Abstract
F1F0 ATP synthases are bidirectional molecular motors that translocate protons across the cell membrane by either synthesizing or hydrolyzing ATP. Alkaliphile ATP synthases are highly adapted, performing oxidative phosphorylation at high pH against an inverted pH gradient (acidin/alkalineout). Unlike mesophilic ATP synthases, alkaliphilic enzymes have tightly regulated ATP hydrolysis activity, which can be relieved in the presence of lauryldimethylamine oxide. Here, we characterized the rotary dynamics of the Caldalkalibacillus thermarum TA2.A1 F1 ATPase (TA2F1) with two forms of single molecule analysis, a magnetic bead duplex and a gold nanoparticle. TA2F1 rotated in a counterclockwise direction in both systems, adhering to Michaelis-Menten kinetics with a maximum rotation rate (Vmax) of 112.4 revolutions/s. TA2F1 displayed 120° unitary steps coupled with ATP hydrolysis. Torque measurements revealed the highest torque (52.4 piconewtons) derived from an F1 molecule using fluctuation theorem. The implications of high torque in terms of extreme environment adaptation are discussed.
Keywords: ATP synthase, enzyme kinetics, enzyme mechanism, F1F0-ATPase, respiratory chain, single molecule biophysics, adaptation, rotation, thermoalkaliphile, torque
Introduction
ATP synthases are membrane-bound rotary molecular motors that are the major enzymes responsible for ATP synthesis in the majority of aerobic life (1, 2). A key feature of the enzyme is the mechanism by which the translocation of protons (or Na+ ions) across the membrane barrier is linked to either synthesizing or hydrolyzing ATP (1–3). Bacterial F1F0-type ATP synthases are two-domain enzymes and are typically composed of eight subunits, α3β3γδϵab2c10–15 (4, 5). The soluble F1 domain is composed of the α3β3γδϵ subunits in which the α- and β-subunits are alternatively arranged in a hexameric ring-shaped catalytic core. ATP hydrolysis and synthesis occur in the catalytic sites that are formed by each of the β-subunits (4, 6). The core rotary shaft is composed of the γ- and ϵ-subunits. The γ-subunit penetrates the center of the α3β3 catalytic hexamer at one end and is associated with the a- and c-subunits of the F0 domain at the other end (7). The F0 domain is composed of ab2c9–15 subunits and is membrane-bound (8). The c-subunits form a ring parallel to the membrane (9); however, although stoichiometry is fixed within a species, it is variable between species (10–15). The a-subunit forms a collar around the c-ring and is thought to conduct ions both into and out of the c-ring in a two-channel model (16–19). The c-ring rotates and, together with a-subunit, conducts protons or Na+ ions across the cell membrane (9, 20). The tightly associated γ- and ϵ-subunits together with the c-ring functionally form the rotor portion of the enzyme, whereas the α3β3δab2 subunits form the stator (4, 5). The coupling of ion translocation through the ac-ring complex to the clockwise rotation of the rotor forces conformational changes within the β-subunits in the α3β3 catalytic hexamer, catalyzing the synthesis of ATP from ADP and inorganic phosphate (21). Conversely, the α3β3 catalytic hexamer can hydrolyze ATP, causing anticlockwise rotation of the γϵc10–15 rotor, resulting in the pumping of either protons or Na+ ions into the bulk phase (21–24). It is also noteworthy that ATP hydrolysis is suppressed in aerobic alkaliphilic organisms (25–27).
When the F1 domain is separated from the F0 domain it can be regarded as an ATPase and is useful for the study of the mechanical properties of ATP/ADP-Pi catalysis and enzyme function (28). Indeed, the rotation of F1 ATPases has been visualized using optical microscopy by attaching a probe to the γ- or ϵ-subunit in the relative area of where the F0 domain would be located had it been attached (23, 28, 29). As this method of study has developed, the type of probe utilized has diversified from actin filaments (23, 30) into gold nanoparticles/nanorods (31, 32), magnetic beads (33, 34), polystyrene beads (35, 36), and fluorescent molecules (37–39). The two most heavily studied bacterial F1 ATPases come from the thermophile Bacillus PS3 (TF1) and the mesophile Escherichia coli (EF1). Both enzymes are capable of ATP hydrolysis activity, rotating stepwise at an ATP concentration-dependent rate in an anticlockwise direction (23, 40). Because the hexameric α3β3 structure harbors three catalytic sites, the basic step size is 120° coupled to either the consumption or production of one ATP molecule (41). For both TF1 and EF1, these 120° steps have been shown to consist of substeps of 80° and 40° and of 85° and 35° substeps, respectively (35, 40, 42). The 80°/85° substeps are proposed to be triggered by ATP binding and ADP release and are referred to as ATP-binding (ATP-waiting) dwells (37), whereas the 40°/35° substeps have been shown to occur after ATP cleavage and release of inorganic phosphate and are referred to as catalytic dwells (43).
Upon catalysis, the generation of torque at the interface between the rotor (γϵ subcomplex) and the stator (α3β3δ subcomplex) results in the rotation of the F1 by using the chemical free energy from ATP hydrolysis. Due to the interconversion of chemical free energy and electrochemical potential via mechanical rotation, torque is an important factor affecting the energy conversion efficiency of a molecular rotor. Stepping torque has been estimated for TF1 (41, 44), EF1 (40), the thermophilic Thermus thermophilus V1 ATPase (TtV1) (44, 45), and the acidophilic Enterococcus hirae V1 ATPase (EhV1) (44), the last of which is proposed to act solely as a V1V0-type ATPase (24, 44, 46). Interestingly, of the F1-type enzymes described here, the torque appears to be higher in the thermophilic TF1 and, according to some reports, lower in EF1. This implies that adaptation to temperature might result in an enzyme with higher torque. In agreement with this, of the V1-type enzymes, the TtV1 torque is greater than that of EhV1; however, the torque difference between TtV1 and EhV1 is much greater than that between TF1 and EF1, suggesting that function may also have a role in defining torque, i.e. ATP synthesis versus hydrolysis (see Table 1).
TABLE 1.
Torque of TA2F1 and other rotary ATPases
| Protein | Torque | Known function | Organism growth conditions | Ref. |
|---|---|---|---|---|
| pNnm | ||||
| TA2F1 | 52 ± 4a,b | ATP synthase | pH 9.5/65 °C | This study |
| EhV1 | 20a,b | ATP hydrolase | pH 5.0/25 °C | 41 |
| TtV1 | 35a,b | ATP Synthase/hydrolase | pH 7.5/65 °C | 45 |
| 33 ± 2a,b | 44 | |||
| TF1 | 41 ± 2a,b | ATP Synthase/hydrolase | pH 7.5/65 °C | This study |
| 39 ± 4a,b | 41 | |||
| EF1 | 30c | ATP Synthase/hydrolase | pH 6.5/35 °C | 40 |
a The values were determined by fluctuation theorem.
b The values are the means ± S.D.
c The value was determined from the angular velocity and frictional drag coefficient of the probe.
In this study, we examine the F1 ATPase from the thermoalkaliphilic Caldalkalibacillus thermarum TA2.A1. Alkaliphile ATP synthases are highly adapted, performing oxidative phosphorylation at high pH against an inverted pH gradient (acidin/alkalineout), thereby challenging Mitchell's chemiosmotic model (25, 27, 47–49). The environmental pressure of an alkaline environment, together with increased membrane permeability to ions at high temperature (50), results in a severe pressure to conserve energy. Unlike the ATP synthases/hydrolases described here, alkaliphilic enzymes have tightly regulated ATP hydrolysis activity (25–27), which can be relieved in the presence of LDAO3 (26, 51–53). In addition to this, both the γ- and ϵ-subunits of the C. thermarum TA2.A1 ATP synthase (TA2F1F0) are proposed to have regulatory roles in ATP hydrolysis (51, 53). Because this enzyme is the antithesis of the EhV1V0 (i.e. it only performs ATP synthesis (26)), it was of significant interest to examine both catalytic mechanism and torque. In addition, due to the thermophilic origin of TA2F1F0, we are also able to directly compare the effect of alkaliphily on torque. Here, we characterized the rotary dynamics of TA2F1 with two forms of single molecule analysis, a magnetic bead and a gold nanoparticle.
Results
Expression of TA2F1 and TA2F1γ2c in E. coli
To examine rotary dynamics in F1 ATPases, the enzyme must be capable of being labeled with a trackable probe such as a 200-nm magnetic bead duplex or a 40-nm gold nanoparticle. For other enzymes such as the TF1 ATPase (23, 31, 34, 42, 54), this was achieved by the introduction of two cysteine residues in the γ-subunit. The F1 portion of the atp operon from C. thermarum TA2.A1, coding for TA2F1 α3β3γδϵ, with a hexahistidine tag at the N terminus of the β-subunit, and the same enzyme including two cysteines introduced at positions 107 and 210 in the γ-subunit (TA2F1γ2c; Fig. 1A) were cloned into the expression plasmid pTrc99A and overexpressed in the unc deletion mutant E. coli DK8. TA2F1 and TA2F1γ2c were extracted from E. coli strain DK8 cytoplasmic fractions and purified via a three-step purification procedure. SDS-PAGE analysis of the purified recombinant mutant enzyme (TA2F1γ2c) revealed five subunits (viz. α, β, γ, δ, and ϵ) that were identical to those of the native F1 purified from the overexpressed WT TA2F1 previously by Keis et al. (51) (Fig. 1B, lane 2). Postpurification, TA2F1γ2c was specifically biotin-modified via the cysteine residues at positions 107 and 210 in the γ-subunit for attachment of rotation probes (Fig. 1B, lane 1). This was confirmed by Western blotting (Fig. 1B, lane 2), revealing that biotin was specifically incorporated into the γ-subunit of TA2F1γ2c.
FIGURE 1.
