Abstract
Mitochondrial ribosomal proteins are important for mitochondrial-encoded protein synthesis and mitochondrial function. In addition to their roles in mitoribosome assembly, several mitochondrial ribosome proteins are also implicated in cellular processes like cell cycle regulation, apoptosis, and mitochondrial homeostasis regulation. Here, we demonstrate that MRPL10 regulates cyclin B1/Cdk1 (cyclin-dependent kinase 1) activity and mitochondrial protein synthesis in mammalian cells. In Drosophila, inactivation of mRpL10 (the Drosophila ortholog of mammalian MRPL10) in eyes results in abnormal eye development. Furthermore, expression of human cyclin B1 suppresses eye phenotypes and mitochondrial abnormality of mRpL10 knockdown Drosophila. This study identified that the physiological regulatory pathway of MRPL10 and providing new insights into the role of MRPL10 in growth control and mitochondrial function.
Keywords: : MRPL10, Drp1, cyclin B1, mitochondrial fission
Introduction
Mitochondria are complex organelles that contain mitochondrial ribosome (mitoribosome) and DNA (mtDNA). Human mtDNA is composed of 16,569 base pairs harboring 37 genes and coding 13 mitochondrial proteins. Human mitoribosome is composed of a small 28 S subunit, which consists of 12S rRNA and 29 proteins, and a large 39 S subunit, which consists of the 16S rRNA and 48 proteins (Sharma et al., 2003; Greber et al., 2014; Kaushal et al., 2014). These mitochondrial ribosomal proteins are encoded by genes in the nucleus, imported into mitochondrial matrix, and involved in mitoribosome assembly for mitochondrial translation. Recent studies showed that besides mitochondrial protein synthesis and energy production, several mitoribosome proteins were also implicated in cellular processes like cell cycle regulation, apoptosis, and mitochondrial homeostatic regulation. For example, the Drosophila mRpL12 (Mitochondrial Ribosomal Protein L12) is required for CycD/Cdk4-induced cell growth (Frei et al., 2005). Human MRPL12 can bind and activate mitochondrial RNA polymerase (POLRMT), which may coordinate mitoribosome biogenesis and transcription in human mitochondria (Nouws et al., 2016). MRPS18-2 is involved in pRb control of the cell cycle through direct interaction with pR (Kashuba et al., 2008). MRPS29 (DAP3) and MRPS30 (PDCD9) are involved in promoting apoptosis in mammalian cells (Cavdar Koc et al., 2001). Additionally, previous studies reported that mutations in mitoribosome genes were associated with mitochondria-related disorders. Mutation in MRPL12 led to growth retardation and neurological deterioration (Serre et al., 2013). MRPL44 gene mutation caused mitochondrial infantile cardiomyopathy (Carroll et al., 2013), while mutation in MRPS16 was associated with mitochondrial respiratory chain disorders (Miller et al., 2004). However, there is still more to learn with regards to mitoribosomes, and the molecular mechanisms of their mutations in the progression of mitochondria-related disorders and other diseases remain elusive.
Until now, one of the best studied mitoribosome conserved region L7/L12 stalk, which contains MRPL7/12, MRPL10, and MRPL11, plays a critical role in the elongation of mitochondrial translation. Previous studies identified that the N-terminal of MRPL10 can interact with the ribosome and the C-terminal can bind to MRPL12, which suggest that MRPL10 act as an anchor to support L7/L12 stalk formation (Koc et al., 2001; Han et al., 2011). A single nucleotide polymorphism rs3209 in MRPL10 associated with early age-related macular degeneration (AMD) was also reported in Chinese Americans (Klein et al., 2013). However, the precise functional relationships between MRPL10 and mitochondrial function are largely unknown.
Cdk1 (cyclin-dependent kinase 1) is a serine/threonine kinase that mediates cell cycle transition from late G2 to mitosis. Cdk1 is also a potent regulator of mitochondrial function and mitochondrial dynamics. Current studies suggest that Cdk1 contributes to mitochondrial segregation via the phosphorylation of mitochondrial fission component Drp1 (at Ser585 rat/Ser 616 human) (Taguchi et al., 2007). Cdk1 kinase activity is repressed through phosphorylation of its inhibitory residue Tyr14 and Tyr15 catalyzed by Myt1 and Weel, and it can be activated through the dephosphorylation of the same residue catalyzed by phosphatase Cdc25C (Kim and Ferrell, 2007; Wurzenberger and Gerlich, 2011). The activation of Cdk1 kinase also needs its interacting protein cyclin B to form the maturation-promoting factor (MPF), which phosphorylates corresponding substrates and control their function (Hara et al., 2012).
In this study, we demonstrate that MRPL10, which encodes a protein of the large subunit of mitoribosome, is required for mitochondrial protein synthesis and mitochondrial activity. Depletion of MRPL10 downregulates Cdk1 kinase activity and leads to mitochondrial fusion via dephosphorylation of Drp1 at Ser616. Moreover, expression of human cyclin B1 in Drosophila suppresses eye defects and mitochondrial abnormality caused by mRpL10 deficiency. Together, our results indicate that MRPL10 plays important roles in mitochondrial function and cell cycling, and identify the possible therapeutic targets for mitoribosome gene-related disorders.
Materials and Methods
Cell culture, transfections, and RNA interference
The HEK293 cells and human retinal pigment epithelial cell line ARPE-19 were grown at 37°C under 5% CO2 in DMEM or DMEM/F12 supplemented with 10% FBS and100 U/mL of penicillin/streptomycin. Two siRNAs were used to target the human MRPL10 mRNA sequences: 5′-GCAGAGGACAAGCUUCUUATT-3′ and 5′-GCGAGA AGGAUUCUGUCAUTT-3′, respectively. A nontargeting siRNA (5′-UUCUCCGAACGUGUCACGUTT-3′) was used as a negative control for all siRNA transfection experiments. Briefly, MRPL10 siRNA-1, MRPL10 siRNA-2, and control siRNA were transfected into HEK293 or ARPE-19 using lipo2000 (Invitrogen), after 24 h cells were transfected again and 72 h post-transfection, cells were harvested and subjected to experiment. For cyclin B1 expression in MRPL10 knockdown cells, cells were first transfected with MRPL10 siRNA-1, MRPL10 siRNA-2, or control siRNA using lipo2000, after 24 h cells were transfected with pcDNA3.1 empty vector or cyclin B1 plasmid.
Quantitation of eye phenotypes
Flies harboring ey-gal4 were obtained from the Bloomington Drosophila Stock Center. mRpL10 RNAi and hCyclin B1 transgenic flies were obtained from TsingHua Fly Center. Flies were grown at 25°C and maintained on standard cornmeal media. The percentage of male and female flies with abnormal eyes was determined (n > 300).
