Abstract
Ehrlichia chaffeensis is an obligatory intracellular bacterium that causes a potentially fatal emerging zoonosis, human monocytic ehrlichiosis. E. chaffeensis has a limited capacity for biosynthesis and metabolism and thus depends mostly on host-synthesized nutrients for growth. Although the host cell cytoplasm is rich with these nutrients, as E. chaffeensis is confined within the early endosome-like membrane-bound compartment, only host nutrients that enter the compartment can be used by this bacterium. How this occurs is unknown. We found that ehrlichial replication depended on autophagy induction involving class III phosphatidylinositol 3-kinase (PtdIns3K) activity, BECN1 (Beclin 1), and ATG5 (autophagy-related 5). Ehrlichia acquired host cell preincorporated amino acids in a class III PtdIns3K-dependent manner and ehrlichial growth was enhanced by treatment with rapamycin, an autophagy inducer. Moreover, ATG5 and RAB5A/B/C were routed to ehrlichial inclusions. RAB5A/B/C siRNA knockdown, or overexpression of a RAB5-specific GTPase-activating protein or dominant-negative RAB5A inhibited ehrlichial infection, indicating the critical role of GTP-bound RAB5 during infection. Both native and ectopically expressed ehrlichial type IV secretion effector protein, Etf-1, bound RAB5 and the autophagy-initiating class III PtdIns3K complex, PIK3C3/VPS34, and BECN1, and homed to ehrlichial inclusions. Ectopically expressed Etf-1 activated class III PtdIns3K as in E. chaffeensis infection and induced autophagosome formation, cleared an aggregation-prone mutant huntingtin protein in a class III PtdIns3K-dependent manner, and enhanced ehrlichial proliferation. These data support the notion that E. chaffeensis secretes Etf-1 to induce autophagy to repurpose the host cytoplasm and capture nutrients for its growth through RAB5 and class III PtdIns3K, while avoiding autolysosomal killing.
Keywords: ATG5, autophagy, BECN1, class III PtdIns3K, Ehrlichia chaffeensis, endosome, Etf-1, infection, LC3, RAB5, type IV secretion effector
Introduction
Ehrlichia chaffeensis, a Gram-negative obligatory intracellular bacterium in the family Anaplasmataceae, primarily infects monocytes and macrophages in mammals and causes the emerging tick-borne zoonosis called human monocytic ehrlichiosis.1-3 This serious and sometimes fatal disease is characterized by fever, headache, myalgia, thrombocytopenia, leukopenia, and elevated liver enzyme levels.3-5
One fundamental virulence factor of microbial pathogens is “nutritional virulence,”6 i.e., the ability to acquire nutrients for pathogen proliferation in competition with hosts and possible other microbes. Many pathogens thrive on extracellular nutrients available in the blood and mucosal surface and on the host cellular lysate. However, E. chaffeensis, as an obligatory intracellular bacterium, acquires nutrients inside host cells which are kept alive until bacteria fully proliferate and mature (reviewed in ref. 7). The host cell-dependency of E. chaffeensis is so extreme that unlike facultative intracellular bacteria such as Legionella pneumophila or Mycobacterium tuberculosis, which can be cultured axenically, E. chaffeensis cannot replicate or even survive outside of host cells for more than a few hours. The intracellular membrane compartment (inclusion) that contains E. chaffeensis has early endosome–like characteristics, including the small GTPase RAB5, a RAB5 effector EEA1 (early endosome antigen 1), TF (transferrin), TFRC (transferrin receptor), and vacuolar-type H+-ATPase, but it lacks late endosomal or lysosomal markers or NADPH oxidase components.8-10 Within this compartment, E. chaffeensis acquires all nutrients for its reproduction to form numerous mature infectious forms. E. chaffeensis has a small genome of 1.176 Mb with a limited capacity for biosynthesis and metabolism.11 Consequently it must depend mostly on host-synthesized nutrients for replication. Although the host cell cytoplasm is rich with these nutrients, it is unlikely that the inclusion membrane is leaky, as ehrlichial inclusions maintain a weakly acidic intralumenal pH.8 It is also unlikely that varieties of active transporters are synthesized and assembled in the proper orientation on the inclusion membrane to import host nutrients during ehrlichial replication. Considering these limitations, it is possible that a novel mechanism has evolved in this group of obligatory intracellular bacteria to acquire nutrients.
Autophagy is an essential and highly regulated eukaryotic cellular homeostatic process that sequesters and digests/recycles intracellular components.12-14 Multiple membrane sources (plasma membrane, ER/endoplasmic reticulum, mitochondria, endosomes, Golgi) and regulatory factors are involved in the process of autophagy to recognize various cargos and activate downstream components of various pathways (reviewed in ref. 15). A number of ATG (autophagy-related) proteins regulate autophagosome biogenesis and maturation (reviewed in refs. 15, 16). In the canonical amino acid starvation-induced pathway in mammalian cells, autophagosome formation is induced by activation of class III PtdIns3K, and an essential component of the PtdIns3K complex, BECN1 (Beclin 1; mammalian ortholog of yeast Vps30/Atg6).17 ATG12, a ubiquitin-like protein that covalently modifies ATG5, and MAP1LC3/LC3 (microtubule-associated protein 1 light chain 3; a mammalian ortholog family of yeast Atg8) are involved in elongation and expansion of autophagosomes.18,19 Biochemical and morphological studies have shown that autophagosomes can fuse with early or late endosomes, forming the amphisome, a hybrid organelle,20-24 although detailed mechanisms of these processes remain, for the most part, unknown. Autophagosomes or amphisomes subsequently fuse with lysosomes to form autolysosomes, where captured substrates are degraded and catabolites are released to the cytosol.15,25,26
Autophagy is an important innate immune response against intracellular infections by bacteria such as Salmonella, Shigella, Listeria, and Mycobacterium.27-31 In contrast, other intracellular bacteria such as Brucella, Francisella, and Coxiella benefit from autophagy.32-34 Brucella abortus-containing double-membrane-enveloped autophagosome formation requires the autophagy initiation proteins ULK1 (unc-51 like autophagy activating kinase 1), BECN1, ATG14, and the class III PtdIns3K, but is independent of ATG5, ATG16L1, ATG4B, ATG7, and MAP1LC3B/LC3B (microtubule-associated protein 1 light chain 3 β).33 In addition, autophagosomes are not required for Brucella replication but are required to complete the intracellular Brucella lifecycle and for cell-to-cell spreading.33 In contrast, Francisella tularensis scavenges intracellular nutrients via ATG5- and LC3-independent noncanonical autophagy.34 Most bacterial factors that induce autophagy for the benefit of bacteria remain unknown.
A tick-borne obligatory intracellular bacterium, Anaplasma phagocytophilum, in the family Anaplasmataceae, replicates in neutrophils in LC3-decorated early autophagosomes that are segregated from the endosomal and lysosomal pathway, thereby gaining access to host cytosolic nutrients while escaping autolysosomal degradation.35-37 However, unlike A. phagocytophilum, E. chaffeensis replicates in the early endosome-like compartment;9 whether or how autophagy is involved in the ehrlichial infection process is unknown.
E. chaffeensis and A. phagocytophilum encode a type IV secretion (T4S) system11,38-41 that mediates the transport of bacterial DNA and/or proteins, referred to as effectors/substrates, across the bacterial membrane into the eukaryotic cell to deregulate or modulate target cell functions.42 Ats-1 (Anaplasma translocated substrate-1), an A. phagocytophilum T4S effector, directly binds BECN1 and induces ER-derived autophagosomes that target and fuse with A. phagocytophilum inclusions to deliver host cytosolic nutrients.37 Other than Ats-1, bacterial factors that induce autophagy to promote bacterial nutrition have not been reported. In this study, we explored roles of autophagy in E. chaffeensis infection and mechanisms by which autophagy is usurped for its replication in the early endosome-like compartment. ECH0825 (here referred to as E. chaffeensis translocated factor 1, Etf-1) is the first experimentally proven T4S effector in the genus Ehrlichia.43 Our findings demonstrate that a unique Etf-1-induced pathway distinct from that of A. phagocytophilum has evolved in E. chaffeensis to co-opt autophagy for proliferation.
Results
E. chaffeensis inclusion membranes are enriched with PtdIns3P and class III PtdIns3K
A RAB5 effector, EEA1, localizes on E. chaffeensis inclusion membranes.9 Because membrane localization of EEA1 is dependent on PtdIns3P (phosphatidylinositol 3-phosphate), we examined whether a 2×FYVE finger protein probe with unique specificity for PtdIns3P in the absence of RAB5 and an ER-localization signal44 is localized to E. chaffeensis replicative inclusions. We used monkey endothelial RF/6A cells, which can be readily infected with E. chaffeensis, are adherent cells with a flattened morphology for unambiguous localization, and can be more effectively transfected than the nonadherent human acute leukemia cell line THP-1.43 Ectopically expressed 2×FYVE-GFP produced large puncta (vesicles) and was localized on ∼80% of E. chaffeensis replicative inclusions at 2 d post-infection (p.i.) (Fig. 1A and C). To further verify the presence of PtdIns3P on E. chaffeensis inclusions, we examined another RAB5 effector, ANKFY1/Rabankyrin-5 (ankyrin repeat and FYVE domain containing 1), the membrane localization of which is also dependent on PtdIns3P.45 GFP-ANKFY1 produced large puncta (vesicles) and accumulated on E. chaffeensis inclusions (Fig. S1). In uninfected cells, GFP-ANKFY1 showed smaller puncta than in infected cells (Fig. S1).
Figure 1.
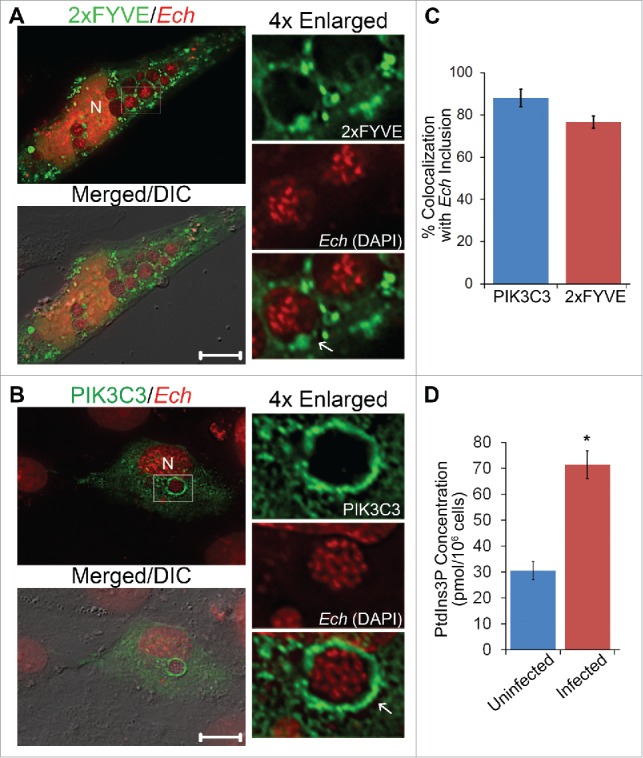
E. chaffeensis inclusion membrane is enriched with PtdIns3P and class III PtdIns3K. (A and B) E. chaffeensis (Ech)-infected RF/6A cells were transfected with plasmids encoding 2×FYVE-GFP or FLAG-PIK3C3/VPS34. At 15 h p.t. (2 d p.i.), cells were fixed and stained with DAPI to indicate E. chaffeensis (pseudocolored in red). PIK3C3 was labeled with mouse anti-FLAG. Merged/DIC, fluorescence image merged with differential interference contrast (DIC) image. Each boxed area is enlarged 4-fold on the right. N, nucleus; scale bars: 10 μm. (C) The percent colocalization of E. chaffeensis inclusions with PIK3C3 or 2×FYVE was determined by counting 10 to 20 inclusions per cell in 5 to 10 cells per experiment from 3 independent experiments. (D) PtdIns3P levels are increased in E. chaffeensis-infected THP-1 cells. Uninfected or E. chaffeensis-infected THP-1 cells (2 × 106 cells) at 1 d p.i. were collected, and PtdIns3P lipids were purified and the amount determined by competitive ELISA. Assays were carried out in triplicate. Data are presented as the mean ± standard deviation. * Significantly different by the Student t test (P < 0.05).
PtdIns3P is the product of activated class III PtdIns3K.46,47 Thus we examined localization of human PIK3C3/VPS34, the catalytic subunit of class III PtdIns3K (mammalian ortholog of yeast Vps34/vacuolar protein sorting 34).48 FLAG-PIK3C3, when transfected at 1 d p.i., was localized on ∼80% of E. chaffeensis replicative inclusions (Fig. 1B and C). Furthermore, cellular PtdIns3P amount was significantly higher in E. chaffeensis-infected THP-1 cells at the beginning and during ehrlichial exponential replication (1 and 2 d p.i., respectively) than in uninfected cells (Fig. 1D, 2 d p.i. data not shown).
E. chaffeensis proliferation requires class III PtdIns3K activation and BECN1, and is enhanced by induction of autophagy with rapamycin
Because PtdIns3P was elevated during E. chaffeensis infection, we next examined whether class III PtdIns3K activation is required for E. chaffeensis endocytosis or proliferation by using 3-methyladenine (3-MA), an inhibitor of class III PtdIns3K.49-51 At a late stage of infection, we used qPCR (quantitative real-time PCR) to analyze the effect, as there were too many bacteria to be counted accurately.43 At the early stage of infection or to analyze binding and entry, a direct counting method by immunofluorescence microscopy was used since qPCR cannot distinguish bound vs. internalized bacteria. Western blot analysis was used to compare knocked-down host protein levels and E. chaffeensis major outer membrane protein P2852 levels relative to controls. All these methods are comparable in quantifying bacteria.53,54 3-MA addition at 0 h did not inhibit E. chaffeensis entry as determined at 2 h p.i. (Fig. 2A). However, when 3-MA was added at 1 h p.i. (immediately after ehrlichial entry) or at 1 d p.i. (at the beginning of exponential growth) and the infected THP-1 cells were continuously incubated in the presence of 3-MA, infection—based on both the percent of infected cells and bacterial numbers per host cell—was greatly inhibited compared with untreated infected cells (Fig. 2B and C). When 3-MA was added to infected cells at 23 h p.i. and incubated for 6 h or 58 h, E. chaffeensis became condensed and were marginalized in enlarged vacuoles (Fig. 2D, panels iv and vi), which was confirmed by staining with an antibody against E. chaffeensis P2852 (Fig. 2E). The vacuoles found in infected cells after 3-MA treatment were specific to E. chaffeensis infection, because similar vacuoles were not seen in uninfected THP-1 cells after 3-MA treatment for 2 d (Fig. S2A). 3-MA did not have direct toxicity on E. chaffeensis: when E. chaffeensis was pretreated with 3-MA and then incubated with THP-1 cells in the absence of 3-MA, there was no inhibitory effect on its ability to infect the cells (Fig. S2B). 3-MA did not have direct toxicity on host mammalian cells, as treatment with 2 mM 3-MA for 1 d had no effect on THP-1 cell viability (Fig. S2A, Table S1). 3-MA does not seem to block intracellular bacterial infection nonspecifically by impairing host cell metabolism, because 3-MA treatment enhances intracellular Mycobacterium growth inside macrophages.31 In addition, 3-MA treatment did not induce lysosomal fusion with ehrlichial inclusions as indicated by an absence of LAMP1 (lysosomal-associated membrane protein 1) colocalization (Fig. S3). The inhibition by 3-MA was reversible, as the inclusions were filled with replicating ehrlichiae at 52 h after 3-MA withdrawal (Fig. 2D, panel v).
Figure 2.

Growth of E. chaffeensis is reversibly inhibited by the class III PtdIns3K inhibitor 3-MA. (A) 3-MA does not inhibit internalization of E. chaffeensis. The percentage of intracellular bacteria vs. total cell-associated bacteria was determined in THP-1 cells incubated with 3-MA or RPMI 1640 medium control (RPMI) at 2 h p.i. by scoring 100 E. chaffeensis bacteria in each group after 2 rounds of immunofluorescence labeling. Data are presented as the mean ± standard deviation of triplicate samples (not significantly different by the Student t test, P > 0.05). (B and C) 3-MA added at 1 h or 1 d p.i. inhibits E. chaffeensis infection. 3-MA was added to THP-1 cells at a final concentration of 2 mM, and infection was assessed at 3 d p.i. by Diff-Quik staining to determine the percentage of infected cells (B) and the number of bacteria per cell (C). Data are presented as the mean ± standard deviation of triplicate assays. *, Significantly different by the Tukey HSD test (P < 0.05). (D) 3-MA reversibly inhibits E. chaffeensis replication in THP-1 cells. i to iii, E. chaffeensis in THP-1 cells without 3-MA treatment at 23, 29, and 81 h p.i., respectively. iv to vi, E. chaffeensis in THP-1 cells treated with 10 mM 3-MA at 23 h p.i. for 6 h (iv) and incubated an additional 52 h with (vi) or without (v) 3-MA. Arrows indicate E. chaffeensis as shown by Diff-Quik staining. (E) Cells in panel (D) (iv) were immunostained with anti-P28. White arrows indicate large vacuoles containing condensed bacteria. Merged/DIC, fluorescence image merged with differential interference contrast (DIC) image. Deconvolution microscopy. Scale bar: 10 μm.
