per milligram of total membrane protein.
Cell membrane integrity assay.
Propidium iodide (PI) staining was used to monitor the membrane integrity of C. glabrata cells growing logarithmically in YNB at pH 6.0 or pH 2.0 for 6 to 8 h (36). Samples (500 μl) of cultures at an initial A600 of 1.0 were incubated with 3 μl PI stock solution (1 mg/ml; Sigma, Shanghai City, China) and kept in the dark for 5 min at 30°C prior to flow cytometry analysis.
Flow cytometry was performed using a FACSCalibur instrument (BD Biosciences, Shanghai, China). The fluorescence emission of PI-labeled cells was measured through a 660/16-nm band-pass filter. At least 20,000 events were analyzed for each sample at a low flow rate. CellQuest software (37) was used for data acquisition and analysis.
Measurement of fluorescence anisotropy.
The membrane fluidity of C. glabrata cells grown in logarithmic phase in YNB at pH 6.0 or pH 2.0 for 6 to 8 h was determined by measuring fluorescence anisotropy using 1,6-diphenyl-1,3,5-hexatriene as a probe. Steady-state fluorescence anisotropy was measured with a spectrofluorimeter (Photon Technology International, Princeton, NJ, USA) with excitation at 360 nm and emission at 450 nm. The fluorescence anisotropy value (r) was calculated as r = (IVV − GIVH)/(IVV + GIVH), where G (G = IHV/IHH) is the correlation factor for instrument polarization, I is the fluorescence intensity, VV indicates a measurement with both polarizers positioned vertically, VH indicates a measurement with the excitation and emission polarizers in the vertical and horizontal positions, respectively, and HV and HH indicate that polarizer positions are the opposite of those of VH and VV, respectively. Fluorescence anisotropy values and membrane fluidity showed a negative correlation, and all analyses were performed in triplicate.
Accession number(s).
Raw reads (Sequence Read Archive [SRA] entries SRX1542155, SRX1542171, SRX1542172, and SRX1542173) were submitted to NCBI under BioProject number PRJNA309480 and SRA study number SRP068794.
RESULTS
CgCrz1p is required for growth at pH 2.0.
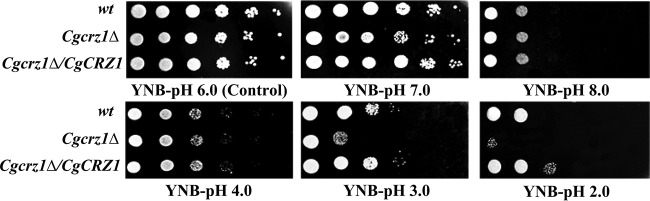
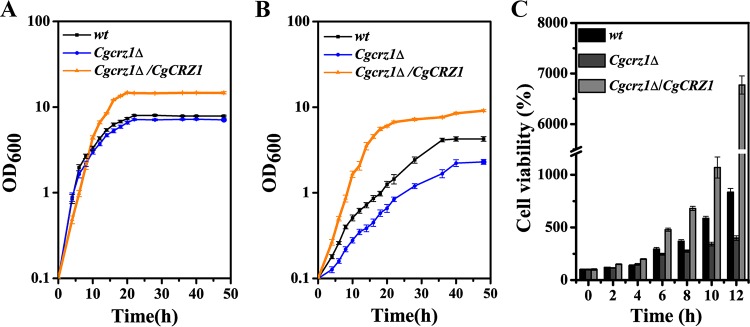
First, we checked whether CgCrz1p was necessary for growth at a low pH. The wild-type, Cgcrz1Δ, and Cgcrz1Δ/CgCRZ1 strains were spotted and grown on YNB at pH values ranging from 2.0 to 8.0. Deletion of CgCRZ1 caused a significant growth defect at pH 2.0, whereas overexpression enhanced growth with respect to that of the wild type (Fig. 1). Growth curves for all three strains were determined at pH 6.0 and pH 2.0 (Fig. 2). Whereas the final biomasses of the wild-type and Cgcrz1Δ strains were similar, that of the overexpression strain increased by 100% at pH 6.0 (Fig. 2A). At pH 2.0, growth of the Cgcrz1Δ strain decreased by 60% and that of the Cgcrz1Δ/CgCRZ1 strain increased by 125% compared to that of the wild type (Fig. 2B). The viability of C. glabrata at pH 2.0 was determined by counting CFU. After 12 h of incubation, the Cgcrz1Δ strain displayed a 50% reduction in viability compared to that of wild-type cells, while the Cgcrz1Δ/CgCRZ1 strain exhibited a 7-fold increase (Fig. 2C; see also Table S5 in the supplemental material). These results strongly suggest that CgCrz1p has a vital role in the growth of C. glabrata at pH 2.0.
FIG 1.
Growth profiles of the wild-type (wt), Cgcrz1Δ, and Cgcrz1Δ/CgCRZ1 strains grown on YNB medium at different pH levels.
FIG 2.
CgCRZ1 is essential for the growth of C. glabrata under low-pH conditions, namely, pH 6.0 (A) and pH 2.0 (B). (C) Cell viability of the Cgcrz1Δ and Cgcrz1Δ/CgCRZ1 mutants at pH 2.0. All data are presented as mean values of three independent experiments.
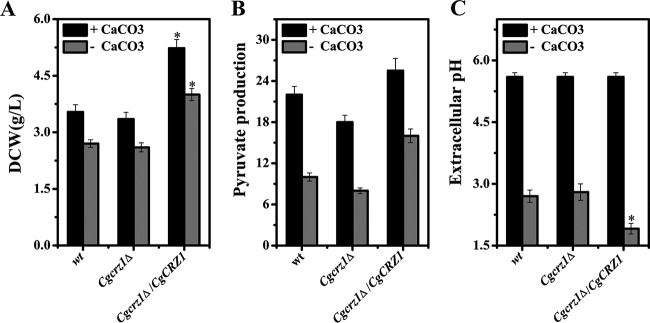
Time course measurements of pyruvate production were performed both with and without pH buffering with CaCO3 (Fig. 3). Upon addition of CaCO3, the pHs of the media were comparable between the mutants and the wild type (Fig. 3). The DCW and pyruvate titer were decreased by 5% and 18%, respectively, in the Cgcrz1Δ strain compared to those in the wild type. In contrast, in the Cgcrz1Δ/CgCRZ1 strain, the DCW and pyruvate titer were increased by 47% and 16%, respectively (Fig. 3). In nonbuffered cultures, the DCW and pyruvate titer were decreased by 15% and 30%, respectively, in the Cgcrz1Δ strain but were increased by 48% and 60%, respectively, in the Cgcrz1Δ/CgCRZ1 strain compared to those of the wild type. The pH dropped by 0.2 in the Cgcrz1Δ strain but rose by 0.8 in the Cgcrz1Δ/CgCRZ1 strain (Fig. 3; see Fig. S2 in the supplemental material). These results suggest that overexpression of CgCrz1p may enhance pyruvate production under acidic stress conditions.