Immunoblotting, immunoprecipitation, and mitochondria separation
Immunoblotting and immunoprecipitation were done essentially as described previously (Xiong et al., 2009). The following primary antibodies were used for immunoblotting: rabbit anti-MRPL10 (HPA021234; Sigma); rabbit anti-Phospho Cdk1, mouse anti-Cdk1, mouse anti-Tom20, and mouse anti-NDUFS3 (ab18, ab47594, ab56783, and ab110246; Abcam); mouse anti-cyclin B1, rabbit anti-Drp1, rabbit anti-Mfn2, and rabbit anti-Opa1 antibodies (4135, 11925, 14739, and 67589; Cell Signaling); and rabbit anti-Fis1 was from Alexis Biochemicals (ALX-210-907-R100). Mitochondria protein were separated with mitochondria fractionation kit (ab65320; Abcam) according to the protocol, and the primary antibody rabbit anti-ND5 (NADH dehydrogenase subunit 5, ab92624; Abcam), rabbit anti-ND2 (NADH dehydrogenase subunit 2, ab102753; Abcam), rabbit anti-Cox II (ab110258; Abcam), rabbit anti-Cox I (ab14705; Abcam), rabbit anti-Cox IV (4850; Cell Signaling), mouse anti-ATP5A (anti-ATP synthase α, ab14748; Abcam), anti-phosphoserine/threonine antibody (ab17464; Abcam), rabbit anti-NDUFA12 antibody (ab192617; Abcam), and mouse anti-NDUFA12 antibody (ab110244; Abcam) were used to detect enriched mitochondrial fractions by immunoblotting or immunoprecipitation. Gray density of immunoblotting results were measured using ImageJ software (http://rsb.info.nih.gov/ij/). Bar graphs depict the signal intensities of protein indicated. The ratio of protein indicated to the loading control were first calculated, and the relative expression is shown as a percentage of the mean using the control sample as reference.
Mitochondrial respiration assay and ATP assay
HEK293 cells were transfected with MRPL10 siRNAs with or without cyclin B1 for 48 h. The cells were harvested and resuspended in phosphate-buffered saline (PBS) supplemented with 25 mM glucose, 1 mM pyruvate, and 2% BSA. After cell count 2 × 106 cells were put in clark oxygen electrode and cellular respiration was measured. ATP level was quantified as previously described using luminescent ATP detection assay kit (ab113849; Abcam). Briefly, lysates from cells or flies were prepared. Protein content was measured by the BCA protein assay reagent, and the substrate buffer was added to the lyophilized ATP substrate to prepare luminescent solution. The samples were mixed with luminescent solution and measured by an illuminometer (Berthold Technologies). The relative expression is shown as a percentage of the mean using the control sample as reference.
Mitochondrial membrane potential assay
Mitochondrial membrane potential assay was performed using TMRE (tetramethyl rhodamine ethyl ester)-mitochondrial membrane potential assay kit (ab113852; Abcam). Briefly, HEK293 cells were transfected with MRPL10 siRNAs twice for 72 h followed by staining with TMRE (100 nM) for 20 min. Post treatment, cells were immediately analyzed using a FACScan flow cytometer (Becton Dickinson, San Jose, CA) with excitation at 488 nm and emission at 530 nm. The results were obtained from three independent experiments.
Immunocytochemistry
Transfected ARPE-19 cells were harvested by washed once with PBS, followed by fixation with 3.7% paraformaldehyde for 10 min and permeabilization with 0.1% Triton X-100. After blocking with 5% BSA for 30 min, cells were incubated with primary antibodies and detected with Alexa-conjugated secondary antibodies. The antibodies used to detect Tom20 was from Abcam (ab56783). Cells were imaged using a confocal microscope (TCS SP5; Leica). Image contrast and brightness were adjusted in Image J.
Mitochondrial complex activity assay
Mitochondrial complex activities were measured using mitochondrial complex I, II, III, IV, and V activity assay kit (Novagen, Merck KGaA, Darmstadt, Germany) according to the manufacturer's instructions. Cells treated with rotenone (5 μM), thenoyltrifluoroacetone (TTFA, 10 μM), antimycin A (2 μM), potassium cyanide (KCN, 100 μM), and oligomycin (10 μM) to inhibit mitochondrial complex I, II, III, IV, and V activities, respectively, were used as positive controls. Briefly, protein content was first measured by the BCA protein assay reagent, and then diluted the concentration to 5 mg/mL in incubation solution. After incubating the sample on plate for 3 h at room temperature, the plate was measured at OD450 at 1 min intervals for 30 min. The activity was expressed as the change in absorbance per minute per amount of sample loaded into the well. The relative expression was shown as a percentage of the mean using the control sample as reference.
mtDNA copy number measurement and real-time PCR
The copy number of mtDNA was measured as previously described (Cotney et al., 2007). Total RNA was isolated using TRIzol reagent (Life Technologies Corporation) following the manufacturer's instruction. cDNA was synthesized from total RNA using Revert Aid™ First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). The following primer pairs were used for PCR assays: human MRPL10, 5′-TGCATTGATGACACCATCCT-3′ (forward) and 5′-GCC GACATGACAGAATCCTT-3′ (reverse); human GAPDH, 5′-TCATCTCTGCCCCCTCTGCT-3′ (forward) and 5′-CGA CGCCTGCTTCACCACCT-3′ (reverse). Drosophila mRpL10, 5′-ACAGCAGCAGGAGAATCCAT-3′ (forward) and 5′-GTG CAGGTTCTGCTTGTGAA-3′ (reverse). Drosophila actin, 5′-ACTTCTGCTGGAAGGTGGAC-3′ (forward) and 5′-ATC CGCAAGGATCTGTATGC-3′ (reverse).
Human GAPDH and Drosophila actin gene expression were used for normalization, respectively. Real-time PCR was performed using SYBR Green Master Mix (Thermo Scientific) and in Bio-Rad Cycler thermal cycler (Bio-Rad). Each cycle was at 95°C for 5 s and at 60°C for 30 s for 40 cycles.
Electron microscopy
For electron microscopy, Drosophila eyes were dissected and fixed in 2% glutaraldehyde for 2 h at 4°C, and then postfixed in 1% osmium tetroxide for 1 h at 4°C. The samples were dehydrated, embedded in plastic, and cut into in 70-nm sections for microscopy. The sections were then poststained with 5% uranyl acetate. Samples were viewed with a Hitachi H-7650 electron microscope.
Statistical analysis
Statistical analysis was performed using Prism 5 software (GraphPad). Two-tailed Student's t-test was used to determine the significance of difference between two groups. One-way analysis of variance with Dunnett's Multiple Comparison Test were used to assess the difference between treatments groups against their respective controls. The data were presented as mean ± standard error of the mean.