PIK3C3 and PtdIns3P are not only essential for endocytic sorting, trafficking, and maturation but also required for autophagy in mammalian cells, and 3-MA is known to inhibit autophagy.48,50,55,56 Localized PtdIns3P production has been linked to the promotion of autophagy.55 For autophagy induction, BECN1 binding to PIK3C3 is essential.57 Given the lack of maturation of ehrlichial inclusions into late endosomes9 and the requirement of class III PtdIns3K activation for ehrlichial proliferation, but not for entry, we examined roles of autophagy in ehrlichial infection by examining the requirement of BECN1. First, we used a siRNA (small interfering RNA) to reduce BECN1 expression in human embryonic kidney HEK293 cells, which are readily transfected and infected with E. chaffeensis.54 E. chaffeensis proliferation was significantly reduced in BECN1 siRNA–transfected cells compared with cells transfected with the control scrambled siRNA (Fig. 3A). Next, we treated infected THP-1 cells with a cell-permeable potent autophagy inhibitor, spautin-1.58 Spautin-1 promotes the degradation of BECN1 by inhibiting 2 USPs (ubiquitin-specific peptidases), USP10 and USP13, which target BECN1.58 Spautin-1 treatment of infected cells significantly decreased E. chaffeensis proliferation (Fig. 3B). These results demonstrated that both class III PtdIns3K activation and BECN1 are required for E. chaffeensis replication, implying that autophagy induction by class III PtdIns3K complex is required for E. chaffeensis replication.
Figure 3.
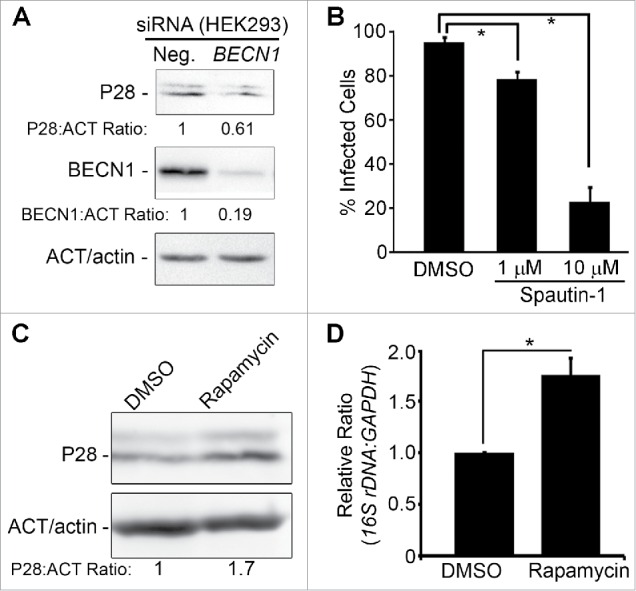
E. chaffeensis infection requires BECN1 and is enhanced by rapamycin. (A) Depletion of BECN1 suppresses E. chaffeensis infection. HEK293 cells were transfected with BECN1 siRNA or control scrambled siRNA (Neg.) for 40 h and then infected with E. chaffeensis for 36 h. Western blotting was performed using anti-P28, ACT/actin, and BECN1. The values under the bands show the relative ratio of band intensities vs. ACT/actin, with the ratios of those from control siRNA set as 1. (B) Spautin-1 inhibits E. chaffeensis growth. E. chaffeensis–infected THP-1 cells were treated at 1 d p.i. with DMSO solvent control or with 1 μM or 10 μM spautin-1 and incubated for an additional 2 d. Infection was assessed at 3 d p.i. by Diff-Quik staining to determine the percent of infected cells. *, Significantly different by the Tukey HSD test (P < 0.05). (C and D) Rapamycin enhances E. chaffeensis infection in THP-1 cells. (C) Western blot analysis with anti-P28. ACT/actin was used as a loading control. The values under the bands show the relative ratio of band intensities normalized against ACT/actin, with the ratio of DMSO (control) set as 1. (D) qPCR of E. chaffeensis 16S rDNA normalized to human GAPDH DNA. *, Significantly different (P < 0.05) by the Student t test.
E. chaffeensis infection was significantly increased in cells treated with rapamycin, the inhibitor of MTOR (mechanistic target of rapamycin [serine/threonine kinase]),59 based on western blot analysis of E. chaffeensis P28 (Fig. 3C) and qPCR of the E. chaffeensis 16S rRNA gene (rDNA, Fig. 3D). Rapamycin treatment under these conditions had no effect on the viability of the infected host cells (Table S1). These results are similar to A. phagocytophilum,35 except that 3-MA treatment does not induce enlarged vacuoles containing A. phagocytophilum, and opposite to intracellular bacteria such as Salmonella and Mycobacterium: 3-MA inhibits intracellular mycobacterium killing by macrophages via autophagy,31 and induction of autophagy by rapamycin, suppresses intracellular bacteria such as Salmonella and Mycobacterium.31,60
ATG5 is required for infection and localizes to E. chaffeensis inclusions
To further investigate the involvement of autophagy induction in E. chaffeensis infection of primary macrophages, the requirement of an autophagy double-membrane initiation protein, ATG5,61 was examined. Homozygous deletion of Atg5 gene results in neonatal lethality in mice.62 Therefore, tissue-specific knockout (TSKO) mice were created by a site-specific recombinase technology using the third exon of Atg5 flanked by LoxP sites in combination with the Cre recombinase driven by tissue-specific promoter.63 Because siRNA transfection of monocytes-macrophages is not efficient, and WT (wild-type) mouse bone marrow-derived macrophages (BMDMs) are readily infected with E. chaffeensis,54 we used BMDMs from atg5flox/flox-Lyz2-Cre mice64 in which Lyz2 promoter-driven Cre is used for myeloid cell-specific KO of Atg5.65 Peritoneal macrophages and BMDMs from these mice lack ATG5.66 E. chaffeensis infection in BMDMs from atg5flox/flox-Lyz2-Cre (atg5 TSKO) mice was markedly reduced compared with WT mice (Fig. 4A), demonstrating the critical role of ATG5 in E. chaffeensis infection of primary macrophages. The viability of BMDMs from atg5 TSKO and WT mice, either uninfected or infected with E. chaffeensis, was >90% at 4 d p.i. (Fig. S4). Moreover, GFP-ATG5 transfected at 1 d p.i. localized to E. chaffeensis inclusions (Fig. 4B). Similar to ectopically expressed GFP-ATG5, immunostaining of E. chaffeensis-infected cells with anti-ATG5 to detect the endogenous protein, showed the localization of endogenous ATG5 in 54 ± 12% of inclusions (n = 75 cells; Fig. S5B).
Figure 4.
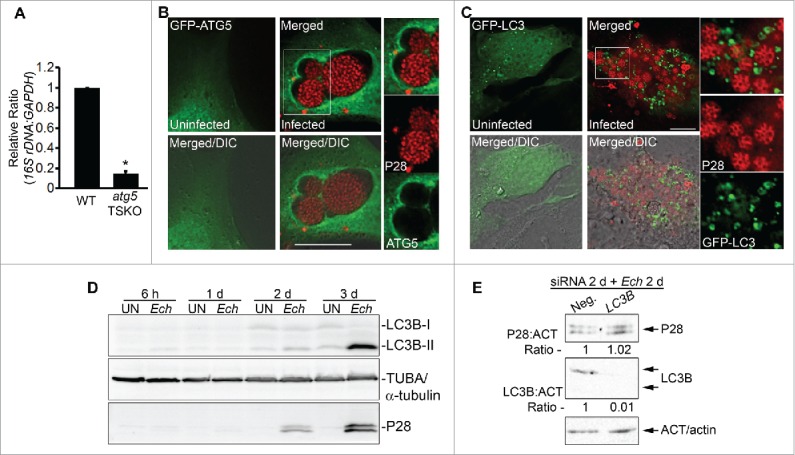
E. chaffeensis infection requires ATG5, and ATG5 but not LC3 localizes to E. chaffeensis inclusions. (A) E. chaffeensis load in macrophages derived from bone marrow of wild-type (WT) and atg5flox/flox-Lyz2-Cre mutant (atg5 TSKO) mice at 7 d p.i. qPCR of E. chaffeensis 16S rDNA was normalized to mouse Gapdh. *, Significantly different (P < 0.05) by the Student t test. (B) ATG5 traffics to E. chaffeensis inclusions. E. chaffeensis-infected cells were transfected at 1 d p.i. and were immunostained with anti-P28 (P28; red) at 17 h p.t. (41 h p.i.). Uninfected RF/6A cells were examined at 17 h p.t. (C) Diffused localization of GFP-LC3 was observed in uninfected RF/6A cells, but puncta were apparent in infected cells. GFP-LC3-transfected RF/6A cells were infected with E. chaffeensis at 1 d p.t. and immunostained with anti-P28 (P28; AF555) at 3 d p.i. and 4 d p.t. (B and C) Each boxed area is enlarged on the right. Merged, merged image; Merged/DIC, fluorescence image merged with DIC image. Scale bars: 10 μm. (D) Conversion of LC3-I to LC3-II occurs at a late stage of infection. HL-60 cells infected with E. chaffeensis (Ech), along with control uninfected cells (UN), were harvested at 6 h, 1 d, 2 d, and 3 d p.i for western blot analysis using rabbit anti-LC3B, rabbit anti-P28, and mouse anti-tubulin. (E) E. chaffeensis infection does not require LC3. RF/6A cells were transfected with LC3B siRNA or control scrambled siRNA (Neg.) for 2 d and incubated with E. chaffeensis for 2 d. Western blotting was performed using anti-P28, anti-ACT/actin, and anti-LC3B. The values under the bands show the relative ratio of band intensities normalized against ACT/actin, with the ratios of those from control siRNA set as 1.
In yeast, Atg5 is involved in the sequestration of the cytosolic precursor of aminopeptidase I and aminopeptidase IV, and the Atg12–Atg5 conjugate localizes only to phagophores (precursors of autophagosomes) and dissociates just before or after completion of autophagic vacuole formation.67 Therefore, ATG5 and ATG12 are not associated with mature autophagosomes. LC3, also called LC3-I (cytosolic form), becomes conjugated to phosphatidylethanolamine (termed LC3-II) and localizes on autophagosomal membranes (appearing as small puncta by fluorescence microscopy) when autophagy is induced.19 Thus LC3-II serves as a marker for canonical autophagosomes. ATG12–ATG5 conjugation is required for the conversion of LC3-I to LC3-II,61 and A. phagocytophilum inclusions are heavily encased by LC3-II.35,37 However, GFP-LC3, as well as endogenous LC3B, did not encase individual E. chaffeensis inclusions (Fig. 4C and S5B). In contrast, both GFP-LC3 and endogenous LC3B were observed as puncta in E. chaffeensis-infected RF/6A cells at 2 to 3 d p.i. (Fig. 4C and S5B). The conversion from LC3B-I to LC3B-II did not increase during exponential E. chaffeensis proliferation but was evident at 3 d p.i. in human promyelocytic leukemia HL-60 cells (Fig. 4D) when infected cells began to rupture because of the heavy bacterial burden, as shown by fluorescence microscopy (Fig. 4C), suggesting that the late onset of LC3B-II conversion may be caused by canonical autophagy: nutritional and ATP depletion that is due to overwhelming ehrlichial replication. Because treatment of E. chaffeensis-infected cells at 2 d p.i. with 10 nM bafilomycin A1 (BAF) for 6 h significantly increased the ratio of endogenous LC3B-II to LC3B-I (Fig. S6A), the basal autophagy flux was not blocked during the exponential growth phase (2 d p.i.) of intracellular E. chaffeensis. In addition, LC3B knockdown with siRNA did not reduce E. chaffeensis infection at 2 d p.i. (Fig. 4E). Thus ATG5 autophagosome localization to E. chaffeensis inclusions was not followed by the LC3B-II localization, a process that is distinct from the development of GFP-LC3-II-loaded autophagosomes routed to A. phagocytophilum inclusions.35,37
E. chaffeensis increases cellular glutamine and glutamate and takes up host-incorporated amino acids
Mammalian cellular free amino acid concentrations (those not incorporated into proteins) are tightly regulated by membrane transporters and the autophagy pathway.68 Given that E. chaffeensis replication requires host cell autophagy proteins (BECN1 and ATG5) and the class III PtdIns3K activation, we examined cellular free amino acid levels in E. chaffeensis-infected cells by targeted metabolomics analysis using LC-MS/MS.69 Compared with uninfected THP-1 cells, l-glutamine and l-glutamate in E. chaffeensis-infected THP-1 cells at 2 d p.i. were notably increased, and cellular glutamine, glutamate, aspartate, and proline were profoundly depleted at 3 d p.i., because of the overwhelming consumption of cellular free amino acids by E. chaffeensis (Tables 1 and S2).
Table 1.
E. chaffeensis infection increases glutamine and glutamate.
| Amino acids | Uninfected (mM) | 48 h Infected (mM) | 72 h Infected (mM) |
|---|---|---|---|
| Glu | 18.08 ± 0.48 | 48.46 ± 9.02 | 1.95 ± 0.14 |
| Gln | 2.44 ± 0.78 | 16.72 ± 7.2 | 0.98 ± 0.26 |
| Asn | 0.21 ± 0.02 | 0.36 ± 0.07 | 0.11 ± 0.02 |
| Pro | 1.34 ± 0.01 | 0.71 ± 0.19 | 0.03 ± 0.01 |
To determine whether host preincorporated amino acids are taken up by E. chaffeensis via autophagy, uninfected THP-1 cells were prelabeled with [3H]-glutamine for 1 d and then infected with E. chaffeensis for 2 d in the absence of [3H]-glutamine in the fresh culture medium. The cells were then incubated with or without 3-MA for an additional 6 h prior to harvesting at 2 d p.i. Host preincorporated l-glutamine was indeed taken up by E. chaffeensis, and 3-MA significantly blocked this uptake (Fig. 5A). The results suggest that class III PtdIns3K activity is critical for E. chaffeensis to have access to host-derived amino acids.
Figure 5.
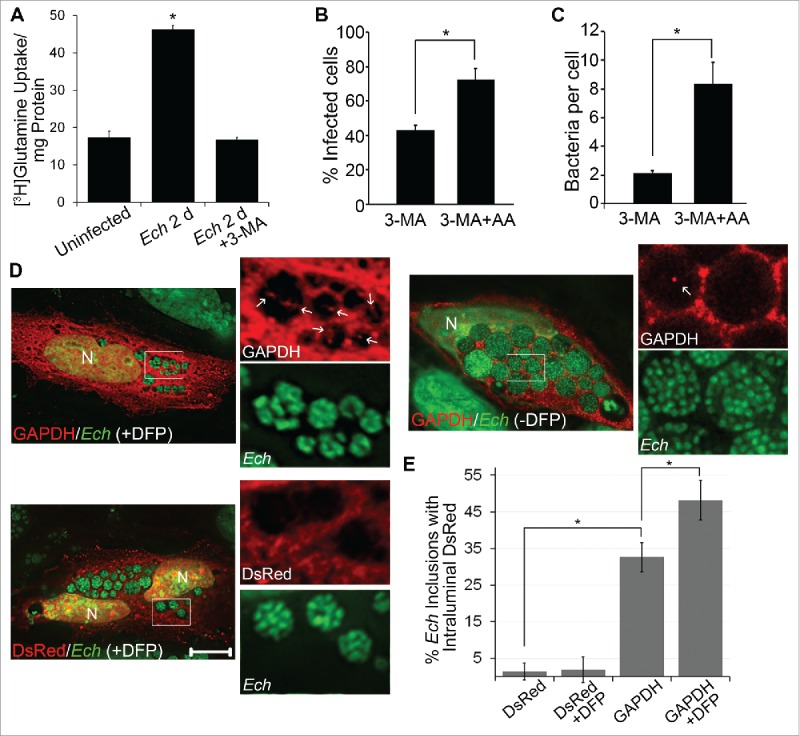
Host preincorporated amino acids are taken up by E. chaffeensis (Ech) in a host class III PtdIns3K-dependent manner. (A) THP-1 cells were prelabeled with [3H]glutamine for 1 d and then infected with E. chaffeensis for 2 d in the absence of [3H]glutamine. Infected cells were treated with 2 mM 3-MA or solvent control for an additional 6 h. E. chaffeensis was purified from infected cells, and the [3H]glutamine incorporated in E. chaffeensis was determined by liquid scintillation counter and normalized to the total protein amount. Data are presented as the mean ± standard deviation of triplicate samples. *, Significantly different (P< 0.05) by the Student t test. (B and C) E. chaffeensis growth inhibition by 3-MA is partially rescued upon supplementation with essential amino acids. E. chaffeensis-infected cells were treated at 1 d p.i. with 3-MA or 3-MA and amino acids (3-MA + AA) for 2 d. The percentage of infected cells (B) and number of bacteria (C) were scored in each group. *, Significantly different (P < 0.05) by the Student t test. (D) DsRed-GAPDH (white arrows) was detected within E. chaffeensis-containing inclusions more with 0.1 μM DFP treatment for 2 h than without treatment before fixation by deconvolution microscopy. DAPI was pseudocolored in green. N, nucleus. Bar: 10 μm. (E) Percentage of inclusions that contain DsRed-GAPDH or DsRed with or without DFP treatment was determined by counting 10 to 20 inclusions per cell in 10 to 20 cells per experiment from 3 independent experiments. *, Significantly different (P < 0.01) by the Student t test.