FIG 3.
Pyruvate production by the wild-type, Cgcrz1Δ, and Cgcrz1Δ/CgCRZ1 strains. Cell growth (A), pyruvate production (B), and extracellular pH (C) were determined after 64 h of culturing. Error bars represent the standard deviations (*, P < 0.05 compared with wild-type cells, as determined by the t test).
Global transcriptome analysis of the Cgcrz1Δ and wild-type strains at pH 2.0.
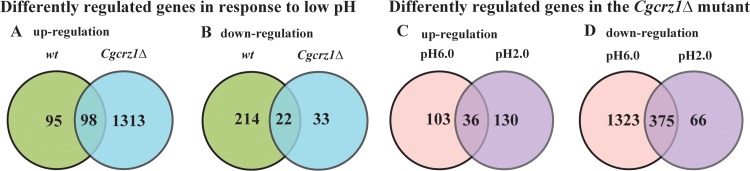
To further elucidate the physiological role of CgCrz1p in C. glabrata, we performed transcriptome sequencing (RNA-seq) analysis of gene expression in both the wild-type and Cgcrz1Δ strains. We first compared wild-type cells growing logarithmically at pH 6.0 and pH 2.0. Transcriptional profiling analysis revealed 429 genes whose expression was significantly modified (>3.0-fold change, with a P of ≤0.05): 193 were upregulated and 236 were downregulated. In the Cgcrz1Δ strain, 1,466 genes displayed a significant change in expression; of these, 1,411 were upregulated and 55 were downregulated. Specifically, a subset of 98 upregulated and 22 downregulated genes were common to both the wild-type and Cgcrz1Δ strains (Fig. 4A and B), indicating a significant overlap. Gene Ontology (GO) analysis of commonly upregulated genes included fatty acid biosynthesis (GO:0005835), arginine metabolism (GO:0006525), inositol phosphate metabolism (GO:0043647), and glycerophospholipid metabolism (GO:0006650) terms, whereas downregulated gene sets encompassed membrane proteins, stress proteins, and cell wall proteins (see Data Sets S1 and S2 in the supplemental material).
FIG 4.
Comparison of genome-wide expression levels in response to acid stress. The Venn diagrams depict the overlap between upregulated (A) and downregulated (B) genes in the wild-type and Cgcrz1Δ strains at pH 2.0 compared with the corresponding strains at pH 6.0. Upregulated (≥3-fold change) (C) and downregulated (D) genes in the Cgcrz1Δ mutant compared with their expression in the wild-type strain.
Transcriptional profiling of cells grown in YNB at pH 6.0 uncovered 139 and 1,698 genes that were up- and downregulated, respectively, in the deletion mutant compared to their level of expression in the wild type. The set of upregulated genes included processes such as glycolysis (GO:0006096), ribosome biogenesis (GO:0042254), and rRNA processing (GO:0006364), whereas downregulated genes were implicated in the cell cycle (GO:0007049), a growth-related pathway (meiosis) (GO:0007067), fatty acid biosynthesis (GO:0005835), sphingolipid metabolism (GO:0006665), inositol phosphate metabolism (GO:0043647), and glycosylphosphatidylinositol (GPI) anchor biosynthesis (GO:0031225), among others. Furthermore, there were 166 up- and 441 downregulated genes at pH 2.0 in the Cgcrz1Δ mutant compared to their levels of expression in the wild type. Among those in the Cgcrz1Δ mutant were genes involved in the cell cycle (GO:0007049), lysine biosynthesis (GO:0009085), and DNA repair (GO:0006281). The set of repressed genes included those coding for proteins implicated in fatty acid biosynthesis (GO:0005835), fatty acid metabolism (GO:0006635), biosynthesis of unsaturated fatty acids (UFA) (GO:0006636), glycerophospholipid metabolism (GO:0006650), inositol phosphate metabolism (GO:0043647), GPI anchor biosynthesis (GO:0031225), ether lipid metabolism (GO:0046485), some amino acid biosynthesis processes (e.g., phenylalanine, tyrosine, and tryptophan biosynthesis), cofactor biosynthesis processes (e.g., α-linolenic acid, pantothenate, coenzyme A, lipoic acid, and biotin), a growth-related pathway (meiosis) (GO:0007067), fungus-type cell wall assembly (GO:0071940), and mitogen-activated protein kinase (MAPK) signal transduction (GO:0007165) (Fig. 4C and D; see Data Sets S3 and S4 in the supplemental material).
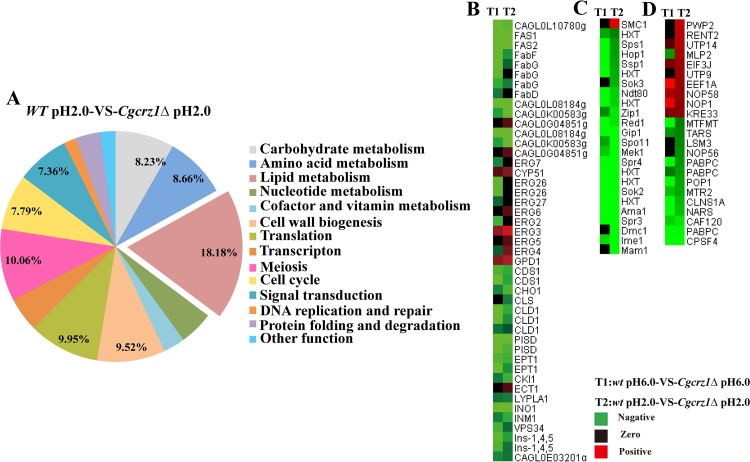
On the basis of the above results, we concluded that lipid metabolism, a growth-related pathway (meiosis), and translation were the three most notable differentially regulated pathways (as mapped in the KEGG database), accounting for 18.81%, 10.06%, and 9.56%, respectively, of all distinctively regulated genes in the Cgcrz1Δ mutant compared to those in the wild type at pH 2.0 (Fig. 5A; see also Data Set S5 in the supplemental material). The next set corresponded to cell wall biogenesis, cell cycle, carbohydrate metabolism, and amino acid metabolism. Some stress response pathways, signal transduction, DNA replication and repair, and protein folding and degradation pathways were also affected (Fig. 5A). Furthermore, heat maps of differentially regulated genes associated particularly with lipid metabolism but also with growth-related (meiosis) and translation pathways showed downregulation (Fig. 5B; Fig. S3). These results suggest that lack of CgCrz1p prominently affects membrane lipid biosynthesis and metabolism in response to low-pH stress (Fig. 6A; see also the detailed description in the supplemental material).