Results
MRPL10 knockdown leads to decreased mitochondrial activity and mitochondrial complexes expression
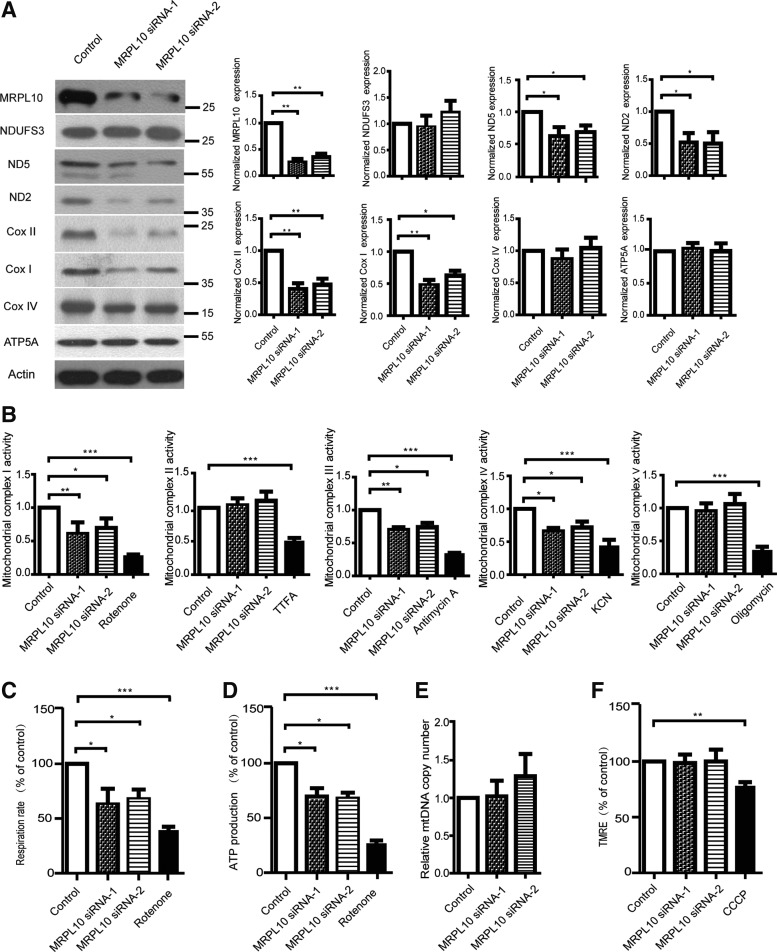
To explore the interaction of MRPL10 and mitochondrial activity in vitro, MRPL10 was knockdown by siRNA in HEK293 cells. As illustrated in Figure 1A, the expression level of mitochondria-encoded protein ND5 and ND2 of mitochondrial complex I, COX I and COX II of mitochondrial complex IV were significantly decreased in MRPL10 knockdown cells, whereas nuclear-encoded mitochondrial proteins, such as NUDFS3, COX IV, and ATP5A had not changed. Consistent with previous findings which showed that mitochondrial translation was decreased globally upon MRPL12 depletion or Mrps34 mutation (Richman et al., 2015; Nouws et al., 2016). Next, mitochondrial complex activity was assayed as shown in Figure 1B. Mitochondrial complex I, III, and IV activity were significantly reduced in MRPL10 knockdown cells, consistent with the observed decrease of mitochondrial complex proteins. Yet the activity of complex II, and V revealed no obvious difference between MRPL10 knockdown and control cells. Moreover, MRPL10 knockdown diminished mitochondrial respiration and intracellular ATP levels (Fig. 1C, D). The mtDNA copy number and mitochondrial membrane potential were not changed upon MRPL10 depletion (Fig. 1E, F), suggesting that MRPL10 knockdown did not affect mtDNA copy number control or mitochondrial depolarization. To verify that the effects observed were due to knockdown of MRPL10, MRPL10 was knocked down first, and transfected with pcDNA3.1 empty vector or human MRPL10 expression plasmid, then the observed effects were detected. The results showed that decreased expression level of mitochondria-encoded protein ND2 and COX II in MRPL10 knockdown cells were returned to normal upon exogenous MRPL10 expression (Supplementary Fig. 1A; Supplementary Data are available online at www.liebertpub.com/dna). Mitochondrial complex I, IV activity, mitochondrial respiration, and intracellular ATP production were also significantly rescued in MRPL10 re-expression cells (Supplementary Fig. 1B–E). Taken together these findings provide evidence that MRPL10 is required for mitochondrial function and respiratory complex proteins synthesis.
FIG. 1.
MRPL10 knockdown leads to decreased mitochondrial activity and mitochondrial complex expression. (A) Several components of mitochondrial respiratory complexes from mitochondria lysates in HEK293 cells with or without MRPL10 knockdown were subjected to SDS-PAGE and immunoblotted with the indicated antibodies. Bar graphs depict the signal intensities of protein indicated. The relative expression is shown as a percentage of the mean using the control sample as reference. (B) Activities of the five mitochondrial respiratory complexes were measured in mitochondria isolated from control or MRPL10 knockdown HEK293 cells. Cells treated with rotenone (5 μM), TTFA (10 μM), antimycin A (2 μM), KCN (100 μM), and oligomycin (10 μM) to inhibit mitochondrial complex I, II, III, IV, and V activities were used as positive controls. (C) Mitchondrial respiration rate was measured using a clark type electrode. Cells treated with mitochondrial complex I inhibitor rotenone (5 μM) were used as positive controls. (D) Comparison of the ATP production in control or MRPL10 knockdown cells. Cells treated with mitochondrial complex I inhibitor rotenone (5 μM) were used as positive controls. (E) The copy number of mtDNA was measured by quantitative PCR analysis. (F) Mitochondrial membrane potential was detected by FACS analysis in control or MRPL10 knockdown HEK293 cells by three independent experiments. Cells treated with CCCP (10 μM) for 2 h were used as positive controls. The results were obtained from three independent experiments. The relative expression is shown as a percentage of the mean using the control sample as reference. All data in this figure represent the mean ± SEM of three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001, KCN, potassium cyanide; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophores; SEM, standard error of the mean; TTFA, thenoyltrifluoroacetone.
