Abstract
The enzyme acid sphingomyelinase-like phosphodiesterase 3B (SMPDL3B) was shown to act as a negative regulator of innate immune signaling, affecting cellular lipid composition and membrane fluidity. Furthermore, several reports identified this enzyme as an off target of the therapeutic antibody rituximab, with implications in kidney disorders. However, structural information for this protein is lacking. Here we present the high resolution crystal structure of murine SMPDL3B, which reveals a substrate binding site strikingly different from its paralogs. The active site is located in a narrow boot-shaped cavity. We identify a unique loop near the active site that appears to impose size constraints on incoming substrates. A structure in complex with phosphocholine indicates that the protein recognizes this head group via an aromatic box, a typical choline-binding motif. Although a potential substrate for SMPDL3B is sphingomyelin, we identify other possible substrates such as CDP-choline, ATP, and ADP. Functional experiments employing structure-guided mutagenesis in macrophages highlight amino acid residues potentially involved in recognition of endogenous substrates. Our study is an important step toward elucidating the specific function of this poorly characterized enzyme.
Keywords: cell surface enzyme, crystal structure, lipid metabolism, sphingomyelinase, substrate specificity, SMPDL3B
Introduction
Sphingomyelins (SM)3 represent a major class of phospholipids found in cellular membranes and are particularly enriched in the outer leaflet of the plasma membrane (1). Removal of the phosphocholine (PC) head group from SM leads to the generation of ceramide sphingolipids, whose resting levels in the plasma membrane are extremely low and which have been shown to alter biophysical properties of the membrane and affect cellular processes (2). This reaction is carried out by enzymes called sphingomyelinases (SMases). In mammals, three SMase families can be distinguished: the alkaline SMase is expressed on microvilli in the intestines, where it metabolizes dietary SM (3); the four neutral SMases are localized in the plasma membrane inner leaflet, Golgi, endoplasmic reticulum, nucleus, or mitochondria (4); and the acid SMase (ASMase, ASM, SMPD1) is found in the lysosome where it serves catabolic purposes and can also be secreted to act on plasma lipoproteins or the cell membrane (5). This latter function occurs in response to numerous stresses and modulates signaling events via ceramide production (5).
ASMase is the founding member of a family of three paralogs, along with the relatively poorly characterized enzymes SMPDL3A and SMPDL3B. Interestingly, SMPDL3A is not a SMase but hydrolyzes various nucleotide triphosphates and diphosphates (6). In this regard, an anti-inflammatory role in purinergic signaling in the extracellular space was proposed for SMPDL3A (6), although it was also reported to be found in lysosomes (7). However, its function is still linked with lipid metabolism, because its expression and secretion is stimulated by cholesterol and liver X receptor ligands in macrophages (6, 8).
On the other hand, SMPDL3B is a glycosylphosphatidylinositol (GPI)-anchored protein (9–11) with reported roles in inflammatory processes, as well as in kidney diseases. A negative regulatory role in innate immune signaling was uncovered in murine macrophages, where the enzyme is found on the cell surface and up-regulated upon inflammatory stimuli (11, 12). Its knockdown results in enhanced responsiveness to Toll-like receptor (TLR) stimulation and Smpdl3b-deficient mice display an increased inflammatory response. Furthermore, deficiency of this protein in macrophages is associated with alterations in the cellular lipid composition and changes in membrane fluidity.
In the kidney disease focal segmental glomerulosclerosis, expression of SMPDL3B on podocytes is reduced; overexpression of the enzyme prevents actin cytoskeleton disruption and apoptosis associated with this condition (13). Rituximab, a therapeutic anti-CD20 monoclonal antibody targeting B cells but also used in certain nephrological conditions, binds to SMPDL3B (13–15) and remediates these defects in a SMPDL3B-dependent manner (13). Rituximab similarly protects podocytes from morphological disruption, decreased proliferation, and SMPDL3B protein loss in xenotransplants (14). In contrast, this protein is elevated in glomeruli from patients with diabetic kidney disease where it binds to the soluble urokinase plasminogen activator receptor on podocytes; SMPDL3B knockdown protects podocytes from apoptosis in this condition (16).
The identification of this enzyme at other locations suggests additional functions. For instance, SMPDL3B has been found in pancreatic zymogen granules, an organelle containing digestive enzymes to be released into the pancreatic duct (17); in saliva exosomes, which are secreted vesicles with various functions (18); in the plasma protein-depleted cerebrospinal fluid (19); and in milk (20).
Although these findings provide clues as to the potential roles of SMPDL3B, its precise function and substrate specificity remain unknown. That it is membrane-anchored, bears similarity to ASMase, and possesses membrane-modulating activities suggests that it may act on lipid substrates. On the other hand, its other homolog, SMPDL3A, is a nucleotide phosphodiesterase. The crystal structures of ASMase and SMPDL3A were recently reported (21–24), but no structural information is as yet available for SMPDL3B.
Here we present the high resolution crystal structure of murine SMPDL3B, which revealed that its substrate binding site is a narrow pocket, quite distinct from its paralogs, a consequence of a unique conformation of a surface loop. The structure of the enzyme in complex with PC showed that this potential product is accommodated inside a narrow pocket surrounded by a “cage” of aromatic side chains, a common feature of choline-binding proteins (25). Although no in vitro enzymatic activity against lipids could be detected, SMPDL3B can cleave other choline-containing substrates including CDP-choline and releases phosphate from ATP and ADP. Functional experiments in murine macrophages further revealed amino acid residues lining the channel leading to the active site of SMPDL3B as potentially important for recognition of endogenous substrate molecules involved in the modulation of macrophage function.
Results
Overall Structure
We determined the crystal structure of full-length murine SMPDL3B, which shares 78% identity with its human counterpart. Our construct excluded the N-terminal signal peptide, as well as the C-terminal GPI attachment signal peptide, which is normally cleaved off to generate a new C terminus, the ω-site of GPI attachment. The ω-site is Ala-431 for human SMPDL3B (10), corresponding to Gly-431 in the murine protein. Although the crystallized construct contained four additional residues beyond the ω-site (residues 19–435), Gly-431 is the last residue visible in the crystal structure. The structure was solved by single-wavelength anomalous diffraction from two zinc ions in the active site and refined to a resolution of 1.14 Å (Table 1).
TABLE 1.