FIG 5.
Statistical analysis of the metabolic pathways in which the identified differentially regulated genes (up- or downregulated in the wild-type and Cgcrz1Δ strains at pH 2.0) are involved. (A) Statistical analysis of metabolic pathways based on KEGG databases (excludes pathways without maps in KEGG databases). Heat maps of differentially regulated genes involved in lipid metabolism (B), growth-related pathway (meiosis) (C), and translation (D) are shown.
FIG 6.
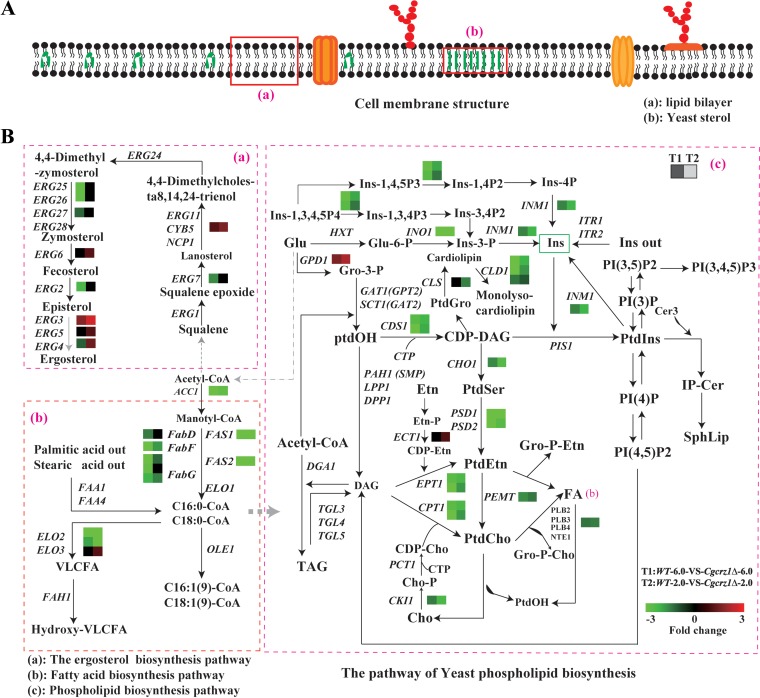
Effect of low pH on the plasma membrane of C. glabrata. (A) Structure of the plasma membrane; (B) measured changes in the expression of genes involved in membrane phospholipid biosynthesis in the Cgcrz1Δ mutant compared to those in the wild-type strain at pH 6.0 (left dashed-line squares) and at pH 2.0 (right dashed-line square). Fold changes in gene expression are indicated by the color scale. Genes not altered are indicated by black boxes. Abbreviations: Ins-1,3,4,5P4, Ins-1,3,4P3, Ins-1,4,5P3, Ins-1,4P2, Ins-3,4P2, and Ins-4P, inositol phosphates; Ins-3-P, inositol 3-phosphate; Ins, inositol; Ins ext, inositol externally added; CoA, coenzyme A; Glu, glucose; Glu-6-P, glucose 6-phosphate; DAG, diacylglycerol; CDP-DAG, cytidine diphosphate diacylglycerol; Gro-3-P, glycerol 3-phosphate; Gro-P-Cho, glycerophosphocholine; Cho, choline; Cho-P, choline phosphate; CDP-Cho, cytidine diphosphate choline; Etn, ethanolamine; Gro-P-Etn, glycerophosphoethanolamine; PtdIns, phosphatidylinositol; PI(3)P, PI(4)P, PI(3,5)P2, PI(4,5)P2, and PI(3,4,5)P3, phosphoinositides; PtdCho, phosphatidylcholine; PtdEtn, phosphatidylethanolamine; PtdOH, phosphatidic acid; PtdSer, phosphatidylserine; SphLip, sphingolipids; IP-Cer, inositol phosphoceramide; TAG, triacylglycerols; VLCFA, very-long-chain fatty acids.
Next, we compared the levels of expression of genes associated with membrane lipid metabolism in the deletion mutant and wild type (Fig. 6A and B; see Table S6 and Fig. S4 and S5 in the supplemental material). We observed the following: (i) a reduction in the transcriptional level of genes involved in fatty acid biosynthesis, such as ACC1, FAS1, FAS2, FabF, and FabG, and (ii) elongation of long-chain fatty acids, such as ELO2 and ELO3. We observed an upregulation of genes associated with ergosterol biogenesis, such as CYP5, ERG3, ERG4, and ERG6, and a downregulation of those involved in terpenoid and sterol biosynthesis (3). We also observed a downregulation of CDS1, PSD1, PSD2, CHO1, CHPT1, and PEMT1, which are involved in phospholipid metabolism, while transcription of phosphatidylinositol biogenesis-associated genes appeared to be unaffected (4). There was also a decrease in the expression of the inositol synthesis genes INO1 and INM1. These results indicate that pathways influencing membrane composition, such as those for fatty acid synthesis and elongation, as well as ergosterol, phospholipid, and inositol biosynthesis, were affected in the deletion mutant. As a consequence, CgCrz1p may regulate cell membrane composition under acid stress conditions.
CgCrz1p regulates plasma membrane composition.