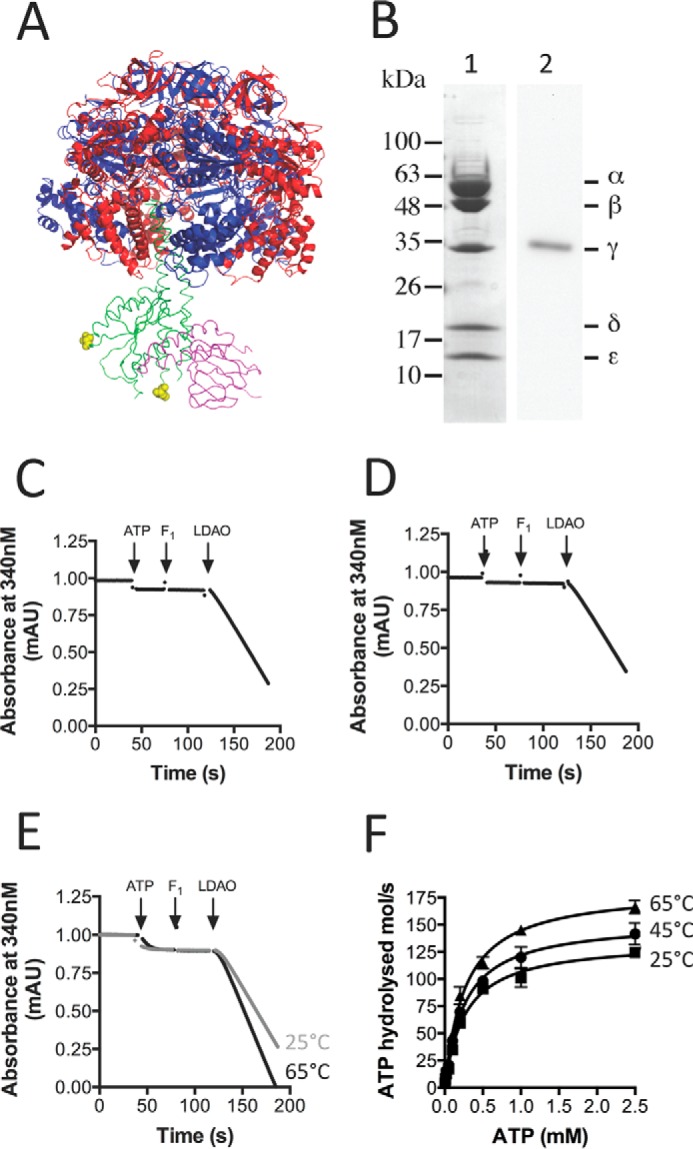
Purification, labeling, and biochemical features of TA2F1γ2c. A, schematic representation of the TA2F1 structure (Protein Data Bank code 2QE7 (53)) showing the relative positions of the two introduced cysteine residues (γH107C/γS210C; in yellow Corey-Pauling-Koltun modeling; hence TA2F1γ2c) located in the γ-subunit (green). The ϵ-subunit is pink, and the α- and β-subunits are shown in blue and red, respectively. This model was constructed and rendered using PyMOL (Schrödinger, LLC). B, lane 1, SDS-PAGE analysis of the purified, biotin-labeled TA2F1γ2c ATPase resolved on a 12% polyacrylamide gel and stained with Coomassie Brilliant Blue. B, lane 2, prior to Western blotting analysis, biotin-labeled TA2F1γ2c ATPase was separated by SDS-PAGE (12%). TA2F1γ2c was subsequently transferred onto a polyvinylidene difluoride membrane and immunoblotted using a streptavidin-alkaline phosphatase conjugate directed against the biotin-modified γ-subunit of TA2F1γ2c. 10 μg of protein was used in all separations. C and D, representative plots showing the effect of 0.1% LDAO on biotin-labeled TA2F1γ2c (C) or TA2F1 WT (D) ATPase activity at 45 °C. E, representative plots showing the effect of 0.1% LDAO on biotin-labeled TA2F1γ2c ATPase activity at 25 (gray line) and 65 °C (black line). Arrows indicate the points during the time traces where various assay components were added. F, effect of ATP concentration and temperature on biotin-labeled TA2F1γ2c ATP hydrolysis activity in the presence of 0.1% LDAO. All measurements were at pH 8.0, and three replicates were performed using 5 μg of protein per measurement with the ATP-regenerating assay described under “Experimental Procedures.” mAU, milliabsorbance units. Error bars represent S.E.
ATP Hydrolysis of TA2F1γ2c-Biotin
A feature of the TA2F1 enzyme is its specific blockage in ATP hydrolysis activity and that this activity can be stimulated up to 30-fold with 0.05% LDAO (26, 51). Like the recombinantly expressed native TA2F1, TA2F1γ2c-biotin was blocked in ATP hydrolysis activity. Neither the WT nor the biotin-modified mutant displayed any measureable ATP hydrolysis activity (Fig. 1, C and D). ATP hydrolysis could be induced in TA2F1γ2c-biotin at both 25 and 45 °C with 0.1% LDAO (Fig. 1, E and F). Per TA2F1γ2c-biotin molecule, this resulted in a maximal specific activity of 124.3 (30.4 units/mg of protein), 141.6 (34 units/mg of protein), and 166.2 mol/s (Fig. 1F) at 25, 45, and 65 °C, respectively. The Km for ATP was 0.27 mm at 25 °C, 0.28 mm at 45 °C, and 0.26 mm at 65 °C. The activities reported here are consistent with those observed previously for TA2F1 by Keis et al. (51) (28.5 units/mg of protein at 45 °C). We have further optimized the assay by introduction of 50 mm KCl to optimize pyruvate kinase activity (data not shown). In this optimized assay system, upon addition of ATP, we observe a small sharp drop indicative of a small amount of ADP present in the ATP. This is followed by a flat line indicating that the TA2F1 ATPase has close to zero ATP hydrolysis activity at 25, 45, and 65 °C (see Fig. 1, C–E, upon ATP introduction).
Rotation Velocity Is Dependent on ATP Concentration
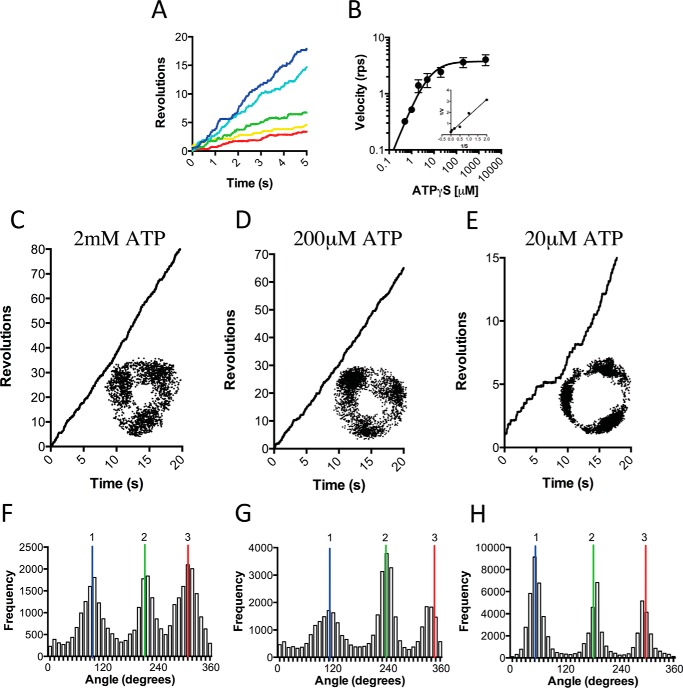
Because TA2F1 does not natively catalyze hydrolysis and we sought to examine TA2F1 rotational dynamics with ATP hydrolysis-driven rotation, we required a system capable of activating F1 ATPase activity. Fortunately, we have two methods to achieve this in the form of LDAO and the use of magnetic beads with magnetic tweezers, the latter of which is well documented to aid in relieving the TF1 ATPase inhibited states (34, 55, 56). Although the use of magnetic beads is a powerful tool, to examine the true maximum rotation speed, a probe with no viscous drag is required. Therefore, to explore the kinetic properties of the TA2F1, a 40-nm gold nanoparticle was utilized. The hydrodynamic friction on a 40-nm gold nanoparticle is practically negligible, allowing the observation of unloaded rotation of TA2F1 to be observed provided the bead is unobstructed and free to rotate in the assay solution (42). Considering our low flow chamber volume and requirement for accessibility to the chamber volume, assays at 65 °C present a significant technical problem. However, because ATP hydrolysis at 25 °C is 73% of that possible at 65 °C (see Fig. 1E), we considered it appropriate to conduct all rotation experiments forthwith at 25 °C. To observe single enzyme rotation, TA2F1γ2c-biotin was immobilized on an NTA-modified glass surface in a flow chamber using hexahistidine tags at the N terminus of the β-subunit, and a 40-nm streptavidin-functionalized gold nanoparticle was attached at the top of the γ-subunit as a rotation probe (Fig. 2A). Initial experiments did not include an ATP regeneration system or LDAO. These experiments revealed very few rotating molecules, each of which only performed two to five revolutions before lapsing into an inactive state. Henceforth, we performed rotation studies using TA2F1γ2c using a 40-nm gold nanoparticle system in the presence of 0.1% LDAO and an ATP regeneration system. Rotation was observed at a variety of ATP concentrations using total internal reflection dark field microscopy (31) recorded at a rate of 10,000 frames per second (fps). In the presence of LDAO, TA2F1 rotated counterclockwise for continuous revolutions that decreased in velocity upon decrease in ATP concentration (Fig. 2, B and C). Above 1 mm, rotation rates were essentially constant, and below 200 μm ATP, the rotation rates were proportional to ATP concentration. This suggests that ATP binding may be rate-limiting at ATP concentrations below 200 μm. Rotation velocities at various ATP concentrations collectively followed Michaelis-Menten kinetics with a Vmax of 112.4 ± 4.3 revolutions s−1 at 2 mm ATP and an apparent ATP Km of 46.85 ± 3.69 μm (Fig. 2C). The second-order rate constant (Kon[ATP]) was determined using 3 × Vmax/Km derived from Fig. 2C and was estimated to be (7.20 ± 0.1) × 106 m−1 s−1 assuming that three ATP molecules are hydrolyzed per revolution of the gold nanoparticle attached to the γ-subunit.
FIGURE 2.
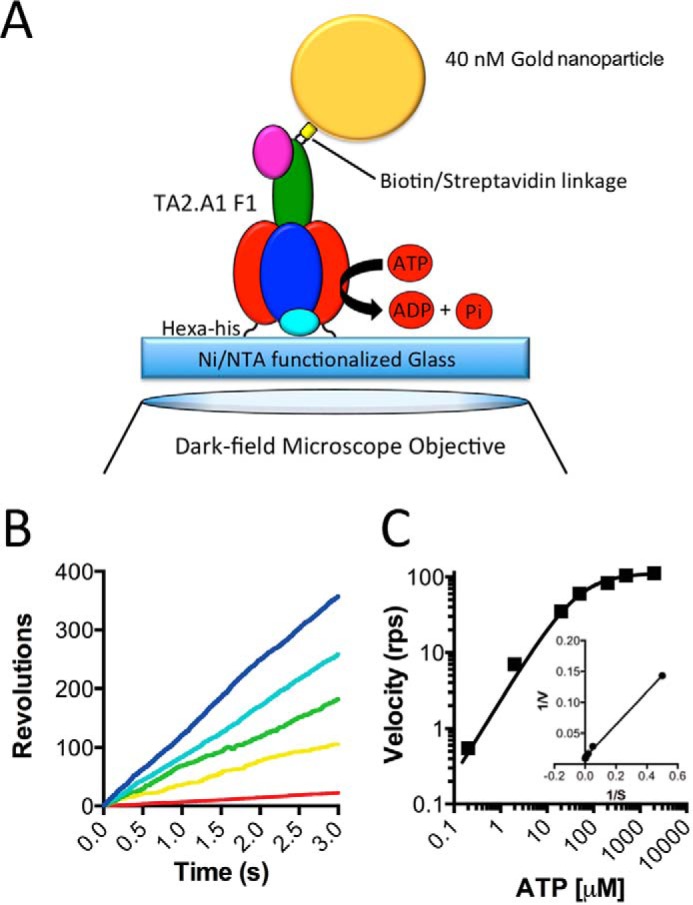
Effect of ATP concentration on TA2F1γ2c rotation rate with a gold nanoparticle. A, schematic representation of the experimental setup for a single molecule rotation assay of the TA2F1γ2c gold nanoparticle probe. The stator α3β3δ subcomplex of TA2F1γ2c was fixed onto the nickel-NTA glass surface with the hexahistidine tag at the N terminus of each β-subunit. A streptavidin-coated 40-nm gold nanoparticle was attached to biotinylated cysteine residues in the γ-subunit rotor (γH107C/γS210C). B and C, effect of ATP concentration on rotation of TA2F1γ2c using the system displayed in A. B, displayed are representative rotation traces recorded at 10,000 fps with a variety of ATP concentrations: 2 mm (blue), 200 μm (cyan), 50 μm (green), 20 μm (yellow), and 2 μm (red). C, displayed are the average rotation rates of 15 molecules at the concentrations of ATP indicated on the graph. Error bars represent S.E. C, inset, Lineweaver-Burke plot of data from C used to define the Vmax of 112.4 ± 4.3 revolutions s−1 (rps) and Km of 46.85 ± 3.69 μm.