Reduced MRPL10 disturbs cyclin B1/Cdk1 activity
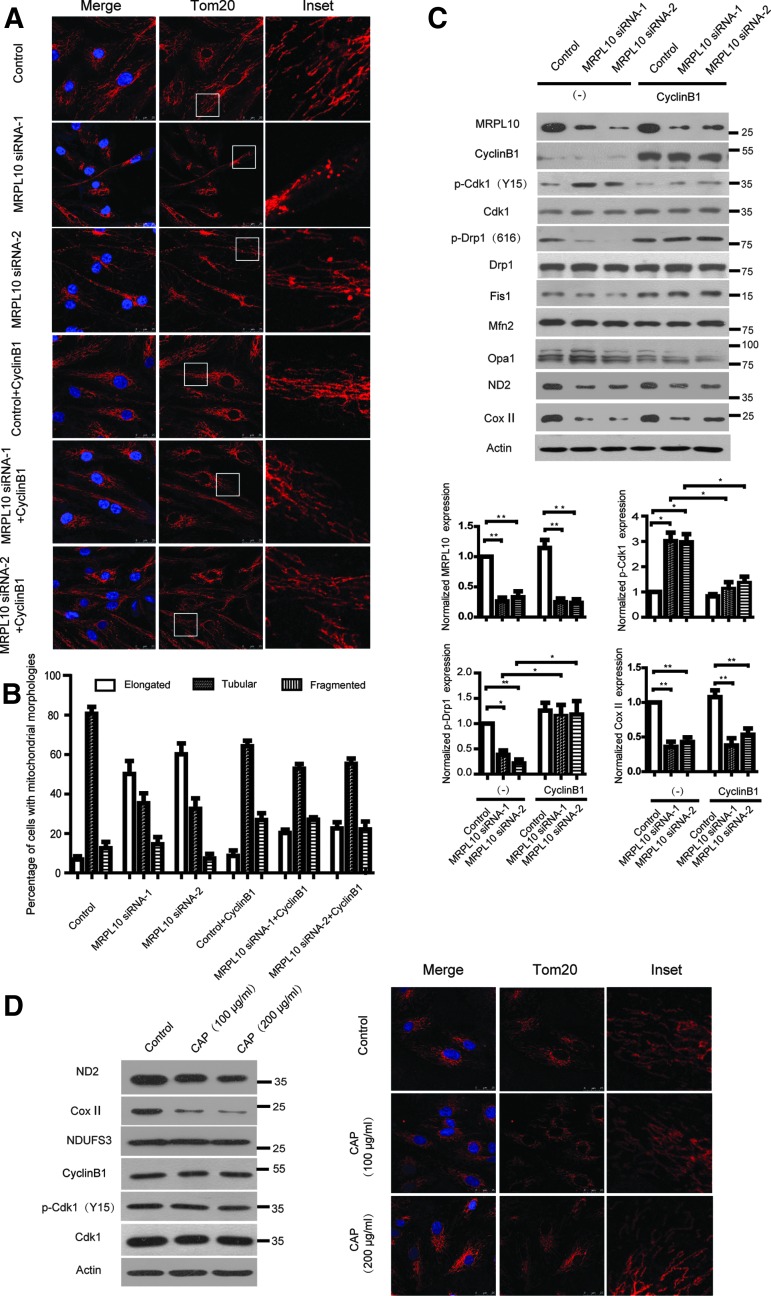
To investigate whether mitochondrial morphology was disrupted by the abnormal mitochondrial function upon MRPL10 depletion, HEK293 (data not shown) and ARPE-19 cells were transfected with MRPL10 siRNA and scramble control, and mitochondrial morphology was assessed by immunostaining using Tom20. Knockdown of MRPL10 led to a significant mitochondrial clustering and elongation (Fig. 2A). The percentage of cells with elongated mitochondria in control, MRPL10 siRNA-1, and MRPL10 siRNA-2 is 6.8% ± 1.6%, 50.2% ± 6.6%, and 60.3% ± 5.5%, respectively (Fig. 2B). To determine whether the alteration of mitochonrial morphology was regulated by mitochondria dynamics-related proteins, the expression of mitochondrial fission proteins Drp1, Fis1, and fusion proteins Mfn2, Opa1 were detected by immunoblotting. Surprisingly, the results suggested that compared with control cells, there was significant decrease in phosphorylation of Drp1 at Ser616 in MRPL10 knockdown cells. In addition, the protein levels of other mitochondria dynamics regulated proteins showed no significant change (Fig. 2C). It has been reported that Drp1 is phosphorylated by cyclin B1/Cdk1 at Serine 616 in humans, which contributes to mitochondrial fission in cycling cells (Taguchi et al., 2007). Next, the expression of cyclin B1 and phosphorylation of Cdk1 on the inhibitory residue Tyr15 were tested, the results showed that Cdk1 Y15 phosphorylation was upregulated in MRPL10 knockdown cells. We hypothesized that cyclin B1 would facilitate mitochondrial normal morphology via phosphorylation of Drp1 at Ser616 in MRPL10 knockdown cells. As anticipated, overexpression of cyclin B1 resulted in significant decrease of Cdk1 Y15 phosphorylation and further increased the percentage of cells with normal tubular mitochondria as seen in controls. While cyclin B1 overexpression in MRPL10 knockdown cells did not restore MRPL10 expression or mitochondrial translation (Fig. 2B, C).
FIG. 2.
Reduced MRPL10 disturbs cyclin B/Cdk1 activity. (A) ARPE-19 cells were transfected with MRPL10 siRNA and Cyclin B1 in different combination and stained with Tom20 (red) and DAPI (blue). Magnified images of the boxed regions are shown in the insets. (B) The number of cells with elongated mitochondrial structures (elongated), tubular mitochondrial (tubular), or fragmented mitochondrial (fragmented) was counted. Mitochondrial morphology was scored as follows: fragmented, mainly small and round; elongated, cluster and long filamentous; and tubulated, long and higher interconnectivity. For each identified object, the major axis length and the roundness index were calculated. Cells were scored with elongated or fragmented mitochondria when >50% of the mitochondria in the image displayed a major axis longer than 5 μm or <2 μm, respectively. The number of cells with the indicated mitochondrial morphology is shown as a percentage. More than 100 cells of three independent experiments were calculated. (C) HEK293 cells were transfected with MRPL10 siRNA and Cyclin B1 in different combination, mitochondrial dynamics related protein from total cell lysates were subjected to SDS-PAGE and immunoblotted with the antibodies indicated. Bar graphs depict the signal intensities of protein indicated. The relative expression is shown as a percentage of the mean using the control sample as reference. (D) ARPE-19 cells were treated with vehicle (Ethanol) or CAP (100 and 200 μg/mL) for 48 h and total cell lysates were subjected to SDS-PAGE and immunoblotted with the antibodies indicated. The cells were also stained with Tom20 (red) and DAPI (blue). Magnified images of the boxed regions are shown in the insets. All data in this figure represent the mean ± SEM of three independent experiments. *p < 0.05, **p < 0.01. CAP, chloramphenicol; Cdk1, cyclin-dependent kinase 1.
To test whether the decreased cyclin B1/Cdk1 activity in MRPL10 knockout cells was due to decreased mitochondrial translation in general, chloramphenicol (CAP) was used to block mitochondrial translation, and the expression of cyclin B1 or Cdk1 Y15 phosphorylation were examined. There was no marked difference on the expression of cyclin B1 or Cdk1 Y15 phosphorylation between control and CAP treatment, the inhibition effect of CAP was evaluated by immunoblotting with ND2 and COX II. Likewise, CAP treatment did not induce the inhibition of mitochondrial fission (Fig. 2D). Moreover, knockdown of the other two mitoribosome protein MRPL7 or MRPL11 also did not cause obvious mitochondrial morphology change (data not shown), which suggest the alteration of cyclin B1/Cdk1 activity and mitochondrial morphology were associated with MRPL10. Together, these results indicate that MRPL10 is essential for maintaining mitochondria fission via the modulation of cyclin B1/Cdk1 activity.