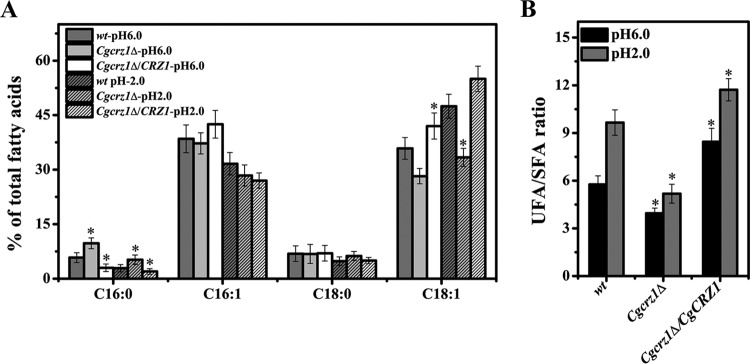
On the basis of the above results, we investigated the membrane compositions of the wild-type, Cgcrz1Δ, and Cgcrz1Δ/CgCRZ1 strains. Compared to those in the wild type at pH 6.0, the proportions of fatty acids in the Cgcrz1Δ strain were 70% higher for C16:0, unchanged for C18:0, and 3% and 20% lower for C16:1 and C18:1, respectively. In the Cgcrz1Δ/CgCRZ1 strain, C16:0 was 48% lower, C16:1 was 10% higher, C18:0 was unchanged, and C18:1 showed an 18% increase (Fig. 7A). At pH 2.0, all fatty acid ratios were substantially different from those in the wild type. In the Cgcrz1Δ strain, C16:0 showed an 80% decrease, C18:0 showed a 10% increase, C16:1 was up by 30%, and C18:1 was down by 30% compared to those in the wild type. In the Cgcrz1Δ/CgCRZ1 strain, C16:0 and C16:1 were 30% and 15% lower, respectively, than in the wild type, C18:0 was unaffected, and C18:1 was up 16% (Fig. 7A). Overall, at pH 6.0 we observed a 31% decrease in the ratio of unsaturated to saturated fatty acids (UFA/SFA) in the Cgcrz1Δ strain and a 6% increase in the Cgcrz1Δ/CgCRZ1 strain with respect to that in the wild type (Fig. 7B). However, at pH 2.0, the ratio of UFA to SFA decreased by 46% in the Cgcrz1Δ strain and increased by 21% in the Cgcrz1Δ/CgCRZ1 strain.
FIG 7.
Effect of CgCrz1p on the proportion of fatty acids in the plasma membrane of C. glabrata. (A) Change in the proportion of fatty acids at pH 6.0 and pH 2.0; (B) change in the UFA/SFA ratio at pH 6.0 and pH 2.0. Error bars represent standard deviations. *, P < 0.05 compared to the corresponding wild-type cells, as determined by the t test.
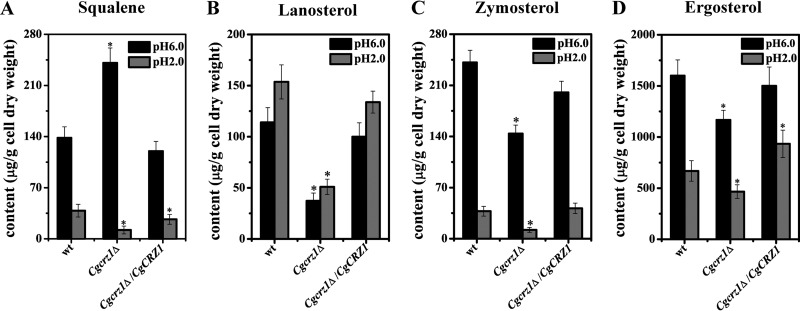
Yeast sterol content was measured to determine whether CgCrz1p could affect the biosynthesis of ergosterol (Fig. 8). At pH 6.0, deletion of CgCRZ1 resulted in a 74% increase in the amount of squalene, while amounts of lanosterol and zymosterol decreased by 60% and 40%, respectively. At pH 2.0, levels of these lipids decreased by 65%, 60%, and 40%, respectively, compared with those in wild-type cells. In the Cgcrz1Δ/CgCRZ1 strain at pH 6.0, levels of squalene, lanosterol, and zymosterol decreased by 13%, 12.7%, and 17%, respectively; at pH 2.0, their concentrations increased by 30% and 40% and decreased by 10%, respectively. In the Cgcrz1Δ strain, the concentration of the ergosterol precursor, an important modulator of the response to environmental variations, decreased by 36% at pH 6.0 and by 30% at pH 2.0 with respect to the levels in the wild type. In the Cgcrz1Δ/CgCRZ1 strain, we observed that the precursor level was 7% lower at pH 6.0 and 40% higher at pH 2.0.
FIG 8.
Effect of CgCrz1p on the sterol content of the plasma membrane of C. glabrata. Sterol analysis was performed at pH 6.0 and pH 2.0. (A) Squalene content; (B) lanosterol content; (C) zymosterol content; (D) ergosterol content. Error bars represent standard deviations. *, P < 0.05 compared to the corresponding wild-type cells, as determined by the t test.
CgCrz1p affects plasma membrane integrity and fluidity.
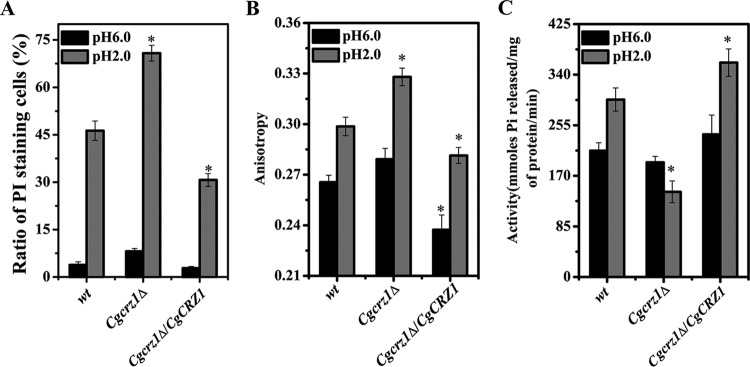
Functional alterations of the plasma membrane can affect the ability of a cell to survive environmental stress. Thus, to investigate the role of CgCrz1p in membrane integrity and rigidity, C. glabrata cells were subjected to acidic (pH 2.0) conditions for 8 h. While we did not observe any differences in the numbers of PI-stained cells at pH 6.0, at pH 2.0 we recorded a 49.7% increase in the number of PI-positive cells in the Cgcrz1Δ strain and a 35.1% decrease in the Cgcrz1Δ/CgCRZ1 strain compared to numbers in the wild type (Fig. 9A; see Fig. S6 in the supplemental material). The membrane fluidity of the Cgcrz1Δ mutant was similar to that of the wild type at pH 6.0 but was 11% higher in the Cgcrz1Δ/CgCRZ1 strain. In contrast, at pH 2.0, membrane fluidity was 9% lower and 6% higher in the Cgcrz1Δ and Cgcrz1Δ/CgCRZ1 strains, respectively, than in the wild type (Fig. 9B). At pH 6.0, H+-ATPase activity was decreased by 9% in the Cgcrz1Δ strain, while it was increased by 13% in the overexpression strain compared to that in the wild type. At pH 2.0, H+-ATPase activity was 52% lower and 20% higher for the Cgcrz1Δ and Cgcrz1Δ/CgCRZ1 strains, respectively (Fig. 9C). In conclusion, these results implicate CgCrz1p in the maintenance of membrane integrity, the regulation of fluidity, and the control of H+-ATPase activity under acidic conditions.