Stepping Behavior and Dwell Time Analysis of TA2F1
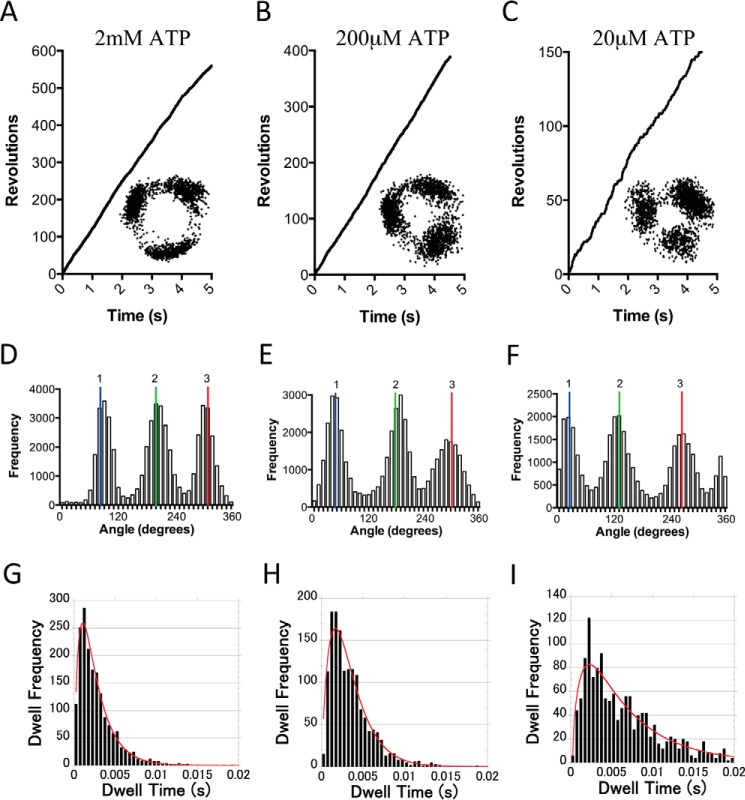
A feature of F1 ATP synthases is the 3-fold symmetry of the catalytic domain of the enzyme. Typical examples of rotation at ATP concentrations of 2 mm, 200 μm, and 20 μm ATP are shown in Fig. 3, A–F. At almost all ATP concentrations, it was possible to observe clear 120° stepwise rotation of TA2F1 under both Vmax and low ATP conditions as shown in the x-y trajectories and distribution of rotary angles (Fig. 3, A–C, insets, and D–F). Under Vmax conditions, each 120° step was completed within 0.2 ms; however, this time period was highly dependent on ATP concentration, underscoring that rotation rate is directly related to the duration of the intervening 120° pauses. At 2 mm ATP, considerably above the Km of 46.85 ± 3.69 μm, these pauses are proposed to represent catalytic dwells. To obtain the time constants and kinetic parameters, the duration of these pauses were examined. Unambiguous analysis was achieved by examination of the first 30 s of traces from 10 molecules distributed over 2 orders magnitude of substrate concentration. These were modeled using a three-consecutive reaction scheme representing ATP binding, hydrolysis, and ADP/Pi release. All distributions of the durations showed a convex shape (Fig. 3, G–I). The average time constants at 2 mm, 200 μm, and 20 μm ATP (Fig. 3, G–I) revealed three time constants that were (i) 2.2 ± 0.2, 2.6 ± 0.3, and 6.2 ± 0.9 ms, respectively; (ii) 0.42 ± 0.04, 0.32 ± 0.04, and 0.68 ± 0.07 ms, respectively; and (iii) 0.42 ± 0.24, 0.78 ± 0.27, and 0.7 ± 0.18 ms, respectively. These time constants correspond to (i) ATP-hydrolysis, (ii) ATP binding, and (iii) ADP/phosphate release. It is unclear from this analysis what time constant corresponds to which reaction step. The second-order rate constant (Kon[ATP]) determined from the distribution of the duration of the ATP-waiting dwells was estimated to be (8.3 ± 0.2) × 106 m−1 s−1, consistent with (7.20 ± 0.1) × 106 m−1 s−1 determined by 3 × Vmax/Km derived from Fig. 2C. To resolve what time constant corresponds to which reaction step, we examined the catalytic mechanism to see whether TA2F1 displayed the same substep behavior as TF1 (35). This is achieved by closely examining rotational stepping behavior around the Km value. Unfortunately, we were unable to observe substepping using the full TA2F1 construct with a gold nanoparticle; however, the 120° steps were clearly broadened at 50 μm ATP (Km = 46.85 ± 3.69 μm; data not shown).
FIGURE 3.
Rotation of TA2F1 reveals three distinct dwell states separated by 120°. A–C, representative rotation traces recorded using gold nanoparticle as a rotation probe at a variety of ATP concentrations: 2 mm (A), 200 μm (B), and 20 μm (C). Insets from A–C show the x-y trajectories of the centroid of the rotating gold nanoparticle from the respective traces. D–F are distributions of rotary angles shown in A–C. G–I are distributions of duration times of dwells at each 120° step composed of dwells from 15 molecules from rotation traces analyzed collectively at the same ATP concentrations as in A–C. 1, 2 and 3 are the three dwell positions separated by 120°. The bin width across all plots was 0.5 ms, and the red curves show the fitting with a triple exponential model: m1 × (exp(−1/m2 × m0) − exp(−1/m3 × m0) − exp(−1/m4 × m0)); m1 = 150, m2 = 0.002, m3 = 0.0005, and m4 = 0.0005. For G, τ1 = 2.2 ± 0.2 ms, τ2 = 0.42 ± 0.04 ms, and τ3 =0.42 ± 0.24 ms (n = 1835). For H, τ1 = 2.6 ± 0.3 ms, τ2 = 0.32 ± 0.04 ms, and τ3 = 0.98 ± 0.27 ms (n = 1500). For I, τ1 = 6.2 ± 0.9 ms, τ2 = 0.68 ± 0.07 ms, and τ3 = 0.7 ± 0.18 ms (n = 1500). Fitting was conducted with KaleidaGraph version 4.5.
Rotation of TA2F1 Using ATP and ATPγS with Magnetic Beads Reveals a Six-dwell Rotation Profile
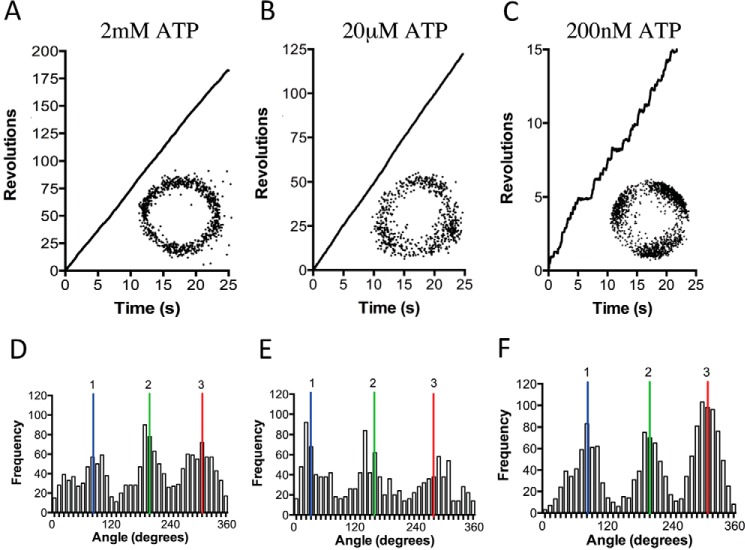
To avoid using LDAO as an activation method, we replaced the gold nanoparticle with magnetic beads, allowing mechanical activation; this is a well documented method to aid in relieving TF1 inhibited states (34, 55, 56). In place of a gold nanoparticle, a ∼200-nm streptavidin-functionalized magnetic bead duplex was attached to the γ-subunit as a rotation probe (Fig. 4A). Initially, rotation was observed at a saturating ATP concentration of 2 mm using phase-contrast microscopy at 30 fps. As with the gold nanoparticle rotation system, in the absence of LDAO or any mechanical stimulation, very few rotating molecules were observed. Each molecule in this system only performed two to five revolutions before lapsing into an inactive state. However, upon a single forced counterclockwise rotation using magnetic tweezers, TA2F1 was capable of ATP hydrolysis (Fig. 4B)-driven rotation at speeds of up to 6.9 revolutions s−1 (Fig. 4, C and D). Rotation was observed at a variety of ATP concentrations that decreased in velocity upon decrease in ATP concentration, obeying Michaelis-Menten kinetics. The maximum rotary velocity measured under Vmax conditions was 6.5 ± 0.4 revolutions s−1 with an apparent ATP Km of 2.72 ± 0.45 μm (Fig. 4, C and D). The suppression of rotation velocity and lowering of apparent Km are a typical feature of using a large, high viscosity probe for rotation observation (31, 33, 44). The second-order rate constant (Kon[ATP]) was determined using 3 × Vmax/Km derived from Fig. 4D and was estimated to be (9.90 ± 0.1) × 106 m−1 s−1, assuming that three ATP molecules are hydrolyzed per revolution of the γ-subunit. This is very much in agreement with observations with the values for ATP of (7.20 ± 0.1) × 106 m−1 s−1 in the same assay system using a gold nanoparticle. However, in the magnetic bead system, these 120° steps were only clearly observed during rotation at very low ATP concentrations (Fig. 5, A–F). Because this is due to the combined effect of camera and probe response times coupled with the high viscous friction of the probe, we changed to using a fast framing camera at 5000 fps to take advantage of the ability to capture stepping. Initially, ATP was used as substrate; however, the 120° steps were simply broadened at 3 μm ATP. In light of this, we attempted to slow the catalytic steps to clarify the stepping behavior. One such method of achieving this is using the slowly hydrolyzable ATP analogue ATPγS (35); however, using this analogue removes the possibility of harnessing the ATP regeneration assay to remove the buildup of Mg-ADP. To limit this unwanted effect, observation times in the rotation assay were limited to 10 min before flushing the flow cell with fresh ATPγS at the desired concentration. Magnetic bead rotation observations at various ATPγS concentrations revealed a maximum rotation velocity of 4.02 ± 0.3 revolutions s−1 under saturating ATPγS concentrations (2 mm; Fig. 6A). From the various ATPγS concentrations tested, TA2F1 has an apparent Km of 5.69 ± 1.33 μm (Fig. 6B). Due to the recording rate being at 5000 fps, discrete 120° steps were observed at most concentrations of ATPγS tested (Fig. 6, C–H). However, at a concentration of 5 μm ATPγS, multistep (six-step) rotary profiles with 50° and 70° substeps were observed (data not shown). Unfortunately, due to the rarity of the events measured and the less than discrete localization, this is a tentative estimate.