Cyclin B1 restores mitochondrial abnormality caused by MRPL10 knockdown
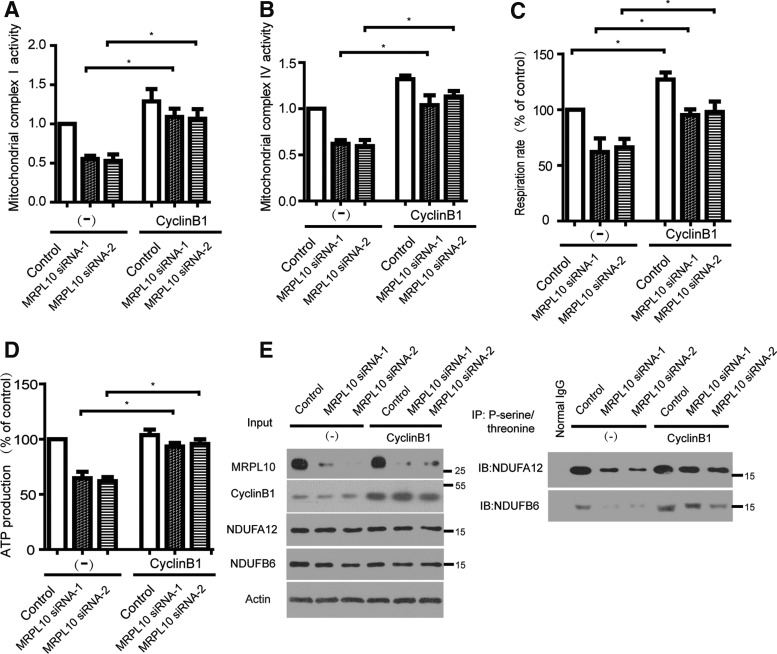
Previous studies have suggested that cyclin B1/Cdk1 was able to increase mitochondrial respiration with enhanced oxygen consumption and ATP generation through upregulation of mitochondrial complex I enzymatic activity (Wang et al., 2014). To further explore whether cyclin B1 facilitates mitochondrial function in MRPL10 knockdown cells, MRPL10 was knocked down by siRNA in HEK293 cells, and then transfected with pcDNA3.1 empty vector or cyclin B1 plasmid. As illustrated in Figure 3A and B, mitochondrial complex I and IV activity were enhanced upon cyclin B1 overexpression in MRPL10 knockdown cells. Expression cyclin B1 also lead to the increase of mitochondrial respiration rate and intracellular ATP levels (Fig. 3C, D). Compared with MRPL10 siRNA-1, MRPL10 siRNA-2 transfected cells, the ATP production was increase up to 163% and 186% in cells expressing cyclin B1. To test whether cyclin B1/Cdk1 regulates mitochondrial activity through phosphorlation of mitochondrial complex I substrates. Mitochondrial proteins from various groups were extracted by immunoprecipitation using a phosphoserine/threonine antibody followed by immunoblotting using antibodies to two mitochondrial complex I subunits NDUFA12 and NDUFB6 (Fig. 3E). Consistent with the decreased activity of cyclin B1/Cdk1 in MRPL10 knockdown cells, the phosphorylation levels of NDUFA12 and NDUFB6 were decreased in MRPL10 knockdown cells, and overexpression of cyclin B1 increased the phosphorylation levels of the two proteins, suggesting that cyclin B1 may restore mitochondrial function via the improvement of mitochondrial respiration in MRPL10 knockdown cells. Thus, based on these results, we conclude that cyclin B1 compensates for MRPL10 depletion through the regulation of mitochondrial function in vitro.
FIG. 3.
Cyclin B restores mitochondrial abnormality caused by MRPL10 knockdown. (A, B) HEK293 cells were transfected with MRPL10 siRNA and Cyclin B1 in different combination. Activities of the mitochondrial respiratory complexes were measured in isolated mitochondria. (C) Mitchondrial respiration rate was measured using a clark type electrode in each group. (D) Comparison of the ATP production in each group. (E) HEK293 cells were transfected with MRPL10 siRNA and Cyclin B1 in different combination for 72 h, mitochondrial proteins were separated and immunoprecipitated using a phosphoserine/threonine antibody followed by immunoblotting using antibodies to NDUFA12 and NDUFB6. Direct immunoblotting of cell lysates with the indicated antibody are shown as the input control (left panel). β-actin is shown as loading control. All data in this figure represent the mean ± SEM of three independent experiments. *p < 0.05.
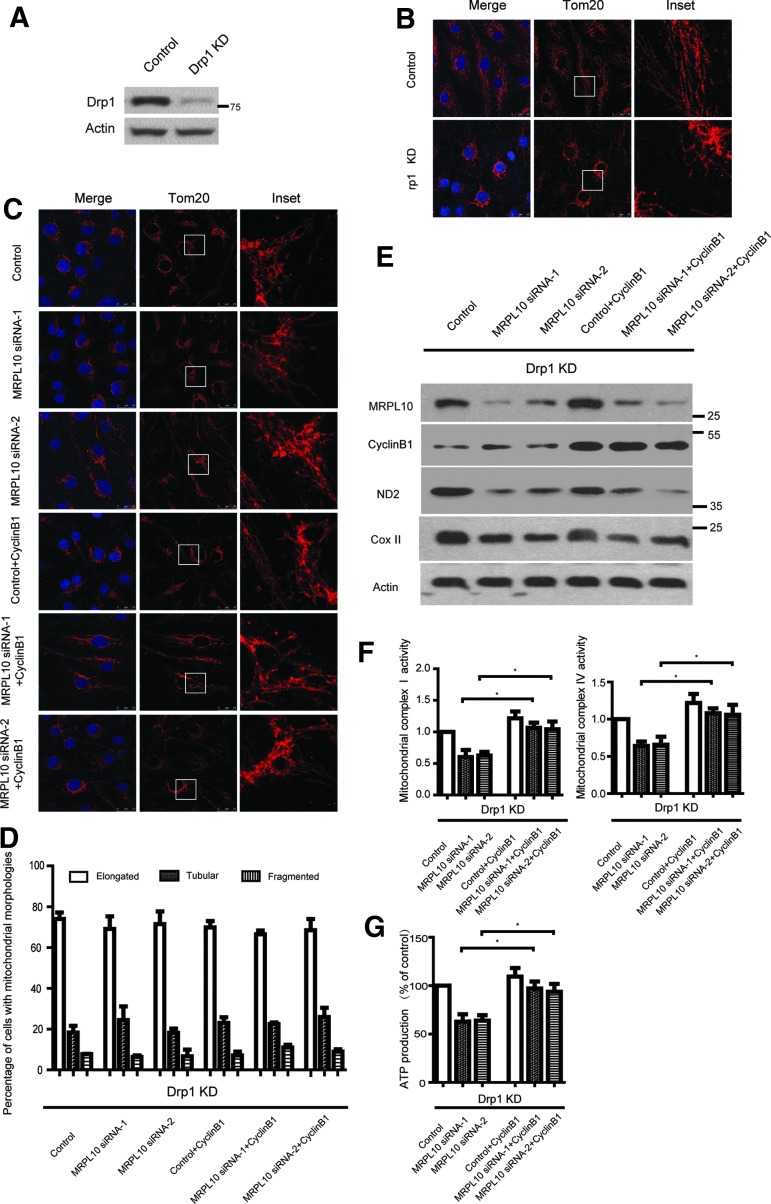
Cyclin B1 rescues respiration defect caused by MRPL10 knockdown independent of mitochondrial morphology
Overexpression of Cyclin B1 rescues both mitochondrial respiration defects and mitochondrial morphology in MRPL10 knockdown cells. To test whether mitochondrial respiration and mitochondrial morphology are connected to each other, Drp1 knockdown cells were established with pSuper vectors expression of small hairpin RNAs designed to knockdown Drp1 (Fig. 4A), and MRPL10 siRNAs along with cyclin B1 expression plasmid were transfected in different combination, mitochondrial morphology was assessed by immunostaining using Tom20. Drp1 and MRPL10 double knockdown led to a significant mitochondrial fusion and enlongation (Fig. 4B), which is similar in Drp1 knockdown cells. Compared with Drp1 knockdown and Drp1and MRPL10 double knockdown cells, overexpression of cyclin B1 didn't increase the percentage of cells with normal tubular mitochondrial (Fig. 4D), or the expression of ND2 and COXII (Fig. 4E). However, cyclin B1 expression still rescues the respiration defect in Drp1 and MRPL10 double knockdown cells (Fig. 4F, G). This result suggest that cyclin B1 rescues mitochondrial respiration defect in MRPL10 knockdown cells independent of mitochondrial morphology.