FIG 9.
Analysis of plasma membrane integrity, fluidity, and H+-ATPase activity. (A) Ratio of PI-stained cells at pH 6.0 and pH 2.0; (B) anisotropy values at pH 6.0 and pH 2.0; (C) H+-ATPase activity at pH 6.0 and pH 2.0. Error bars represent standard deviations. *, P < 0.05 compared to the corresponding wild-type cells, as determined by the t test.
DISCUSSION
The cell membrane is an important barrier that separates a cell from its environment. Hence, changes in its composition and function affect the ability of a microorganism to cope with variations in the surrounding habitat. In this study, deletion of CgCRZ1, the gene encoding the zinc finger transcription factor Crz1p in C. glabrata, resulted in defective growth and altered expression of genes involved in various fatty acid, sterol, and phospholipid biosynthetic pathways when grown under highly acidic conditions. As a result, at pH 2.0, the ergosterol content, the proportion of C18:1, and the UFA/SFA ratio decreased in the Cgcrz1Δ mutant, concomitant with changes in membrane integrity, fluidity, and H+-ATPase activity. Conversely, overexpression of CgCrz1p increased membrane integrity, fluidity, and H+-ATPase activity, which led to enhanced pyruvate titers in the absence of a pH-buffering agent (CaCO3). These results demonstrate that CgCrz1p regulates the composition of membrane lipids during adaptation to acid stress.
CgCrz1p appears to regulate pH homeostasis under acidic conditions. The amino acid sequence of C. glabrata Crz1p shares 34.1% and 27.6% similarity with those of S. cerevisiae and C. albicans, respectively. These orthologous proteins have been associated with various physiological functions. ScCrz1p participates in the acidic and alkaline pH response through regulation of intracellular Ca2+ (21, 22) and, in Na+/Li+ hypertonic saline stress, by increasing the activity of the Na+/Li+-ATPase proton pump (19). Overexpression of ScCrz1p was shown to increase the fermentative capacity of industrial strains (38). CaCrz1/2p are also involved in acidic and alkaline pH regulation but through Rim101 and calcineurin signaling pathways (24, 25). CaCrz2p acts independently of calcineurin but in parallel with the Rim101 pathway during adaptation to high concentrations of Li+ (25). In our hands, deletion of CgCRZ1 did not make C. glabrata more sensitive to salt stress (data not shown). Rather, the deletion mutant exhibited a severe growth defect at pH 2.0. Interestingly, overexpression of CgCRZ1 enhanced cell growth compared to that in the wild type and improved the capacity for pyruvate fermentation. Our results are in contrast to those of a previous study that did not report a role for CgCRZ1 in pH regulation (27).
Deletion of CgCRZ1 altered significantly the expression of 606 genes at pH 2.0. The differentially expressed genes were found to be involved in various metabolic pathways related to acid stress sensitivity. For example, expression of FAS1, ELO2, ERG3, and CDS1, which are involved in fatty acid biosynthesis and elongation and sterol and phospholipid metabolism, decreased. These results are consistent with findings for other fungal species, in which Crz1p-dependent genes were shown to function in lipid and sterol metabolism under various stress conditions (39, 40). Specifically, ERG26, UPC2, and SUR1, which are CRZ1-dependent genes, were repressed at least 3-fold in S. cerevisiae following alkaline pH stress (21). Meanwhile, deletion of Cryptococcus neoformans CRZ1 (CnCRZ1) decreased the transcription of SWH1, OLE1, and AYR1, which are involved in sterol transport, unsaturated fatty acid, and phosphatidic acid biosynthesis, 1.4-, 1.2-, and 2.2-fold, respectively (40). The mRNA levels of MEK1, HXT, SOK2, IME1, and NDT80, all of which are involved in the growth-related pathway (meiosis), were also downregulated 3.2-, 3.6-, 3.5-, 6.7-, and 2.5-fold, respectively. These results are consistent with those of an earlier report demonstrating the role of Crz1p in controlling the cell cycle by regulating cell cycle-related genes (18). HK, PFK, and GAPDH, some genes involved in glycolysis, were downregulated by 4.3-, 1.2-, and 2.6-fold, respectively, and Pyc, Cs, and Aco, some key genes in the tricarboxylic acid (TCA) cycle, were upregulated by 1.0-, 3.3-, and 1.6-fold, respectively. These results might affect the yield of pyruvate and mitochondrial functions, and ATP might be undersupplied, just as the lower membrane H+-ATPase activity suggested. Additionally, SLT2, MAPKK1, CHS1, CHS2, and SED1, which are involved in cell wall integrity and organization, were downregulated 2.5-, 2.7-, 1.8-, 1.8-, and 6.7-fold, respectively, in agreement with the view that Crz1p regulates cell wall biogenesis in S. cerevisiae, C. albicans, and other species (24, 27, 39). Finally, the calcineurin-CgCrz1p pathway may work together with other signal transduction pathways in response to acidic conditions, as suggested by alkaline pH inducing both Rim101 and calcineurin-Crz1p signal transduction in S. cerevisiae and C. albicans (21, 25, 41).