FIGURE 4.
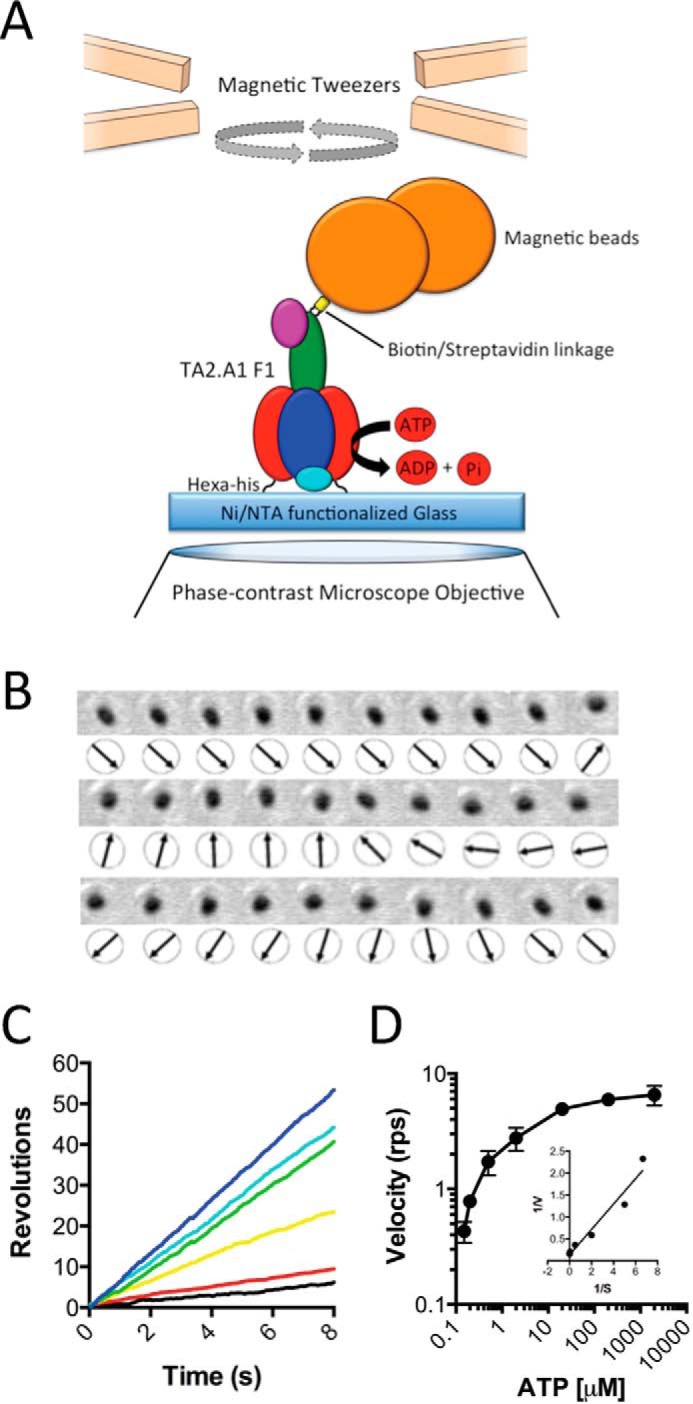
Effect of ATP concentration on TA2F1 rotation rate with magnetic beads. A, schematic representation of the experimental setup for a single molecule rotation assay of TA2F1γ2c with a magnetic bead as a probe. The stator α3β3δ subcomplex of TA2F1γ2c was fixed onto the nickel-NTA glass surface with the hexahistidine tag at the N terminus of each β-subunit. A streptavidin-coated 200-nm magnetic bead was attached to biotinylated cysteine residues in the γ-subunit rotor (γH107C/γS210C). B, frame-by-frame montage of a single rotating magnetic bead directly showing rotation states. C and D, effect of ATP concentration on rotation of TA2F1γ2c using the system displayed in A. C, displayed are representative rotation traces recorded at 30 fps with a variety of ATP concentrations: 2 mm (blue), 200 μm (cyan), 20 μm (green), 2 μm (yellow), 200 nm (red), and 150 nm (black). D, displayed are the average rotation rates of 15 molecules at the concentrations of ATP indicated. Error bars represent S.D. D, inset, Lineweaver-Burke plot of data from D used to define the Vmax of 6.5 ± 0.4 revolutions s−1 (rps) and Km of 2.72 ± 0.45 μm. The rotation assay was conducted as described under “Experimental Procedures.”
FIGURE 5.
Effect of ATP concentration on TA2F1 dwell state distribution with a magnetic bead. A–C, representative rotation traces recorded using a magnetic particle as a rotation probe at a variety of ATP concentrations: 2 mm (A), 20 μm (B), and 200 nm (C). Insets from A–C show the x-y trajectories of the centroid of the rotating particles from the respective traces. D–F show the distributions of rotary angles shown in A–C. 1, 2 and 3 are the three dwell positions separated by 120°. The traces, plots, and trajectories are representative of 15 molecules. The rotation assay was conducted as described under “Experimental Procedures.”
FIGURE 6.
Effect of ATPγS concentration on TA2F1 rotation rate with a magnetic bead. Shown is the effect of ATPγS concentration on rotation of TA2F1γ2c using the system displayed in Fig. 4A. A, displayed are representative rotation traces recorded at 1000 fps with a variety of ATPγS concentrations: 2 mm (blue), 200 μm (cyan), 20 μm (green), 2 μm (yellow), and 1 μm (red). B, displayed are the average rotation rates of 15 molecules at the concentrations of ATP indicated. Error bars represent S.D. B, inset, Lineweaver-burke plot of data from B used to define the Vmax of 4.02 ± 0.3 revolutions s−1 (rps) and Km of 5.61 ± 1.33 μm. C–E, representative rotation traces recorded using a magnetic particle as a rotation probe at a variety of ATPγS concentrations: 2 mm (C), 200 μm (D), and 20 μm (E). Insets from C–E show the x-y trajectories of the centroid of the rotating particles from the respective traces. F–H are distributions of rotary angles shown in C–E. 1, 2 and 3 are the three dwell positions separated by 120°. The traces, plots, and trajectories are representative of 15 molecules. The rotation assay was conducted as described under “Experimental Procedures.”
TA2F1 Torque Is the Highest Estimated Using Fluctuation Theorem
Because the conservation of energy is key to survival at alkaline pH, the stepping torque of TA2F1 was measured to examine the power required to turn the rotor to drive ATP hydrolysis. One of the most accurate methods for this type of analysis is by utilizing fluctuation theorem (FT) (44). FT describes entropy production in small non-equilibrium systems, various colloidal particle systems, and more recently biological systems such as RNA hairpins and the F1 ATPase (44). FT is of particular use as it estimates torque by use of steps isolated from the time courses of rotary angles (Fig. 7A) without the need for an accurate measurement of the frictional drag coefficient of the probe itself. Because this analysis requires perfect 120° stepping motion with close to equal dwell time at each step, we used rotational data of TA2F1 hydrolyzing ATPγS under Vmax conditions. Ten TA2F1 rotation traces recorded in the presence of 2 mm ATPγS were analyzed, revealing a rotary torque of 52.4 ± 4 pNnm (Fig. 7, B and C). For comparison with another ATPase under the same conditions and using the same analysis, we also measured the torque for TF1. In our hands, TF1 generated a torque of 41.4 ± 2.2 pNnm, which is quite comparable with that reported previously (Table 1), strongly supporting the reliability of the estimated torque value reported.
FIGURE 7.
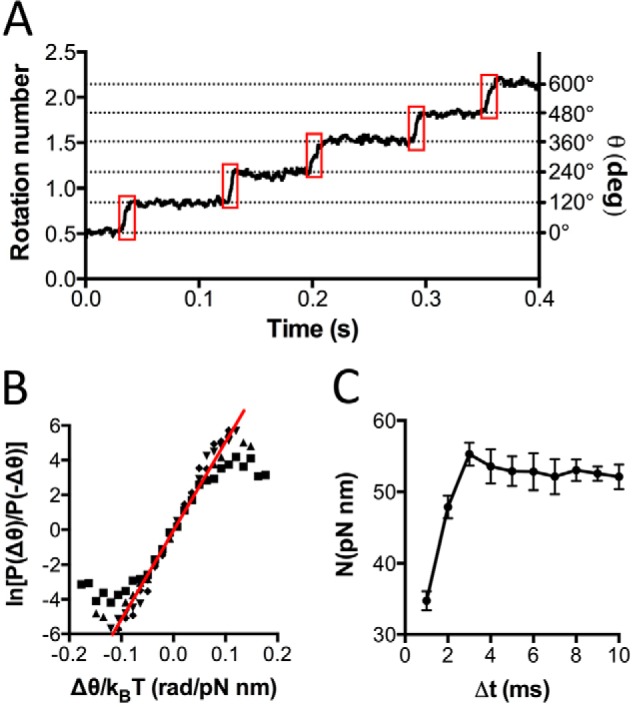
Rotary torque of TA2F1 is the highest determined by fluctuation theorem. A, typical time course of TA2F1γ2c rotation with 2 mm ATPγS filmed at 5000 fps. Boxed in red are the stepping regions of the trace, θ(t), used to determine the torque. The steps were determined by eye. B and C, 10 TA2F1 rotation traces were collectively analyzed. B, ln[P(Δθ)/P(−Δθ)] as a function of Δθ/kBT. The red line shows the average torque of the 10 molecules. The slope revealed a rotary torque of 52.4 ± 4 pNnm/radian (rad) in the case of Δt = 6 ms. C, NFT as a function of Δt for the recording rate of 5000 fps. Torque from 5 to 10 ms was regarded as consistent enough to judge the rotary torque of TA2F1. deg, degrees. Error bars represent S.E.