FIG. 4.
Cyclin B1 rescues respiration defect caused by MRPL10 knockdown independent of mitochondrial morphology. (A) Drp1 were silenced with pSuper vectors, which express small hairpin RNAs designed to knockdown Drp1. Transfected ARPE-19 cells were selected for growth in 1 μg/mL puromycin for 14 days, and the total cell lysates of resistant cells were subjected to SDS-PAGE and immunoblotted with Drp1. (B) Drp1 knockdown cells were stained with Tom20 (red) and DAPI (blue). (C) Drp1 knockdown cells were transfected with MRPL10 siRNA and Cyclin B1 in different combination and stained with Tom20 (red) and DAPI (blue). Magnified images of the boxed regions are shown in the insets. (D) The number of cells with elongated mitochondrial structures (elongated), tubular mitochondrial (tubular), or fragmented mitochondrial (fragmented) was counted. (E) Drp1 knockdown cells were transfected with MRPL10 siRNA and Cyclin B1 in different combination. Protein from total cell lysates were subjected to SDS-PAGE and immunoblotted with the antibodies indicated. (F) Drp1 knockdown cells were transfected with MRPL10 siRNA and Cyclin B1 in different combination. Activities of the mitochondrial respiratory complexes I and IV were measured in isolated mitochondria. (G) ATP production was detected in each group. *p < 0.05.
Cyclin B1 suppresses phenotypes caused by mRpL10 deficiency in Drosophila
Drosophila has been extensively used as a model system to study the function of mitoribosome protein. The amino acid identity between human MRPL10 and its Drosophila homolog mRpL10 is 27%. Drosophila ubiquitous knockdown of mRpL10 showed development delay and stayed in larval stage (not shown). Specific knockdown of mRpL10 in Drosophila eyes conducted by an eye-specific enhancer from the eyeless(ey) gene (Quiring et al., 1994) drives GAL4 to express double-stranded RNAs throughout the eye disc. Compared with control, the eye had pronounced eye defect without pigments lost (Fig. 5A). Interestingly, overexpression of human cyclin B1 can partially suppress the eyes' phenotype of mRpL10 knock down flies (Fig. 5B), the percentage of gross eye in eye specific mRpL10 RNAi Drosophila was 96.4% ± 1.2%, overexpression of cyclin B1 in eye specific mRpL10 RNAi Drosophila resulted in significant decrease of the proportion of flies with gross eye (43.2% ± 10.8%). Concomitantly, most rhabdomeres of photoreceptor cells of eye specific mRpL10 RNAi Drosophila were lost by transmission electron micrograph cross section (Fig. 5C). The percentage of photoreceptor cells with rhabdomeres lost in eye specific mRpL10 RNAi Drosophila was 57.8%, and the proportion decreased to 16.2% upon cyclin B1 overexpression (not shown). Ultrastructure analysis revealed swelling of mitochondrial with disrupted cristae in eyes of mRpL10 RNAi knockdown flies, which was suppressed by cyclin B1 overexpression (Fig. 5D). The knockdown efficiency of mRpL10 was assayed by quantitative real-time PCR (Fig. 5E). Moreover, the decreased mitochondrial complex I activity and ATP production in eye specific mRpL10 RNAi Drosophila were also attenuated by cyclin B1 overexpression (Fig. 5F, G). Together, these results suggest that mRpL10 play an essential role in the formation of compound eye in Drosophila, and human cyclin B1 partially restores mitochondrial function in mRpL10 deficiency Drosophila.
FIG. 5.
Cyclin B1 suppresses phenotypes caused by mRpL10 deficiency in Drosophila. (A) Eye-specific mRpL10 knockdown flies displayed abnormal eyes with reduced size and asymmetric eyes, which can be suppressed by overexpression of Cyclin B1 (ey>mRpL10 RNAi; hCyclin B1). (B) Percentage of flies with gross eye. The percentage of flies with reduced size and abnormal eye was determined in at least 300 flies. The results were obtained from three independent experiments. (C) Transmission electron micrograph images show the rhabdomeres of photoreceptor cells of control, ey-GAL4>mRpL10 RNAi, and hCyclin B1 expression flies. (D) Transmission electron micrograph images show the mitochondrial of each fly group. Black arrowheads indicate mitochondria in the transmission electron microscopy images. The size bar is 1 μm. (E) Real-Time PCR to detect mRpL10 expression in control, ey-GAL4>mRpL10 RNAi, and hCyclin B1 expression flies. The relative expression level is shown as a percentage of the mean using the control sample as reference. Bar graphs quantify the expression level of mRpL10 mRNA. (F) Activities of the mitochondrial respiratory complex I were measured in isolated mitochondria from the eye of each fly group. (G) Comparison of the ATP production in control, ey-GAL4>mRpL10 RNAi, and hCyclin B1 expression flies. The relative expression is shown as a percentage of the mean using the control sample as reference. All data in this figure represent the mean ± SEM of three independent experiments. *p < 0.05, **p < 0.01.
Discussion
This study has identified MRPL10, as a mitoribosome large subunit component, plays an essential role in cell growth via mediating cyclin B1/Cdk1 activity. In addition, we further defined MRPL10 is critical for mitochondrial function and mitochondrial fission by affecting mitochondrial protein synthesis and mitochondria dynamics. This is consistent with previous observation that knockdown of MRPL10 affect mitoribosome assembly (Nouws et al., 2016), and disrupting the mitoribosome intact by actinonin can block cell proliferation due to a mitoribosomal rescue pathway triggered by mitochondrial translation stress (Richter et al., 2013). The study also identifies the role of MRPL10 in the regulation of different signaling pathways, including mitochondrial complexes activity, cyclin B1/Cdk1 signaling, and mitochondria fission, which may help us to understand the mutual connection among these pathways. Moreover, overexpression of cyclin B1 restores mitochondrial dysfunction and promotes mitochondrial respiration in MRPL10 knockdown cells, suggesting the possible therapeutic targets for disorders of mitochondrial protein synthesis.
