Summary
Sedentary plant‐parasitic nematodes (PPNs) induce and maintain an intimate relationship with their host, stimulating cells adjacent to root vascular tissue to re‐differentiate into unique and metabolically active ‘feeding sites’. The interaction between PPNs and their host is mediated by nematode effectors. We describe the discovery of a large and diverse family of effector genes, encoding C‐TERMINALLY ENCODED PEPTIDE (CEP) plant hormone mimics (RrCEPs), in the syncytia‐forming plant parasite Rotylenchulus reniformis. The particular attributes of RrCEPs distinguish them from all other CEPs, regardless of origin. Together with the distant phylogenetic relationship of R. reniformis to the only other CEP‐encoding nematode genus identified to date (Meloidogyne), this suggests that CEPs probably evolved de novo in R. reniformis. We have characterized the first member of this large gene family (RrCEP1), demonstrating its significant up‐regulation during the plant–nematode interaction and expression in the effector‐producing pharyngeal gland cell. All internal CEP domains of multi‐domain RrCEPs are followed by di‐basic residues, suggesting a mechanism for cleavage. A synthetic peptide corresponding to RrCEP1 domain 1 is biologically active and capable of up‐regulating plant nitrate transporter (AtNRT2.1) expression, whilst simultaneously reducing primary root elongation. When a non‐CEP‐containing, syncytia‐forming PPN species (Heterodera schachtii) infects Arabidopsis in a CEP‐rich environment, a smaller feeding site is produced. We hypothesize that CEPs of R. reniformis represent a two‐fold adaptation to sustained biotrophy in this species: (i) increasing host nitrate uptake, whilst (ii) limiting the size of the syncytial feeding site produced.
Keywords: effector, evolution, plant‐parasitic nematode, plant peptide hormone
Introduction
Sedentary plant‐parasitic nematodes (PPNs) induce and maintain an intimate relationship with their host. These obligate biotrophs stimulate root cells to re‐differentiate into unique and metabolically active ‘feeding sites’, from which they feed exclusively for a number of weeks until their life cycle is completed. On selection of the initial feeding cell, the motile nematodes become sedentary and are thus committed to that site. They must maintain the biotrophic interaction for several weeks; if the plant dies or the feeding site ceases to function and to provide nutrients, the nematode will also die. This obligate dependence on host survival, which, in some cases, can extend over many nematode generations, is a strong selection pressure against killing the host.
There are several genera of sedentary PPNs that impose varying degrees of host damage, induce feeding sites of different ontogeny and in which parasitism has evolved independently (van Megen et al., 2009). The multinucleate feeding sites of the most widely studied sedentary PPNs can be divided into two general classes: (i) the cluster of individual giant cells of the root‐knot nematodes, each formed from a single cell by multiple rounds of mitosis in the absence of cytokinesis (de Almeida Engler et al., 1999); (ii) the syncytial feeding sites of the cyst, reniform and false‐root‐knot nematodes, formed by partial cell wall dissolution and subsequent protoplast fusion of multiple adjacent cells (Grymaszewska and Golinowski, 1998; Holtmann et al., 2000; Jones and Payne, 1977; Rahman Razak and Evans, 1976; Robinson et al., 1998; Sobczak and Golinowski, 2011). Syncytia of cyst nematodes are typically initiated from a single pericycle, procambial or inner cortical cell. Increased cytoplasmic density and proliferation of organelles are accompanied by widening of existing plasmodesmata, followed by local dissolution of the cell walls between the syncytium and neighbouring cells and fusion of protoplasts. In the early stages of feeding site development, the syncytium grows by continually incorporating neighbouring cells which have been stimulated to divide (Sobczak and Golinowski, 2011). The syncytia of reniform and false‐root‐knot nematodes form by similar cell wall dissolution and share many of the same characteristics. For Rotylenchulus reniformis, the initial syncytial cell is typically an endodermal cell (Robinson et al., 1998) and the feeding site subsequently extends around the root as a single, curved cell layer (Jones and Dropkin, 1975).
Much of the work to understand the molecular basis of the relationship between parasites and their hosts focuses on parasite proteins secreted during the interaction, termed ‘effectors’. Although effectors of PPNs can originate from a range of tissues, the vast majority are produced in pharyngeal gland cells and are delivered into the host via a hollow, protractible stylet. A number of classes of effectors have been described to date (reviewed in Hewezi, 2015; Mitchum et al., 2013). The most relevant to this work are those which ‘mimic’ post‐translationally modified plant peptide hormones in both sequence and function (Bobay et al., 2013; Mitchum et al., 2012; Wang et al., 2005). Two such classes have been identified to date. The first (Wang et al., 2005) is the CLAVATA3/ESR (CLE)‐like peptides which are present in all sedentary PPNs (Mitchum et al., 2012), regardless of their origin of parasitism. More recently, genes encoding C‐TERMINALLY ENCODED PEPTIDES (CEPs) have been identified in a single lineage, the giant cell‐forming root‐knot nematodes (Bird et al., 2015; Bobay et al., 2013), although neither the gland cell expression nor biological activity of the peptides has been reported.
CEPs were discovered in plants relatively recently (Ohyama et al., 2008), and are an ancient peptide hormone common to all vascular land plants. More than 900 CEP genes have been identified to date (Ogilvie et al., 2014; Roberts et al., 2013), although only those from Arabidopsis (Delay et al., 2013; Ohyama et al., 2008; Roberts et al., 2013; Tabata et al., 2014) and Medicago truncatula (Bobay et al., 2013; Imin et al., 2013; Mohd‐Radzman et al., 2015) have been characterized in detail. CEP precursor proteins contain a signal peptide for secretion, followed by at least one CEP domain, and are processed in the apoplast into biologically active 15‐amino‐acid peptides. These short peptides are most typically modified with hydroxyprolines in positions 4 and 11 (Ohyama et al., 2008), although variants with hydroxylation of the proline (Pro) at position 7 and/or arabinosylation of Pro‐11 have been identified (Mohd‐Radzman et al., 2015). The use of synthetic functional CEP peptides, corresponding to domains within genes of interest, has allowed rapid advances in the field (Bobay et al., 2013; Delay et al., 2013; Imin et al., 2013; Ohyama et al., 2008; Tabata et al., 2014). The current paradigm suggests that CEP genes are up‐regulated by nitrogen starvation (Imin et al., 2013) and that the active peptides move in the xylem to the leaves, where they initiate an as yet unidentified descending signal which results in increased and compensatory up‐regulation of nitrate transporters on a whole‐root scale. Two Arabidopsis leucine‐rich repeat receptor kinases (LRR‐RKs; CEPR1 and CEPR2) have been identified as the shoot‐expressed receptors for several CEPs (Tabata et al., 2014). CEPs are clearly multifunctional, as one of their first described roles was the modulation of plant architecture (Ohyama et al., 2008). Over‐expression of CEP genes or exogenous application of CEP domain peptides to Arabidopsis results in suppression of the rate of root cell division, as evidenced by reduced primary root elongation (Delay et al., 2013). This can be rationalized by systemic up‐regulation of nitrate transporters, presumably as an adaptive local response to reduce plant growth where nitrate is limiting, and thus where CEPs are up‐regulated. The function of CEPs in PPNs remains unclear. Over‐expression of MtCEP1 or exogenous application of the CEP domain peptide to Md. truncatula produces periodic, circumferential root swellings which are phenotypically similar to the galls induced in this plant by the root‐knot nematode Meloidogyne hapla (Imin et al., 2013); however, similar activity has not been described for the M. hapla peptides.