The plasma membrane shields yeast cells from environmental changes. Alteration of the plasma membrane composition represents a way of modulating the response to variations in pH (42). On the basis of KEGG pathway and heat map analyses, our results indicate that membrane lipid biosynthetic and metabolic pathways were affected by the deletion of CgCRZ1, which is known to play a vital role in acid tolerance (11, 43–47). Deletion of CgCRZ1 decreased the proportion of C18:1, the UFA/SFA ratio, and the ergosterol content at pH 2.0. In contrast, all these parameters were increased upon CgCRZ1 overexpression. As a result, the integrity and fluidity of the C. glabrata cell membrane were significantly altered. This is in agreement with previous reports finding that (i) increasing the UFA content leads to a higher UFA/SFA ratio (11, 43, 48) and higher membrane fluidity (43, 44) and (ii) inhibiting ergosterol synthesis and increasing membrane permeability allow acids to enter microbial cells (48). To maintain pH homeostasis and cope with acidic conditions, microorganisms have developed a number of strategies. These include manipulating the lengths and saturation of fatty acid chains (43, 44, 49), the contents and types of sterols (48), and the presence of specific phospholipids, such as phosphatidic acid (45), cardiolipin (47), and some inositol phosphates (46). Changes in lipid composition of the Cgcrz1Δ mutant might affect the activity of membrane proteins essential for barrier function (50, 51). The deletion of CgCRZ1 might have the following effects: (i) a reduction of CgYPS1 mRNA levels, thus affecting Pma1p and pH homeostasis under acidic conditions (52, 53), (ii) a decrease in ELO3 mRNA levels, thus affecting biosynthesis and maturation of very-long-chain (C26) fatty acids that control Pma1p biogenesis (54), and (iii) a decrease in the activity of membrane H+-ATPase, which pumps protons out of yeast cells at a low pH (55). Our results suggest that manipulating the plasma membrane lipid composition is a novel strategy for improving growth under acid stress conditions. Furthermore, these results may aid in the development of other low-pH-tolerant microorganisms for use in industrial-scale fermentation of organic acids.
Supplementary Material
ACKNOWLEDGMENTS
We declare no conflicts of interest.
We are sincerely grateful to Karl Kuchler for the generous gift of the C. glabrata ATCC 2001 and ATCC 55 strains.
This work was supported by the National Natural Science Foundation of China (grants 31270079 and 21422602).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02186-16.
REFERENCES
- 1.Yonehara T, Miyata R. 1994. Fermentative production of pyruvate from glucose by Torulopsis glabrata. J Ferment Bioeng 78:155–159. doi: 10.1016/0922-338X(94)90255-0. [DOI] [Google Scholar]
- 2.Chen X, Wu J, Song W, Zhang L, Wang H, Liu L. 2015. Fumaric acid production by Torulopsis glabrata: engineering the urea cycle and the purine nucleotide cycle. Biotechnol Bioeng 112:156–167. doi: 10.1002/bit.25334. [DOI] [PubMed] [Google Scholar]
- 3.Chen X, Xu G, Xu N, Zou W, Zhu P, Liu L, Chen J. 2013. Metabolic engineering of Torulopsis glabrata for malate production. Metab Eng 19:10–16. doi: 10.1016/j.ymben.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Zhang D, Liang N, Shi Z, Liu L, Chen J, Du G. 2009. Enhancement of α-ketoglutarate production in Torulopsis glabrata: redistribution of carbon flux from pyruvate to α-ketoglutarate. Biotechnol Bioprocess Eng 14:134–139. doi: 10.1007/s12257-008-0169-2. [DOI] [Google Scholar]
- 5.Schugerl K. 2000. Integrated processing of biotechnology products. Biotechnol Adv 18:581–599. doi: 10.1016/S0734-9750(00)00051-3. [DOI] [PubMed] [Google Scholar]
- 6.Yanez R, Marques S, Girio FM, Roseiro JC. 2008. The effect of acid stress on lactate production and growth kinetics in Lactobacillus rhamnosus cultures. Process Biochem 43:356–361. doi: 10.1016/j.procbio.2007.12.014. [DOI] [Google Scholar]
- 7.Sanchez C, Neves AR, Cavalheiro J, dos Santos MM, Garcia-Quintans N, Lopez P, Santos H. 2008. Contribution of citrate metabolism to the growth of Lactococcus lactis CRL264 at low pH. Appl Environ Microbiol 74:1136–1144. doi: 10.1128/AEM.01061-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klein-Marcuschamer D, Stephanopoulos G. 2008. Assessing the potential of mutational strategies to elicit new phenotypes in industrial strains. Proc Natl Acad Sci U S A 105:2319–2324. doi: 10.1073/pnas.0712177105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hida H, Yamada T, Yamada Y. 2007. Genome shuffling of Streptomyces sp. U121 for improved production of hydroxycitric acid. Appl Microbiol Biotechnol 73:1387–1393. doi: 10.1007/s00253-006-0613-1. [DOI] [PubMed] [Google Scholar]
- 10.Hua Q, Joyce AR, Fong SS, Palsson BO. 2006. Metabolic analysis of adaptive evolution for in silico-designed lactate-producing strains. Biotechnol Bioeng 95:992–1002. doi: 10.1002/bit.21073. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Qin Y, Zhao L, Chen J, Liu L. 2011. Physiological characteristics of a low-pH tolerant strain Torulopsis glabrata RT-6. Wei Sheng Wu Xue Bao 51:340–345. (In Chinese.) [PubMed] [Google Scholar]
- 12.Estruch F. 2000. Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol Rev 24:469–486. doi: 10.1111/j.1574-6976.2000.tb00551.x. [DOI] [PubMed] [Google Scholar]
- 13.Grey M, Brendel M. 1994. Overexpression of the SNQ3/YAP1 gene confers hyper-resistance to nitrosoguanidine in Saccharomyces cerevisiae via a glutathione-independent mechanism. Curr Genet 25:469–471. doi: 10.1007/BF00351788. [DOI] [PubMed] [Google Scholar]