X-ray data collection and structure refinement statistics
The values in parentheses refer to the highest resolution shell. Anomalous resolution refers to the resolution limit for substructure determination by SAD. Zn-SAD, zinc single wavelength anomalous diffraction; ASU, asymmetric unit; CMCF, Canadian Macromolecular Crystallography Facility; CLS, Canadian Light Source; MacCHESS, Macromolecular Diffraction Facility at the Cornell High Energy Synchrotron Source.
| Parameters | SMPDL3B | SMPDL3B with PC | Crystal used for Zn-SAD |
|---|---|---|---|
| PDB code | 5KAR | 5KAS | |
| Data collection | |||
| Beamline | F1, MacCHESS | A1, MacCHESS | 08ID-1, CMCF, CLS |
| Wavelength (Å) | 0.91790 | 0.97700 | 1.28206 |
| Space group | P 2 | P 2 | P 2 |
| Unit cell dimensions (Å) | 50.89, 47.03, 93.86 | 50.90, 47.03, 93.87 | 50.98, 46.00, 93.77 |
| Unit cell angles (°) | 90, 98.07, 90 | 90, 98.07, 90 | 90, 97.87, 90 |
| Resolution range (Å) | 31.99–1.14 (1.18–1.14) | 31.28–1.62 (1.68–1.62) | 50.00–1.72 (1.78–1.72) |
| Anomalous resolution (Å) | 2.81 | ||
| Total reflections | 874,062 | 341,710 | 265,057 |
| Unique reflections | 147,638 (8,714) | 50,662 (2,654) | 38,784 (2,258) |
| Multiplicity | 5.9 (2.7) | 6.7 (3.9) | 6.8 (4.8) |
| Completeness (%) | 92.9 (56.0) | 90.5 (51.3) | 84.8 (50.0) |
| I/σ(I) | 17.4 (0.7) | 12.8 (2.4) | 17.9 (1.7) |
| Wilson B-factor (Å2) | 13.57 | 20.28 | |
| Rmerge (%) | 8.6 (>100) | 11.1 (56.9) | 7.9 (88.7) |
| CC½ (%) | (38) | (83) | (81) |
| Refinement | |||
| Reflections used in refinement | 147,420 (8,531) | 50,343 (2,646) | |
| Reflections used for Rfree | 7,396 (425) | 2,537 (130) | |
| Rwork (%) | 13.46 (35.46) | 16.92 (23.51) | |
| Rfree (%) | 15.18 (35.88) | 19.32 (26.76) | |
| Protein molecules/ASU | 1 | 1 | |
| Number of non-hydrogen atoms | 4,019 | 3,887 | |
| Macromolecules | 3,380 | 3,344 | |
| Ligands, carbohydrates | 143 | 140 | |
| Solvent | 496 | 403 | |
| Protein residues | 412 | 412 | |
| RMS bonds (Å) | 0.009 | 0.005 | |
| RMS angles (°) | 1.07 | 0.8 | |
| Ramachandran favored (%) | 96 | 96 | |
| Ramachandran allowed (%) | 4.2 | 4.1 | |
| Ramachandran outliers (%) | 0 | 0 | |
| Rotamer outliers (%) | 0.27 | 0.55 | |
| Clashscore | 1.88 | 1.62 | |
| Average B-factor (Å2) | 20.07 | 29.69 | |
| Macromolecules | 17.63 | 27.48 | |
| Ligands, carbohydrates | 33.11 | 53.57 | |
| Solvent | 32.96 | 39.73 | |
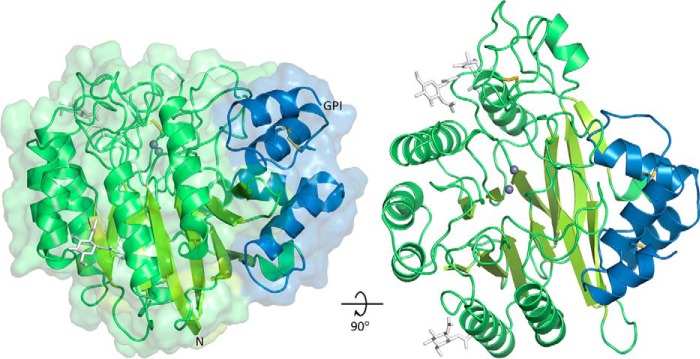
The enzyme comprises a catalytic domain affixed with a small C-terminal subdomain (CTD) (Fig. 1). The catalytic domain consists of two central mixed β-sheets of six strands each, surrounded by eight α-helices. This architecture classifies SMPDL3B in the calcineurin-like phosphoesterase structural superfamily (PFAM code PF00149). One end of the β-sandwich has a wide opening, against which the CTD, consisting of a globular cluster of four α-helices, is packed up. The fourth helix of the CTD is terminated by Gly-431. The CTD appears to be a defining feature of the ASMase-like proteins because it is also found in ASMase (5) and SMPDL3A (6), but not in other phosphoesterases. It displays lower sequence conservation than the catalytic domain, and its exact function is unknown, but it may enhance the stability of these proteins in their lysosomal and extracellular environments.
FIGURE 1.
Crystal structure of SMPDL3B. The catalytic domain is shown in green, and the C-terminal subdomain is in blue. Zinc ions are represented by gray spheres. N-Linked glycans (white sticks) are only partially displayed for clarity. Disulfide bonds are shown as yellow sticks. The N terminus and the putative C-terminal ω-site of GPI anchor attachment are indicated.
The protein fold is stabilized by a total of three disulfide bridges: two in the CTD and a third at the base of a large loop in the catalytic domain. Although murine SMPDL3B has five potential sites of N-linked glycosylation, only three glycans are discernable in the structure. The two other sites are located on the surface of the protein, suggesting that they potentially can be glycosylated. Human SMPDL3B shares only two of these five sites and has a third potential one, also on the surface of the protein.
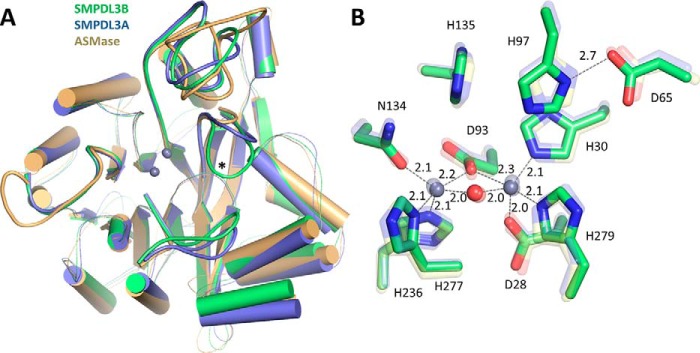
Structural superimposition of SMPDL3B with the ASMase catalytic domain and with SMPDL3A (Fig. 2A) shows that the overall fold and all secondary structure elements are well conserved within this subfamily. The main differences arise in the lengths and compositions of several loops on the “top” face of the proteins, which harbors the active site.
FIGURE 2.
Structural comparison of SMPDL3B, SMPDL3A, and ASMase. A, superimposition of the ASMase family of proteins. SMPDL3B is shown in green, SMPDL3A is in blue (PDB code 5FC6), and ASMase is in yellow (PDB code 5HQN). Loops on the face of the protein containing the active site that diverge between the family members are displayed as thick lines. Loop L9–10 of SMPDL3B (described later) is marked with an asterisk. SMPDL3B shares 28% identity with ASMase (root mean square deviation of 0.851 Å over 284 alpha carbons) and 38% identity with SMPDL3A (root mean square deviation of 0.606 Å over 295 alpha carbons). The saposin domain of ASMase is not shown. B, comparison of the SMPDL3B active site with that of SMPDL3A and ASMase. Active site residues of SMPDL3B are shown as green sticks. The active site of SMPDL3A (transparent blue sticks) and ASMase (transparent yellow sticks) are overlaid. Zinc ions are represented by gray spheres. The bridging water molecule is displayed as a red sphere. Distances are indicated (Å).