Discussion
Here, we have conducted the first full biophysical interrogation of a thermoalkaliphilic F1 ATPase in single molecule analysis with simultaneous comparison with biochemical methods. In this study, we comprehensively demonstrate that the TA2F1 is a unidirectional rotary molecular motor. In previous studies with TA2F1, the small amount of hydrolysis activity observed was likely due to the presence of α3β3 incomplete complex (51). However, the fully intact TA2F1 used here does not natively hydrolyze ATP. However, in agreement with previous studies (26, 49, 51, 52), TA2F1 hydrolyzes ATP upon either mechanical or chemical induction. In TF1 studies, the requirement for mechanical activation was proposed to be due to inhibition from a classical feedback loop of product inhibition by Mg-ADP (57); however, rotation was always observable without activation mechanically or with LDAO. Furthermore, TF1 biochemically measured ATP hydrolysis activity was only enhanced 3–4-fold with LDAO (58). In contrast, TA2F1 is activated from an inactive state with LDAO or mechanical activation, the mechanisms by which remain to be fully described. Interestingly, the Km and Kon values obtained from bulk-phase and rotation experiments do not agree. The estimation of Km revealed by gold nanoparticle rotation was 5.95-fold lower than that determined by the bulk-phase biochemical assay, and the estimation of Kon reflects this. However, this is unsurprising when considering that single molecule analysis specifically selects active molecules, whereas the bulk-phase assay does not, providing an ensemble measurement. The lowering of Km is a known hallmark for competitive inhibition, suggesting that inactive molecules are likely due to ADP inhibition. In support of this, it is well known that ATP hydrolysis in TF1 is regulated by Mg-ADP inhibition to prevent wasteful ATP consumption (57), a phenomenon yet to be explored in TA2F1. In TF1 studies, the difference between single molecule results and biochemical results has long been attributed to Mg-ADP inhibition. Under single molecule conditions using a 40-nm gold nanoparticle, TF1 has a maximal rotation velocity of 200 revolutions s−1; however, in the ATP-regenerating assay under Vmax conditions, only 270 ATP/s/molecule were consumed. Because three ATP molecules are consumed per revolution, bulk biochemistry results would imply only 90 revolutions s−1 are possible; however, TF1 is 55% faster according to single molecule data (59). In line with this, according to TA2F1 biochemical data, 124.3 ATP/s/molecule are consumed; therefore a maximum velocity of 41.44 revolutions s−1 is expected at 25 °C. However, TA2F1 single molecule results show a maximal velocity of 112.4 revolutions s−1, a 63% faster Vmax velocity than that derived from biochemical data. This observation is strikingly close to the biochemical versus single molecule discrepancies reported for the TF1 and EhV1 (24, 44, 59).
At almost all ATP and ATPγS concentrations tested above or below Km, TA2F1 rotation profiles exhibited three dwells separated by 120° (Figs. 3 and 6) when recorded at 5000 fps. Upon using the slowly hydrolyzable substrate ATPγS, six substeps were able to be resolved close to ATPγS Km. This is in close agreement with observations of two other F1 ATPases, TF1 and EF1, that have both been reported to rotate with six dwells per revolution, close to their respective Km values (35, 40). This observation together with the lack of six-step rotation observed in the EhV1 ATPases (24) and structural differences between F1 and V1 ATPases (60, 61) implies that the interactions between rotor and stator determine the presence or absence of substeps upon enzyme rotation. However, we report the angular spacing of these 50° and 70° substeps conservatively due to insufficient quality and quantity of the molecules observed. In addition, it was observed that in almost all cases, one of the six substeps was masked showing the “six-step profile.”
Conservation of energy is a constant challenge under thermoalkaliphilic conditions. In light of this, it is unsurprising that the stepping torque derived using fluctuation theorem for TA2F1 is very high (52.4 ± 4 pNnm); however, it was unexpected that it would be 20% higher than that for TF1 (44). Although TF1 and other F1 ATPases studied in rotation studies come from organisms that grow at neutral (E. coli and Bacillus PS3) and acidic (E. hirae) pH, the C. thermarum TA2.A1 F1F0 is highly adapted to function at alkaline pH (Fig. 8A) (48, 53, 62). Given the difference in torque between TF1 and TA2F1 (Fig. 8B) despite both enzymes originating from organisms that grow optimally at 65 °C and that the internal pH of both organisms is similar at 7.5–8.0 (63, 64), we suggest that the differences in torque across species may be more related to evolutionary adaptation to external pH than temperature (Fig. 8, A and B). It is also noteworthy that both TA2F1 and TF1 torque was measured at 25 °C, ruling out temperature as a potential evolutionary pressure on torque. Even if the torque of TF1 measured at 65 °C was the same as that of EF1 measured at 25 °C, TA2F1 has 13 pNnm greater torque than TF1.
FIGURE 8.
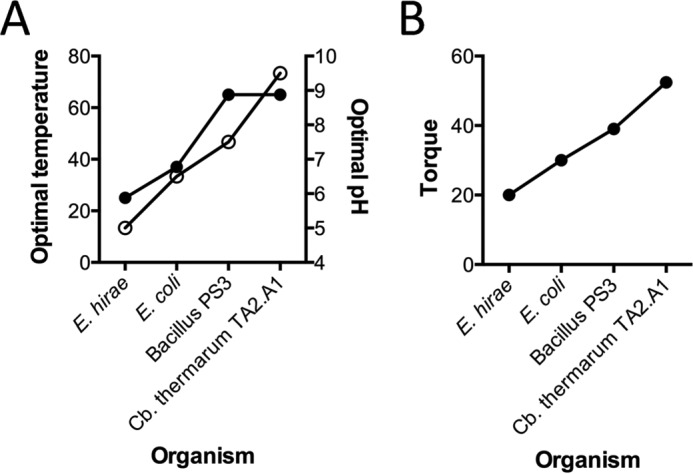
Comparison of bacterial growth conditions in relation to ATPase torque. Shown is a comparison of mild acidophilic (E. hirae), mesophilic (E. coli), thermophilic (Bacillus PS3), and thermoalkaliphilic (C. thermarum TA2.A1) A, optimal temperature and pH of growth conditions (temperature, solid symbols, left y axis; pH, empty circles, right y axis). B, torque of the ATPase. The torque value here for EF1 was derived using a method similar to that used for TF1 and TA2F1 in this study. For methodology details and appropriate references, see Table 1.
Notwithstanding, a higher torque implies a much stiffer rotor. One of the key features of alkaliphilic adaptation is a large c-subunit ring rotor. In the case of C. thermarum TA2.A1, this is a 13-mer (15) with an F1 torque of 52 pNnm in comparison with the E. coli 10-mer (65) with an F1 torque of 30 pNnm (40) and the E. hirae 10-mer (46, 66) with a V1 torque of 20 pNnm (44). This ion coupling ratio is critical in understanding the power of the molecular motor because although velocity and torque are important for determining enzyme efficiency the gearing (i.e. the number of protons coupled to a single revolution event) is also important when considering purpose and efficiency and that C. thermarum TA2.A1 must partially maintain internal pH by synthesis of ATP. However, it is clear that differences in torque cannot be fully ascribed to the number of c-subunits because E. coli and E. hirae both have 10 c-subunits, but E. coli has a 10 pNnm greater torque (40, 44, 65, 66). This strongly suggests that although c-subunit ring size may have a mechanical influence it is the native direction of catalysis (i.e. TA2F1 is a synthase, and EhF1 is a hydrolase) that has the greater mechanical influence. This seems logical given that there is no torque without resistance; the greater the resistance, the greater the torque. Because we are examining TA2F1 in hydrolysis, it is rotating against its native function; hence, one would expect more resistance.
However, in the field of ATPase dynamics, there are several methods by which to estimate torque values for enzymes (Table 1) (28, 40, 44, 67–70). Early EF1 torque measurements were estimated to be 50 pNnm using the drag on the curvature of an actin filament during rotation (67). However, more recently, when rotation of EF1 was assessed using gold nanorods and analysis of torque was conducted using a “power stroke” drag force method, the torque was estimated to be 63 pNnm (68). Clearly, different experimental methods and systems can result in different torque estimations. Here, we use a very conservative method of analysis, “fluctuation theorem,” to directly compare TA2F1 stepping torque with that of TF1 and other enzymes that have been previously analyzed with similar methodology, thus allowing a very reliable comparison between enzymes. Although the stepping torque measured here reflects the shape of rotation potential, future measurement of stall torque will reflect the thermodynamic efficiency. Interestingly, the complete E. hirae V1V0 ATPase has a higher torque than V1 (44). This could be due to a “tighter” overall structure influenced by V1-V0 contacts and/or the influence of specific subunit interactions, i.e. the observation of reinforced torque by genetic fusion of the F-subunit to the D-subunit in T. thermophilus V1 (70). It remains to be examined whether this is a V-type ATPase feature or applies more generally. Future studies will focus on what features dictate torque, activity of the TA2F1F0 complete with the c13-ring, the specific role of ADP inhibition, the substep size, and the mechanical roles of the ϵ- and/or γ-subunits in TA2F1F0 regulation of hydrolysis.
Experimental Procedures
Bacterial Strains, Plasmids, and Growth Conditions
E. coli DH5α (71) was used for all cloning experiments, and E. coli DK8 (72) lacking the ATP synthase genes encoding the unc operon (Δatp) was used to overproduce the TA2F1 (63, 73). For cloning and overexpression of TA2F1 enzyme, the plasmid pTrc99A (Amersham Biosciences) was used. To overproduce F1 ATPases, transformants of E. coli DK8 were routinely grown in 2× YT medium (74) containing 2 g/liter glucose and 100 μg/ml ampicillin at 37 °C with shaking at 200 rpm.