Next, we examined whether the simultaneous addition of essential amino acids and 3-MA to infected cells would abrogate the 3-MA-induced inhibition of E. chaffeensis replication. Our result showed that E. chaffeensis growth inhibition by 3-MA was partially rescued (Fig. 5B and C). Taken together, E. chaffeensis dysregulates the homeostasis of cellular free amino acid levels and raises cellular free glutamine and glutamate levels during exponential growth, and E. chaffeensis within membrane-bound inclusions has the ability to take up free amino acids from host cells in a class III PtdIns3K activity-dependent manner.
To demonstrate that host cytosolic molecules are indeed delivered into E. chaffeensis inclusions via autophagy, the autophagy cargo protein human GAPDH70 was ectopically expressed in E. chaffeensis-infected cells. E. chaffeensis has surface proteases that are required for its replication.71,72 Compared with untreated cells (Fig. 5D), brief treatment with diisopropylfluorophosphate (DFP), an irreversible serine protease inhibitor, increased the presence of DsRed-GAPDH inside E. chaffeensis inclusions (Fig. 5E), supporting autophagic delivery of host cytoplasmic molecules into the inclusion lumen.
E. chaffeensis induces autophagy independently of MTOR, ULK1, PRKA/AMPK, and ubiquitination pathways
Macroautophagy induction by amino acid starvation is initiated by the activation of ULK, which is mainly regulated by MTOR kinase complex or by PRKA/AMPK (protein kinase, AMP-activated) independently from MTOR activity.73,74 MTOR inhibition leads to dephosphorylation of ULK1 at Ser757,74 and of RPS6 (ribosomal protein S6) at Ser240/244.75 PRKA is a heterotrimeric protein composed of α (PRKAA), β, and γ subunits. The γ subunit detects shifts in the AMP-to-ATP ratio and activates PRKA through phosphorylation of the α subunit at Thr172.76 We therefore investigated whether E. chaffeensis induces host autophagy through these well-known signaling pathways using phosphorylation-specific antibodies that demonstrate the activation status of ULK1, PRKA, and RPS6. Western blotting results showed that tricyclic benzonaphthyridinone (Torin-1; a potent inhibitor of MTOR complex 1 [MTORC1] or MTORC2)77 or rapamycin treatment activated ULK1 by dephosphorylation at Ser757 and RPS6 at Ser240/244., but not PRKAA at Thr172 (Fig. S6B). However, infection of E. chaffeensis in THP-1 cells for 1 d did not alter the activation status of ULK1, RPS6, or PRKA (Fig. S6B).
Aggregated proteins, overabundant proteins, and certain microbial proteins can induce selective autophagy through ubiquitination that binds LC3 via SQSTM1/p62 (sequestosome 1).78-80 The autophagic sequestration of invading bacteria is an important innate immune mechanism. Intracellular Salmonella, Mycobacterium, Streptococcus, and Legionella-containing vacuoles or bacteria are ubiquitinated, and subsequent binding of SQSTM1/p62 delivers the bacteria to autolysosomes for degradation.81-85 E. chaffeensis inclusions were not ubiquitinated as monoclonal antibody FK2 that recognize both mono- and poly-ubiquitinated proteins did not label the inclusions (Fig. S7). Taken together, our observations suggest that E. chaffeensis induces autophagy independent of MTOR, ULK1 or PRKA, and ubiquitination-induced signaling pathways.
Ectopic expression of a T4S effector, Etf-1, enhances E. chaffeensis infection, and Etf-1 traffics to E. chaffeensis inclusions
We have previously shown that the T4S effector Etf-1 is produced and secreted at the start of ehrlichial exponential growth, which is required for E. chaffeensis replication in THP-1 cells, since bacteria amount was reduced while affinity-purified anti-Etf-1 IgG was delivered into infected cells using the Chariot protein delivery system at 1 d p.i.43 Here, we examined whether Etf-1 has additional effects on E. chaffeensis infection. E. chaffeensis infection was significantly enhanced in cells transfected with Etf-1-GFP compared with control GFP-transfected cells as determined by qPCR (Fig. 6A). This growth-stimulating activity of Etf-1-GFP was specific to E. chaffeensis because Etf-1-GFP did not enhance infection by A. phagocytophilum (Fig. S8A), which replicates in autophagosomes.35
Figure 6.
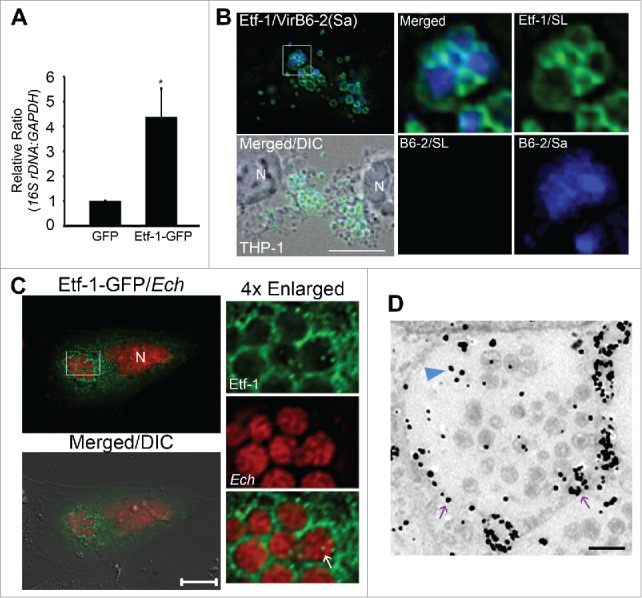
Etf-1 promotes E. chaffeensis infection and traffics to E. chaffeensis inclusions. (A) HEK293 cells transfected with Etf-1-GFP or with GFP alone (control) were infected with E. chaffeensis at 1 d p.t. qPCR was performed at 2 d p.i. The 16S rDNA/GAPDH ratio for GFP-transfected HEK293 cells was set as 1. Data are presented as the mean ± standard deviation of triplicate assays. *, Significantly different (P < 0.05) by the Student t test. (B) Native Etf-1 localizes on the cytoplasmic side of E. chaffeensis inclusions. N, nucleus. The plasma membrane of infected THP-1 cells was selectively permeabilized with SLO and labeled with anti-Etf-1 (Etf-1/SL) and anti-VirB6-2 (B6-2/SL). After the first round of staining, all cell membranes were permeabilized with saponin (Sa), and the cells were stained again with anti-VirB6 (B6-2/Sa). Secondary antibodies with distinct fluorochromes were used for VirB6-2 labeling before (red) and after (blue) saponin treatment. (C) E. chaffeensis (Ech) inclusions are enveloped by Etf-1-GFP. E. chaffeensis-infected RF/6A cells were transfected with Etf-1-GFP at 1 d p.i., and treated with 0.1 μM DFP for 2 h prior to fixation at 16 h p.t. (40 p.i.). DAPI was used to stain DNA in host cell nuclei and E. chaffeensis DNA and pseudocolored in red. N, nucleus. Merged/DIC, fluorescence image merged with DIC image. Boxed area was enlarged 4-fold on the right. The white arrow indicated the presence of Etf-1-GFP inside E. chaffeensis-containing inclusions. Scale bars: 10 μm. (D) Immunogold labeling of Etf-1-GFP in E. chaffeensis-infected RF/6A cells. Silver-enhanced anti-GFP immunogold labeling of Etf-1-GFP detected on the inclusion membrane (purple arrows) or inside the inclusions (blue arrowhead). Scale bar: 2 μm.
Although our previous study shows that Etf-1 targets mitochondria, it also localizes in part to E. chaffeensis inclusions.43 To determine whether Etf-1 is present on cytoplasmic side of the inclusion membrane, we selectively permeabilized the plasma membrane of host cells using streptolysin O (SLO).86 SLO is a thiol-activated protein toxin that binds cholesterol to form membrane-penetrating channels in a temperature-dependent manner.87 Cytoplasmic Etf-1 was labeled with anti-Etf-1 after SLO treatment (Fig. 6B). In contrast, the intrainclusion ehrlichial protein VirB6-240 was not labeled with anti-VirB6-2 in SLO-treated cells (Fig. 6B), indicating the inability of SLO to permeabilize E. chaffeensis inclusions. Saponin treatment permeabilizes all cholesterol-containing membranes. Both Etf-1 and VirB6-2 could be labeled with their respective antibodies and colocalized in saponin-permeabilized cells (Fig. 6B). This result indicates that native Etf-1 is present on the cytoplasmic face of inclusions. Furthermore, ectopically expressed Etf-1-GFP at 2 d p.i. was recruited to E. chaffeensis inclusions, and Etf-1-GFP could be observed inside bacteria-containing inclusions (56.8 ± 4.8% of E. chaffeensis inclusions as quantitated from 10 to 20 cells each in 3 independent experiments, Fig. 6C). Similarly, immunogold labeling showed Etf-1-GFP on the E. chaffeensis inclusion membranes or inside inclusions (Fig. 6D). Etf-1 targeting to bacteria-containing inclusions was specific to E. chaffeensis, as Etf-1-GFP was not present on A. phagocytophilum inclusions (Fig. S8B).
Ectopically expressed Etf-1 activates class III PtdIns3K, induces autophagosomes, and clears mutant HTT
Ectopic expression of Etf-1 activated class III PtdIns3K in the absence of any other ehrlichial molecule, as indicated by a significantly higher amount of cellular PtdIns3P in Etf-1-transfected cells (Fig. 7A). When GFP-ATG5 or GFP-LC3 alone was ectopically expressed, they each showed a few weak and small puncta in uninfected cells (Fig. 4B and C). Coexpression of Etf-1-DsRed with GFP-ATG5 or GFP-LC3 induced numerous puncta, and >80% of Etf-1 puncta colocalized with ATG5 or LC3 (Fig. S9). BAF treatment for 16 h produced enlarged autophagosomes, in which Etf-1 and GFP-ATG5 or GFP-LC3 colocalized (Fig. 7B and C). This indicates that Etf-1 is sufficient in inducing autophagosomes in the absence of any other ehrlichial molecules.
Figure 7.
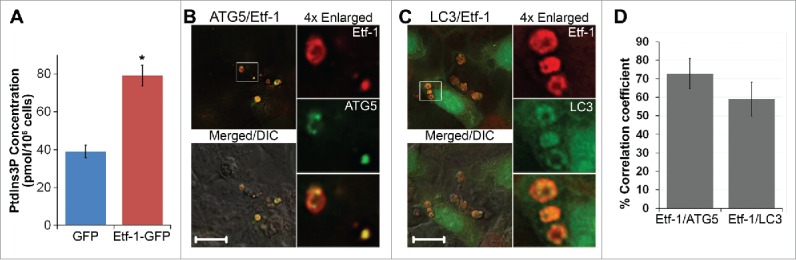
Etf-1 activates PtdIns3K and colocalizes with ATG5 and LC3. (A) RF/6A cells were transfected with Etf-1-GFP or GFP control plasmids, and the PtdIns3P amount was determined by competitive ELISA at 2 d p.t. Data are presented as the mean ± standard deviation of triplicate assays. *, Significantly different by the Tukey HSD test (P < 0.05). (B and C) Etf-1 colocalizes with ATG5 and LC3. HEK293 cells cotransfected with GFP-ATG5 and Etf-1-DsRed (B) or GFP-LC3 and Etf-1-DsRed (C) were treated at 1 d p.t. with 10 nM BAF for 16 h prior to fixation. Merged/DIC, fluorescence image merged with DIC image. Scale bars: 10 μm. (D) The percentage colocalization of Etf-1 with ATG5 or LC3 was analyzed using Pearson correlation coefficients with ImageJ software. Results were average values of 10 to 20 cells per group ± standard deviation from 3 independent experiments.
Eukaryotic proteasomes can cleave only very poorly (if at all) within polyglutamine sequences such as polyglutamine tracts in HTT (huntingtin) protein.88 Expansions of polyglutamine tracts in HTT causes aggregation of polyglutamine peptides, leading to a neurodegenerative genetic disorder, Huntington disease.89 NPEPPS/PSA (aminopeptidase, puromycin sensitive; EC 3.4.11.14) is a mammalian cytosolic Zn2+ metallopeptidase that can digest aggregation-prone proteins including polyglutamine-containing peptides such as HTT via autophagy, thereby reducing cellular toxicity.90 NPEPPS overexpression can enhance macroautophagy.91 In the yeast cytoplasm-to-vacuole targeting pathway, the precursor form of aminopeptidase I is sequestered in phagophores in an Atg5-dependent manner67 and is targeted to the vacuole (the yeast equivalent of mammalian lysosomes).92 We found that ectopically expressed NPEPPS-GFP colocalized with Etf-1 puncta (Fig. 8A) and, at 1 d p.i., encased E. chaffeensis inclusions (Fig. 8B). PAQ-22 is a synthetic, noncompetitive NPEPPS inhibitor that does not act as a substrate mimic and thus binds to NPEPPS at a site distinct from the catalytic site.93,94 Compared with the DMSO-treated control cells, E. chaffeensis infection was significantly decreased in PAQ-22-treated cells at 2 d p.i. based on qPCR (Fig. 8C). PAQ-22 was not toxic to the host cells at 100 μM (data not shown).
Figure 8.

NPEPPS-GFP colocalizes with Etf-1 in cotransfected cells, traffics to E. chaffeensis inclusions, and reduces aggregation of Q103-HTT. (A and B) NPEPPS/PSA-GFP colocalizes with Etf-1 in cotransfected cells and surrounds E. chaffeensis inclusions. (A) DH82 cells were sequentially transfected first with Etf-1 and 1 d later with NPEPPS-GFP. At 1 d p.t. with NPEPPS-GFP, cells were immunostained with anti-Etf-1 (AF555). White arrows indicate the colocalization between the 2 proteins. (B) E. chaffeensis-infected RF/6A cells were transfected with NPEPPS-GFP at 1 d p.i. and stained with DAPI at 1 d p.t. (2 d p.i.). Merged/DIC, fluorescence image merged with DIC image. The boxed area is enlarged on the right. Scale bars: 15 μm. (C) PAQ-22 inhibits E. chaffeensis replication in THP-1 cells. E. chaffeensis-infected THP-1 cells were incubated with 0.1% DMSO (control) or 10 or 100 μM PAQ-22. qPCR of E. chaffeensis 16S rDNA normalized to human GAPDH. *, Significantly different by the Tukey HSD test (P< 0.05). (D to F) Etf-1 reduces aggregation of Q103-GFP. (D) RF/6A cells transfected with Q103-GFP alone or cotransfected with Q103-GFP and DsRed control; exposure time, 0.001 sec. RF/6A cells cotransfected with Q103-GFP and Etf-1 were immunostained with anti-Etf-1 (red); exposure time, 0.6 sec. Scale bars: 15 μm. (E) Percentage of RF/6A cells with Q103-GFP aggregation in cells cotransfected with Q103-GFP and vector control or Etf-1. *, Significantly different by the Tukey HSD test (P < 0.05). (F) Relative amount of Q103-GFP in RF/6A cells with or without Etf-1. Western blot analysis was performed using anti-GFP and - ACT/actin IgG, and band intensities were normalized against ACT/actin.
Ectopic expression of exon 1 of aggregation-prone mutant HTT with 103 polyglutamine repeats fused to GFP (Q103-GFP) is toxic to eukaryotic cells, but coexpression with NPEPPS protects cells via autophagy from death induced by Q103-GFP.91 Indeed, ectopic expression of Q103-GFP in RF/6A cells caused striking aggregation of the protein; however, cotransfection with Etf-1, but not DsRed control, significantly reduced aggregation of Q103-GFP (Fig. 8D-E) and reduced the amount of Q103-GFP, which was partially abrogated by 3-MA treatment (Fig. 8F), suggesting that Etf-1 helps clearance of Q103-GFP through enhanced autophagy.
Certain overexpressed cytosolic proteins can be ubiquitinated and degraded by autophagy.78 We investigated whether native or ectopically expressed Etf-1 was ubiquitinated in infected or transfected cells. Based on immunoprecipitation of E. chaffeensis-infected THP-1 cells with anti-Etf-1 IgG followed by western blotting, native Etf-1 was not ubiquitinated (Fig. S10). Immunofluorescence labeling with anti-ubiquitin showed that neither GFP staining nor Etf-1-GFP puncta were ubiquitinated (Fig. S11A, B, and D). As a positive control, strong ubiquitination was detected on Q103-GFP aggregates (Fig. S11C and D).