Active Site and Substrate Recognition
At the core of SMPDL3B are found two zinc ions coordinated by the side chains of seven residues located on loops extending from the central β-sheets (Fig. 2B). Two histidines, His-135 along with the His-97–Asp-65 pair, complete the active site of the enzyme. All of these residues are perfectly conserved in SMPDL3A and ASMase (Fig. 2B), suggesting that all three family members utilize the same general catalytic mechanism for substrate hydrolysis. Briefly, a phosphate-containing substrate would bind in the active site mainly via strong electrostatic interactions between the zinc ions and the phosphate moiety. A zinc-activated nucleophilic water molecule would then attack the phosphate, resulting in departure of the leaving group, protonated by one of the neighboring histidines (21–24). The nucleophilic water molecule coordinated midway between the two zinc ions is clearly visible in the high-resolution structure of SMPDL3B (Fig. 2B).
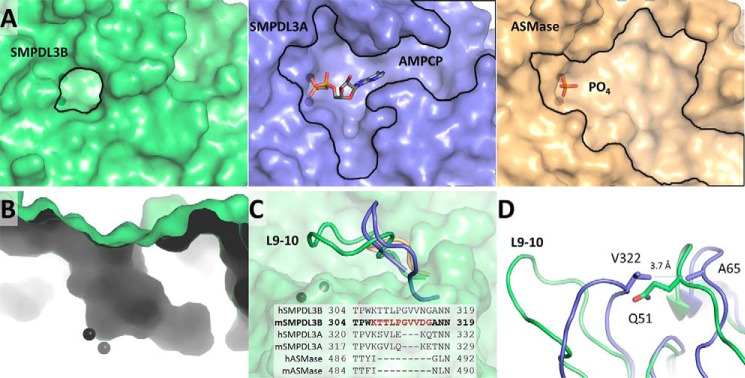
Although the three phosphodiesterases likely employ the same catalytic mechanism, the identity of the phosphate-containing substrate varies widely within this subfamily. ASMase hydrolyzes SM to ceramide and PC. The related lipid phosphatidylcholine, bearing the same PC head group, may also serve as substrate to a lower extent (5). On the other hand, SMPDL3A is inactive against SM and instead hydrolyzes various nucleotide triphosphates and diphosphates (6). The physiological substrate of SMPDL3B, however, is not known. Interestingly, despite the overall structures of the three enzymes being highly conserved (Fig. 2A), the shapes of the regions surrounding their active sites are markedly different (Fig. 3A). The structure of ASMase in complex with sulfate (24) displays a wide depression emanating from the sulfate and zinc ions at the active site. In contrast, murine SMPDL3A (22) accommodates its nucleotide substrate in a roughly T-shaped cleft, which restricts the type of molecules that can enter its active site. Human SMPDL3A displays a similar T-shaped configuration (21). In all cases, the regions adjacent to the active site of these proteins are surface-exposed depressions.
FIGURE 3.
Region around active site. A, the pocket at the active site of SMPDL3B (green). Zinc ions are represented by black spheres. The substrate binding site of SMPDL3A in complex with a nucleotide (blue, PDB code 5FC6) and ASMase bound to phosphate (yellow, PDB code 5HQN) are shown for comparison. B, boot-shaped cavity at the active site of SMPDL3B. C, the loop L9–10 of SMPDL3B (green) is compared with the corresponding loops of SMPDL3A (blue) and ASMase (yellow). Sequences of the loops of human and mouse homologs are aligned. D, conformational differences between the L9–10 loop of SMPDL3B and SMPDL3A are highlighted.
Strikingly, the structure of SMPDL3B revealed a relatively closed off active site with a narrow boot-shaped cavity accessible through a small circular opening on the protein surface (Fig. 3, A and B). The active site zinc ions are located at the heel of the boot, indicating that substrates must reach deep into this cavity. The unique nature of the SMPDL3B substrate binding site is mainly due to a loop insertion between strand β9 and strand β10 (L9–10), which assumes a distinct conformation relative to its paralogs (Fig. 3C) and forms the “tongue” of the boot-shaped substrate-binding cavity. In ASMase, this insertion is essentially absent, leading to its more expansive substrate binding site. In SMPDL3A, this region is shorter by three residues and is pinned back by hydrophobic interactions between Val-322 at the tip of the loop and Ala-65 on a neighboring loop (Fig. 3D). This creates space for the accommodation of nucleobases in this region of SMPDL3A. In SMPDL3B, however, the sequence of L9–10 differs from SMPDL3A, and this loop is oriented toward the substrate binding site, leaving a small, 9 Å opening for substrate entry.
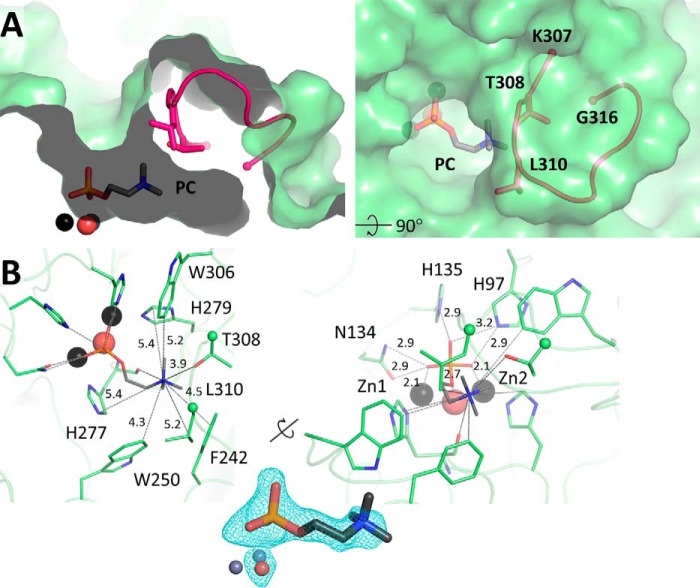
The shape of the substrate binding site provides clues as to the nature of the phosphate-containing substrate of this enzyme. Given that SMPDL3B could potentially act on lipids at the membrane surface (11), we wondered whether it can accommodate the small PC head group of SM. To assess this, we crystallized the protein in presence of PC and found that it is indeed located in the substrate binding cavity (Fig. 4A). Its phosphate group forms numerous electrostatic interactions with the zinc ions and several active site residues as observed previously in structures of ASMase and SMPDL3A (Fig. 4B). The choline moiety extends into the cavity and interacts with the backbone carbonyl of His-277 and the side chains of Leu-310 and Thr-308, which emanate from L9–10 of SMPDL3B. This space is occupied by a glycerol molecule from the crystallization condition in the apo structure. Additionally, the choline moiety is surrounded by five aromatic residues: Phe-242, Trp-250, Trp-306, His-277, and His-279, the latter two of which also participate in zinc coordination. This arrangement of several aromatic residues within 4–5 Å of the cationic quaternary amine has been termed a “cation-π box” and is a common choline recognition motif (25).
FIGURE 4.