We have identified a large family of CEP genes in the reniform nematode R. reniformis. This sedentary semi‐endoparasite, which has a host range encompassing more than 300 plant species, induces a syncytial feeding site and does not cause galling of infected roots. Second‐stage juveniles (J2s) hatch from eggs in the soil and then develop to vermiform adult females and males via non‐motile, non‐feeding J3 and J4 stages. Only the female nematode invades the host root and becomes sedentary, with the posterior body remaining outside and subsequently swelling to a kidney shape (Robinson et al., 1998). We present evidence to suggest that CEPs in R. reniformis originated independently from those in both plants and the root‐knot nematodes. Rotylenchulus reniformis CEPs (RrCEPs) are characterized by unique features; unlike CEPs from all other organisms, those cloned from R. reniformis contain one intron per domain sequence, regardless of the number of tandem domains that are present. We characterize one member of the RrCEP family in detail and demonstrate that it is highly up‐regulated during the biotrophic infection phase of the life cycle and expressed in the effector‐producing pharyngeal gland cell. We show that this gene, RrCEP1, encodes a functional CEP domain which significantly up‐regulates a host nitrate transporter. This domain also inhibits root elongation and limits feeding site expansion for a non‐CEP‐containing, syncytial‐forming cyst nematode. CEP effectors of R. reniformis may therefore represent a two‐fold adaptation to sustained biotrophy by: (i) increasing host nitrate uptake, whilst (ii) limiting the size of the syncytial feeding site produced.
Results and Discussion
Rotylenchulus reniformis contains a large and diverse family of CEP genes
Transcripts containing CEP‐like domains, identified in unpublished R. reniformis next‐generation sequencing (NGS) data (ERA PRJEB8325 and SRR949271), were used to design primers to amplify sequences of interest. Using a primer pair targeting a single CEP‐like gene, multiple polymerase chain reaction (PCR) products were generated from genomic DNA; these were cloned and sequenced. Cloned genomic sequences which encoded complete open reading frames, were unique at the protein level (or, where identical, contained considerably different introns) and were different in total gene length were deemed to arise from unique loci and used to construct a preliminary phylogeny of the gene family (Fig. 1 and Dryad accession doi:10.5061/dryad.q8h75). The level of sequence diversity within Fig. 1 is higher than that likely to arise as a result of allelic variation, and thus the 12 cloned genomic sequences included were named sequentially.
Figure 1.
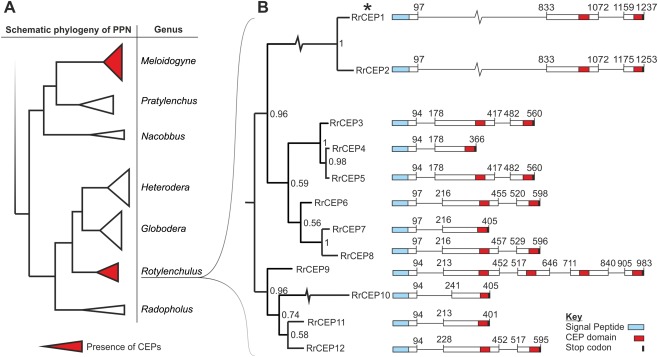
Phylogenetic analysis and genetic structure of the C‐terminally‐encoded peptide (CEP) gene family in Rotylenchulus reniformis. (A) Schematic diagram of the local phylogenetic neighbourhood of sedentary plant‐parasitic nematodes, including the only two CEP‐containing genera. Rotylenchulus reniformis has a distant phylogenetic relationship and an independent origin of biotrophic parasitism to the only other CEP‐containing nematodes, with many intermediate species that lack CEPs (van Megen et al., 2009). (B) Using a mid‐point re‐routed Bayesian phylogeny, all CEP sequences cloned to date from R. reniformis genomic DNA can be grouped into several distinct clades based on an alignment of complete genomic sequences. A representative genetic structure of the sequences in each clade identifies a large variation in CEP domain number and arrangement. Despite the diversity, CEP domain motifs are highly similar between clades. Unlike CEPs from all other organisms (Bobay et al., 1994; Delay et al., 2013), those of R. reniformis all contain at least one intron. Numbers correspond to base position; signal peptides (blue), CEP domains (red) and stop codons (black) are indicated. *Indicates RrCEP1 for further study.
Analysis of the R. reniformis transcriptomic data showed that the full complement of the cloned CEP sequences is not represented in the assembled transcriptome and, similarly, that not all sequences present in the transcriptome were cloned. Given this disparity, it is therefore likely that we have identified only a subset of what is a large and diverse family of CEP‐encoding genes in R. reniformis. Adopting a conservative approach, for their first identification in R. reniformis, we focus on only the cloned genes and their deduced amino acid sequences with the aim of characterizing their function and providing a basis for further study. As for plant and other nematode CEPs, all R. reniformis CEP genes (RrCEPs) encode a signal peptide, followed by at least one CEP domain. According to the grouping system proposed by both Delay et al. (2013) and Roberts et al. (2013), all RrCEPs conform to group I. By comparing group I CEP domain sequences between kingdoms (Fig. 2), those of R. reniformis conform well to the expected characteristics indicative of bona fide CEPs.
Figure 2.
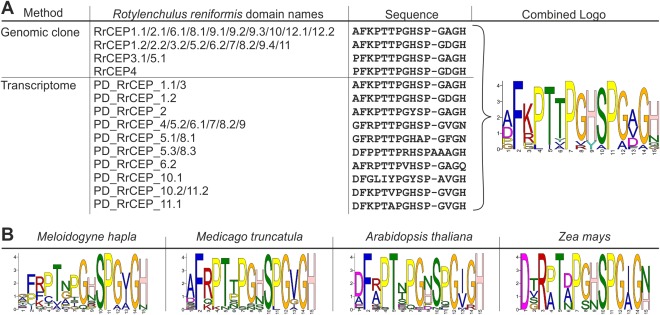
C‐terminally‐encoded peptide (CEP) domain alignments between kingdoms. (A) A combination of all unique Rotylenchulus reniformis CEP domains from the 12 cloned genes displayed in Fig. 1 and all unique putative domains (PDs) present in a de novo transcriptome assembly. According to the grouping system proposed by both Delay et al. (2013) and Roberts et al. (2013), all R. reniformis CEPs conform to group I. (B) For comparison, logo plots are shown of all unique CEP domains from the root‐knot nematode Meloidogyne hapla, and all group I CEP domains from the dicotyledons Medicago truncatula and Arabidopsis thaliana and the monocotyledon Zea mays.
RrCEPs are additionally characterized by unique attributes and probably evolved de novo in R. reniformis
CEP precursor proteins in R. reniformis can contain several CEP domains in tandem (n max = 4), where the final domain is followed directly by a stop codon (Fig. 1). Although multiple domains are common in plant CEPs, and indeed occur in other nematode peptide hormone mimics, e.g. CLEs of the potato cyst nematode Globodera rostochiensis (Lu et al., 2009), they are unique among nematode CEPs. All CEP genes cloned from R. reniformis contain one intron per domain sequence, regardless of the number of tandem domains. Introns range in size from <100 to >700 base pairs (Fig. 1). This is highly unusual as all of the other several hundred CEPs identified to date, from plant or animal origin, are encoded on a single exon (Bobay et al., 2013; Delay et al., 2013; Ogilvie et al., 2014). The biological significance of introns in RrCEPs (particularly in multi‐domain CEP effectors), or of single exon genes in every other genus, is unclear. There is no evidence that the intron structure of R. reniformis CEPs introduces additional variation through alternative splicing.