- 14.Hussain M, Lenard J. 1991. Characterization of PDR4, a Saccharomyces cerevisiae gene that confers pleiotropic drug resistance in high-copy number: identity with YAP1, encoding a transcriptional activator. Gene 101:149–152. doi: 10.1016/0378-1119(91)90238-7. [DOI] [PubMed] [Google Scholar]
- 15.Schnell N, Entian KD. 1991. Identification and characterization of a Saccharomyces cerevisiae gene (PAR1) conferring resistance to iron chelators. Eur J Biochem 200:487–493. doi: 10.1111/j.1432-1033.1991.tb16209.x. [DOI] [PubMed] [Google Scholar]
- 16.Schnell N, Krems B, Entian KD. 1992. The PAR1 (YAP1/SNQ3) gene of Saccharomyces cerevisiae, a c-jun homologue, is involved in oxygen metabolism. Curr Genet 21:269–273. doi: 10.1007/BF00351681. [DOI] [PubMed] [Google Scholar]
- 17.Wu J, Chen X, Cai L, Tang L, Liu L. 2015. Transcription factors Asg1p and Hal9p regulate pH homeostasis in Candida glabrata. Front Microbiol 6:843. doi: 10.3389/fmicb.2015.00843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thewes S. 2014. Calcineurin-Crz1 signaling in lower eukaryotes. Eukaryot Cell 13:694–705. doi: 10.1128/EC.00038-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mendizabal I, Rios G, Mulet JM, Serrano R, de Larrinoa IF. 1998. Yeast putative transcription factors involved in salt tolerance. FEBS Lett 425:323–328. doi: 10.1016/S0014-5793(98)00249-X. [DOI] [PubMed] [Google Scholar]
- 20.Ouedraogo JP, Hagen S, Spielvogel A, Engelhardt S, Meyer V. 2011. Survival strategies of yeast and filamentous fungi against the antifungal protein AFP. J Biol Chem 286:13859–13868. doi: 10.1074/jbc.M110.203588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Serrano R, Ruiz A, Bernal D, Chambers JR, Arino J. 2002. The transcriptional response to alkaline pH in Saccharomyces cerevisiae: evidence for calcium-mediated signalling. Mol Microbiol 46:1319–1333. doi: 10.1046/j.1365-2958.2002.03246.x. [DOI] [PubMed] [Google Scholar]
- 22.Brandao RL, Rosa JCC, Nicoli JR, Almeida MVS, Carmo APD, Queiros HT, Castro IM. 2014. Investigating acid stress response in different Saccharomyces strains. J Mycol 2014:178274. doi: 10.1155/2014/178274. [DOI] [Google Scholar]
- 23.Araki Y, Wu H, Kitagaki H, Akao T, Takagi H, Shimoi H. 2009. Ethanol stress stimulates the Ca2+-mediated calcineurin/Crz1 pathway in Saccharomyces cerevisiae. J Biosci Bioeng 107:1–6. doi: 10.1016/j.jbiosc.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 24.Karababa M, Valentino E, Pardini G, Coste AT, Bille J, Sanglard D. 2006. CRZ1, a target of the calcineurin pathway in Candida albicans. Mol Microbiol 59:1429–1451. doi: 10.1111/j.1365-2958.2005.05037.x. [DOI] [PubMed] [Google Scholar]
- 25.Kullas AL, Martin SJ, Davis D. 2007. Adaptation to environmental pH: integrating the Rim101 and calcineurin signal transduction pathways. Mol Microbiol 66:858–871. doi: 10.1111/j.1365-2958.2007.05929.x. [DOI] [PubMed] [Google Scholar]
- 26.Miyazaki T, Yamauchi S, Inamine T, Nagayoshi Y, Saijo T, Izumikawa K, Seki M, Kakeya H, Yamamoto Y, Yanagihara K, Miyazaki Y, Kohno S. 2010. Roles of calcineurin and Crz1 in antifungal susceptibility and virulence of Candida glabrata. Antimicrob Agents Chemother 54:1639–1643. doi: 10.1128/AAC.01364-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y-L, Konieczka JH, Springer DJ, Bowen SE, Zhang J, Silao FGS, Bungay AAC, Bigol UG, Nicolas MG, Abraham SN, Thompson DA, Regev A, Heitman J. 2012. Convergent evolution of calcineurin pathway roles in thermotolerance and virulence in Candida glabrata. G3 (Bethesda) 2:675–691. doi: 10.1534/g3.112.002279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou J, Dong Z, Liu L, Du G, Chen J. 2009. A reusable method for construction of non-marker large fragment deletion yeast auxotroph strains: a practice in Torulopsis glabrata. J Microbiol Methods 76:70–74. doi: 10.1016/j.mimet.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 29.Liu LM, Li Y, Li HZ, Chen J. 2004. Manipulating the pyruvate dehydrogenase bypass of a multi-vitamin auxotrophic yeast Torulopsis glabrata enhanced pyruvate production. Lett Appl Microbiol 39:199–206. doi: 10.1111/j.1472-765X.2004.01563.x. [DOI] [PubMed] [Google Scholar]
- 30.Xu G, Liu L, Chen J. 2012. Reconstruction of cytosolic fumaric acid biosynthetic pathways in Saccharomyces cerevisiae. Microb Cell Fact 11:24. doi: 10.1186/1475-2859-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marumo K, Aoki Y. 1990. Discriminant analysis of cellular fatty acids of Candida species, Torulopsis glabrata, and Cryptococcus neoformans determined by gas-liquid chromatography. J Clin Microbiol 28:1509–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo Q, Wu J, Wu M. 2014. Enhanced acetoin production by Bacillus amyloliquefaciens through improved acetoin tolerance. Process Biochem 49:1223–1230. doi: 10.1016/j.procbio.2014.05.005. [DOI] [Google Scholar]
- 33.Tian H-C, Zhou J, Qiao B, Liu Y, Xia J-M, Yuan Y-J. 2010. Lipidome profiling of Saccharomyces cerevisiae reveals pitching rate-dependent fermentative performance. Appl Microbiol Biotechnol 87:1507–1516. doi: 10.1007/s00253-010-2615-2. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura K, Niimi M, Niimi K, Holmes AR, Yates JE, Decottignies A, Monk BC, Goffeau A, Cannon RD. 2001. Functional expression of Candida albicans drug efflux pump Cdr1p in a Saccharomyces cerevisiae strain deficient in membrane transporters. Antimicrob Agents Chemother 45:3366–3374. doi: 10.1128/AAC.45.12.3366-3374.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Viegas CA, Sebastiao PB, Nunes AG, Sa-Correia I. 1995. Activation of plasma-membrane H(+)-ATPase and expression of PMA1 and PMA2 genes in Saccharomyces cerevisiae cells grown at supraoptimal temperatures. Appl Environ Microbiol 61:1904–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Branco P, Viana T, Albergaria H, Arneborg N. 2015. Antimicrobial peptides (AMPs) produced by Saccharomyces cerevisiae induce alterations in the intracellular pH, membrane permeability and culturability of Hanseniaspora guilliermondii cells. Int J Food Microbiol 205:112–118. doi: 10.1016/j.ijfoodmicro.2015.04.015. [DOI] [PubMed] [Google Scholar]