Phosphocholine bound in the active site of SMPDL3B. A, PC (sticks) in the active site of SMPDL3B, in proximity to loop L9–10 (pink). B, residues that interact with the choline group of PC are shown as sticks (left). Distances between the choline nitrogen atom and the closest protein non-hydrogen atoms are indicated (Å). Residues that form contacts with the phosphate portion of PC, including zinc ions and water, are shown as sticks (right). Interatomic distances are indicated (Å). The electron density Fo − Fc simulated annealing omit map around PC, and the nucleophilic water molecule is contoured at 3σ.
The dimensions of the PC group just about match the size of the substrate binding site opening, suggesting that it may have to adjust slightly to allow PC access to the buried active site. Comparison of the PC-bound structure with the apo form, however, revealed no conformational changes, although binding of PC seems to increase the disorder in L9–10 as interpreted from higher crystal temperature factors (an increase by 18–64% in alpha carbon temperature factors normalized to the entire protein). Thus, L9–10 may have some malleability to allow entry of small substrates like PC, or it can perhaps move out of the way altogether to accommodate larger substrates. To assess these possibilities, we turned to mutagenesis and enzymatic assays.
Enzymatic Activity in Vitro
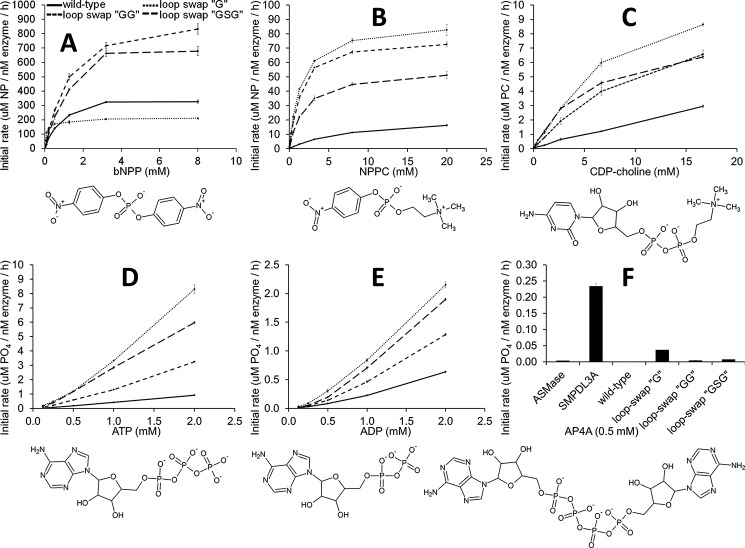
It had been previously shown that SMPDL3B possesses phosphodiesterase activity against the artificial generic substrate bis(4-nitrophenyl) phosphate (bNPP) (11). Based on its relatedness to ASMase and its effects on cellular lipid composition and membrane biophysical properties (11), we initially sought to study the activity of the purified enzyme in vitro against PC-containing lipids. Interestingly, no head group hydrolysis was detected with the following substrates: SM in Triton X-100 micelles, SM in neutral liposomes (also comprising phosphatidylcholine and cholesterol), the soluble micelle-forming lipids sphingosylphosphorylcholine (lyso-SM), and platelet-activating factor. These assays, however, were carried out with the soluble form of the protein used for crystallization that lacks the GPI moiety responsible for anchoring SMPDL3B to the cell surface (9, 10, 11). Thus, it may be that the membrane tethered form of the enzyme is capable of hydrolyzing lipids. On the other hand, SMPDL3B was able to cleave CDP-choline (Fig. 5C), an essential building block for the biosynthesis of PC-containing phospholipids in the Kennedy pathway (26). Additionally, as with SMPDL3A, we found that SMPDL3B can release phosphate from ATP and ADP (Fig. 5, D and E), but not from AMP.
FIGURE 5.
In vitro enzymatic activity against various substrates. A–F, the activity profiles of purified SMPDL3B are shown for the wild type protein (solid line), as well as for loop swaps of L9–10 by glycine, GG, or GSG (dashed lines). The chemical structures of substrates are depicted. In F, activity of purified murine ASMase (24) and SMPDL3A (22) is also shown. The values are the means and standard deviations of triplicates representative of one of two experiments.
To assess the role of L9–10 in substrate binding, this loop was replaced by a glycine residue, GG, or GSG. However, none of these modifications enabled hydrolysis of lipid substrates. Notably, shortening of L9–10 did increase activity against CDP-choline, as well as bNPP, and the artificial substrate p-nitrophenylphosphorylcholine (NPPC) (Fig. 5, A–C), suggesting that it does regulate access to the active site. In theory, CDP-choline could be recognized by the protein either with its choline group positioned at the bottom of the cavity, as in the PC bound structure (Fig. 4A), or in the reverse orientation with the nucleotide moiety occupying that location, as occurs with SMPDL3A substrates (21, 22). The latter would require a stable rearrangement of L9–10 because the cytidine moiety from CDP-choline is too large to fit in the observed substrate cavity.
To ascertain between these possibilities, the enzyme was challenged with diadenosine tetraphosphate (AP4A), a substrate with nucleosides at both ends of a tetraphosphate group. Purified SMPDL3A can cleave this molecule, but SMPDL3B is completely inactive against it (Fig. 5F). Moreover, shortening of L9–10 did not allow hydrolysis of AP4A (although the G loop swap did have a very low rate of activity). This suggests that L9–10 cannot stably rearrange into a different conformation to accommodate larger substrates like nucleosides. In the case of CDP-choline, its PC moiety would be bound to the active site as observed in the PC bound structure, and for ATP/ADP, only the terminal phosphate would enter the active site. Thus, L9–10 appears to act as a relatively fixed selectivity filter that through slight “breathing motions” can allow entry of smaller substrates like PC and isolated phosphate ends.