RAB5 is required for E. chaffeensis infection and traffics to E. chaffeensis inclusions
The small GTPase RAB5 regulates endosome maturation to late endosomes, thereby regulating fusion of LC3-decorated autophagosomes with late endosomes to form intermediary compartments prior to fusion with lysosomes.21 RAB5 also regulates autophagy upstream of LC3 conjugation.95-97 Given that E. chaffeensis replicates in inclusions that contain endogenous RAB5,9 we first examined whether endogenous RAB5 is required for E. chaffeensis infection. There are 3 RAB5 isoforms expressed in human cells, namely RAB5A, 5B, and 5C, which share similar subcellular localization and regulatory functions in the early endocytic pathway.98 RAB5 isoforms are at least partially redundant because downregulation of expression of any single RAB5 isoform using siRNA has no effect on EGF or transferrin internalization, but downregulation of all 3 isoforms has a significant effect.99 Similarly, silencing of all 3 RAB5 isoforms is required to block AKT activation after insulin treatment.100 With this in mind, we tested the role of RAB5 in E. chaffeensis infection by silencing expression of all 3 RAB5 isoforms using siRNA. Western blotting results showed that E. chaffeensis infection was significantly reduced by RAB5 knockdown compared with the control scrambled-siRNA transfection (Fig. 9A). Next, we examined whether exogenous RAB5 traffics to already established E. chaffeensis inclusions by transfecting GFP-RAB5. All 3 GFP-RAB5 isoforms produced puncta (vesicles) and accumulated on E. chaffeensis inclusions when transfected at 15 h p.i., and quantitation from 10 to 20 cells each in 4 independent experiments showed RAB5A positive E. chaffeensis inclusions were 94.0 ± 5.5%. RAB5 delivery appeared to occur largely via fusion of the E. chaffeensis inclusions with RAB5-containing vesicles, suggesting the recruitment and retention of RAB5 endosomes during E. chaffeensis replication and expansion of the inclusion (Fig. 9B to D). In uninfected cells, GFP-RAB5 showed smaller diffuse puncta than in infected cells (RAB5A shown in Fig. 9E).
Figure 9.
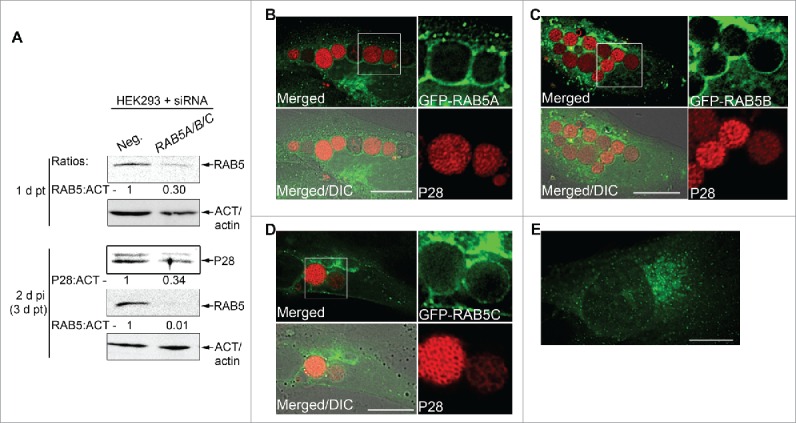
E. chaffeensis infection requires RAB5, and GFP-RAB5 traffics to the E. chaffeensis inclusion membrane. (A) E. chaffeensis infection requires RAB5. RF/6A cells were transfected with control scrambled siRNA (Neg.) or RAB5A/B/C siRNAs for 1 d, and then infected with E. chaffeensis for 2 d. Western blotting was performed using anti-P28, -ACT/actin, or -RAB5 IgG. The values under the bands show the relative ratio of band intensities normalized against ACT/actin, with the ratios of those from control siRNA set as 1. (B to D) E. chaffeensis-infected RF/6A cells at 1 d p.i. were transfected with GFP-RAB5A, GFP-RAB5B, or GFP-RAB5C. At 15 h p.t. (39 h p.i.), cells were subjected to immunofluorescence labeling with rabbit anti-P28 (AF555). Merged, merged images; Merged/DIC, fluorescence image merged with DIC image. Each boxed area is enlarged on the right. (E) Control uninfected RF/6A cells transfected with GFP-RAB5A. Scale bars: 15 μm.
Etf-1 interacts with RAB5, PIK3C3, and BECN1
Given that endogenous Etf-1 and RAB5 localized on the cytoplasmic face of E. chaffeensis inclusions and that both Etf-1 and RAB5-containing vesicles targeted E. chaffeensis inclusions, we examined whether Etf-1- and RAB5-containing vesicles interact. Furthermore, since most cellular BECN1 exists in a complex with PIK3C3, a RAB5 effector,57,101 we examined whether the BECN1-PIK3C3 complex contains RAB5 and Etf-1. With reciprocal immunoprecipitation of E. chaffeensis-infected THP-1 cell lysates using antibodies against Etf-1, RAB5, BECN1, or PIK3C3 followed by western blotting, we found that native Etf-1 and the 3 endogenous human proteins formed a multimeric complex in the infected cells (Fig. 10). Very small amount of BECN1 and PIK3C3 bound to RAB5 in uninfected cells, whereas infection highly upregulated this complex formation. Furthermore, endogenous PIK3C3 and RAB5 proteins were consistently upregulated in E. chaffeensis-infected cells compared with uninfected cells (Fig. 10).
Figure 10.
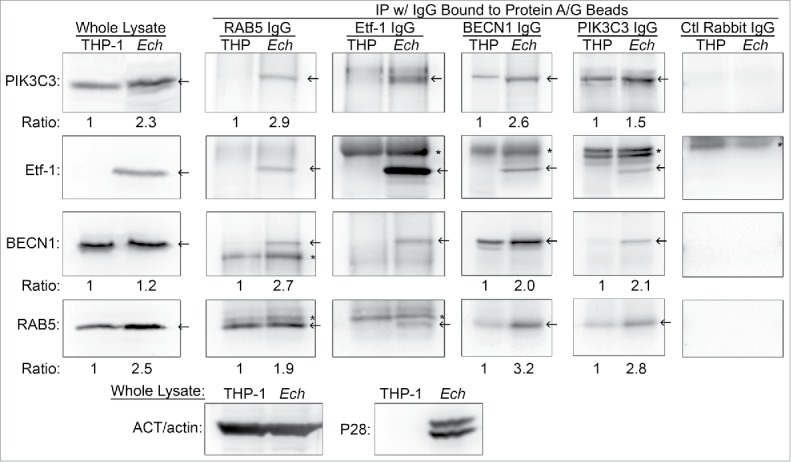
Etf-1 interacts with the RAB5-class III PtdIns3K complex. Co-immunoprecipitation of native Etf-1 with endogenous RAB5, BECN1, and PIK3C3/VPS34. Uninfected (THP) or E. chaffeensis-infected (Ech) THP-1 cells at 2 d p.i. were lysed in modified lysis buffer and immunoprecipitated (IP) with rabbit anti-Etf-1, BECN1, or PIK3C3, or mouse anti-RAB5 IgG cross-linked to protein A/G-magnetic beads for 2 h. Bound proteins were eluted and subjected to western blotting. Arrows indicate the target proteins. *, IgG heavy or light chains. The number under the figure is the density ratio relative to ACT/actin, with uninfected cells set as 1. The absence of a number indicates an infinite ratio (i.e., the protein was absent in the control). Images were representative of 3 experiments with similar results.
In vitro affinity isolation showed that glutathione S-transferase (GST)-RAB5A fusion protein, but not GST, pulled down native Etf-1 from E. chaffeensis-infected THP-1 cell lysates (Fig. S12A). In addition, recombinant Etf-1 interacted with endogenous RAB5, PIK3C3, and BECN1 in uninfected THP-1 cells (Fig. S12B). These results suggest that E. chaffeensis inclusions recruit Etf-1, RAB5, and major components of the class III PtdIns3K complex through protein-protein interactions.
GTP-bound RAB5 interacts with Etf-1 and the BECN1-class III PtdIns3K complex, and promotes infection
RAB GTPase cycles between GTP- and GDP-bound forms and acts via RAB effectors.102,103 RAB effectors such as PIK3C3 and ANKFY1 bind to RAB-GTP but dissociate from RAB-GDP.45,95 RAB5AS34N (dominant-negative RAB5A/RAB5A-DN), a GDP-bound form of RAB5 that sequesters RAB5 guanine nucleotide exchange factor (RAB5-GEF) and thus prevents RAB5 activation.104 In RAB5-regulated autophagy, RAB5-DN decreases LC3-puncta formation and increases ATG5-containing structures (autophagosome precursors).97 To analyze whether RAB5 GTPase cycle is involved in Etf-1 interaction with RAB5 and the BECN1-class III PtdIns3K complex, and Etf-1-induced autophagy, Etf-1-HA and GFP vector control, GFP-RAB5A (WT), GFP-RAB5A-DN, or GFP-RAB5AQ79L (constitutively active RAB5A/RAB5A-CA) were cotransfected into HEK293 cells, and the cell lysates were immunoprecipitated with anti-HA. Western blot analysis showed that although Etf-1-GFP bound RAB5A (WT), RAB5A-DN and RAB5A-CA, PIK3C3 and BECN1 interacted with Etf-1 only in the presence of RAB5A (WT) and RAB5A-CA (Fig. 11A). Fluorescence microscopy showed enlarged GFP-RAB5A-CA endosomes that colocalized with Etf-1 puncta (Fig. 11B). Colocalization analysis between GFP-RAB5A-CA with Etf-1 showed that the Pearson correlation coefficient of 24 ± 2 %, which was increased to 49 ± 5% when cells were treated with rapamycin for 16 h before harvesting at 2 d post-transfection (p.t.), and both were significantly greater (P < 0.01) vs. GFP control (12 ± 2 %) by the Student unpaired t test. Taken together, Etf-1 interacts with the RAB5-BECN1-PIK3C3, autophagy master regulator complex via GTP-bound RAB5, to induce autophagy.
Figure 11.
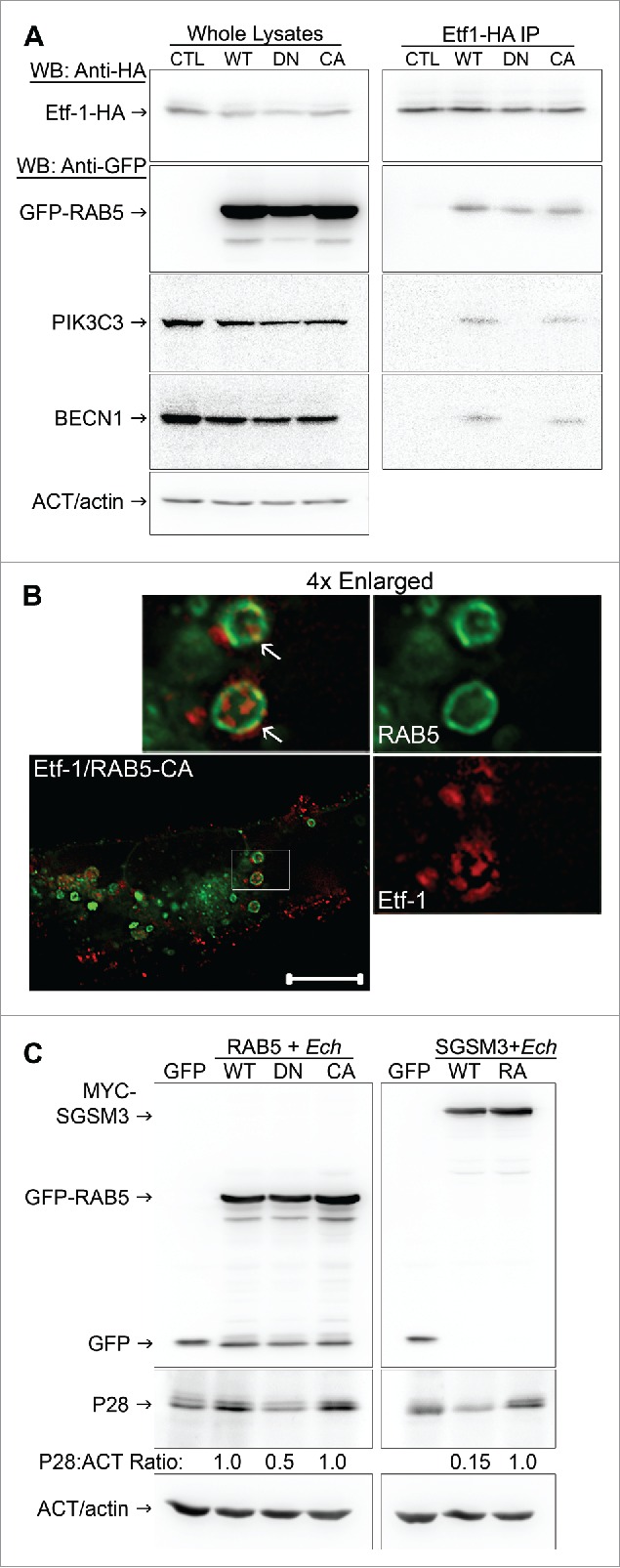
RAB5-GTP is required for E. chaffeensis infection. (A) HEK293 cells were cotransfected with plasmids expressing Etf-1-HA and GFP (CTL) or GFP-RAB5A (WT, DN, or CA mutant). At 2 d p.t., samples were lysed and immunoprecipitated with mouse anti-HA cross-linked on protein G-sepharose beads for 2 h. Images were representative of 3 experiments with similar results. (B) RF/6A cells cotransfected with Etf-1-DsRed and GFP-RAB5A-CA for 2 d. White arrows indicate the colocalization between the 2 proteins. The boxed area was enlarged 4-fold. Scale bar: 10 μm. (C) HEK293 cells were transfected with GFP, GFP-RAB5 (WT, DN, or CA mutant), SGSM3/RABGAP5 (WT), or SGSM3R165A mutant (RA) and then infected with E. chaffeensis at 1 d p.t. for 2 d. Samples were examined by western blotting, and the ratios of P28: ACT/actin were quantified and compared with those of RAB5 WT or SGSM3R165A groups, which were arbitrarily set as 1. Images were representative of 3 experiments with similar results.
Overexpression of GFP-RAB5A-DN impaired E. chaffeensis infection compared with cells transfected with GFP-RAB5A (WT) or GFP-RAB5A-CA (Fig. 11C), indicating that activated RAB5 (the GTP-bound state of RAB5) is indeed critical for infection. RABs have a low intrinsic rate of GTP hydrolysis, which is controlled by GAPs/GTPase activating proteins. Thus we assessed the effects of a RAB5-specific GAP, SGSM3/RABGAP5 (small G protein signaling modulator 3),105 on E. chaffeensis infection. SGSM3 overexpression at 1 d p.i. profoundly impaired E. chaffeensis replication compared with overexpression of SGSM3R165A (SGSM3-RA), a catalytic site mutant105 (Fig. 11C), confirming the critical role of GTP-bound RAB5 in E. chaffeensis replication. Taken together, Etf-1 induces autophagy by recruiting the BECN1-class III PtdIns3K complex via GTP-bound RAB5, to promote E. chaffeensis replication.
Discussion
Autophagy is induced in response to a variety of infections and is a critical innate immune mechanism for clearing intracellular infection.106,107 Thus, what is distinctive for the obligatory intracellular pathogen E. chaffeensis, compared with most other intracellular pathogens, is that infection-induced autophagy does not kill the pathogen; on the contrary, pathogen growth is autophagy dependent. Ehrlichial infection significantly increases cellular PtdIns3P levels at early stage of infection, and dramatic inhibition of ehrlichial growth results from the inhibition of autophagy induction by 3-MA or spautin-1 treatment, or knockdown of ATG5 or BECN1. The inhibition is not due to reduced viability of the host cells, or due to direct toxicity to bacteria. Conversely autophagy induction by rapamycin enhanced infection. How does host cellular autophagy facilitate ehrlichial proliferation? Gln is a primary energy and carbon source of E. chaffeensis that cannot utilize glucose.53 The present study implies that autophagy supplies Gln for E. chaffeensis, because it acquires host preincorporated Gln in a class III PtdIns3K activity–dependent manner, and growth inhibition by autophagy inhibition was reversed by excess amino acid supplementation. Moreover, free Gln and Glu, which are the primary amino acids generated by the autophagic degradation of eukaryotic cytoplasm,108 are also the primary amino acids that increased during exponential E. chaffeensis growth (Table 1).
Our data revealed lack of involvement of MTOR-ULK1-PRKA and ubiquitination pathways in ehrlichial infection. Instead, our data indicate that E. chaffeensis Etf-1, which is secreted at an early stage of infection, is responsible in inducing autophagy by recruiting PIK3C3 and BECN1, because ectopic expression of Etf-1 is sufficient in increasing cellular PtdIns3P levels; and ectopically expressed Etf-1 induces vesicles that colocalize with ATG5 and LC3.
RAB GTPases are the central regulators of membrane trafficking and organellar identity in eukaryotic cells.104 RAB5 is found in nascent phagosomes and early endosomes that lack the microbicidal capacity required to kill invading pathogens; this requires subsequent maturation into late endosomes via the transition from RAB5 to RAB7 and lysosomal fusion. Consequently several intracellular pathogens prevent or delay the RAB5 to RAB7 transition. For example, M. tuberculosis localizes in the RAB5-positive endocytic compartment by reducing PtdIns3P in the phagosomal membrane, thereby interfering with phagosome maturation.109,110 Listeria monocytogenes GAPDH binds and carries out ADP-ribosylation of RAB5A and impairs the GDP/GTP exchange and thereby blocks phagosome maturation.111 Maturation arrest of RAB5-positive E. chaffeensis inclusions is distinct from these examples, as PtdIns3P is highly enriched and the blockade seems to be due to RAB5 GTPase inactivation, based on the ability of SGSM3 overexpression to overcome the blockade. In agreement with this observation, the stable presence of the RAB5 effectors EEA1, PIK3C3, and ANKFY1 on E. chaffeensis–containing inclusions and the inhibition of E. chaffeensis infection by overexpression of RAB5-DN or SGSM3 suggest that RAB5 is mostly locked in the GTP-bound state on E. chaffeensis inclusions.