An insight into the genomic organization of RrCEPs was gained during the cloning and sequencing process. A genomic DNA fragment was cloned that encoded two RrCEPs in tandem. The two CEP gene models, in the same orientation, overlap, yet exist in different reading frames (Fig. S1, see Supporting Information). Precedent exists for this type of genomic organization in the bacterial EcoKI DNA methyltransferase (Roberts et al., 2012). This trimeric protein comprises two modification subunits (M) and one sequence specificity subunit (S). The 3′ end of the gene encoding the M subunit overlaps the 5′ end of the S subunit by one nucleotide where translation from the two different open reading frames is translationally coupled (Roberts et al., 2012). Further investigation will be required to determine whether these RrCEPs are similarly translationally coupled, and whether or not both are produced as full‐length functional proteins. These genes are identical in deduced coding sequence, with the exception that the first does not contain a full‐length CEP domain. The second is already represented in the phylogeny at 100% amino acid identity; thus, neither gene was additionally included in our analysis.
With the exception of the 15‐amino‐acid CEP domain itself, R. reniformis CEPs share no sequence similarity with any other CEPs from plants or animals. Despite the inherent difficulty associated with the limited phylogenetic signal and functional sequence constraints of CEPs, R. reniformis CEPs probably arose independently of other plant and animal CEPs. This is supported by the lack of sequence similarity outside of the CEP domain, the unusual genetic structure seen within R. reniformis and the fact that the only other known CEP‐encoding nematode genus (Meloidogyne) is distantly related to R. reniformis with an independent origin of biotrophy (van Megen et al., 2009). These two genera are separated by many CEP‐absent intermediate species. Contrary to CLEs, there is no evidence of CEP‐like sequences in available cyst nematode genomes (to which R. reniformis is basal), suggesting that these sequences arose after the split from the last common ancestor between R. reniformis and cyst nematodes.
Rotylenchulus reniformis CEP1 is highly expressed in a large secretory pharyngeal gland cell during plant–nematode biotrophic interactions
As transcripts corresponding to the deduced coding sequence of RrCEP1 were present in the transcriptome assembly, this gene was studied further. RrCEP1 is highly up‐regulated during the biotrophic phase of the plant–nematode interaction (Fig. 3A, P < 0.001). Using a complementary digoxigenin‐labelled DNA probe for in situ hybridization (de Boer et al., 1998), the RrCEP1 transcript was localized in the single large secretory pharyngeal gland cell (Fig. 3B,C). No such staining pattern was seen with the non‐complementary negative control DNA probe (Fig. 3D). Contrary to other PPNs, adult female R. reniformis apparently contain only a single large pharyngeal gland cell (Robinson et al., 1998), consistent with the images presented here. This indicates that the protein encoded by RrCEP1 is likely to be secreted in planta during the biotrophic interaction.
Figure 3.
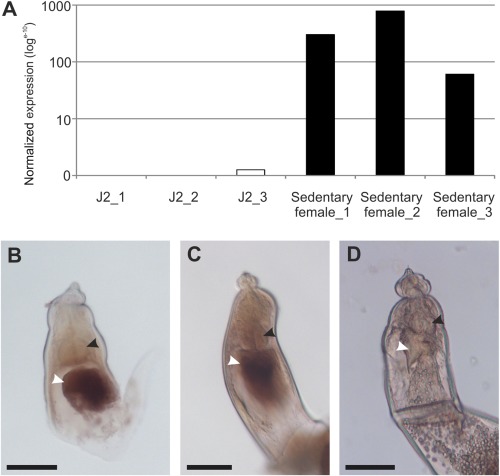
Rotylenchulus reniformis C‐terminally‐encoded peptide (CEP) temporal and spatial expression. (A) RrCEP1 is significantly up‐regulated during the sedentary parasitic phase of the life cycle compared with the second‐stage juvenile (n = 3, P < 0.001). Normalized expression data from each of three independent RNAseq libraries is shown for juvenile and sedentary female stages. (B, C) In situ hybridization identifies R. reniformis CEP1 expression in a large secretory gland cell (white arrow) posterior to the metacorpal bulb (black arrow) of mature females. (D) No such staining is observed with the negative control in situ hybridization probe. Scale bars indicate 40 μm.
Plant peptide hormones function in the apoplast, where they bind to transmembrane LRR‐RKs (Matsubayashi, 2014). The exact site of delivery in planta for R. reniformis CEPs is at present unclear; the gland cells of PPNs deliver proteins both inside the feeding site cytoplasm (Replogle et al., 2011) and directly into the apoplast (Vieira et al., 2011). CLE peptide precursors of the plant‐parasitic cyst nematode Heterodera glycines are apparently delivered into the feeding site cytoplasm, where they can be detected by immunolocalization (Wang et al., 2010). The canonical signal peptide is cleaved on translocation into the gland cell endoplasmic reticulum within the nematode, but the CLE pro‐peptide is then secreted from the plant cell into the apoplast by a post‐translational trafficking mechanism (Wang et al., 2010). It is not clear how this functions, but a putative second ‘cryptic’ signal peptide, present in the N‐terminal region of the variable domain (VDI), has been implicated. A similar cryptic signal peptide sequence was identified within the precursor region of R. reniformis CLE pro‐peptides, although trafficking was not investigated (Wubben et al., 2015). Utilizing the same approach, it is possible to identify cryptic signal peptides within R. reniformis CEP precursors (Fig. S2, see Supporting Information) that have a strikingly high degree of conservation with the equivalent region in RrCLE1. However, further work is required to determine the route to the apoplast, which may be different from that used to deliver root‐knot nematode CEPs to their site of action. All CEP genes identified from both M. hapla and Meloidogyne incognita encode only a signal peptide, followed directly by the 15‐amino‐acid CEP domain (Bird et al., 2015). As for some other pharyngeal gland‐derived root‐knot nematode effectors (Vieira et al., 2011), these peptides are presumably delivered directly through the stylet into the apoplast, with post‐translational modifications occurring in the nematode. The CLE‐peptide effectors of M. hapla (Bird et al., 2015) similarly lack the pro‐domains that characterize both plant CLEs and those of cyst (Mitchum et al., 2012) and reniform (Wubben et al., 2015) nematodes, suggesting a more general divergence in effector delivery.
Amino acids directly following CEP domains are highly non‐random in R. reniformis and plants
Ultimately, for CEPs to function, they require proteolytic processing to release the active domain from a pro‐protein. Like many plant peptide hormones, CEPs in plants are processed in the plant apoplast. Di‐basic residues adjacent to active domain motifs of plant pro‐peptide hormones are common in CLEs (DeYoung and Clark, 2001), PHYTOSULFOKINEs (PSKs) (Yang et al., 2001) and RAPID ALKALINIZATION FACTOR peptides (RALFs) (Matos et al., 2008); indeed, for AtRALF1, a double arginine is required for correct processing and function (Matos et al., 2008). For all cloned R. reniformis CEPs with more than one CEP domain, internal domains are directly followed by a double arginine as part of the consensus sequence RRLM (Fig. 4A). The AtS1P subtilisin which cleaves AtRALF23 has the canonical cleavage site RRIL (Srivastava et al., 2009). Therefore, the presence of single and di‐basic residues directly following the internal domains was also assessed for plant CEPs that contain more than one CEP domain. From 66 plant CEP genes across monocotyledonous and dicotyledonous species, amino acids directly following internal CEP domains are highly non‐random (n = 141, P < 0.0001) and are significantly enriched for both single and di‐basic residues (Fig. 4B,C), perhaps indicating that this is a common feature of all multi‐domain CEPs, not just those in animals.
Figure 4.