Enzymatic Activity in Cells
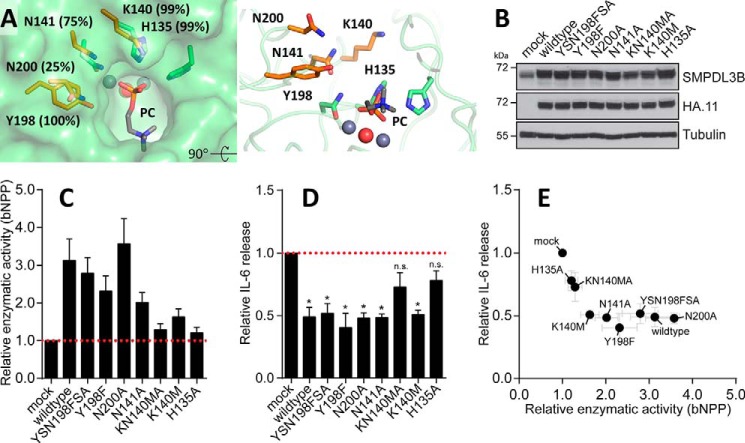
The observed activity on nucleotides, including CDP-choline, and the absence of knowledge of the precise physiological substrate lead us to examine other residues in addition to the aromatic cage and the L9–10 loop that may interact with the substrate. The walls of the channel leading to the active site are lined with amino acids involved in catalysis but also with four other residues (Lys-140, Asn-141, Tyr-198, and Asn-200; Fig. 6A), two of which, Lys-140 and Tyr-198, are highly conserved in vertebrates. We introduced mutations in these residues, as well as in His-135, one of the conserved amino acids contacting the substrate phosphate group. These mutations were designed to avoid disrupting local protein structure, but could potentially abrogate polar contacts with the physiological substrate. SMPDL3B was reported to negatively regulate TLR signaling in murine macrophages, affecting the release of pro-inflammatory cytokines from these cells (11). To test the impact of the mutants in such an experimental system, we generated a series of stable RAW264.7 macrophage cell lines, overexpressing an empty vector control (mock), wild type, or mutant forms of SMPDL3B. Western blot analysis revealed that introduction of all SMPDL3B constructs led to similarly increased protein expression as compared with the endogenous level observed in control cells (mock) (Fig. 6B). The cell lines were subsequently stimulated with the TLR ligand LPS and evaluated for cell-associated enzymatic activity toward the experimental substrate bNPP, as well as for release of the TLR-induced cytokine IL-6. Overexpression of wild type SMPDL3B led to an increase in cell-associated enzymatic activity toward bNPP (Fig. 6C) and concomitantly to a significant reduction of IL-6 release in comparison with control (Fig. 6D), confirming its negative impact on TLR-dependent processes. Individual mutants differed in their ability to hydrolyze the experimental substrate bNPP, with K140M/N141A and H135A almost completely failing to increase detectable activity (Fig. 6C). Interestingly, these are also the only two mutants tested that failed to significantly reduce LPS-induced IL-6 release from RAW264.7 cells in this experimental system (Fig. 6, D and E). The molecular position of His-135, Lys-140, and Asn-141 in relation to the PC group in the structure, along with the functional data, suggest that these residues are indeed important features for recognition and binding of endogenous substrates affecting the activity of macrophages.
FIGURE 6.
Functional impact of SMPDL3B mutants on LPS-induced IL-6 release in macrophages. A, residues lining the wall of the channel leading to the active site are displayed in orange along with their degree of conservation in vertebrates. Other adjacent amino acids, including the catalytic machinery, are in green. B, Western blot analysis for SMPDL3B, HA, and tubulin in samples from RAW264.7 macrophages stably transduced with an empty vector (mock) or HA-tagged wild type or mutant murine SMPDL3B constructs as indicated. C and D, RAW264.7 macrophages stably transduced with an empty vector (mock) or HA-tagged wild type or mutant murine SMPDL3B constructs were stimulated with 100 ng/ml LPS for 8 h. The cells were probed for enzymatic activity by incubation with bNPP (C), and supernatants were analyzed for IL-6 release by ELISA (D). Experimental data were normalized to mock and represent means ± S.E. from five independent biological replicates. Statistical significance was tested using a one sample t test against a hypothetical mean value of 1. *, p ≤ 0.05; n.s., not significant. E, relative enzymatic activity and relative IL-6 release shown in C and D plotted against each other.
Discussion
The overall structure of SMPDL3B determined here is similar to that of its paralogs SMPDL3A and ASMase. All three possess a highly conserved active site that hydrolyzes phosphate-containing substrates via a zinc-activated water molecule. Despite this, differences in protein localization and the shape of the substrate binding site appear to confer functional differences in these proteins. ASMase has an additional N-terminal saposin domain, a small membrane- and lipid-binding module that enables it to act on its SM substrate on the outer leaflet of the plasma membrane, on intralysosomal vesicles, on lipoprotein particles in the circulation, and at other locations (5). Additionally, ASMase has a relatively wide and accessible substrate binding site with optimal activity coupled to the conformation and position of the saposin domain relative to the catalytic domain (24). Conversely, SMPDL3A is not known to be membrane-interacting and has a T-shaped substrate binding site. These attributes alter its substrate specificity to soluble nucleoside-diphosphates and triphosphates and implicate it in purinergic signaling pathways (6).
In contrast to its paralogs, SMPDL3B is constitutively attached to the cell membrane exoplasmic face via a GPI anchor (9–11), and the structure reveals that its substrate binding site is different from either paralog. An extended and conformationally distinct loop, L9–10, closes off a large portion of the surface near the active site, leaving a small restricted opening. This configuration of the active site might suggest an altogether different substrate specificity for SMPDL3B from either ASMase or SMPDL3A. However, the point of GPI attachment of SMPDL3B is located on the “top” face of the enzyme (Fig. 1) and places the active site opening in proximity to the membrane lipid head groups. Moreover, previous studies implicated SMPDL3B in TLR signaling, where its action on macrophage membranes modulated lipid composition, affecting the levels of SM, other PC-based lipids, and ceramide, concomitantly altering membrane fluidity (11).
This notion is further bolstered by the structure of SMPDL3B bound to PC. The small molecule head group of SM binds in the tight cavity of the active site, and the chemical surroundings are appropriate for choline recognition as multiple aromatic residues around the quaternary ammonium group establish cation-π interactions with it. A more anionic environment would have been expected for phosphoethanolamine or other cationic or polar head groups, which would be inappropriate for choline because it cannot form hydrogen bonds or salt bridges with protein residues.
The difference in conformation of L9–10 from the corresponding loops in SMPDL3A or ASMase and that it was slightly more mobile in the presence of PC raised the possibility of a switch-like regulatory role for this segment. Although replacement of L9–10 does increase activity on some substrates tested, there is no indication that a stable conformational change occurs, at least in solution. Instead, the conformation of L9–10 appears to be more or less fixed, allowing entry of only appropriately sized substrates through small breathing motions. Alternatively, the conformation of L9–10 may be impacted at the membrane by presence of lipid head groups, because this loop is on the side of the protein likely to face the membrane. This could also apply to the other loops that diverge between SMDPL3B and its paralogs and that are found on the same face (Fig. 2A).
Based on the crystal structure, we identified amino acid residues potentially required for recognition of endogenous substrates, and evaluated their importance for SMPDL3B-dependent effects in a cellular model of LPS-induced IL-6 release. In addition to His-135, a conserved residue mediating interactions with the phosphate group, we found that a mutant affecting both Lys-140 and Asn-141, two amino acids situated in the channel leading to the active site of SMPDL3B, failed to significantly reduce LPS-induced IL-6 release. The location of these amino acids relative to PC seen in the structure, along with the functional information, makes it likely that these residues indeed specifically participate in substrate recognition.