Construction of pTA2F1γ2c (γH107C/γS210C) and TA2F1 α3β3γδϵ Complexes
In a previous study, the genes coding for the C. thermarum TA2.A1 F1 ATPase were cloned into the expression vector pTrc99A, yielding the plasmid pTrcF1 (75). To substitute the native histidine 107 and serine 210 in the γ-subunit of TA2F1 to cysteine residues, double site-directed mutagenesis was carried out using the plasmid pTrcF1 as template with a three-fragment strategy based on the overlap extension PCR method (76). To construct pTA2F1γ2c, an 837-bp PCR product was generated using primers Tg-cysF and Tg-cysMutR, and a 1492-bp PCR product was generated using primers Tg-cysMutF2 and Tg-cysR. A 331-bp PCR product that overlapped the 3′- and 5′-ends of the 837- and 1492-bp PCR products, respectively, was generated using primers Tg-cysMutF1 and Tg-cysMutR2. The 837- and 331-bp PCR products were then joined by overlap PCR to generate a 1145-bp PCR product using primers Tg-cysF and Tg-cysMutR2. Lastly, the 1145- and 1492-bp PCR products were joined by overlap PCR to generate a 2615-bp fragment using primers Tg-cysF and Tg-cysR and digested with AflII and AfeI, and the resulting 2292-bp fragment was cloned with a 6817-bp fragment from pTrcF1 digested with the same enzymes (75). This new plasmid was designated pDMTA11. All PCRs were conducted with the Phusion high fidelity PCR kit (New England Biosciences) according to the manufacturer's instructions. Mutant inserts in both plasmids were confirmed to be correct by DNA sequencing, and the recombinant plasmid was transformed into E. coli DK8. Primer sequences and specific details are displayed in Table 2.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′–3′)a | Description or mutations | Direction |
|---|---|---|---|
| Tg-cysF | ATCTGTTCTACTCCGGTGTGCG | 5′ external primer upstream from an AflII restriction site | Forward |
| Tg-cysR | AGGAAACGCTGGATTTTGCGGG | 3′ external primer downstream from an AfeI restriction site | Reverse |
| Tg-cysMutF | GAGGAGCGttgTCAGTCCAAAGAC | γH107C | Forward |
| Tg-cysMutR | TGGACTGAcaaCGCTCCTCGATC | γH107C | Reverse |
| Tg-cysMutF2 | CGAACCGGACtGTGAATCGGTC | γS210C | Forward |
| Tg-cysMutR2 | GACCGATTCACaGTCCGGTTCG | γS210C | Reverse |
a Lowercase letters indicate specific changes in the primer for site-directed mutagenesis.
Expression and Purification of TA2F1 and TA2F1γ2c
The procedure applied was a modified hybrid of that in McMillan et al. (48) and Keis et al. (51). DK8 harboring plasmid pTrcF1 or pDMTA11 was grown to an A600 of 0.4. The culture was induced with 1 mm isopropyl β-d-thiogalactopyranoside, and incubation continued for 4 h. Cells were harvested, washed with precooled resuspension buffer (50 mm Tris (pH 8.0), 5 mm MgCl2), and resuspended in the same buffer. Phenylmethylsulfonyl fluoride (PMSF) was added to 0.1 mm, and the cells were disrupted by two passages through a French pressure cell at 20,000 p.s.i. Pancreatic DNase I was added to 0.1 mg/ml, and the mixture was kept on ice for 30 min. Lysate was cleared of debris by centrifugation at 8000 × g for 10 min, and the inverted membrane vesicles were pelleted from the supernatant at 180,000 × g for 1 h at 4 °C. To extract TA2F1 or TA2F1γ2c, the supernatant was heated to 60 °C for 15 min to precipitate contaminating E. coli proteins, and the precipitate was pelleted from the supernatant at 180,000 × g for 30 min at 4 °C. The supernatant was then adjusted to 500 mm NaCl and 10 mm imidazole before application to a 20-ml column of nickel-Sepharose High Performance (GE Healthcare) that was previously washed with water and equilibrated with buffer A (50 mm Tris (pH 8.0), 2 mm MgCl2, 10% glycerol, 500 mm NaCl) containing 10 mm imidazole. To remove contaminating proteins, the resin was washed with buffer A containing 30 mm imidazole, and TA2F1, TA2F1γ2c, or TA2F1ϵ2c was eluted with buffer A supplemented with 200 mm imidazole. The eluted TA2F1, TA2F1γ2c, or TA2F1ϵ2c was pooled and concentrated to 10–30 mg/ml using Amicon Ultra centrifugal filter devices (molecular weight cutoff, 50,000). The concentrated sample was then applied to a Superdex 200 10/300 (GE Healthcare) gel filtration column and fractionated with 50 mm Tris-HCl (pH 8.0), 2 mm MgCl2, 100 mm NaCl buffer. The eluted TA2F1- or TA2F1γ2c-containing fractions (a peak between 370 and 380 kDa) were pooled and concentrated to 10 mg/ml using Amicon Ultra centrifugal filter devices (molecular weight cutoff, 50,000) in 50 mm Tris-HCl (pH 8.0) containing 2 mm MgCl2, 100 mm NaCl, and 10% glycerol.
Labeling
TA2F1γ2c preparations were treated with a 5-fold molar excess of tris(2-carboxyethyl)phosphine for 2 h at 4 °C to reduce the two cysteines on the γ-subunit. This was then buffer-exchanged by gel filtration through a Nap5 column (GE Healthcare) into a potassium phosphate buffer (pH 7.0) containing 5% (v/v) glycerol, 5 mm MgCl2, and a 2-fold molar excess of tris(2-carboxyethyl)phosphine. After reduction, a 10-fold molar excess of biotin-PEAC5-maleimide was added to the reduced TA2F1 and incubated for 1.5 h at room temperature. Unreacted biotin-PEAC5-maleimide was removed by gel filtration through a Nap10 column (GE Healthcare) and buffer-exchanged into 50 mm Tris-HCl (pH 8.0) containing 2 mm MgCl2, 100 mm NaCl, and 10% glycerol.
SDS-PAGE and Immunoblotting
TA2F1 and TA2F1γ2c preparations were routinely analyzed on 12% sodium dodecyl sulfate (SDS)-polyacrylamide gels using the buffer system of Laemmli (77). Polypeptide bands were visualized using Coomassie Brilliant Blue. During immunoblotting, proteins were separated by 12% SDS-PAGE followed by blotting onto a polyvinylidene difluoride (PVDF) membrane. Detection was achieved using a streptavidin-alkaline phosphatase conjugate (Roche Applied Science) directed against the biotin-modified γ-subunit of TA2F1γ2c. The antibody-specific bands were visualized using the 4-nitro blue tetrazolium chloride/5-bromo-4-chloro-3-indolyl phosphate, 4-toluidine detection system (Roche Applied Science).
Protein Assay
Protein concentrations were determined using a bicinchoninic acid (BCA) protein assay kit (Sigma) with bovine serum albumin as the standard.
Biochemical Assays
ATP hydrolysis activity was measured using a spectrophotometric ATP-regenerating assay at 25 and 45 °C similar to that used in McMillan et al. (48, 52). The assay mixture contained 100 mm Tris·Cl (pH 8.0), 1.5 mm MgCl2, 3 mm phosphoenolpyruvate, 50 mm KCl, 0.25 mm NADH, 5 units/ml pyruvate kinase, 6.4 units/ml lactate dehydrogenase, and variable concentrations of sodium ATP (Sigma) or lithium ATPγS (Roche Applied Science). For measurements at 65 °C, thermophilic enzymes, 10 units/ml pyruvate kinase HI1 and 6.4 units/ml lactate dehydrogenase 2, were used (Funakoshi Ltd.). The reaction was initiated by the addition of enzyme or ATP, as indicated, into 1 ml of assay mixture, and the rate of NADH oxidation was monitored continuously at 340 nm using a Jasco V-660 spectrophotometer. Approximately 5–10 μg (as indicated) of recombinant TA2F1 or TA2F1γ2c subunit complexes was used for assays.
Single Molecule Rotation Assays
Single molecule rotation of TA2F1γ2c was measured as described previously using either a ∼200-nm magnetic bead duplex (high load probe) or a 40-nm gold nanoparticle (low load probe) (33, 44, 55, 78). Briefly, a flow cell was constructed using a Ni2+-NTA-modified glass surface and an uncoated cover glass (Matsunami Glass Ltd., Japan), and biotinylated TA2F1γ2c subunit complexes were immobilized on the Ni2+-NTA surface via a hexahistidine tag introduced at the N terminus of the β-subunit. After 5 min, non-immobilized TA2F1γ2c subunit complexes were removed using buffer A (100 mm Tris (pH 8.0), 1.5 mm MgCl2, 50 mm KCl) followed by a 5-min incubation in 20 mg/ml bovine serum albumin in buffer A to prevent nonspecific interactions of probes with the surface. Either ∼0.2-μm high load streptavidin-functionalized magnetic bead duplexes (Seradyn Inc., Indianapolis, IN) or a low load 40-nm streptavidin-functionalized gold nanoparticle (British BioCell International, UK) were subsequently attached to the biotinylated cysteine residues in the TA2F1 γ-subunit in buffer A and incubated for 10 min. Non-bound probe was then removed from the flow cell using buffer A. Observation of TA2F1γ2c rotation was initiated after the infusion of either buffer A containing ≥0.2 mm sodium ATP/lithium ATPγS or buffer B (100 mm Tris (pH 8.0), 300 μm MgCl2, 50 mm KCl) containing <0.2 mm ATP/sodium ATP/lithium ATPγS. In line with previous studies, 3 mm phosphoenolpyruvate and 2.5 units/ml pyruvate kinase were used to prevent accumulation of ADP in solution; where necessary, 0.1% LDAO was also included in the assay mixture. When using magnetic beads, the rotation of the duplex attached to the TA2F1γ2c γ-subunit was observed using bright field or phase-contrast microscopy and, when necessary, manipulated with home-built magnetic tweezers (33) controlled with custom-made software (Celery, Library) for activation of “stalled” enzyme. Images were recorded on an inverted microscope (IX-71, Olympus, Japan) at 30 fps (FC300M, Takenaka Systems, Japan) for all characterization experiments with the exception of the six-step rotation observation that was recorded using a high speed complementary metal oxide semiconductor camera at 5000 fps (FASTCAM-1024PCI, Photron). When using gold nanoparticle, the rotation of the duplex attached to the TA2F1γ2c γ-subunit was observed using a modified inverted microscope (IX-71) using total internal reflection dark field microscopy (31) with a lens capable of total internal reflection microscopy (APON 60XOTIRF, Olympus) (44). Images were recorded with a high speed complementary metal oxide semiconductor camera at 10,000 fps (FASTCAM-1024PCI).
Torque Measurements
Stepping torque (N) was determined from the rotation trajectories of magnetic duplex beads (∼0.2 μm in diameter) attached to TA2F1γ2c by using the FT (44). For continuous rotation of TA2F1, the time evolution of θ(t) can be described by the Langevin equation (Equations 1 and 2).
| (Eq. 1) |
| (Eq. 2) |
where Γ is the frictional drag coefficient, ξ is a random force representing thermal noise, kB is the Boltzmann constant, and T is the room temperature (298 K). N is torque and is assumed to be a constant value as described previously (44). Observation of rotation of TA2F1γ2c was performed as described above except that observations were recorded at 5000 fps, and 2 mm lithium ATPγS was used (FASTCAM-1024PCI). Only enzymes exhibiting clear and even dwell times of three steps (120°) were used for analysis where the area on each trace used for analysis was the period moving between one 120° step and the next. Because the rotary motor proteins rotate in one direction, the following expression is derived from FT.
| (Eq. 3) |
where θ(t) is the rotary angle of the bead, Δθ = θ(t + Δt) − θ(t), and P(Δθ) is the probability distribution of Δθ. For molecules continuously rotating for at least 4 s, P(Δθ) was calculated for the case Δt = 5 ms, and then ln[P(Δθ)/P(−Δθ)] versus Δθ/kBT was plotted. The slope of the resulting plot corresponds to the torque. The torque of each molecule was defined as the maximum value obtained from the FT analysis when using a 4-s moving window with windows starting at 1-ms intervals.