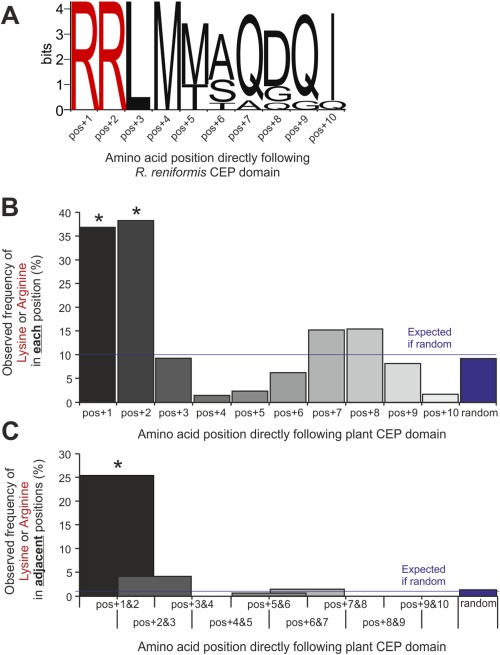
Non‐random single or di‐basic residues directly following nematode and plant C‐terminally‐encoded peptide (CEP) domains. (A) For all Rotylenchulus reniformis CEP sequences that contain more than one domain, the final domain is followed directly by a stop codon. However, all internal CEP domains, irrespective of domain sequence, are directly followed by a double arginine residue (n = 10). From 66 plant CEP genes across monocotyledonous and dicotyledonous species, amino acids directly following internal CEP domains are highly non‐random (n = 141, χ 2, *P < 0.0001) and are significantly enriched for single (B) and di‐basic (C) residues.
Given that single and di‐basic residues are enriched directly following internal plant CEP domains, it is clear that the host machinery required to process nematode multi‐domain CEPs is already present in the apoplast, irrespective of their route to that compartment. Both animals and plants utilize di‐basic residues as cleavage sites; it is therefore possible that nematode CEPs are either processed in the animal and released as functional peptides or secreted as pro‐peptides and processed in the plant apoplast.
Rotylenchulus reniformis CEP1 encodes a biologically active peptide hormone
The current paradigm suggests that CEPs in plants have evolved as part of an adaptive response to nitrogen stress (Tabata et al., 2014); therefore, the ability of a CEP domain encoded by RrCEP1 to up‐regulate the expression of an Arabidopsis nitrate transporter (NRT2.1) was tested. Synthetic peptides corresponding to the Arabidopsis CEP5 domain (AtCEP5, positive control) and to the first domain in RrCEP1 (RrCEP1.1) with hydroxyprolines in positions 4 and 11 were exogenously applied to Arabidopsis seedlings in plant growth medium. A randomized RrCEP1.1 peptide acted as a negative control. RrCEP1.1 significantly up‐regulated NRT2.1 expression (Fig. 5, P < 0.05, n = 4), a response similar to, but lower in magnitude than, that induced by the endogenous plant CEP (AtCEP5), thus indicating that a CEP effector of R. reniformis encodes a biologically active peptide hormone. No effect was observed with the randomized RrCEP1.1. Up‐regulation of host nitrate transport provides a plausible benefit to both plant and animal.
Figure 5.
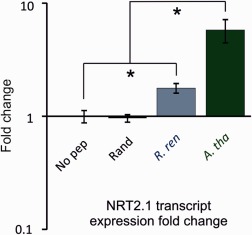
Exogenous application of synthetic Rotylenchulus reniformis CEP1.1 domain peptide up‐regulates nitrate transporter AtNRT2.1 mRNA. Synthetic peptides corresponding to the RrCEP1.1 (R. ren) and AtCEP5 (A. tha) C‐terminally‐encoded peptide (CEP) domains at 1 μm significantly increase the transcript abundance of Arabidopsis NRT2.1 compared with the no‐peptide control (No pep) (n = 3–4, *P < 0.05). No such effect is observed with an equimolar concentration of a randomized RrCEP1.1 peptide (Rand).
The dual roles of CEPs
CEPs also inhibit root proliferation by limiting the rate of cell division (Delay et al., 2013). Utilizing the well‐established system of reduction in Arabidopsis primary root length as a proxy for the inhibition of cell division, we were able to demonstrate that the RrCEP1.1 domain, the M. hapla CEP2 (MhCEP2) and the Md. truncatula CEP1.1 (MtCEP1.1) inhibit cell division, similarly to Arabidopsis CEP5 (Fig. 6A,B, P < 0.001 accounting for multiple t‐tests, n = 21–27). No such inhibition was observed with a randomized RrCEP1.1 peptide (Fig. 6A). In addition, the plant response to both RrCEP1.1 and AtCEP5 is dose dependent, as expected for a bona fide hormone ligand (Fig. 6C). No effect of increasing concentration was observed with the randomized RrCEP1.1 (Fig. 6C). Substituting canonical hydroxyprolines in positions 4 and 11 with Pro markedly reduced the magnitude of the host response for both AtCEP5 and RrCEP1.1 (Fig. S3, see Supporting Information). A similar requirement of hydroxyprolines in positions 4 and 11 of CEP peptides has been described previously (Delay et al., 2013, Imin et al., 2013), but it is not clear whether CEP domains lacking this hydroxylation are affected in receptor binding (Tabata et al., 2014), signal transduction or folding (Bobay et al., 2013). The inclusion of both RrCEP1.1 and AtCEP5 at equimolar concentrations produced the same effect as AtCEP5 alone (Fig. 6C), suggesting that RrCEP1.1 with hydroxyprolines in positions 4 and 11 is not refractory and does not block the CEP receptor when in competition with endogenous plant CEPs, but functions similarly to reduce the cell division rate.
Figure 6.
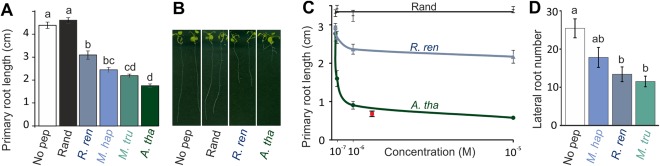
Exogenous application of synthetic Rotylenchulus reniformis CEP1.1 domain peptide affects root development in a dose‐dependent manner. (A) Synthetic peptides corresponding to the C‐terminally‐encoded peptide (CEP) domains of RrCEP1.1 (R. reniformis, R. ren), AtCEP5 (Arabidopsis thaliana, A. tha), MhCEP2 (Meloidogyne hapla, M. hap) and MtCEP1.1 (Medicago truncatula, M. tru) (1 μm) significantly reduce the primary root length of A. thaliana compared with a randomized RrCEP1.1 (Rand) or a no‐peptide control (No pep) at 15 days post‐germination (n = 21–27, lower case letters indicate homogeneous subsets, P < 0.001 accounting for multiple t‐tests; error bars indicate standard error of the mean). (B) Representative primary root length at 15 days post‐germination. (C) Increasing concentrations of both RrCEP1.1 and AtCEP5 increase the magnitude of the response. No response is observed for the randomized peptide, even at 10 μm. Combined application of RrCEP1.1 and AtCEP5 at equimolar concentrations (red, 1 μm each) does not reduce the effect of AtCEP5, suggesting that R. reniformis CEPs are not refractory and do not block the CEP receptor when in competition with endogenous plant CEPs, but function similarly to reduce the cell division rate. (D) RrCEP1.1 and MtCEP1.1, but not MhCEP2, reduce the lateral root number on Medicago truncatula compared with wild‐type (n = 16–23, lower case letters indicate homogeneous subsets, P < 0.002 accounting for multiple t‐tests; error bars indicate standard error of the mean).