Our results suggest that inactivation of RAB5 GTPase on inclusions not only protects E. chaffeensis from a microbicidal mechanism and expands inclusions via homotypic fusion but also enhances “RAB5-regulated autophagy” as a result of the increase in GTP-bound RAB5. Indeed, several previous studies point to the critical role of RAB5-GTP in early autophagosome development/maturation. 1) Ectopic expression of dominant-negative RAB5 or 3-MA treatment blocks the progression of early ATG5-positive phagophores to form LC3-positive autophagosomes.97 2) Upon growth factor restriction, the class 1A phosphoinositide-3-kinase catalytic subunit β (PIK3CB/p110β) dissociates from the growth factor receptor and binds RAB5.95 Such interaction blocks the RAB5 GTPase-activating protein PIK3R1/p85α (class 1A phosphoinositide-3-kinase regulatory subunit 1) from binding to RAB5, which consequently increases the amount of GTP-bound RAB5 and enhances the RAB5-PIK3C3 interaction to promote autophagy.95 3) Hepatitis C virus NS4B protein forms a complex with RAB5 and PIK3C3 and induces RAB5-GTP-promoted autophagy, which is required for virus replication.96 4) HTT binds RAB5 via the RAB5 effector F8A1/HAP40 (coagulation factor VIII-associated 1), and degradated by autophagy.112 Similar to E. chaffeensis infection, such RAB5-regulated autophagy is independent from MTOR signaling.95,97
The present study revealed host cytoplasmic molecules (amino acids, GAPDH, and Etf-1-GFP) are delivered into ehrlichial inclusions. It is still unclear how host cytoplasmic molecules enter into ehrlichial inclusions. Unlike most other intracellular bacteria that are killed by autophagy, E. chaffeensis inclusions lack ubiquitination, LC3, and late endosome or lysosomal markers, although they do contain ATG5, PIK3C3, PtdIns3P, and RAB5. Although the classical view has been that autophagic and endocytic pathways converge at the phagolysosomal and lysosomal levels, autophagosomes have also been found to undergo fusion with earlier components of the endocytic pathway.21,22,24,113 The presence of ATG5 suggests that E. chaffeensis inclusions are large amphisomes, and Etf-1-, RAB5-, and ATG5-containing small vesicles might also be considered amphisomes. Multivesicular bodies (MVBs), morphologically distinctive endosomes, internally accumulate small membrane vesicles (60 to 80 nm), which contain cytosolic cargo molecules.21,114 Morphological evidence suggests that MVBs are the main endocytic fusion partner of the autophagosome, forming the amphisome.21,115 It is tempting to speculate that fusion of these vesicles with ehrlichial inclusions or invagination of the ehrlichial inclusion membrane would deliver host cytosolic cargo into the inclusion lumen. PIK3C3 and PtdIns3P are essential for the generation of the internal vesicles of MVBs, and RAB5 promotes the formation of PtdIns3P on endosomes116 and phagosomes.117 Most 2×FYVE labeling is associated with the internal membranes of MVBs, which have low levels of the late endosome and lysosome markers like lysosomal membrane glycoproteins and lyso-bisphosphatidic acid.44,118 PtdIns3P accumulates within ECVs (endosomal carrier vesicles) and MVBs, and PtdIns3P signaling regulates receptor sorting into ECVs/MVBs.44,119 Indeed, our results with 2×FYVE-GFP-transfected cells also showed strong intralumenal PtdIns3P in large vesicles docked onto E. chaffeensis inclusions, suggesting that these vesicles are MVBs (Fig. 1A, arrow). Inhibition of PtdIns3K blocks formation of intralumenal vesicles in MVBs, which results in swollen vesicles.120 Reversible vacuolation of ehrlichial inclusions in 3-MA-treated cells (Fig. 2D, 2E, and S2) suggests the inclusions also have the feature of MVBs.
Although autolysosomal degradation does not ensue in ehrlichial inclusions (to protect the bacteria), serine protease may be involved in autophagic cargo degradation, because treatment with DFP, an irreversible serine protease inhibitor, increased the presence of DsRed-GAPDH inside E. chaffeensis inclusions. E. chaffeensis has surface proteases that are required for its replication.71,72 Host cytosolic NPEPPS, which is incorporated into Etf-1 autophagosomes and targeted to E. chaffeensis inclusions, is also expected to aid degradation of autophagic cargo. Ectopic expression of Etf-1 facilitates the class III PtdIns3K-dependent degradation of Q103-GFP that is degraded by NPEPPS via autophagy,91 supporting the involvement of NPEPPS in Etf-1 autophagosome cargo degradation.
ANKFY1 plays a role in homotypic early endosomal fusion, micropinocytosis, and retromer-based transport.45,121 Because GFP-ANKFY1121 and transferrin8 colocalize to E. chaffeensis inclusions, macropinocytosis and endocytosis also directly deliver some extracellular nutrients to E. chaffeensis in inclusions.
Taken together, several aspects are unusual in this form of pathogen effector-induced autophagy: 1) autophagy induction is essential for bacterial replication, as it provides nutrients to intracellular bacteria; 2) autophagosomes form proximally to inclusions, but their maturation is arrested, so autophagy cannot function as an innate immune mechanism to clear the infection; 3) a T4S effector interacts with autophagy proteins to induce autophagosome formation and homes to the bacterial inclusions; and 4) canonical starvation-induced MTOR inhibition and ULK activation are not involved. Although this is similar to A. phagocytophilum Ats-1-induced autophagy,37 there are clear differences between the 2 bacteria: 1) RAB5 is involved in E. chaffeensis-induced autophagy, but not in A. phagocytophilum-induced autophagy; 2) Etf-1 and Ats-1 promote distinct autophagy nucleation complexes derived from early endosomes and ER, respectively; 3) E. chaffeensis inclusions appear as early amphisomes without LC3-II localization, whereas A. phagocytophilum inclusions are early autophagosomes with distinct LC3-II localization;37 and 4) Etf-1 and Ats-1, despite their 21% amino acid sequence identity, selectively target and promote E. chaffeensis and A. phagocytophilum growth,37 respectively. How the 2 molecules recruit distinct autophagy membrane precursors and recognize the parent bacterial inclusions remains to be studied.
Materials and methods
Bacteria and cell culture
The E. chaffeensis Arkansas strain1 was cultured in THP-1 cells (ATCC, TIB-202),122 and the A. phagocytophilum HZ strain123 was cultured in HL-60 cells (ATCC, CCL-240) in RPMI 1640 medium (Mediatech, 10-040-CV) supplemented with 10% fetal bovine serum (Atlanta Biologicals, S12450) and 2 mM l-glutamine (Gibco, 25030). RF/6A cells (ATCC, CRL-1780) were cultured in advanced minimal essential medium (Gibco, 12492) supplemented with 10% fetal bovine serum and 2 mM l-glutamine. The method of synchronous culture of E. chaffeensis in HL-60 cells was similar to those described previously.9,43 Host cell-free E. chaffeensis was used to initiate infection as previously described.9,43 HEK293 cells (ATCC, CRL-1573) and canine histiocytic leukemia DH82 cells were cultured in DMEM (Dulbecco's minimal essential medium; Mediatech, 10-013-CV) supplemented with 10% fetal bovine serum and 2 mM l-glutamine.1 Cultures were incubated at 37°C under 5% CO2 in a humidified atmosphere.
Cloning and cell transfection
Full-length Etf-1 that had been codon-optimized for mammalian expression43 was cloned into pEGFP-N1 (Clontech, 6085-1) and pDsRed-N1 (Clontech, 632412) to create plasmids encoding Etf-1-GFP and Etf-1-DsRed fusion proteins. pEtf-1-GFP was further modified to create pEtf-1-HA. Full-length RAB5A was cloned into pET-41a(+) (Novagen, 70556-3) for GST-RAB5A expression. A double-FYVE finger (2×FYVE) of HGS (HGF-regulated tyrosine kinase substrate) was cloned from mouse cDNA as described44 into vector pEGFP-N1 to create plasmid 2×FYVE-GFP for mammalian expression. pEGFP-C1-ATG5 (pEGFP-C1-hApg5) was obtained from Addgene (plasmid 22952, deposited by Dr. Noboru Mizushima).18 Dominant negative GFP-RAB5AS34N and constitutive active GFP-RAB5AQ79L mutants were constructed from GFP-RAB5A by using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, 200519). RF/6A, HEK293, and DH82 cells were transfected using Fugene HD according to the manufacturer's instructions (Promega, E2311).
Antibodies
Antibodies against A. phagocytophilum outer membrane protein P44 (mAb 5C11),124 dog anti-E. chaffeensis,122 rabbit anti-E. chaffeensis P28,52 ACT/actin (Sigma, A2066), Etf-1,43 BECN1 (Cell Signaling Technology, 3738), PIK3C3/VPS34 (Cell Signaling Technology, 4263), ATG5 (Abcam ab78073), MAP1LC3B/LC3B (NovusBio, NB100-2220), poly-ubiquitinated proteins (Enzo Life Sciences, FK1, BML-PW8805), mono- and poly-ubiquitinated proteins (Enzo Life Sciences, FK2, BML-PW8810), mouse anti-TUBA/α-tubulin (Santa Cruz Biotechnology, sc-5286), MYC/c-Myc (BioLegend, 901501), FLAG M2 (Sigma, F1804), RAB5,125 GFP (mAb Clone B-2; Santa Cruz Biotechnology, sc-9996), anti-VirB6-2,40 Alexa Fluor (AF) 350-conjugated goat anti-mouse IgG (Invitrogen, A-21049), AF555-conjugated goat anti-mouse IgG (Invitrogen, A-21427), and AF488-conjugated goat anti-rabbit IgG (Invitrogen, A-11008) or anti-mouse IgG (Invitrogen, A-11029) were used.
Immunofluorescence assay
E. chaffeensis-infected RF/6A cells at 1 d p.i. were transfected with a plasmid encoding Etf-1-GFP, GFP-RAB5A, GFP-RAB5B, GFP-RAB5C, GFP-ANKFY1, FLAG-PIK3C3, 2×FYVE-GFP, GFP-ATG5, or NPEPPS-GFP.91 Localization was determined at 15 to 17 h (or 24 h for NPEPPS-GFP) p.t. Cells transfected with GFP-LC319 were infected 1 d p.t., and localization was determined at 3 d p.i. Briefly, cells were fixed with 4% paraformaldehyde (PFA) and incubated with rabbit anti-P28, followed by secondary antibodies AF488-conjugated goat anti-rabbit IgG in PGS (phosphate-buffered saline [PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4] supplemented with 0.5% bovine serum albumin [Sigma, A2153], 0.1% gelatin [Sigma, G8150], and 0.1% saponin [Sigma, S4521]). For cells transfected with GFP, Etf-1-GFP, 2×FYVE-GFP, or NPEPPS-GFP, DAPI (4',6-diamidino-2-phenylindole; Invitrogen, D1306) was used to label DNA in the host cell nucleus and E. chaffeensis. Localization of PIK3C3 in transfected cells was determined by immunofluorescence assay with mouse anti-FLAG, followed by AF488-conjugated goat anti-mouse IgG and DAPI.
Localization of native ATG5, LC3, or ubiquitinated proteins was performed with uninfected or E. chaffeensis-infected RF/6A cells cultured on coverslips in a 6-well plate at 2 d p.i. Cells were fixed in 4% PFA and labeled with rabbit anti-ATG5 or -LC3B IgG or with mAb FK2 against mono- and poly-ubiquitinated proteins, followed by AF555-conjugated goat anti-rabbit or -mouse IgG. DAPI was used to label DNA in the host cell nucleus and E. chaffeensis.
Localization of Etf-1 in the host cytoplasm of E. chaffeensis-infected THP-1 cells was determined using SLO (Sigma, S0149) to selectively permeabilize the plasma membrane as described.37 After SLO treatment, cells were immediately fixed in 4% PFA and incubated with rabbit anti-Etf-1 (1:100) and mouse anti-VirB6-2 (1:50), followed by incubation with AF488-conjugated goat anti-rabbit IgG and AF555-conjugated goat anti-mouse IgG in PBS supplemented with 0.5% bovine serum albumin. After washing with PBS, cells were further labeled with mouse anti-VirB6-2 and AF350-conjugated goat anti-mouse IgG diluted in PGS to permeabilize all membranes.43
To examine the localization of DsRed-GAPDH in E. chaffeensis-infected RF/6A cells, infected cells cultured on coverslips in a 12-well plate at 1 d p.i. were transfected with pDsRed-N1 control or pDsRed-GAPDH plasmids using Fugene HD. At 1 d p.t., cells were treated with 0.1 μM DFP (Sigma, D0879) for 2 h and then fixed in 4% PFA and incubated with DAPI to stain bacteria.
To examine the colocalization of Etf-1-DsRed with GFP-ATG5 or GFP-LC3, RF/6A or HEK293 cells at ∼80% confluence were used for transfection. Cells were treated with or without 10 nM BAF (Enzo, BML-CM110-0100) for 16 h at 1 d p.t., prior to fixation in PFA. To examine the distribution of NPEPPS-GFP and Etf-1, sequential transfections were performed. Etf-1 was first transfected into DH82 cells, and NPEPPS-GFP was transfected 1 d later. At 1 d p.t. with NPEPPS-GFP, cells were fixed and incubated with rabbit anti-Etf-1 and AF555-conjugated goat anti-rabbit IgG. To examine the effect of Etf-1 on the degradation of Q103-GFP, Q103-GFP was first transfected into RF/6A cells and, at 16 h p.t., Etf-1-DsRed or DsRed was transfected into RF/6A cells. At 2 d p.t., cells were fixed and incubated with rabbit anti-Etf-1 and AF555-conjugated goat anti-rabbit IgG. Images were captured with a DeltaVision Deconvolution microscope system (GE Healthcare, Marlborough, MA). For western blot analysis, cells were transfected with Q103-GFP or cotransfected with Q103-GFP and Etf-1. At 16 h p.t., the cells were treated with or without 2 mM 3-MA (Sigma, M9281) for 2 d.
Image analysis
Colocalization analysis was performed with softWoRx software from DeltaVision microscope on a single z-section by counting 10 to 20 inclusions per cell in 5 to 20 cells per experiment from 3 independent experiments to obtain percentage colocalization of E. chaffeensis-inclusions with various markers including 2×FYVE, PIK3C3, GAPDH, and Etf-1. Colocalization of Etf-1 with RAB5, ATG5, or LC3 was analyzed by Coloc 2 in the NIH Image J software package for the Pearson correlation coefficient from 10 to 20 cells per group from 3 independent experiments.126 Results were presented as average colocalization percentage or Pearson coefficients (r × 100) ± standard deviation, and data were analyzed by the Student unpaired t test for statistical significance.
3-MA, amino acid supplementation, rapamycin, and PAQ-22 treatment
To determine the effect of 3-MA on E. chaffeensis internalization into THP-1 cells, E. chaffeensis was incubated with 1.5 × 105 THP-1 cells at an multiplicity of infection (MOI) of 1,500:1 for 2 h in the presence of 2.5 mM 3-MA. The cells were washed to remove bacteria that were unbound and then were fixed with 4% PFA. The extracellular and intracellular bacteria were distinguished by immunofluorescence assay using rabbit anti-P28 and AF555-conjugated goat anti-rabbit IgG before permeabilization and rabbit anti-P28 and AF488-conjugated goat anti-rabbit IgG after permeabilization as described.54
THP-1 cells (1.5 × 105) were incubated with E. chaffeensis at an MOI of 50:1. 3-MA at a final concentration of 2 mM was added to E. chaffeensis-infected THP-1 cells at 1 h and 1 d p.i., and E. chaffeensis infection was assessed at 3 d p.i. To determine the reversibility of 3-MA inhibition of E. chaffeensis replication, THP-1 cells were infected with E. chaffeensis for 23 h, and then a portion of cells was incubated with 10 mM 3-MA for 6 h or 58 h. The cells that were treated with 3-MA for 6 h were washed to remove 3-MA, and the incubation was continued for an additional 52 h. Bacterial infection was assessed by Diff-Quik staining (Thermo Scientific, 9990700) and immunofluorescence staining with anti-P28.
For amino acid supplementation, 3 × 105 THP-1 cells were infected with E. chaffeensis in 1 ml complete RPMI 1640 medium. At 1 d p.i., cells were treated with complete RPMI 1640 medium with 2 mM 3-MA, or complete RPMI 1640 medium with 2 mM 3-MA plus 5× MEM essential amino acids solution (Invitrogen, 11130-051), and E. chaffeensis infection was assessed at 3 d p.i. THP-1 cells at 1.5 × 105 were preincubated with 0.1 μM rapamycin (Sigma, R0395) or 0.1% DMSO (control) for 2.5 h, and E. chaffeensis was added at an MOI of 50:1 in the continued presence of rapamycin or DMSO at 2 d p.i. Bacterial infection was assessed by western blotting using antibodies against P28 and ACT/actin and by qPCR using specific primers for E. chaffeensis 16S rDNA, with data normalized against the level of expression of human GAPDH.127
To study the effect of PAQ-22 (Wako Pure Chemical Industries, 165-23581) on bacterial infection, 3 × 105 E. chaffeensis–infected THP-1 cells were incubated with 10 or 100 μM of PAQ-22 or 0.1% DMSO (control) for 2 d.