Author Contributions
D. G. G. M. conceived the study, performed the experiments, analyzed the data, and wrote the article. R. W. and H. U. provided critical insight and experimental/analytical knowledge necessary for the magnetic bead and gold rotation assay platforms, respectively. G. M. C. and H. N. provided critical insight into the broader interpretation of results in the view of both physiology and mechanical mechanism.
Acknowledgment
We thank Dr. Kengo Adachi for the use of Labview analysis software.
This work was supported by long term Japan Society for the Promotion of Sciences International Fellowship P14383 (to D. G. G. M.). The authors declare that they have no conflicts of interest with the contents of this article.
- LDAO
- lauryldimethylamine oxide
- TA2F1
- C. thermarum TA2.A1 F1 ATPase
- TF1
- thermophile Bacillus PS3 F1 ATPase
- EF1
- E. coli F1 ATPase
- TtV1
- T. thermophilus V1 ATPase
- EhV1
- E. hirae V1 ATPase
- pNnm
- piconewton nanometer
- NTA
- nitrilotriacetic acid
- fps
- frames per second
- ATPγS
- adenosine 5′-O-(thiotriphosphate)
- FT
- fluctuation theorem
- biotin-PEAC5-maleimide
- N-6-(biotinylamino)hexanoyl-N′-[2-(N-maleimido)ethyl]piperazone.
References
- 1. Yoshida M., Muneyuki E., and Hisabori T. (2001) ATP synthase—a marvellous rotary engine of the cell. Nat. Rev. Mol. Cell Biol. 2, 669–677 [DOI] [PubMed] [Google Scholar]
- 2. Dimroth P., von Ballmoos C., and Meier T. (2006) Catalytic and mechanical cycles in F-ATP synthases. Fourth in the Cycles Review Series. EMBO Rep. 7, 276–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Abrahams J. P., Leslie A. G., Lutter R., and Walker J. E. (1994) Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature 370, 621–628 [DOI] [PubMed] [Google Scholar]
- 4. Weber J., and Senior A. E. (1997) Catalytic mechanism of F1-ATPase. Biochim. Biophys. Acta 1319, 19–58 [DOI] [PubMed] [Google Scholar]
- 5. Morales-Rios E., Montgomery M. G., Leslie A. G., and Walker J. E. (2015) Structure of ATP synthase from Paracoccus denitrificans determined by X-ray crystallography at 4.0 Å resolution. Proc. Natl. Acad. Sci. U.S.A. 112, 13231–13236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cingolani G., and Duncan T. M. (2011) Structure of the ATP synthase catalytic complex (F1) from Escherichia coli in an autoinhibited conformation. Nat. Struct. Mol. Biol. 18, 701–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stock D., Leslie A. G., and Walker J. E. (1999) Molecular architecture of the rotary motor in ATP synthase. Science 286, 1700–1705 [DOI] [PubMed] [Google Scholar]
- 8. Deckers-Hebestreit G., Greie J., Stalz W., and Altendorf K. (2000) The ATP synthase of Escherichia coli: structure and function of F0 subunits. Biochim. Biophys. Acta 1458, 364–373 [DOI] [PubMed] [Google Scholar]
- 9. von Ballmoos C., Meier T., and Dimroth P. (2002) Membrane embedded location of Na+ or H+ binding sites on the rotor ring of F1F0 ATP synthases. Eur. J. Biochem. 269, 5581–5589 [DOI] [PubMed] [Google Scholar]
- 10. Müller D. J., Dencher N. A., Meier T., Dimroth P., Suda K., Stahlberg H., Engel A., Seelert H., and Matthey U. (2001) ATP synthase: constrained stoichiometry of the transmembrane rotor. FEBS Lett. 504, 219–222 [DOI] [PubMed] [Google Scholar]
- 11. Pogoryelov D., Yu J., Meier T., Vonck J., Dimroth P., and Muller D. J. (2005) The c15 ring of the Spirulina platensis F-ATP synthase: F1/F0 symmetry mismatch is not obligatory. EMBO Rep. 6, 1040–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vonck J., von Nidda T. K., Meier T., Matthey U., Mills D. J., Kühlbrandt W., and Dimroth P. (2002) Molecular architecture of the undecameric rotor of a bacterial Na+-ATP synthase. J. Mol. Biol. 321, 307–316 [DOI] [PubMed] [Google Scholar]
- 13. Pogoryelov D., Reichen C., Klyszejko A. L., Brunisholz R., Muller D. J., Dimroth P., and Meier T. (2007) The oligomeric state of c rings from cyanobacterial F-ATP synthases varies from 13 to 15. J. Bacteriol. 189, 5895–5902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Meier T., Yu J., Raschle T., Henzen F., Dimroth P., and Muller D. J. (2005) Structural evidence for a constant c11 ring stoichiometry in the sodium F-ATP synthase. FEBS J. 272, 5474–5483 [DOI] [PubMed] [Google Scholar]
- 15. Matthies D., Preiss L., Klyszejko A. L., Muller D. J., Cook G. M., Vonck J., and Meier T. (2009) The c13 ring from a thermoalkaliphilic ATP synthase reveals an extended diameter due to a special structural region. J. Mol. Biol. 388, 611–618 [DOI] [PubMed] [Google Scholar]
- 16. Allegretti M., Klusch N., Mills D. J., Vonck J., Kühlbrandt W., and Davies K. M. (2015) Horizontal membrane-intrinsic α-helices in the stator a-subunit of an F-type ATP synthase. Nature 521, 237–240 [DOI] [PubMed] [Google Scholar]
- 17. Angevine C. M., and Fillingame R. H. (2003) Aqueous access channels in subunit a of rotary ATP synthase. J. Biol. Chem. 278, 6066–6074 [DOI] [PubMed] [Google Scholar]
- 18. Steed P. R., and Fillingame R. H. (2009) Aqueous accessibility to the transmembrane regions of subunit c of the Escherichia coli F1F0 ATP synthase. J. Biol. Chem. 284, 23243–23250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pogoryelov D., Nikolaev Y., Schlattner U., Pervushin K., Dimroth P., and Meier T. (2008) Probing the rotor subunit interface of the ATP synthase from Ilyobacter tartaricus. FEBS J. 275, 4850–4862 [DOI] [PubMed] [Google Scholar]
- 20. Pogoryelov D., Krah A., Langer J. D., Yildiz Ö., Faraldo-Gómez J. D., and Meier T. (2010) Microscopic rotary mechanism of ion translocation in the Fo complex of ATP synthases. Nat. Chem. Biol. 6, 891–899 [DOI] [PubMed] [Google Scholar]
- 21. Suzuki T., Ueno H., Mitome N., Suzuki J., and Yoshida M. (2002) F0 of ATP synthase is a rotary proton channel. Obligatory coupling of proton translocation with rotation of c-subunit ring. J. Biol. Chem. 277, 13281–13285 [DOI] [PubMed] [Google Scholar]
- 22. Wächter A., Bi Y., Dunn S. D., Cain B. D., Sielaff H., Wintermann F., Engelbrecht S., and Junge W. (2011) Two rotary motors in F-ATP synthase are elastically coupled by a flexible rotor and a stiff stator stalk. Proc. Natl. Acad. Sci. U.S.A. 108, 3924–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Noji H., Yasuda R., Yoshida M., and Kinosita K. Jr. (1997) Direct observation of the rotation of F1-ATPase. Nature 386, 299–302 [DOI] [PubMed] [Google Scholar]
- 24. Minagawa Y., Ueno H., Hara M., Ishizuka-Katsura Y., Ohsawa N., Terada T., Shirouzu M., Yokoyama S., Yamato I., Muneyuki E., Noji H., Murata T., and Iino R. (2013) Basic properties of rotary dynamics of the molecular motor Enterococcus hirae V1-ATPase. J. Biol. Chem. 288, 32700–32707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hicks D. B., and Krulwich T. A. (1990) Purification and reconstitution of the F1F0-ATP synthase from alkaliphilic Bacillus firmus OF4. Evidence that the enzyme translocates H+ but not Na+. J. Biol. Chem. 265, 20547–20554 [PubMed] [Google Scholar]
- 26. Cook G. M., Keis S., Morgan H. W., von Ballmoos C., Matthey U., Kaim G., and Dimroth P. (2003) Purification and biochemical characterization of the F1Fo-ATP synthase from thermoalkaliphilic Bacillus sp. strain TA2.A1. J. Bacteriol. 185, 4442–4449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hoffmann A., and Dimroth P. (1990) The ATPase of Bacillus alcalophilus. Eur. J. Biochem. 194, 423–430 [DOI] [PubMed] [Google Scholar]
- 28. Noji H., Häsler K., Junge W., Kinosita K. Jr, Yoshida M., and Engelbrecht S. (1999) Rotation of Escherichia coli F1-ATPase. Biochem. Biophys. Res. Commun. 260, 597–599 [DOI] [PubMed] [Google Scholar]
- 29. Kato-Yamada Y., Noji H., Yasuda R., Kinosita K. Jr., and Yoshida M. (1998) Direct observation of the rotation of e subunit in F1-ATPase. J. Biol. Chem. 273, 19375–19377 [DOI] [PubMed] [Google Scholar]
- 30. Sielaff H., Rennekamp H., Engelbrecht S., and Junge W. (2008) Functional halt positions of rotary FOF1-ATPase correlated with crystal structures. Biophys. J. 95, 4979–4987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ueno H., Nishikawa S., Iino R., Tabata K. V., Sakakihara S., Yanagida T., and Noji H. (2010) Simple dark-field microscopy with nanometer spatial precision and microsecond temporal resolution. Biophys. J. 98, 2014–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Spetzler D., York J., Daniel D., Fromme R., Lowry D., and Frasch W. (2006) Microsecond time scale rotation measurements of single F1-ATPase molecules. Biochemistry 45, 3117–3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hirono-Hara Y., Ishizuka K., Kinosita K. Jr, Yoshida M., and Noji H. (2005) Activation of pausing F1 motor by external force. Proc. Natl. Acad. Sci. U.S.A. 102, 4288–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Watanabe R., Okuno D., Sakakihara S., Shimabukuro K., Iino R., Yoshida M., and Noji H. (2012) Mechanical modulation of catalytic power on F1-ATPase. Nat. Chem. Biol. 8, 86–92 [DOI] [PubMed] [Google Scholar]
- 35. Shimabukuro K., Yasuda R., Muneyuki E., Hara K. Y., Kinosita K. Jr, and Yoshida M. (2003) Catalysis and rotation of F1 motor: cleavage of ATP at the catalytic site occurs in 1 ms before 40° substep rotation. Proc. Natl. Acad. Sci. U.S.A. 100, 14731–14736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sakaki N., Shimo-Kon R., Adachi K., Itoh H., Furuike S., Muneyuki E., Yoshida M., and Kinosita K. Jr. (2005) One rotary mechanism for F1-ATPase over ATP concentrations from millimolar down to nanomolar. Biophys. J. 88, 2047–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yasuda R., Masaike T., Adachi K., Noji H., Itoh H., and Kinosita K. Jr. (2003) The ATP-waiting conformation of rotating F1-ATPase revealed by single-pair fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. U.S.A. 100, 9314–9318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Börsch M., Turina P., Eggeling C., Fries J. R., Seidel C. A., Labahn A., and Gräber P. (1998) Conformational changes of the H+-ATPase from Escherichia coli upon nucleotide binding detected by single molecule fluorescence. FEBS Lett. 437, 251–254 [DOI] [PubMed] [Google Scholar]