Exploring CEP loss of function in R. reniformis is technically intractable; the efficacy of nematode gene knockdown by RNA interference (RNAi) is complicated by large gene families and exacerbated by redundancy, whereas the lack of a known host CEP receptor for the inhibition of cell division phenotype (Tabata et al., 2014) precludes the use of host mutants in the CEP response. During the induction of cyst nematode syncytia, a cell division event occurs in adjacent cells prior to their incorporation into the expanding feeding site (de Almeida Engler and Gheysen, 2013; Golinowski et al., 1996). Treatment of infected Arabidopsis roots with the mitotic inhibitor oryzalin prevented the division of neighbouring cells, resulting in smaller, narrower syncytia than in untreated roots (de Almeida Engler et al., 1999). Therefore, it might be expected that syncytium size is reduced in a CEP‐rich environment. The application of CEPs in a CEP‐negative state is required, given that the application of CEPs in an already CEP‐positive environment (R. reniformis infection of wild‐type Arabidopsis), either by exogenous application or over‐expression, is unlikely to be informative. Thus, CEP gain of function was explored in the cyst nematode species Heterodera schachtii. Heterodera schachtii forms a syncytial feeding site that involves cell division prior to incorporation, is able to infect Arabidopsis and, based on current knowledge, does not contain CEPs. These nematodes were therefore allowed to infect an Arabidopsis host in the presence of exogenous CEP peptide. When either RrCEP1.1 or AtCEP5 was included in the growth medium, syncytia induced by H. schachtii were significantly smaller than those induced in roots with no peptide (Fig. 7A,B, P < 0.0002, n = 45–59). This may indicate that an apparent cost associated with the production of CEPs to up‐regulate nitrate transporter expression is a reduced ability to create a large feeding site. Consistent with this, formation of the simple R. reniformis syncytium does not involve hyperplasia of cells (Robinson, 2007) and additional lateral cell layers do not appear to be incorporated (Jones and Dropkin, 1975). The syncytium typically comprises just a single layer of cells in the pericycle (Agudelo et al., 2005). Importantly, this apparent ‘trade‐off’ would not be valid for the other CEP‐containing nematode group (the root‐knot nematodes), as progression through the cell cycle to cytokinesis is inhibited in the normal formation of their feeding sites, the giant cells. An important consideration for both groups of CEP‐containing nematodes is the number of individuals required to produce a host response.
Figure 7.
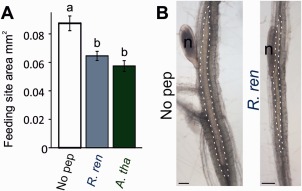
Allowing non‐C‐terminally‐encoded peptide (CEP)‐containing nematodes to infect in a CEP‐rich environment is detrimental to the size of the feeding site produced. (A) The size of the feeding site produced by Heterodera schachtii on Arabidopsis thaliana at 15 days post‐infection is significantly reduced when induced in the presence of 1 μm exogenous RrCEP1.1 (R. ren) or AtCEP5 (A. tha) (n = 45–59, lower case letters indicate homogeneous subsets, P < 0.0005 accounting for multiple t‐tests; error bars indicate standard error of the mean). (B) Representative syncytial size comparison between no‐peptide control (No pep) and RrCEP1.1 (n, nematode; syncytial boundaries are indicated by the broken white line).
Relative to the effects of AtCEP5, both NRT2.1 transcript up‐regulation and inhibition of root growth are similarly smaller in magnitude in response to RrCEP1.1. Although RrCEP1.1 and AtCEP5 differ by only three amino acids, CEPs in both A. thaliana and R. reniformis are within large gene families. It will be interesting to explore whether other R. reniformis CEPs are more effective at either of the dual roles for this particular plant species, or on any of its >300 host species (Robinson et al., 1998). RrCEP1.1, but not MhCEP2, modified root architecture in an additional host species, Md. truncatula (Fig. 6D), yet both were functional in A. thaliana, providing an indication that nematode CEPs may vary in efficacy depending on the host.
Rotylenchulus reniformis has one of the shortest life cycles amongst sedentary PPNs (Robinson et al., 1998), where eggs deposited by females hatch and, because of their lack of macro‐mobility, often re‐infect the same host, resulting in a single plant actively supporting thousands of closely related individuals. Thus, the presence of CEPs in this syncytial‐forming species may represent an adaptation to obligate dependence on host survival of individuals and their progeny for up to 10 generations (Barker et al., 1994; Karam et al., 2006). We hypothesize that CEPs of R. reniformis may contribute to a reduced syncytial size, and increase host nitrate uptake, thus reducing the overall drain on the host during infection.
Experimental Procedures
Nematode growth and collection
Rotylenchulus reniformis (Linford & Oliveira) was maintained on cotton plants (Gossypium hirsutum cv. Coker 201) growing in a mix of 2 : 1 loam soil : sand in a glasshouse at 25–27 ºC with a 16‐h day length. Sedentary females were extracted from washed roots at 8–10 weeks post‐infection. Clean root tissue was cut into approximately 2‐cm lengths and subjected to a short 2‐s burst in a Waring blender. The contents were then passed over a series of sieves and nematodes collected from the 150‐ and 63‐μm sieves were cleaned by sucrose (40%, w/v) centrifugation. Nematodes were extensively washed in tap water and individual young adult females were collected and flash frozen in liquid nitrogen. J2s were hatched from eggs collected from infected roots. Washed root tissue was blended as above for 10 s and homogenized tissue was passed over 150‐, 63‐ and 25‐μm sieves. Eggs were collected from the 25‐μm sieve, cleaned by sucrose centrifugation, washed and sterilized for 20 min in an appropriate volume of hexadecyltrimethylammonium bromide (CTAB, 0.5 mg/mL, Sigma, Dorset, UK) containing 0.1% v/v chlorhexidine digluconate (Sigma) and 0.01% v/v Tween‐20, followed by three washes in sterile tap water. Eggs were allowed to hatch at room temperature in water over a 25‐μm mesh. J2 nematodes were collected daily from under the 25‐μm mesh, cleaned as described above and flash frozen.
RNAseq and transcriptome assembly
Total RNA was extracted from three biological replicate samples each of sedentary female and J2 nematodes using an RNeasy Mini kit (Qiagen, Manchester, UK) according to the manufacturer's instructions with an on‐column DNase I digestion. RNA quality was determined using a Bioanalyser (Agilent Technologies, Santa Clara, CA, USA). Samples with an RNA Integrity Number (RIN) > 8 were employed for library construction and sequencing using the service provided by The Genome Analysis Centre (TGAC, Norwich, UK), as described previously (Eves‐van den Akker et al., 2014a). Raw reads, trimmed of adapter sequences and low‐quality bases (q < 30), were assembled de novo using Trinity (Grabherr et al., 2011) with default parameters.
Identification and cloning of R. reniformis CEP genes
Rotylenchulus reniformis transcripts encoding CEP‐like sequences were identified from the assembled transcriptome by regular expression search as described previously (Eves‐van den Akker et al., 2014a). Three transcripts encoding CEP‐like sequences were identified (Dryad accession doi:10.5061/dryad.q8h75). Forward (ATGAAATTGACCTTAATTTTGATGCT) and reverse (TCAATGTCCATCTCCAGGAGAATG) oligonucleotide primers were designed to clone the genomic DNA sequence corresponding to comp45145_c2_seq7. DNA was isolated from J2s, collected as above and extracted as described previously (Eves‐van den Akker et al., 2014b). Sequences of interest were amplified using Phusion Hi‐fidelity proofreading polymerase, following the manufacturer's instructions (New England Biolabs, Hitchin, Herts, UK). 5′ Adenosines were added to PCR products by incubation at 72 ºC with the addition of 1 µL BioTaq DNA polymerase (Bioline, London, UK), and ligated into the pGEM‐T Easy vector (Promega, Southampton, UK). Positive clones were confirmed by sequencing. Amino acid sequences were deduced by similarity to sequences in the transcriptome assembly.