PtdIns3P assay
Uninfected or E. chaffeensis-infected THP-1 cells (∼2 × 106 cells) at 1 d p.i. were collected. Alternatively, RF/6A cells (1 × 106 cells) were transfected with EGFP (1 μg) or Etf-1-EGFP plasmids (5 μg) by electroporation (100 V) in 0.2-cm cuvettes. Cells were cultured in a T75 flask for 1 d for the Etf-1-transfected groups, before being collected. PtdIns3P lipids were purified according to the manufacturer's instructions (PtdIns3P Mass ELISA kit; Echelon Biosciences, K-3300). Cells were precipitated with 0.5 M trichloroacetic acid (Sigma, T0699), lipids were extracted with MeOH:CHCl3 (2:1), and acidic lipids were extracted with MeOH:CHCl3:12 M HCl (80:40:1). The supernatant (∼2.25 mL) was mixed with 0.75 mL of CHCl3 and 1.35 mL of 0.1 M HCl, and the organic (lower) phase was collected and vacuum dried. The PtdIns3P amount was measured with the competitive ELISA using a SpectraMax Plus 384 microplate reader (Molecular Devices, Sunnyvale, CA).
Utilization of [3H]l-glutamine-labeled THP-1 proteins for synthesis of E. chaffeensis proteins
THP-1 cells (2 × 106) were grown for 24 h in RPMI medium (with 10% FBS and 2 mM glutamine) containing 10 μCi/ml of [3H] l-glutamine (PerkinElmer, NET551250UC), washed, resuspended in fresh growth medium, and infected with E. chaffeensis for 2 d.128 One group of infected cells was treated with 2 mM 3-MA for 6 h. E. chaffeensis was purified from infected cells by sonication and differential centrifugation as described,128 and uninfected THP-1 cells were processed in parallel as control for potential host protein contamination. The protein amount of purified samples was determined, and the incorporation of [3H]glutamine into E. chaffeensis proteins was determined with a liquid scintillation counter, with data normalized to the total protein amount.
siRNA transfection
RF/6A cells were transfected with BECN1 siRNA (Ambion, AM16104) or control siRNA (Santa Cruz Biotechnology; 100 pmol/4 × 104 cells), or HEK293 cells were transfected with LC3B siRNA (Dharmacon, L-012846-00-0005) or RAB5A/B/C siRNA (33 pmol each) (Sigma custom oligos as described in ref. 129) using Lipofectamine 2000 (Invitrogen, 11668-019). E. chaffeensis was added to cells at 1 or 2 d p.t. At 2 d p.i., cells were harvested and subjected to western blotting using antibodies against BECN1, LC3B, RAB5, and E. chaffeensis P28. Band intensities were determined using a Fujifilm Multi Gauge software (GE Healthcare).
E. chaffeensis-infection in bone marrow-derived macrophages from wild-type and atg5flox/flox-Lyz2-Cre mice
BMDMs with atg5 knockout were isolated from atg5flox/flox-Lyz2-Cre mice (atg5 TSKO),64 in which the Cre recombinase driven by Lyz2 promoter was used to target myeloid cells to knockout the third exon of Atg5 flanked by LoxP sites.65 Monocytes were derived from bone marrow of wild type (WT) and atg5 TSKO mice, and macrophages differentiated from the monocytes were incubated with E. chaffeensis as described.54 The infected samples were collected at 7 d p.i and subjected to DNA isolation using the QIAamp DNA blood mini kit (Qiagen, 51106); qPCR was performed using primers specific for E. chaffeensis 16S rDNA and mouse Gapdh.130 The protocol for using animals in this study was approved by the Institutional Laboratory Animal Care and Use Committee.
Effect of Etf-1-GFP, RAB5, or SGSM3 on Ehrlichia infection
Etf-1-GFP or GFP control was transfected with Fugene HD into HEK293 or RF/6A cells during exponential growth. At 8 h p.t., E. chaffeensis or A. phagocytophilum, which were freshly isolated as described,43 were added to infect transfected cells.37,43 Bacteria that were not internalized were removed at 1 d p.i., and cells were harvested at 2 d p.i. Bacterial numbers in each sample were determined by qPCR using specific primers for E. chaffeensis 16S rDNA and normalized against the level of expression of human GAPDH.127 Samples were subjected to western blotting using antibodies against A. phagocytophilum P44 and ACT/actin.
HEK293 cells cultured in a 12-well plate were transfected with plasmids encoding pEGFP, pEGFP-RAB5 (WT, DN, or CA), pcDNA3.1-MYC-SGSM3 (RABGAP5),105 or pcDNA3.1-MYC-SGSM3R165A,105 and infected with E. chaffeensis at 1 d p.t. for 2 d. Cells were lysed in 1× SDS sample buffer, sonicated, and examined by western blotting using antibodies against EGFP (for RAB5), MYC/c-Myc (for SGSM3), and E. chaffeensis P28.
Co-immunoprecipitation
Uninfected or E. chaffeensis-infected THP-1 cells at 2 d p.i. were lysed in modified lysis buffer (25 mM Tris, pH 7.4, 150 mM NaCl, 1% NP40 [USB, 19638], 5% glycerol, and 1% protease inhibitor cocktail III [Calbiochem, 539134]) for 15 min and immunoprecipitated for 2 h with rabbit anti-Etf-1, -BECN1, or -PIK3C3, or mouse anti-RAB5 IgG that was cross-linked to protein A/G-magnetic beads (Pierce, 88805). Normal rabbit (Santa Cruz Biotechnology, sc-2027) or mouse IgG (Santa Cruz Biotechnology, sc-2025) was used as a negative control for immunoprecipitation. Bound proteins were eluted by 2× SDS sample buffer and subjected to western blotting.
HEK293 cells expressing codon-optimized Etf-1-HA and GFP-RAB5 (WT, DN, or CA) or GFP control were lysed in modified lysis buffer as described above. After centrifugation, each supernatant fraction was incubated with mouse anti-HA mAb cross-linked on protein G-sepharose beads (BioLegend, 900801) for 2 h. After extensive washing with lysis buffer, bound proteins were dissociated from the beads with 2× SDS sample buffer, and western blotting was performed.
Affinity isolation assay
A total of 100 μg GST or GST-RAB5 purified and bound to nickel-agarose resin (Sigma, P6611) was used to affinity isolate 100 μg of lysate from E. chaffeensis-infected THP-1 cells (lysis buffer: 43 mM Na2HPO4, 14.7 mM KH2PO4, 137 mM NaCl, 27 mM KCl, pH 7.3, 1% [v/v] Triton X-100 [Sigma, T8787] and 1% protease inhibitor cocktail III) that were precleared with nickel-agarose resin. Western blotting with rabbit anti-Etf-1 was carried out as described.43
Affinity isolation using rEtf-1 (C-terminal 250 residues) containing an N-terminal 6×His-tag43 was performed as described with minor modifications.131 Purified and nickel-agarose-bound rEtf-1 (0.5 mg) was added to lysates from 3 × 107 uninfected THP-1 cells precleared with nickel-agarose. After extensive washing with phosphate buffer (50 mM sodium phosphate, pH 8.0, 0.3 M NaCl) with 1% (v/v) NP40, the bound proteins were eluted with phosphate buffer containing 250 mM imidazole and precipitated with 10% trichloroacetic acid. After centrifugation, the pellet was neutralized with 1.5 M Tris-HCl (pH 8.8) and dissolved in 2× SDS-PAGE loading buffer. Western blotting was performed with rabbit anti-RAB5, rabbit anti-PIK3C3, and mouse anti-BECN1.
Free amino acid analysis by LC-MS/MS
Amino acids were extracted using boiling water as described.69 The resulting extract (200 μl per sample) was loaded onto a 3-kDa Amicon Ultra 0.5-ml filtering device. The samples were spun at 14,000 × g for 45 min at 4°C. Amino acids were separated and quantified using known quantities of amino acid standards that were run before and after samples by LC-MS/MS as described (AB Sciex QTRAP 5500, Foster City, CA and Agilent 1290 LC Infinity II LC System, Santa Clara, CA)69 with minor modifications. A 40-μl aliquot of extract was added to a vial containing 860 μl of nano-pure water and 100 μl of 10 mM hydrochloric acid, and 10 μl of the diluted sample was injected onto the column. The declustering potentials of glutamate, phenylalanine, and proline were modified to 150 V, 150 V, and 160 V, respectively, to avoid saturation of the mass spectrometer detector.
Immunogold labeling
The immunogold labeling method with preembedding gold enhancement was performed as described.132 In brief, cells were fixed in 4% PFA in 0.1 M sodium phosphate buffer (pH 7.4) for 2 h at room temperature and permeabilized in the same buffer containing 0.25% saponin for 30 min. After blocking for 30 min in the same buffer containing 0.1% saponin, 10% bovine serum albumin, 10% normal goat serum (Vector Laboratories, S-1000), and 0.1% cold-water fish skin gelatin (Sigma, G7765) (blocking solution), specimens were incubated with mouse anti-GFP or control mouse IgG overnight, followed by incubation with colloidal gold (1.4 nm in diameter; Nanoprobes, 2001) conjugated to goat anti-mouse IgG in blocking solution for 2 h. After treatment with a silver enhancement kit (SilverEnhance EM; Nanoprobes, 2012) for 2 min at room temperature to intensify the signal, the specimens were postfixed in 1% OsO4 solution containing 1.5% potassium ferrocyanide, dehydrated in a series of graded ethanol solutions, and embedded in epoxy resin. Ultra-thin sections were collected, stained with uranyl acetate and lead citrate, and observed with a Hitachi H7600 (Tokyo, Japan) transmission electron microscope.
Supplementary Material
Abbreviations
- AF
Alexa Fluor
- ANKFY1/Rabankyrin-5
ankyrin repeat and FYVE domain containing 1
- ATG
autophagy-related
- Ats-1
Anaplasma translocated substrate-1
- BECN1
Beclin 1
- CA
constitutively active
- DFP
diisopropylfluorophosphate
- DN
dominant negative
- EEA1
early endosome antigen 1
- Etf-1
Ehrlichia translocated factor-1
- GFP
green fluorescent protein
- HA
hemagglutinin
- HTT
huntingtin
- 3-MA
3-methyladenine
- LAMP1
lysosomal-associated membrane protein 1
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- MAP1LC3/LC3
microtubule-associated protein 1 light chain 3
- MOI
multiplicity of infection
- MTOR
mechanistic target of rapamycin (serine/threonine kinase)
- MVBs
multivesicular bodies
- p.i.
post-infection
- p.t.
post-transfection
- PtdIns3K
phosphatidylinositol 3-kinase
- PtdIns3P
phosphatidylinositol 3-phosphate
- Q103
exon 1 of mutant HTT with 103 glutamine repeats
- qPCR
quantitative polymerase chain reaction
- siRNA
small interfering RNA
- PIK3C3
phosphatidylinositol 3-kinase catalytic subunit type 3
- SGSM3/RABGAP5
small G protein signaling modulator 3
- RAB5-CA
RAB5Q79L
- RAB5-DN
RAB5S34N
- SLO
streptolysin O
- TF
transferrin
- TFRC
transferrin receptor
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. John Brumell at the University of Toronto, Toronto, Canada, for providing plasmids encoding GFP-RAB5A, GFP-RAB5B, and GFP-RAB5C; Dr. Jean Gruenberg at University of Geneva, Switzerland, for monoclonal antibody against RAB5; Dr. Francis Barr at University of Oxford, Oxford, England for providing plasmids pcDNA3.1-MYC-SGSM3 (RABGAP-5) and pcDNA3.1-MYC-SGSM3R165A; and Dr. Junying Yuan, Harvard Medical School, Boston, MA, for providing spautin-1. FLAG-PIK3C3/Vps34 plasmid was kindly provided by Dr. Tamotsu Yoshimori at the Research Institute for Microbial Diseases, Osaka University, Japan. Plasmid pEGFP-C1-LC3, which was kindly provided by Dr. T. Yoshimori, through Dr. Jean Celli at the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Hamilton, MT. Plasmid pEGFP-C1-ATG5 (pEGFP-C1-hApg5, Addgene plasmid # 22952) was a gift from Dr. Noboru Mizushima, and mice with the atg5flox/flox-Lyz2-Cre mutation were kindly provided by Dr. N. Mizushima at the Tokyo Medical Dental University, Tokyo, Japan, through Dr. Herbert W. Virgin, Washington University School of Medicine. The plasmid NPEPPS/PSA-GFP was kindly provided by Dr. Eric Reits, University of Amsterdam, Netherlands. HTT-exon-1-Q103-GFP was a kind gift from Dr. Krill Bersuker, Stanford University, Stanford, CA. Plasmid pEGFP-C1-ANKFY1/Rabankyrin-5 was a kind gift from Dr. Steve Caplan, University of Nebraska Medical Center, Omaha, NE.
We are grateful to the Ohio State University Targeted Metabolomics Laboratory (metabolomics.osu.edu) for access to the LC-MS/MS equipment, which is funded by Translational Plant Sciences Targeted Investment in Excellence.
Funding
This work was supported by a National Institutes of Health grant, R01 AI054476.