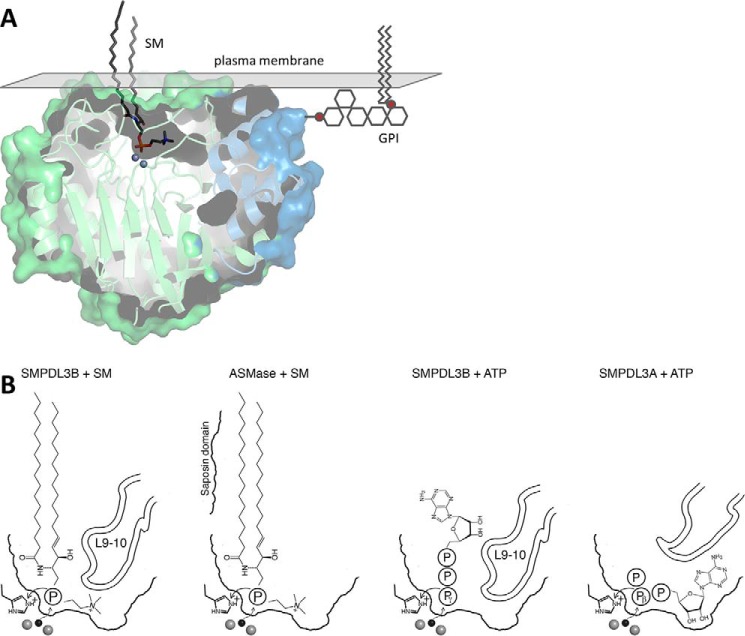
Taken together, these findings suggest that SM with its small PC head group is a viable candidate substrate for SMPDL3B. Manual modeling of SM based on previous docking of SM onto ASMase (23, 24) shows that in both cases, SM is positioned similarly and can insert into the active site with few clashes (Fig. 7A). With SMPDL3B, only the headgroup of SM would be stably bound, whereas with ASMase, portions of the ceramide moiety may also interact with the protein with the aid of is accompanying saposin domain. Although we did not observe any in vitro hydrolysis of SM in detergent micelles or liposomes or of other soluble lipids even when the loop L9–10 is replaced, this is likely because the protein requires proper attachment to membranes for substrate hydrolysis to occur.
FIGURE 7.
Proposed binding modes of potential substrates. A, SM (dark gray sticks) was manually placed in the substrate binding site using bound PC as a guide. A GPI anchor (stylized gray lines) is attached to the putative C-terminal ω-site. The surface of the plasma membrane is represented by a gray plane. B, comparison of potential substrate positioning in SMPDL3B and ASMase for SM and in SMPDL3B and SMPDL3A for nucleotide triphosphates. Zinc ions (gray spheres) and the nucleophilic water molecule (black sphere) are shown.
Alternatively, the physiological substrate could in fact be a non-lipid molecule. We found that the enzyme can hydrolyze CDP-choline, the activated metabolite of PC used in lipid biosynthesis, in vitro. Even though this molecule is usually found intracellularly, it might encounter SMPDL3B under specific physiological conditions, such as inflammatory tissue damage or digestive processes (27). However, the affinity and activity rate of the protein on CDP-choline are rather low (Fig. 5C). We also found activity against ATP and ADP. In all of these cases, however, the means by which substrate molecules are hydrolyzed would be different from SMPDL3A, which also acts on nucleoside diphosphates and triphosphates (Fig. 7B). In SMPDL3A, during the hydrolysis of ATP or ADP, the phosphate preceding the terminal phosphate (β-phosphate for ATP, α-phosphate for ADP) must be coordinated by the zinc ions for attack by the nucleophilic water molecule, and the terminal phosphate (γ-phosphate for ATP, β-phosphate for ADP) is the leaving group because of the constraints imposed by the position of the protonating histidine side chain and orientation of the nucleobase in the T-shaped substrate binding site (22). Conversely, in SMPDL3B, because of the block imposed by L9–10 preventing accommodation of large moieties like complex glycolipids or nucleobases inside the cavity, ATP or ADP phosphate release likely proceeds in a “tail-first” manner in which the terminal phosphate contacts the zinc ions in the active site for nucleophilic attack, while the rest of the molecule remains in the channel leading to them and is the leaving group upon hydrolysis. The suitability of nucleotides as physiological substrates of SMPDL3B, however, is unclear because of their weak, millimolar Km (Fig. 5, D and E) and relatively low activity rate, about 10-fold slower than SMPDL3A at neutral pH (22).
In summary, the structure of murine SMPDL3B highlights molecular details important for catalysis and substrate recognition. This knowledge will further contribute to the understanding of which substrates are bound and hydrolyzed under physiological conditions and, as a consequence, how SMPDL3B influences cellular properties such as lipid composition or inflammatory signaling. The tight cavity forming its substrate binding site, differing from its paralogs, also provides a promising feature for the identification of enzyme-specific inhibitors that could be exploited for modulation of SMPDL3B-dependent functions in relevant experimental systems, including immunity and kidney biology.
Experimental Procedures
Constructs
For expression in Sf9 insect cells, murine SMPDL3B (residues 19–435; UniProt no. P58242) was subcloned into a derivative of pFastBac 1 (Invitrogen). The vector contained the melittin signal peptide MKFLVNVALVFMVVYISYIYA followed by a hexahistidine tag DRHHHHHHKL. Swaps of the loop L9–10 consisted of residues 307–316 (KTTLPGVVDG) replaced by a glycine, GG, or GSG. Expression constructs for mammalian cells were based on a derivative of the entry vector pEntry1A (Invitrogen) encoding the signal peptide of influenza HA followed by an HA tag and the cDNA for murine SMPDL3B (residues 19–456) as reported previously (11). Individual point mutations corresponding to H135A, K140M, N141A, K140M/N141A, Y198F, N200A, and Y198F/N200A were introduced by site-directed mutagenesis using the QuikChange II system (Agilent). For retroviral gene transduction, cDNAs were shuttled to a derivative of pMSCV-GW containing a blasticidin resistance gene using the Gateway LR Clonase (Invitrogen) system. All constructs and mutants were verified by sequencing.
Protein Production
Soluble SMPDL3B was expressed as a secreted protein in Sf9 cells and purified as previously described for SMPDL3A (22). The protein eluted as a monomer in size exclusion chromatography.
Crystallization and Data Collection
Soluble SMPDL3B at 10 mg ml−1 in buffer (15 mm Tris-HCl, pH 7.5, 100 mm NaCl) was mixed with an equal volume of well solution consisting of 0.2 m NH4NO3 and 25% PEG 3350. Crystals were grown by hanging drop vapor diffusion at 22 °C. For anomalous diffraction experiments, the crystals were soaked in well solution supplemented with 10 mm ZnCl2. PC-bound crystals were obtained by preincubating and crystallizing the protein with 20 mm PC. Diffraction data were collected at 100 K on Beamline 08ID-1 at the Canadian Macromolecular Crystallography Facility, Canadian Light Source, and on Beamlines MacCHESS A1 and F1, Cornell High Energy Synchrotron Source. The data were processed with HKL2000 (28).
Structure Determination and Refinement
The structure of SMPDL3B was solved by zinc single-wavelength anomalous diffraction using Autosol in Phenix (29, 30) and manually rebuilt in Coot (31). The PC-bound structure was obtained by molecular replacement using Phaser (32) in Phenix. Refinement was performed with phenix.refine (33). Data processing and refinement statistics are presented in Table 1. Structural images were generated with PyMOL (PyMOL Molecular Graphics System, version 1.3; Schrödinger).