Phylogenetic and motif analyses
Cloned genomic sequences which encoded complete open reading frames, were unique at the protein level (or, where identical, contained considerably different introns) and were different in total gene length were deemed to arise from unique loci and were used to construct a preliminary phylogeny of the gene family. Complete genomic sequences were aligned and refined using MUSCLE (Edgar, 2004), and used to construct a Bayesian phylogeny of the gene family employing 300 000 generations in TOPALi (Milne et al., 2009). Based on the above criteria, highly similar sequences were collapsed into a single representative displayed in Fig. 1. CEP domain motif plots and di‐basic residue plots were generated using MEME (Bailey et al., 2009). The putative ‘cryptic signal peptide’ was identified by analysing a 50‐base‐pair sliding window across the protein sequence of RrCEP1 using SignalP v4.0 (Petersen et al., 2011)
Differential expression and in situ hybridization
Trimmed RNAseq reads were mapped back to the transcriptome using tophat2 (Kim et al., 2013). Read counts were TMM normalized, and differentially expressed transcripts were identified using the trinity wrapper scripts for EdgeR, specifying a minimum P value of 0.001 and a minimum fold change of four. In situ hybridization was carried out with sedentary‐stage adult females of R. reniformis, fixed and extracted from the roots of cotton plants according to previously described methods (Eves‐van den Akker et al., 2014b). Cut nematodes were treated with proteinase K (2 mg/mL) for 1 h at room temperature and then the in situ hybridization protocol was continued from the dehydration step, as described by de Boer et al. (1998). Single‐stranded, digoxygenin‐labelled hybridization probes, 140 bp in length, were synthesized as described previously (Eves‐van den Akker et al., 2014b) using forward (CTGGACGGCTTTTAGTGCAC) and reverse (CAGTCAGAACGCGTGCAAAA) primers to generate sense and anti‐sense probes, respectively.
Plant growth and root analysis
Surface‐sterilized seeds of Arabidopsis thaliana (Col‐0) were stratified for 1–2 days and sown onto 9‐cm‐diameter Petri plates containing 25 mL of half‐strength Murashige and Skoog (MS) medium adjusted to pH 5.7 and solidified with 1% phytagel. Where indicated, synthetic peptides corresponding to AtCEP5 (DFR‐hyP‐TTPGHS‐hyP‐GIGH), MtCEP1.1 (AFQ‐hyPTTPGNS‐hyPGVGH), MhCEP2 (AFR‐hyPTAPGHS‐hyPGVGH), RrCEP1.1 (AFK‐hyP‐TTPGHS‐hyP‐GAGH) or its randomized version RrCEP1.1rand (TA‐hyP‐GTHGP‐hyP‐SFAKGH) at 1 mm in 50 mm sodium phosphate buffer, pH 6, were added to the medium immediately before pouring to achieve the desired final concentration. Plates were held vertically in a growth chamber (Microclima, Snijders Scientific, Tilburg, The Netherlands) at 20 ºC with a 16‐h day length at 15 μmol/m2/s photosynthetically active radiation and 70% humidity. Plates were imaged using an HP scanner and primary root lengths were calculated from saved images using Image‐Pro Analyser v7 (MediaCybernetics, Rockville, MD, USA). Significant differences in root lengths between treatments were determined by analysis of variance (ANOVA) with a Tukey post hoc test. Medicago truncatula seedlings were prepared as above, but grown on Fahraeus medium with no nitrogen (according to Imin et al., 2013) under 11 μmol/m2/s of photosynthetically active radiation.
Nematode infection and feeding site measurement
Seedlings of Arabidopsis were grown as described above with the addition of 1% sucrose to the medium. J2s of the beet cyst nematode H. schachtii were hatched from cysts, sterilized as described previously (Davies et al., 2012) and then resuspended in 0.01% Tween‐20 at a concentration of approximately one nematode/µL. At 12 days post‐imbibition, three root tips per plant were each inoculated with 20 µL of nematode suspension with two plants per plate. Infection points were covered with GF/A paper (Whatman, Maidstone, Kent, UK) for 2 days to facilitate invasion. All root lengths harbouring syncytia at 15 days post‐infection (dpi) were excised, mounted on microscope slides and images were captured using an Olympus, Southend, Essex, UK BH2 microscope with a MicroPublisher 3.3 RTV camera and Q‐Capture Pro software (QImaging, Surrey, BC, Canada). Syncytium sizes were estimated from the projected cross‐sections and measured using Image‐Pro Analyser v7 (MediaCybernetics). At least 45 individual syncytia per treatment were measured for plants growing on unsupplemented medium or on plates containing 1 μm AtCEP5 or 1 μm RrCEP1.1.
RNA extraction, cDNA synthesis and quantitative reverse transcriptase‐polymerase chain reaction (qRT‐PCR) analysis
Total RNA was isolated at 16 days post‐imbibition from the pooled roots of six Arabidopsis seedlings per treatment using an RNeasy Plant Mini Kit (Qiagen) according to the manufacturer's instructions. An on‐column DNase treatment was carried out. cDNA was prepared from 500 ng of RNA using SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA, USA) and an anchored oligo(dT) primer according to the manufacturer's instructions. qRT‐PCR was carried out on the resulting cDNA using Brilliant III SYBR® Green Master Mix (Agilent Technologies) to determine the expression level of the high‐affinity nitrate transporter NRT2.1 (At1g08090). qRT‐PCR conditions were as follows: 95 ºC for 3 min, followed by 40 cycles of 95 ºC for 30 s, 60 ºC for 30 s and 72 ºC for 30 s. Fluorescence data were collected at the end of the annealing phase. Primer pairs for NRT2.1 (FOR, 5′‐TCTTTTGGGTCCCCGTTACG‐3′; REV, 5′‐CGCCAGGCAAAAACCAATCA‐3′) and Elongation Factor 1‐α At5g60390 (FOR, 5′‐GACAGGCGTTCTGGTAAGGA‐3′; REV, 5′‐GCTTGGTTGGGGTCATCTTA‐3′) had amplification efficiencies of 90%–110%. Relative expression was determined by the efficiency‐corrected ΔΔCT method from a minimum of three biological replicates per treatment, each with three technical replicates and using elongation factor for normalization.
Accession numbers
Raw RNAseq reads are available under SRA accession PRJEB8325. Transcripts encoding CEP‐like sequences identified in the transcriptome, all cloned CEP sequences and the CEP phylogeny .tre file are available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.q8h75
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Insight into the genomic organization of Rotylenchulus reniformis C‐terminally‐encoded peptides (CEPs). Schematic diagram to show the organization of two RrCEP genes cloned in a tandem array as a single amplification product.
Fig. S2 Amino acid features of RrCEP1.1. Using a 50‐bp sliding window across the full length of RrCEP1.1, two putative signal peptide cleavage sites are identified (SignalP v4.0): one at the N‐terminus (blue) and a second prior to the first (internal) C‐terminally‐encoded peptide (CEP) domain. Signal peptide cleavage sites were also identified in equivalent positions for the other RrCEPs. CEP domains are highlighted in red, di‐basic residues in orange and a conserved KND motif of unknown function present prior to CEP domains in green.
Fig. S3 Comparison of the activity of RrCEP1.1 and AtCEP5 with and without hydroxyprolines. Substituting canonical hydroxyprolines in positions 4 and 11 with proline markedly reduced the magnitude of the host response for both AtCEP5 and RrCEP1.1 (Student's t‐test, n = 36–40, error bars indicate standard error of the mean).