References
- [1]. Dawson JE, Anderson BE, Fishbein DB, Sanchez JL, Goldsmith CS, Wilson KH, Duntley CW. Isolation and characterization of an Ehrlichia sp. from a patient diagnosed with human ehrlichiosis. J Clin Microbiol 1991; 29:2741–5; PMID:1757540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2]. Anderson BE, Dawson JE, Jones DC, Wilson KH. Ehrlichia chaffeensis, a new species associated with human ehrlichiosis. J Clin Microbiol 1991; 29:2838–42; PMID:1757557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3]. Maeda K, Markowitz N, Hawley RC, Ristic M, Cox D, McDade JE. Human infection with Ehrlichia canis, a leukocytic rickettsia. N Engl J Med 1987; 316:853–6; PMID:3029590; http://dx.doi.org/10.1056/NEJM198704023161406 [DOI] [PubMed] [Google Scholar]
- [4]. Paddock CD, Childs JE. Ehrlichia chaffeensis: a prototypical emerging pathogen. Clin Microbiol Rev 2003; 16:37–64; PMID:12525424; http://dx.doi.org/10.1128/CMR.16.1.37-64.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5]. Walker DH, Ismail N, Olano JP, McBride JW, Yu XJ, Feng HM. Ehrlichia chaffeensis: a prevalent, life-threatening, emerging pathogen. Trans Am Clin Climatol Assoc 2004; 115:375–82; discussion 82-4; PMID:17060980 [PMC free article] [PubMed] [Google Scholar]
- [6]. Abu Kwaik Y, Bumann D. Microbial quest for food in vivo: 'nutritional virulence' as an emerging paradigm. Cell Microbiol 2013; 15:882–90; PMID:23490329; http://dx.doi.org/10.1111/cmi.12138 [DOI] [PubMed] [Google Scholar]
- [7]. Rikihisa Y. Molecular pathogenesis of Ehrlichia chaffeensis infection. Annu Rev Microbiol 2015; 69:283–304; PMID:26488275; http://dx.doi.org/10.1146/annurev-micro-091014-104411 [DOI] [PubMed] [Google Scholar]
- [8]. Barnewall RE, Rikihisa Y, Lee EH. Ehrlichia chaffeensis inclusions are early endosomes which selectively accumulate transferrin receptor. Infect Immun 1997; 65:1455–61; PMID:9119487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9]. Mott J, Barnewall RE, Rikihisa Y. Human granulocytic ehrlichiosis agent and Ehrlichia chaffeensis reside in different cytoplasmic compartments in HL-60 cells. Infect Immun 1999; 67:1368–78; PMID:10024584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10]. Lin M, Rikihisa Y. Degradation of p22phox and inhibition of superoxide generation by Ehrlichia chaffeensis in human monocytes. Cell Microbiol 2007; 9:861–74; PMID:17087735; http://dx.doi.org/10.1111/j.1462-5822.2006.00835.x [DOI] [PubMed] [Google Scholar]
- [11]. Hotopp JC, Lin M, Madupu R, Crabtree J, Angiuoli SV, Eisen JA, Seshadri R, Ren Q, Wu M, Utterback TR, et al. . Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet 2006; 2:e21; PMID:16482227; http://dx.doi.org/10.1371/journal.pgen.0020021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12]. De Duve C, Wattiaux R. Functions of lysosomes. Annu Rev Physiol 1966; 28:435–92; PMID:5322983; http://dx.doi.org/10.1146/annurev.ph.28.030166.002251 [DOI] [PubMed] [Google Scholar]
- [13]. Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat?Cell Biol 2010; 12:814–22; PMID:20811353; http://dx.doi.org/10.1038/ncb0910-814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14]. Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell 2011; 147:728–41; PMID:22078875; http://dx.doi.org/10.1016/j.cell.2011.10.026 [DOI] [PubMed] [Google Scholar]
- [15]. Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol 2013; 14:759–74; PMID:242021801009 1109; http://dx.doi.org/10.1038/nrm3696 [DOI] [PubMed] [Google Scholar]
- [16]. Mizushima N, Yoshimori T, Ohsumi Y. The role of atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 2011; 27:107–32; PMID:21801009; http://dx.doi.org/10.1146/annurev-cellbio-092910-154005 [DOI] [PubMed] [Google Scholar]
- [17]. Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell 2008; 19:5360–72; PMID:18843052; http://dx.doi.org/10.1091/mbc.E08-01-0080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18]. Mizushima N, Sugita H, Yoshimori T, Ohsumi Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J Biol Chem 1998; 273:33889–92; PMID:9852036; http://dx.doi.org/10.1074/jbc.273.51.33889 [DOI] [PubMed] [Google Scholar]
- [19]. Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 2000; 19:5720–8; PMID:11060023; http://dx.doi.org/10.1093/emboj/19.21.5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20]. Dunn WA, Jr. Studies on the mechanisms of autophagy: formation of the autophagic vacuole. J Cell Biol 1990; 110:1923–33; PMID:2351689; http://dx.doi.org/10.1083/jcb.110.6.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21]. Berg TO, Fengsrud M, Stromhaug PE, Berg T, Seglen PO. Isolation and characterization of rat liver amphisomes. Evidence for fusion of autophagosomes with both early and late endosomes. J Biol Chem 1998; 273:21883–92; PMID:9705327; http://dx.doi.org/10.1074/jbc.273.34.21883 [DOI] [PubMed] [Google Scholar]
- [22]. Liou W, Geuze HJ, Geelen MJ, Slot JW. The autophagic and endocytic pathways converge at the nascent autophagic vacuoles. J Cell Biol 1997; 136:61–70; PMID:9008703; http://dx.doi.org/10.1083/jcb.136.1.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23]. Ahlberg J, Marzella L, Glaumann H. Uptake and degradation of proteins by isolated rat liver lysosomes. Suggestion of a microautophagic pathway of proteolysis. Lab Invest 1982; 47:523–32; PMID:6755063 [PubMed] [Google Scholar]
- [24]. Morvan J, Kochl R, Watson R, Collinson LM, Jefferies HB, Tooze SA. In vitro reconstitution of fusion between immature ?autophagosomes and endosomes. Autophagy 2009; 5:676–89; PMID:19337031; http://dx.doi.org/10.4161/auto.5.5.8378 [DOI] [PubMed] [Google Scholar]
- [25]. Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol 2007; 9:1102–9; PMID:17909521; http://dx.doi.org/10.1038/ncb1007-1102 [DOI] [PubMed] [Google Scholar]
- [26]. Klionsky DJ, Eskelinen EL, Deretic V. Autophagosomes, phagosomes, autolysosomes, phagolysosomes, autophagolysosomes … wait, I'm confused. Autophagy 2014; 10:549–51; PMID:24657946; http://dx.doi.org/10.4161/auto.28448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27]. Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol 2009; 10:1215–21; PMID:19820708; http://dx.doi.org/10.1038/ni.1800 [DOI] [PubMed] [Google Scholar]
- [28]. Yoshikawa Y, Ogawa M, Hain T, Yoshida M, Fukumatsu M, Kim M, Mimuro H, Nakagawa I, Yanagawa T, Ishii T, et al. . Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat Cell Biol 2009; 11:1233–40; PMID:19749745; http://dx.doi.org/10.1038/ncb1967 [DOI] [PubMed] [Google Scholar]
- [29]. Dupont N, Lacas-Gervais S, Bertout J, Paz I, Freche B, Van Nhieu GT, Van Der Goot FG, Sansonetti PJ, Lafont F. Shigella phagocytic vacuolar membrane remnants participate in the cellular response to pathogen invasion and are regulated by autophagy. Cell Host Microbe 2009; 6:137–49; PMID:19683680; http://dx.doi.org/10.1016/j.chom.2009.07.005 [DOI] [PubMed] [Google Scholar]
- [30]. Zheng YT, Shahnazari S, Brech A, Lamark T, Johansen T, Brumell JH. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol 2009; 183:5909–16; PMID:19812211; http://dx.doi.org/10.4049/jimmunol.0900441 [DOI] [PubMed] [Google Scholar]
- [31]. Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 2004; 119:753–66; PMID:15607973; http://dx.doi.org/10.1016/j.cell.2004.11.038 [DOI] [PubMed] [Google Scholar]
- [32]. Gutierrez MG, Vazquez CL, Munafo DB, Zoppino FC, Beron W, Rabinovitch M, Colombo MI. Autophagy induction favours the generation and maturation of the Coxiella-replicative vacuoles. Cell Microbiol 2005; 7:981–93; PMID:15953030; http://dx.doi.org/10.1111/j.1462-5822.2005.00527.x [DOI] [PubMed] [Google Scholar]
- [33]. Starr T, Child R, Wehrly TD, Hansen B, Hwang S, Lopez-Otin C, Virgin HW, Celli J. Selective subversion of autophagy complexes facilitates completion of the Brucella intracellular cycle. Cell Host Microbe 2012; 11:33–45; PMID:22264511; http://dx.doi.org/10.1016/j.chom.2011.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34]. Steele S, Brunton J, Ziehr B, Taft-Benz S, Moorman N, Kawula T. Francisella tularensis harvests nutrients derived via ATG5-independent autophagy to support intracellular growth. PLoS Pathog 2013; 9:e1003562; PMID:23966861; http://dx.doi.org/10.1371/journal.ppat.1003562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35]. Niu H, Yamaguchi M, Rikihisa Y. Subversion of cellular autophagy by Anaplasma phagocytophilum. Cell Microbiol 2008; 10:593–605; PMID:17979984; http://dx.doi.org/10.1111/j.1462-5822.2007.01068.x [DOI] [PubMed] [Google Scholar]
- [36]. Niu H, Rikihisa Y. Ats-1: a novel bacterial molecule that links ?autophagy to bacterial nutrition. Autophagy 2013; 9:787–8; PMID:23388398; http://dx.doi.org/10.4161/auto.23693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37]. Niu H, Xiong Q, Yamamoto A, Hayashi-Nishino M, Rikihisa Y. Autophagosomes induced by a bacterial Beclin 1 binding protein facilitate obligatory intracellular infection. Proc Natl Acad Sci U S A 2012; 109:20800–7; PMID:23197835; http://dx.doi.org/10.1073/pnas.1218674109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38]. Ohashi N, Zhi N, Lin Q, Rikihisa Y. Characterization and transcriptional analysis of gene clusters for a type IV secretion machinery in human granulocytic and monocytic ehrlichiosis agents. Infect Immun 2002; 70:2128–38; PMID:11895979; http://dx.doi.org/10.1128/IAI.70.4.2128-2138.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39]. Cheng Z, Wang X, Rikihisa Y. Regulation of type IV secretion apparatus genes during Ehrlichia chaffeensis intracellular development by a previously unidentified protein. J Bacteriol 2008; 190:2096–105; PMID:18192398; http://dx.doi.org/10.1128/JB.01813-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40]. Bao W, Kumagai Y, Niu H, Yamaguchi M, Miura K, Rikihisa Y. Four VirB6 paralogs and VirB9 are expressed and interact in Ehrlichia chaffeensis-containing vacuoles. J Bacteriol 2009; 191:278–86; PMID:18952796; http://dx.doi.org/10.1128/JB.01031-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41]. Rikihisa Y, Lin M. Anaplasma phagocytophilum and Ehrlichia chaffeensis type IV secretion and Ank proteins. Curr Opin Microbiol 2010; 13:59–66; PMID:20053580; http://dx.doi.org/10.1016/j.mib.2009.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42]. Backert S, Meyer TF. Type IV secretion systems and their effectors in bacterial pathogenesis. Curr Opin Microbiol 2006; 9:207–17; PMID:16529981; http://dx.doi.org/10.1016/j.mib.2006.02.008 [DOI] [PubMed] [Google Scholar]
- [43]. Liu H, Bao W, Lin M, Niu H, Rikihisa Y. Ehrlichia type IV secretion effector ECH0825 is translocated to mitochondria and curbs ROS and apoptosis by upregulating host MnSOD. Cell Microbiol 2012; 14:1037–50; PMID:22348527; http://dx.doi.org/10.1111/j.1462-5822.2012.01775.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44]. Gillooly DJ, Morrow IC, Lindsay M, Gould R, Bryant NJ, Gaullier JM, Parton RG, Stenmark H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J 2000; 19:4577–88; PMID:10970851; http://dx.doi.org/10.1093/emboj/19.17.4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45]. Schnatwinkel C, Christoforidis S, Lindsay MR, Uttenweiler-Joseph S, Wilm M, Parton RG, Zerial M. The Rab5 effector Rabankyrin-5 regulates and coordinates different endocytic mechanisms. PLoS Biol 2004; 2:E261; PMID:15328530; http://dx.doi.org/10.1371/journal.pbio.0020261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46]. Patki V, Virbasius J, Lane WS, Toh BH, Shpetner HS, Corvera S. Identification of an early endosomal protein regulated by phosphatidylinositol 3-kinase. Proc Natl Acad Sci U S A 1997; 94:7326–30; PMID:9207090; 18215151http://dx.doi.org/10.1073/pnas.94.14.7326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47]. Simonsen A, Lippe R, Christoforidis S, Gaullier JM, Brech A, Callaghan J, Toh BH, Murphy C, Zerial M, Stenmark H. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature 1998; 394:494–8; PMID:9697774; 18215151http://dx.doi.org/10.1038/28879 [DOI] [PubMed] [Google Scholar]
- [48]. Backer JM. The regulation and function of Class III PI3Ks: novel roles for Vps34. Biochem J 2008; 410:1–17; PMID:18215151; http://dx.doi.org/10.1042/BJ20071427 [DOI] [PubMed] [Google Scholar]
- [49]. Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3'-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem 2000; 275:992–8; PMID:10625637; http://dx.doi.org/10.1074/jbc.275.2.992 [DOI] [PubMed] [Google Scholar]
- [50]. Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci U S A 1982; 79:1889–92; PMID:69516467569 2238; http://dx.doi.org/10.1073/pnas.79.6.1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51]. Lindmo K, Stenmark H. Regulation of membrane traffic by ?phosphoinositide 3-kinases. J Cell Sci 2006; 119:605–14; PMID:16467569; http://dx.doi.org/10.1242/jcs.02855 [DOI] [PubMed] [Google Scholar]
- [52]. Ohashi N, Zhi N, Zhang Y, Rikihisa Y. Immunodominant major outer membrane proteins of Ehrlichia chaffeensis are encoded by a polymorphic multigene family. Infect Immun 1998; 66:132–9; PMID:942384925425236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53]. Cheng Z, Lin M, Rikihisa Y. Ehrlichia chaffeensis proliferation begins with NtrY/NtrX and PutA/GlnA upregulation and CtrA degradation induced by Proline and Glutamine Uptake. MBio 2014; 5:e02141; PMID:25425236; http://dx.doi.org/10.1128/mBio.02141-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54]. Mohan Kumar D, Yamaguchi M, Miura K, Lin M, Los M, Coy JF, Rikihisa Y. Ehrlichia chaffeensis uses its surface protein EtpE to bind GPI-anchored protein DNase X and trigger entry into mammalian cells. PLoS Pathog 2013; 9:e1003666; PMID:24098122; http://dx.doi.org/10.1371/journal.ppat.1003666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55]. Jaber N, Dou Z, Chen JS, Catanzaro J, Jiang YP, Ballou LM, Selinger E, Ouyang X, Lin RZ, Zhang J, et al. . Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc Natl Acad Sci U S A 2012; 109:2003–8; PMID:22308354; http://dx.doi.org/10.1073/pnas.1112848109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56]. Vieira OV, Bucci C, Harrison RE, Trimble WS, Lanzetti L, Gruenberg J, Schreiber AD, Stahl PD, Grinstein S. Modulation of Rab5 and Rab7 recruitment to phagosomes by phosphatidylinositol 3-kinase. Mol Cell Biol 2003; 23:2501–14; PMID:12640132; http://dx.doi.org/10.1128/MCB.23.7.2501-2514.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57]. Kihara A, Kabeya Y, Ohsumi Y, Yoshimori T. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep 2001; 2:330–5; PMID:11306555; http://dx.doi.org/10.1093/embo-reports/kve061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58]. Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, Cai Y, Norberg HV, Zhang T, Furuya T, et al. . Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell 2011; 147:223–34; PMID:21962518; http://dx.doi.org/10.1016/j.cell.2011.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59]. Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 1991; 253:905–9; PMID:1715094; 20637199http://dx.doi.org/10.1126/science.1715094 [DOI] [PubMed] [Google Scholar]
- [60]. Homer CR, Richmond AL, Rebert NA, Achkar JP, McDonald C. ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in Crohn's disease pathogenesis. Gastroenterology 2010; 139;1630–41, 41 e1-2; PMID:20637199; http://dx.doi.org/10.1053/j.gastro.2010.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61]. Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol 2001; 152:657–68; PMID:11266458; http://dx.doi.org/10.1083/jcb.152.4.657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62]. Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature 2004; 432:1032–6; PMID:15525940; http://dx.doi.org/10.1038/nature03029 [DOI] [PubMed] [Google Scholar]
- [63]. Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al. . Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006; 441:885–9; PMID:16625204; http://dx.doi.org/10.1038/nature04724 [DOI] [PubMed] [Google Scholar]
- [64]. Zhao Z, Fux B, Goodwin M, Dunay IR, Strong D, Miller BC, Cadwell K, Delgado MA, Ponpuak M, Green KG, et al. . Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe 2008; 4:458–69; PMID:18996346; http://dx.doi.org/10.1016/j.chom.2008.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65]. Cross M, Mangelsdorf I, Wedel A, Renkawitz R. Mouse lysozyme M gene: isolation, characterization, and expression studies. Proc Natl Acad Sci U S A 1988; 85:6232–6; PMID:3413093; 17700057http://dx.doi.org/10.1073/pnas.85.17.6232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66]. Zhao Z, Thackray LB, Miller BC, Lynn TM, Becker MM, Ward E, Mizushima NN, Denison MR, Virgin HW, 4th. Coronavirus replication does not require the autophagy gene ATG5. Autophagy 2007; 3:581–5; PMID:17700057; http://dx.doi.org/10.4161/auto.4782 [DOI] [PubMed] [Google Scholar]
- [67]. George MD, Baba M, Scott SV, Mizushima N, Garrison BS, Ohsumi Y, Klionsky DJ. Apg5p functions in the sequestration step in the cytoplasm-to-vacuole targeting and macroautophagy pathways. Mol Biol Cell 2000; 11:969–82; PMID:10712513; http://dx.doi.org/10.1091/mbc.11.3.969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68]. Chen R, Zou Y, Mao D, Sun D, Gao G, Shi J, Liu X, Zhu C, Yang M, Ye W, et al. . The general amino acid control pathway regulates mTOR and autophagy during serum/glutamine starvation. J Cell Biol 2014; 206:173–82; PMID:25049270; http://dx.doi.org/10.1083/jcb.201403009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69]. Cocuron JC, Anderson B, Boyd A, Alonso AP. Targeted metabolomics of Physaria fendleri, an industrial crop producing hydroxy fatty acids. Plant Cell Physiol 2014; 55:620–33; PMID:24443498; http://dx.doi.org/10.1093/pcp/pcu011 [DOI] [PubMed] [Google Scholar]