Previous studies have detected cyclin B1/Cdk1, which regulates nuclear envelope dynein recruitment and centrosome separation during mitosis, plays important roles in cell cycle transition from late G2 to mitosis (Lindqvist et al., 2007; Santamaria et al., 2007; Baffet et al., 2015). Inactivation of cyclin B1/Cdk1 results in development arrest in multiple species. In Drosophila melanogaster, inactivation of Cdk1 or cyclin B1 results in size defective, developmental program arrest and die during embryonic stage, indicating a critical role for the cyclin B1/Cdk1 in cell mitosis. Cdk1 knockout mice show embryonic development arrest around the blastocyst stage (Diril et al., 2012). Cdk1 kinase plays its roles via transferring phosphate from ATP to substrates, and the identification of its phosphorylation targets is important for the understanding of cell division process.
Recent studies have identified some components of mitochondrial electron transport chain as potential cyclin B1/Cdk1 phosphorylation targets. Cyclin B1/Cdk1 can mediate mitochondrial complex I activation and enhanced mitochondrial ATP generation via phosphorylation of its mitochondrial targets, which provide sufficient energy for cell cycle progression (Wang et al., 2014). The major antioxidant enzyme of mitochondrial manganese superoxide dismutase (MnSOD) is mediated by the phosphorylation of cyclin B1/Cdk1 at Ser106, which promotes MnSOD activity and improves mitochondrial function (Candas et al., 2013). Cyclin B1/Cdk1 can also phosphorylate mitochondrial P53 at Ser315 residue and enhance mitochondrial ATP production (Nantajit et al., 2010). Together, these studies indicate the critical role of cyclin B1/Cdk1 in the regulation of mitochondrial activity through phosphorylation of mitochondrial substrates. However, it is still poorly understood how MRPL10 controls cyclin B1/Cdk1 activity, one possible speculation is that different stress conditions induced by MRPL10 knockdown might affect cyclin B1/Cdk1 or its upstream regulators, which subsequently leads to adaptive metabolic response, blockage of cell proliferation, and cell survival. Some evidence support this notion, the activation of AMPK induced by the decrease of ATP production causes cell cycle arrest via the phosphorylation of p53 and the regulation of Cyclin E (Jones et al., 2005; Mandal et al., 2005). Interestingly, in Drosophila mRpL12 mutant cells also have reduced growth rates. Furthermore, mRpL12 is required for CycD/Cdk4-induced cell growth, and CycD/Cdk4 overexpression can stimulate mitochondrial activity. Our results also demonstrate that cyclin B1 overexpression enhances mitochondrial complexes activity and promotes mitochondrial respiration in MRPL10 knockdown cells or mRpL10 RNAi Drosophila. Results from these studies reveal mitochondrial ribosomal proteins are required for cell proliferation through the regulation of different cell cycle-dependent kinases and its regulatory partner cyclins. Moreover, proper regulation of Cdks or cyclins expression may facilitate adaptive metabolic response and cell survival to mitochondrial dysfunction.
Mitochondrial fission are essential for tissue development and cell cycle progression, mitochondrial fission related protein Drp1 is important for proper mitochondrial distribution in polarized cells (Detmer and Chan, 2007; Ishihara et al., 2009). Furthermore, Drp1 is phosphorylated by Cdk1 (at Ser585 rat/Ser 616 human) and contribute to mitochondrial segregation, which help the production and delivery of mitochondria to daughter cells in cycling cells (Taguchi et al., 2007). Importantly, the induced formation of hyperfused mitochondria through Drp1 inhibition causes cell cycle blockage and cell quiescence (Mitra et al., 2009). Downregulation of another mitochondrial fission related protein Fis1 also leads to the reduction of cyclins and G2/M cell cycle arrest (Lee et al., 2014). These studies suggest the crosstalk between mitochondrial fission/fusion proteins and cell cycle regulators and their synchronization regulation in cell cycle progression and mitochondrial activity in dividing cells. In this study our results indicate that MRPL10 depletion decreases the phosphorylation of Drp1 at Ser616 by the inhibition of cyclin B1/Cdk1 activity, which lead to a significant mitochondrial clustering and fusion, suggesting the important roles of mitochondrial ribosomal proteins in mitochondrial homeostasis. Interestingly, MRPS29 (DAP3) knockdown can induce mitochondrial fragmentation, indicating that different mitochondrial ribosomal proteins may have multiple effects on the regulation of mitochondrial homeostasis (Nouws et al., 2016). The importance of mitochondrial fragmentation or excess mitochondrial fusion induced by the knockdown of distinct mitochondrial ribosomal proteins need to be tested, whether it is a rescue pathway for mitochondrial translation stress resistance or just a byproduct is still unknown. Further studies aimed at elucidating physiologic significance of the Drp1 phosphorylation and the molecular mechanisms of MRPL10 along with other mitoribosome proteins are needed, and results from these studies may help us to find novel therapeutic strategies for mitochondria-related disorders caused by mutations in mitoribosome genes.
Supplementary Material
Acknowledgment
This work was supported by Chinese clinical rehabilitation research foundation of Xiangya Hospital-Sinobioway (xywm2015 I 31). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the article.
Disclosure Statement
No competing financial interests exist.