Acknowledgements
This work was supported by the Biotechnology and Biological Sciences Research Council (BBSRC) and The James Hutton Institute through a PhD studentship to SE‐vdA. The James Hutton Institute receives funding from the Scottish Government Rural and Environment Science and Analytical Services Division. SE‐vdA is supported by BBSRC grant BB/M014207/1. We thank Dr Martin Wubben (USDA‐ARS, East Mississippi State, MS, USA) for kindly providing R. reniformis.
References
- Agudelo, P. , Robbins, R.T. , Stewart, J.M. , Bell, A. and Robinson, A.F. (2005) Histological observations of Rotylenchulus reniformis on Gossypium longicalyx and interspecific cotton hybrids. J. Nematol. 37, 444–447. [PMC free article] [PubMed] [Google Scholar]
- de Almeida Engler, J. and Gheysen, G. (2013) Nematode‐induced endoreduplication in plant host cells: why and how? Mol. Plant–Microbe Interact. 26, 17–24. [DOI] [PubMed] [Google Scholar]
- de Almeida Engler, J. , De Vleesschauwer, V. , Burssens, S. , Celenza, J.L. , Inzé, D. , Van Montagu, M. , Engler, G. and Gheysen, G. (1999) Molecular markers and cell cycle inhibitors show the importance of cell cycle progression in nematode‐induced galls and syncytia. Plant Cell, 11, 793–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, T.L. , Boden, M. , Buske, F.A. , Frith, M. , Grant, C.E. , Clementi, L. , Ren, J. , Li, W.W. and Noble, W.S. (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker, K. , Konning, S. and Walters, S. (1994) Effect of soil type on the reproductive potential of Meloidogyne incognita and Rotylenchulus reniformis on cotton and related effects on crop maturity. J. Nematol. 26, 91–126. [PMC free article] [PubMed] [Google Scholar]
- Bird, D.M. , Jones, J.T. , Opperman, C.H. , Kikuchi, T. and Danchin, E.G. (2015) Signatures of adaptation to plant parasitism in nematode genomes. Parasitology, 142, S71–S84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobay, B.G. , DiGennaro, P. , Scholl, E. , Imin, N. , Djordjevic, M.A. and Bird, D.M. (2013) Solution NMR studies of the plant peptide hormone CEP inform function. FEBS Lett. 587, 3979–3985. [DOI] [PubMed] [Google Scholar]
- de Boer, J.M. , Yan, Y. , Smant, G. , Davis, E.L. and Baum, T.J. (1998) In situ hybridization to messenger RNA in Heterodera glycines . J Nematol. 30, 309–312. [PMC free article] [PubMed] [Google Scholar]
- Davies, L.J. , Lilley, C.J. , Knox, J.P. and Urwin, P.E. (2012) Syncytia formed by adult female Heterodera schachtii in Arabidopsis thaliana roots have a distinct cell wall molecular architecture. New Phytol. 196, 238–246. [DOI] [PubMed] [Google Scholar]
- Delay, C. , Imin, N. and Djordjevic, M.A. (2013) CEP genes regulate root and shoot development in response to environmental cues and are specific to seed plants. J. Exp. Bot. 64, 5383–5394. [DOI] [PubMed] [Google Scholar]
- DeYoung, B.J. and Clark, S.E. (2001) Signaling through the CLAVATA1 receptor complex. Plant Mol. Biol. 46, 505–513. [DOI] [PubMed] [Google Scholar]
- Edgar, R.C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eves‐van den Akker, S. , Lilley, C. , Danchin, E. , Rancurel, C. , Cock, P. , Urwin, P. and Jones, J.T. (2014a) The transcriptome of Nacobbus aberrans reveals insights into the evolution of sedentary endoparasitism in plant‐parasitic nematodes. Genome Biol. Evol. 6, 2181–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eves‐van den Akker, S. , Lilley, C.J. , Jones, J.T. and Urwin, P.E. (2014b) Identification and characterisation of a hyper‐variable apoplastic effector gene family of the potato cyst nematodes. PLoS Pathog. 10, e1004391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golinowski, W. , Grundler, F. and Sobczak, M. (1996) Changes in the structure of Arabidopsis thaliana during female development of the plant‐parasitic nematode Heterodera schachtii . Protoplasma, 194, 103–116. [Google Scholar]
- Grabherr, M.G. , Haas, B.J. , Yassour, M. , Levin, J.Z. , Thompson, D.A. , Amit, I. , Adiconis, X. , Fan, L. , Raychowdhury, R. , Zeng, Q. , Chen, Z. , Mauceli, E. , Hacohen, N. , Gnirke, A. , Rhind, N. , di Palma, F. , Birren, B.W. , Nusbaum, C. , Lindblad‐Toh, K. , Friedman, N. and Regev A. (2011) Full‐length transcriptome assembly from RNA‐Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grymaszewska, G. and Golinowski, W. (1998) Structure of syncytia induced by Heterodera schachtii Schmidt in roots of susceptible and resistant radish (Raphanus sativus L. var. oleiformis). Acta. Soc. Bot. Pol. 67, 207–216. [Google Scholar]
- Hewezi, T. (2015) Cellular signaling pathways and posttranslational modifications mediated by nematode effector proteins. Plant Physiol. 169, 1018–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtmann, B. , Kleine, M. and Grundler, F.M.W. (2000) Ultrastructure and anatomy of nematode‐induced syncytia in roots of susceptible and resistant sugar beet. Protoplasma, 211, 39–50. [Google Scholar]
- Imin, N. , Mohd‐Radzman, N.A. , Ogilvie, H.A. and Djordjevic, M.A. (2013) The peptide‐encoding CEP1 gene modulates lateral root and nodule numbers in Medicago truncatula . J. Exp. Bot. 64, 5395–5409. [DOI] [PubMed] [Google Scholar]
- Jones, M. and Dropkin, V. (1975) Cellular alterations induced in soybean roots by three endoparasitic nematodes. Physiol. Plant Pathol. 5, 119–124. [Google Scholar]
- Jones, M.G.K. and Payne, H.L. (1977) Structure of syncytia induced by phytoparasitic nematode Nacobbus aberrans in tomato roots, and possible role of plasmodesmata in their nutrition. J. Cell Sci. 23, 299–313. [DOI] [PubMed] [Google Scholar]
- Karam, F. , Lahoud, R. , Masaad, R. , Daccache, A. , Mounzer, O. and Rouphael, Y. (2006) Water use and lint yield response of drip irrigated cotton to the length of irrigation season. Agric. Water Manage. 85, 287–295. [Google Scholar]
- Kim, D. , Pertea, G. , Trapnell, C. , Pimentel, H. , Kelley, R. and Salzberg, S.L. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, S.‐W. , Chen, S. , Wang, J. , Yu, H. , Chronis, D. , Mitchum, M.G. and Wang, X. (2009) Structural and functional diversity of CLAVATA3/ESR (CLE)‐like genes from the potato cyst nematode Globodera rostochiensis . Mol. Plant–Microbe Interact. 22, 1128–1142. [DOI] [PubMed] [Google Scholar]
- Matos, J.L. , Fiori, C.S. , Silva‐Filho, M.C. and Moura, D.S. (2008) A conserved dibasic site is essential for correct processing of the peptide hormone AtRALF1 in Arabidopsis thaliana . FEBS Lett. 582, 3343–3347. [DOI] [PubMed] [Google Scholar]
- Matsubayashi, Y. (2014) Posttranslationally modified small‐peptide signals in plants. Annu. Rev. Plant Biol. 65, 385–413. [DOI] [PubMed] [Google Scholar]