- [70]. Sneve ML, Overbye A, Fengsrud M, Seglen PO. Comigration of two autophagosome-associated dehydrogenases on two-dimensional polyacrylamide gels. Autophagy 2005; 1:157–62; PMID:16874067; http://dx.doi.org/10.4161/auto.1.3.2037 [DOI] [PubMed] [Google Scholar]
- [71]. Ge Y, Rikihisa Y. Surface-exposed proteins of Ehrlichia chaffeensis. Infect Immun 2007; 75:3833–41; PMID:17517859; http://dx.doi.org/10.1128/IAI.00188-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72]. Kumagai Y, Matsuo J, Hayakawa Y, Rikihisa Y. Cyclic di-GMP signaling regulates invasion of Ehrlichia chaffeensis into human monocytes. J Bacteriol 2010; 192:4122–33; PMID:20562302; http://dx.doi.org/10.1128/JB.00132-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73]. Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL. U.K. induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol 2013; 15:741–50; PMID:23685627; http://dx.doi.org/10.1038/ncb2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74]. Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 2011; 13:132–41; PMID:21258367; http://dx.doi.org/10.1038/ncb2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75]. Ferrari S, Bandi HR, Hofsteenge J, Bussian BM, Thomas G. Mitogen-activated 70K S6 kinase. Identification of in vitro 40 S ribosomal S6 phosphorylation sites. J Biol Chem 1991; 266:22770–5; PMID:193928219150980 [PubMed] [Google Scholar]
- [76]. Hawley SA, Davison M, Woods A, Davies SP, Beri RK, Carling D, Hardie DG. Characterization of the AMP-activated protein kinase kinase from rat liver and identification of threonine 172 as the major site at which it phosphorylates AMP-activated protein kinase. J Biol Chem 1996; 271:27879–87; PMID:8910387; 19150980http://dx.doi.org/10.1074/jbc.271.44.27879 [DOI] [PubMed] [Google Scholar]
- [77]. Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem 2009; 284:8023–32; PMID:19150980; http://dx.doi.org/10.1074/jbc.M900301200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78]. Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci U S A 2008; 105:20567–74; PMID:19074260; http://dx.doi.org/10.1073/pnas.0810611105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79]. Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007; 282:24131–45; PMID:17580304; http://dx.doi.org/10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- [80]. Ichimura Y, Kumanomidou T, Sou YS, Mizushima T, Ezaki J, Ueno T, Kominami E, Yamane T, Tanaka K, Komatsu M. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem 2008; 283:22847–57; PMID:18524774; http://dx.doi.org/10.1074/jbc.M802182200 [DOI] [PubMed] [Google Scholar]
- [81]. Fujita N, Morita E, Itoh T, Tanaka A, Nakaoka M, Osada Y, ?Umemoto T, Saitoh T, Nakatogawa H, Kobayashi S, et al. . ?Recruitment of the autophagic machinery to endosomes during infection is mediated by ubiquitin. J Cell Biol 2013; 203:115–28; PMID:24100292; http://dx.doi.org/10.1083/jcb.201304188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82]. Alonso S, Pethe K, Russell DG, Purdy GE. Lysosomal killing of Mycobacterium mediated by ubiquitin-derived peptides is enhanced by autophagy. Proc Natl Acad Sci U S A 2007; 104:6031–6; PMID:17389386; http://dx.doi.org/10.1073/pnas.0700036104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83]. Purdy GE, Russell DG. Ubiquitin trafficking to the lysosome: keeping the house tidy and getting rid of unwanted guests. Autophagy 2007; 3:399–401; PMID:17457035; http://dx.doi.org/10.4161/auto.4272 [DOI] [PubMed] [Google Scholar]
- [84]. Barnett TC, Liebl D, Seymour LM, Gillen CM, Lim JY, Larock CN, Davies MR, Schulz BL, Nizet V, Teasdale RD, et al. . The globally disseminated M1T1 clone of group A Streptococcus evades autophagy for intracellular replication. Cell Host Microbe 2013; 14:675–82; PMID:24331465; http://dx.doi.org/10.1016/j.chom.2013.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85]. Khweek AA, Caution K, Akhter A, Abdulrahman BA, Tazi M, ?Hassan H, Majumdar N, Doran A, Guirado E, Schlesinger LS, et al. . A bacterial protein promotes the recognition of the Legionella pneumophila vacuole by autophagy. Eur J Immunol 2013; 43:1333–44; PMID:23420491; http://dx.doi.org/10.1002/eji.201242835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86]. Campbell AM, Kessler PD, Fambrough DM. The alternative carboxyl termini of avian cardiac and brain sarcoplasmic reticulum/endoplasmic reticulum Ca(2+)-ATPases are on opposite sides of the membrane. J Biol Chem 1992; 267:9321–5; PMID:153362915068806 [PubMed] [Google Scholar]
- [87]. Bhakdi S, Tranum-Jensen J, Sziegoleit A. Mechanism of membrane damage by streptolysin-O. Infect Immun 1985; 47:52–60; PMID:388073015068806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88]. Venkatraman P, Wetzel R, Tanaka M, Nukina N, Goldberg AL. Eukaryotic proteasomes cannot digest polyglutamine sequences and release them during degradation of polyglutamine-containing proteins. Mol Cell 2004; 14:95–104; PMID:15068806; http://dx.doi.org/10.1016/S1097-2765(04)00151-0 [DOI] [PubMed] [Google Scholar]
- [89]. Raspe M, Gillis J, Krol H, Krom S, Bosch K, van Veen H, Reits E. Mimicking proteasomal release of polyglutamine peptides initiates aggregation and toxicity. J Cell Sci 2009; 122:3262–71; PMID:19690053; http://dx.doi.org/10.1242/jcs.045567 [DOI] [PubMed] [Google Scholar]
- [90]. Bhutani N, Venkatraman P, Goldberg AL. Puromycin-sensitive aminopeptidase is the major peptidase responsible for digesting polyglutamine sequences released by proteasomes during protein degradation. EMBO J 2007; 26:1385–96; PMID:17318184; http://dx.doi.org/10. 1038,/sj.emboj.760 1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91]. Menzies FM, Hourez R, Imarisio S, Raspe M, Sadiq O, Chandraratna D, O'Kane C, Rock KL, Reits E, Goldberg AL, et al. . Puromycin-sensitive aminopeptidase protects against aggregation-prone proteins via autophagy. Hum Mol Genet 2010; 19:4573–86; PMID:20829225; 14985334http://dx.doi.org/10. 1093/hmg/ddq385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92]. Harding TM, Morano KA, Scott SV, Klionsky DJ. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol 1995; 131:591–602; PMID:7593182; 14985334http://dx.doi.org/10. 1083/jcb.131.3.591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93]. Komoda M, Kakuta H, Takahashi H, Fujimoto Y, Kadoya S, Kato F, Hashimoto Y. Specific inhibitor of puromycin-sensitive aminopeptidase with a homophthalimide skeleton: identification of the target molecule and a structure-activity relationship study. Bioorg Med Chem 2001; 9:121–31; PMID:11197332; 14985334http://dx.doi.org/10. 1016,/ S0968,–0896(00) 00231–5 [DOI] [PubMed] [Google Scholar]
- [94]. Kakuta H, Koiso Y, Nagasawa K, Hashimoto Y. Fluorescent bioprobes for visualization of puromycin-sensitive aminopeptidase in living cells. Bioorg Med Chem Lett 2003; 13:83–6; PMID:12467622; 14985334http://dx.doi.org/10. 1016,/ S0960,–894X(02) 00845–4 [DOI] [PubMed] [Google Scholar]
- [95]. Dou Z, Pan JA, Dbouk HA, Ballou LM, DeLeon JL, Fan Y, Chen JS, Liang Z, Li G, Backer JM, et al. . Class IA PI3K p110beta subunit promotes autophagy through Rab5 small GTPase in response to growth factor limitation. Mol Cell 2013; 50:29–42; PMID:23434372; 14985334http://dx.doi.org/10. 1016/j.molcel.2013.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96]. Su WC, Chao TC, Huang YL, Weng SC, Jeng KS, Lai MM. Rab5 and class III phosphoinositide 3-kinase Vps34 are involved in hepatitis C virus NS4B-induced autophagy. J Virol 2011; 85:10561–71; PMID:21835792; 14985334http://dx.doi.org/10. 1128,/JVI. 00173–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97]. Ravikumar B, Imarisio S, Sarkar S, O'Kane CJ, Rubinsztein DC. Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease. J Cell Sci 2008; 121:1649–60; PMID:18430781; 14985334http://dx.doi.org/10. 1242/jcs.025726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98]. Bucci C, Lutcke A, Steele-Mortimer O, Olkkonen VM, Dupree P, Chiariello M, Bruni CB, Simons K, Zerial M. Co-operative regulation of endocytosis by three Rab5 isoforms. FEBS Lett 1995; 366:65–71; PMID:7789520; 14985334http://dx.doi.org/10. 1016,/ 0014–5793(95)00477-Q [DOI] [PubMed] [Google Scholar]
- [99]. Huang F, Khvorova A, Marshall W, Sorkin A. Analysis of clathrin-mediated endocytosis of epidermal growth factor receptor by RNA interference. J Biol Chem 2004; 279:16657–61; PMID:14985334; http://dx.doi.org/10.1074/jbc.C400046200 [DOI] [PubMed] [Google Scholar]
- [100]. Su X, Lodhi IJ, Saltiel AR, Stahl PD. Insulin-stimulated Interaction between insulin receptor substrate 1 and p85alpha and activation of protein kinase B/Akt require Rab5. J Biol Chem 2006; 281:27982–90; PMID:16880210; http://dx.doi.org/10.1074/jbc.M602873200 [DOI] [PubMed] [Google Scholar]
- [101]. Christoforidis S, Miaczynska M, Ashman K, Wilm M, Zhao L, Yip SC, Waterfield MD, Backer JM, Zerial M. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nature cell biology 1999; 1:249–52; PMID:10559924; http://dx.doi.org/10.1038/12075 [DOI] [PubMed] [Google Scholar]
- [102]. Pfeffer SR. GTP-binding proteins in intracellular transport. Trends Cell Biol 1992; 2:41–6; PMID:14731525; http://dx.doi.org/10.1016/0962-8924(92)90161-F [DOI] [PubMed] [Google Scholar]
- [103]. Bourne HR. Do GTPases direct membrane traffic in secretion? Cell 1988; 53:669–71; PMID:2836065; http://dx.doi.org/10.1016/0092-8674(88)90081-5 [DOI] [PubMed] [Google Scholar]
- [104]. Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol 2009; 10:513–25; PMID:19603039; http://dx.doi.org/10.1038/nrm2728 [DOI] [PubMed] [Google Scholar]
- [105]. Haas AK, Fuchs E, Kopajtich R, Barr FA. A GTPase-activating protein controls Rab5 function in endocytic trafficking. Nat Cell Biol 2005; 7:887–93; PMID:16086013; http://dx.doi.org/10.1038/ncb1290 [DOI] [PubMed] [Google Scholar]
- [106]. Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature 2011; 469:323–35; PMID:21248839; http://dx.doi.org/10.1038/nature09782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107]. Deretic V. Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol Rev 2011; 240:92–104; PMID:21349088; http://dx.doi.org/10.1111/j.1600-065X.2010.00995.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108]. Lin TC, Chen YR, Kensicki E, Li AY, Kong M, Li Y, Mohney RP, Shen HM, Stiles B, Mizushima N, et al. . Autophagy: resetting glutamine-dependent metabolism and oxygen consumption. Autophagy 2012; 8:1477–93; PMID:22906967; http://dx.doi.org/10.4161/auto.21228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109]. Fratti RA, Chua J, Vergne I, Deretic V. Mycobacterium tuberculosis glycosylated phosphatidylinositol causes phagosome maturation arrest. Proc Natl Acad Sci U S A 2003; 100:5437–42; PMID:12702770; http://dx.doi.org/10.1073/pnas.0737613100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110]. Puri RV, Reddy PV, Tyagi AK. Secreted acid phosphatase (SapM) of Mycobacterium tuberculosis is indispensable for arresting phagosomal maturation and growth of the pathogen in guinea pig tissues. PLoS One 2013; 8:e70514; PMID:23923000; http://dx.doi.org/10.1371/journal.pone.0070514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111]. Alvarez-Dominguez C, Madrazo-Toca F, Fernandez-Prieto L, ?Vandekerckhove J, Pareja E, Tobes R, Gomez-Lopez MT, Del Cerro-Vadillo E, Fresno M, Leyva-Cobián F, et al. . Characterization of a Listeria monocytogenes protein interfering with Rab5a. Traffic 2008; 9:325–37; PMID:18088303; http://dx.doi.org/10.1111/j.1600-0854.2007.00683.x [DOI] [PubMed] [Google Scholar]
- [112]. Pal A, Severin F, Lommer B, Shevchenko A, Zerial M. Huntingtin-HAP40 complex is a novel Rab5 effector that regulates early endosome motility and is up-regulated in Huntington's disease. J Cell Biol 2006; 172:605–18; PMID:16476778; http://dx.doi.org/10.1083/jcb.200509091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113]. Tooze J, Hollinshead M, Ludwig T, Howell K, Hoflack B, Kern H. In exocrine pancreas, the basolateral endocytic pathway converges with the autophagic pathway immediately after the early endosome. J Cell Biol 1990; 111:329–45; PMID:2166050; http://dx.doi.org/10.1083/jcb.111.2.329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114]. Eskelinen EL. Maturation of autophagic vacuoles in Mammalian cells. Autophagy 2005; 1:1–10; PMID:16874026; http://dx.doi.org/10.4161/auto.1.1.1270 [DOI] [PubMed] [Google Scholar]
- [115]. Fader CM, Colombo MI. Autophagy and multivesicular bodies: two closely related partners. Cell Death Differ 2009; 16:70–8; PMID:19008921; http://dx.doi.org/10.1038/cdd.2008.168 [DOI] [PubMed] [Google Scholar]
- [116]. Shin HW, Hayashi M, Christoforidis S, Lacas-Gervais S, Hoepfner S, Wenk MR, Modregger J, Uttenweiler-Joseph S, Wilm M, Nystuen A, et al. . An enzymatic cascade of Rab5 effectors regulates ?phosphoinositide turnover in the endocytic pathway. J Cell Biol 2005; 170:607–18; PMID:16103228; http://dx.doi.org/10.1083/jcb.200505128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117]. Vieira OV, Botelho RJ, Rameh L, Brachmann SM, Matsuo T, Davidson HW, Schreiber A, Backer JM, Cantley LC, Grinstein S. Distinct roles of class I and class III phosphatidylinositol 3-kinases in phagosome formation and maturation. J Cell Biol 2001; 155:19–25; PMID:11581283; http://dx.doi.org/10.1083/jcb.200107069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118]. Kobayashi T, Stang E, Fang KS, de Moerloose P, Parton RG, ?Gruenberg J. A lipid associated with the antiphospholipid syndrome regulates endosome structure and function. Nature 1998; 392:193–7; PMID:9515966; http://dx.doi.org/10.1038/32440 [DOI] [PubMed] [Google Scholar]
- [119]. Petiot A, Faure J, Stenmark H, Gruenberg J. PI3P signaling regulates receptor sorting but not transport in the endosomal pathway. J Cell Biol 2003; 162:971–9; PMID:12975344; http://dx.doi.org/10.1083/jcb.200303018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120]. Fernandez-Borja M, Wubbolts R, Calafat J, Janssen H, Divecha N, Dusseljee S, Neefjes J. Multivesicular body morphogenesis requires phosphatidyl-inositol 3-kinase activity. Curr Biol 1999; 9:55–8; PMID:9889123; http://dx.doi.org/10.1016/S0960-9822(99)80048-7 [DOI] [PubMed] [Google Scholar]
- [121]. Zhang J, Reiling C, Reinecke JB, Prislan I, Marky LA, Sorgen PL, Naslavsky N, Caplan S. Rabankyrin-5 interacts with EHD1 and Vps26 to regulate endocytic trafficking and retromer function. Traffic 2012; 13:745–57; PMID:22284051; http://dx.doi.org/10.1111/j.1600-0854.2012.01334.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122]. Barnewall RE, Rikihisa Y. Abrogation of gamma interferon-induced inhibition of Ehrlichia chaffeensis infection in human monocytes with iron-transferrin. Infect Immun 1994; 62:4804–10; PMID:7927758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123]. Rikihisa Y, Zhi N, Wormser GP, Wen B, Horowitz HW, Hechemy KE. Ultrastructural and antigenic characterization of a granulocytic ehrlichiosis agent directly isolated and stably cultivated from a patient in New York state. J Infect Dis 1997; 175:210–3; PMID:8985223; http://dx.doi.org/10.1093/infdis/175.1.210 [DOI] [PubMed] [Google Scholar]
- [124]. Kim HY, Rikihisa Y. Characterization of monoclonal antibodies to the 44-kgdalton major outer membrane protein of the human granulocytic ehrlichiosis agent. J Clin Microbiol 1998; 36:3278–84; PMID:9774579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125]. Cavalli V, Vilbois F, Corti M, Marcote MJ, Tamura K, Karin M, Arkinstall S, Gruenberg J. The stress-induced MAP kinase p38 regulates endocytic trafficking via the GDI:Rab5 complex. Mol Cell 2001; 7:421–32; PMID:11239470; http://dx.doi.org/10.1016/S1097-2765(01)00189-7 [DOI] [PubMed] [Google Scholar]
- [126]. Dunn KW, Kamocka MM, McDonald JH. A practical guide to evaluating colocalization in biological microscopy. Am J Physiol Cell Physiol 2011; 300:C723–42; PMID:21209361; http://dx.doi.org/10.1152/ajpcell.00462.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127]. Cheng Z, Kumagai Y, Lin M, Zhang C, Rikihisa Y. Intra-leukocyte expression of two-component systems in Ehrlichia chaffeensis and Anaplasma phagocytophilum and effects of the histidine kinase inhibitor closantel. Cell Microbiol 2006; 8:1241–52; PMID:16882029; http://dx.doi.org/10.1111/j.1462-5822.2006.00704.x [DOI] [PubMed] [Google Scholar]
- [128]. Hatch TP. Competition between Chlamydia psittaci and L cells for host isoleucine pools: a limiting factor in chlamydial multiplication. Infect Immun 1975; 12:211–20; PMID:1095493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129]. Chen PI, Kong C, Su X, Stahl PD. Rab5 isoforms differentially regulate the trafficking and degradation of epidermal growth factor receptors. J Biol Chem 2009; 284:30328–38; PMID:19723633; http://dx.doi.org/10.1074/jbc.M109.034546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130]. Miura K, Rikihisa Y. Liver transcriptome profiles associated with strain-specific Ehrlichia chaffeensis-induced hepatitis in SCID mice. Infect Immun 2009; 77:245–54; PMID:19001077; http://dx.doi.org/10.1128/IAI.00979-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131]. Nougayrede JP, Foster GH, Donnenberg MS. Enteropathogenic Escherichia coli effector EspF interacts with host protein Abcf2. Cell Microbiol 2007; 9:680–93; PMID:17064289; http://dx.doi.org/10.1111/j.1462-5822.2006.00820.x [DOI] [PubMed] [Google Scholar]
- [132]. Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, ?Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol 2009; 11:1433–7; PMID:19898463; http://dx.doi.org/10.1038/ncb1991 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