- 39. Börsch M., and Wrachtrup J. (2011) Improving FRET-based monitoring of single chemomechanical rotary motors at work. Chemphyschem 12, 542–553 [DOI] [PubMed] [Google Scholar]
- 40. Bilyard T., Nakanishi-Matsui M., Steel B. C., Pilizota T., Nord A. L., Hosokawa H., Futai M., and Berry R. M. (2013) High-resolution single-molecule characterization of the enzymatic states in Escherichia coli F1-ATPase. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20120023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ueno H., Minagawa Y., Hara M., Rahman S., Yamato I., Muneyuki E., Noji H., Murata T., and Iino R. (2014) Torque generation of Enterococcus hirae V-ATPase. J. Biol. Chem. 289, 31212–31223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yasuda R., Noji H., Yoshida M., Kinosita K. Jr, and Itoh H. (2001) Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature 410, 898–904 [DOI] [PubMed] [Google Scholar]
- 43. Watanabe R., Iino R., and Noji H. (2010) Phosphate release in F1-ATPase catalytic cycle follows ADP release. Nat. Chem. Biol. 6, 814–820 [DOI] [PubMed] [Google Scholar]
- 44. Hayashi K., Ueno H., Iino R., and Noji H. (2010) Fluctuation theorem applied to F1-ATPase. Phys. Rev. Let. 104, 218103. [DOI] [PubMed] [Google Scholar]
- 45. Imamura H., Takeda M., Funamoto S., Shimabukuro K., Yoshida M., and Yokoyama K. (2005) Rotation scheme of V1-motor is different from that of F1-motor. Proc. Natl. Acad. Sci. U.S.A. 102, 17929–17933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Murata T., Yamato I., Kakinuma Y., Leslie A. G., and Walker J. E. (2005) Structure of the rotor of the V-Type Na+-ATPase from Enterococcus hirae. Science 308, 654–659 [DOI] [PubMed] [Google Scholar]
- 47. Dimroth P., and Cook G. M. (2004) Bacterial Na+- or H+-coupled ATP synthases operating at low electrochemical potential. Adv. Microb. Physiol. 49, 175–218 [DOI] [PubMed] [Google Scholar]
- 48. McMillan D. G., Keis S., Dimroth P., and Cook G. M. (2007) A specific adaptation in the a subunit of thermoalkaliphilic F1FO-ATP synthase enables ATP synthesis at high pH but not at neutral pH values. J. Biol. Chem. 282, 17395–17404 [DOI] [PubMed] [Google Scholar]
- 49. Guffanti A. A., and Krulwich T. A. (1994) Oxidative phosphorylation by ADP + Pi-loaded membrane vesicles of alkaliphilic Bacillus firmus OF4. J. Biol. Chem. 269, 21576–21582 [PubMed] [Google Scholar]
- 50. Albers S. V., Van de Vossenberg J. L., Driessen A. J., and Konings W. N. (2001) Bioenergetics and solute uptake under extreme conditions. Extremophiles 5, 285–294 [DOI] [PubMed] [Google Scholar]
- 51. Keis S., Stocker A., Dimroth P., and Cook G. M. (2006) Inhibition of ATP hydrolysis by thermoalkaliphilic F1Fo-ATP synthase is controlled by the C terminus of the ϵ subunit. J. Bacteriol. 188, 3796–3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. McMillan D. G., Keis S., Berney M., and Cook G. M. (2009) Nonfermentative thermoalkaliphilic growth is restricted to alkaline environments. Appl. Environ. Microbiol. 75, 7649–7654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stocker A., Keis S., Vonck J., Cook G. M., and Dimroth P. (2007) The structural basis for unidirectional rotation of thermoalkaliphilic F1-ATPase. Structure 15, 904–914 [DOI] [PubMed] [Google Scholar]
- 54. Noji H. (1998) The rotary enzyme of the cell: the rotation of F1-ATPase. Science 282, 1844–1845 [DOI] [PubMed] [Google Scholar]
- 55. Itoh H., Takahashi A., Adachi K., Noji H., Yasuda R., Yoshida M., and Kinosita K. (2004) Mechanically driven ATP synthesis by F1-ATPase. Nature 427, 465–468 [DOI] [PubMed] [Google Scholar]
- 56. Sambongi Y., Iko Y., Tanabe M., Omote H., Iwamoto-Kihara A., Ueda I., Yanagida T., Wada Y., and Futai M. (1999) Mechanical rotation of the c subunit oligomer in ATP synthase (F0F1): direct observation. Science 286, 1722–1724 [DOI] [PubMed] [Google Scholar]
- 57. Hirono-Hara Y., Noji H., Nishiura M., Muneyuki E., Hara K. Y., Yasuda R., Kinosita K. Jr., and Yoshida M. (2001) Pause and rotation of F1-ATPase during catalysis. Proc. Natl. Acad. Sci. U.S.A. 98, 13649–13654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Paik S. R., Jault J.-M., and Allison W. S. (1994) Inhibition and inactivation of the F1 adenosinetriphosphatase from Bacillus PS3 by dequalinium and activation of the enzyme by lauryl dimethylamine oxide. Biochemistry 33, 126–133 [DOI] [PubMed] [Google Scholar]
- 59. Kohori A., Chiwata R., Hossain M. D., Furuike S., Shiroguchi K., Adachi K., Yoshida M., and Kinosita K. Jr. (2011) Torque generation in F1-ATPase devoid of the entire amino-terminal helix of the rotor that fills half of the stator orifice. Biophys. J. 101, 188–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Svergun D. I., Konrad S., Huss M., Koch M. H., Wieczorek H., Altendorf K., Volkov V. V., and Grüber G. (1998) Quaternary structure of V1 and F1 ATPase: significance of structural homologies and diversities. Biochemistry 37, 17659–17663 [DOI] [PubMed] [Google Scholar]
- 61. Kasho V. N., and Boyer P. D. (1989) Vacuolar ATPases, like F1F0-ATPases, show a strong dependence of the reaction velocity on the binding of more than one ATP per enzyme. Proc. Natl. Acad. Sci. U.S.A. 86, 8708–8711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Murata T., Yamato I., Kakinuma Y., Shirouzu M., Walker J. E., Yokoyama S., and Iwata S. (2008) Ion binding and selectivity of the rotor ring of the Na+-transporting V-ATPase. Proc. Natl. Acad. Sci. U.S.A. 105, 8607–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kalamorz F., Keis S., McMillan D. G., Olsson K., Stanton J. A., Stockwell P., Black M. A., Klingeman D. M., Land M. L., Han C. S., Martin S. L., Becher S. A., Peddie C. J., Morgan H. W., Matthies D., et al. (2011) Draft genome sequence of the thermoalkaliphilic Caldalkalibacillus thermarum strain TA2.A1. J. Bacteriol. 193, 4290–4291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Soga N., Kinosita K. Jr., Yoshida M., and Suzuki T. (2011) Efficient ATP synthesis by thermophilic Bacillus FoF1-ATP synthase. FEBS J. 278, 2647–2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ballhausen B., Altendorf K., and Deckers-Hebestreit G. (2009) Constant c10 ring stoichiometry in the Escherichia coli ATP synthase analyzed by cross-linking. J. Bacteriol. 191, 2400–2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Murata T., Arechaga I., Fearnley I. M., Kakinuma Y., Yamato I., and Walker J. E. (2003) The membrane domain of the Na+-motive V-ATPase from Enterococcus hirae contains a heptameric rotor. J. Biol. Chem. 278, 21162–21167 [DOI] [PubMed] [Google Scholar]
- 67. Pänke O., Cherepanov D. A., Gumbiowski K., Engelbrecht S., and Junge W. (2001) Viscoelastic dynamics of actin filaments coupled to rotary F-ATPase: torque profile of the enzyme. Biophys. J. 81, 1220–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hornung T., Ishmukhametov R., Spetzler D., Martin J., and Frasch W. D. (2008) Determination of torque generation from the power stroke of Escherichia coli F1-ATPase. Biochim. Biophys. Acta 1777, 579–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yasuda R., Noji H., Kinosita K. Jr, and Yoshida M. (1998) F1-ATPase is a highly efficient molecular motor that rotates with discrete 120° steps. Cell 93, 1117–1124 [DOI] [PubMed] [Google Scholar]
- 70. Kishikawa J., Seino A., Nakanishi A., Tirtom N. E., Noji H., Yokoyama K., and Hayashi K. (2014) F-subunit reinforces torque generation in V-ATPase. Eur. Biophys. J. 43, 415–422 [DOI] [PubMed] [Google Scholar]
- 71. Taylor R. G., Walker D. C., and McInnes R. R. (1993) E. coli host strains significantly affect the quality of small scale plasmid DNA preparations used for sequencing. Nucleic Acids Res. 21, 1677–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Klionsky D. J., Brusilow W. S., and Simoni R. D. (1984) In vivo evidence for the role of the e subunit as an inhibitor of the proton-translocating ATPase of Escherichia coli. J. Bacteriol. 160, 1055–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Peddie C. J., Cook G. M., and Morgan H. W. (1999) Sodium-dependent glutamate uptake by an alkaliphilic, thermophilic Bacillus strain, TA2.A1. J. Bacteriol. 181, 3172–3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sambrook J., and Green M. R. (2012) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 75. Stocker A., Keis S., Cook G. M., and Dimroth P. (2005) Purification, crystallization, and properties of F1-ATPase complexes from the thermoalkaliphilic Bacillus sp. strain TA2.A1. J. Struct. Biol. 152, 140–145 [DOI] [PubMed] [Google Scholar]
- 76. Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., and Pease L. R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59 [DOI] [PubMed] [Google Scholar]
- 77. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 78. Watanabe R., Iino R., Shimabukuro K., Yoshida M., and Noji H. (2008) Temperature-sensitive reaction intermediate of F1-ATPase. EMBO Rep. 9, 84–90 [DOI] [PMC free article] [PubMed] [Google Scholar]