- 37.McKenna SL, Cotter TG. 2000. Inhibition of caspase activity delays apoptosis in a transfected NS/0 myeloma cell line. Biotechnol Bioeng 67:165–176. [PubMed] [Google Scholar]
- 38.Panadero J, Hernandez-Lopez MJ, Prieto JA, Randez-Gil F. 2007. Overexpression of the calcineurin target CRZ1 provides freeze tolerance and enhances the fermentative capacity of baker's yeast. Appl Environ Microbiol 73:4824–4831. doi: 10.1128/AEM.02651-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshimoto H, Saltsman K, Gasch AP, Li HX, Ogawa N, Botstein D, Brown PO, Cyert MS. 2002. Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J Biol Chem 277:31079–31088. doi: 10.1074/jbc.M202718200. [DOI] [PubMed] [Google Scholar]
- 40.Adler A, Park Y-D, Larsen P, Nagarajan V, Wollenberg K, Qiu J, Myers TG, Williamson PR. 2011. A novel specificity protein 1 (SP1)-like gene regulating protein kinase C-1 (Pkc1)-dependent cell wall integrity and virulence factors in Cryptococcus neoformans. J Biol Chem 286:20977–20990. doi: 10.1074/jbc.M111.230268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H, Liang Y, Zhang B, Zheng W, Xing L, Li M. 2011. Alkaline stress triggers an immediate calcium fluctuation in Candida albicans mediated by Rim101p and Crz1p transcription factors. FEMS Yeast Res 11:430–439. doi: 10.1111/j.1567-1364.2011.00730.x. [DOI] [PubMed] [Google Scholar]
- 42.Dupont S, Lemetais G, Ferreira T, Cayot P, Gervais P, Beney L. 2012. Ergosterol biosynthesis: a fungal pathway for life on land? Evolution 66:2961–2968. doi: 10.1111/j.1558-5646.2012.01667.x. [DOI] [PubMed] [Google Scholar]
- 43.Yang X, Hang X, Zhang M, Liu X, Yang H. 2015. Relationship between acid tolerance and cell membrane in Bifidobacterium, revealed by comparative analysis of acid-resistant derivatives and their parental strains grown in medium with and without Tween 80. Appl Microbiol Biotechnol 99:5227–5236. doi: 10.1007/s00253-015-6447-y. [DOI] [PubMed] [Google Scholar]
- 44.Wu C, Zhang J, Wang M, Du G, Chen J. 2012. Lactobacillus casei combats acid stress by maintaining cell membrane functionality. J Ind Microbiol Biotechnol 39:1031–1039. doi: 10.1007/s10295-012-1104-2. [DOI] [PubMed] [Google Scholar]
- 45.Young BP, Shin JJH, Orij R, Chao JT, Li SC, Guan XL, Khong A, Jan E, Wenk MR, Prinz WA, Smits GJ, Loewen CJR. 2010. Phosphatidic acid is a pH biosensor that links membrane biogenesis to metabolism. Science 329:1085–1088. doi: 10.1126/science.1191026. [DOI] [PubMed] [Google Scholar]
- 46.Kooijman EE, King KE, Gangoda M, Gericke A. 2009. Ionization properties of phosphatidylinositol polyphosphates in mixed model membranes. Biochemistry 48:9360–9371. doi: 10.1021/bi9008616. [DOI] [PubMed] [Google Scholar]
- 47.MacGilvray ME, Lapek JD Jr, Friedman AE, Quivey RG Jr. 2012. Cardiolipin biosynthesis in Streptococcus mutans is regulated in response to external pH. Microbiology 158:2133–2143. doi: 10.1099/mic.0.057273-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kodedova M, Sychrova H. 2015. Changes in the sterol composition of the plasma membrane affect membrane potential, salt tolerance and the activity of multidrug resistance pumps in Saccharomyces cerevisiae. PLoS One 10:e0139306. doi: 10.1371/journal.pone.0139306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu P, Chernyshov A, Najdi T, Fu Y, Dickerson J, Sandmeyer S, Jarboe L. 2013. Membrane stress caused by octanoic acid in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 97:3239–3251. doi: 10.1007/s00253-013-4773-5. [DOI] [PubMed] [Google Scholar]
- 50.Gruenberg J. 2012. An MBoC favorite: functional interactions between sphingolipids and sterols in biological membranes regulating cell physiology. Mol Biol Cell 23:2402–2402. doi: 10.1091/mbc.E12-03-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spanova M, Zweytick D, Lohner K, Klug L, Leitner E, Hermetter A, Daum G. 2012. Influence of squalene on lipid particle/droplet and membrane organization in the yeast Saccharomyces cerevisiae. Biochem Biophys Acta 1821:647–653. doi: 10.1016/j.bbalip.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bairwa G, Kaur R. 2011. A novel role for a glycosylphosphatidylinositol-anchored aspartyl protease, CgYps1, in the regulation of pH homeostasis in Candida glabrata. Mol Microbiol 79:900–913. doi: 10.1111/j.1365-2958.2010.07496.x. [DOI] [PubMed] [Google Scholar]
- 53.Bairwa G, Rasheed M, Taigwal R, Sahoo R, Kaur R. 2014. GPI (glycosylphosphatidylinositol)-linked aspartyl proteases regulate vacuole homoeostasis in Candida glabrata. Biochem J 458:323–334. doi: 10.1042/BJ20130757. [DOI] [PubMed] [Google Scholar]
- 54.Gaigg B, Toulmay A, Schneiter R. 2006. Very long-chain fatty acid-containing lipids rather than sphingolipids per se are required for raft association and stable surface transport of newly synthesized plasma membrane ATPase in yeast. J Biol Chem 281:34135–34145. doi: 10.1074/jbc.M603791200. [DOI] [PubMed] [Google Scholar]
- 55.Malinsky J, Opekarova M, Tanner W. 2010. The lateral compartmentation of the yeast plasma membrane. Yeast 27:473–478. doi: 10.1002/yea.1772. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.