References
- Baffet A.D., Hu D.J., and Vallee R.B. (2015). Cdk1 activates pre-mitotic nuclear envelope dynein recruitment and apical nuclear migration in neural stem cells. Dev Cell 33, 703–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candas D., Fan M., Nantajit D., Vaughan A.T., Murley J.S., Woloschak G.E., et al. (2013). CyclinB1/Cdk1 phosphorylates mitochondrial antioxidant MnSOD in cell adaptive response to radiation stress. J Mol Cell Biol 5, 166–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C.J., Isohanni P., Poyhonen R., Euro L., Richter U., Brilhante V., et al. (2013). Whole-exome sequencing identifies a mutation in the mitochondrial ribosome protein MRPL44 to underlie mitochondrial infantile cardiomyopathy. J Med Genet 50, 151–159 [DOI] [PubMed] [Google Scholar]
- Cavdar Koc E., Ranasinghe A., Burkhart W., Blackburn K., Koc H., Moseley A., et al. (2001). A new face on apoptosis: death-associated protein 3 and PDCD9 are mitochondrial ribosomal proteins. FEBS Lett 492, 166–170 [DOI] [PubMed] [Google Scholar]
- Cotney J., Wang Z., and Shadel G.S. (2007). Relative abundance of the human mitochondrial transcription system and distinct roles for h-mtTFB1 and h-mtTFB2 in mitochondrial biogenesis and gene expression. Nucleic Acids Res 35, 4042–4054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detmer S.A., and Chan D.C. (2007). Functions and dysfunctions of mitochondrial dynamics. Nat Rev Mol Cell Biol 8, 870–879 [DOI] [PubMed] [Google Scholar]
- Diril M.K., Ratnacaram C.K., Padmakumar V.C., Du T., Wasser M., Coppola V., et al. (2012). Cyclin-dependent kinase 1 (Cdk1) is essential for cell division and suppression of DNA re-replication but not for liver regeneration. Proc Natl Acad Sci U S A 109, 3826–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei C., Galloni M., Hafen E., and Edgar B.A. (2005). The Drosophila mitochondrial ribosomal protein mRpL12 is required for Cyclin D/Cdk4-driven growth. EMBO J 24, 623–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber B.J., Boehringer D., Leitner A., Bieri P., Voigts-Hoffmann F., Erzberger J.P., et al. (2014). Architecture of the large subunit of the mammalian mitochondrial ribosome. Nature 505, 515–519 [DOI] [PubMed] [Google Scholar]
- Han M.J., Cimen H., Miller-Lee J.L., Koc H., and Koc E.C. (2011). Purification of human mitochondrial ribosomal L7/L12 stalk proteins and reconstitution of functional hybrid ribosomes in Escherichia coli. Protein Expr Purif 78, 48–54 [DOI] [PubMed] [Google Scholar]
- Hara M., Abe Y., Tanaka T., Yamamoto T., Okumura E., and Kishimoto T. (2012). Greatwall kinase and cyclin B-Cdk1 are both critical constituents of M-phase-promoting factor. Nat Commun 3, 1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N., Nomura M., Jofuku A., Kato H., Suzuki S.O., Masuda K., et al. (2009). Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat Cell Biol 11, 958–966 [DOI] [PubMed] [Google Scholar]
- Jones R.G., Plas D.R., Kubek S., Buzzai M., Mu J., Xu Y., et al. (2005). AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell 18, 283–293 [DOI] [PubMed] [Google Scholar]
- Kashuba E., Yurchenko M., Yenamandra S.P., Snopok B., Isaguliants M., Szekely L., et al. (2008). EBV-encoded EBNA-6 binds and targets MRS18-2 to the nucleus, resulting in the disruption of pRb-E2F1 complexes. Proc Natl Acad Sci U S A 105, 5489–5494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushal P.S., Sharma M.R., Booth T.M., Haque E.M., Tung C.S., Sanbonmatsu K.Y., et al. (2014). Cryo-EM structure of the small subunit of the mammalian mitochondrial ribosome. Proc Natl Acad Sci U S A 111, 7284–7289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.Y., and Ferrell J.E., Jr. (2007). Substrate competition as a source of ultrasensitivity in the inactivation of Wee1. Cell 128, 1133–1145 [DOI] [PubMed] [Google Scholar]
- Klein R., Li X., Kuo J.Z., Klein B.E., Cotch M.F., Wong T.Y., et al. (2013). Associations of candidate genes to age-related macular degeneration among racial/ethnic groups in the multi-ethnic study of atherosclerosis. Am J Ophthalmol 156, 1010–1020.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koc E.C., Burkhart W., Blackburn K., Moyer M.B., Schlatzer D.M., Moseley A., et al. (2001). The large subunit of the mammalian mitochondrial ribosome. Analysis of the complement of ribosomal proteins present. J Biol Chem 276, 43958–43969 [DOI] [PubMed] [Google Scholar]
- Lee S., Park Y.Y., Kim S.H., Nguyen O.T., Yoo Y.S., Chan G.K., et al. (2014). Human mitochondrial Fis1 links to cell cycle regulators at G2/M transition. Cell Mol Life Sci 71, 711–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist A., van Zon W., Karlsson Rosenthal C., and Wolthuis R.M. (2007). Cyclin B1-Cdk1 activation continues after centrosome separation to control mitotic progression. PLoS Biol 5, e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal S., Guptan P., Owusu-Ansah E., and Banerjee U. (2005). Mitochondrial regulation of cell cycle progression during development as revealed by the tenured mutation in Drosophila. Dev Cell 9, 843–854 [DOI] [PubMed] [Google Scholar]
- Miller C., Saada A., Shaul N., Shabtai N., Ben-Shalom E., Shaag A., et al. (2004). Defective mitochondrial translation caused by a ribosomal protein (MRPS16) mutation. Ann Neurol 56, 734–738 [DOI] [PubMed] [Google Scholar]
- Mitra K., Wunder C., Roysam B., Lin G., and Lippincott-Schwartz J. (2009). A hyperfused mitochondrial state achieved at G1-S regulates cyclin E buildup and entry into S phase. Proc Natl Acad Sci U S A 106, 11960–11965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nantajit D., Fan M., Duru N., Wen Y., Reed J.C., and Li J.J. (2010). Cyclin B1/Cdk1 phosphorylation of mitochondrial p53 induces anti-apoptotic response. PloS One 5, e12341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouws J., Goswami A.V., Bestwick M., McCann B.J., Surovtseva Y.V., and Shadel G.S. (2016). Mitochondrial ribosomal protein L12 is required for POLRMT stability and exists as two forms generated by alternative proteolysis during import. J Biol Chem 291, 989–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiring R., Walldorf U., Kloter U., and Gehring W.J. (1994). Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans. Science 265, 785–789 [DOI] [PubMed] [Google Scholar]
- Richman T.R., Ermer J.A., Davies S.M., Perks K.L., Viola H.M., Shearwood A.M., et al. (2015). Mutation in MRPS34 compromises protein synthesis and causes mitochondrial dysfunction. PLoS Genet 11, e1005089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter U., Lahtinen T., Marttinen P., Myohanen M., Greco D., Cannino G., et al. (2013). A mitochondrial ribosomal and RNA decay pathway blocks cell proliferation. Curr Biol 23, 535–541 [DOI] [PubMed] [Google Scholar]
- Santamaria D., Barriere C., Cerqueira A., Hunt S., Tardy C., Newton K., et al. (2007). Cdk1 is sufficient to drive the mammalian cell cycle. Nature 448, 811–815 [DOI] [PubMed] [Google Scholar]
- Serre V., Rozanska A., Beinat M., Chretien D., Boddaert N., Munnich A., et al. (2013). Mutations in mitochondrial ribosomal protein MRPL12 leads to growth retardation, neurological deterioration and mitochondrial translation deficiency. Biochim Biophys Acta 1832, 1304–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M.R., Koc E.C., Datta P.P., Booth T.M., Spremulli L.L., and Agrawal R.K. (2003). Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell 115, 97–108 [DOI] [PubMed] [Google Scholar]
- Taguchi N., Ishihara N., Jofuku A., Oka T., and Mihara K. (2007). Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J Biol Chem 282, 11521–11529 [DOI] [PubMed] [Google Scholar]
- Wang Z., Fan M., Candas D., Zhang T.Q., Qin L., Eldridge A., et al. (2014). Cyclin B1/Cdk1 coordinates mitochondrial respiration for cell-cycle G2/M progression. Dev Cell 29, 217–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurzenberger C., and Gerlich D.W. (2011). Phosphatases: providing safe passage through mitotic exit. Nat Rev Mol Cell Biology 12, 469–482 [DOI] [PubMed] [Google Scholar]
- Xiong H., Wang D., Chen L., Choo Y.S., Ma H., Tang C., et al. (2009). Parkin, PINK1, and DJ-1 form a ubiquitin E3 ligase complex promoting unfolded protein degradation. J Clin Invest 119, 650–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.