In Vitro Enzymatic Activity Assays
Proteins for activity assays were purified as described above, but the final buffer included 10 μm ZnCl2. To detect hydrolysis of the choline-containing substrates SM in Triton X-100 micelles, SM in neutral liposomes (also comprising phosphatidylcholine and cholesterol), sphingosylphosphorylcholine (lyso-SM), platelet-activating factor and CDP-choline, the Amplex Red sphingomyelinase assay (Thermo Fisher Scientific) was slightly modified. Proteins at 5–25 nm for CDP-choline or up to 10 μm otherwise were incubated with substrate in buffer (20 mm Tris-HCl, pH 7.5, 100 mm NaCl) for 1 h at 37 °C. The reaction was terminated at 95 °C for 5 min, and an equal volume of the second step solution was added as recommended. The second step was carried out at 37 °C, and the change in fluorescence (560-nm excitation and 590-nm emission) after 30 min was used to quantify product formation with the help of a PC standard curve. To detect hydrolysis of bNPP and NPPC, proteins at 1–50 nm were incubated with substrate in buffer (100 nm NaCl, Tris-HCl, pH 7.5, at 20 mm for NPPC or 100 mm for bNPP) for 30 min at 37 °C. NaOH was added to 100 mm before measuring absorbance at 405 nm. Product formation was quantified with a p-nitrophenol standard curve. Enzymatic activity on ATP, ADP, AMP, and AP4A was measured by a slightly modified Biomol Green phosphate detection method (Enzo Life Sciences). Proteins at 10 nm to 1 μm were incubated with substrate in buffer (20 mm Tris-HCl, pH 7.5, 100 mm NaCl) for 30 min at 37 °C; four volumes of Biomol Green reagent were added followed by incubation for 20 min and absorbance measurement at 620 nm. Product formation was quantified with a phosphate standard curve.
Mammalian Cell Culture and Retroviral Gene Transduction
HEK293T and RAW264.7 cells (ATCC) were maintained in DMEM (Gibco) supplemented with 10% FCS (Gibco) and antibiotics (100 units/ml penicillin and 100 μg/ml streptomycin; Gibco) at 37 °C, 5% CO2. Stable expression constructs were introduced to RAW264.7 cells by retroviral gene transduction. Briefly, HEK293T cells were transiently transfected with packaging plasmids pGAG-POL, pVSV-G, pADVANTAGE, and the individual pMSCV expression constructs using PolyFect (Qiagen). 24 h after transfection, the medium was replaced. 48 h after transfection, the virus-containing supernatants were harvested, filtered through 0.45-μm polyethersulfone filters (GE Healthcare), supplemented with 5 μg/ml Polybrene (Sigma), and added to 40–60% confluent RAW264.7 cells. 24 h after infection, the cells were replated in fresh medium; 48 h after infection, transduced cells were selected by addition of 25 μg/ml blasticidin (InvivoGen).
Western Blot Analysis
Protein was extracted from stably transduced RAW264.7 cells using radioimmune precipitation assay buffer (10 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS, 1 mm EDTA, Complete protease inhibitor mixture (Roche), and 250 u/ml Benzonase (Novagen)). Protein concentration in cleared lysates was determined using the Bradford protein assay (Bio-Rad) and normalized using radioimmune precipitation assay buffer. Samples were mixed with Laemmli sample buffer, resolved by SDS-PAGE, and analyzed by Western blot. Custom rabbit anti-SMPDL3B antibodies raised against recombinant murine SMPDL3B (amino acids 1–281) were reported previously (11). Mouse monoclonal anti-HA.11 antibodies were from Covance; anti-tubulin ones were from Abcam. HRP-conjugated secondary antibodies were from Jackson ImmunResearch.
LPS Stimulation, Enzymatic Activity Measurement, and IL-6 release from RAW264.7 Cells
RAW264.7 cells stably transduced with empty vector, wild type, or mutant SMPDL3B constructs were harvested in serum-free DMEM medium and counted. In technical triplicates, 5 × 104 cells in 100 μl of DMEM/well were seeded in tissue culture-treated 96-well plates (Sarstedt) and adhered for 2 h at 37 °C, 5% CO2. The cells were stimulated for 8 h with 100 ng/ml ultrapure LPS from Escherichia coli 0111:B4 (InvivoGen). Supernatants were harvested and analyzed for mouse IL-6 by ELISA (eBioscience) according to the manufacturer's instructions. In parallel, the cells were gently washed with PBS and incubated with 100 μl/well of 1 mm bNPP (Sigma) in isotonic TBS buffer (G-Biosciences) at 37 °C, 5% CO2 as described previously (11). 80 μl from each well were transferred to new 96-well plates and combined with 20 μl of 2 m NaOH/well. Generation of p-nitrophenol from bNPP by phosphodiesterase activity was determined as absorbance at 405 nm in a microplate reader (SpectraMax).
Author Contributions
A. G., L. X. H., K. I., G. S. F., and B. N. designed the experiments. A. G. performed structural biology work. L. X. H. carried out mammalian cell experiments. A. G. and L. X. H. performed enzymatic work. K. I. conducted insect cell culture. A. G. and L. X. H. wrote the manuscript. All authors reviewed and approved the final version of the manuscript.
Acknowledgments
We thank Dr. Guennadi Kozlov for crystallographic data collection. This work is based upon research conducted at the Cornell High Energy Synchrotron Source, which is supported by the National Science Foundation and the National Institutes of Health/National Institute of General Medical Sciences under National Science Foundation award DMR-1332208, using the Macromolecular Diffraction facility at the Cornell High Energy Synchrotron Source, which is supported by award GM-103485 from the National Institute of General Medical Sciences of the National Institutes of Health. We also thank Dr. Shaun Labiuk for crystallographic data collection performed at Beamline 08ID-1 at the Canadian Light Source, which is supported by the Canada Foundation for Innovation, Natural Sciences and Engineering Research Council of Canada, the University of Saskatchewan, the Government of Saskatchewan, Western Economic Diversification Canada, the National Research Council Canada, and the Canadian Institutes of Health Research.
This work is supported in part by the Austrian Academy of Sciences (to G. S.-F.). The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (codes 5KAR and 5KAS) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- SM
- sphingomyelin
- PC
- phosphocholine
- SMase
- sphingomyelinase
- ASMase
- acid sphingomyelinase
- GPI
- glycosylphosphatidylinositol
- TLR
- Toll-like receptor
- CTD
- C-terminal subdomain
- bNPP
- bis(4-nitrophenyl) phosphate
- NPPC
- p-nitrophenylphosphorylcholine
- AP4A
- diadenosine tetraphosphate
- PDB
- Protein Data Bank.