- van Megen, H. , van den Elsen, S. , Holterman, M. , Karssen, G. , Mooyman, P. , Bongers, T. , Holovachov, O. , Bakker, J. and Helder, J. (2009) A phylogenetic tree of nematodes based on about 1200 full‐length small subunit ribosomal DNA sequences. Nematology, 11, 927–950. [Google Scholar]
- Milne, I. , Lindner, D. , Bayer, M. , Husmeier, D. , McGuire, G. , Marshall, D.F. and Wright, F. (2009) TOPALi v2: a rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi‐core desktops. Bioinformatics, 25, 126–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchum, M.G. , Wang, X. , Wang, J. and Davis, E.L. (2012) Role of nematode peptides and other small molecules in plant parasitism. Annu. Rev. Phytopathol. 50, 175–195. [DOI] [PubMed] [Google Scholar]
- Mitchum, M.G. , Hussey, R.S. , Baum, T.J. , Wang, X. , Elling, A.A. , Wubben, M. and Davis, E.L. (2013) Nematode effector proteins: an emerging paradigm of parasitism. New Phytol. 199, 879–894. [DOI] [PubMed] [Google Scholar]
- Mohd‐Radzman, N.A. , Binos, S. , Truong, T.T. , Imin, N. , Mariani, M. and Djordjevic, M.A. (2015) Novel MtCEP1 peptides produced in vivo differentially regulate root development in Medicago truncatula . J. Exp. Bot. 66, 5289–5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogilvie, H.A. , Imin, N. and Djordjevic, M.A. (2014) Diversification of the C‐TERMINALLY ENCODED PEPTIDE (CEP) gene family in angiosperms, and evolution of plant‐family specific CEP genes. BMC Genomics, 15, 870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama, K. , Ogawa, M. and Matsubayashi, Y. (2008) Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC‐MS‐based structure analysis. Plant J. 55, 152–160. [DOI] [PubMed] [Google Scholar]
- Petersen, T.N. , Brunak, S. , von Heijne, G. and Nielsen, H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods, 8, 785–786. [DOI] [PubMed] [Google Scholar]
- Rahman Razak, A. and Evans, A. (1976) An intracellular tube associated with feeding by Rotylenchulus reniformis on cowpea root. Nematologica, 22, 182–189. [Google Scholar]
- Replogle, A. , Wang, J. , Bleckmann, A. , Hussey, R.S. , Baum, T.J. , Sawa, S. , Davis, E.L. , Wang, X. , Simon, R. and Mitchum, M.G. (2011) Nematode CLE signaling in Arabidopsis requires CLAVATA2 and CORYNE. Plant J. 65, 430–440. [DOI] [PubMed] [Google Scholar]
- Roberts, G.A. , Chen, K. , Cooper, L.P. , White, J.H. , Blakely, G.W. and Dryden, D.T. (2012) Removal of a frameshift between the hsdM and hsdS genes of the EcoKI Type IA DNA restriction and modification system produces a new type of system and links the different families of Type I systems. Nucleic Acids Res. 40, 10 916–10 924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, I. , Smith, S. , De Rybel, B. , Van Den Broeke, J. , Smet, W. , De Cokere, S. , Mispelaere, M. , De Smet, I. and Beeckman, T. (2013) The CEP family in land plants: evolutionary analyses, expression studies, and a role in Arabidopsis shoot development. J. Exp. Bot. 64, 5371–5381. [DOI] [PubMed] [Google Scholar]
- Robinson, A.F. (2007) Reniform in US cotton: when, where, why, and some remedies. Annu. Rev. Phytopathol. 45, 263–288. [DOI] [PubMed] [Google Scholar]
- Robinson, A.F. , Inserra, R.N. , Caswell‐Chen, E.P. , Vovlas, N. and Troccoli, A. (1998) Rotylenchulus species: identification, distribution, host ranges, and crop plant resistance. Nematropica, 27, 127–180. [Google Scholar]
- Sobczak, M. and Golinowski, W. (2011) Cyst nematodes and syncytia In: Genomics and Molecular Genetics of Plant–Nematode Interactions (Jones J., Gheysen G. and Fenoll C., eds.), Springer, Dordrecht, The Netherlands pp. 61–82.
- Srivastava, R. , Liu, J.X. , Guo, H. , Yin, Y. and Howell, S.H. (2009) Regulation and processing of a plant peptide hormone, AtRALF23, in Arabidopsis. Plant J. 59, 930–939. [DOI] [PubMed] [Google Scholar]
- Tabata, R. , Sumida, K. , Yoshii, T. , Ohyama, K. , Shinohara, H. and Matsubayashi, Y. (2014) Perception of root‐derived peptides by shoot LRR‐RKs mediates systemic N‐demand signaling. Science, 346, 343–346. [DOI] [PubMed] [Google Scholar]
- Vieira, P. , Danchin, E.G.J. , Neveu, C. , Crozat, C. , Jaubert, S. , Hussey, R.S. , Engler, G. , Abad, P. , de Almeida‐Engler, J. , Castagnone‐Sereno, P. and Rosso, M.N. (2011) The plant apoplasm is an important recipient compartment for nematode secreted proteins. J. Exp. Bot. 62, 1241–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Lee, C. , Replogle, A. , Joshi, S. , Korkin, D. , Hussey, R. , Baum, T.J. , Davis, E.L. , Wang, X. and Mitchum, M.G. (2010) Dual roles for the variable domain in protein trafficking and host specific recognition of Heterodera glycines CLE effector proteins. New Phytol. 187, 1003–1017. [DOI] [PubMed] [Google Scholar]
- Wang, X.H. , Mitchum, M.G. , Gao, B.L. , Li, C.Y. , Diab, H. , Baum, T.J. , Hussey, R.S. and Davis, E.L. (2005) A parasitism gene from a plant‐parasitic nematode with function similar to CLAVATA3/ESR (CLE) of Arabidopsis thaliana . Mol. Plant Pathol. 6, 187–191. [DOI] [PubMed] [Google Scholar]
- Wubben, M.J. , Gavilano, L. , Baum, T.J. and Davis, E.L. (2015) Sequence and spatiotemporal expression analysis of CLE‐motif containing genes from the reniform nematode (Rotylenchulus reniformis Linford & Oliveira). J. Nematol. 47, 159. [PMC free article] [PubMed] [Google Scholar]
- Yang, H. , Matsubayashi, Y. , Nakamura, K. and Sakagami, Y. (2001) Diversity of Arabidopsis genes encoding precursors for phytosulfokine, a peptide growth factor. Plant Physiol. 127, 842–851. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Insight into the genomic organization of Rotylenchulus reniformis C‐terminally‐encoded peptides (CEPs). Schematic diagram to show the organization of two RrCEP genes cloned in a tandem array as a single amplification product.
Fig. S2 Amino acid features of RrCEP1.1. Using a 50‐bp sliding window across the full length of RrCEP1.1, two putative signal peptide cleavage sites are identified (SignalP v4.0): one at the N‐terminus (blue) and a second prior to the first (internal) C‐terminally‐encoded peptide (CEP) domain. Signal peptide cleavage sites were also identified in equivalent positions for the other RrCEPs. CEP domains are highlighted in red, di‐basic residues in orange and a conserved KND motif of unknown function present prior to CEP domains in green.
Fig. S3 Comparison of the activity of RrCEP1.1 and AtCEP5 with and without hydroxyprolines. Substituting canonical hydroxyprolines in positions 4 and 11 with proline markedly reduced the magnitude of the host response for both AtCEP5 and RrCEP1.1 (Student's t‐test, n = 36–40, error bars indicate standard error of the mean).