References
- 1. Slotte J. P. (2013) Biological functions of sphingomyelins. Prog. Lipid Res. 52, 424–437 [DOI] [PubMed] [Google Scholar]
- 2. Castro B. M., Prieto M., and Silva L. C. (2014) Ceramide: a simple sphingolipid with unique biophysical properties. Prog. Lipid Res. 54, 53–67 [DOI] [PubMed] [Google Scholar]
- 3. Wu J., Hansen G. H., Nilsson A., and Duan R. D. (2005) Functional studies of human intestinal alkaline sphingomyelinase by deglycosylation and mutagenesis. Biochem. J. 386, 153–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Airola M. V., and Hannun Y. A. (2013) Sphingolipid metabolism and neutral sphingomyelinases. Handb. Exp. Pharmacol. 215, 57–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kornhuber J., Rhein C., Müller C. P., and Mühle C. (2015) Secretory sphingomyelinase in health and disease. Biol. Chem. 396, 707–736 [DOI] [PubMed] [Google Scholar]
- 6. Traini M., Quinn C. M., Sandoval C., Johansson E., Schroder K., Kockx M., Meikle P. J., Jessup W., and Kritharides L. (2014) Sphingomyelin phosphodiesterase acid-like 3A (SMPDL3A) is a novel nucleotide phosphodiesterase regulated by cholesterol in human macrophages. J. Biol. Chem. 289, 32895–32913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sleat D. E., Jadot M., and Lobel P. (2007) Lysosomal proteomics and disease. Proteomics Clin. Appl. 1, 1134–1146 [DOI] [PubMed] [Google Scholar]
- 8. Noto P. B., Bukhtiyarov Y., Shi M., McKeever B. M., McGeehan G. M., and Lala D. S. (2012) Regulation of sphingomyelin phosphodiesterase acid-like 3A gene (SMPDL3A) by liver X receptors. Mol. Pharmacol. 82, 719–727 [DOI] [PubMed] [Google Scholar]
- 9. Cortes L. K., Vainauskas S., Dai N., McClung C. M., Shah M., Benner J. S., Corrêa I. R. Jr., VerBerkmoes N. C., and Taron C. H. (2014) Proteomic identification of mammalian cell surface derived glycosylphosphatidylinositol-anchored proteins through selective glycan enrichment. Proteomics 14, 2471–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Masuishi Y., Nomura A., Okayama A., Kimura Y., Arakawa N., and Hirano H. (2013) Mass spectrometric identification of glycosylphosphatidylinositol-anchored peptides. J. Proteome Res. 12, 4617–4626 [DOI] [PubMed] [Google Scholar]
- 11. Heinz L. X., Baumann C. L., Köberlin M. S., Snijder B., Gawish R., Shui G., Sharif O., Aspalter I. M., Müller A. C., Kandasamy R. K., Breitwieser F. P., Pichlmair A., Bruckner M., Rebsamen M., Blüml S., et al. (2015) The lipid-modifying enzyme SMPDL3B negatively regulates innate immunity. Cell Rep. 11, 1919–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Köberlin M. S., Snijder B., Heinz L. X., Baumann C. L., Fauster A., Vladimer G. I., Gavin A. C., and Superti-Furga G. (2015) A conserved circular network of coregulated lipids modulates innate immune responses. Cell 162, 170–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fornoni A., Sageshima J., Wei C., Merscher-Gomez S., Aguillon-Prada R., Jauregui A. N., Li J., Mattiazzi A., Ciancio G., Chen L., Zilleruelo G., Abitbol C., Chandar J., Seeherunvong W., Ricordi C., et al. (2011) Rituximab targets podocytes in recurrent focal segmental glomerulosclerosis. Sci. Transl. Med. 3, 85ra46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tasaki M., Shimizu A., Hanekamp I., Torabi R., Villani V., and Yamada K. (2014) Rituximab treatment prevents the early development of proteinuria following pig-to-baboon xeno-kidney transplantation. J. Am. Soc. Nephrol. 25, 737–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Takahashi Y., Ikezumi Y., and Saitoh A. (2016) Rituximab protects podocytes and exerts anti-proteinuric effects in rat adriamycin-induced nephropathy independent of B-lymphocytes. Nephrology (Carlton) 10.1111/nep.12737 [DOI] [PubMed] [Google Scholar]
- 16. Yoo T. H., Pedigo C. E., Guzman J., Correa-Medina M., Wei C., Villarreal R., Mitrofanova A., Leclercq F., Faul C., Li J., Kretzler M., Nelson R. G., Lehto M., Forsblom C., Groop P. H., et al. (2015) Sphingomyelinase-like phosphodiesterase 3b expression levels determine podocyte injury phenotypes in glomerular disease. J. Am. Soc. Nephrol. 26, 133–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rindler M. J., Xu C. F., Gumper I., Smith N. N., and Neubert T. A. (2007) Proteomic analysis of pancreatic zymogen granules: identification of new granule proteins. J. Proteome Res. 6, 2978–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ogawa Y., Miura Y., Harazono A., Kanai-Azuma M., Akimoto Y., Kawakami H., Yamaguchi T., Toda T., Endo T., Tsubuki M., and Yanoshita R. (2011) Proteomic analysis of two types of exosomes in human whole saliva. Biol. Pharm. Bull. 34, 13–23 [DOI] [PubMed] [Google Scholar]
- 19. Thouvenot E., Urbach S., Dantec C., Poncet J., Séveno M., Demettre E., Jouin P., Touchon J., Bockaert J., and Marin P. (2008) Enhanced detection of CNS cell secretome in plasma protein-depleted cerebrospinal fluid. J. Proteome Res. 7, 4409–4421 [DOI] [PubMed] [Google Scholar]
- 20. Picariello G., Ferranti P., Mamone G., Klouckova I., Mechref Y., Novotny M. V., and Addeo F. (2012) Gel-free shotgun proteomic analysis of human milk. J. Chromatogr. A. 1227, 219–233 [DOI] [PubMed] [Google Scholar]
- 21. Lim S. M., Yeung K., Trésaugues L., Ling T. H., and Nordlund P. (2016) The structure and catalytic mechanism of human sphingomyelin phosphodiesterase like 3a: an acid sphingomyelinase homologue with a novel nucleotide hydrolase activity. FEBS J. 283, 1107–1123 [DOI] [PubMed] [Google Scholar]
- 22. Gorelik A., Illes K., Superti-Furga G., and Nagar B. (2016) Structural basis for nucleotide hydrolysis by the acid sphingomyelinase-like phosphodiesterase SMPDL3A. J. Biol. Chem. 291, 6376–6385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xiong Z. J., Huang J., Poda G., Pomès R., and Privé G. G. (2016) Structure of human acid sphingomyelinase reveals the role of the saposin domain in activating substrate hydrolysis. J. Mol. Biol. 428, 3026–3042 [DOI] [PubMed] [Google Scholar]
- 24. Gorelik A., Illes K., Heinz L. X., Superti-Furga G., and Nagar B. (2016) Crystal structure of mammalian acid sphingomyelinase. Nat. Commun. 7, 12196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cheng J., Goldstein R., Gershenson A., Stec B., and Roberts M. F. (2013) The cation-pi box is a specific phosphatidylcholine membrane targeting motif. J. Biol. Chem. 288, 14863–14873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fagone P., and Jackowski S. (2013) Phosphatidylcholine and the CDP-choline cycle. Biochim. Biophys. Acta 1831, 523–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cornell R. B., and Ridgway N. D. (2015) CTP:phosphocholine cytidylyltransferase: Function, regulation, and structure of an amphitropic enzyme required for membrane biogenesis. Prog. Lipid Res. 59, 147–171 [DOI] [PubMed] [Google Scholar]
- 28. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 29. Terwilliger T. C., Adams P. D., Read R. J., McCoy A. J., Moriarty N. W., Grosse-Kunstleve R. W., Afonine P. V., Zwart P. H., and Hung L. W. (2009) Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr. 65, 582–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., and Adams P. D. (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 [DOI] [PMC free article] [PubMed] [Google Scholar]